Abstract
Background
Since colorectal cancer (CRC) is one of the most common malignant tumors worldwide, we aimed to identify the role of sex determining region Y (SRY)-box 18 (SOX18) in CRC.
Methods
RT-PCR and immunohistochemistry were employed to detect the expression of SOX18 in CRC samples. We then identified the effect of SOX18 on cell proliferation, cell cycle, and apoptosis by cell counting kit-8 (CCK-8), flow cytometry, and annexin V/PI staining, respectively. The effect of silencing SOX18 expression in CRC development was evaluated by using a xenograft mouse model.
Results
First, we found that SOX18 was overexpressed in CRC tissues and cell lines and that SOX18 levels in CRC tissues were positively associated with advanced clinical stages, vascular invasion, and lymph node metastasis. Furthermore, patients with higher expression of SOX18 had a lower survival rate. Overexpression of SOX18 significantly promoted cell proliferation, promoted S cell cycle progression, and inhibited cell apoptosis. Conversely, downregulation of SOX18 clearly weakened cell proliferation, induced G0/G1 cell cycle phase arrest, and gave rise to cell apoptosis. The results showed that shSOX18 significantly inhibited tumor growth and weight. Ki67 expression was also decreased by SOX18 silencing treatment.
Conclusion
Our study indicates that SOX18 may have a carcinogenic effect on CRC, which might provide novel insights into CRC prevention and treatment.
Keywords: sex determining region Y, prognosis, biomarker, CRC, cell apoptosis, cell cycle
Introduction
CRC is a common malignant cancer worldwide, and its high mortality makes it the second leading cause of cancer-related deaths due to its poor prognosis after cancer progression.1 In China, CRC mortality rate has been ranked fifth in malignant tumor mortality, seriously endangering human health.2,3 The occurrence of colon cancer is closely related to diet, environment, heredity, and other factors.4 The prevention and treatment of colon cancer has received increasing attention in recent years. When the balance between the oncogene and the tumor suppressor gene is disrupted, the expression of the oncogene that controls cell proliferation is sustained or upregulated and the tumor suppressor gene is not expressed or is inactivated; thus, the cancerous cell escapes the control of the body’s immune mechanism and forms a tumor.5 This leads to the deterioration and metastasis of cells. The detection of cancer-related genes is of clinical significance in predicting the occurrence, development, therapeutic effect, and prognosis of colon cancer.6–8
The sex determining region Y (SRYSOX18) is a member of the Sox transcription factor subfamily which plays an important role in the development of blood and lymphatic vessels. Mutations of the gene or abnormal expression are related to tumorigenesis and tumor development. Recently, studies reported that abnormal expression of SOX18 was observed in prostate cancer,9 breast cancer,10 non-small-cell lung carcinoma,11 non-melanoma skin cancer,12 and ovarian cancer.13 The role of SOX18 in CRC remains unknown.
Previously, we found abnormal SOX18 expression in CRC tissues, and patients with higher expression of SOX18 had a lower survival rate. SOX18 also regulated CRC cell growth in vitro and in vivo. The present study aimed at exploring SOX18 expression in CRC samples and the correlation between SOX18 and CRC prognosis.
Materials and methods
Patients and tissue samples
Sixty samples of CRC tissues and benign tissues surgically removed from patients at The First Affiliated Hospital of Gannan Medical University were collected from 2013 to 2016. Adjacent normal tissues were also collected as negative controls. Preoperative clinical and pathological follow-up data were completed by all patients. Ethical approval for the study was provided by the Ethics Committee of The First Affiliated Hospital of Gannan Medical University. Written informed consent was obtained from the study subjects.
IHC
Tissue sections were initially deparaffinized, rehydrated, and then heated in EDTA (pH 8.0); antigen retrieval was performed in 10 mM citrate buffer for 5 minutes at 100°C. The incubation with SOX18 antibody (1:200; cat. no ab109194, Abcam, Cambridge, MA, USA) or Ki67 (1:400; cat. no Ab833, Abcam, USA) was performed at room temperature for 1 hour, followed by incubation with biotin-labeled secondary antibodies. Slides were then developed using DAB solution and counterstained with hematoxylin staining (BASO, Zhuhai, China). Immunohistochemical signals were calculated with positive staining cells under a microscope (Olympus Corporation, Tokyo, Japan) with magnification of 200×.
