Abstract
A cute myeloid leukemia is a disease of the elderly (median age at diagnosis, 65–70 years). The prognosis of older acute myeloid leukemia patients is generally poor. While genetic markers have become important tools for risk stratification and treatment selection in young and middle-aged patients, their applicability in very old patients is less clear. We sought to validate existing genetic risk classification systems and identify additional factors associated with outcomes in intensively treated patients aged ≥75 years. In 151 patients who received induction chemotherapy in the AMLCG-1999 trial, we investigated recurrently mutated genes using a targeted sequencing assay covering 64 genes. The median number of mutated genes per patient was four. The most commonly mutated genes were TET2 (42%), DNMT3A (35%), NPM1 (32%), SRSF2 (25%) and ASXL1 (21%). The complete remission rate was 44% and the 3-year survival was 21% for the entire cohort. While adverse-risk cytogenetics (MRC classification) were associated with shorter overall survival (P=0.001), NPM1 and FLT3-ITD mutations (present in 18%) did not have a significant impact on overall survival. Notably, none of the 13 IDH1-mutated patients (9%) reached complete remission. Consequently, the overall survival of this subgroup was significantly shorter than that of IDH1-wildtype patients (P<0.001). In summary, even among very old, intensively treated, acute myeloid leukemia patients, adverse-risk cytogenetics predict inferior survival. The spectrum and relevance of driver gene mutations in elderly patients differs from that in younger patients. Our data implicate IDH1 mutations as a novel marker for chemorefractory disease and inferior prognosis. (AMLCG-1999 trial: clinicaltrials.gov identifier, NCT00266136)
Introduction
The incidence of acute myeloid leukemia (AML) is highest among the elderly, with the median age at diagnosis being around 70 years.1 The prognosis of older AML patients is generally considered poor. In this age group, comorbidities, poor performance status and a reluctance of physicians and patients to use treatment regimens perceived as toxic often lead to the decision to avoid induction chemotherapy in favor of less intensive therapies.2–6 On the other hand, data from patient registries and randomized trials suggest that intensive induction therapy can result in prolonged overall survival (OS), at least in a subset of elderly patients aged 70–79 years, and may even be beneficial in selected octogenarians.4,7–10 However, it remains unclear which genetic and clinical factors are relevant to identify those elderly patients most likely to benefit from, and least likely to be harmed by, induction chemotherapy.11
Advances in the field of molecular genetics and the development of next-generation sequencing expanded our knowledge of recurrently mutated genes in AML and their role in disease pathophysiology, and led to refined risk classifications in younger patients.1,12,13 However, elderly patients were underrepresented or excluded in these studies, and the question thus arises whether the spectrum and prognostic relevance of gene mutations are similar in very old AML patients. Some studies of established genetic risk factors indicate that there may be important differences between elderly and younger patients.12,14,15 For example, the FLT3-ITD mutation is a well-recognized adverse prognostic factor in young adults, whereas its impact is reduced or absent in elderly patients.1,7,14,16,17 Therefore, comprehensive genetic analyses in cohorts of very old patients are needed to clarify the relevance of distinct gene alterations in this subset of patients.
To identify prognostic factors associated with clinical outcomes in elderly AML patients, we studied 151 patients aged ≥75 years who received intensive induction therapy. The mutational spectrum in 64 recurrently mutated AML genes was analyzed by targeted next-generation sequencing. We then studied associations of genetic alterations with other known prognostic factors, patients’ characteristics, and outcomes. Our aim was to define subsets of patients who may benefit from intensive induction therapy. Furthermore, we evaluated the prognostic relevance of the British Medical Research Council (MRC) 2010 and European LeukemiaNet (ELN) 2017 risk classification schemes in this subset of patients.1,18
Methods
Patients, treatment, karyotype and molecular analyses
We studied 151 patients aged ≥75 years with newly diagnosed AML or high-risk myelodysplastic syndromes (10% – <20% bone marrow blasts; n=3), diagnosed according to World Health Organization criteria, who had suitable bone marrow or peripheral blood specimens for genetic analysis. All patients received intensive induction treatment during the German AML Cooperative Group AMLCG-1999 randomized, multicenter, phase III trial (clinicaltrials.gov identifier, NCT00266136) between 1999 and 2011. Participants aged ≥60 years were randomized to receive a first induction course with high-dose cytarabine and mitoxantrone (HAM) or with standard-dose cytarabine, daunorubicin and 6-thioguanine (TAD-9). A second HAM induction course was administered from day 21 only if ≥5% residual blasts were present in a bone marrow aspirate taken on day 15. Another cycle of TAD-9 was given as consolidation therapy, followed by monthly cytarabine-based maintenance chemotherapy (Online Supplementary Data). Karyotype analyses were performed centrally and results were classified according to the 2010 MRC classification.18
Molecular analysis encompassed sequencing of 64 genes recurrently mutated in AML. We analyzed either known mutational hotspots or the entire coding sequence using an amplicon-based approach (Haloplex, Agilent, Boeblingen, Germany) as described previously.12 The median sequencing coverage of the target region across all samples was 520-fold, and 98.8% of the target region was covered at >30-fold. Gene alterations with variant allele frequencies of ≥2% were classified as driver mutations, variants of unknown significance or germline polymorphisms.12 The Catalogue Of Somatic Mutations In Cancer (COSMIC), The Cancer Genome Atlas (TCGA) and the Single Nucleotide database (dbSNP) served as databases to compare and classify the mutational results.19–21 NPM1,22 FLT3-ITD23 and CEBPA24 mutations were additionally tested using polymerase chain reaction followed by Sanger sequencing and/or fragment analysis.
Written informed consent for inclusion in the clinical trial and genetic analyses was provided by all patients. All study protocols were in accordance with the Declaration of Helsinki and approved by the institutional review boards of each participating center.
Statistical analysis
Associations among gene mutations, and between gene mutations and patients’ pretreatment features, were analyzed using the Fisher exact test for categorical variables and the Wilcoxon rank-sum test for continuous variables. The Kaplan-Meier method was used to calculate estimated survival probabilities, with the log-rank test evaluating differences between survival distributions. A multivariate Cox regression model including known risk factors [MRC risk category and Eastern Cooperative Oncology Group performance status (ECOG PS)], and gene mutations that showed a univariate association with OS (at P<0.10), was used to identify factors associated with survival. Statistical analyses were performed using SPSS version 24.0 (Chicago, IL, USA). All statistical tests are two-sided, and a P value of ≤0.05 was considered statistically significant.
