Abstract
Recently, LOC285194 has shown a potential tumor-suppressor function in several types of human cancers, but its function in non-small cell lung cancer (NSCLC) remains unknown. This study intended to investigate the biological role of LOC285194 and its clinical significance in NSCLC. LOC285194 was detected by qRT-PCR, and its correlation with clinicopathological features of NSCLC was analyzed. The expression of LOC285194 was knocked down or ectopically expressed in lung cancer cells (A549 and H1299) and tumor cell growth, migration and invasion in vitro were investigated. In addition, the interaction of LOC285194 and target proteins was assessed by RNA pulldown and RNA immunoprecipitation in vitro. The results revealed that the expression of LOC285194 was significantly lower in tumor tissues when compared with the corresponding non-tumor tissues (P<0.001). Its expression was correlated with the tumor size (P=0.027). Kaplan-Meier analysis revealed that patients with lower LOC285194 expression had worse disease-free survival and overall survival rates (P<0.05). RNA protein interaction analysis revealed that p53 was the direct binding target of LOC285194 in NSCLC. Bioinformatics analyses suggested that depletion of LOC285149 could affect its antitumor function through the KRAS/BRAF/SMEK pathway. Our findings indicated that LOC285194 was a novel non-coding prognostic indicator and contributed to tumor suppression by targeting p53 in NSCLC, suggesting that it may be a non-coding target for NSCLC gene therapy.
Keywords: LOC285194, p53, lncRNA, NSCLC, tumor suppressor
Introduction
Lung cancer is the leading cause of cancer-related deaths worldwide. The five-year survival of patients diagnosed with NSCLC remains poor (1). Currently, researchers and physicians are committed to the development of molecular targets for cancer. With the increasing understanding of the molecular basis of lung cancer, some targeted treatments have been revealed to be effective in clinical practice (2). However, the targeted therapies for other common somatic mutations such as p53 and KRAS in lung cancer are still far from optimal. Therefore, the identification of the potential targets for lung cancer is crucial for the development of novel molecular-targeting agents.
Long non-coding RNAs (lncRNAs) are non-protein-coding transcripts longer than 200 nucleotides, which represent novel biomarkers for prognosis (3). The expression of lncRNAs is frequently dysregulated in cancer (4) and participates in the proliferation, survival, migration, and invasion of cancer cells by modulating transcriptional, post-transcriptional, and epigenetic molecular events. Studies have revealed that targeting cancer-associated lncRNAs can impair cancer cell growth and metastasis (5). Since lncRNAs are critical for the carcinogenic process, they could serve as novel therapeutic targets for cancer treatment (6).
p53 is one of the most common sites of genetic alterations of NSCLC (7). p53 mutations have been associated with lung cancer progression and treatment responses (8), however the molecular mechanism underlying their tumor-suppressing effects requires further studies (9). Recently, the tumor-suppressor gene p53 was found to be associated with lncRNAs (10). lncRNA LOC285194 has been revealed to inhibit the growth of tumor cells as a tumor suppressor in osteosarcoma (11). Liu et al (12) demonstrated that LOC285194 was a transcriptional target of p53, and that its ectopic expression functioned as an inhibitor of tumor cell growth in vitro and in vivo. Whether LOC285194 is associated with the loss of p53 tumor suppression in lung cancer has not been studied.
In the present study, we first focused on LOC285194 expression patterns in NSCLC tissues and cell lines as compared to that in normal tissues or cell lines. Further functional experiments revealed that LOC285194 may act as a tumor-suppressing non-coding RNA through its interaction with LOC285194 and p53 in NSCLC cells. Finally, based on examinations of the relationship between the expression levels of LOC285194 in tumor tissues and the clinicopathological features of clinical samples, LOC285194 may be a potential non-coding predictor for diagnosis.
Materials and methods
Analysis of TCGA data
RNA sequencing data and the accompanying clinical data were downloaded from the lung adenocarcinoma (LUAD) dataset of the TCGA sequencing project (13) and used for signaling pathway analysis. The survival data for LOC285194 and p53 were obtained from cBioPortal (http://www.cbioportal.org/) (14,15). Overall survival was analyzed by Kaplan-Meier curves and log-rank tests for all LUAD patients. P-values <0.05 were considered to indicate a statistically significant result.
Patients and tissue samples
A total of 56 paired NSCLC tissues and adjacent non-tumor tissues were obtained from the Affiliated Cancer Hospital and Institute of Guangzhou Medical University (Guangzhou, China) between January 2006 and January 2011. All specimens were resected by surgery, and a pathological examination confirmed the diagnosis of lung adenocarcinoma. Patients with tuberculosis, diabetes, pneumonia, and COPD were excluded from the study. None of patients had received preoperative chemotherapy or radiotherapy. The tissues were cut into specimens 0.5 cm × 0.5 cm in size and immediately placed in cryogenic vials with 1.5 ml RNAlater (Thermo Fisher Scientific, Inc., Waltham, MA, USA). NSCLC and normal tissues were stored at −80°C until total RNA was extracted. This study was performed in accordance with a protocol approved and reviewed by the Medical Ethics Committee of the Affiliated Cancer Hospital and Institute of Guangzhou Medical University.
Cell lines and culture conditions
Five human NSCLC adenocarcinoma cell lines (A549, H1299, PC9, H460 and Calu3) and a normal human bronchial epithelial cell line (16HBE) were obtained from American Type Culture Collection (ATTC; Manassas, VA, USA). All cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen GmbH, Karlsruhe, Germany), supplemented with 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 g/ml streptomycin, and left to grow at 37°C in a 5% CO2 atmosphere.
RNA Preparation, reverse transcription and quantitative real-time PCR
Total RNAs were extracted with TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Carlsbad, CA, USA), following the manufacturer's protocol. The reverse transcription reactions were performed by Reverse Transcriptase M-MLV (Takara Biotechnology Co., Ltd., Dalian, China), and the samples were incubated for 60 min at 42°C, 15 min at 72°C, and stored at −20°C. For real-time PCR, 1 µl of diluted RT products were mixed with 10 µl of SYBR® Premix Ex Taq™ (Takara Biotechnology Co., Ltd.), 0.5 µl of forward and reverse primers, and 4 µl of nuclease-free water for a final volume of 20 µl, according to the manufacturer's instructions. The primers used in the present study were 5′-TGTGCCTGTTTGACCTCTGA-3′ (forward) and 5′-AGGAAGGATAAAAGACCGACCA-3′ (reverse). The qPCR reactions were run on the ABI-7500 real-time PCR system using the following conditions: 95°C for 30 sec, followed by 45 cycles at 95°C for 3 sec and 60°C for 34 sec. The relative expression of LOC285194 was calculated using the comparative cycle threshold (CT) (2−∆∆CT), with small nucleolar RNA U6 as the internal control to normalize the data.
