Interdigitating dendritic cell sarcoma (IDCS) is a rare neoplasm considered to derive from a dendritic cell. Recent studies have shown that B- or T-lymphoblastic leukemia/lymphomas can develop clonally related histiocytic/dendritic cell (H/DC) neoplasms, such as histiocytic sarcoma, Langerhans cell sarcoma, and IDCS.1–3 Considering the cell types involved, these H/DC neoplasms experience lineage conversion from lymphoid neoplasms into myeloid neoplasms. One explanation of this phenomenon is that the origins of lymphoblastic leukemia/lymphomas are immature precursor lymphoid cells, which might retain lineage plasticity. However, mature B-cell lymphomas, such as follicular lymphoma and small lymphocytic lymphoma, can also develop H/DC neoplasms with identical genetic changes.4,5 These observations indicate lineage plasticity even in lineage-committed mature B-cell neoplasms, and the precise mechanism of such lineage conversion is still unknown. Diffuse large B-cell lymphoma (DLBCL) is a common subtype of aggressive B-cell non-Hodgkin lymphoma, which sometimes harbors MYC gene rearrangement. Herein, we describe the first case of DLBCL with MYC translocation that subsequently developed into clonally related IDCS. We performed a comprehensive genomic analysis using time-sequential samples to elucidate clonal development and driver event of lineage conversion from B-lymphoid into myeloid lineage.
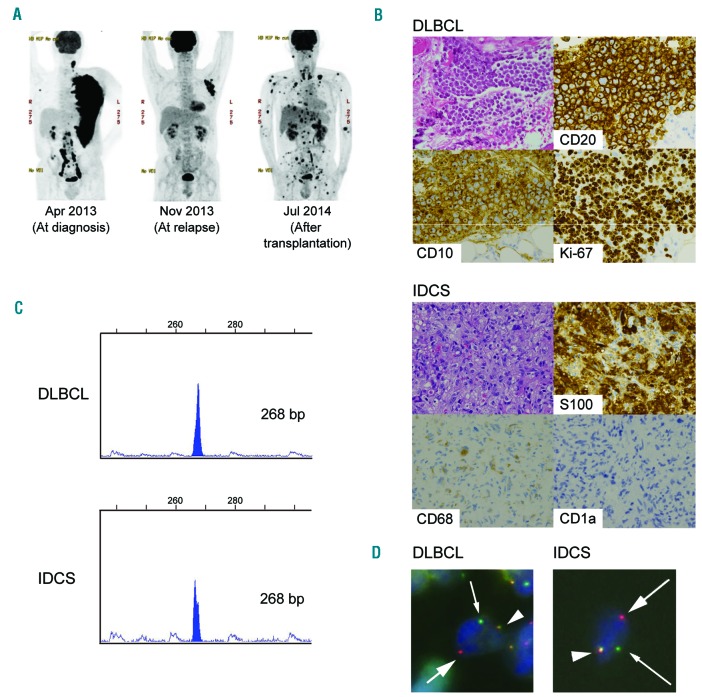
A 52-year-old man with human immunodeficiency virus type 1 (HIV-1) infection presented with rapidly enlarging axillary lymph nodes (Figure 1A, left). Immunohistochemical analysis of the left axillary lymph node biopsy revealed that the tumor cells were positive for CD10, CD20, BCL6, MYC, and MKI67 (their index was more than 95%), but negative for BCL2 or EBER (Figure 1B, top). Fluorescent in situ hybridization (FISH) analysis revealed the presence of MYC translocation. We diagnosed the tumor as MYC positive DLBCL and performed first-line chemotherapy. After a transient complete remission state, DLBCL relapsed in the same lesion (Figure 1A, center), and the patient received salvage chemotherapies, including allogeneic bone marrow transplantation. However, an FDG-PET/CT scan taken after transplantation showed strong FDG accumulation in multiple organs, whereas FDG uptake at the original lesion site disappeared (Figure 1A, right). Upper gastrointestinal endoscopy on day 35 after transplantation showed multiple submucosal tumors, and immunohistochemical analysis of the gastric tumor biopsy revealed that tumor cells were strongly positive for S100, fascin, and CD163 and focally positive for CD68, but negative for lysozyme, CD1A, langerin, CD20, CD21, myeloperoxidase, or PAX5 (Figure 1B, bottom). We diagnosed the tumor as IDCS with histiocytic differentiation. In addition, this tumor showed no evidence of DLBCL, which was evident from negativity for PAX5 and CD20. To investigate the clonal relationship of DLBCL and subsequently developed IDCS, genetic studies were performed in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the Graduate School of Medicine, Kyoto University, and Kobe City Medical Center General Hospital. A written informed consent was obtained for genetic studies. We performed immunoglobulin heavy chain (IGH) gene rearrangement analysis by PCR of DNAs extracted from DLBCL and IDCS specimens, which revealed identical peaks in both tumors (Figure 1C). FISH experiments performed on paraffin section using MYC (8q24) break apart probes revealed that both DLBCL and IDCS cells had split MYC signals (Figure 1D). These results indicated that both tumors from distinct lineages had a clonal relationship. Detailed clinical course and experimental methods are described in the Online Supplementary Information.
