Abstract
Understanding factors that regulate zygotic genome activation (ZGA) is critical for determining how cells are reprogrammed to become totipotent or pluripotent. There is limited information regarding how this process occurs physiologically in early mammalian embryos. Here, we identify a mediator complex subunit, MED13, as translated during mouse oocyte maturation and transcribed early from the zygotic genome. Knockdown and conditional knockout approaches demonstrate that MED13 is essential for ZGA in the mouse, in part by regulating expression of the embryo-specific chromatin remodeling complex, esBAF. The role of MED13 in ZGA is mediated in part by interactions with E2F transcription factors. In addition to MED13, its paralog, MED13L, is required for successful preimplantation embryo development. MED13L partially compensates for loss of MED13 function in preimplantation knockout embryos, but postimplantation development is not rescued by MED13L. Our data demonstrate an essential role for MED13 in supporting chromatin reprogramming and directed transcription of essential genes during ZGA.
Keywords: mediator complex, transcription, translation, zygote, oocyte-to-embryo transition, preimplantation embryo, MED13, MED13L
Summary Sentence
MED13 is required for activating transcription of essential genes during the maternal to zygotic transition in the mouse.
Introduction
The “oocyte-to-embryo transition” (OET) encompasses a series of tightly orchestrated developmental events that convert the prophase I-arrested, germinal vesicle-intact oocyte (GV oocyte) through maturation to metaphase II (MII) and then fertilization to become a totipotent, transcriptionally active cleavage stage embryo. Because GV oocytes are transcriptionally quiescent once fully grown, most of these events depend on cytoplasmic factors generated and stored during the oocyte growth phase. A key process regulating the OET is temporally controlled polyadenylation and recruitment to polysomes of stored dormant maternal mRNAs that encode proteins critical for the success of preimplantation embryo development [1, 2]. These proteins include requisite components for DNA replication such as CDC6 and ORC6L, proteins required for chromatin remodeling such as SIN3A, histone H3.3 and histone H3 methyltransferase ASH2L, and mRNA-degrading enzymes including DCP1A and DCP2 [3–9]. Waiting for oocyte maturation to begin generating these key proteins accomplishes at least two goals: (1) limited oocyte resources are not wasted generating proteins not yet needed, and (2) lack of these proteins prevents precocious DNA replication, chromatin reprogramming, and mRNA degradation prior to fertilization.
There is limited information regarding how embryonic gene transcription is initiated during the OET in mammals. Data from zebrafish and Drosophila have led to a model of embryonic genome activation in which “pioneer” transcription factors bind specific DNA sequence motifs in gene-regulatory regions of genes important during early genome activation. DNA occupancy by these transcription factors leads to recruitment of chromatin remodeling complexes, nucleosome repositioning and histone modifications, and subsequent recruitment of RNA polymerase to the transcription start sites of these genes [10]. In the mouse, embryonic genome activation begins at the one-cell (1C; zygote) stage; hence, the term “zygotic genome activation” (ZGA) is used in this species. During this first wave of genome activation, there is promiscuous transcription of numerous retrotransposons, but relatively few protein-coding genes are transcribed [11, 12]. Many critical factors driving zygotic transcription are maternally encoded [13]. For example, tripartite motif-containing protein 24 (TRIM24) is a maternally generated general transcription regulator located in the oocyte cytoplasm that translocates into the zygote pronuclei where it supports the first wave of transcription [14]. In contrast to the dearth of information regarding factors that support the onset of genome activation in pronuclear stage embryos, recent reports have described several transcription factors essential for the major wave of transcription that occurs in the two-cell (2C) stage embryo, including DUX, ZSCAN, YAP1, and NFYα (reviewed in [15]).
The mediator complex is an extremely large, multiprotein complex that serves to link regulatory information from transcription factors to RNA polymerase II and is conserved from yeast to human (reviewed in [16]). A key role for mediator is to facilitate assembly of the preinitiation complex, in part by serving as an adapter between DNA-binding transcription factors at enhancers and RNA polymerase II at promoters. Mediator is comprised of three modules (head, middle, and tail) that make up the “core” mediator complex and a kinase domain that is variably associated with the core complex. The kinase domain was initially thought to serve only as a transcriptional repressor, but more recent studies have demonstrated that it also functions in context-dependent transcriptional activation (reviewed in [17]). Transcriptional activation may be carried out via kinase domain-mediated phosphorylation of enhancer-associated transcription factors or coactivators [18]. The kinase domain has four protein components: MED13, mediator complex subunit 12 (MED12), cyclin C (CCNC), and cyclin-dependent kinase 8 (CDK8). Three of the kinase domain proteins have paralogs (MED12L, MED13L, and CDK19) that can replace their cognate proteins in the kinase domain but do not appear to have complete functional redundancy [19].
Fully grown GV oocytes and MII eggs are transcriptionally quiescent, so mediator function is not required at these stages, but mediator would be essential for RNA polymerase II-mediated transcription of embryonic genes after fertilization. We reasoned that components of the mediator complex would be good candidates to serve as proteins recruited for translation during oocyte maturation and/or egg activation as part of the developmentally programmed ZGA pathway. Here, we show using both knockdown and conditional knockout approaches that the mediator complex component MED13 is a critical upstream regulator of ZGA, and that its paralog, MED13L, can partially compensate for its function.
Materials and methods
Animals and chemicals
The following mouse strains were used: CF-1 (Envigo, Indianapolis, IN); B6SJLF1/J, C57Bl6/J, and Zp3-Cre [20] (Jackson Laboratories, Bar Harbor, ME); Med13f/f [21]; and Hspa2-Cre [22]. Mice were housed in a temperature-controlled environment under a 12 h light:12 h dark cycle. All animal procedures complied with National Institutes of Health animal care guidelines under an approved protocol. Chemicals were purchased from Sigma-Aldrich (St. Louis, MO) unless otherwise indicated. Antibodies are described in detail in Supplemental Table S1.
Plasmid construction and in vitro transcription
A 1.1-kb region of the Fam208a promoter (–1001 to +101 relative to transcription start site) was amplified with primers 5ʹGTCGAACTCGAGTAAATACTTTGGATGAAATTAG3ʹ and 5ʹGCTAACAAGCTTCGCACTCCAGCCACAGAGACG3ʹ using mouse genomic DNA as the template. The fragment was digested with XhoI and HindIII and inserted into pGL4.10[luc2] (Promega), resulting in F208wt. This promoter region contains two E2F consensus binding sequences (TTTCCCCGGGC and TTTGGCGGG). Site-directed mutagenesis was conducted to sequentially mutate the upstream site to TTTCCaattGC and the downstream site to TTTttaaGG using QuickChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, California). The resulting plasmid was named F208mut.
The coding regions of mouse Smarca4 and Smarcc1 were amplified from pMX-Brg1 (Addgene plasmid #25855) and pMX-Baf155 (Addgene plasmid #25856), gifts of Hans Schöler [23]. The primers used were 5ʹGGGCTTGTCGACGCCACCATGTCTACTCCAGACCCACCCTTTG3ʹ and 5ʹGCTCAATCTAGATTCAGTCTTCCTCACTGCCACTTCCTG3ʹ (Smarca4) and 5ʹ GGCTTCTCGAGGCCACCATGGCCGCGACAGCGGGTGGCG3ʹ and 5ʹ GGTCAAACTAGTCTACTTACACATGCCGTTTGGTG3ʹ (Smarcc1). The PCR products were ligated into the Sal1 and Xba1 sites of a modified pIVT plasmid [24]. Following DNA sequence confirmation, in vitro transcription was carried out using AmpliCap-Max T7 High Yield Message Maker Kit (CELLSCRIPT, Madison, WI).
Oocyte and embryo collection, culture, and microinjection
GV-intact oocytes were collected from female mice 48 h following injection with 5 IU equine CG (Calbiochem, San Diego, CA). The cumulus cells were removed mechanically and the oocytes were cultured in minimal essential medium alpha (MEMα; Invitrogen, Carlsbad, CA) containing 2.5 μM milrinone to prevent GV breakdown. Following microinjection, oocytes were matured for 16 h in MEMα containing 5% fetal calf serum. Metaphase II-arrested eggs were collected from the oviducts of female mice superovulated using 5 IU equine CG followed 48 h later with 5 IU human CG (hCG) (Calbiochem) at 13–14 h following hCG administration. Cumulus cells were removed using 0.1% hyaluronidase. One-cell embryos were collected from superovulated, mated females at 15–16 h following hCG administration. The collection medium was either Hepes-buffered Whitten medium [25] or Leibovitz L-15 medium (Invitrogen) supplemented with 1% fetal calf serum. Microinjections were performed using a Leica DMI 6000B inverted microscope equipped with a XenoWorks micromanipulator system and PrimeTech PMM-150FU piezo drill (Sutter Instruments, Novato, CA). One-cell embryo injections were completed prior to 18 h following hCG injection. GV-intact oocytes and embryos were injected into the cytoplasm with 5–10 pl of a 1.5-mM pipette concentration of the following morpholino oligonucleotides (5ʹ to 3ʹ): Scr-MO, CCTCTTACCTCAGTTACAATTTATA; Med13UTR-MO, GCCACAACCCACCATCCGCCATTAC; Med13-MO, CGTTCGACACGAAGGAGGAACTCAT; or Med13L-MO, TCATGGTCCTCCGCGAGCCC (GeneTools, Philomath, OR). Smarca4 and Smarcc1 cRNAs were microinjected with Med13-MO at a final pipette concentration of 0.25 pg/pl each. Embryos were cultured in KSOM medium (EMD Millipore, Billerica, MA; cat. no. MR-106-D) in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2.
RNA isolation and real-time RT-PCR
For most experiments, total RNA was isolated from 50 oocytes, eggs, or embryos using the PicoPure RNA Isolation kit (Thermo Fisher Scientific, Waltham, MA). Enhanced GFP (eGFP) cRNA was transcribed in vitro from pIVT-eGFP [26], and 1 ng was added to each sample prior to RNA isolation as an internal control. Real-time RT-PCR was performed as previously described [27], using cDNA from two oocytes or embryos per reaction. All E2f transcripts were amplified from the same three sets of GV oocytes to enable comparison of Ct values. Relative gene expression was calculated using the ΔCt method [28] with eGFP expression for normalization. For testing embryos for the presence or absence of Med13 floxed exons 7–8, total RNA was isolated as above from a pool of 10 morula stage embryos from two mice/group, N = 2 pools/genotype. Real-time RT-PCR was performed using cDNA from one morula per reaction. Expression of Med13 exons 7–8 was calculated relative to expression of Med13 exons 29–30 in the same sample using the ΔCt method [28]. Primer sequences are provided in Supplemental Table S2.