Plasmid constructs and transfection of NCSCL cells
The LOC285194 shRNA sequence was synthesized and cloned into pLVX-Puro (Clontech; Takara Biotechnology Co., Ltd.). The pLVX Puro and package plasmids were then transfected into 293T cells. Next, the 293T cell line generated the lentivirus particles, and a supernatant containing the virus was centrifuged to concentrate the virus. The virus was then used to transduce the cell lines of interest: A549 and H1299. The expression level of LOC285194 was assessed by qPCR.
Cell proliferation assays
Cells were digested using the common passage method to cause cell suspension, and stably-transfected plasmids and empty vector cells (3000 cells/well) were seeded in 96-well plates. When harvested, the cells were stained with methylthiazolyldiphenyl-tetrazolium bromide (MTT) and dissolved in dimethyl sulphoxide (DMSO). Cell growth was assessed every 12 h. All experiments were performed in triplicate.
Flow cytometric analysis of apoptosis
Stably-transfected A549 and H1299 cells and empty vectors were harvested by trypsinization. Following the standard FITC-Annexin V and propidium iodide (PI) double staining procedure, the cells were analyzed by flow cytometry (BD Accuri C6; BD Biosciences, San Jose, CA, USA). The percentage of apoptotic cells was assessed (GuavaSoft 3.2). All of the samples were assayed in triplicate.
Cell invasion assay
After 48 h transfection, 1×105 cells in serum-free media were placed into the upper chamber pre-coated with Matrigel (8.0 µm; BD Biosciences). The chambers were then incubated for 0, 24 and 48 h, respectively, in culture medium with 10% FBS in the lower chambers before cell counting. The cells on the upper surface were removed with a cotton swab, whereas the invaded cells on the lower surface were fixed with 4% paraformaldehyde and stained with 0.05% crystal violet. Finally, the invaded cells were counted using a inverted microscope (BDS200) in triplicate. The experiments were independently repeated in triplicate.
Wound healing assay
The cells were seeded in 6-well plates and cultured until 100% confluence. A scratch was performed using 200-µl pipette tips, and then the serum-free medium was replaced. The scratch healing process was observed at 0, 24 and 48 h after incubation.
RNA pull down and protein analysis of the LOC285194-associated protein
The lncRNA cDNA sequence was cloned to the pGMT vector, and the linear cDNA was transcribed in vitro using a MEGAscript T7 Transcription kit (cat. no. AM1333; Life Technologies; Thermo Fisher Scientific, Inc.), following the manufacturer's instructions. Protein products (8–10 µg/µl; 50–60 µg total) were separated with 1% SDS-PAGE on a 10% gel and subsequently transferred overnight onto a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA) using SDS transfer buffer (Bio-Rad Laboratories, Inc., Hercules, CA, USA). The membrane was blocked for 1 h by a western-blocking reagent (Bio-Rad Laboratories, Inc., Hercules, CA, USA) at room temperature prior to protein detection by specific monoclonal p53 antibody at 1:1,000 dilution (cat. no. ab131442; Abcam, Cambridge, MA, USA) overnight at 4°C. This was followed by incubation with a horseradish peroxidase-conjugated anti-mouse secondary antibody (1:1,000; cat. no. 6120-05; SouthernBiotech, Birmingham, AL, USA). The Amersham™ ECL™ Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences, Shanghai, China) was used to visualize the blots, following the manufacturer's protocol, with a 5–10 min exposure to SuperRX X-ray film (Fujifilm Investment Co., Ltd., Shanghai, China).
RNA immunoprecipitation
An EZ-Magna RIP™ kit (EMD Millipore, Bedford, MA, USA) was used (according to the manufacturer's instructions) to perform RNA-binding protein immunoprecipitation. The anti-p53 antibody co-precipitated RNAs were purchased from Abcam and the primers used for the detection of LOC285194 were: H-LOC285194-F forward, 5′-CCTGTGCCTGTTTGACCTCT-3′ and reverse, 5′-CTGGTTTGCAGTTTGGCCTC-3′; LOC285194 P2 forward, 5′-CCCTCTTGTAGAGCCACAGG-3′ and reverse, 5′-CGAACACTGGCATTCATTGAGGG-3′; LOC285194 P3 forward, 5′-CAGTTCCTCAAATTTGACCCC-3′ and reverse, 5′-TTTGAAGGTTTTCCACATGG-3′.
Western blot analysis
Briefly, the cells were washed with PBS and lysed. Protein products (8–10 µg/µl; 50–60 µg total) were separated using 10% SDS-PAGE and subsequently transferred overnight onto a polyvinylidene difluoride membranes (EMD Millipore). The membranes were blocked for 1 h with a Blotting-Grade Blocker (no. 1706404, Bio-Rad Laboratories, Inc.). The specific monoclonal p53 antibody (diluted 1:1,000; cat. no. ab1101; Abcam) was incubated overnight at 4°C, followed by incubation with a horseradish peroxidase-conjugated anti-mouse secondary antibody (1:1,000; cat. no. 6120-05; SouthernBiotech, Birmingham, AL, USA). Amersham™ ECL™ Prime Western Blotting Detection Reagent (GE Healthcare Life Sciences) was used to visualize the blots. The protein bands were exposed onto SuperRX X-ray film (Fujifilm Investment Co., Ltd.). Anti-GAPDH was used as a loading control (1:1,000; cat. no. ab9485; Abcam, Cambridge, UK).
Statistical analysis
All data were presented as the means ± standard error of the mean (SEM). The mean values of the two groups were compared using the Student's t-test. Differences between the groups were analyzed with a one-way analysis of variance (ANOVA). The survival data were compared using the Kaplan-Meier analysis and log-rank test. SPSS 19 software (IBM Corp., Armonk NY, USA) was used for statistical analysis.
