Summary
Background
The role of microRNAs (miRNAs) in animal models of palatogenesis has been shown, but only limited research has been carried out in humans. To date, no miRNA expression study on tissues or cells from cleft palate patients has been published. We compared miRNA expression in palatal fibroblasts from cleft palate patients and age-matched controls.
Material and Methods
Cultured palatal fibroblasts from 10 non-syndromic cleft lip and palate patients (nsCLP; mean age: 18 ± 2 months), 5 non-syndromic cleft palate only patients (nsCPO; mean age: 17 ± 2 months), and 10 controls (mean age: 24 ± 5 months) were analysed with next-generation small RNA sequencing. All subjects are from Western European descent. Sequence reads were bioinformatically processed and the differentially expressed miRNAs were technically validated using quantitative reverse-transcription polymerase chain reaction (RT-qPCR).
Results
Using RNA sequencing, three miRNAs (hsa-miR-93-5p, hsa-miR-18a-5p, and hsa-miR-92a-3p) were up-regulated and six (hsa-miR-29c-5p, hsa-miR-549a, hsa-miR-3182, hsa-miR-181a-5p, hsa-miR-451a, and hsa-miR-92b-5p) were down-regulated in nsCPO fibroblasts. One miRNA (hsa-miR-505-3p) was down-regulated in nsCLP fibroblasts. Of these, hsa-miR-505-3p, hsa-miR-92a, hsa-miR-181a, and hsa-miR-451a were also differentially expressed using RT-PCR with a higher fold change than in RNAseq.
Limitations
The small sample size may limit the value of the data. In addition, interpretation of the data is complicated by the fact that biopsy samples are taken after birth, while the origin of the cleft lies in the embryonic period. This, together with possible effects of the culture medium, implies that only cell-autonomous genetic and epigenetic differences might be detected.
Conclusions
For the first time, we have shown that several miRNAs appear to be dysregulated in palatal fibroblasts from patients with nsCLP and nsCPO. Furthermore, large-scale genomic and expression studies are needed to validate these findings.
Introduction
Palatogenesis occurs through an ordered sequence of embryological events involving the maxillary and medial nasal processes. This is driven by cellular processes such as migration, proliferation, differentiation, and apoptosis (1). Perturbation of these processes can lead to non-syndromic cleft lip and/or palate (nsCLP and nsCPO) and, complex disorders, which are caused by both genomic and environmental factors (2). Previously, large-scale genome-wide association studies (GWAS) have identified several protein-coding susceptibility genes (3). However, these genes only account for part of the total genomic risk associated with cleft palate (4). Furthermore, more than 80% of the disease-associated genetic loci are found within the non-coding genome (5). The non-coding genome is a major regulator of gene expression and as such also of human disease (6). Therefore, further research into nsCLP/CPO aetiology needs to include the non-coding genome.
MicroRNAs (miRNAs) are part of the non-coding genome and have elicited interest because of their association with a wide range of complex disorders such as cancer, cardiovascular disease, diabetes, and many developmental disorders (7). MiRNAs are small non-coding RNAs, which function as post-transcriptional repressors of gene expression. A mature miRNA pairs with the 3′ untranslated region of its target messenger RNA (mRNA) and recruits an miRNA-induced silencing complex (miRISC) (8). Depending on base complementarity, the miRISC catalyses mRNA cleavage or translational repression and destabilization (9). To date, 35 828 miRNAs in 223 species have been identified, including thousands in humans (10). A wide range of biological processes are regulated by miRNAs, including epithelial to mesenchymal transition, migration, apoptosis, differentiation, and proliferation (11). Hence, miRNAs are also key regulators of embryogenesis, including the morphogenesis of most organs such as the heart, lung, hair, skin, teeth, and limbs (12–15). Genomic alterations affecting miRNA expression and function may therefore act as risk factors for complex congenital disorders such as nsCLP and nsCPO (16, 17).
It has been shown that miRNAs play a crucial role in animal models of palatogenesis (18, 19). Although only four human studies on patients from Asian descent have been carried out, they indicate that miRNAs are also important for human palatogenesis. A Chinese study identified a single nucleotide polymorphism (SNP) within the precursor of miR-140 to be associated with clefting (20). In a Thai patient with nsCPO, a mutation in the 3′ UTR of PDGFRa was shown to disrupt target recognition by miR-140 (21). In a Chinese cohort, an SNP in the binding site for miR-3649 in MSX1 was associated with cleft palate risk (22). Finally, altered miR-496-FGF2, miR-145-FGF5, and miR-187-FGF9 interactions were associated with clefting in 289 nsCLP and 49 nsCPO patients (23). The ultimate aim of this human research was to improve our understanding of the genomic aetiology of nsCLP and CPO and allow more accurate genomic counselling. To the best of our knowledge, no miRNA expression study has been carried out in the developmentally relevant tissues from human cleft palate patients up to now. To assess miRNA expression in cleft palate, we performed a small RNA sequencing study on palatal fibroblasts from cleft palate patients and age-matched controls.
