Multiple myeloma (MM) is a hematologic neoplasm that develops after transformation and excessive growth of plasma cells in the bone marrow. MM is characterized by heterogeneous genetic abnormalities and an extensive range of clinical outcomes.1,2 The marked variability of responses and the limited clinical prognostic value of the current genetic information justify the need to identify new biomarkers of response and develop personalized medicine strategies.3 The recent introduction of next-generation sequencing technologies has considerably advanced our understanding of the biological features of MM.4–6 Accordingly, we used a targeted deep sequencing panel, applying the highest read depth to date, to detect minor subclones and evaluated their impact on response to treatment. We also integrated these data with the clinical features of a very homogeneous cohort of patients to unearth new patterns of genetic alterations and gain new insights into the complexity of the clonal and subclonal architecture of MM.
We analyzed 79 tumor samples from newly diagnosed MM patients enrolled in the GEM10MAS65 clinical trial (registered at www.clinicaltrials.gov as #NCT01237249).7 This phase II study enrolled patients older than 65 years, and randomly assigned them to one of two treatment arms: sequential melphalan/prednisone/bortezomib (MPV) followed by lenalidomide/low-dose dexamethasone (Rd), or alternating MPV with Rd. This clinical trial was carried out in compliance with the Helsinki Declaration: it was approved by the appropriate research Ethics Committees and all participants provided written informed consent before inclusion. The median time to progression of this cohort of patients was 26.4 months and the median follow up was 31.5 months. Plasma cells were enriched from bone marrow aspirates using anti-CD138+ immunomagnetic beads obtaining a purity higher than 85% in all cases and only 20 ng of DNA were used for library construction. The panel screens 77 genes that are frequently mutated in MM, including those related to critical pathophysiological pathways, genes associated with drug resistance, or those targetable by molecular drugs (Online Supplementary Table S1).8–10 The average coverage depth for CD138+ and CD138− samples was 1600 × and 900 ×, respectively. The minimum coverage of the detected variants was 20 ×, and the average cover age was 370 ×. We applied two different bioinformatics pipelines developed by Thermofisher (Ion reporter 5.0) and by the Spanish National Cancer Research Center (Rubioseq, http://rubioseq.bioinfo.cnio.es/).11 We also sequenced 11 germline fractions available to confirm that most of the germinal variants had been correctly excluded by our conservative filter criteria.
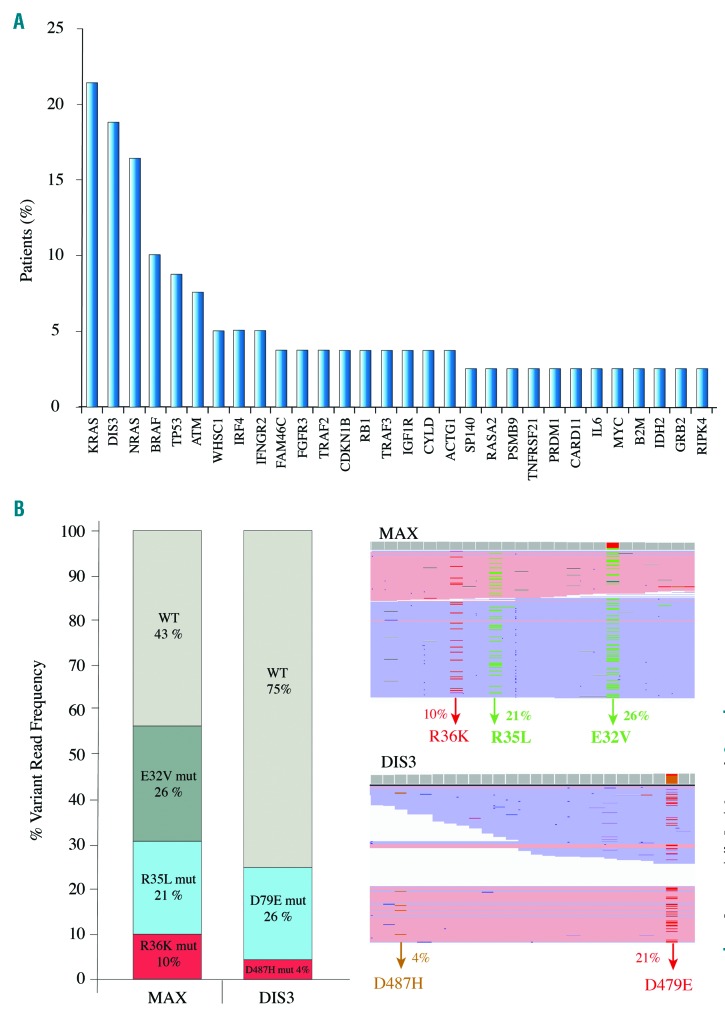
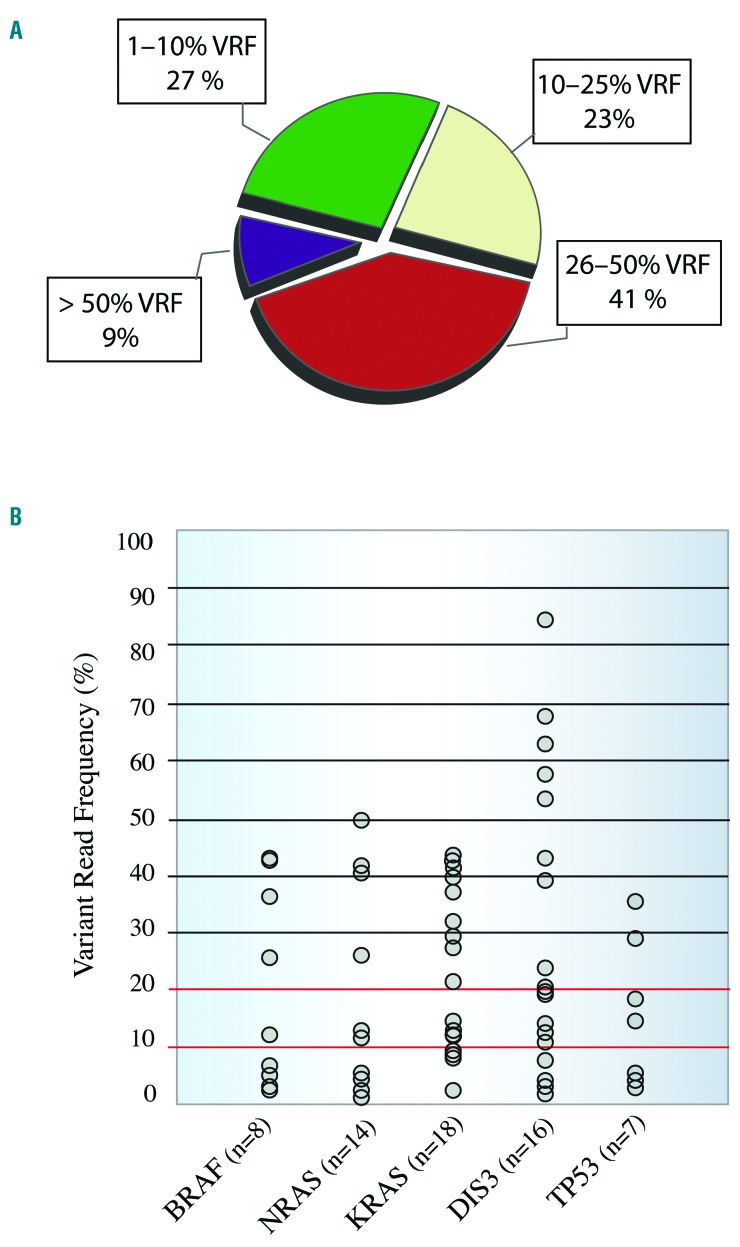
We identified 168 non-synonymous missense/nonsense/stop-loss single variants, of which 81 (48%) were predicted to be pathogenic by SIFT and PolyPhen-2, and 61 (36%) have been described in the COSMIC database. Eighty-five percent of patients harbored at least one mutation, with there being a median of 2.1 mutations per patient. Mutations were detected in 53 genes, although the following five genes accounted for 42% of the total number of mutations: KRAS (21.5% of patients), DIS3 (19%), NRAS (16.5%), BRAF (10.1%) and TP53 (8.8%) (Figure 1A). The RAS pathway was the most recurrently mutated, present in 48% (39/79) of patients. Seventy-two and 100% of KRAS and NRAS mutations, respectively, were detected at the hotspot codons 12, 13 and 61, and the targetable BRAF V600E activating mutation was detected in one patient. KRAS and BRAF mutations were co-existent in three patients, while RAS was always mutated exclusively. NF-κB was the second most fre quently mutated pathway in our cohort, accounting for 15% of all mutations; mutations were found in the genes of 25% of the patients (19/79), including the adapter signaling genes TRAF3 (1 nonsense and 4 missense) and TRAF2 (3 missense). All were predicted to be pathogenic by SIFT and PolyPhen-2. Other pathways that were altered in our cohort were MYC, in 11% of patients (9/79), and Cereblon and Cyclin, each found in 9% of patients (7/79). Multiple mutations in the same gene were observed in 12 patients, affecting DIS3 (n=4), KRAS (n=3), NRAS (n=2), MAX (n=1), TRAF3 (n=1) and TP53 (n=1) genes. The MAX mutations were missense and were located at the known MAX hotspots E32V, R35L and R36K. All three MAX mutations were found to be subclonal, with variant read frequencies (VRF) of 26%, 21% and 10%, respectively. Interestingly, due to the proximity between mutations it was possible to confirm that each variant was located on a different sequencing read, with no read containing more than one mutation (Figure 1B). Since this patient had a diploid karyotype, the entire tumor was affected by the mutations in the MAX gene, but with a parallel evolution of the three different subclones. The same finding was made in one of the patients who harbored multiple mutations in DIS3 (Figure 1B); in this case, the two D487H and D479E hotspot mutations were subclonal, with a VRF of 4% and 21%, respectively, and presented exclusively in different reads. Our cohort of patients revealed a broad VRF landscape, with 50% of variants (85/170) having a VRF below 25% and 27% (46/170) having a VRF below 10% (Figure 2A). The VRF of most of the mutations in KRAS, BRAF, NRAS and TP53 was lower than 40%, while that of DIS3 mutations showed a broader range (2% to 85%) (Figure 2B). The DIS3 gene had the largest number of low VRF mutations (5/20), all of which were considered to be pathogenic by SIFT and PolyPhen-2, and four of which were located in hotspots of the catalytic RNB domain (Online Supplementary Figure S1D).
Figure 1.
Mutated genes and parallel clonal evolution. (A) Fifty-four genes were mutated in 79 newly diagnosed MM patients. KRAS, DIS3, NRAS, BRAF and TP53 mutations accounted for 42% of all the mutations. (B) MAX (right and up) and DIS3 (left and down) showed concomitant mutations within the same patient. The proximity between subclonal mutations enabled us to detect that variants were in different sequencing reads, indicating a possible parallel subclonal evolution.
Figure 2.
Clonal and subclonal heterogeneity and variant read frequency distributions. (A) Fifty percent of mutations (85/168) had a variable read frequency (VRF) <25%, and almost half of them (46/168) had a VRF <10%. These values highlight the importance of detecting variants at very low frequencies in newly diagnosed patients. (B) Distribution of the detected variants in the most frequently mutated genes. The number of patients harboring the mutations is indicated below the name of the gene. DIS3 was the only gene in which a mutation VRF >50% was found.
