Abstract.
Given the increased need for consistent quantitative image analysis, variations in radiomics feature calculations due to differences in radiomics software were investigated. Two in-house radiomics packages and two freely available radiomics packages, MaZda and IBEX, were utilized. Forty regions of interest (ROIs) from 40 digital mammograms were studied along with 39 manually delineated ROIs from the head and neck (HN) computed tomography (CT) scans of 39 patients. Each package was used to calculate first-order histogram and second-order gray-level co-occurrence matrix (GLCM) features. Friedman tests determined differences in feature values across packages, whereas intraclass-correlation coefficients (ICC) quantified agreement. All first-order features computed from both mammography and HN cases (except skewness in mammography) showed significant differences across all packages due to systematic biases introduced by each package; however, based on ICC values, all but one first-order feature calculated on mammography ROIs and all but two first-order features calculated on HN CT ROIs showed excellent agreement, indicating the observed differences were small relative to the feature values but the bias was systematic. All second-order features computed from the two databases both differed significantly and showed poor agreement among packages, due largely to discrepancies in package-specific default GLCM parameters. Additional differences in radiomics features were traced to variations in image preprocessing, algorithm implementation, and naming conventions. Large variations in features among software packages indicate that increased efforts to standardize radiomics processes must be conducted.
Keywords: radiomics, texture analysis, head and neck CT, mammography, software packages
1. Introduction
The texture of a medical image refers to the coarseness, consistency, and arrangement of pixels within the image. Radiologists typically make qualitative assessments regarding a patient’s condition based on the texture and spatial patterns they perceive within the image.1 With large databases of medical images available for researchers and radiologists to analyze, high-throughput computing through the conversion of medical images into mineable data (i.e., radiomics) is possible, which may offer additional insight into a patient’s underlying pathophysiology.2–4 Since the development of several systematic and large-scale texture analysis-based computer-aided diagnosis (CAD) schemes began at the University of Chicago in the early 1980s, investigators have used texture analysis over the decades to develop various automated detection, diagnosis, and segmentation strategies. By the mid 1980s, the University of Chicago had set the foundation for CAD schemes used to detect lung nodules in digital chest radiographs as well as microcalcifications in mammography images.5–8 While these early investigations resulted in a relatively large number of false positives per patient, it was shown that radiologists’ ability to detect lesions significantly improved when these texture analysis-based CAD schemes were used.9,10 Since these earlier investigations, programs using texture analysis have extended to examine and make clinical predictions concerning additional tissues such as the colon, liver, and brain.11–13 Because of the growing promise texture analysis and radiomics have shown and the large amounts of imaging data available, investigators have become more interested in quantifying texture to increase the amount of information that can be extracted from medical images and to limit variability among radiologists.
Many research groups have developed in-house and freely available radiomics software packages to allow for the advancement of radiomics research. These packages, however, are often used with a one-size-fits-all approach without considering the underlying mechanisms embedded in the algorithms that may result in variations among packages or differences in the images to which the algorithms might be applied. Such variations could be caused by differences in preprocessing, differences in the algorithms used to calculate features or differences in algorithm implementation. In addition, often a radiomics package is used to analyze images of one specific imaging modality, anatomic location, or tissue type, although the software is designed to analyze another type of image. Radiomics features have been shown to vary substantially based on differences in image acquisition parameters, reconstruction algorithms, and gating techniques, and these differences may be exacerbated when computed with different packages.2,14–23
A number of studies have noticed this lack of harmonization among radiomics research and have called for greater standardization.18,24,25 Hatt et al.26 conducted a review of studies involving texture analysis of positron emission tomography (PET) images and identified sources of discrepancies in these studies that need to be addressed to achieve more reproducible radiomics research. Due to the growing demand for harmonized radiomics research, the imaging biomarker standardization initiative (IBSI), composed of 55 researchers from 19 institutions in eight countries, aims to standardize the computation of radiomics features as well as any potential image processing required before feature extraction. Through this initiative, a number of recommendations have been made regarding feature calculation.27 The IBSI has compared radiomics methods across collaborators using a small digital phantom with limited size and gray-level range as well as a single computed tomography (CT) scan from a patient with lung cancer with five different realistic image processing configurations in order to achieve greater standardization among these institutions.28,29 In addition, a multi-institutional study developed by the quantitative imaging network investigated the sensitivity of quantitative radiomics descriptors of lung nodules when computed by different research institutions. The variability of feature values, however, was compounded by differences in the nodule segmentation methods and other institution-specific factors, whereas the dependence of the variability in features due to image-specific parameters (e.g., tissue type, imaging modality, and image-acquisition settings) was not discussed.30 Therefore, the purpose of this study was to compare two in-house radiomics software packages to two freely available software packages using clinical images of various anatomic regions and imaging modalities and to determine the sources of these variations in clinical data.
2. Methods and Materials
2.1. Medical Imaging Data
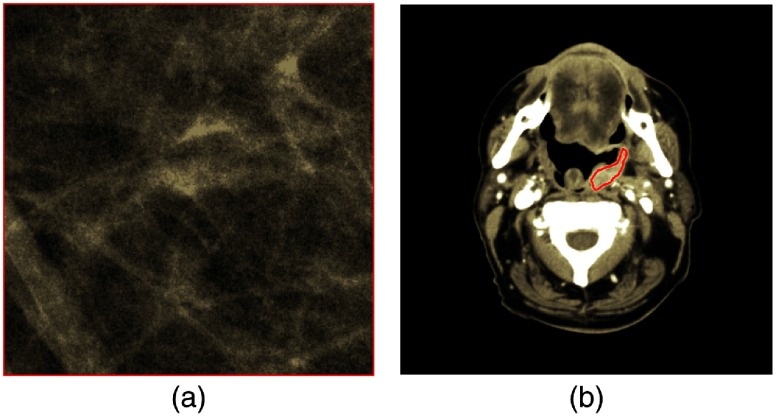
Cranial-caudal (CC) digital mammography and head and neck (HN) CT images were obtained through the Human Imaging Research Office under institutional review board approval.24 Image parameters and patient information are shown in Table 1. Pixel information was extracted from a single region of interest (ROI) in each image. Mammography ROIs () contained normal breast parenchyma, whereas HN CT ROIs contained manually segmented tumor (mean number of pixels: 1102; range: 174 to 2819) with example ROIs from each database shown in Fig. 1.
