Abstract
Readily accessible samples such as urine or blood are seemingly ideal for differentiating and stratifying patients; however, it has proven a daunting task to identify reliable biomarkers in such samples. Noncoding RNA holds great promise as a source of biomarkers distinguishing physiologic wellbeing or illness. Current methods to isolate and characterize RNA molecules in urine are limited. In this proof of concept study, we present a method to extract and identify small noncoding RNAs in urine. Initially, quantitative reverse transcription PCR was applied to confirm the presence of microRNAs in total RNA extracted from urine. Once the presence of micro RNA in urine was confirmed, we developed a method to scale up RNA extraction to provide adequate amounts of RNA for next generation sequence analysis. The method described in this study is applicable to detecting a broad range of small noncoding RNAs in urine; thus, they have wide applicability for health and disease analyses.
Keywords: noncoding RNA, extracellular RNA in urine, ovarian cancer, small RNA next generation sequencing, tRNA fragments
1. Introduction
Physiologically representative and accessible samples such as saliva, blood or urine have long been expected to provide a source of biomarkers with high potential for characterizing conditions of health and disease. These types of samples are referred to as liquid biopsies and may harbor circulating cells, protein, DNA, and RNA biomarkers (1). RNA is one component within these samples that was initially ignored due to its propensity for rapid degradation by ribonucleases (RNases). However, with the identification of microRNAs and their notable stability in physiologic samples, RNA has come to the forefront of readily accessible molecules for the discovery of novel biomarkers (2–4).
Noncoding RNAs (ncRNAs) found in liquid biopsies include, but are not limited to, ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), tRNA fragments (tRFs) and microRNAs. The wide variety of functions of extracellular ncRNAs are currently under investigation. For example, the tRFs, although having the unfortunate name of tRNA fragments, are actively processed from mature tRNAs and their function is beginning to be elucidated (5). While the microRNAs are already known to function in most if not all biological processes. Circulating microRNAs as serum biomarkers of health and disease have been robustly explored (6–8); however, the microRNA repertoire in urine is less studied. The majority of the early studies of RNA in urine were focused on prostate, bladder and kidney disease because these tissues would directly contribute extracellular vesicles (EVs) to urine (9,10). In many of the studies of EVs in urine, they are referred to as exosomes due to their size of approximately 40–100 nm. The term EV refers to any extracellular vesicle including but not limited to the microvesicles, ectosomes and exosomes (11). The misconception that extracellular RNAs (exRNAs) must be associated with EVs persists despite recent studies that show exRNAs may be associated with protein and lipid complexes independent of vesicles (12–14). These findings suggest that exRNA in urine may provide useful biomarkers of physiological relevance to many diseases not limited to those involving urologic disease (15).
Urine based biomarkers would be ideal for many studies because of the accessible nature of urine. Urine is readily collected in many animal models, as well as in the veterinary setting and for human health assessment. A urine-based RNA biomarker for ovarian cancer would be particularly useful due to the inaccessible nature of the ovaries and the ability of this cancer to metastasize with few symptoms (16–18). Studies of microRNAs in ovarian cancer have delineated a set of differentially expressed microRNAs that are expected to regulate key tumor suppressors such as BRCA1 (19). Such oncomirs would be expected to be over expressed in biological fluids from cancer patients and have been studied in tissue, serum, ascites fluid and urine from ovarian cancer patients (20–24). microRNAs associated with extracellular vesicles (EVs) are also found in urine from healthy volunteers (25,26). It is possible to isolate EVs from large amounts of urine by a combined method of filtration and ultracentrifugation (25,26); however, a rapid method to isolate total RNA from small amounts of urine is needed. In this article, we present a proof of concept study of the isolation of total RNA from urine allowing either quantitative reverse transcription-PCR (qRT-PCR) or next generation sequencing (NGS).
2. Materials and methods
2.1. Participants and Sample Collection
The Gynecologic Tissue and Fluid Bank (GTFB), at the University of Colorado Anschutz Medical Campus, collected urine from women undergoing gynecologic surgery. Urine was obtained from women with ovarian cancer under an IRB approved protocol (COMIRB Protocol 11–0626). Samples were provided as 1 ml aliquots of urine with information on patient age, stage and histology of the ovarian cancer. Urine samples were collected during surgery and centrifuged prior to freezing at −80 °C for long term storage. All samples were de-identified and data was analyzed under a second IRB approved protocol at the University of Minnesota (study number: 1610E97724).
2.2. RNA extraction and Quantitative RT-PCR analysis of microRNA
Urine was defrosted once and aliquoted in 100 μl aliquots and refrozen following the addition of 700 μl of Qiazol Reagent (Qiagen, Valencia, CA). Total RNA was isolated from 100 μl of urine using the miRNeasy Mini Kit (Qiagen). The isolation procedure followed the miRNeasy protocol with a few clarifications explained in more detail in Supplemental Table 1. The final elution volume for total RNA from the spin column was the minimum required, 30 μl RNase free water, to allow the maximum concentration of RNA per μl for downstream applications. The concentration and quality of extracted RNA were assessed by spectrophotometry on the NanoDrop 1000 (Thermo Scientific, Waltham, MA). However, the concentration of RNA obtained is much lower than the expected accuracy for the NanoDrop 1000; thus, in the following steps, we ignored concentration estimates and simply used the maximum template volume allowed in the protocol. For example, in the Miscript II kit (Qiagen) for a 20 μl reaction it is possible to reverse transcribe a volume of 12 μls of total RNA for cDNA preparation. Amplifiable RNA extraction and cDNA preparation were confirmed by a positive result for qRT-PCR of a small RNA compared to the cDNA water control. qRT-PCR was conducted using the miScript SYBR Green reagent (Qiagen) with a custom primer for miR-29a-3p 5′-cccTAGCACCATCTGAAATCGGTTA or miR-146a-5p 5′-ggT GAG AAC TGA ATT CCA TGG GTT. We also used the RNU6B primer available from Qiagen for qRT-PCR assays (RNU6B_13). Although in this study, we did not use an internal control for calibration of qRT-PCR results, it is important to mention that an appropriate internal control for exRNA studies is currently controversial. Several helpful studies have examined this controversy and may be of use to those working in this field (27–29).
