Abstract
For cells to grow faster they must increase their protein production rate. Microorganisms have traditionally been thought to accomplish this increase by producing more ribosomes to enhance protein synthesis capacity, leading to the linear relationship between ribosome level and growth rate observed under most growth conditions previously examined. Past studies have suggested that this linear relationship represents an optimal resource allocation strategy for each growth rate, independent of any specific nutrient state. Here we investigate protein production strategies in continuous cultures limited for carbon, nitrogen and phosphorus, which differentially impact substrate supply for protein versus nucleic acid metabolism. Unexpectedly, we find that at slow growth rates, Escherichia coli achieves the same protein production rate using three different strategies under the three different nutrient limitations. Under phosphorus (P) limitation, translation is slow due to a particularly low abundance of ribosomes, which are RNA-rich and thus particularly costly for phosphorous-limited cells. Under nitrogen (N) limitation, translation elongation is slowed by processes including ribosome stalling at glutamine codons. Under carbon (C) limitation, translation is slowed by accumulation of inactive ribosomes not bound to messenger RNA. These extra ribosomes enable rapid growth acceleration during nutrient upshift. Thus, bacteria tune ribosome usage across different limiting nutrients to enable balanced nutrient-limited growth while also preparing for future nutrient upshifts.
Resource allocation during growth is a fundamental challenge faced by all cells1–4. For example, with a fixed resource budget, cells must balance production of the machinery that makes proteins (ribosomes, transfer RNAs, translation factors) with the production of the proteins themselves. This balance is generally represented by the RNA/protein ratio (R/P ratio)5,6. The R/P ratio captures protein production capacity, as > 95% of total RNA is devoted to translation (ribosomal RNAs and tRNAs5,7). In single-celled organisms such as E. coli, previous studies have demonstrated that there is a linear relationship between R/P ratio and growth rate, with faster growth rates requiring more protein production capacity and therefore higher R/P ratios5,6,8,9. Production of ribosomes is costly as each contains 52 protein subunits and three large rRNAs10,11; hence, it is advantageous for the cell to saturate ribosomes with substrates. In this efficient ribosome scenario, the ribosome level should be fixed and independent of nutrient conditions for any growth rate, with the only way to increase protein synthesis rate being to increase the number of ribosomes1,5,12. One surprise for such a seemingly optimized system is that multiple studies have demonstrated that at slow growth rates E. coli accumulates inactive ribosomes13. There are two possible explanations for the presence of inactive ribosomes. First, it is possible that E. coli translation is constrained in such a way that it cannot function when ribosome levels drop too low13. Alternatively, E. coli could regulate ribosome production independently of growth rate. Here we settle this debate by showing that E. coli ribosome production and usage differ across nutrient conditions.
Results
Phosphate-limited cells achieve the same growth rate with fewer ribosomes than C- or N-limited cells.
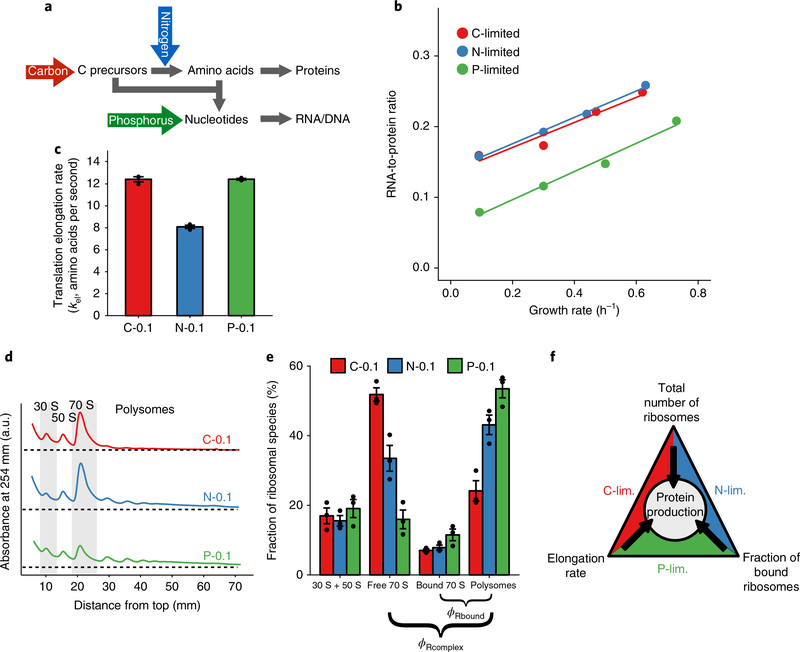
To determine the generality of the relationship between growth rate and ribosome content, we examined how the R/P ratio changes as a function of growth under different nutrient limitations. We measured R/P ratios in E. coli under glucose (C, carbon), ammonia (N, nitrogen) and phosphate (P, phosphorus) limitations over a range of different growth rates in chemostats (Fig. 1a). Surprisingly, P-limited cells consistently exhibited lower R/P ratios than C-limited or N-limited cells, with a roughly 2-fold difference at the slowest growth rate tested (0.1 h−1, Fig. 1b). Whereas most bacterial physiology studies have been performed with batch cultures, chemostat cultures reach a steady state due to limitation of a specific nutrient. To determine if our findings are specific to chemostat-grown cultures we also determined the R/P ratios for batch cultures with different growth rates. These results confirmed previous findings that the R/P ratios of C- and N-limited cells follow the same trend regardless of whether they are grown in batch or chemostat conditions (Supplementary Fig. 1a,b and Supplementary Table 2). Measured protein levels were similar in all cells regardless of growth rate or nutrient limitation (Supplementary Fig. 1c,d). Moreover, the rRNA fraction of total RNA remained similar across different nutrient limitations at the same growth rate and decreased as growth was slowed (Supplementary Fig. 1e,f). Since the nutrient-specific changes in R/P ratio cannot be explained by changes in rRNA fraction, and the P-limited cells with the lowest R/P ratios also have the lowest rRNA fraction, our data collectively suggest that P-limited cells produce protein at the same rate as C/N-limited cells using fewer ribosomes.
Fig. 1 |. RNa-to-protein ratio is both growth-rate and nutrient dependent.
a, Schematic flow of nutrients for biomass formation. Carbon (C) and nitrogen (N) combine to make amino acids. Amino acids combine with carbon precursors and phosphorus (P) to make nucleic acids. b, RNA-to-protein ratios for chemostat cultures under C, N and P limitations at different growth rates. Each data point shows the mean value from three technical replicates. c, Translation elongation rates (amino acids per second) as measured by the lacZ induction assay after correction for translation initiation. The bar height represents mean values with error bars as s.e.m. from three biological replicates. d, Polysome profiles of cells grown under C, N and P limitations in chemostats at a dilution rate of 0.1 h−1. Five independent experiments were repeated, with a single representative curve shown. e, Quantification of ribosomes in the form of subunits (30 S + 50 S), free 70 S, mRNA-bound 70 S (one ribosome on one mRNA) and polysomes (multiple ribosomes on one mRNA). The bar height represents mean values with error bars as s.e.m. from three biological replicates. f, Cells adapt to different nutrient limitations using different strategies of translational regulation that achieve the same protein production rate.