Materials and Methods
Palatal fibroblast isolation and culture
Samples from 13 Dutch children with nsCLP (8 patients, mean age: 18 months±2 months), nsCPO (5 patients, mean age 17 ± 2 months), and 10 age-matched control individuals (mean age: 24 ± 5 months) were used. Control individuals had no congenital malformations. All children were treated in the Cleft Palate Craniofacial Unit of the Radboud University Medical Centre. Sample collection was approved by the Central Ethical Committee on Research Involving Human Subjects of the Netherlands (CCMO nr Poz.o544C). A 3 mm biopsy of the palatal mucosa from the hard palate was obtained during primary surgical palate closure at about 1 cm from the medial edge of the cleft. In control individuals, the biopsy was obtained during tonsillectomy at about 1 cm from the median of the hard palate. In order to have sufficient RNA yield, fibroblasts from the tissue were cultured, expanded, and frozen in liquid nitrogen in a highly standardized procedure as described previously (24). Fibroblasts, all with passage number 3, were thawed and cultured in a T75 bottle in Dulbecco’s Modified Eagle’s Medium (Sigma-Aldrich) containing 10% fetal calf serum, 100 U/ml penicillin, and 100 μg/ml streptomycin (all from Invitrogen) for RNA isolation. The fibroblasts displayed typical spindle-shaped fibroblast morphology.
Small RNA isolation
Upon reaching 90% confluency, the fibroblasts were harvested with a cell scraper. Lysis was performed with QIAzol reagent (Qiagen) and homogenization was achieved by pipetting up and down vigorously. Cell lysates were stored at −80°C. Small RNAs were isolated using the miRNeasy Mini Kit (Qiagen) according to the manufacturer’s protocol. The yield was quantified by the NanoDrop ND-2000 (Thermo Scientific) and quality control was done using the 2200 TapeStation System (Agilent Technologies).
Small RNA sequencing
The small RNA fraction (400 ng) of each sample was used for individual sequencing. Bar-coded small RNA libraries were generated according to the manufacturers’ protocols using the Ion Total RNA-Seq Kit v2 and the Ion Xpress™ RNA-Seq bar coding kit (Life Technologies). The size distribution and yield of the bar coded libraries were assessed using the 2200 TapeStation System with Agilent D1K ScreenTapes (Agilent Technologies). Sequencing templates were prepared using the Ion PI™ Hi-Q™ OT2 200 Kit on an Ion OneTouch 2 system and enriched on an Ion OneTouch ES system (Life Technologies). Sequencing was performed on the Ion Proton system using the Ion PI Chip v3 and the Ion PI™ Hi-Q™ Sequencing 200 Kit (Life Technologies) according to the manufacturers’ protocols.
In silico miRNA expression analysis
Sequence reads were analysed on the CLC Genomics Workbench v9.0 (http://www.clcbio.com/genomics/). Adapters were trimmed and sequence reads were counted. Annotation was done using default parameters by aligning counted reads to known Homo sapiens miRNA sequences from miRBase version 21 (25). Identified miRNAs with a read count below 10 were excluded because of possible sequencing errors. Expression was normalized to the total read count. Groups were compared and P-values were calculated using the β-binomial test (26). Correction for multiple testing was done using the false discovery rate procedure from Benjamini–Hochberg. A value of P < 0.05 was considered significant.
