An outbreak of HIV among people who inject drugs in Scotland follows similar recent outbreaks in Greece, Romania, Ireland, and the United States. Phylodynamic analysis demonstrates the infections are tightly linked genetically and the effective reproductive number remains above 1.
Keywords: HIV, phylodynamic, network, transmission, people who inject drugs
Abstract
Background
Harm reduction has dramatically reduced HIV incidence among people who inject drugs (PWID). In Glasgow, Scotland, <10 infections/year have been diagnosed among PWID since the mid-1990s. However, in 2015 a sharp rise in diagnoses was noted among PWID; many were subtype C with 2 identical drug-resistant mutations and some displayed low avidity, suggesting the infections were linked and recent.
Methods
We collected Scottish pol sequences and identified closely related sequences from public databases. Genetic linkage was ascertained among 228 Scottish, 1820 UK, and 524 global sequences. The outbreak cluster was extracted to estimate epidemic parameters.
Results
All 104 outbreak sequences originated from Scotland and contained E138A and V179E. Mean genetic distance was <1% and mean time between transmissions was 6.7 months. The average number of onward transmissions consistently exceeded 1, indicating that spread was ongoing.
Conclusions
In contrast to other recent HIV outbreaks among PWID, harm reduction services were not clearly reduced in Scotland. Nonetheless, the high proportion of individuals with a history of homelessness (45%) suggests that services were inadequate for those in precarious living situations. The high prevalence of hepatitis C (>90%) is indicative of sharing of injecting equipment. Monitoring the epidemic phylogenetically in real time may accelerate public health action.
(See the Editorial commentary by Poon and Dearlove, on pages 1854–7.)
People who inject drugs (PWID) are at risk of acquiring human immunodeficiency virus (HIV) from sharing injecting equipment and from high-risk sexual activity while under the influence of drugs or in exchange for drugs [1]. There are 9–22 million PWID worldwide of whom 1–5 million have HIV [2].
Major outbreaks of HIV were identified among PWID in Scotland in the 1980s [3–5], along with other parts of northern [3, 6] and southern Europe [5]. A major outbreak in Edinburgh in 1983 associated with extensive needle sharing [5] led to 50% of PWID becoming infected [3]. This epidemic was closely linked to similar ones in Dundee and Dublin [3], but few HIV cases were seen among PWID in Glasgow at the time [7]. Rapid introduction in the United Kingdom of needle exchange in 1986, followed by other harm reduction measures [8], dramatically decreased spread of HIV within this population. Since the mid-1990s, HIV diagnoses among PWID in Glasgow have averaged 10 per year [9]. Similarly in the rest of Western Europe, incidence had declined in accordance with public health responses [10].
However, in 2011 there were reports of outbreaks of HIV among PWID in Greece [11], Romania [12], and Ireland [13]. Prior to this, HIV incidence among PWID in Greece and Ireland had been similar to the United Kingdom, around 0.1% [2, 14, 15]. In Greece, fewer than 20 infections per year were reported among PWID between 2001 and 2010, but in 2011 this surged to 256 cases, accounting for one-quarter of all new HIV diagnoses that year [16]. The epidemics in Greece [11] and Ireland [13] followed an economic crisis, which led to increases in homelessness. The recession of 2008 resulted in funding cuts to opiate substitution therapy and needle exchange programs in Greece and Romania [17]. In parallel, the surge in injection of stimulant-based new psychoactive substances, which are typically injected more frequently than heroin thus increasing the risk of needle sharing, contributed to the outbreaks in Romania [12] and Ireland [13].
In 2015, a significant rise in HIV diagnoses among PWID was noted in Glasgow. Data from Scotland’s Needle Exchange Surveillance Initiative showed that HIV prevalence among PWID increased from 0.3% in 2011–2012 to 1.9% in 2015–2016 [18]. Routine sequencing to test for drug resistance revealed many were HIV subtype C, a subtype rarely observed among PWID in the United Kingdom [19, 20], suggesting a common source for the outbreak.
Reconstruction of the transmission network through contact tracing is difficult for HIV because of the time delay between infection and diagnosis, the low transmission rate, and the challenge of collecting information pertaining to sexual and drug-taking behaviors. Phylogenetic analysis of viral sequences provides an alternative and independent route to reconstructing transmission networks [21]. As viral sequences are generated as a component of routine clinical care in the United Kingdom, we conducted a phylogenetic analysis to investigate whether PWID cases were related, when infections had been acquired, and whether the strain was spreading into the wider community and elsewhere in the United Kingdom. Results informed the shape and intensity of the public health response.