Results
Patients’ characteristics
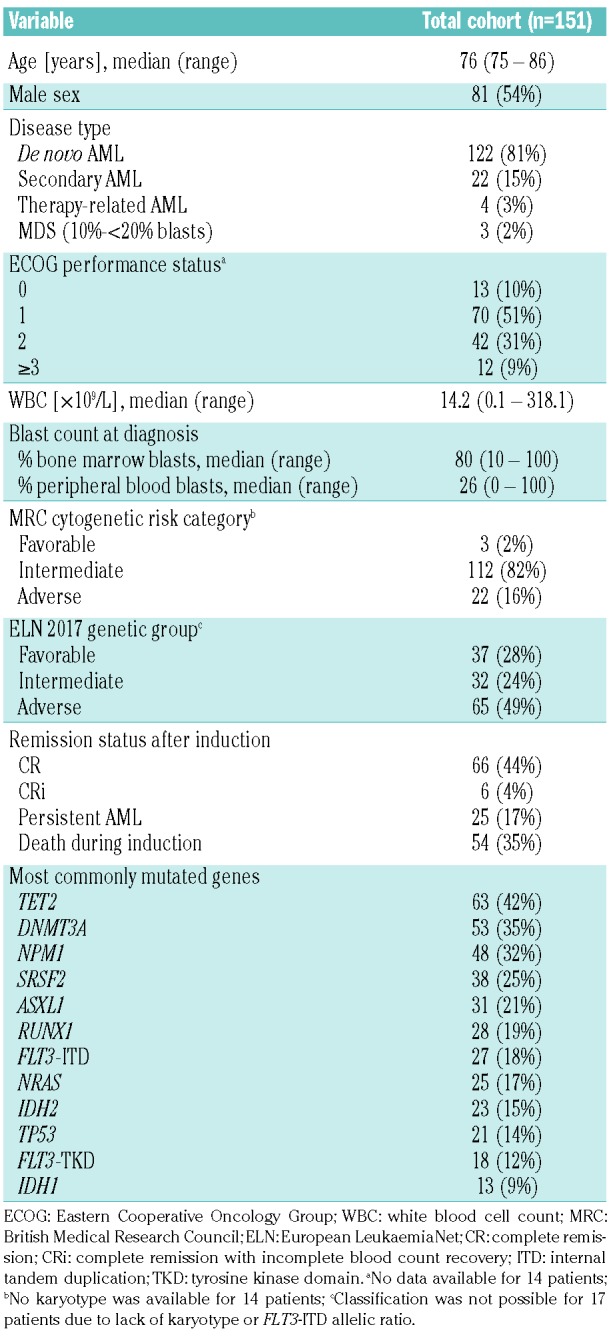
We identified 151 patients aged ≥75 years treated in the AMLCG-1999 trial for whom suitable material for genetic analyses was available. Details of this trial have been published elsewhere.25 The patients’ baseline characteristics are shown in Table 1. The median age of the subjects was 76 years (range, 75–86). Eighty-one percent of patients had a clinical diagnosis of de novo AML, 15% had secondary AML and 3% had therapy-related AML. Three patients (2%) had high-risk myelodysplastic syndromes (10-<20% bone marrow blasts). Most patients (90%) had an ECOG PS of ≤2. Among 137 patients with cytogenetic data, 82% belonged to the intermediate-risk group according to the MRC 2010 classification, including 52% with cytogenetically normal AML. Only three patients (2%) had favorable cytogenetics, and 16% fell in the MRC adverse-risk group.
Table 1.
Patients’ baseline characteristics.
Treatment outcomes
In the overall cohort, the rates of complete remission (CR) and CR with incomplete blood count recovery (CRi) were 44% and 4%, respectively. The median event-free survival (EFS) was 1.7 months. The median relapse-free survival (RFS) for patients achieving a remission was 12 months, and the median OS was 6.0 months (Online Supplementary Figure S1). The 3-year OS rate for the entire study population was 21%, and survival was similar for patients aged 75–79 or 80–86 years (P=0.3) (Online Supplementary Figure S2A). Patients with an ECOG PS of 3 or 4 had shorter OS compared to patients with an ECOG PS of 0–2 (P=0.036) (Online Supplementary Figure S2B), mostly because of an increased risk of early death within 60 days from starting treatment (26% for patients with an ECOG PS of 0–1, 40% for those with a PS of 2, and 75% for patients with an ECOG PS of 3 or 4). Patients randomized to HAM induction tended to have longer OS compared to patients randomized to TAD induction (median OS: TAD, 3.1 months versus HAM, 7.8 months; P=0.09) (Online Supplementary Results and Online Supplementary Figure S3).
Impact of cytogenetics on survival
Since only three patients had favorable-risk cytogenetics according to the MRC classification, the favorable-and intermediate-risk groups were analyzed jointly. Patients in the favorable-or intermediate-risk categories had a non-significantly higher remission rate than adverse-risk patients (CR/CRi, 51% versus 27%; P=0.2). The EFS, RFS, and OS of favorable-and intermediate-risk patients were significantly longer than those for adverse-risk patients (EFS: P=0.001; RFS: P=0.006; OS: P=0.001) (Figure 1A and Online Supplementary Figure S4).
Figure 1.
