Abstract
The dynamic and polymicrobial oral microbiome is a direct precursor of diseases such as dental caries and periodontitis, two of the most prevalent microbially induced disorders worldwide. Distinct microenvironments at oral barriers harbour unique microbial communities, which are regulated through sophisticated signalling systems and by host and environmental factors. The collective function of microbial communities is a major driver of homeostasis or dysbiosis and ultimately health or disease. Despite different aetiologies, periodontitis and caries are each driven by a feedforward loop between the microbiota and host factors (inflammation and dietary sugars, respectively) that favours the emergence and persistence of dysbiosis. In this Review, we discuss current knowledge and emerging mechanisms governing oral polymicrobial synergy and dysbiosis that have both enhanced our understanding of pathogenic mechanisms and aided the design of innovative therapeutic approaches for oral diseases.
Diverse microorganisms inhabit the oral cavity1,2 and in many cases are unique to this niche as they have evolved an exquisite specificity for oral colonization3. Within the oral cavity, there are distinct microenvironments such as the hard non-shedding surfaces of the teeth and the epithelial surfaces of the mucosal membranes (FIG. 1). These surfaces are exposed to a fluid phase of saliva, or if subgingival, to gingival crevicular fluid (GCF). The microbial communities that grow on these surfaces are also distinct, and any one site contains ~50 species — a subset of ~1,000 species that are capable of oral colonization4,5. Tissue-specific tropisms are often defined by specificity and avidity of adherence, which is a feature of many successful oral colonizers, providing resistance to the mechanical shearing forces of fluid flow and mastication. Primary colonizers of oral surfaces are predominantly facultative anaerobes such as streptococci and Actinomyces species. Within the confines of the subgingival area, reduced oxygen tensions favour population shifts with increasing abundance of strict anaerobes such as Bacteroidaceae spp. and spirochaetes. In addition to microbial composition, the spatial and structural organization (biogeography) of natural microbial communities is being increasingly recognized as essential for physical and metabolic interspecies interactions that can be antagonistic or cooperative6,7. Microorganisms on tooth surfaces tend to form multispecies biofilm communities that are often embedded in a matrix of extracellular polymeric substances (EPS). By contrast, the shedding, more transient epithelial surfaces necessitate a specialized colonization strategy, and although organisms do form biofilms on these surfaces, there is less time for biofilm maturation than with abiotic or tooth surfaces. In addition, bacteria penetrate and grow within epithelial tissues and even intracellularly. Most of the time, a homeostatic balance exists between the host and microbial communities, and the resident microbiota is thought to compete with and exclude exogenous pathogens as a component of ecosystem stability, as well as contribute to normal tissue and immune system development8,9. Although gingivitis is an almost inevitable consequence of prolonged accumulation of biofilms (also known as plaque) on tooth surfaces, it is a controlled immune-inflammatory state (BOX 1) that does not permanently compromise the integrity of the tissues supporting the teeth. Host saliva also contributes to ecosystem stability by buffering the oral environment, providing nutrition to the community and delivering antimicrobial factors that are antagonistic to exogenous species. Nonetheless, under particular conditions, the host-community interaction becomes dysbiotic and site-specific diseases involving the teeth or gums (gingivae) can follow10–12. The accessibility of the oral ecosystems has facilitated the characterization of microbial communities that are associated with health or disease at distinct oral sites. In this Review, we discuss the mechanisms by which oral microbial communities develop and become functionally specialized. We examine the progression of polymicrobial communities towards pathogenicity with a particular emphasis on the induction of immune responses that are ineffective, uncontrolled and destructive, and on cariogenic biofilm development stimulated by host diet. Finally, the opportunities for therapeutic intervention directed towards interfering with acidogenic biofilm development or the subversion of the immune response are explored.
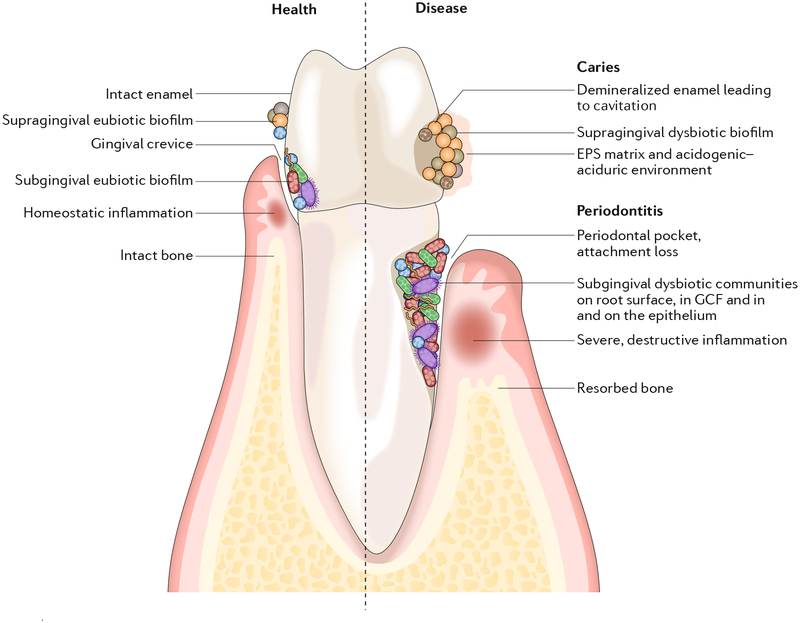
Fig. 1 |. Biogeography of oral microbiota colonization in the diverse habitats of the oral cavity.
Microbial colonization occurs on all available surfaces, and microorganisms can also penetrate epithelial tissues and cells. The microbiota assembles into biofilm communities on the abiotic and biotic surfaces. In health (left), eubiotic biofilms maintain a homeostatic balance with the host. In disease (right), caries and periodontitis ensue when biofilms become dysbiotic, resulting in increased levels and duration of low pH challenge and the induction of destructive inflammatory responses, respectively. EPS, extracellular polymeric substance; GCF, gingival crevicular fluid.
Box 1 | Models of microbiota-induced periodontitis.
Gingivitis is highly prevalent in the human population. An immune-inflammatory state characterizes this condition, and neutrophils are continuously recruited into the gingival tissues. Mild periodontal inflammation can therefore be seen as a normal and controlled state that may prevent, or at least does not contribute to, tissue destruction. Disruption of this equilibrium is necessary for the onset of destructive inflammation and permanent tissue damage. The widespread occurrence and clinical variety of destructive periodontal conditions has complicated attempts to construct an overarching model of disease initiation and progression. Additionally, experimentally tractable animal and in vitro models by definition do not fully recapitulate the human situation, although different aspects of the disease can be productively addressed by in vitro and animal models. the recently formulated polymicrobial synergy and dysbiosis (PsD) hypothesis is consistent with human microbiome studies and mechanistic studies in animal models and with the ‘ecological plaque’ hypothesis24,99. according to the ecological plaque concept, environmental factors (for example, inflammation, pH, redox potential and nutrient availability) drive the selection and enrichment of specific pathogenic bacteria, also known as pathobionts36,45. The PSD model proposes that disease is not initiated by individual causative pathogens but rather by a synergistic polymicrobial community, within which specific constituents, or combinations of functional genes, fulfil distinct roles that converge to shape and stabilize a dysbiotic microbiota, which perturbs host homeostasis11. Disease is caused by reciprocally reinforced interactions between such physically and metabolically integrated polymicrobial communities and a dysregulated host inflammatory response. Thus, the PsD model bypasses the ‘chicken-or-the-egg’ question on whether dysbiosis initiates inflammation or vice versa but rather places an emphasis on the continuous cyclic process in which dysbiosis and inflammation are reciprocally reinforced and constitute the actual driver of periodontitis.
The oral microbiota and disease
Several processes underlie the transition of a microbial community to a state of dysbiosis. Alterations in host immune competence or diet can affect the community composition and the metatranscriptional landscape, with increases in the production of virulence factors. As a community develops, microbial metabolism and by-products of the host immune response can cause changes to the local environment that facilitate the outgrowth or over-representation of microorganisms associated with a dysbiotic state. The microbiota associated with a healthy state is thus considered more generalist, whereas the disease-associated microbiota is influenced by ‘specialist’ microorganisms that possess metabolic functions and an elevated virulence potential that are largely absent in health13. Once a community has transitioned to a dysbiotic state, the structural stability of functionally specialized components6,14,15 will allow the condition to persist for an extended period of time, and oral diseases such as periodontitis and dental caries (FIG. 1) are often chronic and slowly progressing (although acute onset of both diseases can be triggered under particular host-compromising conditions).
Supragingival communities and dental caries.
In dental caries, overexposure to dietary carbohydrates and host factors promotes the production of EPS and acidic metabolites, in addition to causing the accumulation of acidogenic and aciduric microorganisms. Excess fermentable carbohydrate thus drives the transition to a pathogenic biofilm community7,16,17. If sugar consumption is low and infrequent, the microbial communities on teeth remain stable and, despite being able to produce acids that demineralize enamel, the episodic pH decrease can be readily neutralized by saliva, which restores and maintains the mineralization of enamel17. However, with frequent exposure to fermentable carbohydrates, microorganisms become embedded in an EPS-rich biofilm matrix while constantly producing acids that are physically protected from rapid buffering by saliva. Localized regions of low pH within biofilms formed on tooth surfaces18,19 continue to select for aciduric microorganisms17. If the biofilm is not removed and frequent sugar consumption continues, a prolonged and repeated state of acidification ensues (which can be exacerbated by dysfunction in salivary secretion or composition), disrupting the homeostatic mineral balance towards enamel demineralization. Mutans streptococci (especially Streptococcus mutans) and lactobacilli have long been recognized as pathogens that are associated with caries; however, more recent molecular analyses have revealed the existence of a pathogenic community that includes non-streptococcal bacteria (for example, Bifidobacterium spp., Scardovia spp. and Actinomyces spp.) and fungi (for example, Candida albicans)20–23. The microbial composition can vary depending on the different sites of the tooth surface. These microorganisms interact with each other in a dynamic and concerted polymicrobial synergy to form a cariogenic biofilm (that is, a biofilm that can cause caries) within which the community changes as caries progress from early onset (initial demineralization) to deeper lesions with dentin exposure7.
Subgingival communities and periodontal disease.
In periodontal diseases, polymicrobial communities induce a dysregulated and destructive host response through an overall mechanism referred to as polymicrobial synergy and dysbiosis24 (Box 1). Conventional culture-based approaches identified a pathogenic triad of Porphyromonas gingivalis, Tannerella forsythia and Treponema denticola25,26 (the red complex), and culture-independent molecular studies have extended the list of candidate pathogens to include the Gram-positive bacteria Filifactor alocis and Peptoanaerobacter stomatis; Gram-negative members of the Firmicutes phylum (Dialister spp., Megasphaera spp. and Selenomonas spp.); species in the genera Prevotella, Desulfobulbus and Synergistes; and many others1,2,5,27,28. Indeed, contrary to the situation in the gastrointestinal tract, periodontal diseases are associated with increases in diversity of the microbiome, thought to be the consequence of additional nutrients derived from host tissue damage and increasing physical space as the gingival crevice deepens. However, it is important to note that in vivo studies show substantial variation in the microbiomes among individuals with periodontitis and even between sites in the same individual29–32. Although human studies do not provide insights into disease mechanisms, the integration of metagenomic and metatranscriptomic data indicates that, rather than there being a discrete pathogenic cohort of organisms, a particular set of gene functions is required to induce dysbiosis.
The oral microbiota in cancer and other systemic diseases.