Cell culture
NCM460 human colon epithelial cells were obtained from the Cell Engineering Research Center of The Fourth Military Medical University (Xi’an, Shanxi, China) and cultured in RPMI-1640 (Gibco) containing 10% FBS, 10 ng/mL EGF, 1% insulin (First Biological and Chemical Medication Co., LTD, Shanghai, China), 5 µg/mL hydrocortisone (The Third Pharmaceutical Company, Beijing, China), and 1% penicillin/streptomycin, at 37°C in a humidified atmosphere of 5% CO2. The use of the NCM460 cell line was approved by the Ethics Committee of The First Affiliated Hospital of Gannan Medical University. Human colon cancer cells (HT-29, HCT-116, SW620, and OUMS23) were purchased from the SUER Bio-Technique Co. Ltd. in Shanghai, China and cultured in DMEM (St Louis, MO, USA) containing 10% FBS and 1% penicillin-streptomycin at 37°C in a humidified atmosphere of 5% CO2.
Cell transfection
HT-29 and SW620 cells were seeded onto 6-well culture plates at a density of 3×105 cells/well. SOX18, SOX18 shRNA, and the respective negative controls (all purchased from Shanghai GenePharma Co., Ltd, Shanghai, China) were then transfected into cells at 50%–60% confluence using Lipofectamine™ 2000 (Invitrogen, Shanghai, China), following the manufacturer’s protocol. The targeted sequences for shRNA-SOX18-1 and shRNA-SOX18-2 were 5′-GCGGCCCTTCGTGGAGGAAGC-3′ and 5′-GGCTGCGCGTGCAGCACTTGC-3′, respectively. After 48 hours, transfected cells were collected and processed for subsequent experiments.
CCK-8 assay
Cell viability was evaluated by the CCK-8 assay. Briefly, 48 hours after transfection, HT-29 and SW620 cells were seeded at a density of 4×103 cells/well in 96-well plates and incubated for 0, 24, 48, and 72 hours. Subsequently, 20 µL of CCK-8 was added to each well, and the plates were incubated for 1 hour. The optical density was read at 450 nm using a microplate reader (Thermo, Fisher Scientific, Waltham, MA, USA). All experimental concentrations were assessed in triplicate.
Colony formation assay
The colony formation assay was used to assess the clonogenic ability of transfected cells. CRC cells (2×103/well) were trypsinized in a single-cell suspension and seeded in 6-well dishes. Cells were maintained in RPMI-1640 supplemented with 10% FBS for approximately two weeks. Visible colonies were fixed in 4% paraformaldehyde for 4 hours at 37°C and stained with 0.5% crystal violet for 2 hours at 37°C (Beyotime Institute of Biotechnology, Haimen, China). Colony numbers were counted under a light microscope (Olympus Corporation, Japan).
Cell cycle analysis
Flow cytometry was employed for the analysis of the cell cycle. After transfection, cells were harvested and fixed overnight in ice-cold 70% ethanol (stored at -20°C). Afterwards, cells were washed with PBS prior to re-suspension in DNA-staining solution [40 µg/mL PI, 250 µg/mL RNase in PBS with 2 mM EDTA] for 30 minutes at 37°C. Analysis was performed using a flow cytometer (FACSCalibur, BD Biosciences).
Apoptosis analysis
The apoptosis levels of HT-29 and SW620 cells transfected with SOX18, shSOX18, or negative control RNA were measured by flow cytometry. HT-29 and SW620 cells transfected with SOX18 or negative control RNA were then treated with cisplatin (30 µg/mL). The detection procedures for overall cell apoptosis were similar to those already described up to the ethanol fixation step, with slight modifications in the cell numbers. Briefly, ethanol-fixed cells (1×106 cells/well) were harvested in centrifuge tubes, washed and detached in 2 PBS with 2 mM EDTA, and centrifuged at 15,000 g for 5 minutes. Cells were then stained with 250 µL of hypotonic fluorochrome solution (50 µg PI, 0.1% sodium citrate, and 0.1% Triton X-100 in PBS) with RNase A (100 U/mL; BD PharMingen) for 30 minutes in the dark at room temperature. Measurements were made using a flow cytometer (BD Influx; BD Biosciences, Franklin Lakes, NJ, USA). The B3 quadrant represented viable cells; B2 and B4 quadrants represented apoptotic cells.
