Abstract
The association of primary Sjögren’s syndrome (pSS) with Major Histocompatibility Complex (MHC) alleles is quintessential of MHC-disease associations. Indeed, although disease associations with classical HLA class I and II alleles/haplotypes are amply documented, further dissection is often prevented by the strong linkage disequilibrium across the entire MHC complex. Here we study the association of pSS, not with HLA genes, but with the non-conventional MHC encoded class I gene, MICA (MHC class I chain-related gene A). MICA is selectively expressed within epithelia, and is the major ligand for the activatory receptor, NKG2D, both attributes relevant to pSS’ etiology. MICA-pSS association was studied in two independent (French and UK) cohorts representing a total of 959 cases and 1,043 controls. MICA*008 allele was shown to be significantly associated with pSS (pcor=2.61 × 10−35). A multivariate logistic regression showed that this association was independent of all major known MHC-linked risk loci/alleles, as well as other relevant candidate loci that are in linkage disequilibrium with MICA*008 i.e. HLA-B*08:01, rs3131619 (T), MICB*008, TNF308A, HLA-DRB1*03:01 and HLA-DRB1*15:01 (P = 1.84 × 10−04). Furthermore, independently of the MICA*008 allele, higher levels of soluble MICA proteins were detected in sera of pSS patients compared to healthy controls. This study hence defines MICA as a new, MHC-linked, yet HLA-independent, pSS risk locus and opens a new front in our understanding of the still enigmatic pathophysiology of this disease. The fact that the soluble MICA protein is further amplified in MICA*008 carrying individuals, might also be relevant in other auto-immune diseases and cancer.
Introduction
Primary Sjögren's syndrome (pSS) is a systemic autoimmune disease with a population prevalence of 0.05 to 0.6%, and a 14:1 female to male ratio (1–3). pSS could be defined as an exocrinopathy where the dysfunction of salivary and lacrimal glands respectively lead to xerostomia and keratoconjunctivitis sicca (4). Like most complex autoimmune diseases, pSS is believed to be caused by an interplay between environmental factors (infection, stress…) and host’s genome. Among the latter, MHC (also known in man as the "Human Leukocyte Antigen"; HLA) genes are major players in susceptibility to pSS, as is the case for most, if not all, autoimmune diseases (5). Indeed, a number of pSS associated HLA loci - including risk and protective alleles - have been reported in the past three decades (6). HLA class II DRB1 and DQB1 have been extensively studied, with HLA-DRB1*15:01 (also called HLA-DR2) and HLA-DRB1*03:01 (also known as HLA-DR3) alleles defining the strongest reported associations in Caucasians (7,8). In the MHC class I region, associations are well known with HLA-B*008 (first reported in 1975) and HLA-A*024 (9,10). In the class III region, SNPs in TNF and more recently NCR3/NKp30 have also been reported as pSS risk factors (11–13). More recently, two genome-wide association studies (GWAS) identified SNPs with high predictive values in the HLA region. The first study was performed in a Han Chinese population. Amongst the 15 associated SNPs uncovered in the HLA region, two reached genome-wide significance, namely rs9271588 (HLA-DQA1 region) and rs4282438 (HLA-DPB1 region) (14). The second study examined patients and controls of European descent. The authors identified 160 variants in the HLA region with three peaks accounting for most of the association: rs115575857 (HLA-DQB1 region), rs3129770 (HLA-DQA1 region) and rs3131619 (MHC class I chain-related gene A (MICA) region) (15). The vicinity between MICA and rs3131619 (51.2 kb apart) begs investigating the relevance of MICA’s coding sequence per se in susceptibility to pSS.
The MIC gene family encodes a distinct family of MHC class I genes within the HLA complex (16–18). MICA is distant of only 46 kb from the HLA-B locus. It encodes a stress-induced, membrane-bound, single chain (no dimerization with ß2-microglobulin) glycoprotein recognized by NKG2D expressing CD8+ αß, ɣδ T-cells and/or NK cells (18). MIC proteins appear to be specifically expressed in epithelial/mucosal tissues (19) although they do show a wider transcription pattern (20). By triggering the cytolysis mediated by NKG2D-bearing cells, MICA participates in the first line of defense against pathogens and cancer. As MICA-NKG2D interaction has been shown to increase inflammatory cytokine production and proliferation of certain subsets of T cells, its implication in autoimmunity seems plausible (18,20). MICA is highly polymorphic, with over 100 alleles identified to date and such with only few thousands individuals sequenced (21,22) (http://hla.alleles.org; date last accessed February 28, 2017). This is to be compared to close to 16,000 alleles for HLA genes, but uncovered after sequence analyses of several million individuals (http://hla.alleles.org; date last accessed February 28, 2017). Nevertheless, and as of now, after the classical HLA (A/B/C/DR/DP/DQ) genes, MICA is the most polymorphic human and hence MHC gene. The polymorphism of MICA comprises a mixture of coding SNPs throughout the molecule and an exon 5 (transmembrane domain) embedded triplet repeat microsatellite (GCTn). With regards to the latter, seven different GCT (alanine) repeats have been described: alleles carrying four alanine repeats are dubbed A4, those with 5, A5, 6 (A6), 7 (A7), 8 (A8), 9 (A9) and 10 (A10). An additional allele is defined by the A5 allele with a guanine insertion after the second of the five trinucleotide repeats. This causes a frameshift mutation leading to a premature intradomain stop codon. This particular allele is called A5.1. Several studies have found associations between MICA alleles and autoimmune diseases (23–25), although many have suffered from the well-known caveats of MHC-disease association studies i.e the MHC-wide strong linkage disequilibrium (LD) (5) and the lack of replication in independent cohorts (26), respectively. Finally, and in addition to above-mentioned circumstantial evidences for the potential relevance of MICA in pSS pathophysiology, the recent identification of the SNP rs3131619, located in close proximity to MICA and showing genome-wide significance in a pSS GWAS (15), further compelled us to address the following two questions directly: (1) are MICA alleles associated with pSS? (2) if yes, is this a primary association or merely due to linkage disequilibrium with the already known HLA class I, class II or other closely linked loci? The study presented here – incidentally the first analysis of MICA association with pSS - provides answers to both questions.
Results
The distribution of MICA alleles was first studied in a discovery cohort (French Assessment of Systemic Symptoms and Evolution in patients with pSS; ASSESS) of 347 pSS patients and 553 unrelated healthy subjects (Table 1), upon full-length sequencing of exons encoding the extracellular domains of the MICA glycoprotein. The MICA*008 allele showed a significant association with pSS as its frequency was 46.8% in patients and 31.6% in controls (OR = 1.90; 95% CI 1.56–2.31; P = 9.37 × 10−09). In order to validate this association, the microsatellite marker of MICA (this is a coding microsatellite within MICA’s trans-membrane encoding exon) was analyzed in a replication cohort from United Kingdom (UK primary Sjögren’s syndrome registry; UKPSSR) composed of 612 pSS patients and 490 controls (hereafter called replication cohort). The microsatellite allele MICA A5.1 - which tags MICA*008 (25) (99.77% correlation in Europeans, data not shown) - was also found to be significantly more frequent in patients than in controls in this cohort (OR = 2.28; 95% CI 1.92–2.70; P = 1.54 × 10−20), confirming the genuine association of this MICA allele with pSS (Supplementary Material, Table S1).
Table 1.
MICA allele frequencies in patients and controls of the discovery cohort
| Patients (n = 347) |
Controls (n = 553) |
||||||
|---|---|---|---|---|---|---|---|
| Allele (TMmicrosatellite marker) | n | % | n | % | Pa | OR | 95% Cl |
| MICA*008 (A5.1) | 325 | 46.8 | 350 | 31.6 | 9.37 × 10−09 | 1.90 | 1.56–2.31 |
| MICA*002 (A9) | 60 | 8.6 | 123 | 11.1 | ns | ||
| MICA*004 (A6) | 66 | 9.5 | 150 | 13.6 | ns | ||
| MICA*009 (A6) | 47 | 6.8 | 121 | 10.9 | ns | ||
| MICA*010 (A5) | 39 | 5.6 | 52 | 4.7 | ns | ||
| MICA*018 (A4) | 25 | 3.6 | 56 | 5.1 | ns | ||
| MICA*007 (A4) | 25 | 3.6 | 50 | 4.5 | ns | ||
| MICA*012 (A4) | 17 | 2.4 | 20 | 1.8 | ns | ||
| MICA*027 (A5) | 12 | 1.7 | 26 | 2.4 | Ns | ||
| MICA*001 (A4) | 15 | 2.2 | 22 | 2.0 | Ns | ||
| MICA*017 (A9) | 16 | 2.3 | 33 | 3.0 | Ns | ||
| MICA*016 (A5) | 18 | 2.6 | 31 | 2.8 | Ns | ||
| MICA*011 (A6) | 4 | 0.6 | 20 | 1.8 | Ns | ||
| MICA*019 (A5) | 16 | 2.3 | 32 | 2.9 | Ns | ||
| MICA*006 (A6) | 4 | 0.6 | 5 | 0.5 | Ns | ||
| MICA*015 (A9) | 3 | 0.4 | 2 | 0.2 | Ns | ||
| Other alleles | 2 | 0.3 | 13 | 1.2 | Ns | ||
Chi square test (Pearson) with Bonferroni correction (P-values were multiplied by 17).