Immunofluorescent staining, microscopy, and quantification
For MED13 staining, embryos were fixed in ice-cold methanol for 10 min, then blocked overnight at 4°C in blocking buffer (PBS, 3 mg/ml BSA, 0.01% Tween-20) supplemented with 10% fetal calf serum. Embryos were then incubated in primary anti-MED13 antibody (Santa Cruz, Dallas, TX, sc-5369; 10 μg/ml) for 1 h at room temperature (RT) followed by Alexa Fluor 488 anti-goat IgG (Thermo Fisher Scientific, cat# A-11055; 2 μg/ml) for 1 h at RT. For γH2AX staining, embryos were fixed in 4% paraformaldehyde for 30 min, permeabilized in 0.1% Triton X-100 for 20 min, then incubated in blocking buffer for 1 h. Embryos were incubated in primary γH2AX antibody (Bioworld Technology, St Louis Park, MN; BS4760; 5 μg/ml) at 4°C overnight followed by Alexa Fluor 555 anti-rabbit antibody (Thermo Fisher Scientific, cat# A-21429; 2 μg/ml) for 1 h at RT. For MED13L staining, embryos were fixed in 2% paraformaldehyde for 40 min, permeabilized as above, then washed in blocking buffer. Embryos were incubated in primary MED13L antibody (Bethyl Laboratories, Montgomery, TX, A302-421A; 10 μg/ml) at 4°C overnight followed by Alexa Fluor 555 anti-rabbit antibody (Thermo Fisher Scientific, cat# A-21429; 4 μg/ml) for 1 h at RT. For NANOG staining, embryos were fixed in 4% paraformaldehyde for 30 min, permeabilized in 0.1% Triton X-100 for 20 min, then incubated in blocking buffer for 1 h. Embryos were incubated in primary NANOG antibody (R&D Systems, Minneapolis, MN, cat# AF2729; 10 μg/ml) at 4°C overnight followed by Alexa Fluor 647 anti-goat IgG (Thermo Fisher Scientific, cat# A-21447; 4 μg/ml) for 1 h at RT. Terminal deoxynucleotidyl transferase dUTP Nick-End Labeling (TUNEL) was performed as described previously [29] using the In Situ Cell Death Detection Kit, Fluorescein (Sigma-Aldrich) except that 1.5 μg/mL DAPI was in the mounting medium. Peroxide-treated embryos served as positive controls [29]. For some experiments, zona pellucidae were removed immediately prior to fixation using acid Tyrode medium.
For cell counts, blastocysts were fixed 120 h after hCG injection in 2% paraformaldehyde for 40 min, permeabilized as above, then washed in blocking buffer. Embryos were incubated in primary anti-CDX2 antibody (Abcam, Cambridge, MA; ab157524; 1 μg/ml) at 4°C overnight followed by primary OCT-3/4 antibody (Santa Cruz, Dallas, TX; sc-8628; 10 μg/ml) for 1 h at RT. The embryos were then washed and incubated with Alexa Fluor 488 donkey anti-mouse (Thermo Fisher Scientific, cat# A-21202; 4 μg/ml) and Alexa Fluor 555 donkey anti-goat (Thermo Fisher Scientific, cat# A-21432; 4 μg/ml) for 1 h at RT.
All oocytes and embryos were mounted in Vectashield containing 1.5 μg/mL DAPI (Vector Laboratories, Burlingame, CA), and slides were scanned using a Zeiss LSM 510 UV confocal microscope. Immunofluorescence signals were quantified using ImageJ software. For EU, EdU, and γH2AX staining, a region of interest was drawn around each nucleus and the average pixel intensity was determined. The values for each nucleus were then averaged to generate an intensity level for each embryo. Intensity was expressed relative to the average intensity of the control embryos for each independent experiment. Quantification of CDX2, OCT4, and NANOG staining intensity was done from confocal z-stack scans through entire blastocysts followed by automated measurement of signal intensity in each slice using ImageJ. The total image intensity was calculated by multiplying the overall average intensity by the number of images in the stack. Quantification of blastocyst inner cell mass (OCT4-positive), trophectoderm (CDX2-positive), and total cells (DAPI) was performed manually after scanning a z-stack of images through the full depth of each embryo. Total cell numbers were counted after encoding nuclear depth using a rainbow look-up table as described previously [30].
Immunoblot analysis
Immunoblot analyses for cyclin B1 and actin were performed as described previously [31].
DNA replication, transcription, and luciferase assays
Embryos were cultured in KSOM with 10 μM 5-ethynyl-2ʹ-deoxyuridine (EdU) or 1 mM ethynyl uridine (EU) for 1 h and fixed in 4% paraformaldehyde in PBS for 30 min at RT. Edu or EU incorporation into DNA or RNA, respectively, was detected using Click-iT EdU Alexa Fluor 488 Imaging Kit or Click-iT RNA Alexa Fluor 488 Imaging Kit (both from ThermoFisher, Waltham, MA) according to the manufacturer's protocols. Fluorescence was detected on a Zeiss confocal microscope (Zeiss LSM 510 UV), and fluorescence intensity was quantified using ImageJ software [32] as described previously [33]. For luciferase assays, 1–2 pl of 50 000 copies/pl plasmid DNA was microinjected into the male pronucleus using a pressure-controlled digital microinjector (Sutter Instruments). The injected plasmid DNA was a 1:1 mixture (13.3 ng/μl each) of TK-ren with either F208wt or F208mut firefly luciferase plasmids. Plasmid DNA injections were performed 25 h following hCG administration; for doubly injected embryos, this was 9 h following morpholino injections. Following plasmid injection, the embryos were cultured in KSOM containing 2 μg/ml aphidicolin to inhibit DNA replication. Luciferase assays were performed 24 h later using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. Firefly luciferase activity was normalized using renilla luciferase activity from the same embryo.
RNA sequencing and analysis
Total RNA was isolated from 4 independent collections of 40 embryos/group using the PicoPure RNA Isolation Kit (Thermo Fisher Scientific) and amplified using Ovation RNA-Seq System V2 (NuGEN, San Carlos, CA). mRNA libraries were constructed from 400 ng cDNA using Illumina TruSeq RNA Sample Prep Kits, version 2. The cDNA was first fragmented using a Covaris E210 focused ultrasonicator. Library amplification was performed using 12 cycles to minimize the risk of overamplification. Unique barcode adapters were applied to each library. Libraries were pooled in equimolar ratio and sequenced together on an Illumina HiSeq 2000 with version 3 flow cells and sequencing reagents. At least 15 million 101-base read pairs were generated for each individual library. Data were processed using RTA 1.13.48 and CASAVA 1.8.2 (Illumina, San Diego, CA). Data were deposited in the NCBI GEO database, accession number GSE90710.
To identify differentially expressed transcripts, raw sequence reads were mapped to mouse cDNA sequences using BWA (ver. 0.7.12) [34], with the default setting of its BWA-MEM algorithm paired-end read mapping protocol. The cDNA sequence library used for the mapping included all transcripts in the mm10 GENCODE comprehensive gene annotation (http://www.gencodegenes.org/). The mapping was done separately for each replicate. Differentially expressed transcripts were identified using the improved version of EpiCenter [35], which considers both read coverage depth and within-group variation of read counts for testing significance of gene expression change between groups. Specifically, we first filtered out all reads that were not mapped in-pair to the reference cDNA sequences, and then counted reads mapped to each cDNA sequence. Read counts in each biological replicate were then normalized so that the average number of total mapped reads was same across all replicates in both groups. To increase the reliability of the log ratio data used in statistical testing, any transcript with a mean of normalized counts less than 100 in both groups was removed from further analysis. This filter also allowed us to focus on genes with more than minimal levels of expression. The significance cutoff was set at 5% false discovery rate (FDR) based on the Benjamini–Hochberg method. The generation of heatmap plots of all differentially expressed transcripts, and hierarchical clustering analysis of seven RNAseq samples were carried out using R (https://www.r-project.org) statistical language with the gplots package.
Differentially expressed transcripts were submitted to DAVID (https://david.ncifcrf.gov) for gene enrichment analysis in all gene ontology (GO) terms and pathways. This was done separately for upregulated and downregulated transcripts. The analysis of transcription of repeats was performed separately. In this analysis, all raw reads were mapped to the whole mouse genome reference (mm10) using Tophat2 [36], and EpiCenter was used to do comparison analysis of transcription levels of repeat regions between the two groups. All repeat regions and repeat types were based on the annotation from the UCSC rmsk table downloaded from the UCSC genome browser. The cutoff for differentially transcribed repeat regions was set at 5% FDR based on the Benjamini–Hochberg method.
Differential motif enrichment analysis of promoter sequences
The 1-kb promoter sequences for the 1054 downregulated genes and 664 upregulated genes were extracted using the mouse reference gene annotation (mm10). The total numbers of unique promoter sequences obtained were 923 and 654, respectively; these sequences were referred to as foreground sequences. For each promoter sequence in each dataset, we randomly selected 10 promoter sequences of the same length (1 kb) with similar C/G content from the entire mouse genome to serve as the matched background sequences. We then scanned each set of the foreground promoter sequences and the corresponding background sequences [37] for putative transcription factor-binding sites using motif models from TRANSFAC, JASPAR, and UniPROBE [38–40]. A total of ∼1300 motif models were used in the analysis. For each motif model, we counted the number of foreground sequences and background sequences containing at least one predicted site for the motif. Motif enrichment analysis was then carried out on the resultant 2 × 2 contingency table using Fisher exact test.