RNA isolation and qRT-PCR
Gene expression was evaluated by qRT-PCR (Trans-Start Top Green qPCR SuperMix, TransGen Biotech Co., LTD, Beijing, China). Cells were seeded in 6-well plates at a density of 5×105 cells/well, cultured overnight, and treated with either SOX18 or shRNA-SOX18 for 48 hours. Total RNA was extracted from cells using TRIzol reagent (Invitrogen) according to the manufacturer’s protocol, and 2 µg were used for reverse-transcription with the TransCript One-step gDNA Removal and cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China). Expression of SOX18 in HT-29 and in SW620 cells was detected by qRT-PCR with the following cycling parameters: 5 minutes at 95°C, followed by 40 cycles at 95°C for 15 seconds, 60°C for 30 seconds, and 72°C for 30 seconds. The primers used for the amplification of the indicated genes were designed using the Primer Express Software (Applied Biosystems, Foster City, CA, USA): 5′-CGCGTGTATGTTTGGTTC-3′ and 5′-ATGTAACCCTGGCAACTC-3′ for SOX18 and 5′-ACCRCGAAGACTGTGGATGG-3′ and 5′-TCAGCTC AGGGATGACCTTG-3′ for GAPDH. The relative expression level was calculated as follows: Relative expression level = 2−ΔΔCt, where ΔCt = Ct (gene of interest) − Ct (housekeeping gene).14 GAPDH was used as the housekeeping gene. All reactions were performed in triplicate.
Western blotting analysis
Total protein lysates were resolved by 10% SDS-PAGE and transferred to polyvinyl difluoride membranes (EMD Millipore, Burlington, MA, USA). After blocking with 5% non-fat dry milk in Tris-buffered saline containing 0.1% Tween-20 (TBS-T) for 30 minutes, membranes were washed four times in TBS-T and incubated overnight at 4°C with primary antibodies anti-SOX18 or anti-GAPDH at 1:1,000 (ab66145 and ab8245, respectively; Cambridge, MA, USA). After extensive washing, membranes were incubated with horseradish peroxidase-linked goat polyclonal anti-rabbit IgG secondary antibody, at a dilution of 1:2,000, for 1 hour at room temperature. Immunoreactivity was detected by enhanced chemiluminescence (ECL Kit, Pierce Biotechnology, Waltham, MA, USA) and captured on a radiographic film. Expression of GAPDH served as loading control.
In vivo xenograft experiments
Male BALB/c nude mice (6-week-old; n=6) were purchased from Beijing (HFK Bioscience Co. Ltd. Beijing, China) and maintained under pathogen-free conditions, with the approval of the Ethics Committee of The First Affiliated Hospital of Gannan Medical University. For tumor propagation analysis, 1×107 HT-29 tumor cells were subcutaneously injected into BALB/c nude mice. The tumor volume was calculated using the formula v=πab2/6 (where v is volume, a is tumor length, and b is tumor width) at the indicated time points. Tumor weight was measured at week four post-injection. Animal experiments were performed in accordance with the relevant guidelines and regulations of the Animal Care and Use Committees at The First Affiliated Hospital of Gannan Medical University.
Statistical analysis
All results are presented as the mean ± SD of three independent experiments, and the data were processed with SPSS 13.0 software. Pearson’s χ2 tests were used for the analysis of the correlation between clinicopathological features and SOX18 expression in CRC patients. Kaplan–Meier survival analysis was used for the analysis of the survival rate, and the P-value was calculated by the log-rank test. Data for multiple comparisons were subjected to one-way ANOVA and chi-square tests. P<0.05 was considered statistically significant.
Results
Increased expression of SOX18 in CRC patients and cell lines
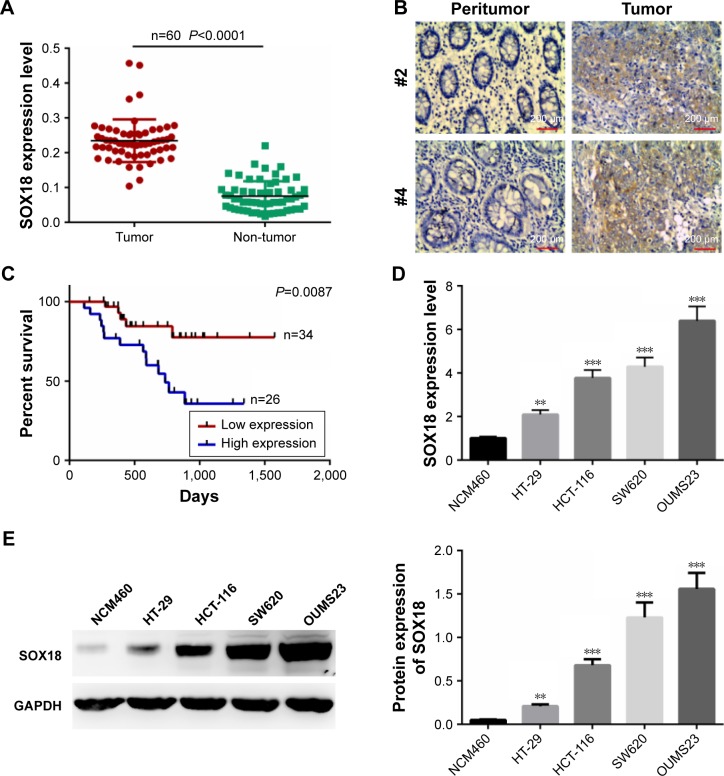
We first analyzed the expression of SOX18 in CRC tissues and adjacent non-tumor tissues by qRT-PCR. We found that the expression of SOX18 was significantly increased in CRC tissues compared to that in normal tissues (P<0.0001, Figure 1A). The expression of SOX18 in CRC tissues and non-tumor tissues was also examined by IHC with pairs of CRC samples. As shown in Figure 1B, the expression of SOX18 in CRC tissues was higher than that in non-tumor tissues. To further evaluate the clinicopathological signifi-cance of SOX18 levels in patients with CRC, the 60 samples from patients were divided into two subgroups based on the mean value of SOX18 expression: low SOX18 group (34 cases) and high SOX18 group (26 cases). As displayed in Table 1, SOX18 levels in CRC tissues were positively associated with advanced clinical stages, vascular invasion, and lymph node metastasis. Furthermore, patients with higher expression of SOX18 had a lower survival rate (Figure 1C).
