Abstract
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors worldwide, and current treatments exhibit limited efficacy against advanced HCC. The majority of cancer-related deaths are caused by metastasis from the primary tumor, which indicates the importance of identifying clinical biomarkers for predicting metastasis and indicating prognosis. Patient-derived cells (PDCs) may be effective models for biomarker identification. In the present study, a wound healing assay was used to obtain 10 fast-migrated and 10 slow-migrated PDC cultures from 36 HCC samples. MicroRNA (miRNA) signatures in PDCs and PDC-derived exosomes were profiled by microRNA-sequencing. Differentially expressed miRNAs between the low- and fast-migrated groups were identified and further validated in 372 HCC profiles from The Cancer Genome Atlas (TCGA). Six exosomal miRNAs were identified to be differentially expressed between the two groups. In the fast-migrated group, five miRNAs (miR-140-3p, miR-30d-5p, miR-29b-3p, miR-130b-3p and miR-330-5p) were downregulated, and one miRNA (miR-296-3p) was upregulated compared with the slow-migrated group. Pathway analysis demonstrated that the target genes of the differentially expressed miRNAs were significantly enriched in the ‘focal adhesion’ pathway, which is consistent with the roles of these miRNAs in tumor metastasis. Three miRNAs, miR-30d, miR-140 and miR-29b, were significantly associated with patient survival. These findings indicated that these exosomal miRNAs may be candidate biomarkers for predicting HCC cell migration and prognosis and may guide the treatment of advanced HCC.
Keywords: exosome, microRNA, migration, prognosis, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is one of the leading causes of cancer-related mortality worldwide (1). Due to its insidious onset, lack of apparent symptoms in the early stage, and quick progression, HCC usually is advanced when diagnosed. The efficacy of traditional treatments, such as chemotherapy and radiotherapy, is not satisfactory, and emerging therapeutics, including targeted therapy and immunotherapy, have achieved limited success in the treatment of advanced HCC thus far (2). Cancer metastasis is the major cause of treatment failure (2), exerting a marked negative effect on the cure and survival of patients with HCC. Numerous studies have investigated metastasis-associated markers as potential prognostic indicators and candidate therapeutic targets. As one of the most well-studied microRNAs (miRNAs/miRs) in HCC, the overexpression of miR-21 was previously reported to promote cell proliferation, metastasis and invasion, and to be an indicator of poor prognosis (3). Knockdown of miR-21 was also demonstrated to significantly inhibit cancer cell migration in vitro (4).
Exosomes are small membrane vesicles (50–150 nm diameter) that are released by most cell types and are present in various body fluids, including plasma, urine, saliva, breast milk and malignant effusions. Exosomes are able to envelop proteins and miRNAs within their double-membrane structure and transfer contents from donor cells to recipient cells (5,6). The release of exosomes from primary tumors into the circulatory system has been demonstrated in various models (7), and numerous studies have reported that exosomal miRNAs can contribute to cancer progression and metastasis (8). Accumulating research is investigating the use of exosome contents as biomarkers for patient diagnosis, treatment and drug resistance.
Exosomes are also present in the supernatant of cultured cells, including tumor cells (9). Despite the ubiquitous use of commercial cancer cell lines, it is often questioned whether these cell lines are able to model the biological processes of tumors effectively, as commercial cell lines tend to lose their original tumor characteristics due to repeated passaging (10). Additionally, it has been previously demonstrated that patient-derived cells (PDCs) inherit the complexity and genetic diversity of original tumors, as they are directly derived from fresh tumor tissues (11). Therefore, PDCs were selected for use in the present study, as they may be of a superior representative value compared with conventional cell line models. To the best of our knowledge, this is the first use of PDCs for investigation of exosomal miRNAs in HCC.
miRNA expression profiling has proven useful in diagnosing and monitoring the development and progression of tumors. Reverse transcription-quantitative polymerase chain reaction (PCR) and microarrays are the classical methods for miRNA expression analysis; however, these only detect a limited number of known miRNAs. In recent years, with the rapid development of next-generation sequencing, miRNA sequencing (miRNA-Seq) has offered increased specificity and sensitivity in miRNA profiling (12); in particular, it is able to identify novel miRNAs, which enables rapid profiling and further investigation of miRNAs.
Given the current limited efficacy of treatments for advanced HCC, biomarkers for liver metastasis as potential prognostic indicators are worth extensive investigation. Although current research is investigating the applications of exosomes as biomarkers in the detection, diagnosis and treatment monitoring in various cancers, little has been done to evaluate metastasis-associated exosomal biomarkers in HCC. Therefore, the aim of the present study was to identify differentially expressed exosomal miRNAs in PDCs grouped by the migration rate, explore the pathway enrichment of miRNA-targeted genes and verify the association between the expression level of miRNAs and patient survival using data from The Cancer Genome Atlas (TCGA). To the best of our knowledge, the present study is the first to investigate metastasis-related exosomal miRNA biomarkers using HCC PDC models.
Materials and methods
PDC culture
HCC tissues were collected from the Eastern Hepatobiliary Surgery Hospital (Shanghai, China) with informed consent obtained from the 36 patients for PDC culture and further research from August 2013 to June 2015. These patients with a median age of 49 years (range, 37–74) consisted of 30 males and 6 females (Table I). PDCs were established and cultured by 3D Medicine Inc. (Shanghai, China) using standard procedures. In brief, fresh tissues from partial tumor of HCC patients were washed with Dulbeccos modified Eagles medium (DMEM)/F12 medium (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) to remove excess blood. The tissues were minced into 2-mm pieces and incubated with DMEM/F12 medium supplemented with 5% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.). When the cells reached 80% confluence, they were trypsinized and prepared for subculture. Subsequently, the medium was changed every 3 days.
Table I.
Clinical information of the patients with hepatocellular carcinoma.
| Clinical information | All patients (n=36) | Fast-migrating group (n=10) | Low-migrating group (n=10) | P-value |
|---|---|---|---|---|
| Sex | ||||
| Male | 30 | 8 | 9 | 1.000 |
| Female | 6 | 2 | 1 | |
| Age (years) | ||||
| <40 | 5 | 3 | 1 | 0.582 |
| >40 | 31 | 7 | 9 | |
| Hepatitis B surface antigen | ||||
| Positive | 32 | 9 | 9 | 1.000 |
| Negative | 4 | 1 | 1 | |
| Histological grade | ||||
| Well/moderately differentiated | 0 | 0 | 0 | 1.000 |
| Poorly differentiated | 35 | 10 | 10 | |
| Undifferentiated | 1 | 0 | 0 | |
| Number of lesions | ||||
| Solitary | 12 | 5 | 1 | 0.266 |
| Multiple | 9 | 3 | 4 | |
| NA | 15 | 2 | 5 | |
| Microvascular invasion | ||||
| M0 | 5 | 1 | 2 | 0.915 |
| M1 | 8 | 1 | 3 | |
| M2 | 8 | 1 | 4 | |
| NA | 15 | 7 | 1 | |
NA, not available.