Real-time quantitative polymerase chain reaction
cDNA was generated from 400 ng small RNA using the miScript II RT Kit (Qiagen) according to the manufacturer’s protocol. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was performed in duplicate in a total reaction volume of 10 μl, containing 5 μl SYBR Green PCR Master Mix (Qiagen), 1 μl of cDNA, and 2 μl of RNAse-free water. For U6, 2 μl of a 5 μM forward and reverse primer solution was used. For the miRNAs, 1 μl of 5μM forward primer and 1μl of 10× miScript Universal Primer (Qiagen) were used. cDNA amplification was performed in the Rotor-Gene Q (Qiagen) with an initial activation step at 95°C for 15 minutes followed by 40 cycles of 94°C for 15 seconds, 55°C for 30 seconds, and 70°C for 30 seconds. The primers for U6, hsa-miR-505-3p, hsa-miR-181a-5p, hsa-miR-29c-5p, hsa-miR-92b-5p, hsa-miR-451a, and hsa-miR-3182 are shown in Table 1. Expression was calculated from the mean Ct values of the duplicates from each sample using the delta Ct method (2−ΔCt). Samples were repeated if the difference between the duplicates was greater than 0.5. U6 snRNA was used as the internal control. Groups were compared using a two sample t-test. A value of P < 0.05 was considered significant.
Table 1.
List of primers used. miRNA, microRNA.
| miRNA (hsa-) | Forward primer (5′–3′) | Reverse primer (5′ –3′) |
|---|---|---|
| U6 snRNA | GCTTCGGCAGCACATATACTAAAAT | CGCTTCACGAATTTGCGTGTCAT |
| miR-505-3p | CGT CAA CAC TTG CTG GTT TCC T | |
| miR-181a-5p | AAC ATT CAA CGC TGT CGG TGA GT | |
| miR-29c-5p | TGA CCG ATT TCT CCT GGT GTT C | |
| miR-92b-5p | AGG GAC GGG ACG CGG TGC AGT G | |
| miR-451a | AAA CCG TTA CCA TTA CTG AGT T | |
| miR-3182 | GCT TCT GTA GTG TAG TC | |
| mir-93-5p | CAAAGTGCTGTTCGTGCAGGTAG | |
| mir-18a-5p | TAAGGTGCATCTAGTGCAGATAG | |
| mir-92a-3p | TATTGCACTTGTCCCGGCCTGT | |
| mir-549a | TGACAACTATGGATGAGCTCT |
Results
The patients were clinically diagnosed by the multidisciplinary cleft palate craniofacial unit and clinical phenotype information about the patients and controls is shown in Table 2. Gender distribution within the sample represents male:female prevalence of nsCLP and nsCPO. The age of the controls is slightly higher due to the timing of the tonsillectomy.
Table 2.
Sample phenotype. BCLP, bilateral CLP; UCLP, unilateral CLP; UCLA, unilateral cleft lip and alveolus.
| Controls | nsCLP | nsCPO | |
|---|---|---|---|
| Mean age+/−SD | 24 ± 5 months | 18 months ± 2 months | 17 ± 2 months |
| Gender | 5 females | 1 female | 5 females |
| 5 males | 7 males | ||
| Phenotype | 2 BCLP complete | 3 complete | |
| 3 UCLP complete left | 2 soft palate | ||
| 2 UCLP complete right | |||
| 1 UCLA left with soft palate |
Expression of canonical miRNAs
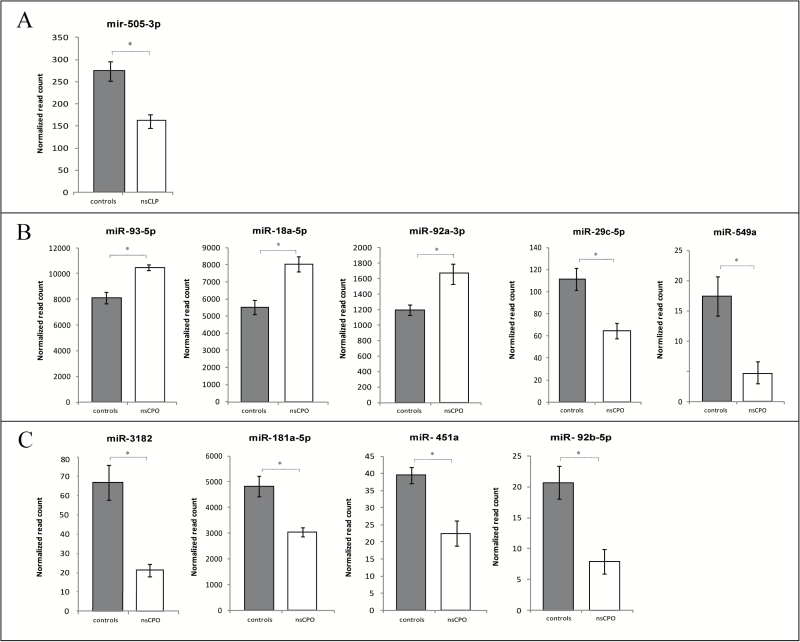
