Abstract
Background.
Rapid diagnostic tests (RDTs) account for more than two-thirds of malaria diagnoses in Africa. Deletions of the Plasmodium falciparum hrp2 (pfhrp2) gene cause false-negative RDT results and have never been investigated on a national level. Spread of pfhrp2-deleted P. falciparum mutants, resistant to detection by HRP2-based RDTs, would represent a serious threat to malaria elimination efforts.
Methods.
Using a nationally representative cross-sectional study of 7,137 children under five years of age from the Democratic Republic of Congo (DRC), we tested 783 subjects with RDT-/PCR+ results using PCR assays to detect and confirm deletions of the pfhrp2 gene. Spatial and population genetic analyses were employed to examine the distribution and evolution of these parasites.
Results.
We identified 149 pfhrp2-deleted parasites, representing 6.4% of all P. falciparum infections country-wide (95% confidence interval 5.1–8.0%). Bayesian spatial analyses identified statistically significant clustering of pfhrp2 deletions near Kinshasa and Kivu. Population genetic analysis revealed significant genetic differentiation between wild-type and pfhrp2-deleted parasite populations (GST = .046, p ≤ .00001).
Conclusions.
Pfhrp2-deleted P. falciparum is a common cause of RDT-/PCR+ malaria among asymptomatic children in the DRC and appears to be clustered within select communities. Surveillance for these deletions is needed, and alternatives to HRP2-specific RDTs may be necessary.
Keywords: rapid diagnostic tests, false-negative, diagnostic resistance, histidine-rich protein 2, pfhrp3, hrp2, hrp3, RDT, deletion, malaria.
BACKGROUND
Malaria-related mortality has fallen by 60% in Africa since 2000 [1]. One of the cornerstones of malaria control programs is the use of RDTs instead of traditional microscopy for diagnosis. From 2008 to 2014, annual sales of RDTs increased from 46 million to 314 million units, representing a $151 million annual investment funded largely by multilateral and bilateral donors [2]. In 2014, RDTs accounted for 71% of all diagnostic testing employed to evaluate suspected malaria cases in Africa [1]. In the Democratic Republic of the Congo (DRC) and throughout Africa, RDTs have become the primary mode of malaria diagnosis.
Most widely used RDTs rely on the detection of histidine-rich protein 2 (HRP2), an antigen specific to Plasmodium falciparum. HRP2-based RDTs are known to generate false-positive results in the setting of persistent circulating HRP2 antigen after antimalarial treatment and false-negative results in individuals with low levels of parasitemia beneath the assay’s threshold for detection, typically around 200 parasites/µL for most commercially available RDTs [2–4]. Recently, however, false-negative results have been reported in individuals infected with P. falciparum parasites harboring a deletion of the pfhrp2 gene [5–15]. Several of these reports also identified co-existing deletions of the P. falciparum histidine-rich protein 3 (pfhrp3) gene, which produces an antigen that exhibits some cross-reactivity with select HRP2-based RDTs. Understanding the distribution and evolution of these mutant parasites is a priority for the World Health Organization, which recently hosted a Technical Consultation on P. falciparum hrp2/3 gene deletions and drafted interim guidance for investigating false-negative RDTs [16, 17]. It is unknown whether reliance on HRP2-based RDTs to guide treatment is exerting evolutionary pressure favoring the spread of this mutation.
Such a finding could have significant implications for malaria control and elimination efforts in Africa. To date, pfhrp2-deleted mutant parasites have not been investigated using country-wide population-based surveys. Here, we use samples from a large, nationally representative cross-sectional study of children younger than 5 years of age in the DRC to determine the prevalence, spatial distribution, and population genetics of these deletions.
METHODS
Study Population
Samples were collected from children under 5 years of age as part of the 2013–2014 DRC Demographic and Health Survey (DHS) [18]. The majority of children were asymptomatic at the time of sample collection. As previously described, heel- or finger-prick blood from each participant was analyzed by light microscopy for parasites, applied to an RDT targeting the P. falciparum HRP2 antigen (SD BIOLINE Malaria Ag P.f., Standard Diagnostics, Gyeonggi-do, Republic of Korea), and used to prepare dried blood spots (DBS) [18, 19]. The SD BIOLINE Malaria Ag P.f. RDT is P. falciparum-specific and meets current procurement criteria recommended by the World Health Organization, with high panel detection scores of 95 and 99 (on a 100-point scale) at 200 and 2,000 parasites/µL, respectively [2]. Three diagnostic methods were performed for each child: RDT, microscopy, and polymerase chain reaction (PCR). After extracting DNA from the DBS using established methods [20], a real-time PCR assay with a limit of detection of 100 parasites/µL was employed to identify P. falciparum infection by amplifying a region of the single-copy pfldh gene as previously described [19]. This study was approved by the University of Kinshasa School of Public Health and the University of North Carolina Institutional Review Boards. Informed consent was obtained from a parent or responsible adult for all subjects.