Increasing evidence supports an association between the oral microbiome and oral cancers such as oral squamous cell carcinoma. A mechanistic understanding for such an association may arise from the ability of many oral microorganisms to alter the inflammatory microenvironment and to interfere with host signalling pathways that control cell viability, proliferation and differentiation33–38. Remarkably, the influence of the oral microbiome can extend beyond the oral cavity, and systemic conditions such as coronary artery disease, preterm delivery of low-birthweight neonates and rheumatoid arthritis are associated with the oral microbiome39–41. In the case of rheumatoid arthritis, the enzyme peptidylarginine deiminase PPAD — produced uniquely by P. gingivalis — can citrullinate host proteins, which may then induce autoantibody production42.
Polymicrobial synergy
Microorganisms within communities often interact synergistically to enhance colonization, persistence or pathogenicity. The concept of polymicrobial synergy among members of the periodontal microbiome has been established in vivo; experiments in animal models consistently show elevated pathogenicity with combinations of organisms compared with monospecies infection10,43,44. Such polymicrobial synergy can arise from several classes of interspecies interactions including one organism providing a substratum for the attachment and colonization of another, nutritional cross feeding, and the co-ordinated metabolism of complex substrates10,45–48. Physical interactions among organisms and the diffusion of soluble factors can modulate virulence gene expression and nososymbiocity45,49. Interdependence among community members has led to functional specialization, with different species contributing discrete sets of community-essential genes. Similarly, in cariogenic communities, synergistic interactions occur between acidogenic and aciduric bacteria during the different stages of biofilm development and acidification7. Recently, inter-kingdom synergies with Candida albicans were found to enhance biofilm nososymbiocity, leading to the onset of severe caries in vivo. Furthermore, mechanistic studies have indicated complex signalling and cross-feeding interactions combined with elevated EPS production7,50,51.
The notion that commensalism or pathogenicity represent a fixed duality has in recent years been replaced by the concept of a fluid continuum45,52–55, and the oral microbiome provides many illustrative examples. Oral streptococci of the Mitis group (including Streptococcus gordonii, Streptococcus sanguinis, Streptococcus parasanguinis, Streptococcus oralis and Streptococcus mitis) were once considered strict commensals in the oral cavity, a view that is now considered as too restrictive56. These microorganisms are abundant pioneer colonizers of tooth surfaces, in large part owing to the expression of numerous adhesins for receptors in the salivary pellicle that coats tooth surfaces3,57. Monoinfection of animal models with Mitis streptococci does not induce substantial pathology56,58,59. However, mixed infections of S. gordonii and P. gingivalis result in increased alveolar bone loss compared with infection with P. gingivalis alone60; S. gordonii is considered an accessory pathogen in this context56. However, the interaction between S. gordonii and P gingivalis is nuanced and multidimensional. Sensing of the streptococcal metabolite 4-amino benzoate (pABA) by P. gingivalis increases the activity of the tyrosine kinase Ptk1, and the resulting protein tyrosine phosphorylation-dependent signalling converges on the FimA and Mfa1 fimbrial adhesins61. Although this primes P. gingivalis for attachment, the pathogenic potential of this organism is diminished61. Engagement of the Mfa1 adhesin with streptococcal SspA or SspB surface proteins initiates community development and activates the tyrosine phosphatase Ltp1. Dephosphorylation of Ptk1 by Ltp1 suppresses adhesin production and ultimately constrains the degree of community accretion62,63 (Fig. 2). Interspecies communication based on physical contact increases the pathogenic potential of P gingivalis. Moreover, communication is bidirectional, and as a keystone pathogen, P gingivalis increases the nososymbiocity of communities in which it resides and drives pathogenicity even at low abundances (discussed below).
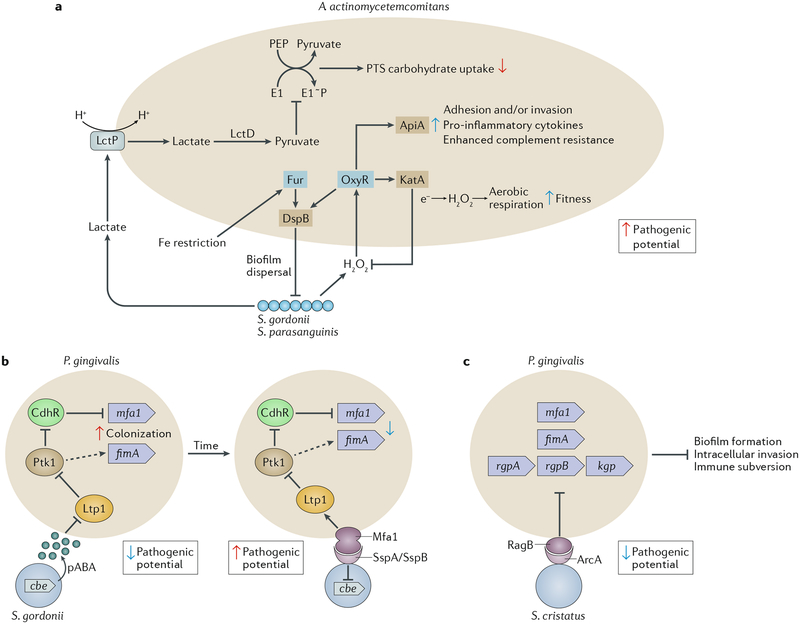
Fig. 2 |. Interactions among bacterial species that affect nososymbiocity.
Oral bacteria interact through multiple pathways that can be separated both spatially and temporally. a | Streptococcus gordonii and Streptococcus parasanguinis produce hydrogen peroxide, to which Aggregatibacter actin-omycetemcomitans responds by activation of the OxyR transcriptional regulator; consequently, transcription of both apiA and katA is elevated. Higher levels of the ApiA surface protein increases complement resistance and will also potentially induce intracellular invasion and pro-inflammatory cytokine production. KatA (a catalase) degrades the hydrogen peroxide produced by both streptococci and neutrophils, thus protecting A. actino-mycetemcomitans from oxidative damage64,66,154–156. OxyR also regulates production of dispersin B (DspB), an enzyme that degrades the biofilm matrix and facilitates dispersal of A. actinomycetemcomitans65. Hydrogen peroxide increases the bioavailability of oxygen, and, in response, A. actinomycetemcomitans shifts from a primarily fermentative to a respiratory metabolism, an interaction referred to as cross respiration157. Respiratory metabolism enhances the growth and fitness of A. actinomycetemcomitans in vivo. Transport of streptococcal lactate into A. actinomycetemcomitans through the proton-driven lactate permease (LctP) leads to conversion to pyruvate by lactate dehydrogenase (LctD). Pyruvate suppresses autophosphorylation of E1, which then decreases uptake of phosphotransferase system (PTS) carbohydrates such as glucose155. Preferential utilization of lactate through this carbon resource partitioning gives a competitive advantage to A. actinomycetemcomitans in the presence of organisms that can metabolize glucose more efficiently. Communities of A. actinomycetemcomitans and S. gordonii also become restricted for iron. The Fur transcriptional regulator of A. actinomycetemcomitans responds to iron limitation and induces upregulation of the gene encoding DspB, which will release A. actinomycetemcomitans from biofilms158. A. actinomycetemcomitans responses to both oxidative stress and iron restriction thus involve DspB activity and re-localization, and in vivo A. actinomycetemcomitans maintains an optimal distance from streptococci in communities that are synergistically virulent65. b | Interactions between Porphyromonas gingivalis and S. gordonii resulting from metabolite (4-amino benzoate (pABA)) perception (left) and direct contact (right). pABA secreted by S. gordonii inactivates the P. gingivalis tyrosine phosphatase Ltp1. Dephosphorylation and inactivation of the tyrosine kinase Ptk1 is thus reduced. Ptk1 phosphorylates and inactivates the transcription factor CdhR, which is a repressor of the mfa1 gene. Ptk1 activity also converges on expression of the fimA gene. Expression of both fimbrial adhesins is increased, and in this mode P. gingivalis is primed for attachment to S. gordonii. However, nososymbiocity is reduced, and pABA-treated P. gingivalis are less pathogenic in animal models. Engagement of Mfa1 with the streptococcal SspA or SspB surface protein increases Ltp1 and reverses information flow through the Ltp1-Ptk1 axis. In addition, Mfa1-SspA/SspB binding suppresses expression of chorismate binding enzyme (Cbe), which is responsible for pABA production. Prolonged physical interaction between P. gingivalis and S. gordonii leads to increased nososymbiocity, and dual infection of animal models causes more alveolar bone loss than P. gingivalis infection alone60–63,159,160. c | Streptococcus cristatus arginine deiminase (ArcA) interacts with the P. gingivalis surface protein RagB. Signal transduction results in downregulation of genes encoding the FimA and Mfa1 component fimbriae along with the arginine-specific (RgpA or RgpB) and lysine-specific (Kgp) gingipain proteinases. Adhesion, biofilm formation, epithelial cell invasion and degradation of cytokines are consequently reduced and nososymbiocity is suppressed72–74. Part a adapted with permission from REF56, Wiley-VCH.
Both S. gordonii and S. parasanguinis are accessory pathogens to Aggregatibacter actinomycetemcomitans — a keystone pathogen in localized aggressive periodontitis64–66 (Fig. 2). Mitis group streptococci have also been shown to enhance the virulence of C. albicans by promoting fungal tissue invasion and increasing the severity of mucosal infection67,68. However, consistent with the notion that pathogenic potential is context-dependent, S. gordonii, S. sanguinis and S. oralis are antagonistic towards S. mutans69–71 and may help protect against caries (Fig. 3). A related streptococcal species, Streptococcus cristatus, is antagonistic towards P. gingivalis and suppresses virulence gene expression72,73 (Fig. 2). S. cristatus can reduce the pathogenicity of P. gingivalis in animal models of disease74, and the two organisms are negatively correlated in human oral biofilm samples75. Host epithelial cells are an additional factor in community nososymbiocity as they provide an interactive interface for colonizing microorganisms; interactions between colonizers and the host epithelia affect inflammatory responses11,76. Streptococci are also major components of microbial communities that are associated with oral epithelia77 and can restrain pro-inflammatory responses and stimulate the expression of antimicrobial peptides (for example, β-defensins)43,78,79. Thus, on the epithelial interface, oral streptococci can act as homeostatic commensals and help maintain a host-community equilibrium. Interestingly, although homeostatic commensals can induce the expression of antimicrobial proteins that preferentially target periodontitis-associated bacteria80, the reverse is also true. Indeed, P gingivalis activates Notch 1 signalling in oral epithelial cells, leading to the production of PLA2-IIA (also known as PLA2G2A), an antimicrobial protein that can promote dysbiosis81. Polymicrobial infections involving functionally specialized organisms within an interactive communication network are thus inherently more complex than infections caused by single species, which complicates the study of pathogenicity and the development of diagnostic and treatment options.
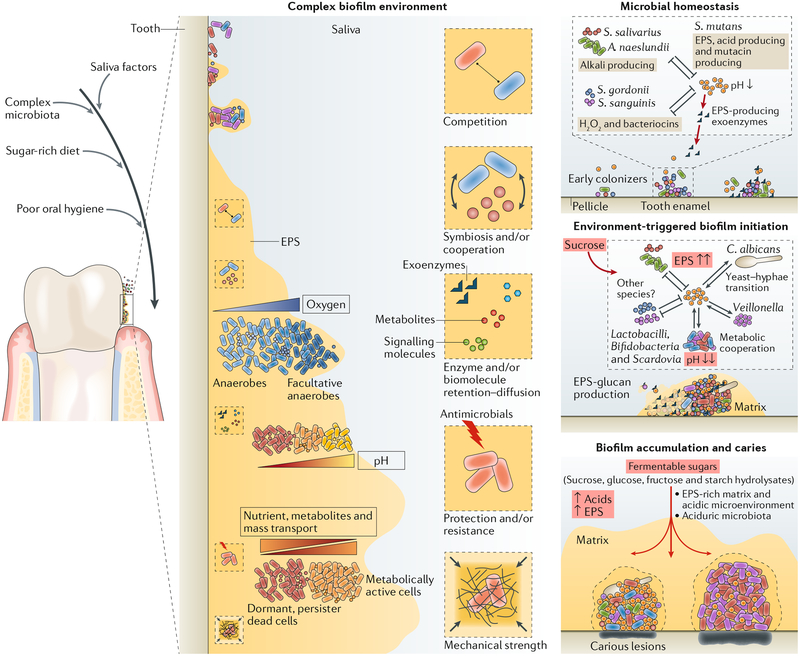
Fig. 3 |. Diet-microbiota interactions trigger the assembly of cariogenic biofilm microenvironment.