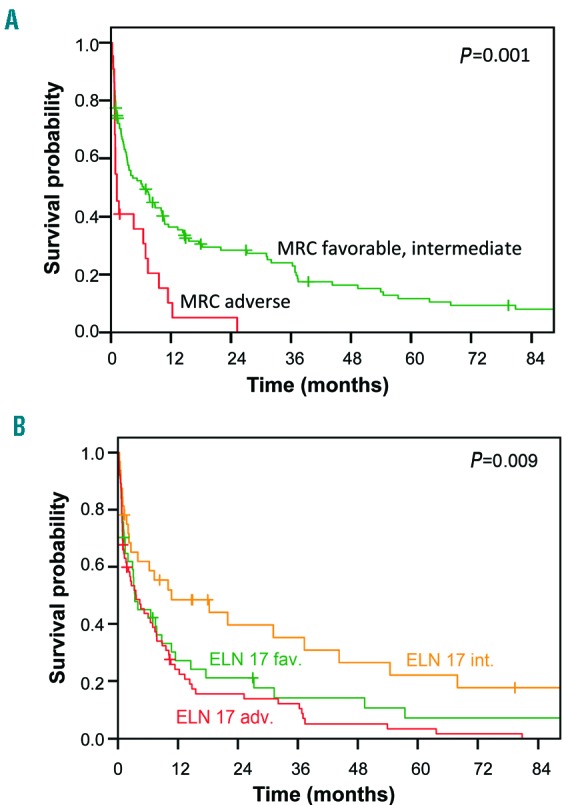
Overall survival according to the MRC and ELN risk classifications. (A) Overall survival for patients in the favorable-and intermediate-risk groups (green) compared to the adverse-risk group (red) according to the MRC cytogenetic risk category. (B) Overall survival for patients in the favorable-(green), intermediate-(orange) and adverse-risk groups (red) according to the ELN 2017 genetic category.
Gene mutations and patients’ pretreatment characteristics
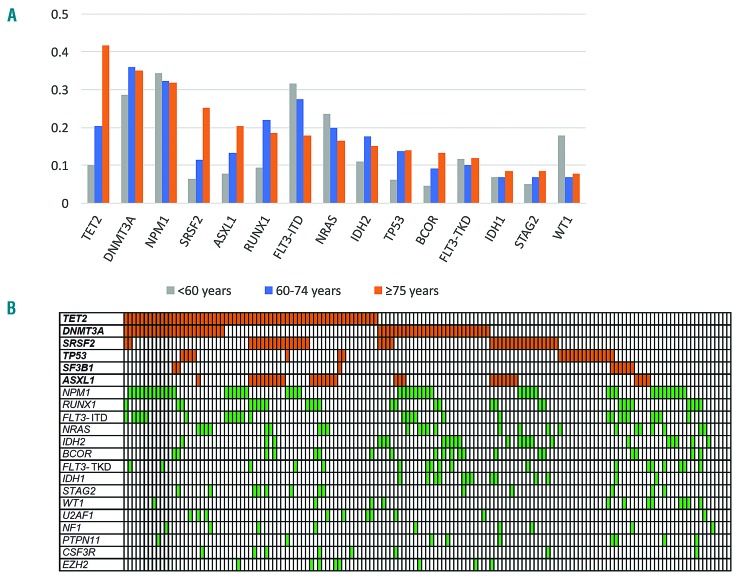
We identified a total of 622 driver mutations affecting 64 genes, with a median number of four mutated genes per patient (range, 1–10 mutations/patient) (Online Supplementary Figure S5). Older age (81–86 years versus 75–80 years) was not associated with a higher number of driver gene mutations (P=0.5, data not shown). The most frequently mutated genes in this age group were TET2 (63/151, 42%), DNMT3A (53/151, 35%), NPM1 (48/151, 32%), FLT3 (45/151, 30%), SRSF2 (38/151, 25%), ASXL1 (31/151, 21%), and RUNX1 (28/151, 19%) (Table 1, Figure 2A). Compared to previously studied patients aged <60 or 60–74 years12, those aged ≥75 years had a higher frequency of TET2, SRSF2 and ASXL1 mutations. TP53 mutations occurred in 14% of patients (21/151) and were strongly associated with complex karyotypes (P<0.001). Eighty-three percent of patients (126/151) harbored one or more mutations in TET2, DNMT3A, SRSF2, ASXL1, TP53 or SF3B1 - genes known to be involved in age-associated clonal hematopoiesis (Figure 2B).26,27 RUNX1 mutations were positively associated with mutations in SRSF2 and ASXL1 (SRSF2, P=0.02; ASXL1, P=0.01), while ASXL1 and DNMT3A mutations were inversely correlated with each other (P=0.003). NPM1 mutations were mutually exclusive with RUNX1 mutations, and only two patients had co-mutations in NPM1 and ASXL1 (P<0.001) (Figure 2B).
Figure 2.
Genetic landscape of old acute myeloid leukemia patients. (A) Driver gene mutations in 151 AML patients ≥75 years of age at primary diagnosis. The bar chart shows the 15 most commonly mutated genes in the 151 AML patients aged ≥75 years compared to 664 patients aged <60 and 60–74 at primary diagnosis (data from Metzeler et al.12). (B) Heatmap showing associations between different driver gene mutations. Each column represents one patient.
Association between gene mutations, therapy response and survival
The number of mutated genes per patient was not associated with OS in a comparison of patients with one to three (62/151, 41%), four to seven (80/151, 53%) or eight to ten (9/151, 6%) mutated genes (P=0.6) (Online Supplementary Figure S6).
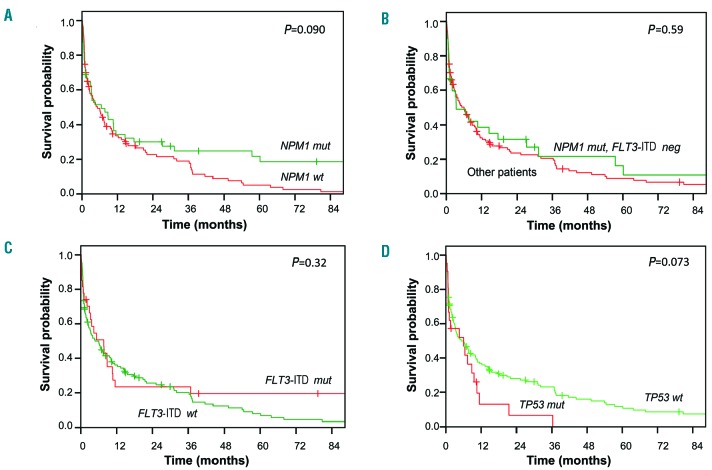
Univariate analyses of the associations between the most common gene mutations and CR rate and OS are shown in Online Supplementary Table S1. NPM1 mutations, which have been shown to be associated with a higher CR rate in younger AML patients, were not associated with response to induction treatment in our cohort. Patients with NPM1 mutations had a significant longer EFS (P=0.027) (Online Supplementary Figure S7A) and tended to have a longer RFS (P=0.081) (Online Supplementary Figure S7B) and OS (P=0.090) (Figure 3A) than wildtype patients. Among patients who achieved CR, those with mutated NPM1 had a significantly longer OS (calculated from the day of achieving CR) than NPM1-wildtype patients (P=0.037) (Online Supplementary Figure S7C). Within the subgroup of patients with cytogenetically normal AML, there was no difference in OS between NPM1-mutated and -wildtype patients (P=0.4, data not shown). Likewise, the OS of NPM1mutated/FLT3-ITDwildtype patients - a known favorable-risk subgroup, at least in younger patients - was similar to that of the rest of the cohort (P=0.59) (Figure 3B).