Detection of pfhrp2 Deletions
We performed PCR to identify the presence or absence of pfhrp2 deletions among the subset of P. falciparum parasites with RDT-/PCR+ results. We included both microscopy-positive and -negative samples because excluding samples with insufficient parasite density for microscopic diagnosis could lead to an underestimate of the true pfhrp2-deleted parasite prevalence. We employed PCR assays targeting pfhrp2 exon 1 and exon 2, using both 72°C and 60°C elongation temperatures based on reports of improved PCR sensitivity for AT-rich targets using lower elongation temperatures [21]. Additionally, we tested RDT+/PCR+ samples from select provinces to evaluate the occurrence of RDT-positivity due to HRP3 alone (expressed by pfhrp2-deleted parasites with an intact pfhrp3 gene). PCR methods are described in the Supplementary Material.
Confirmation of P. falciparum Infection
For the subset of samples in which PCR assays failed to amplify either region of the pfhrp2 gene, we performed additional PCR assays targeting other single-copy genes to confirm that sufficient P. falciparum DNA persisted in each sample for amplification, including the pfhrp3 gene as described below and/or the P. falciparum beta tubulin (Pf β-tubulin) gene (see Supplementary Materials). Samples with successful amplification of pfhrp3 or Pf β-tubulin were deemed to contain pfhrp2-deleted P. falciparum. The presence of amplifiable DNA was further confirmed for a subset of samples using PCR assays for multiple flanking microsatellite loci as described below.
Detection of Coexisting pfhrp3 Deletions
We performed PCR assays to detect co-existing complete pfhrp3 deletions among pfhrp2-deleted P. falciparum parasites. We employed assays targeting pfhrp3 exon 2, followed by assays for exon 1 as described in the Supplementary Material. Samples with negative results for these assays were deemed to contain coexisting pfhrp2 and pfhrp3 deletions.
Prevalence and Geographical Distribution of pfhrp2-Deleted P. falciparum
We calculated the prevalence of P. falciparum infections among children younger than 5 years of age, using PCR-positive infections (determined previously by the initial real-time PCR assay targeting pfldh) as the numerator and the total number tested as the denominator. We calculated the proportion of pfhrp2-deleted P. falciparum infections overall and by province, using pfhrp2 deletions as the numerator and total PCR-positive P. falciparum cases as the denominator. All calculations utilized sample weights generated by the DHS to account for complex sampling design. Locations of pfhrp2-deleted isolates were mapped using R software (R Core Team, Vienna, Austria) and assessed for clustering by province using a Bayesian clustering technique as previously described [22]. We allowed for 20% of the study population to be included in the scanning window, consistent with other scan-based cluster methods. For the prior distributions for the null and non-null configurations, we specified conjugate gamma priors, such that null relative risks ranged between 0.95 and 1.05, and non-null relative risks ranged between 0 and 4 with 95% probability. Finally, we specified a prior probability of no clusters as 0.95 (consistent with a null hypothesis of no clustering). See Supplementary Material for details.
Microsatellite Analysis
In order to explore whether evolutionary selective pressure favoring pfhrp2-deleted parasites was present (e.g. selective treatment based on HRP2-based RDT results), we performed additional testing on a subset of pfhrp2-deleted parasites. We chose to test all pfhrp2-deleted parasites identified in Kinshasa, North Kivu, and South Kivu Provinces. Additionally, an equal number of randomly selected, geographically matched HRP2-positive control samples were included in our testing to achieve a 1:1 ratio of deleted to control samples in each province, resulting in a frequency-matched sample that was analyzed without regard to individually matched pairs. We performed PCR to amplify six previously described microsatellite regions flanking the pfhrp2 gene and employed capillary electrophoresis (Eton Biosciences, San Diego, CA) to analyze the amplicons [6]. Primers and reaction conditions are described in Supplementary Table 1. Capillary electrophoresis chromatograms were visualized and scored for amplicon length using GeneMapper v. 4.1 (Applied Biosystems, Foster City, CA). We considered peaks with an intensity of less than 200 relative fluorescent units (RFU) as background signal and analyzed only the dominant peak at each locus. We adjusted amplicon lengths according to the difference between P. falciparum 3D7 controls and their theoretical fragment lengths from the consensus 3D7 (v3) sequence in PlasmoDB [23].
