Abstract
Objective
To determine the effects of the disease-modifying therapies, glatiramer acetate (GA) and dimethyl fumarate (DMF), on the gut microbiota in patients with MS.
Methods
Participants with relapsing MS who were either treatment-naive or treated with GA or DMF were recruited. Peripheral blood mononuclear cells were immunophenotyped. Bacterial DNA was extracted from stool, and amplicons targeting the V4 region of the bacterial/archaeal 16S rRNA gene were sequenced (Illumina MiSeq). Raw reads were clustered into Operational Taxonomic Units using the GreenGenes database. Differential abundance analysis was performed using linear discriminant analysis effect size. Phylogenetic investigation of communities by reconstruction of unobserved states was used to investigate changes to functional pathways resulting from differential taxon abundance.
Results
One hundred sixty-eight participants were included (treatment-naive n = 75, DMF n = 33, and GA n = 60). Disease-modifying therapies were associated with changes in the fecal microbiota composition. Both therapies were associated with decreased relative abundance of the Lachnospiraceae and Veillonellaceae families. In addition, DMF was associated with decreased relative abundance of the phyla Firmicutes and Fusobacteria and the order Clostridiales and an increase in the phylum Bacteroidetes. Despite the different changes in bacterial taxa, there was an overlap between functional pathways affected by both therapies.
Interpretation
Administration of GA or DMF is associated with differences in gut microbial composition in patients with MS. Because those changes affect critical metabolic pathways, we hypothesize that our findings may highlight mechanisms of pathophysiology and potential therapeutic intervention requiring further investigation.
MS is believed to result from interactions between underlying genetic predisposition and environmental exposures.1,2 Environmental risk factors, including vitamin D levelse1 (links.lww.com/NXI/A86), viral exposures,e2 smoking,e3 and obesity,e4,e5 are related to MS onset and disease course. However, most of the environmental risk factors in MS remain unexplained. In consideration of additional potential mediators, the gut is a natural site of investigation, given that it is a major locus of environmental interaction and home to a large portion of the human immune system. In addition, immunologic development and patterning, dysregulated in MS and other autoimmune diseases, is heavily influenced by resident commensal microbes, the microbiota. Gut microbial composition is shaped by both genetics and environmental exposures that begin in utero, providing a potential pathogenic link between these factors and autoimmune diseases such as MS. Studies of experimental allergic encephalomyelitis (EAE) in mice have demonstrated reduced disease severity with administration of oral antibiotics3 and established that the presence of commensal flora is required for EAE induction.4
Several groups have begun to study the gut microbiota in patients with MS to identify a potential disease-associated signature.5–9 However, initial studies were limited by small numbers of participants and potentially confounded by the effects of disease-modifying therapies (DMTs). Separating changes in microbial composition related to MS disease state from changes induced by DMTs is crucial to further understand the role of gut microbiota in MS. We recently reported structural and functional changes in gut microbiota in untreated patients with MS compared with healthy controls10; in the current study, we began to examine the effects of DMT.
Therapy-induced changes in microbial composition may contribute toward efficacy by favoring microbes with anti-inflammatory properties, for example by influencing T-cell differentiation toward regulatory phenotypes or promoting the production of regulatory cytokines. This could manifest either as a “reversal” of observed differences in baseline gut microbiota associated with the MS disease state or in unrelated changes that nonetheless promote immune tolerance over inflammation in MS. Some changes may be neutral and still others may be detrimental, contributing to incomplete efficacy. Changes to individual taxa may be less important than the overall reshaping of the microbial community.
This cross-sectional study was designed to define the effects of 2 commonly prescribed DMTs on gut microbial composition in patients with MS. Glatiramer acetate (GA) is administered by subcutaneous injection. It is a random copolymer of alanine, lysine, glutamic acid, and tyrosine with widespread immunomodulatory effects including effects on antigen presentation and polarization of naive T cells away from TH1 and TH17 and toward TH2 and regulatory phenotypes with accompanying changes in cytokine profilese6 (links.lww.com/NXI/A86). Efficacy in MS was established by several clinical trialse7–e9 and it has been U.S. Food and Drug Administration (FDA) approved for the treatment of MS since 1996. Dimethyl fumarate (DMF) is an orally administered fumaric acid ester. Immunological effects relevant to MS include antiproliferative effects, inhibition of the NF-KB pathway, promotion of the heme oxygenase pathway, and anti-inflammatory effects related to T-cell differentiation and cytokine production.e10 Efficacy was established by 2 large clinical trials,e11,e12 and U.S. FDA approval was granted for MS in 2013.
In this study, we demonstrate that these 2 immunomodulatory therapies distinctly alter gut microbial composition in patients with MS.
Methods
Participants were recruited at the Corinne Dickinson Goldsmith Center for Multiple Sclerosis at Mount Sinai (New York, NY) between August 2013 and December 2015 and at the University of California San Francisco Multiple Sclerosis Center (San Francisco, CA) between July 2013 and September 2016 as part of the MS Microbiome Consortium. Potential participants aged 18–65 years were approached by MS Center clinicians during clinical visits. Patients with MS were eligible to participate if they met the McDonald 2010 criteria for relapsing-remitting MS or qualified as having clinically isolated syndrome along with abnormal brain MRI and were either naive to MS DMT or stable on GA or DMF for at least 3 months. Participants could not have taken antibiotics within 3 months or high-dose corticosteroids within 1 month of enrollment. Additional exclusions were a diagnosis of diabetes or inflammatory bowel disease, recent gastroenteritis, and treatment with an immunosuppressant medication for any condition in the 3 months preceding enrollment.
Clinical data
Clinical details including demographic information, detailed MS disease history, medical history, and height and weight were recorded. The Expanded Disability Status Scale was completed by the treating MS clinician or abstracted from the electronic medical record by one of the study investigators (I.K.S. and R.B.). Statistics were performed using SPSS 23.
Microbiota
Sample collection
Research coordinators reviewed detailed instructions regarding stool collection with all participants and provided them stool kits to be completed at home. Care was taken to match the kits and instructions at both institutions using a dry swab technique (BD #220135; Becton, Dickinson and Company, Franklin Lakes, NJ). Participants were instructed to collect stool samples using the first bowel movement of the day on 2 separate days, preferably 2 days within the same week. They were then instructed to place each completed kit between 2 freezer packs inside a thermal envelope and ship immediately or keep in their home freezer if there was to be a short delay in shipping. A prepaid return shipping label for overnight shipping was provided to minimize transit time. Participants were also instructed to send samples only in the early part of the week to avoid samples arriving over the weekend. On receipt, samples were immediately placed at −80°C until bacterial DNA extraction.