The finding that P-limited cells make the same amount of protein with fewer ribosomes, suggests that C/N-limited E. coli cells do not use ribosomes with optimal efficiency and their ‘extra’ ribosomes do not reflect a biophysical limitation. Since RNA accounts for two-thirds of the mass of bacterial ribosomes10,14, producing fewer ribosomes under P limitation makes sense as a way for cells to deal with a limitation that preferentially reduces an elemental substrate needed to make RNA but not protein. This lower ribosome level may be a direct consequence of low phosphate resulting in limited nucleotide pools, as deletion of genes involved in phosphate sensing or storage, phoB15 or ppk16, did not alter the R/P ratio (Supplementary Fig. 1g,h).
N-limited ribosomes translate slowly while C-limited cells accumulate more mRNA-free ribosomes.
Why do C/N-limited cells accumulate so many ribosomes if P-limited cells can achieve the same protein synthesis rates with fewer ribosomes? One possibility is that the ribosomes in these cells translate slowly. We thus used a lacZ induction assay to compare the translation elongation rates of slow-growing C-, N- and P-limited cells (0.1 h−1)17,18. We observed a reduced elongation rate in N-limited cells compared to C- and P-limited cells but no difference between C- and P-limited cells (Fig. 1c and Supplementary Fig. 2). Thus, N-limited cells may need higher ribosome numbers to compensate for their slow translation elongation, but something else must explain the elevated ribosome numbers in C-limited cells.
To characterize ribosome pools we performed polysome profiling, which separates ribosome species using a sucrose gradient19. Regardless of the growth condition, all cells exhibited similar fractional pools of dissociated 30 S and 50 S subunits (Fig. 1d). In contrast, the fraction of 70 S monosomes was significantly larger in C/N-limited cells than in P-limited cells (Fig. 1d). Since growth rate is proportional to protein synthesis rate, growth rate can be estimated by the product of the number of active ribosomes and the translation elongation rate. However, using the assumption that all 70 S monosomes are active yielded very different growth rate estimates for C-, N- and P-limited cells (Table 1 and Supplementary Information), which is inconsistent with the fact that these cells are growing at the same rate and have the same protein content. This inconsistency suggested that a fraction of the 70 S ribosomes may not be active.
Table 1|.
Growth rate estimation from parameters measured
| Sample | R/P ratio | rRNa frac. | ϕRcomplex | ϕRbound | kel (amino acids per second) | Estimated growth rate from all 70 S (h–1) | Estimated growth rate from bound 70 S (h–1) | Actual growth rate (h–1) |
|---|---|---|---|---|---|---|---|---|
| C-0.1 | 0.16 | 0.64 ± 0.03 | 0.83 ± 0.02 | 0.31 ± 0.03 | 12.4 ± 0.2 | 0.29 ± 0.02 | 0.09 ± 0.01 | 0.09 ± 0.00 |
| N-0.1 | 0.16 | 0.59 ± 0.06 | 0.84 ± 0.02 | 0.51 ± 0.03 | 8.1 ± 0.1 | 0.17 ± 0.01 | 0.09 ± 0.01 | 0.09 ± 0.00 |
| P-0.1 | 0.08 | 0.51 ± 0.03 | 0.81 ± 0.03 | 0.65 ± 0.04 | 12.4 ± 0.1 | 0.12 ± 0.01 | 0.08 ± 0.01 | 0.09 ± 0.01 |
Estimation of growth rate using measurements from R/P ratio, fraction of rRNA in total RNA (rRNA frac.), elongation rates (kel) and fractions of different ribosomal species (ϕRcomplex and ϕRbound). All values are mean values. R/P ratio values are derived from three technical replicates and the rest of the parameters are from three biological replicates with the s.e.m. shown. The propagated error is shown for growth rate estimates.
The mass of a single mRNA is small relative to the mass of a ribosome, such that 70 S monosomes could represent either mRNAs with only one ribosome per transcript or inactive ‘free’ ribosomes that are not associated with an mRNA. To distinguish free and mRNA-bound 70 S monosomes, we used their differential sensitivity to high potassium levels (170 mM)20. High potassium causes free ribosomes to shift to a lower density but does not shift the density of mRNA-bound monosomes20. We thus designed a high-resolution ‘free-ribosome profiling’ method to resolve this density shift (Supplementary Fig. 3). As controls, we confirmed that our assay detects the potassium-dependent shift of 70 S monosomes induced by puromycin (Supplementary Fig. 3a), which releases elongating ribosomes from their associated mRNAs21. There was no potassium-dependent shift detected in fast-growing cells that lack free ribosomes (Supplementary Fig. 3a). As a second validation of our free-ribosome profiling method we confirmed that free ribosome pools decreased as growth rate increased across all three nutrient limiting conditions (Supplementary Fig. 3b).
By combining traditional and free-ribosome polysome profiling, we quantified the relative fractions of all ribosome species in slow-growing C-, N- and P-limited E. coli. The fraction of free monosomes was roughly 3-fold greater in the C-limited cells than in the P-limited cells, while the fraction of mRNA-bound monosomes remained relatively constant across nutrient limitations (Fig. 1e). The accumulation of free 70 S monosomes in C- and N-limited cells appears to be independent of a previously described RaiA-dependent mechanism for ribosome storage as deletion of raiA had no impact on R/P ratios or polysome profiles (Supplementary Fig. 4). Importantly, revising our protein synthesis rate estimates to account for the fraction of inactive 70 S monosomes correctly yielded similar values for all cells, regardless of nutrient limitation (Table 1). These results both validate our experimental measurements and suggest that, in different nutrient states, E. coli differentially tune ribosome number, elongation rate and active fraction to produce proteins at the same rates (Fig. 1f).
Quantitative modelling describes three different strategies of ribosome dynamics to achieve the same protein production rate.
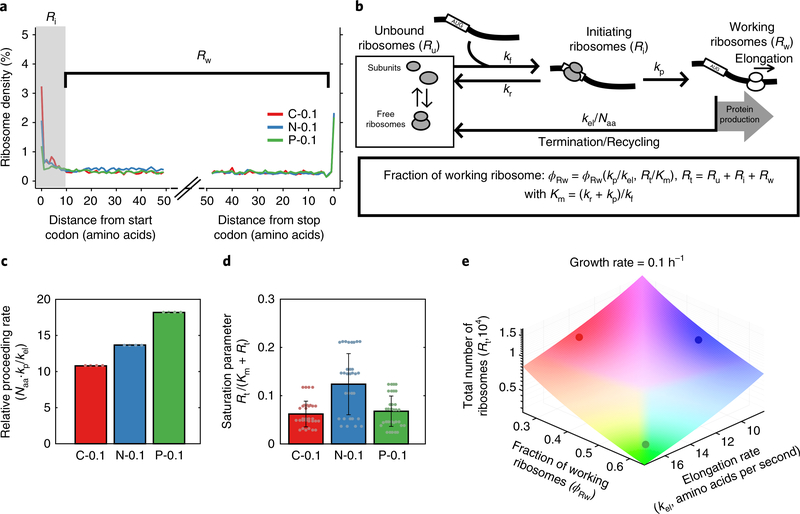
To better understand nutrient-dependent ribosome dynamics, we probed translation in slow-growing C-, N- and P-limited cells by ribosome profiling. Analysis of ribosome densities as a function of distance from the start and stop codons revealed higher ribosome occupancy near the start codon for C- and N-limited ribosomes (Fig. 2a). Ribosome density thereafter was similar, with no decrease in ribosome density between the first and second halves of genes (Fig. 2a and Supplementary Fig. 5a), suggesting that there is little to no aborted translation after the first few codons.
