Abstract
Stem cells must maintain proliferation during tissue development, repair and homeostasis, yet avoid tumor formation. In Drosophila, neural stem cells (neuroblasts) maintain proliferation during embryonic and larval development and terminate cell cycle during metamorphosis. An important question for understanding how tissues are generated and maintained is: what regulates stem cell proliferation versus differentiation? We performed a genetic screen which identified nucleostemin 3 (ns3) as a gene required to maintain neuroblast proliferation. ns3 is evolutionarily conserved with yeast and human Lsg1, which encode putative GTPases and are essential for organism growth and viability. We found NS3 is cytoplasmic and it is required to retain the cell cycle repressor Prospero in neuroblast cytoplasm via a Ran-independent pathway. NS3 is also required for proper neuroblast cell polarity and asymmetric cell division. Structure-function analysis further shows that the GTP-binding domain and acidic domain are required for NS3 function in neuroblast proliferation. We conclude NS3 has novel roles in regulating neuroblast cell polarity and proliferation.
Keywords: Nucleostemin, Prospero, Neuroblast, Subcellular localization, Proliferation
1. Introduction
Stem cells have the unique capacity to maintain their undifferentiated state while rapidly producing lineage-specific, differentiating daughter cells. Towards the end of development, stem cells may enter a reversible, non-dividing G0 state, known as quiescence, or permanently exit cell cycle to undergo differentiation, senescence, or apoptosis. Determining the mechanisms that maintain stem cell proliferation may help illuminate mechanisms that impair development or cause tumor-igenesis.
Drosophila neuroblasts (NBs) have emerged as a widely used model system for studying stem cell proliferation, self-renewal, and cell cycle exit (Doe, 2008; Egger et al., 2008; Homem and Knoblich, 2012; Homem et al., 2015; Maurange and Gould, 2005). NBs delaminate from the neuroectoderm and immediately proceed to undergo asymmetric cell division to self-renew and generate neurons or glia. Most NBs in the thorax and central brain enter quiescence at the end of embryogenesis, and subsequently re-enter the cell cycle and proliferate after larval hatching. Based on their cell lineage, two types of NBs are distinguishable within the nervous system: type I and type II. The majority of NBs are categorized as type I NBs, which asymmetrically divide to self-renew and produce a smaller ganglion mother cell (GMC) that further divides to produce two neurons or glia. Type I NBs can also switch to type 0 during late embryonic development to produce a single neuronal daughter cell, instead of a GMC (Baumgardt et al., 2014, 2009; Bertet et al., 2014; Karcavich and Doe, 2004). In addition, there are eight type II NBs in each bilateral brain lobe that asymmetrically divide to self-renew and generate a smaller intermediate neural progenitors (INPs) that each undergo multiple rounds of asymmetric cell division to self-renew and produce GMCs. The latter type of division allows NBs to rapidly expand their lineage within a limited proliferation window (Bayraktar et al., 2010; Bello et al., 2008; Boone and Doe, 2008; Bowman et al., 2008; Walsh and Doe, 2017).
Proliferative embryonic NBs contain a set of transcription factors that promote stem cell attributes (asymmetric cell division, self-renewal, survival, and proliferation); this set of factors include the basic helix-loop-helix transcription factors Deadpan (Dpn) and Asense (Ase), the Snail family gene Worniu (Wor), the cell cycle regulator Cyclin E (CycE), the Sox family members SoxN and Dichaete (D), and the early temporal genes Hunchback (Hb), Krupple (Kr), and POU domain protein family genes (Nub/Pdm2) (Bahrampour et al., 2017; Lai and Doe, 2014; Lai et al., 2012). Proliferative embryonic and larval type I NBs also express the differentiation-promoting Prospero (Pros) transcription factor, but Pros protein is not active due to its segregation into the cytoplasm. Pros is an atypical homeodomain transcription factor with similar functions to its mammalian ortholog, Prox1, to repress progenitor-specific genes, repress cell cycle genes, and promote cell cycle exit and differentiation (Doe et al., 1991; Dyer et al., 2003; Elsir et al., 2012; Vaessin et al., 1991). Although high levels of nuclear Pros triggers cell cycle exit and differentiation (Bayraktar et al., 2010), low levels of Pros induce NB quiescence via repression of ase, wor, and cycE, but not dpn (Lai and Doe, 2014). NB cortical polarity is also essential for maintaining proliferation. The atypical protein kinase C (aPKC) is localized to the NB apical cortex and is required to restrict the Miranda/Pros complex to the basal cortex, where it is partitioned into the newborn GMC at mitosis (Atwood and Prehoda, 2009). Loss of aPKC results in delocalization of Miranda/Pros from the cortex, nuclear import of Pros, and NB cell cycle arrest (Rolls et al., 2003).
Here we use a forward genetic screen to identify essential genes that are expressed in NBs and required to maintain NB proliferation. We identify the highly conserved Lsg1 family gene nucleostemin 3 (ns3) as being required for maintaining larval NB proliferation. Moreover, we find that cytoplasmic retention of Pros is regulated by NS3, independent of Ran-mediated nucleo-cytoplasmic trafficking, and knocking down ns3 leads to the disruption of NB asymmetric cell division. We further show that NS3 requires the GTP-binding domain to be excluded from the nucleus, and the GTP-binding domain and acidic domain act together to retain Pros in the cytoplasm.
2. Materials and methods
2.1. Fly stocks
The following flies were used: (1) wor-gal4 (Lai et al., 2012); (2) wor-gal4, ase-gal80, UAS‐mCD8:GFP (Carney et al., 2013; Joy et al., 2015); (3) UAS-Dcr-2; wor-gal4; UAS‐mCD8:GFP (Carney et al., 2013; Joy et al., 2015); (4) UAS-nls-nes+-gfp (FBst0007032); (5) UAS-ns3-RNAi (FBst0036626); (6) UAS-pros-RNAi (FBst0042538); (7) tub-Gal80ts (FBst0007019). RNAi lines used in the screening experiments are listed in Table S1.
2.2. NS3 rescue constructs
UAS‐HA:ns3WT, UAS‐HA:ns3ΔA, UAS‐HA:ns3ΔG and UAS‐HA:ns3ΔB were generated by using PCR to amplify each individual fragment which was later assembled directly in EcoRI/NotI digested pUAST-attB with NEBuilder (New England BioLabs, Ipswich, MA). Primers used in PCR and fragments used for each construct are listed in Table S2. All constructs were verified by sequencing (Sequetech, Mountain View, CA) and inserted into attP40 (FBst0036304) via PhiC31 integrase-mediated transgenesis (BestGene Inc, Chino Hills, CA). To prevent RNAi knockdown of the rescue constructs, we mutated the DNA sequence in the acidic domain targeted by ns3 RNAi from GCCAGGTATGTGCTTAAAGACTAC to GCACGATACGTATTAAAGGATTAC.
2.3. Dissections, antibody staining, fixation and confocal microscopy
The following primary antibodies were used: rabbit anti-atypical PKC (1:1000) (Santa Cruz Biotechnology), rabbit anti-Asense (Cheng-Yu Lee, Univ. of Michigan, Ann Arbor, MI), rat anti-Dpn (1:100) (Abcam), guinea pig anti-Dpn (1:2000) (Jim Skeath, Washington Univ., St Louis, MO), mouse anti-Elav (1:50) (9F8A9, DSHB), chicken anti-GFP (1:500) (Aves Labs), mouse anti-Lamin (ADL67.10, DSHB), guinea pig anti-Miranda (1:500) (Doe lab), rabbit anti-PH3 (EMD Millipore), mouse anti-Pros purified antibody (1:500) (MR1A, Abcam), and rat anti-Worniu (1:100) (Abcam). Fluorophore-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories (West Grove, PA). Texas Red™-X Phalloidin (ThermoFisher Scientific) and DAPI (Sigma-Aldrich) were applied after secondary antibody staining by following manufacturer’s protocol. Larvae were raised at 29°C until third instar, and the brains were dissected, fixed and stained by following published procedures (Lai and Doe, 2014; Lai et al., 2012). Images were taken with Zeiss confocal microscope LSM710, processed with open source software Fiji (Schindelin et al., 2012) and assembled in the software Adobe Photoshop or Illustrator (Adobe Systems Inc., San Jose, CA). All statistical analyses in the figures used one-tail t-test included in Microsoft Excel (Microsoft Corp., Redmond, WA).