CI: confidence interval; ns: not significant (P ≥ 0.05); OR: odds ratio.
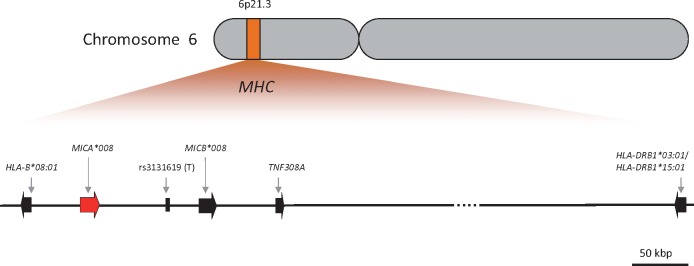
Because the HLA region displays a high degree of LD and some pSS associated genes/markers are already known within this chromosomal segment, using proxy SNPs, we tested in addition to these well-known MHC-linked pSS susceptibility loci, HLA-DRB1*03:01 and HLA-DRB1*15:01, the following MICA neighboring risk alleles for association with pSS: HLA-B*08:01 (telomeric to MICA), rs3131619(T) (T allele, centromeric to MICA), MICB*008 (centromeric to MICA), TNF308A (centromeric to MICA) (Fig. 1). No significant deviation (p ≤ 0.001) from Hardy-Weinberg equilibrium was observed for any tested gene (or allele) in controls. Table 2 summarizes the statistical analysis with respect to presence/absence of potential risk alleles in different patients’ sub-groups. The following six sets of comparisons were performed for both cohorts (i.e. discovery cohort and replication cohort) as well as in the combined data set (meta-analysis): (I) all pSS cases (irrespective of their anti-SSA/SSB auto-antibody status) versus healthy controls, (II) pSS cases positive for anti-SSA antibodies only versus healthy controls, (III) pSS cases positive for anti-SSA and anti-SSB antibodies versus healthy controls, (IV) pSS cases negative for anti-SSA and anti-SSB antibodies versus healthy controls, (V) pSS cases positive for anti-SSA antibodies only versus negative for anti-SSA and anti-SSB, and (VI) pSS cases positive for anti-SSA and anti-SSB antibodies versus negative for anti-SSA and anti-SSB antibodies. In both cohorts, when considering all patients (again, irrespective of their auto-antibody status) versus healthy controls, with the exception of HLA-DRB1*15:01, all tested loci/alleles were found to be strong susceptibility markers (all P-value < 10−34 in meta-analysis) (Table 2). Of note, in both controls and cases, the frequencies of all tested markers were higher in the replication cohort than in the discovery cohort, confirming the frequency gradient of HLA markers across Europe (27). In both cohorts, the association of HLA-B*08:01, MICA*008, rs3131619(T), MICB*008, TNF308A and HLA-DRB1*03:01 was stronger in the subset of patients with both disease-specific anti-SSA and anti-SSB autoantibodies than in all pSS patients, with OR (meta) of 5.19 versus 3.57, 2.83 versus 2.24, 5.98 versus 3.96, 5.22 versus 3.54, 3.65 versus 2.57 and 6.01 versus 3.90 for HLA-B*08:01, MICA*008, rs3131619(T), MICB*008, TNF308A and HLA-DRB1*03:01, respectively (Table 2). Moreover, these markers were statistically more significant in the same subset with P-values (meta) of 3.41 × 10−69 versus 9.13 × 10−51, 1.13 × 10−37 versus 2.61 × 10−35, 4.94 × 10−81 versus 3.91 × 10−58, 2.76 × 10−67 versus 3.03 × 10−48, 4.94 × 10−49 versus 2.20 × 10−34 and 1.42 × 10−82 versus 1.20 × 10−57 for HLA-B*08:01, MICA*008, rs3131619 (T), MICB*008, TNF308A and HLA-DRB1*03:01, respectively. In the subset of patients with only anti-SSA antibodies, the disease association was stronger as compared to all pSS patients only for HLA-DRB1*15:01 (OR (meta) of 2.14 vs. 1.54 and P-values (meta) of 3.01 × 10−08 vs. 6.50 × 10−06). Only weak associations of HLA-B*08:01, MICA*008, MICB*008, TNF308A and HLA-DRB1*03:01 could be observed in the subset of patients without anti-SSA and anti-SSB antibodies. Finally, the same loci/alleles showed a weak association with anti-SSA and anti-SSB positivity but not with isolated anti-SSA positivity (Table 2).
Figure 1.
Schematic chromosomal localization of the tested alleles/loci. This figure represents a simplified MHC map centered on genes of interest in this manuscript.
Table 2.
Association analysis of various MHC-linked genetic markers with pSS in discovery and replication cohorts
| Discovery cohort |
Replication cohort |
Meta-analysis |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases % | Controls % | Pa | OR (95%CI) | Cases % | Controls % | Pa | OR (95%CI) | Cases % | Controls % | Pa | OR (95%CI) | ||
| n = | 347 | 553 | 612 | 490 | 959 | 1043 | |||||||
| All pSS cases vs. healthy controls | HLA-B*08:01 b | 21.5 | 7.7 | 1.81 × 10−16 | 3.28 (2.47–4.37) | 33.9 | 13.6 | 3.52 × 10−27 | 3.27 (2.63–4.06) | 29.4 | 10.5 | 9.13 × 10−51 | 3.57 (3.01–4.24) |
| MICA*008c | 46.8 | 32.1 | 2.45 × 10−09 | 1.86 (1.53–2.27) | 62.2 | 42.1 | 5.28 × 10−20 | 2.26 (1.9–2.68) | 56.6 | 36.8 | 2.61× 10 − 35 | 2.24 (1.97–2.54) | |
| rs3131619 | 22.2 | 7.5 | 2.79 × 10−18 | 3.5 (2.63–4.66) | 34.7 | 12.4 | 1.78 × 10−32 | 3.74 (2.99–4.68) | 30.2 | 9.8 | 3.91× 10−58 | 3.96 (3.33–4.71) | |
| MICB*008 | 21 | 8.6 | 5.00 × 10−13 | 2.82 (2.13–3.72) | 31.6 | 11.5 | 8.05 × 10−30 | 3.68 (2.92–4.63) | 27.7 | 10 | 3.03× 10 − 48 | 3.54 (2.98–4.22) | |
| TNF308A | 24.2 | 13.9 | 2.11 × 10−07 | 1.97 (1.55–2.52) | 36.9 | 17.7 | 1.35 × 10−22 | 2.73 (2.23–3.34) | 32.3 | 15.7 | 2.20 × 10−34 | 2.57 (2.21–2.99) | |
| HLA-DRB1*15:01 d | 13.3 | 10.8 | Ns | 21.1 | 14.7 | 8.04 × 10−04 | 1.55 (1.24–1.94) | 18.2 | 12.7 | 6.50 × 10−06 | 1.54 (1.3–1.83) | ||
| HLA-DRB1*03:01 e | 23.1 | 8.2 | 6.79 × 10−18 | 3.34 (2.53–4.41) | 34.6 | 12.1 | 4.34 × 10−33 | 3.82 (3.05–4.78) | 30.4 | 10.1 | 1.20 × 10−57 | 3.9 (3.28–4.64) | |
| SSA+/B- cases % | Controls % | SSA+/B−cases % | Controls % | SSA+/B−cases % | Controls % | ||||||||
| n = | 91 | 553 | 120 | 490 | 211 | 1043 | |||||||
| SSA+/B-f cases vs. healthy controls | HLA-B*08:01 b | 20.9 | 7.7 | 1.40 × 10−07 | 3.17 (2.08 − 4.83) | 28.8 | 13.6 | 1.00 × 10−07 | 2.57 (1.84–3.59) | 25.4 | 10.5 | 6.45 × 10−16 | 2.91 (2.24–3.78) |
