Abstract
Aims
The contribution of resident stem or progenitor cells to cardiomyocyte renewal after injury in adult mammalian hearts remains a matter of considerable debate. We evaluated a cell population in the adult mouse heart induced by myocardial infarction (MI) and characterized by an activated Nkx2.5 enhancer element that is specific for multipotent cardiac progenitor cells (CPCs) during embryonic development. We hypothesized that these MI-induced cells (MICs) harbour cardiomyogenic properties similar to their embryonic counterparts.
Methods and results
MICs reside in the heart and mainly localize to the infarction area and border zone. Interestingly, gene expression profiling of purified MICs 1 week after infarction revealed increased expression of stem cell markers and embryonic cardiac transcription factors (TFs) in these cells as compared to the non-mycoyte cell fraction of adult hearts. A subsequent global transcriptome comparison with embryonic CPCs and fibroblasts and in vitro culture of MICs unveiled that (myo-)fibroblastic features predominated and that cardiac TFs were only expressed at background levels.
Conclusions
Adult injury-induced reactivation of a cardiac-specific Nkx2.5 enhancer element known to specifically mark myocardial progenitor cells during embryonic development does not reflect hypothesized embryonic cardiomyogenic properties. Our data suggest a decreasing plasticity of cardiac progenitor (-like) cell populations with increasing age. A re-expression of embryonic, stem or progenitor cell features in the adult heart must be interpreted very carefully with respect to the definition of cardiac resident progenitor cells. Albeit, the abundance of scar formation after cardiac injury suggests a potential to target predestinated activated profibrotic cells to push them towards cardiomyogenic differentiation to improve regeneration.
Keywords: Myocardial infarction, Nkx2.5 cardiac enhancer, Cardiac resident progenitor cells, Myofibroblasts, Postnatal cardiac repair
1. Introduction
Recently, limited regenerative capacity of the postnatal mammalian heart under physiological and pathophysiological circumstances was postulated.1 However, this capacity is meager and the underlying mechanism remains poorly understood. Proliferation of pre-existing cardiomyocytes after dedifferentiation and cytokinesis appeared to be the major source for newly formed myocardium.2 However, the extent to which resident or circulating stem/progenitor cells contribute to cardiomyocyte renewal post injury in the adult heart remains a matter of considerable debate.3–5 Based on surface antigens, specific gene expression (e.g. the embryonic developmental marker Isl1) or other abilities, several putative cardiomyogenic cardiac stem or progenitor cell populations have been described as primitive cells residing in niches of embryonic, fetal and adult mammalian hearts.6
The transcription factor (TF) Nkx2.5 is a key factor guiding embryonic heart development.7,8 In the developing heart of the Nkx2.5 cardiac enhancer-eGFP (NkxCE-GFP) transgenic mouse specific multipotent cardiac progenitor cells (CPCs) are clearly marked by GFP expression under the regulatory control of a 2.1 kb Nkx2.5 enhancer.8 Remarkably, Nkx2.5 gene expression in these CPCs is low compared to cardiomyocytes.9,10 While the expression domain of Nkx2.5 marks several tissues, including the thyroid, pharynx, stomach and spleen,11 the 2.1 kb Nkx2.5 enhancer driven GFP expression is specific to the embryonic heart.8 NkxCE-GFP-positive CPCs are able to differentiate into cardiomyocytes, smooth muscle cells, endothelial cells and fibroblasts in vitro.8,9 Recently, it was shown that GFP-expressing cells with an activated Nkx2.5 enhancer were re-activated in the neonatal uninjured heart that have the potential to differentiate into cardiomyocytes and smooth muscle cells in vitro and in vivo.10 However, by the age of 3 weeks, no such GFP-expressing cells are detectable in uninjured NkxCE-GFP mouse hearts.10 Using the same transgenic NkxCE-GFP mouse, we sought to investigate a cell population re-emerging after myocardial infarction (MI) in the adult mouse heart (> 8 weeks) also marked by the activated cardiac-specific Nkx2.5 enhancer (NkxCE) element. We hypothesized that MI-induced cells might reflect cardiomyogenic properties of their embryonic or neonatal counterparts and by this constitute a potential regenerative cardiac resident stem/progenitor cell population. We further characterized their cellular origin, localization in the heart and identity. Interestingly, phenotypic and molecular characterization, including a direct global transcriptomic comparison with embryonic cardiac progenitors and fibroblasts, revealed that the cell population identified after MI resembles (myo-)fibroblasts rather than a cardiac stem/progenitor cell population with cardiomyogenic properties. Our study represents a detailed comparison of a putative postnatal cardiac resident stem/progenitor cell population with their corresponding embryonic counterparts and fibroblasts and suggests that a re-expression of embryonic, stem or progenitor cell features in the adult injured heart must be interpreted very carefully with respect to the definition of cardiac resident progenitor cells. However, the abundance of scar formation after cardiac injury/disease suggests a potential to target fibrotic cells, especially predestinated subpopulations, to push them towards cardiomyogenic differentiation to improve regeneration.
2. Methods
2.1 Transgenic mouse strains
Mice were housed in an accredited facility in compliance with the European Community Directive related to laboratory animal protection (2010/63/EU). All procedures implemented in mice were approved by the German Heart Center and the local legislation on protection of animals (Commission on Animal Protection, Regierung von Oberbayern, Munich, Germany, 55.2-1-55-2532-141-14, 55.2-1-54-2532.3-17-13) and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
All transgenic mouse lines have been previously described in detail. The NkxCE-GFP mouse was described by Wu et al.8 The pNTK-mouse was also developed in the laboratory of Sean Wu.12 Mice from the C57Bl/6 J strain (The Jackson Laboratories, Bar Harbor, ME, USA, Stock No 000664) were used as wild-type controls.
All animal organ or embryo extractions were performed in accordance with the European regulations for animal care and handling (2010/63/EU). For extraction of embryos or organs mice were pre-anaesthetized in a closed chamber with 5% isofluorane [2-chloro-2-(difluoromethoxy)-1, 1, 1-trifluoro-ethane] in oxygen and then euthanized by cervical dislocation.
2.2 Induction of MI by left anterior descending artery ligation
Adult mice (NkxCE-GFP, pNTK-ROSAYFP) older than 8 weeks were anesthetized using midazolam (5.0 mg/kg), medetomidine (0.5 mg/kg) and fentanyl (0.05 mg/kg) via intraperitoneal injection (i.p.), then intubated and ventilated with oxygen enriched air using a small animal respirator (Mouse Ventilator MiniVent Type 845; Harvard apparatus, Holliston, MA, USA). Buprenorphine (0.05 mg/kg) was administered subcutaneously prior to surgery for analgesia. The left anterior descending artery (LAD) was permanently ligated using an 8/0 monofilament suture (Prolene, Johnson & Johnson Medical GmbH, Ethicon, Norderstedt, Germany). Narcotics were antagonized by subcutaneous injection of flumazenil (0.5 mg/kg) and atipamezol (2.5 mg/kg). Sham-operated control mice underwent the same surgical procedure except that the suture around the coronary artery was not tied. Preemptive analgesia was provided by buprenorphine (0.05 mg/kg BW s.c.) three times per day for at least 72 h. After different periods mice were pre-anaesthetized in a closed chamber with 5% isofluorane [2-chloro-2-(difluoromethoxy)-1, 1, 1-trifluoro-ethane] in oxygen and euthanized by cervical dislocation. Hearts were extracted and subsequent analyses were performed (described under the respective sections).
For the thymosin β4 (TB4) in vivo stimulation experiments NkxCE-GFP mice were primed with TB4 (12 mg/kg BW i.p.) daily for 7 days before inducing MI. As a control group NkxCE-GFP mice were injected with phosphate buffered saline (PBS) (i.p.) daily for 7 days before inducing MI. Then, in both groups, MI was induced by LAD ligation as described earlier. Surgery was performed in a blinded fashion. The surgeon did not know which mouse belonged to which group. After MI mice achieved additional doses of TB4 and PBS (for 4 days). Mice were imaged by in vivo magnetic resonance imaging (MRI) 1 week after induction of MI to evaluate left ventricular function (LVEF) and infarct size (%LV) (see Section 2.11). Subsequently, mice were pre-anaesthetized in a closed chamber with 5% isofluorane [2-chloro-2-(difluoromethoxy)-1, 1, 1-trifluoro-ethane] in oxygen and then euthanized by cervical dislocation. Hearts were harvested, digested and prepared for flow cytometry analysis (see Section 2.5).
2.3 Heterotopic cervical heart transplantation
Hetereotpoic heart transplantation was performed as described previously.13 Briefly the animals were anesthetized by i.p. injection of midazolam (5.0 mg/kg), medetomidine (0.5 mg/kg) and fentanyl (0.05 mg/kg). Buprenorphine (0.05 mg/kg) was administered subcutaneously prior to surgery. After removing the donor heart the graft was stored on ice in cardioplegic solution (6 h) until implantation. After preparing the recipient animal, the graft was cervically implanted and anastomoses were secured with circumferential 8–0 silk ligatures. Normally, the heart developed sinus rhythm within 1 min. Narcotics were antagonized by subcutaneous injection of flumazenil (0.5 mg/kg) and atipamezol (2.5 mg/kg). Preemptive analgesia was provided by 0.05 mg/kg buprenorphine three times per day for at least 72 h. After 1 week of reperfusion mice were pre-anaesthetized in a closed chamber with 5% isofluorane [2-chloro-2-(difluoromethoxy)-1, 1, 1-trifluoro-ethane] in oxygen and then euthanized by cervical dislocation. Donor hearts and recipient original hearts were extracted and subsequent flow cytometry analysis was performed.
2.4 Induction of pressure overload-induced cardiac hypertrophy by transverse aortic constriction
Transverse aortic constriction (TAC) was performed to create pressure overload-induced cardiac hypertrophy and heart failure. When compared to a complete occlusion of the left anterior descending (LAD) coronary artery, TAC provides a more gradual time course in the development of heart failure.14 Adult mice (> 8 weeks) were anaesthetized by i.p. injection of midazolam (5.0 mg/kg), medetomidine (0.5 mg/kg) and fentanyl (0.05 mg/kg), intubated and ventilated with a rodent ventilator (Harvard Apparatus, 0.2 mL tidal volume, 150 breaths per minute). Buprenorphine (0.05 mg/kg) was administered subcutaneously prior to surgery. Aortic banding was performed by ligating the aorta between the innominate artery and left common carotid artery with a 27-gauge needle using a 5–0 silk suture.15 Narcotics were antagonized by subcutaneous injection of flumazenil (0.5 mg/kg) and atipamezol (2.5 mg/kg). Preemptive analgesia was provided by buprenorphine (0.05 mg/kg BW s.c.) three times per day for at least 72 h. For heart explantation mice were pre-anaesthetized in a closed chamber with 5% isofluorane [2-chloro-2-(difluoromethoxy)-1, 1, 1-trifluoro-ethane] in oxygen and then euthanized by cervical dislocation. Hearts were extracted after 1 week or 6 weeks and flow cytometry analysis was performed.
2.5 Flow cytometry
Mouse hearts (MI, TAC, HTX, sham, ctr without operation) were extracted as outlined above. As described previously,16 hearts were enzymatically digested to obtain single cell suspension. By filtration steps cardiac cell suspensions get depleted from cardiomyocytes (size discrimination). Dead cells were stained with propidium iodide solution (2 µg/ml, Sigma-Aldrich, St. Louis, MO, USA) or 4, 6-diamidino-2-phenylindole (DAPI; 1 µg/ml, BioLegend, SanDiego, CA).
Whole blood from infarcted NkxCE-GFP mice was obtained before harvesting the hearts 1 week post-MI. Bone marrow cells from infarcted NkxCE-GFP mice were obtained by flushing tibiae and femora of donor mice with PBS 1 week post-MI.
