Abstract
Gastric cancer is currently the fourth most common cancer and the third leading cause of cancer-associated mortality worldwide. Studies have identified that certain biomarkers contribute to the prognosis, diagnosis and treatment of gastric cancer. However, the biomarkers of gastric cancer are rarely used clinically. Therefore, it is imperative to define novel molecular networks and key genes to guide the further study and clinical treatment of gastric cancer. In the present study, raw RNA sequencing data and clinicopathological information on patients with gastric cancer were downloaded from The Cancer Genome Atlas, and a weighted gene co-expression network analysis was conducted. Additionally, functional enrichment and protein-protein interaction analyses were implemented to further examine the significant modules. As a result, 16 modules of highly correlated genes were acquired and colour coded, and the yellow module containing 174 genes associated with chemotherapy resistance and prognosis in gastric cancer was further analysed. The biological processes of the yellow module were primarily associated with cell adhesion, vasculature development and the regulation of cell proliferation. In addition, the Kyoto Encyclopedia of Genes and Genomes pathways primarily involved the transforming growth factor-β signalling pathway, the cellular tumour antigen p53 signalling pathway, extracellular matrix-receptor interactions and focal adhesions. Notably, survival analysis and cell verification confirmed that high expression of GLIS family zinc finger 2 is significantly associated with chemoresistance and a worse prognosis in gastric cancer, and that this high expression is likely to be an important biomarker for the guidance of clinical treatment and prognostic evaluation.
Keywords: GLIS family zinc finger 2, chemoresistance, prognosis, gastric cancer, weighted gene co-expression network analysis
Introduction
Gastric cancer is currently the fourth most common cancer, after lung cancer, breast cancer and colon cancer, and the third leading cause of cancer-associated mortality worldwide (1,2). A previous study identified various biomarkers for gastric cancer, including cancer antigen 19-9, erb-b2 receptor tyrosine kinase 2 and vascular endothelial growth factor, certain of which may predict clinical prognosis and therapeutic effect or serve as a hallmark of diagnosis (3). However, biomarkers of gastric cancer are rarely used clinically due to a lack of specificity or sensitivity. Thus, it is imperative to identify novel molecular biomarkers that are able to predict the clinical outcome of gastric cancer, which may help to further the study of gastric cancer pathogenesis and facilitate personalised treatment.
With the development and maturation of third-generation sequencing technology, network approaches have been used to study the progression of various diseases, bridging the gaps between individual genes and the occurrence and development of disease (4,5). However, identification of key biomarkers remains a challenge. At present, although there are numerous studies based on bioinformatics approaches, including screening for differentially expressed genes (DEGs), similar expression patterns between genes have not been a focus (6). Weighted gene co-expression network analysis (WGCNA), an unbiased algorithm, elucidates the higher-order associations between genes or between gene sets and clinical features based on their co-expression relationships and delineates modules of biologically-associated genes (7). WGCNA has been widely applied to screen key biomarkers associated with clinical characteristics, including tumour grade, metastasis and prognosis, among different tumour types and even among different diseases or species (8–11). The present study utilized the unbiased strategy of WGCNA to study a module of genes significantly associated with chemotherapy resistance and prognosis in gastric cancer, and GLIS family zinc finger 2 (GLIS2) was identified as a previously unreported biomarker of gastric cancer by integrating the data analysis with cellular verification in vitro.
Materials and methods
Dataset acquisition and pre-processing
A flow diagram of the study is presented in Fig. 1. The raw data on patients with gastric cancer containing RNA sequencing (RNA_seq) and clinical information were obtained from The Cancer Genome Atlas (TCGA) repository website (https://cancergenome.nih.gov/). The R ‘limma’ Bioconductor package (12) was adopted to screen the DEGs between normal gastric and tumour tissues based on the following criteria: Fold change (FC), |log2(FC)|>1; and false discovery rate (FDR) <0.05. To minimize noise in the DEG dataset, in strict accordance with the analysis method on the WGCNA official website (https://labs.genetics.ucla.edu/horvath/htdocs/CoexpressionNetwork/Rpackages/WGCNA/), genes with too many missing values and unknown names were checked for and excluded, and genes for which the names corresponded to multiple probes were deleted. The samples were clustered to exclude the obvious outliers (7,13). The microarray dataset of filtered probe sets was constructed using the remaining genes and samples. In addition, clinical variables, including age and gender, pathological variables [including drug response, tumour (T) stage and tumour grade] and survival information were compiled for the WGCNA analysis. The DEGs were normalized by log2 transformation prior to the subsequent analyses. A total of two investigators separately collated and analysed the data, and a joint decision was reached in cases of disagreement.
