Abstract
An 82-residue-long chimeric peptide was synthesised by solid phase peptide synthesis (SPPS), following the Fmoc protocol. Microwave (MW) radiation-assisted synthesis was compared to standard synthesis using low loading (0.20 mmol/g) of polyethylene glycol (PEG) resin. Similar synthetic difficulties were found when the chimeric peptide was obtained via these two reaction conditions, indicating that such difficulties were inherent to the sequence and could not be resolved using MW; by contrast, the number of coupling cycles and total reaction time became reduced whilst crude yield and percentage recovery after purification were higher for MW radiation-assisted synthesis.
Keywords: solid phase peptide synthesis, chimeric peptide, coupling, microwave radiation, Fmoc protocol
1. Introduction
Bruce Merrifield developed solid phase peptide synthesis (SPPS) in 1963; this involves a solid support where the peptide becomes elongated and excess reagents and by-products being withdrawn through simple filtration after every reaction step (incorporating α-amino and deprotection) [1,2]. Significant advances have been made in SPPS; however, this methodology has involved difficulties regarding synthesis in some cases, these being very important concerning difficult peptide sequences having severe inefficiencies regarding acylation and deprotection reactions depending on peptide sequence of amino acid (aa) residues. Peptide sequences containing much hydrophobic aa, bulky side chains or bulky protecting groups are more prone to such inefficiencies, thereby producing peptide chain deletion or truncation, in turn, is the product of peptide-resin and/or secondary structure (mostly β-sheet) aggregation causing steric hindrance [3]. These associations produce low peptide-resin complex solvation and low reagent accessibility to reaction sites. Such complications could be significant regarding long peptide synthesis because there is the high likelihood of interactions between adjacent chains and steric hindrance; moreover, long peptides require more reaction steps regarding coupling and deprotection reactions, thereby increasing the probability of incomplete reactions [4,5,6].
Polystyrene-polyethylene glycol (PS-PEG), polyethylene glycol-acrylamide (PEGA) [7] and polyethylene glycol (PEG) [8] resins have been used to overcome such synthesis aggregation problems, usually involving great swelling and high solvation directing reagent diffusion toward reaction sites; also, low loading resins have been used to reduce the probability of association.
Chaotropic agents and pseudo-prolines have been used to avoid these associations and conventional heat and microwave (MW) radiation have been used for disrupting these associations and accelerating synthesis [6,9]. However, such strategies do not ensure such difficulties being overcome because target molecule synthesis also depends on its aa composition which could involve significant steric hindrance and intra-molecular interactions which would lead to inefficient synthesis.
SPPS protocols highly standardised let produce peptides of high purity, and, average peptide of 50 residues of long, but, also well know that shorter peptide could be very problematic because the sequence composition and longer peptide could be synthesized by this stepwise synthesis like Ribonuclease A was made first time in 1971 [9,10,11]. Two main techniques were introduced to try overcoming the cumulative effect of stepwise synthesis of long peptide sequences and small proteins; protected peptide fragments and chemical ligation. Condensation of the protected peptide was the method commonly used in the 80’s, and can be performed in solution or solid phase; however, this approach provides limited utility because of the racemisation, low solubility and longer reactions times of the protected peptides [12,13]. Chemical ligation (Native chemical ligation, peptide hydrazides, click reaction, a-Ketoacid-hydroxylamine ligation and Staudinger ligation) is a reaction chemoselective, which do not have racemisation problems and better solubility than the approach of the protected peptide; however, both require the purification steps of intermediaries and the final product [14]. On the other hand, stepwise synthesis has let produce these macromolecules by standard and MW-assisted SPPS, and these only require the purification of the final product which simplifies the process; but, MW radiation not only accelerate coupling and deprotection reactions, moreover, could accelerate byproducts formation if the reaction conditions are not controlled [15,16,17,18].
This article reports the synthesis of an 82-residue-long linear peptide chimera (82KNSFSLGENPNANPGGVIKHMRFHADYQAPFLGGGYGGEVLYHVPLAGVYRSLKKQLEGGMTDVNRYRYSNNYEEQPHISGG1, derived from four protein fragments which are relevant to Plasmodium falciparum invasion of target cells [19]) and its evaluation regarding two reaction conditions (standard and MW-assisted) involving a low loading (0.20 mmol/g) PEG resin (Rink Amide ChemMatrix).
