Abstract
A cute myeloid leukemia is a malignant disease of immature myeloid cells. Despite significant therapeutic effects of differentiation-inducing agents in some acute myeloid leukemia subtypes, the disease remains incurable in a large fraction of patients. Here we show that SK053, a thioredoxin inhibitor, induces differentiation and cell death of acute myeloid leukemia cells. Considering that thioredoxin knock-down with short hairpin RNA failed to exert antiproliferative effects in one of the acute myeloid leukemia cell lines, we used a biotin affinity probe-labeling approach to identify potential molecular targets for the effects of SK053. Mass spectrometry of proteins precipitated from acute myeloid leukemia cells incubated with biotinylated SK053 used as a bait revealed protein disulfide isomerase as a potential binding partner for the compound. Biochemical, enzymatic and functional assays using fluorescence lifetime imaging confirmed that SK053 binds to and inhibits the activity of protein disulfide isomerase. Protein disulfide isomerase knockdown with short hairpin RNA was associated with inhibition of cell growth, increased CCAAT enhancer-binding protein α levels, and induction of differentiation of HL-60 cells. Molecular dynamics simulation followed by the covalent docking indicated that SK053 binds to the fourth thioredoxin-like domain of protein disulfide isomerase. Differentiation of myeloid precursor cells requires the activity of CCAAT enhancer-binding protein α, the function of which is impaired in acute myeloid leukemia cells through various mechanisms, including translational block by protein disulfide isomerase. SK053 increased the levels of CCAAT enhancer-binding protein α and upregulated mRNA levels for differentiation-associated genes. Finally, SK053 decreased the survival of blasts and increased the percentage of cells expressing the maturation-associated CD11b marker in primary cells isolated from bone marrow or peripheral blood of patients with acute myeloid leukemia. Collectively, these results provide a proof-of-concept that protein disulfide isomerase inhibition has potential as a therapeutic strategy for the treatment of acute myeloid leukemia and for the development of small-molecule inhibitors of protein disulfide isomerase.
Introduction
Acute myeloid leukemia (AML), the most prevalent acute leukemia among adults, is a malignancy of myeloid lineage cells characterized by the inhibition of cell differentiation leading to accumulation of abnormal white blood cells.1 The use of differentiation-inducing agents, such as all-trans retinoic acid and arsenic trioxide, for the treatment of acute promyelocytic leukemia has brought remarkable therapeutic effects.2,3 However, not all patients with acute promyelocytic leukemia benefit from differentiation treatment and there has been no such significant progress in the treatment of other types of AML.4 The development of new therapeutic agents exerting anti-leukemic effects by targeting unique cellular mechanisms of differentiation is still, therefore, a pressing need of clinical importance.5 It is particularly desirable to develop differentiation-promoting compounds that induce terminal differentiation of leukemic cells leading to cell cycle arrest followed by cell death, and obviate overt cytotoxicity.
A critical transcription factor involved in the development and differentiation of myeloid lineage cells is CCAAT enhancer-binding protein α (C/EBPα). In C/EBPα-deficient mice granulocyte differentiation is blocked,6 and C/EBPα expression in bipotential myeloid progenitors is sufficient to induce granulocytic development.7 Dysregulation of C/EBPα activity is frequently observed in AML patients. Lack of, aberrant or suboptimal C/EBPα activity can result from genomic mutations in the CEBPA gene,8 transcriptional suppression originating from promoter hypermethylation, or functional inactivation by phosphorylation.9 A translational block that occurs in cells experiencing endoplasmic reticulum stress has also been reported as a mechanism leading to C/EBPα downregulation at the mRNA level.10 Various mechanisms such as loss of Ca2+ homeostasis, inhibition of disulfide bond formation, oxidative stress, or hypoxia, lead to endoplasmic reticulum stress, which triggers the unfolded protein response. The role of the unfolded protein response is to restore protein homeostasis and normal endoplasmic reticulum function. Accordingly, this response has been reported to be upregulated in a significant percentage of patients with AML and to be associated with a more favorable course of the disease.10
We have previously developed SK053, a peptidomimetic inhibitor of thioredoxin that exerts cytostatic/cytotoxic effects and endoplasmic reticulum stress-mediated apoptosis in tumor cells.11 Conspicuously, we have observed that AML cells incubated with SK053 undergo growth arrest followed by differentiation into more mature myeloid stages and cell death. We, therefore, employed RNA sequencing and a biotin affinity probe-labeling approach to identify the molecular mechanism of the differentiation-promoting effects of SK053, revealing protein disulfide isomerase (PDI) as a druggable target for AML treatment.
Methods
A detailed description of the methods used can be found in the Online Supplementary Data file.
Culture conditions for the acute myeloid leukemia cell lines
NB4, MOLM14, HS-5 and HL-60 cells were cultured in RPMI 1640 (Sigma-Aldrich) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone), 100 mg/mL streptomycin and 100 U/mL penicillin (P/S, Sigma-Aldrich). KG1 cells were cultured in IMDM (Gibco) supplemented with 20% heat-inactivated fetal bovine serum (Hyclone) and P/S. HeLa cells were cultured in DMEM (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Hyclone) and P/S. All cells were cultured at 37°C in a fully humidified atmosphere of 5% CO2.
Acute myeloid leukemia cell differentiation assays
Myeloid differentiation was assessed by staining cytospun cells with May-Grünwald-Giemsa as well as in flow cytometry by examining the surface expression of CD11b. A conventional semi-quantitative microscopic nitroblue tetrazolium (NBT; Sigma-Aldrich) assay was used to determine the production of superoxide anion upon stimulation with phorbol myristate acetate in differentiated AML cells. A modified quantitative NBT assay was established and implemented by dissolving the blue formazan particles from equal cell numbers in sodium hydroxide, followed by measurement of absorbance at 620 nm using an ASYS UVM 340 microplate reader (Biochrom, Cambridge, UK).
RNA extraction and quantitative real-time polymerase chain reaction
Total RNA was extracted from HL-60 cells using an RNA purification kit (EurX) and relative gene expression was quantified using a LightCycler II 480 Real-Time PCR System (Roche, Basel, Switzerland), LightCycler Probes Master and hydrolysis probes [Universal Probe Library (UPL), Roche] according to the manufacturer’s recommendations.