| MICA*008 c | 46.2 | 32.1 | 1.47 × 10−03 | 1.81 (1.32–2.49) | 66.7 | 42.1 | 6.25 × 10−11 | 2.75 (2.04–3.69) | 57.8 | 36.8 | 7.37 × 10−15 | 2.35 (1.9–2.91) | |
| rs3131619 | 20.3 | 7.5 | 2.73 × 10−07 | 3.13 (2.05–4.79) | 30.8 | 12.4 | 2.83 × 10−11 | 3.13 (2.24–4.36) | 26.3 | 9.8 | 1.26 × 10−19 | 3.26 (2.52–4.23) | |
| MICB*008 | 20.9 | 8.6 | 3.29 × 10−06 | 2.8 (1.85–4.24) | 26.3 | 11.5 | 1.31 × 10−08 | 2.83 (1.99–4.01) | 23.9 | 10.0 | 4.18 × 10−15 | 2.89 (2.22–3.77) | |
| TNF308A | 22.0 | 13.9 | 3.41 × 10−02 | 1.74 (1.18–2.57) | 34.2 | 17.7 | 1.20 × 10−07 | 2.42 (1.77–3.31) | 28.9 | 15.7 | 6.99 × 10−10 | 2.19 (1.72–2.78) | |
| HLA-DRB1*15:01 d | 17.0 | 10.8 | ns | 28.8 | 14.7 | 1.91 × 10−06 | 2.34 (1.68–3.26) | 23.7 | 12.7 | 3.01 × 10−08 | 2.14 (1.65–2.78) | ||
| HLA-DRB1*03:01 e | 22.0 | 8.2 | 9.05 × 10−08 | 3.14 (2.08–4.74) | 29.2 | 12.1 | 4.52 × 10−10 | 2.98 (2.12–4.18) | 26.1 | 10.1 | 1.82 × 10−18 | 3.15 (2.43–4.08) | |
| SSA+/B+ cases % | Controls % | SSA+/B+ cases % | Controls % | SSA+/B+ cases % | Controls % | ||||||||
| n = | 118 | 553 | 347 | 490 | 465 | 1043 | |||||||
| SSA+/B+ cases vs. healthy controls | HLA-B*08:01 b | 32.2 | 7.7 | 4.72 × 10−25 | 5.71 (4.01–8.11) | 39.6 | 13.6 | 1.47 × 10−33 | 4.18 (3.3–5.3) | 37.7 | 10.5 | 3.41 × 10−69 | 5.19 (4.28–6.3) |
| MICA*008 c | 53.8 | 32.1 | 1.92 × 10−09 | 2.46 (1.85–3.28) | 65.1 | 42.1 | 1.28 × 10−19 | 2.56 (2.1–3.14) | 62.3 | 36.8 | 1.13 × 10−37 | 2.83 (2.41–3.32) | |
| rs3131619 | 33.9 | 7.5 | 1.84 × 10−28 | 6.3 (4.44–8.94) | 41.4 | 12.4 | 3.92 × 10−41 | 4.97 (3.9–6.34) | 39.5 | 9.8 | 4.94 × 10−81 | 5.98 (4.92–7.27) | |
| MICB*008 | 31.4 | 8.6 | 8.58 × 10−21 | 4.85 (3.43–6.86) | 37.3 | 11.5 | 2.72 × 10−37 | 4.81 (3.75–6.18) | 35.8 | 10.0 | 2.76 × 10−67 | 5.22 (4.29–6.36) | |
| TNF308A | 36.4 | 13.9 | 1.76 × 10−15 | 3.54 (2.59–4.86) | 41.8 | 17.7 | 1.06 × 10−26 | 3.35 (2.68–4.18) | 40.4 | 15.7 | 4.94 × 10−49 | 3.65 (3.06–4.35) | |
| HLA-DRB1*15:01 d | 11.9 | 10.8 | ns | 19.6 | 14.7 | ns | 17.6 | 12.7 | 2.07 × 10-03 | 1.48 (1.19-1.83) | |||
| HLA-DRB1*03:01 e | 36.0 | 8.2 | 1.15 × 10−29 | 6.28 (4.46–8.83) | 41.6 | 12.1 | 8.93 × 10−43 | 5.16 (4.05–6.59) | 40.2 | 10.1 | 1.42 × 10−82 | 6.01 (4.95–7.29) | |
| Cases % | Controls % | Pa | OR (95%CI) | Cases % | Controls % | Pa | OR (95%CI) | Cases % | Controls % | Pa | OR (95%CI) | ||
| n = | 131 | 553 | 73 | 490 | 204 | 1043 | |||||||
| SSA-/B- cases vs. healthy controls | HLA-B*08:01b | 13.4 | 7.7 | 2.46 × 10−02 | 1.85 (1.22–2.82) | 24.7 | 13.6 | 3.27 × 10−03 | 2.08 (1.37–3.17) | 17.4 | 10.5 | 4.22 × 10−04 | 1.81 (1.35–2.42) |
| MICA*008 c | 40.8 | 32.1 | 5.00 × 10−02 | 1.46 (1.11–1.93) | 54.8 | 42.1 | 2.83 × 10−02 | 1.66 (1.17–2.36) | 45.8 | 36.8 | 4.28 × 10−03 | 1.45 (1.17–1.8) | |
| rs3131619 | 14.1 | 7.5 | 4.98 × 10−03 | 2.02 (1.34–3.05) | 20.5 | 12.4 | ns | 16.4 | 9.8 | 7.13 × 10−04 | 1.8 (1.33–2.42) | ||
| MICB*008 | 12.6 | 8.6 | ns | 24.0 | 11.5 | 2.32 × 10−04 | 2.42 (1.58–3.71) | 16.7 | 10.0 | 4.95 × 10−04 | 1.81 (1.35–2.44) | ||
| TNF308A | 16.0 | 13.9 | ns | 28.8 | 17.7 | 1.00 × 10−02 | 1.88 (1.27–2.79) | 20.6 | 15.7 | Ns | |||
| HLA-DRB1*15:01d | 11.1 | 10.8 | ns | 15.8 | 14.7 | ns | 12.7 | 12.7 | Ns | ||||
| HLA-DRB1*03:01e | 13.4 | 8.2 | ns | 21.2 | 12.1 | 1.80 × 10−02 | 1.95 (1.26–3.03) | 16.2 | 10.1 | 2.25 × 10−03 | 1.72 (1.28–2.33) | ||
| SSA+/B- cases % | SSA-/B- cases % | SSA+/B- cases % | SSA-/B- cases % | SSA+/B- cases % | SSA-/B- cases % | ||||||||
| n = | 91 | 131 | 120 | 73 | 211 | 204 | |||||||
| SSA+/B- cases vs. SSA-/B- cases | HLA-B*08:01 b | 20.9 | 13.4 | ns | 28.8 | 24.7 | ns | 25.4 | 17.4 | 3.68 × 10−02 | 1.61 (1.15–2.26) | ||
| MICA*008 c | 46.2 | 40.8 | ns | 66.7 | 54.8 | ns | 57.8 | 45.8 | 3.85 × 10−03 | 1.62 (1.23–2.13) | |||
| rs3131619 | 20.3 | 14.1 | ns | 30.8 | 20.5 | ns | 26.3 | 16.4 | 3.68 × 10−03 | 1.82 (1.29–2.55) | |||
| MICB*008 | 20.9 | 12.6 | ns | 26.3 | 24 | ns | 23.9 | 16.7 | Ns | ||||
| TNF308A | 22.0 | 16.0 | ns | 34.2 | 28.8 | ns | 28.9 | 20.6 | 3.87 × 10−02 | 1.57 (1.14–2.16) | |||
| HLA-DRB1*15:01 d | 17.0 | 11.1 | ns | 28.8 | 15.8 | 2.56 × 10−02 | 2.16 (1.28–3.65) | 23.7 | 12.7 | 3.18 × 10−04 | 2.13 (1.47–3.07) | ||
| HLA-DRB1*03:01 e | 22.0 | 13.4 | ns | 29.2 | 21.2 | ns | 26.1 | 16.2 | 3.45 × 10-03 | 1.83 (1.3-2.57) | |||
| SSA+/B+ cases % | SSA-/B- cases % | SSA+/B+ cases % | SSA-/B- cases % | SSA+/B+ cases % | SSA-/B- cases % | ||||||||
| n = | 118 | 131 | 347 | 73 | 465 | 204 | |||||||
| SSA+/B+ cases vs. SSA-/B- cases | HLA-B*08:01 b | 32.2 | 13.4 | 3.17 × 10−06 | 3.08 (1.97–4.82) | 39.6 | 24.7 | 4.64 × 10−03 | 2.01 (1.34–3.01) | 37.7 | 17.4 | 1.18 × 10−12 | 2.88 (2.16–3.84) |
| MICA*008 c | 53.8 | 40.8 | 2.64 × 10−02 | 1.69 (1.18–2.41) | 65.1 | 54.8 | ns | 62.3 | 45.8 | 1.58 × 10−07 | 1.95 (1.54–2.47) | ||
| rs3131619 | 33.9 | 14.1 | 1.41 × 10−06 | 3.12 (2.01–4.84) | 41.4 | 20.5 | 1.51 × 10−05 | 2.74 (1.78–4.21) | 39.5 | 16.4 | 6.53 × 10−16 | 3.33 (2.48–4.46) | |
| MICB*008 | 31.4 | 12.6 | 3.15 × 10−06 | 3.14 (1.99–4.96) | 37.3 | 24 | 6.18 × 10−03 | 1.99 (1.32–3) | 35.8 | 16.7 | 2.97 × 10−12 | 2.88 (2.15–3.86) | |
| TNF308A | 36.4 | 16.0 | 1.36 × 10−06 | 3 (1.97–4.59) | 41.8 | 28.8 | 2.41 × 10−02 | 1.78 (1.2–2.62) | 40.4 | 20.6 | 1.40 × 10−11 | 2.62 (1.99–3.44) | |
| HLA-DRB1*15:01 d | 11.9 | 11.1 | ns | 19.6 | 15.8 | ns | 17.6 | 12.7 | ns | ||||
| HLA-DRB1*03:01 e | 36.0 | 13.4 | 2.49E-08 | 3.65 (2.34–5.69) | 41.6 | 21.2 | 2.74 × 10−05 | 2.65 (1.73–4.05) | 40.2 | 16.2 | 4.82E-17 | 3.49 (2.6–4.68) | |
Chi square test (Pearson) with Bonferroni correction (P-values were multiplied by 7). CI: confidence interval; OR: odds ratio.