Table 1.
Patient and scan characteristics.
| Mammography | HN CT | ||
|---|---|---|---|
| Number of scans | 40 | 39 | |
| Number of ROIs | 40 | 39 | |
| Peak kilovoltage (kVp) | 24 () | 29 () | 120 () |
| 26 () | 30 () | 140 () | |
| 27 () | 31 () | ||
| 28 () | 32 () | ||
| Slice thickness (mm) | NA | 2.5 () | |
| 3.0 () | |||
| Mean pixel spacing (range) (mm) | 0.1 (0.1 to 0.1) | 0.536 | |
| (0.424 to 0.688) | |||
| Scanner manufacturer and model | GE Senographe 2000D | Philips brilliance 16 () | |
| Philips brilliance 64 () Philips iCT 256 () | |||
| Siemens biograph 64 () | |||
Fig. 1.
Example ROIs depicting (a) a mammography ROI and (b) an HN ROI containing contoured tumor.
2.2. Radiomics Software
Four radiomics software packages were utilized for this study. Two packages had been developed in-house by independent research labs at the University of Chicago (A1 and A2),21,31–34 and two were freely available packages, MaZda v4.6 (Institute of Electronics, Technical University of Lodz, Poland)35–38 and IBEX v1.0 beta (The University of Texas MD Anderson Cancer Center).39 The two packages from outside our institution were chosen because they were freely available at the initiation of this study, and they had been cited in a number of recent publications. Each package was capable of calculating several classifications of radiomics features including first-order histogram features, fractal features, Fourier features, and gray-level run length matrix features; however, only first-order histogram features and second-order gray-level co-occurrence matrix (GLCM) features were common among all four packages as shown in Table 2.
Table 2.
Number of directionally independent features per feature category that can be calculated by each radiomics package.
| Feature category | A1 | A2 | MaZda | IBEX |
|---|---|---|---|---|
| Shape | 73 | 18 | ||
| First-order histogram | 22 | 18 | 9 | 24 |
| Intensity histogram Gaussian fit | 5 | |||
| Absolute gradient | 5 | |||
| Run-length matrix | 5 | 11 | ||
| Neighborhood intensity difference | 5 | |||
| Co-occurrence matrix | 14 | 14 | 11 | 22 |
| Autoregressive model parameters | 5 | |||
| Wavelet | 20 | |||
| Fractal | 5 | 25 | ||
| Fourier | 17 | 22 | ||
| Laws | 84 |
In an abstract from the IBSI, the union of all features across all packages was compared;28 however, in the current study, only the features shared by all four packages with the same naming conventions were compared. These common features are shown in Table 3.
Table 3.
First- and second-order radiomics features common among all four packages.
| First-order histogram features | Second-order GLCM features |
|---|---|
| Maximum | Entropy |
| Minimum | Contrast |
| Mean | Sum average |
| Standard deviation | Sum variance |
| Skewness | Sum entropy |
| Kurtosis | Difference entropy |
2.3. Sources of Feature Variation: GLCM Parameters
GLCM features first were calculated using the package-specific default GLCM parameters unique to each package. These parameters included the gray-level limits, the dimensions of the GLCM, and the directions used in the final average of the GLCM feature values; the distance between neighboring pixel values was 1 for all packages, and the GLCM features were all normalized by the number of pixels within the ROI. Next, these parameters were modified to allow for the greatest possible consistency among packages. The package-specific default along with the consistent GLCM parameters is shown in Table 4. Due to limitations in the customizability of the MaZda interface, the gray-level limits and the number of directions could not be modified to match the other packages. Features were calculated by each software package for each ROI of each image.
Table 4.
Package-specific default GLCM parameters and GLCM parameters that were modified to maximize consistency among radiomics packages.
| GLCM parameter | A1 | A2 | MaZda | IBEX |
|---|---|---|---|---|
| Package-specific default | ||||
| Gray-level limits | [Min PV, Max PV] | [1,4096] | [Min PV, Max PV] | |
| Number of gray levels | 3001 | 64 | 256 | (Max PV – Min PV) |
| Number of directions | 4 | 8 | 4 | 8 |
| Consistenta GLCM parameters | ||||
| Gray-level limits | [Min PV, Max PV] | [Min PV, Max PV] | 1 to 4096b | [Min PV, Max PV] |
| Number of gray levels | 64 | 64 | 64 | 64 |
| Number of directions | 8 | 8 | 4b | 8 |
PV = pixel value.
Parameters were modified to maximize consistency across packages.
These parameters could not be modified.
2.4. Sources of Feature Variation: Algorithm Implementation
Differences in algorithm implementation and ROI processing were investigated to determine sources of variation among software packages. While the underlying mathematical equations used in all software packages are expected to be mathematically the same, the interpretation and implementation of these formulas may vary from one package to another. Packages A2, MaZda, and IBEX cited Haralick,40 whereas A1 cited publications by Felipe41 and the Handbook of Computer Vision Applications41 for the equations used for feature calculation; however, the algorithmic implementation of these equations may vary due to differences in notation, equation representation, and implementation strategies. Furthermore, the Handbook of Computer Vision Applications cites the Haralick paper for its equations after modifying the notation and imposing some corrections and conditions on the equations used. The source code for the packages, excluding MaZda since it was not available, was investigated for differences between algorithm implementation.