Conditional knockout mouse breeding and embryo development assays
For litter size analysis, Med13f/f females were mated to Med13f/f males (three pairs) and Med13f/f; ZP3-Cre females were mated to Med13f/▵ males (seven females, five males) for 6 months. The cages were monitored daily, and litter sizes from each group were recorded. For parthenogenetic activation, MII eggs were collected from superovulated females and then activated for 6 h in 10 mM SrCl2 in Ca2+-free CZB medium [41] containing 5 μg/ml cytochalasin B or 2 μg/ml cytochalasin D to prevent second polar body emission. In vivo-fertilized 1C embryos were collected from either spontaneously cycling or superovulated females mated to males overnight and found to be vaginal plug-positive the following morning. All embryos were cultured in KSOM at 37°C in a humidified atmosphere of 5% CO2, 5% O2, and 90% N2 and development was monitored daily. Postimplantation embryo development was assayed by mating Med13f/f or Med13f/f;ZP3-Cre females to Med13f/▵;Hspa2-Cre males. The females were sacrificed on E12.5, and the number of viable and nonviable implantation sites was recorded.
Statistical analysis
All analyses were performed using GraphPad Prism software. Comparisons of two groups were performed using two-tailed T-tests, with Welch correction in case of unequal standard deviations. Multiple group results were analyzed using ANOVA with Holm-Sidak multiple comparisons test. A two-tailed Fisher exact test was used to compare numbers of viable and nonviable implantation sites. Detailed information is included in the figure legends.
Results
To determine which mediator complex components were recruited for translation during oocyte maturation, we utilized a published microarray dataset that identified transcripts associated with polysome fractions isolated from mouse oocytes at different stages of maturation [1]. Three kinase domain proteins were the most highly recruited mediator mRNAs. Med13 was increased more than 11-fold, whereas Med13l and Med12l were increased about 9- and 8-fold, respectively, suggesting that these mRNAs were strongly recruited for translation during oocyte maturation. Relative to its expression in oocytes, Med13 mRNA levels remained relatively stable through the 2C stage (Figure 1A). To determine if Med13 was transcribed very early from the embryonic genome, we measured Med13 mRNA expression in late 2C-stage embryos that were cultured beginning at the 1C stage with or without α-amanitin to block transcription [42]. Med13 mRNA levels were reduced to about one-third of that in controls by α-amanitin treatment (Figure 1B), indicating that the majority of the mRNA present in late 2C-stage embryos was a result of new transcription from the embryonic genome. Consistent with these findings, MED13 immunoreactivity increased markedly between the GV oocyte and MII egg stages, and was detected in the cytoplasm of 1C embryos, but increased in 2C- and 4C-stage embryos where it localized to the nuclei (Figure 1C).
Figure 1.
MED13 knockdown inhibits preimplantation embryo development. (A) Expression of Med13 mRNA in oocytes and preimplantation embryos relative to that in GV oocytes. N = 3. (B) Expression of Med13 mRNA in late 2C embryos cultured without (C, control) or with α-amanitin (α-am) starting at the 1C stage to block zygotic transcription. N = 3; *P ≤ 0.05, T-test. (C) MED13 protein in oocytes, eggs, and 1C, 2C, and 4C-stage embryos. Bar = 20 μm. N = 3, total of 11–25/group. (D) MED13 protein in 4C-stage embryos following microinjection at the 1C stage using the indicated morpholino. Bar = 20 μm. Graph indicates relative signal intensity in nuclei. N = 3, total of 47–53/group; *P < 0.05, T-test. (E) Representative DIC images of embryos 3 days following microinjection at the 1C stage using the indicated morpholino. (F) Percentage of embryos to reach the various preimplantation embryo stages following microinjection at the 1C stage with the indicated morpholino. Bar = 80 μm. N = 4, 16–37 1C embryos/group for each replicate; *P ≤ 0.05 compared to Scr-MO control at the same time point, ANOVA with Holm-Sidak multiple comparisons test. All graphs show mean ± s.e.m. Abbreviations used in this and all subsequent figures: ooc, GV-intact oocyte; egg, MII-arrested egg; 1C, one-cell embryo; 2C, two-cell embryo; 4C, four-cell embryo; 8C, eight-cell embryo; mo, morula; bl, blastocyst.
These findings led us to hypothesize that MED13 is a very early regulator of ZGA in mouse preimplantation embryos. To test this hypothesis, we knocked down MED13 protein in 1C embryos by microinjecting morpholino oligonucleotides targeting the Med13 translation start site (Med13-MO) approximately 4 h after fertilization in vivo. This procedure reduced MED13 protein levels as indicated by immunofluorescence analysis (Figure 1D). Med13-MO-injected embryos cleaved to 2C, but many became arrested after this point, with only ∼40% of the embryos developing to the four-cell (4C) stage, and none beyond 4C (Figure 1E and F). In contrast, when 1C embryos were microinjected with the MED13-MO starting 6 h after fertilization in vivo, when pronuclei had already formed, there was far less impact on preimplantation embryo development, with 78% (54/69) progressing to the morula stage, though only 13% (9/69) developed into blastocysts. Embryos injected with a scrambled control morpholino (Scr-MO) served as negative controls. We generated capped, polyadenylated mRNA encoding full-length MED13 protein, with or without either V5, GFP, or mCherry tags on either end, but following microinjection of these mRNAs into 1C embryos, we were unable to observe expression of the exogenous mRNA despite no difficulties in expressing many other positive control proteins. For this reason, we could not perform a standard rescue experiment to rule out off-target effects of the Med13-MO. As one alternative approach, we tested the effect of microinjecting a morpholino that targeted a region of the Med13 5ʹ UTR (Med13UTR-MO). Like the Med13-MO-injected embryos, the Med13UTR-MO-injected embryos had significantly arrested development starting at the 4C stage, though a few developed into morulae (Figure 1F). These results suggested that translation of MED13 after fertilization was essential for preimplantation embryo development.
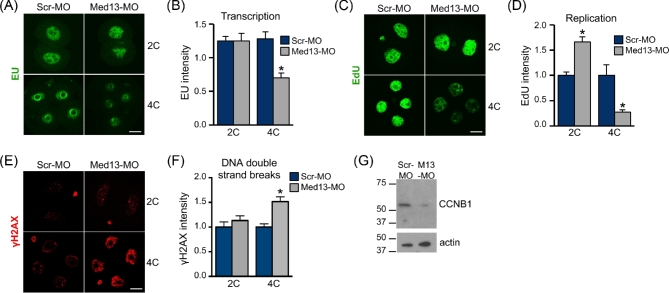
To determine whether MED13 knockdown caused embryo development arrest because of a global failure of transcription, total transcriptional activity in Med13-MO-injected embryos was measured using the uridine analog 5-EU [43]. Despite the finding that many of the embryos arrest at the 2C stage, there was no detectable difference in overall transcriptional activity of MED13 knockdown embryos until the 4C stage (Figure 2A and B). These findings suggested that MED13 knockdown did not disrupt global function of the mediator complex. Another explanation for embryo developmental arrest was a failure of DNA replication. DNA replication was measured using EdU [44]. MED13 knockdown embryos at the 2C stage had significantly increased EdU staining intensity compared to Scr-MO-injected embryos indicating successful DNA replication, whereas 4C embryos had very low EdU incorporation (Figure 2C and D). In addition, Med13-MO-injected embryos had evidence of increased DNA double strand breaks at the 4C stage as indicated by γH2AX nuclear staining (Figure 2E and F). To determine if the Med13-MO-injected embryos arrested before or after entering M phase, we performed immunoblot analysis of cyclin B1 protein. Cyclin B1 was dramatically lower in Med13-MO-injected 2C embryos as compared to controls (Figure 2G). We conclude from these experiments that a failure of DNA replication cannot explain the developmental arrest that occurs at the 2C stage, and that loss of MED13 causes cell cycle arrest prior to M phase.
Figure 2.
MED13 knockdown inhibits embryo transcription and cell cycle progression. One-cell stage embryos were microinjected with the indicated morpholino, then cultured to the 2C or 4C stage. (A) EU staining as an indicator of transcription levels. (B) Quantification of EU staining. N = 3, total of 23–24 embryos/group; *P ≤ 0.05, T-test. (C) EdU staining as an indicator of DNA replication. (D) Quantification of EdU staining. N = 3, total of 20–24 embryos/group; *P ≤ 0.05, T-test. (E) Gamma H2AX staining as an indicator of DNA double strand breaks. (F) Quantification of γH2AX staining. N = 4, total of 14–36 embryos/group; *P ≤ 0.05, T-test. (G) Immunoblot analysis of cyclin B1 in late 2C-stage embryos (upper panel). Actin was used as a loading control (lower panel). Numbers indicate apparent Mr × 10−3. Lysate of 20 embryos in each lane. Blot is representative of N = 3 independent replicates. All graphs show mean ± s.e.m. All bars = 20 μm.
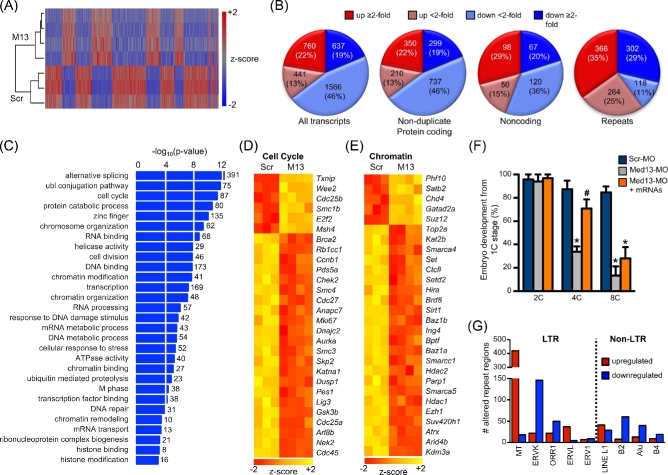
Based on findings that many mediator subunits regulate transcriptional activation or repression of specific developmental pathways [45], we tested the idea that MED13 functions to regulate the transcription of a subset of genes during ZGA. This was done by microinjecting either Scr-MO or Med13-MO at the very early 1C stage and then performing RNA sequencing (RNAseq) following culture to the late 2C stage. Of the four independent replicates per group, one replicate from the Scr-MO group failed quality control, leaving three Scr-MO and four Med13-MO replicates for analysis. Unsupervised hierarchical analysis revealed clearly distinct sorting by treatment group (Figure 3A). Analysis of these data using the GENCODE comprehensive dataset [46] and a FDR of 5% revealed significant changes in transcription of protein-coding genes, noncoding RNAs, and repetitive elements (Figure 3B; see also Supplemental Table S3). Compared to the Scr-MO controls, expression of 3404 transcripts was altered in the Med13-MO-injected embryos, with 1201 (35%) upregulated and 2203 (65%) downregulated. Almost twice as many genes were downregulated, suggesting that MED13 is required for transcriptional activation at more gene loci than for transcriptional repression. Of note, there was no evidence of positive TUNEL staining in either injection group at the late 2C stage (data not shown), and the RNAseq data revealed no alterations in transcription or splicing of Trp53 or its downstream target Cdkn1a (encoding p21/WAF1/CIP1) in the knockdown embryos. These findings indicate that the potential off-target effect of p53-induced apoptosis had not occurred [47].