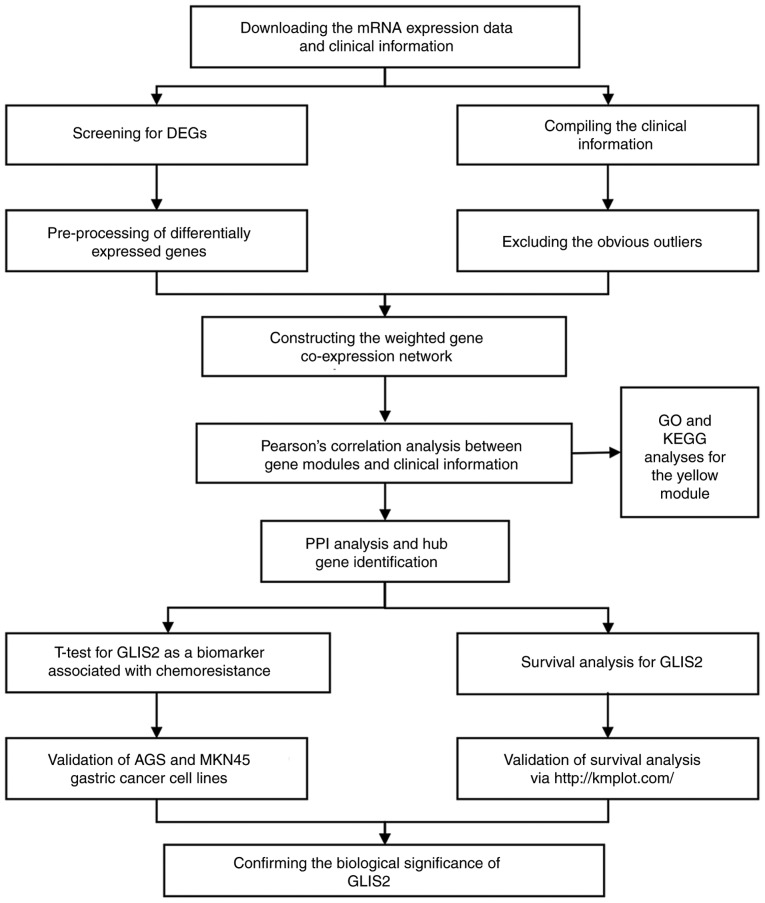
Figure 1.
Flow diagram of the present study. GO, gene ontology; KEGG, Kyoto Encyclopaedia of Genes and Genomes; PPI, protein-protein interaction; DEG, differentially-expressed gene; GLIS2, GLIS family zinc finger 2.
WGCNA construction
Construction of a weighted co-expression network represents an effective method for identifying modules and for defining the intra-module connectivity. In the current study, WGCNA was performed on the microarray data of filtered probe sets using the R ‘wgcna’ Bioconductor package (13). Each paired gene-gene association was correlated utilizing the absolute value of the Pearson product moment correlation (a gene co-expression similarity measure); the absolute value represents the co-expression similarity. An adjacency matrix was constructed utilizing a ‘soft’ power adjacency function, where the ‘soft’ power indicates that the resulting adjacency measures the connection strength. Clusters of co-expressed genes were designated by hierarchical cluster analysis following subtraction of the topological overlap measure of similarity of 1 (7,10).
The modules of WGCNA are groups of highly correlated genes. In network terminology, modules are sets of genes with high topological overlap. To obtain the potential associations between the co-expressed gene clusters and clinical variables, a single column of vectors termed the module eigengenes (MEs) (7) was used. The MEs were generated by reserving the first principal component of a given module and were representative of the gene expression profiles in a module. Since each ME contains the majority of the variance in the original data, it represents a summary measure for the overall co-expression network. The consistency between the expression of a particular gene and the ME expression is termed the module membership. This measure of co-expression network centrality is decided by calculating the Pearson correlation coefficient between each individual gene and the ME. Further details on WGCNA theory and algorithm have been previously published (7,10).
Key module identification and analysis
To define the pivotal module, the correlations between the MEs and clinical features were calculated. Furthermore, functional enrichment analysis and protein-protein interaction (PPI) analysis was conducted on the genes in the yellow module to examine the probable mechanism underlying the impact of the genes on chemotherapy resistance and the prognosis of gastric cancer. The Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david-d.ncifcrf.gov/) was used to analyse the enriched biological process (BP) terms and pathways (14). The top 15 most significant BP terms (P≤0.05) are presented. Only those KEGG pathways with P≤0.05 and ≥10 enriched genes were considered to be significant. In addition, the PPI networks were constructed using STRING (https://string-db.org/).
Cell culture and plasmid transfection
The human gastric cancer cell lines AGS and MKN45 were purchased from the American Type Culture Collection (Manassas, VA, USA). All the cell lines were maintained in RPMI 1640 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with 10% foetal bovine serum (ScienCell Research Laboratories, Inc., San Diego, CA, USA) and 2 mM penicillin and streptomycin, and were cultured in a humidified 37°C incubator at 5% CO2.
The GLIS2 overexpression plasmids were constructed and cloned into the CV061 vector (Shanghai GeneChem Co., Ltd., Shanghai, China) between the EcoRI and HindIII sites. The primer sequence used for GLIS2 was: 5′-ACGGGCCCTCTAGACTCGAGATGCACTCCCTGGACGAGCCGCTCG-3′. AGS and MKN45 cells were transfected with GLIS2 overexpression plasmids using Lipofectamine® 3000 (Invitrogen; Thermo Fisher Scientific, Inc.). According to the manufacturer's instructions, 1×105 gastric cancer cells were inoculated onto a 24-well plate and 1 µg/ml of the plasmid was added, and the transfected cells were incubated in a 5% CO2 incubator at 37°C for 48 h. Subsequent treatments were performed when the cells had grown to 80–90% confluence.