Antibody staining of single cell suspensions from infarcted NkxCE-GFP hearts was performed 1 week post-MI. The cardiomyocyte-depleted cell fractions were incubated with first antibodies (CD45, CD31, CD90, Sca1; all primary antibodies are specified in Supplementary material online, Table S2) overnight at 4°C. Alexa 555-conjugated secondary antibodies (1:2000) matching to the species and subclasses of the primary antibodies were subsequently used to label the target cells (e.g. anti-rabbit Alexa Fluor 555 and anti-rat Alexa Fluor 555; both Abcam, Cambridge, UK).
Flow cytometry data (cell counting) were acquired using a BD LSRFortessaTM cell analyzer equipped with three lasers (blue, red and violet) (BD Biosciences, San Jose, CA) and the BD FACSDiva software version 8.0.1 (BD Biosciences). Results were analyzed by the FlowJo 7.6.5 software (Tree Star, Ashland, OR, USA).
Cell sorting for RNA extraction was performed using a FACS ARIATM Illu flow cytometer (BD Biosciences, San Jose, CA) and the BD FACSDiva software version 6.1.2 (BD Biosciences).
Cell sorting for in vitro cultivation of MICs was performed using a MoFloTM Legacy flow cytometer (Beckman Coulter, Brea, CA) and the Summit software version 4.3 (Beckman Coulter).
2.6 Histological analysis of cardiac sections (immunofluorescent, Movat’s penatchrome stain)
One week after MI or heterotopic heart transplantation hearts were perfused with PBS and then harvested [mice were pre-anaesthetized in a closed chamber with 5% isofluorane (2-chloro-2-(difluoromethoxy)-1, 1, 1-trifluoro-ethane] in oxygen and euthanized by cervical dislocation). Hearts were fixed with 4% paraformaldehyde for 12 h at 4°C. Then, hearts were embedded in optimum cutting temperature compound (OCT) for frozen sections and stored at −80°C for further use (immunohistochemical staining or Movat’s stain). OCT-embedded hearts were sectioned with a cryotome (Cryostat, LEICA CM1850; 8 µm per section).
2.7 In situ 2-photon-microscopy
Infarcted hearts of NkxCE-GFP mice were harvested 1 week after MI {mice were pre-anaesthetized in a closed chamber with 5% isofluorane [2-chloro-2-(difluoromethoxy)-1, 1, 1-trifluoro-ethane] in oxygen and euthanized by cervical dislocation}. Hearts were cross-sectioned with a scalpel to expose the infarct area. No fixation or staining procedure was performed. Hearts were analyzed in situ. Images were acquired with a TrimScope (LaVision Biotech, Bielefeld, Germany) connected to an upright Olympus microscope, equipped with a MaiTai Laser (Spectra-Physics, Munich, Germany) and a x20 water immersion objective (numerical aperture 0.95, Olympus, Tokyo, Japan). Scans were performed using a wavelength of 800 nm in a frame of 558 μm × 558 μm with 1030 × 1030 pixels, and a z-step of 3 μm in a range of approximately 30 μm.
2.8 Micro arrays (mRNA)
For MicroArray analysis, total RNA samples from NkxCE-GFP+ MICs (1 week post MI), murine ES-derived CPCs (in vitro differentiated for 7 days), murine cardiac fibroblasts (CFs) and tail-tip fibroblasts (TTFs) (cultivated for several passages < 4) were used (each n = 3). First, the integrity of the total RNA was assessed through analysis on an Agilent Bioanalyzer 2100 (Agilent Technologies, Böblingen, Germany). Samples with a RIN less than seven were excluded from further analyses. A two-round amplification and labelling was done with the TargetAmp-Pico Labelling Kit (Epicentre, Illumina, CA, USA) starting with 500 pg total RNA and finally Biotin-aRNA was analyzed with the Mouse WG-6 v2.0 Expression BeadChip (Illumina, CA, USA). BeadChips were scanned using the Illumina Bead Array Reader. Subsequently, data were analyzed bioinformatically.
2.9 In vitro cultivation of MICs including stimulation by various substances
Infarcted NkxCE-GFP mice were euthanized after 1 week {pre-anaesthetized in a closed chamber with 5% isofluorane [2-chloro-2-(difluoromethoxy)-1, 1, 1-trifluoro-ethane] in oxygen first and then euthanized by cervical dislocation}, hearts were extracted, single-cell suspension was prepared, and 20 GFP+ MICs were sorted by fluorescence-activated cell sorting (FACS) directly into collagen-coated 96 well plates. Cells were analyzed as follows: cells were fixed with 4% paraformaldehyde and nuclei were counterstained with DAPI to count the cells. Doubling times were calculated. MICs were further subjected to stimulation by several substances: ActivinA (recombinant hu/mu/rat ActivinA, 100 ng/ml and 500 ng/ml; R&D Systems, MN, USA, according to Chen et al.12), bone morphogenic protein 4 (BMP4) (recombinant mu BMP4, 100 ng/ml and 500 ng/ml; R&D Systems, according to Chen et al.12), TB4 (100 ng/ml and 500 ng/ml; RegenRX, Maryland, USA; ), A83-01, a small molecule ALK4, 5, 7 inhibitor (1 µM and 10 µM; Tocris-Bioscience, Bristol, UK; according to Chen et al.12). In addition, MICs were stained according to standard immunocytochemical protocols (primary antibodies are specified in Supplementary material online, Table S2; fluorochrome-conjugated secondary antibodies were used for detection).
2.10 Epigenetic analysis of the Nkx2.5 genomic locus
Previously published Hi-C,17,18 DNA methylation,18–20 RNA-seq18,20 and ChIP-seq20,21 data were reanalyzed. Principal component analysis (PCA) was performed on Hi-C data using the HiC-Explorer.22 RNA-seq and ChIP-seq data were mapped and genome wide read coverage was calculated. DNA methylation data were mapped using Bismark.23 Coverage tracks were calculated using deepTools24 and the HiC-Explorer22 was used for visualization.
2.11 MRI
Mice were imaged by in vivo MRI using a 7 T GE/Agilent (GE Healthcare, Munich, Germany/Agilent, Böblingen, Germany) MR901 at 1 week after induction of LAD ligation to evaluate LVEF and infarct size (%LV). Mice were pre-anaesthetized in a closed chamber with 3% isofluorane in oxygen. Anaesthesia was then maintained at 1.5% isofluorane in oxygen.
2.12 Data analysis and statistics
Data are presented as mean values ± standard error of the mean (S.E.M.). Statistical differences were evaluated using the unpaired Student’s t-test or the Mann-Whitney-U test. Comparison of several groups was performed by one way ANOVA or Kruskal–Wallis test on ranks including appropriate post-hoc tests. A value of P < 0.05 was considered to be statistically significant. In all figures, statistical significance is indicated as follows: *means P < 0.05 and ** means P < 0.01.
2.13 Data availability
The datasets generated and/or analysed during the current study will be available in the GEO repository from NCBI (https://www.ncbi.nlm.nih.gov/gds).
See Supplementary material online for an extended methods section.
3. Results
3.1 Identification of an NkxCE positive cell population after MI
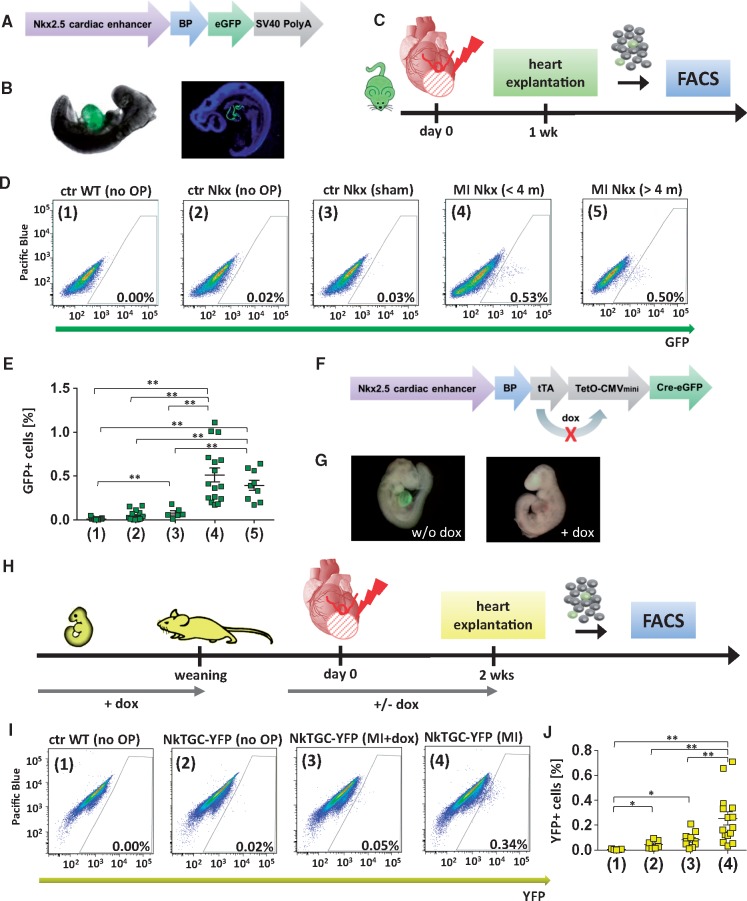
NkxCE-GFP transgenic mice (Figure 1A) specifically express eGFP in the developing heart (Figure 1B). GFP fluorescence in the heart of NkxCE-GFP embryos peaks around E9.5, fades after E11.5 and vanishes in the uninjured adult heart until postnatal week three.7,8,10 After inducing MI by permanent ligation of the LAD (Figure 1C) in adult mice (>8 weeks), approximately 0.51 ± 0.08% GFP+ cells reemerged in the injured heart after one week (vs. the nucleated, cardiomyocyte-depleted ventricular cell fraction). In adult uninjured controls (ctr Nkx, no OP) and sham operated animals (ctr Nkx, sham), 0.02% GFP+ cells were found (Figure 1D and E), confirming that no GFP-positive cells with an activated NkxCE exist in hearts of mice older than 3 weeks.10 Interestingly, the reemerging GFP+ cell population was also found in older (>4 months) murine hearts post-injury (Figure 1D and E). Even in mice older than 1 year, reemerging GFP+ cells were detectable after MI (see Supplementary material online, Figure S1A and B); no significant difference was detected by age.
Figure 1.
