Abstract
Because phenotypic innovations must be genetically heritable for biological evolution to proceed, it is natural to consider new mutation events as well as standing genetic variation as sources for their birth. Previous research has identified a number of single-nucleotide polymorphisms that underlie a subset of adaptive traits in organisms. However, another well-known class of variation, genomic structural variation, could have even greater potential to produce adaptive phenotypes, due to the variety of possible types of alterations (deletions, insertions, duplications, among others) at different genomic positions and with variable lengths. It is from these dramatic genomic alterations, and selection on their phenotypic consequences, that adaptations leading to biological diversification could be derived. In this review, using studies in humans and other mammals, we highlight examples of how phenotypic variation from structural variants might become adaptive in populations and potentially enable biological diversification. Phenotypic change arising from structural variants will be described according to their immediate effect on organismal metabolic processes, immunological response and physical features. Study of population dynamics of segregating structural variation can therefore provide a window into understanding current and historical biological diversification.
Keywords: genomic structural variation, copy number variation, adaptive variant, evolution, nonpathogenic phenotype, segregating polymorphisms
Introduction
Single-nucleotide polymorphisms (SNPs) introduce only single DNA base pair (bp) variation, yet they have been shown to directly alter a number of non-disease phenotypes in mammals, including hair thickness [1], muscle mass [2–4] and locomotion [5], among others. Attempts to determine how SNPs affect complex trait phenotypes is now standard practice, especially in the field of human genetics [6]. Genomic structural variants (SVs) comprise another class of phenotype-shaping genetic variation that has emerged in the past decade, due in large part to the development of high-resolution technologies, such as array comparative genomic hybridization, and advanced computational tools to analyze next-generation sequence data. SVs (used broadly in this review to indicate any structural change to the length or organization of a chromosome of ≥1 bp) encompass a variety of chromosomal alterations such as deletions, insertions, duplications, inversions, translocations and transposable elements. A special subset of SVs, copy number variants (CNVs), refers to loci that differ in integer allelic copy number between individuals and was among the first classes of SVs observed to be polymorphic at many loci in human genomes [7, 8]. Unlike SNPs, which only involve substitution of one nucleotide-pair for another, SVs involve a local sequence gain or loss or reorganization. SNP analysis itself may be considered a saturated mutagenesis experiment in humans, whereby across the whole human population statistically every genomic position that does not affect viability has likely been mutagenized. With structural variation, however, this is almost certainly not the case, due to the enormous breadth of possibility for individual events, including of different SV types, genomic positions and lengths. Thus, SVs have the potential to markedly affect organismal phenotype owing to the range of possible consequences that include gene dosage variation, open reading frame alterations, transcription factor binding site modifications and gain or loss of functional genomic elements.
General surveys of SVs, particularly CNVs, have been conducted in humans [9–12] and other mammalian species [13–27]. In humans, SVs have been shown to affect more base pairs in an average individual than SNPs [28], and CNVs in particular contribute a sizeable amount (nearly 18%) of the variation in lymphoblastoid cell gene expression [29]. Additional studies in humans [30] and mice [31, 32] have confirmed this expression effect in various tissues and cell types, with one study in mice suggesting that CNVs can modify not only gene expression levels but also affect the timing of expression [33]. Multi-allelic CNVs (loci that have more than two segregating structural alleles) have been shown in humans to account for nearly 90% of the variation in gene dosage between individuals, though they represent only a small proportion of all CNVs [34]. Interestingly, copy number of certain genes can rise to >50 in some human populations [35], suggesting positive selective pressure on paralogs acting cumulatively or neofunctionalization of the duplicates, or at least reduced purifying selection. Iskow et al. [36] provide a table of many CNV genes that show apparent evidence of positive selection in humans, yet the functional output of variable copy number is often unclear. Studies in humans have also examined the genomic extent of variable pseudogenes (copies of genes, usually truncated into exon-only sequences relative to their ‘parent’ sequences) [37], and have found a subset that encode chimeric transcripts that might have the potential for cellular function [38]. Studies in humans have also examined the relationship between CNV genes (compared with all genes) and microRNAs; one study found an enrichment for both the number of microRNAs that target CNV genes as well as the number of microRNA binding sites within CNV genes [39]. Another study found that genes targeted by copy-number variable microRNAs tended to have larger expression variability and a greater likelihood of differential expression between tissues and developmental stages [40].
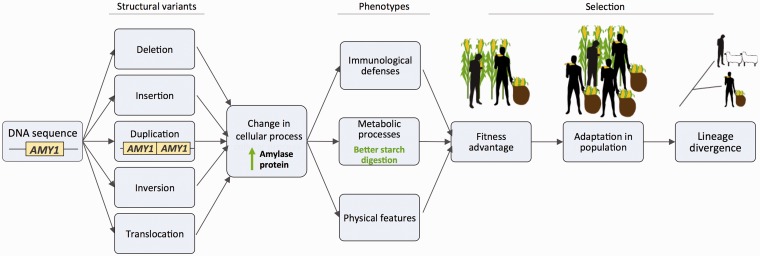
Although general survey studies of SVs are highly informative, finding differences within species at thousands of loci, these studies do not pinpoint specific SVs as leading to particular phenotypes. The vast majority of genomic studies involving particular SVs have focused on disease-associated phenotypes, especially in humans (reviewed by Weischenfeldt et al. [41]). Yet, given the large number of SV events observed in humans that do not appear to cause disease [11, 42], nonpathogenic SVs either alone or aggregated together may have functional impact on phenotype, as changing genomic arrangements may have wide-ranging regulatory or protein consequences. This may be true even if the alteration is small compared with overall genome size, but whether and how this occurs is poorly understood. However, the focus of this review is the growing body of studies that have analyzed how nonpathogenic genomic diversity, associated with specific SVs of large functional effect, shapes organismal phenotype. These studies give insight into the genomic processes that may be shaping species’ interactions with their own members or with their environment. If a particular genomic alteration can produce a phenotypic innovation that facilitates a fitness advantage to the individuals possessing the variation, this enables positive selection on the variant and may set the stage for adaptation in the population. Through selection in a lineage, these alterations could ultimately lead to fixation of the trait in the species and thus alter its evolutionary trajectory. Since this type of genetic variation has also existed throughout biological history, by studying it, we might gain insight into its contribution to phenotypic divergence in past evolution events (see Figure 1 for a model). A greater understanding of the role of SVs in adaptive diversification might also help us to identify potential present-day diversification events. Analysis of segregating (polymorphic) SVs can, therefore, provide insight into the biological diversification potential of a population or a species.
Figure 1.
Model of biological diversification mediated through adaptive structural variants. AMY1 represents the human salivary amylase gene on chromosome 1 that has been shown to undergo adaptive duplication [43]. This schematic model represents DNA sequence that can undergo structural sequence mutation to alter phenotype that produces lineage divergence of a population through positive selective pressure. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)
In this review, we will demonstrate that naturally occurring genetic variation in humans and other mammals, arising particularly from SVs, is a source of phenotypic potential that could directly enhance organismal fitness in any of three classes of phenotypes: (i) metabolic processes, (ii) immunological response, and (iii) physical features. This categorization provides a powerful framework for how to conceive of evolutionary adaptation, and can likely be applied to other groups of organisms besides mammals. Although connections and interrelationships exist between these three classes of phenotypic adaptation, their partition provides a helpful way to conceive of the processes of evolutionary adaptation.