Figure 3.
Overall survival according to gene mutations. (A) NPM1 mutations: overall survival in NPM1-mutated (green) compared to NPM1-wildtype patients (red). (B) NPM1-mutated/FLT3-ITD-negative patients: overall survival in NPM1-mutated/FLT3-ITD-wildtype patients (green) compared to other patients (red). (C) FLT3-ITD mutations: overall survival in FLT3-ITD-mutated (red) compared to FLT3-ITD-wildtype patients (green). (D) TP53 mutations: overall survival in TP53-mutated patients (red) compared to TP53-wildtype patients (green). “Mut” denotes mutated and “wt” wildtype.
FLT3-ITD mutations had no impact on EFS, RFS, or OS in the entire cohort [EFS: P=0.54 (Online Supplementary Figure S8A); RFS: P=0.78 (Online Supplementary Figure S8B); OS: P=0.32 (Figure 3C)] or in patients with cytogenetically normal AML (P=0.26) (Online Supplementary Figure S8C). Since the FLT3-ITD allelic ratio is an important prognosticator recognized in the ELN 2017 classification, we also explored its impact on OS. A high FLT3-ITD allelic ratio (≥0.5) was not associated with a shorter OS compared to a low allelic ratio (<0.5) or FLT3-ITD wildtype (OS: P=0.53) (Online Supplementary Figure S8D). In the context of co-mutated NPM1, there was also no impact on OS of a high FLT3-ITD allelic ratio compared to a low allelic ratio or to NPM1mutated/FLT3-ITDwildtype patients (OS: P=0.4) (Online Supplementary Figure 7D). Patients with TP53 mutations showed trends towards shorter EFS (P=0.079) (Online Supplementary Figure S9A) and OS (P=0.073) (Figure 3D), and had a significantly shorter RFS (P=0.041) (Online Supplementary Figure S9B), compared to TP53-wildtype patients. TP53-mutated patients who achieved CR had a significantly shorter OS (calculated from the day of achieving CR) compared to that of TP53-wildtype patients in CR (P=0.005) (Online Supplementary Figure S9C).
IDH1 mutations: impact on event-free and overall survival, association with patients’ pretreatment characteristics and other gene mutations
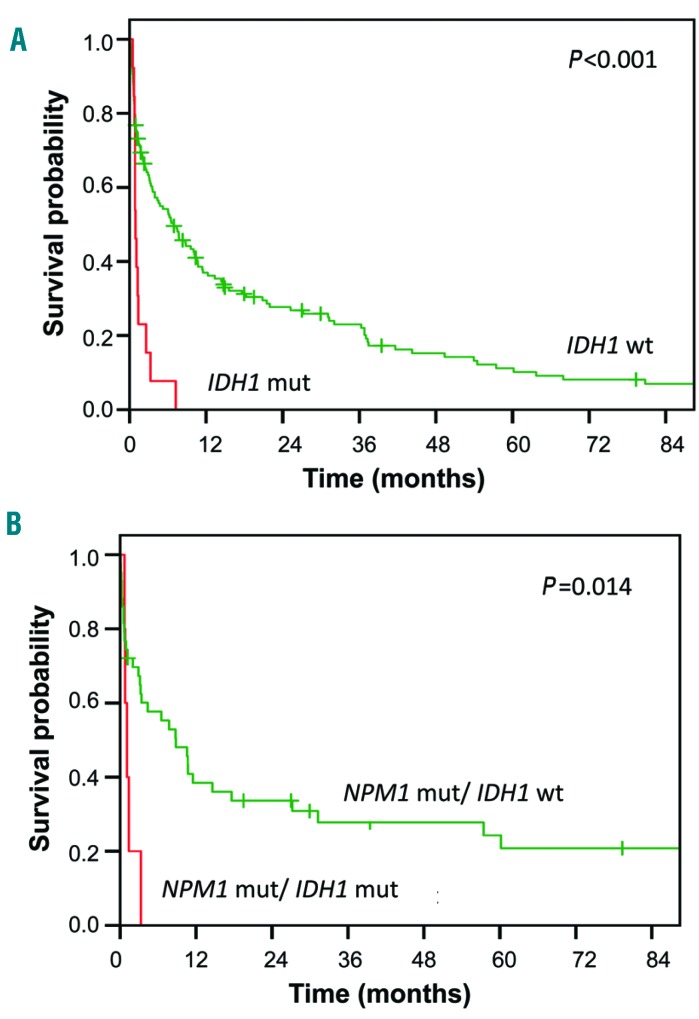
In our cohort of old AML patients, the only gene significantly associated with OS in univariate analysis was IDH1 (Online Supplementary Table S1). None of the IDH1 codon R132-mutated patients (13/151, 9%) reached CR/CRi (P<0.001). Consequently, IDH1-mutated patients had a significantly shorter EFS (P<0.001) (Online Supplementary Figure S10A) and OS (P≤0.001) Figure 4A) than IDH1-wildtype patients.
Figure 4.
IDH1 mutations and survival. (A) Overall survival in IDH1-mutated patients (red line) compared to IDH1-wildtype patients (green line). (B) Overall survival in NPM1-mutated/IDH1-mutated patients (red line) compared to NPM1-mutated/IDH1-wildtype patients (green line). Mut: mutated; wt: wildtype.