Genotyping of HLA-B*008 was performed using proxies rs6457374 and rs2844535.
MICA*008 was typed using proxy MICA A5.1.
Genotyping of HLA-DRB1*15:01 was performed using proxy rs3129860).
HLA-DRB1*03:01 was typed using proxy rs2187668.
antibody status of patients are presented in Supplementary Material, Table S2.
Linkage disequilibrium (LD) between the tested loci was evaluated with Haploview software. MICA showed LD with HLA-B, rs3131619, MICB and TNF (D’ of respectively 0.95, 0.75, 0.86 and 0.63 in discovery cohort; D’ of, respectively, 0.96, 0.78, 0.87 and 0.80 in replication cohort). Significant LD was also observed between HLA-B, rs3131619, MICB and TNF (Supplementary Material, Fig. S1). Given this high degree of LD observed between the different markers, to statistically assess the independent effect of MICA, association of MICA*008 (presence/absence in controls and patients) was evaluated using a multiple logistic regression model including HLA-B*08:01, rs3131619(T), MICB*008, TNF308A and HLA-DRB1*03:01 alleles as co-variables. For HLA-DRB1, we included only the risk allele HLA-DRB1*03:01 in the multivariate model because it is the strongest risk allele of this gene and because HLA-DRB1*15:01 was not significant in univariate analysis (data not shown). In both cohorts MICA*008 showed a significant effect (pdiscovery = 0.029; preplication = 0.001; pmeta = 1.84 × 10−04) on pSS susceptibility independently of HLA-B*08:01, rs3131619(T), MICB*008, TNF308A and HLA-DRB1*03:01 (Table 3). Similar results were obtained when including sex as a co-variate in the multivariate model (Supplementary Material, Table S4). To further account for potential LD we restricted the analysis to subgroups of individuals without HLA-B*08:01, rs3131619(T), MICB*008, TNF308A, and HLA-DRB1*03:01 risk alleles. We found that MICA*008 was associated with pSS in the discovery cohort (pdiscovery = 0.032), replication cohort (preplication = 0.002) and meta-analysis (pmeta = 1.33 × 10−04) (Table 4). The distribution of the haplotype HLA-B*08:01+/MICA*008+/rs3131619(T)+/MICB*008+/TNF308A+/HLA-DRB1*03:01+ was found to be significantly more frequent in patients than in controls in all tested cohorts (pdiscovery = 5.01 × 10−19; preplication = 3.79 × 10−26; pmeta = 2.49 × 10−51; Supplementary Material, Table S5). Finally, to further characterize the genuine association of MICA*008 with pSS, independently of HLA-DRB1*03:01, we performed a haplotype association analysis on the subset of HLA-DRB1*03:01 negative patients and controls. The two haplotypes HLA-B8+/MICA*008+/rs115146037(T)+/MICB*008+/TNF308A+ and HLA-B*08:01-/MICA*008+/rs115146037(T)-/MICB*008-/TNF308A- were associated with the disease (pmeta = 1.4×10−6 and pmeta = 0.0023, respectively; Supplementary Material, Table S6).
Table 3.
Multiple logistic regression analysis in discovery and replication cohorts
| Discovery cohort |
Replication cohort |
Meta-analysisd |
||||
|---|---|---|---|---|---|---|
| P | OR (95%CI) | P | OR (95%CI) | P | OR (95%CI) | |
| HLA-B*08:01a | 0.180 | 1.63 (0.80–3.31) | 0.732 | 1.12 (0.60–2.09) | 0.347 | 1.25 (0.78–1.99) |
| MICA*008b | 0.029 | 1.45 (1.04–2.03) | 0.001 | 1.78 (1.25–2.53) | 1.84 × 10−04 | 1.58 (1.24–2.01) |
| rs3131619(T) | 0.073 | 2.05 (0.94–4.47) | 0.625 | 1.22 (0.55–2.68) | 0.076 | 1.64 (0.95–2.84) |
| MICB*008 | 0.300 | 0.70 (0.36–1.37) | 0.210 | 1.48 (0.80–2.75) | 0.829 | 1.05 (0.67–1.63) |
| TNF308A | 0.356 | 0.80 (0.50–1.28) | 0.622 | 0.89 (0.57–1.40) | 0.372 | 0.86 (0.63–1.19) |
| HLA-DRB1*03:01c | 5.96 × 10−09 | 3.16 (2.14–4.65) | 2.48 × 10−12 | 3.62 (2.53–5.20) | 8.22 × 10−20 | 3.38 (2.6–4.39) |
CI: confidence interval; OR: odds ratio.
Only variables showing a P < 0.05 in univariate logistic regression were used for this model.
Genotyping of HLA-B*008 was performed using proxies rs6457374 and rs2844535.
MICA*008 was typed using proxy MICA A5.1.
HLA-DRB1*03:01 was typed using proxy rs2187668.
The analysis was controlled for a cohort effect by including the cohort as a co-variate in the model.
Table 4.
Logistic regression analysis in subgroups of individuals without HLA-B*08:01, TNF308A, rs3131619(T), MICB*008 and HLA-DRB1*03:01 risk alleles
| Discovery cohort (n = 140 cases vs.
352 controls) |
Replication cohort (n = 133 cases vs.
279 controls) |
Meta-analysisb (n = 273 cases vs. 631
controls) |
||||
|---|---|---|---|---|---|---|
| P | OR (95%CI) | P | OR (95% CI) | P | OR (95% CI) | |
| MICA*008a | 0.032 | 1.54 (1.04–2.28) | 0.002 | 1.99 (1.29–3.07) | 2.13 × 10−04 | 1.73 (1.29–2.31) |
CI: confidence interval; OR: odds ratio.
MICA*008 was typed using proxy MICA A5.1.
The analysis was controlled for a cohort effect by including the cohort as a co-variate in the model.
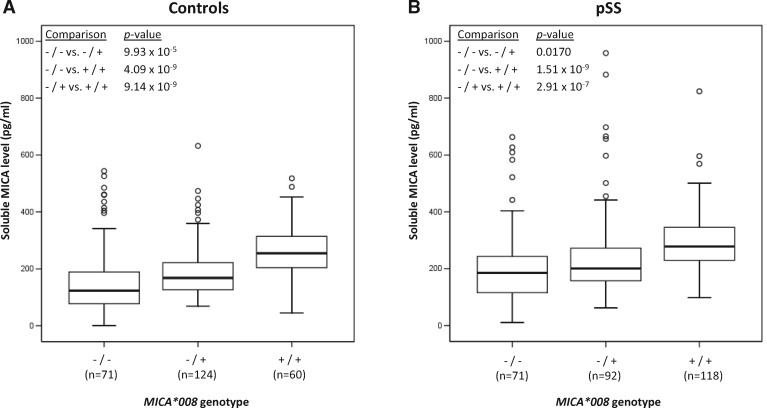
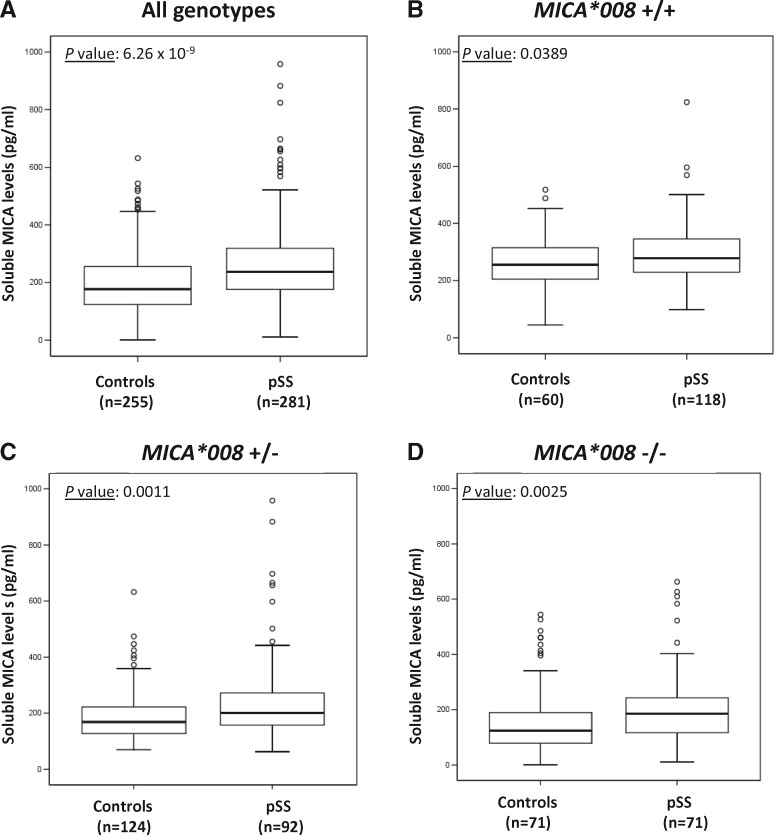
To assess the functional consequences of this genetic association, we measured the level of soluble MICA protein (sMICA) in the serum of a subset of pSS patients of the replication cohort (n = 281) and compared them to healthy blood donors (n = 255). When analyzing controls and pSS patients separately, the presence of the MICA*008 allele was correlated with a higher level of sMICA in a dose-dependent manner, i.e. ([sMICA] in MICA*008-/-) < ([sMICA] in MICA*008+/-) < ([sMICA] in MICA*008+/+) (Fig. 2). Moreover, independently of the genotype, there was significantly more sMICA in the serum of pSS patients compared to healthy controls (Fig. 3). Finally, we assessed the association of high-level sMICA (> 182.925 pg/ml) with pSS, in different patients’ sub-groups. Similarly to what was observed within the same subgroups for the genetic association analyses (see above), there were more subjects with high level sMICA in the patients’ groups compared to controls (OR = 2.94; 95% CI 2.05–4.22; P = 4.34 × 10−09) and this difference was amplified when the analysis was restricted to the pSS cases with only anti-SSA antibodies (OR = 3.19; 95% CI 1.62–6.27; P = 7.79 × 10−04) or pSS cases with anti-SSA and anti-SSB antibodies (OR = 4.25; 95% CI 2.66–6.81; P = 1.63 × 10−09) (Table 5). In addition, pSS was weakly associated with high-level sMICA in patients with anti-SSA and anti-SSB antibodies compared to patients without anti-SSA and/or anti-SSB antibodies (OR = 2.53; 95% CI 1.13–5.67; P = 0.024) (Table 5). Finally, the association of soluble MICA levels was shown to be stronger than, and independent of, the HLA-DRB1*03:01 allele (Supplementary Materials, Figs S3 and S4).