The adjusted amplicon lengths were then used to create haplotypes for each sample consisting of each of the six microsatellite loci. We defined haplotypes in the form of repeats per microsatellite locus rather than fragment lengths. We calculated the number of repeats in each amplicon by dividing the difference between our adjusted amplicon length and the minimum theoretical amplicon length, assuming zero repeats, by the repeat unit length. Only the dominant haplotype for each sample was included in population genetic analyses.
Population Genetic Analysis
The unbiased expected heterozygosity by locus for pfhrp2-deleted and non-deleted parasites was calculated using GenAlEx version 6.5 and plotted in R [24, 25]. To visualize potential relationships between individual parasites, we generated a matrix of pairwise genetic distances between parasites using GenAlEx for haploid simple-sequence repeats, with missing data interpolated by inserting the average genetic distances for each population-level pairwise contrast [24, 25]. We then created a neighbor-joining tree using the APE package in R and visualized the tree using Cytoscape [26, 27].
To explore genetic differentiation between parasite populations, we calculated the observed GST for pfhrp2-deleted parasites versus controls overall and by province. GST, a measure of genetic distance between populations that can incorporate multiple genetic markers, was calculated using the formula of Nei at each locus before averaging over all loci [28]. We used permutation testing to determine the statistical significance of observed GST values; population labels (pfhrp2-deletion vs. positive control) were permuted 99,999 times and the position of the observed value in this ordered list was converted to an empirical p-value.
Data Analysis
We made comparisons using Fisher’s exact test for categorical variables, the Wilcoxon rank-sum test for non-normally distributed continuous variables, and the t-test for normally distributed continuous variables. Statistical analyses were performed using SAS 9.4 (SAS Institute, Cary, NC) and R software.
RESULTS
Prevalence and Geographical Distribution of pfhrp2-Deleted P. falciparum
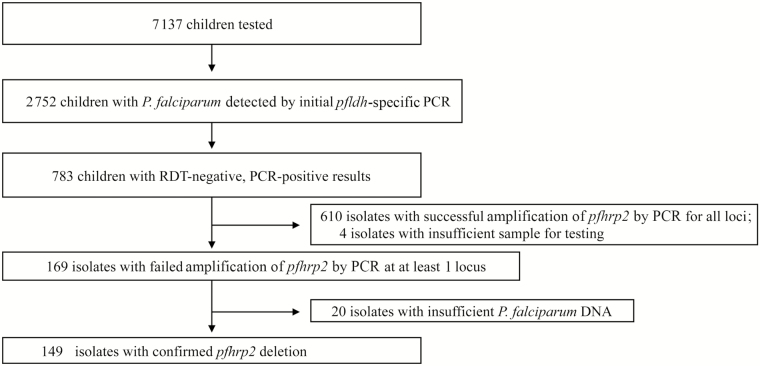
We identified 149 P. falciparum isolates with a deletion of the pfhrp2 gene, representing a country-wide prevalence of 6.4% (95% CI 5.1 – 8.0) among the 2,752 children diagnosed with P. falciparum infection by real-time PCR and 19.0% of the 783 RDT-/PCR+ parasites tested. Figure 1 and Supplementary Figure 1 outline the outcome of PCR assays employed to detect deletions of the pfhrp2 gene and to confirm the presence of P. falciparum DNA. Of the 149 pfhrp2-deleted P. falciparum isolates, only 5 (3.4%) had co-existing complete pfhrp3 deletions (Supplementary Figure 1). We did not identify any pfhrp2 deletions among the 107 RDT+/PCR+ parasites detected in children from Kinshasa, North Kivu, or South Kivu Provinces. The characteristics of children diagnosed with P. falciparum infection by PCR and the subset with pfhrp2-deleted P. falciparum are described in Table 1. We include previously published results of the initial P. falciparum ldh (pfldh) real-time PCR for reference.[18, 19] Pfhrp2-deleted parasites had lower parasite densities than wild-type parasites (p < .0001), with mean pfldh cycle threshold values of 36.1 (standard deviation [SD] 3.2) and 32.7 (SD 3.6), respectively. Children infected with pfhrp2-deleted P. falciparum were significantly younger than those infected with wild-type parasites (p = .0007) and more likely to be microscopy-negative (p < .0001) and afebrile (p = .0004). There was no significant difference in gender between the groups (p = .92).