Wound healing assay
Cells in the logarithmic growth phase were seeded on 6-well plates (5×105 cells/well) and incubated for 24 h to obtain a 90% confluent monolayer. The wound healing assay was performed by creating scratches using 200-µl pipette tips, followed by gentle washing three times with phosphate-buffered saline (PBS). Cells were cultivated in serum-free medium for 24 h, and the images of the wound gaps were captured at 0, 6 and 24 h after the wound was made. Wound healing ability was determined by measuring the change in the scraped area using Image-Pro Plus 6.0 software (Media Cybernetics, Inc., Rockville, MD, USA). Each experiment was repeated three times.
Exosome isolation
Exosomes were collected from PDCs during passages 10–15. Briefly, when the cells reached 80% confluence, the culture medium was replaced with fresh serum-free DMEM/F12, and the cells were cultured for an additional 48 h. Subsequently, the supernatants were centrifuged at 1,000 × g for 5 min and 4,000 × g for 5 min to remove cellular debris, and then collected in 50-ml tubes for storage at −80°C. Exosomes were isolated using a 3D Medicine exosome isolation kit (CFDA license no. Hu min xie bei 20170019). Briefly, supernatants were brought to room temperature, passed through a 0.45-µm filter, and then a 0.22-µm filter. 3D-TC reagent was added to the supernatants at a 1:2 ratio and mixed by inverting the tubes several times. The mixture was incubated overnight at 4°C, and centrifuged at 4,700 × g for 30 min at 4°C to obtain the precipitated exosomes. The isolated exosomes were resuspended in 200 µl PBS.
Transmission electron microscopy
Exosomes were fixed with 2% glutaraldehyde (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany). One drop of the sample was loaded onto Formvar® coated copper grids (Agar Scientific, Ltd., Stansted, UK), and was dried at room temperature for 10 min. The grids were washed with ultrapure water three times and negatively stained with 2.5% uranyl acetate (pH 7.0; SPI-Chem; Structure Probe, Inc., West Chester, PA, USA), followed by methyl cellulose/uranyl acetate (pH 4.0; Sigma-Aldrich; Merck KGaA). Exosome particles were visualized using an H-600 electron microscope (Hitachi, Ltd., Tokyo, Japan) at 100 kV accelerating voltage.
Western blot analysis
Cellular protein was extracted from PDCs using radioimmunoprecipitation assay lysis buffer (Thermo Fisher Scientific, Inc.). Exosomal protein was extracted from exosomes using 3D Medicine protein lysis buffer. The protein lysate was then centrifuged at 4°C for 15 min, and the supernatant was quantified using a BCA kit (Pierce; Thermo Fisher Scientific, Inc.). Protein (70 µg/well) was electrophoresed by SDS-PAGE on 10% gels and transferred to a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA, USA). The membranes were blocked using 3% bovine serum albumin (BSA; Thermo Fisher Scientific, Inc.) at 4°C for 12 h and then incubated with CD9 antibody (diluted at 1:1,000; cat. no. sc-13118; Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Horseradish peroxidase-conjugated anti-mouse IgG was used as the secondary antibody (diluted at 1:5,000; cat. no. sc-2380; Santa Cruz Biotechnology, Inc.). Bands were visualized using enhanced chemiluminescence (Thermo Fisher Scientific, Inc.). Comparison of the grey signal intensity bands on the western blots were performed using software Adobe Photoshop CS6. Each experiment was repeated three times.
Exosomal and cellular RNA isolation
RNA from exosomes was isolated using miRNeasy Serum/Plasma kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturers protocol. Briefly, 700 µl QIAzol was added to 200 µl exosomes, vortexed and incubated. Then 90 µl chloroform was added to the mixture for further vortexing and incubation, and the mixture was centrifuged at 4°C for 15 min. The upper aqueous phase was transferred to a new tube, and two volumes of 100% ethanol were added. Following vortexing, the solution was loaded to a spin column and centrifuged at 8,000 × g for 15 sec. The flow-through was discarded, and the spin column was rinsed twice with wash buffer, air-dried and the recovered exosomal RNA was eluted using 15 µl RNase-free water. Total RNA from PDCs was isolated using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturers protocol. RNA quality and concentration were analyzed using the NanoDrop ND-100 Spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
Small RNA library construction and sequencing
Small RNA library preparation for high throughput sequencing was performed using the protocols and reagents from the NEBNext Small RNA Library Prep Set for Illumina (New England BioLabs, Inc., Ipswich, MA, USA). The small RNA library prepared from PDCs was fractionated by DNA length on a 2% TAE agarose gel for the excision of the 150-bp band. The small RNA library was purified using NucleoSpin Gel and PCR Clean-Up kit (Qiagen, Shanghai, China). The purified library was analyzed for purity using an Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., Santa Clara, CA, USA) and then subjected to single-ended strand-specific sequencing on the Illumina HiSeq X10 platform (Illumina, Inc., San Diego, CA, USA).
Sequence mapping and small RNA identification
Following trimming of adaptor sequences, reads were mapped to hg19 using the Burrows-Wheeler Aligner 0.7.12-r1039 and a number of reads mapped to miRNAs from miRBase v21 were calculated. The expression levels were normalized using DESeq2 version 1.20.0. The principal component analysis (PCA) and the contribution of each gene to the dominant direction were performed as described by Huang et al (13). miRNAs that met all the following conditions were considered differentially expressed: i) P<0.05 between two groups; ii) the absolute value of contribution was >0.03; and iii) the average expression level of groups with higher mean expression was >100 reads per kilobase per million mapped reads (RPKM).
Statistical analysis
The data were presented as the mean ± SD of three independent experiments. The different migration rates between the fast and slow groups were assessed by Students t-test. For differentially expressed miRNAs, the Students t-test was performed to calculate the P-value. Pearsons correlation analysis of miRNA expression between PDCs and their exosomes was performed. The target genes of differentially expressed miRNAs were analyzed using miRTarBase (http://mirtarbase.mbc.nctu.edu.tw) instead of frequently used computational tools, such as TargetScan, miRanda, PicTar and DIANA-microT. The overall survival (OS) in patients with HCC was analyzed using the Kaplan-Meier method and log-rank test. Fishers test and Chi-square test were used to analyze the clinical characteristic differences between the fast and slow groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Characterization of PDCs
HCC tissues obtained from surgical resection were cultured to establish PDCs. All cell lines were free of contamination by bacteria or mycoplasma. Short tandem repeat (STR) analysis revealed that all PDCs were derived from the corresponding tissue samples and the STR loci profiles were all unique when compared with each other (data not shown). The growth rate was estimated by the passage time, and the PDCs with a passage time of >20 days were excluded. Considering the potential similarity of PDCs derived from the same patient, all selected PDCs were derived from different patients. In total, 36 PDC cultures were selected for further use in migration assays.