Fig. 2 |. A macroscopic model reveals different ribosome dynamics that achieve the same growth rate.
a, Averaged A-site ribosome counts within the first and last 50 codons of the transcripts from ribosome profiling analysis. Ribosomes bound to the first 10 codons are defined as ‘initiating ribosomes’ (Ri) and those bound to the rest of the transcripts except the stop codon are ‘working ribosomes’ (Rw) (n = 1 independent biological sample). b, A macroscopic model of ribosome dynamics. kf, forward rate (s−1); kp, proceeding rate (s−1); kel, elongation rate (amino acids per second); Naa, number of amino acids in an average protein of E. coli; kr, aborted translation rate (s−1); Rt, total number of ribosomes. c, The relative proceeding rate under C, N and P limitations at a growth rate of 0.1 h−1. The bar height represents mean values with error bars as standard deviation from all the possible combinations of experimental measurements. d, The saturation parameter under C, N and P limitations at a growth rate of 0.1 h−1. The bar height represents mean values with error bars as standard deviation from all the possible combinations of experimental measurements. e, The relationship between elongation rate (kel), fraction of working ribosomes (ϕRw) and total number of ribosomes (Rt) that leads to the same growth rate at 0.1 h−1. Coloured dots indicate the values for C-, N- and P-limited wild-type cells.
To gain more mechanistic insights into the differences in translation across nutrient limitations, we developed a macroscopic mathematical model of ribosome dynamics (Fig. 2b, Supplementary Fig. 5b-e and details in the Supplementary Information). This macroscopic model incorporates the ribosome profiling data and models ribosomes as three species: unbound ribosomes, initiating ribosomes and working ribosomes. Unbound ribosomes (Ru) can bind to an mRNA with a free ribosomal binding site (with rate constant kf) to become initiating ribosomes (Ri). The initiating ribosomes can proceed to elongation (with rate constant kp) to become working ribosomes (Rw) or can abort translation (with rate constant kr). Working ribosomes elongate to finish translation and become unbound (with rate constant kel/Naa), where Naa is the length of an average protein in amino acids. We defined the fraction of ribosomes bound to the first 10 codons as initiating ribosomes since the ribosome footprint size is ~10 codons22,23. To validate our model we used it to calculate elongation rates, which closely agreed with those we measured experimentally (Supplementary Fig. 6a).
Analysis of our model indicated that the system can be characterized by two main dimensionless parameters (Supplementary Information): the ‘relative proceeding rate’, defined as the ratio between the rate of ribosomes proceeding from initiation to elongation (kp, s−1) and the rate of elongation (kel /Naa, s−1); and the ‘saturation parameter’ (Rt /(Km + Rt)), reflecting the degree of saturation of ribosome binding sites on mRNAs, where Rt is the total ribosome number and Km = (kr + kp)/kf (Fig. 2c,d and Supplementary Fig. 6b,c). Fitting the measured ribosome densities, translation elongation rates and pool sizes of ribosome species to the macroscopic model revealed that P-limited cells have the highest relative proceeding rate while C-limited cells have the lowest saturation parameter, and N-limited cells have the highest saturation parameter (Fig. 2c,d and Supplementary Fig. 6b-e). Thus, C-, N- and P-limited cells produce proteins at the same rate using three different strategies: P-limited cells have few ribosomes that are mostly active and elongate rapidly, N-limited cells have more ribosomes but fewer are active and they elongate slowly, and C-limited cells have many ribosomes, which elongate rapidly, but even fewer are bound to mRNA (Fig. 2e and Supplementary Fig. 6f).
N-limited ribosomal regulation is mediated by RelA, the ppGpp alarmone synthase.
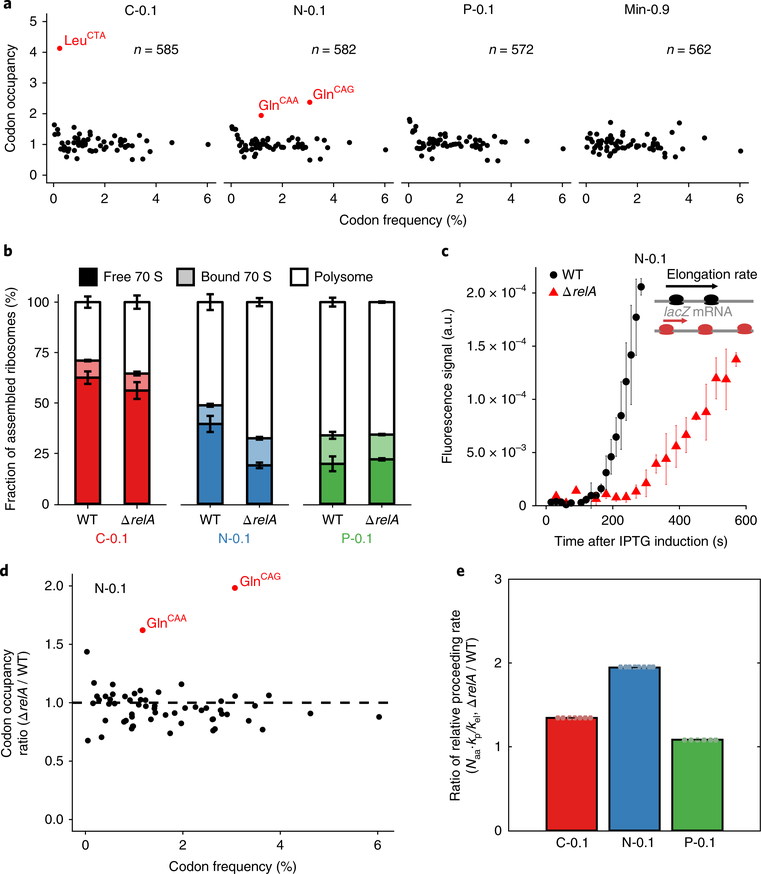
Our ribosome profiling data provides additional insight into the molecular basis of nutrient-specific ribosome regulation through analysis of codon occupancies. Codon-specific ribosome stalling leads to increased codon occupancy and is a hallmark of insufficient pools of the corresponding charged tRNAs. P-limited cells exhibited no elevated codon frequencies, consistent with the efficient ribosome usage of these cells (Fig. 3a). In contrast, both C- and N-limited cells exhibited significant codon-specific stalling. Under N limitation, ribosomes stalled at both of the two glutamine-encoding codons. This result is consistent with previous studies indicating that glutamine is the most strongly depleted amino acid pool under N limitation and serves as an intracellular sensor for extracellular nitrogen levels24–26. Since glutamine codons account for 4.4% of all predicted open reading frame (ORF) codons in E. coli, the ~2-fold increase in glutamine codon occupancy indicates that additional mechanisms also contribute to the ~30% reduction in translation elongation rate observed under N limitation. Under C limitation, we observed elevated occupancy of the Leu-CTA codon, which was surprising as there is no known intracellular carbon sensor that controls translation. E. coli has six leucine codons decoded by five leucine tRNA species27,28. Leu-CTA is the rarest Leu codon, accounting for only 0.4% of all predicted ORF codons.