Figure 1.
Expression of SOX18 in CRC tissues and cell lines.
Notes: (A) mRNA levels of SOX18 in 60 paired CRC samples and normal non-tumor tissues, as assessed by qRT-PCR. (B) Protein expression of SOX18 in CRC tissues and normal tissues determined by immunohistochemistry. Magnification: 200×. (C) Survival rates of patients with CRC with high and low SOX18 levels by Kaplan–Meier survival analysis. (D) mRNA levels of SOX18 in human colon epithelial cells (NCM460) and CRC cells (HT-29, HCT-116, SW620, and OUMS23) detected by qRT-PCR. (E) Protein levels of SOX18 in human colon epithelial cells (NCM460) and CRC cells (HT-29, HCT-116, SW620, and OUMS23) detected by Western blotting. Data are presented as mean ± SD. **P<0.01, ***P<0.001 vs NCM460 cell line.
Abbreviations: CRC, colorectal cancer; SOX18, region, sex determining region Y (SRY)-box 18; qRT-PCR, quantitative real-time PCR.
Table 1.
Association between clinicopathological features and SOX18 expression in 60 patients with CRC
| Characteristics | Number of patients | SOX18 Low expression (≤medium) | SOX18 High expression (≥medium) | P-value |
|---|---|---|---|---|
| Number | 60 | 34 | 26 | |
| Ages (years) | 0.660 | |||
| <60 | 25 | 15 | 10 | |
| ≥60 | 35 | 19 | 16 | |
| Gender | 0.944 | |||
| Female | 32 | 18 | 14 | |
| Male | 28 | 16 | 12 | |
| Location | 0.651 | |||
| Left | 32 | 19 | 13 | |
| Right | 28 | 15 | 13 | |
| Tumor size | 0.944 | |||
| ≤3 | 28 | 16 | 12 | |
| >3 | 32 | 18 | 14 | |
| AJCC stage | <0.001 | |||
| I | 21 | 18 | 3 | |
| II | 14 | 10 | 4 | |
| III | 21 | 5 | 16 | |
| IV | 4 | 1 | 3 | |
| Differentiation | 0.472 | |||
| Well | 18 | 10 | 8 | |
| Moderately | 27 | 15 | 12 | |
| Poorly | 15 | 9 | 6 | |
| Vascular invasion | 0.002 | |||
| Yes | 28 | 10 | 18 | |
| No | 32 | 24 | 8 | |
| Depth of invasion | 0.036 | |||
| T1 | 8 | 6 | 2 | |
| T2 | 11 | 8 | 3 | |
| T3 | 24 | 13 | 11 | |
| T4 | 17 | 7 | 10 | |
| Lymph node metastasis | 0.001 | |||
| N0 | 26 | 21 | 5 | |
| N1 | 20 | 10 | 10 | |
| N2 | 14 | 3 | 11 | |
| Distant metastasis | 0.015 | |||
| M0 | 46 | 30 | 16 | |
| M1 | 14 | 4 | 10 |
Abbreviations: AJCC stage, American Joint Committee on Cancer; CRC, colorectal cancer; SOX18, sex determining region Y (SRY)-box 18.
In addition, mRNA and protein levels of SOX18 in human colon epithelial cells (NCM460) and CRC cells (HT-29, HCT-116, SW620, and OUMS23) were also detected. As presented in Figure 1D and E, expression of SOX18 increased remarkably in CRC cell lines compared to that in human colon epithelial cells (P<0.01). The results indicate that SOX18 might act as an oncogene in CRC.