Figure 3.
MED13 knockdown impairs transcription of genes required for the OET, including cell cycle and chromatin-remodeling proteins. One-cell stage embryos were microinjected with the indicated morpholino, then cultured to the late 2C stage. RNA sequencing was performed on independent pools of 40 embryos per group. (A) Unsupervised hierarchical clustering of gene expression differences in Scr-MO (Scr)- and Med13-MO (M13)-injected embryos. (B) Numbers of altered transcripts of the indicated types in Med13-MO-injected embryos relative to Scr-MO-injected embryos. (C) Highly significant GO biological categories of genes downregulated in Med13-knockdown embryos. Numbers to right of bars indicate number of genes in category. (D) Selected GO cell cycle genes significantly altered in Med13-knockdown embryos. (E) Selected GO chromatin modification genes significantly altered in Med13-knockdown embryos. (F) Percentage of embryos to reach the indicated preimplantation embryo stages following microinjection at the 1C stage with the either Scr-MO, Med13-MO, or Med13-MO and both Smarca4 and Smarcc1 mRNAs. N = 3, 8–12 1C embryos/group for each replicate; *P ≤ 0.05 compared to Scr-MO control at the same time point, #P ≤ 0.05 compared to MED13-MO at the same time point, ANOVA with Holm-Sidak multiple comparisons test. Graph shows mean ± s.e.m. (G) Graph showing total number of chromosomal regions either up- or downregulated for each of the indicated repetitive element classes.
GO pathway analysis of the altered genes (5% FDR) was performed separately for both upregulated and downregulated genes. A nonredundant set of highly significant pathways (Benjamini–Hochberg adjusted P ≤ 0.05) was manually sorted into categories. The categories for genes upregulated in response to MED13 depletion were in general far less significantly altered than the downregulated genes, but included nucleotide binding, cancer, signaling, and cytoskeleton (see Supplemental Table S4). The downregulated gene categories included RNA processing, protein catabolism, cell cycle, transcription, chromatin modification, and DNA repair (Figure 3C; see also Supplemental Table S5). These specific downregulated gene categories and their high degree of significance suggested that MED13 was preferentially involved in activating transcription of genes that control pathways previously identified to be critical for the success of the OET [48]. Notably, the list of significantly altered cell cycle genes included a large number of genes either documented as or anticipated to be critical regulators of preimplantation embryo development, including Wee2, Brca2, Ccnb1, Chek2, Cdc25a, and Cdc45 (Figure 3D). Similarly, the significantly downregulated chromatin modifying genes included several identified as essential for preimplantation embryo development, including Smarca4 (previously Brg1), Hira, and Hdac1 [7, 49, 50] (Figure 3E). Furthermore, Smarca4 and another significantly downregulated gene, Smarcc1 (previously Baf155), encode subunits of the BAF (Brg/Brahma-associated factors) ATP-dependent chromatin remodeling complex known as esBAF, which is required for postimplantation embryo development as well as embryonic stem cell self-renewal and pluripotency [51–53].
To determine whether the reductions in mRNAs encoding the esBAF components SMARCA4 and SMARCC1 were responsible for the embryo arrest phenotype in MED13-knockdown embryos, we microinjected mRNAs encoding both of these proteins into zygotes along with the MED13-MO. This manipulation doubled the success of development of MED13-knockdown embryos to the 4C stage, but did not promote their development beyond this point (Figure 3F). These findings indicate that MED13 is a critical upstream regulator of the OET in part via its effect on transcription of esBAF components.
Consistent with previous reports [11, 12], we found that long-terminal repeats (LTRs) were highly abundant in the RNAseq datasets, comprising >75% of the repetitive element transcripts expressed in 2C embryos in both the SCR-MO and MED13-MO groups. Depletion of MED13 caused significant alterations in specific repeat classes. There was a dramatic increase in the number of mouse transcript (MT)-type LTRs as a result of upregulation of MT transcripts from many different chromosomal regions; only a few MT regions were downregulated (Figure 3G). Given the roles of dicer 1 and argonaute 2 in degrading MT retrotransposons in oocytes [54, 55], this finding is consistent with our RNAseq data showing downregulation of Dicer1 by 1.6-fold and Ago2 by 1.9-fold. In contrast, two other LTR transcript types, ERVK and ORR1, as well as several non-LTR transcript types, were downregulated in more repeat regions following MED13 depletion.
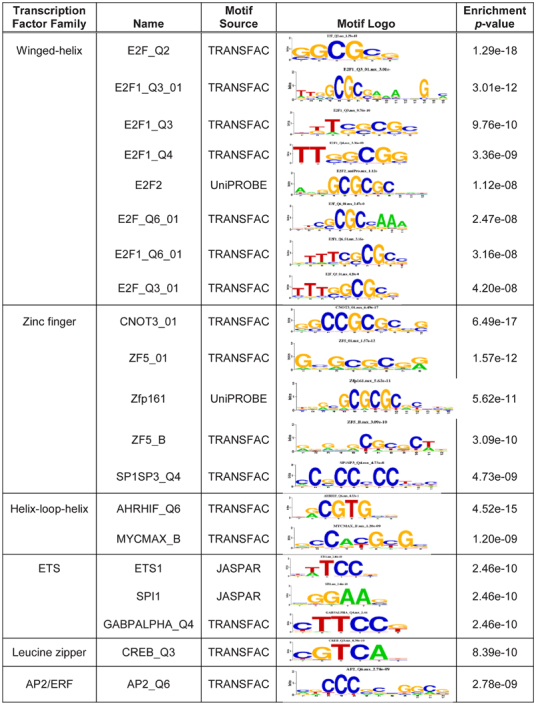
We next asked whether MED13 was interacting with specific transcription factors to regulate expression of the altered mRNAs. We analyzed the promoter sequences of protein-coding genes identified as significantly altered in our RNAseq dataset; promoters of upregulated and downregulated genes were analyzed separately. For each promoter from an altered gene, 10 additional promoters with similar GC content were randomly selected to serve as controls. This analysis identified several transcription factor-binding motifs that were strongly associated with the promoters of significantly downregulated genes (Table 1). In particular, multiple motifs that bind E2F family winged-helix transcription factors were identified as highly enriched. Indeed, E2f1, E2f2, and E2f5 transcripts were relatively abundant in fully grown GV oocytes (Supplemental Table S6), suggesting the presence of the corresponding proteins in early embryos.
Table 1.
Top 20 significantly enriched motifs in the promoters of genes downregulated in MED13-knockdown embryos.
|
To determine whether the E2F-binding sites in gene promoters were necessary for transcriptional regulation by MED13, we performed luciferase assays using the promoter region of Fam208a. This gene was selected because in MED13-knockdown embryos it was downregulated by 4.1-fold from the RNAseq assay, was validated as a downregulated gene by quantitative real-time PCR (down 3.1-fold ± 0.08 s.e.m.), and contained two predicted E2F-binding sites in the proximal promoter. A plasmid construct was generated in which the region from –1000 to +100 relative to the transcription start site of Fam208a was cloned in front of firefly luciferase (F208wt; Figure 4A). A second construct was generated with the same promoter region but the two predicted E2F sites were mutated (F208mut; Figure 4A). A plasmid containing renilla luciferase driven by the TK promoter (TK-ren) was used as a control for the pronuclear microinjection procedure. The control TK-ren plasmid was mixed at a 1:1 molar ratio with either the F208wt or F208mut plasmids, and the mixture was microinjected into the male pronucleus of each 1C embryo. The embryos were then cultured for 24 h in the presence of aphidocolin to prevent DNA replication and allow efficient transcription of the microinjected plasmids (Figure 4B) [56]. The firefly and renilla luciferase signals were then read from each embryo and expressed as a ratio (firefly luciferase/renilla luciferase). Mutation of the E2F sites caused luciferase activity to decrease to about 25% of that from the wild-type construct, indicating that the E2F-binding sites were required for efficient luciferase transcription from the Fam208a promoter (Figure 4C). We next tested whether this promoter required MED13 for efficient transcription. Early 1C-stage embryos were microinjected with either Med13-MO or Scr-MO, cultured for 9 h to allow protein knockdown, and then the same embryos were injected with the luciferase constructs into the male pronucleus (Figure 4D). MED13-knockdown embryos had significantly lower transcription from the F208wt plasmid than the Scr-MO-injected embryos and similar transcription levels to that from F208mut plasmid in MED13-knockdown embryos (Figure 4E). In fact, transcription of F208wt in MED13-knockdown embryos was similar to that of the F208mut plasmid in Scr-MO-injected embryos. These findings indicated that the E2F-binding sites were required for efficient MED13-mediated transcription from this promoter.
Figure 4.