The clinical information of the patients is presented in Table I. The 21 PDCs with a passage time of ≤7 days were classified as having a fast growth rate, and the 15 PDCs with a passage time of >7 days were classified as having a slow growth rate. Hepatitis B virus (HBV) DNA integration in PDCs was examined by polymerase chain reaction (PCR) amplification of the virus gene, and 20 PDCs tested positive, while 5 tested negative for HBV. Serological analysis for HBV antigen and antibody of the derived patient was also performed. HBV surface antigen was detected in 32 patients, indicating a chronic HBV infection in 89% of patients. Edmondson-Steiner grading indicated poor differentiation in the majority of the patients. Quantity and distribution of microvascular invasion (MVI) was evaluated and classified as M0-MVI for no visible MVI, M1-MVI for low-risk MVI, and M2-MVI for high-risk MVI. Other clinical information, including TNM stage and prognosis, was unavailable.
Group enrollment of PDCs by migration rate
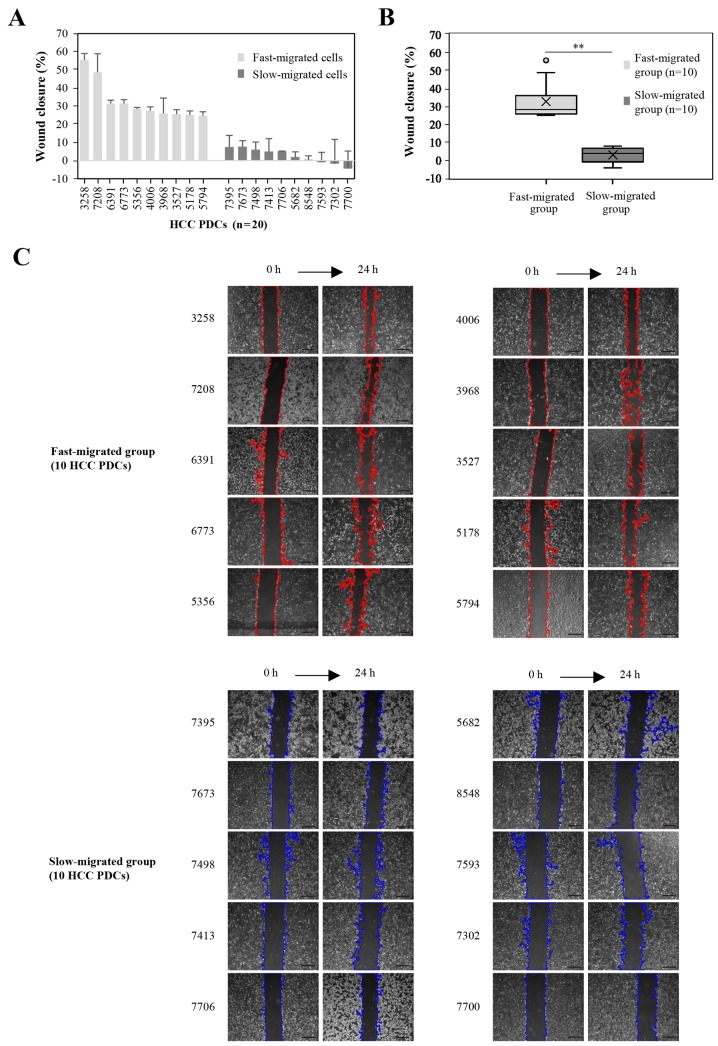
To identify miRNAs that may play a role in the metastasis of liver tumors, cell migration rates were determined using an in vitro wound healing assay. The migration rate was quantified as the wound closure percentage at 24 h after the scratch wound was made. Among the 36 PDCs, the top 10 and bottom 10 PDCs according to the migration rate were designated as the fast- and slow-migrated groups, respectively. No association was observed between PDC migration rate and patient clinical characteristics, such as the growth rate and HBV infection status in the patients. The wound closure percentages were all ≥24.8% in the fast-migrated group, and ≤7.5% in the slow-migrated group. Statistically, the mean and median values were 32.5 and 28.0% in the fast-migrated group, and 2.7 and 3.6% in the slow-migrated group (Fig. 1), respectively, with a significant difference in wound closure between the groups (P=1.8×10−7). In order to exclude the influence of the proliferation differences of HCC PDCs on the cellular migration, the 20 HCC PDCs in the fast-migrated group and the slow-migrated group were resuscitated and detected in serum-free medium. When cells were cultivated in serum-free medium for 24 h, the results revealed that the proliferation rates in the fast-migrated group and the slow-migrated group were not significant (data not shown).
Figure 1.
Migration rate between the fast- and slow-migrated PDC groups. Among the 36 PDC samples, the wound closure percentage of the top 10 and bottom 10 PDCs according to migration rate are presented as (A) individuals and (B) groups. (C) Representative images of wound-healing assays at 0 and 24 h. Upper, top 10 PDCs in migration rate (the edges of the wound-healing scratches are marked red). Lower, bottom 10 PDCs in migration rate (the edges of wound-healing scratches are marked blue). Data represent at least three independent experiments. **P<0.01. Scale bar, 100 µm. PDCs, patient-derived cells. The numbers in (A) are representative of the different PDCs.
Identification of PDC-derived exosomes
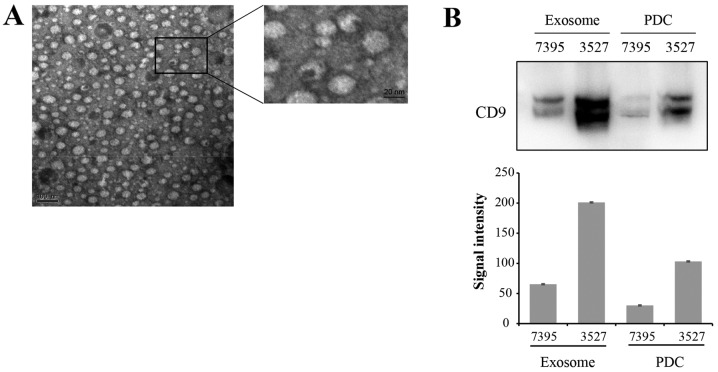
Exosomes were isolated from the supernatant of PDCs. Fig. 2A presents PDC-derived exosomes visualized by electron microscopy. Exosomes appeared as typical spherical vesicles ranging from 50–150 nm in diameter. Protein expression of known exosome marker CD9 was confirmed by western blot analysis in PDC-derived exosomes and in the PDCs (Fig. 2B upper image). The gray signal intensities of CD9 were presented in Fig. 2B (lower image). The CD9 blot produced two protein bands, with the upper band presumably due to glycosylation modification.
Figure 2.
Characterization of PDC-derived exosomes. (A) Representative electron micrograph of PDC-derived exosomes revealed a typical size distribution of 50–150 nm in diameter. Scale bars, 100 nm and 20 nm. (B) Western blot (upper image) and the gray signal intensities (lower image) of exosome marker CD9 in PDCs and PDC-derived exosomes were shown. PDCs, patient-derived cells.