Results
LOC285194 is downregulated in cancer cell lines and tissues
First, we aimed to investigate whether LOC285194 was detectable and aberrantly expressed in NSCLC and bronchial epithelial cell lines. Among the five NSCLC cell lines, the expression level of LOC285194 was lower in these selected NSCLC cell lines when compared with normal bronchial epithelial cells (HBE) (P<0.05; Fig. 1A). Furthermore, we examined the expression of LOC285194 in NSCLC cancer tissues and adjacent normal tissues. We detected that LOC285194 expression was significantly downregulated in both the lung adenocarcinoma and the squamous tumor tissues when compared to the adjacent normal tissues (P<0.001; Fig. 1B).
Figure 1.
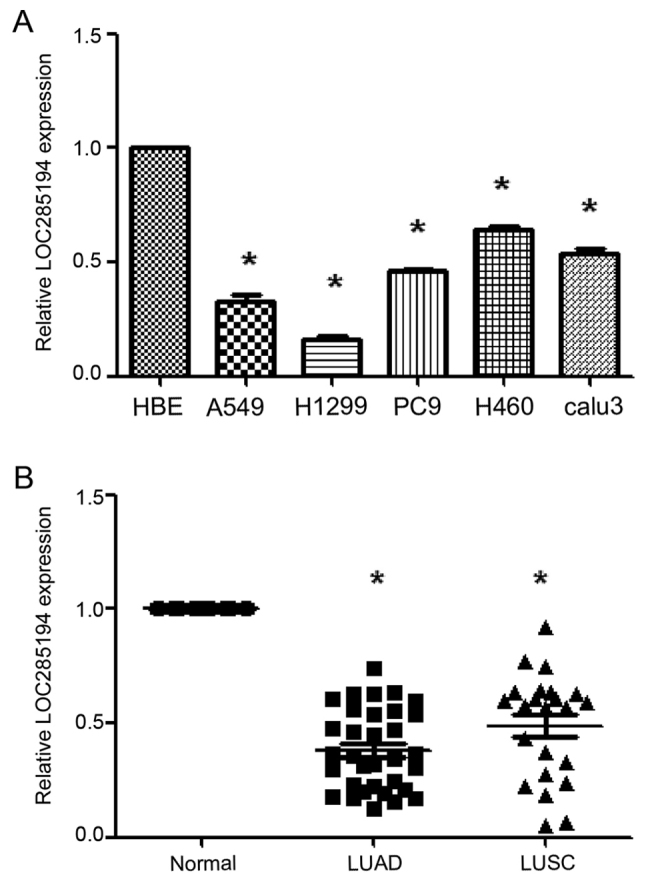
Quantitative real-time polymerase chain reaction analysis of LOC285194 expression in NSCLC. (A) LOC285194 expression levels were determined by qPCR in five lung cancer cell lines (A549, H1299, PC9, H460 and Calu3 cells) and normal bronchial epithelial cell line (HBE). Data are presented as the mean ± SEM (n=3). (B) LOC285194 was significantly downregulated in 56 NSCLC tissues when compared to the adjacent normal tissues. The ∆∆Ct method was used to measure the relative LOC285194 expression, which was normalized by the U6 expression level. *P<0.05 indicated a significant difference from the control.
LOC285194 expression and clinicopathological factors in NSCLC
The clinicopathological data of 56 patients are shown in Table I. We divided the 56 patients into low or high expression groups (average ∆∆Ct expression value of 0.44), with the median expression level of LOC285194 as a cutoff. As indicated in Table I, the low LOC285194 group was significantly associated with increased tumor size (P=0.027), but no significant association was found between LOC285194 expression and other clinicopathological data including sex, age, tumor location, histological subtype, lymph node metastasis, distant organ metastases, and TNM stage (P>0.05; Table I). Thus, our results indicated that the reduced expression of LOC285194 was correlated with tumor growth in NSCLC.
Table I.
LOC285194 expression and clinicopathological characteristics of 56 NSCLC patients.
| LOC285194 subgroupsa | |||
|---|---|---|---|
| Low | High | P-value | |
| Sex | 0.576 | ||
| Male | 19 | 18 | |
| Female | 10 | 9 | |
| Age | 0.202 | ||
| <60 | 14 | 17 | |
| ≥60 | 15 | 10 | |
| Location | 0.169 | ||
| LUL | 4 | 5 | |
| LLL | 3 | 9 | |
| RUL | 13 | 10 | |
| RML | 3 | 1 | |
| RLL | 6 | 2 | |
| Histology | 0.595 | ||
| LUAD | 16 | 15 | |
| LUSC | 13 | 12 | |
| Size | 0.027c | ||
| T1-T2 | 12 | 19 | |
| T3-T4 | 17 | 8 | |
| Lymph node | 0.469 | ||
| Negative | 18 | 18 | |
| Positive | 11 | 9 | |
| Metastasis | 0.535 | ||
| Negative | 26 | 25 | |
| Positive | 3 | 2 | |
| TNM stageb | 0.485 | ||
| I+ II | 16 | 16 | |
| III+ IV | 13 | 11 | |
Median expression level of LOC285194 was used as a cutoff to divide the 56 patients into LOC285194 low and high subgroups.
Tumor stage was defined according to the 7th TNM criteria.