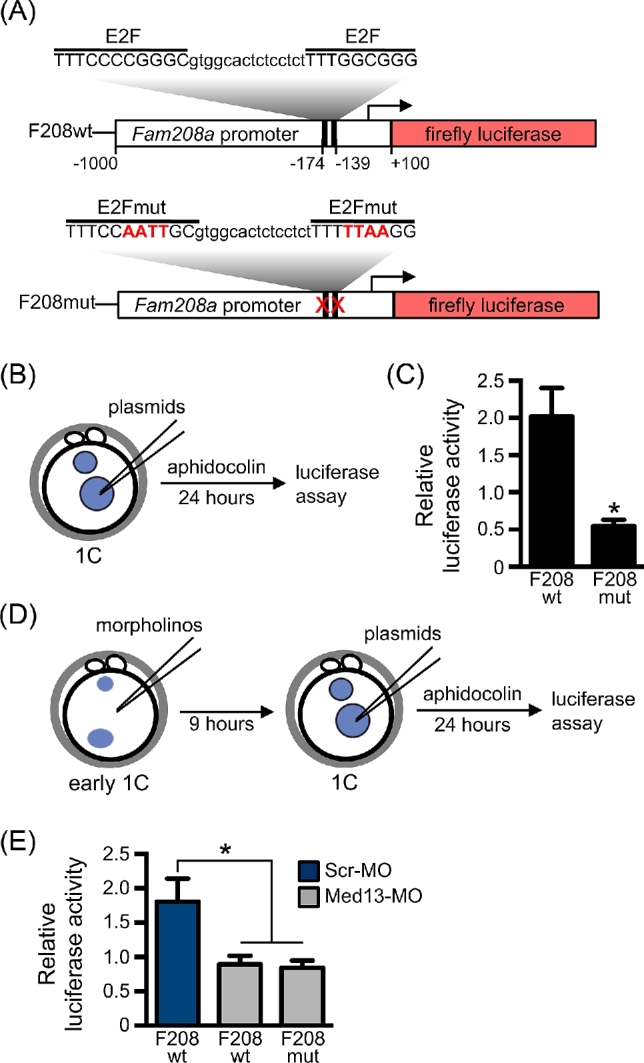
E2F transcription factor binding motif mediates MED13-dependent gene transcription. (A) Schematic diagram of luciferase plasmids showing wild-type and mutated E2F-binding motifs in the Fam208a promoter. Numbers indicate position relative to TSS (arrow). (B) Schematic of experimental design for plasmid injections. (C) Relative luciferase activity following plasmid injection alone using F208wt or F208mut plasmids. N = 4, total of 23–26 embryos/group; *P ≤ 0.05, T-test. (D) Schematic of experimental design for morpholino and plasmid injections. (E) Relative luciferase activity in embryos first injected with the indicated morpholino and then 9 h later with the indicated plasmid. N = 6, total of 21–22 embryos/group; *P ≤ 0.05 compared to F208wt plasmid/Scr-MO group, ANOVA with Holm-Sidak multiple comparisons test. All graphs show mean ± s.e.m.
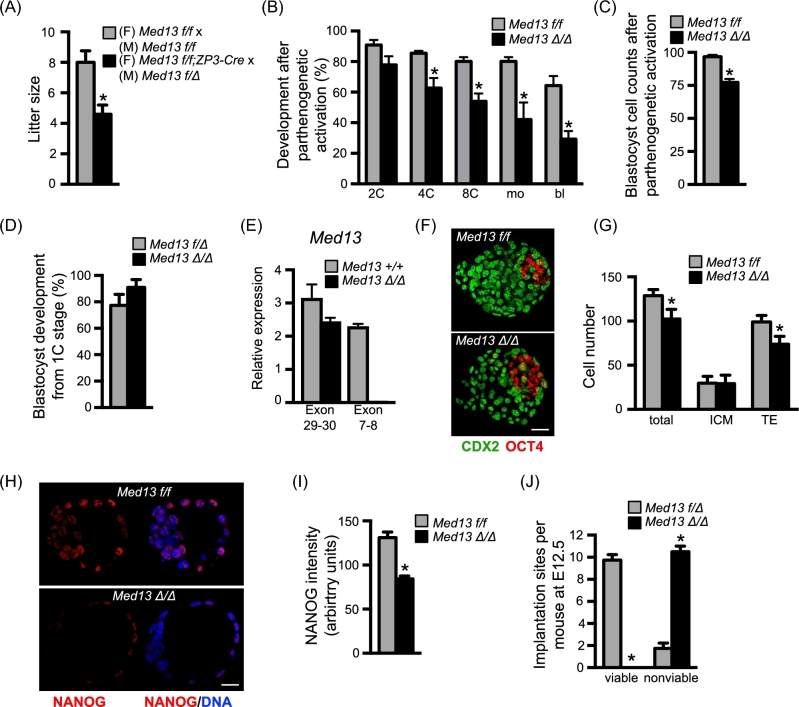
Based on the above findings, we predicted that oocytes completely lacking MED13 would not undergo successful preimplantation embryo development unless rescued by a wild-type Med13 allele (Med13+) at fertilization. We generated female mice carrying the ZP3-Cre transgene and homozygous for the Med13-floxed allele [21] (Med13f/f;ZP3-Cre); ovulated eggs of these females will carry two copies of a loss-of-function Med13 allele (Med13Δ/Δ). These mice were mated to Med13f/Δ males, and the resulting offspring were genotyped. As anticipated, the litter sizes were reduced as compared to those of controls (Figure 5A), and 100% of the offspring of Med13f/f;ZP3-Cre females were Med13f/Δ, indicating that Med13Δ/Δ embryos did not survive to delivery. To determine the efficiency of preimplantation embryo development in embryos lacking MED13, we collected Med13Δ/Δ and Med13f/f eggs and parthenogenetically activated them to avoid generation of heterozygous embryos. Of note, parthenogenetic activation, particularly when performed with strontium to induce repetitive calcium oscillations and cytochalasin to maintain a diploid chromosome complement, results in embryos practically indistinguishable from normally fertilized embryos until the blastocyst stage or later [57–60]. Activated Med13Δ/Δ eggs, unlike Med13-MO-injected 1C embryos, developed beyond the 4C stage but had far lower development to the morula and blastocyst stages than activated Med13f/f eggs (Figure 5B). Furthermore, the total number of cells in the Med13Δ/Δ embryos that did reach the blastocyst stage was significantly reduced (Figure 5C). The abnormal but improved development of Med13Δ/Δ eggs following parthenogenetic activation compared to Med13-MO-injected 1C embryos suggested that Med13Δ/Δ eggs had partially compensated for loss of MED13 during oocyte growth within the ovarian follicle.
Figure 5.
Postimplantation developmental arrest of Med13 knockout embryos. (A) Average litter size from breeding pairs of indicated genotypes. N = 10 litters/group, *P < 0.05, T-test. (B) Metaphase II eggs of the indicated genotype were parthenogenetically activated and allowed to develop in vitro. Graph represents average percentage of embryos to develop to the indicated stages. N = 3, total of 51–55 1C embryos/group; P < 0.05 compared to same stage, ANOVA with Holm-Sidak multiple comparisons test. (C) Total cell numbers in parthenogenetically activated embryos of each genotype that reached the blastocyst stage. N = 27 embryos/group, *P < 0.0001, T-test. (D) In vivo fertilized, 1C-stage embryos of the indicated genotypes were collected and allowed to develop in vitro. Graph represents percentage of embryos to develop to the blastocyst stage. N = 3 breeding pairs per group, 8–13 1C embryos collected per female. (E) Relative expression of Med13 mRNA encoding exons 29–30 and 7–8. Graph shows median ± range. N = 2 replicates. (F) Representative immunofluorescent staining of blastocyst stage embryos of the indicated genotypes that developed from the 1C stage in vitro. (G) Graph of cell numbers in the blastocysts from (E). Inner cell mass (ICM), trophectoderm (TE). N = 3, total of 13–16 embryos/group; *P < 0.05, T-test. (H) Representative immunofluorescent staining of blastocyst stage embryos of the indicated genotypes that developed from the 1C stage in vitro. Red channel on left, merged image on right. (I) Graph of NANOG staining intensity in the blastocysts from (G). N = 3, total of 23–31 embryos/group; *P < 0.0001, T-test with Welch correction. (J) Females carrying oocytes with normal amounts of MED13 (Med13f/f) or lacking MED13 (Med13f/f;ZP3-Cre) were mated to males carrying sperm lacking MED13 (Med13f/Δ;Hspa2-Cre) to generate either Med13f/Δ or Med13Δ/Δ embryos. Graph shows numbers of viable and nonviable implantation sites on E12.5. N = 4 females/group; *P < 0.0001, Fisher exact test. All graphs except panel E show mean ± s.e.m. All bars = 20 μm.
To better characterize the preimplantation development phenotype of embryos lacking MED13, we generated male mice in which Med13 was conditionally deleted only in sperm using the Hspa2-Cre transgene [22]. This transgene efficiently deleted the targeted allele because when Med13f/f;Hspa2-Cre males were mated to wild-type females, 100% of the offspring were Med13+/Δ (31/31 pups from three litters). Med13f/Δ;Hspa2-Cre males were then mated to either Med13f/f or Med13f/f;ZP3-Cre females to generate Med13f/Δ or Med13Δ/Δ embryos. In contrast to the parthenogenetically activated embryos, the in vivo-derived Med13Δ/Δ 1C embryos developed efficiently in vitro to the blastocyst stage (Figure 5D). Because of this difference in phenotype, we performed RT-PCR and confirmed efficient deletion of the floxed exons (7–8) in the Med13Δ/Δ embryos (Figure 5E). The total number of cells in Med13Δ/Δ blastocysts was reduced compared to controls, with the difference in cell numbers explained by a reduction in the number of trophectoderm cells as indicated by CDX2 labeling (Figure 5F and G). Although the expression levels of both CDX2 and OCT4 were no different in the two groups of blastocysts, expression of NANOG was reduced by ∼40% in Med13Δ/Δ embryos as compared to Med13f/f as indicated by a consistent reduction in immunofluorescence intensity (Figure 5H and I). To determine how far Med13Δ/Δ embryos developed, the same mating was done and maternal weights were followed from E0.5 to E15.5. All pregnant females carrying Med13Δ/Δ embryos lost weight between E8.5 and E12.5, indicating that their pregnancies were disrupted by this time. Examination of E12.5 uteri for the number of resorption sites indicated that Med13Δ/Δ embryos implanted as frequently as control Med13f/Δ embryos but did not survive to E12.5 (Figure 5J).