16S rRNA amplicon sequencing and data analysis
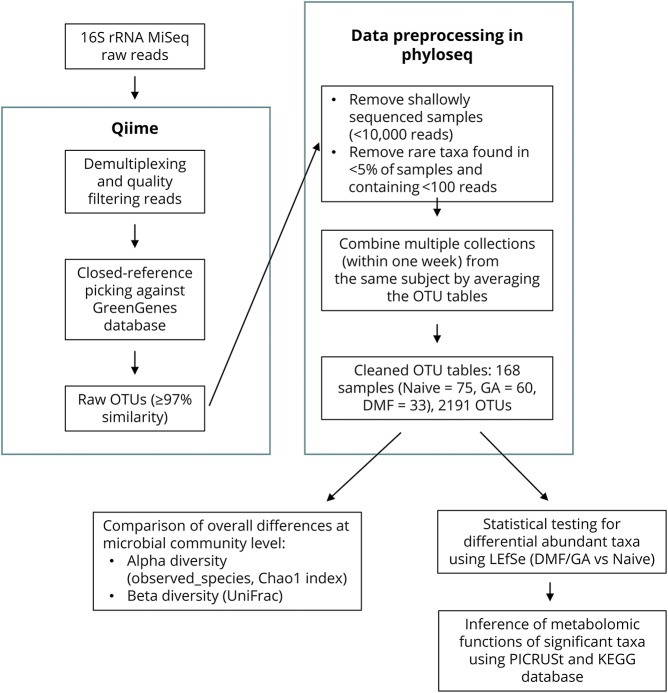
The workflow for microbiota sequencing and analysis is outlined in figure 1. DNA was extracted from samples using the PowerSoil DNA Isolation Kit (MoBio #12888) according to the manufacturer's instructions. For each sample, PCR targeting the V4 region of the bacterial/archaeal 16S rRNA gene was completed in triplicate using the 515/806 primer pair, and amplicons were sequenced using Illumina MiSeq (150 bp paired-end) sequencing primers, and procedures were described in the Earth Microbiome Project standard protocol.11
Figure 1. Flow chart explaining the bioinformatic pipeline and statistical methods (see Methods for details).
Raw reads were clustered into Operational Taxonomic Units (OTUs) using closed-reference SortMeRNA12 method at 97% identity based on the GreenGenes database v13.8 using Quantitative Insights Into Microbial Ecology (QIIME) v1.9.13 Taxonomy was assigned to the retained OTUs based on the GreenGenes reference sequence, and the GreenGenes tree was used for all downstream phylogenetic community comparisons. Samples with less than 10,000 sequences per sample were filtered out. OTUs were filtered to retain only OTUs present in at least 5% of the samples and containing at least 100 reads. Several very rare (relative abundance <5 × 10−5) OTUs including OTUs 4295564 (k__Bacteria; p__[Thermi]; c__Deinococci; o__Thermales; f__Thermaceae; g__Thermus; s__), 112194 (k__Bacteria; p__Proteobacteria; c__Alphaproteobacteria; o__Rickettsiales; f__Rickettsiaceae; g__Rickettsia; s__), and 4319519 (k__Bacteria; p__Spirochaetes; c__Spirochaetes; o__[Borreliales]; f__[Borreliaceae]; g__Borrelia; s__) were identified as potential contaminants and were removed. Samples collected from the same participant within 1 week were combined to 1 sample by averaging the OTU tables.
After filtering, samples were rarefied to 10,000 sequences per sample, and alpha and beta diversities were computed using QIIME v1.9. Unweighted UniFrac14 was used as the distance metric to perform principal coordinate analysis and to generate the corresponding plots. Alpha-diversity differences across the 3 treatment groups were tested by the non–parametric Kruskal-Wallis rank test. Beta diversity differences were assessed by comparing the within-group and between-group unweighted UniFrac distances using a non–parametric 2-sample t-test based on 1,000 Monte Carlo permutations, followed by a Bonferroni correction for multiple testing. Differential abundance analysis was performed using linear discriminant analysis effect size (LEfSe).15 Briefly, the non–parametric Kruskal-Wallis rank test was first applied to detect differentially abundant taxa. Significant taxa (p < 0.05) were then used to build a linear discriminant analysis (LDA) model, which estimated the effect size associated with a treatment group. p < 0.05 and LDA score >2 were used as the criteria to determine differentially changed taxa after treatment compared with samples from those who were treatment-naive.
To infer metagenomic functions (and predict metabolic functions from the predicted gene content) of the bacteria significantly altered by DMTs, the software package phylogenetic investigation of communities by reconstruction of unobserved states (PICRUSt) v1.1.116 was applied to all OTUs (2,191 OTUs) after quality filtering as described above, following the recommended pipeline of normalizing OTUs by copy number (to account for copy number differences of the 16S rRNA gene among taxa), predicting functions using the Kyoto Encyclopedia of Genes and Genomes orthologs, and assembling predicted Kyoto Encyclopedia of Genes and Genomes orthologs into functional pathways. Differentially changed metabolomic pathways between the treated and naive groups were identified using the non–parametric Kruskal-Wallis rank test, followed by the Benjamini-Hochberg correction for multiple testing with a false discovery rate ≤15%, reflecting the exploratory nature of this piece of the study.
Immunophenotyping
Whole blood was collected for immunophenotyping (only at Mount Sinai because of the requirement for fresh samples) and immediately processed by Mount Sinai's Human Immune Monitoring Core. Specimens were stained with a preoptimized T-cell antibody cocktail and acquired within 3 hours using a BD LSR Fortessa (BD, San Jose, CA). A minimum of 500,000 events were recorded from each sample to accurately assess minor cell populations. Compensation was performed with unstained cells and BD compensation beads. FlowJo 9.4 software (Treestar Inc, San Carlos, CA) was used for postacquisition analysis. Statistical analysis was performed using unpaired 2 tailed t-test (GraphPad Software, La Jolla, CA).
Standard protocol approvals, registrations, and patient consents
The research was approved by the Institutional Review Boards at Mount Sinai and UCSF, and written informed consent was obtained from all participants.
Data availability statement
Anonymized data are available and will be shared on request from any qualified investigator.