Figure 1.
Clinical findings in a patient with diffuse large B-cell lymphoma (DLBCL) and interdigitating dendritic cell sarcoma (IDCS). (A) FDG PET/CT scans at diagnosis (left), at relapse (center), and after transplantation (right). (B) Hematoxylin-eosin staining (original magnification ×200) and immunostaining of tumors. (C) DLBCL and IDCS cells show identical clonal peaks according to PCR for the immunoglobulin heavy chain (IGH) gene. Primers for IGH variable framework region 2 and joining region were used. (D) Fluorescent in situ hybridization using MYC break apart probes demonstrates splitting of MYC gene signals (arrows) in DLBCL and IDCS cells.
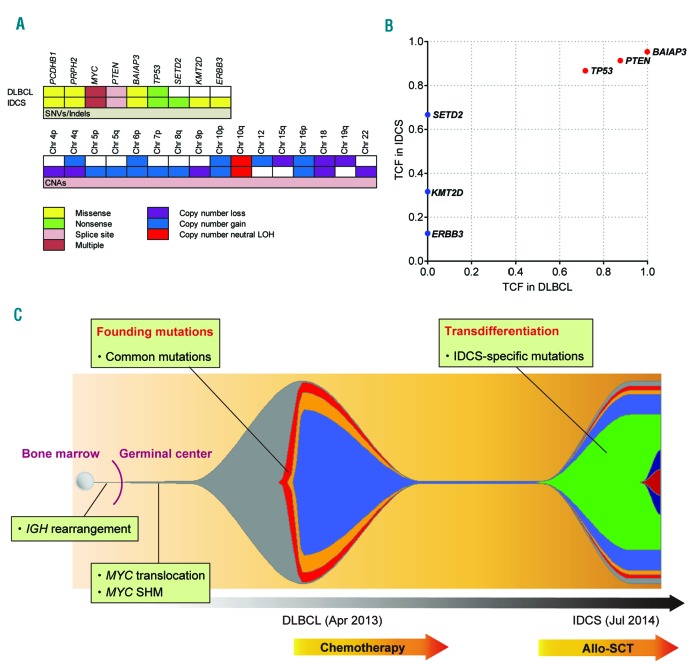
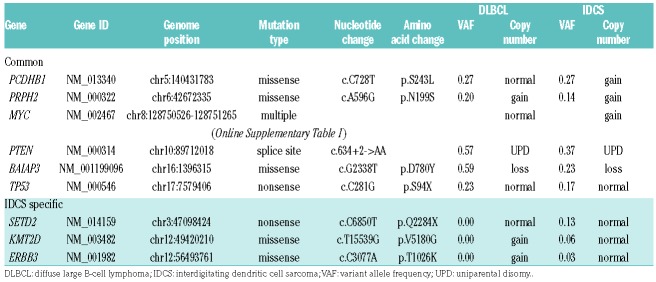
Next, we performed whole-exome sequencing (WES), using samples of DNA from DLBCL, IDCS, and normal tissue (buccal cells, as control). Overall, the average depth for WES was 142× and 93% of the exome targets covered more than 30×. In total, 37 somatic non-synonymous mutations in nine genes were identified and validated by amplicon sequencing (Figure 2A, Table 1, and Online Supplementary Table S1). Of these mutations, 34 were present in both DLBCL and IDCS, whereas three were specific to IDCS. We found a total of 29 mutations in MYC exon 2, all of which were common to both tumors. We also analyzed copy number alterations using sequencing depth and allele frequencies of heterozygous single nucleotide polymorphisms and revealed complex karyotype-like abnormalities, including uniparental disomy of chromosome 10q in both tumors (Figure 2A and Online Supplementary Figure S1). Therefore, biallelic inactivation of the PTEN gene was evident due to a splice site mutation and uniparental disomy of chromosome 10q in both tumors. Next, to clarify the clonal architecture of both tumors, we calculated the tumor cell fraction of each mutation based on the variant allele frequency, involved copy number, and estimated tumor purity (Figure 2B). We found that major clones of both tumors contained all common mutations, whereas some IDCS-specific mutations were subclonal. IDCS-specific mutations in genes such as SETD2, KMT2D, and ERBB3 were putative drivers that enforced clonal evolution from DLBCL to IDCS (Figure 2C).