Metabolic processes
Adaptations at the molecular level that affect fundamental internal physiological processes in cells and tissues, provide possibly the most direct examples of ongoing evolution in a population of organisms. Although many of the early survey studies of structural variation gave clues as to the general metabolic processes affected by SVs, a direct link between an SV and altered internal physiology has been difficult to establish. Studies looking at how overall patterns of SVs, particularly CNVs, affect metabolic variation have identified changes to extracellular processes and signal transduction, such as the NF-κB and MAPK signaling pathways [44, 45]. A literature review by He et al. [46] examined the associations between CNV regions, particularly CYP2D6, that coincide with pharmacologic targets and how those regions can modulate drug response/toxicity. Other studies have shown an enrichment of SVs in olfactory genes in humans [47–49], and a study in cattle identified a deletion polymorphism of olfactory receptor genes including BTA5 [50]. Taken together, these findings suggest CNVs can broadly affect many types of internal metabolite responses that may affect organismal phenotype, leading to overall fitness differences.
Several examples of SVs affecting specific metabolic processes with the potential to positively alter fitness consequences have emerged (Table 1). One classic example is the correlation between copy number of the salivary amylase gene, AMY1 and starch digestion. In the landmark study, our lab was able to positively correlate copy number of AMY1 in humans to salivary amylase protein level [43], and found that individuals from regions with high-starch diets had, on average, more copies of AMY1 than those from populations with low-starch diets. Based on these findings, we have hypothesized that the observed directional selection of AMY1 copy number confirms an adaptive benefit: the more AMY1 copies—and higher AMY1 enzyme levels—a person has, the more rapidly one can break down starch after a meal and thus more efficiently use the sugar source available from a high-starch diet. Because of agricultural production, which tends to yield high-starch food, possessing a greater ability to biochemically break down starch would enable an individual to better use the energy-richness of sugar that farming provides. This would explain why AMY1 was shown to be abundant in various farming populations. More recently, a study demonstrated variable copy number of pancreatic amylase in dogs, from the gene AMY2B [51]. The authors hypothesized that the increase in amylase copy number was owing to metabolic selection occurring in dogs during the domestication process, adapting away from the carnivorous diets of wolves to the high-starch diets given to them by humans.
Table 1.
Metabolic process phenotypes observed from structural variants in mammals
| Phenotype | Organism | SV type, extent | Genetic element | Study |
|---|---|---|---|---|
| Increase in salivary amylase production | Human | Duplication, full | AMY1 | Perry (2007) [43] |
| Increase in pancreatic amylase production | Dog | Duplication, full | AMY2B | Axelsson (2013) [51] |
| Bone mineral density increases | Human | Deletion, full | APC | Chew (2012) [52] |
| Lack of urinary testosterone | Human | Deletion, full | UGT2B17 | Jakobsson (2006) [53] |
| Increase in testosterone and estradiol in serum | Human | Deletion, full | UGT2B17 | Yang (2008) [54] |
| Increase in production traits | Cow | Duplication, full | APOL3 and FABP2 | Bickhart (2012) [55] |
| Increase in total merit | Cow | Duplication, full | BTA18 | Seroussi (2010) [56] |
| Higher monounsaturated fatty acid proportion | Cow | Deletion, partial | SREBP-1 | Hoashi (2007) [57]; Cecchinato (2012) [58] |
| In females, more offspring and higher recombination rate | Human | Inversion, partial | 900kb located in 17q21.31 locus | Stefansson (2005) [59] |
| Increase in bull fertility | Cow | Duplication, full | TSPY | Hamilton (2012) [60] |
| Increase in breeding values of sires | Cow | Duplication, full | KIAA1683 | Glick (2011) [61] |
A few other examples in humans of SVs operating to modulate metabolic activities have also emerged. One study was able to calculate that Caucasian individuals with a single copy loss of the tumor suppressor APC locus had a net increase in bone mineral density of the forearm (∼8%), spine (13%) and hip (13%) [52]. In a study of men from Sweden and Korea, urinary testosterone concentrations were shown to correlate with UGT2B17 allele count. UGT2B17 codes for an enzyme involved in the metabolism of steroids (among other substrates), and men who had a homozygous deletion of the gene had no or negligible amounts of urinary testosterone compared with men of other genotypes [53]. In another study, homozygous deletion of UGT2B17 was significantly associated with higher serum concentrations of both testosterone and estradiol in Chinese men [54].
SVs have also been shown to modulate metabolic activities in domesticated cattle. From ‘artificial’ selection on beef breeds, genes involved in lipid transport and metabolism, including APOL3 and FABP2, are highly duplicated, suggesting a link between CNVs and production traits [55]. In Holstein cattle in particular, close associations have been reported between copy number within the BTA18 telomeric region and total merit, as well as fat and protein production levels [56]. In Japanese Black cattle, a deletion in an intron of the transcription factor gene SREBP-1 was associated with beef having a higher (1.3%) monounsaturated fatty acid level and a lower (−1.6C) melting point of intramuscular fat [57]. A follow-up study confirmed higher fat content in cattle carrying the non-deletion (longer) allele [58].
An evolutionarily interesting phenotype found to be affected by structural variation is fertility. A study in humans identified a 900 kb inversion on chromosome 17 that exists in two distinct structural segments termed H1 and H2 (now more recently shown to exist in multiple haplotypes [62]), with different population frequencies [59]. In what turned out to be a provocative interpretation, the researchers claimed that the inverted H2 segment is undergoing positive selection in an Icelandic population, owing to their data indicating that carrier females tended to have more children, on average, than noncarriers, and have an overall higher recombination rate. More offspring and the ability to generate more genetic diversity of haplotypes through recombination ‘shuffling’ could be regarded as having adaptive potential owing to likely fitness increases. These inversion polymorphisms, along with other examples, are discussed in more detail in this same issue, page XXX [63]. Increased fertility resulting from structural variation has also been tentatively shown in domesticated cattle, with a positive correlation between copy number of TSPY (testes-specific protein Y-encoded) and bull fertility [60], as well as the number of copies of a specific variant (V1) of the uncharacterized gene KIAA1683 associating with breeding values of sires [61].
Immunological response
The ability to resist predation from harmful microorganisms or having a reduced susceptibility to a particular disease presents a clear fitness advantage. A classic example of immunological adaptation in humans, in this case due to balancing selection, is seen in people who lack a full complement of the α-globin genes, in areas of the world where malaria has high prevalence. Individuals typically have four copies in a diploid genome (αα/αα). For most individuals, deletion of two copies, either in the form (–/αα) or (-α/-α), causes the blood disorder thalassemia and results in a form of anemia, while deletion of three copies results in very severe anemia (zero copies is embryonically lethal). However, the normal complement of four copies increases susceptibility to severe malaria infection; therefore, when living in endemic malaria environments, having two copies of the allele is immunologically advantageous, as these individuals can often avoid the worst malarial and anemic outcomes [64, 65].
Another popular example of purported immunological adaptation in humans is that derived from the copy number of the CCL3L1 gene. The product of this gene is a ligand for the C-C chemokine receptor-5 (CCR5), an important cell-surface protein that acts as a co-receptor for HIV entry into cells. In a controversial paper, Gonzalez et al. (2005) made the claim that humans with a copy number (which varies from 0 to at least 14) lower than the population average had greater susceptibility to HIV-1/AIDS infection [66]. A later study claimed that a higher copy number of CCL3L in rhesus macaques accounted for an appreciable amount (18%) of the variance of time of progression to simian-AIDS, with lower copy number associated with faster disease progression [67]. These studies led some to conclude that higher CCL3L1 copy number may confer some protection against HIV-1 susceptibility, with a meta-analysis of available data supporting this conclusion [68]. However, conflicting findings from various studies make it difficult to determine the overall benefits versus risks of copy number variation for this gene. Some research groups had difficulty replicating the associations between low copy number and increased HIV-1 susceptibility, including a study in a Zimbabwean population [69] and a study in a North American population [70], among others. One study found some evidence that the presence of low CCL3L1 copy number served as slight protection against anemia [71]. Another study found that for people already at a reduced risk for a rare inflammatory disease, Kawasaki disease (KD), due to a deletion in CCR5, individuals who also carried a high copy number of CCL3L1 had even more significantly reduced risk of developing KD [72]. Difficulty interpreting together specific findings such as these should serve as reminders for the research community of the potential danger of broadening particular biological findings from a certain cohort or population to other groups, given the complexities of epistatic interactions resulting from various haplotypes and genetic backgrounds. Usher and McCarroll in this same issue [73] review some of these difficulties.