METHODS
Study Population
Since 2005, the West of Scotland specialist virology center has routinely carried out baseline sequencing of all new HIV diagnoses. The HIV-1 protease and reverse transcriptase regions (HXB2 positions 2253 to 3549) were amplified using primers described previously [22] with Expand reverse transcriptase (and the Expand High Fidelity polymerase chain reaction (PCR) System (Roche) and the following program: reverse transcription PCR (RT-PCR) 42°C for 45 minutes; first-round PCR (2 minutes at 94°C; 10 cycles of 15 seconds at 94°C; 30 seconds at 55°C; 1 minute 30 seconds at 72°C; 25 cycles of 15 seconds at 94°C; 30 seconds at 55°C; 1 minute 30 seconds at 72°C + 5 seconds/cycle; 10 minutes at 72°C) and nested PCR (2 minutes at 94°C; 10 cycles of 15 seconds at 94°C; 30 seconds at 55°C; 1 minute 30 seconds at 72°C; 25 cycles of 15 seconds at 94°C; 30 seconds at 55°C; 1 minute 30 seconds at 72°C + 5 seconds/cycle; 10 minutes at 72°C). Sanger sequencing was performed on ABI3130xl using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems). Alignment and base-calling was performed using the online sequence analysis software RECall [23]. REGAv3 was used to subtype sequences and detect drug resistance mutations [24]. All subtype C sequences were extracted from the laboratory database for further analysis. At each stage (extraction through to PCR) and for each patient, negative controls were included in each assay to detect contamination. If evidence for contamination was observed, all patient samples in that run were re-extracted. For each weekly run a simple phylogenetic tree was constructed to detect contamination occurring at the sequencing stage. Any cases of sequence identity in the same batch were repeated from the extraction stage.
Avidity testing was used to classify infections as recent or older than 4 months. The assay is a modification of the Genscreen HIV1/2 version 2 (Bio-Rad) [25] and testing has been performed on HIV diagnoses since 2012. Clinical and epidemiological information was obtained through the UK National Health Service clinical portal, a virtual electronic patient record that contains clinical notes on interactions with tertiary services and test results. Data retrieved included hepatitis C status, viral load, date of last HIV negative test, sex, risk group, age, nationality, and history of drug use, incarceration, and homelessness.
Background Sequences
The UK HIV Drug Resistance Database (UKRDB) receives pol sequences obtained for routine clinical surveillance and submitted by participating laboratories. Resistance data are linked to demographic and clinical patient data held in the UK Collaborative HIV Cohort study (UK CHIC) database [26] and the national HIV/AIDS Reporting System (HARS) database held at Public Health England [27]. After linking sequences to epidemiological data, the data were anonymized. In the 2014 release of the database (sequences up to mid-2013), sequences were available for around 60% of the infected population and >80% of patients diagnosed since 2005. Of 63163 sequences in the UKRDB, 15864 sequences (25.1%) were classified as subtype C by REGAv3 [24]. Epidemiological data contributed by Public Health England and Health Protection Scotland included year of birth, gender, and self-reported most likely route of infection (PWID, heterosexual sex, men who have sex with men [MSM], mother to child, blood product, or unknown).
The Los Alamos National Laboratory (LANL) HIV database compiles all published HIV sequences, including 11658 non-UK subtype C pol sequences (accessed 8 August 2014). We used Geneious to BLAST Scottish sequences against UKRDB and LANL sequences, selecting the 10 closest matches for each individual Scottish sequence for analysis [28]. As the same sequence is hit multiple times, this procedure limits the size of the final alignments. Final alignments comprised 228 sequences from Scotland, 1820 from the UKRDB, and 524 from LANL (2572 in total).
Genetic Linkage and Phylodynamic Analysis
Sequences were stripped of 44 sites associated with drug resistance based on the 2013 International AIDS Society list [29]. We reconstructed genetic clusters by calculating genetic distances between pairs of sequences under a TN93 substitution model. Each sequence was represented in the network by a node and nodes were linked if their pairwise distance was below a certain genetic distance threshold. At thresholds 1%–2.5%, the same PWID outbreak cluster stood out (n = 104, see Results), with all sequences from Scotland. We selected the tightest threshold because 1% is consistent with recent and rapid transmission [30]. Ten percent of outbreak sequences were submitted to Genbank (accession numbers MG76186:MG761826). Sequences from the outbreak were further analyzed using the Bayesian birth-death skyline model in BEAST2 [31, 32]. The birth-death skyline is well suited to analyzing outbreaks because, unlike coalescent models, it does not assume low sample fraction within an infinite population size; instead, sample fraction is explicitly parameterized. Furthermore, the birth-death skyline estimates the effective reproductive number (Re), the average number of infections originating from each individual, directly yielding epidemiologically relevant results. Substitution models (TN93, GTR+G+I) and clock models (strict, uncorrelated lognormal) were compared and a GTR+G+I model with an uncorrelated lognormal clock was selected based on its Bayes factor. Chain samples were run for 500000000 generations in triplicate. Prior distributions for Re and the rate of becoming noninfectious were extracted from a previous analysis of the UK epidemic [31], and priors for the origin of the tree and the sampling proportion were based on our knowledge of the UK epidemic. The origin of the tree was set with a maximum value of 30 years and the sampling proportion was set as 0 until 2005 (the date of the first outbreak sequence) then with a normal distribution with mean = 0.65 and SD = 0.05. Because sampling fraction, Re, and time to becoming noninfectious are correlated in the birth-death skyline model, at least one must be set with a narrow prior [31].
