Abstract
Cyanobacterial natural products offer new possibilities for drugs and lead compounds but many factors can inhibit the production of sufficient yields for pharmaceutical processes. While Escherichia coli and Streptomyces sp. have been used as heterologous expression hosts to produce cyanobacterial natural products, they have not met with resounding success largely due to their inability to recognize cyanobacterial promoter regions. Recent work has shown that the filamentous freshwater cyanobacterium Anabaena sp. strain PCC 7120 recognizes various cyanobacterial promoter regions and can produce lyngbyatoxin A from the native promoter. Introduction of Anabaena sigma factors into E. coli might allow the native transcriptional machinery to recognize cyanobacterial promoters. Here, all 12 Anabaena sigma factors were expressed in E. coli and subsets were found to initiate transcription from several cyanobacterial promoters based on transcriptional fusions to the chloramphenicol acetyltransferase (CAT) reporter. Expression of individual Anabaena sigma factors in E. coli did not result in lyngbyatoxin A production from its native cyanobacterial gene cluster, possibly hindered by deficiencies in recognition of cyanobacterial ribosomal binding sites by native E. coli translational machinery. This represents an important step toward engineering E. coli into a general heterologous expression host for cyanobacterial biosynthetic gene cluster expression.
Keywords: Cyanobacteria, secondary metabolites, Anabaena sp. strain PCC 7120, promoter
Expression of cyanobacterial sigma factors in Escherichia coli.
ABBREVIATIONS
- BGC
biosynthetic gene clusters
- CAT
chloramphenicol acetyltransferase
- DNA
deoxyribonucleic acid
- ILV
indolactam V
- Km
kanamycin
- LB
Lysogeny broth
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- LTXA
lyngbyatoxin A
- NMVT
N-methyl-L-valyl-L-tryptophanol
- NRPS
nonribosomal peptide synthetase
- PKS
polyketide synthase
- RiPP
ribosomal peptide natural products
- RNA
ribonucleic acid
- Sp
spectinomycin
- UV-vis
Ultraviolet-visible
INTRODUCTION
Facing a scarcity of new anti-infective drugs entering the market in an era of increasing microbial drug resistance, pharmaceutical discovery efforts have recently returned to screening microbial sources for bioactive compounds, especially within marine environments (Cooper and Shlaes 2011: 32; Gerwick and Fenner 2013: 800–6; Newman and Cragg 2016: 629–61; Singh, et al.2011: 401–12). Within this effort, cyanobacterial natural products represent a remarkably underexplored and rich source of novel, complex secondary metabolites including terpenoids, alkaloids, polyketides and nonribosomal peptides with antibacterial, anticancer, antifungal, antiviral, antiprotozoal, molluscicidal and protease inhibition activities (Calteau et al.2014: 977; Ehrenreich, Waterbury and Webb 2005: 7401–13; Niedermeyer 2015: 1309–25; Pattanaik and Lindberg 2015: 269–93; Tan 2007: 954–79; Welker and Von Döhren 2006: 530–63). Many of these compounds exhibit potent and varied biological activities and serve as appealing drug leads (Tan 2007: 954–79). Although hundreds of cyanobacterial bioactive metabolites have been isolated in adequate yields for initial characterization, drug discovery and development efforts have not been pursued for a variety of practical impediments (Gerwick and Fenner 2013: 800–6; Tan 2007: 954–79). Characterization sufficient for marketing, biological trials and production of synthetic structural analogues require compounds in much greater yields than what can be isolated from field collections. Filamentous cyanobacterial strains known to produce natural products can be slow-growing, with doubling times ranging from 12 h to multiple days. Axenic isolation and identification of compound-producing strains can be challenging, and even when axenic strains can be isolated, they may cease to produce the desired compound during long-term culture (Vestola et al.2014: E1909–E17). Moreover, genetic engineering and synthetic biology approaches are often prohibitory because producing strains are commonly genetically intractable. A viable solution to bypass the aforementioned problems is to use a heterologous expression host to produce cyanobacterial natural products.
Attempts at heterologous expression of cyanobacterial natural product biosynthetic gene clusters (BGCs) in bacterial hosts have encountered mixed success. Using an Escherichia coli host, the cyanobacterial ribosomal peptide natural products (RiPPs) patellamides A and C (Long et al.2005: 1760–5; Schmidt et al.2005: 7315–20) and the microviridins (Ziemert et al.2008: 7756–9) were produced from their native promoters. Yields for the patellamides were low at approximately 20 μg/L (Schmidt et al.2005: 7315–20), but the microviridins were produced at titers in the range of the native producers, up to 7280 μg/L (Ziemert et al.2008: 7756–9). In the case of the microviridins, the presence of multiple incorrectly processed analogues was observed in addition to the expected products. The expression of more complex natural products from nonribosomal peptide synthetase (NRPS) and polyketide synthase (PKS) pathways has encountered minimal success. An attempt at expressing the NRPS/PKS hybrid barbamide A in the actinobacterium Streptomyces venezuelae led to the production of 4-O-demethylbarbamide A, a closely related product (Kim et al.2012: 5824–7). Yields of this product were very low (<1 μg/L), possibly due to differences in codon usage between cyanobacterial DNA compared to the high % GC content of actinobacteria (∼45% G + C vs. ∼70% G + C, respectively). Analogously, attempts to produce lyngbyatoxin A (LTXA) in Streptomyces coelicolor A3(2) proved unsuccessful, likely due to premature transcript termination within ltxA, the first gene in the BGC (Jones et al.2012: 1243–51). This termination may also have been a result of the large difference in the G + C % between the producing cyanobacterium and S. coelicolor. In contrast, LTXA was successfully produced in E. coli in high yield (25.6 mg/L) using promoter exchange, as the native cyanobacterial promoter(s) were not recognized by the E. coli host (Ongley et al.2013: 1888–93). Taking a different approach, the genetically tractable freshwater filamentous heterocystous cyanobacterium Anabaena sp. strain PCC 7120 (hereafter, Anabaena) was recently assessed as a host for cyanobacterial natural product expression (Videau et al.2016: 978–88). Successful production of LTXA was achieved when expressed from the native BCG promoters at titers comparable to the native producer, Moorea producens, at 174.9 ng per mg of dry cell mass. To show its general utility as a heterologous host, several promoters from cyanobacterial BGCs present in unicellular and filamentous marine cyanobacteria were recognized and expressed by the Anabaena transcriptional machinery (Videau et al.2016: 978–88). In this report, we detail our attempts to engineer E. coli as a general heterologous host for the expression of cyanobacterial natural products by heterologously expressing sigma factors from Anabaena.