P<0.05. LUL, left upper lobe; LLL, left lower lobe; RUL, right upper lobe; RML, right middle lobe; RLL, right lower lobe; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
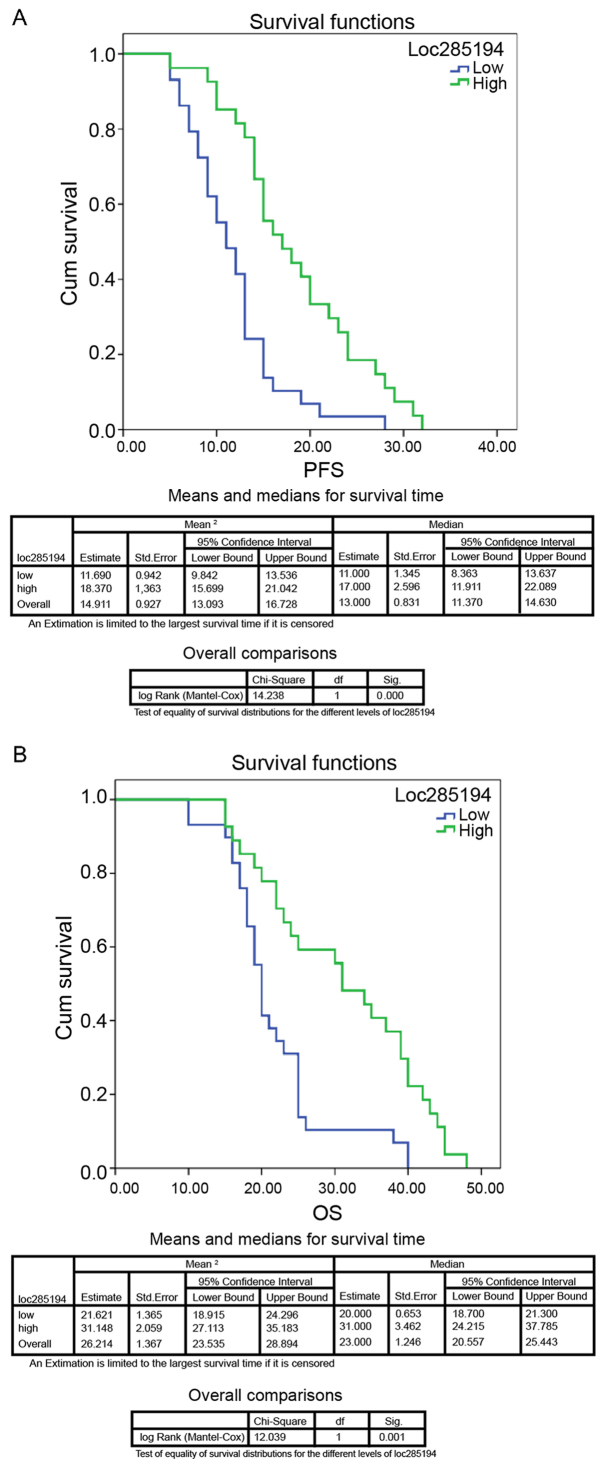
Kaplan-Meier analysis revealed that patients with lower LOC285194 expression levels had a significantly worse progression-free survival (PFS) (P<0.01; Fig. 2A) and overall survival (OS) (P<0.01; Fig. 2B) than those with high LOC285194 expression. Moreover, multivariate Cox proportional hazard regression analysis revealed that low LOC285194 expression was an independent prognostic factor in NSCLC patients (P<0.01; Table II). Additionally, the survival data from cBioPortal also revealed that the deletion of LOC285194 led to poor patient survival compared to patients with normal LOC285194 expression, similar to the loss of p53 in NSCLC patients compared with patients without a loss of p53 expression. These data inferred that LOC285194 expression was associated with prognosis after lobectomy and shared the prognostic pattern of p53 loss in NSCLC populations.
Figure 2.
Dowregulation of LOC285194 is associated with poor prognosis in NSCLC. (A and B) Kaplan-Meier PFS analysis revealed that patients with low expression of LOC285194 had a significantly lower PFS rate (P<0.001) and OS rate (P<0.001) compared with patients with a high level of LOC285194 expression.
Table II.
Multivariate Cox regression analysis of the 5-year progression-free survival and overall survival of 56 NSCLC patients.
| PFS | OS | |||||
|---|---|---|---|---|---|---|
| Variables | HR | 95% CI | P-value | HR | 95% CI | P-value |
| Age | ||||||
| (<60/≥60 years) | 0.177 | 0.085–0.368 | <0.001a | 0.241 | 0.118–0.490 | <0.001a |
| Sex | ||||||
| (Male/Female) | 0.427 | 0.201–0.908 | 0.027a | 0.681 | 0.334–1.388 | 0.291 |
| Location | ||||||
| (LUL/LLL/RUL/RML/RLL) | 1.138 | 0.887–1.458 | 0.309 | 1.250 | 0.968–1.614 | 0.087 |
| Histology | ||||||
| (LUAD/LUSC) | 1.427 | 0.116–17.499 | 0.781 | 3.253 | 0.251–42.207 | 0.367 |
| Size | ||||||
| (T1+T2/T3+T4) | 1.132 | 0.594–2.160 | 0.706 | 0.821 | 0.439–1.536 | 0.821 |
| Lymph node | ||||||
| (Absence/Presence) | 0.819 | 0.287–2.341 | 0.710 | 0.385 | 0.125–1.185 | 0.096 |
| Metastasis | ||||||
| (Absence/Presence) | 0.734 | 0.212–2.539 | 0.625 | 1.194 | 0.353–4.039 | 0.775 |
| Stage | ||||||
| (I+II/III+IV) | 6.535 | 0.521–82.002 | 0.146 | 5.304 | 0.375–74.950 | 0.217 |
| LOC285194 | ||||||
| (Low/High) | 0.153 | 0.073–0.318 | <0.001a | 0.199 | 0.094–0.423 | <0.001a |
P<0.05. HR, hazard ratio; CI, confidence interval.
LOC285194 inhibits proliferation and promotes apoptosis of lung cancer cells in vitro
To further study the effect of LOC285194 in NSCLC, lentiviral vectors of pLVX-LOC285194 and shLOC285194 were produced and then separately transfected into A549 and H1299 cells. The qPCR results confirmed that the expression level of LOC285194 was significantly knocked down or overexpressed in both cells transfected with shLOC285194 or pLVX-LOC285194 (Fig. 3A and B). We also found that the ectopic expression of LOC285194 inhibited the proliferation ability of both A549 and H1299 cells (Fig. 3D), while the knockdown of LOC285194 did not increase the proliferation ability of H1299 cells (Fig. 3C). Additionally, the flow cytometric analysis of A549 and H1299 cells revealed that overexpression of LOC285194 induced apoptosis in comparison with the control cells (Fig. 4A and B).
Figure 3.