2.4. EdU incorporation
Immediately following dissections, brains were incubated in PBS containing 100 μg/mL EdU (ThermoFisher Scientific) at room temperature for 2hr with rocking. After standard fixation and antibody staining, EdU was detected by following the manufacturer’s protocol.
2.5. Cellular localization of NLS-NES analysis
We used the “Plot Profile” functionality of Fiji to measure the intensity of GFP-NLS-NES and Deadpan signal across the diameter of 5 NBs, whose cell cortex was outlined by Phalloidin staining. We then used R software to normalize the intensity and NB length to the scale 0–1, and smoothing splines were fit to the 5 data sets for each genotype. The R code is available upon request.
3. Results
3.1. An RNAi screen identifies genes required to maintain larval neuroblast proliferation
We used an RNAi-based approach to screen for genes that, when knocked down in larval brains, led to a loss of larval NB proliferation without decreasing NB numbers (i.e. specifically leading to quiescence or permanent cell cycle arrest). We screened 28 essential genes that we previously found to have enriched expression in NBs (Carney et al., 2012). For each gene, we used the NB-specific worniu-gal4 transgene to drive expressions of a single copy of each UAS-RNAi transgene in NBs and membrane-tethered GFP to mark the affected NB lineage. NBs were identified by staining for Deadpan (Dpn), which marks both proliferating and quiescent NBs (Lai and Doe, 2014), and we assessed NB cell cycle progression via a 2-hr pulse of EdU.
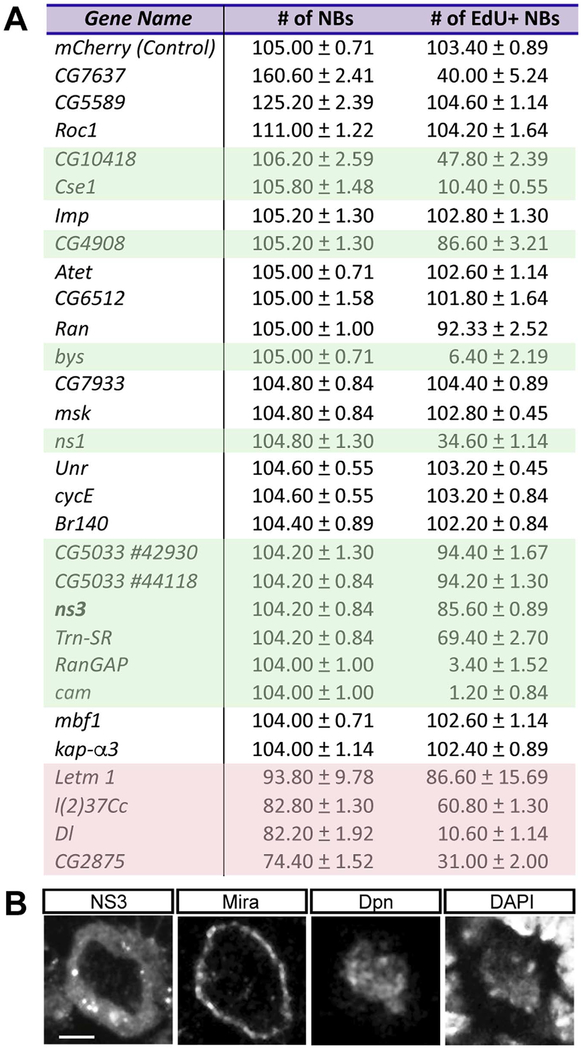
In wild type larval brains there are ~105 NBs per lobe, and all are labeled by a 2hr pulse of EdU (Fig. 1A, top row). In contrast, knocking down expression of 11 genes led to a significant reduction in EdU + NBs without decreasing NB number (Fig. 1A, green highlighted); this represents induction of NB cell cycle exit. Knockdown of an additional 4 genes showed reduced NB number (Fig. 1A, red shading); these are likely to be genes required to prevent NB death or differentiation, and while interesting, we do not focus on this class of genes here. We conclude that loss of function of 11 genes results in NB cell cycle exit without inducing differentiation or death.
Fig. 1.
RNAi screen identifies candidate genes to promote NB proliferation. (A) The 28 NB-enriched genes assayed for RNAi phenotypes. The number of total NBs and EdU+ NBs in the L3 brain were quantified for each RNAi gene knockdown. Green shading indicates RNAi lines where no significant change in NB number (p > 0.05) but a decrease in EdU+ NBs (p < 0.05) indicating premature NB cell cycle exit. Red shading indicates RNAi lines that lead to a reduction in total NB number, due to NB death or differentiation. n = 5 brain lobes from 5 larvae for each gene. Genotype: UAS-Dcr-2; wor-gal4; UAS‐mCD8:GFP/UAS‐gene RNAi. (B) A representative neuroblast in the L3 brain. wor-gal4 was used to drive expression of UAS‐ns3:YFP (FBst0050769). The neuroblast was stained with membrane marker Mira, NB marker Dpn, and DNA marker DAPI. Scale bar: 5 μm. Genotype: wor‐gal4/UAS‐ns3:YFP.
Among the 11 candidate genes, we were particularly interested in the conserved Lsg1 family member, called nucleostemin 3 (ns3) in Drosophila. The yeast and human ortholog, Lsg1, is cytoplasmic and promotes release of the nuclear export adapter from the large ribosomal subunit for ribosome maturation (Hedges et al., 2005; Kallstrom et al., 2003; Reynaud et al., 2005; Weis et al., 2014). In Drosophila, NS3 is also cytoplasmic in all cell types examined including larval NBs (Fig. 1B), and required in serotonergic neurons to regulate organismal growth (Hartl et al., 2013; Kaplan et al., 2008), yet little is known its roles in Drosophila nervous system development. Here, we investigate the role of NS3 in regulating NB proliferation.
3.2. Reduced NS3 leads to proliferation arrest in type I NBs
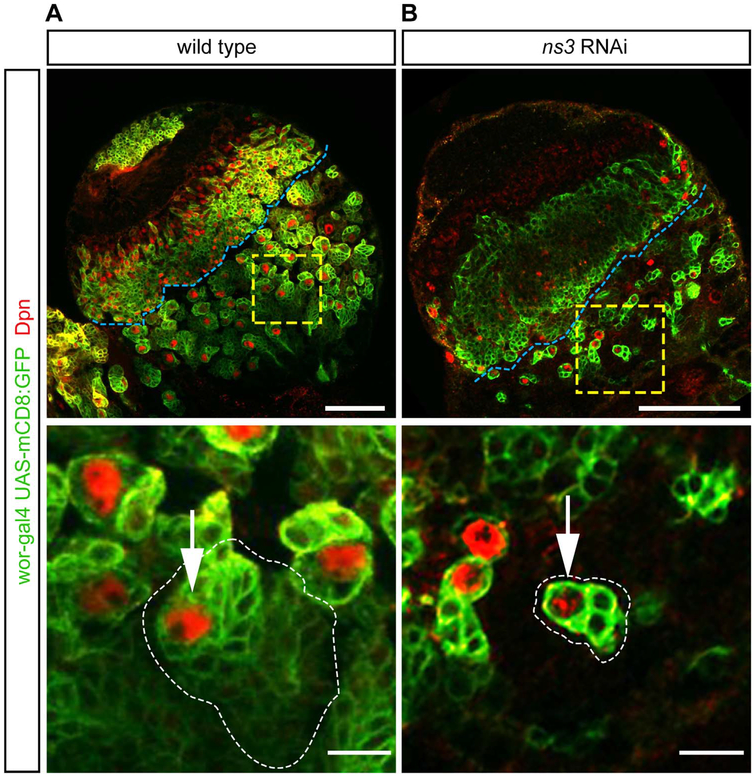
Wild type larval NBs produce a clone of neuronal progeny that can be visualized by expression of GFP in the NB (worniu-gal4 UAS‐ mCD8:GFP) followed by perdurance of the GFP in the most recently born progeny (Fig. 2A, green). We found that ns3 RNAi NBs had fewer neuronal progeny (Fig. 2B), despite normal NB numbers. Loss of NS3 could lead to NB cell cycle arrest or quiescence; the persistence of Dpn+ NBs rules out apoptosis or differentiation.