Figure 1.
Study population and PCR results. Abbreviations: PCR, polymerase chain reaction; RDT, rapid diagnostic test.
Table 1.
Characteristics of Children Infected With P. falciparum
| Characteristic | All subjects | All P. falciparum infections* | pfhrp2-deleted P. falciparum | p** |
|---|---|---|---|---|
| N (weighted prevalence [95% CI]) | 7,137 | 2752 (34.8 [32.3 – 37.4]) | 149 (6.4 [5.1 – 8.0]) | — |
| Age in months, median (IQR) | 32 (19 – 46) | 36 (23 – 48) | 31 (14 – 45) | 0.0007 |
| Female sex, n (weighted proportion) | 3568 (49.9) | 1340 (49.0) | 67 (48.4) | 0.92 |
| Microscopy positive, n (weighted proportion) | -- | 1695 (58.7) | 18 (9.7) | <0.0001 |
| Fever in last 2 weeks, n (weighted proportion)& | 2094 (0.32) | 934 (39.8) | 27 (18.5) | 0.0004 |
| Province, n (weighted prevalence [95% CI]) | ||||
| Kinshasa | 362 | 92 (25.6 [19.9,32.1]) | 20 (21.9 [13.9,32.8]) | + |
| Kwango | 308 | 67 (19.4 [13.3,27.4]) | 9 (12.8 [5.4,27.4]) | -- |
| Kwilu | 360 | 70 (18.9 [14.0,24.9]) | 6 (3.0 [1.1,8.2]) | -- |
| Mai-Ndombe | 272 | 116 (44.0 [31.9,56.8]) | 4 (9.6 [2.6,29.6]) | -- |
| Kongo Central | 321 | 148 (43.1 [31.6,55.4]) | 6 (4.2 [1.5,11.7]) | -- |
| Equateur | 198 | 57 (23.6 [11.4,42.5]) | 3 (2.8 [0.4,18.7]) | -- |
| Mongala | 255 | 74 (36.6 [28.3,45.7]) | 5 (4.4 [0.9,19.0]) | -- |
| Nord-Ubangi | 231 | 143 (61.5 [46.6,74.6]) | 5 (5.3[1.7,15.9]) | -- |
| Sud-Ubangi | 276 | 86 (29.5 [23.6,36.2]) | 7 (12.4 [5.4,26.1]) | -- |
| Tshuapa | 205 | 56 (26.3 [16.4,39.5]) | 5 (8.1 [2.7,22.1]) | -- |
| Kasai | 310 | 118 (34.5 [20.9,51.2]) | 2 (1.3 [0.3,5.1]) | -- |
| Kasai-Central | 327 | 155 (50.9 [42.1,59.7]) | 3 (3.4 [1.2,9.2]) | -- |
| Kasai-Oriental | 255 | 106 (36.4 [22.8,52.6]) | 3 (3.2 [0.9,11.1]) | -- |
| Lomami | 351 | 221 (63.6 [52.7,73.3]) | 5 (1.8 [0.5,5.9]) | -- |
| Sankuru | 194 | 68 (26.4 [12.2,48.0]) | 6 (14.8 [5.6,33.8]) | -- |
| Haut-Katanga | 236 | 81 (31.7 [19.1,47.8]) | 3 (3.5 [1.1,10.5]) | -- |
| Haut-Lomami | 261 | 99 (42.8 [34.0,52.2]) | 3 (3.7 [1.0,12.5]) | -- |
| Lualaba | 176 | 87 (44.9 [30.8,59.9]) | 2 (5.7 [1.1,25.5]) | -- |
| Tanganyka | 245 | 157 (61.7 [45.1,75.9]) | 3 (1.7 [0.3,9.5]) | -- |
| Maniema | 349 | 182 (50.6 [38.0,63.1]) | 13 (4.6 [2.1,10.0]) | -- |
| Nord-Kivu | 426 | 72 (12.2 [8.5,17.3]) | 12 (15.5 [7.5,29.4]) | + |
| Bas-Uele | 180 | 109 (68.2 [54.9,79.1]) | 1 (0.2[0,1.8]) | -- |
| Haut-Uele | 170 | 87 (57.5 [46.6,67.7]) | 0 (0[NA]) | -- |
| Ituri | 255 | 122 (43.7 [30.6,57.8]) | 9 (11.5 [6.4,19.7]) | + |
| Tshopo | 225 | 106 (45.3 [36.3,54.6]) | 6 (4.5 [1.5,12.3]) | -- |
| Sud-Kivu | 389 | 73 (14.2 [9.8,20.0]) | 8 (18.3 [7.1,39.9]) | + |
* Diagnosed by real-time PCR targeting the pfldh gene during initial screening, as previously described [19].