Western blotting
For the western blotting assay, cells were lysed using radioimmunoprecipitation assay and phenylmethylsulfonyl fluoride buffers (100:1; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), and the lysates were collected by centrifugation at 12,000 × g for 12 min at 4°C. The protein concentration was determined using a bicinchoninic acid protein assay kit (Sigma-Aldrich; Merck KGaA). Equal amounts of protein lysates (30 µg) were electrophoresed on 10% SDS-polyacrylamide gels and transferred to methanol-activated polyvinylidene fluoride (PVDF) membranes. The PVDF membranes were blocked with 5% non-fat dried milk in Tris-buffered saline (pH 7.4) containing 0.1% Tween-20 (TBST) for 1 h, and incubated with the primary GLIS2 (Abcam, Cambridge, UK; cat. no. ab28462; diluted 1:750) and GAPDH antibodies (Abcam; cat. no. ab8245; diluted 1:750) in TBST at 4°C overnight. The membranes were subsequently washed three times with TBST and incubated with a secondary Alexa Fluor-conjugated anti-rabbit antibody (Abcam; cat. no. ab150077; goat anti-rabbit IgG H&L; diluted 1:5,000) for 1 h at room temperature. Signal detection was performed for 5 min using an enhanced chemiluminescence reaction (Dalian Meilun Biotechnology Co., Ltd., Dalian, China). The GAPDH antibody was used to normalize protein expression.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). RT was performed (on 500 ng of RNA) using the PrimeScript RT Reagent kit (Takara Biotechnology Co., Ltd., Dalian, China), and the RT-qPCRs were performed using SYBR Premix Ex Taq (Takara Biotechnology Co., Ltd.), according to the manufacturer's protocol. The following primers were used: GLIS2 forward, 5′-CTTCGGGGAGGCTGGATT-3′ and reverse, 5′-GGTGATACTCAGCTTCAGGTCG-3′; and GAPDH forward, 5′-AATCCCATCACCATCTTCCAG-3′ and reverse, 5′-GAGCCCCAGCCTTCTCCAT-3′. The relative expression of the genes (GLIS2 and GAPDH) was calculated using the 2−ΔΔCq method (15).
MTT assay
Fluorouracil (5-FU), cisplatin and doxorubicin were purchased from Selleck Chemicals (Houston, TX, USA). Cells (6×103 per well) were seeded in 96-well plates and incubated for 48 h. 5-FU, cisplatin and doxorubicin were added at final concentrations of 0, 2, 4, 8, 16 and 32 µmol/l with dimethyl sulfoxide (DMSO) alone serving as a control. The cells were incubated in a 5% CO2 incubator at 37°C for 24 h. The medium was removed, and the cells were cultured in fresh growth medium containing 0.5 mg/ml MTT for 4 h under the above conditions. The supernatant was removed, and 100 µl DMSO was added to dissolve the formazan crystals. The absorption was measured at 490 nm, and the results were expressed as the half-maximal inhibitory concentration (IC50), the concentration of chemotherapeutic drug required for 50% inhibition in vitro.
Statistical analysis and visualization
The majority of the visualizations were generated using R version 3.3.1 (https://www.r-project.org/) except for the KEGG network and PPI visualizations, for which the ClueGO (16) and STRING online tools were used. Survival analysis and clinical correlation analysis was performed using TCGA expression and clinical data on gastric cancer, setting the median expression value of GLIS2 as a cut-off value between high and low expression. The Kaplan-Meier method was used to estimate survival, and a log-rank test was used to assess differences between the survival curves. Furthermore, the prognostic role of GLIS2, including its role in predicting overall survival and disease-free survival, was validated using online tools (http://kmplot.com/analysis/) (17). Statistical analyses were implemented using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA). All data are presented as the mean ± standard deviation. Student's t-tests were used for comparisons between groups, except for the analysis of clinicopathological features, for which the two-sample t-test was used. P<0.05 was considered to indicate a statistically significant difference.
Results
DEG screening and pre-processing
As presented in the flow chart (Fig. 1), public RNA_seq datasets were downloaded from TCGA repository website, containing 238 gastric cancer samples and 33 normal tissue samples. Based on the criteria of |log2(FC)|>1 and FDR<0.05, a total of 5,630 DEGs were screened out. Prior to constructing the microarray dataset of filtered probe sets, 204 low-quality DEGs and 26 cancer samples were excluded (data not shown). To facilitate the subsequent WGCNA analysis, the clinical information was compiled and converted. To obtain an overview of the clinical information, hierarchical cluster analysis was performed (data not shown).
Co-expression network construction
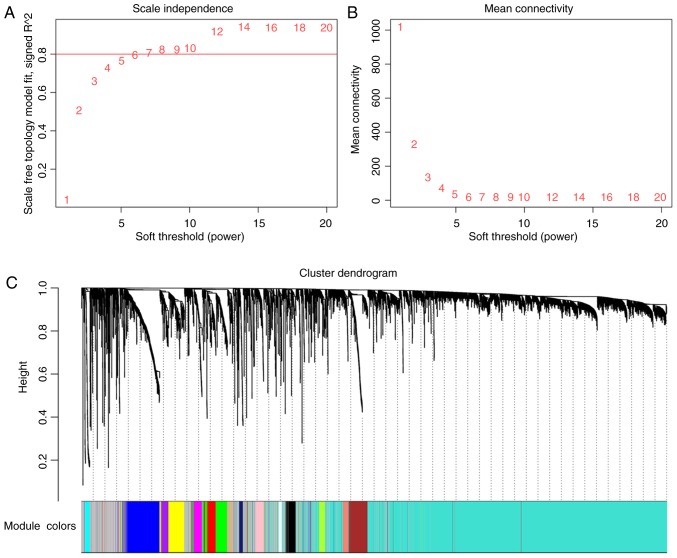
The remaining 5,426 most varying and most connected genes from 212 gastric cancer samples were obtained for construction of the co-expression network. As presented in Fig. 2, 16 distinct gene modules with high topological overlap were identified when the soft threshold power β was set to 6. The number of genes included in the modules ranged between 36 (light cyan) and 3,068 (turquoise). Each module was assigned a different colour to distinguish between them, and the grey module was reserved for genes that were identified as not being co-expressed.
Figure 2.