2.3. RNA precipitation, RNA quality assessment and Illumina Mi-seq methods
Urine is a complex liquid containing EVs, protein, nucleic acids and many other metabolites (30). Tamm-Horsfell is a protein in urine that is known to form networks that trap EVs (31). In the study presented here we extracted RNA from six 100 μl aliquots of urine and then combined these in the following step to reduce the concentration of contaminating substances such as Tamm-Horsfell protein. Total RNA from 600 μl of urine (in 100 μl aliquots) was extracted using the above protocol. The 30 μl aliquots of purified RNA were combined into 1 tube for a total of 180 μls. This was extracted with addition of GenElute LPA as described by the manufacturer (Sigma-Aldrich, St. Louis, MO) as previously described (32). The use of LPA as a carrier is required because alternatives such as glycogen or yeast tRNA are isolated from biological sources and maybe contaminated with small ncRNA (32). After LPA addition, the samples were extracted with low pH phenol (Ambion 9710) and chloroform:isoamyl alcohol (49:1) with a standard ethanol precipitation using 3 M NaAcetate pH 5.2 (protocol included, in Supplemental Table 1).
2.4. Library preparation and Illumina sequencing analysis
RNA was forwarded to the UCD Genomics and Microarray Core for library construction. In the core facilities, RNA was assessed for quality on the Agilent Bioanalyzer 2100 using the Eukaryote Total RNA Pico Chip (Agilent Technol., Palo Alto, CA). RNA libraries were constructed using a volume of 5 μls of total RNA rather than the recommended concentration of RNA need to prepare the Illumina HiSeq libraries. The TruSeq Small RNA kit uses a 3′ adapter modified to target microRNAs and other small RNAs that have a 3′ hydroxyl group. Enriching for RNA with a 3’hydroxyl allows the detection of RNA that has been enzymatically cleaved by Dicer or other RNA processing enzymes. Small RNA template libraries were sequenced using NGS technology on the Illumina HiSeq2000 platform at the University of Colorado’s Genomics and Sequencing Core Facility.
2.5. miR-seq data analysis
The microRNA sequence reads were identified for known and novel microRNA sequences using the program miRDeep. We calculated the expression of the microRNA variants based on normalized read counts and tested for significant differences using ANOVA in R. Galaxy cutadapt was used for each fastq file to remove adapters. Then fastq files were imported into CLC Genomics workbench to identify and count unique small RNAs using two databases as reference (miRBase Release 19 and Homo sapiens GRCh37.57 ncrna). The individual data files of trimmed reads have been uploaded to the publicly available data base at https://doi.org/10.5281/zenodo.801484.
Additional data sets were searched from publicly available data at NCBI on the SRA website (33).
3. Results
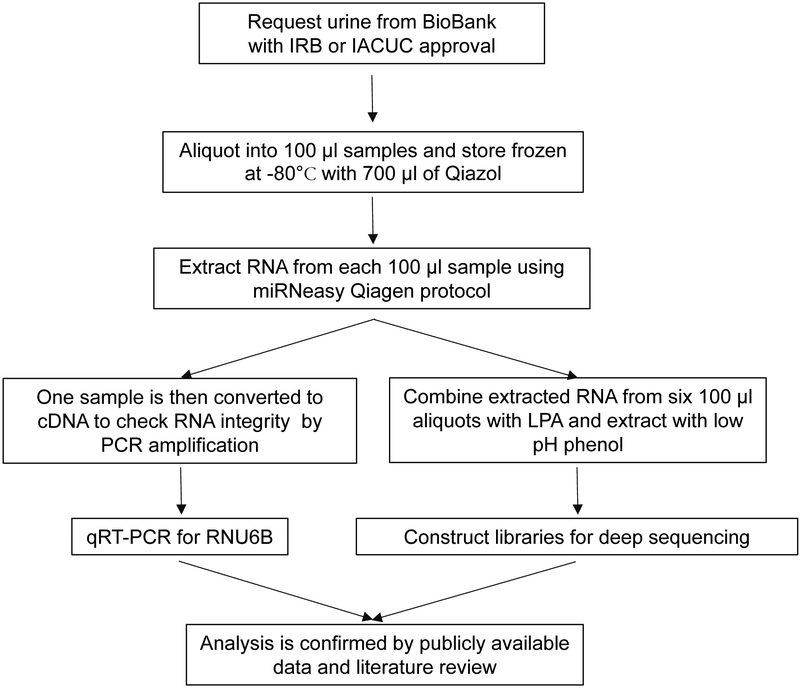
In order to identify the repertoire of ncRNAs in urine samples, we developed methods to obtain sufficient RNA for NGS. The amount of total RNA obtained from a one hundred microliter aliquot of urine provided amplifiable RNA for qRT-PCR (Table 1), however, not enough RNA for NGS. In order to collect adequate amounts of RNA for NGS, one milliliter samples of urine were obtained from the GTFB at the University of Colorado and divided into 100 μl aliquots. Each aliquot was then stored at −80 °C with 700 μl of the Phenol reagent, Qiazol for at least one hour (Fig. 1). A final concentration step using low pH phenol was used to combine six samples into one, thus allowing for an increase in total RNA extracted from the same urine sample (Fig. 1).
Table 1.
Total RNA was extracted from eight urine samples and analyzed by qRT-PCR, bioanalysis and RNA-seq. The age, stage and histology for each patient sample is included.
| Sample # | Age, stage | Histology | RNA quality (Detectable RNA) | RNA quality (RIN) | Total reads in millions |
|---|---|---|---|---|---|
| 1 | 72, IIIC | mucinous adenocarcinoma | U6, miR-29a | 2.5 | 2.6 |
| 2 | 63, IIIC | high grade serous | U6, miR-29a, miR-146a | 2.2 | 49.6 |
| 3 | 48, IIIC | high grade serous | U6, miR-29a, miR-146a | 2.6 | 6.2 |
| 4 | 67, IV | high grade serous | U6 | 2.5 | 2.7 |
| 5 | 72, IV | high grade serous | U6 | 1 | 1.6 |
| 6 | 57, IIIC | high grade serous | U6, miR-29a, miR-146a | 2.5 | 5.8 |
| 7 | 48, IIC | high grade serous | U6 | Less than 1.0 | 4.8 |
| 8 | 72, IIIC | high grade serous | U6 | 1.1 | 2.5 |
Fig. 1.
Suggested workflow for ncRNA extraction and analysis from urine obtained from patients or animal models. Notes of specific clarification of the method are included in Supplemental Table 1 (a–d).
RNA extracted from eight samples was first assessed for the presence of small RNAs by qRT-PCR for either U6, miR-29a or miR-146a. If the sample were positive by qRT-PCR, the sample was then submitted for library preparation regardless of its RNA integrity number (RIN). Later, the total number of reads were compared across the eight samples and it was found that the total number of reads varied widely (Table 1).