Figure 2.
Correlation between soluble MICA levels and MICA*008 genotype. The x-axis shows the MICA*008 genotype status: -/- corresponds to a subject without the MICA*008 allele, +/- to a MICA*008 heterozygous subject and +/+ to a MICA*008 homozygous subject. The y-axis shows the concentration of soluble MICA in pg/ml. The number of independent samples in each group is shown in parentheses. The horizontal line in the box-plots corresponds to the median values of each tested group. The box covers the 25th to 75th percentiles and the whiskers extend to the highest and lowest value within 1.5 times the interquartile range. Points outside the whiskers are outliers. Comparison of median values between groups was tested with the Mann-Whitney’s test. Plots were generated with the SPSS software (release 12.0.1; SPSS, USA).
Figure 3.
Correlation between soluble MICA levels and pSS disease. The x-axis shows the disease status and the y-axis the concentration of soluble MICA in pg/ml. The number of independent samples in each group is shown in parentheses. The horizontal line in the box-plots corresponds to the median values of each tested group. The box covers the 25th to 75th percentiles and the whiskers extend to the highest and lowest value within 1.5 times the interquartile range. Points outside the whiskers are outliers. Comparison of median values between groups was tested with the Mann-Whitney’s test. Plots were generated with the SPSS software (release 12.0.1; SPSS, USA).
Table 5.
Logistic regression analysis of high sMICA levels in various categories of pSS patients and controls
| Category 1 (% high sMICA levels)a | Category 2 (% high sMICA levels)a | P | OR (95%CI) | |
|---|---|---|---|---|
| All pSS cases (n = 281) vs. healthy controls (n = 255) | 73.0% | 47.8% | 4.34 × 10−09 | 2.94 (2.05–4.22) |
| SSA+/B- cases (n = 51) vs. healthy controls (n = 255) | 74.5% | 47.8% | 7.79 × 10−04 | 3.19 (1.62–6.27) |
| SSA+/B+ cases (n = 147) vs. healthy controls (n = 255) | 79.6% | 47.8% | 1.63 × 10−09 | 4.25 (2.66–6.81) |
| SSA-/B- cases (n = 33) vs. healthy controls (n = 255) | 60.6% | 47.8% | ns | – |
| SSA+/B- cases (n = 51) vs. SSA-/B- cases (n = 33) | 74.5% | 60.6% | ns | – |
| SSA+/B+ cases (n = 147) vs. SSA-/B- cases (n = 33) | 79.6% | 60.6% | 0.024 | 2.53 (1.13–5.67) |
the cutoff value to discriminate high from low sMICA levels was 182.925 pg/mL as established by ROC curve analysis (Supplementary Material, Fig. S2). CI: confidence interval; OR: odds ratio.
Discussion
This is the first report investigating the association of MICA with pSS. Genetic associations of MICA in several autoimmune/auto-inflammatory diseases such as rheumatoid arthritis, Behçet’s disease or yet systemic lupus erythematosus have been previously reported (25,28,29). However, with the possible exception of Behçet’s disease (linked to another MICA allele, MICA*009, harboring the A6 transmembrane repeat and incidentally also an epithelial/mucosal disease) (25,30,31), these studies most often concluded to a secondary association with MICA because of LD with other susceptibility/protective loci, or were not able to conclude as to whether MICA or the neighboring loci in LD, was the primary causal locus. In the present study, by extensively taking into account the genotypes of all major and/or potential pSS susceptibility genes/alleles - i.e. HLA-B*08:01, MICA*008, rs3131619(T), MICB*008, TNF308A, HLA-DRB1*03:01 and HLA-DRB1*05:01—an independent association of MICA*008 with pSS is evidenced. The fact that the P-value for MICA*008 association is higher in univariate (P = 2.61 × 10−35, Table 2) than in multivariate analysis (P = 5 × 10−5; Table 3) confirms that there is an LD between MICA*008 and other markers of this region, but does not question the independent association with pSS.
MICA*008 is the most frequent MICA allele in nearly all populations studied (21,32–36). It is moreover a peculiar allele, as it differs from other MICA alleles by a single nucleotide insertion within the transmembrane-encoding exon (25,16). This leads to a truncated protein devoid of any cytoplasmic tail and a shortened transmembrane segment. Due to this structural specificity, it was suggested that the well-known process of proteolytic shedding of MICA is more likely to happen for the MICA*008 allele than for other MICA variants (37). Our results here show for the first time a clear dose dependent effect of the MICA*008 genotype of a subject—irrespective of the disease status - on the amount of sMICA in his/her serum (Fig. 2).sMICA has been implicated both in cancer and autoimmunity through NKG2D receptor dis-regulation (38–41). Secretion of MICA protein in the soluble or exosomal form has been shown to be a major immune escape strategy in human tumors (39,42). Higher amounts of sMICA were also reported in serum of rheumatoid arthritis patients, where they might stimulate autoreactive T cells (41). Here we describe a genotype independent association of high level of sMICA with pSS (Fig. 3 and Table 5) which suggests a possible role of sMICA in the pathophysiology on pSS. The release of sMICA, which is amplified by the genetic association with the MICA*008 allele (Fig. 2), may therefore, as described in other conditions (39,43–45), trigger a systemic down-regulation of the NKG2D receptor on NK and CD8 T cells or paradoxically augment the cytotoxicity of these cells as recently shown for an NKG2D ligand in mouse (46).
Besides the increased production of a soluble MICA*008, at least four different other mechanisms alone or in various combinations, could also explain the contribution of MICA*008 to pSS (1) a differential binding of the allele to NKG2D as this has been recently documented for other MIC alleles (18) and indirectly shown in man in a transplant setting where the effect of LD was totally eliminated (as donors and recipients were 100% allele matched for all major HLA loci) (47,48); (2) a distinct subcellular targeting of MICA*008, as this has been clearly shown - in an experimental set-up - in polarized epithelial cells (49); the case for pSS lesions, (3) an escape from viral evasion mechanisms due to the lack of cytoplasmic tail in this particular allele (versus all other MICA alleles which are full-length), a known target of viral immunoevasins (50,51). It is of note that a viral etiology for pSS is intermittently put forward (like for many auto-immune diseases); and in some cases documented (52); (4) a last, circumvoluted explanation, is the presentation to the T-cell receptor, of MICA*008 specific antigenic peptides by HLA-B*08:01. This mechanism, which could be relevant not only to pSS but to other tissue specific auto-immune diseases (Bahram et al. unpublished), could (in this case) reconcile the tissue specificity of pSS (through restricted MICA expression), the linkage disequilibrium (here MICA*008 and HLA-B*08:01), and the ubiquitous expression of HLA-B*08:01 (like any classical MHC-I). Whatever the actual mechanism, this genetic study puts the MIC-NKG2D axis at the heart of pSS pathophysiology and corroborates recent functional studies hinting to the involvement of NK cells in the induction phase of pSS (12).
In conclusion, our results demonstrate that MICA is a novel, independent, HLA-linked disease susceptibility locus for pSS. As such, MICA should be included in investigational genetic profiling of pSS patients and the MICA-NKG2D axis carefully examined in the context of the complex pathophysiology of disease. We additionally show a dose dependent expression of sMICA - a phenomenon relevant to many other diseases as well - further increased in pSS patients, where it might affect NK and T cell activity via modulation of NKG2D signaling. In order to fully decipher the pathophysiological mechanism involving MICA*008, additional functional studies are needed. By using CRISPR/Cas9 for instance, the specific deletion of MICA alleles in relevant cell lines or patient cells could be informative.