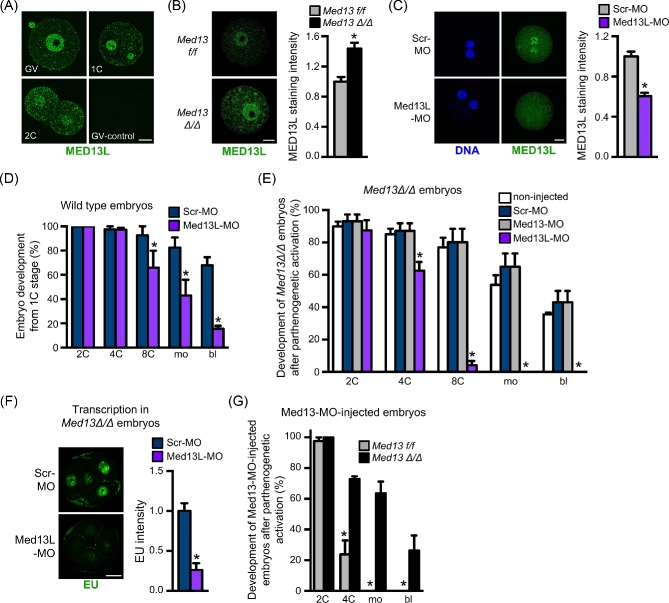
We hypothesized that the successful preimplantation embryo development and implantation of Med13Δ/Δ embryos, in contrast to the early cleavage stage development arrest observed in the Med13-MO-injected embryos, might be explained by compensatory upregulation of the MED13 paralog, MED13L. The ZP3-Cre transgene is expressed and excises floxed alleles during the relatively long phase of oocyte growth within the ovarian follicle [20], so there is a significant period of time when oocytes are transcriptionally active and could in theory compensate for a deleted protein. In wild-type mice, MED13L is expressed in GV oocytes, 1C, and 2C embryos, with predominant localization in the nuclei (Figure 6A). To determine if MED13L was upregulated in oocytes lacking MED13, we compared MED13L protein expression in Med13f/f and Med13Δ/Δ oocytes. MED13L was upregulated ∼1.4-fold in Med13Δ/Δ oocytes (Figure 6B). We next tested whether MED13L, like MED13, was required for successful preimplantation embryo development. Wild-type GV stage oocytes were injected with morpholino oligonucleotides targeting the Med13l translation start site (Med13L-MO) and then matured to MII and parthenogenetically activated. The resulting embryos had reduced levels of MED13L protein at the late 1C stage, documenting efficacy of the Med13L-MO (Figure 6C). We then injected wild-type 1C embryos with Med13L-MO approximately 4 h after fertilization in vivo and monitored preimplantation development. Med13L-MO-injected embryos had reduced efficiency of development to the blastocyst stage as compared to Scr-MO-injected embryos (Figure 6D), though most embryos arrested at the 8C-morula stage rather than at the 2C–4C stage like Med13-MO-injected embryos, and a few developed to the blastocyst stage. These findings suggested that MED13 and MED13L were both required for successful preimplantation embryo development, and that their functions did not overlap completely.
Figure 6.
MED13L compensates for lack of MED13 during preimplantation embryo development. (A) Immunofluorescent staining of MED13L protein in wild-type GV oocytes, 1C- and 2C-stage embryos. GV-control staining done with no primary antibody. (B) Immunofluorescent staining and quantification (arbitrary units) of MED13L in GV oocytes of the indicated genotype. N = 3, total of 30/group; *P < 0.05, T-test. (C) Immunofluorescent staining and quantification (arbitrary units) of MED13L in parthenogenetically activated 1C embryos following microinjection of the indicated morpholino at the GV stage. N = 3, total of 18/group; *P < 0.05, T-test. (D) Percentage of wild-type embryos to reach the various preimplantation embryo stages following microinjection at the 1C stage with the indicated morpholino. N = 3, total of 61–70 1C embryos/group; *P < 0.05 compared to Scr-MO control at same time point, ANOVA with Holm-Sidak multiple comparisons test. (E) Percentage of embryos derived from parthenogenetically activated eggs to reach the various preimplantation embryo stages following microinjection at the 1C stage with the indicated morpholino. N = 4, 15–24 1C embryos/group for each replicate; *P < 0.05 compared to Scr-MO control at same time point, ANOVA with Holm-Sidak multiple comparisons test. (F) Representative images of Med13Δ/Δ embryos microinjected with the indicated morpholino and stained with EU at the 4C stage as an indicator of transcription levels. Graph shows quantification of staining intensity. N = 3–6 embryos/group; *P < 0.05, T-test. (G) Percentage of embryos derived from parthenogenetically activated eggs of the indicated genotypes to reach the various preimplantation embryo stages following microinjection at the 1C stage with Med13-MO. Graph represents average percentage of embryos to develop to the indicated stages. N = 3, total of 56–59 1C embryos/group; *P < 0.05 compared to same stage, ANOVA with Holm-Sidak multiple comparisons test. All graphs show mean ± s.e.m. All bars = 20 μm.
To determine if MED13L was compensating for MED13 functions in Med13Δ/Δ early cleavage stage embryos, Med13Δ/Δ GV oocytes were microinjected with Scr-MO or Med13L-MO, matured in vitro, and then parthenogenetically activated. As a control for morpholino specificity, additional Med13Δ/Δ oocytes were injected with the Med13-MO, which should not affect their development unless off-target effects are present. Med13Δ/Δ oocytes injected with Scr-MO and Med13-MO developed to the blastocyst stage as well as noninjected Med13Δ/Δ oocytes (Figure 6E). However, Med13Δ/Δ oocytes injected with Med13L-MO arrested at the 2–4C stage, much like wild-type 1C embryos injected with Med13-MO (compare Figure 6E to Figure 1F). These embryos also had a marked reduction in transcriptional activity at the 4C stage, similar to wild-type embryos injected with MED13-MO (Figure 6F). To further test the existence of a compensating mechanism and to test for off-target effects of the Med13-MO, Med13f/f, and Med13Δ/Δ, metaphase II stage eggs were parthenogenetically activated and then injected with Med13-MO and allowed to develop in vitro. Consistent with our previous observations using wild-type embryos (Figure 1F), Med13-MO-injected Med13f/f embryos arrested at the 2C to 4C stage (Figure 6G). In contrast, Med13-MO-injected Med13Δ/Δ embryos developed to the blastocyst stage, confirming that the Med13-MO does not have significant off-target effects. Taken together, these findings suggest that compensatory upregulation of MED13L in early cleavage stage embryos allows Med13Δ/Δ embryos to survive long enough to form blastocysts that implant, but that during preimplantation or early postimplantation development, MED13 has critical functions that are not compensated by MED13L.
Discussion
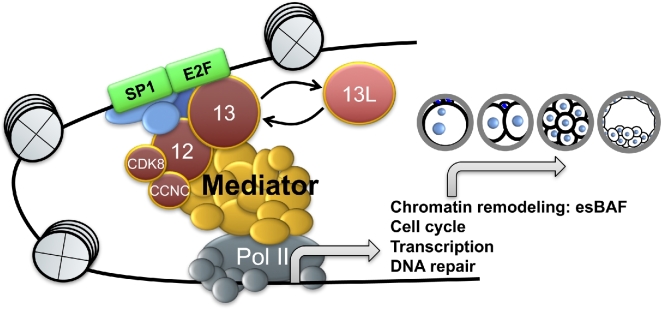
The data reported here demonstrate that MED13 is essential for driving the OET in the mouse. Maternally generated Med13 mRNA is recruited for translation during oocyte maturation. The resulting protein specifically supports transcription of a set of genes required for key aspects of the OET, including chromatin remodeling, RNA processing, protein catabolism, cell cycle control, and DNA repair [48]. This function does not appear to be a result of global effects on transcription. Instead, MED13 interacts directly or indirectly with specific transcription factors including those of the E2F family to direct expression of a new set of genes from the zygotic genome, including esBAF components, which facilitate reprogramming of the zygote into a totipotent, developmentally competent preimplantation embryo (Figure 7).
Figure 7.
Schematic illustrating function of MED13 and MED13L during the OET. MED13 serves as a crucial link between transcription factors (e.g., E2F family and SP1), coactivators, and the main mediator complex. The mediator complex recruits RNA polymerase II and its accessory factors, resulting in transcription of specific gene classes required for preimplantation embryo development. In the absence of MED13, its paralog MED13L partially compensates for this function.
Initial reports indicated that the mediator kinase domain serves as a transcriptional repressor by interfering with core mediator-RNA polymerase II interactions; however, later studies reported that it also functions in transcriptional activation [16]. For example, CDK8 phosphorylates the transactivation domain of STAT1 to both up- and downregulate transcription of genes important for the interferon signaling pathway [61]. CDK8 also promotes transcriptional elongation of proximally paused HIF1A target genes in response to hypoxia [62]. Here, we demonstrate that MED13 is required for both transcriptional activation and repression during ZGA based on our identification of both up- and downregulated transcripts in MED13-knockdown embryos.
Because a global decrease in transcription was not detected in 2C-stage MED13-knockdown embryos, despite the fact that ∼60% of these embryos arrested, it is unlikely that MED13 is required for transcription of most genes at this stage. Similarly, the 2C-stage embryo development arrest could not be explained by a failure of DNA replication. However, an increase in DNA double strand breaks was clearly evident in 4C-stage MED13-knockdown embryos. This process probably began at the 2C stage based on the significant downregulation of genes involved in DNA repair pathways detected by RNAseq, and may have contributed to the 2C-stage arrest. Although it would be satisfying to explain the 2C-stage arrest based on disruption of one or just a few specific gene targets, the degree to which pathways critical for the OET were disrupted in the MED13-knockdown embryos (Supplemental Table S5) prevents us from drawing such a conclusion. Instead, we attribute the cell cycle arrest phenotype to the combined effects of disrupting expression of cell cycle, DNA repair, and chromatin remodeling protein components, many of which have been shown previously to be required for successful preimplantation embryo development. For example, Nfyb, which encodes an essential subunit of the NF-Y transcription factor complex that establishes open chromatin at specific promoter regions [63, 64], was downregulated 1.9-fold in MED13-knockdown embryos. NF-Y was shown recently to regulate chromatin opening in mouse 2C embryos during ZGA [65]. The partial rescue of MED13-knockdown embryo development by overexpressing esBAF components highlights the importance of MED13 function upstream of this critical regulator of preimplantation development and pluripotency.