Western blot
HL-60 cells were washed twice with ice-cold phosphate-buffered saline, lysed in RIPA buffer (50 mmol/L Tris base, 150 mmol/L NaCl, 1% NP-40, 0.25% sodium deoxycholate, and 1 mmol/L EDTA) or cytoplasmic/nuclear fractions of proteins were extracted using an NE-PER Kit (Pierce). Immunoblotting was done via standard procedures using the antibodies according to the manufacturer’s instructions.
Isolation of primary acute myeloid leukemia blasts and evaluation of the anti-leukemic effects of SK053
The study was approved by the Institutional Review Board of the Medical University of Lublin (approval n. KE-0254/71/2010) and the Medical University of Warsaw (approval n. KB 182/2011) and was conducted according to the Declaration of Helsinki. Each patient signed written informed consent to the procedures. Leukemic cells were obtained from the bone marrow of patients with AML and isolated by 1.077 g/mL Histopaque (Sigma-Aldrich) density gradient centrifugation. Cells were seeded in IMDM culture medium, supplemented with 10% heat-inactivated fetal bovine serum at a density 5×105 cells/mL and incubated for 72 h with SK053. Cytotoxic effects, growth and differentiation upon incubation with SK053 were evaluated with methods described above. 7-AAD staining was used to discriminate dead cells. Additionally, anti-CD33 antibody (BD Biosciences, cat. n. 551378) was used to identify myeloid cells among all peripheral blood mononuclear cells. The patients’ basic characteristics are shown in Online Supplementary Table S11.
Statistical analysis
Data were analyzed using Microsoft Excel 2010 and GraphPad Prism 6.0 for Windows (GraphPad Software Inc., La Jolla, CA, USA) software. The results were analyzed for significance using a two-tailed Student t-test or one-way ANOVA with a Dunnett post-hoc test. Statistical significance was defined as P values <0.05.
Results
SK053 induces differentiation of acute myeloid leukemia cells
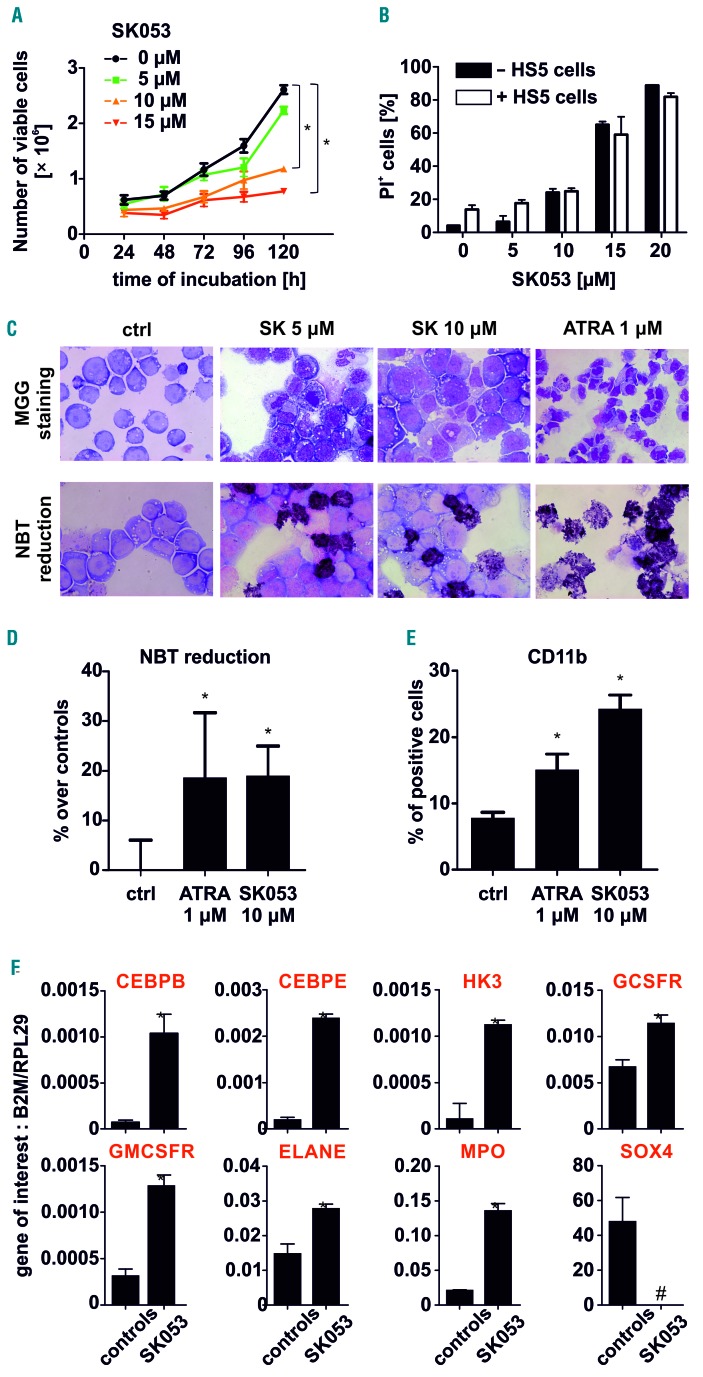
HL-60 acute promyelocytic leukemia cells were incubated for 24 to 120 h with increasing concentrations of SK053 and cell growth as well as cytotoxic effects were determined by counting viable cells and flow cytometry. SK053 inhibited cell growth in a time- and concentration-dependent manner (Figure 1A). We observed similar cytotoxic effects of SK053 against HL-60 cells grown in cell culture medium alone or on the top of bone marrow stromal cells (HS5) mimicking the interaction between AML blasts and the bone marrow niche (Figure 1B). HL-60 cells incubated with SK053 also showed features of more mature myelopoietic stages with a decreased nucleus/cytoplasm ratio and a greater reduction of NBT (Figure 1C), corresponding to NADPH oxidase activity. An NBT reduction assay (Figure 1D) as well as increased surface levels of CD11b measured in flow cytometry (Figure 1E) confirmed the differentiation-promoting effects of SK053. Quantitative real-time polymerase chain reaction analysis revealed that incubation of HL-60 cells with SK053 resulted in increased levels of transcripts for differentiation-associated genes, such as differentiation-driving transcription factors (CEBPB, CEBPE), early markers of neutrophil differentiation (HK3) and genes expressed by mature granulocytes (CSF3R encoding G-CSFR, CSF2R encoding GM-CSFR, ELANE and MPO) (Figure 1F). Cytostatic/cytotoxic and differentiation-promoting effects of SK053 were also observed in NB4, another acute promyelocytic leukemia cell line, as well as in non-acute promyelocytic leukemia AML cell lines, such as MOLM14 and KG1 (Online Supplementary Figure S1).