The libraries of ncRNAs in the urine from eight samples consisted of a wide range of RNA molecules including, but not limited to, ribosomal RNAs (rRNA), microRNAs and tRNA fragments (tRFs). The combined NGS results from all eight samples are shown in Fig. 2A, which is derived from the total annotated reads of ncRNA and shown as the percent of microRNAs, tRFs and other ncRNAs for all libraries. In Fig. 2B the three categories are shown as percentage of total annotated reads in each individual library.
Fig. 2.
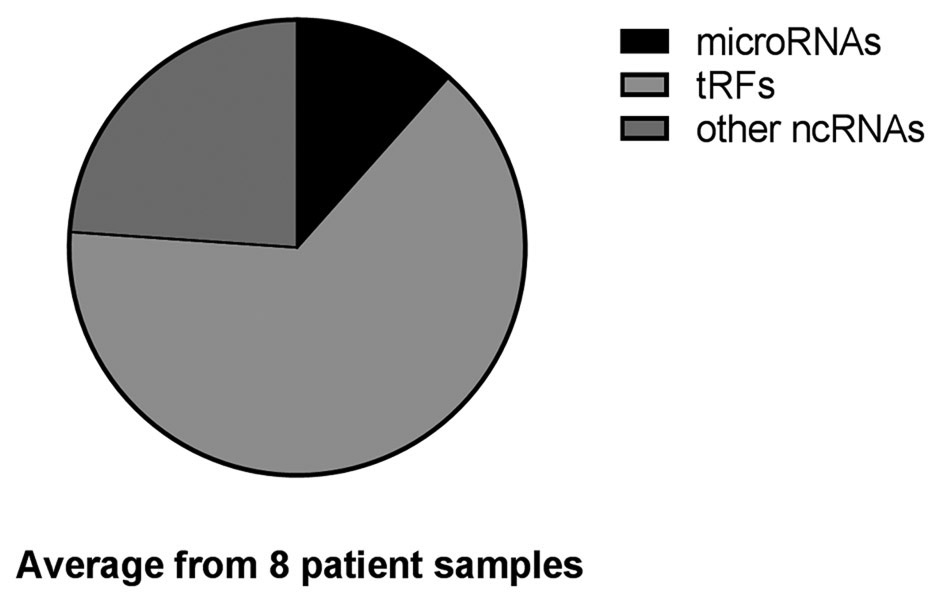
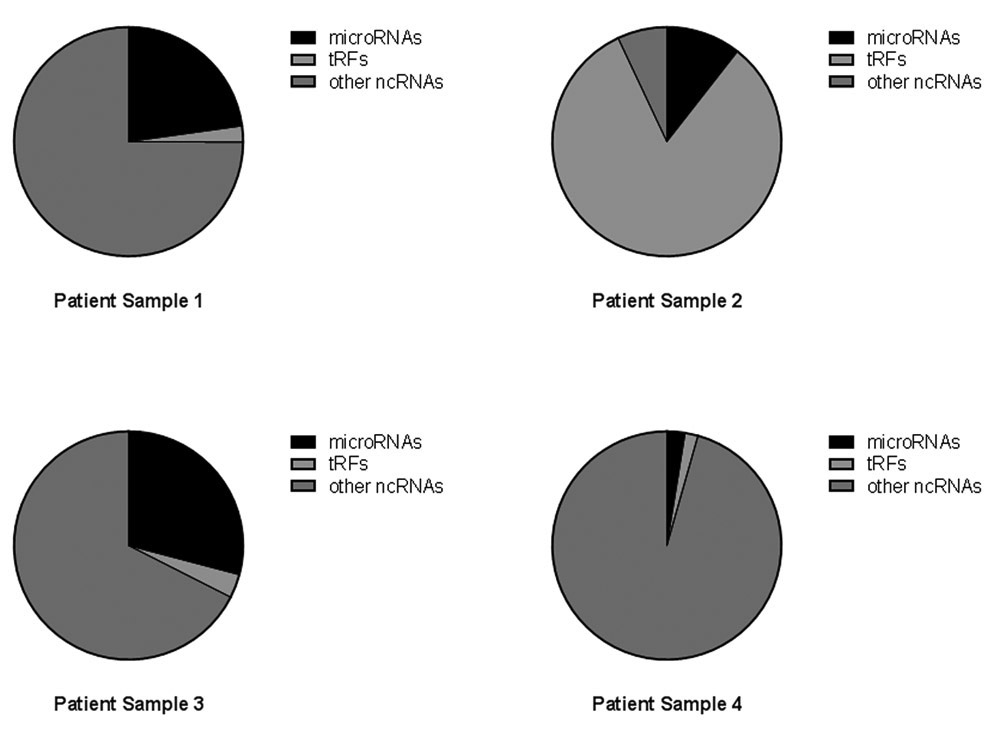
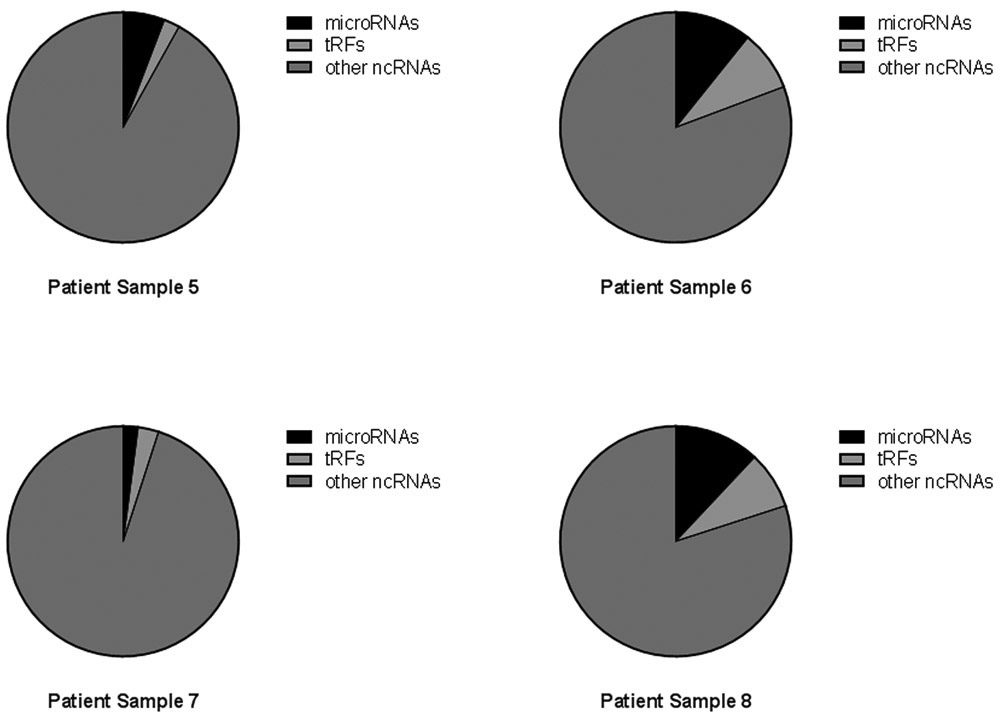
Percentage of noncoding RNA. Reads from urine classified as microRNA, tRFs or other ncRNAs. (A) Total reads from 8 urine samples summarized by percent of reads from each sample, microRNAs (black), tRFs (light grey) and other noncoding RNAs (dark grey). (B) The percentage of reads for each individual sample again with microRNAs (black), tRFs (light grey) and other noncoding RNAs (dark grey).