Composition of cellular and exosomal small RNAs
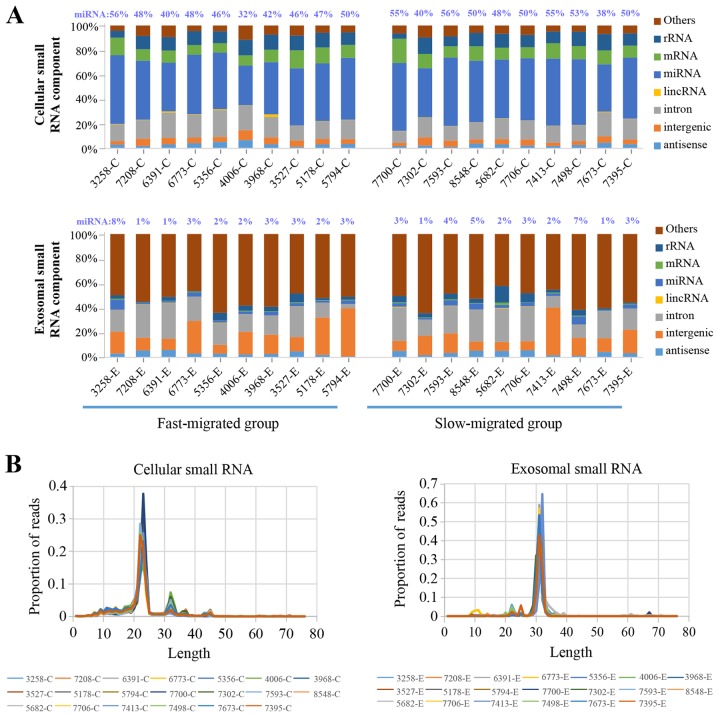
Deep sequencing of small RNA generated an average of 13 million and 9 million total reads for PDC and PDC-derived exosome samples, respectively. Of the total reads, >97% were successfully mapped to the reference genome and the sequencing data was adequate for further analysis.
In PDC samples, miRNA was the most abundant class of small RNAs, representing 47.5% of the entire cellular small RNA component. The percentage of miRNAs among all small RNAs varied from 32.3 to 55.9% from sample to sample, with no significant difference between the fast- and slow-migrated groups (Fig. 3A). By contrast, in exosome samples miRNAs accounted for only 3.0% of all small RNA molecules. Similarly, no significant difference in the exosome miRNA proportion was observed between the two groups. The most abundant class of small RNAs present in the exosomes was unannotated small RNA (Fig. 3A).
Figure 3.
Composition of cellular and exosomal small RNAs. (A) miRNAs represent 47.5 and 3.0% of the cellular and exosomal small RNA repertoires, respectively. (B) Read length distribution produced a predominant peak at 23 and 32 nucleotides in cellular and exosomal small RNAs, respectively. Small RNA is a class of non-coding RNA molecules with a length of ~20–30 nucleotides. miRNA, microRNA.
Considering the marked difference in the composition of the small RNA population between cellular and exosomal samples, the read length distribution of the samples was investigated. The PDC samples produced a predominant peak at 23 nucleotides and an additional peak at 32 nucleotides, while exosomes had a major peak at 32 nucleotides, and tiny peaks from 22–25 nucleotides (Fig. 3B). miRNAs are 21–25 nucleotides, which corresponded to the major peak in cellular small RNAs and the minor peaks in the exosomal components. The distribution of read length corresponded to the percentile of miRNAs present in cellular and exosomal RNA samples.
Differentially expressed exosomal miRNAs are associated with migration rate
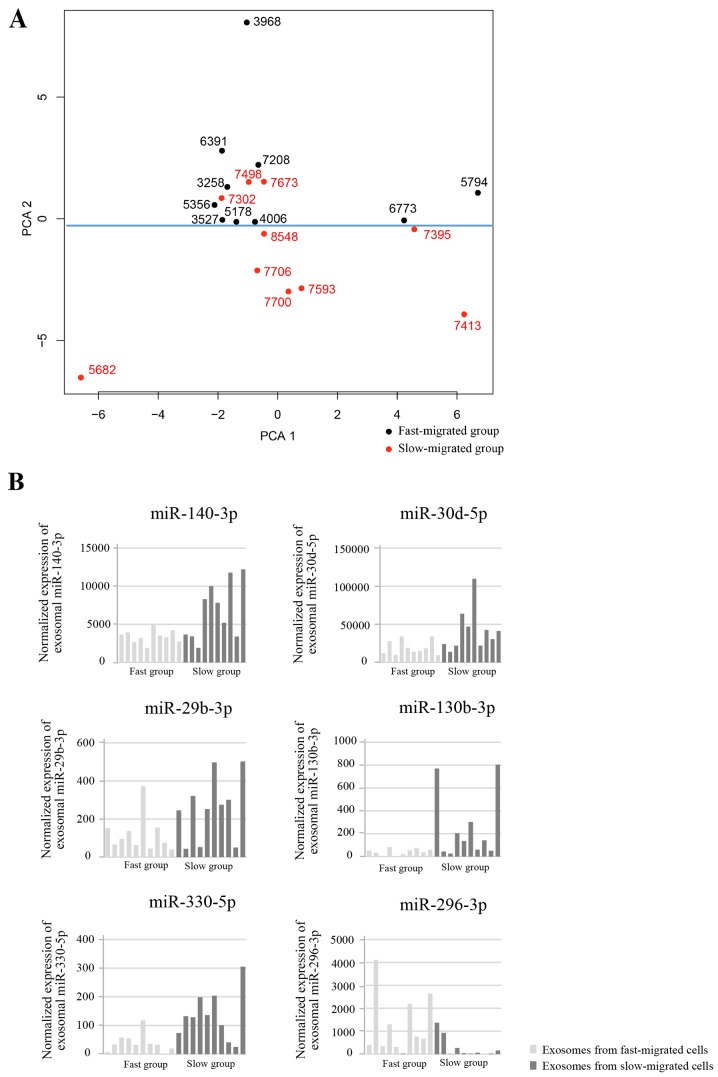
Next it was also investigated whether specific miRNAs were upregulated or downregulated in the fast-migrated group compared with the slow-migrated group. PCA was performed and the contribution of each gene to the dominant direction was calculated. The unsupervised PCA clustered the samples into two groups (separated by the blue line in Fig. 4A), which generally accorded with the migration rate of each sample. To focus on the miRNAs with relatively high expression, miRNAs with mean expressions of <100 RPKM were removed. Each miRNA with a Students t-test result of P<0.05 between groups, and an absolute value of contribution of >0.03 was considered to be differentially expressed. Six exosomal miRNAs were differentially expressed between the fast- and slow-migrated groups. In the fast-migrated group, five miRNAs (miR-140-3p, miR-30d-5p, miR-29b-3p, miR-130b-3p and miR-330-5p) were downregulated, and one miRNA (miR-296-3p) was upregulated (Fig. 4B). The detailed parameters for the six miRNAs are listed in Table II.
Figure 4.
Differentially expressed exosomal miRs between the fast- and slow-migrated groups. (A) Unsupervised principal component analysis clustered the samples into two groups (separated by the blue line). Ten samples with a fast migration rate and 7 out of 10 samples with a slow migration rate were naturally clustered into two groups. (B) Six exosomal miRs were differentially expressed between fast- and slow-migrated groups. In the fast-migrated group, five of them were downregulated (miR-140-3p, miR-30d-5p, miR-29b-3p, miR-130b-3p and miR-330-5p) and one (miR-296-3p) was upregulated compared with the low-migrated group. miR, microRNA.