Fig. 2.
ns3 RNAi mutant NBs in the central brain have reduced lineage sizes. Single slice confocal images of L3 wild type and RNAi brain lobes; NBs are identified by Dpn and NBs and their lineages are marked with GFP (wor-gal4, UAS-mCD8:GFP). (A) Wild type NBs produce large clones of neuronal progeny. Genotype: UAS-Dcr-2; wor-gal4; UAS-mCD8:GFP/UAS-mCherry-RNAi. (B) ns3 RNAi NBs show reduced numbers of neuronal progeny. Genotype: UAS-Dcr-2; wor-gal4; UAS-mCD8:GFP/UAS-ns3-RNAi. The central brain / optic lobe boundary is indicated by a blue line. Representative NBs (white arrows) are boxed in yellow and enlarged in the bottom row. Scale bars: 50 μm top row, 10 μm bottom row.
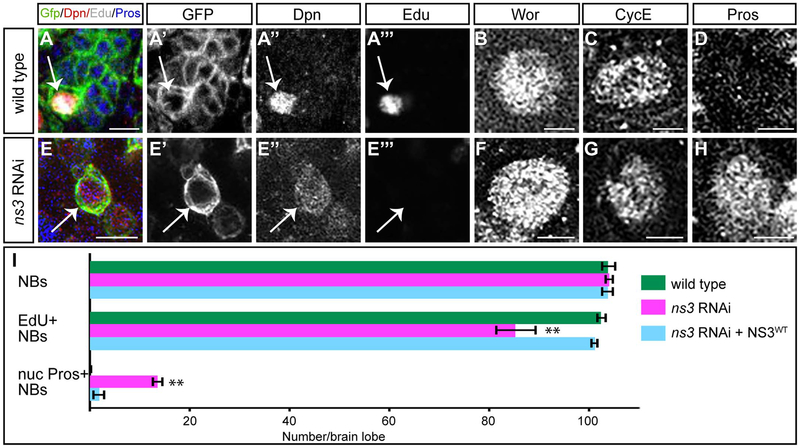
We used cell cycle progression, molecular markers, and morphological criteria to determine whether reduced NS3 resulted in NB cell cycle arrest or quiescence. Wild type NBs can be labeled by a pulse of EdU, are Dpn+ Wor+ CycE+, and lack nuclear Pros (Fig. 3A–D). In contrast, quiescent NBs are Dpn+ Wor− CycE− nuclear Pros+, and extend a neurite-like process (Chell and Brand, 2010; Lai and Doe, 2014; Tsuji et al., 2008). We found that ns3 RNAi NBs did not match either profile. They did not incorporate EdU like quiescent NBs but were Dpn+ Wor+ CycE+ nuclear Pros+ (Fig. 3E–H), but they failed to extend a neurite-like process (Fig. 2B, 3E′). These results suggest that reduced NS3 leads to NB cell cycle arrest, but not entry into quiescence. Importantly, overexpression of full length NS3 can fully rescue the ns3 RNAi phenotype (Fig. 3I), showing NS3 is responsible for the cell cycle arrest phenotype.
Fig. 3.
Reduced NS3 leads to proliferation arrest in type I NBs. (A-D) Wildtype type I NBs (white arrows) in the L3 brain, identified by wor-gal4 UAS- mCD8:GFP (A′) and Dpn (A”). They can be labeled by a pulse of EdU (A′”), are Wor+ (B) and CycE+ (C), and have no detectable nuclear Pros (D). The Wor, CycE and Pros images were obtained from different confocal stacks. Genotype: UAS-Dcr-2; wor-gal4; UAS-mCD8:GFP/UAS-mCherry-RNAi. (E-H) ns3 RNAi type I NBs (white arrows) in the L3 brain, identified by wor-gal4 UAS- mCD8:GFP (E′) and Dpn (E”). They are not labeled by a pulse of EdU (E′”), but are Wor+ (F) and CycE+(G), and have nuclear Pros (H). The Wor, CycE and Pros images were obtained from different confocal stacks. Genotype: UAS-Dcr-2; wor-gal4; UAS-mCD8:GFP/UAS-ns3-RNAi. (I) Quantifications. n = 5 brain lobes. Error bars indicate s.d. * *: p < 0.001. Scale bars: 10 μm (A, E) and 5 μm (B-D, F-H).
To further test whether ns3 RNAi leads to larval NB proliferation arrest or quiescence, we transiently knocked down NS3 and assayed for NB arrest followed by NB re-entry into the cell cycle, a hallmark of quiescent NBs. We used temperature-sensitive Gal80 (Gal80ts) to temporally control the expression of Gal4-induced ns3 RNAi. We raised the ns3 RNAi larvae for two days at 29°C (where Gal80ts is inactive) to allow expression of ns3 RNAi to induce NB proliferation arrest, followed by 2 days at 23°C (where Gal80ts is active) to restore NS3 levels. We found that NBs remain cell cycle arrested (26.0 ± 2.9 NBs, n = 6 brain lobes), despite recovery of NS3 expression. We conclude that permanent or transient reduction of NS3 in type I NBs results in NB cell cycle arrest, but not entry into quiescence.
3.3. NS3 promotes Pros nuclear export in type I NBs through a Ran-independent mechanism
We noted that ns3 RNAi NBs had nuclear Pros, which has been shown to arrest the NB cell cycle (Lai and Doe, 2014). This led us to investigate how NS3 promotes Pros nuclear export. Pros contains a canonical nuclear localization signal (NLS) to drive nuclear import (Hirata et al., 1995). Additionally, Pros harbors a nuclear export signal (NES) near its conserved homeodomain (Demidenko et al., 2001). The competition between NLS-driven import and NES-driven export determines the subcellular localization of Pros, however, the underlying mechanisms remain unclear (Bi et al., 2005, 2003; Demidenko et al., 2001; Ryter et al., 2002).
Most NLS/NES containing proteins require Ran-dependent transport across the nuclear membrane (Terry et al., 2007; Yudin and Fainzilber, 2009). Ran cycling is expedited by a steep concentration gradient between cytoplasmic Ran GTPase activating protein (RanGAP) and nuclear RanGTP exchange factor (RanGEF) (Steggerda and Paschal, 2002). In Drosophila, Ran, RanGAP, and RanGEF (Bj1) are all enriched in NBs (Carney et al., 2013; Joy et al., 2015), and the loss of any of these genes induces Pros nuclear localization (Joy et al., 2015, data not shown). Moreover, knocking down Ran or RanGAP also halted cell cycle progression without altering the quantity of NBs (Joy et al., 2015; Fig. 1B). These results led us to investigate whether NS3 has a novel role in Ran-mediated nuclear transport and functions together with Ran to control the nuclear-cytoplasmic localization of proteins with both an NLS and NES, including Pros.