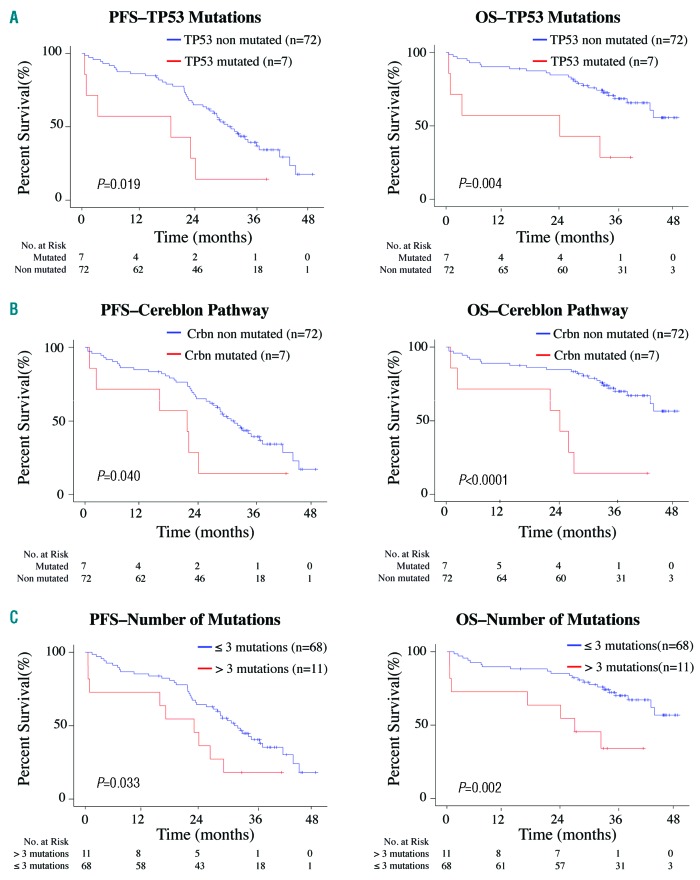
We also evaluated the impact on survival of each individual mutated gene, and of genes grouped according to their pathways. No significant impact on survival was observed for KRAS, NRAS, BRAF or DIS3 genes or for the RAS and NF-κB pathways. The existence of multiple mutations in the same gene or the number of subclonal mutations (<40% VRF) did not affect patients’ survival. However, TP53 mutations were associated with a negative impact on progression-free survival and overall survival (P=0.019 and P=0.004, respectively) (Figure 3A). Patients with mutations in the Cereblon pathway had significantly shorter progression-free survival and overall survival (P=0.040 and P<0.0001, respectively) (Figure 3B). We also noted a negative impact on progression-free survival and overall survival as a function of the number of mutations: patients with more than three mutations (>1 mutation per 61 kb of DNA analyzed) relapsed and died prematurely (P=0.033 and P=0.002, respectively) (Figure 3C). We performed multivariate survival analysis using a Cox regression model with the Akaike Information Criterion technique for the selection of demographic and cytogenetic variables. The variables selected to build the model were International Staging System score, Eastern Cooperative Oncology Group score and the patient’s age. We found a significant negative impact on survival for TP53 mutations (progression-free survival: hazard ratio, 3.68; 95% confidence interval: 1.45–9.29, P=0.005; overall survival: hazard ratio, 9.07; 95% confidence interval: 2.78–9.57, P=0.0002).
Figure 3.
Negative impact of genetic alterations on progression-free survival (left) and overall survival (right). (A) Mutations in TP53. (B) Mutations in the Cereblon pathway. (C) More than three mutations (1 mutation/61 kb of DNA).
The genes and pathways found to be most frequently mutated in this study are consistent with those in previous reports (KRAS 21% versus 21%, NRAS 18% versus 16%, BRAF 7% versus 8% and TP53 8% versus 8%).6,9 The exception was DIS3, for which the mutational incidence increased from 9% to 19% due to the deep coverage which enabled the detection of a higher number of subclonal mutations (5/20). In addition, the VRF of mutated RAS genes (<50% in all cases) supports the hypothesis that newly diagnosed MM patients who are not exposed to selective pressure by a therapy are more likely to harbor subclonal than clonal RAS mutations.5 The detection of minor subclones has allowed the identification of a subgroup of patients with poor outcome who would not have been identified by their cytogenetic profile or clinical manifestation, including those negative for del(17p) but positive for TP53 mutations.12 Considering the detection threshold in whole-genome and whole-exome sequencing studies, very useful genetic information might be being overlooked. Additionally, the recent discovery that Cereblon is essential for the anti-MM activity of immunomodulatory drugs13–15 is especially relevant here because patients enrolled in the GEM10MAS65 trial were all treated with lenalidomide, suggesting that downstream genes such as IKZF1, IKZF2, IRF4 and MYC should be additionally studied when biomarkers of lenalidomide resistance need to be identified.