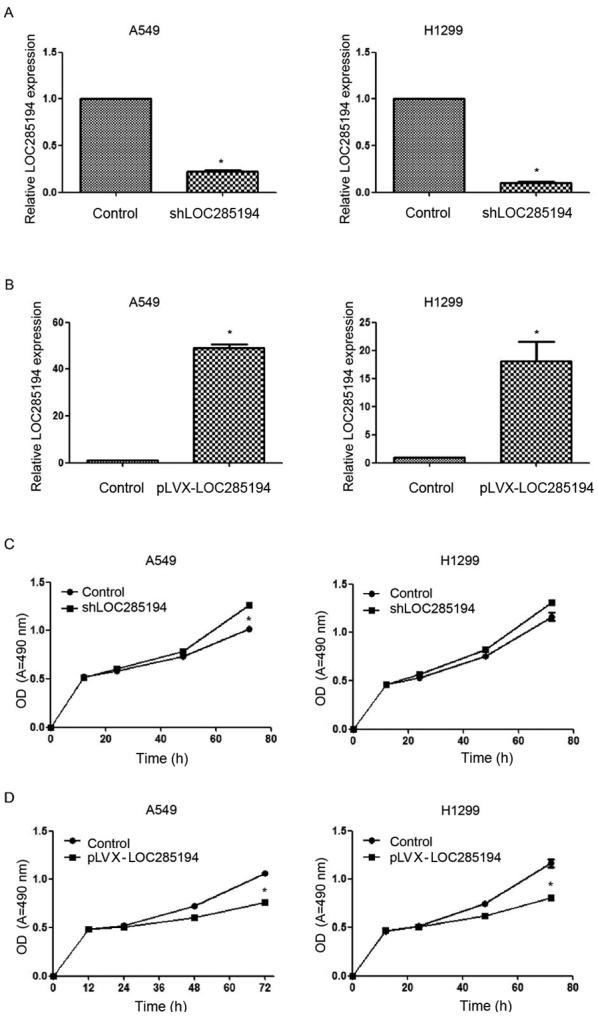
Overexpression of LOC285194 inhibits cell proliferation and induces apoptosis of NSCLC cells. (A and B) LOC285194 expression levels were evaluated by qPCR in pLVX-LOC285194- and shLOC285194-transfected A549 and H1299 cells. The expression level of LOC285194 was significantly knocked down in both A549 and H1299 cells transfected with shLOC285194 and overexpressed in both cell lines transfected with pLVX-LOC285194. (C) The effect of LOC285194 knockdown on the proliferation of A549 and H1299 cell lines was determined by MTT assay. (D) The effect of LOC285194 ectopic expressed on the proliferation of A549 and H1299 cell lines was determined by MTT assay. *P<0.05.
Figure 4.
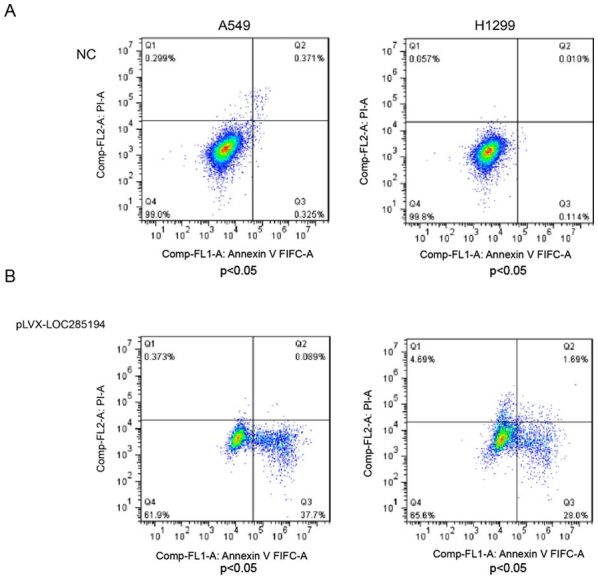
The apoptotic impact of A549 and H1299 cells stably overexpressing LOC285194. (A and B) The apoptotic impact of A549 and H1299 cells stably overexpressing LOC285194 was detected by flow cytometry. The flow cytometric analysis of A549 and H1299 cells revealed that overexpression of LOC285194 induced apoptosis in comparison with the control cells. Data were presented as the mean ± SD from three independent experiments.
Effect of LOC285194 on cell migration and invasiveness
To determine whether LOC285194 was involved in the migration of tumor cells and facilitated cell invasion in vitro, we evaluated cancer cell migration and invasion by wound healing assay and matrigel invasion assay. Knockdown of LOC285194 expression by shRNA increased A549 and H1299 cell migration and invasion (Fig. 5A and B). Ectopic expression of LOC285194 led to significantly decreased migration and invasion abilities of A549 cells. (P<0.05; Fig. 6A and B). Collectively, these results revealed that the alteration of LOC285194 expression could affect NSCLC cell migration and invasion abilities compared with the control group.
Figure 5.
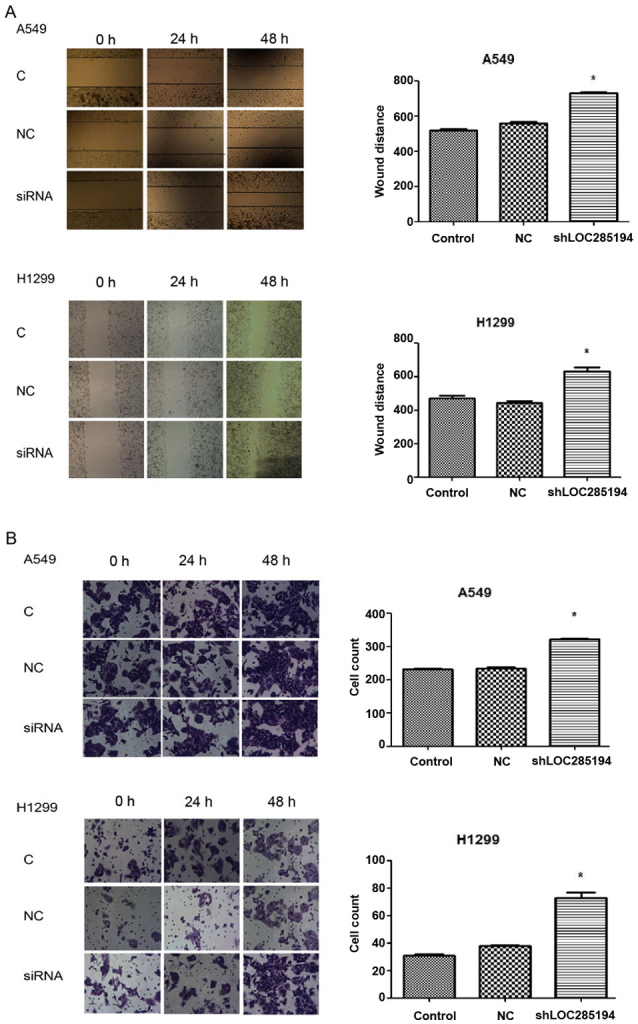
The migration and invasion effects of shLOC285194 on A549 and H1299 cells. (A) The migration capacity of A549 and H1299 cells was increased by shLOC285194 (*P<0.05) at 0, 24 and 48 h after the creation of wounds. (B) The invasion capacity of A549 and H1299 cells was increased by shLOC285194 (*P<0.05) at 0, 24 and 48 h.
Figure 6.