** Comparing pfhrp2-deleted to non-deleted P. falciparum.
&Fever in the last 2 weeks based on survey response.
+Provinces with geographical clustering of pfhrp2-deleted P. falciparum by Bayesian spatial cluster analysis.
Abbreviations: CI, confidence interval; IQR, interquartile range; NA, not applicable.
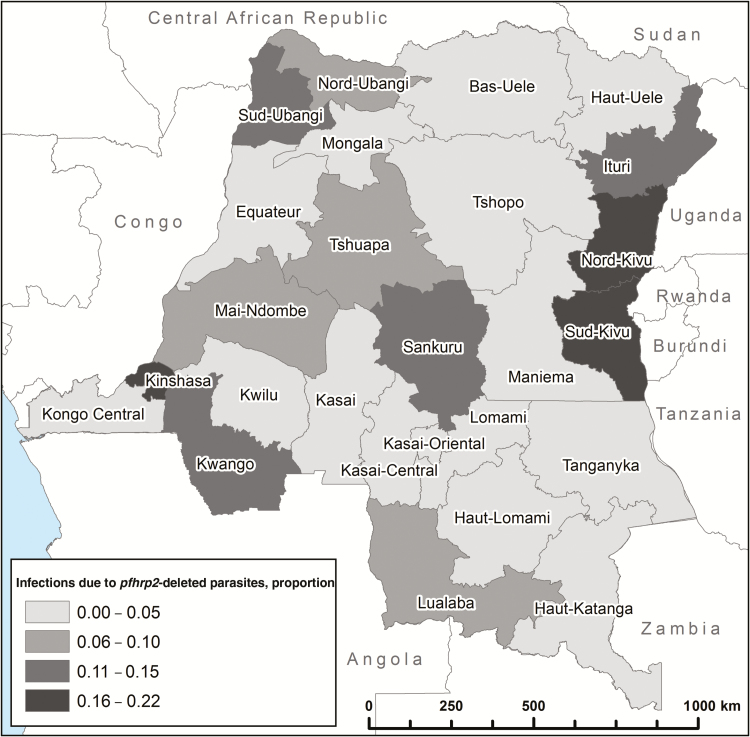
Pfhrp2-deleted parasites were present in nearly all provinces but were significantly clustered in the provinces surrounding the capital Kinshasa in the southwest and Lake Kivu in the northeast (Table 1 and Figure 2). They were especially common in Kinshasa, where they were responsible for more than one of every five P. falciparum infections. A Bayesian spatial cluster analysis supported significant geographical clustering of pfhrp2-deleted parasites in Kinshasa Province (posterior probability of cluster membership = 0.99) and three provinces in the northeast near Lake Kivu (posterior probabilities of 0.69 in South Kivu, 0.70 in North Kivu, and 0.69 in Ituri Provinces [Supplementary Figure 2]). A sensitivity analysis using different prior probabilities for no clustering (0.50) and different scanning windows (up to 50% of the study population) did not change the results meaningfully (Supplementary Figures 3 and 4, Supplementary Material). The pattern of deletions (i.e. complete deletion of the gene, partial deletion of exon 1, or partial deletion of exon 2) is depicted in Supplementary Figure 5. Because spatial clustering suggests the possibility of person-to-person spread and/or expansion of the mutant parasite population, we attempted to obtain genetic evidence for spread and selective pressures using microsatellite analyses.
Figure 2.
Distribution of pfhrp2-deleted P. falciparum parasites.