To remove the effects of ROI preprocessing and GLCM parameter variability for each package, feature functions were extracted from the two in-house packages as well as IBEX such that only the functions used to calculate the individual features were investigated. These functions were used to calculate features directly on a single mammographic image. Individual feature values were compared across packages to determine differences in algorithm implementation. These equations were extracted after GLCM construction such that differences in the GLCMs across packages did not affect the resultant feature values.
2.5. Statistical Analysis
After verifying that data were not normally distributed using the Shapiro–Wilk test, nonparametric repeated measures Friedman tests were used to test for significance among features calculated by the software packages. The null hypothesis is that all features calculated using each software packages are the same and sampled from the same population. Significance was assessed at the level using Bonferroni correction to account for the 12 features evaluated ().
The intraclass correlation coefficient (ICC) was used to assess the agreement of radiomics feature values among packages with package-specific and consistent GLCM parameters using the two-way mixed effect model illustrating the absolute agreement of the feature values across packages [i.e., ICC(A,1)].42 The ICC quantifies the absolute agreement between the sets of data by comparing the variability in feature values across software packages to the variability in values across patients. ICC values are stratified to indicate “excellent” (), “good” (), “moderate” (), or “poor” () agreement.43
3. Results
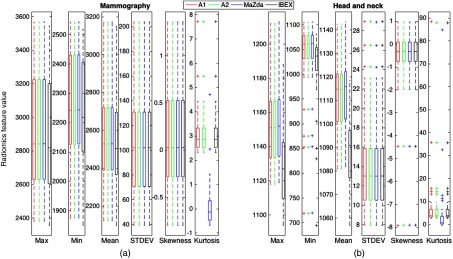
First-order gray-level features and second-order GLCM features were generated using the 40 mammography and 39 HN CT ROIs as input for each of the four radiomics software packages. Boxplots depicting the distributions of the calculated features among all four packages are shown in Fig. 2.
Fig. 2.
Distribution of first-order features calculated on (a) the mammography ROIs and (b) the HN CT ROIs. Boxes extend from the first to the third quartile with the median represented by the centerline. Outliers are indicated by +.
ICCs and the -values for differences across packages for each feature are shown in Table 5. While first-order features showed significant differences, the ICCs for all first-order mammography features besides kurtosis demonstrated excellent agreement. This indicates that while systematic biases are introduced due to differences in each of the packages resulting in significant differences, the magnitude of these biases are small relative to the feature values themselves. Therefore, the ICC still reflected excellent agreement in these features among packages. Among HN CT ROIs, maximum showed good agreement, and mean showed moderate agreement, whereas the remaining first-order features all showed excellent agreement.
Table 5.
-Values resulting from the nonparametric Friedman tests comparing radiomics features across packages and ICCs illustrating agreement in features among packages. Second-order features were calculated using package-specific default GLCM parameters.
| Mammography | HN CT | |||
|---|---|---|---|---|
| Feature | -Value | ICC | -Value | ICC |
| Max | 0.999 | 0.755 | ||
| Min | 0.996 | 0.975 | ||
| Mean | 0.997 | 0.614 | ||
| Standard deviation | 1.000 | 1.000 | ||
| Skewness | 0.917 | 1.000 | 1.000 | |
| Kurtosis | 0.297 | 0.989 | ||
| GLCM contrast | 0.001 | 0.193 | ||
| GLCM entropy | 0.001 | 0.003 | ||
| GLCM sum entropy | 0.002 | 0.004 | ||
| GLCM sum average | ||||
| GLCM sum variance | 0.002 | |||
| GLCM difference entropy | 0.004 | 0.006 | ||
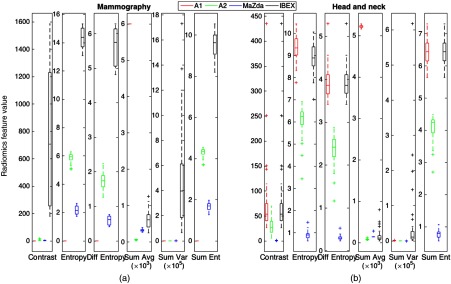
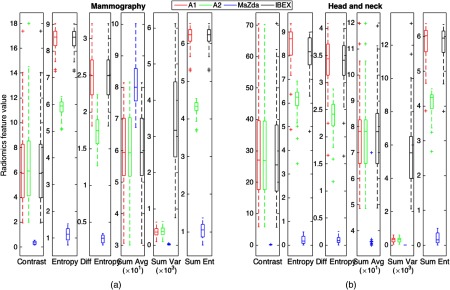
Second-order GLCM features were calculated using the package-specific default GLCM parameters with the distributions of feature values shown in Fig. 3.
Fig. 3.
Distribution of second-order features for (a) the mammography ROIs and (b) the HN CT ROIs using the package-specific default GLCM parameters outlined in Table 3.
All features differed significantly among the four packages for both mammography and HN CT ROIs.