Materials and Methods
Patients and controls
The discovery cohort was composed of 347 pSS patients from the French nationwide ASSESS cohort (French Assessment of Systemic Symptoms and Evolution in patients with pSS) and 553 control individuals. The replication cohort was composed of 612 pSS patients from the UK primary Sjögren’s syndrome registry (UKPSSR, United Kingdom) and 490 controls. All patients fulfilled the American-European Consensus Criteria for Sjögren's Syndrome (53). Principal component analysis (PCA) based on 47 Ancestry Informative Markers (AIMs) was used to detect population outliers in the discovery cohort. After outlier removal, the discovery cohort consisted of a total of 347 pSS patients. For the replication cohort, we performed a PCA analysis with 29 independent genetic markers from a previous GWAS (15). No outlier was detected by this analysis. Both cohorts used EIGENSTRAT for PCA and outlier detection (54). The controls were all self-reported unrelated healthy individuals of European descent originating from the same geographical area as patients. All patients and controls have provided written informed consents and the research protocol was approved by relevant institutional review boards.
Genotyping
Full-length sequencing of MICA and MICB extracellular domains (α1, α2 and α3) encoding exons was performed as previously described (22,55). Briefly, fragments spanning exons 2 to 5 of MICA and MICB were PCR amplified using the Expand Long Template PCR System (Roche, Germany), following manufacturer's recommendations. After purification with the QIAquick PCR Purification Kit (QIAGEN, Germany), the PCR products were directly sequenced with the BigDye Terminator v3.1 Cycle sequencing kit and run on an 96 capillary ABI3730XL Genetic Analyzer (Life Technologies, USA). Sequences were analyzed using Seqscape software (Life Technologies, USA). Final MICA and MICB genotypes were assigned using an in-house developed software compiling sequence data and MICA-transmembrane (TM) genotypes.
Genotyping of the MICA-TM microsatellite polymorphism was performed as previously described (25). The TM coding region was amplified with a forward primer labeled at the 5′ end with 6-Carboxyfluorescein (FAM) (5′-CCTTTTTTTCAGGGAAAGTGC-3′) and a reverse primer (5′-CCTTACCATCTCCAGAAACTGC-3′). To determine the number of the triplet repeats in the TM region of the MICA gene, PCR products were electrophoresed on a 16 capillary ABI3130xl Genetic Analyzer and their sizes were determined using the Genemapper software v4.0 (Life Technologies, USA). MICA-TM genotypes (MICA A4, A5, A5.1, A6 or A9) were determined by comparing the sizes of the obtained fragments with controls of known genotypes.
Genotyping of SNPs rs3131619 and rs1800629 (TNF308A), as well as SNP proxies for HLA-B*008 (rs6457374 and rs2844535) (estimated correlation of 98%, data not shown) (56–58), HLA-DRB1*15:01 (rs3129860) (56,59) and HLA-DRB1*03:01 (rs2187668) (56,59) was carried out by the Taqman method using pre-designed primer sets (Life Technologies, USA). PCR reactions were performed following manufacturer’s recommendations in 10 µl final reaction volumes with 15 ng of genomic DNA as template. Cycling conditions were as follows: one cycle at 95 °C for 10 min, 40 cycles at 92 °C for 15 s, 60 °C for 1 min. End-point fluorescence intensities were read using an ABI7000 Real-Time PCR System and analyzed by the Sequence Detection Software v1.2 (Life Technologies, USA).
Quantification of soluble MICA (sMICA)
sMICA levels were measured in patients’ and controls’ sera with an in-house developed sandwich enzyme-linked immunosorbent assay (ELISA) using two monoclonal mouse antibodies for capture (A13-C485B10 and A9-C255A9 at 2 µg/ml and 0.2 µg/ml, respectively) and one biotinylated monoclonal mouse antibody for detection (A15-C199B9 at 60 pg/ml). Coating of MaxiSorp ELISA plates (ThermoFisher Scientific) was performed in PBS at +4 °C overnight. After three washing steps with PBS, the wells were blocked with 200 µl of 10% BSA in PBS for 1 h at room temperature. All the following steps were carried out at room temperature with PBS/0.05% Tween 20/10% BSA used as a diluent for all the reagents and sera. The plates were washed three times with PBS/0.05% Tween 20 between incubation steps. After blocking, the plates were incubated with 100 µl of sera, standards and controls for 2 h, followed by incubation with 100 µl biotinylated detection antibody for 1 h. Then the plates were incubated during 1h with 100 µl of a 5000-fold dilution of streptavidin poly-HRP (ThermoFisher Scientific) per well. The reactions were finally revealed using TMB Ultra (Thermofisher Scientific) at 100 µl/well for 15 min and stopped with 100 µl of 1M HCl. The absorbance was measured at 450 nm.
The sensitivity and allelic specificity of the ELISA has previously been validated using a panel of 16 purified MICA and 6 MICB alleles and the assay was shown to be MICA-specific. For quantification, standard curves were generated by 2.5-fold serial dilutions of purified MICA*008 and MICA*018 alleles (showing respectively the highest and lowest sensitivity of all tested alleles in the ELISA) with starting concentration of 1 ng/ml and seven calibration points. In addition, each microplate contained four internal control samples that were generated by pooling several human sera and spiking them with different amounts of MIC proteins. Spiking recovery served as a reference for the quality of each experiment. In addition, the spiking controls also served for normalization of the data. The spiking controls and human sera were diluted 10-fold. Each sample was tested in duplicate.
Statistical analysis
Comparing distributions of genotypes between cases and controls was performed using the Pearson’s Chi-square test. Odds ratios (OR) and 95% confidence intervals (95% CI) were calculated for the tested alleles in the pSS patients groups vs. control groups. The P-values were adjusted with the Bonferroni correction by multiplying the P-values by the number of tests conducted. Corrected P-values below 0.05 were considered statistically significant. PLINK (60) was used to test for Hardy–Weinberg equilibrium. LD between markers was calculated using Haploview program (61) as normalized value of pairwise LD (D’=D/Dmax). Univariate analysis and logistic regression analysis were performed using SPSS (release 12.0.1; SPSS, USA). Only markers displaying P < 0.05 in univariate analysis were included in the multiple logistic model. For fitting the model, the status of patient or control was used as a binary dependent variable, and the following independent predictor variables, expressed as presence/absence, were used: MICA*008, HLA-B*08:01, TNF308A, rs3131619 (T), MICB*008, and HLA-DRB1*03:01. Results of logistic regression were reported in terms of odds ratio (OR) with 95% confidence interval (CI). The meta-analysis was performed by combining data of both cohorts. In multivariate analyses presented in Tables 2 and 3 and Supplementary Material, Table S4, the cohort was included as a covariate in the model.
Concentrations of sMICA were calculated with a five-parameter logistic curve analysis using the MICA*008 allele as a reference for the standard curve (GraphPad Prism; GraphPad Software, Inc., USA). Data were expressed as mean of duplicates for each serum. Comparison of the ELISA results between patient and control groups was statistically analyzed using Mann-Whitney’s nonparametric, unpaired, two-tailed test (SPSS release 12.0.1; SPSS, USA). The significance level was set to P-value < 0.05. The comparative analyses were validated to be equivalent when using the MICA*016 allele as a reference for standard curves. ROC curve analysis was performed with SPSS release 12.0.1 (SPSS, USA) and the Youden index was used to select the optimum cutoff values for the ELISA (62).
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We would like to thank Ben Hargreaves (Newcastle upon Tyne Hospitals) for data extraction. This work is published under the framework of the LABEX TRANSPLANEX [ANR-11-LABX-0070_TRANSPLANTEX] The authors thank all the investigators of the ASSESS cohort, Valérie Devauchelle-Pensec, Philippe Dieudé, Jean-Jacques Dubost, Anne-Laure Fauchais, Vincent Goeb, Eric Hachulla, Claire Larroche, Véronique Le Guern, Jacques Morel, Aleth Perdriger, Xavier Puéchal, Stéphanie Rist, Damien Sène, and Olivier Vittecoq, The authors also thank Joëlle Benessiano and all staff members from the Centre de Ressources Biologiques, Hôpital Bichat, Assistance Publique-Hôpitaux de Paris for their help in centralizing and managing biologic data collection from the French ASSESS cohort, Karine Inamo, Stanie Gaete, Djilali Batouche, Domitille Molinari from the Unité de Recherche Clinique Paris-Sud, Assistance Publique-Hôpitaux de Paris and Mickael Randrianandrasana, Isabelle Pane, Adeline Abbe, Gabriel Baron, Philippe Ravaud from the Centre d’Epidémiologie clinique, Hôpital Hôtel Dieu, Assistance Publique-Hôpitaux de Paris, for clinical data collection and management.
Conflict of Interest statement. SB is the scientific founder and a (minority) shareholder in BIOMICA SAS.