Fig. 3 |. Deletion of relA disrupts translation regulation under nitrogen limitation.
a, A-site codon occupancy under different growth conditions from ribosome profiling. Occupancy was calculated as the ratio between measured and expected counts for each gene based on codon frequency. The average of this ratio is plotted (with number of genes specified). Codons that have higher than average ratio by 2.5 standard deviations are highlighted. b, Fraction of assembled (70 S) ribosomes in wild type and Δ relA under C, N and P limitations at a growth rate of 0.1 h−1. The bar height represents mean values with error bars as s.e.m. from three biological replicates. c, lacZ induction assay for wild type and ΔrelA under nitrogen limitation at a growth rate of 0.1 h−1. The lag time measures when the first functional LacZ is produced and is inversely proportional to the elongation rate. Mean values from three biological replicates are shown with error bars as s.e.m. d, Ratio of codon occupancy between Δ relA and wild type (WT) under nitrogen limitation at a growth rate of 0.1 h−1 (n = 1 independent biological sample). e, Ratio of the relative proceeding rate (Naa·kp/kel) between Δ relA and wild type across different conditions at a growth rate of 0.1 h−1. The bar height represents mean values with error bars as standard deviation from all the possible combination of experimental measurements.
In addition to clarifying the link between metabolism and translation for N limitation, the observation that ribosomes stall at specific codons under C/N limitation suggested a molecular mechanism for nutrient-specific translation regulation (Fig. 3a). In bacteria, insufficient charged tRNA pools activate the stringent response, which is mediated by the accumulation of the cellular alarmone, ppGpp29,30. ppGpp is known to regulate rRNA transcription in vivo. Its role in translation is less well understood but ppGpp has been shown in vitro to inhibit translation factors such as EF-Tu and IF-2 by competing with GTP31,32. To test how ppGpp accumulation might affect translation in vivo, we induced ppGpp synthesis by treating batch-grown E. coli with serine hydroxamate (SHX). SHX is a serine analogue that competitively inhibits serine tRNA synthetase to yield uncharged serine tRNA and thereby activate the RelA ppGpp synthase33. SHX treatment increased the pool of mRNA-free 70 S monosomes, and this effect was completely dependent on relA (Supplementary Fig. 7a,b). Thus, inducing ppGpp by activating RelA alters translation by increasing the fraction of inactive free 70 S ribosomes.
RelA is primarily required for the accumulation of ppGpp under N limitation but not C limitation30. Consistently, deletion of relA had little impact on the accumulation of free ribosomes under C limitation, but significantly reduced free ribosome pools under N limitation to levels similar to those observed under P limitation (Fig. 3b and Supplementary Fig. 7c). We could not probe the role of ppGpp in C-limited ribosome accumulation because, unlike N limitation, C limitation elevates ppGpp through spoT, which is essential34. N-limited ΔrelA cells also contained more polysomes than wild type (Fig. 3b), suggesting that these cells had a higher fraction of elongating ribosomes. This result was initially confusing because the wild type and ΔrelA N-limited cells were grown at the same growth rate and maintained the same R/P ratio (Supplementary Fig. 7d). We thus measured the rate of translation elongation and found that while N-limited ΔrelA cells have higher fractions of translating ribosomes, their ribosomes elongate more slowly (Fig. 3c), resulting in the same rate of protein production as wild type.
To understand how RelA influences ribosome dynamics we performed ribosome profiling. N-limited ΔrelA cells exhibited even more pronounced ribosome stalling at both glutamine codons than wild type N-limited cells, while relA deletion had no effect on P- or C-limited cells (Fig. 3d and Supplementary Fig. 8). Fitting the ΔrelA cells measurements to our ribosome dynamics model revealed that ΔrelA cells specifically increase the relative proceeding rate under N limitation, but display no effect on relative proceeding rate under P or C limitation and no effect on the saturation parameter in any condition (Fig. 3e and Supplementary Fig. 6b,c). These results suggest that in N-limited cells, RelA-dependent ppGpp serves to restrict ribosome function by regulating the translation initiation rate and/or the frequency at which ribosomes transition from initiation to elongation. Furthermore, we speculate that in the absence of RelA more ribosomes attempt to elongate, which exacerbates the depletion of charged tRNA pools, leading to increased stalling and a slower translational elongation rate.
Extra ribosomes may facilitate growth acceleration during nutrient upshift.
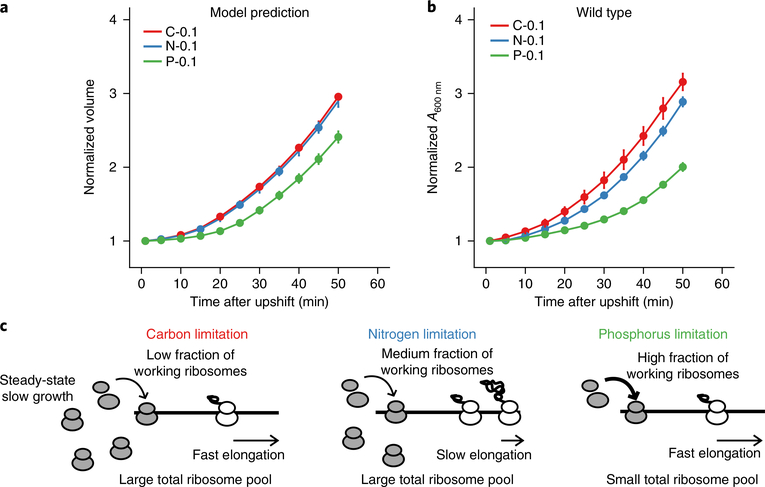
While cells can modulate different aspects of ribosome dynamics to achieve the same protein production rate, what benefits might be served by the inefficient translation system used by C-limited cells where many ribosomes are inactive? Since free ribosomes accumulate the most at the slowest growth rates, we hypothesized that our findings could reflect a trade-off between steady-state growth rate and the ability to respond to a fluctuating environment. Such transitions could include rapidly and safely slowing growth when nutrients become depleted and rapidly increasing growth rate when nutrients are replenished. In this scenario, cells may benefit more by optimizing their ability to rapidly use new nutrients, for example to outcompete their neighbours or maximally use a transient pulse of nutrients, than by optimizing a steady-state growth rate when nutrient levels are low.