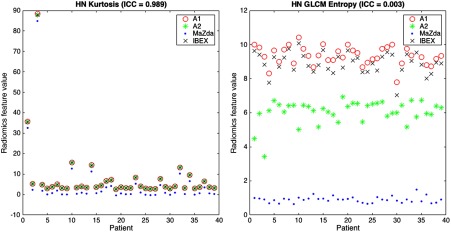
All second-order features for both mammography and HN CT ROIs showed poor agreement; however, HN features tended to show slightly higher ICC values. Plots showing values for the kurtosis and GLCM entropy across the 39 HN CT ROIs are shown in Fig. 4, which demonstrate excellent and poor agreement across packages, respectively. In the scatter plot depicting the feature distributions for kurtosis, the differences in feature values among packages for each patient are small relative to the variation in feature values among patients. In addition, the differences in feature values introduced by differences among radiomics packages were consistent, resulting in significant differences when using Friedman tests; however, because the bias was small compared to the variation among patients, the ICC is close to 1 reflecting excellent agreement. In contrast, for GLCM entropy, the differences in feature values for each patient across packages are large and systematic, resulting in significant differences and an ICC value near 0 reflecting poor agreement.
Fig. 4.
Scatter plots illustrating the agreement of features across packages. HN kurtosis showed excellent agreement () because the variability in feature values among packages is much less than the variability in feature values among patients, whereas HN GLCM entropy showed poor agreement (). Because of the consistent bias introduced in the feature distributions, HN kurtosis is still significantly different when calculated using different radiomics packages despite the strong agreement reflected by the ICC for HN kurtosis.
3.1. Variation in Image Importation and Preprocessing
In both the A1 and A2 packages, the raw images are imported with no preprocessing or normalization applied before features are calculated. MaZda, however, applies a default normalization such that the value of each pixel is increased by one. The MaZda user manual35 states that this is done to keep consistency with the equations presented by Haralick and Shapiro.44 In addition, MaZda was not designed to accommodate negative pixel values, and the resulting pixel values are greatly dependent on whether or not the image was stored as signed or unsigned because of the way signed and unsigned images differ in their bit allocation. In comparison, pixel values in IBEX are not dependent on if the image was stored as signed or unsigned, but negative pixel values are truncated at zero while nonnegative pixel values retain their original value. In addition, IBEX imports images using the RescaleSlope and RescaleIntercept tags from the DICOM header in the following manner:
| (1) |
The RescaleSlope tag for a standard CT scan typically has a value of , resulting in the value of each pixel in the image being reduced by 24. These trends can be seen in the boxplots for the min, max, and mean in Fig. 2. MaZda, on the contrary, does not consider any information contained in the DICOM header, resulting in fundamentally different results than when analyzed in IBEX. Differences in image importation are summarized in Table 6.
Table 6.
Differences in image importation characteristics.
| A1 | A2 | MaZda | IBEX | |
|---|---|---|---|---|
| Imported image dependent on image being signed or unsigned | ✓ | |||
| Capable of importing negative pixel values | ✓ | ✓ | ||
| Capable of performing calculations using negative pixel values without manual preprocessing | ✓ | ✓ | ✓ | |
| Capable of performing calculations using negative pixel values with manual preprocessing | ✓ | ✓ | ✓ | ✓ |
| Uses DICOM header in preprocessing | ✓ |
3.2. Variations in Algorithm Implementation
First- and second-order feature values for the single mammography image when feature functions were extracted from the packages A1, A2, and IBEX are shown in Table 7.
Table 7.
Feature values for a single mammography image when feature algorithms are extracted from packages A1, A2, and IBEX.
| Feature | A1 | A2 | IBEX |
|---|---|---|---|
| Max | 3161 | 3161 | 3161 |
| Min | 2123 | 2123 | 2123 |
| Mean | 2545.7 | 2545.7 | 2545.7 |
| Standard deviation | 152.6 | 152.6 | 152.6 |
| Skewness | 0.439 | 0.439 | 0.439 |
| Kurtosis | 2.967 | 2.967 | 2.967 |
| GLCM contrast | |||
| GLCM entropy | |||
| GLCM sum entropy | |||
| GLCM sum average | |||
| GLCM sum variance | |||
| GLCM difference entropy |
When calculated by isolating feature functions from preprocessing steps, most features show strong agreement among packages. Sum variance is shown to greatly differ between A1 and IBEX; however, A1 references Jahne et al.45 for this equation, which incorporates the value of the sum average in its calculation, whereas IBEX references Haralick et al.,40 which instead incorporates the value of the sum entropy. It is stated in Jahne et al.45 that this discrepancy is thought to be a typographical error. In addition, while skewness and kurtosis among A1, A2, and IBEX appear to be relatively similar, A1 uses a bias correction that could result in large discrepancies for smaller images.
Differences in values for GLCM entropy, sum entropy, and difference entropy between package A2 and the other two packages arise from different entropy definitions. While A1 and IBEX use a logarithm with base 2 in this calculation, A2 uses a natural logarithm. When these features from A2 are scaled by a ratio incorporating the two logarithm bases, the values of the entropy, sum entropy, and difference entropy agree with values calculated by packages A1 and IBEX to within four significant digits.
3.3. Variations in Naming Conventions
While some features with a common name have different algorithmic implementations in different software packages, other features use the same equation (and potentially the same implementation) but are known by a number of different names. The individual features analyzed in this study were those that had common naming conventions among software packages. This feature set might have been larger had common naming conventions been used to describe common mathematical calculations. As an example, the literature, as well as the notes in some published Matlab functions, shows that GLCM energy can also be referred to as “uniformity,” “uniformity of energy,” and “angular second moment.”44,46 The same Matlab functions refer to GLCM contrast as “variance” or “inertia.” Also, the GLCM homogeneity used in A1 was identified as identical to the inverse difference moment outlined in Haralick;40 however, the homogeneity in A1 uses the absolute value of the involved differences44 rather than the square of that difference. Despite the underlying code differing greatly in one package, the computed GLCM absolute value and GLCM difference average feature values were identical for all patients, indicating that these features may be equivalent; however, variations in naming conventions may be difficult to identify when both feature names and algorithm implementation differ among software packages. Finally, it can be seen in Fig. 2 that the kurtosis calculated by MaZda is exactly three less than the kurtosis calculated by the remaining packages for each ROI. This is because MaZda instead calculates the kurtosis that exceeds that of a Gaussian distribution, i.e., the excess kurtosis, which has a value of about three. This discrepancy in naming convention is not explicit in MaZda’s interface.