The overall repertoire of noncoding RNAs varied across samples, with a predominant number of reads falling into the “other ncRNAs” category, although one sample had a higher percentage of tRFs (Fig. 2B, sample 2). The percent of microRNAs varied widely from a few percent to more than 25% (Supplemental Table 2). To better understand the classification of noncoding RNA in the urine samples, we queried the eight samples for the five most highly expressed noncoding RNAs in each library and limited the analysis to those RNAs that were annotated in the GRCH37.57. ncrna data base (Table 2).
Table 2.
The top 5 ncRNAs in each of the urine samples as determined by percent of total annotated reads. The Ensembl genome number from GRCH37.57. ncrna is included as well as a sequence name when available.
| Patient Samples #╲Top5 ncRNAs | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | ENST00000466130 (rRNA pseudo gene) | miR-10b | ENST00000474885 ENST00000459522 (5.8S ribosomal 1) | ENST00000493956 (rRNA pseudo gene) | ENST00000489202 (rRNA pseudo gene) |
| 2 | ENST00000483476 (tRF) | ENST00000486830 (tRF) | miR-10b | miR-30a | miR-10a |
| 3 | ENST00000474885 ENST00000459522 (5.8S ribosomal 1) | miR-10b | ENST00000489202 (rRNA pseudo gene) | miR-10a | ENST00000493956 (rRNA pseudo gene) |
| 4 | ENST00000466130 (rRNA pseudo gene) | ENST00000474885 ENST00000459522 (5.8S ribosomal 1) | ENST00000496481 (rRNA pseudo gene) | ENST00000493956 (rRNA pseudo gene) | ENST00000492060 (rRNA pseudo gene) |
| 5 | ENST00000493956 (rRNA pseudo gene) | ENST00000479524 (rRNA pseudo gene) | ENST00000474870 (rRNA pseudo gene) | ENST00000474075 (rRNA pseudo gene) | ENST00000463737 (rRNA pseudo gene) |
| 6 | ENST00000466130 (rRNA pseudo gene) | ENST00000474885 ENST00000459522 (5.8S ribosomal 1) | ENST00000483476 (tRF) | ENST00000486830 (tRF) | miR-10b |
| 7 | ENST00000493956 (rRNA pseudo gene) | ENST00000479524 (rRNA pseudo gene) | ENST00000463737 (rRNA pseudo gene) | ENST00000476674 (rRNA pseudo gene) | ENST00000459949 (rRNA pseudo gene) |
| 8 | ENST00000493956 (rRNA pseudo gene) | ENST00000479524 (rRNA pseudo gene) | miR-10b | ENST00000466130 (rRNA pseudo gene) | ENST00000474870 (rRNA pseudo gene) |
The most highly expressed small RNAs were generally fragments related to rRNAs and pseudo rRNAs. Very little is known about the function of extracellular rRNAs and pseudo rRNAs; however, they have been proposed to be processed in response to stress, including oxidative stress (34,35).
Among the noncoding RNAs, microRNAs are currently the most well studied group in urine. Overall analysis of the microRNAs in the eight samples revealed that miR-10b was by far the most highly expressed microRNA in all eight samples followed by miR-10a in five of the eight samples. Other microRNAs that were second to the highest in at least one patient were miR-22 and miR-30a. Interestingly, a microRNA often associated with oncogenesis, miR-21, was only in the top five in one sample (Table 3).
Table 3.
The top 5 microRNAs in each of the urine samples as determined by percent of total annotated reads.
| Patient Sample #╲Top5 microRNAs | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | miR-10b | miR-22 | miR-10a | miR-205 | miR-30a |
| 2 | miR-10b | miR-30a | miR-10a | miR-21 | miR-22 |
| 3 | miR-10b | miR-10a | miR-22 | miR-205 | miR-30d |
| 4 | miR-10b | miR-10a | miR-30a | miR-22 | miR-205 |
| 5 | miR-10b | miR-10a | miR-92a-1 | miR-92a-2 | miR-203a |
| 6 | miR-10b | miR-10a | miR-22 | miR-205 | miR-30d |
| 7 | miR-10b | miR-10a | miR-30a | miR-204 | miR-4454 |
| 8 | miR-10b | miR-10a | miR-30a | miR-204 | miR-22 |
The most highly expressed microRNA in this study, miR-10b, is not well studied in ovarian cancer, although it has recently been implicated in metastasis in other cancer sites (36). Based on a literature review we expected to find miR-146a and miR-29a in urine due to their previous association with ovarian cancer and previous findings of their presence in exRNA samples (23,24,37,38). Neither of these microRNAs were highly expressed in any of the eight samples as detected by NGS.
A large group of reads in the NGS studies derived from tRNAs and belonged to the group of functional noncoding RNAs known as tRFs (39). The small RNA reads in this study were mapped against the known human tRNAs using the GtRNAdb data base (40,41). The most highly expressed tRFs in the eight samples are reported in Table 4.
Table 4.