RESULTS
The Drug-Resistant Subtype C Outbreak Has Not Been Observed Outside Scotland
All Scottish subtype C sequences were included in the phylogenetic analysis (n = 228), alongside 1820 sequences from the UKRDB and 524 from LANL (2572 in total). Of 2572 sequences, 501 (19.5%) linked to at least 1 other in the network using a genetic distance cutoff of 1%.
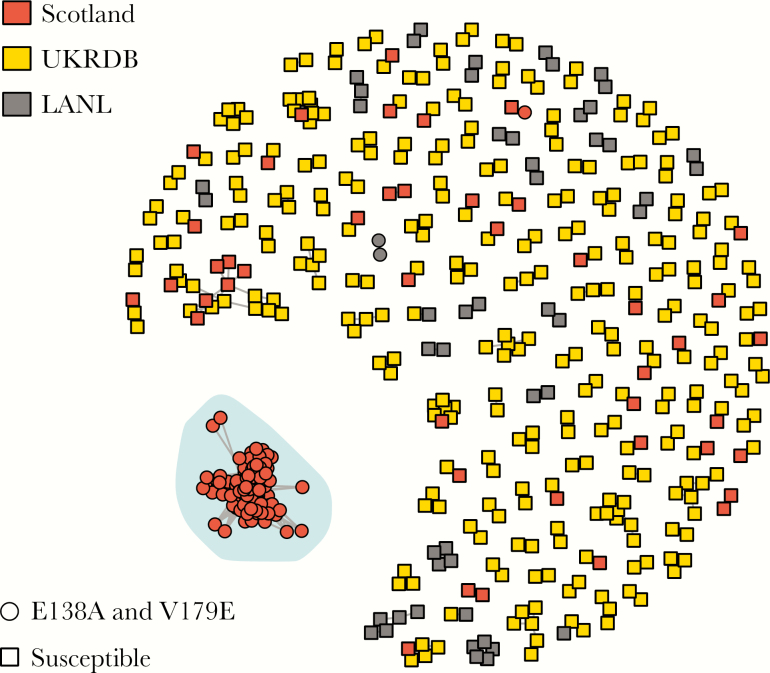
Within the network, a tight cluster of 104 sequences stood out (Figure 1). All sequences within the cluster were less than 1% genetic distance from at least 1 other sequence in the cluster. Mean genetic distance was <1% with 7 patients with pol sequences exactly identical to another, 2 pairs, and 1 triad. All sequences originated from Scotland and contained E138A and V179E. Thus we have not yet observed evidence of spread of this strain outside Scotland. E138A is a common polymorphic accessory mutation weakly selected in patients receiving etravirine and rilpivirine that reduces susceptibility to these 2 antiretrovirals by around 2-fold. V179E is a nonpolymorphic mutation weakly selected by nevirapine and efavirenz and associated with resistance to nevirapine, efavirenz, and etravarine. In the UKRDB, which includes sequences sampled in Scotland until mid-2013, E138A is found in 1648/15864 of subtype C sequences (10.39%) and V179E is found in 50 (0.32%). Only 41 sequences in the UKRDB contained both mutations (0.25%), of which 26 were from the present outbreak. Among the remaining 15 sequences containing both mutations, 12 were not closely related to the outbreak and were not included in the phylogenetic analysis and 3 were included in the analysis but did not link to the outbreak. Between 2014 and mid-2016, 5 nonoutbreak HIV diagnoses were made among PWID in Scotland.
Figure 1.
Genetic distance network of relatedness at 1%. Of 2572 sequences analyzed, only those linked to at least 1 other sequence at 1% are shown (501 in total). Sequences are colored by origin: Scotland, UK HIV Drug Resistance Database (UKRDB) or Los Alamos National Database (LANL). Node shapes are determined by drug susceptibility of viruses. One large cluster, highlighted and circled, stood out due to its size (104 sequences), its concentration of drug-resistant sequences, and its Scottish origin.
Spread of HIV Among Scottish PWID Has Been Recent and Rapid
Sequences from the outbreak cluster (n = 104) were selected for analysis using the birth-death skyline models in BEAST2 to estimate growth through time and to better infer the origin of the cluster. Runs converged within 5000000 generations with effective sample size values above 200.
The common ancestor to the cluster was dated as mid-2003 (2003.4; 95% highest posterior density [HPD], 2001.8–2005.0), while the oldest outbreak sequence was from a female PWID diagnosed in 2005. Five patients were diagnosed in 2008–2009 (4.8%), 27 were diagnosed between 2010 and 2013 (26.0%), and 71 patients were diagnosed after 2014 (68.3%). All were diagnosed in Scotland and all those with a risk group reported were PWID.