2. Results
Loading resin was initially reduced from 0.60 mmol/g to 0.20 mmol/g to minimise the probability of synthesis difficulties caused by aggregation between adjacent chains (β-sheet secondary structure) during peptide elongation over the solid support.
Cleavage controls were made every 11 residues coupled, approximately, to evaluate synthesis progress. A single signal for the 11th residue was observed in the chromatogram during the first standard synthesis control (i.e., its ion peak at 1228.53 Da); this was also observed in the MW-assisted strategy (Table 1, Figure S1). A single signal for the 22nd residue was observed in the chromatogram for the second control of both synthesis conditions (i.e., molecular ion peak at 2628.16 Da; Table 1, Figure S2). Synthesis inefficacy regarding peptide elongation thus did not occur up to this point; SPPS peptides of this length are usually obtained without difficulties because such length is not long enough for adjacent interaction between peptide chains [19].
Table 1.
Results of controls and chimeric peptide for standard and microwave (MW)-assisted synthesis.
| Residue Control | Molecular Weight (Da) § | Standard | Microwave-Assisted | ||
|---|---|---|---|---|---|
| (m/z) a | tR (min) | (m/z) a | tR (min) | ||
| 11 | 1228.53 | 1229.52 | 14.20 | 1230.01 | 14.43 |
| 22 | 2628.16 | 2628.29 | 17.17 | 2628.06 | 17.59 |
| 33 | 3887.86 | 3831.50 (−57.81) b 3887.90 |
19.72 | 3831.93 (−57.52) b 3889.61 |
19.85 |
| 3889.31 | 21.18 | 3889.45 | 21.29 | ||
| 3888.07 4055.84 (+166.53) c |
22.43 | 3889.18 4058.04 (+168.59) c |
22.26 | ||
| 44 | 5065.51 | 4939.21 (−128. 02) 4996.03 (−71.20) 5066.91 |
27.36 | 4938.52 (−128.34) 4995.93 (−70.93) 5067.40 |
27.50 |
| 5067.23 | 28.60 | 5066.86 | 28.64 | ||
| 5065.90 5178.52 (+111.29) c |
29.87 | 5066.98 5178.61 (+111.75) c |
29.42 | ||
| 57 | 6348.07 | 6221.41 (−127.59) 6234.78 (−114.54) 6350.06 |
28.17 | 6235.14 (−114.91) 6351.36 |
27.97 |
| 6291.44 (−57.88) 6349.32 |
29.05 | 6350.05 | 28.66 | ||
| 6349.47 6632.30 (+282.98) |
29.41 | ND | 29.16 | ||
| 66 | 7467.68 | ND | 28.65 | ND | 28.73 |
|
7469.35 7580.21 (+111.86) c |
29.51 | 7467.84 | 29.54 | ||
| 82 e | 9051.42 | 8146.32 (−928.97) 9075.29 (+23.87) d |
26.61 27.73 |
9075.99 (+24.57) d | 26.79 |
§ Molecular weight calculated (Table S1). a The values in bold letters marked for m/z of standard and MW-assisted synthesis correspond to molecular ion of the target molecule in the main peak of chromatogram (control residues 11th and 22nd) or the peak of fraction recollected. b ion peak corresponds to a glycine deletion. c Values calculated for multiple ter-butylations (t-Bu = 56.1 Da, 2t-Bu = 112.2 Da, 3t-Bu = 168.3Da; Table S2). d Value calculated for peak ion plus sodium. e Here fractions did not recollect. Note: the values in parentheses correspond to differences with the expected molecular weight.