Genetic Origins of pfhrp2-Deleted P. falciparum
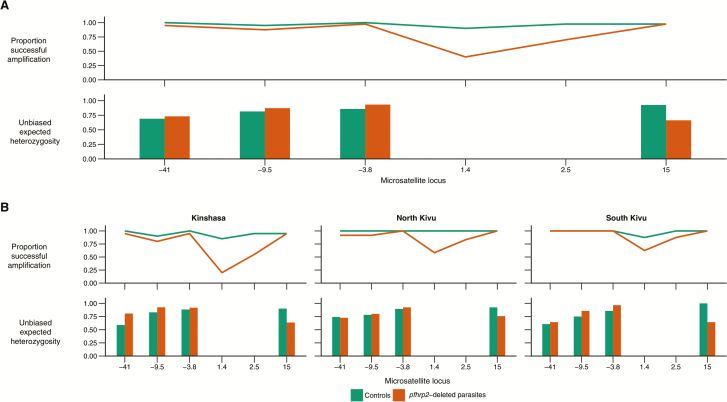
Failure to amplify microsatellite loci 1.4kb and 2.5kb downstream in pfhrp2-deleted parasites suggests that the predominant gene deletion pattern spans at least several kilobases and includes regions downstream from the pfhrp2 gene (Figure 3). This pattern of amplification failure was most common in pfhrp2-deleted parasites from Kinshasa, but it was also observed in a subset of parasites from North and South Kivu. These findings, in conjunction with the distribution of partial versus complete gene deletions (Supplementary Figure 5), suggest that there are multiple deletion genotypes circulating in the DRC.
Figure 3.
Flanking microsatellites. Analyses revealed frequent failure to amplify flanking microsatellite loci downstream from the pfhrp2 gene and reduced expected heterozygosity at a single locus in pfhrp2-deleted parasites overall (panel A) and by province (panel B).
We did not observe clear evidence of a selective sweep (Figure 3), although there appeared to be loss of heterozygosity at a single microsatellite locus 15kb downstream from the pfhrp2 gene (unbiased expected heterozygosity of 0.66 among pfhrp2-deleted parasites and 0.93 among controls). Amplification failure among pfhrp2-deleted parasites prevented similar analyses at the two proximal downstream loci.
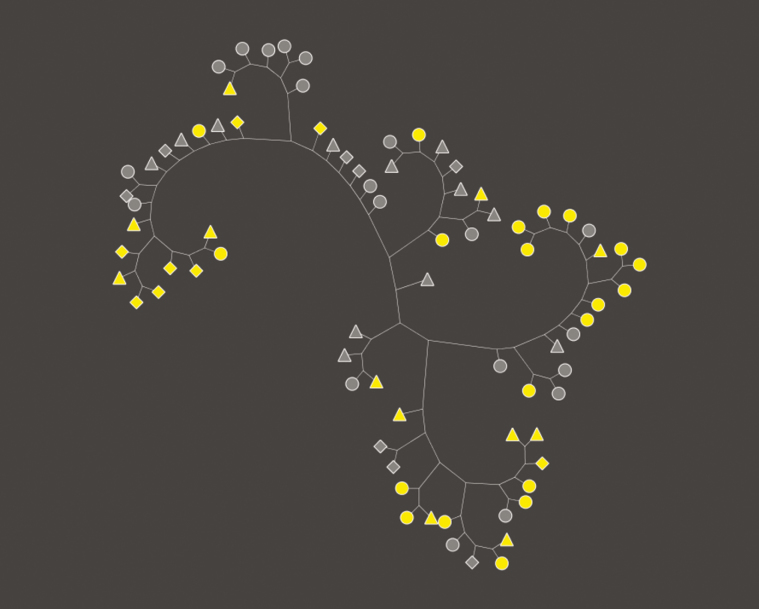
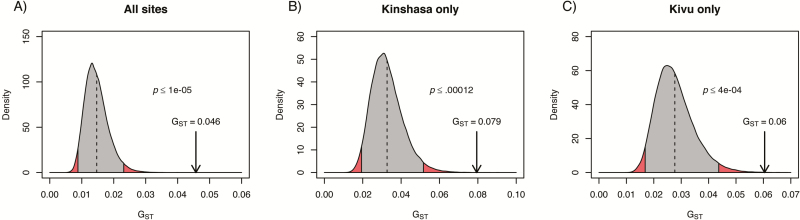
Initial analysis of haplotypes using an unrooted neighbor-joining tree revealed three distinct clusters of pfhrp2-deleted parasites, one containing primarily isolates from Kinshasa Province, another containing a mixture of isolates from North and South Kivu, and a third containing a mixture of isolates from all three provinces (Figure 4). Because analysis of individual loci did not provide sufficient discriminatory power to distinguish parasite populations (Supplementary Table 2), we explored the genetic relatedness of pfhrp2-deleted parasites and non-deleted controls using a permutation test based on the statistic GST. This analysis revealed significant genetic differentiation between the two populations (GST = .046, p ≤ .00001, Figure 5). Inclusion of the parasites’ geographical origins in the analysis confirmed significant differentiation between pfhrp2-deleted and non-deleted parasites in both Kinshasa (GST = .079, p ≤ .00012) and Kivu (GST = .06, p ≤ .0004).