Analysis of network topology for various soft thresholding powers. (A) The scale-free fit index (y-axis) as a function of the soft thresholding power (x-axis). (B) The mean connectivity (degree, y-axis) as a function of the soft thresholding power (x-axis). (C) Clustering dendrogram of genes, with dissimilarity based on topological overlap, together with assigned module colours.
Key module identification
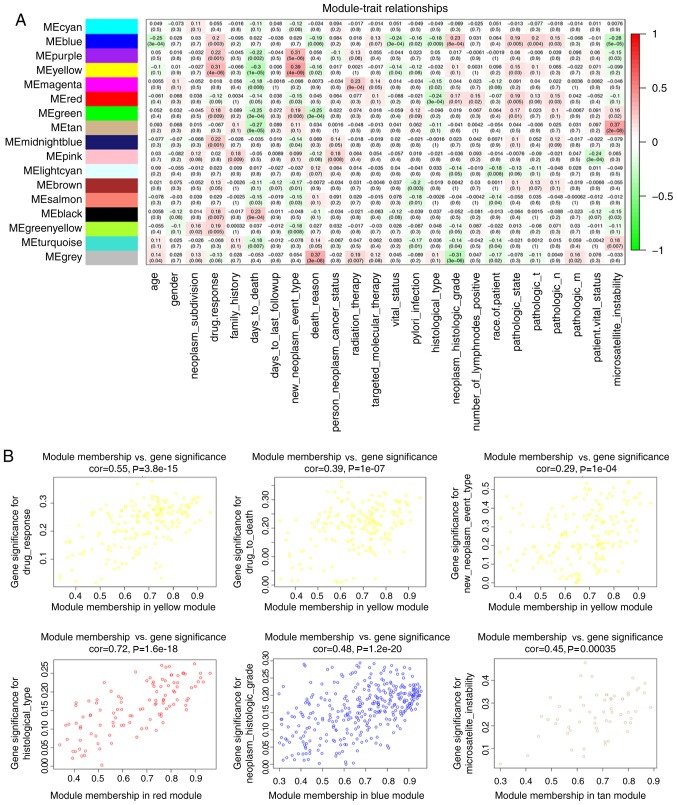
The yellow module had the highest correlation with drug response (r=0.31; P<0.001) and days to mortality (r=−0.3; P<0.001) among the module-trait relationships (Fig. 3A), and was selected for study in the subsequent analyses due to its clinical importance. The yellow module was also associated with new tumour type (including locoregional recurrence, distant metastasis and new primary tumour; r=0.39; P<0.001). It was additionally identified that the red module was associated with histological type (r=−0.24; P<0.001), the blue module was associated with tumour histological grade (r=0.23; P<0.001), and the tan module was associated with microsatellite instability (r=0.37; P<0.001). In addition, the intra-modular connectivity was calculated for each gene based on its Pearson correlation with all other genes in the module (Fig. 3B), to help prove the importance of these modules.
Figure 3.
Module-trait associations. (A) Each cell contains the corresponding correlation and P-value. (B) The scatterplot of gene significance for biological trait vs. module membership in different modules. ME, module eigengene; cor, correlation.
Biological importance of the modules
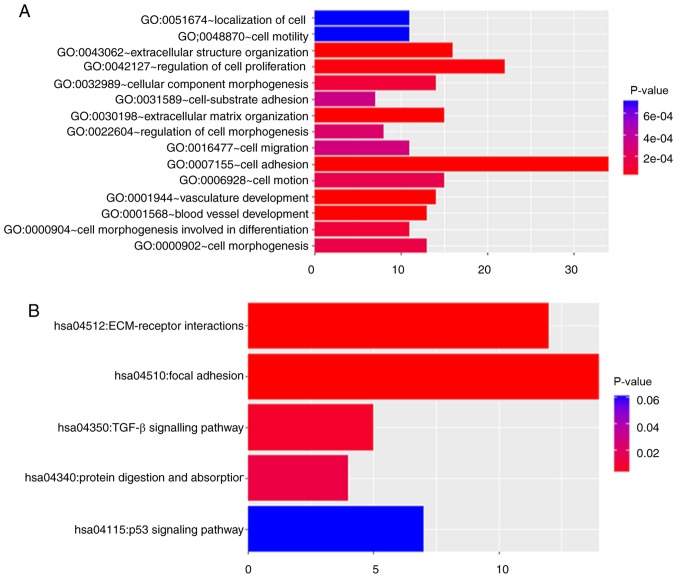
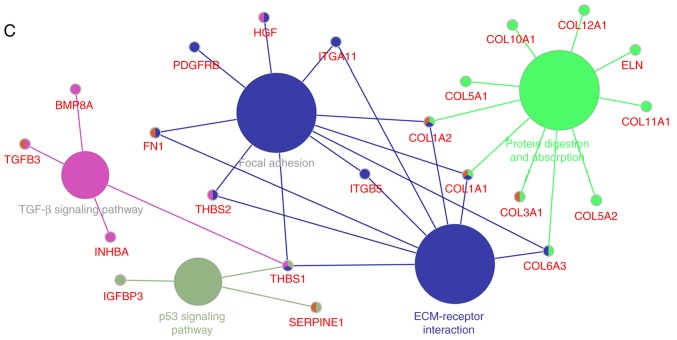
To investigate the biological importance of the yellow module, 174 genes in the yellow module were functionally enriched using DAVID for BP analysis and KEGG pathways analyses. The BPs of the yellow module were primarily enriched in a number of aspects, including ‘cell adhesion’ (P<0.001), ‘vasculature development’ (P<0.001), and ‘regulation of cell proliferation’ (P<0.001. Fig. 4A). In addition, the DAVID-based enrichment analysis identified KEGG pathways for the 174 genes in the yellow module (Fig. 4B and C). Among them, certain tumorigenesis-associated signalling pathways were present, including the TGF-β (P=0.006) and p53 (P=0.036) signalling pathways. In particular, cell adhesion-associated signalling pathways, including extracellular matrix (ECM)-receptor interactions (P<0.001) and focal adhesion (P<0.0001), were observed to serve a key role in the regulation of drug resistance in gastric cancer.