Figure 1.
SK053 induces differentiation of AML cells. (A) HL-60 cells were incubated with various concentrations of SK053 for up to 5 days. Each day the number of dead cells was evaluated with trypan blue exclusion (mean ± SD from 6 experiments). (B) Co-culture of HL-60 cells with universal bone marrow stromal cells (HS5) did not impair SK053 cytotoxicity. HL-60 cells were seeded in a transwell system on top of HS5 cells and incubated with SK053 for 72 h. Next, HL-60 cells were analyzed for viability using propidium iodide (PI) staining in flow cytometry (data are shown as mean percentages of PI-positive cells ± SD from 3 experiments); *P<0.05 in one-way ANOVA with the Dunnett post-hoc test. (C) May-Grünwald-Giemsa (MGG) staining (upper panel) and nitroblue tetrazolium (NBT) (lower panel) in HL-60 cells incubated with SK053 for 120 h. All-trans retinoic acid (ATRA) served as a positive control. (D) Semi-quantitative colorimetric NBT reduction assay in HL-60 cells incubated for 5 days with 10 mM SK053 (data are shown as mean increases, in %, over controls ± SD for 3 experiments); *P<0.05 in one-way ANOVA with the Dunnett post-hoc test. (E) Mean percentage ± SD of HL-60 cells expressing CD11b myeloid marker was determined in flow cytometry in 3 experiments; *P<0.05 in one-way ANOVA with the Dunnett post hoc test. (F) Real-time quantitative polymerase chain reaction analysis of HL-60 cells incubated for 5 days with 10 μM SK053; results are presented as mean target-to-reference ratio ± SD of three experiments; *P<0.05 vs. controls, two-tailed Student t test, #P<0.05 vs. controls, one-tailed Student t test.
SK053 induces gene expression profile associated with stress response and differentiation
To get further insight into the mechanisms of SK053 activity in AML cells, we sequenced and analyzed transcriptomes of controls and SK053-treated HL-60 cells using next-generation sequencing. Analysis of transcriptomes of controls and HL-60 cells incubated with SK053 for 48 h (Online Supplementary Figure S2) revealed differential expression of genes (Online Supplementary Figure S3A, Online Supplementary Table S1) that, in gene ontology analysis, clustered into two major groups: (i) biological processes (Online Supplementary Figure S3B, Online Supplementary Table S2) involving the stress response (e.g. genes encoding calreticulin or proteasome subunits), regulation of apoptosis (e.g. genes encoding Bcl-2-related protein, CFLAR or TNFRSF10B) and myeloid cell differentiation (CBFB, JAK2 or PIR) and (ii) molecule function (Online Supplementary Figure S3C, Online Supplementary Table S3) associated with changes in oxidoreductases activity [e.g. NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, ATP synthase or cytochrome c]. Furthermore, a 5-day incubation of HL-60 cells with SK053 induced the expression of genes regulating myeloid cell differentiation (JUN, CSF1, ID2) and cell proliferation (BCL6, PNMT) (Online Supplementary Figure S4A-C, Online Supplementary Tables S4-S6). Immunoblotting of the lysates of HL-60 cells confirmed the results of the next-generation sequencing analysis showing that SK053 induces endoplasmic reticulum stress and the unfolded protein response (Online Supplementary Figure S5).
SK053 binds to and inhibits protein disulfide isomerase
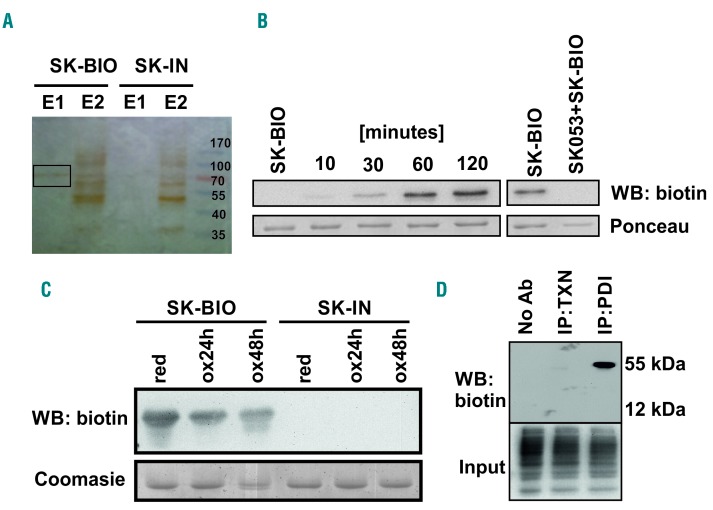
We then used lentiviral particles encoding short hairpin RNA (shRNA) to target thioredoxin in HL-60 cells. Intriguingly, thioredoxin knock-down with shRNA failed to inhibit the growth of HL-60 cells (Online Supplementary Figure S6A). Subsequently, we used two other shRNA sequences targeting thioredoxin (Online Supplementary Figure S6B) and assessed the effects of thioredoxin knockdown in HL-60 and MOLM14 cells. While both hairpins effectively suppressed the growth of MOLM14 cells, only one shRNA slightly inhibited and the other had no effect on the growth of HL-60 cells (Online Supplementary Figure S6C,D). These observations indicate that inhibition of thioredoxin might not be responsible for the differentiation-inducing effects of SK053 in HL-60 cells. Therefore, using a biotin affinity probe-labeling approach followed by mass spectrometry, we sought to identify other potential intracellular SK053 targets in these cells. Incubation of HL-60 cells for 4 h with SK-BIO, an active, biotinylated SK053 analog (see Online Supplementary Figure S8 for the general structures of compound used herein), followed by pulldown with avidin-coated beads and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), revealed a prominent band of ~50-70 kDa molecular weight in silver-stained gels (Figure 2A). An inactive, biotinylated analog of SK053 that lacks the electrophilic double bond (referred to as SK-IN, Online Supplementary Figure S8) failed to precipitate any protein. The ~50-70 kDa protein was identified in mass spectrometry to be a protein disulfide isomerase (PDI), a product of the P4HB gene (Online Supplementary Figure S9A-D, Online Supplementary Table S7). SDS-PAGE followed by the detection of biotin revealed covalent binding of SK-BIO to recombinant human (rhu) PDI (Figure 2B). Oxidation of rhuPDI, which decreases the number of sulfhydryl groups, reduced binding of SK-BIO to PDI indicating a thioldependent interaction (Figure 2C). Moreover, in a competition assay, pre-incubation of rhuPDI with SK053 prevented binding of SK-BIO, indicating the same binding site for both compounds (Figure 2B).