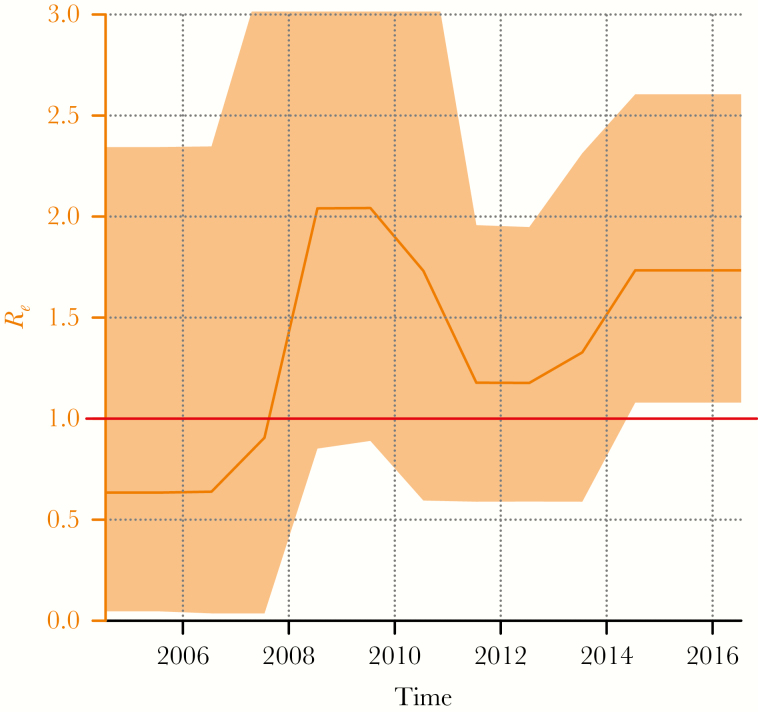
The birth death skyline infers the effective reproductive number Re (the average number of transmissions for each individual). Importantly, Re has remained above 1 throughout the course of the outbreak (Figure 2), implying that the number of cases would be expected to continue to rise. Mean Re was estimated as 1.5 (95% HPD
Figure 2.
Reproductive number (Re) inferred from the birth-death skyline plot. The line across the plot marks Re = 1, the threshold above which an infection will continue to spread.
, 0.1–3.9) over the course of the outbreak, rising to 1.8 (HPD, 1.1–2.6) during the last 2 years. At its highest point, in 2009, Re exceeded 2. Sample fraction was estimated as 0.66 (HPD, 0.46 –0.81).
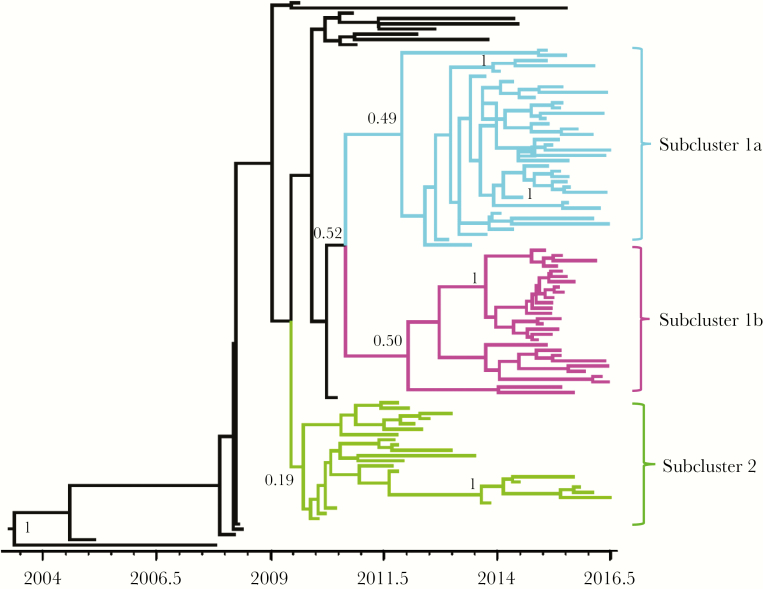
The distance between internal nodes in the tree approximates the upper bound on time between transmission events [33]. Based on the time-resolved trees, the average transmission interval was estimated as 6.7 months for the outbreak as a whole (Supplementary information). Looking at the phylogeny in more detail, it divided into 3 subclusters: 1a, 1b, and 2, originating in 2011, 2012, and 2010, respectively (Figure 3). Subclusters 1a and 1b had a higher density of recent transmission events, but there was no difference in transmission intervals between subclusters based on an analysis of 1000 trees from the posterior distribution (Supplementary information), indicating that while subclusters 1a and 1b are most active at present, the transmission dynamics within all 3 subclusters have been similar. The origin of subclusters 1a and 1b correlated with an increase in Re (Figure 2).
Figure 3.
Time-resolved phylogeny for the outbreak cluster comprising 104 sequences from Scotland with drug-resistant mutations E138A and V179E. The outbreak subdivided into 3 subclusters.
Demographic and Clinical Characteristics of Outbreak Members
Among the 104 individuals in the outbreak diagnosed by mid-2016, 102 (98.1%) reported injecting drugs. Mean age was 38.4 (SD = 6.5), 63/103 (61.2%) were men and 40 (38.8%) were women, 99/100 were white British (1 mixed race), 38/96 (39.6%) had a recorded history of incarceration, and 41/92 (44.6%) reported having ever been homeless; 96/98 (98.0%) were confirmed to be HCV antibody positive, with 6 not tested, suggesting wide-spread sharing of injecting equipment; 68/96 (70.8%) had ongoing HCV infection with a positive HCV antigen or PCR result.