Copy number of beta-defensin genes, the peptides of which are highly antimicrobial agents, has been implicated in variable immune response in humans. A cluster of at least seven of these genes is known to have diploid copy numbers ranging between two to seven. One study examined meiotic transmission of this region and identified the rate of copy number change in the germ line to be about 0.7% per gamete, making this cluster one of the highest-frequency recurrent CNVs known to date [74]. A study involving 67 populations revealed an unusually high frequency of DEFB103-expressing copies in East Asia, which corresponds to the geographical location of both historical and modern influenza epidemics, which the authors suggest may be the result of selection for resistance to influenza [75]. One older study showed a positive correlation of DEFB4 genomic copy number in humans and the corresponding levels of its mRNA transcript, implicating copy number of this gene as having important immune consequences [76]. This correlation was shown in a cervical cancer study demonstrating that women with five or more copies of DEFB4 had a lower likelihood of developing cervical cancer compared with those with four or less copies [77]. However, as noted above, it is important to be cautious when drawing broad conclusions from highly specific disease studies, especially when involving immune system modulation, as immune-related SVs that appear protective in some contexts might be detrimental in others. A classic example illustrating this is the homozygous 32 bp internal deletion within CCR5 that has been found to be protective against HIV-1 infection [78], yet also increases the risk of symptomatic West Nile virus infection [79]. In the case of DEFB4, a study in Chinese individuals showed that increased copy number led to greater susceptibility of the autoimmune diseases systemic lupus erythematosus (SLE) and ANCA-associated small vasculitis [80]. In a different study, individuals possessing five or more copies of the complement component C4 and three or more copies of C4A, compared with having lower copy numbers, had some protection against developing SLE [81]. Being aware of these nuances when synthesizing multiple disease studies from participants with different genetic backgrounds and then attempting to associate SVs with disease susceptibility is an important challenge for researchers in which to gain mastery.
Several other findings in humans suggest that immunological diversity in general may still be underappreciated. One study in an Irish cohort was able to demonstrate that individuals that carried more FLG intragenic tandem repeats have lower risk of having the inflammatory skin disease eczema [82]. In relation to the human immunoglobulin heavy-chain locus, specifically that of the variable chain, a study using a hydatidiform mole BAC clone resource was able to add over 1 Mb of additional sequence for alternative assembly to the reference genome in the region 14q32.33, including identification of eight CNV-containing haplotypes from a panel of nine genomes [83]. One study surveying the TRIM5 locus, a region encoding a retroviral restriction factor, found this region to be duplicated in one Han Chinese woman (out of 72 individuals) with 12 copies of TRIM genes beyond the study’s other individuals [84]. Additionally, deletion at G176 of TRIM5α was found to be associated with reduced susceptibility to HIV-1 infection, with the truncated protein having enhanced antiviral activity [85]. These studies indicate that the human immune response may be even more diverse than previously known, and suggest that further study be done across populations of different ancestry.
Several other studies have examined how SVs interface with immune system function in nonhuman mammals. A study involving rhesus macaques found that higher copy number of KIR2DL4, an immunoglobulin-like receptor gene expressed in natural killer (NK) cells, resulted in increased interferon-gamma production from NK cells and better retainment of CD4+ T cells during SIV infection, which may slow disease progression [86]. A study of Angus cattle identified nearly 300 CNVs that associated with increased resistance to gastrointestinal nematodes; by network analysis, many of these variants fell into regions annotated as immune-related, including the WC1 gene, the product of which is expressed on certain T cells in cattle [87]. Another study in cattle showed that two genes related to pathogen and parasite resistance, CATHL4 and ULBP17, respectively, were highly duplicated in one indicine cow compared with taurine cows [55].
It is apparent from these findings (see Table 2 for a summary) that immune system modulation via structural variation might confer fitness advantages. If selective pressures are strong enough, they could become adaptive or even fixed in a population, thereby permanently altering how its members interact with the environment. For example, if individuals are capable of fending off (or are adapted to) previous external immunological threats, they may be able to explore ecological niches previously unavailable, such as new food sources (plants or animals) that have natural microorganisms associated with them that previously prevented their consumption. Also, mutualistic microbial interactions could eventually result if animals were not susceptible to the potentially negative effects of the microbes’ colonization in the gut or other bodily location.
Table 2.
Immunological response phenotypes observed from structural variants in mammals
| Phenotype | Organism | SV type, extent | Genetic element | Study |
|---|---|---|---|---|
| Reduced susceptibility to malaria infection | Human | Deletion, full | α-globin genes | Flint (1986) [64]; May (2007) [65] |
| Reduced susceptibility to HIV-1/AIDS | Human | Duplication, full | CCL3L1 | Gonzalez(2005) [66]; Liu (2010) [68] |
| Slower time progression to simian-AIDS | Rhesus macaque | Duplication, full | CCL3L1 | Degenhardt (2009) [67] |
| Slight protection against anemia | Human | Deletion, full | CCL3L1 | Carpenter (2012) [71] |
| Reduced risk for Kawasaki disease | Human | Deletion, partial ; Duplication, full | CCR5 CCL3L1 | Burns (2005) [72] |
| Reduced likelihood of developing cervical cancer | Human | Duplication, full | DEFB4 | Abe (2013) [77] |
| Protective against HIV-1 infection | Human | Deletion, partial | CCR5 | Liu et al. (1996) [78] |
| Reduced risk for developing systemic lupus erythematosus | Human | Duplication, full | C4 and C4A together | Yang (2007) [81] |
| Reduced risk of having eczema | Human | Duplication, full | FLG intragenic repeats | Brown (2012) [82] |
| Reduced susceptibility to HIV-1 infection | Human | Deletion, partial | TRIM | Nakajima (2009) [85] |
| Increased interferon-gamma production; better retainment of CD4+ T cells | Rhesus macaque | Duplication, full | KIR2DL4 | Hellmann (2013) [86] |
| Increased resistance to gastrointestinal nematodes | Cow | Duplication, full | WC1 and ∼300 other genes | Hou (2012) [87] |
| Increased resistance to pathogens and parasites | Cow | Duplication, full | CATHL4 and ULBP17 | Bickhart (2012) [55] |
Physical features
Unlike the many examples in humans involving metabolic or immunological response alterations resulting from SVs, very few examples [88, 89] of nonpathogenic physical feature differences caused by SVs have been documented. This could be due to the bias in human studies toward disease phenotypes or to a broader issue of the definition of what is pathological in an organismal context, especially in humans where disease research is more common. Nonetheless, examples in other mammals of seemingly nonpathogenic physical feature alterations are abundant, suggesting that future work should be done investigating human variation in physical features arising from SVs. Coat color variation is a prime example of physical feature variation in nonhuman mammals. SVs involving several genes have been repeatedly linked to different coat coloration outcomes (Table 3). One common example involves the Agouti signaling protein (ASIP) gene whose product serves to regulate mammalian pigmentation by acting as an inverse agonist at melanocortin receptors. Melanocortin-1 receptor (MC1R) is a G protein-coupled receptor whose activation promotes the production of the dark pigment eumelanin; when antagonized by ASIP, cells instead produce the light pigment phaeomelanin [90]. Many studies have identified SVs of ASIP and MC1R that led to multiple pigmentation outcomes. For these instances, the SV was located in a genomic position sufficient to alter either gene regulation or affect the gene product, a change that led directly to coat coloration differences. While many of these examples are due to artificial selection from human breeders, some instances could be suggestive of potential fitness advantages gained by the affected individuals in natural habitats given the right circumstances. For example, rabbits with a deletion in the reading frame of MC1R have altered coat coloration to either black or red, depending on which deletion they carry [91]. Similar darkening of coat color has been observed in both deer mice [92] and gray squirrels [93] and resulted from an SV in ASIP and MC1R, respectively. These color changes could theoretically provide enhanced concealment from predators in terrestrial habitats, especially at night. As well, darkening of coat color may provide other advantages, such as social dominance or mating behavior preferences, as has been seen in a sheep population [125].