The top 5 tRFs in each of the urine samples as determined by percent of total annotated reads.
| Patient Sample #╲Top5 tRF | 1 | 2 | 3 | 4 | 5 |
|---|---|---|---|---|---|
| 1 | Glu | Gly | Lys | Val | His |
| 2 | Gly | Glu | Val | Lys | His |
| 3 | Gly | Glu | Val | Lys | His |
| 4 | Glu | Gly | Lys | Val | His |
| 5 | Glu | Gly | Lys | Val | His |
| 6 | Gly | Glu | Val | Lys | His |
| 7 | Gly | Glu | Lys | Val | His |
| 8 | Gly | Glu | Lys | Val | His |
Highly expressed tRFs in the urine included the tRNA halves as well as a large number of variants differing by only a few bases. One tRF variant of the tRNA halve for tRNA-Glu-CTC (miR-2476) was of particular interest because it had been previously reported as a microRNA in the cow. The miR-2476 had not been previously described in human samples; further, it has been removed from the microRNA data base because of its location near a tRNA, tRF5-Glu-CTC (42). The tRF5-Glu-CTC variant that was previously called miR-2476 is missing a guanine at the sixth base (Table 5A, named tRF5-Glu 1Gv). We examined several NGS studies available at GenBank to determine if this variant of tRF5-Glu-CTC had been observed in other human studies. We queried the publicly available data for the presence of tRF5-Glu 1Gv as well as the most prevalent variant from our urine analysis called tRF5-Glu 2Gv (Table 5B). One study of particular interest, where both variants were observed, was a comparison of the RNA cargo in exosomes isolated from the media of ovarian cancer cells, ovarian cancer associated adipocytes, normal adipocytes, cancer associated fibroblasts and normal fibroblasts (33).
Table 5.
The expression of tRF5-Glu-CTC and its variants in urine and human samples. (A) The variants detected by NGS in all eight urine samples. The average in all eight samples is given followed by the range of expression. The length in base pairs refers to the length of the variant. The entire length of tRNA-Glu-CTC-1-1 is included for comparison. (B) The two most prevalent variants of tRF5-Glu-CTC are presented here along with the SRA accession number and type of sample.
| Sequence of variants | Average in 8 tumors (range) |
Length in base pairs |
|---|---|---|
| TCCCTG-TGGTCTAGTGGTTAGGATTCGGC | 2926 (14, 22967) | 29 |
| TCCCTGGTGGTCTAGTGGTTAGGATTCGGC | 4555 (51, 35617) | 30 |
| TCCCTGGTGGTCTAGTGGTTAGGATTCGGCG | 4044 (17, 31848) | 31 |
| TCCCTGGTGGTCTAGTGGTTAGGATTCGGCGC | 7064 (37, 49667) | 32 |
| TCCCTGGTGGTCTAGTGGTTAGGATTCGGCGCT | 7326 (144, 40615) | 33 |
| TCCCTGGTGGTCTAGTGGTTAGGATTCGGCGCTC | 2079 (70, 12402) | 34 |
| TCCCTGGTGGTCTAGTGGTTAGGATTCGGCGCTCT | 850 (58, 4303) | 35 |
| TCCCTGGTGGTCTAGTGGTTAGGATTCGGCGCTCTC | 452 (10, 2463) | 36 |
| tRNA-Glu-CTC-1–1 | ||
| TCCCTGGTGGTCTAGTGGTTAGGATTCGGCGCTCTCACCGCCGCGGCCCGGGTTCGATTCCCGGTCAGGGAA | ||
The variants of tRF5-Glu (both 1Gv and 2Gv) were found in all 13 samples, with 2Gv being more highly expressed in most samples. The increased expression of the 2Gv is similar to our finding in urine from ovarian cancer patients (Table 5A). Both variants were highly expressed in cancer adipocytes as compared to the other samples. The ovarian carcinoma derived cell line A2780 had increased expression compared to the other three cell lines (43). The top five tRFs from this study were also analyzed for their expression in data bases available at NCBI in the SRA collection (Supplemental Table 3). In this expanded analysis tRF5-Glu was by far the most highly expressed tRF from urinary exosomes. The use of publicly available data to verify the existence of specific variants of tRFs provides a readily available resource to the research community for follow-up studies of NGS analysis. Confirmation of variant expression by their presence in additional publicly available samples may guide the choice for further study of specific exRNAs.
4. Discussion
Liquid biopsies utilizing novel and robust ncRNA targets hold great potential for the development of new biomarkers (4,44). High throughput sequencing studies have fueled the search for novel ncRNA biomarkers from accessible samples including serum, plasma and urine (3,24). The unexpected stability of exRNA and new technologies to deep sequence RNA have led to an explosion of new biomarker studies focusing on ncRNAs in accessible samples (1–4). RNA is a preferred molecule for biomarker discovery in liquid biopsies due to its representation of the physiological state of the organism (44,45). RNA as a potential biomarker in urine was recognized as early as the 1970s when tRNAs were detected at increased levels in urine from cancer patients (46). In these early studies, it was not possible to distinguish the specific noncoding RNA in urine, while it is now feasible with the advent of RNA-seq technologies and the bioinformatics tools to analyze the vast amount of information.
Methods to extract exRNA from accessible samples are widely ranging and varied in the ability to capture amplifiable RNA (11,47). It was initially thought that RNA was only stable in urine if it were protected as cargo in exosomes from bladder and urinary tract cells or from the kidney if there is damage. Thus, studies of exRNA in urine exosomes were initially expected to favor the isolation of ncRNA from the bladder. However, it is now increasingly accepted that exRNA in urine can also come from other tissue sites as well as tumors (48–50).
Many of the recent studies aimed at identifying exRNA in urine through NGS protocols are focused on the ncRNA found in EVs and including the subset known as exosomes (47). The extraction of RNA from urinary extracellular vesicles requires a large volume of urine to ensure that enough RNA will be extracted for downstream applications. The suggested volume of urine for NGS biomarker discovery protocols ranges anywhere from 5 mls to 250 mls of urine (25–27,47). Furthermore, the collection of exosomes requires either ultracentrifugation or treatment with a proprietary method such as the Exosome RNA Isolation kit from Norgen (Norgen Biotek Corporation, Thorold, ON, Canada) to concentrate the exosomes (27). Studies of the repertoire of exRNA associated with EVs and specifically exosomal RNA provide a partial understanding of the exRNA present in urine. Methods to extract total RNA from urine are also needed.
Methods designed to conduct NGS on total RNA extracted from urine are less common than exosomal studies. One such study of total RNA extracted from the urine of male goats utilized 500 μl samples successfully to accomplish NGS (51). Other studies based on the extraction of total RNA from a small volume of urine did not attempt NGS, rather these methods were used for microRNA Array studies or qRT-PCR and required as little as 50 μls for analysis (52). The methods we describe in this article require similar volumes of urine and allow the isolation and analysis of total RNA by both qRT-PCR and NGS. Once biomarkers are discovered in pilot studies such as these, it will be possible to scale up to larger studies and develop methods with direct clinical utility.