In control fibroblasts, nsCLP fibroblasts and nsCPO fibroblasts 264, 261, and 266 miRNAs were identified, respectively. By comparing the sequencing data of the three groups, a number of differentially expressed miRNAs were found. In nsCLP fibroblasts, 28 miRNAs were significantly down-regulated whereas 11 were up-regulated. Two had a fold change of more than 2 (hsa-miR-155-3p and hsa-miR-135b-5p) and four miRNAs of more than −2 (hsa-mir-505-5p, hsa-miR-145-5p, hsa-miR-380-5p, and hsa-miR-641-5p). After correction for multiple testing, only one miRNA was significantly down-regulated in nsCLP fibroblasts (hsa-miR-505-3p), as shown in Figure 1A. In nsCPO fibroblasts, 33 miRNAs were significantly down-regulated and 11 were up-regulated. Three had a fold change of more than 2 (hsa-miR-301a-5p, hsa-miR-155-3p, and hsa-miR-135-5p) and nine miRNAs had a fold change of more than −2 (hsa-miR-424-3p, hsa-miR-549a, hsa-miR-451a, hsa-miR-1537-3p, hsa-miR-641, hsa-miR-204-5p, hsa-miR-410-3p, hsa-miR-598-3p, and hsa-miR-29b-2-5p). After correction for multiple testing, three miRNAs were significantly up-regulated in nsCPO fibroblasts (hsa-miR-93-5p, hsa-miR-18a-5p, and hsa-miR-92a-3p) and two were down-regulated (hsa-miR-29c-5p and hsa-miR-549a), as shown in Figure 1B.
Figure 1.
Small RNA sequencing data. A = canonical miRNAs in nsCLP, B = canonical miRNAs in nsCPO, C = isomiRs in nsCPO. *P < 0.05. miRNA, microRNAs; nsCLP, non-syndromic cleft lip palate; nsCPO, non-syndromic cleft palate only.
Expression of isomiRs
Because a single miRNA locus can give rise to multiple distinct mature length variants (isomiRs), these were also analysed. Both 5′and 3′ isomiRs with either nucleotide addition or trimming were included. In nsCLP fibroblasts, an additional 28 isomiRs were significantly down-regulated and two were up-regulated, of which three had a fold change of more than −2 (hsa-miR-9-5p, hsa-miR-320b-3p, and hsa-miR-1-3p). After correction for multiple testing, none was significant. In nsCPO fibroblasts, an additional 3 isomiRs were significantly up-regulated and 20 were down-regulated, of which three had a fold change of more than −2 (hsa-miR-3182, hsa-miR-548c-5p, and hsa-miR-497-3p). After correction for multiple testing, four were significantly down-regulated (hsa-miR-3182, hsa-miR-181a-5p, hsa-miR-451a, and hsa-miR-92b-5p), as shown in Figure 1C.
Quantitative real-time PCR
The significantly differentially expressed miRNAs were analysed again using qRT-PCR. Only the canonical miRNAs were analysed. In all patient cells, down- or up-regulated expression was consistent with the RNA sequencing results. In nsCLP fibroblasts, hsa-miR-505-3p was significantly down-regulated, as shown in Table 3. However, it was down-regulated with a fold change of −5.79 compared to −1.49 in the RNA sequencing. In nsCPO fibroblasts, only hsa-miR-92a, hsa-miR-181a, and hsa-miR-451a were significantly different, as shown in Table 3. All had a larger fold change than in the RNA sequencing. No accurate melt curve or amplification signal for mir-3182 could be detected for the qPCR.
Table 3.
Fold change and P-values of differentially expressed miRNAs. CLO, cleft lip only; CLP, cleft lip palate; FDR, false discovery rate; miRNA, microRNA; qPCR, quantitative PCR.
