Abstract
The nutrient sensing protein, SIRT1 influences aging and nutritional interventions such as caloric restriction in animals, however, the role of SIRT1 in human aging remains unclear. Here, the role of SIRT1 single-nucleotide polymorphisms (SNPs) and serum-induced SIRT1 protein expression (a novel assay that detects circulating factors that influence SIRT1 expression in vitro) were studied in the Concord Health and Ageing in Men Project (CHAMP), a prospective cohort of community dwelling men aged 70 years and older. Serum-induced SIRT1 expression was not associated with age or mortality, however participants within the lowest quintile were less likely to be frail (odds ratio (OR) 0.34, 95% confidence interval (CI) 0.17–0.69, N = 1,309). Serum-induced SIRT1 expression was associated with some markers of body composition and nutrition (height, weight, body fat and lean % mass, albumin, and cholesterol) but not disease. SIRT1 SNPs rs2273773, rs3740051, and rs3758391 showed no association with age, frailty, or mortality but were associated with weight, height, body fat and lean, and albumin levels. There were some weak associations between SIRT1 SNPs and arthritis, heart attack, deafness, and cognitive impairment. There was no association between SIRT1 SNPs and the serum-induced SIRT1 assay. SIRT1 SNPs and serum-induced SIRT1 expression in older men may be more closely associated with nutrition and body composition than aging and age-related conditions.
Keywords: Sirtuin, SIRT1, Polymorphism, Frailty, Mortality, Body composition
SIRT1 is an NAD+-dependent deacetylase with key roles in aging and the responses to caloric restriction through its regulation of downstream targets such as PGC1α, FOXO, p53, and NF-κB (1,2). Caloric restriction is associated with increased activity and/or expression of SIRT1 (3,4), whereas increased SIRT1 activity generated by genetic or pharmaceutical interventions has been associated with delayed aging and delayed onset of metabolic disorders in animal models (1,2,5).
SIRT1 is a protein found in nuclei of most tissues, which limits the biological significance of blood measurements of SIRT1. Therefore, de Cabo and colleagues developed a bioassay to determine circulating factors that regulate the expression of SIRT1 protein (3,6). Rats and humans undergoing caloric restriction have increased levels of circulating factors that induce SIRT1 expression, which is consistent with the conclusion that caloric restriction delays aging at least in part by increasing SIRT1 expression (3,7). However, in a pilot study of older men (n = 159), we found that those men with the lowest levels of serum-induced SIRT1 protein expression were less frail and had higher body lean (muscle) mass. We proposed that serum-induced SIRT1 expression might primarily reflect nutritional intake, with higher levels found when food intake is low, either as a result of illness or caloric restriction (8). On the other hand, direct measurement of circulating SIRT1 protein in 200 older subjects found that frailty was associated with lower levels of SIRT1 protein (9).
An alternative option to determine the role of SIRT1 in humans is from the study of SIRT1 single-nucleotide polymorphisms (SNPs). There have been several reports studying the association between various SIRT1 SNPs with longevity (10–12) as well as a number of different diseases (13,14). As yet there is no consensus about whether SIRT1 SNPs influence aging and hence susceptibility to age-related conditions and diseases in humans.
The aims of the present study were to investigate the relationship between serum-induced SIRT1 expression and age, frailty, mortality, and age-related diseases (in a much larger cohort than our previous pilot study (8)) and to determine whether there are any associations between serum-induced SIRT1 expression and nutritional and body composition biomarkers. Genotyping assays were undertaken to assess whether selected SIRT1 SNPs are associated with age, frailty, mortality, and age-related diseases. Finally, we determined whether there is any relationship between SIRT1 SNPs and markers of nutrition and body composition and with the serum-induced SIRT1 expression assay. These were studied utilizing an established cohort, the Concord Health and Ageing in Men Project (CHAMP) (8,15–18).
Method
Participants
CHAMP is a longitudinal study of health and aging in a population of Australian men aged 70 years and older that commenced in 2005 and has been described previously (eg, (8,15–18)). The CHAMP study population comprises of 1,705 men aged 70 years and older living in an urban region (the Local Government Areas of Burwood, Canada Bay, and Strathfield) near Concord Hospital in Sydney, Australia. The sampling frame was the New South Wales Electoral Roll, on which registration is compulsory in Australia. The only exclusion criterion was living in a residential aged care facility. Eligible men were sent a letter describing the study and, if they had a listed telephone number, were telephoned about 1 week later. Of the 2,815 eligible men with whom contact was made, 1,511 participated in the study (54%). An additional 194 eligible men living in the study area heard about the study from friends or the local media and were recruited before receiving a letter, yielding a total cohort of 1,705 subjects. All participants gave written informed consent. The study was approved by the Sydney South West Area Health Service Human Research Ethics Committee, Concord Repatriation General Hospital, Sydney, Australia.