Results
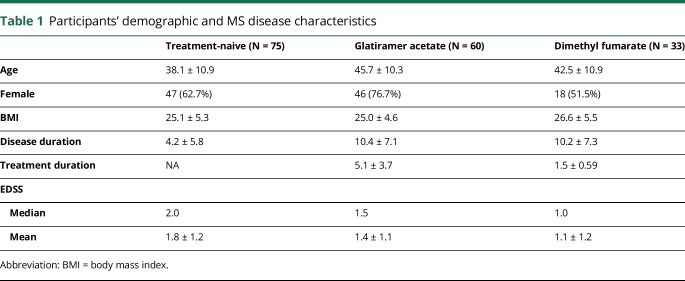
The study enrolled a total of 186 participants who met the inclusion criteria. Of these, 168 (90.3%) had at least 2 samples whose DNA passed quality control measures and were ultimately included. This includes 75 participants who were treatment-naive, 33 treated with DMF, and 60 treated with GA as outlined in figure 1. Immunophenotyping data were available only for a subset of the participants recruited at Mount Sinai (n = 30 naive, n = 26 DMF, and n = 12 GA). Clinical characteristics of study participants, broken down by treatment group, are outlined in table 1 (microbiota) and table e-1 (links.lww.com/NXI/A89, immunophenotyping).
Table 1.
Participants' demographic and MS disease characteristics
Immunophenotyping profiles validated the expected effects of DMF and GA
Peripheral blood mononuclear cells were collected from 68 participants recruited at Mount Sinai (treatment-naive n = 30, DMF n = 26, and GA n = 12) and analyzed by FACS to identify CD3, CD4, and CD8 populations. The surface markers CCR4 and CCR6 were used to further classify CD4 T cells as Th17 (CCR4+CCR6+)e13 (links.lww.com/NXI/A86) or Th2 (CCR4+CCR6−).e14
DMF treatment was associated with reduced percentage of CD3, CD4, and CD8 T cells as previously describede15 (links.lww.com/NXI/A86). DMF treatment was also associated with a reduced percentage of CCR4+ CCR6+ T cells (t-test, p < 0.0001), as well as a milder reduction of CCR4+CCR6− T cells (t-test, p = 0.02). Treatment with GA did not have any effect on total CD3 or CD4 percentage but was associated with a reduced percentage of CCR4+ CCR6+ T cells compared with treatment-naive patients (t-test, p = 0.02), which was not significantly different from the effect of DMF on those cells (t-test, p = 0.2) (figure e-1, links.lww.com/NXI/A87).
Administration of DMF or GA results in distinct alterations to gut microbial composition in patients with MS
16S amplicon sequencing was performed on the Illumina MiSeq platform to assess changes in the fecal microbiome of patients with MS after different treatments. In total, 49,679,344 raw reads passed the quality filtering in QIIME and were clustered into OTUs based on the GreenGenes database at a 97% sequence identity. We further applied stringent filters to remove samples with low sequencing reads and rare taxa (see Methods for detail) and then combined multiple collections within a short time interval from the same participant to counteract temporal variations (figure 1). This resulted in 8,542,763 high-quality reads (on average 49,628 ± 31,746 reads per participant) mapping to 2,191 unique OTUs. At the phylum level, the microbiota of all patients with MS was comprised predominately of Firmicutes (45.9%) and Bacteroidetes (41.0%) and to a lesser extent of Proteobacteria (8.7%), Actinobacteria (2.0%), and Verrucomicrobia (2.0%), consistent with recent reports on microbiota profiles identified in patients with MS or healthy participants on a Western diet5,6,17,18 (figure 2).
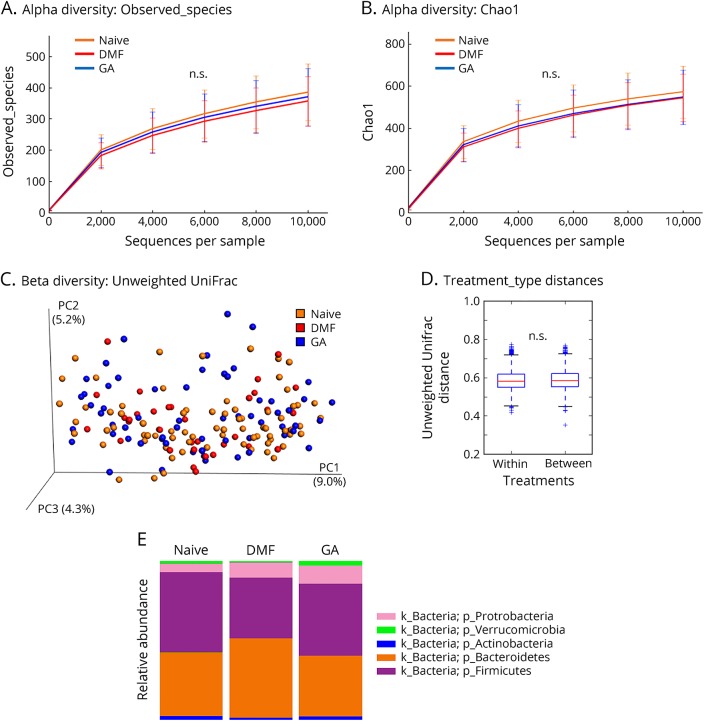
Figure 2. Overall community differences in the gut microbiota of treated or untreated patients with MS.
(A and B) Rarefaction curves plotting alpha diversity indices (A: Observed_species; B: Chao1) vs the number of sequences for each treatment group. n.s.: not statistically significant by the Kruskal-Wallis sum-rank test. (C) Principal coordinate plot based on the unweighted UniFrac distance metric. The percentage of variability explained by the first 3 principal components was plotted. Each colored point represents a participant from one of the therapy groups (Naive, DMF, or GA). (D) No obvious beta diversity differences were observed across groups as demonstrated by similar within-group and between-group distances. n.s.: not statistically significant by a non–parametric 2-sample t-test based on 1,000 Monte Carlo permutations, followed by a Bonferroni correction for multiple testing. (E) Mean relative abundance of prevalent microbes (>1% in any therapy group) at the phylum level. DMF = dimethyl fumarate; GA = glatiramer acetate; k = kingdom; p = phylum.