Figure 2.
Whole exome sequencing data and clonal architecture of DLBCL and IDCS. (A) Somatic mutations and copy number alterations (CNAs) in DLBCL and IDCS. Chr: chromosome; LOH: loss of heterozygosity; SNV: single nucleotide variant. (B) Distribution of tumor cell fraction (TCF) of each mutation. (C) Scheme summarizing clonal evolution from DLBCL to IDCS. Each color represents a distinct mutation. The timeline is indicated below. SHM: somatic hypermutation; allo-SCT: allogeneic stem cell transplantation.
Table 1.
Non-synonymous somatic mutations in diffuse large B-cell lymphoma and interdigitating dendritic cell sarcoma.
Recent comprehensive genetic studies have identified many genetic drivers of DLBCL and classified the disease into some specific molecular subgroups.6,7 In our case, we identified genetic alterations in known targets such as TP53 mutation, biallelic inactivation of the PTEN gene, and somatic hypermutation (SHM) of the MYC gene, in addition to the MYC translocation. Our case was classified into germinal center B cell-like DLBCL by Hans algorithm, did not show the typical genomic characteristics of a specific molecular subgroup based on the mutation profiles of DLBCL,7 and was characterized by the MYC translocation and mutations which are associated with poorer survival.6 Though MYC translocation is also a feature of Burkitt lymphoma, our case did not show the genetic hallmarks of Burkitt lymphoma other than MYC translocation such as mutations in ID3 and TCF3.8 IDCS is an extremely rare disease, and only ~100 cases have been described in literature. Although TP53 and BRAF mutations have been reported in several cases,9 the molecular basis of this neoplasm largely remains unknown due to the lack of comprehensive genomic studies. WES of IDCS demonstrated a mutational profile similar to that of DLBCL, with mutations in the tumor suppressor genes TP53 and PTEN and multiple mutations in MYC. In addition, we found IDCS-specific mutations in genes such as SETD2, KMT2D, and ERBB3. SETD2 and KMT2D are associated with histone modification and are frequent targets of somatic mutations in cancers, including lymphomas.6 Deregulated epigenetic modifications due to these mutations may act as driver events in IDCS development. Furthermore, somatic mutations of ERBB3, which encodes a member of the epidermal growth factor receptor family of receptor tyrosine kinases, are common in colorectal and gastric cancers, but rare in hematological malignancies. The function of this gene remains elusive.10 Of note, IDCS as well as DLBCL harbored IGH rearrangement, MYC SHM, and even MYC translocation, which had not ever been identified in IDCS in literature. Although these novel genetic lesions may be involved in IDCS pathogenesis, further genetic studies investigating larger cohorts are needed to establish IDCS genetic landscape properly.
DLBCL and IDCS originate from two distinct (B-lymphoid and myeloid) lineages, and there are several possible pathways that can mediate lineage conversion. Here, we consider this lineage conversion process as transdifferentiation defined as direct conversion of a lineage-committed cell to another type of differentiated cell. This hypothesis is supported by the characteristic genetic alterations common to both tumors, such as IGH rearrangement, MYC SHM, and MYC translocation, which helped us to determine the developmental stage of the common ancestor B cell. Firstly, the origin of both tumors can be traced back to the bone marrow because they harbored clonal IGH gene rearrangement, which is considered to occur as a recombinase-mediated event in the bone marrow at the pre-B cell stage. Secondly, the shared MYC translocation and MYC SHM in both tumors indicated that the putative common ancestor cell had already been differentiated to the germinal center B cell stage because these genetic changes are known to be caused by the activity of activation-induced cytidine deaminase in the germinal center.11,12 Based on these findings, we propose that this lineage conversion was indeed transdifferentiation as the putative common ancestor cell had already been fully differentiated at the mature B cell stage and lineage conversion occurred thereafter (Figure 2C).