Figure 4.
Gene ontology (GO) functional analysis and KEGG pathway enrichment analysis for genes in the yellow module. The x-axis indicates the number of genes, and the y-axis indicates the GO and KEGG pathway terms. The-log10 (P-value) of each term is coloured according to the legend. (A) Biological process enrichment analyses. (B) KEGG pathway enrichment analysis. (C) KEGG pathway visualization. GO, gene ontology; KEGG, Kyoto Encyclopaedia of Genes and Genomes; ECM, extracellular matrix; p53, cellular tumour antigen p53.
Hub gene identification
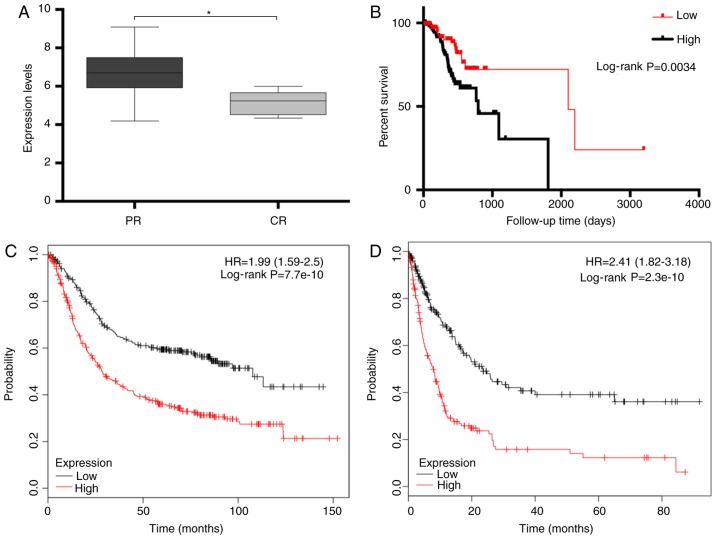
The present study proceeded to develop the PPI network and investigate the hub genes of the yellow module (Table I). To identify novel proteins and pathways modulated by the pivotal genes, GLIS2 was selected as a candidate gene for further analysis and verification subsequent to comparing the data and reviewing the literature. Analysis of the TCGA dataset demonstrated that high expression of GLIS2 was significantly associated with tumour chemotherapeutic resistance (P=0.031; Fig. 5A). Furthermore, high GLIS2 positivity in gastric cancer samples corresponded to a significantly worse prognosis (P=0.003; Fig. 5B). Similar results were obtained using the online tools to verify the prognostic role of GLIS2. Patients with higher GLIS2 positivity had significantly shorter overall survival (P<0.001; Fig. 5C) and disease-free survival (P<0.001; Fig. 5D) times. In addition, the association between GLIS2 and other clinicopathological characteristics was evaluated in patients with gastric cancer (Table II). The expression of GLIS2 was significantly associated with histological type (P=0.039), histological grade (P=0.02), pathological stage (P=0.013), T stage (P=0.002) and microsatellite instability (P<0.001).
Table I.
Hub genes (n=30) contained in the yellow module.
| Gene symbol | Gene name |
|---|---|
| COL1A1a | Collagen type I α1 chain |
| COL5A1a | Collagen type V α1 chain |
| COL1A2a | Collagen type I α2 chain |
| FN1a | Fibronectin 1 |
| COL3A1a | Collagen type III α1 chain |
| COL6A3a | Collagen type VI α3 chain |
| COL5A2a | Collagen type V α2 chain |
| THBS1a | Thrombospondin 1 |
| SPARCa | Secreted protein acidic and cysteine rich |
| COL11A1a | Collagen type XI α1 chain |
| ABCA1 | ATP binding cassette subfamily A member 1 |
| ADAMTSL1 | ADAMTS like 1 |
| BET3L | Trafficking protein particle complex 3 like |
| BTBD19 | BTB domain containing 19 |
| ADAM12 | ADAM metallopeptidase domain 12 |
| AEBP1 | AE binding protein 1 |
| BGN | Biglycan |
| C1QTNF5 | C1q and TNF related 5 |
| ADAMTS12 | ADAM metallopeptidase with thrombospondin type 1 motif 12 |
| ANTXR1 | ANTXR cell adhesion molecule 1 |
| BICC1 | BicC family RNA binding protein 1 |
| C1QTNF6 | C1q and TNF related 6 |
| ADAMTS2 | ADAM metallopeptidase with thrombospondin type 1 motif 2 |
| ARHGAP22 | Rho GTPase activating protein 22 |
| BMP1 | Bone morphogenetic protein 1 |
| CCDC71L | Coiled-coil domain containing 71 like |
| ADAMTS3 | ADAM metallopeptidase with thrombospondin type 1 motif 3 |
| ATP10A | ATPase phospholipid transporting 10A (putative) |
| BMP8A | Bone morphogenetic protein 8a |
| CD248 | CD248 molecule |
Top ten hub genes.
Figure 5.
(A) Expression level of GLIS2 in cancer tissues. (B) Prognostic value of GLIS2 at different expression levels among gastric cancer patients. Survival analysis of patients with gastric cancer was performed based on GLIS2 expression, as assessed with online tools. (C) Overall survival time; (D) disease-free survival time. *P<0.05. HR, hazard ratio; CI, confidence interval; GLIS2, GLIS family zinc finger 2; PR, partial response to chemotherapy; CR, complete response to chemotherapy.