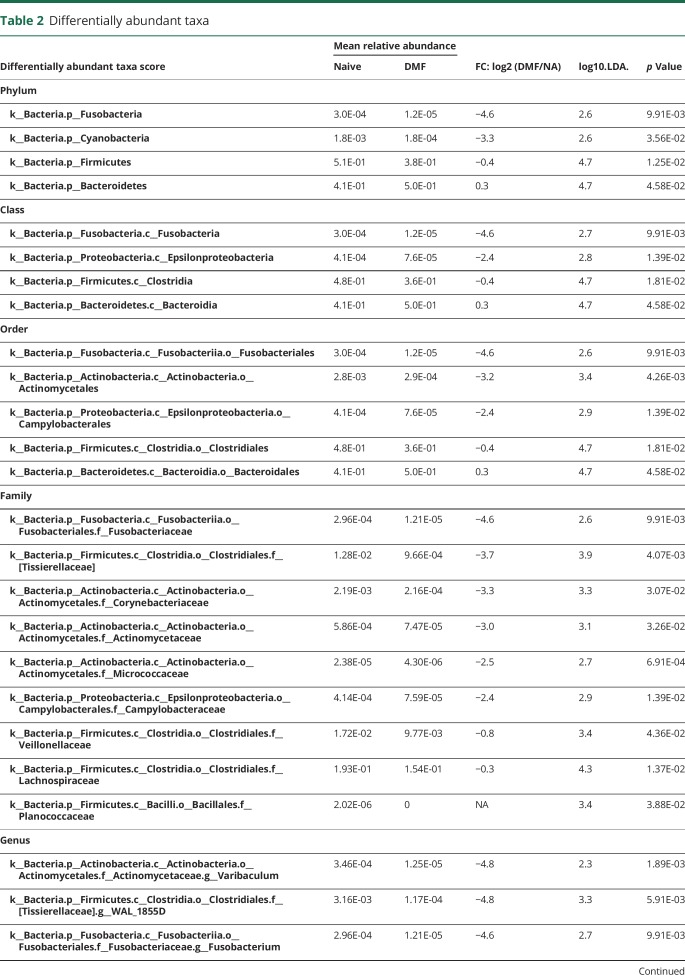
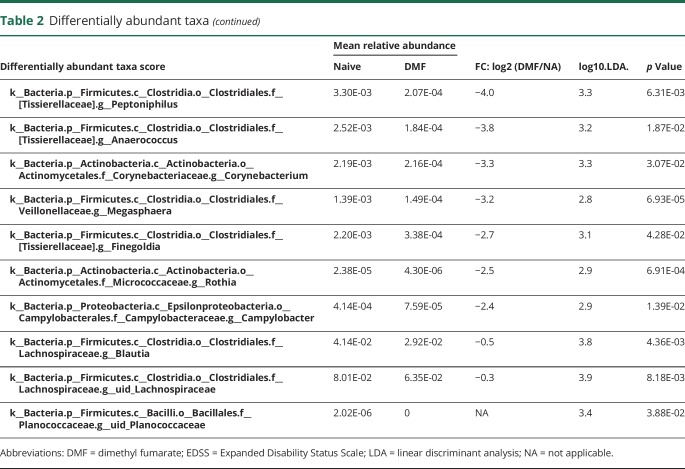
Because our sample collections were made at 2 geographically distinct sites, we first established that there were no community-level differences between the 2 sites (figure e-2, links.lww.com/NXI/A88). Because there were more women than men in our study and the proportion of women differed by therapy, we also performed diversity analyses between sexes. Although no alpha diversity differences were detected, we detected small within-sex and between-sex beta diversity differences (p = 0.01, effect size = 0.6%) that are not noticeable on a principal coordinate analysis plot (figure e-2). As shown in figure 2, administration of DMF or GA did not result in overall changes to microbial community structure as measured by alpha diversity (richness of taxa within samples by observed_species or richness and evenness of taxa within samples by chao1) or beta diversity (diversity between different samples by unweighted UniFrac. However, LEfSe analysis identified 13 differentially abundant bacterial genera in patients with MS on DMF and 14 differentially abundant genera in patients on GA, as compared to those who were treatment-naive (table 2). Of interest, GA led to a reduction in the relative abundance of 7 genera and an increase of 7 genera, whereas all the 13 genera affected by DMF treatment showed decreased relative abundance (figure 3). Among the taxonomic units decreased by DMF, the most dramatic effect was mapped to the order Clostridiales (figure 4), thereby suggesting potential importance of this taxon.
Table 2.
Differentially abundant taxa
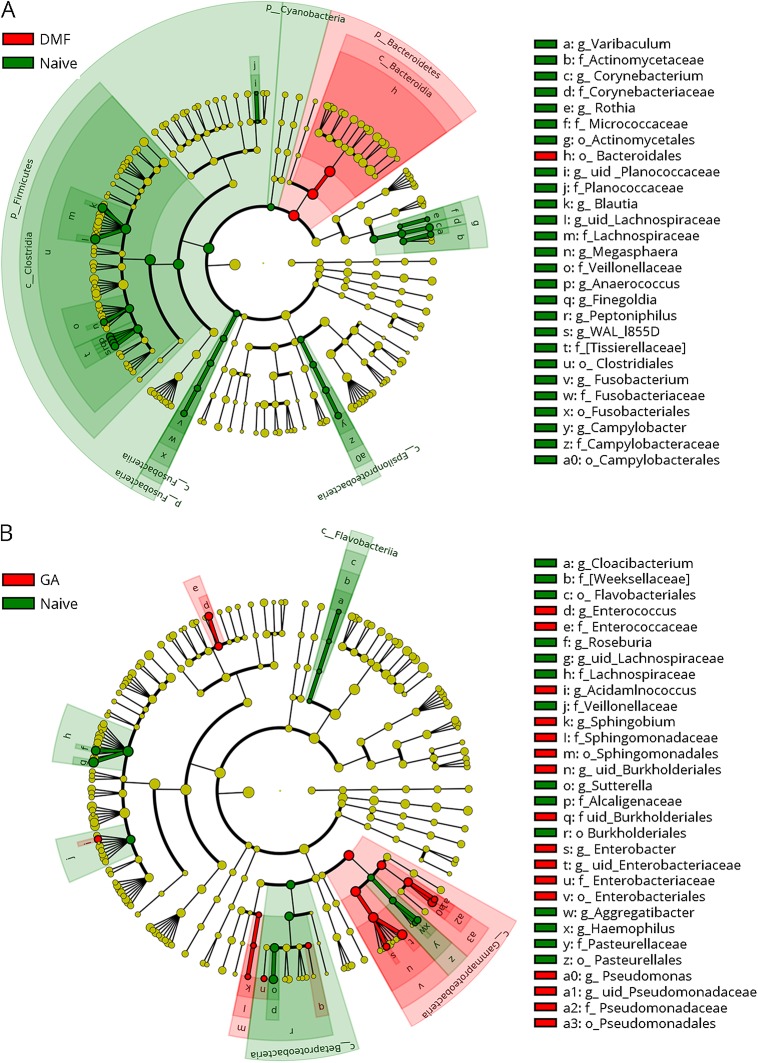
Figure 3. Cladogram indicating the effect of disease-modifying therapies on the phylogenetic structure of MS patients' microbiota.