Figure 2.
SK053 binds to protein disulfide isomerase. (A) HL-60 cells were incubated with 100 μM SK-BIO and lysed. Proteins binding SK-BIO were precipitated with avidin-coated beads and eluted with buffers E1 and E2 as described in the Methods section. Next, the samples were separated by SDS-PAGE. A band obtained with buffer E1 (black rectangle) was excised from silver-stained gel and analyzed by mass spectrometry. An inactive biotinylated SK053 analog (SK-IN) was used as a negative control. A representative result of a series of experiments is presented. (B) Recombinant human (rhu) PDI was incubated with a 10× molar excess of SK-BIO for 10–120 min at 37°C (left) or preincubated for 1 h with a 10× molar excess of SK053 followed by 1 h incubation with a 10× molar excess of SK-BIO (right). Next, proteins were separated by SDS-PAGE followed by western blotting (WB) using anti-biotin monoclonal antibody. Membranes stained with Ponceau red served as loading controls. A representative result of a series of experiments is presented. (C) Reduced (red) and oxidized (ox) rhuPDI were incubated with SK-BIO followed by immunoblotting for biotin. Gels stained with Coomasie blue served as loading controls. SK-IN was used as a negative control. A representative result of a series of experiments is presented. (D) The lysates of HL-60 cells were incubated with SK-BIO followed by biotin immunoprecipitation and western blotting. Biotin detection in total cell lysates (input) was used as a loading control. Lysate incubated with protein G agarose beads in the absence of antibodies was used as a negative control (No Ab). A representative result of a series of experiments is presented. IP: immunoprecipitation; TXN: thioredoxin.
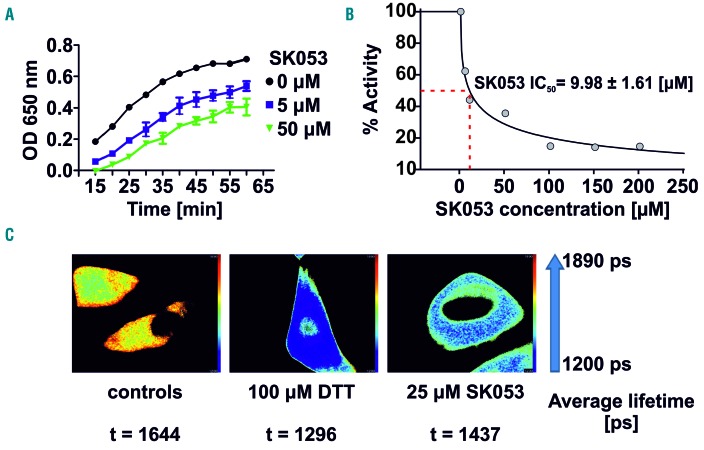
Immunoprecipitation of PDI and thioredoxin from the lysates of HL-60 cells cultured with SK-BIO revealed the presence of biotinylated PDI but not thioredoxin (Figure 2D), indicating that in HL-60 cells PDI is a preferential target for the compound. Further mass spectrometry revealed that SK053 binds to the catalytic cysteines localized in the fourth thioredoxin-like domain of PDI (Online Supplementary Figure S9E). Furthermore, the molecular dynamics simulation followed by covalent docking of SK053 to the fourth thioredoxin-like domain of human PDI (pdb|4ekz) indicated that the binding of the truncated form of SK053 (a molecule without the leaving group, Online Supplementary Figure S8D) is equally likely to occur by forming a covalent bond with either sulfur from Cys397 or Cys400 (Online Supplementary Figure S10, Online Supplementary Movie 1). Next, we sought to determine whether binding of SK053 translates into inhibition of PDI activity. A turbidimetric assay of PDI-mediated insulin reduction revealed that SK053 efficiently blocks the enzymatic activity of rhuPDI with an IC50 value of 9.98 μM (Figure 3A,B). To further investigate whether SK053 can inhibit PDI activity in cells, we transfected HeLa cervical cancer cells with a plasmid encoding an endoplasmic reticulum-tuned fluorescent redox-responsive probe (roGFPiE)12 and measured the response of this probe using fluorescence lifetime imaging in a time-correlated single-photon counting mode. The fluorescent lifetime of the probe declined in SK053-treated HeLa cells, reflecting the reductive shift in the dithiol-disulfide steady state and indicating SK053-mediated inhibition of PDI activity in the endoplasmic reticulum of living cells (Figure 3C).
Figure 3.
SK053 inhibits enzymatic activity of protein disulfide isomerase. (A) A turbidimetric assay of recombinant human PDI-mediated insulin disulfide reduction (mean ± SD of 6 experiments). (B) Determination of the IC50 value for SK053 with SigmaPlot software. (C) Fluorescence lifetime imaging (FLIM) of HeLa cells transfected with plasmid encoding an endoplasmic reticulum-tuned fluorescent redox-responsive probe (roGFPiE) tracking the activity of the PDI in cells; 100 μM dithiothreitol (DTT), a reducing agent, was used as a positive control. A representative result of a series of experiments is presented.