Blood tests were performed at the Diagnostic Pathology Unit of Concord RG Hospital, which is a National Australian Testing Authority-accredited pathology service, using a MODULAR Analytics system (Roche Diagnostics, Castle Hill, Australia). Body composition was measured by dual x-ray absorptiometry (DEXA) using a Discovery-W scanner (Hologic, Bedford, MA). Frailty was defined according to the Cardiovascular Health Study frailty criteria of weight loss, weakness, exhaustion, slowness, and low activity as described (8,16). Diseases, smoking, and alcohol consumption were determined by self-report. Mortality data were collected via telephone contact every 4 months and through access to the NSW Registry of Births, Deaths and Marriages. After 8 years of follow-up, 616 deaths had been recorded.
The outcome of aging was indirectly inferred from chronological age, frailty, and age-related diseases.
Serum-Induced SIRT1 Expression Bioassay
An indirect enzyme-linked immunosorbent assay was designed to measure the amount of SIRT1 protein produced by SK Hep1 cells (American Type Tissue Culture Collection, Manassas, VA) due to factors present in human serum as described (8). The sensitivity of the SIRT1 enzyme-linked immunosorbent assay was 0.125 ng/mL. The mean interassay coefficient of variance was 15.3% (5%–18%). The mean intra-assay coefficient of variance was 7.9% (4%–12%). Linearity was confirmed over a range of dilutions. In each assay, the internal pooled human serum control lysate was spiked with human recombinant SIRT1 to determine percentage recovery.
SIRT1 SNPs
The SNPs rs2273773, rs3740051, and rs3758391 were selected from published human studies on the SIRT1 gene and its association with aging, age-related disorders, and longevity (10–14). Primers were designed by the LightScanner primer design software version 1.0.R84 (Idaho Technology; Supplementary Table 1). Buffy coat genomic DNA samples were outsourced to Macrogen (Seoul, Korea) for genotyping using the standard company protocols with Taqman probes and the ABI PRISM 7900HT Real-time PCR system.
Statistics
All results are presented as mean ± SD and statistical tests were considered significant with a p value less than .05. Only participants with complete data sets were used for each analysis. The correlation between age and serum-induced SIRT1 expression was assessed using the Pearson’s product correlation coefficient. The relationships between quintiles of serum-induced SIRT1 expression and frailty were evaluated using analysis of variance and χ2 test. Log rank and Kaplan–Meier survival analyses were performed according to serum-induced SIRT1 protein expression. Genotypic and allelic frequencies were analyzed using χ2 test or Fisher probability test statistics. Dominant and recessive models were considered. Bonferroni correction for multiple comparisons was used to generate target p values for statistical significance. Statistical Package for the Social Sciences SPSS (version 23.0’ IBM SPSS Statistics Software) and SigmaPlot (version 11.0, Systat Software Germany) were used for analyses.
Results
Serum-Induced SIRT1 Expression and Age, Frailty, Mortality, and Age-Related Disease
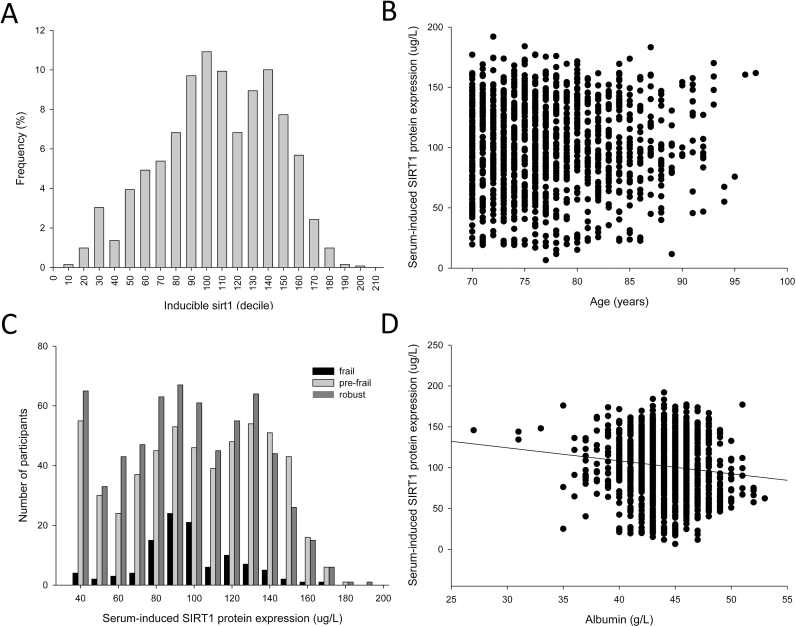
The bioassay measurements were completed for serum samples from 1,318 participants (77.0 ± 5.5 years, age range: 70–97 years). There was a bimodal distribution of concentrations but there was no relationship with the age of the participants or their frailty status (Figure 1A–C). Kaplan–Meier survival analysis performed according to quintiles of serum-induced SIRT1 expression showed no relationship with survival (Supplementary Figure 1). Serum-induced SIRT1 expression levels were not influenced by self-reported history of disease apart from osteoporosis (Supplementary Figure 2).