Promoter recognition is governed by sigma factors, which form complexes with RNA polymerase to initiate specific transcription from a promoter (Feklístov et al.2014: 357–76). In our previous experiments, Anabaena was able to initiate transcription from promoter regions from other cyanobacterial BGCs (Videau et al.2016: 978–88). Anabaena encodes 12 sigma factors in its genome, which show low amino acid identity to those of E. coli (26%–59%, Table 1) (Imamura and Asayama 2009: 65–87; Kaneko et al.2001: 205–13). The Anabaena sigma factors are organized into four groups: Group 1 sigma factors (also known as principal sigma factors) are essential for expression of constitutive genes and cell viability; Group 2 sigma factors are structurally similar to group 1, but not required for cell viability; Group 3 sigma factors are structurally different from the other groups and are expressed during stress responses; Group 4 sigma factors are known as extracytoplasmic function (ECF) sigma factors. Previous research on Anabaena sigma factors has primarily focused on their roles in heterocyst differentiation, the production of a secondary cell type utilized for nitrogen fixation (Muro-Pastor and Hess 2012: 548–57). The Group 2 sigma factors sigC (all1692) and sigE (alr4249) are upregulated in developing heterocyst cells and are required for proper nitrogen fixation while sigG (alr3280) is most likely involved in envelope stress given the similarity of Alr3280 to SigG from Nostoc punctiforme ATCC 29133 (Bell, Lee and Summers 2017: 179–94). Each of these sigma factors has a role in regulating expression of heterocyst-specific genes required for differentiation and heterocyst function (Aldea, Mella-Herrera and Golden 2007: 8392–6; Ehira and Miyazaki 2015: 587–603; Khudyakov and Golden 2001: 6667–75; Mella-Herrera et al.2011: 1823–32). Additional studies have shown that the Group 3 sigma factor SigJ (alr0277) is required for resistance to high light and desiccation conditions in Anabaena (Srivastava et al.2017: 287–97; Yoshimura et al.2007: 13–24).
Table 1.
Anabaena sigma factors used in this study. Percent amino acid identity/similarity compared to E. coli K-12 substr. MG1655 as determined by BLAST analysis (blastp algorithm). The E. coli sigma factor used for comparison is listed in parentheses. *Identified using PSI-BLAST. Sigma factor adapted from Bell, Lee and Summers (2017).
| Sigma factor | Group | Mass (kDa) | % Amino acid identity/similarity to E. coli σ factors |
|---|---|---|---|
| All5263 (SigA) | 1 | 46.7 | 59/77 (RpoD) |
| All7615 (SigB) | 2 | 38.4 | 44/68 (RpoD) |
| Alr3800 (SigB2) | 37.9 | 46/73 (RpoD) | |
| All7608 (SigB3) | 38.0 | 41/69 (RpoD) | |
| All7179 (SigB4) | 36.4 | 42/68 (RpoD) | |
| All1692 (SigC) | 47.4 | 48/71 (RpoD) | |
| Alr3810 (SigD) | 37.6 | 46/69 (RpoD) | |
| Alr4249 (SigE) | 45.4 | 45/69 (RpoD) | |
| All3853 (SigF) | 3 | 30.0 | 26/50 (RpoD) |
| Alr0277 (SigJ) | 30.0 | 28/48 (FliA) | |
| Alr3280 (SigG) | 4 | 25.2 | 34/52 (RpoE) |
| All2193 (SigI) | 22.6 | 26/44 (RpoE)* |
Due to complications arising from codon usage in Streptomyces sp. and the lack of broad recognition and transcription from native cyanobacterial promoters in E. coli, neither bacterium is a suitable general host for cyanobacterial natural product expression (Jones et al.2012: 1243–51; Kim et al.2012: 5824–7; Ongley et al.2013: 1888–93). Because promoter exchange has facilitated the expression of some cyanobacterial natural products in E. coli strains created for heterologous expression (specifically those with activating enzymes like 4’- phosphopantetheinyl transferases integrated into their genomes, e. g., E. coli strain BAP1 (Pfeifer et al.2001: 1790–2)), generating an E. coli strain with the ability to recognize a wide range of cyanobacterial promoters would allow it to serve as a more general expression host. Predicated on the success of Anabaena in recognizing native promoters from diverse cyanobacterial BGCs (Videau et al.2016: 978–88), here we individually overexpressed the 12 sigma factors from Anabaena in E. coli strain BAP1 and assessed the ability of each to recognize various cyanobacterial promoters. This represents an important step toward developing E. coli into a fast-growing, genetically tractable host for the expression of cyanobacterial BGCs.