Within this elderly cohort of patients, IDH1-mutated patients showed a trend towards older age (P=0.08) (Online Supplementary Table S2). Furthermore, IDH1-mutated patients tended to have higher platelet counts (P=0.20) and lower WBC counts (P=0.10) than IDH1-wildtype patients. There was no difference in peripheral blood or bone marrow blast counts between IDH1-wildtype and IDH1-mutated patients. IDH1 mutations were associated with cytogenetically normal AML (P=0.069): 10/13 IDH1-mutated patients had a normal karyotype, whereas one patient had a 7q deletion, one had trisomy 8, and karyotype was unknown for one patient. Further information on clinical characteristics of the 13 IDH1-mutated patients are shown in Online Supplementary Table S3. IDH1 and IDH2 mutations were mutually exclusive (P=0.2), and IDH2 mutations, found in 15% of patients (23/151), had no impact on OS (P=0.5). TET2 mutations were mutually exclusive with IDH1 mutations (P=0.001), and with mutated IDH2 (P=0.002).
Five IDH1-mutated patients also carried NPM1 mutations. While patients with NPM1 mutations overall tended to have favorable OS (Figure 3A), IDH1 mutations had a dominant negative prognostic effect even when co-occurring with mutated NPM1 (Figure 4B). Four IDH1-mutated patients had simultaneous FLT3-TKD mutations, an association that almost reached statistical significance (P=0.051). There was no association between IDH1 and FLT3-ITD mutations (P=0.5).
Multivariate analysis of prognostic factors in old, intensively treated acute myeloid leukemia patients
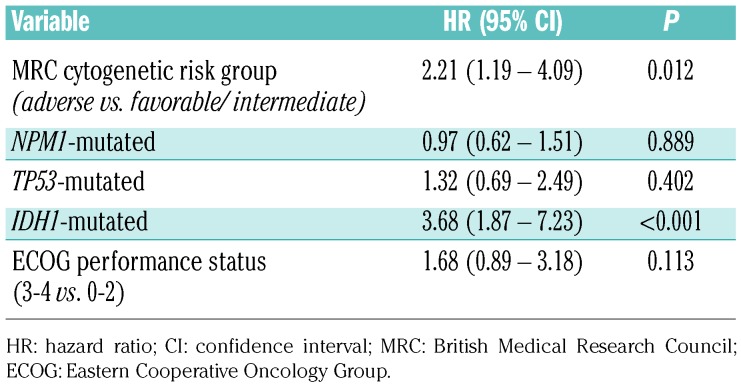
We used a multivariate Cox regression model to identify the most relevant predictors of OS in this cohort of intensively treated AML patients ≥75 years (Table 2). In this multivariate model, using the MRC classification, ECOG PS, and IDH1, NPM1 and TP53 mutations as covariates, only IDH1 mutations and the MRC adverse cytogenetic risk category had significant negative impacts on OS. There was a trend towards worse OS for patients with poor ECOG PS.
Table 2.
Multivariate analysis for overall survival.
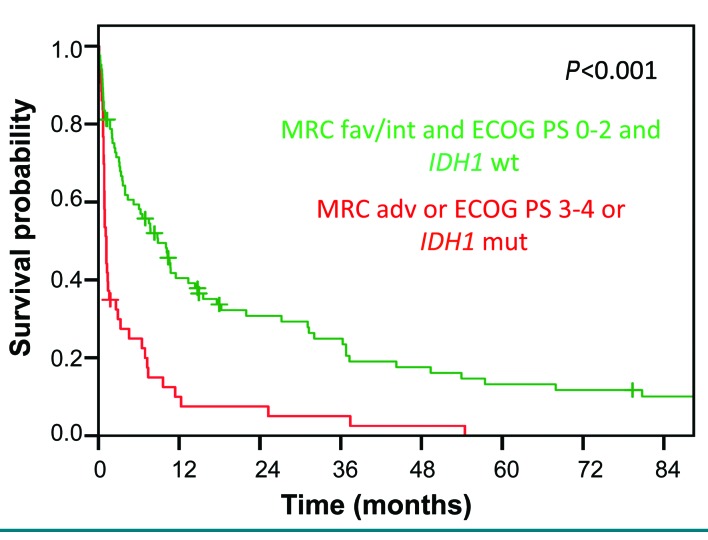
Patients who were characterized by wildtype IDH1, favorable or intermediate cytogenetics and an ECOG PS of 0-2 had a significantly longer median OS than patients who had at least one high-risk feature (mutated IDH1, adverse cytogenetics, and/or performance status ≥3) (3-year OS, 25% versus 5%; P<0.001) (Figure 5).
Figure 5.
Impact of Eastern Cooperative Oncology Group performance status, Medical Research Council classification and IDH1 mutations on overall survival. Overall survival in patients with an ECOG performance status of 0-2, favorable or intermediate MRC category and IDH1-wildtype compared to patients with an ECOG performance status of 3–4 or MRC adverse category or IDH1 mutation.
Association between the European LeukemiaNet 2017 and Medical Research Council classifications and outcomes
When we applied the novel ELN 2017 genetic risk classification to our cohort of very old patients, more patients were assigned to the favorable-and adverse-risk categories, and fewer patients to the intermediate-risk group, compared to the MRC classification (Table 1). Surprisingly, patients classified as ELN-2017 intermediate-risk had longer OS (median, 10.7 months) compared to both the favorable- and adverse-risk groups (median, 3.4 months and 3.6 months, respectively; P=0.009) (Figure 1B). Furthermore, in the MRC adverse-risk subgroup, all patients died within 2 years, whereas seven ELN-2017 adverse-risk patients survived for 3 years or longer.
Discussion
Elderly AML patients are frequently underrepresented in clinical trials evaluating induction chemotherapy schedules, as well as in studies of the genetic basis of the disease.12,28–30 For example, the widely-used MRC cytogenetic risk categories were derived from a cohort of patients aged 16–59 years.18 This selection bias contrasts with the epidemiology of AML, which is mostly a disease of elderly patients. While many study groups have excluded older patients from trials involving induction chemotherapy, and less-intensive regimens, including the hypomethylating agents, decitabine and azacitidine, are now widely used in this age group, some studies suggest that a subgroup of old patients may benefit from induction chemotherapy.7,8,10 There is, therefore, a vital need to identify factors that are associated with outcomes and that could support therapeutic decision-making in this difficult-to-treat patient cohort.