In the oral microbial community on tooth surfaces, social interactions begin with primary colonizers that can rapidly attach and then co-adhere with later colonizers. Microorganisms can interact physically and metabolically to determine the initial biofilm community. Both antagonistic and cooperative interactions can occur, and these dynamically change according to the host diet, and other factors such as salivary dysfunction, fluoride exposure and oral hygiene. In particular, dietary sucrose provides a substrate for extracellular polysaccharide production and synthesis of organic acids by acidogenic microorganisms. The extracellular matrix, which also contains other biomolecules (extracellular DNA (eDNA) and bacterial or host-derived proteins), provides a multi-functional scaffold for spatial organization, mechanical coherence and interbacterial interactions. The matrix can trap or sequester substances, which, in combination with diffusion-modifying properties, can generate a variety of chemical and protective microenvironments. Biofilms thus become persistently adhered to the surface and recalcitrant to antimicrobial action. S. mutans has a key pathogenic role as an EPS-matrix producer, acidogenic and aciduric organism. With frequent dietary sugar exposure, continued bacterial metabolism of carbohydrates and reduced accessibility to salivary buffering systems causes the microenvironment within the matrix to become increasingly and constantly acidic. As the biofilm accumulates, the microenvironment also becomes progressively anaerobic (hypoxic). In a feedforward loop, microbial diversity decreases as an aciduric microbiota predominates. If the biofilm is not removed, persistent low-pH conditions at the tooth-biofilm interface shift the demineralization-remineralization balance towards net mineral loss from the tooth enamel, leading to the development of a carious lesion. EPS, extracellular polymeric substance. Adapted with permission from REF7, Cell Press.
Host diet and biofilms in dental caries
Dental caries is a polymicrobial biofilm disease driven by diet-microbiota interactions that cause the destruction of the mineralized tooth tissue7,16,17,82 (FIG. 1). Oral microorganisms are required for the formation of dental caries, but not sufficient, as the formation of pathogenic biofilms is dependent on frequent consumption of dietary sugars by the host7,16,22. Other host and behavioural factors (for example, poor oral hygiene, salivary flow and composition, and enamel defects) and inadequate fluoride exposure also contribute to caries development7,16,17,82. Early (primary) colonizers associated with dental health, such as Mitis streptococci, have substantial ecological advantages over cariogenic organisms when the diet of the host is not rich in dietary sugars. These organisms can bind more avidly to salivary-pellicle-coated teeth, show more rapid growth and antagonize pathogens through multiple mechanisms, including the production of alkali, bacteriocins and hydrogen peroxide83–85, helping to maintain microbial homeostasis and stability (FIG. 3). However, when ecological perturbations exceed thresh-olds, interspecies competition is altered, thus triggering pathogenic processes. Specifically, the balance between commensals and pathogens can be disrupted by overexposure to fermentable carbohydrates. Sucrose is particularly cariogenic as the component hexoses (glucose and fructose) are used to synthesize EPS (glucans and fructans) and are efficiently fermented to produce organic acids (such as lactic acid), which greatly influence the structure and composition of dental bio-films7,17,18. EPS provide binding sites for adhesion to the tooth surface and co-adhesion between bacterial cells, and the microbial communities become embedded in a polymeric matrix that provides cohesion, protection and stability. Such structural organization, together with environmental acidification, promotes microbial shifts towards acidogenic and aciduric organisms and new interspecies interactions7,16,17. However, organisms within dental biofilms must manage a wide range of stresses and large nutrient fluctuations to persist and contribute to the onset of caries (FIG. 3). Thus, diet can modulate both the ecology of the oral microbiota and polymicrobial synergies by providing a highly structured and localized acidic microenvironment. In turn, this acidic microenvironment shapes the composition and metabolic activity of the community in a manner that promotes caries development.
Polymicrobial interactions and acidogenesis.
Caries development is a consequence of dietary sugar-driven biofilm accumulation and localized acidification that causes deleterious microbial community shifts and disrupts tooth-enamel mineral homeostasis. This ecological plaque (biofilm) hypothesis has provided a logical and a tractable model for caries microbial pathogenesis16 and is supported by microbiome-based studies that reveal microbial composition shifts during the transition from health to the various stages of caries16,17,20–22,86–88. Overall, the microbiota becomes dominated by increasingly acidogenic and aciduric organisms, including mutans and non-mutans streptococci, actinomyces, lactobacilli, bifidobacteria and Scardovia spp., which can synergize to enhance EPS production and promote further acidification of the biofilm milieu with frequent sugar exposure. This increased acidification is accompanied by loss of diversity and a reduction in the levels and metabolic activity of beneficial bacteria, which preferentially grow at neutral pH. However, the community composition in advanced caries lesions may reflect the exposure of dentin, an important microenvironment change that allows proteolytic bacteria to thrive89. Other genera commonly found in cariogenic communities, including Propionibacterium, Corynebacterium, Granulicatella and certain strains of Leptotrichia, exhibit high saccharolytic potential and produce acids21,31,90. Although acid-sensitive species survival is disrupted in acidic microenvironments, microorganisms that use lactate as a carbon source (for example, Veillonella spp.)91,92 will benefit from decreases in pH, which may help inhibit the feedforward loop of increasing acidification. Some bacteria found in cariogenic biofilms are not acidogenic-aciduric, such as Prevotella spp., Atopobium spp. and Gram-negative bacteria21,31, and their contribution to the biofilm community or to caries pathogenesis remains to be elucidated. Intriguingly, C. albicans can be detected in higher numbers, often together with S. mutans, in cariogenic plaque from toddlers with severe caries20,93. C. albicans can interact syn-ergistically with S. mutans and colonize tooth surfaces in the presence of dietary sucrose through EPS-mediated interactions. Exoenzymes, termed glucosyltransferases, secreted by S. mutans, bind to the Candida surface and synthesize glucans in situ using sucrose as a substrate. The glucans formed on the surface enhance bacterial-fungal co-adhesion and embed the microorganisms in an EPS-rich matrix, promoting mixed-biofilm accumulation50,94. Within biofilms, the microorganisms cooperate by providing substrates or metabolites (cross feeding), providing growth-stimulating factors, enhancing EPS production and maintaining an aciduric environment51,95. In a rodent model, cross-kingdom interactions enhance biofilm virulence when fed a sugar-rich diet, leading to the onset of extensive carious lesions similar to those found clinically50,94. Whether other Candida species or additional fungi are associated with the cariogenic microbiome in childhood caries remains to be determined. Altogether, the available evidence indicates that synergistic polymicrobial interactions that are triggered by host dietary sugars drive the development of caries, which can be exacerbated by salivary dysfunction, inadequate fluoride exposure and poor oral hygiene.
The role of the extracellular matrix.
Although early studies of caries focused on the composition of oral microbial communities, the importance of the extracellular matrix in collective microbial behaviour and virulence is being increasingly recognized as essential for the biofilm lifestyle7,96. The structural and biochemical properties of the matrix provide the emergent properties of biofilms, including surface adhesion, social interactions and antimicrobial tolerance96. The major components of the matrix are EPS, including exopolysaccharides such as glucans, extracellular DNA (eDNA), lipoteichoic acid, amyloid-like proteins, glycoproteins and host proteins7. The microorganisms that are embedded in the matrix of cariogenic biofilms are cohesive and adherent, making these biofilms difficult to remove from surfaces. Furthermore, the extracellular matrix provides protection against antimicrobials. The extracellular matrix also enables chemical or nutrient gradients to form, including pH and redox gradients, thereby affecting the behaviour and survival of microorgan-isms7,96. Moreover, the matrix can limit the diffusion of charged buffering ions, whereas uncharged sugars such as glucose and sucrose can readily diffuse into biofilms and these can be metabolized into acids7. Conversely, extracellular matrix glucans provide an endogenous source of sugars and can directly trap protons to help the accumulation of acids within biofilms7,18,19,97. These properties may help explain how localized acidification within biofilms occurs in the presence of buffering saliva, shear forces and the near-alkaline environment of the oral cavity. Thus, the extracellular matrix provides a multi-functional platform for the organization of cells into a cohesive multicellular ecosystem that promotes adherence and spatially localizes acid metabolites. Additionally, acids produced within biofilms are sheltered from saliva, which impedes rapid neutralization and helps create a cariogenic microenvironment. This sheltering effect potentiates the ability of acids to demineralize enamel and cause caries. The ability of S. mutans to reduce the pH to levels that are toxic to some microorganisms and create a cohesive EPS-rich environment that can facilitate colonization of other organisms and provide protection is compatible with a keystone or specialist role (FIG. 3). Establishing a localized pathogenic habitat through simultaneous matrix and acid production that promotes a dysbiotic and aciduric community is consistent with the variable levels of the bacterium in plaque during caries development (<1–30% of microbial composition)16,21,22,31,86,87,90,98, although further mechanistic in vivo studies are needed to validate these concepts.
Altogether, dental caries can be conceptually defined as a host-diet-dependent pathological process that relies not only on ecological shifts and polymicrobial acidogenesis but also on the biofilm extracellular milieu within which organisms interact and acids accumulate. This evolving view of ecological flux and concerted polymicrobial synergies within a structured and protected environment, together with salivary and behavioural factors, has direct implications for the development of new and more effective antibiofilm therapeutics.
Inflammation and dysbiosis in periodontitis
A common theme that links caries and periodontal disease is a central axis of microbial community interactions with the host (diet in the case of caries and inflammation in the case of periodontal disease). Additionally, the structure of microbial communities (that is, species composition and abundance of individual species) changes from health to disease. In periodontal disease, the rise of newly dominant species, rather than the appearance of new species (such as exogenous pathogens that are absent in health), has been observed1. Thus, species or genera that dominate disease-associated polymicrobial communities are also found in health but at markedly reduced relative abundance, as predicted by the ecological plaque hypothesis, according to which changes in environmental conditions may favour the outgrowth of pathogens (now defined as pathobionts) beyond a threshold that can instigate periodontitis99. This concept gained experimental support through recent animal model studies showing that anti-inflammatory treatments not only inhibit periodontitis in mice, rats and rabbits but also diminish the periodontal bacterial burden and reverse dysbiosis100–105. Conversely, the bacterial load of subgingival biofilms from individuals with periodontitis accumulates with increasing clinical inflammation1.
Therefore, inflammation appears to be an important ecological change that can drive the outgrowth of periodontitis-associated microorganisms through tissue destruction that releases nutrients (for example, degraded collagen, haem-containing compounds, sources of amino acids and iron, respectively)28,106. These nutrients can be carried via the inflammatory exudate (that is, the GCF) into the gingival crevice to foster the growth of subgingival proteolytic and asaccharolytic bacteria with iron-acquisition capacity (FIG. 4). Accordingly, in situ community-wide transcriptomic analyses of periodontitis-associated subgingival biofilms revealed elevated expression of proteolysis-related genes and genes for peptide transport and acquisition of iron as well as genes for the synthesis of lipopolysaccharides that could further increase the pro-inflammatory potential of the microbial community107. Thus, a subset of species, termed inflammophilic pathobionts106, may selectively expand at the expense of those that fail to adapt to the new environmental conditions, thereby creating a dysbiotic imbalance in the community28–106. In support of this notion, the addition of serum, haemoglobin or hemin to generated oral multispecies biofilms in vitro induces the selective outgrowth of organisms that can act as pathobionts, which additionally upregulate virulence genes including those encoding proteases, haemo-lysins and proteins involved in hemin transport108. This remodelling of the original eubiotic biofilm into a dysbiotic one also enhances the ability of the biofilm to induce pro-inflammatory cytokines by host cells108, thus mimicking the in vivo setting where dysbiosis leads to inflammation.