HIV avidity was tested on 87/104 (83.7%) patients and 49/104 has a previous negative test result; 24/87 (27.6%) had low avidity results indicating that infection had occurred within the last 4 months. Five additional patients had a date of last negative HIV test less than a year previous to their diagnosis. Three patients had antibody levels too low for avidity testing indicating acute seroconversion, confirmed with negative western blot and BiSpot results. Therefore, in total at least 32/87 (36.8%) had HIV for less than a year at diagnosis, consistent with the short terminal branch lengths in the phylogenetic tree (Figure 3).
DISCUSSION
This recent outbreak in Scotland is the latest in a series of rapid transmissions of HIV among PWID: in Greece [11], Romania [12], Ireland [13], and Indiana, United States [34]. Prior to these outbreaks, HIV incidence among PWID in these regions had been fairly static since the epidemics of the 1980s. From 2001 to 2014, there were 10–20 new cases per year in Scotland with 5–10 new cases around Glasgow [35]. The Scottish outbreak now comprises over 100 linked cases.
Phylodynamic analysis demonstrated how rapidly the virus was transmitted, with average transmission intervals around 6 months, similar to MSM [33] and shorter than heterosexuals [36] in the United Kingdom. In contrast to the more commonly presented R0, which represents the number of onward transmissions in a fully susceptible population, Re is the number of secondary infections for the current frequency of susceptibles [37]. The number of secondary infections has averaged 1.5 since the outbreak originated around 2003, reaching 2 at its peak in 2009. In contrast, HIV diagnoses among PWID did not reach a peak until 2015 in Scotland, remained around 20 per year between 2008 and 2010, not reaching a peak until 2015 [38]. UK estimates of Re for heterosexuals and MSM are below 1, and just above 1, respectively [39]. Previous estimates of Re among PWID have ranged as high as 21.7 in Lithuania [40]. For the United Kingdom, estimates of Re do not exist for PWID, but Re was consistently above 1 for this outbreak, indicating that spread was ongoing in 2016. This number is specific to this outbreak and should not be extrapolated to reflect HIV transmission among PWID in the United Kingdom in general. The outbreak subdivided into subclusters, indicating independent concurrent transmission chains. All 3 transmission chains showed evidence of recent transmission events, and had equally short transmission intervals.
Genetic distance within the outbreak was extremely low, with multiple sets of identical partial pol sequences, a phenomenon observed in cases of rapid transmission [41]. While in part due to the short region of the virus sequenced, such low divergence demonstrates how rapidly the virus spread in this group. The potential for multiple partners during acute infection leads to low genetic diversity within PWID transmission networks. The recent outbreak among Greek PWID similarly displayed low diversity and high clustering [11], reminiscent of the spread of near identical subtype A variants throughout Eastern European and Russian PWID in the 1990s [42]. Full genome sequencing, currently being undertaken, may disentangle the order of transmissions within the outbreak.
The outbreak was in part detected because of 2 resistance mutations, E138A and V179E, repeatedly identified in subtype C viruses, which had not previously associated with PWID infection in the United Kingdom. This prompted a search through the UKRDB for the mutations and for related sequences. At present, despite the UKRDB collecting sequences from all HIV resistance tests, sequences are used for research purposes and not as a systematic surveillance tool. Genetic analysis confirmed the strain was unique to Scotland and is not transmitting elsewhere in the United Kingdom. In the United Kingdom it is rare to find large clusters from a single region [43], and this is now the largest cluster of HIV among PWID seen in the United Kingdom since the 1980s. However, we did not have samples from the rest of the United Kingdom as recent as those from Scotland. The most recent Scottish sequence was sampled in 2016, whereas the most recent UKRDB sequence was sampled in 2013. It is possible that the outbreak has spread outside Scotland but that we have not captured it.
No published PWID outbreaks have reported transmission of resistance mutations, although preliminary results from Saskatchewan, Canada, demonstrated the G190 mutation disproportionately affected Aboriginal PWID [44]. Earlier studies found a higher prevalence of resistance mutations among PWID than among those infected sexually [45]. Suboptimal treatment adherence among this group may provide an explanation for this phenomenon [46]. Another possibility is that blood to blood transmission could enable transmission of lower-fitness viruses unable to establish infection through sexual transmission [47].