Upper: DMF vs Naive. Lower: GA vs Naive. Only significant taxa (p-value < 0.05, LDA score >2) identified by LEfSe were displayed. Differential abundant taxa are marked by colored shades: red indicates enrichment in the therapy groups (DMF or GA), and green indicates enrichment in the naive group. Circles represent phylogenetic levels from kingdom to genus from inside out. Each dot represents a taxon and its diameter is proportional to the taxon's effect size. c = class; DMF = dimethyl fumarate; f = family; g = genus; GA = glatiramer acetate; k = kingdom; LEfSe = linear discriminant analysis effect size; LDA = linear discriminant analysis; o = order; p = phylum; uid = unidentified, according to the GreenGenes database.
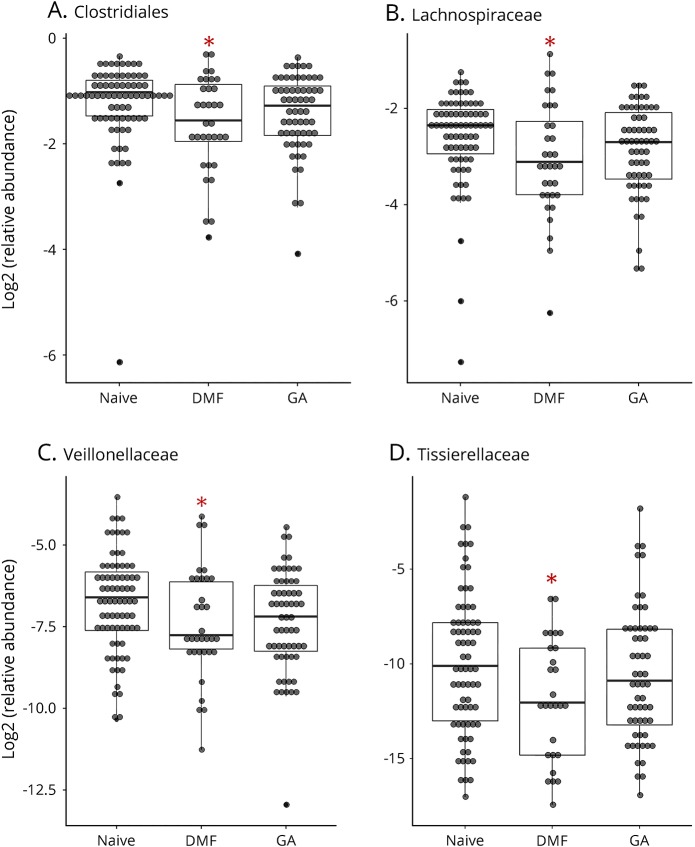
Figure 4. Relative abundances of the order Clostridiales and its family members are differentially changed by therapy.
Relative abundances of the order Clostridiales (A) and its family members are differentially changed by therapies (B: Lachnospiraceae; C: Veillonellaceae; D: Tissierellaceare). Asterisk (*) indicates significant change compared with naive as determined by the LEfSe analysis (see also in table 2). The inner bisecting line indicates the median. The lower and upper edges of the box represent the 25th and the 75th percentiles, respectively. LEfSe = linear discriminant analysis effect size.
Inference of functional consequences of alterations in gut microbial composition induced by DMF and GA
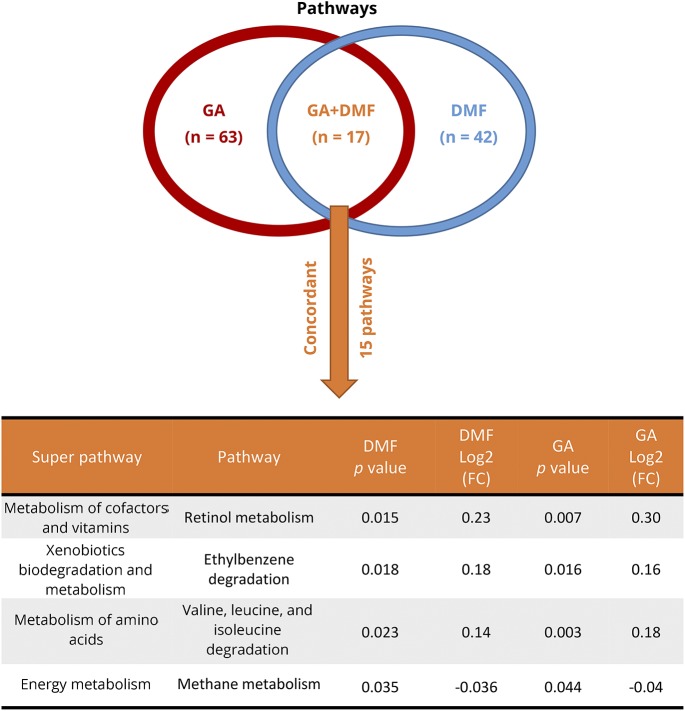
PICRUSt was used to evaluate potential functional implications of these changes in gut microbial profiles.16 We noted significant changes in 42 pathways in DMF-treated participants (30 increased and 12 decreased) and in 63 pathways in GA-treated participants (36 increased and 27 decreased). Despite the different route of administration and the different effect on bacterial taxa, there was an overlap of 17 pathways between the 2 therapies. GA and DMF had a concordant effect on 15 of those pathways, which included pathways affecting vitamin, amino acid, energy, and xenobiotic metabolism. More specifically, among the common concordant pathways affected were retinol (vitamin A), methane, and ethylbenzene metabolism, as well as valine, leucine, and isoleucine degradation (figure 5).
Figure 5. Functional implications of gut microbial profile changes.