Identification of NkxCE-positive cells after myocardial infarction (MICs). Vector construct of the Nkx2.5 cardiac enhancer GFP (NkxCE-GFP) mouse (A). (B) Cardiac-specific GFP-fluorescence in an NkxCE-GFP embryo (E9.5). Left picture shows whole-mount in situ GFP-fluorescence, whereas the right picture shows a cryo-section stained for GFP (green) (blue: DAPI). (C) MI was induced by ligation of the LAD in adult NkxCE-GFP mice; heart explantation after 1 week. Single-cell suspension was cardiomyocyte-depleted via several filter steps. Quantification of GFP+ cells by FACS. (D) Amount of GFP+ MICs detected by FACS (exemplary plots). (1) Ctr WT (no OP): Wild type mouse (C57Bl/6) without surgery. (2) Ctr Nkx (no OP): NkxCE-GFP mouse without surgery. (3) Ctr Nkx (sham): NkxCE-GFP mouse with sham operation. (4) MI Nkx (<4 m): NkxCE-GFP mouse with MI (< 4 months). (5) MI Nkx (>4 m): NkxCE-GFP mouse with MI (>4 months). (E) About 0.5% GFP+ MICs were detected in mice with MI but not in the control groups. [groups see D., n(1) = 19, n(2) = 15, n(3) = 6, n(4) = 16, n(5) = 9]. **P < 0.01 (Kruskal–Wallis, postHoc: Dunn’s method). (F) Vector construct of the transgenic NkxCE-tTA-Cre-GFP (NkTGC) mouse. (G) Left panel shows cardiac-specific whole-mount in situ YFP-fluorescence in an NkTGC x Rosa26YFP embryo (E9.5) (without doxycycline substitution in the dam’s drinking water). Right panel shows no YFP-fluorescence in an E9.5-embryo substituted with dox (+ dox) in the dam’s drinking water during embryonic development. (H) MI was induced by LAD-ligation in adult NkTGC x Rosa26YFP (NkTGC-YFP) mice substituted with dox from embryonic development until weaning indicating no YFP-fluorescence in their hearts. As a control, half of the MI-surgery group received dox-enriched drinking water from 1 week before LAD-ligation to monitor the ‘leakiness’ of the system; heart explantation 14 days after MI. Quantification of YFP+ cells by FACS. (I) Amount of YFP+ cells detected by FACS (exemplary plots). Groups: (1) Ctr WT (no OP): Wild type mouse (C57Bl/6) without surgery. (2) Ctr NkTGC-YFP (no OP): NkTGC-YFP mouse without surgery. (3) Ctr NkTGC-YFP (MI+Dox): NkTGC-YFP mouse with MI (achieved dox by drinking water from 1 week before MI until heart explantation). (4) NkTGC-YFP (MI): NkTGC-YFP mouse with MI. (J) About 0.25% of YFP+ cells were detected in mice with MI (group 4) but not in the control groups [groups see I., n(1) = 5, n(2) = 7, n(3) = 10, n(4) = 15]. *P < 0.05; **P < 0.01 (Kruskal–Wallis, postHoc: Dunn’s method). FACS, fluorescence-activated cell sorting.
To exclude that reemerging GFP-expression was an artifact of the NkxCE-GFP transgene, a second transgenic mouse strain carrying the same NkxCE element but combined with a doxycycline (dox) responsive (tet-off) element to create a NkxCE-tetO-eGFP-Cre mouse model, hereafter called NkTGC mouse10,12 (Figure 1F), underwent MI surgery. Since the eGFP-Cre fusion protein weakly fluoresces (see Supplementary material online, Figure S1C), a Rosa26YFP reporter mouse strain was crossed with the NkTGC mouse. The NkTGC-Rosa26YFP transgene labels NkxCE-positive cells and their derivatives selectively and irreversibly. Mouse embryos (NkTGC-Rosa26YFP) without dox supplementation showed cardiac-specific YFP-fluorescence, whereas dox administration to the gravid dam suppressed YFP fluorescence in embryonic hearts (Figure 1G). For these experiments, mice were treated with dox from conception (in the dam’s drinking water, 1 mg/ml) until weaning at 4 weeks of age, preventing cardiac progenitor cell labelling during embryonic development. After weaning, mice incepted regular drinking water. As a control, half of the mice of the MI-surgery group received dox in drinking water (1 mg/ml) from one week before LAD-ligation until heart explantation to monitor the number of YFP+ cells after MI under dox suppression (Figure 1H). Two weeks after MI, approximately 0.25 ± 0.05% YFP+ cells, indicating an active NkxCE, were detected in the MI group (4) without dox (Figure 1I and J).
We were able to reproduce GFP/YFP induction in NkxCE-GFP mice and a second independent mouse model (NkTGC) after MI.
3.2 Localization of re-emerging NkxCE-positive cells after MI
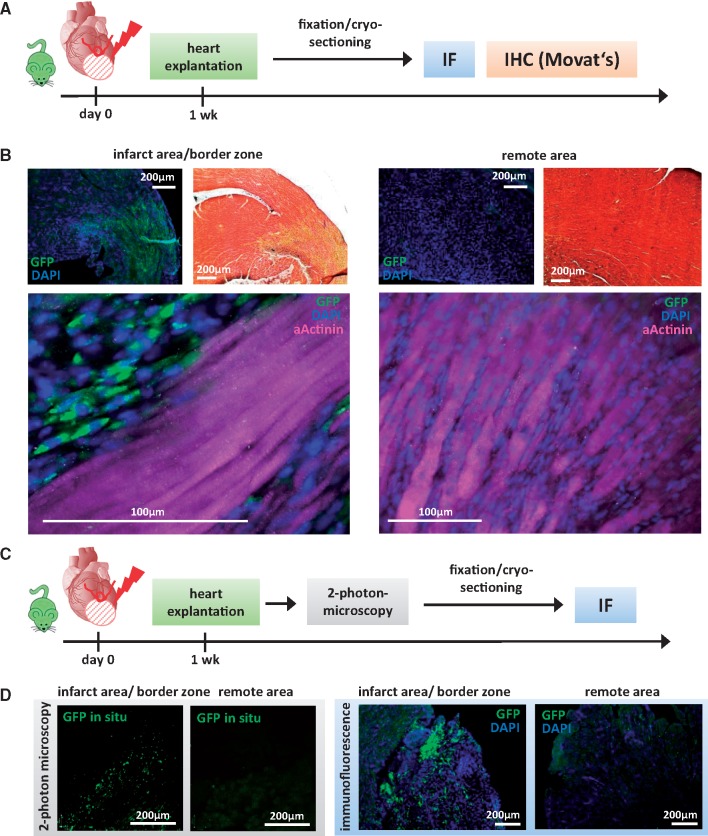
Next, we sought to evaluate the localization of MI-induced GFP-positive cells (MICs). One week after MI, NkxCE-GFP mouse hearts were cryosectioned for various stainings (Figure 2A and B, Supplementary material online, Figure S2A–C). GFP+ MICs were detected in the infarction area and border zone but not in remote myocardium (Figure 2B, Supplementary material online, Figure S2A–C). To achieve clear differentiation between the infarction area and unaffected areas, adjacent sections underwent Movat’s pentachrome staining to stain cardiomyocytes and collagen fibers. GFP+ MICs clearly overlap with collagen fibers in the infarction area and the border zone (Figure 2B, upper panel; Supplementary material online, Figure S2A–C). To exclude autofluorescence related to fixation- or staining processes, 2-photon-microscopy-analysis was employed to detect fluorescence in situ (hearts extracted and analyzed in the absence of fixation or staining; Figure 2C and D). GFP+ cells were again detected in infarction- and border zone areas but not in remote myocardium (Figure 2D, left panel). The same hearts were then fixed, cryo-sectioned and stained for GFP, confirming that MICs are located exclusively in the infarction area and border zone (Figure 2D, right panel).
Figure 2.
Localization of NkxCE-GFP+ MICs after MI. (A) MI was induced by LAD-ligation in NkxCE-GFP mice (n = 7). After 1 week, hearts were explanted, fixed, cryo-sectioned and subjected to immunoflusorescent or histological staining (IHC-Movat’s). (B) Sections were stained for GFP (green) (DAPI = nuclei, blue). Adjacent sections were stained with Movat’s pentachrome (yellow: collagen fibers; red: muscle). Left upper panels: GFP+ MICs were observed in the infarct area and border zone overlapping with Movat’s yellow stained areas. Right upper panels: in remote myocardium no GFP+ MICs were detected. Lower panels: Stainings for GFP (green) and αActinin (cardiomyocytes, magenta) (DAPI = nuclei, blue). (C) One week after MI was induced in NkxCE-GFP mice (n = 4), hearts were explanted and analyzed in situ under a 2-photon-microscope for GFP+ MICs. Then, hearts were fixed, cryo-sectioned and subjected to IF. (D) Left panel shows the results of 2-photon-microscopy. In situ GFP+ cells (green) are visible in the infarct area and border zone. No GFP+ MICs were detected in the remote myocardium. Right panel: IF confirmed the results of 2-photon-microscopy.
3.3 Origin of NkxCE-GFP+ MICs
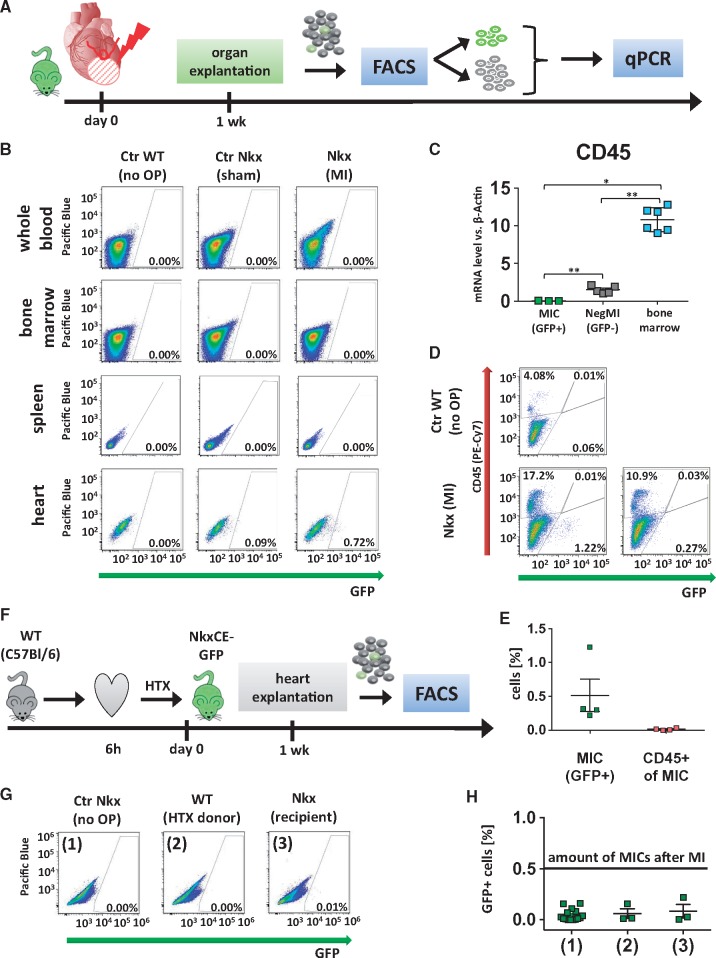
Next, we asked whether re-emerging GFP+ MICs originate from cardiac-resident cells or secondarily migrate into the heart as derivatives of the hematopoietic system in response to injury. While GFP+ MICs were clearly detected in infarcted hearts, flow cytometric analysis detected no GFP-positive cells in corresponding whole blood, bone marrow or spleen samples one week after MI (Figure 3A and B). These data suggest that MICs do not originate from the hematopoietic compartment. To further exclude hematopoietic origin, the pan-hematopoietic cell surface antigen CD45 was analyzed in sorted GFP+ MICs as compared to corresponding GFP-negative cells and murine bone marrow by qPCR (Figure 3C). GFP+ MICs did not show notable CD45 expression as compared to GFP-negative cells or bone marrow. Additionally, we analyzed CD45 expression using flow cytometry in cardiac cells harvested from infarcted NkxCE-GFP hearts (Figure 3D and E) and found, in accordance with our gene expression data, very few CD45-positive MICs.
Figure 3.