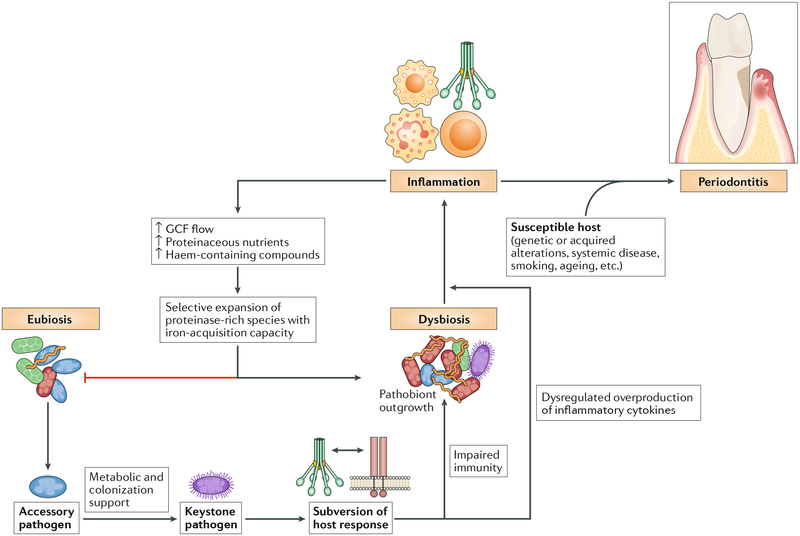
Fig. 4 |. Reciprocally reinforced interactions between dysbiosis and inflammation drive chronic periodontitis.
Colonization by keystone pathogens (for example, Porphyromonas gingivalis) aided by accessory pathogens (for example, Streptococcus gordonii) leads to impaired innate host defence and promotion of inflammation (for example, by subverting complement-Toll-like receptor (TLR) crosstalk in neutrophils and other myeloid cells)60,102,117,119. These alterations contribute to the emergence of dysbiosis (quantitative and compositional changes in the periodontal microbiota). Inflammation worsens dysbiosis by increasing the flow of gingival crevicular fluid (GCF), which, as a result of inflammatory tissue destruction, carries degraded collagen and haem-containing compounds into the gingival crevice, where dysbiotic communities develop. These molecules are selectively used by proteolytic and asaccharolytic bacteria with iron-acquisition capacity. By contrast, health-associated (eubiotic) species cannot capitalize on the new environmental conditions and are outcompeted. This imbalance drives dysbiosis, which further exacerbates inflammation, culminating in periodontitis in susceptible individuals. The ability of inflammation and dysbiosis to positively reinforce each other in a self-sustained feedforward loop may contribute to the chronicity of periodontitis.
In addition to iron and amino acid transport, another enhanced metabolic change detected in developing dysbiosis of the human periodontal microbiome involves the transport of potassium ions109, which become more concentrated in the GCF with increased periodontal disease110. Interestingly, elevating the concentration of potassium in an ex vivo dental plaque biofilm model causes compositional and phenotypic changes in the microbial community, resulting in enhanced production of pro-inflammatory cytokines and a decrease in the production of human β-defensin 3 in gingival epithelial cells111. Another by-product of inflammation is nitrate, which can be used as an electron acceptor for anaerobic respiration by Entero-bacteriaceae to outcompete fermenting microorganisms during colitis-associated dysbiosis112. In a murine model of spontaneous periodontal inflammation (due to deficiency of growth arrest-specific gene 6 (GAS6)), microbial dysbiosis was associated with the selective expansion of nitrate reductase-expressing Proteobacteria, which can use the elevated nitrate in the periodontal environment of Gas6−/− mice113. Potassium and nitrate thus appear to be inflammation-related environmental cues that can contribute to the remodelling of the oral microbiome from a eubiotic community to a dysbiotic one.
Whether dysbiosis is a cause or a consequence of inflammatory disease has been the subject of debate114,115. In periodontitis, there appears to be a reciprocal cause-and-effect relationship between dysbiosis and inflammation. As inflammation fuels the selective growth of dysbiotic communities and dysbiosis exacerbates inflammation28,105–108,111,113, it is likely that neither destructive inflammation nor dysbiosis could fully develop without interactions between these two processes. In conclusion, inflammation acts as a reciprocal ecological driver of dysbiosis, and this inflammation-dysbiosis interplay appears to generate a self-sustained feedforward loop that drives periodontitis (FIG. 4).
Subversion of the host response
Uncoupling bactericidal activity from inflammation.
The crucial importance of inflammation for the development of dysbiosis creates a biological conundrum — dysbiotic communities need inflammation to acquire nutrients but also must downregulate the host immune response for their protection. Although immunosuppression is a common microbial evasion strategy116, this tactic would not be a viable option for inflammophilic bacteria as it would create a non-inflammatory environment that starves the bacteria of essential nutrients. Periodontitis-associated bacteria have resolved this paradox by manipulating the host response in a manner that uncouples inflammation from bactericidal activity, as exemplified by the action of the keystone pathogen P. gingivalis115. In this context, P. gingivalis can benefit the entire microbial community by impairing the bactericidal activity of innate leukocytes while promoting their inflammatory responses60,102,117–120 (FIG. 4). The importance of this keystone function was demonstrated in mice, where this dual subversive action disrupts host-microorganism homeostasis and contributes to the emergence of a dysbiotic microbiota and the development of periodontitis102,117.
Manipulation of complement, Toll-like receptor signalling and cytokines.
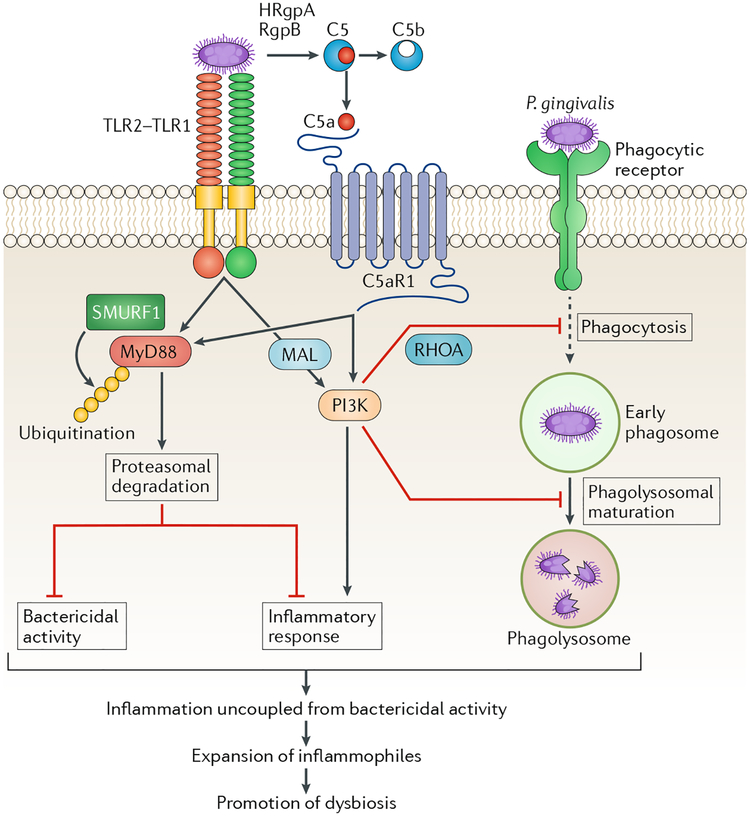
The complement C5a receptor 1 (C5aR1) and Toll-like receptor 2 (TLR2) are at the core of the immune-subversive action of P gingivalis117,119. In human or mouse neutrophils, P. gingivalis initiates C5aR1-TLR2 crosstalk signalling that separates a host-protective TLR2-MyD88 pathway from a TLR2-MyD88-adaptor-like (MAL; also known as TIRAP)-PI3K pathway, which blocks phagocytosis and promotes inflammation117 (FIG. 5). P. gingivalis can also bypass MyD88 to induce pro-inflammatory and anti-phagocytic TLR2-PI3K signalling in macrophages; intriguingly, even within cells that do phagocytose P. gingivalis, PI3K signalling suppresses phagolysosomal maturation and thus promotes intracellular survival118 (FIG. 5). Pharmacological inhibition of either C5aR1 or TLR2, or key downstream signalling intermediates, blocks P. gingivalis-induced dysbiosis and periodontitis in mice102,117. The capacity of P gingivalis to induce TLR2-dependent inflammation while bypassing MyD88, which undergoes ubiquitylation and proteasomal degradation117, is unusual given that prototypical TLR2 agonists (for example, the Pam3CSK4 lipopeptide) activate TLR2 in a strictly MyD88-dependent manner117,118. Consistent with the redundant role of MyD88 in P. gingivalis-induced inflammation, this bacterium induces inflammatory bone loss in mice regardless of the presence or absence of MyD88, whereas the presence of TLR2 is absolutely necessary118.
Fig. 5 |. P. gingivalis induces dysbiosis by impairing innate host defences while promoting inflammatory responses in phagocytic cells.
Porphyromonas gingivalis expresses cell-surface molecules that activate the Toll-like receptor 2 (TLR2)-TLR1 complex and secretes enzymes (HRgpA and RgpB gingipains) that act on the complement component C5 to generate high local concentrations of C5a, a ligand of complement C5a receptor 1 (C5aR1). The bacterium can thus co-activate C5aR1 and TLR2 in phagocytic cells such as neutrophils and macrophages. In both of these myeloid cell types, P gingivalis can bypass MyD88 and thus prevent the associated bactericidal activity117,161, which in neutrophils is possibly mediated by downstream activation of IRAK4-dependent granule exocytosis162. In neutrophils, the inactivation of MyD88 involves its ubiquitylation via the E3 ubiquitin ligase SMURF1 and subsequent proteasomal degradation. Although MyD88-dependent inflammation is blocked by P. gingivalis, this organism induces PI3K-dependent inflammatory cytokines in both neutrophils and macrophages118,120. Similarly, in both cell types, P. gingivalis-induced activation of PI3K leads to inhibition of phagocytosis117,118. In neutrophils, this activity is mediated by the ability of PI3K to suppress RhoA GTPase and actin polymerization117. Intriguingly, even within those macrophages that do manage to phagocytose P. gingivalis bacteria, PI3K signalling suppresses phago-lysosomal maturation, thereby preventing pathogen destruction118. These tactics compromise innate immunity and promote inflammation that leads to the selective expansion of inflammophilic pathobionts. Conversely, inhibition of C5aR1, TLR2 or PI3K reverses dysbiotic inflammation and periodontitis in mice102,117. MAL, MyD88-adaptor-like.