We designed our study to focus on a cohort of very old patients (aged 75 years or older) included in a trial of induction chemotherapy. In this age group, the benefits of induction chemotherapy and potential clinical and genetic markers for therapy success or failure are not well defined. We found that, even among very old patients, favorable-and intermediate-risk cytogenetics remain associated with a relatively favorable OS in a comparison with adverse-risk cytogenetics. This finding is in agreement with that of a retrospective analysis by Heiblig and colleagues.31 In their study, older age (≥75 years in a cohort with an age range of 70–93 years) was also a strong prognostic factor in terms of OS whereas according to our own data, higher age (80–86 years versus 75–79 years) did not associate with OS, potentially due to the fact that only very fit octagenarians were enrolled in a trial of induction chemotherapy. Patients in the cohort reported by Heiblig and colleagues received three different treatment modalities: intensive chemotherapy, less intensive treatment (i.e. low-dose AraC, azacitidine or decitabine) or best supportive care only.31 The median OS in patients aged ≥75 years, regardless of therapy type, was 4.3 months with a 3-year OS rate of 14%, compared to a median OS of 10 months and a 3-year OS rate of 20% in patients aged <75 years. In patients ≥75 years of age who all received intensive chemotherapy, Heiblig and colleagues reported a 3-year OS rate of 24%, which is comparable to the 3-year OS rate of 21% found in our retrospective analysis. Importantly, the analysis by Heiblig and colleagues also showed that long-term survival beyond 3 years was exceedingly rare in patients treated with hypomethylating agents, while survival times exceeding 10 years were observed in intensively treated patients.
Our previous analysis of the mutational landscape in 664 intensively treated AML patients included 288 patients ≥60 years of age at primary diagnosis.12 We reported that the mutational spectrum in older AML patients differed from that in younger patients (<60 years). In this study, we extend this finding to very old patients aged ≥75 years. The five most frequently mutated genes were TET2, DNMT3A, NPM1, SRSF2 and ASXL1.12 The high incidence of TET2, DNMT3A, SRSF2, ASXL1, TP53 and SF3B1 mutations in our patients is in agreement with reports showing that these genes are frequently mutated in age-associated clonal hematopoiesis.26,32 Overall, 83% (126/151) of AML patients ≥75 years carried at least one mutation in one of these six genes.
Recently, Baldus and colleagues published data on the genetic and epigenetic landscape in 93 elderly AML patients (65–90 years) enrolled in a Study Alliance Leukemia (SAL) registry.33 Similar to our results, they identified a high frequency of mutations in DNMT3A, TET2, SRSF2, ASXL1 and RUNX1. Strikingly, NPM1 mutations were much less common (16.1% of patients) than in our cohort (32%). This difference remains unexplained, since there are no obvious differences regarding, for example, the proportion of patients with de novo versus secondary AML which could affect the frequency of NPM1 mutations.
We unexpectedly identified IDH1 mutations as the strongest genetic predictor of shorter survival in this age group. The prognostic value of IDH1 and IDH2 mutations in AML has been studied by multiple groups but is still controversial.34,35 While some analyses found no impact on OS,36 others demonstrated associations with poor37–40 or favorable prognosis.28 A recently published meta-analysis of 33 studies found that IDH1 and IDH2 mutations, analyzed jointly, do not affect OS or EFS.34 IDH1 mutations, when analyzed separately, are associated with lower CR rates, shorter EFS and shorter OS. A negative impact of IDH1 mutations on outcomes has also been described in younger NPM1-mutated/FLT3-ITD-negative patients.37,39 These findings are generally compatible with the results of our study. This meta-analysis also revealed that IDH2-mutated patients had a longer OS than IDH2-wildtype patients, while in our study there was no impact of IDH2 mutations on OS (P=0.53). This discrepancy may be explained by our focus on very old AML patients, among whom the prognostic relevance of gene mutations may differ from that among younger patients. It remains unclear why mutations in two related genes with apparently similar functional consequences21,41,42 might have varying or even opposite impacts on OS. The poor outcome of old AML patients with mutated IDH1 needs to be validated in additional cohorts. If the negative prognostic effect of IDH1 mutations in old patients is confirmed in other studies, these patients should not be considered candidates for induction chemotherapy. Targeted inhibitors of the mutant IDH1 enzyme, which are currently being tested in clinical trials, may become a preferred therapeutic approach for these patients in the future.
In our multivariate analysis, results of cytogenetic analysis (classified according to the MRC system) were a strong predictor of outcomes. Compared to the MRC risk categories, application of the new ELN-2017 genetic risk classification led to an expansion of the favorable- and adverse-risk categories (Table 1). The inclusion of RUNX1, ASXL1 and TP53 mutations in the ELN adverse-risk category, in particular, affects the risk classification of this cohort of very old patients in whom these mutations are common. Many patients who were classified as intermediate-risk based on cytogenetic results alone according to the MRC recommendations are now re-classified into the adverse-risk group. At least in our cohort, however, the MRC cytogenetic risk classification appears to provide better prognostic stratification compared to the ELN-2017 system. Thus, further validation of the ELN-2017 classification in larger cohorts of elderly patients seems warranted to define its applicability in this age group.