Funding
French National Research Agency (ANR), Genomax, Strasbourg School of Medicine Next Generation Sequencing center, the Institut Universitaire de France (IUF), the Partenariat Hubert Curien GUNDISHAPUR by the ministères des Affaires étrangères et du développement international (MAEDI) and Enseignement supérieur et de la Recherche (MESR), the Institut de Recherche pour le développement (IRD), the Centre National pour la Recherche Scientifique (CNRS) and the EU-funded (ERDF) INTERREG V project n°3.2 TRIDIAG, Medical Research Council, UK, French Ministry of Health (PHRC N°2006-AOM06133).
References
- 1. Brito-Zerón Pilar, Acar-Denizli Nihan, Zeher Margit, Rasmussen Astrid, Seror Raphaele, Theander Elke, Li Xiaomei, Baldini Chiara, Gottenberg Jacques-Eric, Danda Debashish, et al. (2016) Influence of geolocation and ethnicity on the phenotypic expression of primary Sjogren's syndrome at diagnosis in 8310 patients: a cross-sectional study from the Big Data Sjogren Project Consortium. Ann. Rheum. Dis., doi:10.1136/annrheumdis2016-209952. [DOI] [PubMed] [Google Scholar]
- 2. Gøransson L.G., Haldorsen K., Brun J.G., Harboe E., Jonsson M.V., Skarstein K., Time K., Omdal R. (2011) The point prevalence of clinically relevant primary Sjogren's syndrome in two Norwegian counties. Scand. J. Rheumatol., 40, 221–224. [DOI] [PubMed] [Google Scholar]
- 3. Maldini C., Seror R., Fain O., Dhote R., Amoura Z., De Bandt M., Delassus J.L., Falgarone G., Guillevin L., Le Guern V., et al. (2014) Epidemiology of primary Sjogren's syndrome in a French multiracial/multiethnic area. Arthritis Care Res., 66, 454–463. [DOI] [PubMed] [Google Scholar]
- 4. Nocturne G., Mariette X. (2013) Advances in understanding the pathogenesis of primary Sjogren's syndrome. Nat. Rev. Rheumatol., 9, 544–556. [DOI] [PubMed] [Google Scholar]
- 5. Trowsdale J., Knight J.C. (2013) Major histocompatibility complex genomics and human disease. Annu. Rev. Genomics Hum. Genet., 14, 301–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cobb B.L., Lessard C.J., Harley J.B., Moser K.L. (2008) Genes and Sjogren's syndrome. Rheum. Dis. Clin. North Am., 34, 847-868. vii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jean S., Quelvennec E., Alizadeh M., Guggenbuhl P., Birebent B., Perdriger A., Grosbois B., Pawlotsky P.Y., Semana G. (1998) DRB1*15 and DRB1*03 extended haplotype interaction in primary Sjogren's syndrome genetic susceptibility. Clin. Exp. Rheumatol., 16, 725–728. [PubMed] [Google Scholar]
- 8. Kang H.I., Fei H.M., Saito I., Sawada S., Chen S.L., Yi D., Chan E., Peebles C., Bugawan T.L., Erlich H.A., et al. (1993) Comparison of HLA class II genes in Caucasoid, Chinese, and Japanese patients with primary Sjogren's syndrome. J. Immunol., 150(8 Pt 1), 3615–3623. [PubMed] [Google Scholar]
- 9. Loiseau P. et al. (2001) HLA class I and class II are both associated with the genetic predisposition to primary Sjogren syndrome. Hum. Immunol., 62, 725–731. [DOI] [PubMed] [Google Scholar]
- 10. Gershwin M.E., Terasaki I., Graw R., Chused T.M. (1975) Increased frequency of HL-A8 in Sjogren's syndrome. Tissue Antigens, 6, 342–346. [DOI] [PubMed] [Google Scholar]
- 11. Gottenberg J.E., Busson M., Loiseau P., Dourche M., Cohen-Solal J., Lepage V., Charron D., Miceli C., Sibilia J., Mariette X. (2004) Association of transforming growth factor beta1 and tumor necrosis factor alpha polymorphisms with anti-SSB/La antibody secretion in patients with primary Sjogren's syndrome. Arthritis Rheum., 50, 570–580. [DOI] [PubMed] [Google Scholar]
- 12. Rusakiewicz S., Nocturne G., Lazure T., Semeraro M., Flament C., Caillat-Zucman S., Sène D., Delahaye N., Vivier E., Chaba K., et al. (2013) NCR3/NKp30 contributes to pathogenesis in primary Sjogren's syndrome. Sci. Transl. Med., 5, 195ra196.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bolstad A.I., Le Hellard S., Kristjansdottir G., Vasaitis L., Kvarnström M., Sjöwall C., Johnsen S.J., Eriksson P., Omdal R., Brun J.G., et al. (2012) Association between genetic variants in the tumour necrosis factor/lymphotoxin alpha/lymphotoxin beta locus and primary Sjogren's syndrome in Scandinavian samples. Ann. Rheum. Dis., 71, 981–988. [DOI] [PubMed] [Google Scholar]
- 14. Li Y., Zhang K., Chen H., Sun F., Xu J., Wu Z., Li P., Zhang L., Du Y., Luan H., et al. (2013) A genome-wide association study in Han Chinese identifies a susceptibility locus for primary Sjogren's syndrome at 7q11.23. Nat. Genet., 45, 1361–1365. [DOI] [PubMed] [Google Scholar]
- 15. Lessard C.J., Li H., Adrianto I., Ice J.A., Rasmussen A., Grundahl K.M., Kelly J.A., Dozmorov M.G., Miceli-Richard C., Bowman S., et al. (2013) Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjogren's syndrome. Nat. Genet., 45, 1284–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bahram S., Bresnahan M., Geraghty D.E., Spies T. (1994) A second lineage of mammalian major histocompatibility complex class I genes [see comments]. Proc. Natl Acad. Sci. U S A, 91, 6259–6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bahram S. (2000) MIC genes: from genetics to biology. Adv. Immunol., 76, 1–60. [DOI] [PubMed] [Google Scholar]
- 18. Carapito R., Bahram S. (2015) Genetics, genomics, and evolutionary biology of NKG2D ligands. Immunol. Rev., 267, 88–116. [DOI] [PubMed] [Google Scholar]
- 19. Groh V., Bahram S., Bauer S., Herman A., Beauchamp M., Spies T. (1996) Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc. Natl Acad. Sci. U S A, 93, 12445–12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schrambach S., Ardizzone M., Leymarie V., Sibilia J., Bahram S. (2007) In vivo expression pattern of MICA and MICB and its relevance to auto-immunity and cancer. PloS One, 2, e518.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fodil N., Pellet P., Laloux L., Hauptmann G., Theodorou I., Bahram S. (1999) MICA haplotypic diversity. Immunogenetics, 49, 557–560. [DOI] [PubMed] [Google Scholar]
- 22. Fodil N., Laloux L., Wanner V., Pellet P., Hauptmann G., Mizuki N., Inoko H., Spies T., Theodorou I., Bahram S. (1996) Allelic repertoire of the human MHC class I MICA gene. Immunogenetics, 44, 351–357. [DOI] [PubMed] [Google Scholar]
- 23. Zhou X., Wang J., Zou H., Ward M.M., Weisman M.H., Espitia M.G., Xiao X., Petersdorf E., Mignot E., Martin J., et al. (2013) MICA, a gene contributing strong susceptibility to ankylosing spondylitis. Ann. Rheum. Dis., 73,1552–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Feng B.J., Sun L.D., Soltani-Arabshahi R., Bowcock A.M., Nair R.P., Stuart P., Elder J.T., Schrodi S.J., Begovich A.B., Abecasis G.R., et al. (2009) Multiple Loci within the major histocompatibility complex confer risk of psoriasis. PLoS Genet., 5, e1000606.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mizuki N., Ota M., Kimura M., Ohno S., Ando H., Katsuyama Y., Yamazaki M., Watanabe K., Goto K., Nakamura S., Bahram S., Inoko H. (1997) Triplet repeat polymorphism in the transmembrane region of the MICA gene: a strong association of six GCT repetitions with Behcet disease. Proc. Natl Acad. Sci. U S A, 94, 1298–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kirino Y., Remmers E.F. (2015) Genetic architectures of seropositive and seronegative rheumatic diseases. Nat. Rev., 11, 401–414. [DOI] [PubMed] [Google Scholar]
- 27. Heath S.C., Gut I.G., Brennan P., McKay J.D., Bencko V., Fabianova E., Foretova L., Georges M., Janout V., Kabesch M., et al. (2008) Investigation of the fine structure of European populations with applications to disease association studies. Eur. J. Hum. Genet., 16, 1413–1429. [DOI] [PubMed] [Google Scholar]
- 28. Martinez A., Fernandez-Arquero M., Balsa A., Rubio A., Alves H., Pascual-Salcedo D., Martin-Mola E., de la Concha E.G. (2001) Primary association of a MICA allele with protection against rheumatoid arthritis. Arthritis Rheum., 44, 1261–1265. [DOI] [PubMed] [Google Scholar]