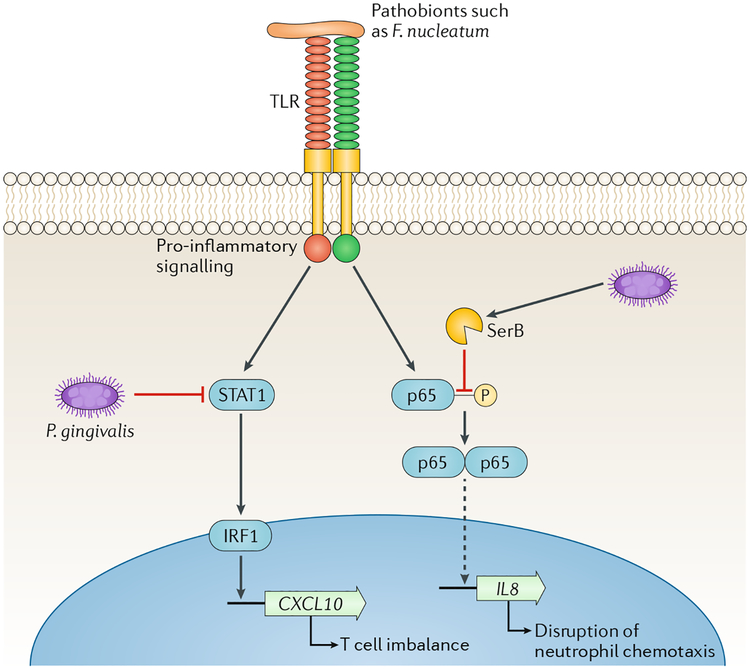
The ability of P. gingivalis to subvert the host response in a manner that benefits the entire microbial community is also supported by the localized chemokine paralysis concept. The expression of IL-8 (also known as CXCL8) by the junctional gingival epithelium, adjacent to the tooth-associated biofilm, is a homeostatic feature of the healthy periodontium as it generates a chemotactic gradient for neutrophil recruitment to the gingival crevice121,122. Consistent with its keystone role, P. gingivalis selectively suppresses expression of IL-8 and T helper 1 (TH1) cell-biasing chemokines (CXCL9, CXCL10 and CXCL11), even in the presence of otherwise stimulatory pathobionts such as Fusobacterium nucleatum123,124. Within epithelial cells, P. gingivalis secretes the serine phosphatase SerB, which dephosphorylates the p65 subunit of the NF-κB transcription factor125. Translocation of NF-κB-p65 homodimers into the nucleus is consequently reduced and IL8 transcription is diminished (Fig. 6). The suppression of TH1-associated chemokine expression by P. gingivalis is mediated through inhibition of the STAT1-IRF1 pathway in epithelial cells, neutrophils and monocytes124 (FIG. 6). Collectively, these chemokine-paralyzing phenomena, even if transient as suggested by in vivo observations102, could have a debilitating effect on immune surveillance during the development of a dysbiotic biofilm. Indeed, defective neutrophil recruitment into the gingival crevice could allow overgrowth of pathobionts, whereas disruption of TH1-biasing chemokines may perturb the balance of protective and destructive immunity in the periodontium126, thereby contributing to increased nososymbiocity.
Fig. 6 |. Localized chemokine paralysis.
Oral pathobionts such as Fusobacterium nucleatum are recognized by Toll-like receptors (TLRs) on epithelial cell surfaces, which leads to the activation of pro-inflammatory signalling pathways. The keystone pathogen Porphyromonas gingivalis can manipulate these pathways and cause a targeted and precise reduction in the production of specific chemokines. Inactivation of STAT1 by P gingivalis leads to reduced expression of CXCL10, which is controlled by the IRF1 transcription factor124. Intracellularly, P. gingivalis secretes SerB, a serine phosphatase that specifically dephosphorylates the serine 536 residue of the p65 NF-κB subunit, thus inhibiting formation and nuclear translocation of NF-κB-p65 homodimers. Transcription of the IL8 gene is reduced and the IL-8 neutrophil gradient is disrupted125. These chemokine paralysis activities will be localized to tissue adjacent to, or containing, P. gingivalis, and in animal models supersede the effects of community pathobionts163. The continuous recalibration of host cell signalling pathways also limits the temporal extent of the phenomenon, which may contribute to the cyclical nature of periodontal tissue destruction. Adapted with permission from REF10, Cell Press.
P. gingivalis as a community activist.
Consistent with the notion that P gingivalis has important roles in initiating dysbiosis, a longitudinal metatranscriptomic analysis of microbial communities from stable or disease-progressing sites showed that, among those bacteria commonly associated with the red complex, only P. gingivalis expressed virulence factors at healthy sites that progressed to disease109. By contrast, T. denticola and T. forsythia virulence gene expression was upregulated at later times when tissue breakdown was clinically observed109. These findings suggest that, at early stages, P. gingivalis acts as a keystone pathogen that contributes to the dysbiotic process, leading to disease progression, whereas T. denticola and T. forsythia may contribute to the nososymbiocity of the microbial community once homeostasis is disrupted, thereby acting as pathobionts that accelerate disease progression. Although not an essential mechanism for dysbiosis (for example, host genetic deficiencies can cause dysbiosis and periodontitis in the absence of P. gingivalis127), P. gingivalis is likely an important risk factor in periodontitis. By manipulating host immunity and separating bactericidal from inflammatory activities117,118,128, P. gingivalis can enhance the adaptive fitness of the entire microbial community in a nutritionally favourable and disease-promoting inflammatory environment (FIG. 4).
Novel approaches to prevention and treatment
Oral biofilms harbour complex polymicrobial communities involving interspecies interactions with host, diet and immunity that control dysbiosis and explain nososymbiocity. The lack of a single obvious target for therapeutic intervention and the potential for transfer of antibiotic resistance genes and physical protection provided by EPS complicate treatment options, and conventional antimicrobial elimination has proved difficult. The presence of a fluid phase that can inactivate bioactive molecules and the difficulty of accessing different oral sites where the disease occurs, combined with poor retention of topically delivered agents, pose additional challenges. These unique conditions are hurdles but also opportunities for the development of innovative drug delivery and effective therapies that could target these complex biological traits and protected oral niches.
For caries prevention, multitargeted therapeutic strategies may be required to prevent pathogenic biofilm accumulation or disrupt established biofilms and decrease dissolution of the enamel mineral. Although supragingival biofilms can be mechanically dislodged by manual or powered toothbrushing, these approaches do not remove biofilms completely, particularly in areas that cannot be easily accessed such as sulcal and interproximal surfaces. Furthermore, subpopulations at high risk of caries, such as young children, elderly adults and those with disabilities, lack the dexterity for optimal brushing techniques, necessitating new or adjunctive therapies. Current antimicrobials do not target important structural and functional traits of biofilms or drug tolerance mechanisms, resulting in limited clinical efficacy to prevent caries7,129,130. Conversely, fluoride can decrease the rate of enamel demineralization and enhance remineralization but has limited biofilm-killing effects (although fluoride can disrupt acid production and acid tolerance by cariogenic bacteria131). In this context, prospective therapeutic strategies should be developed to specifically target the biofilm matrix, the acidic pH microenvironment and the polymicrobial synergies associated with acidogenesis and to facilitate the action of remineralizing agents or mechanical removal. Likewise, enhanced strategies to deliver and retain bioactive agents at the sites where pathogenic biofilms develop (for prevention) or that can penetrate the complex structure of biofilms (for disruption) are needed.
A number of approaches to manipulate biofilm microbial communities have recently emerged, including pH modulation. The simplest strategy that has found clinical applicability involves the use of arginine as a prebiotic-like agent83,132,133. l-Arginine can be metabolized by arginolytic oral species (for example, S. gordonii and Actinomyces spp.) to produce alkali. This alkali can counter the biofilm acidification process and modulate pH homeostasis within oral biofilms, preventing overgrowth of acidogenic-aciduric bacteria and enhance the anti-caries activity of oral care products. Alternatively, bacteria naturally found in the oral cavity such as Streptococcus dentisani or Streptococcus A12 can display dual probiotic action by inhibiting the growth of cariogenic species and by pH modulation through their arginolytic activities50. Conversely, a new generation of antimicrobial peptides consisting of a broad-spectrum, novispirin-derived ‘killing’ region attached to a species-specific peptide pheromone can enhance the targeting specificity against caries pathogens such as S. mutans and increase the abundance of commensal streptococci51. Once a cariogenic biofilm is established, more aggressive measures are needed. These include synergistic combinations of antimicrobial action with biofilm matrix degradation or enhanced physical (mechanical) disruption techniques such as shear-generating high-velocity water sprays129,134–136, particularly in difficult-to-reach and protected retentive sites. Other potential biofilm-specific targeting approaches, including surface modification, antibiofilm coatings and small molecules identified from chemical libraries or in silico screening, have been reviewed elsewhere129.
New drug delivery nanotechnologies have emerged that can penetrate biofilms more effectively and expedite drug release in response to acidic pH, which could enhance the efficacy of current and prospective chemical modalities that target cariogenic biofilms129,137–140. Another approach employs pH-dependent catalytic nanoparticles that generate free radicals from hydrogen peroxide only at acidic pH values and simultaneously degrade the biofilm matrix and kill embedded bacteria with high efficacy under cariogenic conditions. Topical applications of catalytic nanoparticles with low concentrations of hydrogen peroxide prevented the onset of severe caries (including on interproximal and sulcal surfaces) without cytotoxic effects in vivo137. Thus, stimuli or environment-triggered technologies can enhance the selectivity of drug activation or delivery within biofilms, targeting the matrix and the embedded bacteria to eradicate the pathological niches with precision and efficacy without affecting the surrounding host tissues and the commensal microbiota.
The self-sustained feedforward loop between dysbiosis and inflammation discussed earlier (FIG. 4) appears to underlie the chronicity of periodontitis and suggests that host modulation is an effective adjunctive therapy to standard treatment (mechanical debridement to remove the disease-associated biofilm). Targeted inhibition of implicated inflammatory pathways may break the reciprocal cause-and-effect relationship between dysbiosis and inflammation, arrest further development of the disease and promote an environment conducive to inflammation resolution and periodontal tissue repair. Host modulation is indirectly an antimicrobial approach as inflammation control should limit the nutrient supply (inflammatory tissue breakdown products) that sustains dysbiosis, thereby restoring ecological conditions that favour microbiotas compatible with periodontal health. Preclinical interventions in animal models, ranging from mice to non-human primates, have targeted distinct but interconnected signalling pathways involving inflammation initiation (for example, inflammatory cell recruitment103), mechanisms that amplify and propagate inflammation (for example, complement141 and specific pro-inflammatory cytokines142) and pathways that promote inflammation resolution143. In terms of anti-cytokine therapy, antibody-mediated blocking of IL-23, which drives aggressive periodontitis associated with leukocyte adhesion deficiency, led to resolution of inflammatory periodontal lesions in both mouse and human disease104,144. Furthermore, new nanotechnologies for drug delivery, regenerative tissue engineering and biomaterials are emerging for the treatment of periodontal diseases145,146.
Future development should focus on achieving maximal in vivo efficacy and targeting specificity with minimal toxicity and long-term therapeutic effects (compared with current treatments). These, combined with safety and efficacy clinical trials, affordable manufacturing and the development of practical formulations, will determine whether any of the current preclinical strategies could be used for the treatment of human periodontitis and caries.
Conclusions and perspectives
In both dental caries and periodontitis, the nososymbiocity of the polymicrobial communities involved in disease is largely regulated by host factors, predominantly dietary sugars and inflammation. In periodontitis, inflammation and dysbiosis co-develop in a reciprocally reinforced manner, and their interplay develops to become the driver of periodontitis in susceptible individuals (FIG. 4). In caries, diet-microbiota interactions help assemble a persistent tooth-associated biofilm that provides protection to resident microorganisms. The biofilm is also where polymicrobial acidogenesis creates an aciduric environment that disrupts enamel tissue homeostasis, favouring demineralization, which drives caries onset and progression (exacerbated by salivary dysfunction, inadequate fluoride exposure and poor oral hygiene).