Despite access to injecting equipment, HIV still poses a significant risk to PWID. The identification of a unique strain facilitated its detection in Scotland during this outbreak, but real-time monitoring may help accelerate public health action. British Columbia recently deployed a real-time phylogeny response, where monthly reports were generated detailing cluster growth [48]. This analysis revealed a highly active cluster that expanded by 11 cases in 3 months. Members of the cluster were contacted to ensure linkage to care and partner notification and subsequently no further cases linked to those members were diagnosed. In the case of the Scottish outbreak, real-time phylogenetic monitoring could have brought the cluster to attention sooner. At present, all UKRDB analyses are conducted with anonymized data, while Poon et al identified subjects to reach out to them [48]. Use of nonanonymized HIV data for phylogenetic analyses is avoided in some jurisdictions because of the criminalization of HIV transmission. An anonymized version of Poon’s system can also be imagined, in which the background of sequences for comparison is anonymous but data are available for the patient being seen at that moment [49]. If the patient’s sequence were to cluster with 2 or more recent sequences, that patient could be selected for early initiation of treatment and preexposure prophylaxis could be offered to their partners. The advantage of Poon’s method is that all members of the cluster can be retrospectively contacted whereas under the anonymized system, only patients diagnosed after the first few in a cluster would be identified. Overall, advances such as avidity testing and real-time phylogenetic analysis can be used to improve our understanding of outbreaks to better target public health responses.
Many PWID involved in the outbreak had experienced homelessness. Scotland’s Needle Exchange Surveillance Initiative emphasized this point: almost 90% (20/23) of PWID from Glasgow who tested positive for HIV in 2015–16 had a history of homelessness, three-quarters of whom had been homeless within the last 6 months [18]. The situation in Scotland differs from that in other PWID outbreaks, however, because harm reduction services (injecting equipment provision, opiate substitution therapy) were available in Scotland postrecession. Indeed, Glasgow operates one of the most active injecting equipment provision service in Europe, distributing over 1 million syringes per year [18]. In contrast, in Indiana, neither needle exchange nor HIV testing were available at the time of the outbreak [34]. Nonetheless, the association observed with homelessness suggests that harm reduction services available in Glasgow may have been difficult to access for those in precarious living situations, often with chaotic lifestyles.
This outbreak may have been due to a change in circumstances, but it may result from the unfortunate introduction of HIV into a group of connected but previously uninfected PWID, such as was the case in Sweden in 2006 [50] and in Indiana in 2015 [34]. The high prevalence of hepatitis C among PWID in this outbreak (>90%) is indicative of widespread injecting equipment sharing. In contrast, in Romania and Greece, multiple strains and networks were uncovered [11, 12]; these outbreaks resulted from the reduced availability of harm reduction services. The Scottish outbreak is being managed through education of the population at risk and service providers, improved addiction services, increasing provision of needle exchange (eg, greater evening availability), improving accessibility of HIV testing, and outreach services to support early treatment and retention. Further research is needed to demonstrate whether homelessness, or other behavioral factors, played a role in the outbreak.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. The authors would like to acknowledge Denise Kuhnert for proving advice on the skyline model, and would like to thank the clinical teams for their input.
Financial support. This work was supported by the Bill and Melinda Gates Foundation and by NIH (grant number GM110749) through the Pangea-HIV Consortium.
Potential conflicts of interest. M. R. C. is currently supported by a grant from Gilead to the University of California, San Diego for work on hepatitis C. This funding was received after the present work was completed and did not have any influence on this work. All other authors report no conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Presented in part: Conference on Retroviruses and Opportunistic Infections, Seattle, 2017; and HIV Dynamics and Evolution, Isle of Skye, Scotland, 2017.
References
- 1. United Nations Office on Drugs and Crime. World Drug Report 2005 Volume 1: Analysis. Geneva: United Nations, 2005. [Google Scholar]
- 2. Des Jarlais DC, Kerr T, Carrieri P, Feelemyer J, Arasteh K. HIV infection among persons who inject drugs: ending old epidemics and addressing new outbreaks. AIDS 2016; 30:815–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown AJ, Lobidel D, Wade CM, et al. The molecular epidemiology of human immunodeficiency virus type 1 in six cities in Britain and Ireland. Virology 1997; 235:166–77. [DOI] [PubMed] [Google Scholar]
- 4. Holmes EC, Zhang LQ, Robertson P, et al. The molecular epidemiology of human immunodeficiency virus type 1 in Edinburgh. J Infect Dis 1995; 171:45–53. [DOI] [PubMed] [Google Scholar]