| miRNA (hsa-) | Fold change (RNAseq) | P-value (FDR) | Fold change (qPCR) | P-value |
|---|---|---|---|---|
| CLP (n = 8) | ||||
| miR-505-3p | −1.49 | 0.001 | −5.79 | 0.004 |
| CPO (n = 5) | ||||
| miR-93-5p | 1.29 | 0.002 | 1.25 | 0.329 |
| miR-18a-5p | 1.46 | 0.011 | 1.47 | 0.214 |
| miR-92a-3p | 1.40 | 0.011 | 1.81 | 0.041 |
| miR-29c-5p | −1.73R | 0.031 | −2.19 | 0.062 |
| miR-549a | −3.77 | 0.041 | −2.24 | 0.082 |
| miR-3182 | −3.14 | 0.002 | ||
| miR-181a-5p | −1.59 | 0.022 | −2.59 | 0.027 |
| miR-451a | −1.76 | 0.031 | −3.53 | 0.029 |
| miR-92b-5p | −2.61 | 0.031 | −1.32 | 0.174 |
Discussion
We studied the expression of miRNAs in palatal fibroblasts from cleft palate patients and control subjects. NsCLP and nsCPO fibroblasts were analysed separately as both disorders are believed to have a different aetiology (2). Several miRNAs were found to be dysregulated in nsCPO while only one was dysregulated in nsCLP fibroblasts.
For nsCPO, only hsa-miR-18a, hsa-miR-92a, and hsa-miR-93 have been linked with cleft palate in mice (19), while 92a was the only one confirmed by both RNAseq and qRT-PCR. Furthermore, miR-93 has also shown to be stably methylated in murine secondary palate development (27). Both hsa-miR-18a and hsa-miR-92a belong to the miR-17-92 cluster located in the third intron of a ~7 kb primary transcript known as C13orf25 on human chromosome 13q31.3. It encodes six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92a-1) with highly conserved sequences and organization. Up-regulation in mouse palatal mesenchymal cells induces proliferation (28). Ancient gene duplications have given rise to two miR-17-92 cluster paralogs in mammals: the miR-106b-25 cluster (located on human chromosome 7) and the miR-106a-363 cluster (located on the X chromosome). Hsa-miR-93 is part of the miR-106b-25 cluster. Deletion of the cluster in mice, alone or in combination with its paralog cluster miR-106b-25, leads to several developmental defects, including cleft palate with greatly reduced cell proliferation in the palatal shelves (19). Our data, however, show that the three miRNAs are up-regulated in fibroblasts of nsCPO patients. MiRNAs of this cluster or its paralogs may, therefore, have a dose-dependent effect, similar to miR-140 (18). Perturbation of precise expression levels may, therefore, lead to disturbed proliferation in the palatal shelves and cleft palate.
The remaining miRNAs, of which only hsa-miR-92a, hsa-miR-181a, and hsa-miR-451a were confirmed by both RNAseq and qRT-PCR, have not been linked with cleft palate before. However, during E12–E14 of embryonic development in mice, several of the miRNAs are expressed in mouse palatal shelves in a specific temporal pattern (29). Only miR-29c, miR-3182, and miR-549a were not found in this microarray study. Hsa-miR-92b, hsa-miR-181a, hsa-miR-451a, and hsa-miR-29c have been studied in cancer, and regulate proliferation, migration, epithelial-to-mesenchymal transition, and apoptosis (30–39). These cellular processes are also crucial for palatogenesis and perturbation hereof may therefore lead to cleft palate. The functions and associations of hsa-mir-3182 and hsa-miR-549a have not been documented yet.
For nsCLP, hsa-miR-505 has not been linked with cleft palate. It does, however, suppress proliferation, regulate migration, and increase apoptosis in several cancer cell lines and endothelial cells (40–42). Dysregulation of hsa-miR-505 during palatogenesis may thus effect these cellular processes important for correct palate formation.
No previous miRNA expression study has been carried out in tissues or cells from human cleft palate patients and thus the results represent novel findings. Two studies from the same research group analysed miRNA expression in plasma of nsCL and nsCP patients (43, 44). Using a microarray screening method, 305 miRNAs in plasma of nsCL patients and 241 miRNAs in plasma of nsCP patients were differentially expressed (fold change ≥2). No overlap is seen with our identified differentially expressed miRNAs except for miR-17-3p which also belongs to the miR-17-92 cluster and is also up-regulated. However, it is unclear whether this is after correction for multiple testing. Furthermore, previous studies have shown that miRNAs have different functions and expression patterns depending on the tissue in which they are expressed (45). It is, therefore, more interesting to study miRNA expression in the developmentally relevant tissues. The palatal fibroblasts used in this study are derived from the palatal ectomesenchyme within the developing maxillary and medial nasal processes during embryonic development. Conditional disruption of miRNA expression in mice within either the palatal ectomesenchyme or ectoderm leads to cleft lip and or palate (46–50). As such, correct and specific miRNA expression is necessary in both ectomesenchyme and ectoderm-derived palatal cells for normal palate development. As we limited our analysis to fibroblasts, it will be interesting to include keratinocytes in future studies.