Figure 1.
The frequency distribution of serum-induced SIRT1 expression (A) and its relationship with age (B) (n = 1,318) and frailty (C) (n = 1,289). There was overrepresentation of robust and prefrail subjects with low levels of serum-induced SIRT1 expression. (D) The relationship between serum albumin and the serum-induced SIRT1 expression assay. There was a significant relationship (Pearson’s Product Moment correlation −0.12, p < .0001).
There was no difference in the serum-induced SIRT1 expression levels between the robust, prefrail, and frail groups (Table 1) but the distributions of the SIRT1 results appeared to differ between groups (Figure 1C) as noted in our pilot study (8). Frail participants were less likely (odds ratio (OR) 0.34, 95% confidence interval (CI) 0.17–0.69) and robust participants more likely (OR 1.31, 95% CI 1.00–1.72) to have a serum-induced SIRT1 expression result in the lowest quintile.
Table 1.
Relationship Between Serum-Induced SIRT1 Expression and Frailty
| N | Mean | ANOVA | |
|---|---|---|---|
| Robust | 636 | 99.7 ± 36.5 | |
| Prefrail | 548 | 104.1 ± 38.8 | p = .1, ns |
| Frail | 105 | 102.3 ± 26.8 | |
| Q1 | Q2–Q5 | OR (95% CI) | |
| Frail | 9 | 96 | 0.34 (0.17–0.69, p < .002) |
| Robust + prefrail | 254 | 930 | |
| Robust | 144 | 492 | 1.31 (1.00–1.72, p = .049) |
| Frail + prefrail | 119 | 534 |
Note: ANOVA = analysis of variance; CI = confidence interval; OR = odds ratio. The OR is calculated from participants in quintile 1 (lowest serum- induced SIRT1 expression) vs participants in the other four quintiles.
Serum-Induced SIRT1 Expression and Markers of Body Composition and Nutrition
Participants with serum-induced SIRT1 expression in the lowest quintile had anthropometric and blood results (Table 2) that were consistent with increased nutritional intake compared with the rest of the participants. These parameters included higher body weight, height, fat mass, albumin, cholesterol, and hemoglobin levels. Participants with serum-induced SIRT1 expression in the highest quintile had blood test results consistent with reduced nutritional intake including lower albumin and increased creatinine levels. A scatter plot of albumin versus serum-induced SIRT1 expression confirmed a strong negative correlation (Figure 1D, Pearson’s correlation p < .0001). Other biochemical and hematological parameters were not associated with SIRT1 expression (data not shown).
Table 2.
Relationship Between Quintiles of Serum-Induced SIRT1 Expression and Markers of Nutrition and Body Composition
| Q1 | Q2 | Q3 | Q4 | Q5 | p Value | ||
|---|---|---|---|---|---|---|---|
| Q1 vs Rest | Q5 vs Rest | ||||||
| BMI | 27.93 ± 4.04 | 27.66 ± 3.94 | 27.65 ± 3.77 | 27.44 ± 4.00 | 28.10 ± 4.18 | ns | ns |
| Height | 1.70 ± 0.07 | 1.69 ± 0.08 | 1.68 ± 0.07 | 1.68 ± 0.07 | 1.68 ± 0.13 | 0.002 | ns |
| Weight | 80.74 ± 12.86 | 79.04 ± 13.53 | 78.41 ± 12.28 | 78.03 ± 13.12 | 79.68 ± 13.46 | 0.030 | ns |
| DEXA % fat | 29.05 ± 5.53 | 27.96 ± 5.35 | 27.66 ± 5.79 | 28.10 ± 6.04 | 28.11 ± 5.64 | 0.006 | ns |
| DEXA % lean | 70.95 ± 5.53 | 72.04 ± 5.35 | 72.34 ± 5.80 | 71.90 ± 6.04 | 71.89 ± 5.64 | 0.006 | ns |
| Albumin | 44.76 ± 2.75 | 43.95 ± 2.95 | 43.76 ± 2.51 | 43.56 ± 2.58 | 43.55 ± 2.85 | < 10–5 | 0.02 |
| Creatinine | 94.53 ± 24.95 | 95.47 ± 26.54 | 97.21 ± 33.85 | 94.19 ± 25.01 | 102.64 ± 68.61 | ns | 0.008 |
| Hemoglobin | 145.28 ± 13.69 | 141.86 ± 17.21 | 140.64 ± 20.63 | 141.26 ± 15.70 | 141.45 ± 15.44 | 0.001 | ns |
| Cholesterol | 4.65 ± 0.99 | 4.67 ± 1.06 | 4.56 ± 0.94 | 4.35 ± 0.93 | 4.43 ± 0.92 | 0.03 | ns |
| LDL | 2.59 ± 0.90 | 2.58 ± 0.99 | 2.47 ± 0.85 | 2.32 ± 0.83 | 2.40 ± 0.84 | 0.02 | ns |
Note: DEXA = dual x-ray absorptiometry; LDL = low-density lipoprotein.