MATERIALS AND METHODS
General bacterial methods
All E. coli strains were routinely grown in liquid Lysogeny Broth, Miller (LB) or on plates solidified with 1.5% agar. Cultures were incubated at 37°C and shaken at 200 rpm unless otherwise stated. For plasmid selection and maintenance in E. coli, kanamycin (Km) and spectinomycin (Sp) were used at final concentrations of 50 μg mL−1 and 100 μg mL−1, respectively. Cultures were supplemented with a final concentration of 20 mM glucose to repress spurious protein expression and 100 μM isopropyl β-D-1-thiogalactopyranoside (IPTG) to induce protein expression. All UV-vis spectroscopy was performed using a BioSpectrometer kinetic (Eppendorf, Hauppauge, NY).
Plasmid construction
All plasmids and primers used in this study are listed in Tables S1 and S2 (Supporting Information), respectively. Primestar GXL polymerase (CloneTech; Mountain View, CA) was used for PCR per the manufacturer's instructions. Sanger sequencing was performed at the Center for Genome Resources and Biocomputing at Oregon State University (Corvallis, OR). Plasmids pJEE002, pJEE003, pJEE004, pBJP0041, pBJP0043, pBJP0045, pBJP0047, pBJP0049, pBJP0051, pBJP0053, pBJP0055 and pBJP0057 are plasmids based on pCOLADuet-1 (EMD Millipore; Burlington, MA) for expressing the Anabaena polyhistidine epitope-tagged sigma factors alr3800, alr3280, all5263, alr0277, all1692, all2193, alr3810, all3853, alr4249, all7179, all7608 and all7615, respectively. The coding regions of alr3800, alr3280, all5263, alr0277, all1692, alr3810, all3853, all7179 and all7608 were amplified by PCR from Anabaena genomic DNA using the primer pairs 3800-BamHI-Fwd/3800-HindIII-Rev, 3280-BamHI-Fwd/3280-HindIII-Rev, 5263-BamHI-Fwd/5263-HindIII-Rev, alr0277-BamHI-Fwd/alr0277-HindIII-Rev, all1692-BamHI-Fwd/all1692-HindIII-Rev, alr3810-BamHI-Fwd/alr3810-HindIII-Rev, all3853-BamHI-Fwd/all3853-HindIII-Rev, all7179-BamHI-Fwd/all7179-HindIII-Rev and all7608-BamHI-Fwd/all7608-HindIII-Rev, respectively. The PCR products were digested with BamHI and HindIII and cloned into the same sites in pCOLADuet-1 to create pJEE0002, pJEE0003, pJEE0004, pBJP0041, pBJP0043, pBJP0047, pBJP0049, pBJP0053 and pBJP0055, respectively. The coding regions of alr4249, and all7615 were amplified by PCR from Anabaena genomic DNA using the primer pairs alr4249-BamHI-Fwd/alr4249-SalI-Rev, and all7615-BamHI-Fwd/all7615-SalI-Rev. The PCR products were digested with BamHI and SalI and cloned into the same sites in pCOLADuet-1 to create pBJP0051 and pBJP0057. The coding region of all2193 was amplified by PCR from Anabaena genomic DNA using the primer pair all2913-BamHI-Fwd/all2913-PstI-Rev, digested with BamHI and PstI, and cloned into the same sites in pCOLADuet-1 to yield pBJP0045.
Plasmid pKNW132 is a mobilizable shuttle vector based on pPJAV361 (Videau et al.2016: 978–88) containing the full −1 to −1000 of PltxA fused to the start codon of the cat gene. PltxA was amplified by PCR from pPJAV562 (Videau et al.2016: 978–88) using the primer pair PltxA-XhoI-F and KNWPltxA-CAT-R and cat was amplified from pPJAV562 using the primer pair KNWCAT-F and CAT-SacI-R. The products were fused by overlap extension PCR (Higuchi, Krummel and Saiki 1988: 7351–67), cloned into the SmaI site of pPJAV361, and directionality was verified by PCR to create pKNW132.
In vitro chloramphenicol acetyltransferase (CAT) assay
Each of the sigma factor expression plasmids (pJEE002, pJEE003, pJEE004, pBJP0041, pBJP0043, pBJP0045, pBJP0047, pBJP0049, pBJP0051, pBJP0053, pBJP0055 and pBJP0057) was transformed into chemically competent E. coli BAP1 cells containing pPJAV562, pPJAV575, pPJAV576, pPJAV577, pPJAV578 or pKNW132. Positive transformants were selected on LB agar plates containing Km, Sp and glucose (1% w/v) and grown at 37°C overnight. Colonies were inoculated into 2 mL LB media supplemented with Km, Sp and glucose (20 mM) and grown at 37°C for 14–16 h. Cultures were then diluted 1:100 in 2 mL of LB supplemented with Km, Sp and glucose (20 mM) and grown at 37°C at 200 rpm for approximately 3.5 h until the OD600 reached ∼0.6. The cultures were equilibrated at 28°C for 15 min and then IPTG was added to a final concentration of 0.1 mM to induce sigma factor protein expression, and all cultures were grown at 28°C for an additional 16 h at 200 rpm.