The polymicrobial communities in caries and periodontitis display sophisticated structural and functional integration that confers a quasi-organismal status to these entities. This interdependence among constituent members of oral polymicrobial communities lends support to the ‘black queen hypothesis’147, according to which functions that are energetically costly can be discarded as dispensable by ‘cheater’ organisms provided that they are not entirely lost from the community (that is, they are retained by a subset of community members (‘helpers’) that benefit the community). This hypothesis offers a theoretical framework for the emergence of keystone or specialist pathogens that contribute an indispensable public service to the microbial community (for example, formation of a matrix or subversion of host immunity). Whereas in classic infectious diseases overt pathogens employ strategies to overcome colonization resistance as a prerequisite to cause disease, keystone or specialist pathogens exploit the inherent metabolic and/or colonization properties of their commensal neighbours (accessory pathogens) or help create a selective environment in which they increase their virulence and the nososymbiocity of the community.
In a state of compromised homeostasis, the expansion of pathobionts or changes in microenvironment marks a potential tipping point for full-blown development of nososymbiocity (in that restoration of host-microbiota homeostasis is unlikely to occur without treatment intervention). Multitargeted approaches to counter cariogenic biofilm establishment by disrupting both biofilm matrix and acidification, together with mechanical removal, could help re-establish the healthy microbiota and concomitantly enhance the efficacy of remineralizing agents to prevent and treat caries. These approaches are essential given that necessary dietary changes to control caries entail behavioural modifications of at-risk populations that have proved difficult to achieve. Strategies to control other environmental variables that drive and perpetuate dysbiosis (for example, inhibition of inflammation in periodontitis) or to restore immune function in immunodeficient individuals should arrest disease development and promote host-microbiota homeostasis148–151. Moreover, approaches to block the synergistic mechanisms that drive nososymbiocity (for example, targeting key interspecies interactions, acidogenesis or host signalling pathways that are exploited by keystone pathogens to subvert immunity) may also help control polymicrobial biofilm-associated diseases60,72,117. These strategies, combined with enhanced drug delivery and retention approaches, could substantially help control these prevalent and costly oral diseases.
Advances in omics technologies and improved database curation and bioinformatics have improved the identification of active microbial social networks and their products (genes, proteins and metabolites)37,152. However, to move beyond correlations and begin to address causation, further refinement of these technologies is needed. Integration of omics data combined with microorganism phenotype-pathogenicity association, together with complementary in vivo polymicrobial models153, may be a powerful strategy to identify additional disease-related microorganisms, their virulence properties and synergistic interactions that modulate components of the microbiota and host immunity within the conceptual framework presented here. In turn, a more complete set of therapeutic targets can be revealed, providing better opportunities to develop highly precise and efficacious therapeutic approaches.
Acknowledgements
The work of the authors is supported by US National Institutes of Health grants DE01111, DE012505, DE017921 and DE0130585 (R.J.L.); DE018023, DE025220 and DE025848 (H.K.); and DE015254, DE024153, DE024716, DE026152 and AI068730 (G.H.).
Glossary
- Subgingival
Relating to the area under the gum margins.
- Gingival crevicular fluid
(GCF). A serum exudate that contains immune and inflammatory mediators, along with large numbers of neutrophils recruited along a chemokine gradient.
- Extracellular polymeric substances
(EPS). Extracellular biomolecules including exopolysaccharides, fibrous and globular proteins in addition to extracellular enzymes, lipids and nucleic acids.
- Gingivitis
Mild and reversible inflammation of the gum often accompanied by bleeding upon toothbrushing. Tissue destruction does not occur. Gingivitis results from an accumulation of the plaque biofilm around the gingival margin and resolves after removal of the plaque.
- Acidogenic
organisms capable of producing acidic metabolites and reducing environmental pH.
- Dysbiosis
An imbalanced interaction that can be among bacteria in a community or between the microbiome and the host, and is detrimental to the host. The imbalance can be in the amount and/or the influence of individual microbial species relative to their abundance or influence in health. Alternatively, the imbalance can be caused by a poorly controlled immune response.
- Periodontitis
An episodic, slowly progressing inflammatory disease of the periodontal tissues that usually occurs in adults, although aggressive, rapidly progressing forms exist and can occur in adolescents.
- Dental caries
A polymicrobial and diet-dependent disease that is characterized by the development of pathogenic biofilms (dental plaque) within which acid production from bacterial metabolism of dietary carbohydrates causes demineralization of the mineralized tooth tissues (enamel, dentin and cementum), eventually leading to the clinical onset of cavitation or tooth decay.
- Aciduric
Organisms capable of growth at acidic pH levels that are often toxic to other bacteria.
- Polymicrobial synergy
Interactions among organisms that increase microbial fitness in the local environment.
- Dentin
Calcified tissue, predominantly hydroxyapatite, forming the bulk of the tooth, which is beneath and is softer (less mineralized with more organic material) than enamel.
- Periodontal diseases
A collection of conditions in which poorly controlled inflammatory responses induced by the microbiota cause destruction of the supporting structures of the tooth.
- Red complex
The triad of P gingivalis, T forsythia and T denticola — organisms that are often isolated together and were classically considered to be the predominant pathogens in chronic periodontitis.
- Gingival crevice
The compartment between the tooth root and the gingival (gum) tissue. The gingival crevice deepens into a periodontal pocket as periodontal disease progresses and tissue is destroyed.
- Citrullinate
Post-translational modification of a protein involving deamination of arginine by the enzyme peptidylarginine deiminase PPAD to produce citrulline.
- Cross feeding
The utilization of a metabolic by-product of one organism as a nutrient source by another organism.
- Nososymbiocity
The potential for a microbial community to contribute to disease; this recognizes the community rather than a single species as the aetiological agent.
- Salivary pellicle
A layer of salivary proteins and glycoproteins adsorbed to the enamel surface and to which adhesins of initial colonizers of the oral surface can attach. Pellicle can also contain molecules of microbial origin and those derived from epithelial cells.
- Accessory pathogen
organisms that act synergistically with more pathogenic species (keystone pathogens or pathobionts) to elevate community nososymbiocity. Accessory pathogens can provide an attachment substratum for colonization and metabolic support, and can increase virulence gene expression in other organisms through physical interactions or small-molecule-dependent communication.
- Keystone pathogen
Species that exert an influence on their communities that is disproportionate relative to their abundance and therefore form the ‘keystone’ of the community’s structure.
- Homeostatic commensals
Species that act to maintain a host-microbiota equilibrium by mitigating the action of more pathogenic species. Mechanisms include reducing the impact of pathogens on host cell signalling pathways or production of metabolites that favour a homeostatic inflammatory response.
- Homeostasis
A state of equilibrium or stability in a system that is maintained by adjusting physiological processes to counteract external changes.
- Ecological plaque (biofilm) hypothesis
A model encompassing microbiological, biochemical and ecological properties of oral biofilms and their association with disease. The model also accommodates host-derived changes (for example, frequent dietary sugar exposure) that trigger changes in the nososymbiocity of biofilm communities.
- Saccharolytic potential
The ability of an organism to metabolize carbohydrates.
- Emergent properties
Novel structures, activities, patterns and properties that arise during self-organization into complex systems. In the context of biofilm communities, these include surface adhesion and interbacterial cohesion, spatial organization, physical and social interactions, chemical heterogeneity and increased tolerance to antimicrobials.
- Pathobionts
organisms that are generally benign or commensal within an indigenous community but transition to pathogenic upon the breakdown of host-microbiota homeostasis (for example, as a result of antibiotic treatment, tissue damage, dietary shifts and especially immune deficiencies). These conditions promote pathobiont outgrowth and disrupt the symbiotic microbiota, causing further dysbiosis and inflammation.
- Asaccharolytic
A property of organisms incapable of breaking down carbohydrates for energy and thus reliant on the degradation of proteins and the generation of amino acids for metabolic energy and growth.
- Inflammophilic
A property of bacteria that thrive on inflammation and utilize inflammatory tissue breakdown products for nutrition.
- Hemin
An iron-containing porphyrin compound released from red blood cells; exploited by bacteria to obtain iron for growth.
- Localized chemokine paralysis
Precise and targeted suppression of specific chemokines by microbial community participants, superseding the otherwise stimulatory activity of other community inhabitants.
- Black queen hypothesis
A theory of reductive evolution to account for co-dependency.
Footnotes
Competing interests
The authors declare no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Reviewer information
Nature Reviews Microbiology thanks G. Belibasakis, B. Keijser and other anonymous reviewers for their contributions to the peer review of this work.
References
- 1.Abusleme L et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7, 1016–1025 (2013).This study documents alterations in subgingival microbial communities that underpin the development of periodontitis and describes the relationship between clinical inflammation and the disease-associated microbiome.
- 2.Griffen AL et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6, 1176–1185 (2012).This landmark study establishes the complexity of the periodontal microbial community and the demarcation between health and disease.
- 3.Rosan B & Lamont RJ Dental plaque formation. Microbes Infect. 2, 1599–1607 (2000). [DOI] [PubMed] [Google Scholar]
- 4.Aas JA, Paster BJ, Stokes LN, Olsen I & Dewhirst FE Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol 43, 5721–5732 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dewhirst FE et al. The human oral microbiome. J. Bacteriol 192, 5002–5017 (2010).References 4 and 5 are the basis of our current understanding of the diversity of the oral microbiome.
- 6.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE & Borisy GG Biogeography of a human oral microbiome at the micron scale. Proc. Natl Acad. Sci. USA 113, E791–E800 (2016).This imaging study provides the foundation for concepts of oral microbial biogeography.
- 7.Bowen WH, Burne RA, Wu H & Koo H Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26, 229–242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kamada N, Chen GY, Inohara N & Nunez G Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol 14, 685–690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamada N, Seo SU, Chen GY & Nunez G Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol 13, 321–335 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Lamont RJ & Hajishengallis G Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med 21, 172–183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajishengallis G & Lamont RJ Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur. J. Immunol 44, 328–338 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baker JL, Bor B, Agnello M, Shi W & He X Ecology of the oral microbiome: beyond bacteria. Trends Microbiol. 25, 362–374 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dabdoub SM, Ganesan SM & Kumar PS Comparative metagenomics reveals taxonomically idiosyncratic yet functionally congruent communities in periodontitis. Sci. Rep 6, 38993 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suwannakul S, Stafford GP, Whawell SA & Douglas CW Identification of bistable populations of Porphyromonas gingivalis that differ in epithelial cell invasion. Microbiology 156, 3052–3064 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Valm AM et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl Acad. Sci. USA 108, 4152–4157 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsh PD & Zaura E Dental biofilm: ecological interactions in health and disease. J. Clin. Periodontol 44 (Suppl. 18), 12–22 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Takahashi N & Nyvad B The role of bacteria in the caries process: ecological perspectives. J. Dent. Res 90, 294–303 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Xiao J et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLOS Pathog. 8, e1002623 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo L, McLean JS, Lux R, He X & Shi W The well-coordinated linkage between acidogenicity and aciduricity via insoluble glucans on the surface of Streptococcus mutans. Sci. Rep 5, 18015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hajishengallis E, Parsaei Y, Klein MI & Koo H Advances in the microbial etiology and pathogenesis of early childhood caries. Mol. Oral Microbiol 32, 24–34 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mira A, Simon-Soro A & Curtis MA Role of microbial communities in the pathogenesis of periodontal diseases and caries. J. Clin. Periodontol 44 (Suppl. 18), S23–S38 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Tanner ACR, Kressirer CA, Rothmiller S, Johansson I & Chalmers NI The caries microbiome: implications for reversing dysbiosis. Adv. Dent. Res 29, 78–85 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Eriksson L, Lif Holgerson P, Esberg A & Johansson I Microbial complexes and caries in 17-year-olds with and without Streptococcus mutans. J. Dent. Res 97, 275–282 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Hajishengallis G & Lamont RJ Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol 27, 409–419 (2012).This paper questions the primary importance of individual pathogens, such as the red complex bacteria, and proposes that periodontitis is initiated by a synergistic polymicrobial community within which different species, or specific gene combinations thereof, mediate distinct roles that converge to shape and stabilize a dysbiotic and disease-provoking microbiota.