The much larger number of downregulated genes than those upregulated in MED13-knockdown embryos was mainly explained by changes in transcription of protein-coding genes. In fact, when repetitive elements were evaluated separately, there were more upregulated transcripts than downregulated. By far, the most abundant repetitive elements present in oocytes and 2C embryos were of the LTR class, which represented more than 75% of repeat transcripts [11, 12, 66]. The MT-type LTRs accounted for the vast majority of upregulated repeat regions in the MED13-knockdown embryos. MT transcripts are degraded in oocytes by the combined activities of the RNAse III enzyme, dicer 1, and the catalytic component of the RNA-induced silencing complex, argonaute 2 [54, 55]. Our findings that transcripts encoding both of these proteins were significantly downregulated, coincident with a dramatic increase in MT transcripts, suggest that as in oocytes, they are involved in degrading MT transcripts in preimplantation embryos.
Structural and biochemical analyses indicate that MED12 serves as the central core to which MED13 and CCNC-CDK8 are bound, and that both MED13 and MED12 tether the kinase domain to the core mediator complex via interactions with several subunits (reviewed in [17]). These findings suggest that CCNC-CDK8 interactions with core mediator depend strictly on the presence of an intact 4-subunit kinase module. In contrast, there is evidence in Drosophila that the homologs of MED12 and MED13 may function independently of CCNC and CDK8 in regulating transcriptional responses important for specific developmental pathways [67–69]. Our data do not delineate whether MED13 is functioning independently from or as part of an intact kinase module in the mouse embryo. Based on the presence of mRNA transcripts, all of the kinase module components are expressed in mouse zygotes. There is a requisite role for CDK8 as early as the zygote stage because homozygous null zygotes (generated via a gene-trap strategy) do not survive [70]. The specific function of CDK8 in zygotes is unknown, but this requisite CDK8 functional activity could be supported by interactions with residual, maternally generated MED13 protein still present in the MED13-knockdown embryos.
E2F transcription factors appear critical for activating transcription of genes required for the OET. This is not surprising given that E2Fs have important roles in somatic cells regulating transcription of genes needed for mitotic cell cycle progression, including components of the DNA replication machinery [71]. Cell cycle-related genes were highly enriched in the set of downregulated genes in Med13-MO-injected embryos, suggesting that MED13 serves as an important activator of E2F-mediated gene transcription in early embryos and that it could serve this function in somatic cells as well. In support of this idea, there is evidence for an interaction between the mediator kinase domain subunit CDK8 and E2F1 in human colorectal cancer cells [72]. E2Fs are divided into two categories based on their predominant effects on the cell cycle. In general, “activating” E2Fs, including E2F1, E2F2, and E2F3, activate transcription of genes important for cell proliferation, whereas “repressive” E2Fs, including E2F4 and E2F5, promote cell cycle exit and cellular differentiation [71]. It was previously reported that MED13L interacts with E2Fs to promote growth inhibition in cancer cells; this effect was at least partially mediated by E2F5 [73]. As the activating E2Fs, E2F1 and/or E2F2 are likely responsible for interacting with MED13 to promote expression of the genes downregulated in Med13-MO-injected embryos.
Phenotypic differences between genetic mutants and “morphants,” in which protein levels are knocked down, are often observed and can be explained by compensatory activation of functional protein networks rather than off-target effects [74]. We found that MED13L can compensate for MED13 function during the OET sufficiently to support embryo development to the blastocyst stage. This is not surprising given their high homology (52% amino acid identity, 67% similarity), and the observation that MED13L can substitute for MED13 to form a kinase module complex with MED12 and CCNC-CDK8 [19]. There is some evidence for functional compensation by other mediator kinase domain paralogs. For example, both MED12 and MED12L bind the transcription factor SOX10 in similar ways, suggesting that both proteins could mediate SOX10-based transcriptional responses during myelination [75]. Similarly, CDK8 and CDK19 both directly interact with the histone arginine methyltransferase, PRMT5, resulting in repression of gene transcription in mammalian cells [76]. However, the embryonic lethal knockout mouse phenotypes observed for Med12 [77], Cdk8 [70], and Med13 (reported here) indicate that there is not complete functional redundancy for any of the kinase module paralogs. Our future studies will focus on determining additional critical factors that, like MED13, direct transcription of genes essential for reprogramming the zygote into a totipotent embryo.
Supplementary data
Supplementary data are available at BIOLRE online.
Supplemental Table S1. Antibody table.
Supplemental Table S2. Primers used for real-time RT-PCR.
Supplemental Table S3. Significantly different genes in late 2C-stage embryos following microinjection with either Scr-MO or Med13-MO (FDR < 5).
Supplemental Table S4. GO categories of genes upregulated in MED13-knockdown embryos.
Supplemental Table S5. GO categories of genes downregulated in MED13-knockdown embryos.
Supplemental Table S6. Expression of E2F family transcription factors in oocytes by qPCR.
Acknowledgments
We thank Tom Kunkel and David Fargo for advice throughout this project; Paula Stein, Richard Schultz, Guang Hu, and Jurrien Dean for critical review of the manuscript; Eric Olson (UT Southwestern) for providing the Med13-floxed mice; and Mitch Eddy (NIEHS) for providing the Hspa2-Cre mice.
Author contributions: Conception and experimental design: Y-LM, AG, MLB, WH, LL, CJW. Acquisition of data: Y-LM, AG, YZ, EPB, WNJ, MLB. Analysis and interpretation of data: Y-LM, AG, WH, LL, CJW. Drafting the manuscript: YLM, CJW. Editing and final approval of the submitted manuscript: all authors.
Conflict of Interest: The authors have declared that no conflict of interest exists.
Footnotes
Grant Support: This work was supported by National Natural Science Foundation of China (Grant No. 31471350 to Y-LM) and the Intramural Research Program of the National Institutes of Health, National Institutes of Environmental Health Sciences (1ZIAES102985 to CJW, 1ZIAES101765 to LL).
Accession Number: RNA sequencing data were deposited in the NCBI GEO database, accession number GSE90710.
References
- 1. Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev 2011; 25(7):755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Potireddy S, Vassena R, Patel BG, Latham KE. Analysis of polysomal mRNA populations of mouse oocytes and zygotes: dynamic changes in maternal mRNA utilization and function. Dev Biol 2006; 298(1):155–166. [DOI] [PubMed] [Google Scholar]
- 3. Ma J, Flemr M, Strnad H, Svoboda P, Schultz RM. Maternally recruited DCP1A and DCP2 contribute to messenger RNA degradation during oocyte maturation and genome activation in mouse. Biol Reprod 2013; 88(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shao GB, Chen JC, Zhang LP, Huang P, Lu HY, Jin J, Gong AH, Sang JR. Dynamic patterns of histone H3 lysine 4 methyltransferases and demethylases during mouse preimplantation development. In Vitro Cell Dev Biol Anim 2014; 50(7):603–613. [DOI] [PubMed] [Google Scholar]
- 5. Lemaitre JM, Bocquet S, Terret ME, Namdar M, Ait-Ahmed O, Kearsey S, Verlhac MH, Mechali M. The regulation of competence to replicate in meiosis by Cdc6 is conserved during evolution. Mol Reprod Dev 2004; 69(1):94–100. [DOI] [PubMed] [Google Scholar]
- 6. Murai S, Stein P, Buffone MG, Yamashita S, Schultz RM. Recruitment of Orc6l, a dormant maternal mRNA in mouse oocytes, is essential for DNA replication in 1-cell embryos. Dev Biol 2010; 341(1):205–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lin CJ, Koh FM, Wong P, Conti M, Ramalho-Santos M. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell 2014; 30(3):268–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wiekowski M, Miranda M, Nothias JY, DePamphilis ML. Changes in histone synthesis and modification at the beginning of mouse development correlate with the establishment of chromatin mediated repression of transcription. J Cell Sci 1997; 110(Pt 10):1147–1158. [DOI] [PubMed] [Google Scholar]
- 9. Jimenez R, Melo EO, Davydenko O, Ma J, Mainigi M, Franke V, Schultz RM. Maternal SIN3A regulates reprogramming of gene expression during mouse preimplantation development. Biol Reprod 2015; 93(4):89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ostrup O, Andersen IS, Collas P. Chromatin-linked determinants of zygotic genome activation. Cell Mol Life Sci 2013; 70(8):1425–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Evsikov AV, de Vries WN, Peaston AE, Radford EE, Fancher KS, Chen FH, Blake JA, Bult CJ, Latham KE, Solter D, Knowles BB. Systems biology of the 2-cell mouse embryo. Cytogenet Genome Res 2004; 105(2–4):240–250. [DOI] [PubMed] [Google Scholar]
- 12. Peaston AE, Evsikov AV, Graber JH, de Vries WN, Holbrook AE, Solter D, Knowles BB. Retrotransposons regulate host genes in mouse oocytes and preimplantation embryos. Dev Cell 2004; 7(4):597–606. [DOI] [PubMed] [Google Scholar]
- 13. Hamatani T, Ko M, Yamada M, Kuji N, Mizusawa Y, Shoji M, Hada T, Asada H, Maruyama T, Yoshimura Y. Global gene expression profiling of preimplantation embryos. Human Cell 2006; 19(3):98–117. [DOI] [PubMed] [Google Scholar]
- 14. Torres-Padilla ME, Zernicka-Goetz M. Role of TIF1alpha as a modulator of embryonic transcription in the mouse zygote. J Cell Biol 2006; 174(3):329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Jukam D, Shariati SAM, Skotheim JM. Zygotic genome activation in vertebrates. Dev Cell 2017; 42(4):316–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Conaway RC, Conaway JW. The Mediator complex and transcription elongation. Biochim Biophys Acta 2013; 1829(1):69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Clark AD, Oldenbroek M, Boyer TG. Mediator kinase module and human tumorigenesis. Crit Rev Biochem Mol Biol 2015; 50(5):393–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Petrenko N, Jin Y, Wong KH, Struhl K. Mediator undergoes a compositional change during transcriptional activation. Mol Cell 2016; 64(3):443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Daniels D, Ford M, Schwinn M, Benink H, Galbraith M, Amunugama R, Jones R, Allen D, Okazaki N, Yamakawa H. Mutual exclusivity of MED12/MED12L, MED13/13L, and CDK8/19 paralogs revealed within the CDK-mediator kinase module. J Proteomics Bioinform 2013; 2:2. [Google Scholar]