The liquid cultures of E. coli BAP1 from the above paragraph were stored at 4°C to prevent changes in CAT abundance, and the cells were pelleted by centrifugation at 15,000 × g for 1 min and stored at −20°C until use. The cell pellets were resuspended in 300 μL of 100 mM Tris base (pH 7.8). Cell suspensions were sonicated on ice with 2 pulses at 30% amplitude for 10 s each using a Qsonica Q55 sonicator and centrifuged at 21,000 × g for 20 min at 4°C. The protein concentration of the supernatant was determined by measuring the OD280 with a BioSpectrometer kinetic using a G1.0 microcuvette. Supernatants were diluted to a final concentration of 2 μg protein/μL in cold 100 mM Tris base (pH 7.8) and 5 μL was placed in the appropriate wells of a 96-well polystyrene clear bottom plate (Costar 3370, Corning, Tewksbury, MA) followed by a brief, 15 s centrifugation step. An assay master mix containing 8.5 mL 100 mM Tris (pH 7.8), 280 μL Acetyl CoA (5 mM), 280 μL 2.5 mM 5 5'-Dithiobis-2-nitrobenzoic acid, 140 μL chloramphenicol (0.3% w/v) was assembled and mixed by a brief vortex. A portion of the master mix (105 μL) was aliquoted into each well of a 96-well polypropylene V-bottom plate (Costar 3363, Corning). The plates were placed in a Sciclone ALH3000 workstation (Perkin Elmer, Waltham, MA) and 95 μL of the master mix was transferred from the V-bottom plate to the clear bottom 96-well plate containing the diluted E. coli supernatant and the resulting solution was mixed by gentle pipetting three times. The clear bottom plate was then moved to a Synergy 4 (BioTek, Winooski, VT) preequilibrated at 25°C. Each well was read for absorbance at 412 nm every 30 s for a total of 30 min. The experiments were performed in biological triplicate and technical duplicate. The data were then analyzed using the GraphPad Prism software (GraphPad, La Jolla, CA). CAT activity in units was determined by comparison to a standard curve created using varying activities of purified CAT (Sigma-Aldrich, St. Louis, MO).
Attempted production and extraction of LTXA in E. coli
Either pCOLADuet-1 or sigma factor containing plasmids were individually co-transformed into chemically competent E. coli BAP1 cells with the plasmid pPJAV500 harboring PltxA-ltxABC (Videau et al.2016: 978–88). Positive colonies were selected and overnight cultures grown as described above. 50 μL of the overnight cultures was inoculated into three 250 mL Erlenmeyer flasks containing either 50 mL of LB media, Terrific Broth (TB) media or M9 minimal media supplemented with Km and Sp. The cultures were grown at 37°C to an OD600 of 0.6–0.8 (approximately 4–7 h) and then IPTG was added to a final concentration of 0.1 mM. The cultures were grown for an additional 1 h at 28°C followed by growth at 18°C for 24 h. Half of each culture (25 mL) was transferred to a 50-mL conical tube and pelleted at 6000 × g at 4°C for 10 min. The supernatant was aspirated and the pellet frozen at −80°C until use. The remaining half of the cultures continued to grow at 18°C for an additional 48 h and were then harvested as above. Frozen pellets were defrosted on ice, resuspended in 10 mL of methanol, and sonicated in a Branson 3510 sonicator for 10 min followed by 10 min on ice, six times. Cell debris was cleared by centrifugation at 6000 × g for 10 min at 4°C and the supernatant was concentrated in vacuo. Extracts were dissolved in methanol at a concentration of 10 mg mL−1, particulates were removed by filtration through a 0.2 μm nylon syringe filter, and 10 μL was injected for analysis by LC-MS/MS as previously described (Videau et al.2016: 978–88).
Western blot analysis
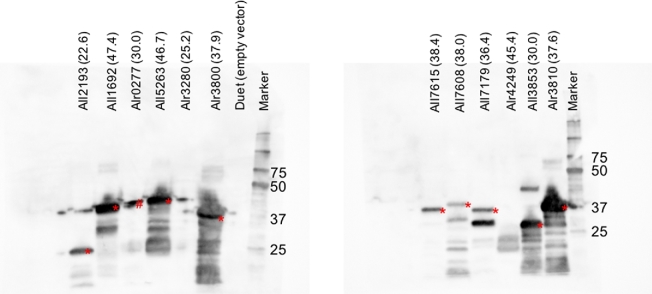
The pre-dilution supernatants of E. coli BAP1 cells harboring expression plasmids containing each of the 12 sigma factors or the empty control vector (pCOLADuet-1) and PltxA-cat (pPJAV562) from the paragraph entitled ‘In vitro chloramphenicol acetyltransferase (CAT) assay’ were immediately frozen at −20°C. The supernatants were then defrosted on ice and diluted to a final protein concentration of 7.7 mg/mL (Fig. 3) or 5.5 mg/mL (Fig. S1, Supporting Information) and 30 μL was mixed with 10 μL 4x SDS loading buffer (Sambrook and Russell 2001: 2344). Aliquots (30 μL) of E. coli lysates containing 173 μg (Fig. 3) or 123 μg (Fig. S3, Supporting Information) of protein were subjected to resolution via SDS-PAGE electrophoresis using a precast miniProtean TGX 4%–15% gradient gel (Bio-Rad, Hercules, CA) followed by electrophoretic transfer onto an Immobilon P polyvinylidene difluoride membrane (EMD Millipore). Polyhistidine epitope-tagged sigma factors were detected with primary mouse anti-histidine tag antibodies (Bio-Rad) and secondary goat anti-mouse horseradish peroxidase (Bio-Rad) conjugated antibodies followed by chemiluminescent detection (Clarity Western ECL Substrate, Bio-Rad) and imaging with a ChemiDoc MP (Bio-Rad).