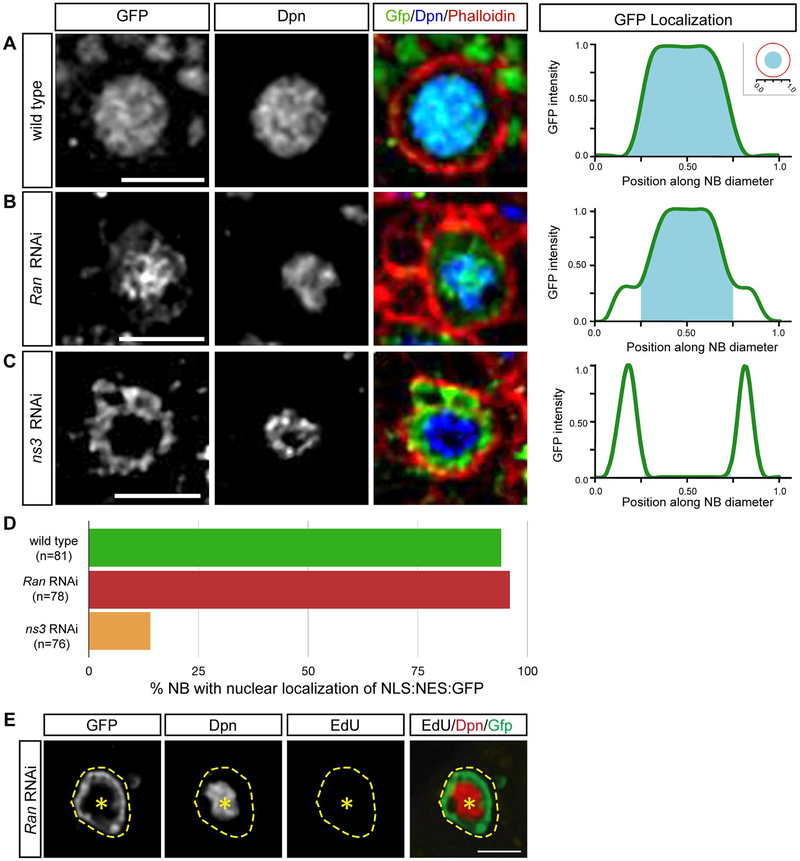
To test this hypothesis, we determined the role of NS3 and Ran in the nucleo-cytoplasmic localization of a dual NLS/NES GFP reporter, NLS: NES:GFP (Kusano et al., 2001). In wild type NBs, NLS: NES:GFP was enriched in the nucleus, showing that the NLS was dominant over the NES (Fig. 4A, quantified in D). Similarly, Ran RNAi NBs showed nuclear localization of NLS: NES:GFP, showing that Ran was not required for nuclear localization of NLS: NES:GFP (Fig. 4B, quantified in D). In contrast, ns3 RNAi larval NBs localized NLS: NES:GFP to the cytoplasm, showing that NS3 was required for nuclear import of the NLS: NES:GFP protein (Fig. 4C, quantified in D). We can be sure the Ran RNAi was effective, because it arrested the NB cell cycle (Fig. 4E). Interestingly, this is the opposite of the role of NS3 in preventing nuclear localization of Pros (see Discussion). Nevertheless, the finding that Ran RNAi and the NS3 RNAi had opposite phenotypes in this assay strongly suggests that NS3 is acting in in a Ran-independent pathway.
Fig. 4.
NS3 regulates Pros localization via a Ran-independent pathway. (A-D) Localization of NLS:NES:GFP (green line) in wild type (A), Ran RNAi (B), and ns3 RNAi (C) L3 brain NBs. The NB nucleus marked by Dpn (cyan area); pixel intensity trace in right panels. Quantification (D) from 5 brains, n = number of NBs. Genotype: (A) wor-gal4 UAS-NLS:NES:GFP; UAS-mCherry-RNAi; (B) wor-gal4 UAS-NLS:NES:GFP; UAS-Ran-RNAi; (C) wor-gal4 UAS-NLS:NES:GFP; UAS-ns3-RNAi. Scale bars: 10 μm (E) Confocal images of a Ran RNAi L2 central brain NB lineage; NB and its lineage are marked with GFP (wor-gal4, UAS-mCD8:GFP), and NB is identified by Dpn. NB is EdU-negative. Genotype: (E) UAS-Dcr-2; wor-gal4; UAS-mCD8:GFP/UAS-Ran-RNAi. Scale bars: 10 μm.
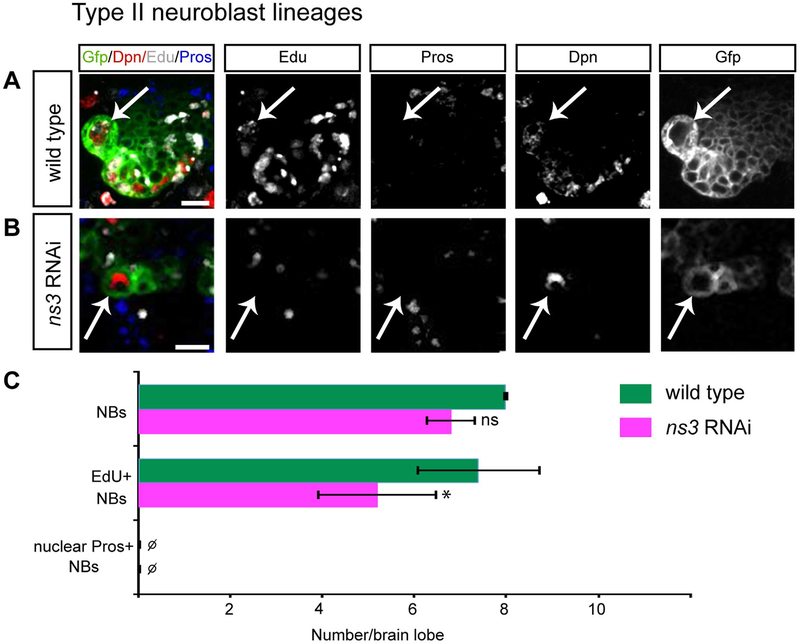
3.4. Reduced NS3 leads to proliferation arrest in Prospero-negative type II NBs
To test whether ns3 RNAi induces NB proliferation arrest via nuclear import of Pros, we examined type II larval brain NBs. Type II NBs have undetectable levels of Pros, which is thought to allow newborn INPs to remain proliferative (Bayraktar et al., 2010). If nuclear Pros is a required step in ns3 RNAi induced NB arrest, the type II NBs should show no ns3 RNAi phenotype. We used a type II NB-specific driver (wor-gal4, ase-gal80) to express ns3 RNAi and drive expression of GFP in each type II NB lineage. In wild type, the type II NBs were Dpn+ Pros-negative and could be labeled by a pulse of EdU, showing that they are proliferative (Fig. 5A, quantified in C). In contrast, ns3 RNAi type II NBs were Dpn+ nuclear Pros-negative, but failed to efficiently incorporate EdU (Fig. 5B, quantified in C). We conclude NS3 knockdown can produce NB proliferation arrest using a Pros-independent pathway. This does not rule out a role for nuclear Pros in type I NB proliferation arrest, but it does show that at least one additional mechanism must be used to block proliferation in ns3 RNAi type II neuroblasts.
Fig. 5.
Reduced NS3 leads to proliferation arrest in type II NBs. Type II NBs (white arrows) in the L3 brain, identified by Dpn and wor-gal4 ase-gal80 UAS-GFP. (A) Wild type NBs are labeled by a pulse of EdU. Genotype: UAS-Dcr-2; wor-gal4 ase-gal80; UAS-mCD8:GFP/UAS-mCherry-RNAi. (B) ns3 RNAi NBs are not labeled by a pulse of EdU. Genotype: UAS-Dcr-2; wor-gal4 ase-gal80; UAS-mCD8:GFP/UAS-ns3-RNAi. (C) Quantifications. n = 5 brain lobes. Error bars indicate s.d. *: p < 0.05. Scale bars: 10 μm.