3.4. Variations in GLCM Parameters
When GLCM parameters were modified to those shown in Table 3 to maximize the consistency among radiomics packages, the resulting feature distributions are shown in Fig. 5. Compared with the distributions shown in Fig. 3, the ranges in the feature values are dramatically reduced for both the mammography and HN images, indicating greater agreement among packages. When using the modified GLCM parameters for both mammography and HN CT ROIs, ICCs increased (Table 8) compared with those calculated using default GLCM parameters but remained less than 0.33 indicating poor agreement. In addition, all second-order features were still significantly different across packages for both mammography and HN CT ROIs.
Fig. 5.
Distribution of second-order features for (a) the mammography ROIs and (b) the HN CT ROIs using the consistent GLCM parameters outlined in Table 3. Feature distributions show greater agreement when using the package-specific default GLCM parameters shown in Fig. 3.
Table 8.
-Values and ICC values for second-order features when using GLCM parameters modified to maximize consistency both with and without MaZda included. While all features differed significantly when MaZda was not included in the analysis, agreement improved when MaZda was not included.
| Mammography | HN CT | ||||
|---|---|---|---|---|---|
| Feature | -Value | ICC | -Value | ICC | |
| With MaZda | GLCM contrast | 0.327 | 0.315 | ||
| GLCM entropy | 0.006 | 0.021 | |||
| GLCM sum entropy | 0.008 | 0.017 | |||
| GLCM sum average | 0.309 | 0.208 | |||
| GLCM sum variance | 0.015 | ||||
| GLCM difference entropy | 0.043 | 0.041 | |||
| Without MaZda | GLCM contrast | 0.998 | 0.989 | ||
| GLCM entropy | 0.061 | 0.281 | |||
| GLCM sum entropy | 0.055 | 0.387 | |||
| GLCM sum average | 1.000 | 0.994 | |||
| GLCM sum variance | 0.023 | ||||
| GLCM difference entropy | 0.330 | 0.387 | |||
Because MaZda limited the GLCM parameters that could be customized, the analysis was repeated excluding MaZda. When using the modified GLCM parameters, all second-order features still showed significant differences for both mammography and HN CT ROIs. While ICCs, excluding MaZda, increased for every feature, only two features (GLCM contrast and sum average) for both mammography and HN CT ROIs increased in value to exceed 0.9 indicating excellent agreement.
4. Discussion
This study demonstrated dramatic differences in computed radiomics features values among the four packages due to various sources of discrepancy. These sources of variation among packages include differences in image importation and preprocessing, algorithm implementation, as well as GLCM and feature-specific parameters. While many first-order features showed relatively good agreement across packages, nearly all features significantly differed. All second-order features showed very poor agreement and differed significantly when using package-specific default GLCM parameters. Therefore, when these radiomics features are used for predictive modeling, computer-aided diagnosis, or image segmentation, for example, the results could greatly differ depending on the software being used. Subsequently, the results from studies that use one particular package potentially may not correlate with studies that rely on a different package, and if the same package is used, results may still not agree if feature parameters (e.g., GLCM parameters) are not consistent across these studies.
The exchange and comparison of radiomics software may allow for a standardization of these software packages resulting in more translatable radiomics-based research. For example, it was found by comparing the feature values across packages that package A1 had an error in the normalization of the GLCMs before feature calculation. Therefore, software errors may be revealed through a comparison of results and code from one institution to another, and it may be more likely to address previously unknown errors. Use of a standard set of “calibration” cases and reporting of the resulting feature values such as those provided by the IBSI could serve as a tool by which to validate and commission new radiomics software.27 In addition, an editorial from Vallières et al.47 summarizes the elements of the radiomics workflow that may also result in variations and practices that could be used when incorporating radiomics into research. For example, the radiomics ontology offers a means of consistently reporting aspects of the radiomics workflow including radiomics features, segmentation algorithms, and image filters. This editorial refers to the responsible research and innovation website for guidelines regarding the effective reporting of research methods and results.
A number of studies have recognized and reported the need for standardizing the radiomics pipeline.18,24–26 Because of this, the IBSI worked toward standardizing radiomics research by compiling an extensive manual of recommended feature definitions and image processing protocols. The collection of 19 institutions included in the IBSI used these recommendations to iteratively modify the feature extraction process when using a shared digital phantom and eventually a CT scan from a single lung cancer patient. Features were considered standardized if 50% of the contributors produced the same feature value. Through this process, agreement was achieved for 99.4% and 96.4% of features extracted from the digital phantom and CT scan, respectively.28,29 While the institutions included in the IBSI have increased the homogeneity of their radiomics workflow, this study illustrates that the field would benefit from a broader standardization effort that captures institutions using both propriety in-house radiomics packages as well as freely available open-source packages. The IBSI has established an important role in the standardization of feature definitions and radiomics algorithms. The goal of this study was not to duplicate the IBSI effort or offer recommendations outside of those established by the IBSI but rather to quantify the differences in radiomics features computed from real-world clinical images by radiomics software packages that have been the basis for numerous publications. The findings provide additional support for the goals that the IBSI seeks to achieve and quantifies the sources of variation that are highlighted here and also by the IBSI.