In conclusion, this is the first prospective study addressing the role of targeted sequencing in a highly homogeneous series of elderly, newly diagnosed MM patients treated within a clinical trial. Despite the limited number of samples included in the study, the next-generation sequencing strategy used here was capable of detecting mutations in purified plasma cells in most patients. The affordability of next-generation sequencing together with the small sample requirement offers an excellent opportunity to introduce personalized medicine protocols into clinical practice and also to intensify the search for new biomarkers and therapeutic targets to improve treatment selection and survival of MM patients.
Supplementary Material
Footnotes
Funding: this work was partially supported by a CRIS foundation grant, Red Temática de Investigación Cooperativa en Cáncer (RTICC) (RD12/0036/0061), Joan Rodés grant (JR 14/00016), Contratos Predoctorales de Formación en Investigación en Salud i-PFIS (IFI 14/00008) and Fondo de Investigación Sanitaria PI15/01484 awarded to Joaquín Martínez López
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Avet-Loiseau H, Durie BGM, Cavo M, et al. Combining fluorescent in situ hybridization data with ISS staging improves risk assessment in myeloma: an International Myeloma Working Group collaborative project. Leukemia. 2013;27(3):711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyd KD, Ross FM, Chiecchio L, et al. A novel prognostic model in myeloma based on co-segregating adverse FISH lesions and the ISS: analysis of patients treated in the MRC Myeloma IX trial. Leukemia. 2012;26(2):349–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kumar SK, Rajkumar SV, Dispenzieri A, et al. Improved survival in multiple myeloma and the impact of novel therapies. Blood. 2008; 111(5):2516–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolli N, Avet-Loiseau H, Wedge DC, et al. Heterogeneity of genomic evolution and mutational profiles in multiple myeloma. Nat Commun. 2014;5:2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lohr JG, Stojanov P, Carter SL, et al. Widespread genetic heterogeneity in multiple myeloma: implications for targeted therapy. Cancer Cell. 2014;25(1):91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walker BA, Boyle EM, Wardell CP, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015; 33(33):3911–3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mateos M-V, Martínez-López J, Hernández M-T, et al. Sequential vs alternating administration of VMP and Rd in elderly patients with newly diagnosed MM. Blood. 2016;127(4):420–425. [DOI] [PubMed] [Google Scholar]
- 8.Kortüm KM, Langer C, Monge J, et al. Longitudinal analysis of 25 sequential sample-pairs using a custom multiple myeloma mutation sequencing panel (M(3)P). Ann Hematol. 2015;94(7):1205–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kortuem KM, Braggio E, Bruins L, et al. Panel sequencing for clinically oriented variant screening and copy number detection in 142 untreated multiple myeloma patients. Blood Cancer J. 2016;6(2):e397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bolli N, Li Y, Sathiaseelan V, et al. A DNA target-enrichment approach to detect mutations, copy number changes and immunoglobulin translocations in multiple myeloma. Blood Cancer J. 2016;6(9):e467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubio-Camarillo M, Gómez-López G, Fernández JM, Valencia A, Pisano DG. RUbioSeq: a suite of parallelized pipelines to automate exome variation and bisulfite-seq analyses. Bioinforma Oxf Engl. 2013;29(13):1687–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kortüm KM, Langer C, Monge J, et al. Targeted sequencing using a 47 gene multiple myeloma mutation panel (M 3 P) in -17p high risk disease. Br J Haematol. 2015; 168(4):507–510.sup [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu YX, Braggio E, Shi C-X, et al. Identification of cereblon-binding proteins and relationship with response and survival after IMiDs in multiple myeloma. Blood. 2014; 124(4):536–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu YX, Braggio E, Shi C-X, et al. Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood. 2011;118(18):4771–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kortum KM, Mai EK, Hanafiah NH, et al. Targeted sequencing of refractory myeloma reveals a high incidence of mutations in CRBN and Ras pathway genes. Blood. 2016;128(9):1226–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.