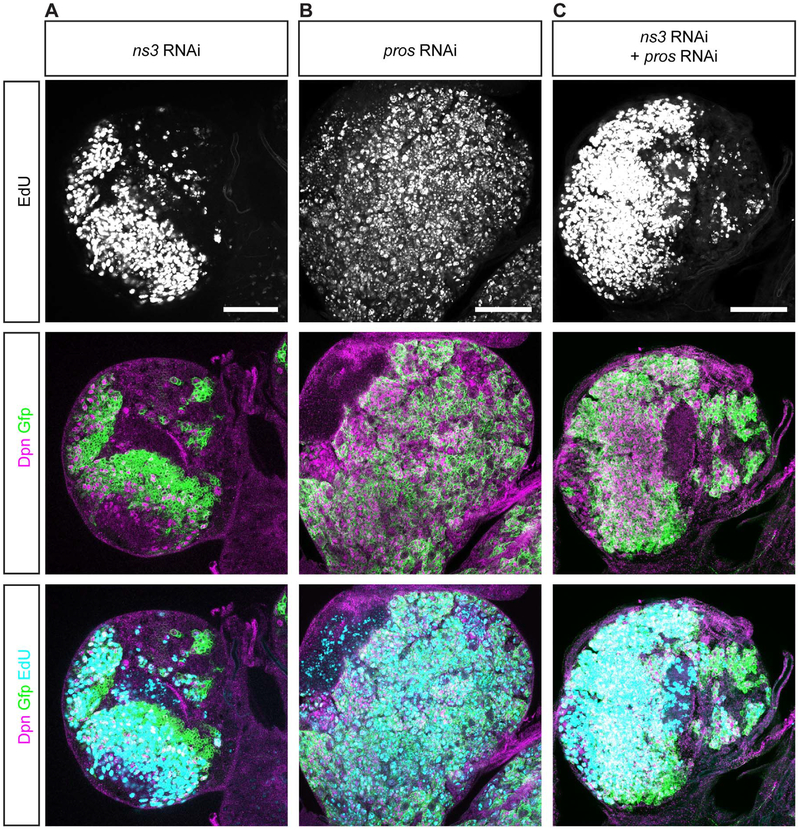
To determine if ns3 RNAi also acts via a Pros-independent mechanism in type I NBs, we generated ns3 pros double RNAi knockdown larvae. If NS3 acts via a Pros-independent mechanism, we would expect that ns3 pros double RNAi knockdown would match the ns3 single RNAi phenotype. We found that the ns3 pros double RNAi knockdown phenotype was intermediate: few proliferating NBs in ns3 RNAi brains (Fig. 6A), many proliferating NBs in pros RNAi brains (Fig. 6B), and an intermediate level of proliferating NBs in the ns3 pros double RNAi brains (Fig. 6C). The intermediate phenotype could be due to (a) partial knockdown of both NS3 and Pros proteins, resulting in expansion of the small number of “escaper” NBs that maintain proliferation in ns3 RNAi brains, or (b) less effective ns3 RNAi knockdown due to an additional UAS-transgene in the double versus the single knockdown experiment (the extra UAS-transgene may titrate Gal4 levels, thereby reducing expression of all UAS-transgenes). Thus, this experiment did not allow us to determine whether proliferation arrest of ns3 RNAi type I NBs is Pros-dependent or Pros-independent; both may contribute to the phenotype (see Discussion). Nevertheless, based on our findings that ns3 RNAi can arrest proliferation of type II NBs in the absence of detectable Pros, we favor the model that NS3 acts via a Pros-independent pathway to maintain NB proliferation in all larval NBs.
Fig. 6.
ns3 pros double knockdown shows intermediate phenotype between ns3 or pros single knockdown. Single slice confocal images of single or double knockout L3 brains; NBs are identified by Dpn and NBs and their lineages are marked with GFP (wor-gal4, UAS-mCD8:GFP). Representative brains are shown (n = 5 brains per genotype). (A) ns3 RNAi brain has fewer EdU+ cells. Genotype: UAS-Dcr-2; wor-gal4; UAS-mCD8:GFP/UAS-ns3-RNAi. (B) pros RNAi brain has many more EdU+ cells. Genotype: UAS-Dcr-2; wor-gal4/UAS-pros-RNAi; UAS-mCD8:GFP. (C) ns3 pros double RNAi brain has intermediate number of EdU+ cells. Genotype: UAS-Dcr-2; wor-gal4/UAS-pros-RNAi; UAS-mCD8:GFP/UAS-ns3-RNAi. Scale bar: 50 μm.
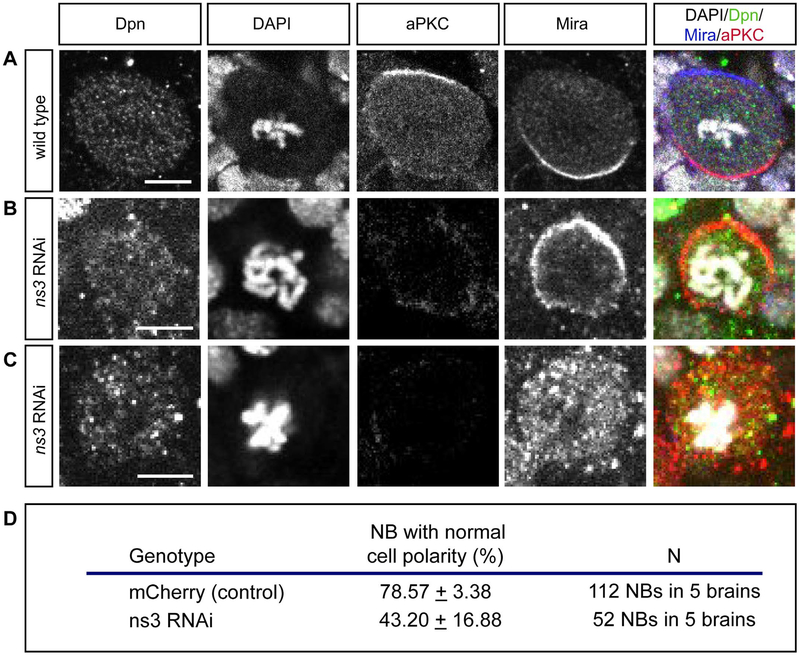
3.5. NS3 is required to establish NB cortical asymmetry
If NS3 acts via a Pros-independent mechanism, regulating NB cortical polarity is a good possibility, as loss of cortical polarity has been shown to lead to both NB proliferation arrest (Rolls et al., 2003) and nuclear Pros localization (Ikeshima-Kataoka et al., 1997; Shen et al., 1997). Wild type larval NBs segregate Miranda and its cargo protein Pros to the basal cortex during mitosis, and aPKC to the apical cortex, where it excludes the Miranda:Pros complex via phosphorylation of Miranda (Atwood and Prehoda, 2009). Loss of functional aPKC results in delocalization of Miranda, NB cell cycle arrest (Rolls et al., 2003) and premature Pros nuclear localization (unpublished results). We confirm that wild type mitotic NBs have strictly non-overlapping cortical crescents of aPKC and Miranda (Fig. 7A, quantified in D). In contrast, ns3 RNAi mitotic NBs show a significant loss of cortical polarity, with aPKC mostly undetectable and Miranda uniform cortical or cytoplasmic (Fig. 7B,C, quantified in D). These phenotypes show that NS3 is required for normal NB asymmetric cell division, and the Miranda delocalization phenotype is consistent with failure to segregate Pros out of the NB during asymmetric division, which may lead to abnormal nuclear Pros in the NB following cell division (see Discussion). Our results support a model in which NS3 regulates NB cortical polarity to maintain NB proliferation; note that this does not exclude a role for NS3-regulated Pros nuclear export in maintaining type I NB proliferation.
Fig. 7.
ns3 RNAi NBs have abnormal cortical polarity. (A) Wild type pro-metaphase NB in the L3 brain, identified by Dpn and condensed chromatin (DAPI). aPKC and Mira asymmetrically segregate to the apical and basal cortex, respectively. Genotype: wor-gal4; UAS-mCherry-RNAi. (B,C) ns3 RNAi pro-metaphase NBs in L3 brain lack detectable aPKC and show either uniform cortical Miranda (Mira; B) or cytoplasmic Mira (C). Genotype: wor-gal4; UAS-ns3-RNAi. Scale bars: 5 μm. (D) Quantification of the percent of mitotic NBs that display proper asymmetric segregation of aPKC and Mira.