We extended our mathematical model of ribosome dynamics to predict cellular growth dynamics during nutrient upshift. This modelling supported the hypothesis that the larger free ribosome pools of C/N-limited cells should enable them to increase their growth rates more quickly than P-limited cells during nutrient upshift (Fig. 4a, Supplementary Fig. 9 and details in the Supplementary Information)4,35. We experimentally tested this prediction by measuring the growth rates of slow-growing (0.1 h−1) E. coli immediately after being shifted to rich media (Luria-Bertani media (LB) + 0.4% glucose)36. As predicted, C/N-limited cells increased their growth rates significantly faster than P-limited cells (Fig. 4b). Thus, the distinct translation strategies employed under different nutrient conditions may represent nutrient-specific adaptations, with P-limited cells optimizing for current steady-state growth under slow growth conditions, and C- and N-limited cells favouring the ability to rapidly recover growth (Fig. 4c).
Fig. 4 |. Extra ribosomes confer growth advantage during nutrient upshift.
a, Theoretically predicted growth curves from the macroscopic model. b, Experimental nutrient-upshift growth curves in LB + 0.4% glucose for wild-type cells from C-, N- and P-limited chemostats at a growth rate of 0.1 h−1. The initial absorbance at 600 nm and standard deviation of three biological replicates for each condition are 0.141 ± 0.008 (C), 0.107 ± 0.017 (N) and 0.154 ± 0.008 (P). Each data point shows the mean value from three biological replicates with error bars as standard deviation. c, Model for nutrient-dependent ribosome usage: under C or N limitation, the total ribosome pool is high while under P limitation the total pool is low. C-limited cells elongate fast but have a low fraction of working ribosomes. N-limited cells elongate slower but have a higher fraction of working ribosomes than C-limited cells. Under P limitation the low supply of ribosomes leads to both a high fraction of working ribosomes and fast elongation to meet the protein production demand.
Discussion
In previous studies, E. coli were found to vary R/P ratio with growth rate independently of the specific nutrient limitation used to produce a given growth rate5,9. While pioneering physiology studies acknowledged the possibility that such growth laws could depend on the specific growth state of the cell6,8, the reproducibility of the correlation between R/P ratio and growth rate has often been considered to be more general1,9. Meanwhile, transcriptional analysis in Saccharomyces cerevisiae suggested that the primary determinant of the response to a wide range of stresses was the cellular growth rate rather than the specific stressor37. Together, these studies suggested that the primary regulator of microbial physiology is growth rate. However, our findings demonstrate that, at the same growth rate, E. coli exhibit significantly different translation strategies across nutrient limitations; at the lowest growth rate tested, P-limited cells produced the same amount of protein with roughly half as many ribosomes as C/N-limited cells. P-limited cells also exhibited smaller inactive 70 S monosome pools and higher relative proceeding rates than C/N-limited cells. Furthermore, while C/N-limited cells have similar ribosome numbers, they also display differences in free ribosome pools, translational elongation rates and sensitivity to the loss of the ppGpp synthase RelA. Thus, our results suggest that the extra ribosomes of C/N-limited cells do not reflect essential constraints, but rather reflect a selectively beneficial adaptation.
Our findings also implicate ppGpp as a key mediator of ribosome activity that specifically modulates the transition from translation initiation to elongation. ppGpp is a well-characterized cellular alarmone that senses translational activity and inhibits rRNA transcription30,38. However, ppGpp can competitively inhibit GTP-dependent enzymes, including the translation initiation factor IF-2 that is required for the transition to elongation39. Since ppGpp can also be produced by SpoT and spoT relA mutants cannot survive in the conditions of our experiments40, the effect of complete loss of ppGpp remains unclear. Nevertheless, the phenotypes we observe in the absence of RelA provide in vivo support that ppGpp can affect translation in addition to affecting ribosome production. Our modelling suggests that ppGpp affects translation by altering the relative proceeding rate, which can be influenced by both the translation initiation rate and the frequency with which ribosomes transition from translation initiation to elongation. One attractive candidate target for ppGpp is IF-2, as a previous in vitro study demonstrated that ppGpp inhibits IF-2 function32 and IF-2 has been implicated in both initiation and the transition to elongation. Since ppGpp can also affect other GTP-binding proteins involved in translation31,41, future studies on bacterial translation will need to address both how RelA-dependent ppGpp interferes with ribosome function and whether other sources of ppGpp are important translational modulators in other contexts.
In native environments such as the mammalian gut, bacteria such as E. coli are faced with feast and famine cycles induced by feeding cycles42. Here we show at the same growth rate that the R/P ratio in E. coli does not reflect the optimization of steady-state growth but, instead, may reflect the ability of cells with higher inactive ribosome pools to rapidly accelerate growth during nutrient repletion. These findings suggest that E. coli may improve their fitness by sacrificing their maximal growth rate in nutrient-poor periods in return for the ability to respond to a changing environment, including rapidly accelerating growth in nutrient-rich periods. Our macroscopic ribosome dynamics model highlights how cells can tune total ribosome number, the fraction of working ribosomes and the rate of translational elongation to achieve the same total protein production rate while balancing other constraints such as reduced amino acid availability or the need to rapidly accelerate growth. This strategy could help explain recent reports of the suboptimality of protein allocation for E. coli in the presence of poor carbon sources43–46, the suboptimal expression levels of essential genes in Bacillus subtilis47 and excess ribosome production in S. cerevisiae48,49. Future studies will address the consequences of adaptation strategies in dynamic conditions, for example the generality of optimizing growth rate transitions at the expense of steady-state growth.
Methods
Cell strains and growth conditions.
E. coli strain NCM3722 was grown in batch or continuous cultures. To achieve different growth rates, different carbon or nitrogen sources were provided in batch culture, whereas dilution rates ranging from 0.1 h−1 to 0.7 h−1 were used in continuous (chemostat) cultures. The chemostat (Sixfors, HT) volume was 300 ml with oxygen and pH probes to monitor the culture. pH was maintained at 7.2 ± 0.1 and the aeration rate was set at 4.5 l/h. 40 mM MOPS media (M2120, Teknova) was used with glucose (0.4%, Sigma G8270), ammonia (9.5 mM NH4Cl, Sigma A9434) and phosphate (1.32 mM K2HPO4, Sigma P3786) added separately. For C- and N-limiting media, glucose and ammonia concentrations were reduced by 5-fold (0.08% and 1.9 mM, respectively). The P-limiting medium contains 0.132 mM K2HPO4. ΔrelA mutant was generated by P1 transduction from the Keio collection50 into E. coli strain NCM3722.