Origin of NkxCE-GFP+ MICs. (A) One week after MI was induced in NkxCE-GFP mice (n = 4), hearts and spleens were explanted. Whole blood and bone marrow were also extracted. GFP+ cells were quantified by FACS analysis. GFP+ and GFP-negative cells from heart samples were isolated and subjected to qPCR. (B) No GFP+ cells were detected in whole blood, bone marrow or spleens. However, in the corresponding hearts between 0.5% and 1% of MICs were present (exemplary FACS plots). Groups: Ctr WT (no OP): Wild-type mouse (C57Bl/6) without surgery. Ctr Nkx (sham OP): NkxCE-GFP mouse with sham operation. MI Nkx: NkxCE-GFP mouse with MI. (C) No CD45 gene expression was detected in MICs by qPCR, whereas GFP-negative cells from corresponding hearts (NegMI) showed low levels of CD45 and bone marrow served as positive control (n = 6 per group). *P < 0.05; ** P < 0.01; (Kruskal–Wallis, postHoc: Dunn’s method). (D) Additionally, 1 week after MI, single-cell suspensions of infarcted hearts were stained for CD45 (exemplary plots). For Nkx (MI), one example with a high and one with a low number of GFP+ MICs is depicted. No GFP+ MICs were stained for CD45. (E) Summarized FACS results (see D, n = 4 per group). GFP+ MICs were detected not stained positive for CD45. (F) To verify if MICs originate from the heart itself hearts of C57Bl/6 wild type mice were harvested and transplanted into NkxCE-GFP recipients after 6 h of ischemia. Hearts were explanted after 1 week and GFP+ cells were quantified by FACS (n = 3). (G) No relevant number of GFP+ cells was detected either in transplanted WT or in transgenic recipient hearts (exemplary FACS plots). Groups: (1) Ctr Nkx (no OP): Nkx2.5-GFP mouse without surgery. (2) WT (HTX donor): Cervically transplanted heart of a wild type mouse (C57Bl/6). (3) Nkx (recipient): Heart of the Nkx2.5CE-GFP recipient mouse. (H) Summarized FACS results of cervical heart transplantation. The black line shows the number of GFP+ MICs detected after MI (groups see G). FACS, fluorescence-activated cell sorting.
To underline our assumption above and to exclude the possibility that MICs migrated from extra-cardiac origins, two assays employing heterotopic transplantation of ischemic hearts were performed.13 First, NkxCE-GFP hearts were ectopically transplanted into wild type C57Bl/6 recipients after 6 h of global cold ischemia. A similar number of GFP+ cells (0.41% ± 0.3%) in the transplanted transgenic donor hearts like post MI was detected by flow cytometry (see Supplementary material online, Figure S3A–D), indicating that global ischemia reperfusion (I/R) injury leads to the appearance of NkxCE-GFP+ cells as seen after MI. These GFP+ cells appearing in transgenic donor hearts must stem from the transplanted heart itself, since they were grafted into wild type recipients. To fully exclude MIC extra-cardiac origin, the transplantation assay was performed in reverse (i.e. wild-type donor heart and transgenic recipient). It would be reasonable to expect that if MICs migrate to the heart from extra-cardiac sources, they would be attracted by the ischemic stimulus of the ectopically-transplanted wild type heart. However, when hearts from C57Bl/6 wild type mice were transplanted into NkxCE-GFP recipients, no GFP+ cells were detected in donor organs one week after transplantation (Figure 3F–H). We thus conclude that MICs are resident to injured hearts and do not migrate from extra-cardiac sources.
3.4 Characterization of NkxCE-GFP+ MICs
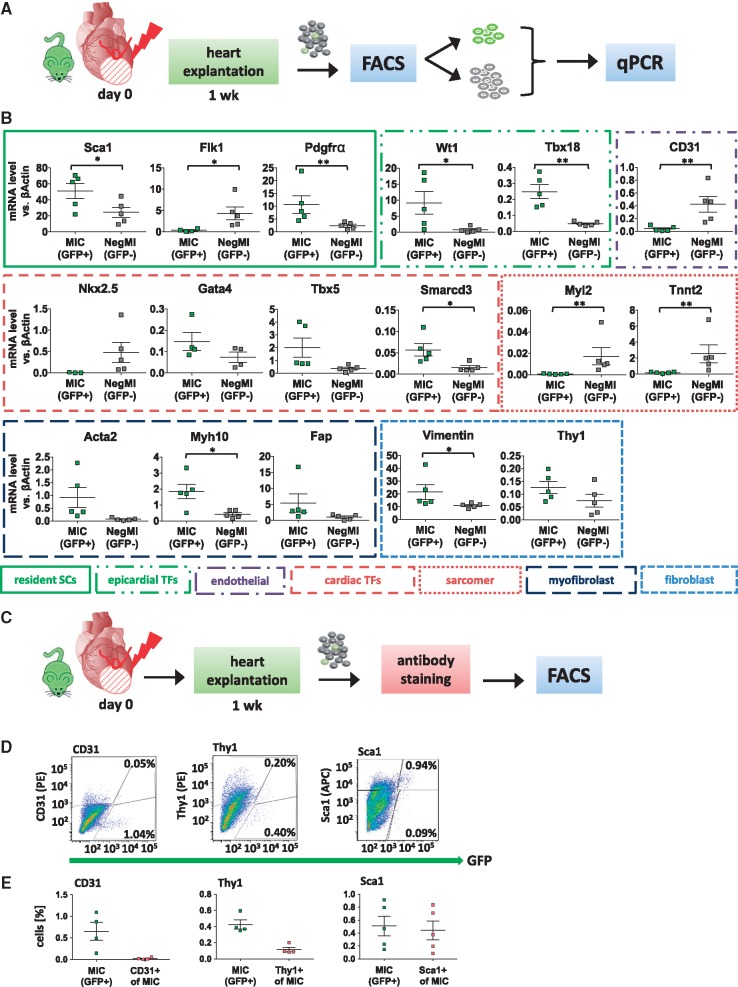
As NkxCE-GFP-positive cells clearly show cardiomyogenic properties during embryonic development8 and to a lesser extent one week after birth,10 we hypothesized that adult MICs might share cardiomyogenic properties with their embryonic or neonatal counterparts. Thus, we characterized MICs by gene expression analysis (qPCR) as compared to respective GFP-negative cells (Figure 4A and B). Gene expression profiling of MICs revealed a mixed phenotype with significantly upregulated genes typical of resident mesenchymal stem cells (Sca1, Pdgfrα), epicardial progenitor cells (EPDCs; Wt1, Tbx18) and cardiac developmental markers (Gata4, Tbx5, Smarcd3). As expected,9,10Nkx2.5 expression was lower in MICs (potential resident progenitor cells) compared to the GFP-negative cell fraction which, although depleted from cardiomyocytes by filtration steps, still might contain some residual cardiomyocytes. Simultaneously, high expression of fibroblast (e.g. Vimentin, Thy1) and myofibroblast markers (e.g. Myh10, Acta2, Fap) was found in GFP+ MICs as compared to the GFP-negative cell fraction (Figure 4B).
Figure 4.
Characterization of NkxCE-GFP+ MICs after MI. (A) One week after MI was induced in NkxCE-GFP mice hearts were explanted, GFP+ and GFP- cells were sorted and qPCR was performed. (B) qPCR analysis of a panel of cardiac resident stem cell marker (Sca1, Flk1, Pdgfrα), epicardial developmental marker (Wt1, Tbx18), cardiac developmental TFs (Nkx2.5, Gata4, Tbx5, Smarcd3), sarcomeric proteins (Myl2, Tnnt2), endothelial marker (CD31), myofibroblast marker (Acta2, Myh10, Fap) and fibroblast/mesenchymal marker (Vimentin, Thy1) was performed in GFP+ MICs and GFP-negative cells (NegMI). n = 5 per group. *P < 0.05; **P < 0.01. Stem cells; TFs (C) One week after MI single cell suspensions of NkxCE-GFP mice were stained with fluorochrome-conjugated antibodies for Sca1, CD31 and Thy1 and quantified by FACS (n = 4/5). (D) Virtually no GFP+ MICs stained for CD31. A subset of GFP+ cells stained for Thy1, and nearly all MICs stained for Sca1 (exemplary FACS plots). (E) Summarized staining results corresponding to D. FACS, fluorescence-activated cell sorting.
Gene expression results were verified by antibody staining and subsequent flow cytometric analysis (Figure 4C–E). GFP+ MICs did not show significant expression of the endothelial marker CD31 (Figure 4D and E). However, about 25% of GFP+ MICs were positive for Thy1 (CD90) and nearly 80% were positive for the mesenchymal stem cell marker Sca1 (Figure 4D and E).
Additionally, we performed immunofluorescent staining on cryosections of infarcted NkxCE-GFP hearts for select markers one week after MI (see Supplementary material online, Figures S4 and S5). We co-stained for GFP and Sca1 or Fap (see Supplementary material online, Figure S4A and B). Additionally, GFP+ cells were found to co-express Thy1 (CD90), Myh10 (SMemb), Wt1, Col1, αSMA and Vimentin (see Supplementary material online, Figures S4C–H and S5A). Moreover, MICs showed phospho-Histone H3 nuclear staining, indicating that cells were mitotically active (see Supplementary material online, Figure S5B). No CD31-positive staining was detected in GFP+ cells (see Supplementary material online, Figure S5C). Taken together, MICs exhibit a distinct fibroblast/myofibroblast expression signature rather than a cardiac progenitor cell signature.
3.5 Global transcriptome of NkxCE-GFP+ MICs
To further examine the question of whether MICs represent a cardiac resident stem/progenitor cell population, a detailed comparative microarray analysis was performed. The global gene expression profile of GFP+ MICs was compared to that of in vitro differentiated (day 7) GFP+ cells (i.e. either CPCs or very early cardiomyocytes) from an NkxCE-GFP ES cell line, cultivated CFs from adult uninjured mouse hearts and adult TTFs (Figure 5A, Supplementary material online, Figure S6A–D).
Figure 5 .
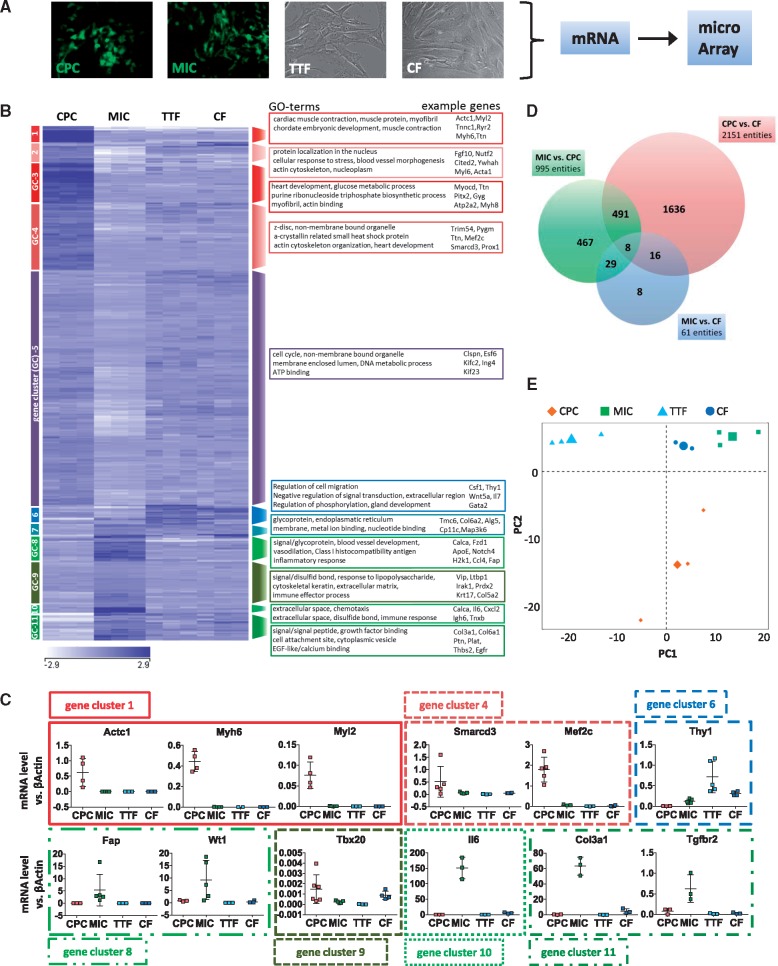
Global transcriptome characterization of NkxCE-GFP+ MICs as compared to select cell types. (A) CPCs (in vitro differentiated CPCs from murine NkxCE-GFP embryonic stem cells), MICs , mouse TTFs and mouse CFs underwent comparative microarray-based transcriptional profiling (n = 3). (B) Global heat map was divided into 11 GC and select GO terms are provided with associated exemplary genes. (C) Select gene expression was confirmed by qPCR (n = 5). (D) The Venn diagram shows 2151 differentially-regulated mRNAs/entities between CPCs and CFs, whereas only 61 mRNAs/entities are differentially regulated between MICs and CFs (fold change ≥1.5; P ≤ 0.05). (E) PCA clusters MICs in close proximity to CFs (upper right corner).