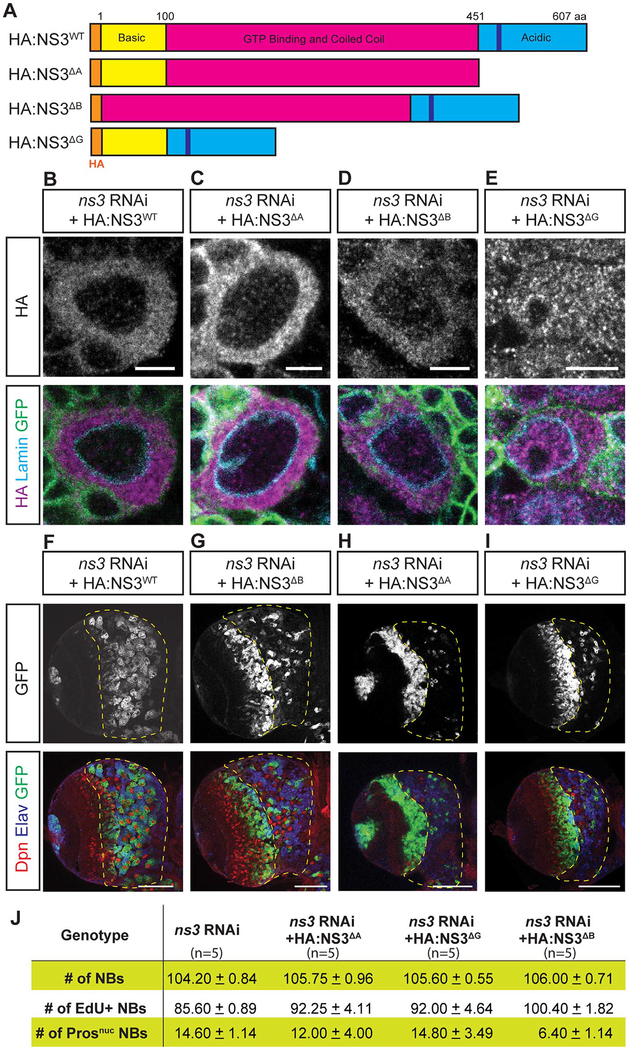
3.6. Defining the NS3 domains required for promoting NB proliferation
To understand how NS3 promotes NB proliferation, we performed a structure-function analysis to identify the NS3 protein domains required for this function. NS3 consists of three major domains, the N-terminal basic domain, the GTP binding and coiled-coil domain, and the C-terminal acidic domain (Fig. 8A). The GTP binding domain functions as GTPase and is essential for global body growth (Hartl et al., 2013), yet less is understood about the functional significance of both the basic and acidic domains. To determine the function of each domain, we depleted endogenous NS3 using RNAi and rescued with one of the following constructs: acidic domain-deleted NS3 (NS3ΔA); the GTP-binding domain-deleted NS3 (NS3ΔG); or the basic domain deleted NS3 (NS3ΔB) (Fig. 8A). We also mutated the DNA sequence in the acidic domain to prevent RNAi from targeting the rescue constructs (see Methods).
Fig. 8.
The GTP-binding domain and acidic domain together is essential to promote NB proliferation. (A) Schematics of NS3 and the rescue constructs. The DNA sequence in the acidic domain is mutated to avoid RNAi knocked-down (see Methods). (B-E) L3 brain NBs. The NB-specific driver wor-gal4 was used to drive UAS-ns3-RNAi, the indicated UAS-HA:NS3 rescue constructs, and UAS-mCD8:GFP. HA staining shows NS3 localization, GFP marks the NB membrane, and Lamin marks the nuclear envelope. Genotype: UAS-Dcr-2; wor-gal4/UAS-NS3 rescue constructs; UAS-mCD8:GFP/UAS-ns3-RNAi. Scale bars: 5 μm. (F-J) A single focal plane of L3 brain. ns3-RNAi and the rescue constructs (F-I) were concurrently expressed by wor-gal4, and the cells were marked by UAS-mCD8:Gfp. The central brain region was outlined with yellow dash lines. Quantification results (J) from 5 brain lobes. Genotype: UAS-Dcr-2; wor-gal4/UAS-NS3 rescue constructs; UAS-mCD8:GFP/UAS-ns3-RNAi. Scale bars: 50 μm.
We co-expressed ns3-RNAi and each of the four rescue constructs in larval NBs and determined the subcellular localization of the rescue constructs. We found that NS3ΔA, and NS3ΔB were located in cytoplasm similar to wild type NS3 localization (Fig. 8B–D); in contrast, the NS3ΔG protein lacking the GTP-binding domain was evenly distributed in both the cytoplasm and the nucleus (Fig. 8E). Expression of the rescue constructs in wild type (without ns3 RNAi) did not generate any phenotypes (data not shown). We then assayed the ability of each rescue construct to restore proliferation to ns3 RNAi larval NBs (Fig. 8F–J, quantified in J). Partial rescue was obtained by the NS3ΔB construct (Fig. 8G). In contrast, the NS3ΔA and NS3ΔG constructs failed to rescue the ns3 RNAi phenotype (Fig. 8H,I). We conclude that the NS3 acidic domain and GTP-binding domain are essential for maintaining larval NB proliferation.
4. Discussion
Our genetic screen identified 11 genes, including ns3, that are required to prevent premature NB cell cycle exit. These genes include RNA or ribonucleoprotein binding proteins (CG10418, ns1, ns3 and CG5033), nuclear-cytoplasmic transport machinery (Ran, RanGAP, Trn-SR, and Cse-1), an ATPase (CG4908), a calcium signal transducer (Calmodulin), and a nuclear protein (bys) (Fig. 1A). Our results indicate that multiple pathways are required to promote cell cycle progression. It is unknown yet how each gene contributes to the prevention of NB cell cycle arrest. For example, Calmodulin is highly enriched in NBs (Kovalick and Beckingham, 1992), and we have shown it is required to maintain NB proliferation. Calmodulin is activated by calcium binding and subsequently activates target kinases or phosphatases, but how these targets maintain NB proliferation remains to be explored.
Here we focused on ns3. This gene is not expressed at detectable levels in the embryonic CNS (BDGP in situ homepage: http://insitu.fruitfly.org/cgi-bin/ex/report.pl?ftype=10&ftext=FBgn0266284). Consistent with this observation, ns3 homozygous mutant embryos showed no detectable NB proliferation defect, nor did overexpression of ns3 in otherwise wild type embryos produce a NB proliferation phenotype (data not shown). ns3 homozygous mutants failed to grow after larval hatching, possibly due to non-neuronal phenotypes or background lethal mutations, preventing us from analyzing larval NBs. Mosaic clonal analysis of ns3 mutants in larval NBs did not reveal a NB proliferation phenotype (data not shown), possibly because the strongest available allele is not a null allele (Hartl et al., 2013).
Lsg1, the ortholog of NS3, is a highly-conserved protein and is found in Plantae, Fungi and Animalia, suggesting widespread importance. Lsg1 was identified as required to promote the release of nuclear export adapter from the large ribosomal unit for ribosome final maturation (Hedges et al., 2005; Kallstrom et al., 2003; Reynaud et al., 2005; Weis et al., 2014). NS3 is also required for ribosome biogenesis in the cytoplasm and regulates the synthesis of ribosomal proteins RpL13 and Rps6 (Hartl et al., 2013). Therefore, we had originally hypothesized that NS3 might inhibit the NB cell cycle by down-regulating NB-specific genes that are required for NB proliferation or self-renewal. However, we observed normal expression of many NB-specific markers (Fig. 3), which suggests a distinct mechanism may operate in place of, or in parallel to, a cell growth pathway.
In Drosophila, NS3 functions non-autonomously to regulate insulin signaling in dopaminergic neurons to control whole body growth, and expression of insulin downstream effector Akt1 in ns3 mutants is sufficient to rescue the developmental defect (Kaplan et al., 2008). We found that the cell cycle defect of ns3 RNAi NBs can be fully rescued by overexpressing NS3 in NBs, showing that NS3 functions cell-autonomously to promote cell cycle progression. Interestingly, reactivation of quiescent NBs and cell cycle progression of proliferating NBs both require Akt1 (Chell and Brand, 2010; Cheng et al., 2011; Sousa-Nunes et al., 2011), and it would be important to know if Akt1 is also downstream of ns3 pathway in NBs and if ns3 RNAi compromises production or activation of Akt1.