Overexpression of SOX18 promoted cell proliferation and suppressed cell apoptosis
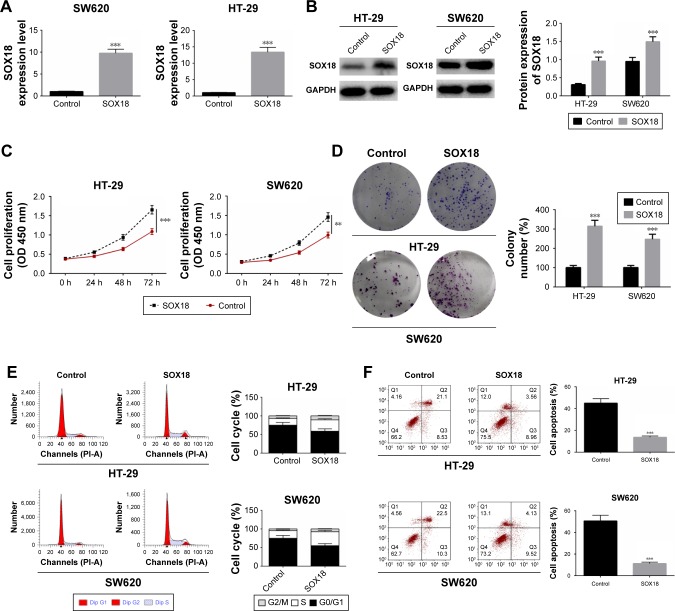
To explore the biological role of SOX18 in CRC, HT-29 and SW620 cells were transfected with SOX18 overexpression or negative control plasmids. After transfection for 48 hours, the expression of SOX18 in HT-29 and SW620 cells was detected by qRT-PCR and Western blot (Figure 2A and B). To induce effective cell apoptosis, HT-29 and SW620 cells transfected with SOX18 overexpression or negative control plasmids were treated with cisplatin. Cell proliferation was assessed by CCK-8 and colony formation assays. Cell cycle and apoptosis were determined by flow cytometry and annexin V/PI staining, respectively. As shown in Figure 2C and D, cell viability and colony formation were significantly promoted by SOX18 overexpression compared to the cells in control groups (P<0.01). Upregulation of SOX18 also accelerated the progression from G0/G1 to S phase in CRC cells (Figure 2E). Annexin V/PI staining showed that SOX18 over-expression effectively inhibited cell apoptosis in HT-29 and SW620 cells (Figure 2F). These results reveal that SOX18 has a carcinogenic role in CRC.
Figure 2.
Effect of SOX18 on cell proliferation, cell cycle, and apoptosis.
Notes: (A, B) HT-29 and SW620 cells were transfected with SOX18 overexpression or negative control plasmids and then treated with cisplatin. The transfection efficiency was examined by qRT-PCR and Western blot. (C, D) Cell proliferation was determined by CCK-8 and cell colony formation. (E) Cell cycle was analyzed by flow cytometry. (F) Cell apoptosis was measured in HT-29 and SW620 cells transfected with SOX18 overexpression or negative control by staining with annexin V/PI. Data are presented as mean ± SD. **P<0.01, ***P<0.001 vs Control.
Abbreviations: CCK-8, cell counting kit-8; PI, propidium iodide; SOX18, sex determining region Y (SRY)-box 18.
Downregulation of SOX18 inhibited cell proliferation and induced cell apoptosis
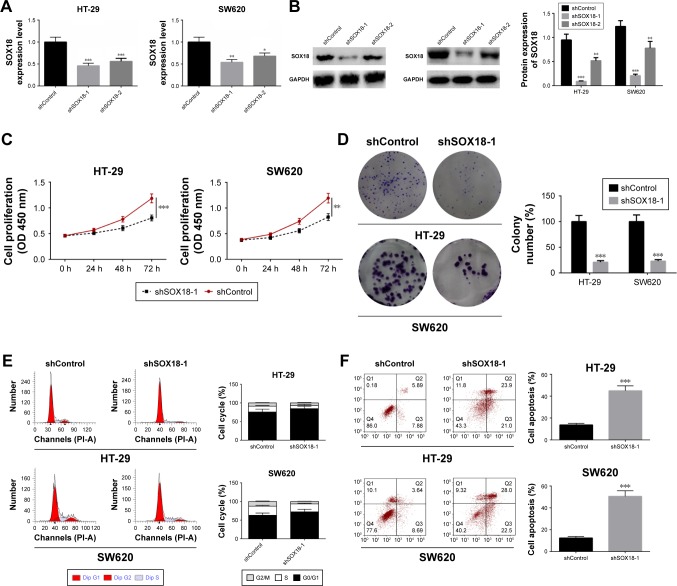
To explore further the function of SOX18 in CRC, HT-29 and SW620 cells were transfected with shSOX18-1, shSOX18-2, or negative control (shControl). Forty-eight hours after transfection, SOX18 expression in HT-29 and SW620 cells was detected by qRT-PCR and Western blot. Compared to control groups, cells transfected with shSOX18-1 and shSOX18-2 clearly downregulated SOX18 (P<0.05), especially shSOX18-1 (Figure 3A and B). Therefore, this sequence was selected for subsequent experiments. CCK-8 and colony formation assays indicated that cell proliferation was significantly reduced by shSOX18 transfection in CRC cells (P<0.01; Figure 3C and D). Cell cycle was then evaluated by flow cytometry, and the results indicate that silencing of SOX18 expression effectively induced G0/G1 phase arrest compared to CRC cells transfected with shControl (Figure 3E). Results of the annexin V/PI staining revealed that cell apoptosis was increased 1.67 and 2.33 fold in HT-29 and SW620 cells transfected with shSOX18, respectively (P<0.01; Figure 3F). The results demonstrate that downregulation of SOX18 can determine progression of CRC.