Three signals appeared in the chromatogram for the third control of the 33rd residue regarding crude peptide synthesised by both reaction conditions (Table 1, Figure S3). Fractions from every peak were collected and analysed by MALDI-TOF MS in each case. Minor peak tR (19.72 min. standard and 19.85 min. MW-assisted) for both reaction conditions was likely due to Gly deletion (residue 23 or 24). The tR of 21.18 min. standard and 21.29 min. MW-assisted were for the 33-residue-long elongated peptide (3887.86 Da). The higher tR (22.43 and 22.26 min. standard and MW-assisted, respectively) ter-butylated cleavage-derived species caused to ter-butyl group junction with the peptide chain; this group came from the aa chain previously protected by t-Boc group. This frequently occurs regarding peptide synthesis cleavage and could be overcome by improving cleavage conditions (time, volume and/or scavenger) [20]. It is worth stressing that the highest intensity peak for both reaction conditions occurred for the 33-residue-long target molecule.
Three signals were observed in the chromatograms for the fourth control of the 44th residue regarding both reaction conditions; they were for deletion species, the 44-residue-long target molecule and the ter-butylated species (Table 1, Figure S4). The intensity of the peak containing the target molecule was similar to that for deletion species; such decrease in peak intensity (compared to 33-residue-long control) was produced by the accumulation of incompleted acylations and deprotections caused by aggregation, thereby hampering synthesis (observed in both strategies). Similar peak intensity results were obtained for the control for the 57th residue (Figure S5). Decreased intensity regarding control for the 66th residue for both standard and MW-assisted synthesis was observed; two signals appeared in the chromatograms for the 66-residue-long target molecule. During the synthesis, inefficiencies accumulations caused a decrease in intensity for the peak that contained the target molecule (Figure S6).
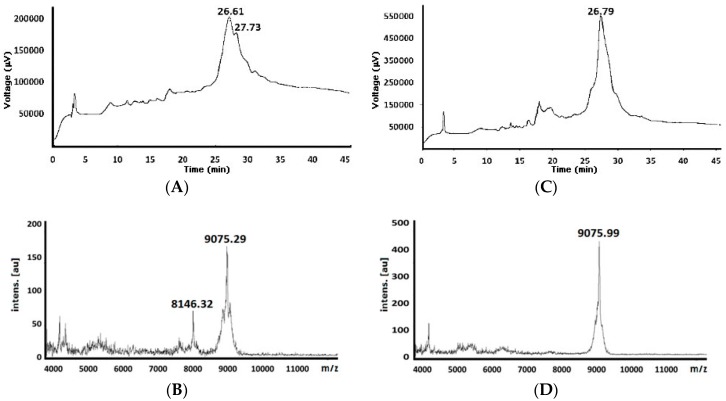
The 82-residue-long chimeric peptide was obtained (82KNSFSLGENPNANPGGVIKHMRFHADYQAPFLGGGYGGEVLYHVPLAGVYRSLKKQLEGGMTDVNRYRYSNNYEEQPHISGG1) by both strategies; the broad signal was observed in the chromatograms which the target peptide (Figure 1); these chromatograms did not have significant differences regarding both reaction conditions. Total synthesis reaction time was 9720 min (162 h) for the standard strategy compared to 2268 min (37.8 h) for the MW-assisted strategy, giving a 4.26:1 ratio. Shorter MW-assisted reaction time was due to less time being required for every coupling and deprotection reaction step enabling this macromolecule to be obtained fourfold faster than in standard conditions.
Figure 1.
Chromatographic profiles and mass spectra for crude peptide’s 82nd residue (theoretical 9051.42 Da) obtained by standard (A) and (B) (Chromatogram and mass spectra, respectively) and MW-assisted reaction conditions (C) and (D) Chromatogram and mass spectra, respectively).
Greater crude peptide yield was obtained by MW-assisted strategy (13.06% compared to 9.98% for the standard condition, Table 2).
Table 2.