The heat map in Figure 5B depicts differentially expressed genes (n = 1548; fold change ≥1.5; P ≤ 0.05). Gene ontology (GO) analysis of gene clusters (GCs) 1-4, upregulated in CPCs, revealed GO terms such as cardiac muscle contraction, heart development, myofibril and blood vessel morphogenesis (Figure 5B), whereas GO-analysis of upregulated genes in MICs, mainly GC 8, 10 and 11, showed GO terms such as extracellular region, extracellular space, inflammatory response and blood vessel development.
Select genes from different clusters were validated by qPCR (Figure 5C). Sarcomeric marker expression (GC 1: Myh6, Myl2, Actc1) was clearly upregulated in ES-derived CPCs as compared to fibroblasts or MICs. This extends to TFs important in cardiac development (e.g. Smarcd3/Baf60c, Mef2c; GC 4). The fibroblast marker Thy1 (GC 6) was mainly detected in fibroblasts. However, MICs also expressed a substantial amount of Thy1. Genes that are mainly associated with myofibroblasts (e.g. Fap, Wt1, Il6, Col3a1, Tgfbr2; GCs 8, 10, 11) were expressed at the highest levels in MICs.25–27 MICs also expressed a certain level of Tbx20, which was demonstrated to be highly expressed in cardiac fibroblasts.28
Additionally, expression of different cardiac TFs, sarcomeric markers, mesenchymal stem cell markers, epicardial developmental TFs, smooth muscle cell and myofibroblast markers were compared between cell types (see Supplementary material online, Figure S6E). While stem cell (e.g., Sca1, but not c-Kit) and fibroblast/myofibroblast markers were highly expressed in MICs (e.g., Postn, Acta2), cardiac developmental marker expression was significantly increased in CPCs (e.g., Nkx2.5, Gata4, Tbx5 and c-Kit).
We also studied candidate microRNA expression by qPCR in MICs as compared to CPCs and adult fibroblasts (CFs and TTFs; Supplementary material online, Figure S6F and G). We found that CPCs showed upregulation of typical cardiac microRNAs (e.g. miR-1 and miR-133, Supplementary material online, Figure S6F) that are nearly undetectable in fibroblasts and MICs. In contrast, characteristic fibroblast/myofibroblast microRNAs (miR-21, miR-2929–31) were only expressed in fibroblasts and MICs but not in CPCs (see Supplementary material online, Figure S6G).
Finally, GO terms related to cardiac muscle, extracellular matrix, oxidative stress and inflammation were selected, their correlated genes were chosen from significantly differentially expressed genes, and focus heat maps were created (see Supplementary material online, Figure S7A–E). In all cases, except for inflammation, MICs clustered with TTFs and CFs, whereas CPCs showed different expression patterns. For inflammation, MICs exhibited the greatest number of upregulated genes.
A Venn diagram of differentially expressed genes shows that CPCs possess the most distinct gene expression as compared to CFs and MICs (Figure 5D). However, MICs appeared largely similar to CFs as only 61 genes were differentially expressed as compared to 2151 (CPCs vs. CFs) or 995 (CPCs vs. MICs) genes (fold change ≥1.5; P ≤ 0.05). PCA supports this finding (Figure 5E). Principal component 1 (PC1) and PC2 explain 58.5% of the observed variability. MICs were found in close vicinity to CFs, whereas CPCs appeared further away.
In summary, the extensive cell type-specific comparison supported our earlier data demonstrating that MICs resemble cardiac fibroblasts and have lost their cardiomyogenic capacity as cardiac stem/progenitor cells.
3.6 In vitro culture of NkxCE-GFP+ MICs and characterization
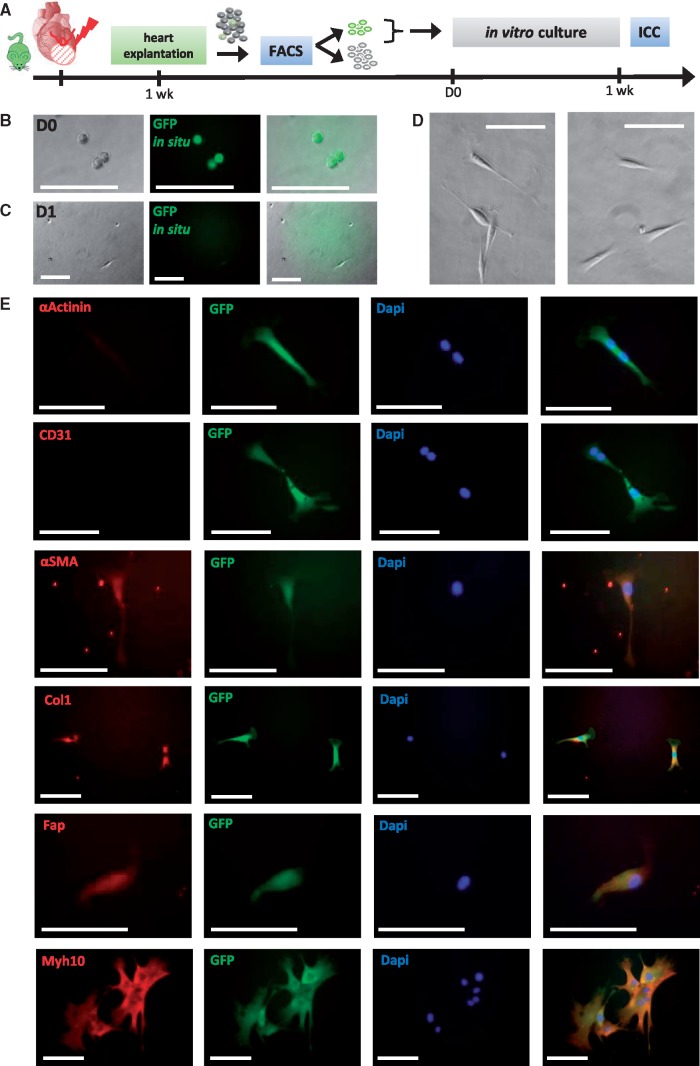
GFP+ MICs were cultured in vitro after FACS sorting (Figure 6A) to further characterize their morphology, growth behavior and differentiation potential. Initially, MICs showed clearly visible intracellular GFP fluorescence, which faded over time (Figure 6B–D). After attachment to the cell-culture dish, MIC cell morphology was typical of fibroblasts (Figure 6C and D). MICs were flat and spindle-shaped and adhered to the plastic cell culture plates. After prolonged culture, MICs exhibited an elongated, stellate shape with branched cytoplasm32 (e.g., Figure 6E, lower panel). No clonal growth was observed what discriminates MICs from colony forming units fibroblasts33 or the clonogenic self-renewing Pdgfrα-positive stem/progenitor cells from adult mouse hearts described by Noseda et al.34 MICs were cultured up to 2 weeks. However, they only proliferate slowly as shown by DAPI staining and subsequent cell counting (see Supplementary material online, Figure S8A–C). Their doubling time is about 6 days when seeded in a density of 20 cells per well (96 well plate). This is shorter than for primary cardiac fibroblasts from healthy hearts which exhibit doubling times of 11 days (50 or 200 cells per well, data not shown). However, Beltrami et al.35 postulate a markedly shorter doubling time of only 1.67 days for their adult cardiac resident c-Kit+ progenitor cells.
Figure 6.
In vitro culture of NkxCE-GFP+ MICs isolated after MI. (A) Hearts from infarcted NkxCE-GFP mice were explanted 1 week after MI (n = 6). 20 GFP+ MICs were directly sorted to collagen-coated 96-well-plates. Immunocytochemical staining was performed after 1 week. (B) Directly after sorting (D0) GFP+ MICs exhibit a round shape and in situ GFP fluorescence (no antibody). Scale bars: 100 µm. (C) One day later (D1), in situ GFP-fluorescence has faded and 3 days after sorting (D), GFP-fluorescence was no longer visible. Morphologically, cells appeared fibroblast-like. Scale bars: 100µm. (E) Immunocytochemical staining 1 week after plating. αActinin and CD31 were not detected. However, fibroblast/myofibroblast markers like αSMA, Col1, Fap, and Myh10 were detected in in vitro cultivated MICs. Scale bars: 100 µm
Next, the effect of mitogenic compounds on MIC proliferation behavior was evaluated. Therefore, 20 GFP-positive MICs were individually seeded into single collagen-coated wells. Without any treatment, each well contained 1.5 ± 4.0 cells after 1 week on average and 3.3 ± 4.2 cells after 2 weeks (P < 0.001; Supplementary material online, Figure S8C). In subsequent assays, sorted MICs were stimulated by ActivinA, BMP4, A83-017,12 and TB436 (see Supplementary material online, Figure S8D and E). To rule out position-specific effects in 96 well plates, untreated MICs at the same positions on a second 96 well plate were used as control. Compared to untreated cells, BMP4, TB4 and the lowest concentration of A83-01 (1 µM)-induced significant proliferation of MICs (see Supplementary material online, Figure S8E).
Immunocytochemical staining of cultivated MICs showed similar gene expression as MICs in infarcted heart sections. During the first week after plating, MICs did not exhibit sarcomeric markers like αActinin or endothelial markers like CD31. However, they were positive for fibroblast and myofibroblast markers like αSMA, Col1, Fap and Myh10 (Figure 6E). Anti-GFP-antibody was still able to detect GFP in most cells after one week, even if GFP was not visible in situ via fluorescent microscopy. However, immunocytochemical staining after prolonged culture (2–3 weeks) revealed that MICs still expressed mesenchymal markers like Vimentin but did not differentiate into cardiomyocytes, since they did not express sarcomeric proteins like αActinin, αMHC or Tnni3 (see Supplementary material online, Figure S8F and G). Additionally, no spontaneous beating activity was observed for MICs.
In summary, MICs were successfully cultured in vitro and select compounds were able to enhance their proliferation. However, no cardiac differentiation was observed. The fibroblast phenotype observed in vivo was confirmed in vitro.
3.7 Chromatin organization at the Nkx2.5 genomic locus in different cell types
Next, we asked if myogenic differentiation of MICs might be inhibited by differences in the chromatin structure between embryonic CPCs and adult MICs. We initially addressed whether the chromatin organization or status is responsible to restrict Nkx2.5 gene expression despite a putative activation of the NkxCE in MICs.