Functional implications of the gut microbial profile changes were assessed by applying the package phylogenetic investigation of communities by reconstruction of unobserved states on whole microbial communities of patients taking either GA or DMF. Forty-two pathways were found to be affected in participants treated with DMF (30 upregulated and 12 downregulated) and 63 pathways in participants treated with GA (36 upregulated and 27 downregulated). Despite the different route of administration and the different effect on bacterial taxa, there was an overlap of 17 pathways that were changed by both therapies, 15 of which had a concordant effect. These included pathways of vitamins, amino acids, energy and xenobiotic metabolism, and more specifically the metabolism of retinol (vitamin A), methane, and ethylbenzene, as well as valine, leucine, and isoleucine degradation. DMF = dimethyl fumarate; GA = glatiramer acetate.
Discussion
Defining the effects of DMT on gut microbial composition is critical for the inclusion of treated MS patients in microbiota studies and interpretation of any findings. This has been established in several other disease states, for example, in separating effects of metformin from those of the type 2 diabetes disease statee16 (links.lww.com/NXI/A86). Many other medications, from non–steroidal anti-inflammatory drugse17 to proton pump inhibitors,e18 have been shown to affect the gut microbiota, and in this study, we demonstrate that MS therapies do as well. Although we acknowledge that definitive effects are best established through longitudinal studies of samples collected before and subsequent to the start of DMT, cross-sectional studies allow for preliminary evaluation of these ideas and generation of hypotheses for further study.
The phylum Bacteroidetes was increased in relative abundance in our DMF cohort compared with the treatment-naive cohort. This difference was mainly driven by an increase in the genus Bacteroides that narrowly missed statistical significance. This result is interesting in the context of other studies that have looked at this commensal group in MS and suggested a potential protective effect. A small study in pediatric MS showed relative depletion of Bacteroides compared with healthy controls.8 From a mechanistic standpoint, oral administration of purified polysaccharide A produced by Bacteroides fragilis was shown to mitigate EAE in an IL-10-dependent fashion.19 Lipid 654, a lipodipeptide generated by several commensal Bacteroidetes species, reaches the systemic circulation and is reduced in the serum of patients with MS compared with healthy controls.20 In an adoptive transfer model of EAE, low-level Lipid 654 was administered to induce toll-like receptor 2 tolerance and was found to attenuate EAE and decrease macrophage activation and TH17 cells in the CNS.21 Our results suggest that the observed effects of DMF on gut microbiota might be relevant to its anti-inflammatory effects in MS.
A decrease in the genus Prevotella in patients with MS was noted in several small studies; however, several of these may have been confounded by treatment with DMT, given the inclusion of a significant number of treated patients (particularly interferon [IFN]).6,7 One study found an increase in Prevotella associated with treatment (pooled group including IFN and GA), suggesting a possible mechanistic association for this genus with DMTs.5 In addition, human-derived Prevotella histicola suppresses EAE in a CD4+FoxP3+Treg–dependent fashion.22 In our cohort, we noted no change in the relative abundance of Prevotella in treated participants compared with those who were treatment-naive, suggesting that this genus is less affected by either GA or DMF than by IFN. Another possible explanation for observed differences in the effects of DMT on Prevotella in our study compared with others relates to our use of the V4 hypervariable region as compared to the use of V3–V5 in the aforementioned studies. However, previous comparisons of these variations in the technique have demonstrated relatively consistent results, making this difference unlikely to fully account for this discordance.23–25
We noted the genus Sutterella to be 2.4-fold less abundant in participants treated with GA as compared to those who were treatment-naive, with no difference in relative abundance with DMF. An increase in Sutterella in treated compared with untreated patients with MS was previously reported; however, the treatment group included patients treated with either IFN or GA.5 Suggesting that IFN may in fact be the driving factor, an additional study has noted an increase in Sutterella in IFN-treated MS patients.26 In a spontaneous EAE model, specimens from mice who received transplanted MS fecal material were noted to be relatively depleted in Sutterella.27 Given that the EAE course worsened in mice transplanted with fecal material from patients with MS, compared with those transplanted from a healthy twin, this finding might be pathologically significant. However, in this experiment, 3 of the 5 MS twins received treatment with IFN, potentially confounding interpretation of these results. Further work is needed to understand whether Sutterella is associated with MS pathology, whether there are differential effects of DMTs on the abundance of this genus, and finally whether such potential effects on Sutterella abundance have any clinical significance.
In our study, the relative abundance of members of the order Clostridiales (phylum Firmicutes, class Clostridia) was decreased. Members of 2 particular Clostridial families, Lachnospiraceae and Veillonellaceae, were decreased by both DMF and GA. Of interest, we previously noted increased relative abundance of the genus Megasphaera (family Veillonellaceae) in untreated MS compared with healthy controls.10 DMF seems to “reverse” this. Clostridial organisms have received significant attention in demyelinating disorders. An increased frequency of immunoreactivity to epsilon toxin–producing Clostridium perfringens type B was observed in patients with MS, suggesting this organism as contributory to MS pathology, given the potentially toxic effects at the blood-brain barrier.28 A subsequent study demonstrated the ability of epsilon toxin to destroy oligodendrocytes and induce demyelination.29 An overabundance of C perfringens has also been noted in a small study of patients with neuromyelitis optica.30 In addition to the described potential effects of Clostridial members on inflammation, Lachnospiraceae was demonstrated to impair oligodendrocyte differentiation in cultured cells and was associated with impaired myelination in mice.31 Clostridial taxa therefore have potential for pathologic relevance in MS and the mechanism of action of DMTs both with respect to inflammation and neurodegeneration/neuroprotection. In concordance with our results regarding other Clostridial family members, DMF was noted to inhibit the log-phase growth of C perfringens in vitro, and investigators hypothesized that this may be contributory to the drug's mechanism of action.32 A decrease in species belonging to Clostridia clusters was previously found in patients with MS compared with controls; however, this majority of participants in this study were treated with DMTs.7 A decrease in the relative abundance of Clostridial members could be common to several MS DMTs.
DMF resulted in a decrease in the relative abundance of many taxa, with Bacteroidetes members as the only significant increase. The methodology used here to calculate relative abundance values precludes conclusions about the absolute numbers of microbes present. However, fumarates have known antimicrobial properties and in fact DMF was previously used as an antimicrobial preservative for upholstery.33 Of interest, previous work in EAE suggests that a reduction in gut bacterial load might be of benefit.3 Compared with mice who received no antibiotics or intraperitoneal antibiotics, those who received oral antibiotics before EAE induction demonstrated reduced EAE severity that correlated to an increase in FoxP3+Treg cells in distal peripheral lymph nodes. Further, costimulation of harvested lymphocytes from distal nodes resulted in decreased production of proinflammatory cytokines such as IFN gamma and increased production of anti-inflammatory cytokines such as IL-10. Additional studies are needed to evaluate this potential effect of DMF as its generalized antimicrobial properties may be a contributing factor to efficacy in MS.