- 25.Holt SC & Ebersole JL Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 38, 72–122 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Socransky SS & Haffajee AD Periodontal microbial ecology. Periodontol. 2000 38, 135–187 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Dewhirst FE The oral microbiome: critical for understanding oral health and disease. J. Calif. Dent. Assoc 44, 409–410 (2016). [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz PI, Hoare A & Hong BY Subgingival microbiome shifts and community dynamics in periodontal diseases. J. Calif. Dent. Assoc 44, 421–435 (2016). [PubMed] [Google Scholar]
- 29.Moore WE et al. The microflora of periodontal sites showing active destructive progression. J. Clin. Periodontol 18, 729–739 (1991). [DOI] [PubMed] [Google Scholar]
- 30.Diaz PI Microbial diversity and interactions in subgingival biofilm communities. Front. Oral Biol 15, 17–40 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Simon-Soro A et al. Microbial geography of the oral cavity. J. Dent. Res 92, 616–621 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Nowicki EM et al. Microbiota and metatranscriptome changes accompanying the onset of gingivitis. mBio 9, e00575–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitmore SE & Lamont RJ Oral bacteria and cancer. PLOS Pathog. 10, e1003933 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Atanasova KR & Yilmaz O Looking in the Porphyromonas gingivalis cabinet of curiosities: the microbium, the host and cancer association. Mol. Oral Microbiol 29, 55–66 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sahingur SE & Yeudall WA Chemokine function in periodontal disease and oral cavity cancer. Front. Immunol 6, 214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cugini C, Klepac-Ceraj V, Rackaityte E, Riggs JE & Davey ME Porphyromonas gingivalis: keeping the pathos out of the biont. J. Oral Microbiol 5, 19804 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi N Oral microbiome metabolism: from “who are they?” to “what are they doing?”. J. Dent. Res 94, 1628–1637 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Han YW & Wang X Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J. Dent. Res 92, 485–491 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hajishengallis G Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol 15, 30–44 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar PS Oral microbiota and systemic disease. Anaerobe 24, 90–93 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Maddi A & Scannapieco FA Oral biofilms, oral and periodontal infections, and systemic disease. Am. J. Dent 26, 249–254 (2013). [PubMed] [Google Scholar]
- 42.Potempa J, Mydel P & Koziel J The case for periodontitis in the pathogenesis of rheumatoid arthritis. Nat. Rev. Rheumatol 13, 606–620 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Ebersole JL et al. The periodontal war: microbes and immunity. Periodontol. 2000 75, 52–115 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Chukkapalli SS et al. Global TLR2 and 4 deficiency in mice impacts bone resorption, inflammatory markers and atherosclerosis to polymicrobial infection. Mol. Oral Microbiol 32, 211–225 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hajishengallis G & Lamont RJ Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. 24, 477–489 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murray JL, Connell JL, Stacy A, Turner KH & Whiteley M Mechanisms of synergy in polymicrobial infections. J. Microbiol 52, 188–199 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michie KL, Cornforth DM & Whiteley M Bacterial tweets and podcasts #signaling# eavesdropping#microbialfightclub. Mol. Biochem. Parasitol 208, 41–48 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Short FL, Murdoch SL & Ryan RP Polybacterial human disease: the ills of social networking. Trends Microbiol. 22, 508–516 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stacy A, McNally L, Darch SE, Brown SP & Whiteley M The biogeography of polymicrobial infection. Nat. Rev. Microbiol 14, 93–105 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hwang G et al. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLOS Pathog. 13, e1006407 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim D et al. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci. Rep 7, 41332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen YE, Fischbach MA & Belkaid Y Skin microbiota-host interactions. Nature 553, 427–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Casadevall A The pathogenic potential of a microbe. mSphere 2, e00015–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lin D & Koskella B Friend and foe: factors influencing the movement of the bacterium Helicobacter pylori along the parasitism-mutualism continuum. Evol. Appl 8, 9–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nelson PG & May G Coevolution between mutualists and parasites in symbiotic communities may lead to the evolution of lower virulence. Am. Nat 190, 803–817 (2017). [DOI] [PubMed] [Google Scholar]
- 56.Whitmore SE & Lamont RJ The pathogenic persona of community-associated oral streptococci. Mol. Microbiol 81, 305–314 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nobbs AH, Lamont RJ & Jenkinson HF Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev 73, 407–450(2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee SF Oral colonization and immune responses to Streptococcus gordonii: potential use as a vector to induce antibodies against respiratory pathogens. Curr. Opin. Infect. Dis 16, 231–235 (2003). [DOI] [PubMed] [Google Scholar]
- 59.Xie E et al. Oral delivery of a novel recombinant Streptococcus mitis vector elicits robust vaccine antigen-specific oral mucosal and systemic antibody responses and T cell tolerance. PLOS ONE 10, e0143422 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Daep CA, Novak EA, Lamont RJ & Demuth DR Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect. Immun 79, 67–74 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuboniwa M et al. Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat. Microbiol 2, 1493–1499 (2017).This study establishes multidimensional communication between two organisms in the oral community that separately either enhance or suppress nososymbiocity.
- 62.Wright CJ et al. Characterization of a bacterial tyrosine kinase in Porphyromonas gingivalis involved in polymicrobial synergy. Microbiologyopen 3, 383–394 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu C, Miller DP, Wang Y, Merchant M & Lamont RJ Structure-function aspects of the Porphyromonas gingivalis tyrosine kinase Ptk1. Mol. Oral Microbiol 32, 314–323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramsey MM & Whiteley M Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc. Natl Acad. Sci. USA 106, 1578–1583 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stacy A et al. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc. Natl Acad. Sci. USA 111, 7819–7824 (2014).This study reveals that the spatial proximity of organisms can depend on the collective outcome of synergistic and antagonistic interactions.
- 66.Duan D, Scoffield JA, Zhou X & Wu H Fine-tuned production of hydrogen peroxide promotes biofilm formation of Streptococcus parasanguinis by a pathogenic cohabitant Aggregatibacter actinomycetemcomitans. Environ. Microbiol 18, 4023–4036 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bertolini MM et al. Candida-streptococcal mucosal biofilms display distinct structural and virulence characteristics depending on growth conditions and hyphal morphotypes. Mol. Oral Microbiol 30, 307–322 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu H, Jenkinson HF & Dongari-Bagtzoglou A Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol. Oral Microbiol 29, 99–116 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cheng X et al. Plasticity of the pyruvate node modulates hydrogen peroxide production and acid tolerance in multiple oral Streptococci. Appl. Environ. Microbiol 84, e01697–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thurnheer T & Belibasakis GN Streptococcus oralis maintains homeostasis in oral biofilms by antagonizing the cariogenic pathogen Streptococcus mutans. Mol. Oral Microbiol 33, 234–239 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Redanz S et al. Live and let die: hydrogen peroxide production by the commensal flora and its role in maintaining a symbiotic microbiome. Mol. Oral Microbiol https://doi.org/10.1111/omi.12231 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ho MH, Lamont RJ & Xie H Identification of Streptococcus cristatus peptides that repress expression of virulence genes in Porphyromonas gingivalis. Sci. Rep 7, 1413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ho MH, Lamont RJ & Xie H A novel peptidic inhibitor derived from Streptococcus cristatus ArcA attenuates virulence potential of Porphyromonas gingivalis. Sci. Rep 7, 16217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xie H, Hong J, Sharma A & Wang BY Streptococcus cristatus ArcA interferes with Porphyromonas gingivalis pathogenicity in mice. J. Periodontal. Res 47, 578–583 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang BY, Wu J, Lamont RJ, Lin X & Xie H Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. J. Clin. Microbiol 47, 3902–3906 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mans JJ, Hendrickson EL, Hackett M & Lamont RJ Cellular and bacterial profiles associated with oral epithelium-microbiota interactions. Periodontol. 2000 52, 207–217 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Colombo AV, Silva CM, Haffajee A & Colombo AP Identification of oral bacteria associated with crevicular epithelial cells from chronic periodontitis lesions. J. Med. Microbiol 55, 609–615 (2006). [DOI] [PubMed] [Google Scholar]
- 78.Kreth J, Giacaman RA, Raghavan R & Merritt J The road less traveled - defining molecular commensalism with Streptococcus sanguinis. Mol. Oral Microbiol 32, 181–196 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shah SA et al. The making of a miscreant: tobacco smoke and the creation of pathogen-rich biofilms. NPJ Biofilms Microbiomes 3, 26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ghosh SK et al. Conceptual perspectives: bacterial antimicrobial peptide induction as a novel strategy for symbiosis with the human host. Front. Microbiol 9, 302 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Al-Attar A et al. Activation of Notch-1 in oral epithelial cells by P. gingivalis triggers the expression of the antimicrobial protein PLA2-IIA. Mucosal Immunol. 11, 1047–1059 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pitts NB et al. Dental caries. Nat. Rev. Dis. Primers 3, 17030 (2017). [DOI] [PubMed] [Google Scholar]
- 83.Liu YL, Nascimento M & Burne RA Progress toward understanding the contribution of alkali generation in dental biofilms to inhibition of dental caries. Int. J. Oral Sci 4, 135–140 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Merritt J & Qi F The mutacins of Streptococcus mutans: regulation and ecology. Mol. Oral Microbiol 27, 57–69 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Qi F & Kreth J Methods to study antagonistic activities among oral bacteria. Methods Mol. Biol 1537, 203–218 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Gross EL et al. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLOS ONE 7, e47722 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Johansson I, Witkowska E, Kaveh B, Lif Holgerson P & Tanner AC The microbiome in populations with a low and high prevalence of caries. J. Dent. Res 95, 80–86 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Teng F et al. Prediction of early childhood caries via spatial-temporal variations of oral microbiota. Cell Host Microbe 18, 296–306 (2015). [DOI] [PubMed] [Google Scholar]
- 89.Takahashi N & Nyvad B Ecological hypothesis of dentin and root caries. Caries Res. 50, 422–431 (2016). [DOI] [PubMed] [Google Scholar]
- 90.Richards VP et al. Microbiomes of site-specific dental plaques from children with different caries status. Infect. Immun 85, e00106–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Knapp S et al. Natural competence is common among clinical isolates of Veillonella parvula and is useful for genetic manipulation of this key member of the oral microbiome. Front. Cell. Infect. Microbiol 7, 139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mashima I & Nakazawa F The influence of oral Veillonella species on biofilms formed by Streptococcus species. Anaerobe 28, 54–61 (2014). [DOI] [PubMed] [Google Scholar]
- 93.Xiao J et al. Candida albicans and early childhood caries: a systematic review and meta-analysis. Caries Res. 52, 102–112 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Falsetta ML et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun 82, 1968–1981 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sztajer H et al. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 8, 2256–2271 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Flemming HC et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol 14, 563–575 (2016). [DOI] [PubMed] [Google Scholar]
- 97.Hwang G et al. Simultaneous spatiotemporal mapping of in situ pH and bacterial activity within an intact 3D microcolony structure. Sci. Rep 6, 32841 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Jiang W et al. Pyrosequencing analysis of oral microbiota shifting in various caries states in childhood. Microb. Ecol 67, 962–969 (2014). [DOI] [PubMed] [Google Scholar]
- 99.Marsh PD Are dental diseases examples of ecological catastrophes? Microbiology 149, 279–294 (2003).This paper proposes that environmental factors drive the selection and enrichment of specific oral pathogenic bacteria, with implications for both dental caries and periodontitis.