Our finding that in patients aged 75 or older, the prognosis of ELN-2017 favorable-risk patients was not better, and may in fact be worse, than that of the intermediate-risk group suggests that favorable molecular markers established in younger patients (e.g., mutated NPM1 without or with a low FLT3-ITD allelic ratio, biallelic CEBPA mutations) have weaker prognostic relevance in elderly patients. Indeed, in our cohort, the NPM1-mutated/FLT3-ITD-negative genotype did not associate with favorable outcomes. NPM1 mutations alone were associated with significantly longer EFS, but there were only trends towards longer RFS and OS. Among patients who achieved CR after induction therapy, those with mutated NPM1 had a significantly longer OS, suggesting that these patients might have benefited from cytarabine-based consolidation and prolonged monthly maintenance therapy as used in the AML-CG 1999 trial. In contrast, the MRC adverse-risk group, defined by cytogenetic alterations only, had significantly shorter OS compared to the cytogenetic favorable- and intermediate-risk groups. In contrast to the ELN-2017 adverse-risk group, no MRC adverse-risk patient survived beyond 2.1 years. Thus, karyotype rather than gene mutations appears to be the major factor defining the prognosis in very old, intensively treated AML patients.43
One limitation of our study is that all patients were treated with the same treatment modality - intensive chemotherapy. Consequently, we were unable to study interactions between genetic alterations and type of treatment (intensive versus not intensive) with regard to patients’ outcomes. However, we found that patients randomized to the more intensive induction regimen (HAM) tended to have longer OS compared to those randomized to TAD. While this non-significant difference may be due to the play of chance, the finding that better outcomes were observed in the more intensive arm supports the conclusion that induction chemotherapy is tolerable for selected patients in this age group. Two randomized phase III trials in newly diagnosed AML patients aged ≥65 years compared treatment with the hypomethylating agents, azacitidine and decitabine, to alternative therapies (best supportive care, low-dose cytarabine, or intensive chemotherapy).44,45 The median OS of azacitidine-treated patients was 10.4 months, and that of decitabine-treated patients was 7.7 months, compared to a median OS of 6.0 months in our study. Obviously, these cohorts of patients cannot be compared directly because of their different age ranges and inclusion criteria. While the lower age limit of our analysis was higher than that in the trials of hypmethylating agents (75 versus 65 years), our cohort represents highly selected patients who were, despite their advanced age, deemed fit enough to undergo induction chemotherapy. Notwhitstanding this obvious limitation, it is noteworthy that the 3-year OS rate in our cohort was nominally higher than the rates in both the trials of hypomethylating agents. In our study, 24 patients (21%) were still alive 3 years after primary diagnosis, whereas two patients (1%) in the azacitidine and no patient in the decitabine trial were still alive at 3 years. Thus, at least a subset of AML patients may benefit from intensive induction therapy in terms of sustained remissions and long-term survival, even at the age of 75 years or above. Nevertheless, the longer median OS achieved in the trials of hypomethylating agents suggests that these agents may be the preferred choice for many elderly patients.
In clinical practice, the choice between intensive, potentially curative chemotherapy or a palliative therapeutic approach in old AML patients should be guided by an evidence-based, individualized assessment of the potential risks (e.g., treatment-related mortality) and benefits of intensive chemotherapy. In our study, favorable- and intermediate-risk cytogenetics, IDH1-wildtype status and good performance status characterized a subset of old AML patients who may achieve long-term survival after induction chemotherapy. These patients might be candidates for intensive therapy in the absence of medical contraindications. On the other hand, patients with adverse-risk cytogenetics, IDH1 mutations or poor performance status did not benefit from intensive induction therapy and might fare better with alternative strategies such as hypomethylating agents, or novel targeted agents that have recently been approved or are currently under development.46 In summary, our results show that clinicians should consider intensive chemotherapy as a therapeutic option even for selected, very old AML patients.
Supplementary Material
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/103/11/1853
References
- 1.Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hiddemann W, Kern W, Schoch C, et al. Management of acute myeloid leukemia in elderly patients. J Clin Oncol. 1999;17(11):3569–3576. [DOI] [PubMed] [Google Scholar]
- 3.Appelbaum FR, Gundacker H, Head DR, et al. Age and acute myeloid leukemia. Blood. 2006;107(9):3481–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Juliusson G, Billstrom R, Gruber A, et al. Attitude towards remission induction for elderly patients with acute myeloid leukemia influences survival. Leukemia. 2006;20(1):42–47. [DOI] [PubMed] [Google Scholar]
- 5.Burnett AK, Milligan D, Prentice AG, et al. A comparison of low-dose cytarabine and hydroxyurea with or without all-trans retinoic acid for acute myeloid leukemia and high-risk myelodysplastic syndrome in patients not considered fit for intensive treatment. Cancer. 2007;109(6):1114–1124. [DOI] [PubMed] [Google Scholar]
- 6.Rao AV. Fitness in the elderly: how to make decisions regarding acute myeloid leukemia induction. Hematology Am Soc Hematol Educ Program. 2016;2016(1):339–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wetzler M, Mrozek K, Kohlschmidt J, et al. Intensive induction is effective in selected octogenarian acute myeloid leukemia patients: prognostic significance of karyotype and selected molecular markers used in the European LeukemiaNet classification. Haematologica. 2014;99(2):308–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lowenberg B, Zittoun R, Kerkhofs H, et al. On the value of intensive remission-induction chemotherapy in elderly patients of 65+ years with acute myeloid leukemia: a randomized phase III study of the European Organization for Research and Treatment of Cancer Leukemia Group. J Clin Oncol. 1989;7(9):1268–1274. [DOI] [PubMed] [Google Scholar]
- 9.Juliusson G, Swedish AMLG. Most 70- to 79-year-old patients with acute myeloid leukemia do benefit from intensive treatment. Blood. 2011;117(12):3473–3474. [DOI] [PubMed] [Google Scholar]
- 10.Juliusson G, Antunovic P, Derolf A, et al. Age and acute myeloid leukemia: real world data on decision to treat and outcomes from the Swedish Acute Leukemia Registry. Blood. 2009;113(18):4179–4187. [DOI] [PubMed] [Google Scholar]