Therefore, we reanalyzed previously published18 Hi-C, ChiP-seq and RNA-seq data (see Supplementary material online, Figure S9A and B). We included tripotent embryonic NkxCE-GFP CPCs as well as fetal, early postnatal and adult cardiomyocytes. Unfortunately, we were not able to isolate MICs from infarcted mouse hearts suitable for epigenetic analysis. Since also published epigenetic data are lacking for cardiac fibroblasts, we included human lung fibroblasts (IMR90) as a representative fibroblast cell line. Remarkably, we found that the Nkx2.5 gene and its flanking regulatory regions are located within an active compartment (A compartment) in all assessed cell types. In addition, in adult cardiomyocytes (expression of Nkx2.5) and fibroblasts (no expression of Nkx2.5), the upstream region of the Nkx2.5 gene, containing the enhancer element (NkxCE) used in this study, show low levels of CpG methylation and enrichment for H3K4me1, which is characteristic for regulatory elements. In cardiomyocytes, these elements were also decorated by H3K27ac and thus represent likely active enhancers. This active signature is weaker in fibroblasts and thus an additional stimulus is likely necessary for true enhancer activation allowing gene expression. In summary, we conclude that the activation of Nkx2.5 gene expression in fibroblasts and cardiomyocytes is not primarily regulated by epigenetic mechanisms. Further studies are required to shed light on these mechanisms. Unfortunately, it is impossible to draw a final conclusion about an epigenetic roadblock for myogenic differentiation in MICs.
3.8 Function of NkxCE-GFP+ MICs in the infarcted heart
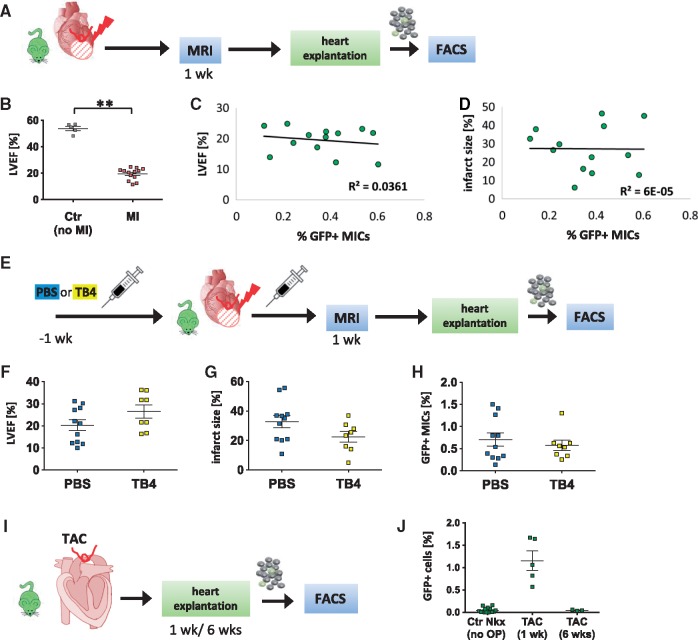
Since only approximately 0.5% of MICs were detected in infarcted mouse hearts, technical challenges made it difficult to extensively study their roles in vivo. Thus, correlations between left ventricular ejection fraction (LVEF) and infarct size (%LV; both parameters as measured by in vivo MRI) and the corresponding amount of GFP+ MICs were evaluated (Figure 7A). One week after MI, a significant reduction in LVEF was detected (P < 0.001). Mean LVEF was 19.5 ± 1.2% in MI mice compared to 53.8 ± 1.6% in healthy controls (Figure 7B). However, one week after MI, no significant correlations between the number of MICs and LVEF (R2 = 0.0361) or infarct size (R2 = 0.00006; Figure 7C and D) were detected.
Figure 7 .
Functional analysis of NkxCE-GFP+ MICs after MI and TAC. (A) Functional measurements were performed 1 week after MI on NkxCE-GFP mice by MRI. Hearts were then explanted and analyzed by FACS. (B) Left ventricular ejection fraction (LVEF) [%] of infarcted NkxCE-GFP mice (MI, n = 13) is significantly reduced compared to control mice (Ctr – no MI, n = 5), **P < 0.01 (t-test). (C) and (D) No correlation was observed between LVEF [%] or infarct size [% of LV], respectively, of infarcted NkxCE-GFP mice and corresponding amounts of GFP+ MICs (n = 13). (E) NkxCE-GFP mice were primed with TB4 or PBS (ctr) for 1 week before induction of MI. One week post injury MRI was performed, followed by heart explantation and FACS analysis (PBS: n = 11; TB4: n = 8). (F) LVEF [%] was slightly increased in TB4-treated mice compared to PBS-treated mice 1 week post-MI whereas infarct size [%LV] and amount of GFP+ MICs were rather reduced in TB4-treated mice (G) and (H). (I) One (n = 5) or 6 weeks (n = 3) after TAC was applied to NkxCE-GFP mice hearts were explanted and GFP+ cells were quantified by FACS. (J) A similar number of GFP+ cells as in infarcted NkxCE-GFP hearts emerged after 1 week. Six weeks after TAC, GFP+ cells have disappeared.
The MIC population shares similarities with previously described Wt1-positive EPDCs. EPDCs alone did not contribute significantly to cardiomyogenesis after MI but TB4 priming/treatment was able to stimulate EPDCs and contribute to cardiac regeneration after MI in mice.36,37 TB4 is an anti-inflammatory agent and is well-known for promoting wound healing, angiogenesis, cardiomyocyte survival and cardiac repair after MI in mice.37 Since we verified in vitro stimulation of MIC-proliferation by TB4 (see Supplementary material online, Figure S8E), we also treated NkxCE-GFP mice with TB4 to stimulate MIC-proliferation in vivo. According to established protocols,36,38 NkxCE-GFP mice were primed with TB4 (12 mg/kg) daily for seven days before inducing MI and received additional doses after MI. Control mice were injected with PBS only (Figure 7E). One week post-MI, functional measurements using MRI were performed (LVEF, %LV). Additionally, re-emerging GFP+ MICs were quantified by FACS after MRI (Figure 7E). TB4-treated mice exhibited a weak trend to higher LVEF and smaller %LV as compared to control mice (LVEF, P = 0.076; %LV, P = 0.097; Figure 7F and G). TB4 appeared beneficial after MI by slightly reducing infarct size and preserving LVEF.36,37 Interestingly, the number of GFP+ MICs was reduced by TB4 treatment (Figure 7H). Although not significant, the reduction in MICs that showed an inflammatory signature after MI (see Supplementary material online, Figure S7E) could be due to TB4’s anti-inflammatory functionality.
In summary, no correlation was observed between MICs and LVEF or infarction size. TB4 treatment may preserve cardiac function in the early phase after MI and is associated with a trend towards a reduced number of MICs.
3.9 Mechanism of activation of NkxCE-GFP+ MICs
Finally, we sought to identify the mechanism of GFP+ MIC activation. MI is an event of local ischemia and subsequent myocardial cell death followed by oxidative stress (hypoxia) and inflammatory reactions in the injured myocardium.27,39 This ‘inflammatory’ phase lasts approximately 3–4 days in mice.27 When inflammation is suppressed, reparation begins with concomitant fibroblast activation to maintain structural integrity.27
In support of these post-injury processes, the time course of MIC appearance after MI was analyzed (see Supplementary material online, Figure S10A and B). As early as three days post-injury, 0.17 ± 0.07% of GFP+ MICs were detectable. This coincides with the end of the inflammatory phase after MI and induction of the reparative phase accompanied by fibroblast activation and transdifferentiation into myofibroblasts.27 The number of MICs increased beginning five days post-MI, peaking after two weeks. Following the reparative phase, scar maturation begins approximately 2–2.5 weeks after MI, in which reparative cells (myofibroblasts) are deactivated or undergo apoptosis.27 This coincides with the time course of vanishing MICs. Six weeks (42 days) after MI, only a small proportion (0.2 ± 0.03%) of MICs was still identifiable (see Supplementary material online, Figure S10B).
I/R injuries maintain the same time course of myofibroblast activation, like persisting ischemia (MI).40 Therefore, global I/R injury simulated by heterotopic heart transplantation after 6 h ex vivo was applied to NkxCE-GFP mice. This induced GFP+ MICs to a similar proportion as was observed post-MI (see Supplementary material online, Figure S3A–D).
Since MICs first appear during the inflammatory phase (< 4 days; Supplementary material online, Figure S10B), we hypothesized that they might be activated by hypoxia or inflammatory signals. Thus, NkxCE-GFP mice underwent TAC surgery to simulate cardiac pressure overload (Figure 7I, Supplementary material online, Figure 11A–F). This non-ischemic heart failure model with a more gradual time course than acute ischemic injuries (MI or I/R injury) induces transient inflammatory and subsequent fibrotic responses accompanied by the appearance of myofibroblasts. However, even if reactive oxygen species are induced, hypoxia or cardiomyocyte cell death does not play a major role after TAC. Interestingly, we observed GFP+ MICs one week after TAC-surgery in NkxCE-GFP mice (1.15± 0.22%; Figure 7J), which disappeared after six weeks.
In summary, we demonstrated that MICs appear as early as three days after MI, and we observed GFP+ MICs after I/R-injury. Additionally, a non-ischemic pressure-overload model of heart failure also induced GFP+ MICs, indicating that inflammation might be the trigger for MICs as opposed to hypoxia.
4. Discussion
Using an NkxCE-GFP transgenic mouse strain, a rare GFP+ cell population appearing after MI (MICs) was identified in adult mouse hearts. Since NkxCE-positive cells physiologically appear exclusively during embryonic cardiac development and, to a lesser extent, in neonatal hearts, we hypothesized that MICs might represent a cardiac resident stem/progenitor cell population activated by MI that harbour cardiomyogenic properties. MICs localized to the infarction area and border zone. An extra-cardiac origin was excluded. Gene expression profiling of purified GFP+ MICs revealed that MI triggers an increase in stem cell marker and embryonic cardiac TF expression in these cells as compared to GFP-negative cells. Cardiac resident stem cell populations are frequently defined by cardiac embryonic or mesenchymal gene expression.41 However, we found several fibroblast and myofibroblast markers to be highly expressed in MICs. We detailed by microarray-based genome-wide transcriptional profiling with ES cell-derived CPCs as well as adult cardiac and TTFs that cardiac developmental markers were only expressed at background levels in GFP+ MICs, similar to their expression in cardiac fibroblasts. Expression of cardiac TFs has been recently demonstrated in cardiac fibroblasts.28 Interestingly, MICs exhibited a distinct (myo-)fibroblast profile and several inflammatory markers. Bioinformatic analysis brought MICs in close vicinity to cardiac fibroblasts, supporting that MICs may only be a subset of activated fibroblasts/myofibroblasts after injury. Under in vitro culture conditions, MICs adopted a fibroblast-like phenotype with minor proliferative ability. In contrast, stem cells usually have a short doubling time.35 Moreover, no clonal growth was observed for MICs, which is one of the key aspects of stem cells.6 Select compounds like A83-0112 or TB436 were able to increase the proliferative capacity of MICs in vitro. However, even after prolonged culture, MICs did not differentiate into cardiomyocytes, as has been reported for comparable adult cardiac resident stem cell populations.42,43
We were able to show that MICs appeared 3 days after MI coinciding with the occurrence of myofibroblasts after MI in the reparative phase, as evidenced by others.27 GFP+ MICs were also observed after I/R-injury where myofibroblasts follow a similar time-course of activation as after MI.40 In addition, a non-ischemic pressure-overload model of heart failure also induced GFP+ MICs indicating that inflammation may be the trigger for MIC induction, rather than hypoxia. This non-ischemic heart failure model with a more gradual time course than acute ischemic injuries (MI, I/R-injury) also induces a transient inflammatory and subsequent fibrotic response accompanied by the appearance of myofibroblasts with a very similar time course to that of acute injuries.44
Our results suggest that GFP+ MICs probably represent a small subset of activated fibroblasts induced by the inflammatory environment in myocardial injuries. It has already been reported previously that enhancer elements might be activated by injury to trigger gene expression in injury sites.45
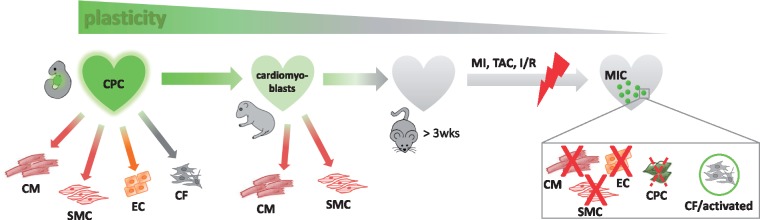
4.1 Changing plasticity of NkxCE-positive cells from development to adulthood
During embryonic cardiac development, NkxCE-positive progenitor cells are multipotent, giving rise to the main cardiac lineages like cardiomyocytes, endothelial cells, smooth muscle cells and also fibroblasts.8,9 Chen et al. and Serpooshan et al. recently found that hearts of neonatal NkxCE-GFP-mice re-activate around 5–15% GFP-positive cells (among non-myocytes).7,10 FACS-purified NkxCE-GFP+ cells from neonatal hearts appear to lack the ability to spontaneously differentiate into cardiomyocytes while retaining only limited capacity for smooth muscle cell differentiation.7 In co-culture experiments with cardiomyocytes, smooth muscle cells and endothelial cells NkxCE-GFP+ cells from neonatal hearts were able to differentiate mainly in cardiomyocytes or smooth muscle cells instead of endothelial cells supporting a commitment to myogenic lineages (cardiomyoblasts).10 However, re-activated MICs from adult infarcted NkxCE-GFP mouse hearts (>8 weeks) were clearly distinct from their multipotent embryonic and neonatal counterparts. Somehow, MICs exhibit features of an embryonic gene program, albeit without the ability to fully revert back to an embryonic phenotype.