Table II.
Screened parameters for the six differentially expressed exosomal microRNAs.
| miRNA | PC2 value | Average fast | Average slow | P-value (t-test) |
|---|---|---|---|---|
| hsa-miR-140-3p | −0.058 | 3413 | 6781 | 0.02 |
| hsa-miR-30d-5p | −0.042 | 19531 | 41768 | 0.037 |
| hsa-miR-29b-3p | −0.037 | 121 | 254 | 0.047 |
| hsa-miR-130b-3p | −0.033 | 41 | 254 | 0.048 |
| hsa-miR-330-5p | −0.032 | 39 | 135 | 0.006 |
| hsa-miR-296-3p | 0.044 | 1276 | 287 | 0.046 |
Pathway analysis
To reveal the potential roles of the differentially expressed miRNAs in HCC, their target genes were analyzed using miRTarBase (http://mirtarbase.mbc.nctu.edu.tw). The prediction principle for computational tools predominantly depends on the complementarity between miRNA seed region and the 3-untranslated region of their target genes. The majority of miRNAs imperfectly bind to the complementary sites, thus various computational methods have been generated for miRNA target prediction, and the resulting lists of candidate target genes from different algorithms often do not overlap. However, miRTarBase only includes experimentally validated miRNA-target interactions, which provides more reliable data for subsequent pathway analysis. To further reveal the potential roles of these miRNAs, the miRNAs and their target genes were analyzed using the miRTarBase database. In summary, 1652 non-repetitive genes were potential targets of the six differentially expressed miRNAs. Pathway enrichment analysis was performed using DAVID 6.8 (https://david.ncifcrf.gov/) for Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome analysis.
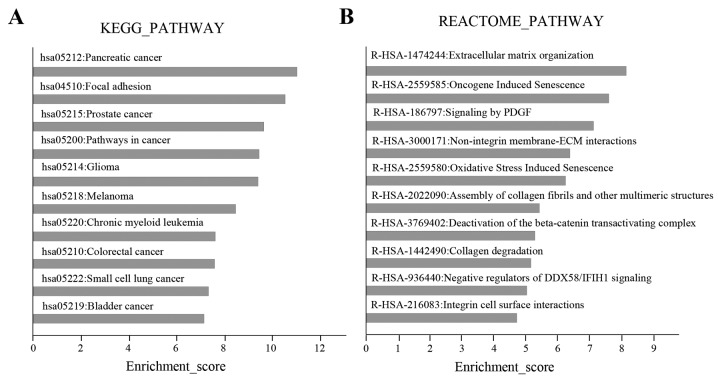
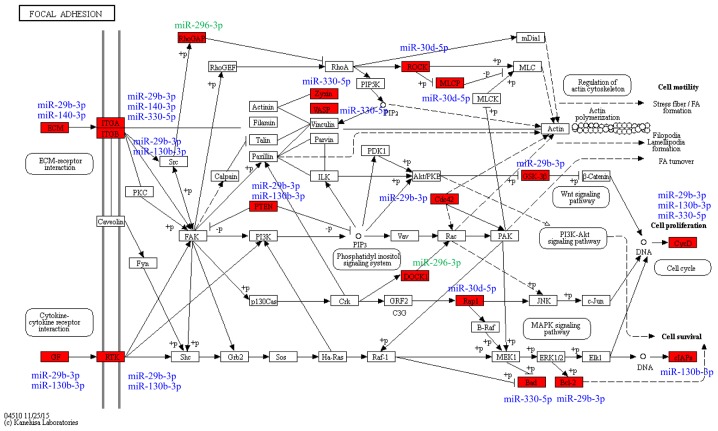
In KEGG pathway analysis, the top 10 pathways were predominantly cancer-associated pathways, including ‘pancreatic cancer’, ‘prostate cancer’ and ‘pathways in cancer’. The second highest scoring pathway was ‘focal adhesion’ (Fig. 5A), which is involved in the regulation of cell migration (14). In the Reactome analysis, the highest scoring pathway was ‘extracellular matrix (ECM) organization’, which was consistent with the ‘focal adhesion’ pathway identified by KEGG analysis. Other Reactome pathways, including ‘signaling by PDGF’ and ‘non-integrin membrane-ECM interactions’ were also associated with cell migration (Fig. 5B). The genes associated with the ‘focal adhesion’ pathway that were predicted as targets of the six exosomal miRNAs were comprised of upstream cytokine-receptor interaction molecules, including ECM components (COL1A1, COL5A2, LAMA and LAMC1), growth factors (PDGFC, VEGFA and IGF1), receptor tyrosine kinases (ERBB2 and PDGFRA) and integrins (ITGA5 and ITGB1), and downstream factors (PTEN and PIK3CA), cell proliferation genes (CCND1 and CCND2), cell survival genes (BCL2 and BAD) The targets of the six differentially expressed miRNAs in the ‘focal adhesion’ pathway and their associated miRNAs are presented in Fig. 6.
Figure 5.
Pathway analysis of target genes of the differentially expressed miRNAs. The 10 highest scoring pathways in (A) KEGG and (B) Reactome analysis.
Figure 6.
Molecules associated with ‘focal adhesion’ pathway. Red block, genes and gene families. Blue and green, downregulated and upregulated miRNAs in the fast-migrated group, respectively. KEGG, Kyoto Encyclopedia of Genes and Genomes.
Validation of migration-associated miRNAs using TCGA data
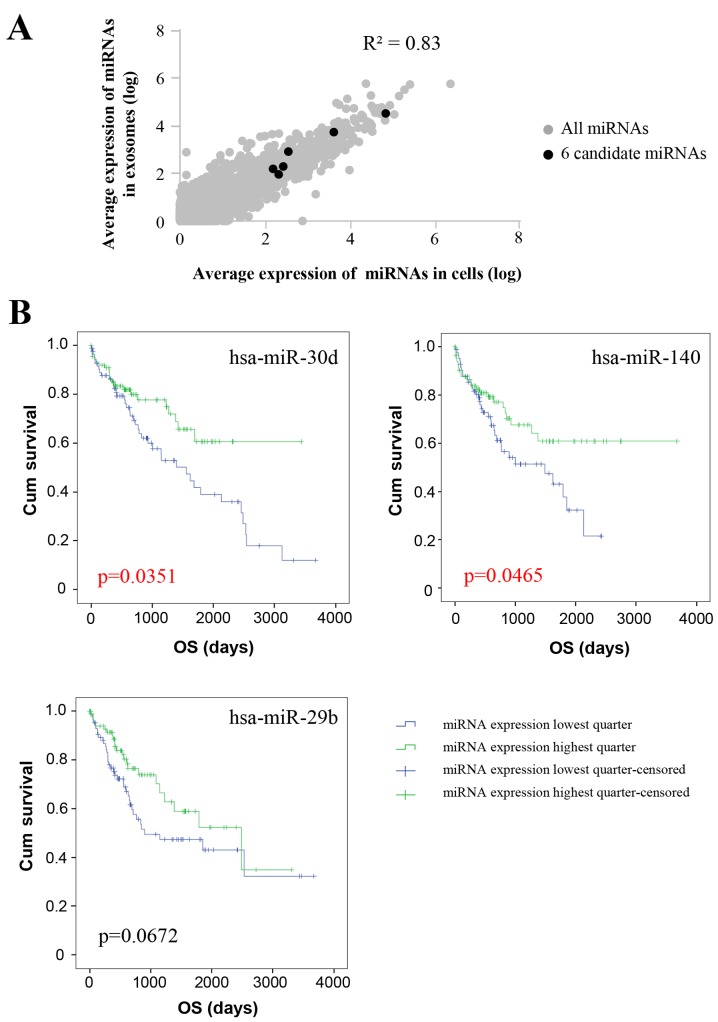
The expression of individual miRNAs was compared between exosomes and their corresponding PDCs. Generally, the association analysis highlighted a strong correlation (R2=0.83) between normalized miRNA expression in PDCs and exosomes, which indicated that exosomes reflect the miRNA composition of the derived cells. As for the six differentially expressed miRNAs, normalized exosomal miRNA levels were also correlated with the miRNA levels of PDCs as well (Fig. 7A). Considering the average miRNA expression levels were evaluated above, the expression levels of the 6 miRNAs in individual PDC samples and the paired exosome samples were further analyzed, and a similar trend of high correlation of miRNA expression between PDCs and their corresponding exosomes was observed (data not shown).