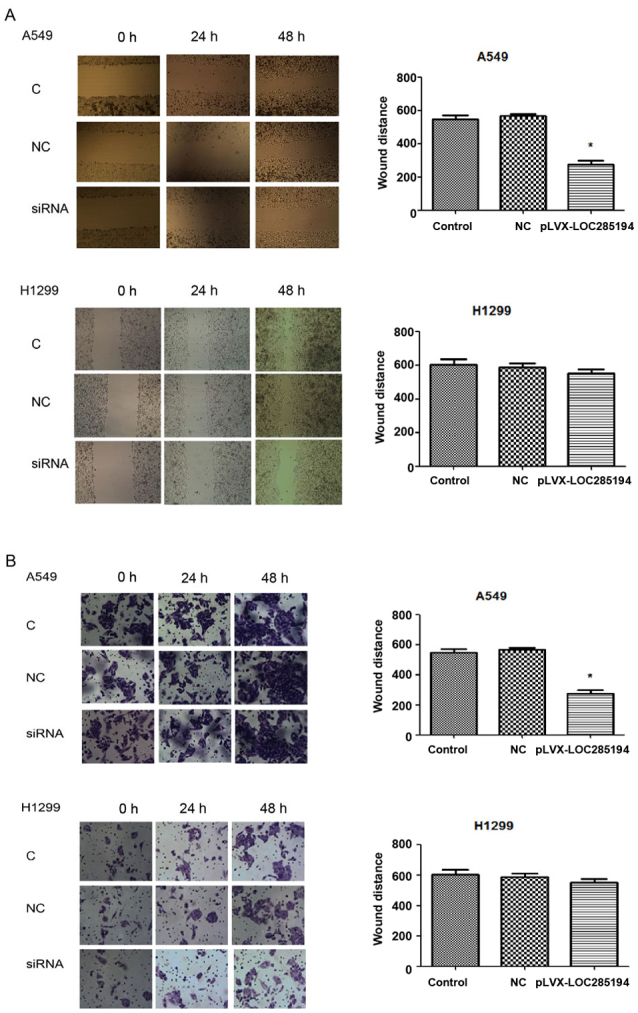
The migration and invasion effects of pLVX-LOC285194 on A549 and H1299 cells. (A) Τhe migration capacity of A549 and H1299 cells was decreased by pLVX-LOC285194 (*P<0.05) at 0, 24 and 48 h after the creation of wounds. (B) The invasion capacity of A549 and H1299 cells was decreased by pLVX-LOC285194 (*P<0.05) at 0, 24 and 48 h.
LOC285194 binds to the tumor supressor p53 protein
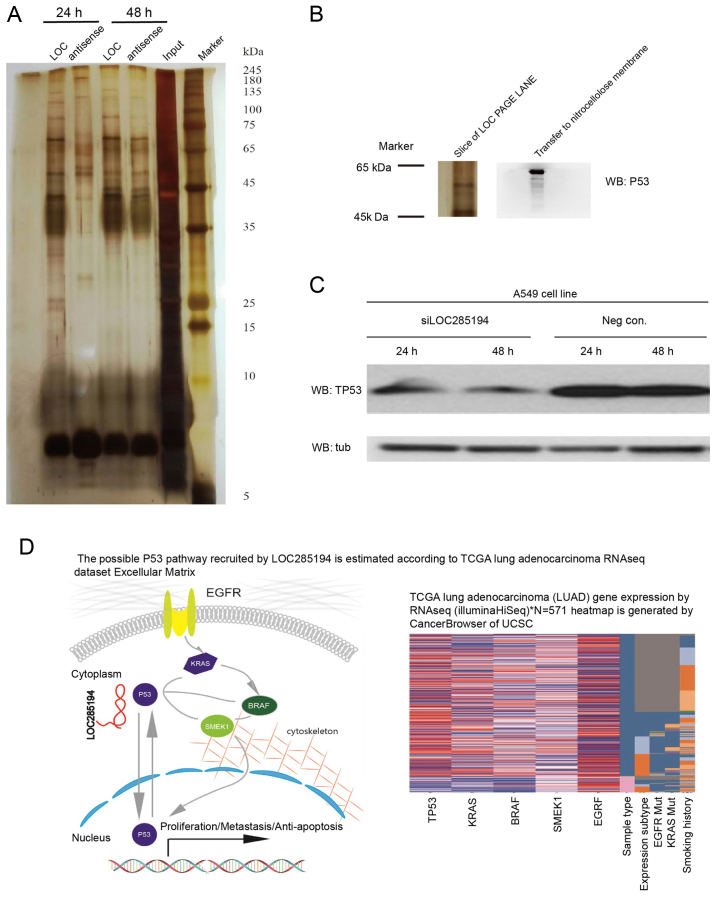
Since lncRNAs may perform their molecular functions by binding to specific proteins and regulate cell growth by modulating the p53 pathway (10), we hypothesized that LOC285194 may inhibit tumor growth and metastasis through this mechanism. The RNA pull-down experiment and western blot analysis were performed to identify proteins that are associated with LOC285194 (Fig. 7A). In brief, we resolved the RNA-enriched proteins on an SDS-PAGE gel, cut out the bands specific to p53, and subjected them to western blot analysis (Fig. 7B). We then detected p53 expression in LOC285194 siRNA-interfering NSCLC cells. The results revealed that the expression of p53 was low as shown in Fig. 7C. By analyzing the LUAD dataset from the TCGA sequencing project by UCSC cancer browser, we discovered that the p53 expression pattern was correlated with KRAS, BRAF and SMEK, but not EGFR and PI3K. These data confirmed that LOC285194 functions as a tumor suppressor non-coding RNA by interfering with EGFR/KRAS/BRAF signaling in NSCLC (Fig. 7D).
Figure 7.
LOC285194 binds to p53 and affects KRAS/BRAF/SMEK signaling. (A and B) Biotinylated LOC285194 or antisense RNA was incubated with total-cell extracts (A549 cells). The associated proteins were resolved in a gel. Western blot analysis of the slice of page at the p53 molecular weight confirms the interaction. (C) The expression of the p53 protein was detected by western blotting. (D) The possible p53 pathway recruited by LOC285194 is estimated by Cancer Browser of UCSC. TCGA lung adenocarcinoma (LUAD) gene expression by RNA-seq (Illumina HiSeq), N=571.