- 11.Krug U, Gale RP, Berdel WE, et al. Therapy of older persons with acute myeloid leukaemia. Leuk Res. 2017;60:1–10. [DOI] [PubMed] [Google Scholar]
- 12.Metzeler KH, Herold T, Rothenberg-Thurley M, et al. Spectrum and prognostic relevance of driver gene mutations in acute myeloid leukemia. Blood. 2016;128(5):686–698. [DOI] [PubMed] [Google Scholar]
- 13.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferrara F, Criscuolo C, Riccardi C, et al. FLT3 mutations have no prognostic impact in elderly patients with acute myeloid leukemia and normal karyotype. Am J Hematol. 2009;84(8):532–535. [DOI] [PubMed] [Google Scholar]
- 15.Mrozek K, Marcucci G, Nicolet D, et al. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. J Clin Oncol. 2012;30(36):4515–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andersson A, Johansson B, Lassen C, Mitelman F, Billstrom R, Fioretos T. Clinical impact of internal tandem duplications and activating point mutations in FLT3 in acute myeloid leukemia in elderly patients. Eur J Haematol. 2004;72(5):307–313. [DOI] [PubMed] [Google Scholar]
- 17.Whitman SP, Maharry K, Radmacher MD, et al. FLT3 internal tandem duplication associates with adverse outcome and gene- and microRNA-expression signatures in patients 60 years of age or older with primary cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. Blood. 2010;116(18):3622–3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimwade D, Hills RK, Moorman AV, et al. Refinement of cytogenetic classification in acute myeloid leukemia: determination of prognostic significance of rare recurring chromosomal abnormalities among 5876 younger adult patients treated in the United Kingdom Medical Research Council trials. Blood. 2010;116(3):354–365. [DOI] [PubMed] [Google Scholar]
- 19.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cancer Genome Atlas Research N. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368(22):2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnittger S, Schoch C, Kern W, et al. Nucleophosmin gene mutations are predictors of favorable prognosis in acute myelogenous leukemia with a normal karyotype. Blood. 2005;106(12):3733–3739. [DOI] [PubMed] [Google Scholar]
- 23.Kiyoi H, Naoe T, Yokota S, et al. Internal tandem duplication of FLT3 associated with leukocytosis in acute promyelocytic leukemia. Leukemia Study Group of the Ministry of Health and Welfare (Kohseisho). Leukemia. 1997;11(9):1447–1452. [DOI] [PubMed] [Google Scholar]
- 24.Benthaus T, Schneider F, Mellert G, et al. Rapid and sensitive screening for CEBPA mutations in acute myeloid leukaemia. Br J Haematol. 2008;143(2):230–239. [DOI] [PubMed] [Google Scholar]
- 25.Krug U, Berdel WE, Gale RP, et al. Increasing intensity of therapies assigned at diagnosis does not improve survival of adults with acute myeloid leukemia. Leukemia. 2016;30(6):1230–1236. [DOI] [PubMed] [Google Scholar]
- 26.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patel JP, Gonen M, Figueroa ME, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kihara R, Nagata Y, Kiyoi H, et al. Comprehensive analysis of genetic alterations and their prognostic impacts in adult acute myeloid leukemia patients. Leukemia. 2014;28(8):1586–1595. [DOI] [PubMed] [Google Scholar]
- 30.Nazha A, Ravandi F. Acute myeloid leukemia in the elderly: do we know who should be treated and how¿ Leuk Lymphoma. 2014;55(5):979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heiblig M, Le Jeune C, Elhamri M, et al. Treatment patterns and comparative effectiveness in elderly acute myeloid leukemia patients (age 70 years or older): the Lyon-university hospital experience. Leuk Lymphoma. 2017;58(1):110–117. [DOI] [PubMed] [Google Scholar]
- 32.Bullinger L, Dohner K, Dohner H. Genomics of acute myeloid leukemia diagnosis and pathways. J Clin Oncol. 2017;35(9):934–946. [DOI] [PubMed] [Google Scholar]
- 33.Silva P, Neumann M, Schroeder MP, et al. Acute myeloid leukemia in the elderly is characterized by a distinct genetic and epigenetic landscape. Leukemia. 2017;31(7): 1640–1644. [DOI] [PubMed] [Google Scholar]
- 34.Xu Q, Li Y, Lv N, et al. Correlation between isocitrate dehydrogenase gene aberrations and prognosis of patients with acute myeloid leukemia: a systematic review and meta-analysis. Clin Cancer Res. 2017;23(15): 4511–4522. [DOI] [PubMed] [Google Scholar]
- 35.Green CL, Evans CM, Hills RK, Burnett AK, Linch DC, Gale RE. The prognostic significance of IDH1 mutations in younger adult patients with acute myeloid leukemia is dependent on FLT3/ITD status. Blood. 2010;116(15):2779–2782. [DOI] [PubMed] [Google Scholar]
- 36.DiNardo CD, Ravandi F, Agresta S, et al. Characteristics, clinical outcome, and prognostic significance of IDH mutations in AML. Am J Hematol. 2015;90(8):732–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paschka P, Schlenk RF, Gaidzik VI, et al. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J Clin Oncol. 2010;28(22):3636–3643. [DOI] [PubMed] [Google Scholar]
- 38.Yamaguchi S, Iwanaga E, Tokunaga K, et al. IDH1 and IDH2 mutations confer an adverse effect in patients with acute myeloid leukemia lacking the NPM1 mutation. Eur J Haematol. 2014;92(6):471–477. [DOI] [PubMed] [Google Scholar]
- 39.Boissel N, Nibourel O, Renneville A, et al. Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: a study by the Acute Leukemia French Association group. J Clin Oncol. 2010;28(23):3717–3723. [DOI] [PubMed] [Google Scholar]
- 40.Abbas S, Lugthart S, Kavelaars FG, et al. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood. 2010;116(12): 2122–2126. [DOI] [PubMed] [Google Scholar]
- 41.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Im AP, Sehgal AR, Carroll MP, et al. DNMT3A and IDH mutations in acute myeloid leukemia and other myeloid malignancies: associations with prognosis and potential treatment strategies. Leukemia. 2014;28(9):1774–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grimwade D, Walker H, Harrison G, et al. The predictive value of hierarchical cytogenetic classification in older adults with acute myeloid leukemia (AML): analysis of 1065 patients entered into the United Kingdom Medical Research Council AML11 trial. Blood. 2001;98(5):1312–1320. [DOI] [PubMed] [Google Scholar]
- 44.Dombret H, Seymour JF, Butrym A, et al. International phase 3 study of azacitidine vs conventional care regimens in older patients with newly diagnosed AML with >30% blasts. Blood. 2015;126(3):291–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kantarjian HM, Thomas XG, Dmoszynska A, et al. Multicenter, randomized, open-label, phase III trial of decitabine versus patient choice, with physician advice, of either supportive care or low-dose cytarabine for the treatment of older patients with newly diagnosed acute myeloid leukemia. J Clin Oncol. 2012;30(21):2670–2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wei AH, Tiong IS. Midostaurin, enasidenib, CPX-351, gemtuzumab ozogamicin, and venetoclax bring new hope to AML. Blood. 2017;130(23):2469–2474. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.