Table 3.
Physical feature phenotypes observed from structural variants in mammals
| Phenotypes | Organism | SV type, extent | Genetic element | Study |
|---|---|---|---|---|
| Olive color skin | Human | Duplication, partial | Promoter region of MATP | Graf (2007) [88] |
| Short stature | Human | Deletion, partial and full | Multiple loci | Dauber (2011) [89] |
| Red coat or black coat | Rabbit | Deletion, partial | MC1R | Fontanesi (2006) [91] |
| Darker coat | Deer mouse | Deletion, partial | ASIP | Kingsley (2009) [92] |
| Black coat | Gray squirrel | Deletion, partial | MC1R | McRobie (2009) [93] |
| White coat | Sheep | Duplication, full | ASIP and ITCH promoter | Norris (2008) [94] |
| Darker coat | Sheep | Deletion, partial | ASIP | Gratten (2010) [95] |
| Grey coat | Sheep | Duplication, full | ASIP | Fontanesi (2011) [96] |
| White/tan coat | Goat | Duplication, full | ASIP | Fontanesi (2009) [97] |
| Black coat | Alpaca | Deletion, partial | ASIP | Feeley (2011) [98] |
| Black coat | Horse | Deletion, partial | ASIP | Rieder (2001) [99] |
| Saddle tan coat | Dog | SINE insertion, partial | ASIP | Dreger (2011) [100] |
| Black coat | Rabbit | Insertion, partial | ASIP | Fontanesi (2010) [101] |
| Red coat | Pig | Insertion, partial | MC1R | Kijas (2001) [102] |
| Yellow coat | Raccoon dog | Deletion, partial | 5′ UTR of MC1R | Han (2012) [103] |
| Brindle coat | Rabbit | Deletion, partial | MC1R | Fontanesi (2010) [104] |
| White coat | Horse | Deletion, partial | KIT | Holl (2010) [105] |
| White coat | Horse | Deletion, partial | KIT | Haase (2011) [106] |
| Tobiano coat | Horse | Inversion, partial | 100 kb downstream of KIT | Brooks (2007) [107] |
| White coat | Pig | Duplication, partial and full | KIT | Moller (1996) [108] |
| Grey-roan coat | Pig | Deletion, partial | KIT | Fontanesi (2010) [109] |
| White coat or white-spotted | Pig | Duplication, full | KIT | Rubin (2012) [110] |
| Color sidedness | Cow | Translocation, partial | KIT locus on chr 6 with locus on chr 29 | Durkin (2012) [111] |
| Red coat | Cow | Deletion, partial | MSHR | Joerg (1996) [112] |
| Dilute-colored coat | Cow | Deletion, partial | PMEL | Schmutz (2013) [113] |
| Splashed white coat | Horse | Insertion, partial | promoter of MITF | Hauswirth (2012) [114] |
| Black coat | Dog | Deletion, partial | CBD103 | Candille (2007) [115] |
| Brown coat | Pig | Deletion, partial | TYRP1 | Ren (2011) [116] |
| Furnishings trait | Dog | Insertion, partial | 3′ UTR of RSPO2 | Cadieu (2009) [117] |
| Rex hair | Rabbit | Deletion, partial | LIPH | Diribarne (2011) [118] |
| More muscular | Cow | Deletion, partial | MSTN | Grobet (1997) [119] |
| More muscular and faster | Dog | Deletion, partial | MSTN | Mosher (2007) [120] |
| More muscular | Horse | SINE insertion, partial | promoter region of MSTN | Petersen (2013) [121] |
| Greater body mass at birth | Cow | Insertion, partial | 3′ UTR of Nfix | Zhou (2012) [122] |
| Increased body proportions | Pig | Duplication, full | seven genes individually | Chen (2012) [123] |
| Greater body mass | Rabbit | Deletion, partial | IGF2 | Fontanesi (2012) [124] |
Physical feature variation resulting from structural variation can also change hair texture in mammals. In a large study of various dog breeds, the presence of an insertion within the 3′ untranslated region (UTR) of RSPO2, which codes for the signaling protein R-spondin-2, was associated with the ‘furnishings’ trait, a distinct facial hair pattern marked by a moustache and eyebrows [117]. In rabbits, deletion of a single nucleotide in an exon of the lipase member H (LIPH) gene is associated with the ‘rex’ hair phenotype (short, soft down hair) [118]. In a follow-up study, it was found that this single-nucleotide deletion, compared with wild-type, reduced by 3-fold the abundance of the LIPH mRNA transcript at both the fetal and adult stages, and reduced the lipase activity of the protein [126]. It is unclear whether this variation has immediate adaptive potential, yet in the wild, if hair-length shortening allows rabbits to sense the external environment differently (temperature, moisture content, etc.), it could alter how they interact with the environment, such as affecting foraging times. Note that this hypothetical SV-induced behavior change could eventually alter whole ecosystem balance if there was any time-dependence to the system—illustrating that given the right selective circumstances, SVs might play an even wider role in the adaptation process beyond that in a single, directly affected species.
Changes in an organism’s body morphology arising from SVs are also seen in several mammalian species (Table 3). A classic example of this is the so-called ‘double-muscled’ phenotype. In domesticated cattle, notable increases in muscle mass have been shown to result from a short deletion in the coding sequence of MSTN, the gene encoding the secreted muscle inhibitory factor myostatin [119]. This phenotype was also observed in dogs carrying a homozygous 2 bp deletion in an exon of MSTN, which leads to a premature stop codon. Even dogs carrying only one copy of this deletion were found, on average, to be statistically more muscular than wild-type dogs and faster in competitive racing events [120]. Similarly, muscle fiber typing studies in Quarter horses showed that insertion of a 5′ SINE in the promoter region of MSTN is associated with altered muscle fiber proportions that favor an optimal alternative gait; this change presumably accounts for the observed strong signal of selection at that locus in the horse lineage [121]. Other examples of genomic alterations leading to body size variation have been found in mammals. In cattle, a short insertion in the Nfix gene, which encodes site-specific binding proteins, is thought to be associated with greater body mass at birth [122]. In pigs, seven CNVs were found as candidate genes for a large number of body morphology phenotypes in categories such as measurement proportions, fat and muscle content and body weight, among others [123]. Similar to pigs, rabbits can also be evaluated for weight, which may be relevant in commercial settings. One study showed an association between a small deletion in the insulin-like growth factor-2 (IGF2) gene and larger weight [124]. Collectively, these body morphology studies stand out as examples of particular fitness enhancements resulting from SVs, owing to the variations directionally increasing animals’ physical presence.