Discussion
Lung cancer is one of leading causes of cancer-related deaths worldwide, with very high incidence and mortality rates (16). Early detection, diagnosis, and treatment are vital to the management of lung cancer patients. The metastasis of distant organs contributes to the decline in the quality of life and to poor disease prognosis. Increasing evidence suggests that lncRNAs may be involved in the regulation of cancer invasion and metastasis. Some lncRNAs are more aberrantly expressed in cancer tissues than in normal tissues (17,18), and thus some of them can be used as novel molecular markers for tumor diagnosis and treatment. Other studies have revealed that lncRNAs play a key role in EGFR exon 19 deletions (19), drug resistance (20), and chemical carcinogenesis (21–24). Additionally, lncRNAs such as MALAT1 can be used as markers of lung cancer, but with low sensitivity (25).
lncRNA LOC285194, also known as antisense RNA LSAMP3, is a long non-coding RNA located at the 3q13.31 loci with a length of 2 kb. It has been revealed to be a tumor suppressor (11), inhibiting the growth of osteosarcoma cells. Liu et al (12) revealed that LOC285194 was a transcriptional target of p53, and that the ectopic expression of LOC285194 inhibited the growth of tumor cells in vitro and in vivo. Qi et al (26) determined that LOC285194 expression was downregulated in colon cancer specimens compared to normal tissues. Similar findings were found in pancreatic ductal adenocarcinoma (27).
In the present study, we first investigated the relationship between LOC285194 expression and the prognosis of NSCLC and found that the expression of LOC285194 was significantly lower in lung cancer tissues than in the adjacent lung tissues. Similar to previous studies, the lack of LOC285194 expression in patients with lung adenocarcinoma led to poor prognosis compared to patients with normal expression (28). In addition, the ectopic expression of LOC285194 significantly inhibited the growth, migration, and invasion abilities of lung cancer cells in vitro. Inversely, the knockdown of LOC285194 promoted cell growth, migration, and invasion. These results revealed that LOC285194 may play an important role in the progression of lung cancer.
The presence of TP53 tumor suppressor gene mutation occurs in 50–80% of human cancers. However, it remains unclear whether the lack of its related tumor suppressor lncRNA can also promote tumor occurrence and development. Recent studies reported that TP53 was involved in tumorigenesis and progression through interaction with lncRNAs. For example, Zhang et al (29) found that lncRNA TUG1 was the direct transcriptional target of p53 genes. Moreover, Han et al (30) confirmed that the lncRNA PANDAR was the direct transcriptional target of the p53 gene in NSCLC. Recently, the two binding sites of the HOTAIR promoter region were reported to be bound by p53, with p53 binding inhibiting the transcription of HOTAIR mRNA. When the p53 gene was overexpressed in A549 cells, the expression of lncRNA HOTAIR was downregulated and the cell proliferation rate and invasion abilities were decreased (31). In the present study, we revealed that LOC285194 can interact with p53 and interfere with lung cancer cell carcinogenesis. By analyzing the LUAD dataset of the TCGA sequencing project and the corresponding clinical data and incorporating the UCSC cancer browser for visualization, we discovered that the p53 expression pattern was correlated with KRAS, BRAF, and SMEK but not with EGFR and PI3K. Therefore, we proposed that in the NSCLC model, the loss of p53 by LOC285194 recruitment may impair antitumor function through the KRAS/BRAF/SMEK pathway.
The main difference between this study and previous research (12) is that we studied the function of LOC285194 on lung cancer cells, and confirmed the direct binding of LOC285194 and p53 through molecular biology experiments. The related research of this study demonstrated that LOC285194 could be used as a biomarker for prognosis (32).
Our results revealed that LOC285194 may be a tumor suppressor that may regulate p53. However, further experi-ments are warranted, such as the western blot analysis of the KRAS/BRAF/SMEK pathway downstream proteins. RNA-targeting drugs based on RNAi and lipid nanoparticles (LNPs) are under development and have been tested in humans (33). Restoring the function of the p53 protein by targeting lncRNAs could lead to a new method of treatment for malignant tumors.
To the best of our knowledge, no study has been previously published with respect to the association between LOC285194 expression levels and lung cancer prognosis. Our results revealed that LOC285194 expression was significantly decreased in NSCLC tissues and cell lines. Lower expression of LOC285194 was associated with poor prognosis. LOC285194 may play a role in suppressing the progression of lung cancer by recruiting p53. Thus, approaches for targeting LOC285194 to block NSCLC proliferation are needed for further investigation.
Acknowledgements
We would like to extend our sincere gratitude to Jianzhao and Wenfan Fu for the collection of lung cancer samples and to Xiaobing Le for investigating the bioinformatics analysis.
Glossary
Abbreviations
- lncRNA
long non-coding RNA
- RNA-seq
RNA sequencing
- TCGA
The Cancer Genome Atlas
- LUAD
lung adenocarcinoma
- NSCLC
non-small cell lung cancer
- p53
tumor protein p53
- qPCR
quantitative real-time polymerase chain reaction
- SDS-PAGE
sodium dodecyl sulfate/polyacrylamide gel electrophoresis
- OS
overall survival
- PFS
progression-free survival
Funding
The present study was supported by the Youth Foundation of Guangzhou Medical University (2015A21).
Availability of data and materials
The datasets used during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
JH designed and supervised the project. XS took part in the fund raising, experimental design, data acquisition, and article writing. HZh performed the majority of the experiments and drafted the manuscript. JL and JC designed and supervised the in vitro functional study. HZo, YZ and QD constructed the LOC285194 overexpressed and knocked down stable cell lines and performed the RNA pull-down and western blot experiments. MH confirmed the pathological diagnosis. HZo supervised the RNA pull-down and western blot experiments and also performed the bioinformatics analysis. AC supervised the results. KS performed the preliminary experiments. JS and XZ contributed in the design of surgical approaches and were major contributors in revising the manuscript. All authors were involved in the conception of the study, read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
All clinical data have been reviewed and approved by the Medical Ethics Committee of the Affiliated Cancer Hospital and Institute of Guangzhou Medical University (no. 2016-170).
Patient consent for publication
Not applicable.