- 5. Robertson JR, Bucknall AB, Welsby PD, et al. Epidemic of AIDS related virus (HTLV-III/LAV) infection among intravenous drug abusers. Br Med J (Clin Res Ed) 1986; 292:527–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Op de Coul EL, Prins M, Cornelissen M, et al. Using phylogenetic analysis to trace HIV-1 migration among western European injecting drug users seroconverting from 1984 to 1997. AIDS 2001; 15:257–66. [DOI] [PubMed] [Google Scholar]
- 7. Davies AG, Cormack RM, Richardson AM. Estimation of injecting drug users in the city of Edinburgh, Scotland, and number infected with human immunodeficiency virus. Int J Epidemiol 1999; 28:117–21. [DOI] [PubMed] [Google Scholar]
- 8. Stimson GV. AIDS and injecting drug use in the United Kingdom, 1987–1993: the policy response and the prevention of the epidemic. Soc Sci Med 1995; 41:699–716. [DOI] [PubMed] [Google Scholar]
- 9. Public Health England, Health Protection Scotland, Public Health Wales, Public Health Agency Northern Ireland. Shooting up: infections among people who inject drugs in the UK, 2016. London: Public Health England, 2017. [Google Scholar]
- 10. Hamers FF, Batter V, Downs AM, Alix J, Cazein F, Brunet JB. The HIV epidemic associated with injecting drug use in Europe: geographic and time trends. AIDS 1997; 11:1365–74. [DOI] [PubMed] [Google Scholar]
- 11. Sypsa V, Paraskevis D, Malliori M, et al. Homelessness and other risk factors for HIV infection in the current outbreak among injection drug users in Athens, Greece. Am J Public Health 2015; 105:196–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Botescu A, Abagiu A, Mardarescu M, Ursan M.. HIV/AIDS among injecting drug users in Romania: Report of a recent outbreak and initial response policies, 2016. European Monitoring Centre for Drugs and Drug Addiction, Lisbon, November 2012. Available at http://www.emcdda.europa.eu/publications/ad-hoc/2012/romania-hiv-update_en. [Google Scholar]
- 13. Giese C, Igoe D, Gibbons Z, et al. Injection of new psychoactive substance snow blow associated with recently acquired HIV infections among homeless people who inject drugs in Dublin, Ireland, 2015. Euro Surveill 2015; 20:30036. [DOI] [PubMed] [Google Scholar]
- 14. Yin Z, Brown AE, Hughes G, Nardone A, Gill ON, Delpech V.. HIV in the United Kingdom: 2014 Report. London: Public Health England, 2014. [Google Scholar]
- 15. Public Health England, Health Protection Scotland, Public Health Wales, Public Health Agency Northern Ireland. Shooting up: infections among people who inject drugs in the UK, 2014. London: Public Health England, 2015. [Google Scholar]
- 16. Fotiou A, Micha K, Paraskevis D, Terzidou M, Malliori M, Hatzakis A. An updated report for the EMCDDA on the recent outbreak of HIV infections among drug injectors in Greece. In: European Monitoring Centre for Drugs and Drug Addiction, ed. Athens, Greece, 2012. Available at http://www.emcdda.europa.eu/system/files/ publications/752/HIV_update_Greece_2012_400439.pdf. [Google Scholar]
- 17. Hedrich D, Kalamara E, Sfetcu O, et al. Human immunodeficiency virus among people who inject drugs: is risk increasing in Europe?Euro Surveill 2013; 18:20648. [DOI] [PubMed] [Google Scholar]
- 18. University of the West of Scotland, Health Protection Scotland, Glasgow Caledonian University, West of Scotland Specialist Virology Centre. The Needle Exchange Surveillance Initiative (NESI): Prevalence of HCV and injecting risk behaviours among people who inject drugs (PWID) attending injecting equipment provision services (IEPs) in Scotland, 2008/2009–2015/2016. Glasgow, UK:Health Protection Scotland, 2016. [Google Scholar]
- 19. The UK Collaborative Group on HIV Drug Resistance. The increasing genetic diversity of HIV-1 in the UK, 2002–2010. AIDS 2014; 28:773–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ragonnet-Cronin M, Lycett SJ, Hodcroft EB, et al. ; United Kingdom HIV Drug Resistance Database Transmission of non-B HIV subtypes in the United Kingdom is increasingly driven by large non-heterosexual transmission clusters. J Infect Dis 2016; 213:1410–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grenfell BT, Pybus OG, Gog JR, et al. Unifying the epidemiological and evolutionary dynamics of pathogens. Science 2004; 303:327–32. [DOI] [PubMed] [Google Scholar]
- 22. Alexander CS, Dong W, Chan K, et al. HIV protease and reverse transcriptase variation and therapy outcome in antiretroviral-naive individuals from a large North American cohort. AIDS 2001; 15:601–7. [DOI] [PubMed] [Google Scholar]
- 23. Woods CK, Brumme CJ, Liu TF, et al. Automating HIV drug resistance genotyping with RECall, a freely accessible sequence analysis tool. J Clin Microbiol 2012; 50:1936–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pineda-Peña AC, Faria NR, Imbrechts S, et al. Automated subtyping of HIV-1 genetic sequences for clinical and surveillance purposes: performance evaluation of the new REGA version 3 and seven other tools. Infect Genet Evol 2013; 19:337–48. [DOI] [PubMed] [Google Scholar]
- 25. Shepherd SJ, McAllister G, Kean J, et al. Development of an avidity assay for detection of recent HIV infections. J Virol Methods 2015; 217:42–9. [DOI] [PubMed] [Google Scholar]
- 26. UK Collaborative HIV Cohort Steering Committee. The creation of a large UK-based multicentre cohort of HIV-infected individuals: the UK Collaborative HIV Cohort (UK CHIC) Study. HIV Med 2004; 5:115–24. [DOI] [PubMed] [Google Scholar]