Figure 3.
Effect of silencing SOX18 on cell proliferation, cell cycle, and apoptosis.
Notes: (A, B) HT-29 and SW620 cells were transfected with shSOX18 or negative control (shControl). The transfection efficiency was examined by qRT-PCR and Western blot. (C, D) Cell proliferation was determined by CCK-8 and cell colony formation. (E) Cell cycle was analyzed by flow cytometry. (F) Cell apoptosis was measured in HT-29 and SW620 cells transfected with shSOX18 or negative control by staining with annexin V/PI. Data are presented as mean ± SD. *P<0.05, **P<0.01, ***P<0.001 vs shControl.
Abbreviations: CCK-8, cell counting kit-8; PI, propidium iodide; SOX18, sex determining region Y (SRY)-box 18.
Tumor-suppressive effect of shSOX18 in vivo
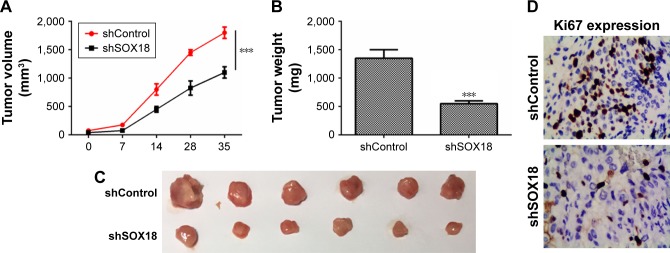
The biological effects of SOX18 on CRC development were addressed by performing a xenograft mouse model. HT-29 cells were transfected with shControl or shSOX18 and then implanted subcutaneously into nude mice. Tumor growth was examined every 7 days. It was observed that downregulation of SOX18 notably delayed tumor growth in vivo (Figure 4A). At 35 days post-implantation, nude mice were sacrificed, and tumors were harvested and weighed. As shown in Figure 4B and C, silencing of SOX18 expression clearly decreased tumor size and weight (P<0.001). Finally, the expression of Ki67, an important biological index of cell proliferation, was determined by IHC. The expression of Ki67 was significantly decreased in the shSOX18 group compared to that in the shControl group (Figure 4D). The results prove that SOX18 can promote CRC development in vivo.
Figure 4.
The role of SOX18 on CRC progression in vivo.
Notes: (A) Tumor growth curves were established by measuring tumor volume every 7 days, for 28 days after injection. (B, C) Tumor weights isolated from nude mice in each treatment group were determined on day 28 after injection. (D) Expression of Ki67 in tumor tissues was assessed by IHC. ***P<0.001 vs shControl.
Abbreviations: CRC, colorectal cancer; SOX18, sex determining region Y (SRY)-box 18.