Control synthesis of standard and MW-assisted strategies.
| Strategy | Resin (g) | Obtained (mg) | % Yield | Purified Quantity (mg) | Amount of Peptide Obtained in the Purification (mg) | % Purification Yield | |
|---|---|---|---|---|---|---|---|
| Theoretical | Experimental | ||||||
| Standard | 0.507 | 905.70 | 90.42 | 9.98 | 10.58 | 0.97 | 9.16 |
| MW-assisted | 0.501 | 905.80 | 118.32 | 13.06 | 11.13 | 1.41 | 12.67 |
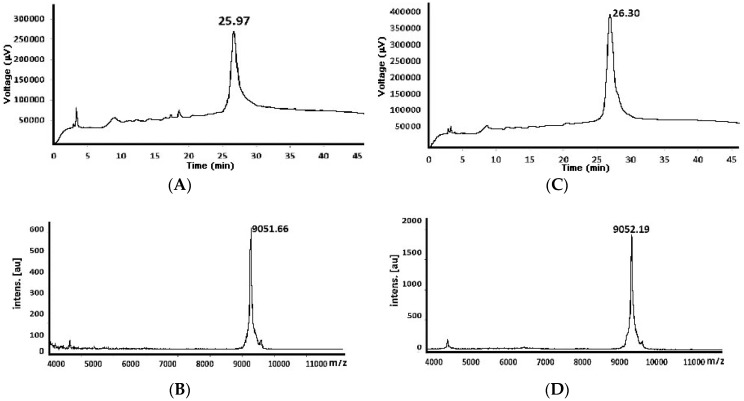
The 82-residue-long chimeric peptide synthesised by these two strategies was purified (Figure 2); the MW-assisted SPPS had a higher recovery percentage (12.67%). All this demonstrated that although the MW did not totally avoid inefficient synthesis; it was good enough to improve chimeric peptide yield and purity.
Figure 2.
Chromatographic profiles and mass spectrum of the purified peptide’s 82nd residue (MW theoretical 9051.42 Da) obtained by standard (A) and (B) (Chromatogram and mass spectra, respectively) and MV-assisted reaction conditions (C) and (D) Chromatogram and mass spectra, respectively).
In a previous study, the chimera obtained here by MW-assisted synthesis was evaluated in murine BALB/C mice against malaria (Plasmodium berghei ANKA and yoelii 17XL) and this shows protection induce capacity [21].
3. Discussion
The obtained chimera peptide or 82-mer is remarkable, and how is shown in the results the standard strategy also lets obtained the molecule, but MW-assisted approach produces a better product in yield with less time. Here is important highlight that the stepwise synthesis is a tool important to obtain long peptide and with the advances obtained every day, this technique is gain more field of development. In another side, chemical ligation let also obtained these types of molecules, but require not only personal prepared but also some special equipment. Besides, chemical ligation has some disadvantages (Table 3) as longer times the reaction is required, and, not always let’s obtain peptide bond. Native Chemical Ligation requires a cysteine or alanine residue in a strategic position to obtain the long peptide or protein, also, all techniques of ligation require an additional step of separation between reagents and product. These are the reasons of what is essential follow investigating in stepwise to obtain this long peptide, and in the literature, some landmark peptides like ribonuclease A (124 residues) and the peptide of human immunodeficiency virus (HIV)-1 (Tat of 86 residues) are good are examples of the potential of SPPS [22,23,24].
Table 3.
Advantages and disadvantages of chemical ligation.
| Advantages | Disadvantages | Ref. |
|---|---|---|
|
|
[22] |
4. Materials and Methods
4.1. Reagents and Solvents
The following reagents, 9-fluorenylmethoxycarbonyl (Fmoc) protected aa, O-(Benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TBTU), N,N′-dicyclohexylcarbodiimide (DDC) and 1-hydroxybenzotrazole (HOBt), were purchased from AAPPTEC (Louisville, KY, USA), ChemMatrix resin (0.60 mmol/g) from PCAS BiomMatrix Inc. (Saint-Jean-sur-Richelieu, QC, Canada), N,N-diisopropylethylamine (DIPEA), triisopropylsilane (TIS), Triton X-100, α-cyano-4-hydroxycinnamic acid (HCCA), methanol and absolute ethanol from Sigma-Aldrich (St. Louis, MO, USA), piperidine, pyridine, phenol, potassium cyanide, isopropyl alcohol (IPA), acetonitrile (ACN), N-methyl-2-pyrrolidone (NMP), trifluoroacetic acid (TFA) and diethyl ether from Panreac (Barcelona, Spain), acetic anhydride (Ac2O), ninhydrin and ethanedithiol (EDT) from Merck (Boston, MA, USA) and dichloromethane (DCM) from The Dow Chemical Company (Bogotá, Colombia).