Figure 3.
Western blots of Anabaena sigma factors expressed from pCOLADuet-1 at 28°C in the soluble fraction of E. coli BAP1 cell lysate. The name of the sigma factor expressed is listed above each lane and the number in parentheses indicates the calculated molecular weight for each protein in kDa. Expected bands are denoted by an red asterisk (*). The bands denoted by a red pound sign (#) are due to overflow from the neighboring lane. The numbers to the right of the lane labeled ‘Marker’ indicate the size of the Precision Prestained Protein Ladder (Bio-Rad) in kDa. The data presented here are from one representative experiment.
Statistical analyses
All data were analyzed using the GraphPad Prism software (GraphPad; La Jolla, CA). Unpaired, two-tailed t-tests were performed between groups. Cutoffs for significance were set at P ≤ 0.0001. Graphs were made using Microsoft Excel.
RESULTS AND DISCUSSION
An ideal heterologous expression host is a strain that grows quickly, is genetically tractable and requires minimal genetic modification of native BGCs for expression. While E. coli grows quickly and has many tools available for genetic manipulation, promoters from cyanobacterial BGCs are only weakly recognized and high-level expression of BGCs typically requires promoter exchange. Promoter exchange is possible for BGCs containing only a few genes but is prohibitive for large BGCs that may contain internal or divergent promoters. Rather than map all promoters in every desirable BGC and exchange them, it would be far simpler to express sigma factors that recognize the promoters and initiate transcription in E. coli. As previous work in Anabaena showed that its transcriptional machinery recognizes promoters from diverse cyanobacterial BGCs, the 12 sigma factors (Table 1) were cloned into an E. coli expression vector (pCOLADuet-1) and assessed for their ability to activate transcription from a cyanobacterial promoter region. We focused our initial investigation on the ltxA promoter (PltxA), which we defined as the 1000 bp upstream of the translational start site of ltxA, the first gene in the lyngbyatoxin BGC (Edwards and Gerwick 2004: 11432–3). The E. coli BAP1 strain (a BL21(DE3) derivative) was used in this study because it contains the T7 RNA polymerase and the Bacillus subtilus 4’-phosphopantetheinyl transferase (sfp) gene downstream of the T7 promoter integrated into the genome (Pfeifer et al.2001: 1790–2). The Sfp protein converts the inactive (apo) form of NRPS and PKS proteins to the active (holo) form through posttranslational attachment of a phosphopantetheinyl arm to a conserved serine residue (Beld et al.2014: 61–108). This modification is required when heterologously expressing NRPS- and PKS-containing BGCs.
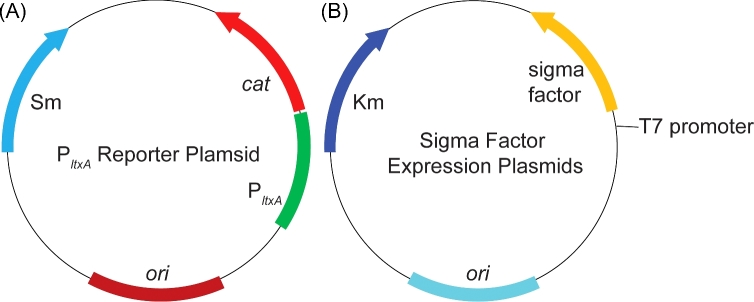
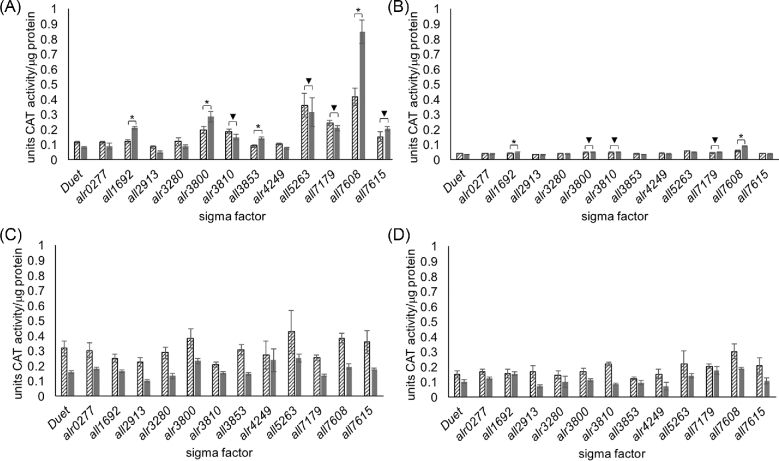
To obtain a direct measurement of CAT activity in E. coli expressing Anabaena sigma factors, an enzymatic assay was adapted from previous work to be compatible with a 96-well plate reader (Videau et al.2016: 978–88). Expression of CAT from cell lysates of E. coli BAP1 cells harboring the PltxA-cat reporter plasmid (Fig. 1A) and a sigma factor expression plasmid (Fig. 1B) were measured in units of CAT activity standardized per μg of protein (Fig. 2A). Negative controls included cultures that were not induced with IPTG, and cultures containing pPJAV562 and an empty pCOLADuet-1. Analysis of the data shows that this in vitro assay provides highly reproducible measurements with low error. The expression of four sigma factors (all1692, alr3800, all3853 and all7608) resulted in a statistically significant increase in levels of CAT activity, which is directly related to cat transcription, compared to both the corresponding uninduced controls and the negative control empty expression vector. Expression of all7608 resulted in highest CAT activity 0.85 ± 0.08 units of CAT activity per μg of protein. There was also a significant amount of CAT activity in E. coli BAP1 cells expressing alr3810, all5263, all7179 and all7615 compared to the empty vector control, but this seemed to be due to low levels of leaky expression of the sigma factor as there was no significant increase between the uninduced and induced samples containing these promoters and, in the cases of alr3810, all5263 and all7179, there was a decrease in activity in the induced sample. This can be best explained because addition of IPTG induces over-expression of sfp and the sigma factor, which could result in decreased CAT protein synthesis.