This investigation included a few limitations that introduced a degree of uncertainty in these results while also indicating areas that may require attention while working toward standardizing radiomics research. The source code for MaZda was not available, making it difficult to investigate the underlying mechanics and isolating the components such as preprocessing and algorithm implementation. To facilitate reproducible research, freely available radiomics packages may want to increase the transparency of their methods by making the source code available to the public for comparison. Also, MaZda does not allow for automated ROI processing, so for robust prediction models that include hundreds or thousands of images, manual feature extraction could take several hours and introduce a high degree of human error. IBEX also did not inherently allow for automated feature extraction for multiple images while also altering feature parameters; however, the IBEX source code could be used to create an automated feature extraction function. Radiomics packages developed in the future should consider automating the feature extraction process while allowing the user to customize the feature calculation parameters such as those involved in constructing the GLCMs. Furthermore, when publishing findings obtained from radiomics research, any relevant material required to reproduce the work, such as feature definitions or GLCM parameters, should be included in paper appendices or supplemental material.
Additional limitations of this study that hindered comparisons among packages included the inconsistency in computed radiomics features and the inability of some packages to calculate features in three-dimensions (3-D). Of the hundreds of features that could be calculated among the various packages, the only feature classes all four software packages had in common were the first-order histogram features and GLCM features, and only six features from each class were common among packages. This illustrates that features should be translatable across radiomics software packages using the feature definitions supplied by the IBSI.27 These definitions should also be used to allow for calculations in both two- and three-dimensions. For instance, because some packages included in this study could not compute 3-D features, comparison to IBSI harmonization data was not possible.27
Future work should incorporate additional radiomics packages to further test the variability of the resultant feature values. The first- and second-order features used in this study were chosen because they were the only 12 features that all four packages had in common; however, using additional package combinations could allow for a larger number of studied features.
Additional studies should directly investigate the effects of analyzing images from various imaging modalities such as magnetic resonance imaging (MRI) or PET. Radiomics packages may have been developed to process a particular type of image from a specific imaging modality. MaZda was originally developed to extract features from MRI scans with a particular range of pixel values, whereas A1 was originally developed to study lung CT scans in Hounsfield Units. Therefore, the package-specific default GLCM parameters for A1 used a gray-level limit of to 1500, whereas the gray-level limit for MaZda was determined automatically based on the bit depth of the MRI image. Investigating images from additional imaging modalities and additional tissue types could offer insight into how different packages behave under various circumstances. Future work could also include studying the effects of using various radiomics packages to accomplish a particular clinical task such as classifying patients with a particular disease.
5. Conclusion
An analysis of the variability in four radiomics software packages was performed to determine sources of discrepancies in computed radiomics features among packages. Inconsistencies in image importation, algorithm implementation, and GLCM parameters were investigated. The vast majority of features demonstrated significant differences in computed values across packages; however, most first-order features showed excellent agreement based on ICC. Second-order features had relatively poor agreement among packages as assessed by ICC. When GLCM parameters were modified to provide greater consistency across packages, ICCs increased but only showed agreement for two features (GLCM contrast and sum average). Investigators should therefore use caution when adopting new radiomics packages and incorporating them into their research, ensuring the software used is appropriate for the images being studied and fully disclosing the underlying calculation parameters so that results from one radiomics-based study may be translatable to other studies. Additional collaboration with groups such as the IBSI should be conducted to achieve greater harmonization of radiomics methods with direct clinical application across a greater number of institutions.
Acknowledgments
The authors are grateful to Roger Engelmann, M.S. and Li Lan M.S. for interface design and to Lauren Nowosatka and Prerana Mitta for help with calculations. The authors would like to thank Kristen Wroblewski, the University of Chicago Department of Health Studies, for her guidance in statistical analysis.
Biographies
Joseph J. Foy is a PhD candidate at the University of Chicago studying in the graduate program in medical physics. His research focuses on the variability in the radiomics and texture analysis workflow, and he is working to help standardize radiomics research across institutions to allow for greater clinical implementation. In particular, his research has focused on the variation in radiomics features due to differences in radiomics software packages, image acquisition parameters, and reconstruction methods.
Kayla R. Robinson is a graduate student in medical physics at the University of Chicago. Her primary research interests include quantitative texture analysis for risk assessment in medical imaging. Her research focuses on the applications of texture descriptors to improve early detection and risk assessment of breast cancer in women, and improvements in screening image analysis.
Hui Li has been working on quantitative imaging analysis on medical images for over a decade. His research interests include breast cancer risk assessment, diagnosis, prognosis, response to therapy, understanding the relationship between radiomics and genomics, and their future roles in precision medicine with both conventional and deep learning approaches.
Maryellen L. Giger is the A. N. Pritzker professor of radiology and the committee on medical physics at The University of Chicago, and a member of the National Academy of Engineering. Her research interests involve the investigation of computer-aided diagnosis and machine learning methods for the assessment of risk, diagnosis, prognosis, and response to therapy of breast cancer on multimodality (mammography, ultrasound, and magnetic resonance) breast images, and data mining for cancer discovery.
Hania Al-Hallaq investigates the use of medical images to: (1) inform treatment selection, (2) guide treatment positioning, and (3) assess treatment response following radiotherapy. She collaborates with Samuel Armato to study lung texture in CT scans, to test whether clinical symptoms correlate with radiomics changes in CT images. Her research background in texture analysis and clinical background as a clinical radiotherapy physicist has allowed her to contribute significantly to translational cancer research.
Samuel G. Armato III is an associate professor of radiology and the committee on medical physics at The University of Chicago. His research interests involve the development of computer-aided diagnostic methods for thoracic imaging, including automated lung nodule detection and analysis in CT scans, semiautomated mesothelioma tumor response assessment, image-based techniques for the assessment of radiotherapy-induced normal tissue complications, and the automated detection of pathologic change in temporal subtraction images.