SIRT1 SNPs and Frailty, Mortality, and Age-Related Diseases
The frequency of the alleles for the three SNPs are shown in Supplementary Table 2. SNP rs3758391 was not in Hardy–Weinberg equilibrium confirming another study (19), which also reported a paucity of the CT heterozygotes.
The distribution of the polymorphisms was not significantly different between the frail, prefrail, and robust groups whether analyzed by allele or genotype (Supplementary Table 3). There was a weakly positive association between the distribution of the rs3740051 alleles and the prefrail phenotype (p = .04) but this is not significant when adjusted for multiple comparisons. In addition, there were no statistically significant relationships between SNPs and mortality (Supplementary Figure 3) or age (data not shown).
Associations were observed between some SIRT1 SNPs and the diseases and conditions evaluated in the CHAMP database (Supplementary Table 4). Both rs3740051 and rs3758391 were associated at the allele and genotype level with arthritis. Other weak associations included rs3758391 with hearing impairment and heart attack and rs3740051 with back pain. The statistical significance of these associations is limited by multiple comparisons and none remain significant following Bonferroni correction. The relationship between the SNPs and MMSE was also evaluated (Table 3). There was an association between rs2273773 and MMSE with the CC genotype having a lower score (24.8 ± 3.5) than the TT genotype (27.2 ± 2.9).
Table 3.
The Relationship Between SIRT1 SNPs and MMSE Scores
| rs2273773 | N | CC | CT | TT | CC Dominant p Value | CC Recessive p Value |
| 1,454 | 24.8 ± 3.5 | 26.73.4 | 27.2 ± 2.9 | .01 | .01 | |
| rs3740051 | N | AA | AG | GG | AA Dominant p Value | AA Recessive p Value |
| 1,440 | 27.2 ± 30. | 26.8 ± 3.4 | 25.9 ± 2.3 | ns | ns | |
| rs3758391 | N | CC | CT | TT | CC Dominant p Value | CC Recessive p Value |
| 1,458 | 27.0 ± 3.1 | 27.1 ± 3.1 | 27.4 ± 2.4 | ns | ns |
Note: The p value for the CC dominant (CC and CT vs TT; AA and AG vs GG) and CC recessive (CC vs CT and TT, AA vs AG and GG) models are provided and are equivalent to the TT recessive and dominant models.
SIRT1 SNPs and Markers of Nutrition and Body Composition
Given the associations between serum-induced SIRT1 expression and markers of body composition and nutrition, we also studied the relationships between these parameters and the SIRT1 SNPs. There were many associations between the three SIRT1 SNPs and weight, height, and body mass index and the DEXA measurements of body fat and body lean (Table 4) and were strongest for rs2273773. The only consistent association between the SNPs and blood tests was for albumin, which was significantly associated with all three SNPs (Supplementary Table 5).
Table 4.