- 27. PublicHealth England. National HIV/AIDS Reporting System (HARS), 2008. Available at https://www.gov.uk/guidance/hiv-surveillance-systems. Accessed 15 May 2017. [Google Scholar]
- 28. Kearse M, Moir R, Wilson A, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012; 28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Johnson VA, Calvez V, Gunthard HF, et al. Update of the drug resistance mutations in HIV-1: March 2013. Top Antivir Med 2013; 21:6–14. [PMC free article] [PubMed] [Google Scholar]
- 30. Smith DM, May SJ, Tweeten S, et al. A public health model for the molecular surveillance of HIV transmission in San Diego, California. AIDS 2009; 23:225–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stadler T, Kühnert D, Bonhoeffer S, Drummond AJ. Birth-death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV). Proc Natl Acad Sci U S A 2013; 110:228–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bouckaert R, Heled J, Kühnert D, et al. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 2014; 10:e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med 2008; 5:e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Conrad C, Bradley HM, Broz D, et al. ; Centers for Disease Control and Prevention (CDC) Community outbreak of HIV infection linked to injection drug use of oxymorphone–Indiana, 2015. MMWR Morb Mortal Wkly Rep 2015; 64:443–4. [PMC free article] [PubMed] [Google Scholar]
- 35. Health Protection Scotland. HIV-infected persons, Scotland: Number of cases reported by NHS Board, transmission category, and year of report; to 31 December 2012. Health Protection Scotland, 2013. [Google Scholar]
- 36. Hughes GJ, Fearnhill E, Dunn D, Lycett SJ, Rambaut A, Leigh Brown AJ; UK HIV Drug Resistance Collaboration Molecular phylodynamics of the heterosexual HIV epidemic in the United Kingdom. PLoS Pathog 2009; 5:e1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Amundsen EJ, Stigum H, Røttingen JA, Aalen OO. Definition and estimation of an actual reproduction number describing past infectious disease transmission: application to HIV epidemics among homosexual men in Denmark, Norway and Sweden. Epidemiol Infect 2004; 132:1139–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Public Health England, Health Protection Scotland, Public Health Wales, Public Health Agency Northern Ireland. Shooting up: infections among people who inject drugs in the UK, 2016. Accompanying data tables. London: Public Health England, 2017. [Google Scholar]
- 39. White PJ, Ward H, Garnett GP. Is HIV out of control in the UK? An example of analysing patterns of HIV spreading using incidence-to-prevalence ratios. AIDS 2006; 20:1898–901. [DOI] [PubMed] [Google Scholar]
- 40. Stadler T, Kouyos R, von Wyl V, et al. ; Swiss HIV Cohort Study Estimating the basic reproductive number from viral sequence data. Mol Biol Evol 2012; 29:347–57. [DOI] [PubMed] [Google Scholar]
- 41. Brooks JT, Robbins KE, Youngpairoj AS, et al. Molecular analysis of HIV strains from a cluster of worker infections in the adult film industry, Los Angeles 2004. AIDS 2006; 20:923–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bobkov A, Kazennova E, Khanina T, et al. An HIV type 1 subtype A strain of low genetic diversity continues to spread among injecting drug users in Russia: study of the new local outbreaks in Moscow and Irkutsk. AIDS Res Hum Retroviruses 2001; 17:257–61. [DOI] [PubMed] [Google Scholar]
- 43. Ragonnet-Cronin M, Hodcroft E, Hué S, et al. ; UK HIV Drug Resistance Database Automated analysis of phylogenetic clusters. BMC Bioinformatics 2013; 14:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wong A, Kambo J, Harrigan PR, Poon AF, Joy J.. Large NNRTI-resistant transmission cluster in injection drug users in Saskatchewan. Conference on Retroviruses and Opportunistic Infections, Seattle, Washington, 2015. http://www.croiconference.org/scientific-program/electronic-materials/croi-2015. Accessed 23 March 2018. [Google Scholar]
- 45. Pham QD, Do NT, Le YN, et al. Pretreatment HIV-1 drug resistance to first-line drugs: results from a baseline assessment of a large cohort initiating ART in Vietnam, 2009-10. J Antimicrob Chemother 2015; 70:941–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jordan MR, La H, Nguyen HD, et al. Correlates of HIV-1 viral suppression in a cohort of HIV-positive drug users receiving antiretroviral therapy in Hanoi, Vietnam. Int J STD AIDS 2009; 20:418–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Leigh Brown AJ, Frost SD, Mathews WC, et al. Transmission fitness of drug-resistant human immunodeficiency virus and the prevalence of resistance in the antiretroviral-treated population. J Infect Dis 2003; 187:683–6. [DOI] [PubMed] [Google Scholar]
- 48. Poon AF, Gustafson R, Daly P, et al. Near real-time monitoring of HIV transmission hotspots from routine HIV genotyping: an implementation case study. Lancet HIV 2016; 3:e231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Brooks JI, Sandstrom PA. The power and pitfalls of HIV phylogenetics in public health. Can J Public Health 2013; 104:e348–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Skar H, Axelsson M, Berggren I, et al. Dynamics of two separate but linked HIV-1 CRF01_AE outbreaks among injection drug users in Stockholm, Sweden, and Helsinki, Finland. J Virol 2011; 85:510–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.