Table II.
Clinicopathological characteristics of patients according to GLIS2 expression.
| Variable | No. patients | GLIS2 relative of expression (mean ± standard deviation) | P-value |
|---|---|---|---|
| Age at diagnosis, years | |||
| <60 | 76 | 6.613±0.1318 | 0.053 |
| ≥60 | 157 | 6.299±0.0926 | – |
| Histological type | |||
| Adenocarcinoma | 159 | 6.506±0.0864 | 0.039 |
| Intestinal adenocarcinoma | 78 | 6.174±0.1460 | <0.05 |
| Pathological stage | |||
| I+II | 125 | 6.231±0.1074 | 0.013 |
| III+IV | 104 | 6.617±0.1087 | <0.05 |
| Lymph node status | |||
| N0 | 84 | 6.234±0.1349 | 0.109 |
| N1-3 | 154 | 6.487±0.0899 | – |
| Microsatellite instability | |||
| MSS | 157 | 6.604±0.0801 | 0.000 |
| MSI-L+ MSI-H | 81 | 5.998±0.1494 | <0.05 |
| Family history | |||
| No | 195 | 6.416±0.0859 | 0.682 |
| Yes | 6 | 6.618±0.3467 | – |
| Sex | |||
| Male | 151 | 6.434±0.0910 | 0.523 |
| Female | 87 | 6.334±0.1333 | – |
| Histological grade | |||
| G1+G2 | 81 | 6.154±0.1378 | 0.020 |
| G3+GX | 157 | 6.523±0.0882 | <0.05 |
| T stage | |||
| T1+T2 | 70 | 6.035±0.1581 | 0.002 |
| T3+T4 | 165 | 6.551±0.0828 | <0.05 |
| Metastasis | |||
| M0 | 216 | 6.400±0.0798 | 0.613 |
| M1 | 11 | 6.584±0.3607 | – |
| Drug response | |||
| Partial response | 48 | 6.638±0.1598 | 0.002 |
| Complete response | 6 | 5.158±0.2595 | <0.05 |
| Helicobacter pylori infection | |||
| No | 58 | 5.893±0.1588 | 0.680 |
| Yes | 4 | 6.153±0.6512 | – |
T, tumour; MSS, microsatellite stability; MSI-H, high-level microsatellite instability; MSI-L, low-level microsatellite instability; GLIS2, GLIS family zinc finger 2.
GLIS2 functional verification
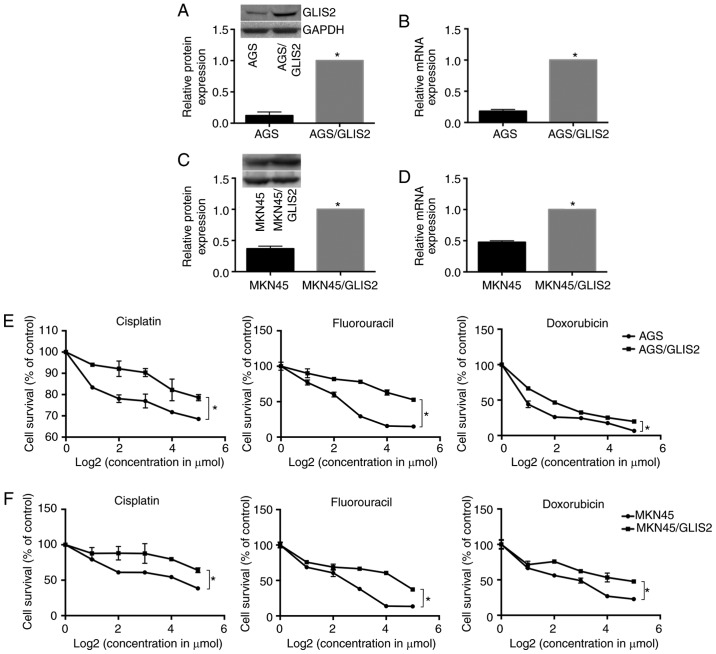
To examine whether the overexpression of GLIS2 enhanced chemotherapy tolerance in gastric cancer, MTT assays were performed. Fig. 6A-D demonstrates that the relative mRNA and protein expression levels of GLIS2 were markedly increased following transfection of GLIS2 plasmids in the AGS and MKN45 cell lines, respectively (P<0.05). Following treatment with different concentrations of three chemotherapeutic agents for 24 h, the results indicated that gastric cancer cells that overexpressed GLIS2 had increased survival in response to cisplatin, fluorouracil and doxorubicin compared with parental cells with lower GLIS2 expression. Specifically, the growth activity of the AGS experimental groups were higher compared with the control group (Fig. 6E; P<0.05), and consistent results were also obtained in the MKN45 cell line (Fig. 6F; P<0.05).
Figure 6.
High expression of GLIS2 promotes chemoresistance. (A) Relative protein expression levels and (B) mRNA expression of GLIS2 in AGS cells. *P<0.05 vs. AGS. (C) Relative protein expression levels and (D) mRNA expression of GLIS2 in MKN45 cells. *P<0.05 vs. MKN45. Growth activity following treatment with different concentrations of cisplatin, 5-FU and doxorubicin was assessed in (E) AGS cells and (F) MKN45 cells; *P<0.05. GLIS2, GLIS family zinc finger 2.