NS3 has physical interactions with insulin/mTOR signaling components Gigas, Thor, Rheb (Ras homolog enriched in brain) and Myc (Vinayagam et al., 2016), and yet little is known how NS3 regulates insulin/mTOR signaling pathways via interacting with these proteins. More work will be needed to understand if ns3 RNAi attenuates insulin/mTOR signaling pathways to induce NB cell cycle arrest. Other genes, including ribosomal proteins, that are affected in ns3 RNAi mutant NBs can be identified in the future by identifying NB-specific RNAs for global transcriptome analysis (Hida et al., 2017; Liu et al., 2015; Miller et al., 2009; Yang et al., 2016).
At the onset of our experiments, it seemed likely that NS3 would function in Ran-dependent transport, because RNAi depletion of NS3, Ran, RanGAP and RanGEF all induce nuclear Pros localization. Surprisingly, we found that the cellular distribution of NLS:NES:GFP reporter was completely different in Ran and ns3 RNAi mutant NBs (Fig. 4), strongly indicating that NS3 and Ran act in the different pathways. This raises the question: why does loss of both Ran and NS3 lead to elevated nuclear Pros? One explanation could be that the Ran pathway acts as a secondary backup mechanism. For example, CRM1 has a pivotal role in relocating proteins “misplaced” in the nucleus, which can frequently occur due to leaky nuclear pore complexes or damage to the nuclear envelope (Fornerod et al., 1997; Fukuda et al., 1997). The Pros NES shows sensitivity to the drug leptomycin B, which is characteristic of CRM1-dependent NESs (Bi et al., 2003; Demidenko et al., 2001). The yeast and human ortholog of NS3, Lsg1, is required for shuttling the nuclear export adapter between cytoplasm and nucleus, and loss of Lsg1 impairs the recycle of nuclear export adapter into nucleus. As such, it is tempting to hypothesize Pros is primarily shuttled to the cytoplasm via the same NS3-dependent nuclear export adapter, and Ran acts as a corrective mechanism. Similarly, it is possible that Pros is regulated by both a Ran-independent and Ran-dependent mechanism.
It is important to note that the Pros NES is currently considered to have an atypical sequence, but the NLS-NES-GFP we used has a canonical NES (Bi et al., 2003; Demidenko et al., 2001). We are beginning to understand NESs are quite diverse and depend not only on the sequence, but also their tertiary structure and physical properties (Xu et al., 2012). Based on the ability to bind CRM1, multiple new consensus sequences have been identified (Kosugi et al., 2008; Xu et al., 2012). Because Pros appears to be CRM1-dependent, we believe the current understanding of what constitutes a consensus sequence simply remains too restrictive, and the use of a canonical NES was not relevant to the Pros localization mechanism.
It has previously been shown that loss of the aPKC cortical polarity protein can lead to NB cell cycle arrest (Rolls et al., 2003). Thus, failure to express, stabilize, or properly localize aPKC to the apical cortex of the mitotic NB appears to be the primary or earliest defect in ns3 RNAi NBs. It is also possible that there are multiple defects in ns3 RNAi NBs that cause cell cycle arrest; in type I NBs one defect is abnormal nuclear localization of Pros, which is known to arrest the NB cell cycle. However, at least one other mechanism is utilized by type II NBs, which do not contain detectable levels of Pros (in wild type or ns3 RNAi NBs). This second mechanism may be failure to properly localize aPKC, or a completely unknown mechanism. Addressing the role of NS3 in type II NBs is an interesting question for the future.
Supplementary Material
Acknowledgements
We thank Judith Eisen, Peter O’Day, Doe lab members, anonymous reviewer 2, and undergraduate researchers in Summer Research Program at the University of Oregon (2014) for comments and support. We also acknowledged Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at the University of Iowa, Department of Biology, Iowa City, IA 52242, to distribute monoclonal antibodies. P.W.J. was supported by NICHD R25 Summer Research Program at the University of Oregon (5R25HD070817–04). S.-L.L. and C.Q.D. were supported by the Howard Hughes Medical Institute.
Footnotes
Appendix A. Supporting information
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.ydbio.2018.04.014.
References
- Atwood SX, Prehoda KE, 2009. aPKC phosphorylates Miranda to polarize fate determinants during neuroblast asymmetric cell division. Curr. Biol 19, 723–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahrampour S, Gunnar E, Jonsson C, Ekman H, Thor S, 2017. Neural lineage progression controlled by a temporal proliferation program. Dev. Cell 43 (332–348), e334. [DOI] [PubMed] [Google Scholar]
- Baumgardt M, Karlsson D, Salmani BY, Bivik C, MacDonald RB, Gunnar E, Thor S, 2014. Global programmed switch in neural daughter cell proliferation mode triggered by a temporal gene cascade. Dev. Cell 30, 192–208. [DOI] [PubMed] [Google Scholar]
- Baumgardt M, Karlsson D, Terriente J, Diaz-Benjumea FJ, Thor S, 2009. Neuronal subtype specification within a lineage by opposing temporal feed-forward loops. Cell 139, 969–982. [DOI] [PubMed] [Google Scholar]
- Bayraktar OA, Boone JQ, Drummond ML, Doe CQ, 2010. Drosophila type II neuroblast lineages keep Prospero levels low to generate large clones that contribute to the adult brain central complex. Neural Dev. 5, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H, 2008. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Dev. 3, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C, Li X, Erclik T, Cavey M, Wells B, Desplan C, 2014. Temporal patterning of neuroblasts controls Notch-mediated cell survival through regulation of Hid or Reaper. Cell 158, 1173–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Jones T, Abbasi F, Lee H, Stultz B, Hursh DA, Mortin MA, 2005. Drosophila caliban, a nuclear export mediator, can function as a tumor suppressor in human lung cancer cells. Oncogene 24, 8229–8239. [DOI] [PubMed] [Google Scholar]
- Bi X, Kajava AV, Jones T, Demidenko ZN, Mortin MA, 2003. The carboxy terminus of Prospero regulates its subcellular localization. Mol. Cell. Biol 23, 1014–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boone JQ, Doe CQ, 2008. Identification of Drosophila type II neuroblast lineages containing transit amplifying ganglion mother cells. Dev. Neurobiol 68, 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman SK, Rolland V, Betschinger J, Kinsey KA, Emery G, Knoblich JA, 2008. The tumor suppressors brat and numb regulate transit-amplifying neuroblast lineages in Drosophila. Dev. Cell 14, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney TD, Miller MR, Robinson KJ, Bayraktar OA, Osterhout JA, Doe CQ, 2012. Functional genomics identifies neural stem cell sub-type expression profiles and genes regulating neuroblast homeostasis. Dev. Biol 361, 137–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney TD, Struck AJ, Doe CQ, 2013. midlife crisis encodes a conserved zinc-finger protein required to maintain neuronal differentiation in Drosophila. Development 140, 4155–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chell JM, Brand AH, 2010. Nutrition-responsive glia control exit of neural stem cells from quiescence. Cell 143, 1161–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng LY, Bailey AP, Leevers SJ, Ragan TJ, Driscoll PC, Gould AP, 2011. Anaplastic lymphoma kinase spares organ growth during nutrient restriction in Drosophila. Cell 146, 435–447. [DOI] [PubMed] [Google Scholar]
- Demidenko Z, Badenhorst P, Jones T, Bi X, Mortin MA, 2001. Regulated nuclear export of the homeodomain transcription factor Prospero. Development 128, 1359–1367. [DOI] [PubMed] [Google Scholar]
- Doe CQ, 2008. Neural stem cells: balancing self-renewal with differentiation. Development 135, 1575–1587. [DOI] [PubMed] [Google Scholar]