Concluding remarks
In this review, we used examples of naturally segregating genetic variation in humans and other mammals, specifically genomic structural variation, as a way to gain insight into potential routes of biological diversification (see Figure 1 for model). If SVs occur in genomic regions (either individually or in combination) that facilitate phenotypic variability, under certain circumstances this could provide a fitness advantage for the organism and its progeny—ultimately giving these variants the opportunity to become adaptive or even fixed in the population under positive selective pressure. Combined with reproductive barriers either in allopatry or sympatry, these SVs could drive biological diversification of a population or species. By classifying potentially adaptive SVs according to their effects on three categories of phenotypes (metabolic processes, immunological response, and physical features), a conceptual framework can be built to better understand how SVs are operating on a population level and shaping evolutionary trajectories.
We have provided multiple examples (though not exhaustive) of nonpathogenic SV-produced phenotypic change that may have adaptive potential either from known or hypothesized selective pressures. In regards to metabolic processes (Table 1), we have seen how selection for increased enzyme activity (in this case from increased copy number of the human salivary amylase gene, AMY1) allows farming populations better energy utilization during digestion of high-starch foods [43]. Also in humans, we have seen that fertility can be potentially increased from an inversion polymorphism [59]. Immunological response alterations provide more evidence of the potential adaptive value of SVs (Table 2). Whether it be from the classic case in humans of balancing selection among the number of the α-globin genes [64, 65], reduced susceptibility to diseases such as eczema [82] or HIV-1 [85] or pathogen and parasite resistance in cattle [55], immune phenotypes from SV alterations can provide observable selective advantage. And while many examples of physical feature alterations caused by SVs (Table 3) come from artificial selection by human breeders to produce more desirable breeds, these examples of coat color diversity, hair texture variation, and body morphology increases, provide a window into the potentially adaptive value of SVs in the wild given the right selective pressures. These extreme examples from artificial selection (e.g., double-muscling in cattle can also result in obstructed labor/dystocia) are still useful in demonstrating the potential of strong or long-term selection acting on a beneficial trait, and can therefore be seen as evidence of the potential of SVs to become adaptive in the wild given the right selective pressures. Similar traits derived through natural selection could help animals more efficiently avoid predators or obtain greater numbers of prey, or otherwise better compete with members of the same population for food and other resources.
If nonpathogenic SVs alter the direction of how a species interacts with its environment, especially if they enable access to previously inaccessible ecological opportunity (the availability of an incompletely filled biological niche [127]), then a special type of evolution known as adaptive radiation may occur. Adaptive radiation can be defined as an ancestral species giving rise to multiple descendant species each adapted to distinct parts of the environment [128], an evolutionary concept that may be among the clearest demonstrations of Darwin’s ‘principle of divergence’ [129]. An often-recognized precondition for adaptive radiation is that of ecological opportunity. The ability of a species to exploit this opportunity is sometimes mediated through the development of a key innovation: an adjustment in a species’ morphological and/or physiological organization [130]. These genetically heritable novel features could enable a species to access the environment in a previously inaccessible way, thereby laying the groundwork for expansion into new a niche and the possibility of an adaptive radiation. Structural variation could theoretically provide a faster route to this process than SNPs due to SVs having potentially larger phenotypic effect-sizes because of affecting chromosomal structure, sometimes of hundreds or thousands of base-pairs in a single event. Therefore, analysis of structural variation in conjunction with adaptive radiation research could yield fruitful insight into evolutionary processes.
We focused the review on differences between individuals of the same species, not fixed genomic differences between species [131], as potential interspecies differences in genomic architecture make it challenging to compare even similar SVs across species. Studies attempting to explain biological adaptation have traditionally focused on fixed differences between species, yet this can result in parsimony difficulties that arise because of historical events that are inherently not directly testable. However, with the availability of high-throughput technologies that can rapidly assess SVs within multiple individuals of the same species, evolutionary questions can now be addressed by analyzing evolution in real-time using genomic alterations still undergoing flux in a species. In this way, analysis of segregating structural variation can provide much insight into the potential for, and constraint of, evolutionary processes, as well as provide a more probabilistic ascertainment of the steps involved in mammalian evolution.
By narrowing the review to non-disease phenotypes (but aware that secondary deleterious consequences might exist that are not readily identifiable), we highlight the potentially adaptive nature of certain SVs. We attempt to provide examples of only genomic structural variation affecting phenotype, though tightly linked SNPs co-segregating on the same haplotype along with the SV may also be responsible in some cases. These data can provide researchers in the field of mammalian evolution, or biological diversification more generally, insight into the nonpathogenic genomic plasticity range of species. Therefore, our summary also serves as a counterpoint to the many published reports on the effects of deleterious SVs that undergo strong purifying selection. This review may be of interest to those involved in agricultural production and maintenance, as awareness of biological enhancements/differences between livestock may alter breeding programs and result in different facility and land-use strategies. Finally, the examples of natural genetic variation given in this review may be potentially valuable to researchers in the emerging field of genomic engineering, as these examples provide proof-of-principle for viable genomic alterations tolerable in organisms, in some cases in regions not well studied experimentally.
Key points.
Structural variation can have wide-ranging nonpathogenic phenotypic consequences in humans and other mammals.
Under the right circumstances, certain structural variants could be under positive selective pressure and therefore adaptive in populations.
Studying currently segregating structural variation allows insight into past and present biological evolution.
Acknowledgements
We would like to thank Anna Lisa Lucido for help in editing earlier drafts of the manuscript, Christina Usher for graphical improvement of Figure 1, and Omer Gokcumen for helpful discussion on the topic.
Funding
This material is based on work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE1144152 (DWR).