Figure 1.
Plasmid maps of (A) pPJAV562, the PltxA-cat reporter plasmid and (B) the sigma factor expression plasmids. Km, kanamycin resistance gene; Sp, spectinomycin resistance gene; cat, chloramphenicol acetyltransferase gene; ori, origin of replication (red represents the pBR322/pDU1 origins, teal represents the ColA origin).
Figure 2.
CAT activity measured from (A) PltxA-cat (pPJAV562), (B) PbarA-cat (pPJAV575), (C) PcurA-cat (pPJAV576) and (D) PpatA-cat (pPJAV578) with either the empty expression vector pCOLADuet-1 or pCOLADuet-1 containing the indicated sigma factor. CAT activity was determined by comparison to a standard curve made with purified CAT, per μg of protein ± standard deviation. The lined bars indicate the uninduced samples while the grey bars indicate the induced sample. (*P < 0.0001 for increased expression in induced samples over uninduced; ▾P < 0.0001 for increased expression of induced sigma factor over induced pCOLADuet-1, but no significant difference between uninduced and induced sample). The data presented here are the aggregate of three biological replicates assayed in technical duplicate.
To determine whether Anabaena sigma factors expressed in E. coli BAP1 are able to initiate transcription from other cyanobacterial promoters, transcriptional fusions of cat with 1000 bp upstream of the BGCs for barbamide A (PbarA; pPJAV575) and curacin A (PcurA; pPJAV576), and 250 bp upstream of patellamide A (PpatA; pPJAV578, not to be confused with the patA gene from Anabaena), were assayed for CAT activity in vitro (Videau et al.2016: 978–88). These promoter regions are derived from the genetically intractable cyanobacterial strains M. producens and Prochloron didemni, and nothing is known about their regulation to date. We observed a low level of activation from the PbarA promoter sequence, which showed a statistically significant increase in CAT activity compared to the uninduced sample and the pCOLADuet-1 induced sample for cells expressing all1692 and all7608 (Fig. 2B). For PbarA/all7608, we observed 10% of the CAT activity (0.089 ± 0.004 units of CAT activity per μg of protein) as observed for the PltxA/all7608 combination. For the PcurA and PpatA promoters (Fig. 2C–D), we did not record any samples in which induction of sigma factor production resulted in an increase in observed CAT activity, despite the fact that these two promoters were found to induce expression of CAT in Anabaena (Videau et al.2016: 978–88). This observation could be due to the fact that the sigma factor responsible for recognizing these promoters is not expressed in the soluble fraction in E. coli BAP1 (Fig. 3). As a postive control we measured the CAT activity from the native promoter (Pcat) in BAP1 cells. In uninduced cultures the measured CAT activity was 6.84 ± 0.90 units of CAT activity per μg of protein, while in induced cultures the measured CAT activity was determined to be 5.86 ± 0.58 units of CAT activity per μg of protein. We also noticed that the addition of IPTG (including the Pcat control) resulted in lower CAT activity for any given pair in almost all cases. We attribute this to the fact that the addition of IPTG triggers the production of three heterologous proteins (the sigma factor, CAT and Sfp). This dispersion of resources (e.g. tRNAs, ribosomes, RNA polymerase) results in a lower amount of CAT synthesis which is reflected in the lower activity observed in the assay.
To verify that the cat expression profiles described for pPJAV562 (PltxA; Fig. 2A) were due to the expression of a sigma factor, a Western blot of all 12 sigma factors expressed in E. coli BAP1 cells was performed (Fig. 3). Most of the sigma factors displayed clear bands at the expected masses from the soluble fraction of the cell lysate, suggesting they were properly expressed and soluble. We noted that a greater amount of soluble sigma factor did not necessarily result in greater amounts of observed CAT activity. For example, All7608 is present at a lower level than Alr3810 but induces 5.7-fold the amount of CAT activity from PltxA (Figs 2A and 3; All7608, 0.85 ± 0.08 vs. Alr3810, 0.15 ± 0.02 units of CAT per μg protein). The lack of CAT activity observed in E. coli BAP1 cells containing pPJAV576 (PcurA) or pPJAV578 (PpatA) could be explained by the fact that Alr0277, Alr3280 and Alr4249 did not appear to be expressed at detectable levels. It is possible that cloning these sigma factors into different expression vectors or screening at different expression temperatures would result in soluble protein that could then be used to screen for promoter recognition. These studies are currently underway. We also noted that most of the sigma factors observed in the Western blot analysis (Fig. 3) had increased expression in the induced vs. uninduced cultures (Alr3800, All1692, Alr3810, All3853, All7608 and All7615). In contrast, All5263 and All7179 had similar expression levels in the uninduced and induced cultures (Fig. S1, Supporting Information), which could explain the high background CAT activity and the lack of statistical significance between the uninduced and induced samples (Fig. 2A).