- 100.Hasturk H et al. Resolvin E1 regulates inflammation at the cellular and tissue level and restores tissue homeostasis in vivo. J. Immunol 179, 7021–7029 (2007). [DOI] [PubMed] [Google Scholar]
- 101.Abe T et al. Local complement-targeted intervention in periodontitis: proof-of-concept using a C5a receptor (cD88) antagonist. J. Immunol 189, 5442–5448 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hajishengallis G et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10, 497–506 (2011).This study substantiates the concept of a keystone pathogen by providing in vivo evidence that a specific microorganism instigates quantitative and qualitative alterations to the commensal microbiota, which is thereby remodelled into a dysbiotic community driving periodontitis.
- 103.Eskan MA et al. The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nat. Immunol 13, 465–473 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moutsopoulos NM et al. Defective neutrophil recruitment in leukocyte adhesion deficiency type I disease causes local IL-17-driven inflammatory bone loss. Sci. Transl Med 6, 229ra240 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lee C-T et al. Resolvin E1 reverses experimental periodontitis and dysbiosis. J. Immunol 197, 2796–2806 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Hajishengallis G The inflammophilic character of the periodontitis-associated microbiota. Mol. Oral Microbiol 29, 248–257 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Duran-Pinedo AE et al. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 8, 1659–1672 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Herrero ER et al. Dysbiotic biofilms deregulate the periodontal inflammatory response. J. Dent. Res 97, 547–555 (2018). [DOI] [PubMed] [Google Scholar]
- 109.Yost S, Duran-Pinedo AE, Teles R, Krishnan K & Frias-Lopez J Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 7, 27 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bang J, Cimasoni G, Rosenbusch C & Duckert A Sodium, potassium and calcium contents of crevicular exudate: their relations to gingivitis and periodontitis. J. Periodontol 44, 770–774 (1973). [DOI] [PubMed] [Google Scholar]
- 111.Yost S, Duran-Pinedo AE, Krishnan K & Frias-Lopez J Potassium is a key signal in host-microbiome dysbiosis in periodontitis. PLOS Pathog. 13, e1006457 (2017).This investigation identifies key metabolic changes in the periodontal microbial community associated with dysbiosis initiation and concludes that disease progression is mediated by the collective virulence of the entire community rather than by the action of a select few pathogens.
- 112.Winter SE & Baumler AJ Dysbiosis in the inflamed intestine: chance favors the prepared microbe. Gut Microbes 5, 71–73 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nassar M et al. GAS6 is a key homeostatic immunological regulator of host-commensal interactions in the oral mucosa. Proc. Natl Acad. Sci. USA 114, E337–E346 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dalal SR & Chang EB The microbial basis of inflammatory bowel diseases. J. Clin. Invest 124, 4190–4196 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hajishengallis G, Darveau RP & Curtis MA The keystone-pathogen hypothesis. Nat. Rev. Microbiol 10, 717–725 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Finlay BB & McFadden G Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782 (2006). [DOI] [PubMed] [Google Scholar]
- 117.Maekawa T et al. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe 15, 768–778 (2014).This study shows that a keystone periodontal pathogen manipulates complement-TLR crosstalk to block bactericidal mechanisms while fostering a nutritionally favourable inflammatory response; this uncoupling of immune bacterial clearance from inflammation promotes dysbiosis and periodontitis.
- 118.Makkawi H et al. Porphyromonas gingivalis stimulates TLR2-PI3K signaling to escape immune clearance and induce bone resorption independently of MyD88. Front. Cell. Infect. Microbiol 7, 359 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang M et al. Microbial hijacking of complement-toll-like receptor crosstalk. Sci. Signal 3, rail (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liang S et al. The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J. Immunol 186, 869–877 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tonetti MS, Cortellini D & Lang NP In situ detection of apoptosis at sites of chronic bacterially induced inflammation in human gingiva. Infect. Immun 66, 5190–5195 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zenobia C et al. Commensal bacteria-dependent select expression of CXCL2 contributes to periodontal tissue homeostasis. Cell. Microbiol 15, 1419–1426 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Darveau RP, Belton CM, Reife RA & Lamont RJ Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun 66, 1660–1665 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Jauregui CE et al. Suppression of T cell chemokines by Porphyromonas gingivalis. Infect. Immun 81, 2288–2295 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Takeuchi H et al. The serine phosphatase SerB of Porphyromonas gingivalis suppresses IL-8 production by dephosphorylation of NF-kappaB RelA/p65. PLOS Pathog. 9, e1003326 (2013).References 123–125 establish the concept of P. gingivalis-induced local ‘chemokine paralysis’, originally shown to affect innate immunity and later expanded to include suppression of T cell-specific chemokines.
- 126.Hajishengallis G Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 35, 3–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Moutsopoulos NM et al. Subgingival microbial communities in leukocyte adhesion deficiency and their relationship with local immunopathology. PLOS Pathog. 11, e1004698 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Darveau RP, Hajishengallis G & Curtis MA Porphyromonas gingivalis as a potential community activist for disease. J. Dent. Res 91, 816–820 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Koo H, Allan RN, Howlin RP, Stoodley P & Hall-Stoodley L Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol 15, 740–755 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liu Y, Ren Z, Hwang G & Koo H Therapeutic strategies targeting cariogenic biofilm microenvironment. Adv. Dent. Res 29, 86–92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marquis RE, Clock SA & Mota-Meira M Fluoride and organic weak acids as modulators of microbial physiology. FEMS Microbiol. Rev 26, 493–510 (2003). [DOI] [PubMed] [Google Scholar]
- 132.Kolderman E et al. L-Arginine destabilizes oral multi-species biofilm communities developed in human saliva. PLOS ONE 10, e0121835 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Nascimento MM et al. The effect of arginine on oral biofilm communities. Mol. Oral Microbiol. 29, 45–54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Pleszczynska M, Wiater A, Janczarek M & Szczodrak J (1-→3)-alpha-D-Glucan hydrolases in dental biofilm prevention and control: a review. Int. J. Biol. Macromol 79, 761–778 (2015). [DOI] [PubMed] [Google Scholar]
- 135.Fabbri S et al. High-velocity microsprays enhance antimicrobial activity in Streptococcus mutans biofilms. J. Dent. Res 95, 1494–1500 (2016). [DOI] [PubMed] [Google Scholar]
- 136.Liu Y et al. Topical delivery of low-cost protein drug candidates made in chloroplasts for biofilm disruption and uptake by oral epithelial cells. Biomaterials 105, 156–166 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gao L et al. Nanocatalysts promote Streptococcus mutans biofilm matrix degradation and enhance bacterial killing to suppress dental caries in vivo. Biomaterials 101, 272–284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Horev B et al. pH-activated nanoparticles for controlled topical delivery of farnesol to disrupt oral biofilm virulence. ACS Nano 9, 2390–2404 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Liu Y et al. Surface-adaptive, antimicrobially loaded, micellar nanocarriers with enhanced penetration and killing efficiency in Staphylococcal biofilms. ACS Nano 10, 4779–4789 (2016).References 137–139 demonstrate the feasibility of using pH-activated nanotechnologies for enhanced drug delivery, release or activation within the biofilm microenvironment to amplify the precision and efficacy of antibiofilm effects.
- 140.Paula AJ & Koo H Nanosized building blocks for customizing novel antibiofilm approaches. J. Dent. Res 96, 128–136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Maekawa T et al. Inhibition of pre-existing natural periodontitis in non-human primates by a locally administered peptide inhibitor of complement C3. J. Clin. Periodontol 43, 238–249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Assuma R, Oates T, Cochran D, Amar S & Graves DT IL-1 and TNF antagonists inhibit the inflammatory response and bone loss in experimental periodontitis. J. Immunol 160, 403–409 (1998). [PubMed] [Google Scholar]
- 143.Hasturk H, Kantarci A & Van Dyke TE Paradigm shift in the pharmacological management of periodontal diseases. Front. Oral Biol 15, 160–176 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Moutsopoulos NM et al. Interleukin-12 and interleukin-23 blockade in leukocyte adhesion deficiency type 1. N. Engl. J. Med 376, 1141–1146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen X et al. Advanced biomaterials and their potential applications in the treatment of periodontal disease. Crit. Rev. Biotechnol 36, 760–775 (2016). [DOI] [PubMed] [Google Scholar]
- 146.Goyal G, Garg T, Rath G & Goyal AK Current nanotechnological strategies for an effective delivery of drugs in treatment of periodontal disease. Crit. Rev. Ther. Drug Carrier Syst 31, 89–119 (2014). [DOI] [PubMed] [Google Scholar]
- 147.Morris JJ, Lenski RE & Zinser ER The black queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3, e00036–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mastellos DC, Ricklin D, Hajishengallis E, Hajishengallis G & Lambris JD Complement therapeutics in inflammatory diseases: promising drug candidates for C3-targeted intervention. Mol. Oral Microbiol 31, 3–17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Van Dyke TE Pro-resolving mediators in the regulation of periodontal disease. Mol. Aspects Med 58, 21–36 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gatej S, Gully N, Gibson R & Bartold PM Probiotics and periodontitis - a literature review. J. Int. Acad. Periodontol 19, 42–50 (2017). [PubMed] [Google Scholar]
- 151.Rosier BT, Marsh PD & Mira A Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J. Dent. Res 97, 371–380 (2017). [DOI] [PubMed] [Google Scholar]
- 152.Garza DR, van Verk MC, Huynen MA & Dutilh BE Towards predicting the environmental metabolome from metagenomics with a mechanistic model. Nat. Microbiol 3, 456–460 (2018). [DOI] [PubMed] [Google Scholar]
- 153.Surana NK & Kasper DL Moving beyond microbiome-wide associations to causal microbe identification. Nature 552, 244–247 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Brown SA & Whiteley M A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J. Bacteriol 189, 6407–6414 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Brown SA & Whiteley M Characterization of the L-lactate dehydrogenase from Aggregatibacter actinomycetemcomitans. PLOS ONE 4, e7864 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ramsey MM, Rumbaugh KP & Whiteley M Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLOS Pathog. 7, e1002012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Stacy A, Fleming D, Lamont RJ, Rumbaugh KP & Whiteley MA Commensal bacterium promotes virulence of an opportunistic pathogen via cross-respiration. mBio 7, e00782–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stacy A, Abraham N, Jorth P & Whiteley M Microbial community composition impacts pathogen iron availability during polymicrobial infection. PLOS Pathog. 12, e1006084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chawla A et al. Community signalling between Streptococcus gordonii and Porphyromonas gingivalis is controlled by the transcriptional regulator CdhR. Mol. Microbiol 78, 1510–1522 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Maeda K et al. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol. Microbiol 69, 1153–1164 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Burns E, Eliyahu T, Uematsu S, Akira S & Nussbaum G TLR2-dependent inflammatory response to Porphyromonas gingivalis is MyD88 independent, whereas MyD88 is required to clear infection. J. Immunol 184, 1455–1462 (2010). [DOI] [PubMed] [Google Scholar]
- 162.Brzezinska AA, Johnson JL, Munafo DB, Ellis BA & Catz SD Signalling mechanisms for Toll-like receptor-activated neutrophil exocytosis: key roles for interleukin-1-receptor-associated kinase-4 and phosphatidylinositol 3-kinase but not Toll/IL-1 receptor (TIR) domain-containing adaptor inducing IFNβ (TRIF). Immunology 127, 386–397 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bainbridge B et al. Role of Porphyromonas gingivalis phosphoserine phosphatase enzyme SerB in inflammation, immune response, and induction of alveolar bone resorption in rats. Infect. Immun 78, 4560–4569 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]