Figure 4.
Neighbor joining tree. Unrooted neighbor joining tree depicting the relationship between dominant haplotypes of pfhrp2-deleted P. falciparum isolates (yellow) and HRP2-positive controls (gray). Shapes indicate the geographical origin of each isolate: Kinshasa (circle), North Kivu (triangle), or South Kivu (diamond).
Figure 5.
Permutation testing for G ST. Value of observed GST between pfhrp2-deleted and HRP2-positive control P. falciparum populations (black arrow) compared with the distribution of GST obtained from 99,999 permutations of population labels. Analysis was carried out for all sites (A), as well as Kinshasa only (B) and Kivu only (North and South combined) (C).
Relationship Between pfhrp2-Deleted Mutants and P. falciparum Prevalence
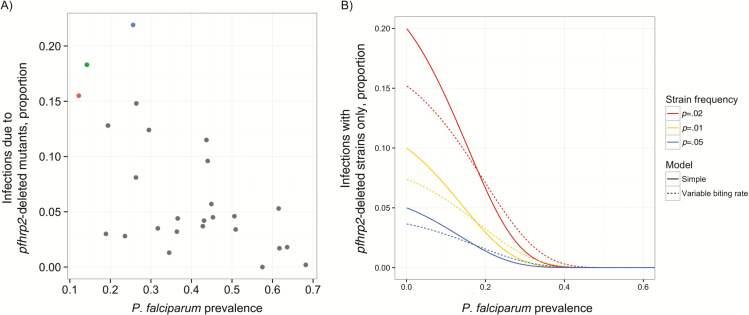
P. falciparum infections are presumed to be more polyclonal in areas of higher prevalence [29, 30]. Accordingly, coinfections with a more diverse population of wild-type parasites could increase the probability of a positive RDT and mask the presence of pfhrp2-deleted parasites. We observed a decline in the frequency of presumed monoclonal infections by parasites harboring a pfhrp2 deletion at higher parasite prevalence, consistent with this hypothesis (Figure 6 and Supplementary Material).
Figure 6.
Relationship between pfhrp2-deleted mutants and overall P. falciparum prevalence. (A) Scatterplot by province comparing the proportion of P. falciparum infections due to pfhrp2-deleted mutants and overall P. falciparum prevalence. Colors correspond to Kinshasa (blue), North Kivu (red), South Kivu (green), and other (gray) Provinces. (B) Modeling results showing the predicted decrease in probability of infection with the pfhrp2-deleted strain only, as a function of prevalence in 0–5 year olds. The underlying strain frequency is held at a constant frequency in this example. Results are shown for a simple model assuming homogeneous force of infection throughout the population, and a more complex model allowing for variation in biting rate between individuals (see Supplementary Material).
DISCUSSION
This is the first nationally representative study to demonstrate the presence and estimate the prevalence of malaria caused by pfhrp2-deleted P. falciparum in asymptomatic children. Our findings suggest that nearly one of every 15 children with falciparum malaria in the DRC are infected by a pfhrp2-deleted mutant. Because most RDTs employed in the DRC are HRP2-based, they will fail to detect these parasites.
Several provinces had a markedly higher prevalence of pfhrp2-deleted mutants. The high prevalence observed in Kivu and Kinshasa is especially concerning because RDTs appear to have been introduced in these regions prior to introduction elsewhere. HRP2-based RDTs have been deployed in humanitarian interventions in Kivu since at least 2005, whereas they were deployed in most remaining regions in approximately 2011 [31–33]. In Kinshasa, HRP2-based RDTs were deployed on a limited basis by at least 2009 and their availability increased 10-fold by 2013 [34]. Geographic and genetic clustering of pfhrp2-deleted mutants raises concern for evolutionary selection in the setting of selective treatment of RDT-positive P. falciparum parasites.