Inhibition of protein disulfide isomerase increases C/EBPa levels and inhibits the growth of acute myeloid leukemia cells
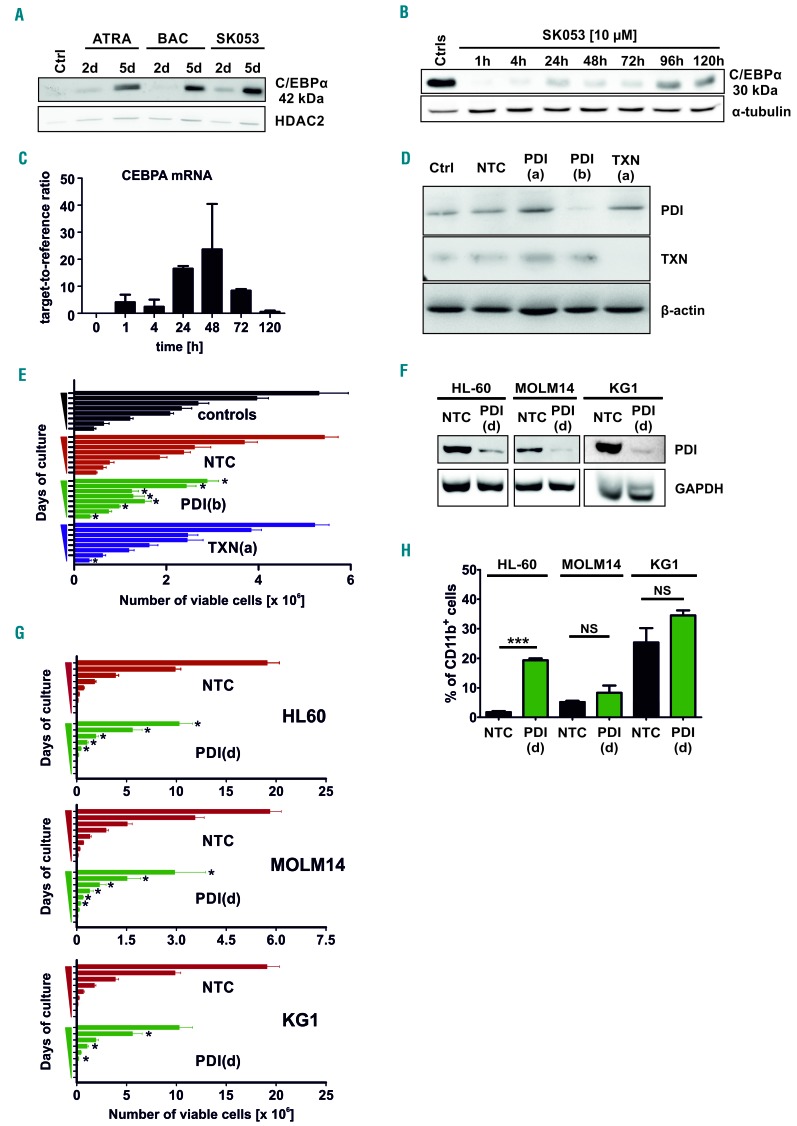
Recently, it was shown that PDI interacts with the stem loop region of mRNA for C/EBPα thereby blocking its translation.13 Considering that C/EBPα is a crucial transcription factor driving myeloid cells differentiation14,15 we investigated the effects of SK053 on C/EBPa levels in AML cells. Incubation of HL-60 cells with SK053 increased the nuclear levels of the 42 kDa C/EBPα isoform (Figure 4A) and induced CEBPA mRNA expression (Figure 4C). Additionally, the oncogenic p30 kDa C/EBPα levels were strongly suppressed in cells incubated with SK053 (Figure 4B). Accordingly, SK053 decreased the levels of SOX4 mRNA (Figure 1G), a direct target of C/EBPα, the expression of which has been shown to correlate inversely with C/EBPα activity, increased self-renewal of leukemic cells as well as a block in blast differentiation.16 shRNA-mediated knock-down of PDI (P4HB) expression significantly suppressed the proliferation of HL-60 cells (Figure 4D,E). Lentiviral transduction of HL-60, MOLM14 and KG1 cells with another shRNA sequence confirmed that PDI knockdown is associated with significant growth suppression (Figure 4F,G) and upregulates C/EBPα protein (Online Supplementary Figure S7A). PDI knock-down also increased the percentage of CD11b+ HL-60 cells as compared with nontargeting controls, but failed to do so in MOLM14 and KG1 cells (Figure 4H). Unexpectedly, SK053 strongly suppressed the growth and induced differentiation of HL-60 cells transduced with two independent shRNA sequences targeting CEBPA (Online Supplementary Figure S7B-D), indicating that the presence of this differentiation-associated transcription factor is not necessary for the antileukemic effects of SK053.
Figure 4.
SK053 increases CEBPA levels in HL-60 cells and protein disulfide isomerase knock-down impairs the growth of acute myeloid leukemia cells and induces differentiation of HL-60 cells. (A) HL-60 cells were incubated for 2 or 5 days with 10 μM SK053. Subcellular fractions were isolated using a NE-PER kit. HDAC2 served as a loading control for the nuclear fraction. A representative result of a series of experiments is presented. (B) HL-60 cells were incubated for 1-120 h with 10 μM SK053, harvested, lysed and total cell lysates were immunoblotted for 30 kDa C/EBP α. α-tubulin levels served as a loading control. A representative result of a series of experiments is presented. For (A) and (B) the controls are HL-60 cells incubated with dimethylsulfoxide at the same concentration as used for SK053-treated group, harvested on day 5. (C) Real-time quantitative polymerase chain reaction of HL-60 cells incubated for the indicated times with 10 μM SK053; results are presented as mean target-to-reference ratio ± SD of three experiments. (D) Western blot showing the efficacy of PDI or thioredoxin (TXN) knock-down 5 days after transduction with shRNA-encoding lentiviruses. PDI(a) and PDI(b) indicate two different shRNA; β-actin (βA) served as a loading control. (E) Growth of HL-60 cells transduced with non-targeting (NTC), TXN [TXN(a)]- or PDI [PDI(b)]-targeting shRNA lentiviral particles. Cells were counted daily in trypan blue from day 1 until day 8. *P<0.05 vs. NTC in a two-tailed Student t-test. For (C) and (D) the controls are non-modified HL-60 cells. (F) Western blot showing the efficacy of PDI knock-down in AML cell lines transduced with PDI-targeting shRNA [PDI(d)]-encoding lentiviruses. GAPDH served as a loading control. (G) Growth of HL-60, MOLM14 and KG1 cells transduced with non-targeting (NTC) or PDI-targeting [PDI(d)] shRNA lentiviral particles. Cells were counted daily in trypan blue from day 1 until day 9. *P<0.05 vs. NTC in a two-tailed Student t-test. (H) Mean percentage ± SD of AML cells modified with non-targeting (NTC) or PDI-targeting [PDI(d)] shRNA, expressing CD11b myeloid marker determined in flow cytometry (n=4 experiments, except for KG1: n=2), ***P<0.0001 vs. NTC, two-tailed Student t-test; NS: not significant.