4.2. Peptide Design
A linear chimeric peptide consisting of four Plasmodium falciparum epitopes separated by Gly-Gly (GG) spacers was selected for studying this long peptide’s synthesis pattern (Scheme 1). The glycine is commonly used as a spacer due to its flexibility, low hindrance [25] and experimentally we have found structural similarity between monomer and polymerised peptides (without G and with G spacer, respectively) platforms. The epitopes were selected bearing previous research in mind [19,26,27,28,29], and the Peptide Companion software (free software of excel version, Spyder Institute’s, Praha, Czech Republic) was used for choosing the order of the chimeric peptide’s epitope [30].
Scheme 1.
Linear chimeric peptide sequence derived from four Plasmodium falciparum proteins (CSP, STARP, MSP-1, and RESA-155; residues marked with letters purple, green, black and blue colour, respectively).
4.3. Peptide Synthesis
The chimeric peptide was synthesised by SPPS using Fmoc/tBu protocol on 0.60 mmol/g Rink Amide ChemMatrix resin (PCAS BiomMatrix Inc., Saint-Jean-sur-Richelieu, QC, Canada), using standard and MW-assisted strategies, briefly described below.
Initially, 1.008 g resin (loading: 0.60 mmol/g) was swollen with 5 mL DCM and NMP for 10 min in each solvent; 0.20 mmol Fmoc-Gly-OH was then added to the resin and reacted for 60 min at room temperature (RT); the aa had been previously activated with TBTU/HOBt/DIPEA (1:1:2 mmol) and dissolved in 5 mL NMP. The resin was rinsed with DMF, IPA, and DCM three times with each solvent for 1 min during this time; the amine group in the resin which did not react was then capped with 10 mL acetylation solution (anhydride acetic/DMF/pyridine; 1:1:1) for 60 min at RT and the resin was rinsed again. A qualitative Kaiser’s test was used at this point for evaluating whether an amine group was present [31]; 0.20 mmol/g resin loading was thus obtained, as described by Boll et al. [32]. Fmoc-Gly coupled resin was used in both aforementioned strategies.
4.3.1. The Standard Strategy
Fmoc-Gly-resin (loading: 0.20 mmol/g) was deprotected with three cycles of 10 mL piperidine/DMF (1:1) for 10 min. and washed with DMF, IPA and DCM for 1 min, three times. The first strategy for all couplings involved using a fivefold excess of Fmoc aa derivative, DDC/HOBT in NMP. The coupling reaction was carried out for 60 min at RT. A second cycle was carried if the Kaiser test indicated incomplete coupling, using a fivefold excess with TBTU/HOBT/DIPEA. The Fmoc group was removed after every aa had been anchored, as previously described here (acylation and deprotection are evaluated by qualitative Kaiser in every reaction step). Peptides were cleaved with 10 mL/g TFA/TIS/H2O/EDT resin (94:1:2.5:2.5) for 3 h at RT; the peptides were then precipitated from the cleavage solution using cold diethyl ether and washed four times with cold ether [33].
4.3.2. The Microwave-Assisted Strategy
All microwave (MW)-assisted strategy couplings and deprotections were performed at 80 °C for 30 s of radiation (10 Watts) and then for 1 min at RT, except for Asp and His aa coupling and Asp aa deprotection which were carried out at RT for 60 min, as described for the standard strategy, to avoid by-product formation (racemisation and aspartimide) [34]. Cleavage involved the same conditions as for standard synthesis.
4.4. Characterization
4.4.1. HPLC Analysis
The crude chimeric peptide and reaction control intermediates were analysed by reversed-phase HPLC using a Merck-Hitachi chromatograph with a Vydac C-18 column (250 mm length, 4.6 mm inner diameter, 5 μm particle size, Merck, Kenilworth, NJ, USA). The samples were dissolved in water and acetonitrile (1:1) with 0.05% TFA and eluted with a 0% to 70% gradient of solution B (0.05% TFA in water [Solution A] and 0.05% TFA in acetonitrile [Solution B]) for 45 min at RT and 1 mL/min. Peak signals were detected at 210 nm.