Nutrient upshift growth measurement.
Cells from chemosats were mixed with 4 × volumes of fresh pre-warmed media and grown in flasks in a 37 °C water bath. Cell growth was monitored every 5 min by checking absorbance at 600 nm in a quartz cuvette (Starna, 16.160-Q-10/Z8.5) using a spectrophotometer (GENESYS™ 20, Thermo Scientific). The LB media (244610, BD) used for upshift was supplemented with 0.4% glucose.
Total RNA measurement.
The method for RNA measurement was adapted from You et al.1. 1.5 ml of cultures were pelleted by centrifugation for 1 min at 13,000g. The pellet was frozen on dry ice and the supernatant was taken to measure absorbance at 600 nm for cell loss. The pellet was then washed twice with 0.6 M HClO4 and digested with 0.3 M KOH for 1 h at 37 °C. The solution was then precipitated with 3 M HClO4 and the supernatant was collected. The pellet was re-extracted again with 0.5 M HClO4. The supernatants were combined and absorbance measured at 260 nm using Tecan Infinite 200 Pro (Tecan Trading AG, Switzerland). Total RNA concentration is the multiplication product of the A260 absorbance value and the extinction coefficient (31 μ g RNA ml–1).
Total protein measurement.
The method for protein measurement is adapted from You et al.1. 1.5 ml of cell cultures were pelleted by centrifugation for 1 min at 13,000g. Cells were washed with 1 ml MOPS buffer once, re-suspended in 200 μl water and left on dry ice. All the supernatant was collected and A600 nm was measured for cell loss. To measure the protein content, samples were thawed, 100 μl of 3 M NaOH was added and the sample was heated for 5 min at 98 °C. The samples were cooled down to room temperature for 5 min before 300 μl of 0.1% CuSO4 were added for biuret assay. The samples were incubated at room temperature for 5 min and centrifuged at 13,000g for 1 min. The supernatant was collected and the absorbance was measured at 555 nm for a 200 μl sample volume in a microplate reader (Synergy HT, BioTek) with software Gen5. Proper dilution of albumin (23209, Thermo) with known concentrations was used to infer the total protein concentration in the cell.
rRNA fraction quantification.
2 ml of culture were pelleted by centrifugation at maximum speed for 1 min. Supernatant was removed and the sample was flash frozen in liquid nitrogen. Total RNA was extracted using a hot-phenol method51. The sample was analysed using a 2100 Bioanalyzer instrument with the RNA 6000 Nano kit (Agilent). The RNA fraction was quantified by integrating the area under each peak in the 2100 Expert software using the Prokaryote Total RNA Nano Assay (Version 2.5). rRNA fraction is defined as the sum of the 16 S and 23 S peak areas divided by the total area. tRNA fraction is defined as the sum of the area between 80 nt and 175 nt divided by the total area.
Polysome profiling and quantification of ribosome fraction.
200 ml of cells were collected from cultures by filtration through 90 mm cellulose acetate membranes with a 0.2 μM pore size (CA029025, Strelitech) at 37 °C, scratched with a clean and pre-warmed stainless steel spatula, and snap frozen in liquid nitrogen. The whole filtration procedure did not take more than 2 min to maintain the original physiological state. Cell pellets were mixed with 650 μl lysis buffer frozen nuggets (20 mM Tris-HCl pH 8.0, 10 mM MgCl2, 100 mM NH4Cl, 0.4% Triton X-100, 0.1% NP-40, 1 mM Chloramphenicol, 100 U ml−1 RNase-free DNase I (04716728001 Roche)) in a pre-chilled 10 ml jar (014620331, Retsch). Pulverization was done by cryomill (Retsch) at 15 Hz for 15 min. The thawed cell lysates were quantified by NanoDrop and 200 μl of lysates with RNA concentration ranging from 80 μ g to 500 μ g were used. For overall polysome quantification, lysates were loaded to 10–55% linear sucrose gradients (20 mM Tris-HCl pH 8.0, 10 mM MgCl2, 100 mM NH4Cl, 300 μM Chloramphenicol) made by GradientMaster (BioComp). The gradients were placed in a SW41Ti bucket and centrifuged in Optima XE-100 Ultracentrifuge (Beckman Coulter) at 35,000 r.p.m. for 2 h at 4 °C. Gradients were fractionated by BioComp Gradient Fractionator and the absorption curves at 254 nm were recorded by a ultraviolet monitor (EM-1, BioRad).
Quantification of the polysome profiles was performed using customized MATLAB codes. First, since chloramphenicol absorbs at the same wavelengths as RNA, we estimated a baseline absorbance using the average of the last 50 readings where no RNA was detected (the bottom of the tubes) and subtracted this background value. The background from the free nucleotides and tRNA was removed by fitting an exponential decay function to the first peak representing the source of non-ribosome signals. Each ribosome peak was picked and quantified by integrating the area underneath the curve.
To quantify different species of ribosomes in the 70 S peak, 100 mM NH4Cl was replaced with 170 mM KCl. Cell lysates were loaded onto 10–30% linear gradient and centrifuged at 35,000 r.p.m. in a SW41Ti bucket at 4 °C for 5 h. Because the two peaks for the free and mRNA-bound 70 S ribosomes are very close in mass and clean separation was not possible, the MATLAB file-exchange package, peakfit (2.0), was used to fit the three overlapped peaks (50 S subunit, mRNA-free 70 S and mRNA-bound 70 S) as three Gaussian distributions. For the free ribosome control, cells were treated with 100 μM puromycin for 5 min and collected. For serine hydroxamate (SHX, Sigma S4503) treatment, cells were grown in MOPS glucose minimal media until A ~ 0.3 and treated with SHX with a final concentration of 1 mg ml−1 for 10 min before collection.
lacZ induction and translational elongation rate measurement.
The method was adapted from Zhu et al52. A final concentration of 5 mM isopropyl-β -D-thiogalactoside (IPTG) (I2481C-25, Gold Biotechnology) was added to cultures. At every 15 s, 1 ml of culture was collected in a tube containing 10 μl of 100 mM chloramphenicol, snap frozen in liquid nitrogen and stored at −20 ˚C before subsequent measurement. After cells were thawed, 400 μl of the sample was added to 100 μl 5 × Z-buffer (0.3 M Na2HPO4.7H2O, 0.2 M NaH2PO4.H2O, 50 mM KCl, 5 mM MgSO4, 20 mM β -mercaptoethanol) and incubated at 37 °C for 10 min. 100 μl of 4 mg ml−1 4-methylumbelliferyl-β -D-galactopyranoside (MUG, 337210010, ACROS Organics) in DMSO was then added to each sample every 10 s to accurately control the reaction time. The samples were incubated at 37 °C in Eppendorf Thermomixer R at 1,400 r.p.m. mixing rate for 30 min to 2 h, depending on the enzyme expression level. The reaction was stopped by addition of 300 μl of 1 M Na2CO3. The tubes were spun down at 16,000g for 3 min to sediment the cell debris. 200 μl of supernatant were taken and the fluorescence measured by a microplate reader (365 nm excitation and 450 nm emission filter).