Disclosures
This study was funded in part by a Team Science Award through the University of Chicago Comprehensive Cancer Center and by NIH T32 EB002103. SGA, HA, MLG, and HL receive royalties and licensing fees for computer-aided diagnosis technology through the University of Chicago. MLG is a stockholder in R2 Technology/Hologic and is a cofounder of and shareholder in Quantitative Insights.
References
- 1.Elsheikh T. M., et al. , “Interobserver and intraobserver variation among experts in the diagnosis of thyroid follicular lesions with borderline nuclear features of papillary carcinoma,” Am. J. Clin. Pathol. 130(5), 736–744 (2008). 10.1309/AJCPKP2QUVN4RCCP [DOI] [PubMed] [Google Scholar]
- 2.Gillies R. J., Kinahan P. E., Hricak H., “Radiomics: images are more than pictures, they are data,” Radiology 278(2), 563–577 (2016). 10.1148/radiol.2015151169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yip S. S., Aerts H. J., “Applications and limitations of radiomics,” Phys. Med. Biol. 61(13), R150–R166 (2016). 10.1088/0031-9155/61/13/R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aerts H. J., et al. , “Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach,” Nat. Commun. 5, 4006 (2014). 10.1038/ncomms5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giger M. L., Doi K., MacMahon H., “Computerized detection of lung nodules in digital chest radiographs,” Proc. SPIE 0767, 384–387 (1987). 10.1117/12.967022 [DOI] [Google Scholar]
- 6.Giger M. L., Doi K., MacMahon H., “Image feature analysis and computer-aided diagnosis in digital radiography. 3. Automated detection of nodules in peripheral lung fields,” Med. Phys. 15(2), 158–166 (1988). 10.1118/1.596247 [DOI] [PubMed] [Google Scholar]
- 7.Chan H. P., et al. , “Image feature analysis and computer-aided diagnosis in digital radiography. 1. Automated detection of microcalcifications in mammography,” Med. Phys. 14(4), 538–548 (1987). 10.1118/1.596065 [DOI] [PubMed] [Google Scholar]
- 8.Doi K., et al. , “Method and system for enhancement and detection of abnormal anatomic regions in a digital image,” US patent 4,907,156 (1990).
- 9.Chan H. P., et al. , “Improvement in radiologists’ detection of clustered microcalcifications on mammograms: the potential of computer-aided diagnosis,” Invest. Radiol. 25(10), 1102–1110 (1990). 10.1097/00004424-199010000-00006 [DOI] [PubMed] [Google Scholar]
- 10.Doi K., “Computer-aided diagnosis in medical imaging: historical review, current status and future potential,” Comput. Med. Imaging Graphics 31(4–5), 198–211 (2007). 10.1016/j.compmedimag.2007.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida H., et al. , “Computerized detection of colonic polyps at CT colonography on the basis of volumetric features: pilot study,” Radiology 222(2), 327–336 (2002). 10.1148/radiol.2222010506 [DOI] [PubMed] [Google Scholar]
- 12.Ganeshan B., et al. , “Dynamic contrast-enhanced texture analysis of the liver: initial assessment in colorectal cancer,” Invest. Radiol. 46(3), 160–168 (2011). 10.1097/RLI.0b013e3181f8e8a2 [DOI] [PubMed] [Google Scholar]
- 13.Wardlaw J. M., White P. M., “The detection and management of unruptured intracranial aneurysms,” Brain 123(3), 205–221 (2000). 10.1093/brain/123.2.205 [DOI] [PubMed] [Google Scholar]
- 14.Oliver J. A., et al. , “Variability of image features computed from conventional and respiratory-gated PET/CT images of lung cancer,” Transl. Oncol. 8(6), 524–534 (2015). 10.1016/j.tranon.2015.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter L. A., et al. , “High quality machine-robust image features: identification in nonsmall cell lung cancer computed tomography images,” Med. Phys. 40(12), 121916 (2013). 10.1118/1.4829514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galavis P. E., et al. , “Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters,” Acta Oncol. 49(7), 1012–1016 (2010). 10.3109/0284186X.2010.498437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng N. M., Fang Y. H., Yen T. C., “The promise and limits of PET texture analysis,” Ann. Nucl. Med. 27(9), 867–869 (2013). 10.1007/s12149-013-0759-8 [DOI] [PubMed] [Google Scholar]
- 18.Leijenaar R. T., et al. , “The effect of SUV discretization in quantitative FDG-PET radiomics: the need for standardized methodology in tumor texture analysis,” Sci. Rep. 5(5), 11075 (2015). 10.1038/srep11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shafiq-Ul-Hassan M., et al. , “Intrinsic dependencies of CT radiomic features on voxel size and number of gray levels,” Med. Phys. 44(3), 1050–1062 (2017). 10.1002/mp.12123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackin D., et al. , “Measuring computed tomography scanner variability of radiomics features,” Invest. Radiol. 50(11), 757–765 (2015). 10.1097/RLI.0000000000000180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cunliffe A., et al. , “Lung texture in serial thoracic computed tomography scans: correlation of radiomics-based features with radiation therapy dose and radiation pneumonitis development,” Int. J. Radiat. Oncol. Biol. Phys. 91(5), 1048–1056 (2015). 10.1016/j.ijrobp.2014.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunliffe A. R., et al. , “Lung texture in serial thoracic CT scans: registration-based methods to compare anatomically matched regions,” Med. Phys. 40(6), 061906 (2013). 10.1118/1.4805110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendel K. R., et al. , “Quantitative texture analysis: robustness of radiomics across two digital mammography manufacturers’ systems,” J. Med. Imaging 5(1), 011002 (2018). 10.1117/1.JMI.5.1.011002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sollini M., et al. , “PET radiomics in NSCLC: state of the art and a proposal for harmonization of methodology,” Sci. Rep. 7, 358 (2017). 10.1038/s41598-017-00426-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nyflot M. J., et al. , “Quantitative radiomics: impact of stochastic effects on textural feature analysis implies needs for standards,” J. Med. Imaging 2, 041002 (2015). 10.1117/1.JMI.2.4.041002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatt M., et al. , “Characterization of PET/CT images using texture analysis: the past, the present… any future?” Eur. J. Nucl. Med. Mol. Imaging 44, 151–165 (2017). 10.1007/s00259-016-3427-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zwanenburg A., et al. , “Image biomarker standardisation initiative (IBSI),” arXiv:1612.07003 (2016).