Protein disulfide isomerase inhibition targets leukemia-initiating cells and is effective in primary acute myeloid leukemia cells
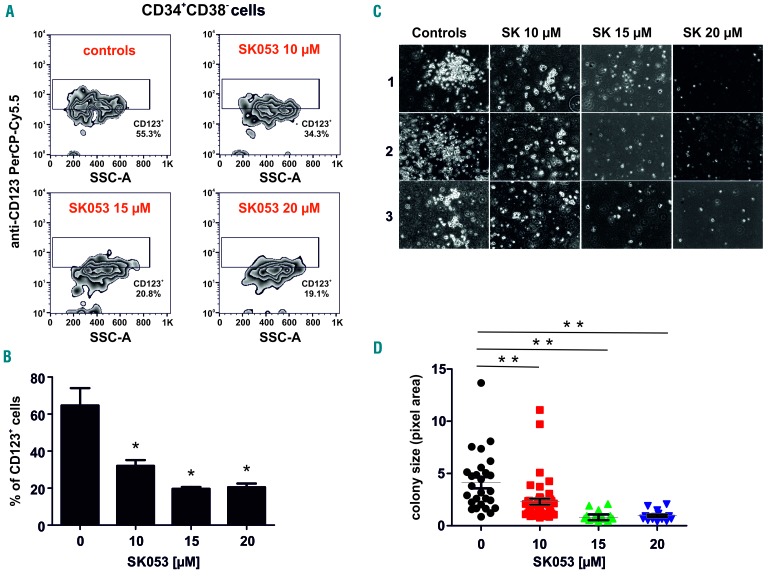
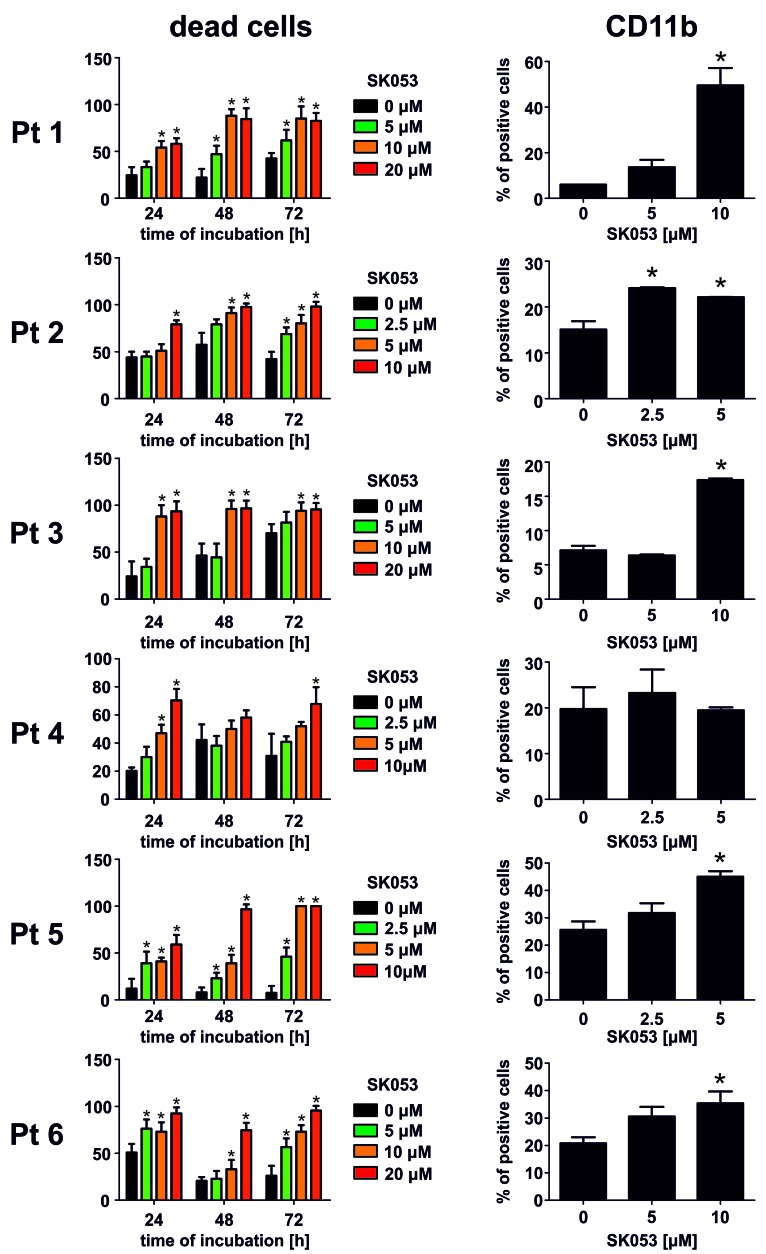
AML is characterized by a small population of self-renewing leukemic stem cells referred to as leukemia-initiating cells that give rise to a large population of immature leukemic blasts.17 Since leukemia-initiating cells are intrinsically resistant to chemotherapeutics and AML frequently relapses due to incomplete elimination of leukemia-initiating cells18 we sought to investigate the effects of SK053 on the survival of CD123+CD34+CD38-leukemia-initiating cell-like cells within the KG1 AML cell line. We observed a concentration-dependent decrease in the percentage of these cells when incubated with SK053 (Figure 5A,B). Moreover, pretreatment of KG1 cells with SK053 decreased the clonogenic potential of these cells in a colony formation assay in vitro (Figure 5C,D). Moreover, SK053 exerted antileukemic effects in primary AML blasts isolated from bone marrow or peripheral blood of AML patients. We observed a decrease in blast survival in all samples from six patients investigated, accompanied by an increased percentage of cells expressing CD11b marker in five out of the six samples (Figure 6).