Association Between SIRT1 SNPs and Body Composition
| rs2273773 | N | CC | CT | TT | CC Dominant p Value | CC Recessive p Value |
| Weight (kg) | 1,615 | 70.82 ± 14.26 | 77.79 ± 12.87 | 79.51 ± 12.64 | 0.02* | 0.02* |
| Height (kg) | 1,610 | 1.67 ± 0.07 | 1.67 ± 0.07 | 1.69 ± 0.07 | 0.00* | ns |
| BMI | 1,607 | 25.39 ± 3.46 | 27.74 ± 3.78 | 27.80 ± 3.93 | ns | 0.03* |
| % fat | 1,610 | 24.56 ± 4.86 | 28.05 ± 5.52 | 28.37 ± 5.64 | ns | 0.02* |
| % lean | 1,610 | 72.02 ± 4.66 | 68.66 ± 5.32 | 68.33 ± 5.40 | ns | 0.02* |
| rs3740051 | N | AA | AG | GG | AA Dominant p Value | AA Recessive p Value |
| Weight (kg) | 1,599 | 79.58 ± 12.60 | 77.59 ± 13.17 | 73.67 ± 14.90 | ns | 0.02* |
| Height (m) | 1,594 | 1.69 ± 0.07 | 1.67 ± 0.07 | 1.67 ± 0.08 | ns | 0.00* |
| BMI | 1,591 | 27.82 ± 3.91 | 27.68 ± 3.89 | 26.29 ± 3.86 | ns | ns |
| % fat | 1,594 | 28.40 ± 5.60 | 28.03 ± 5.64 | 25.33 ± 4.35 | 0.06 | ns |
| % lean | 1,594 | 68.30 ± 5.37 | 68.69 ± 5.44 | 71.30 ± 4.08 | 0.06 | ns |
| rs3758391 | N | CC | CT | TT | CC Dominant p Value | CC Recessive p Value |
| Weight (kg) | 1,617 | 80.25 ± 12.17 | 78.69 ± 12.78 | 77.61 ± 13.96 | 0.04* | 0.004* |
| Height (m) | 1,612 | 1.69 ± 0.07 | 1.69 ± 0.07 | 1.68 ± 0.07 | 0.03* | ns |
| BMI | 1,609 | 28.08 ± 3.78 | 27.57 ± 3.96 | 27.45 ± 4.10 | Ns | 0.01* |
| % fat | 1,613 | 28.60 ± 5.62 | 28.08 ± 5.62 | 27.99 ± 5.58 | Ns | 0.06 |
| % lean | 1,613 | 68.10 ± 5.38 | 68.62 ± 5.41 | 68.70 ± 5.35 | Ns | 0.05* |
Note: BMI = body mass index.
SIRT1 SNPs and Serum-Induced SIRT1 Expression and Ethnicity
There was no relationship between SIRT SNPs and the serum-induced SIRT1 expression bioassay for any of the SNPs (Supplementary Table 6). Although the number of Chinese participants was low, there were differences in the distribution of their SNP genotypes compared to Australian, British and Mediterranean participants. Ethnicity did not influence the serum-induced SIRT1 expression assay (Supplementary Table 7).
Discussion
Much of the research on the biology of aging over the last decades has focused on putative mechanisms for aging. These include cellular processes such as oxidative stress, mitochondrial dysfunction, impaired autophagy, impaired genome maintenance, telomere shortening, and altered gene expression (20). On the other hand, there has also been increasing research that has focused on the mechanisms and cellular pathways associated with interventions that influence aging. The most robust intervention known to influence aging is caloric restriction (21). Recent research has identified four canonical pathways that appear to mediate some of the aging effects of caloric restriction. These include the sirtuins, insulin/IGF1/GH, AMPK and mTOR, and related downstream proteins such as PGC1α and FOXO (22). The sirtuin pathway in particular has received enormous attention because an agonist of SIRT1 called resveratrol was found to delay aging in various laboratory models including yeast, Caenorhabditis elegans, Drosophila, and mice (1,5).
The aim of our study was to evaluate the role of SIRT1 in aging in humans. Two assays were utilized to study SIRT1: the serum-induced SIRT1 expression assay which measures the activity of unidentified circulating factors that stimulate SIRT1 expression in vitro and SNPs in the SIRT1 gene. These were studied in a study of older men (CHAMP) in which we found that men with the lowest results for the serum-induced SIRT1 expression assay were less likely to be frail although the relationship was statistically weak. This result was unexpected because rats and humans undergoing caloric restriction with delayed aging have elevated levels of the serum-induced SIRT1 bioassay (3,6,7) and elevated expression and/or activity of SIRT1 is generally expected to be associated with delayed aging and improved age-related health (1).
To resolve this difference, we previously proposed that the serum-induced SIRT1 expression bioassay reflected nutritional intake rather than healthy aging (8). Therefore, the relationship between the serum-induced SIRT1 expression bioassay and parameters that reflect nutrition was studied. We found an inverse relationship between this assay and markers of nutrition as high levels of the serum-induced SIRT1 expression were associated with parameters such as lower body weight, body fat, and albumin. The results from the in vitro SIRT1 bioassay were not associated with any particular disease except osteoporosis.
Three SIRT1 SNPs were studied in CHAMP. There were no associations between SIRT1 SNPs and age, survival, or frailty. There were weak statistical associations between some of the SNPs with conditions that are common in older people such as arthritis, cognitive impairment, and hearing impairment. There have been other studies of SIRT1 SNPs with disease and these are shown in Table 5. These studies have found associations with cardiometabolic risk factors, diabetes mellitus, chronic obstructive pulmonary disease, cardiovascular disease, depression, cognitive function, and in some studies longevity. The possible association between SIRT1 SNPs with a broad range of diseases in this and other published studies might be consistent with an effect on aging biology influencing susceptibility to many age-related diseases.
Table 5.