Discussion
In recent years, a number of diagnostic strategies and therapeutic methods have been followed to treat CRC. However, the overall survival rate of patients with CRC remains low due to tumor recurrence and metastasis.15,16 Therefore, in order to develop a novel and effective therapeutic approach for CRC treatment, there is an urgent need to fully understand the molecular mechanisms that regulate CRC initiation and progression. SOX18 is a member of the Sox transcription factor subfamily, which plays an important role in the development of blood and lymphatic vessels. Previous studies reported that abnormal expression of SOX18 was observed in a wide variety of cancers.10 Yin et al reported that SOX18 was overexpressed in prostate cancer and that it might regulate the malignant capacity of cells via the upregulation of TCF1, c-Myc, cyclin D1, and MMP-7.9 Ornat et al reported that SOX18 played a crucial role in the development of squamous cell carcinoma and basal cell carcinoma.12 Jethon et al indicated that expression of SOX18 was increased in non-small-cell lung cancer and cytoplasmic SOX18 expression was correlated with poor patient outcome.11 In the present study, we observed that the expression of SOX18 was significantly increased in CRC tissues. Additionally, we discovered that SOX18 expression levels were significantly correlated with AJCC stage, vascular invasion, depth of invasion, lymph node metastasis, and distant metastasis, which demonstrates that SOX18 overexpression is positively correlated with CRC progression. We also found that CRC patients with higher expression of SOX18 present decreased survival rates. Furthermore, it was also noted that expression of SOX18 in CRC cell lines was higher than that in the human colon epithelial cells. Based on the above results, we propose that SOX18 may exert a cancer-promoting role in CRC progression.
Excessive cell proliferation is one of the manifestations of malignant tumors.17 Wang et al demonstrated that SOX18 promoted the proliferation and metastasis of hepatocellular carcinoma cells by regulating focal adhesion and chemokine signaling pathways.18 Therefore, the effects of SOX18 upregulation or downregulation on CRC cell viability were examined by CCK-8 and colony formation assays. Results showed that cell proliferation and clonogenic ability were noticeably inhibited by SOX18 knockdown and significantly promoted by SOX18 overexpression. We then examined whether cell growth inhibition was associated with cell cycle progression and cell apoptosis. Cell cycle and cell apoptosis of CRC cells transfected with SOX18 or shSOX18 were evaluated by flow cytometry and annexin V/PI staining, respectively. In our study, cell cycle arrest and cell apoptosis were markedly induced by shSOX18 in CRC cells. On the contrary, upregulation of SOX18 dramatically promoted cell cycle progression and reduced cell apoptosis ratio. Our results reveal, for the first time, that ectopic expression of SOX18 might give rise to oncogenesis in CRC.
Previous in vivo studies indicated that SOX18 could delay tumor growth in prostate cancer.9 Therefore, we evaluated the biological role of SOX18 in CRC development by performing a xenograft mouse model. Our results revealed that downregulation of SOX18 notably delayed tumor growth, as well as decreased tumor size and weight. The expression of Ki67, a nuclear antigen associated with proliferation, is closely related to the occurrence and development of many tumor diseases.19–21 Melling et al showed that Ki67 acts as an independent prognostic biomarker in CRC.22 In the present study, the expression of Ki67 in tumor tissues of the xeno-graft mouse model was analyzed, and the results showed that silencing of SOX18 effectively inhibited Ki67 expression in CRC tissue. These findings suggest that SOX18 may exert an oncogenic role in CRC progression.
In conclusion, the results of the present study indicate that SOX18 is significantly upregulated in CRC tissues and cell lines compared to that in normal tissues and cells and that SOX18 is associated with CRC cancer risk, survival rate, and cell growth in vitro and in vivo. Our findings suggest that SOX18 may act as a novel diagnostic and prognostic marker and a novel therapeutic target for CRC treatment.
Ethics approval and consent to participate
The use of human tissues was approved by the Ethics Committee of The First Affiliated Hospital of Gannan Medical University and patient consent was obtained.
Abbreviations
- SOX18
sex determining region Y (SRY)-box 18
- IHC
immunohistochemistry
- TCF1
HNF1 homeobox A
- MMP-7
matrix metalloproteinase-7
- AJCC
American Joint Committee on Cancer
- CRC
colorectal cancer
- CCK-8
cell counting kit-8
- qRT-PCR
quantitative real-time PCR
- PI
propidium iodide