In order to determine the full spectrum of exRNA in urine, patient samples were requested from the GTFB at the University of Colorado. These samples had been obtained with informed consent and had all been collected during surgery and handled in a consistent manner prior to storage at −80 °C. Consistent collection and storage of samples is required for all studies of exRNA and the importance of collection and storage has been described previously (11,53). Ovarian cancer is a very difficult disease to detect and frequently is found at late stage; thus, one goal of the GTFB is to provide a resource for studies to discover new biomarkers for patient care. In this pilot study, we analyzed the microRNAs and tRFs and grouped the rRNA fragments with all other noncoding RNAs (Fig. 2). The total tRFs, microRNAs and other noncoding RNAs were first analyzed for all samples by percent of total annotated reads. Combining the reads from all samples gives an overview of the types of RNAs in ovarian cancer patient urine (Fig. 2A). However, the grouping of all reads is misleading as individual samples were very different from each other (Fig. 2B). The variability of classes of RNA in patient samples suggests a potential to discover unique biomarkers in urine. Although this proof of concept project provides methods to identify biomarkers in urine, a larger study with the goal of discovering specific biomarkers for ovarian cancer patients will be required.
The top five most highly expressed exRNAs in most samples were dominated by the group of other noncoding RNAs; however, miR-10b, miR-10a and miR-30a were also found in this group (Table 2). The predominant microRNA was by far miR-10b, which was the top microRNA in all samples (Table 3). miR-10a was also one of the top five in all samples and in many samples miR-22 and miR-30a were frequently observed. miR-21, a microRNA reported to be highly expressed in cancer patients, was only observed among the top five molecules in one sample (Table 3). Although the goal of this pilot study was to develop methods to study RNA biomarkers in patient urine, it will be interesting in future studies to determine if there is a role for miR-10a and 10b in ovarian cancer. A literature review revealed that few previous studies of miR-10 in ovarian cancer have been conducted. However, the role of miR-10 has been elucidated in normal granulosa cells of the ovary and is linked to feedback regulation of the TGF-β signaling pathway (54). We also observed the presence of miR-10a and 10b in publicly available data of normal fibroblasts and cancer associated fibroblasts, where the miR-10 family was equally expressed in both normal and cancer associated fibroblasts (33). A group of microRNAs expressed in ovarian cancer has been suggested to form a signature predictive of poor outcome in ovarian cancer patients (55); however, the only microRNA included in that signature that is also detected in the top five microRNAs in this study was miR-30d (Table 3). The role of microRNAs in ovarian cancer is still being investigated as is the role of tRFs. These exRNAs are expected to provide new physiological relevant biomarkers for ovarian cancer.
exRNA contains a complex variety of noncoding RNAs, including numerous tRFs and their variants whose complexity is just now being realized (35,56–59). The complexity of tRF expression is in part due to the frequent modification of nucleosides in the mature tRNA, which may interfere with tRF detection in high-throughput sequencing studies (60). In addition to high throughput sequencing, additional methods including qRT-PCR and Northern analysis are routinely used to identify tRFs. However, tRFs are derived from pre- and mature tRNAs making them difficult to distinguish by established assays. Honda et al. have developed methods to specifically analyze tRFs using ligation PCR (61). Future studies aimed at applying tRFs as biomarkers in patient samples will require the development of methods specific to tRFs.
In order to confirm the use of NGS of RNA from urine as a potential method of discovering new biomarkers, we chose one tRF, tRF5-Glu-CTC, for further study. We examined publicly available databases of exRNA isolated from exosomes to determine if tRF5-Glu and its variants are detectable in exRNA from extracellular vesicles (Table 5B). Libraries of exRNA isolated from exosomes obtained from ovarian cancer cell lines, cancer associated adipocytes, normal adipocytes, cancer associated fibroblasts and normal fibroblasts were queried for the presence of tRF5-Glu-CTC (33). The predominant variants from the urine samples were also detected in the RNA from exosomes (Table 5). The use of publicly available data to confirm the expression of previously unidentified exRNAs provides an additional resource to study the numerous exRNAs in urine. The methods described here are readily adapted to other species and other disease conditions for the future development of physiologically relevant urine-based biomarkers.
Supplementary Material
| Accession number |
tRF5-Glu 1Gv |
tRF5-Glu 2Gv |
tRF5-Glu 1Gv percentage |
tRF5-Glu 2Gv percentage |
Total reads | |
|---|---|---|---|---|---|---|
| Ovarian cancer cell line A2780 | SRX1550574 | 186 | 1439 | 0.087% | 0.670% | 214630 |
| Ovarian cancer cell line HeyA8 | SRX1550575 | 43 | 105 | 0.015% | 0.037% | 281330 |
| Ovarian cancer cell line OVCA433 | SRX1550576 | 32 | 114 | 0.009% | 0.034% | 338204 |
| Ovarian cancer cell line SKOV3 | SRX1550577 | 5 | 25 | 0.002% | 0.010% | 256003 |
| Ovarian cancer adipocytes OMT924007 | SRX1550585 | 8065 | 10261 | 1.748% | 2.224% | 461313 |
| Ovarian cancer adipocytes OMT916645 | SRX1550586 | 1901 | 1380 | 0.812% | 0.589% | 234100 |
| Cancer associated fibroblasts CAF866652 | SRX1550580 | 20 | 1113 | 0.007% | 0.385% | 288787 |
| Cancer associated fibroblasts CAF869881 | SRX1550581 | 116 | 293 | 0.027% | 0.068% | 427929 |
| Cancer associated fibroblasts CAF888242 | SRX1550582 | 111 | 523 | 0.023% | 0.108% | 485258 |
| Normal ovarian fibroblasts NOF151 | SRX1550578 | 60 | 435 | 0.020% | 0.143% | 305013 |
| Normal ovarian fibroblasts NOF81000 | SRX1550579 | 76 | 254 | 0.023% | 0.078% | 324908 |
| Normal omental adipocytes OMN050312 | SRX1550583 | 1473 | 35 | 0.434% | 0.010% | 339618 |
| Normal omental adipocytes OMN923075 | SRX1550584 | 84 | 97 | 0.031% | 0.036% | 269810 |
| tRF5-Glu 1Gv, tRF5-Glu 1G variant (CCCTG-TGGTCTAGTGGTTAGGATTCGGC) | ||||||
| tRF5-Glu 2Gv, tRF5-Glu most prevalent 2G variant (CCCTGGTGGTCTAGTGGTTAGGATTCGGCGCT) | ||||||
Acknowledgments
Funding for this study was provided by the HERA Foundation to MAS and LTB. Funding was also provided by the Whiteside Institute to LTB.