References
- 1. Kamberov YG, Wang S, Tan J, et al. Modeling recent human evolution in mice by expression of a selected EDAR variant. Cell 2013;152:691–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. McPherron AC, Lee SJ. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci USA 1997;94:12457–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clop A, Marcq F, Takeda H, et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat Genet 2006;38:813–8. [DOI] [PubMed] [Google Scholar]
- 4. Schuelke M, Wagner KR, Stolz LE, et al. Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med 2004;350:2682–8. [DOI] [PubMed] [Google Scholar]
- 5. Andersson LS, Larhammar M, Memic F, et al. Mutations in DMRT3 affect locomotion in horses and spinal circuit function in mice. Nature 2012;488:642–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 2008;9:356–69. [DOI] [PubMed] [Google Scholar]
- 7. Iafrate AJ, Feuk L, Rivera MN, et al. Detection of large-scale variation in the human genome. Nat Genet 2004;36:949–51. [DOI] [PubMed] [Google Scholar]
- 8. Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science 2004;305:525–8. [DOI] [PubMed] [Google Scholar]
- 9. McCarroll SA, Hadnott TN, Perry GH, et al. Common deletion polymorphisms in the human genome. Nat Genet 2006;38:86–92. [DOI] [PubMed] [Google Scholar]
- 10. Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature 2006;444:444–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Conrad DF, Pinto D, Redon R, et al. Origins and functional impact of copy number variation in the human genome. Nature 2010;464:704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mills RE, Walter K, Stewart C, et al. Mapping copy number variation by population-scale genome sequencing. Nature 2011;470:59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sudmant PH, Huddleston J, Catacchio CR, et al. Evolution and diversity of copy number variation in the great ape lineage. Genome Res 2013;23:1373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gokcumen O, Tischler V, Tica J, et al. Primate genome architecture influences structural variation mechanisms and functional consequences. Proc Natl Acad Sci USA 2013;110:15764–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu GE, Hou Y, Zhu B, et al. Analysis of copy number variations among diverse cattle breeds. Genome Res 2010;20:693–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hou Y, Liu GE, Bickhart DM, et al. Genomic characteristics of cattle copy number variations. BMC Genomics 2011;12:127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fontanesi L, Martelli PL, Beretti F, et al. An initial comparative map of copy number variations in the goat (Capra hircus) genome. BMC Genomics 2010;11:639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang J, Jiang J, Fu W, et al. A genome-wide detection of copy number variations using SNP genotyping arrays in swine. BMC Genomics 2012;13:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Doan R, Cohen N, Harrington J, et al. Identification of copy number variants in horses. Genome Res 2012;22:899–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nicholas TJ, Cheng Z, Ventura M, et al. The genomic architecture of segmental duplications and associated copy number variants in dogs. Genome Res 2009;19:491–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nicholas TJ, Baker C, Eichler EE, et al. A high-resolution integrated map of copy number polymorphisms within and between breeds of the modern domesticated dog. BMC Genomics 2011;12:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Berglund J, Nevalainen EM, Molin A-M, et al. Novel origins of copy number variation in the dog genome. Genome Biol 2012;13:R73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fontanesi L, Martelli PL, Scotti E, et al. Exploring copy number variation in the rabbit (Oryctolagus cuniculus) genome by array comparative genome hybridization. Genomics 2012;100:245–51. [DOI] [PubMed] [Google Scholar]
- 24. Guryev V, Saar K, Adamovic T, et al. Distribution and functional impact of DNA copy number variation in the rat. Nat Genet 2008;40:538–45. [DOI] [PubMed] [Google Scholar]
- 25. Quinlan AR, Clark RA, Sokolova S, et al. Genome-wide mapping and assembly of structural variant breakpoints in the mouse genome. Genome Res 2010;20:623–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yalcin B, Wong K, Agam A, et al. Sequence-based characterization of structural variation in the mouse genome. Nature 2011;477:326–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yalcin B, Wong K, Bhomra A, et al. The fine-scale architecture of structural variants in 17 mouse genomes. Genome Biol 2012;13:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Levy S, Sutton G, Ng PC, et al. The diploid genome sequence of an individual human. PLoS Biol 2007;5:e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stranger BE, Forrest MS, Dunning M, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 2007;315:848–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schlattl A, Anders S, Waszak SM, et al. Relating CNVs to transcriptome data at fine resolution: assessment of the effect of variant size, type, and overlap with functional regions. Genome Res 2011;21:2004–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Henrichsen CN, Vinckenbosch N, Zöllner S, et al. Segmental copy number variation shapes tissue transcriptomes. Nat Genet 2009;41:424–9. [DOI] [PubMed] [Google Scholar]
- 32. Cahan P, Li Y, Izumi M, et al. The impact of copy number variation on local gene expression in mouse hematopoietic stem and progenitor cells. Nat Genet 2009;41:430–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chaignat E, Yahya-Graison EA, Henrichsen CN, et al. Copy number variation modifies expression time courses. Genome Res 2011;21:106–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Handsaker RE, Van Doren V, Berman JR, et al. Large multiallelic copy number variations in humans. Nat Genet 2015;47:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sudmant PH, Kitzman JO, Antonacci F, et al. Diversity of human copy number variation and multicopy genes. Science 2010;330:641–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iskow RC, Gokcumen O, Lee C. Exploring the role of copy number variants in human adaptation. Trends Genet 2012;28:245–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Abyzov A, Iskow R, Gokcumen O, et al. Analysis of variable retroduplications in human populations suggests coupling of retrotransposition to cell division. Genome Res 2013;23:2042–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schrider DR, Navarro FCP, Galante PAF, et al. Gene copy-number polymorphism caused by retrotransposition in humans. PLoS Genet 2013;9:e1003242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Felekkis K, Voskarides K, Dweep H, et al. Increased number of microRNA target sites in genes encoded in CNV regions. Evidence for an evolutionary genomic interaction. Mol Biol Evol 2011;28:2421–4. [DOI] [PubMed] [Google Scholar]
- 40. Wu X, Zhang D, Li G. Insights into the regulation of human CNV-miRNAs from the view of their target genes. BMC Genomics 2012;13:707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Weischenfeldt J, Symmons O, Spitz F, et al. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat Rev Genet 2013;14:125–38. [DOI] [PubMed] [Google Scholar]
- 42. Consortium, 1000 Genomes Project. A map of human genome variation from population-scale sequencing. Nature 2010;467:1061–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Perry GH, Dominy NJ, Claw KG, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet 2007;39:1256–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Poptsova M, Banerjee S, Gokcumen O, et al. Impact of constitutional copy number variants on biological pathway evolution. BMC Evol Biol 2013;13:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Orozco LD, Cokus SJ, Ghazalpour A, et al. Copy number variation influences gene expression and metabolic traits in mice. Hum Mol Genet 2009;18:4118–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. He Y, Hoskins JM, McLeod HL. Copy number variants in pharmacogenetic genes. Trends Mol Med 2011;17:244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nozawa M, Kawahara Y, Nei M. Genomic drift and copy number variation of sensory receptor genes in humans. Proc Natl Acad Sci USA 2007;104:20421–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Young JM, Endicott RM, Parghi SS, et al. Extensive copy-number variation of the human olfactory receptor gene family. Am J Hum Genet 2008;83:228–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Waszak SM, Hasin Y, Zichner T, et al. Systematic inference of copy-number genotypes from personal genome sequencing data reveals extensive olfactory receptor gene content diversity. PLoS Comput Biol 2010;6:e1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liu GE, Li RW, Sonstegard TS, et al. Characterization of a novel microdeletion polymorphism on BTA5 in cattle. Anim Genet 2008;39:655–8. [DOI] [PubMed] [Google Scholar]