4.4.2. Mass Spectrometry
A Microflex MALDI-TOF spectrometer (Bruker Daltonics, Billerica, MA, USA) was used for characterising the products; 18.0 μL of an oversaturated α-cyano-4-hydroxycinnamic acid matrix solution was mixed with 2.0 μL peptide solution (5.0 × 10−4 mg/μL), and then 2.5 μL of this mixture was placed in the 96-well plates to be analysed.
4.4.3. Purification of the Chimeric Peptide
The 82-mere peptide obtained by the two strategies were purified on a semi-preparative chromatograph (Merck-Hitachi) using a Vydac C-18 Colum (250 mm length, 22 mm inner diameter, 10 μm particle size). Each peptide (1 mL that contained 10 mg approx.) was injected and eluted with 21–41% gradient of solution B for 60 min at RT and 4.5 mL/min of flow. The peak signals were detected at 210 nm; fractions of these were collected, analysed by MALDI-TOF mass spectrometry and to those that contained the target molecule were chosen to analysed by analysis RP-HPLC (Section 4.4.1). Finally, the fractions selected were joined and lyophilised to obtain the purified peptide (Figure 2).
5. Conclusions
The present study has described the synthesis of a chimeric peptide by SPPS methodology involving standard and MW-assisted strategies for Fmoc/tBu protocol. Both procedures involved some difficulties caused by the sequence itself thereby hampering synthesis of this 82-residue-long molecule. Although the MW-assisted strategy did not totally overcome such difficulties, it had a shorter reaction time, higher crude product yield and higher final product recovery percentage during purification.
Acknowledgments
We would like to thank Jason Garry for reviewing the manuscript and making appropriate corrections.
Supplementary Materials
The following are available online.
Author Contributions
Y.F.V., M.V.M. designed and performed the chimeric peptide synthesis; also wrote the paper. M.E.P. supported the main ideas behind this work. All the authors revised and approved these article.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the chimeric peptide is available by request from the authors.
References
- 1.Mitchell A.R. Bruce Merrifield and solid-phase peptide synthesis: A historical assessment. Biopolym. Pept. Sci. Sect. 2008;90:175–184. doi: 10.1002/bip.20925. [DOI] [PubMed] [Google Scholar]
- 2.Kimmerlin T., Seebach D. ‘100 years of peptide synthesis’: Ligation methods for peptide and protein synthesis with applications to β-peptide assemblies*. 2005;65:229–260. doi: 10.1111/j.1399-3011.2005.00214.x. [DOI] [PubMed] [Google Scholar]
- 3.Coin I., Beyermann M., Bienert M. Solid-phase peptide synthesis: From standard procedures to the synthesis of difficult sequences. Nat. Protoc. 2007;2:3247–3256. doi: 10.1038/nprot.2007.454. [DOI] [PubMed] [Google Scholar]
- 4.De L. Milton R.C., Milton S.C.F., Adams P.A. Prediction of Difficult Sequences in Solid-Phase Peptide Synthesis. J. Am. Chem. Soc. 1990;112:6039–6046. doi: 10.1021/ja00172a020. [DOI] [Google Scholar]
- 5.Furrer J., Piotto M., Bourdonneau M., Limal D., Guichard G., Elbayed K., Raya J., Briand J.P., Bianco A. Evidence of secondary structure by high-resolution magic angle spinning NMR spectroscopy of a bioactive peptide bound to different solid supports. J. Am. Chem. Soc. 2001;123:4130–4138. doi: 10.1021/ja003566w. [DOI] [PubMed] [Google Scholar]
- 6.Paradís-Bas M., Tulla-Puche J., Albericio F. The road to the synthesis of ‘difficult peptides’. Chem. Soc. Rev. 2016;45:631–654. doi: 10.1039/C5CS00680E. [DOI] [PubMed] [Google Scholar]