Abbreviations used:
- ncRNA
noncoding RNA
- exRNA
extracellular RNA
- EVs
extracellular vesicles
- tRNA
transfer RNA
- tRF
tRNA fragment
- rRNA
ribosomal microRNA
- qRT-PCR
quantitative RT-PCR
- NGS
next generation sequencing
- miRNA
microRNA
- RNases
ribonucleases
References
- (1).Perakis S, Speicher MR, Emerging concepts in liquid biopsies, BMC Med 15 (2017) 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, CirculatingmicroRNAs as stable blood-based markers for cancer detection, Proc. Natl.Acad. Sci 105 (2008) 10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Weber JA, Baxter DH, Zhang S, Huang DY, Huang KH, Lee MJ, Galas DJ, Wang K, The microRNA spectrum in 12 body fluids, Clin. Chem 56 (2010) 1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Fernandez-Mercado M, Manterola L, Larrea E, Goicoechea I, Arestin M, Armesto M, Otaegui D, Lawrie CH, The circulating transcriptome as a source of non-invasive cancer biomarkers: concepts and controversies of non-coding and coding RNA in body fluids, J. Cell. Mol. Med 19 (2015) 2307–2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Zhou J, Liu S, Chen Y, Fu Y, Silver AJ, Hill MS, Lee I, Lee YS, Bao X, Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection, J. Gen. Virol 98 (2017) 1600–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Nair VS, Pritchard CC, Tewari M, Ioannidis J, Design and analysis for studying microRNAs in human disease: a primer on-omic technologies, Am. J. Epidemiol 180 (2014) 140–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Velu VK, Ramesh R, Srinivasan A, Circulating microRNAs as biomarkers in health and disease, J. Clin. Diagn Res 6 (2012) 1791–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).de Candia P, Torri A, Pagani M, Abrignani S, Serum microRNAs as biomarkers of human lymphocyte activation in health and disease, Front. Immunol 5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Snowdon J, Boag S, Feilotter H, Izard J, Siemens DR, A pilot study of urinary microRNA as a biomarker for urothelial cancer, Can. Urol. Assoc. J 7 (2013) 28–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Huang X, Liang M, Dittmar R, Wang L, Extracellular microRNAs in urologic malignancies: chances and challenges, Int. J. Mol. Sci 14 (2013) 14785–14799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Nolte EN, Piper MG, Sivaraman S, Skog J, Standardization of sample collection, isolation and analysis methods in extracellular vesicle research, J. Extracell. Vesicles 2 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT, MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins, Nat. Cell Biol 13 (2011) 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma, Proc. Natl. Acad. Sci 108 (2011) 5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Wang K, Zhang S, Weber J, Baxter D, Galas DJ, Export of microRNAs and microRNA-protective protein by mammalian cells, Nucleic Acids Res (2010) gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Matullo G, Naccarati A, Pardini B, MicroRNA expression profiling in bladder cancer: the challenge of next-generation sequencing in tissues and biofluids, Int. J. Cancer 138 (2016) 2334–2345. [DOI] [PubMed] [Google Scholar]
- (16).E. National Academies of Sciences, Medicine, Ovarian Cancers: Evolving Paradigms in Research and Care, National Academies Press, 2016. [PubMed] [Google Scholar]
- (17).Levine DA, Karlan BY, Strauss JF, Evolving approaches in research and care for ovarian cancers: a report from the National Academies of Sciences, Engineering, and Medicine, Jama 315 (2016) 1943–1944. [DOI] [PubMed] [Google Scholar]
- (18).Konecny GE, Winterhoff B, Wang C, Gene-expression signatures in ovarian cancer: promise and challenges for patient stratification, Gynecol. Oncol 141 (2016) 379–385. [DOI] [PubMed] [Google Scholar]
- (19).Shen J, Ambrosone CB, DiCioccio RA, Odunsi K, Lele SB, Zhao H, A functional polymorphism in the miR-146a gene and age of familial breast/ ovarian cancer diagnosis, Carcinogenesis 29 (2008) 1963–1966. [DOI] [PubMed] [Google Scholar]
- (20).Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu C-G, Alder H, MicroRNA signatures in human ovarian cancer, Cancer Res 67 (2007) 8699–8707. [DOI] [PubMed] [Google Scholar]
- (21).Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O’Briant K, Godwin AK, Urban N, Drescher CW, Knudsen BS, Tewari M, Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries, PLoS One 4 (2009) e5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Taylor DD, Gercel-Taylor C, MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer, Gynecol. Oncol 110 (2008) 13–21. [DOI] [PubMed] [Google Scholar]
- (23).Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE, The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform, Gynecol. Oncol 112 (2009) 55–59. [DOI] [PubMed] [Google Scholar]
- (24).Nakamura K, Sawada K, Yoshimura A, Kinose Y, Nakatsuka E, Kimura T, Clinical relevance of circulating cell-free microRNAs in ovarian cancer, Mol. Cancer 15 (2016) 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Cheng L, Sun X, Scicluna BJ, Coleman BM, Hill AF, Characterization and deep sequencing analysis of exosomal and non-exosomal miRNA in human urine, Kidney Int 86 (2014) 433–444. [DOI] [PubMed] [Google Scholar]
- (26).Ben-Dov IZ, Whalen VM, Goilav B, Max KE, Tuschl T, Cell and microvesicle urine microRNA deep sequencing profiles from healthy individuals: observations with potential impact on biomarker studies, PLoS One 11 (2016) e0147249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Crossland RE, Norden J, Bibby LA, Davis J, Dickinson AM, Evaluation of optimal extracellular vesicle small RNA isolation and qRT-PCR normalisation for serum and urine, J. Immunol. Meth 429 (2016) 39–49. [DOI] [PubMed] [Google Scholar]
- (28).Zen K, Zhang CY, Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers, Med. Res. Rev 32 (2012) 326–348. [DOI] [PubMed] [Google Scholar]
- (29).Xiang M, Zeng Y, Yang R, Xu H, Chen Z, Zhong J, Xie H, Xu Y, Zeng X, U6 is not a suitable endogenous control for the quantification of circulating microRNAs, Biochem. Biophys. Res. Commun 454 (2014) 210–214. [DOI] [PubMed] [Google Scholar]
- (30).Lin SY, Linehan JA, Wilson TG, Hoon DS, Emerging utility of urinary cell-free nucleic acid biomarkers for prostate, bladder, and renal cancers, Eur. Urol. Focus (2017) (in press). [DOI] [PubMed] [Google Scholar]