As this system was created with the goal of heterologously expressing cyanobacterial natural products, we tried to express the three gene BGC for LTXA production (ltxA-C) using single Anabaena sigma factors that showed the most promise above. Multiple attempts at producing LTXA using sigma factors Alr3800, All7608, All7179, All1692 and All5263 with PltxA-ltxA-C (pPJAV500) yielded no detectable LTXA or precursors when analyzed by LC–MS/MS, even though these sigma factors activated CAT activity from PltxA in our reporter assay. One reason we did not observe LTXA could be the presence of multiple promoters in the ltx BGC, as previous work in both S. coelicolor and Anabaena suggested the presence of a second promoter upstream of ltxC (Jones et al.2012: 1243–51; Videau et al.2016: 978–88). However, the fact that neither biosynthetic precursor (N-methyl-L-valyl-L-tryptophanol (NMVT) or indolactam V (ILV), Fig. S2, Supporting Information) was observed suggests the absence of a ribosomal binding site (Shine–Dalgarno sequence) that the native E. coli translational machinery could recognize. This is consistent with previous research indicating that the regions upstream of many cyanobacterial genes lack traditional Shine–Dalgarno sequences (Omotajo et al.2015: 604; Zheng et al.2011: 361–73).
In the PltxA-cat plasmid (pPJAV562), the −1 position of PltxA was not fused directly to the ATG start codon of cat, but rather to the -18 position of native cat DNA. These 18 nucleotides (5΄-GGGAGGAGGAAAGCTAAA-3΄) contain a strong E. coli Shine–Dalgarno sequence at positions −10 to −15 (underlined). A consensus Shine–Dalgarno sequence is notably absent in the −1 to −21 region of PltxA (Fig. S3, Supporting Information), although a possible candidate site is located at positions −21 to −25. To investigate the possible influence of these 18 nucleotides on the PltxA-dependent expression of cat, we constructed pKNW132, a plasmid harboring the −1 position of PltxA fused directly to the ATG start codon of cat, which removes the E. coli Shine–Dalgarno sequence. CAT expression was probed during co-expression with all 12 sigma factors. No significant activation of CAT activity was observed and even background expression of CAT activity was severely reduced with pKNW132 in comparison to pPJAV562 (Fig. S4, Supporting Information). The marked decrease in CAT activity following removal of the strong E. coli Shine–Dalgarno sequence suggests that a lack of recognizable ribosomal binding sites in cyanobacterial mRNAs may further compound existing issues with heterologous expression of cyanobacterial gene clusters in E. coli.
In the present work, we assessed the ability of E. coli to recognize and initiate transcription from cyanobacterial promoters using sigma factors from Anabaena. We found the sigma factor All7608 was the most promiscuous in its ability to recruit RNA polymerase to diverse cyanobacterial promoters in E. coli, but other sigma factors are capable of recognition on a promoter specific basis. The lack of ribosomal binding sites recognized by E. coli in cyanobacterial BGCs hindered our ability to produce LTXA from its native gene cluster in E. coli. While future work will address ribosomal recognition of cyanobacterial BGCs, this work demonstrates that E. coli transcriptional machinery can work in tandem with Anabaena sigma factors to recognize and initiate transcription from promoter regions within cyanobacterial BGCs. It also provides a path toward developing an E. coli strain that can recognize a broad range of promoters from cyanobacterial BGCs and serve as a heterologous expression system for compound production and investigations into biosynthetic steps.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSLE online.
Acknowledgements
The authors would like to thank Chaitan Khosla (Stanford University) for the kind gift of E. coli BAP1 cells and William and Lena Gerwick (University of California, San Diego) for the kind gift of the fosmid containing the lyngbyatoxin gene cluster (fos-DE3-86). Taifo Mahmud (Oregon State University) is acknowledged for use of his SpectraMax 190 plate reader. We are indebted to the staff at the OSU High-throughput screening services laboratory for the assistance in adapting the CAT assay to a 96-well plate format and for help in operating the automated equipment. The authors would also like to thank the two anonymous reviewers who helped improve a previous version of this manuscript.
FUNDING
This work was supported by the College of Pharmacy, Oregon State University, a New Investigator Grant from the Medical Research Foundation of Oregon [grant number 1415] and an AREA award grant from the National Institutes of Health [grant number 1R15GM117541-01A1] to Benjamin Philmus, a Professional Development Award from the Oregon State Postdoctoral Association to Patrick Videau, and an Honors Experience Scholarship from the University Honors College and an Undergraduate Summer Research Scholarship from the College of Pharmacy to Kaitlyn Wells.
Conflict of interest. None declared.