- 51. Axelsson E, Ratnakumar A, Arendt M-L, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 2013;495:360–4. [DOI] [PubMed] [Google Scholar]
- 52. Chew S, Dastani Z, Brown SJ, et al. Copy number variation of the APC gene is associated with regulation of bone mineral density. Bone 2012;51:939–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jakobsson J, Ekström L, Inotsume N, et al. Large differences in testosterone excretion in Korean and Swedish men are strongly associated with a UDP-glucuronosyl transferase 2B17 polymorphism. J Clin Endocrinol Metab 2006;91:687–93. [DOI] [PubMed] [Google Scholar]
- 54. Yang T-L, Chen X-D, Guo Y, et al. Genome-wide copy-number-variation study identified a susceptibility gene, UGT2B17, for osteoporosis. Am J Hum Genet 2008;83:663–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bickhart DM, Hou Y, Schroeder SG, et al. Copy number variation of individual cattle genomes using next-generation sequencing. Genome Res 2012;22:778–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Seroussi E, Glick G, Shirak A, et al. Analysis of copy loss and gain variations in Holstein cattle autosomes using BeadChip SNPs. BMC Genomics 2010;11:673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Hoashi S, Ashida N, Ohsaki H, et al. Genotype of bovine sterol regulatory element binding protein-1 (SREBP-1) is associated with fatty acid composition in Japanese Black cattle. Mamm Genome 2007;18:880–6. [DOI] [PubMed] [Google Scholar]
- 58. Cecchinato A, Ribeca C, Maurmayr A, et al. Short communication: Effects of β-lactoglobulin, stearoyl-coenzyme A desaturase 1, and sterol regulatory element binding protein gene allelic variants on milk production, composition, acidity, and coagulation properties of Brown Swiss cows. J Dairy Sci 2012;95:450–4. [DOI] [PubMed] [Google Scholar]
- 59. Stefansson H, Helgason A, Thorleifsson G, et al. A common inversion under selection in Europeans. Nat Genet 2005;37:129–37. [DOI] [PubMed] [Google Scholar]
- 60. Hamilton CK, Verduzco-Gómez AR, Favetta LA, et al. Testis-specific protein Y-encoded copy number is correlated to its expression and the field fertility of Canadian Holstein bulls. Sex Dev 2012;6:231–9. [DOI] [PubMed] [Google Scholar]
- 61. Glick G, Shirak A, Seroussi E, et al. Fine mapping of a QTL for fertility on BTA7 and its association with a CNV in the Israeli holsteins. G3 2011;1:65–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Boettger LM, Handsaker RE, Zody MC, et al. Structural haplotypes and recent evolution of the human 17q21.31 region. Nat Genet 2012;44:881–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Puig M, Casillas S, Villatoro S, Cáceres M. Human inversions and their functional consequences. Brief Funct Genomics 2015;Forthcoming. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Flint J, Hill A V, Bowden DK, et al. High frequencies of alpha-thalassaemia are the result of natural selection by malaria. Nature 1986;321:744–50. [DOI] [PubMed] [Google Scholar]
- 65. May J, Evans JA, Timmann C, et al. Hemoglobin variants and disease manifestations in severe falciparum malaria. JAMA 2007;297:2220–6. [DOI] [PubMed] [Google Scholar]
- 66. Gonzalez E, Kulkarni H, Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 2005;307:1434–40. [DOI] [PubMed] [Google Scholar]
- 67. Degenhardt JD, de Candia P, Chabot A, et al. Copy number variation of CCL3-like genes affects rate of progression to simian-AIDS in Rhesus Macaques (Macaca mulatta). PLoS Genet 2009;5:e1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Liu S, Yao L, Ding D, et al. CCL3L1 copy number variation and susceptibility to HIV-1 infection: a meta-analysis. PLoS One 2010;5:e15778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Larsen MH, Thørner LW, Zinyama R, et al. CCL3L gene copy number and survival in an HIV-1 infected Zimbabwean population. Infect Genet Evol 2012;12:1087–93. [DOI] [PubMed] [Google Scholar]
- 70. Lee EY, Yue FY, Jones RB, et al. The impact of CCL3L1 copy number in an HIV-1-infected white population. AIDS 2010;24:1589–91. [DOI] [PubMed] [Google Scholar]
- 71. Carpenter D, Färnert A, Rooth I, et al. CCL3L1 copy number and susceptibility to malaria. Infect Genet Evol 2012;12:1147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Burns JC, Shimizu C, Gonzalez E, et al. Genetic variations in the receptor-ligand pair CCR5 and CCL3L1 are important determinants of susceptibility to Kawasaki disease. J Infect Dis 2005;192:344–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Usher CL, McCarroll SA. Complex and multi-allelic copy number variations in human disease. Brief Funct Genomics 2015;14:329–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Abu Bakar S, Hollox EJ, Armour JAL. Allelic recombination between distinct genomic locations generates copy number diversity in human beta-defensins. Proc Natl Acad Sci USA 2009;106:853–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hardwick RJ, Machado LR, Zuccherato LW, et al. A worldwide analysis of beta-defensin copy number variation suggests recent selection of a high-expressing DEFB103 gene copy in East Asia. Hum Mutat 2011;32:743–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hollox EJ, Armour JAL, Barber JCK. Extensive normal copy number variation of a beta-defensin antimicrobial-gene cluster. Am J Hum Genet 2003;73:591–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Abe S, Miura K, Kinoshita A, et al. Copy number variation of the antimicrobial-gene, defensin beta 4, is associated with susceptibility to cervical cancer. J Hum Genet 2013;58:250–3. [DOI] [PubMed] [Google Scholar]
- 78. Liu R, Paxton WA, Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 1996;86:367–77. [DOI] [PubMed] [Google Scholar]
- 79. Glass WG, McDermott DH, Lim JK, et al. CCR5 deficiency increases risk of symptomatic West Nile virus infection. J Exp Med 2006;203:35–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhou X-J, Cheng F-J, Lv J-C, et al. Higher DEFB4 genomic copy number in SLE and ANCA-associated small vasculitis. Rheumatology (Oxford) 2012;51:992–5. [DOI] [PubMed] [Google Scholar]
- 81. Yang Y, Chung EK, Wu YL, et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European America. Am J Hum Genet 2007;80:1037–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Brown SJ, Kroboth K, Sandilands A, et al. Intragenic copy number variation within filaggrin contributes to the risk of atopic dermatitis with a dose-dependent effect. J Invest Dermatol 2012;132:98–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Watson CT, Steinberg KM, Huddleston J, et al. Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am J Hum Genet 2013;92:530–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Han K, Lou DI, Sawyer SL. Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet 2011;7:e1002388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nakajima T, Nakayama EE, Kaur G, et al. Impact of novel TRIM5alpha variants, Gly110Arg and G176del, on the anti-HIV-1 activity and the susceptibility to HIV-1 infection. AIDS 2009;23:2091–100. [DOI] [PubMed] [Google Scholar]
- 86. Hellmann I, Letvin NL, Schmitz JE. KIR2DL4 copy number variation is associated with CD4+ T-cell depletion and function of cytokine-producing NK cell subsets in SIV-infected Mamu-A*01-negative rhesus macaques. J Virol 2013;87:5305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Hou Y, Liu GE, Bickhart DM, et al. Genomic regions showing copy number variations associate with resistance or susceptibility to gastrointestinal nematodes in Angus cattle. Funct. Integr. Genomics 2012;12:81–92. [DOI] [PubMed] [Google Scholar]
- 88. Graf J, Voisey J, Hughes I, et al. Promoter polymorphisms in the MATP (SLC45A2) gene are associated with normal human skin color variation. Hum Mutat 2007;28:710–7. [DOI] [PubMed] [Google Scholar]