Figure 7.
Validation of migration-associated miRs in TCGA database. (A) Pearsons correlation analysis revealed a strong association between cellular miRs and their corresponding PDCs. (B) Kaplan-Meier analysis revealed a correlation between the expression levels of the differentially expressed miRs and OS in TCGA database. High expression of miR-30d, miR-140 and miR-29b was associated with improved OS. miR, microRNA; TCGA, The Cancer Genome Atlas; PDC, patient-derived cells; OS, overall survival.
In line with the finding that exosome miRNAs may reflect the miRNA composition of the PDCs that the exosomes were derived from, tumor tissue-derived miRNA levels and the clinical information of liver tumors were obtained from TCGA for validation analysis. Among the 372 available records, only 4 patients were categorized as M1 for definitive distal metastasis; therefore, it was impossible to study the association between miRNA levels and metastasis directly. Considering distal metastasis was positively correlated with poor clinical prognosis, the association between OS and miRNA levels were analyzed. The patients were divided by the expression level of a certain differentially expressed miRNA, with the top quartile and bottom quartile patients set as the low and high miRNA expression groups, respectively. The miRNA expression level and corresponding OS was analyzed using the Kaplan-Meier method (Fig. 7B). Patients with high expression of the three differentially expressed miRNAs, miR-30d, miR-140 and miR-29b, exhibited improved OS (P=0.0351, P=0.0465 and P=0.0672, respectively). Analysis of the clinical characteristics, including tumor stage, pathological T, N and M stage, detected no differences between the low and high miRNA expression groups (Tables III–V), suggesting that the result is not a secondary effect caused by the differences in the clinical characteristics between the two groups. Moreover, we analyzed the difference between the cirrhosis of these patients and survival from TCGA database. The following result revealed that there was no difference between cirrhosis and cum survival (P=0.773), indicating that our result concerning miRNAs and survival is not an epiphenomenon from their differences in underlying cirrhosis (data not shown).
Table III.
Clinical characteristics of HCC patients in the hsa-mir-140 lowest and highest quarters.
| hsa-mir-140 | ||||
|---|---|---|---|---|
| Characteristics | No. of patients | Lowest quarter | Highest quarter | P-value |
| Stage | ||||
| I | 73 | 32 | 41 | 0.240 |
| II | 47 | 23 | 24 | |
| III | 48 | 29 | 19 | |
| IV | 1 | 0 | 1 | |
| NA | 17 | 9 | 8 | |
| Pathologic_T | ||||
| T1 | 75 | 33 | 42 | 0.215 |
| T2 | 49 | 24 | 25 | |
| T3 | 41 | 25 | 16 | |
| NA | 13 | 7 | 6 | |
| Pathologic_N | ||||
| N0 | 125 | 62 | 63 | 0.363 |
| N1 | 2 | 2 | 0 | |
| NX | 47 | 23 | 24 | |
| NA | 12 | 6 | 6 | |
| Pathologic_M | ||||
| M0 | 129 | 64 | 65 | 0.577 |
| M1 | 1 | 0 | 1 | |
| MX | 44 | 23 | 21 | |
| NA | 12 | 6 | 6 | |
NA, not available.
Table V.
Clinical characteristics of HCC patients in the hsa-mir-29b lowest and highest quarters.
| hsa-mir-29b | ||||
|---|---|---|---|---|
| Characteristics | No. of patients | Lowest quarter | Highest quarter | P-value |
| Stage | ||||
| I | 80 | 36 | 44 | 0.536 |
| II | 42 | 20 | 22 | |
| III | 42 | 23 | 19 | |
| IV | 4 | 3 | 1 | |
| NA | 18 | 11 | 7 | |
| Pathologic_T | ||||
| T1 | 83 | 37 | 46 | 0.433 |
| T2 | 46 | 23 | 23 | |
| T3 | 42 | 24 | 18 | |
| TX | 1 | 0 | 1 | |
| NA | 9 | 5 | 4 | |
| Pathologic_N | ||||
| N0 | 121 | 63 | 58 | 0.546 |
| N1 | 3 | 1 | 2 | |
| NX | 52 | 23 | 29 | |
| NA | 10 | 6 | 4 | |
| Pathologic_M | ||||
| M0 | 127 | 63 | 64 | 0.203 |
| M1 | 3 | 3 | 0 | |
| MX | 47 | 22 | 25 | |
| NA | 9 | 5 | 4 | |
NA, not available.
Discussion
In PDC samples, miRNA was the most abundant class of small RNAs, accounting for 47.5% of the entire cellular small RNA population, which is comparable to the proportion in cells and plasma/serum reported in previous studies (15). However, in PDC-derived exosome samples, miRNAs accounted for only 3.0% of the small RNA repertoire, and the length distribution pattern differed from that in PDCs. Notably, similar phenomena were also observed in other studies (15). It was reported miRNAs represented 30 and 2–5% of the total small RNAome in mesenchymal stem cells and cell-derived exosomes, respectively (16). In exosomes collected from cell culture medium, miRNAs accounted for 2–7% of all small RNAs obtained by different isolation methods (17).