- Doe CQ, Chu-LaGraff Q, Wright DM, Scott MP, 1991. The prospero gene specifies cell fates in the Drosophila central nervous system. Cell 65, 451–464. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Livesey FJ, Cepko CL, Oliver G, 2003. Prox1 function controls progenitor cell proliferation and horizontal cell genesis in the mammalian retina. Nat. Genet 34, 53–58. [DOI] [PubMed] [Google Scholar]
- Egger B, Chell JM, Brand AH, 2008. Insights into neural stem cell biology from flies. Philos. Trans. R. Soc. Lond., B, Biol. Sci 363, 39–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsir T, Smits A, Lindström MS, Nistér M, 2012. Transcription factor PROX1: its role in development and cancer. Cancer Metastas-. Rev 31, 793–805. [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW, 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90, 1051–1060. [DOI] [PubMed] [Google Scholar]
- Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E, 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390, 308–311. [DOI] [PubMed] [Google Scholar]
- Hartl TA, Ni J, Cao J, Suyama KL, Patchett S, Bussiere C, Gui DY, Tang S, Kaplan DD, Fish M, et al. , 2013. Regulation of ribosome biogenesis by nucleostemin 3 promotes local and systemic growth in Drosophila. Genetics 194, 101–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges J, West M, Johnson AW, 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24, 567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hida N, Aboukilila MY, Burow DA, Paul R, Greenberg MM, Fazio M, Beasley S, Spitale RC, Cleary MD, 2017. EC-tagging allows cell type-specific RNA analysis. Nucleic Acids Res. 45, e138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata J, Nakagoshi H, Nabeshima Y, Matsuzaki F, 1995. Asymmetric segregation of the homeodomain protein Prospero during Drosophila development. Nature 377, 627–630. [DOI] [PubMed] [Google Scholar]
- Homem CCF, Knoblich JA, 2012. Drosophila neuroblasts: a model for stem cell biology. Development 139, 4297–4310. [DOI] [PubMed] [Google Scholar]
- Homem CCF, Repic M, Knoblich JA, 2015. Proliferation control in neural stem and progenitor cells. Nat. Rev. Neurosci 16, 647–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F, 1997. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 390, 625–629. [DOI] [PubMed] [Google Scholar]
- Joy T, Hirono K, Doe CQ, 2015. The RanGEF Bj1 promotes prospero nuclear export and neuroblast self-renewal. Dev. Neurobiol 75, 485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallstrom G, Hedges J, Johnson A, 2003. The putative GTPases Nog1p and Lsg1p are required for 60S ribosomal subunit biogenesis and are localized to the nucleus and cytoplasm, respectively. Mol. Cell Biol 23, 4344–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan DD, Zimmermann G, Suyama K, Meyer T, Scott MP, 2008. A nucleostemin family GTPase, NS3, acts in serotonergic neurons to regulate insulin signaling and control body size. Genes Dev. 22, 1877–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karcavich R, Doe CQ, 2004. Drosophila neuroblast 7–3 cell lineage: a model system for studying programmed cell death, Notch/Numb signaling, and sequential specification of ganglion mother cell identity. J Comp Neurol. 481, 240–251. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H, 2008. Nuclear export signal consensus sequences defined using a localization-based yeast selection system. Traffic 9, 2053–2062. [DOI] [PubMed] [Google Scholar]
- Kovalick GE, Beckingham K, 1992. Calmodulin transcription is limited to the nervous system during Drosophila embryogenesis. Dev. Biol 150, 33–46. [DOI] [PubMed] [Google Scholar]
- Kusano A, Staber C, Ganetzky B, 2001. Nuclear mislocalization of enzymatically active RanGAP causes segregation distortion in Drosophila. Dev. Cell 1, 351–361. [DOI] [PubMed] [Google Scholar]
- Lai S-L, Doe CQ, 2014. Transient nuclear Prospero induces neural progenitor quiescence. Elife 3, e03363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai S-L, Miller MR, Robinson KJ, Doe CQ, 2012. The Snail family member Worniu is continuously required in neuroblasts to prevent Elav-induced premature differentiation. Dev. Cell 23, 849–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yang CP, Sugino K, Fu CC, Liu LY, Yao X, Lee LP, Lee T, 2015. Opposing intrinsic temporal gradients guide neural stem cell production of varied neuronal fates. Science 350, 317–320. [DOI] [PubMed] [Google Scholar]
- Maurange C, Gould AP, 2005. Brainy but not too brainy: starting and stopping neuroblast divisions in Drosophila. Trends Neurosci. 28, 30–36. [DOI] [PubMed] [Google Scholar]
- Miller MR, Robinson KJ, Cleary MD, Doe CQ, 2009. TU-tagging: cell type-specific RNA isolation from intact complex tissues. Nat. Methods 6, 439–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud EG, Andrade MA, Bonneau F, Ly TB, Knop M, Scheffzek K, Pepperkok R, 2005. Human Lsg1 defines a family of essential GTPases that correlates with the evolution of compartmentalization. BMC Biol. 3, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls MM, Albertson R, Shih H-P, Lee C-Y, Doe CQ, 2003. Drosophila aPKC regulates cell polarity and cell proliferation in neuroblasts and epithelia. J. Cell Biol 163, 1089–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryter JM, Doe CQ, Matthews BW, 2002. Structure of the DNA binding region of prospero reveals a novel homeo-prospero domain. Structure 10, 1541–1549. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. , 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen CP, Jan LY, Jan YN, 1997. Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell 90, 449–458. [DOI] [PubMed] [Google Scholar]
- Sousa-Nunes R, Yee LL, Gould AP, 2011. Fat cells reactivate quiescent neuroblasts via TOR and glial insulin relays in Drosophila. Nature 471, 508–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steggerda SM, Paschal BM, 2002. Regulation of nuclear import and export by the GTPase Ran. Int. Rev. Cytol 217, 41–91. [DOI] [PubMed] [Google Scholar]
- Terry LJ, Shows EB, Wente SR, 2007. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science 318, 1412–1416. [DOI] [PubMed] [Google Scholar]
- Tsuji T, Hasegawa E, Isshiki T, 2008. Neuroblast entry into quiescence is regulated intrinsically by the combined action of spatial Hox proteins and temporal identity factors. Development 135, 3859–3869. [DOI] [PubMed] [Google Scholar]
- Vaessin H, Grell E, Wolff E, Bier E, Jan LY, Jan YN, 1991. prospero is expressed in neuronal precursors and encodes a nuclear protein that is involved in the control of axonal outgrowth in Drosophila. Cell 67, 941–953. [DOI] [PubMed] [Google Scholar]
- Vinayagam A, Kulkarni MM, Sopko R, Sun X, Hu Y, Nand A, Villalta C, Moghimi A, Yang X, Mohr SE, et al. , 2016. An integrative analysis of the InR/PI3K/Akt network identifies the dynamic response to insulin signaling. Cell Rep. 16, 3062–3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh KT, Doe CQ, 2017. Drosophila embryonic type II neuroblasts: origin, temporal patterning, and contribution to the adult central complex. Development 144, 4552–4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis BL, Missbach S, Marzi J, Bohnsack MT, Schleiff E, 2014. The 60S associated ribosome biogenesis factor LSG1–2 is required for 40S maturation in Arabidopsis thaliana. Plant J. 80, 1043–1056. [DOI] [PubMed] [Google Scholar]
- Xu D, Farmer A, Collett G, Grishin NV, Chook YM, 2012. Sequence and structural analyses of nuclear export signals in the NESdb database. Mol. Biol. Cell 23, 3677–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CP, Fu CC, Sugino K, Liu Z, Ren Q, Liu LY, Yao X, Lee LP, Lee T, 2016. Transcriptomes of lineage-specific Drosophila neuroblasts profiled by genetic targeting and robotic sorting. Development 143, 411–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudin D, Fainzilber M, 2009. Ran on tracks—cytoplasmic roles for a nuclear regulator. J. Cell. Sci 122, 587–593. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.