- 7.Renil M., Ferreras M., Delaisse J.M., Foged N.T., Meldal M. PEGA supports for combinatorial peptide synthesis and solid-phase enzymatic library assays. J. Pept. Sci. 1998;4:195–210. doi: 10.1002/(SICI)1099-1387(199805)4:3<195::AID-PSC141>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 8.García-Martín F., Quintanar-Audelo M., García-Ramos Y., Cruz L.J., Gravel C., Furic R., Côté S., Tulla-Puche J., Albericio F. Chemmatrix, a poly (ethylene glycol)-based support for the solid-phase synthesis of complex peptides. J. Comb. Chem. 2006;8:213–220. doi: 10.1021/cc0600019. [DOI] [PubMed] [Google Scholar]
- 9.Behrendt R., White P., Offer J. Advances in Fmoc solid-phase peptide synthesis. J. Pept. Sci. 2016;22:4–27. doi: 10.1002/psc.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li C.H., Yamashiro D. The Synthesis of a Protein Possessing Growth-Promoting and Lactogenic Activities. J. Am. Chem. Soc. 1970;92:7608–7609. doi: 10.1021/ja00729a028. [DOI] [PubMed] [Google Scholar]
- 11.Chandrudu S., Simerska P., Toth I. Chemical methods for peptide and protein production. Molecules. 2013;18:4373–4388. doi: 10.3390/molecules18044373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nishiuchi Y., Inui T., Nishio H., Bodi J., Kimura T., Tsuji F.I., Sakakibara S. Chemical synthesis of the precursor molecule of the Aequorea green fluorescent protein, subsequent folding, and development of fluorescence. Proc. Natl. Acad. Sci. USA. 1998;95:13549–13554. doi: 10.1073/pnas.95.23.13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pan M., Gao S., Zheng Y., Tan X., Lan H., Tan X., Sun D., Lu L., Wang T., Zheng Q., et al. Quasi-Racemic X-ray Structures of K27-Linked Ubiquitin Chains Prepared by Total Chemical Synthesis. J. Am. Chem. Soc. 2016;138:7429–7435. doi: 10.1021/jacs.6b04031. [DOI] [PubMed] [Google Scholar]
- 14.Nilsson B.L., Soellner M.B., Raines R.T. Chemical Synthesis of Proteins. Rev. Lit. Arts Am. 2005;18:91–118. doi: 10.1146/annurev.biophys.34.040204.144700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nutt R.F., Brady S.F., Darke P.L., Ciccarone T.M., Colton C.D., Nutt E.M., Rodkey J.A., Bennett C.D., Waxman L.H., Sigal I.S. Chemical synthesis and enzymatic activity of a 99-residue peptide with a sequence proposed for the human immunodeficiency virus protease. Proc. Natl. Acad. Sci. USA. 1988;85:7129–7133. doi: 10.1073/pnas.85.19.7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kresge N., Simoni R.D., Hill R.L. The solid phase synthesis of ribonuclease A by Robert Bruce Merrifield. J. Biol. Chem. 2006;281:e21. doi: 10.1002/chin.200645260. [DOI] [Google Scholar]
- 17.Pedersen S.L., Tofteng A.P., Malik L., Jensen K.J. Microwave heating in solid-phase peptide synthesis. Chem. Soc. Rev. 2012;41:1826–1844. doi: 10.1039/C1CS15214A. [DOI] [PubMed] [Google Scholar]
- 18.Palasek S.A., Cox Z.J., Collins J.M. Limiting racemization and aspartimide formation in microwave-enhanced fmoc solid phase peptide synthesis. J. Pept. Sci. 2007;13:143–148. doi: 10.1002/psc.804. [DOI] [PubMed] [Google Scholar]
- 19.Patarroyo M.E., Bermúdez A., Alba M.P., Vanegas M., Moreno-Vranich A., Poloche L.A., Patarroyo M.A. IMPIPS: The Immune Protection-Inducing Protein Structure Concept in the Search for Steric-Electron and Topochemical Principles for Complete Fully-Protective Chemically Synthesised Vaccine Development. PLoS ONE. 2015;10:e0123249. doi: 10.1371/journal.pone.0123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biosystems A. Cleavage, Deprotection, and Isolation of Peptides after Fmoc Synthesis. Tech. Bull. 1998:1–12. doi: 10.1038/nrd.2016.29. [DOI] [Google Scholar]