Bollini et al. investigated re-activated Wt1-positive EPDCs after MI and compared them to their Wt1-positive embryonic counterparts.38 Similar to our study, they found that the molecular phenotype of adult re-activated EPDCs is clearly distinct from embryonic Wt1-positive cells. Another publication described a Sca1+/Pdgfrα+ cell population resident to the adult heart that showed a rather similar gene expression profile to MICs (positive for Gata4, Tbx5/20, Mef2c; Nkx2.5 and Isl1 silenced).34 The authors characterized those cells as an incomplete form (forme fruste) of multipotent CPCs that are represented by Pdgfrα-positive cells during cardiac development.6 Isl1-positive cells in the infarcted adult mouse heart were also analyzed.46 During embryogenesis, Isl1 is a marker of second heart field multipotent cardiovascular progenitors.47 However, after MI in the adult heart, no evidence was detected that Isl1-positive cells serve as cardiac progenitor cells,46 also indicating that they lose their cardiogenic potential that they maintained during cardiac development. Similar results were published for c-Kit-positive endogenous stem cells.48,49 In early postnatal murine hearts (P1-3) c-Kit+ cardiac resident stem cells were able to adopt myogenic and vascular fates while partially regenerating the infarction.48 Whereas in adult mice c-kit+ cells rarely (0.03–0.008%) underwent myogenic differentiation49 and mainly adopted vascular fate, indicating a lack of authentic CPCs in adult hearts.48
Our results support decreasing plasticity of cardiac progenitor(-like) cell populations and their potential regenerative properties from embryonic to adult hearts (Figure 8). In embryonic development until the first week after birth, full compensation of injury- or ablation triggered myocardial cell loss has been suggested for mice50,51 coinciding with the decreasing plasticity of NkxCE-positive cells and other progenitor(-like) cell populations.
Figure 8.
Changing plasticity of NkxCE-positive cells from embryonic development to adulthood. During cardiac development, multipotent NkxCE-positive cells are able to build cells of all lineages in the heart. In neonatal hearts few NkxCE-positive cells (cardiomyoblasts) are re-activated and were shown to be able to differentiate into cardiomyocytes and smooth muscle cells. However, MICs induced by MI in adult mouse hearts indeed exhibit a somewhat embryonic gene profile but do not fully revert back to an embryonic phenotype.
5. Conclusion
Here, we show that postnatal cardiac injury (MI, I/R, TAC) reactivates a cardiac-specific Nkx2.5 enhancer element known to specifically mark myocardial progenitor cells during embryonic development. We performed a molecular comparison of MI-induced putative postnatal cardiac resident stem/progenitor cells (MICs) with their embryonic counterparts and cardiac fibroblasts. Although embryonic NkxCE-GFP positive CPCs are able to differentiate into cardiomyocytes, smooth muscle cells and endothelial cells, reemerging MICs resemble activated cardiac fibroblasts and are molecularly clearly distinct from their embryonic counterparts. Our data suggest a decreasing plasticity of cardiac progenitor (-like) cell populations with increasing age. This is in line with recent reports questioning a relevant contribution of putative resident cardiac stem or progenitor cells to postnatal cardiomyocyte renewal after injury.34,38,46,48,49 Thus, a re-expression of embryonic, stem or progenitor cell features in the adult heart must be interpreted very carefully with respect to the definition of cardiac resident progenitor cells. However, the abundance of scar formation after cardiac injury suggests a potential to target fibrotic cells, especially predestinated subpopulations exhibiting embryonic features, to push them towards cardiomyogenic differentiation to improve regeneration.
Supplementary material
Supplementary material is available at Cardiovascular Research online.
Supplementary Material
Acknowledgements
We are grateful to Paul Riley for serving us with the same lot of TB4 that was used for his experiments.37,39
Conflict of interest: none declared.
Funding
M.A.D. and the project are supported by the Rusche Forschungsprojekt (2014) of the Deutsche Stiftung für Herzforschung (DSH) and the Deutsche Gesellschaft für Thorax-, Herz- und Gefäßchirurgie (DGTHG). M.K. is supported by the Deutsches Zentrum für Herz Kreislauf Forschung [DZHK B 15-005, DZHK B 15-039SE] and Deutsche Forschungsgemeinschaft (DFG) – Sachmittelantrag [KR3770/7-1, KR3770/9-1].
Footnotes
Time for primary review: 44 days
References
- 1. Bergmann O, Bhardwaj RD, Bernard S, Zdunek S, Barnabé-Heider F, Walsh S, Zupicich J, Alkass K, Buchholz BA, Druid H, Jovinge S, Frisén J.. Evidence for cardiomyocyte renewal in humans. Science 2009;324:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Senyo SE, Steinhauser ML, Pizzimenti CL, Yang VK, Cai L, Wang M, Wu TD, Guerquin-Kern JL, Lechene CP, Lee RT.. Mammalian heart renewal by pre-existing cardiomyocytes. Nature 2012;493:433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hsieh PC, Segers VF, Davis ME, MacGillivray C, Gannon J, Molkentin JD, Robbins J, Lee RT.. Evidence from a genetic fate-mapping study that stem cells refresh adult mammalian cardiomyocytes after injury. Nat Med 2007;13:970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wu JM, Hsueh YC, Ch'ang HJ, Luo CY, Wu LW, Nakauchi H, Hsieh PC.. Circulating cells contribute to cardiomyocyte regeneration after injury. Circ Res 2015;116:633–641. [DOI] [PubMed] [Google Scholar]
- 5. Eschenhagen T, Bolli R, Braun T, Field LJ, Fleischmann BK, Frisen J, Giacca M, Hare JM, Houser S, Lee RT, Marban E, Martin JF, Molkentin JD, Murry CE, Riley PR, Ruiz-Lozano P, Sadek HA, Sussman MA, Hill JA.. Cardiomyocyte regeneration: a consensus statement. Circulation 2017;136:680–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Noseda M, Abreu-Paiva M, Schneider MD.. The quest for the adult cardiac stem cell. Circ J 2015;79:1422–1430. [DOI] [PubMed] [Google Scholar]
- 7. Chen WP, Wu SM.. Small molecule regulators of postnatal Nkx2.5 cardiomyoblast proliferation and differentiation. J Cell Mol Med 2012;16:961–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu SM, Fujiwara Y, Cibulsky SM, Clapham DE, Lien CL, Schultheiss TM, Orkin SH.. Developmental origin of a bipotential myocardial and smooth muscle cell precursor in the mammalian heart. Cell 2006;127:1137–1150. [DOI] [PubMed] [Google Scholar]
- 9. Li G, Plonowska K, Kuppusamy R, Sturzu A, Wu SM.. Identification of cardiovascular lineage descendants at single-cell resolution. Development 2015;142:846–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Serpooshan V, Liu YH, Buikema JW, Galdos FX, Chirikian O, Paige S, Venkatraman S, Kumar A, Rawnsley DR, Huang X, Pijnappels DA, Wu SM.. Nkx2.5+ Cardiomyoblasts contribute to cardiomyogenesis in the neonatal heart. Sci Rep 2017;7:12590.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ.. Embryonic expression of an Nkx2-5/Cre gene using ROSA26 reporter mice. Genesis 2001;31:176–180. [DOI] [PubMed] [Google Scholar]
- 12. Chen WP, Liu YH, Ho YJ, Wu SM.. Pharmacological inhibition of TGFbeta receptor improves Nkx2.5 cardiomyoblast-mediated regeneration. Cardiovasc Res 2015;105:44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ratschiller T, Deutsch MA, Calzada-Wack J, Neff F, Roesch C, Guenzinger R, Lange R, Krane M.. Heterotopic cervical heart transplantation in mice. J Vis Exp 2015;102:e52907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. deAlmeida AC, van Oort RJ, Wehrens XH.. Transverse aortic constriction in mice. J Vis Exp 2010;38:doi: 10.3791/1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martin TP, Robinson E, Harvey AP, MacDonald M, Grieve DJ, Paul A, Currie S.. Surgical optimization and characterization of a minimally invasive aortic banding procedure to induce cardiac hypertrophy in mice. Exp Physiol 2012;97:822–832. [DOI] [PubMed] [Google Scholar]
- 16. Doppler SA, Werner A, Barz M, Lahm H, Deutsch M-A, Dreßen M, Schiemann M, Voss B, Gregoire S, Kuppusamy R, Wu SM, Lange R, Krane M.. Myeloid zinc finger 1 (Mzf1) differentially modulates murine cardiogenesis by interacting with an Nkx2.5 cardiac enhancer. PLoS One 2014;9:e113775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, Aiden EL.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 2014;159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nothjunge S, Nuhrenberg TG, Gruning BA, Doppler SA, Preissl S, Schwaderer M, Rommel C, Krane M, Hein L, Gilsbach R.. DNA methylation signatures follow preformed chromatin compartments in cardiac myocytes. Nat Commun 2017;8:1667.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, Edsall L, Antosiewicz-Bourget J, Stewart R, Ruotti V, Millar AH, Thomson JA, Ren B, Ecker JR.. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009;462:315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gilsbach R, Preissl S, Gruning BA, Schnick T, Burger L, Benes V, Wurch A, Bonisch U, Gunther S, Backofen R, Fleischmann BK, Schubeler D, Hein L.. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat Commun 2014;5:5288.