Discussion
WGCNA provides an efficient algorithm to reveal the associations between genes and BP among different diseases and species (10,11). Modules, or groups of highly correlated genes, may be a consequence of transcriptional co-activation, which results in the coordinated operation of associated gene networks to accomplish a group of biological functions (13,18). Therefore, a highly connected hub node may be established to control the entire network and the associated biological functions, and the hub node may be used for guiding further study and the clinical treatment of cancer.
Following this approach, raw data for patients with gastric cancer were downloaded from TCGA database and identified 16 gene co-expression modules using WGCNA. Modules that are significantly associated with clinical features have important biological and clinical implications (19). The results of the present study demonstrated that the yellow module was correlated with drug response and days to mortality, the red module was associated with histological type, the blue module was associated with tumour histological grade, and the tan module was associated with microsatellite instability. The yellow module, containing 174 genes, was further functionally enriched, and the PPI network was analysed, as it was apparently associated with chemotherapy resistance and tumour prognosis. Cell adhesion serves an important role in the chemotherapy resistance of gastric cancer, and associated signalling pathways include ECM-receptor interactions and focal adhesion. Previous studies have illustrated that ECM-receptor interactions lead to growth arrest at the G1 phase of the cell cycle and resistance to chemotherapy-induced apoptosis, and the inhibition of focal adhesion may impair DNA repair, resulting in increased residual DNA damage and treatment sensitivity in malignant tumours (20,21). In addition, the present study demonstrated that the TGF-β and p53 signalling pathways are involved in the chemotherapy resistance of gastric cancer. The mechanisms of the TGF-β and p53 signalling pathways in chemoresistance have also been reported and are involved in the induction of cytoskeletal molecular remodelling, cell growth regulation and cytokinesis (22–24).
Relevant hub genes are hypothesized to serve an important role in the biological processes of tissues, and central hub genes in the network are likely to be key drivers of aberrant regulatory mechanisms (25). The hub genes from the yellow module were screened, and it was identified by reviewing the literature that the roles of the majority of the genes in chemoresistance have been revealed. GLIS2 was further validated as a candidate hub gene that has an essential association with the biological behaviour of gastric cancer. GLIS2, a member of the sub-family of Krüppel-like zinc finger proteins, has been linked to the development of nephronophthisis, while few studies have been conducted on the connection between tumorigenesis and GLIS2 (26). In the TCGA dataset, GLIS2 was highly correlated with chemotherapy effects and the prognosis of gastric cancer. To verify this result, a survival analysis online tool was used to demonstrate that low expression of GLIS2 is associated with an improved gastric cancer prognosis. In addition, through gastric cancer cell line culture and MTT assays, it was demonstrated in vitro that high expression of GLIS2 may increase the chemotherapy resistance of gastric cancer cells. Furthermore, the analyses indicated that the expression of GLIS2 was significantly associated with histological type, histological grade, pathological stage, clinical T stage and microsatellite instability. All these results suggested that GLIS2 may serve an important role in the development of gastric cancer. However, whether GLIS2 is an independent prognostic factor requires further study.
The role and mechanism of GLIS2 in tumorigenesis and chemoresistance have rarely been reported. Thirant et al (27,28) reported that the CBFA2/RUNX1 translocation partner 3 (ETO2)-GLIS2 fusion protein controls the transcription of GATA binding proteins 1 and 2 and ERG by directly binding to DNA and upregulating the expression of enhancer elements, and ETO2-GLIS2 is closely associated with a poor prognosis in leukaemia. Guerra et al (29) reported that activation of GLIS2 is associated with the overexpression of trophoblast antigen 2, subsequently driving the transcription factor AP-1, NF-κB and G1/S-specific cyclin-D1 (CCND1) pathways and leading to tumorigenesis. NF-κB signalling activates anti-apoptotic genes and overexpression of drug transporters, and further promotes drug resistance (30). CCND1 is involved in chemoresistance, primarily by increasing the expression of multidrug resistance proteins and regulating the cell cycle (31).
In conclusion, a molecular network was constructed based on gene expression and biological characteristics using the WGCNA approach. Numerous modules and molecular networks associated with clinical manifestations of gastric cancer have been demonstrated. Furthermore, GLIS2 was significantly associated with chemotherapy resistance and the prognosis of gastric cancer. GLIS2 expression is likely to be an important biomarker that is able to guide clinical treatments and prognostic evaluation. However, the present study had certain limitations. First, to determine the validity of this bioinformatics analysis, the expression level of GLIS2 requires further confirmation in different cell types and gastric tissue samples. Second, the mechanisms by which GLIS2 causes chemoresistance require further study. Third, whether GLIS2 overexpression in vivo is an important cause of chemoresistance remains to be confirmed.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- DEGs
differentially expressed genes
- WGCNA
weighted gene co-expression network analysis
- RNA_seq
RNA sequencing
- TCGA
The Cancer Genome Atlas
- FC
fold change
- FDR
false discovery rate
- ME
module eigengene
- PPI
protein-protein interaction
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- BP
biological processes
- KEGG
Kyoto Encyclopaedia of Genes and Genomes
- RT-qPCR
reverse transcription-quantitative polymerase chain reaction
- 5-FU
fluorouracil
- ECM
extracellular matrix
- TGF-β
transforming growth factor-β
- ETO2-GLIS2
CBFA2/RUNX1 translocation partner 3-GLIS family zinc finger 2 fusion protein
- NF-κB
nuclear factor κB
- CCND1
G1/S-specific cyclin-D1
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 81572411).
Availability of data and materials
The datasets used in the present study are available from The Cancer Genome Atlas database (https://cancergenome.nih.gov/).