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bopanna S, Ananthakrishnan AN, Kedia S, Yajnik V, Ahuja V. Risk of colorectal cancer in Asian patients with ulcerative colitis: a systematic review and meta-analysis. Lancet Gastroenterol Hepatol. 2017;2(4):269–276. doi: 10.1016/S2468-1253(17)30004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang J, Xiao J, Feng J, Li K. One-year unplanned readmission after colorectal cancer surgery in Western China. J Invest Surg. 2018:1–5. doi: 10.1080/08941939.2018.1443176. [DOI] [PubMed] [Google Scholar]
- 3.Gu MJ, Huang QC, Bao CZ, et al. Attributable causes of colorectal cancer in China. BMC Cancer. 2018;18(1):38. doi: 10.1186/s12885-017-3968-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong C, Long Z, Yu Y, et al. Dietary factors and polymorphisms in vitamin D metabolism genes: the risk and prognosis of colorectal cancer in northeast China. Sci Rep. 2017;7(1):8827. doi: 10.1038/s41598-017-09356-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kearney MR, Chen EY, Stenzel P, et al. Colorectal cancer-associated spontaneous tumor lysis syndrome: a case report and review of the current literature. J Gastrointest Cancer. 2018 doi: 10.1007/s12029-018-0102-7. In press. [DOI] [PubMed] [Google Scholar]
- 6.Wang B, Yin M, Cheng C, et al. Decreased expression of miR-490-3p in colorectal cancer predicts poor prognosis and promotes cell proliferation and invasion by targeting RAB14. Int J Oncol. 2018;53(3):1247–1256. doi: 10.3892/ijo.2018.4444. [DOI] [PubMed] [Google Scholar]
- 7.Ding N, Li R, Shi W, He C. CENPI is overexpressed in colorectal cancer and regulates cell migration and invasion. Gene. 2018;674:80–86. doi: 10.1016/j.gene.2018.06.067. [DOI] [PubMed] [Google Scholar]
- 8.Liu M, Zhang X, Cai J, et al. Downregulation of TRIM58 expression is associated with a poor patient outcome and enhances colorectal cancer cell invasion. Oncol Rep. 2018;40(3):1251–1260. doi: 10.3892/or.2018.6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin H, Sheng Z, Zhang X, et al. Overexpression of SOX18 promotes prostate cancer progression via the regulation of TCF1, c-Myc, cyclin D1 and MMP-7. Oncol Rep. 2017;37(2):1045–1051. doi: 10.3892/or.2016.5288. [DOI] [PubMed] [Google Scholar]
- 10.Overman J, Fontaine F, Moustaqil M, et al. Pharmacological targeting of the transcription factor SOX18 delays breast cancer in mice. Elife. 2017;6:e21221. doi: 10.7554/eLife.21221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jethon A, Pula B, Olbromski M, et al. Prognostic significance of SOX18 expression in non-small cell lung cancer. Int J Oncol. 2015;46(1):123–132. doi: 10.3892/ijo.2014.2698. [DOI] [PubMed] [Google Scholar]
- 12.Ornat M, Kobierzycki C, Grzegrzolka J, et al. SOX18 Expression in Non-melanoma Skin Cancer. Anticancer Res. 2016;36(5):2379–2383. [PubMed] [Google Scholar]
- 13.Pula B, Kobierzycki C, Solinski D, et al. SOX18 expression predicts response to platinum-based chemotherapy in ovarian cancer. Anticancer Res. 2014;34(8):4029–4037. [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Lee IK, Sung NY, Lee YS, et al. The survival rate and prognostic factors in 26 perforated colorectal cancer patients. Int J Colorectal Dis. 2007;22(5):467–473. doi: 10.1007/s00384-006-0184-8. [DOI] [PubMed] [Google Scholar]
- 16.Sharkas GF, Arqoub KH, Khader YS, et al. Colorectal cancer in Jordan: Survival rate and its related factors. J Oncol. 2017;2017:3180762. doi: 10.1155/2017/3180762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang J, Liu HL, Tao L, et al. Let-7d inhibits colorectal cancer cell proliferation through the CST1/p65 pathway. Int J Oncol. 2018;53(2):781–790. doi: 10.3892/ijo.2018.4419. [DOI] [PubMed] [Google Scholar]
- 18.Wang G, Wei Z, Jia H, Zhao W, Yang G, Zhao H. Knockdown of SOX18 inhibits the proliferation, migration and invasion of hepatocellular carcinoma cells. Oncol Rep. 2015;34(3):1121–1128. doi: 10.3892/or.2015.4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y, Wang N, Zhou X, et al. Prognostic value of ki67 in BCG-treated non-muscle invasive bladder cancer: a meta-analysis and systematic review. BMJ Open. 2018;8(4):e19635. doi: 10.1136/bmjopen-2017-019635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robertson S, Stålhammar G, Darai-Ramqvist E, et al. Prognostic value of Ki67 analysed by cytology or histology in primary breast cancer. J Clin Pathol. 2018;71(9):787–794. doi: 10.1136/jclinpath-2017-204976. [DOI] [PubMed] [Google Scholar]
- 21.Yang C, Zhang J, Ding M, et al. Ki67 targeted strategies for cancer therapy. Clin Transl Oncol. 2018;20(5):570–575. doi: 10.1007/s12094-017-1774-3. [DOI] [PubMed] [Google Scholar]
- 22.Melling N, Kowitz CM, Simon R, et al. High Ki67 expression is an independent good prognostic marker in colorectal cancer. J Clin Pathol. 2016;69(3):209–214. doi: 10.1136/jclinpath-2015-202985. [DOI] [PubMed] [Google Scholar]