Figure 5.
SK053 decreases the percentage of KG1 leukemia-initiating cells and their clonogenic potential in vitro. (A) KG1 cells were incubated for 72 h with the indicated concentrations of SK053, harvested and stained for flow cytometry. Representative density plots from a series of experiments are presented. (B) Mean percentages of CD123+ KG1 cells among CD34+ and CD38− cells ± SD (n=3 experiments) *P<0.05 vs. controls, one-way ANOVA with Dunnett post-hoc test. (C) Representative pictures of the clonogenic assay plates on day 14 after seeding KG1 cells pretreated for 72 h with SK053. (D) Mean clone size (pixel area) ± SEM (n=3 experiments); **P<0.0001 vs. controls, one-way ANOVA with Dunnett post-hoc test.
Figure 6.
SK053 exerts antileukemic effects and induces differentiation of primary leukemic blasts isolated from the bone marrow of patients with acute myeloid leukemia. (Left) Primary AML blasts isolated from six patients (numbered consecutively Pt 1 to Pt 6) were incubated with the indicated concentrations of SK053 for 72 h. Each day the number of dead cells was evaluated with trypan blue exclusion. Graphs present mean percentages of dead cells ± SD (n=6 experiments). *P<0.05 vs. controls, one-way ANOVA with Dunnett post-hoc test. (Right) Primary AML blasts isolated from six patients were incubated with the indicated concentrations of SK053 for 72 h. Percentages of CD11b+ cells among live (7-AAD−) and CD33+ cells were evaluated in flow cytometry. Graphs present mean percentages of dead cells ± SD (n=3 experiments). *P<0.05 vs. controls, one-way ANOVA with Dunnett post-hoc test.
Discussion
In this study we demonstrate that SK053 induces differentiation of AML cells. SK053 has been developed as a thioredoxin inhibitor and was reported to induce endoplasmic reticulum stress and apoptosis of tumor cells.11 Thus, it was to some extent unexpected to observe that SK053 induces cytostatic/cytotoxic effects against HL-60 cells (Figure 1A,B), while thioredoxin knock-down with shRNA turned out to be largely ineffective in suppressing the growth of these cells (Online Supplementary Figure S6C). This observation prompted us to look for an additional cellular target for SK053. Using biotinylated SK053 as a bait in a biotin affinity probe-labeling approach we observed that SK053 binds to PDI (Figure 2, Online Supplementary Figure S9). Molecular docking confirmed the feasibility of this interaction (Online Supplementary Figure S10) and further precipitation, as well as enzymatic and functional analyses, revealed that SK053 binds to and inhibits the activity of human PDI (Figures 2 and 3).
PDI is the original member of a family of PDI proteins that contain a characteristic CXXC motif, with two cysteine residues forming a disulfide bond that is cleaved by oxidoreductases or by thiol-disulfide exchange. Although all PDI family members contain a thioredoxin-like domain, they differ considerably in size, domain composition and enzymatic properties. PDI is involved in the formation and isomerization of disulfide bonds between cysteine residues of polypeptides as they fold.19 Such disulfide modifications participate in post-translational protein control and affect the functions of many proteins. PDI also participates in the maintenance of cellular homeostasis by mediating oxidative protein folding and acts as a chaperone.19 A number of studies indicate that various PDI are highly expressed in multiple cancer types as compared with matched normal tissues and play an important role in supporting cancer progression.19 PDI is induced in tumor cells during the unfolded protein response and has been associated with chemoresistance.20,21 Inhibition of PDI activity with a non-selective inhibitor, bacitracin, enhanced apoptosis triggered by bortezomib or fenretinide.22 Propynoic acid carbamoyl methyl amide (PACMA31), an irreversible PDI inhibitor, exhibited significant antitumor effects in ovarian cancer models23 and potentiated the antitumor effects of sorafenib in hepatocellular carcinoma.24 A structurally different PDI inhibitor (CCF642) was shown to exert anti-myeloma activity associated with the induction of endoplasmic reticulum stress and apoptosis-inducing calcium release.25 All these observations indicate that PDI is a potential drug target in cancer treatment.
Remarkably, Heafliger et al. have shown that PDI binds to the stem loop region of C/EBPα mRNA, thereby blocking its translation.13 Our results indicate that SK053 upregulates C/EBPα levels (Figure 4), downregulates its downstream effector SOX4, and induces differentiation of AML cells (Figure 1F). However, despite C/EBPα upregulation we have observed that SK053 still exerts potent growth-inhibitory effects in HL-60 cells with stably suppressed CEBPA expression induced by two different shRNA sequences (Online Supplementary Figure S7). Thus, it seems unlikely that PDI inhibition with SK053 directly leads to stabilization of CEBPA mRNA. Nonetheless, PDI knockdown with shRNA (Figure 4) exerted cytostatic/cytotoxic effects in HL-60, MOLM14 and KG1 cells and induced differentiation of HL-60 cells. In contrast to HL-60 cells, thioredoxin knock-down in MOLM14 and KG1 cells was also associated with inhibition of cell growth. Thus, it seems that both thioredoxin and PDI are involved in the survival of these cells, and since SK053 targets both enzymes, the antileukemic activity of this compound can be attributed to its dual selectivity. The findings that SK053 significantly decreases the percentage of CD123+CD34+CD38− leukemia-initiating cells and decreases the clonogenic potential of these cells in the colony formation assay in vitro are particularly important. Leukemia-initiating cells seem to have a pivotal role in the relapse of AML and are considered to be resistant to conventional chemotherapy and targeted therapies.26
Altogether, we show that SK053 targets PDI by thiol-dependent interactions, modulates C/EBPα levels and induces differentiation of AML cells. These observations indicate that PDI is a druggable target for differentiation treatment in AML.