Due to ethical constraints, the earliest time point we could analyse miRNA expression was during surgical primary palate closure. Furthermore, the study was performed in cultured fibroblasts and not directly in the tissue samples, because of the small biopsy size. MiRNA expression can be affected by components in the culture medium (e.g. growth factors) (51) and specific signalling pathways during palatogenesis, as highlighted by the specific temporal expression pattern (29, 52). Therefore, our study likely only detected miRNAs whose expression level is changed by cell-autonomous genetic and epigenetic differences related to clefting.
In summary, we used a bias-free approach by performing a whole-transcriptome miRNA expression profiling method. RNA sequencing was used instead of microarray analysis because it is more accurate, absolute miRNA abundance levels are analysed, and it is more sensitive (53). Furthermore, when new miRNAs are annotated in the future, our data set can be re-analysed. The possibility of RNA sequencing to detect splice variants has allowed us to get the additional benefit of analysing isomiRs. These heterogeneous miRNA variants have been widely detected in humans and cooperate to target common biological pathways (54, 55). Including isomiRs, thus provides a more comprehensive analysis and allowed identification of four or more differentially expressed miRNAs (hsa-miR-3182, hsa-miR-181a, hsa-miR-451a, and hsa-miR-92b) in nsCLP fibroblasts. Only the canonical sequences were used for qRT-PCR, which may be the reason that we found only few significant miRNAs. RNA sequencing and qRT-PCR results within same samples sometimes shows a high similarity, but other studies have also found discrepancies (56). In general, both techniques are considered to have a similar accuracy.
Conclusion
This study analysed miRNA expression in palatal fibroblasts from non-syndromic cleft palate patients compared to controls. We found that several miRNAs are significantly up- or down-regulated in cells from patients. Some of these have already been shown to be involved in palatogenesis in animal models indicating a possible biological significant role in humans. The previously unknown cleft palate-associated miRNAs we have identified should be further investigated to confirm their biological significance. Although this study has a quite small sample size, it provides an indication that miRNAs may be dysregulated in fibroblasts of patients with non-syndromic cleft palate. Larger scale genomic expression and functional studies are needed to validate these findings. Although, these findings may not have a direct impact on clinical management, they will improve our understanding of the aetiology of clefting.
Conflict of interest
None to declare.
Acknowledgements
This work was supported by a research grant from the European Orthodontic Society.
References
- 1. Meng L., Bian Z., Torensma R., Von den Hoff J. W. (2009)Biological mechanisms in palatogenesis and cleft palate. Journal of Dental Research, 88, 22–33. [DOI] [PubMed] [Google Scholar]
- 2. Dixon M. J., Marazita M. L., Beaty T. H., Murray J. C. (2011)Cleft lip and palate: understanding genetic and environmental influences. Nature Reviews Genetics, 12, 167–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Böhmer A. C., et al. (2013)Analysis of susceptibility loci for nonsyndromic orofacial clefting in a European trio sample. American Journal of Medical Genetics Part A, 161A, 2545–2549. [DOI] [PubMed] [Google Scholar]
- 4. Khandelwal K. D., van Bokhoven H., Roscioli T., Carels C. E., Zhou H. (2013)Genomic approaches for studying craniofacial disorders. American Journal of Medical Genetics Part C: Seminars in Medical Genetics, 163C, 218–231. [DOI] [PubMed] [Google Scholar]
- 5. Manolio T. A., et al. (2009)Finding the missing heritability of complex diseases. Nature, 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Consortium EP. (2012)An integrated encyclopedia of DNA elements in the human genome. Nature, 489, 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sayed D., Abdellatif M. (2011)MicroRNAs in development and disease. Physiological Reviews, 91, 827–887. [DOI] [PubMed] [Google Scholar]
- 8. Bartel D. P. (2009)MicroRNAs: target recognition and regulatory functions. Cell, 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008)Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight?Nature Reviews Genetics, 9, 102–114. [DOI] [PubMed] [Google Scholar]