To infer translational elongation rate, the square root of the signal in excess of the signal at time zero was plotted. A linear fit was performed on the points after the signal began to increase. The lag time is the x intercept of the line. A previous study has shown that the initiation time remains constant for about 10 s across a wide range of conditions tested52. Therefore, we corrected the elongation times measured by subtracting 10 s from the lag times measured.
Ribosome footprinting and RNA extraction.
The cell collection step was the same as for polysome profiling except that 1 mM of chloramphenicol was used in the sucrose solution. The footprinting and library preparation steps were adapted from Li et al.2 After quantification of RNA concentration with NanoDrop, samples with 500 μ g RNA were digested with 750U MNase (10107921001, Roche) for 1 h at 25 °C before being quenched with 6 mM EGTA. The lysates were then layered onto a 10–55% sucrose gradient and centrifuged. The monosome fraction was collected and snap frozen in liquid nitrogen. No polysome peaks were observed, indicating a thorough digestion. The RNA was isolated using hot phenol and size selected on 15% TBE-Urea PAGE gels run for 1 h at 210 V. Gels were stained with SYBR Gold and visualized using Dark Reader (Clare Chemical Research). RNA fragments with size between 25 nt and 40 nt were extracted using isopropanol precipitation.
Library preparation and sequencing.
Fragments from footprints were dephosphorylated at the 3’ end by T4 Polynucleotide Kinase (M0201, NEB). The repaired fragments were linked to the Universal miRNA Cloning Linker (S1315S, NEB), reverse transcribed (18080044, Thermo) and circularized (CL4111K, Epicentre). rRNA was subtracted from the circularized samples before PCR amplification (M0531L, NEB) and size selection. High-quality PCR samples were checked by Bioanalyzer high-sensitive DNA chip. Deep sequencing was done by Illumina HiSeq 2500 on Rapid flow cells with settings of single end and 75 nt-long read length.
Mapping and sequencing data analysis.
Data manipulation including barcode splitting, linker trimming and mapping were done using Galaxy53. The processed reads were mapped to E coli genome escherichia_coli_k12_nc_000913_3 from the National Center for Biotechnology Information (NCBI) database with the BWA54 short read mapping algorithm. Only the reads between 20 nt and 45 nt that aligned to the coding region were used for further analysis.
To infer the ribosome A-site position, the Python package Plastid55 was used to align the 3’ end of reads to the stop and start codons56, which are known to have higher ribosome densities. We found that the offsets were 12 nt for the stop codon and 15 nt for the start codon. Therefore, we used 11 nt for the A-site position and 14 nt for the P site. The counts were normalized to the total counts in the coding region as reads per million (RPM) and transcripts per million (TPM). Further analysis was done using customized Python and R codes with packages including Plastid, dplyr, tidyr and ggplot2.
Analysis of ribosome profiling data.
TPM was used to identify highly expressed genes for codon occupancy analysis. After assigning each mapped read to the A-site nucleotide, the raw counts were first normalized by the length of their mapped coding region, to yield the count density for each gene. These count densities were then globally normalized to one million counts across all genes within one sample. To determine codon occupancy, transcripts having total counts over 100 TPM and containing more than 200 codons were considered. Ribosome footprint counts for the first and last 40 codons were removed to avoid possible effects from initiation and termination, and counts per codon were recorded for the remaining counts. For each gene and each codon type, the codon occupancy ratio is defined as the ratio of measured counts to expected counts. Expected counts are simply proportional to the frequency of that particular codon in the gene. The final reported codon occupancy ratio is the average of codon occupancy ratios from all genes considered weighted equally.
We calculated ribosome counts along a transcript using RPM. Only transcripts having total counts over 10 TPM and containing more than 100 codons were considered. Total counts after filtering were normalized to one million reads. The selected genes account for more than 75% of the total counts. The ribosome counts are the sum of re-normalized RPM at each position.
To compare the ribosome counts of genes from the first and second halves, the same set of genes and trimming processing for codon occupancy were used. The RPM from the first and second halves of the gene were summed up separately and plotted against each other.
Supplementary Material
Acknowledgements
We thank the members of the Gitai, Wingreen and Rabinowitz laboratories for helpful discussions. We thank G.-W. Li for his support for the ribosome profiling experiments. We thank the Microarray Core Facility at the Lewis-Sigler Institute (D. Sanchez, J. M. Miller, J. Wiggins and W. Wang) for RNA-Seq sample processing and sequencing and the Princeton Proteomics Core Facility (H. Shwe and T. Srikumar) for ribosome profiling sample processing. L. Parsons provided helpful technical support for bioinformatics analysis of the sequencing data. We thank the Botstein lab, particularly S. Silverman, for chemostat operation support and all former members for discussions relating to microbial growth. This work was supported by grants from the National Institutes of Health (DP1AI124669 and R01GM082938) and NSF (PHY-1607612).
Footnotes
Competing interests
The authors declare no competing interests.
Reporting Summary.
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Code availability.
All the codes used for data analysis in this paper are available on request.
Data availability.
All the sequencing data used to reach the conclusion of this paper are available under NCBI BioProject ID PRJNA474410 and SRA accession number SRP149755. All the RNA-to-protein ratio data are available in Supplementary Table 2 as an Excel file.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41564–018-0199–2.
Reprints and permissions information is available at www.nature.com/reprints.