When considering the ultimate influence of DMT on MS gut microbiota, the effects of the overall shaping of the microbial community on downstream functional pathways may be more important than effects on individual genera. PICRUSt analysis may help identify the impact of changes in microbial composition on biological and metabolic pathways and suggest potential mechanisms that link changes in gut microbiota with therapeutic effects.16 One of the common pathways affected by both GA and DMF was retinol (vitamin A) metabolism. Vitamin A metabolites are known to modulate specific functional aspects of the immune response, such as the Th1–Th2-cell balance and the differentiation of Treg and Th17 cells.34 Retinoic acid also modulates lymphocyte trafficking by enhancing integrin a4b7 expression on lymphocytes and imprinting them with gut tropism.35 Of interest, dendritic cells from the gut-associated lymphoid tissue, but not from the spleen, were able to produce retinoic acid from retinol, suggesting that vitamin A plays an important role specifically in the gut-immune system interaction. Another bacterial pathway affected by both therapies was methane metabolism. An increased relative abundance of methane-producing bacteria and exhaled methane in patients with MS compared with healthy controls was reported.5 The role of these pathways in MS pathophysiology and precise effects of therapies will need to be further explored in future studies.
Our study is still limited by the relatively small sample size and cross-sectional nature of sample collection, including unavoidable imperfect matching of groups with respect to age, sex, and disease duration. Also, although recruitment at 2 separate locations is a merit, particularly because careful attention was granted to implement identical methodology and analyses did not demonstrate community-level differences by site, it is possible that unmeasured differences in study groups related to geography confounded our results in some fashion.
Our study results confirm our hypothesis that DMTs for MS have measurable effects on gut microbiota. Future directions will include working with a larger cohort with longitudinal follow-up of individuals who begin new DMTs to assess the effects more precisely and to evaluate questions regarding the relationship of gut microbiota to therapy response and tolerability. Bidirectional relationships have been confirmed in other disease states36–39 and will need to be investigated in MS as well. Effects are likely to be particularly important for orally delivered medications. In addition, lessons learned from these studies may inform our understanding of the pathogenesis of the disease and lead to the exploration of new potential treatment targets.
Acknowledgment
The authors thank all of those who contributed to this project, especially the MS clinicians (Michelle Fabian, Aaron Miller, Fred Lublin, Stephen Krieger, Sylvia Klineova, Aliza Ben-Zacharia, Gretchen Mathewson, and Jennifer Graves) and research coordinators and technicians (Jessica Zheng, Tamjeed Sikder, Cuquita O'Shea, Refujia Gomez, John Morrissey, and Patrick Barba) at Mount Sinai and UCSF who recruited the participants, as well as our MS patients and their families for their participation.
Glossary
- DMF
dimethyl fumarate
- DMT
disease-modifying therapy
- EAE
experimental allergic encephalomyelitis
- FDA
Food and Drug Administration
- GA
glatiramer acetate
- IFN
interferon
- LEfSe
linear discriminant analysis effect size
- LDA
linear discriminant analysis
- OTU
operational taxonomic unit
- PICRUSt
phylogenetic investigation of communities by reconstruction of unobserved states
Author contributions
Conception and design of the study was completed by I. Katz Sand, Y. Zhu, A. Ntranos, R. Knight, B.A.C. Cree, S.E. Baranzini, and P. Casaccia. Acquisition and analysis of data were completed by I. Katz Sand, Y. Zhu, A. Ntranos, J.C. Clemente, R. Brandstadter, Y. Bencosme, E. Cekanaviciute, S. Singh, J. Debelius, R. Knight, S.E. Baranzini, and P. Casaccia. The manuscript was drafted by I. Katz Sand, Y. Zhu, A. Ntranos, and P. Casaccia, with critical editing and approval by all remaining authors.
Study funding
The project was funded by the National MS Society, the United States Department of Defense, and the Friedman Brain Institute.
Disclosure
I. Katz Sand received research support from United States Department of Defense, the National Multiple Sclerosis Society, and the Guthy Jackson Charitable Foundation. Y. Zhu and A. Ntranos report no disclosures. J.C. Clemente is an associate editor for Microbiome and an editor for mBio. E. Cekanaviciute received research support from NIH IRACDA. R. Brandstadter reports no disclosures. E. Crabtree-Hartman received speaker honoraria from Biogen and Genzyme Sanofi; consulted for Teva, Novartis, and Biogen; and served on the speakers bureau for Teva and Biogen. S. Singh reports no disclosures. Y. Bencosme has been employed by Novo Nordisk. J. Debelius received research support from the University of California, San Diego. R. Knight served on the scientific advisory boards of Janssen, CommenSe, and Prometheus; received travel funding from Metagenics, Genentech, and Zurich Insurance Co; served on the editorial boards of Biology Direct, Gut Microbes, AEM, Env Micro, and Genome Biology; is an editor for the ISME Journal; holds a patent for microbiome bases systems, apparatus and methods for monitoring and controlling industrial processes and systems and microbiome bases systems, apparatus and methods for the exploration and production of hydrocarbons; receives publishing royalties from Simon and Schuster and St. Martin's Press; is employed by Biota Technology; consulted for CommenSe and Prometheus; received research support from Janssen (Johnson & Johnson), the NIH, NIDDK, NIH NIAID, DOJ, ONR, USAMRAA, NSF, NIAID/NIH, NIH, NIH-NHLBI, NIH-NIDDK, the Laureate Institute for Brain Research, the Alfred P. Sloan Foundation, the Gerber Foundation, CRISP, and the Robert Wood Johnson Foundation; and received stock or stock options from Biota Technology. B.A.C. Cree served on the scientific advisory board of Akili and consulted for AbbVie, Biogen, EMD Serono, GeNeuro, Novartis, and Sanofi Genzyme. S.E. Baranzini served on the scientific advisory boards of Novartis, EMD Serono, and Sanofi-Aventis; received a gift from Novartis Pharma; received speaker honoraria and travel funding from Novartis; served on the editorial boards of the Multiple Sclerosis Journal, Neurology, and mSystems; has a patent pending for gene expression signature that could identify patients at high risk of developing MS; consulted for Novartis, EMD Serono, and Teva; and received research support from NIH/NINDS, DOD, and NMSS. P. Casaccia received research support from the Department of the Army, NMSS. Full disclosure form information provided by the authors is available with the full text of this article at Neurology.org/NN.