Competing interests
The authors state that they have no competing interests.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Tan CS, Gilligan D, Pacey S. Treatment approaches for EGFR-inhibitor-resistant patients with non-small-cell lung cancer. Lancet Oncol. 2015;16:e447–e459. doi: 10.1016/S1470-2045(15)00246-6. [DOI] [PubMed] [Google Scholar]
- 3.Yarmishyn AA, Kurochkin IV. Long noncoding RNAs: A potential novel class of cancer biomarkers. Front Genet. 2015;6:145. doi: 10.3389/fgene.2015.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: Progress and prospects. Int J Biochem Cell Biol. 2013;45:1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 6.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Semenova EA, Nagel R, Berns A. Origins, genetic landscape, and emerging therapies of small cell lung cancer. Genes Dev. 2015;29:1447–1462. doi: 10.1101/gad.263145.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muller PA, Vousden KH. p53 mutations in cancer. Nat Cell Biol. 2013;15:2–8. doi: 10.1038/ncb2641. [DOI] [PubMed] [Google Scholar]
- 9.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14:359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaudhary R, Lal A. Long noncoding RNAs in the p53 network. Wiley Interdiscip Rev RNA. 2017;8:3. doi: 10.1002/wrna.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasic I, Shlien A, Durbin AD, Stavropoulos DJ, Baskin B, Ray PN, Novokmet A, Malkin D. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 2010;70:160–171. doi: 10.1158/1538-7445.AM10-3400. [DOI] [PubMed] [Google Scholar]
- 12.Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, Wu F, Mo YY. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahman M, Jackson LK, Johnson WE, Li DY, Bild AH, Piccolo SR. Alternative preprocessing of RNA-Sequencing data in The Cancer Genome Atlas leads to improved analysis results. Bioinformatics. 2015;31:3666–3672. doi: 10.1093/bioinformatics/btv377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 17.Yang J, Lin J, Liu T, Chen T, Pan S, Huang W, Li S. Analysis of lncRNA expression profiles in non-small cell lung cancers (NSCLC) and their clinical subtypes. Lung Cancer. 2014;85:110–115. doi: 10.1016/j.lungcan.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Xu G, Chen J, Pan Q, Huang K, Pan J, Zhang W, Chen J, Yu F, Zhou T, Wang Y. Long noncoding RNA expression profiles of lung adenocarcinoma ascertained by microarray analysis. PLoS One. 2014;9:e104044. doi: 10.1371/journal.pone.0104044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Chen W, Chen J, Pan Q, Pan J. LncRNA expression profiles of EGFR exon 19 deletions in lung adenocarcinoma ascertained by using microarray analysis. Med Oncol. 2014;31:137. doi: 10.1007/s12032-014-0137-y. [DOI] [PubMed] [Google Scholar]
- 20.Yang Y, Li H, Hou S, Hu B, Liu J, Wang J. The noncoding RNA expression profile and the effect of lncRNA AK126698 on cisplatin resistance in non-small-cell lung cancer cell. PLoS One. 2013;8:e65309. doi: 10.1371/journal.pone.0065309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao L, Mai A, Li X, Lai Y, Zheng J, Yang Q, Wu J, Nan A, Ye S, Jiang Y. LncRNA-DQ786227-mediated cell malignant transformation induced by benzo(a)pyrene. Toxicol Lett. 2013;223:205–210. doi: 10.1016/j.toxlet.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 22.Yang Q, Zhang S, Liu H, Wu J, Xu E, Peng B, Jiang Y. Oncogenic role of long noncoding RNA AF118081 in anti-benzo[a]pyrene-trans-7,8-dihydrodiol-9,10-epoxide-transformed 16HBE cells. Toxicol Lett. 2014;229:430–439. doi: 10.1016/j.toxlet.2014.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Thai P, Statt S, Chen CH, Liang E, Campbell C, Wu R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am J Respir Cell Mol Biol. 2013;49:204–211. doi: 10.1165/rcmb.2013-0159RC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaplan R, Luettich K, Heguy A, Hackett NR, Harvey BG, Crystal RG. Monoallelic up-regulation of the imprinted H19 gene in airway epithelium of phenotypically normal cigarette smokers. Cancer Res. 2003;63:1475–1482. [PubMed] [Google Scholar]
- 25.Weber DG, Johnen G, Casjens S, Bryk O, Pesch B, Jöckel KH, Kollmeier J, Brüning T. Evaluation of long noncoding RNA MALAT1 as a candidate blood-based biomarker for the diagnosis of non-small cell lung cancer. BMC Res Notes. 2013;6:518. doi: 10.1186/1756-0500-6-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C, Zhou XY, Du X. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. doi: 10.1186/1479-5876-11-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding YC, Yu W, Ma C, Wang Q, Huang CS, Huang T. Expression of long non-coding RNA LOC285194 and its prognostic significance in human pancreatic ductal adenocarcinoma. Int J Clin Exp Pathol. 2014;7:8065–8070. [PMC free article] [PubMed] [Google Scholar]
- 28.Collisson EA, Campbell JD, Brooks AN, Berger AH, Lee W, Chmielecki J, Beer DG, Cope L, Creighton CJ, Danilova L, et al. ; Cancer Genome Atlas Research Network: Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang EB, Yin DD, Sun M, Kong R, Liu XH, You LH, Han L, Xia R, Wang KM, Yang JS, et al. P53-regulated long non-coding RNA TUG1 affects cell proliferation in human non-small cell lung cancer, partly through epigenetically regulating HOXB7 expression. Cell Death Dis. 2014;5:e1243. doi: 10.1038/cddis.2014.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han L, Zhang EB, Yin DD, Kong R, Xu TP, Chen WM, Xia R, Shu YQ, De W. Low expression of long noncoding RNA PANDAR predicts a poor prognosis of non-small cell lung cancer and affects cell apoptosis by regulating Bcl-2. Cell Death Dis. 2015;6:e1665. doi: 10.1038/cddis.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhai N, Xia Y, Yin R, Liu J, Gao F. A negative regulation loop of long noncoding RNA HOTAIR and p53 in non-small-cell lung cancer. OncoTargets Ther. 2016;9:5713–5720. doi: 10.2147/OTT.S110219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi X, Chen Y, Chen AM, Le X, Huang K, Chen J, Wen S, Zeng H, Chen C, Li J. LncRNA TUSC7 affects malignant tumor prognosis by regulating protein ubiquitination: A genome-wide analysis from 10,237 pan-cancer patients. Transl Cancer Res. 2017;6:834–842. doi: 10.21037/tcr.2017.08.19. [DOI] [Google Scholar]
- 33.Zatsepin TS, Kotelevtsev YV, Koteliansky V. Lipid nanoparticles for targeted siRNA delivery - going from bench to bedside. Int J Nanomedicine. 2016;11:3077–3086. doi: 10.2147/IJN.S106625. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used during the present study are available from the corresponding author upon reasonable request.