- 10. Kozomara A., Griffiths-Jones S. (2014)miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Research, 42, D68–D73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mathieu J., Ruohola-Baker H. (2013)Regulation of stem cell populations by microRNAs. Advances in Experimental Medicine and Biology, 786, 329–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harfe B. D., McManus M. T., Mansfield J. H., Hornstein E., Tabin C. J. (2005)The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proceedings of the National Academy of Sciences of the USA, 102, 10898–10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oommen S., et al. (2012)Distinct roles of microRNAs in epithelium and mesenchyme during tooth development. Developmental Dynamics, 241, 1465–1472. [DOI] [PubMed] [Google Scholar]
- 14. Pauli A., Rinn J. L., Schier A. F. (2011)Non-coding RNAs as regulators of embryogenesis. Nature Reviews Genetics, 12, 136–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yi R., et al. (2006)Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nature Genetics, 38, 356–362. [DOI] [PubMed] [Google Scholar]
- 16. Cammaerts S., Strazisar M., De Rijk P., Del Favero J. (2015)Genetic variants in microRNA genes: impact on microRNA expression, function, and disease. Frontiers in Genetics, 6, 186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meola N., Gennarino V. A., Banfi S. (2009)MicroRNAs and genetic diseases. PathoGenetics, 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Eberhart J. K., et al. (2008)MicroRNA Mirn140 modulates Pdgf signaling during palatogenesis. Nature Genetics, 40, 290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang J., et al. (2013)MicroRNA-17-92, a direct Ap-2α transcriptional target, modulates T-box factor activity in orofacial clefting. PLoS Genetics, 9, e1003785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li L., Meng T., Jia Z., Zhu G., Shi B. (2010)Single nucleotide polymorphism associated with nonsyndromic cleft palate influences the processing of miR-140. American Journal of Medical Genetics Part A, 152A, 856–862. [DOI] [PubMed] [Google Scholar]
- 21. Rattanasopha S., Tongkobpetch S., Srichomthong C., Siriwan P., Suphapeetiporn K., Shotelersuk V. (2012)PDGFRa mutations in humans with isolated cleft palate. European Journal of Human Genetics, 20, 1058–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma L., et al. (2014)A miRNA-binding-site SNP of MSX1 is associated with NSOC susceptibility. Journal of Dental Research, 93, 559–564. [DOI] [PubMed] [Google Scholar]
- 23. Li D., et al. (2016)Associations between microRNA binding site SNPs in FGFs and FGFRs and the risk of non-syndromic orofacial cleft. Scientific Reports, 6, 31054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu J., et al. (2008)Cleft palate cells can regenerate a palatal mucosa in vitro. Journal of Dental Research, 87, 788–792. [DOI] [PubMed] [Google Scholar]
- 25. Griffiths-Jones S. (2004)The microRNA registry. Nucleic Acids Research, 32, D109–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baggerly K. A., Deng L., Morris J. S., Aldaz C. M. (2003)Differential expression in SAGE: accounting for normal between-library variation. Bioinformatics, 19, 1477–1483. [DOI] [PubMed] [Google Scholar]
- 27. Seelan R. S., et al. (2014)Methylated microRNA genes of the developing murine palate. MicroRNA, 3, 160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li L., Shi J. Y., Zhu G. Q., Shi B. (2012)MiR-17-92 cluster regulates cell proliferation and collagen synthesis by targeting TGFB pathway in mouse palatal mesenchymal cells. Journal of Cellular Biochemistry, 113, 1235–1244. [DOI] [PubMed] [Google Scholar]
- 29. Mukhopadhyay P., Brock G., Pihur V., Webb C., Pisano M. M., Greene R. M. (2010)Developmental microRNA expression profiling of murine embryonic orofacial tissue. Birth Defects Research Part A: Clinical and Molecular Teratology, 88, 511–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Babapoor S., Fleming E., Wu R., Dadras S. S. (2014)A novel miR-451a isomiR, associated with amelanotypic phenotype, acts as a tumor suppressor in melanoma by retarding cell migration and invasion. PlOS ONE, 9, e107502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen B., et al. (2016)Inhibition of miR-29c promotes proliferation, and inhibits apoptosis and differentiation in P19 embryonic carcinoma cells. Molecular Medicine Reports, 13, 2527–2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Han P., et al. (2017)The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Molecular Cancer, 16, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu Z., et al. (2015)MicroRNA-92b promotes tumor growth and activation of NF-κB signaling via regulation of NLK in oral squamous cell carcinoma. Oncology Reports, 34, 2961–2968. [DOI] [PubMed] [Google Scholar]
- 34. Liu Z., et al. (2015)miR-451a Inhibited cell proliferation and enhanced tamoxifen sensitive in breast cancer via macrophage migration inhibitory factor. BioMed Research International, 2015, 207684. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35. Song H., et al. (2016)miR-92b regulates glioma cells proliferation, migration, invasion, and apoptosis via PTEN/Akt signaling pathway. Journal of Physiology and Biochemistry, 72, 201–211. [DOI] [PubMed] [Google Scholar]