REFERENCES
- Aldea MR, Mella-Herrera RA, Golden JW. Sigma factor genes sigC, sigE, and sigG are upregulated in heterocysts of the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 2007;189:8392–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beld J, Sonnenschein EC, Vickery CR et al. . The phosphopantetheinyl transferases: catalysis of a post-translational modification crucial for life. Nat Prod Rep 2014;31:61–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell N, Lee JJ, Summers ML. Characterization and in vivo regulon determination of an ECF sigma factor and its cognate anti-sigma factor in Nostoc punctiforme. Mol Microbiol 2017;104:179–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calteau A, Fewer D, Latifi A et al. . Phylum-wide comparative genomics unravel the diversity of secondary metabolism in Cyanobacteria. BMC Genomics 2014;15:977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MA, Shlaes D. Fix the antibiotics pipeline. Nature 2011;472:32. [DOI] [PubMed] [Google Scholar]
- Edwards DJ, Gerwick WH. Lyngbyatoxin biosynthesis: sequence of biosynthetic gene cluster and identification of a novel aromatic prenyltransferase. J Am Chem Soc 2004;126:11432–3. [DOI] [PubMed] [Google Scholar]
- Ehira S, Miyazaki S. Regulation of genes involved in heterocyst differentiation in the cyanobacterium Anabaena sp. strain PCC 7120 by a group 2 sigma factor SigC. Life 2015;5:587–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich IM, Waterbury JB, Webb EA. Distribution and diversity of natural product genes in marine and freshwater cyanobacterial cultures and genomes. Appl Environ Microbiol 2005;71:7401–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feklístov A, Sharon BD, Darst SA et al. . Bacterial sigma factors: a historical, structural, and genomic perspective. Annu Rev Microbiol 2014;68:357–76. [DOI] [PubMed] [Google Scholar]
- Gerwick WH, Fenner AM. Drug discovery from marine microbes. Microb Ecol 2013;65:800–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi R, Krummel B, Saiki R. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucl Acids Res 1988;16:7351–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura S, Asayama M. Sigma factors for cyanobacterial transcription. Gene Regul Syst Biol 2009;3:65–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AC, Ottilie S, Eustáquio AS et al. . Evaluation of Streptomyces coelicolor A3(2) as a heterologous expression host for the cyanobacterial protein kinase C activator lyngbyatoxin A. FEBS J 2012;279:1243–51. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Wolk CP et al. . Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res 2001;8:205–13. [DOI] [PubMed] [Google Scholar]
- Khudyakov IY, Golden JW. Identification and inactivation of three group 2 sigma factor genes in Anabaena sp. strain PCC 7120. J Bacteriol 2001;183:6667–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Lee JH, Choi H et al. . Heterologous production of 4-O-demethylbarbamide, a marine cyanobacterial natural product. Org Lett 2012;14:5824–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long PF, Dunlap WC, Battershill CN et al. . Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. ChemBioChem 2005;6:1760–5. [DOI] [PubMed] [Google Scholar]
- Mella-Herrera RA, Neunuebel MR, Kumar K et al. . The sigE gene Is required for normal expression of heterocyst-specific genes in Anabaena sp. strain PCC 7120. J Bacteriol 2011;193:1823–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Pastor AM, Hess WR. Heterocyst differentiation: from single mutants to global approaches. Trends Microbiol 2012;20:548–57. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod 2016;79:629–61. [DOI] [PubMed] [Google Scholar]
- Niedermeyer THJ. Anti-infective natural products from cyanobacteria. Planta Med 2015;81:1309–25. [DOI] [PubMed] [Google Scholar]
- Omotajo D, Tate T, Cho H et al. . Distribution and diversity of ribosome binding sites in prokaryotic genomes. BMC Genomics 2015;16:604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongley SE, Bian X, Zhang Y et al. . High-titer heterologous production in E. coli of lyngbyatoxin, a protein kinase C activator from an uncultured marine cyanobacterium. ACS Chem Biol 2013;8:1888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanaik B, Lindberg P. Terpenoids and their biosynthesis in cyanobacteria. Life 2015;5:269–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer BA, Admiraal SJ, Gramajo H et al. . Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 2001;291:1790–2. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. NY: Cold Spring Harbor Labs Publishing, 2001. [Google Scholar]
- Schmidt EW, Nelson JT, Rasko DA et al. . Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA 2005;102:7315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh RK, Tiwari SP, Rai AK et al. . Cyanobacteria: an emerging source for drug discovery. J Antibiot 2011;64:401–12. [DOI] [PubMed] [Google Scholar]
- Srivastava A, Brilisauer K, Rai AK et al. . Down-regulation of the alternative sigma factor SigJ confers a photoprotective phenotype to Anabaena PCC 7120. Plant Cell Physiol 2017;58:287–97. [DOI] [PubMed] [Google Scholar]
- Tan LT. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry 2007;68:954–79. [DOI] [PubMed] [Google Scholar]
- Vestola J, Shishido TK, Jokela J et al. . Hassallidins, antifungal glycolipopeptides, are widespread among cyanobacteria and are the end-product of a nonribosomal pathway. Proc Natl Acad Sci USA 2014;111:E1909–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Videau P, Wells KN, Singh AJ et al. . Assessment of Anabaena sp. strain PCC 7120 as a heterologous expression host for cyanobacterial natural products: production of lyngbyatoxin A. ACS Synth Biol 2016;5:978–88. [DOI] [PubMed] [Google Scholar]
- Welker M, Von Döhren H. Cyanobacterial peptides—Nature's own combinatorial biosynthesis. FEMS Microbiol Rev 2006;30:530–63. [DOI] [PubMed] [Google Scholar]
- Yoshimura H, Okamoto S, Tsumuraya Y et al. . Group 3 sigma factor gene, sigJ, a key regulator of desiccation tolerance, regulates the synthesis of extracellular polysaccharide in cyanobacterium Anabaena sp. strain PCC 7120. DNA Res 2007;14:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Hu G-Q, She Z-S et al. . Leaderless genes in bacteria: clue to the evolution of translation initiation mechanisms in prokaryotes. BMC Genomics 2011;12:361–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Ishida K, Liaimer A et al. . Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angew Chem Int Ed 2008;47:7756–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.