To identify potential miRNAs that may be involved in the metastasis of liver tumors, a wound healing assay was used to obtain two groups of PDCs with fast or slow cell migration rates. Six exosomal miRNAs were differentially expressed between the fast and slow-migrated groups. Five miRNAs (miR-140-3p, miR-30d-5p, miR-29b-3p, miR-130b-3p and miR-330-5p) were downregulated, and one miRNA (miR-296-3p) was upregulated in the fast-migrated group compared with the slow-migrated group. Although few studies have explored the association between these exosomal miRNAs and tumor metastasis directly, accumulating evidence indicates that these miRNAs may have vital roles in tumor metastasis. For example, exosomal miR-140 was identified as a negative regulator of cell migration in breast cancer via targeting of SRY-box 9 (18). This indicated that miR-140 acts as a potential tumor-suppressor by targeting oncogenes, and this may explain why it was downregulated in the fast-migrated group. Similarly, miR-30d-5p was reported to inhibit tumor cell proliferation and migration by directly targeting cyclin E2 in non-small cell lung cancer (19). Tumor-suppressive miRNA-29s directly regulated lysyl oxidase like 2 expression and inhibited cancer cell migration and invasion in renal cell carcinoma (20). miR-130b-3p was reported to inhibit cell invasion and migration by targeting the Notch ligand gene delta-like 1 in breast carcinoma (21). miR-330-5p regulated tyrosinase and protein disulfide isomerase family A member 3 expression, and suppressed cell proliferation, migration and invasion in cutaneous malignant melanoma (22). As an oncogenic miRNA, the level of miR-296-3p was markedly higher in highly metastatic human prostate cancer cells than in non-metastatic cells (23). However, the roles of miRNAs in tumor metastasis may vary among different studies, in different tumor types, under different physiological and pathological conditions, and through different mechanisms. miR-30d was also demonstrated to promote the metastatic behavior of melanoma cells by directly suppressing the GalNac transferase polypeptide N-acetylgalactosaminyltransferase 7 (24), while miR-296 was reported to inhibit the metastasis and epithelial-mesenchymal transition of colorectal cancer by targeting S100 calcium binding protein A4 (25).
In HCC, specific target genes could not be identified for these six differentially expressed miRNAs. However, the potential targeted genes were enriched in the ‘focal adhesion’ pathway. Focal adhesions are formed of integrin and other adapter proteins, and the dynamic assembly and disassembly of focal adhesion-ECM has a central role in cell dissemination and migration (14), resulting in the ability of cancer cells to metastasize. Focal adhesion kinase was revealed to be associated with the development and progression of HCC (26). Accumulating evidence suggests that miRNAs may regulate tumor migration and metastasis by affecting signal-mediated cytoskeletal and cell matrix adhesion remodeling (23), and the analysis in the present study demonstrated the involvement of ‘focal adhesion’ associated pathways in tumor cell migration and metastasis.
In summary, PDCs, which may be of superior representative value compared with commercial cancer cell line models, were used to investigate exosome miRNAs as biomarkers in the present study. miRNA-Seq was applied for comprehensive screening of differentially expressed exosomal miRNAs, rather than using RT-q PCR or microarray methods. Consequently, six differentially expressed exosomal miRNAs were identified as potential biomarkers for metastasis in HCC, and miRNA-targeted genes were enriched in the ‘focal adhesion’ pathway, supporting the role of miRNA regulation in cell migration and tumor metastasis. Finally, the six miRNA expression levels and corresponding patient survival profiles from TCGA were explored, and the association between the expression levels of miRNAs and patient survival was validated, indicating the potential role of these miRNAs in prognosis. To the best of our knowledge, this is the first study to investigate migration-associated exosomal miRNA biomarkers in HCC PDCs.
Table IV.
Clinical characteristics of HCC patients in the hsa-mir-30d lowest and highest quarters.
| hsa-mir-30d | ||||
|---|---|---|---|---|
| Characteristics | No. of patients | Lowest quarter | Highest quarter | P-value |
| Stage | ||||
| I | 79 | 32 | 47 | 0.103 |
| II | 35 | 18 | 17 | |
| III | 51 | 32 | 19 | |
| IV | 4 | 2 | 2 | |
| NA | 17 | 9 | 8 | |
| Pathologic_T | ||||
| T1 | 82 | 34 | 48 | 0.135 |
| T2 | 40 | 21 | 19 | |
| T3 | 48 | 29 | 19 | |
| TX | 1 | 1 | 0 | |
| NA | 9 | 4 | 5 | |
| Pathologic_N | ||||
| N0 | 125 | 58 | 67 | 0.323 |
| N1 | 3 | 2 | 1 | |
| NX | 50 | 29 | 21 | |
| NA | 8 | 4 | 4 | |
| Pathologic_M | ||||
| M0 | 131 | 66 | 65 | 0.999 |
| M1 | 4 | 2 | 2 | |
| MX | 44 | 22 | 22 | |
| NA | 7 | 3 | 4 | |
NA, not available.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- HCC
hepatocellular carcinoma
- PDCs
patient-derived cells
- miRNA-seq
microRNA sequencing
- miRNA
microRNA
- RPKM
reads per kilobase per million mapped reads
- OS
overall survival
- STR
short tandem repeat
- MVI
microvascular invasion
- PCA
principal component analysis
- ECM
extracellular matrix
Funding
This present study was supported by the National Key Basic Research Program of China (grant no. 2014CB542102) and Science Fund for Creative Research Groups, and the National Natural Science Foundation of China (grant no. 81521091).
Availability of data and materials
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.
Authors contributions
LXY designed the migration rate study, performed the wound healing assay, contributed to the data interpretation and wrote the manuscript; BLZ performed the pathway analysis and wrote the manuscript; YY conceived the study, designed the electron microscopy identification of the exosomes, interpreted the data and wrote the manuscript; MCW analyzed the miRNA data from TCGA database, contributed to the data interpretation and wrote the manuscript; GLL isolated the exosomal and cellular RNA and contributed to the pathway analysis; YG performed the western blot identification of the exosomes and prepared the exosomal samples for transmission electron microscopy; HL collected clinical information of the PDCs and contributed to the data interpretation; CHX conducted the electron microscopy identification of the exosomes and wrote the manuscript; JJX constructed small RNA library, contributed to the quality control of small RNA library and performed miRNA-seq; HQ performed the bioinformatics analysis of the miRNA-seq data, contributed to the data interpretation and wrote the manuscript; XYX isolated the PDC-derived exosomes and performed the migration rate study; ZSC cultured and established the PDCs, performed the wound healing assay and wrote the manuscript; DDZ contributed to the revised manuscript and data interpretation; FGL discussed the hypothesis and contributed to the data interpretation; SGZ conceived the study and led the project; RL conceived the study and participated in its design and coordination. All authors read and approved the manuscript and agree to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Ethics approval and consent to participate
HCC tissues were collected with ethics approval obtained from the Eastern Hepatobiliary Surgery Hospital, and informed consent was obtained from the patients for PDC culture and further research.
Patient consent for publication
Not applicable.