Studies of SIRT1 SNPs, Longevity, and Disease
| SNP | Results | N | Population | Citation |
|---|---|---|---|---|
| rs3758391 | No effect on longevity | 1,026 | German | (12) |
| rs1885472 | ||||
| rs2273773 | ||||
| rs10997870 | ||||
| rs3758391 | rs2758391 associated with cardiovascular mortality and cognitive function | 1,245 | Dutch, Leiden, Netherlands | (13) |
| rs3740051 | ||||
| rs2236319 | ||||
| rs2273773 | ||||
| rs3818291 | ||||
| rs730821 | rs12413112 associated with energy expenditure, insulin sensitivity and blood glucose | 1,013 | Caucasian Tuebingen family Study | (26) |
| rs12413112 | ||||
| rs7069102 | ||||
| rs2273773 | ||||
| rs12778366 | No association with Alzheimer’s disease | 326 | Finnish | (27) |
| rs3740051 | ||||
| rs2236319 | ||||
| rs2273773 | ||||
| rs3758391 | Weak association between rs3758391 and type 2 diabetes | 519 | Mexican | (28) |
| rs3758391 | Both SNPs associated with older age | 482 | Han Chinese | (29) |
| rs4746720 | ||||
| rs10997875 | rs10997875 associated with major depressive disorder | 450 | Japanese | (30) |
| rs2273773 | ||||
| rs4746720 | ||||
| rs12778366 | ||||
| rs3740051 | Associated with diabetic nephropathy | 1304 | Japanese | (14) |
| rs3818291 | ||||
| rs2236319 | ||||
| rs2273773 | ||||
| rs7895833 | rs7069102 associated with cholesterol and rs2273773 with coronary artery calcification | 219 | Japanese hemodialysis patients | (31) |
| rs7069102 | ||||
| rs2273773 | ||||
| rs3758391 | rs4746720 associated with longevity | 500 | Yongfu, Guangxi, China | (11) |
| rs3740051 | ||||
| rs2273773 | ||||
| rs4746720 | ||||
| rs100997870 | ||||
| rs2273773 | rs2273773 and rs7069102 associated with COPD | 200 | Mulga Turkish | (32) |
| rs7069102 | ||||
| rs7895833 | ||||
| rs2273773 | rs2273773 associated with hypertension | 340 | Kazakh Xinjiang, West China | (33) |
| rs4746720 | ||||
| rs7896005 | ||||
| rs144124002 | Associated with cardiovascular disease | 406 | Turkish | (34) |
| rs3758391 | rs3758391 associated with cardiovascular disease | 500 | Iranian | (35) |
| rs369274325 | ||||
| rs10997854 | rs10997854 and rs142194353 associated with blood lipids. No effect on longevity | 392 | Ashkenazi Jewish | (10) |
| rs142194353 | ||||
| rs12778366 | ||||
| rs35706870 | ||||
| rs932658 |
Note: SNP = single-nucleotide polymorphism.
The relationship between the SIRT1 SNPs, nutritional markers, and the serum-induced SIRT1 expression bioassay was studied. It was found that SNPs were associated with anthropometric measures such as height, body mass index, percentages body fat and body lean, and also with albumin but not with other blood measures of nutrition. There have been other reports that SIRT1 SNPs are associated with changes in body composition that might reflect nutrition. For example, Zillikens and colleagues reported that SIRT1 SNPs were associated with body mass index in the Rotterdam study (23), whereas Peeters and colleagues found that they were associated with obesity (24). SIRT1 regulates many processes that influence body metabolism and therefore SIRT1 SNPs may plausibly have an impact on the response to nutrition (1).
There are limitations to this study. The CHAMP cohort is a group of older men and does not provide the opportunity to study women or the relationship between SIRT1 and aging over a broader range of ages. The serum-induced SIRT1 expression assay measures the effects of unidentified circulating factors and until these factors are identified, the biological significance of the assay remains uncertain. The study of the SIRT1 SNPs examined many aging and disease outcomes, therefore the statistical significance of the results is limited by multiple comparisons and the conclusions can only be considered to be hypothesis-generating rather than definitive. Future studies should focus on repeating these investigations in other populations in order to confirm the generalizability of the conclusions. Studies of the association between SNPs and diseases in single populations must be replicated before the results can be considered to be robust. Our conclusion that SIRT1 might be associated with nutritional factors is inferred indirectly on blood and body composition parameters rather than direct measurement of dietary intake. In addition, participants were all aged 70 years or more, so the results do not capture the aging process over a longer timeframe. A major future goal is to identify the circulating factors that underpin the serum-induced SIRT1 expression bioassay.
Overall, the results of these studies of serum-induced SIRT1 expression and SIRT1 SNPs in CHAMP suggest that SIRT1 in humans is more strongly associated with nutritional markers and body composition, than directly with aging per se. Even so, SIRT1 may indirectly influence aging because of the association between nutrition and aging (8,25).
Funding
This study was funded by the National Health and Medical Research Council of Australia (NHMRC#512364), Sydney Medical School Foundation, and the Ageing and Alzheimers Research Institute.