- 36. Tian Y., et al. (2017)HBx promotes cell proliferation by disturbing the cross-talk between miR-181a and PTEN. Scientific Reports, 7, 40089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang H. W., et al. (2016)A regulatory loop involving miR-29c and Sp1 elevates the TGF-β1 mediated epithelial-to-mesenchymal transition in lung cancer. Oncotarget, 7, 85905–85916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhou Z., Wang Z., Wei H., Wu S., Wang X., Xiao J. (2016)Promotion of tumour proliferation, migration and invasion by miR-92b in targeting RECK in osteosarcoma. Clinical Science, 130, 921–930. [DOI] [PubMed] [Google Scholar]
- 39. Zhuang L. K., et al. (2016)MicroRNA-92b promotes hepatocellular carcinoma progression by targeting Smad7 and is mediated by long non-coding RNA XIST. Cell Death & Disease, 7, e2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen S., Sun K. X., Liu B. L., Zong Z. H., Zhao Y. (2016) MicroRNA-505 functions as a tumor suppressor in endometrial cancer by targeting TGF-α. Molecular Cancer, 15, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu L., Qiu C., Li D., Bai G., Liang J., Yang Q. (2016)MicroRNA-505 suppresses proliferation and invasion in hepatoma cells by directly targeting high-mobility group Box 1. Life Sciences, 157, 12–18. [DOI] [PubMed] [Google Scholar]
- 42. Yang Q., et al. (2014)MicroRNA-505 identified from patients with essential hypertension impairs endothelial cell migration and tube formation. International Journal of Cardiology, 177, 925–934. [DOI] [PubMed] [Google Scholar]
- 43. Zou J., Li J., Li J., Ji C., Li Q., Guo X. (2016)Expression profile of plasma microRNAs in nonsyndromic cleft lip and their clinical significance as biomarkers. Biomedicine & Pharmacotherapy, 82, 459–466. [DOI] [PubMed] [Google Scholar]
- 44. Li J., et al. (2016)Assessment of differentially expressed plasma microRNAs in nonsyndromic cleft palate and nonsyndromic cleft lip with cleft palate. Oncotarget, 7, 86266–86279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Spruce T., et al. (2010)An early developmental role for miRNAs in the maintenance of extraembryonic stem cells in the mouse embryo. Developmental Cell, 19, 207–219. [DOI] [PubMed] [Google Scholar]
- 46. Cao H., et al. (2010)MicroRNAs play a critical role in tooth development. Journal of Dental Research, 89, 779–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sheehy N. T., Cordes K. R., White M. P., Ivey K. N., Srivastava D. (2010)The neural crest-enriched microRNA miR-452 regulates epithelial-mesenchymal signaling in the first pharyngeal arch. Development, 137, 4307–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zehir A., Hua L. L., Maska E. L., Morikawa Y., Cserjesi P. (2010)Dicer is required for survival of differentiating neural crest cells. Developmental Biology, 340, 459–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nie X., Wang Q., Jiao K. (2011)Dicer activity in neural crest cells is essential for craniofacial organogenesis and pharyngeal arch artery morphogenesis. Mechanisms of Development, 128, 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Barritt L. C., et al. (2012)Conditional deletion of the human ortholog gene Dicer1 in Pax2-Cre expression domain impairs orofacial development. Indian Journal of Human Genetics, 18, 310–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ikari J., et al. (2015)Effect of culture conditions on microRNA expression in primary adult control and COPD lung fibroblasts in vitro. In vitro Cellular & Developmental Biology - Animal, 51, 390–399. [DOI] [PubMed] [Google Scholar]
- 52. Warner D. R., Mukhopadhyay P., Brock G., Webb C. L., Michele Pisano M., Greene R. M. (2014)MicroRNA expression profiling of the developing murine upper lip. Development, Growth & Differentiation, 56, 434–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marguerat S., Bähler J. (2010)RNA-seq: from technology to biology. Cellular and Molecular Life Sciences, 67, 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Cloonan N., et al. (2011)MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biology, 12, R126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee L. W., et al. (2010)Complexity of the microRNA repertoire revealed by next-generation sequencing. RNA, 16, 2170–2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Git A., et al. (2010)Systematic comparison of microarray profiling, real-time PCR, and next-generation sequencing technologies for measuring differential microRNA expression. RNA, 16, 991–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]