- 29. Yoshida K., Komai K., Shiozawa K., Mashida A., Horiuchi T., Tanaka Y., Nose M., Hashiramoto A., Shiozawa S. (2011) Role of the MICA polymorphism in systemic lupus erythematosus. Arthritis Rheum., 63, 3058–3066. [DOI] [PubMed] [Google Scholar]
- 30. Carapito R., Shahram F., Michel S., Le Gentil M., Radosavljevic M., Meguro A., Abdollahi B.S., Inoko H., Ota M., Davatchi F., Bahram S. (2015) On the genetics of the Silk Route: association analysis of HLA, IL10, and IL23R-IL12RB2 regions with Behcet's disease in an Iranian population. Immunogenetics, 67, 289–293. [DOI] [PubMed] [Google Scholar]
- 31. Hughes T., Coit P., Adler A., Yilmaz V., Aksu K., Düzgün N., Keser G., Cefle A., Yazici A., Ergen A., et al. (2013) Identification of multiple independent susceptibility loci in the HLA region in Behcet's disease. Nat. Genet., 45, 319–324. [DOI] [PubMed] [Google Scholar]
- 32. Romphruk A.V., Naruse T.K., Romphruk A., Kawata H., Puapairoj C., Kulski J.K., Leelayuwat C., Inoko H. (2001) Diversity of MICA (PERB11.1) and HLA haplotypes in Northeastern Thais. Tissue Antigens, 58, 83–89. [DOI] [PubMed] [Google Scholar]
- 33. Tian W., Boggs D.A., Ding W.Z., Chen D.F., Fraser P.A. (2001) MICA genetic polymorphism and linkage disequilibrium with HLA-B in 29 African-American families. Immunogenetics, 53, 724–728. [DOI] [PubMed] [Google Scholar]
- 34. Zhang Y., Lazaro A.M., Lavingia B., Stastny P. (2001) Typing for all known MICA alleles by group-specific PCR and SSOP. Hum. Immunol., 62, 620–631. [DOI] [PubMed] [Google Scholar]
- 35. Zhang Y., Lazaro A.M., Zou Y., Lavingia B., Moraes E.M., Moraes R.J., Stastny P. (2002) MICA polymorphism in South American Indians. Immunogenetics, 53, 900–906. [DOI] [PubMed] [Google Scholar]
- 36. Marin M.L., Savioli C.R., Yamamoto J.H., Kalil J., Goldberg A.C. (2004) MICA polymorphism in a sample of the Sao Paulo population, Brazil. Eur. J. Immunogenet., 31, 63–71. [DOI] [PubMed] [Google Scholar]
- 37. Choy M.K., Phipps M.E. (2010) MICA polymorphism: biology and importance in immunity and disease. Trends Mol. Med., 16, 97–106. [DOI] [PubMed] [Google Scholar]
- 38. Holdenrieder S., Stieber P., Peterfi A., Nagel D., Steinle A., Salih H.R. (2006) Soluble MICA in malignant diseases. Int. J. Cancer, 118, 684–687. [DOI] [PubMed] [Google Scholar]
- 39. Groh V., Wu J., Yee C., Spies T. (2002) Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature, 419, 734–738. [DOI] [PubMed] [Google Scholar]
- 40. Salih H.R., Holdenrieder S., Steinle A. (2008) Soluble NKG2D ligands: prevalence, release, and functional impact. Front. Biosci., 13, 3448–3456. [DOI] [PubMed] [Google Scholar]
- 41. Groh V., Bruhl A., El-Gabalawy H., Nelson J.L., Spies T. (2003) Stimulation of T cell autoreactivity by anomalous expression of NKG2D and its MIC ligands in rheumatoid arthritis. Proc. Natl Acad. Sci. U S A, 100, 9452–9457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kaiser B.K., Yim D., Chow I.T., Gonzalez S., Dai Z., Mann H.H., Strong R.K., Groh V., Spies T. (2007) Disulphide-isomerase-enabled shedding of tumour-associated NKG2D ligands. Nature, 447, 482–486. [DOI] [PubMed] [Google Scholar]
- 43. Ashiru O., Boutet P., Fernández-Messina L., Agüera-González S., Skepper J.N., Valés-Gómez M., Reyburn H.T. (2010) Natural killer cell cytotoxicity is suppressed by exposure to the human NKG2D ligand MICA*008 that is shed by tumor cells in exosomes. Cancer Res., 70, 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chitadze G., Bhat J., Lettau M., Janssen O., Kabelitz D. (2013) Generation of soluble NKG2D ligands: proteolytic cleavage, exosome secretion and functional implications. Scand. J. Immunol., 78, 120–129. [DOI] [PubMed] [Google Scholar]
- 45. Doubrovina E.S., Doubrovin M.M., Vider E., Sisson R.B., O'Reilly R.J., Dupont B., Vyas Y.M. (2003) Evasion from NK cell immunity by MHC class I chain-related molecules expressing colon adenocarcinoma. J. Immunol., 171, 6891–6899. [DOI] [PubMed] [Google Scholar]
- 46. Deng W., Gowen B.G., Zhang L., Wang L., Lau S., Iannello A., Xu J., Rovis T.L., Xiong N., Raulet D.H. (2015) Antitumor immunity. A shed NKG2D ligand that promotes natural killer cell activation and tumor rejection. Science, 348, 136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fuerst D., Neuchel C., Niederwieser D., Bunjes D., Gramatzki M., Wagner E., Wulf G., Glass B., Pfreundschuh M., Einsele H.. et al. (2016) Matching for the MICA-129 polymorphism is beneficial in unrelated hematopoietic stem cell transplantation. Blood, 128, 3169–3176. [DOI] [PubMed] [Google Scholar]
- 48. Carapito R., Jung N., Kwemou M., Untrau M., Michel S., Pichot A., Giacometti G., Macquin C., Ilias W., Morlon A., et al. (2016) Matching for the nonconventional MHC-I MICA gene significantly reduces the incidence of acute and chronic GVHD. Blood, 128, 1979–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Suemizu H., Radosavljevic M., Kimura M., Sadahiro S., Yoshimura S., Bahram S., Inoko H. (2002) A basolateral sorting motif in the MICA cytoplasmic tail. Proc. Natl Acad. Sci. U S A, 99, 2971–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zou Y., Bresnahan W., Taylor R.T., Stastny P. (2005) Effect of human cytomegalovirus on expression of MHC class I-related chains A. J. Immunol., 174, 3098–3104. [DOI] [PubMed] [Google Scholar]
- 51. Seidel E., Khanh Le V.T., Bar-On Y., Tsukerman P., Enk J., Yamin R., Stein N., Schmiedel D., Oiknine Djian E., Weisblum Y.. et al. (2015) Dynamic co-evolution of host and pathogen: HCMV downregulates the prevalent allele MICA *008 to escape elimination by NK Cells. Cell Rep., 10, 968–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Igoe A., Scofield R.H. (2013) Autoimmunity and infection in Sjogren's syndrome. Curr. Opin. Rheumatol., 25, 480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vitali C., Bombardieri S., Jonsson R., Moutsopoulos H.M., Alexander E.L., Carsons S.E., Daniels T.E., Fox P.C., Fox R.I., Kassan S.S., et al. (2002) Classification criteria for Sjogren's syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann. Rheum. Dis., 61, 554–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet., 38, 904–909. [DOI] [PubMed] [Google Scholar]
- 55. Pellet P., Renaud M., Fodil N., Laloux L., Inoko H., Hauptmann G., Debré P., Bahram S., Theodorou I. (1997) Allelic repertoire of the human MICB gene. Immunogenetics, 46, 434–436. [DOI] [PubMed] [Google Scholar]
- 56. de Bakker P.I., McVean G., Sabeti P.C., Miretti M.M., Green T., Marchini J., Ke X., Monsuur A.J., Whittaker P., Delgado M., et al. (2006) A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet., 38, 1166–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Medrano L.M., Dema B., López-Larios A., Maluenda C., Bodas A., López-Palacios N., Figueredo M.Á., Fernández-Arquero M., Núñez C. (2012) HLA and celiac disease susceptibility: new genetic factors bring open questions about the HLA influence and gene-dosage effects. PloS One, 7, e48403.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Conde L., Halperin E., Akers N.K., Brown K.M., Smedby K.E., Rothman N., Nieters A., Slager S.L., Brooks-Wilson A., Agana L., et al. (2010) Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21.32. Nat. Genet., 42, 661–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Taylor K.E., Chung S.A., Graham R.R., Ortmann W.A., Lee A.T., Langefeld C.D., Jacob C.O., Kamboh M.I., Alarcón-Riquelme M.E., Tsao B.P., Moser K.L., et al. (2011) Risk alleles for systemic lupus erythematosus in a large case-control collection and associations with clinical subphenotypes. PLoS Genet., 7, e1001311.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., Sham P.C. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Barrett J.C., Fry B., Maller J., Daly M.J. (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics, 21, 263–265. [DOI] [PubMed] [Google Scholar]
- 62. Youden W.J. (1950) Index for rating diagnostic tests. Cancer, 3, 32–35. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.