- 20. de Vries WN, Binns LT, Fancher KS, Dean J, Moore R, Kemler R, Knowles BB. Expression of Cre recombinase in mouse oocytes: a means to study maternal effect genes. Genesis 2000; 26(2):110–112. [PubMed] [Google Scholar]
- 21. Grueter CE, van Rooij E, Johnson BA, DeLeon SM, Sutherland LB, Qi X, Gautron L, Elmquist JK, Bassel-Duby R, Olson EN. A cardiac microRNA governs systemic energy homeostasis by regulation of MED13. Cell 2012; 149(3):671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Inselman AL, Nakamura N, Brown PR, Willis WD, Goulding EH, Eddy EM. Heat shock protein 2 promoter drives Cre expression in spermatocytes of transgenic mice. Genesis 2010; 48:114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Scholer HR. Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 2010; 141(6):943–955. [DOI] [PubMed] [Google Scholar]
- 24. Winuthayanon W, Bernhardt ML, Padilla-Banks E, Myers PH, Edin ML, Lih FB, Hewitt SC, Korach KS, Williams CJ. Oviductal estrogen receptor alpha signaling prevents protease-mediated embryo death. Elife 2015; 4:e10453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tutuncu L, Stein P, Ord TS, Jorgez CJ, Williams CJ. Calreticulin on the mouse egg surface mediates transmembrane signaling linked to cell cycle resumption. Dev Biol 2004; 270(1):246–260. [DOI] [PubMed] [Google Scholar]
- 26. Igarashi H, Knott JG, Schultz RM, Williams CJ. Alterations of PLCbeta1 in mouse eggs change calcium oscillatory behavior following fertilization. Dev Biol 2007; 312(1):321–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jefferson WN, Chevalier DM, Phelps JY, Cantor AM, Padilla-Banks E, Newbold RR, Archer TK, Kinyamu HK, Williams CJ. Persistently altered epigenetic marks in the mouse uterus after neonatal estrogen exposure. Mol Endocrinol 2013; 27(10):1666–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 2001; 29(9):e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang F, Kooistra M, Lee M, Liu L, Baltz JM. Mouse embryos stressed by physiological levels of osmolarity become arrested in the late 2-cell stage before entry into M phase. Biol Reprod 2011; 85(4):702–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gambini A, Williams CJ. LUTs of blastocyst nuclei for quantification. Mol Reprod Dev 2016; 83(7):575–575. [DOI] [PubMed] [Google Scholar]
- 31. Bernhardt ML, Kim AM, O’Halloran TV, Woodruff TK. Zinc requirement during meiosis I-meiosis II transition in mouse oocytes is independent of the MOS-MAPK pathway. Biol Reprod 2011; 84(3):526–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Meth 2012; 9(7):671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Aoki F, Worrad DM, Schultz RM. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol 1997; 181(2):296–307. [DOI] [PubMed] [Google Scholar]
- 34. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009; 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang W, Umbach DM, Vincent Jordan N, Abell AN, Johnson GL, Li L. Efficiently identifying genome-wide changes with next-generation sequencing data. Nucleic Acids Res 2011; 39(19):e130–e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14(4):R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li L. GADEM: a genetic algorithm guided formation of spaced dyads coupled with an EM algorithm for motif discovery. J Comput Biol 2009; 16(2):317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Knuppel R, Dietze P, Lehnberg W, Frech K, Wingender E. TRANSFAC retrieval program: a network model database of eukaryotic transcription regulating sequences and proteins. J Comput Biol 1994; 1(3):191–198. [DOI] [PubMed] [Google Scholar]
- 39. Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res 2004; 32(90001):D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res 2009; 37(Database):D77–D82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. Reproduction 1989; 86(2):679–688. [DOI] [PubMed] [Google Scholar]
- 42. Braude P, Pelham H, Flach G, Lobatto R. Post-transcriptional control in the early mouse embryo. Nature 1979; 282(5734):102–105. [DOI] [PubMed] [Google Scholar]
- 43. Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc Natl Acad Sci 2008; 105(41):15779–15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci 2008; 105(7):2415–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet 2010; 11(11):761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, Rossier C, Ucla C et al. . GENCODE: producing a reference annotation for ENCODE. Genome Biol 2006; 7(Suppl 1):S4.1–S4.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet 2007; 3:e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Svoboda P, Franke V, Schultz RM. Sculpting the transcriptome during the oocyte-to-embryo transition in mouse. Curr Top Dev Biol 2015; 113:305–349. [DOI] [PubMed] [Google Scholar]
- 49. Bultman SJ, Gebuhr TC, Pan H, Svoboda P, Schultz RM, Magnuson T. Maternal BRG1 regulates zygotic genome activation in the mouse. Genes Dev 2006; 20:1744–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol 2008; 319:110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ho L, Jothi R, Ronan JL, Cui K, Zhao K, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci 2009; 106:5187–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gao X, Tate P, Hu P, Tjian R, Skarnes WC, Wang Z. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci 2008; 105:6656–6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ho L, Ronan JL, Wu J, Staahl BT, Chen L, Kuo A, Lessard J, Nesvizhskii AI, Ranish J, Crabtree GR. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci 2009; 106:5181–5186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Stein P, Rozhkov NV, Li F, Cardenas FL, Davydenko O, Vandivier LE, Gregory BD, Hannon GJ, Schultz RM. Essential role for endogenous siRNAs during meiosis in mouse oocytes. PLoS Genet 2015; 11:e1005013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ. Critical roles for Dicer in the female germline. Genes Dev 2007; 21:682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Forlani S, Bonnerot C, Capgras S, Nicolas JF. Relief of a repressed gene expression state in the mouse 1-cell embryo requires DNA replication. Development 1998; 125:3153–3166. [DOI] [PubMed] [Google Scholar]
- 57. Ma SF, Liu XY, Miao DQ, Han ZB, Zhang X, Miao YL, Yanagimachi R, Tan JH. Parthenogenetic activation of mouse oocytes by strontium chloride: a search for the best conditions. Theriogenology 2005; 64:1142–1157. [DOI] [PubMed] [Google Scholar]
- 58. Petzoldt U, Hoppe PC. Spontaneous parthenogenesis in Mus musculus: comparison of protein synthesis in parthenogenetic and normal preimplantation embryos. Mol Gen Genet 1980; 180:547–552. [DOI] [PubMed] [Google Scholar]
- 59. Henery CC, Kaufman MH. Cleavage rate of haploid and diploid parthenogenetic mouse embryos during the preimplantation period. Mol Reprod Dev 1992; 31:258–263. [DOI] [PubMed] [Google Scholar]
- 60. Liu L, Trimarchi JR, Keefe DL. Haploidy but not parthenogenetic activation leads to increased incidence of apoptosis in mouse embryos. Biol Reprod 2002; 66:204–210. [DOI] [PubMed] [Google Scholar]
- 61. Bancerek J, Poss ZC, Steinparzer I, Sedlyarov V, Pfaffenwimmer T, Mikulic I, Dolken L, Strobl B, Muller M, Taatjes DJ, Kovarik P. CDK8 kinase phosphorylates transcription factor STAT1 to selectively regulate the interferon response. Immunity 2013; 38:250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Galbraith MD, Allen MA, Bensard CL, Wang X, Schwinn MK, Qin B, Long HW, Daniels DL, Hahn WC, Dowell RD, Espinosa JM. HIF1A employs CDK8-mediator to stimulate RNAPII elongation in response to hypoxia. Cell 2013; 153:1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Nardini M, Gnesutta N, Donati G, Gatta R, Forni C, Fossati A, Vonrhein C, Moras D, Romier C, Bolognesi M, Mantovani R. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 2013; 152:132–143. [DOI] [PubMed] [Google Scholar]
- 64. Oldfield AJ, Yang P, Conway AE, Cinghu S, Freudenberg JM, Yellaboina S, Jothi R. Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol Cell 2014; 55:708–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lu F, Liu Y, Inoue A, Suzuki T, Zhao K, Zhang Y. Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 2016; 165:1375–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Evsikov AV, Graber JH, Brockman JM, Hampl A, Holbrook AE, Singh P, Eppig JJ, Solter D, Knowles BB. Cracking the egg: molecular dynamics and evolutionary aspects of the transition from the fully grown oocyte to embryo. Genes Dev 2006; 20:2713–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Janody F, Treisman JE. Requirements for mediator complex subunits distinguish three classes of notch target genes at the Drosophila wing margin. Dev Dyn 2011; 240:2051–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Kuuluvainen E, Hakala H, Havula E, Sahal Estime M, Ramet M, Hietakangas V, Makela TP. Cyclin-dependent kinase 8 module expression profiling reveals requirement of mediator subunits 12 and 13 for transcription of serpent-dependent innate immunity genes in Drosophila. J Biol Chem 2014; 289:16252–16261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gobert V, Osman D, Bras S, Auge B, Boube M, Bourbon HM, Horn T, Boutros M, Haenlin M, Waltzer L. A Genome-wide RNA interference screen identifies a differential role of the mediator CDK8 module subunits for GATA/RUNX-activated transcription in Drosophila. Mol Cell Biol 2010; 30:2837–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Westerling T, Kuuluvainen E, Makela TP. Cdk8 is essential for preimplantation mouse development. Mol Cell Biol 2007; 27:6177–6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 2002; 3:11–20. [DOI] [PubMed] [Google Scholar]
- 72. Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, Kwon EJ, Haigis KM, Naar AM, Dyson NJ. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature 2008; 455:552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Angus SP, Nevins JR. A role for Mediator complex subunit MED13L in Rb/E2F-induced growth arrest. Oncogene 2012; 31:4709–4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rossi A, Kontarakis Z, Gerri C, Nolte H, Holper S, Kruger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 2015; 524:230–233. [DOI] [PubMed] [Google Scholar]
- 75. Vogl MR, Reiprich S, Kuspert M, Kosian T, Schrewe H, Nave KA, Wegner M. Sox10 cooperates with the mediator subunit 12 during terminal differentiation of myelinating glia. J Neurosci 2013; 33:6679–6690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Tsutsui T, Fukasawa R, Shinmyouzu K, Nakagawa R, Tobe K, Tanaka A, Ohkuma Y. Mediator complex recruits epigenetic regulators via its two cyclin-dependent kinase subunits to repress transcription of immune response genes. J Biol Chem 2013; 288:20955–20965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rocha PP, Scholze M, Bleiss W, Schrewe H. Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development 2010; 137:2723–2731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.