Correspondence and requests for materials should be addressed to N.S.W. or Z.G.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.You C et al. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature 500, 301–306 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G-W, Burkhardt D, Gross C & Weissman JS Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scott M, Klumpp S, Mateescu EM & Hwa T Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Syst. Biol. 10, 747–747 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Giordano N, Mairet F, Gouzé J-L, Geiselmann J & de Jong H Dynamical allocation of cellular resources as an optimal control problem: novel insights into microbial growth strategies. PLoS Comput. Biol. 12, e1004802 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dennis PP & Bremer H Modulation of chemical composition and other parameters of the cell at different exponential growth rates. EcoSal Plus https://doi.org/10.1128/ecosal.5.2.3 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Neidhardt FC & Magasanik B Studies on the role of ribonucleic acid in the growth of bacteria. Biochim. Biophys. Acta 42, 99–116 (1960). [DOI] [PubMed] [Google Scholar]
- 7.Baracchini E & Bremer H Determination of synthesis rate and lifetime of bacterial mRNAs. Anal. Biochem. 167, 245–260 (1987). [DOI] [PubMed] [Google Scholar]
- 8.Schaechter M, Maaløe O & Kjeldgaard NO Dependency on medium and temperature of cell size and chemical composition during balanced growth of Salmonella typhimurium. Microbiology 19, 592–606 (1958). [DOI] [PubMed] [Google Scholar]
- 9.Scott M, Gunderson CW, Mateescu EM, Zhang Z & Hwa T Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–1102 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto T, Izumi S & Gekko K Mass spectrometry of hydrogen/deuterium exchange in 70S ribosomal proteins from E. coli. FEBS Lett 580, 3638–3642 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Nomura M & Gourse R Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. 53, 75–117 (1984). [DOI] [PubMed] [Google Scholar]
- 12.Maaloe O & Kjeldgaard NO Control of Macromolecular Synthesis (WA Benjamin, New York, 1966). [Google Scholar]
- 13.Dai X et al. Reduction of translating ribosomes enables Escherichia coli to maintain elongation rates during slow growth. Nat. Microbiol 2, 16231 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewin B Genes VIII (Pearson Education, Upper Saddle River, 2003). [Google Scholar]
- 15.Wanner BL Gene regulation by phosphate in enteric bacteria. J. Cell. Biochem. 51, 47–54 (1993). [DOI] [PubMed] [Google Scholar]
- 16.Ahn K & Kornberg A Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J. Biol. Chem. 265, 11734–11739 (1990). [PubMed] [Google Scholar]
- 17.Schleif R, Hess W, Finkelstein S & Ellis D Induction kinetics of the L-arabinose operon of Escherichia coli. J. Bacteriol. 115, 9–14 (1973). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dalbow DG & Young R Synthesis time of β -galactosidase in Escherichia coli B/r as a function of growth rate. Biochem. J. 150, 13–20 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qin D & Fredrick K Analysis of polysomes from bacteria. Meth. Enzymol. 530, 159–172 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Beller RJ & Lubsen NH Effect of polypeptide chain length on dissociation of ribosomal complexes. Biochemistry 11, 3271–3276 (1972). [DOI] [PubMed] [Google Scholar]
- 21.Nathans D Puromycin inhibition of protein synthesis: incorporation of puromycin into peptide chains. Proc. Natl Acad. Sci. USA 51, 585–592 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steitz JA Polypeptide chain initiation: nucleotide sequences of the three ribosomal binding sites in bacteriophage R17. RNA 224, 957–964 (1969). [DOI] [PubMed] [Google Scholar]
- 23.Ingolia NT, Ghaemmaghami S, Newman JRS & Weissman JS Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doucette CD, Schwab DJ, Wingreen NS & Rabinowitz JD α -Ketoglutarate coordinates carbon and nitrogen utilization via enzyme I inhibition. Nat. Chem. Biol. 7, 894–901 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yuan J et al. Metabolomics‐driven quantitative analysis of ammonia assimilation in E. coli. Mol. Syst. Biol. 5, 302 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ikeda TP, Shauger AE & Kustu S Salmonella typhimurium apparently perceives external nitrogen limitation as internal glutamine limitation. J. Mol. Biol. 259, 589–607 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Subramaniam AR, Pan T & Cluzel P Environmental perturbations lift the degeneracy of the genetic code to regulate protein levels in bacteria. Proc. Natl Acad. Sci. USA 110, 2419–2424 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sørensen MA et al. Over expression of a tRNALeu isoacceptor changes charging pattern of leucine tRNAs and reveals new codon reading. J. Mol. Biol. 354, 16–24 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T & Gerdes K Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat. Rev. Micro 13, 298–309 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Potrykus K & Cashel M (p)ppGpp: Still Magical?. Annu. Rev. Microbiol. 62, 35–51 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Mitkevich VA et al. Thermodynamic characterization of ppGpp binding to EF-G or IF2 and of initiator tRNA binding to free IF2 in the presence of GDP, GTP, or ppGpp. J. Mol. Biol. 402, 838–846 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Milon P et al. The nucleotide-binding site of bacterial translation initiation factor 2 (IF2) as a metabolic sensor. Proc. Natl Acad. Sci. USA 103, 13962–13967 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang M, Sullivan SM, Wout PK & Maddock JR G-protein control of the ribosome-associated stress response protein SpoT. J. Bacteriol. 189, 6140–6147 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gentry DR & Cashel M Mutational analysis of the Escherichia coli spoT gene identifies distinct but overlapping regions involved in ppGpp synthesis and degradation. Mol. Microbiol. 19, 1373–1384 (1996). [DOI] [PubMed] [Google Scholar]
- 35.Pavlov MY & Ehrenberg M Optimal control of gene expression for fast proteome adaptation to environmental change. Proc. Natl Acad. Sci. USA 110, 20527–20532 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kjeldgaard NO, Maaløe O & Schaechter M The transition between different physiological states during balanced growth of Salmonella typhimurium. Microbiology 19, 607–616 (1958). [DOI] [PubMed] [Google Scholar]
- 37.Brauer MJ et al. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell 19, 352–367 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul BJ, Ross W, Gaal T & Gourse RL rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38, 749–770 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Laursen BS, Sørensen HP, Mortensen KK & Sperling-Petersen HU Initiation of protein synthesis in bacteria. Microbiol. Mol. Biol. Rev. 69, 101–123 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao H et al. Residual guanosine 3’,5’-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266, 5980–5990 (1991). [PubMed] [Google Scholar]
- 41.Rojas A-M, Ehrenberg MN, Andersson SGE & Kurland CG ppGpp inhibition of elongation factors Tu, G and Ts during polypeptide synthesis. Mol. Gen. Genet. 197, 36–45 (1984). [DOI] [PubMed] [Google Scholar]
- 42.Zarrinpar A, Chaix A, Yooseph S & Panda S Diet and feeding pattern affect the diurnal dynamics of the gut microbiome. Cell Metab. 20, 1006–1017 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korem Kohanim Y, Levi D, Jona G, Bren A & Alon U A bacterial growth law out of steady-state. 23, 2891–2900 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Erickson DW et al. A global resource allocation strategy governs growth transition kinetics of Escherichia coli. Nature 551, 119–123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mori M, Schink S, Erickson DW, Gerland U & Hwa T Quantifying the benefit of a proteome reserve in fluctuating environments. Nat. Commun 8, 1225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Towbin BD et al. Optimality and sub-optimality in a bacterial growth law. Nat. Commun 8, 14123 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peters JM et al. A comprehensive, CRISPR-based functional analysis of essential genes in bacteria. Cell 165, 1493–1506 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kafri M, Metzl-Raz E, Jona G & Barkai N The cost of protein production. Cell Rep. 14, 22–31 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Metzl-Raz E et al. Principles of cellular resource allocation revealed by condition-dependent proteome profiling. eLife 6, e03528 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baba T et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2, 2006.0008 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ares M Bacterial RNA isolation. Cold Spring Harb. Protoc. 2012, 1024–1027 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Zhu M, Dai X & Wang Y-P Real time determination of bacterial in vivoribosome translation elongation speed based on LacZα complementation system. Nucleic Acids Res. 44, e155 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afgan E et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44, 3–10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li H & Durbin R Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunn JG & Weissman JS Plastid: nucleotide-resolution analysis of next-generation sequencing and genomics data. BMC Genom. 17, 958 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woolstenhulme CJ, Guydosh NR, Green R & Buskirk AR High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell Rep. 11, 13–21 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.