Supplementary Material
References
- 1. Baur JA, Ungvari Z, Minor RK, Le Couteur DG, de Cabo R. Are sirtuins viable targets for improving healthspan and lifespan? Nat Rev Drug Discov. 2012;11:443–461. doi:10.1038/nrd3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sinclair DA, Guarente L. Small-molecule allosteric activators of sirtuins. Annu Rev Pharmacol Toxicol. 2014;54:363–380. doi:10.1146/annurev-pharmtox-010611-134657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cohen HY, Miller C, Bitterman KJ, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi:10.1126/science.1099196 [DOI] [PubMed] [Google Scholar]
- 4. Chen D, Bruno J, Easlon E, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi:10.1101/gad.1650608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baur JA, Pearson KJ, Price NL, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi:10.1038/nature05354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. de Cabo R, Fürer-Galbán S, Anson RM, Gilman C, Gorospe M, Lane MA. An in vitro model of caloric restriction. Exp Gerontol. 2003;38:631–639. [DOI] [PubMed] [Google Scholar]
- 7. Allard JS, Heilbronn LK, Smith C, et al. ; Pennington CALERIE Team. In vitro cellular adaptations of indicators of longevity in response to treatment with serum collected from humans on calorie restricted diets. PLoS One. 2008;3:e3211. doi:10.1371/journal.pone.0003211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Le Couteur DG, Benson VL, McMahon AC, et al. Determinants of serum-induced SIRT1 expression in older men: the CHAMP study. J Gerontol A Biol Sci Med. 2011;66:3–8. doi:10.1093/gerona/glq158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumar R, Mohan N, Upadhyay AD, et al. Identification of serum sirtuins as novel noninvasive protein markers for frailty. Aging Cell. 2014;13:975–980. doi:10.1111/acel.12260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han J, Atzmon G, Barzilai N, Suh Y. Genetic variation in Sirtuin 1 (SIRT1) is associated with lipid profiles but not with longevity in Ashkenazi Jews. Transl Res. 2015;165:480–481. doi:10.1016/j.trsl.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang J, Sun L, Liu M, et al. Association between SIRT1 gene polymorphisms and longevity of populations from Yongfu region of Guangxi. Chinese J Med Genetics. 2013;30:55–59. [DOI] [PubMed] [Google Scholar]
- 12. Flachsbart F, Croucher PJ, Nikolaus S, et al. Sirtuin 1 (SIRT1) sequence variation is not associated with exceptional human longevity. Exp Gerontol. 2006;41:98–102. doi:10.1016/j.exger.2005.09.008 [DOI] [PubMed] [Google Scholar]
- 13. Kuningas M, Putters M, Westendorp RG, Slagboom PE, van Heemst D. SIRT1 gene, age-related diseases, and mortality: the Leiden 85-plus study. J Gerontol A Biol Sci Med Sci. 2007;62:960–965. [DOI] [PubMed] [Google Scholar]
- 14. Maeda S, Koya D, Araki S, et al. Association between single nucleotide polymorphisms within genes encoding sirtuin families and diabetic nephropathy in Japanese subjects with type 2 diabetes. Clin Exp Nephrol. 2011;15:381–390. doi:10.1007/s10157-011-0418-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cumming RG, Handelsman D, Seibel MJ, et al. Cohort profile: the Concord Health and Ageing in Men Project (CHAMP). Int J Epidemiol. 2009;38:374–378. doi:10.1093/ije/dyn071 [DOI] [PubMed] [Google Scholar]
- 16. Le Couteur DG, Blyth FM, Creasey HM, et al. The association of alanine transaminase with aging, frailty, and mortality. J Gerontol A Biol Sci Med Sci. 2010;65:712–717. doi:10.1093/gerona/glq082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Held FP, Blyth F, Gnjidic D, et al. Association rules analysis of comorbidity and multimorbidity: the Concord Health and Aging in Men Project. J Gerontol A Biol Sci Med Sci. 2016;71:625–631. doi:10.1093/gerona/glv181 [DOI] [PubMed] [Google Scholar]
- 18. Hirani V, Naganathan V, Blyth F, et al. Low hemoglobin concentrations are associated with sarcopenia, physical performance, and disability in older Australian men in cross-sectional and longitudinal analysis: the Concord Health and Ageing in Men Project. J Gerontol A Biol Sci Med Sci. 2016;71:1667–1675. doi:10.1093/gerona/glw055 [DOI] [PubMed] [Google Scholar]