The precise mechanism of this lineage conversion remains unknown. However, because lymphoid neoplasms have preceded or coincided with H/DC neoplasms in all reported cases so far, it is reasonable to suppose that additional driver mutations in lymphoid neoplasms drive lineage conversion into H/DC lineage. We found mutations in two tumor suppressor genes, PTEN and TP53, in both DLBCL and IDCS. It has been reported that PTEN is involved in myeloid-lymphoid lineage plasticity and the development of H/DC neoplasms.13 It is also known that TP53 plays a key role in somatic cell differentiation and reprogramming.14 Therefore, mutations in these two tumor suppressor genes might increase the lineage plasticity and susceptibility to transdifferentiation. In addition, histone modification is a major epigenetic regulator and plays a crucial role in somatic cell reprogramming and transdifferentiation.15 Thus, IDCS-specific mutations in the histone modifying genes SETD2 and KMT2D may drive transdifferentiation through altered histone modification. However, this study involves only a single case, and further studies investigating larger cohorts should uncover the frequent driver events of this dynamic evolutionary process.
In summary, our study confirmed the presence of MYC translocation not only in DLBCL but also in subsequently developed IDCS, which indicated transdifferentiation of mature B-cell neoplasm into H/DC neoplasm. A comprehensive genomic analysis revealed novel genomic alterations in IDCS and identified the putative driver events of transdifferentiation.
Supplementary Material
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Feldman AL, Berthold F, Arceci RJ, et al. Clonal relationship between precursor T-lymphoblastic leukaemia/lymphoma and Langerhans-cell histiocytosis. Lancet Oncol. 2005;6(6):435–437. [DOI] [PubMed] [Google Scholar]
- 2.Waanders E, Hebeda KM, Kamping EJ, et al. Independent development of lymphoid and histiocytic malignancies from a shared early precursor. Leukemia. 2016;30(4):955–958. [DOI] [PubMed] [Google Scholar]
- 3.Kato M, Seki M, Yoshida K, et al. Genomic analysis of clonal origin of Langerhans cell histiocytosis following acute lymphoblastic leukaemia. Br J Haematol. 2016;175(1):169–172. [DOI] [PubMed] [Google Scholar]
- 4.Feldman AL, Arber DA, Pittaluga S, et al. Clonally related follicular lymphomas and histiocytic/dendritic cell sarcomas: evidence for transdifferentiation of the follicular lymphoma clone. Blood. 2008;111(12):5433–5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoecker MM, Wang E. Histiocytic/dendritic cell transformation of B-cell neoplasms: pathologic evidence of lineage conversion in differentiated hematolymphoid malignancies. Arch Pathol Lab Med. 2013;137(6):865–870. [DOI] [PubMed] [Google Scholar]
- 6.Reddy A, Zhang J, Davis NS, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171(2):481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmitz R, Young RM, Ceribelli M, et al. Burkitt lymphoma pathogenesis and therapeutic targets from structural and functional genomics. Nature. 2012;490(7418):116–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Liso E, Pennelli N, Lodovichetti G, et al. Braf mutation in interdigitating dendritic cell sarcoma: a case report and review of the literature. Cancer Biol Ther. 2015;16(8):1128–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi MR, Yoo NJ, Lee SH. Oncogenic ERBB3 mutations altering p. Val104 is rare in acute leukemias and non-Hodgkin lymphomas. Eur J Haematol. 2014;92(2):177–178. [DOI] [PubMed] [Google Scholar]
- 11.Robbiani DF, Bothmer A, Callen E, et al. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135(6):1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pasqualucci L, Neumeister P, Goossens T, et al. Hypermutation of multiple proto-oncogenes in B-cell diffuse large-cell lymphomas. Nature. 2001;412(6844):341–346. [DOI] [PubMed] [Google Scholar]
- 13.Carrasco DR, Fenton T, Sukhdeo K, et al. The PTEN and INK4A/ARF tumor suppressors maintain myelolymphoid homeostasis and cooperate to constrain histiocytic sarcoma development in humans. Cancer Cell. 2006;9(5):379–390. [DOI] [PubMed] [Google Scholar]
- 14.Kawamura T, Suzuki J, Wang YV, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460(7259):1140–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin H, Zhao A, Zhang C, Fu X. Epigenetic control of reprogramming and transdifferentiation by histone modifications. Stem Cell Rev. 2016;12(6):708–720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.