Competing interests
All authors affiliated to 3D Medicine Inc., are current or former employees. No potential conflicts of interest were disclosed by the other authors.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Steeg PS. Targeting metastasis. Nat Rev Cancer. 2016;16:201–218. doi: 10.1038/nrc.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu G, Zhang Y, Wei J, Jia W, Ge Z, Zhang Z, Liu X. MicroRNA-21 promotes hepatocellular carcinoma HepG2 cell proliferation through repression of mitogen-activated protein kinase-kinase 3. BMC Cancer. 2013;13:469. doi: 10.1186/1471-2407-13-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ, Hou JH, Fu J, Zeng MS, Yun JP, Wu QL, et al. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res. 2011;13:R2. doi: 10.1186/bcr2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Liu K, Liu Y, Xu Y, Zhang F, Yang H, Liu J, Pan T, Chen J, Wu M, et al. Exosomes mediate the cell-to-cell transmission of IFN-α-induced antiviral activity. Nat Immunol. 2013;14:793–803. doi: 10.1038/ni.2647. [DOI] [PubMed] [Google Scholar]
- 6.Lässer C, Alikhani VS, Ekström K, Eldh M, Paredes PT, Bossios A, Sjöstrand M, Gabrielsson S, Lötvall J, Valadi H. Human saliva, plasma and breast milk exosomes contain RNA: Uptake by macrophages. J Transl Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suetsugu A, Honma K, Saji S, Moriwaki H, Ochiya T, Hoffman RM. Imaging exosome transfer from breast cancer cells to stroma at metastatic sites in orthotopic nude-mouse models. Adv Drug Deliv Rev. 2013;65:383–390. doi: 10.1016/j.addr.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 8.Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, Hergueta-Redondo M, Williams C, García-Santos G, Ghajar C, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kogure T, Patel T. Isolation of extracellular nanovesicle microRNA from liver cancer cells in culture. Methods Mol Biol. 2013;1024:11–18. doi: 10.1007/978-1-62703-453-1_2. [DOI] [PubMed] [Google Scholar]
- 10.van Staveren WC, Solís DY, Hébrant A, Detours V, Dumont JE, Maenhaut C. Human cancer cell lines: Experimental models for cancer cells in situ? For cancer stem cells? Biochim Biophys Acta. 2009;1795:92–103. doi: 10.1016/j.bbcan.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Byrne AT, Alférez DG, Amant F, Annibali D, Arribas J, Biankin AV, Bruna A, Budinská E, Caldas C, Chang DK, et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat Rev Cancer. 2017;17:254–268. doi: 10.1038/nrc.2017.85. [DOI] [PubMed] [Google Scholar]
- 12.Buitrago DH, Patnaik SK, Kadota K, Kannisto E, Jones DR, Adusumilli PS. Small RNA sequencing for profiling microRNAs in long-term preserved formalin-fixed and paraffin-embedded non-small cell lung cancer tumor specimens. PLoS One. 2015;10:e0121521. doi: 10.1371/journal.pone.0121521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang J, Qin H, Yang Y, Chen X, Zhang J, Laird S, Wang CC, Chan TF, Li TC. A comparison of transcriptomic profiles in endometrium during window of implantation between women with unexplained recurrent implantation failure and recurrent miscarriage. Reproduction. 2017;153:749–758. doi: 10.1530/REP-16-0574. [DOI] [PubMed] [Google Scholar]
- 14.van Roosmalen W, Le Dévédec SE, Golani O, Smid M, Pulyakhina I, Timmermans AM, Look MP, Zi D, Pont C, de Graauw M, et al. Tumor cell migration screen identifies SRPK1 as breast cancer metastasis determinant. J Clin Invest. 2015;125:1648–1664. doi: 10.1172/JCI74440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodríguez M, Bajo-Santos C, Hessvik NP, Lorenz S, Fromm B, Berge V, Sandvig K, Linē A, Llorente A. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol Cancer. 2017;16:156. doi: 10.1186/s12943-017-0726-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baglio SR, Rooijers K, Koppers-Lalic D, Verweij FJ, Pérez Lanzón M, Zini N, Naaijkens B, Perut F, Niessen HW, Baldini N, et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res Ther. 2015;6:127. doi: 10.1186/s13287-015-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang YT, Huang YY, Zheng L, Qin SH, Xu XP, An TX, Xu Y, Wu YS, Hu XM, Ping BH, et al. Comparison of isolation methods of exosomes and exosomal RNA from cell culture medium and serum. Int J Mol Med. 2017;40:834–844. doi: 10.3892/ijmm.2017.3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gernapudi R, Yao Y, Zhang Y, Wolfson B, Roy S, Duru N, Eades G, Yang P, Zhou Q. Targeting exosomes from preadipocytes inhibits preadipocyte to cancer stem cell signaling in early-stage breast cancer. Breast Cancer Res Treat. 2015;150:685–695. doi: 10.1007/s10549-015-3326-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen D, Guo W, Qiu Z, Wang Q, Li Y, Liang L, Liu L, Huang S, Zhao Y, He X. MicroRNA-30d-5p inhibits tumour cell proliferation and motility by directly targeting CCNE2 in non-small cell lung cancer. Cancer Lett. 2015;362:208–217. doi: 10.1016/j.canlet.2015.03.041. [DOI] [PubMed] [Google Scholar]
- 20.Nishikawa R, Chiyomaru T, Enokida H, Inoguchi S, Ishihara T, Matsushita R, Goto Y, Fukumoto I, Nakagawa M, Seki N. Tumour-suppressive microRNA-29s directly regulate LOXL2 expression and inhibit cancer cell migration and invasion in renal cell carcinoma. FEBS Lett. 2015;589:2136–2145. doi: 10.1016/j.febslet.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Shui Y, Yu X, Duan R, Bao Q, Wu J, Yuan H, Ma C. miR-130b-3p inhibits cell invasion and migration by targeting the Notch ligand Delta-like 1 in breast carcinoma. Gene. 2017;609:80–87. doi: 10.1016/j.gene.2017.01.036. [DOI] [PubMed] [Google Scholar]
- 22.Su BB, Zhou SW, Gan CB, Zhang XN. MiR-330-5p regulates tyrosinase and PDIA3 expression and suppresses cell proliferation and invasion in cutaneous malignant melanoma. J Surg Res. 2016;203:434–440. doi: 10.1016/j.jss.2016.03.021. [DOI] [PubMed] [Google Scholar]
- 23.Zhou L, Liu F, Wang X, Ouyang G. The roles of microRNAs in the regulation of tumor metastasis. Cell Biosci. 2015;5:32. doi: 10.1186/s13578-015-0028-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gaziel-Sovran A, Segura MF, Di Micco R, Collins MK, Hanniford D, Vega-Saenz de Miera E, Rakus JF, Dankert JF, Shang S, Kerbel RS, et al. miR-30b/30d regulation of GalNAc transferases enhances invasion and immunosuppression during metastasis. Cancer Cell. 2011;20:104–118. doi: 10.1016/j.ccr.2011.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He Z, Yu L, Luo S, Li M, Li J, Li Q, Sun Y, Wang C. miR-296 inhibits the metastasis and epithelial-mesenchymal transition of colorectal cancer by targeting S100A4. BMC Cancer. 2017;17:140. doi: 10.1186/s12885-017-3121-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Panera N, Crudele A, Romito I, Gnani D, Alisi A. Focal adhesion kinase: Insight into molecular roles and functions in hepatocellular carcinoma. Int J Mol Sci. 2017;18:99. doi: 10.3390/ijms18010099. doi: 10.3390/ijms18010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the present study are available from the corresponding author on reasonable request.