- 21.Lozano J.M., Varela Y., Silva Y., Ardila K., Forero M., Guasca L., Guerrero Y., Bermudez A., Alba P., Vanegas M., et al. A large size chimeric highly immunogenic peptide presents multistage plasmodium antigens as a vaccine candidate system against malaria. Molecules. 2017;22:1837. doi: 10.3390/molecules22111837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thapa P., Zhang R.-Y., Menon V., Bingham J.-P. Native Chemical Ligation: A Boon to Peptide Chemistry. Molecules. 2014;19:14461–14483. doi: 10.3390/molecules190914461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frankel A.D., Biancalana S., Hudson D. Activity of synthetic peptides from the Tat protein of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA. 1989;86:7397–7401. doi: 10.1073/pnas.86.19.7397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gutte B., Merrifield R.B. The Synthesis of Ribonuclease A. J. Biol. Chem. 1971;246:1922–1941. [PubMed] [Google Scholar]
- 25.Reddy Chichili V.P., Kumar V., Sivaraman J. Linkers in the structural biology of protein-protein interactions. Protein Sci. 2013;22:153–167. doi: 10.1002/pro.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alba M.P., Salazar L.M., Vargas L.E., Trujillo M., Lopez Y., Patarroyo M.E. Modifying RESA protein peptide 6671 to fit into HLA-DRβ1* pockets induces protection against malaria. Biochem. Biophys. Res. Commun. 2004;315:1154–1164. doi: 10.1016/j.bbrc.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Bermúdez A., Calderon D., Moreno-Vranich A., Almonacid H., Patarroyo M.A., Poloche A., Patarroyo M.E. Gauche+ side-chain orientation as a key factor in the search for an immunogenic peptide mixture leading to a complete fully protective vaccine. Vaccine. 2014;32:2117–2126. doi: 10.1016/j.vaccine.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 28.Bermúdez A., Vanegas M., Patarroyo M.E. Structural and immunological analysis of circumsporozoite protein peptides: A further step in the identification of potential components of a minimal subunit-based, chemically synthesised antimalarial vaccine. Vaccine. 2008;26:6908–6918. doi: 10.1016/j.vaccine.2008.09.071. [DOI] [PubMed] [Google Scholar]
- 29.Bermúdez A., Alba M.P., Vanegas M., Patarroyo M.E. 3D structure determination of STARP peptides implicated in P. falciparum invasion of hepatic cells. Vaccine. 2010;28:4989–4996. doi: 10.1016/j.vaccine.2010.05.025. [DOI] [PubMed] [Google Scholar]
- 30.Krchnák V., Flegelová Z., Vágner J. Aggregation of resin-bound peptides during solid-phase peptide synthesis. Prediction of difficult sequences. Int. J. Pept. Protein Res. 1993;42:450–454. doi: 10.1111/j.1399-3011.1993.tb00153.x. [DOI] [PubMed] [Google Scholar]
- 31.Coin I., Beyermann M., Bienert M. Monitoring solid phase peptide synthesis. Protoc. Exch. 2007:4–6. doi: 10.1038/nprot.2007.461. [DOI] [PubMed] [Google Scholar]
- 32.Boll E., Drobecq H., Ollivier N., Blanpain A., Raibaut L., Desmet R., Vicogne J., Melnyk O. One-pot chemical synthesis of small ubiquitin-like modifier protein-peptide conjugates using bis(2-sulfanylethyl)amido peptide latent thioester surrogates. Nat. Protoc. 2015;10:269–292. doi: 10.1038/nprot.2015.013. [DOI] [PubMed] [Google Scholar]
- 33.Rivera Z., Granados G., Pinto M., Varón D., Carvajal C., Chaves F., Calvo J., Rodríguez R., Guzmán F., Patarroyo M.E. Double dimer peptide constructs are immunogenic and protective against Plasmodium falciparum in the experimental Aotus monkey model. J. Pept. Res. 2002;59:62–70. doi: 10.1046/j.1397-002x.2001.00001_957.x. [DOI] [PubMed] [Google Scholar]
- 34.Murray J.K., Gellman S.H. Parallel synthesis of peptide libraries using microwave irradiation. Nat. Protoc. 2007;2:624–631. doi: 10.1038/nprot.2007.23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.