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu Y-C, Pfenning A, Wang X, ClaussnitzerYaping Liu M, Coarfa C, Alan Harris R, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, David Hawkins R, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Scott Hansen R, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh K-H, Feizi S, Karlic R, Kim A-R, Kulkarni A, Li D, Lowdon R, Elliott GNell, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Abdennur N, Adli M, Akerman M, Barrera L, Antosiewicz-Bourget J, Ballinger T, Barnes MJ, Bates D, Bell RJA, Bennett DA, Bianco K, Bock C, Boyle P, Brinchmann J, Caballero-Campo P, Camahort R, Carrasco-Alfonso MJ, Charnecki T, Chen H, Chen Z, Cheng JB, Cho S, Chu A, Chung W-Y, Cowan C, Athena Deng Q, Deshpande V, Diegel M, Ding B, Durham T, Echipare L, Edsall L, Flowers D, Genbacev-Krtolica O, Gifford C, Gillespie S, Giste E, Glass IA, Gnirke A, Gormley M, Gu H, Gu J, Hafler DA, Hangauer MJ, Hariharan M, Hatan M, Haugen E, He Y, Heimfeld S, Herlofsen S, Hou Z, Humbert R, Issner R, Jackson AR, Jia H, Jiang P, Johnson AK, Kadlecek T, Kamoh B, Kapidzic M, Kent J, Kim A, Kleinewietfeld M, Klugman S, Krishnan J, Kuan S, Kutyavin T, Lee A-Y, Lee K, Li J, Li N, Li Y, Ligon KL, Lin S, Lin Y, Liu J, Liu Y, Luckey CJ, Ma YP, Maire C, Marson A, Mattick JS, Mayo M, McMaster M, Metsky H, Mikkelsen T, Miller D, Miri M, Mukame E, Nagarajan RP, Neri F, Nery J, Nguyen T, O’geen H, Paithankar S, Papayannopoulou T, Pelizzola M, Plettner P, Propson NE, Raghuraman S, Raney BJ, Raubitschek A, Reynolds AP, Richards H, Riehle K, Rinaudo P, Robinson JF, Rockweiler NB, Rosen E, Rynes E, Schein J, Sears R, Sejnowski T, Shafer A, Shen L, Shoemaker R, Sigaroudinia M, Slukvin I, Stehling-Sun S, Stewart R, Subramanian SL, Suknuntha K, Swanson S, Tian S, Tilden H, Tsai L, Urich M, Vaughn I, Vierstra J, Vong S, Wagner U, Wang H, Wang T, Wang Y, Weiss A, Whitton H, Wildberg A, Witt H, Won K-J, Xie M, Xing X, Xu I, Xuan Z, Ye Z, Yen C-A, Yu P, Zhang X, Zhang X, Zhao J, Zhou Y, Zhu J, Zhu Y, Ziegler S, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJM, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai L-H, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M, Kundaje A, Meuleman W, Ernst J, Bilenky M, Yen A, Heravi-Moussavi A, Kheradpour P, Zhang Z, Wang J, Ziller MJ, Amin V, Whitaker JW, Schultz MD, Ward LD, Sarkar A, Quon G, Sandstrom RS, Eaton ML, Wu Y-C, Pfenning AR, Wang X, Claussnitzer M, Liu Y, Coarfa C, Harris RA, Shoresh N, Epstein CB, Gjoneska E, Leung D, Xie W, Hawkins RD, Lister R, Hong C, Gascard P, Mungall AJ, Moore R, Chuah E, Tam A, Canfield TK, Hansen RS, Kaul R, Sabo PJ, Bansal MS, Carles A, Dixon JR, Farh K-H, Feizi S, Karlic R, Kim A-R, Kulkarni A, Li D, Lowdon R, Elliott GNell, Mercer TR, Neph SJ, Onuchic V, Polak P, Rajagopal N, Ray P, Sallari RC, Siebenthall KT, Sinnott-Armstrong NA, Stevens M, Thurman RE, Wu J, Zhang B, Zhou X, Beaudet AE, Boyer LA, De Jager PL, Farnham PJ, Fisher SJ, Haussler D, Jones SJM, Li W, Marra MA, McManus MT, Sunyaev S, Thomson JA, Tlsty TD, Tsai L-H, Wang W, Waterland RA, Zhang MQ, Chadwick LH, Bernstein BE, Costello JF, Ecker JR, Hirst M, Meissner A, Milosavljevic A, Ren B, Stamatoyannopoulos JA, Wang T, Kellis M.. Integrative analysis of 111 reference human epigenomes. Nature 2015;518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ramirez F, Bhardwaj V, Arrigoni L, Lam KC, Gruning BA, Villaveces J, Habermann B, Akhtar A, Manke T.. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun 2018;9:189.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krueger F, Andrews SR.. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 2011;27:1571–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T.. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res 2014;42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tillmanns J, Hoffmann D, Habbaba Y, Schmitto JD, Sedding D, Fraccarollo D, Galuppo P, Bauersachs J.. Fibroblast activation protein alpha expression identifies activated fibroblasts after myocardial infarction. J Mol Cell Cardiol 2015;87:194–203. [DOI] [PubMed] [Google Scholar]
- 26. Braitsch CM, Kanisicak O, van Berlo JH, Molkentin JD, Yutzey KE.. Differential expression of embryonic epicardial progenitor markers and localization of cardiac fibrosis in adult ischemic injury and hypertensive heart disease. J Mol Cell Cardiol 2013;65:108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Prabhu SD, Frangogiannis NG.. The Biological Basis for Cardiac Repair After Myocardial Infarction: from Inflammation to Fibrosis. Circ Res 2016;119:91–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Furtado MB, Costa MW, Pranoto EA, Salimova E, Pinto AR, Lam NT, Park A, Snider P, Chandran A, Harvey RP, Boyd R, Conway SJ, Pearson J, Kaye DM, Rosenthal NA.. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ Res 2014;114:1422–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Thum T, Gross C, Fiedler J, Fischer T, Kissler S, Bussen M, Galuppo P, Just S, Rottbauer W, Frantz S, Castoldi M, Soutschek J, Koteliansky V, Rosenwald A, Basson MA, Licht JD, Pena JT, Rouhanifard SH, Muckenthaler MU, Tuschl T, Martin GR, Bauersachs J, Engelhardt S.. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008;456:980–984. [DOI] [PubMed] [Google Scholar]
- 30. Fiedler J, Thum T.. MicroRNAs in myocardial infarction. Arterioscler Thromb Vasc Biol 2013;33:201–205. [DOI] [PubMed] [Google Scholar]
- 31. Turner NA, Porter KE.. Function and fate of myofibroblasts after myocardial infarction. Fibrogenesis Tissue Repair 2013;6:5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doppler SA, Carvalho C, Lahm H, Deutsch M-A, Dreßen M, Puluca N, Lange R, Krane M.. Cardiac fibroblasts: more than mechanical support. J Thorac Dis 2017;9:S36–S51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chong JJ, Chandrakanthan V, Xaymardan M, Asli NS, Li J, Ahmed I, Heffernan C, Menon MK, Scarlett CJ, Rashidianfar A, Biben C, Zoellner H, Colvin EK, Pimanda JE, Biankin AV, Zhou B, Pu WT, Prall OW, Harvey RP.. Adult cardiac-resident MSC-like stem cells with a proepicardial origin. Cell Stem Cell 2011;9:527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Noseda M, Harada M, McSweeney S, Leja T, Belian E, Stuckey DJ, Abreu Paiva MS, Habib J, Macaulay I, de Smith AJ, Al-Beidh F, Sampson R, Lumbers RT, Rao P, Harding SE, Blakemore AI, Jacobsen SE, Barahona M, Schneider MD.. PDGFRalpha demarcates the cardiogenic clonogenic Sca1+ stem/progenitor cell in adult murine myocardium. Nat Commun 2015;6:6930.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Beltrami AP, Barlucchi L, Torella D, Baker M, Limana F, Chimenti S, Kasahara H, Rota M, Musso E, Urbanek K, Leri A, Kajstura J, Nadal-Ginard B, Anversa P.. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 2003;114:763–776. [DOI] [PubMed] [Google Scholar]
- 36. Smart N, Bollini S, Dube KN, Vieira JM, Zhou B, Davidson S, Yellon D, Riegler J, Price AN, Lythgoe MF, Pu WT, Riley PR.. De novo cardiomyocytes from within the activated adult heart after injury. Nature 2011;474:640–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bock-Marquette I, Saxena A, White MD, Dimaio JM, Srivastava D.. Thymosin beta4 activates integrin-linked kinase and promotes cardiac cell migration, survival and cardiac repair. Nature 2004;432:466–472. [DOI] [PubMed] [Google Scholar]
- 38. Bollini S, Vieira JM, Howard S, Dube KN, Balmer GM, Smart N, Riley PR.. Re-activated adult epicardial progenitor cells are a heterogeneous population molecularly distinct from their embryonic counterparts. Stem Cells Dev 2014;23:1719–1730. [DOI] [PubMed] [Google Scholar]
- 39. Frangogiannis NG, Smith CW, Entman ML.. The inflammatory response in myocardial infarction. Cardiovasc Res 2002;53:31–47. [DOI] [PubMed] [Google Scholar]
- 40. Frangogiannis NG, Michael LH, Entman ML.. Myofibroblasts in reperfused myocardial infarcts express the embryonic form of smooth muscle myosin heavy chain (SMemb). Cardiovasc Res 2000;48:89–100. [DOI] [PubMed] [Google Scholar]
- 41. Furtado MB, Nim HT, Boyd SE, Rosenthal NA.. View from the heart: cardiac fibroblasts in development, scarring and regeneration. Development 2016;143:387–397. [DOI] [PubMed] [Google Scholar]
- 42. Pfister O, Mouquet F, Jain M, Summer R, Helmes M, Fine A, Colucci WS, Liao R.. CD31—but Not CD31+ cardiac side population cells exhibit functional cardiomyogenic differentiation. Circ Res 2005;97:52–61. [DOI] [PubMed] [Google Scholar]
- 43. Matsuura K, Nagai T, Nishigaki N, Oyama T, Nishi J, Wada H, Sano M, Toko H, Akazawa H, Sato T, Nakaya H, Kasanuki H, Komuro I.. Adult cardiac Sca-1-positive cells differentiate into beating cardiomyocytes. J Biol Chem 2004;279:11384–11391. [DOI] [PubMed] [Google Scholar]
- 44. Xia Y, Lee K, Li N, Corbett D, Mendoza L, Frangogiannis NG.. Characterization of the inflammatory and fibrotic response in a mouse model of cardiac pressure overload. Histochem Cell Biol 2009;131:471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kang J, Hu J, Karra R, Dickson AL, Tornini VA, Nachtrab G, Gemberling M, Goldman JA, Black BL, Poss KD.. Modulation of tissue repair by regeneration enhancer elements. Nature 2016;532:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Weinberger F, Mehrkens D, Friedrich FW, Stubbendorff M, Hua X, Muller JC, Schrepfer S, Evans SM, Carrier L, Eschenhagen T.. Localization of Islet-1-positive cells in the healthy and infarcted adult murine heart. Circ Res 2012;110:1303–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Laugwitz KL, Moretti A, Lam J, Gruber P, Chen Y, Woodard S, Lin LZ, Cai CL, Lu MM, Reth M, Platoshyn O, Yuan JX, Evans S, Chien KR.. Postnatal isl1+ cardioblasts enter fully differentiated cardiomyocyte lineages. Nature 2005;433:647–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jesty SA, Steffey MA, Lee FK, Breitbach M, Hesse M, Reining S, Lee JC, Doran RM, Nikitin AY, Fleischmann BK, Kotlikoff MI.. c-kit+ precursors support postinfarction myogenesis in the neonatal, but not adult, heart. Proc Natl Acad Sci U S A 2012;109:13380–13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van Berlo JH, Kanisicak O, Maillet M, Vagnozzi RJ, Karch J, Lin SC, Middleton RC, Marban E, Molkentin JD.. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature 2014;509:337–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sturzu AC, Rajarajan K, Passer D, Plonowska K, Riley A, Tan TC, Sharma A, Xu AF, Engels MC, Feistritzer R, Li G, Selig MK, Geissler R, Robertson KD, Scherrer-Crosbie M, Domian IJ, Wu SM.. Fetal mammalian heart generates a robust compensatory response to cell loss. Circulation 2015;132:109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Porrello ER, Mahmoud AI, Simpson E, Hill JA, Richardson JA, Olson EN, Sadek HA.. Transient regenerative potential of the neonatal mouse heart. Science 2011;331:1078–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.