- 89. Dauber A, Yu Y, Turchin MC, et al. Genome-wide association of copy-number variation reveals an association between short stature and the presence of low-frequency genomic deletions. Am J Hum Genet 2011;89:751–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Lu D, Willard D, Patel IR, et al. Agouti protein is an antagonist of the melanocyte-stimulating-hormone receptor . Nature 1994;371:799–802. [DOI] [PubMed] [Google Scholar]
- 91. Fontanesi L, Tazzoli M, Beretti F, et al. Mutations in the melanocortin 1 receptor (MC1R) gene are associated with coat colours in the domestic rabbit (Oryctolagus cuniculus). Anim Genet 2006;37:489–93. [DOI] [PubMed] [Google Scholar]
- 92. Kingsley EP, Manceau M, Wiley CD, et al. Melanism in peromyscus is caused by independent mutations in agouti. PLoS One 2009;4:e6435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. McRobie H, Thomas A, Kelly J. The genetic basis of melanism in the gray squirrel (Sciurus carolinensis). J Hered 2009;100:709–14. [DOI] [PubMed] [Google Scholar]
- 94. Norris BJ, Whan VA. A gene duplication affecting expression of the ovine ASIP gene is responsible for white and black sheep. Genome Res 2008;18:1282–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Gratten J, Pilkington JG, Brown EA, et al. The genetic basis of recessive self-colour pattern in a wild sheep population. Heredity (Edinb) 2010;104:206–14. [DOI] [PubMed] [Google Scholar]
- 96. Fontanesi L, Dall’Olio S, Beretti F, et al. Coat colours in the Massese sheep breed are associated with mutations in the agouti signalling protein (ASIP) and melanocortin 1 receptor (MC1R) genes. Animal 2011;5:8–17. [DOI] [PubMed] [Google Scholar]
- 97. Fontanesi L, Beretti F, Riggio V, et al. Copy number variation and missense mutations of the agouti signaling protein (ASIP) gene in goat breeds with different coat colors. Cytogenet. Genome Res 2009;126:333–47. [DOI] [PubMed] [Google Scholar]
- 98. Feeley NL, Bottomley S, Munyard KA. Three novel mutations in ASIP associated with black fibre in alpacas (Vicugna pacos). J Agric Sci 2011;149:529–38. [Google Scholar]
- 99. Rieder S, Taourit S, Mariat D, et al. Mutations in the agouti (ASIP), the extension (MC1R), and the brown (TYRP1) loci and their association to coat color phenotypes in horses (Equus caballus). Mamm Genome 2001;12:450–5. [DOI] [PubMed] [Google Scholar]
- 100. Dreger DL, Schmutz SM. A SINE insertion causes the black-and-tan and saddle tan phenotypes in domestic dogs. J Hered 2011;102:S11–8. [DOI] [PubMed] [Google Scholar]
- 101. Fontanesi L, Forestier L, Allain D, et al. Characterization of the rabbit agouti signaling protein (ASIP) gene: transcripts and phylogenetic analyses and identification of the causative mutation of the nonagouti black coat colour. Genomics 2010;95:166–75. [DOI] [PubMed] [Google Scholar]
- 102. Kijas JM, Moller M, Plastow G, et al. A frameshift mutation in MC1R and a high frequency of somatic reversions cause black spotting in pigs. Genetics 2001;158:779–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Han J-I, Yang H, Jeung E-B, et al. Altered expression of melanocortin-1 receptor (MC1R) in a yellow-coloured wild raccoon dog (Nyctereutes procyonoides). Vet Dermatol 2012;23:187–e37. [DOI] [PubMed] [Google Scholar]
- 104. Fontanesi L, Scotti E, Colombo M, et al. A composite six bp in-frame deletion in the melanocortin 1 receptor (MC1R) gene is associated with the Japanese brindling coat colour in rabbits (Oryctolagus cuniculus). BMC Genet 2010;11:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Holl H, Brooks S, Bailey E. De novo mutation of KIT discovered as a result of a non-hereditary white coat colour pattern. Anim Genet 2010;41:196–8. [Google Scholar]
- 106. Haase B, Rieder S, Tozaki T, et al. Five novel KIT mutations in horses with white coat colour phenotypes. Anim Genet 2011;42:337–9. [DOI] [PubMed] [Google Scholar]
- 107. Brooks SA, Lear TL, Adelson DL, et al. A chromosome inversion near the KIT gene and the Tobiano spotting pattern in horses. Cytogenet Genome Res 2007;119:225–30. [DOI] [PubMed] [Google Scholar]
- 108. Moller MJ, Chaudhary R, Hellmén E, et al. Pigs with the dominant white coat color phenotype carry a duplication of the KIT gene encoding the mast/stem cell growth factor receptor. Mamm Genome 1996;7:822–30. [DOI] [PubMed] [Google Scholar]
- 109. Fontanesi L, D’Alessandro E, Scotti E, et al. Genetic heterogeneity and selection signature at the KIT gene in pigs showing different coat colours and patterns. Anim Genet 2010;41:478–92. [DOI] [PubMed] [Google Scholar]
- 110. Rubin C-J, Megens H-J, Barrio AM, et al. Strong signatures of selection in the domestic pig genome. Proc Natl Acad Sci USA 2012;109:19529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Durkin K, Coppieters W, Drögemüller C, et al. Serial translocation by means of circular intermediates underlies colour sidedness in cattle. Nature 2012;482:81–4. [DOI] [PubMed] [Google Scholar]
- 112. Joerg H, Fries HR, Meijerink E, et al. Red coat color in Holstein cattle is associated with a deletion in the MSHR gene. Mamm Genome 1996;7:317–18. [DOI] [PubMed] [Google Scholar]
- 113. Schmutz SM, Dreger DL. Interaction of MC1R and PMEL alleles on solid coat colors in Highland cattle. Anim Genet 2013;44:9–13. [DOI] [PubMed] [Google Scholar]
- 114. Hauswirth R, Haase B, Blatter M, et al. Mutations in MITF and PAX3 cause ‘splashed white’ and other white spotting phenotypes in horses. PLoS Genet 2012;8:e1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Candille SI, Kaelin CB, Cattanach BM, et al. A -defensin mutation causes black coat color in domestic dogs. Science 2007;318:1418–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ren J, Mao H, Zhang Z, et al. A 6-bp deletion in the TYRP1 gene causes the brown colouration phenotype in Chinese indigenous pigs. Heredity (Edinb) 2011;106:862–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Cadieu E, Neff MW, Quignon P, et al. Coat variation in the domestic dog is governed by variants in three genes. Science 2009;326:150–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Diribarne M, Mata X, Chantry-Darmon C, et al. A deletion in exon 9 of the LIPH gene is responsible for the rex hair coat phenotype in rabbits (Oryctolagus cuniculus). PLoS One 2011;6:e19281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Grobet L, Martin LJ, Poncelet D, et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet 1997;17:71–4. [DOI] [PubMed] [Google Scholar]
- 120. Mosher DS, Quignon P, Bustamante CD, et al. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet 2007;3:e79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Petersen JL, Mickelson JR, Rendahl AK, et al. Genome-wide analysis reveals selection for important traits in domestic horse breeds. PLoS Genet 2013;9:e1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Zhou Y, Lan X, Xu Y, et al. Exploring polymorphisms and potential application roles of the bovine Nfix gene in breeding. Genome 2012;55:845–51. [DOI] [PubMed] [Google Scholar]
- 123. Chen C, Qiao R, Wei R, et al. A comprehensive survey of copy number variation in 18 diverse pig populations and identification of candidate copy number variable genes associated with complex traits. BMC Genomics 2012;13:733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Fontanesi L, Mazzoni G, Bovo S, et al. Association between a polymorphism in the IGF2 gene and finishing weight in a commercial rabbit population. Anim Genet 2012;43:651–2. [DOI] [PubMed] [Google Scholar]
- 125. Loehr J, Carey J, Ylönen H, et al. Coat darkness is associated with social dominance and mating behaviour in a mountain sheep hybrid lineage. Anim Behav 2008;76:1545–53. [Google Scholar]
- 126. Diribarne M, Mata X, Rivière J, et al. LIPH expression in skin and hair follicles of normal coat and Rex rabbits. PLoS One 2012;7:e30073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Yoder JB, Clancey E, Des Roches S, et al. Ecological opportunity and the origin of adaptive radiations. J Evol Biol 2010;23:1581–96. [DOI] [PubMed] [Google Scholar]
- 128. Losos JB, Ricklefs RE. Adaptation and diversification on islands. Nature 2009;457:830–6. [DOI] [PubMed] [Google Scholar]
- 129. Glor RE. Phylogenetic insights on adaptive radiation. Annu Rev Ecol Evol Syst 2010;41:251–70. [Google Scholar]
- 130. Mayr E, Schuz E. (eds). Ornithologie als Biologische Wissenschaft: 28 Beitrage als Festschrift zum 60. Geburtstag von Erwin Stresemann (22 November 1949). Heidelberg, Germany: Carl Winter. [Google Scholar]
- 131. Qiu Q, Zhang G, Ma T, et al. The yak genome and adaptation to life at high altitude. Nat Genet 2012;44:946–9. [DOI] [PubMed] [Google Scholar]