- (31).Fernandez-Llama P, Khositseth S, Gonzales PA, Star RA, Pisitkun T, Knepper MA, Tamm-Horsfall protein and urinary exosome isolation, Kidney Int 77 (2010) 736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Bartram AK, Poon C, Neufeld JD, Nucleic acid contamination of glycogen used in nucleic acid precipitation and assessment of linear polyacrylamide as an alternative co-precipitant, Biotechniques 47 (2009) 4. [DOI] [PubMed] [Google Scholar]
- (33).Yeung CLA, Co N-N, Tsuruga T, Yeung T-L, Kwan S-Y, Leung CS, Li Y, Lu ES, Kwan K, Wong K-K, Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1, Nat. Commun 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Li Z, Ender C, Meister G, Moore PS, Chang Y, John B, Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs, Nucleic Acids Res (2012) gks307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Thompson DM, Lu C, Green PJ, Parker R, tRNA cleavage is a conserved response to oxidative stress in eukaryotes, Rna 14 (2008) 2095–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Cao M.-x., Jiang Y.-p., Tang Y.-l., Liang X.-h., The crosstalk between lncRNA and microRNA in cancer metastasis: orchestrating the epithelial-mesenchymal plasticity, Oncotarget 8 (2017) 12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, Lidereau R, Mikaelian I, Mazoyer S, Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers, EMBO Mol. Med 3 (2011) 279–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Yu PN, Yan MD, Lai HC, Huang RL, Chou YC, Lin WC, Yeh LT, Lin YW, Downregulation of miR-29 contributes to cisplatin resistance of ovarian cancer cells, Int. J. Cancer 134 (2014) 542–551. [DOI] [PubMed] [Google Scholar]
- (39).Diebel KW, Zhou K, Clarke AB, Bemis LT, Beyond the ribosome: extratranslational functions of tRNA fragments, Biomark. Insights 11 (2016) 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Chan PP, Lowe TM, GtRNAdb: a database of transfer RNA genes detected in genomic sequence, Nucleic Acids Res 37 (2009) D93–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Lowe TM, Chan PP, tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes, Nucleic Acids Res 44 (2016) W54–W57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Kozomara A, Griffiths-Jones S, miRBase: annotating high confidence micro- RNAs using deep sequencing data, Nucleic Acids Res 42 (2014) D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Eva A, Robbins KC, Andersen PR, Srinivasan A, Tronick SR, Reddy EP, Ellmore NW, Galen AT, Lautenberger JA, Papas TS, Cellular genes analogous to retroviral onc genes are transcribed in human tumour cells, Nature 295 (1982) 116e119. [DOI] [PubMed] [Google Scholar]
- (44).Buschmann D, Haberberger A, Kirchner B, Spornraft M, Riedmaier I, Schelling G, Pfaffl MW, Toward reliable biomarker signatures in the age of liquid biopsies-how to standardize the small RNA-Seq workflow, Nucleic Acids Res 44 (2016) 5995–6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Seifert VA, Clarke BL, Crossland JP, Bemis LT, A method to distinguish morphologically similar Peromyscus species using extracellular RNA and high-resolution melt analysis, Anal. Biochem 508 (2016) 65–72. [DOI] [PubMed] [Google Scholar]
- (46).Borek E, Baliga B, Gehrke CW, Kuo C, Belman S, Troll W, Waalkes TP, High turnover rate of transfer RNA in tumor tissue, Cancer Res 37 (1977) 3362–3366. [PubMed] [Google Scholar]
- (47).Tataruch-Weinert D, Musante L, Kretz O, Holthofer H, Urinary extracellular vesicles for RNA extraction: optimization of a protocol devoid of prokaryote contamination, J. Extracell. Vesicles 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Gonzalez E, Falcon-Perez JM, Cell-derived extracellular vesicles as a platform to identify low-invasive disease biomarkers, Expert Rev. Mol. Diagnostics 15 (2015) 907–923. [DOI] [PubMed] [Google Scholar]
- (49).Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield X, Widmark A, Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer, Br. J. Cancer 100 (2009) 1603–1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Cheng Y, Wang X, Yang J, Duan X, Yao Y, Shi X, Chen Z, Fan Z, Liu X, Qin S, A translational study of urine miRNAs in acute myocardial infarction, J. Mol. Cell. Cardiol 53 (2012) 668–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Longpre KM, Kinstlinger NS, Mead EA, Wang Y, Thekkumthala AP, Carreno KA, Hot A, Keefer JM, Tully L, Katz LS, Seasonal variation of urinary microRNA expression in male goats (Capra hircus) as assessed by next generation sequencing, Gen. Comp. Endocrinol 199 (2014) 1–15. [DOI] [PubMed] [Google Scholar]
- (52).Lorenzen J, Volkmann I, Fiedler J, Schmidt M, Scheffner I, Haller H, Gwinner W, Thum T, Urinary miR-210 as a mediator of acute T-cell mediated rejection in renal allograft recipients, Am. J. Transplant 11 (2011) 2221–2227. [DOI] [PubMed] [Google Scholar]
- (53).Witwer KW, Circulating microRNA biomarker studies: pitfalls and potential solutions, Clin. Chem 61 (2015) 56–63. [DOI] [PubMed] [Google Scholar]
- (54).Jiajie T, Yanzhou Y, Hoi-Hung AC, Zi-Jiang C, Wai-Yee C, Conserved miR-10 family represses proliferation and induces apoptosis in ovarian granulosa cells, Sci. Rep 7 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Bagnoli M, Canevari S, Califano D, Losito S, Di Maio M, Raspagliesi F, Carcangiu ML, Toffoli G, Cecchin E, Sorio R, Development and validation of a microRNA-based signature (MiROvaR) to predict early relapse or progression of epithelial ovarian cancer: a cohort study, Lancet Oncol 17 (2016) 1137–1146. [DOI] [PubMed] [Google Scholar]
- (56).Lee YS, Shibata Y, Malhotra A, Dutta A, A novel class of small RNAs: tRNA-derived RNA fragments (tRFs), Genes Dev 23 (2009) 2639–2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Gebetsberger J, Polacek N, Slicing tRNAs to boost functional ncRNA diversity, RNA Biol 10 (2013) 1798–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Telonis AG, Loher P, Honda S, Jing Y, Palazzo J, Kirino Y, Rigoutsos I, Dissecting tRNA-derived fragment complexities using personalized transcriptomes reveals novel fragment classes and unexpected dependencies, Oncotarget 6 (2015) 24797–24822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Kumar P, Mudunuri SB, Anaya J, Dutta A, tRFdb: a database for transfer RNA fragments, Nucleic Acids Res 43 (2015) D141–D145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM, ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments, Nat. Meth 12 (2015) 879–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Honda S, Loher P, Shigematsu M, Palazzo JP, Suzuki R, Imoto I, Rigoutsos I, Kirino Y, Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers, Proc. Natl. Acad. Sci 112 (2015) E3816–E3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.