Supplementary Material
Acknowledgments
The authors thank Dr. Magdalena Winiarska from the Department of Immunology, MUW and Prof. Andrzej Dziembowski from the Institute of Biochemistry and Biophysics PAS for their critical and helpful review of this work as well as Dr. Tomasz Stoklosa from the Department of Immunology, MUW for advice concerning the clonogenic assays. This work was supported by the Polish National Science Center, grant numbers: 2013/10/E/NZ5/00778 (DN), 2014/15/B/ST6/05082 (DP), Foundation for Polish Science (TEAM to DP), Ministry of Science and Higher Education, grant number: IP2011 038971 (DN) and European Commission Horizon 2020 Programme 692180-STREAMH2020-TWINN-2015 (JG).
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/11/1843
References
- 1.Dohner H, Weisdorf DJ, Bloomfield CD. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–52. [DOI] [PubMed] [Google Scholar]
- 2.Mi JQ, Li JM, Shen ZX, Chen SJ, Chen Z. How to manage acute promyelocytic leukemia. Leukemia. 2012;26(8):1743–1751. [DOI] [PubMed] [Google Scholar]
- 3.Lengfelder E, Hofmann WK, Nowak D. Impact of arsenic trioxide in the treatment of acute promyelocytic leukemia. Leukemia. 2012;26(3):433–442. [DOI] [PubMed] [Google Scholar]
- 4.Coombs CC, Tavakkoli M, Tallman MS. Acute promyelocytic leukemia: where did we start, where are we now, and the future. Blood Cancer J. 2015;5:e304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montalban-Bravo G, Garcia-Manero G. Novel drugs for older patients with acute myeloid leukemia. Leukemia. 2015;29(4): 760–769. [DOI] [PubMed] [Google Scholar]
- 6.Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci USA. 1997;94(2):569–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radomska HS, Huettner CS, Zhang P, Cheng T, Scadden DT, Tenen DG. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol Cell Biol. 1998;18(7):4301–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pabst T, Mueller BU, Zhang P, et al. Dominant-negative mutations of CEBPA, encoding CCAAT/enhancer binding protein-alpha (C/EBPalpha), in acute myeloid leukemia. Nat Genet. 2001;27(3):263–270. [DOI] [PubMed] [Google Scholar]
- 9.Radomska HS, Basseres DS, Zheng R, et al. Block of C/EBP alpha function by phosphorylation in acute myeloid leukemia with FLT3 activating mutations. J Exp Med. 2006;203(2):371–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schardt JA, Weber D, Eyholzer M, Mueller BU, Pabst T. Activation of the unfolded protein response is associated with favorable prognosis in acute myeloid leukemia. Clin Cancer Res. 2009;15(11):3834–3841. [DOI] [PubMed] [Google Scholar]
- 11.Klossowski S, Muchowicz A, Firczuk M, et al. Studies toward novel peptidomimetic inhibitors of thioredoxin-thioredoxin reductase system. J Med Chem. 2012;55(1):55–67. [DOI] [PubMed] [Google Scholar]
- 12.Avezov E, Cross BC, Kaminski Schierle GS, et al. Lifetime imaging of a fluorescent protein sensor reveals surprising stability of ER thiol redox. J Cell Biol. 2013;201(2):337–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haefliger S, Klebig C, Schaubitzer K, et al. Protein disulfide isomerase blocks CEBPA translation and is up-regulated during the unfolded protein response in AML. Blood. 2011;117(22):5931–5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohlsson E, Schuster MB, Hasemann M, Porse BT. The multifaceted functions of C/EBPalpha in normal and malignant haematopoiesis. Leukemia. 2016;30(4):767–775. [DOI] [PubMed] [Google Scholar]
- 15.Pulikkan JA, Tenen DG, Behre G. C/EBPalpha deregulation as a paradigm for leukemogenesis. Leukemia. 2017;31(11): 2279–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang H, Alberich-Jorda M, Amabile G, et al. Sox4 is a key oncogenic target in C/EBPalpha mutant acute myeloid leukemia. Cancer Cell. 2013;24(5):575–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. [DOI] [PubMed] [Google Scholar]
- 18.Jordan CT. Unique molecular and cellular features of acute myelogenous leukemia stem cells. Leukemia. 2002;16(4):559–562. [DOI] [PubMed] [Google Scholar]
- 19.Xu S, Sankar S, Neamati N. Protein disulfide isomerase: a promising target for cancer therapy. Drug Discov Today. 2014;19(3): 222–240. [DOI] [PubMed] [Google Scholar]
- 20.Higa A, Taouji S, Lhomond S, et al. Endoplasmic reticulum stress-activated transcription factor ATF6alpha requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol Cell Biol. 2014;34(10):1839–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tufo G, Jones AW, Wang Z, et al. The protein disulfide isomerases PDIA4 and PDIA6 mediate resistance to cisplatin-induced cell death in lung adenocarcinoma. Cell Death Differ. 2014;21(5):685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovat PE, Corazzari M, Armstrong JL, et al. Increasing melanoma cell death using inhibitors of protein disulfide isomerases to abrogate survival responses to endoplasmic reticulum stress. Cancer Res. 2008;68(13): 5363–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu S, Butkevich AN, Yamada R, et al. Discovery of an orally active small-molecule irreversible inhibitor of protein disulfide isomerase for ovarian cancer treatment. Proc Natl Acad Sci USA. 2012;109(40):16348–16353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Won JK, Yu SJ, Hwang CY, et al. Protein disulfide isomerase inhibition synergistically enhances the efficacy of sorafenib for hepatocellular carcinoma. Hepatology. 2017;66(3): 855–868. [DOI] [PubMed] [Google Scholar]
- 25.Vatolin S, Phillips JG, Jha BK, et al. Novel protein disulfide isomerase inhibitor with anticancer activity in multiple myeloma. Cancer Res. 2016;76(11):3340–3350. [DOI] [PubMed] [Google Scholar]
- 26.Pollyea DA, Jordan CT. Therapeutic targeting of acute myeloid leukemia stem cells. Blood. 2017;129(12):1627–1635. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.