- 28.Zwanenburg A., “EP-1677: multicentre initiative for standardisation of image biomarkers [abstract],” Radiother. Oncol. 123(Suppl.), S914–S915 (2017). 10.1016/S0167-8140(17)32209-0 [DOI] [Google Scholar]
- 29.Hatt M., et al. , “IBSI: an international community of radiomics standardization initiative [abstract],” J. Nucl. Med. 59(Suppl.), 287 (2018). [Google Scholar]
- 30.Kalpathy-Cramer J., et al. , “Radiomics of lung nodules: a multi-institutional study of robustness and agreement of quantitative imaging features,” Tomography 2(4), 430–437 (2016). 10.18383/j.tom.2016.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armato S. G., et al. , “Research imaging in an academic medical center,” Acad. Radiol. 19(6), 762–771 (2012). 10.1016/j.acra.2012.02.002 [DOI] [PubMed] [Google Scholar]
- 32.Li H., et al. , “Computerized texture analysis of mammographic parenchymal patterns of digitized mammograms,” Acad. Radiol. 12(7), 863–873 (2005). 10.1016/j.acra.2005.03.069 [DOI] [PubMed] [Google Scholar]
- 33.Li H., et al. , “Computerized analysis of mammographic parenchymal patterns on a large clinical dataset of full-field digital mammograms: robustness study with two high-risk datasets,” J. Digital Imaging 25(5), 591–598 (2012). 10.1007/s10278-012-9452-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., et al. , “Comparative analysis of image-based phenotypes of mammographic density and parenchymal patterns in distinguishing between BRCA1/2 cases, unilateral cancer cases, and controls,” J. Med. Imaging 1(3), 031009 (2014). 10.1117/1.JMI.1.3.031009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szczypinski P., Strzelecki M., MaZda User’s Manual, Institute of Electronics, Technical University of Lodz, Poland: (2005). [Google Scholar]
- 36.Strzelecki M., et al. , “A software tool for automatic classification and segmentation of 2D/3D medical images,” Nucl. Instrum. Methods Phys. Res. 702(21), 137–140 (2013). 10.1016/j.nima.2012.09.006 [DOI] [Google Scholar]
- 37.Szczypinski P., et al. , “MaZda: a software package for image texture analysis,” Comput. Methods Prog. Biomed. 94(1), 66–76 (2009). 10.1016/j.cmpb.2008.08.005 [DOI] [PubMed] [Google Scholar]
- 38.Szczypinski P., Strzelecki M., Materka A., “MaZda: a software for texture analysis,” in Int. Symp. on Information Technology Convergence (ISITC 2007), pp. 245–249 (2007). 10.1109/ISITC.2007.15 [DOI] [Google Scholar]
- 39.Zhang L., et al. , “IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics,” Med. Phys. 42(3), 1341–1353 (2015). 10.1118/1.4908210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haralick R. M., Shanmugam S., Sinstein I., “Textural features for image classification,” IEEE Trans. Syst. Man Cybern. SMC-3(6), 610–621 (1973). 10.1109/TSMC.1973.4309314 [DOI] [Google Scholar]
- 41.Felipe J. C., Traina A. J. M., Traina C., “Retrieval by content of medical images using texture for tissue identification,” in Proc. 16th IEEE Symp. Computer-Based Medical Systems (2003). 10.1109/CBMS.2003.1212785 [DOI] [Google Scholar]
- 42.McGraw K. O., Wong S. P., “Forming inferences about some intraclass correlation coefficients,” Psychol. Methods 1(1), 30–46 (1996). 10.1037/1082-989X.1.1.30 [DOI] [Google Scholar]
- 43.Koo T. K., Li M. Y., “A guideline of selecting and reporting intraclass correlation coefficients for reliability research,” J. Chiropr. Med. 15(2), 155–163 (2016). 10.1016/j.jcm.2016.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haralick R. M., Shapiro L. G., Computer and Robot Vision, Vol. 1, p. 460, Addison-Wesley Longman Publishing Co., Inc., Boston: (1992). [Google Scholar]
- 45.Jahne B., Haussecker H., Geissler P., Handbook of Computer Vision and Applications, Vol. 2, Academic Press, Inc., Orlando, Florida: (1999). [Google Scholar]
- 46.Karaa W. B. A., Dey N., Biomedical Image Analysis and Mining Techniques for Improved Health Outcomes, IGI Global, Hershey, Pennsylvania: (2016). [Google Scholar]
- 47.Vallièrres M., et al. , “Responsible radiomics research for faster clinical translation,” J. Nucl. Med. 59(2), 189–193 (2018). 10.2967/jnumed.117.200501 [DOI] [PMC free article] [PubMed] [Google Scholar]