Authors' contributions
JG and WS conceived and designed the study. JY was also involved in the design of the study and conducted the data analysis. LT and ZY performed the cell line verifications. KT and GW revised and improved the design of the study and participated in drafting the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric cancer: Descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23:700–713. doi: 10.1158/1055-9965.EPI-13-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yook SH, Oltvai ZN, Barabási AL. Functional and topological characterization of protein interaction networks. Proteomics. 2004;4:928–942. doi: 10.1002/pmic.200300636. [DOI] [PubMed] [Google Scholar]
- 5.Barabási AL, Oltvai ZN. Network biology: Understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 6.Røsok Ø, Sioud M. Discovery of differentially expressed genes: Technical considerations. Methods Mol Biol. 2007;360:115–129. doi: 10.1385/1-59745-165-7:115. [DOI] [PubMed] [Google Scholar]
- 7.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat Appl Genet Mol Biol. 2005;4:Article17. doi: 10.2202/1544-6115.1128. [DOI] [PubMed] [Google Scholar]
- 8.Tian F, Zhao J, Fan X, Kang Z. Weighted gene co-expression network analysis in identification of metastasis-related genes of lung squamous cell carcinoma based on the Cancer Genome Atlas database. J Thorac Dis. 2017;9:42–53. doi: 10.21037/jtd.2017.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Q, Zhao H, Zhang C, Hu T, Wu J, Lin X, Luo D, Wang C, Meng L, Xi L, et al. Gene co-expression network reveals shared modules predictive of stage and grade in serous ovarian cancers. Oncotarget. 2017;8:42983–42996. doi: 10.18632/oncotarget.17785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci USA. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller JA, Horvath S, Geschwind DH. Divergence of human and mouse brain transcriptome highlights Alzheimer disease pathways. Proc Natl Acad Sci USA. 2010;107:12698–12703. doi: 10.1073/pnas.0914257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langfelder P, Horvath S. WGC NA An R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 15.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 16.Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–1093. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han L, Diao L, Yu S, Xu X, Li J, Zhang R, Yang Y, Werner HMJ, Eterovic AK, Yuan Y, et al. The genomic landscape and clinical relevance of A-to-I RNA editing in human cancers. Cancer Cell. 2015;28:515–528. doi: 10.1016/j.ccell.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eke I, Cordes N. Focal adhesion signaling and therapy resistance in cancer. Semin Cancer Biol. 2015;31:65–75. doi: 10.1016/j.semcancer.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 21.Cortesio CL, Boateng LR, Piazza TM, Bennin DA, Huttenlocher A. Calpain-mediated proteolysis of paxillin negatively regulates focal adhesion dynamics and cell migration. J Biol Chem. 2011;286:9998–10006. doi: 10.1074/jbc.M110.187294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cichon MA, Radisky DC. Extracellular matrix as a contextual determinant of transforming growth factor-β signaling in epithelial-mesenchymal transition and in cancer. Cell Adh Migr. 2014;8:588–594. doi: 10.4161/19336918.2014.972788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J, Lee J, Kang W, Chang S, Shin EC, Choi C. TG F-β1 and hypoxia-dependent expression of MKP-1 leads tumor resistance to death receptor-mediated cell death. Cell Death Dis. 2013;4:e521. doi: 10.1038/cddis.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knappskog S, Lønning PE. P53 and its molecular basis to chemoresistance in breast cancer. Expert Opin Ther Targets. 2012;16((Suppl 1)):S23–S30. doi: 10.1517/14728222.2011.640322. [DOI] [PubMed] [Google Scholar]
- 25.Yang Y, Han L, Yuan Y, Li J, Hei N, Liang H. Gene co-expression network analysis reveals common system-level properties of prognostic genes across cancer types. Nat Commun. 2014;5:3231. doi: 10.1038/ncomms4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lichti-Kaiser K, ZeRuth G, Kang HS, Vasanth S, Jetten AM. Gli-similar proteins: Their mechanisms of action, physiological functions, and roles in disease. Vitam Horm. 2012;88:141–171. doi: 10.1016/B978-0-12-394622-5.00007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thirant C, Ignacimouttou C, Lopez CK, Diop M, Le Mouël L, Thiollier C, Siret A, Dessen P, Aid Z, Rivière J, et al. ETO2-GLIS2 hijacks transcriptional complexes to drive cellular identity and self-renewal in pediatric acute megakaryoblastic leukemia. Cancer Cell. 2017;31:452–465. doi: 10.1016/j.ccell.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Thirant C, Lopez C, Malinge S, Mercher T. Molecular pathways driven by ETO2-GLIS2 in aggressive pediatric leukemia. Mol Cell Oncol. 2017;4:e1345351. doi: 10.1080/23723556.2017.1345351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guerra E, Trerotola M, Aloisi AL, Tripaldi R, Vacca G, La Sorda R, Lattanzio R, Piantelli M, Alberti S. The Trop-2 signalling network in cancer growth. Oncogene. 2013;32:1594–1600. doi: 10.1038/onc.2012.151. [DOI] [PubMed] [Google Scholar]
- 30.Li F, Zhang J, Arfuso F, Chinnathambi A, Zayed ME, Alharbi SA, Kumar AP, Ahn KS, Sethi G. NF-κB in cancer therapy. Arch Toxicol. 2015;89:711–731. doi: 10.1007/s00204-015-1470-4. [DOI] [PubMed] [Google Scholar]
- 31.Sewify EM, Afifi OA, Mosad E, Zaki AH, El Gammal SA. Cyclin D1 amplification in multiple myeloma is associated with multidrug resistance expression. Clin Lymphoma Myeloma Leuk. 2014;14:215–222. doi: 10.1016/j.clml.2013.07.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used in the present study are available from The Cancer Genome Atlas database (https://cancergenome.nih.gov/).