- 19. Consiglio CR, Juliana da Silveira S, Monticielo OA, Xavier RM, Brenol JC, Chies JA. SIRT1 promoter polymorphisms as clinical modifiers on systemic lupus erythematosus. Mol Biol Rep. 2014;41:4233–4239. doi:10.1007/s11033-014-3294-3 [DOI] [PubMed] [Google Scholar]
- 20. de Cabo R, Le Couteur DG. The biology of ageing. In: Kasper D, Fauci A, Hauser S, Longo D, Jameson JL, Loscalzo J, eds. Harrisons Principles of Internal Medicine. 19th ed Columbus, OH: McGraw Hill Education; 2015:94e. [Google Scholar]
- 21. Mercken EM, Carboneau BA, Krzysik-Walker SM, de Cabo R. Of mice and men: the benefits of caloric restriction, exercise, and mimetics. Ageing Res Rev. 2012;11:390–398. doi:10.1016/j.arr.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Solon-Biet SM, Mitchell SJ, de Cabo R, Raubenheimer D, Le Couteur DG, Simpson SJ. Macronutrients and caloric intake in health and longevity. J Endocrinol. 2015;226:R17–R28. doi:10.1530/JOE-15-0173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zillikens M, van Meurs J, Rivadeneira F, et al. SIRT1 genetic variation is related to BMI and risk of obesity. Diabetes. 2009;58:2828–2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peeters AV, Beckers S, Verrijken A, et al. Association of SIRT1 gene variation with visceral obesity. Hum Genet. 2008;124:431–436. doi:10.1007/s00439-008-0567-8 [DOI] [PubMed] [Google Scholar]
- 25. Miller SL, Wolfe RR. The danger of weight loss in the elderly. J Nutr Health Aging. 2008;12:487–491. [DOI] [PubMed] [Google Scholar]
- 26. Weyrich P, Machicao F, Reinhardt J, et al. SIRT1 genetic variants associate with the metabolic response of Caucasians to a controlled lifestyle intervention–the TULIP Study. BMC Med Genet. 2008;9:100. doi:10.1186/1471-2350-9-100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Helisalmi S, Vepsäläinen S, Hiltunen M, et al. Genetic study between SIRT1, PPARD, PGC-1alpha genes and Alzheimer’s disease. J Neurol. 2008;255:668–673. doi:10.1007/s00415-008-0774-1 [DOI] [PubMed] [Google Scholar]
- 28. Cruz M, Valladares-Salgado A, Garcia-Mena J, et al. Candidate gene association study conditioning on individual ancestry in patients with type 2 diabetes and metabolic syndrome from Mexico City. Diabetes Metab Res Rev. 2010;26:261–270. doi:10.1002/dmrr.1082 [DOI] [PubMed] [Google Scholar]
- 29. Zhang WG, Bai XJ, Chen XM. SIRT1 variants are associated with aging in a healthy Han Chinese population. Clin Chim Acta. 2010;411:1679–1683. doi:10.1016/j.cca.2010.06.030 [DOI] [PubMed] [Google Scholar]
- 30. Kishi T, Yoshimura R, Kitajima T, et al. SIRT1 gene is associated with major depressive disorder in the Japanese population. J Affect Disord. 2010;126:167–173. doi: 10.1016/j.jad.2010.04.003 [DOI] [PubMed] [Google Scholar]
- 31. Shimoyama Y, Mitsuda Y, Tsuruta Y, Suzuki K, Hamajima N, Niwa T. SIRTUIN 1 gene polymorphisms are associated with cholesterol metabolism and coronary artery calcification in Japanese hemodialysis patients. J Ren Nutr. 2012;22:114–119. doi:10.1053/j.jrn.2011.10.025 [DOI] [PubMed] [Google Scholar]
- 32. Kalemci S, Edgunlu TG, Kara M, Turkcu UO, Cetin ES, Zeybek A. Sirtuin gene polymorphisms are associated with chronic obstructive pulmonary disease in patients in Muğla province. Kardiochir Torakochirurgia Pol. 2014;11:306–310. doi:10.5114/kitp.2014.45682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhong XL, Miao HJ, Fang ZM, et al. The effect of SIRT1 gene polymorphisms on ambulatory blood pressure of hypertensive patients in the Kazakh population. Genet Test Mol Biomarkers. 2015;19:561–565. doi:10.1089/gtmb.2015.0111 [DOI] [PubMed] [Google Scholar]
- 34. İzmirli M, Göktekin Ö, Bacaksız A, Uysal Ö, Kılıç Ü. The effect of the SIRT1 2827 A > G polymorphism, resveratrol, exercise, age and occupation in Turkish population with cardiovascular disease. Anatol J Cardiol. 2015;15:103–106. doi:10.5152/akd.2014.5214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mohtavinejad N, Nakhaee A, Harati H, Poodineh J, Afzali M. SIRT1 gene is associated with cardiovascular disease in the Iranian population. Egypt J Med Human Genet. 2014;16:117–122. doi: 10.1016/j.ejmhg.2014.11.005 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.