References
- 1.Olsson TT, Barcellos LF, Alfredsson L. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat Rev Neurol 2017;13:25–36. [DOI] [PubMed] [Google Scholar]
- 2.Hedström AK, Alfredsson L, Olsson T. Environmental factors and their interactions with risk genotypes in MS susceptibility. Curr Opin Neurol 2016;29:293–298. [DOI] [PubMed] [Google Scholar]
- 3.Ochoa-Reparaz J, Mielcarz DW, Ditrio LE, et al. . Role of gut commensal microflora in the development of experimental autoimmune encephalomyelitis. J Immunol 2009;183:6041–6050. [DOI] [PubMed] [Google Scholar]
- 4.Berer K, Mues M, Koutrolos M, et al. . Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011;479:538–541. [DOI] [PubMed] [Google Scholar]
- 5.Jangi S, Gandhi R, Cox LM, et al. . Alterations of the human gut microbiome in multiple sclerosis. Nat Commun 2016;7:12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JJ, Chia N, Kalari KR, et al. . Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep 2016;6:28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyake SS, Kim S, Suda W, et al. . Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV clusters. PLoS One 2015;10:e0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tremlett HH, Fadrosh DW, Faruqi AA, et al. . Gut microbiota in early pediatric multiple sclerosis: a case-control study. Eur J Neurol 2016;23:1308–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantarel BL, Waubant E, Chehoud C, et al. . Gut microbiota in multiple sclerosis: possible influence of immunomodulators. J Investig Med 2015;63:729–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cekanaviciute E, Yoo BB, Runia TF, et al. . Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Natl Acad Sci USA 2017;114:10713–10718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caporaso JG, Lauber CL, Walters WA, et al. . Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012;6:1621–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kopylova EE, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012;28:3211–3217. [DOI] [PubMed] [Google Scholar]
- 13.Caporaso JG, Kuczynski J, Stombaugh J, et al. . QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 2005;71:8228–8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segata NN, Izard J, Waldron L, et al. . Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Langille MG, Zaneveld J, Caporaso JG, et al. . Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 2013;31:814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schnorr SL, Candela M, Rampelli S, et al. . Gut microbiome of the Hadza hunter-gatherers. Nat Commun 2014;5:3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Filippo C, Cavalieri D, Di Paola M, et al. . Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA 2010;107:14691–14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ochoa-Reparaz J, Mielcarz DW, Wang Y, et al. . A polysaccharide from the human commensal Bacteroides fragilis protects against CNS demyelinating disease. Mucosal Immunol 2010;3:487–495. [DOI] [PubMed] [Google Scholar]
- 20.Farrokhi V, Nemati R, Nichols FC, et al. . Bacterial lipodipeptide, Lipid 654, is a microbiome-associated biomarker for multiple sclerosis. Clin Transl Immunol 2013;2:e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anstadt EJ, Fujiwara M, Wasko N, Nichols F, Clark RB. TLR tolerance as a treatment for central nervous system autoimmunity. J Immunol 1950;197:2110–2118. [DOI] [PubMed] [Google Scholar]
- 22.Mangalam AA, Shahi SK, Luckey D, et al. . Human gut-derived commensal bacteria suppress CNS inflammatory and demyelinating disease. Cel Rep 2017;20:1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res 2007;35:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, DeSantis TZ, Andersen GL, Knight R. Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Res 2008;36:e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soergel DA, Dey N, Knight R, Brenner SE. Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J 2012;6:1440–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Castillo Alvarez F, Pérez Matute P, Colina Lizuain S, et al. . Intestinal microbiota in multiple sclerosis: influence of treatment with interferon B-1b. Presented at the European Committee for Treatment and Research in Multiple Sclerosis (ECTRIMS) 2016; London, United Kingdom.
- 27.Berer K, Gerdes LA, Cekanaviciute E, et al. . Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Natl Acad Sci USA 2017;114:10719–10724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rumah KR, Linden J, Fischetti VA, Vartanian T. Isolation of Clostridium perfringens type B in an individual at first clinical presentation of multiple sclerosis provides clues for environmental triggers of the disease. PLoS One 2013;8:e76359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Linden JR, Ma Y, Zhao B, et al. . Clostridium perfringens epsilon toxin causes selective death of mature oligodendrocytes and central nervous system demyelination. MBio 2015;6:e02513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cree BA, Spencer CM, Varrin-Doyer M, Baranzini SE, Zamvil SS. Gut microbiome analysis in neuromyelitis optica reveals overabundance of Clostridium perfringens. Ann Neurol 2016;80:443–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gacias M, Gaspari S, Santos PM, et al. . Microbiota-driven transcriptional changes in prefrontal cortex override genetic differences in social behavior. ELife 2016;5:pii: e13442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rumah KR, Vartanian TK, Fischetti VA. Oral multiple sclerosis drugs inhibit the in vitro growth of epsilon toxin producing gut bacterium, Clostridium perfringens. Front Cell Infect Microbiol 2017;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schad K, Nobbe S, French LE, Ballmer-Weber B. Sofa dermatitis [in German]. J Dtsch Dermatol Ges 2010;8:897–899. [DOI] [PubMed] [Google Scholar]
- 34.Kim CH. Regulation of FoxP3 regulatory T cells and Th17 cells by retinoids. Clin Dev Immunol 2008;2008:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwata M, Hirakiyama A, Eshima Y, Kagechika H, Kato C, Song SY. Retinoic acid imprints gut-homing specificity on T cells. Immunity 2004;21:527–538. [DOI] [PubMed] [Google Scholar]
- 36.Wilson ID, Nicholson JK. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res 2017;179:204–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Das A, Srinivasan M, Ghosh TS, Mande SS. Xenobiotic metabolism and gut microbiomes. PLoS One 2016;11:e0163099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol 2016;14:273–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Taguer M, Maurice CF. The complex interplay of diet, xenobiotics, and microbial metabolism in the gut: implications for clinical outcomes. Clin Pharmacol Ther 2016;99:588–599. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Anonymized data are available and will be shared on request from any qualified investigator.