There are several lines of evidence confirming that our inability to detect the pfhrp2 gene was not simply due to insufficient DNA, an important consideration in this type of study [35]. The presence of amplifiable DNA in pfhrp2-deleted samples was confirmed by PCR assays for multiple single-copy genes (pfldh, pfhrp3, and/or Pf β-tubulin) and, for a subset, six microsatellite loci flanking the pfhrp2 gene. We successfully amplified pfhrp3 in 96.1% of pfhrp2-deleted P. falciparum isolates with sufficient sample remaining for testing and multiple flanking microsatellite loci in all pfhrp2-deleted parasites included in the population genetic analysis. For those considering similar analyses, useful recommendations for streamlining the process of identifying pfhrp2-deleted P. falciparum are now available [36].
Only a small fraction (3.4%) of pfhrp2-deleted parasites had coexisting pfhrp3 deletions, a finding that contrasts with reports from South America [36]. Because pfhrp3 produces an antigen (HRP3) that shares epitopes with HRP2, pfhrp2-deleted parasites with an intact pfhrp3 gene can, in theory, trigger a positive HRP2-based RDT. This phenomenon may afford select RDTs a “fail-safe” against pfhrp2-deleted parasites. By examining RDT-/PCR+ parasites, it is possible that we may have underestimated the true prevalence of pfhrp2-deleted parasites. However, we expect this effect to be small given that we did not detect any additional pfhrp2-deleted parasites during testing of RDT+/PCR+ parasites from Kinshasa, North Kivu, and South Kivu Provinces.
We observed a notable improvement in our ability to amplify pfhrp2 exon 1/2 using a lower extension temperature during PCR cycling. This finding likely reflects impaired Taq polymerase extension of the AT-rich intron at higher temperatures due to DNA melting [21]. Future assays employed to detect pfhrp2-deleted parasites, especially those that target the AT-rich intron between exons 1 and 2, should be tested at lower extension temperatures to avoid unintentional misclassification based on false-negative PCR results.
Population genetic analyses of flanking microsatellites confirmed that parasites with a pfhrp2 deletion were genetically distinct from wild-type parasites in both Kinshasa and Kivu. While we did not observe evidence of a selective sweep, the loss of heterozygosity observed at a single locus downstream raises the possibility of a “soft selective sweep” in the population, with multiple genetic origins of the deletion due to multiple breakpoints. Soft selective sweeps have been observed in analyses of pfmdr1 duplication, implicated in antimalarial drug resistance, in Southeast Asia [37]. Additionally, recent analyses of beneficial copy number variation mutations in P. falciparum did not reveal the typical long flanking haplotypes required to produce “hard” selective sweeps, which are usually observed around point mutations [38].
Although we cannot draw definitive conclusions about whether selective pressure is responsible for the pfhrp2 deletion, the ability to evade detection by a widespread diagnostic test would provide significant evolutionary advantages for the P. falciparum parasite. Clonal selection of pfhrp2-deleted parasites could jeopardize recent gains in malaria control, already threatened by the rising tide of resistance to artemisinin-based combination therapies in Southeast Asia and to insecticides [1, 39].
Pfhrp2-deleted parasites may be easier to detect in areas of low transmission intensity because polyclonal infections are less likely to occur—i.e. co-infection with wild-type parasites could trigger a positive RDT and mask the presence of a pfhrp2-deleted parasite [36]. The inverse relationship we observed between the prevalence of pfhrp2-deleted mutants and overall P. falciparum prevalence is consistent with this hypothesis. If correct, this hypothesis implies that RDT failure due to pfhrp2 deletions will become more common as transmission declines.
There are several limitations to this study. First, our ability to explore clinical differences between pfhrp2-deleted and wild-type P. falciparum malaria is restricted by limited clinical data. Ongoing studies of symptomatic malaria in the DRC may provide valuable insights into the clinical manifestations of infection with pfhrp2-deleted parasites. In the present study, however, subjects were enrolled in a large, country-wide survey and not selected based on symptoms of malaria. As a result, we cannot draw conclusions about the relative virulence of pfhrp2-deleted parasites. While the lower parasite densities by real-time PCR and the lower proportion of microscopy-positivity and fever among pfhrp2-deleted P. falciparum cases (Table 1) may simply reflect our ability to detect deletions in less complex infections as noted above, these findings also raise the possibility that there may be a fitness cost to the parasite. Our finding that children infected with pfhrp2-deleted parasites were younger than those infected with wild-type parasites could reflect the immature immune system of younger children, who may mount a weaker response to these less fit parasites. The function of pfhrp2 is not well understood, but detoxification of heme and other possibilities have been suggested [40]. While parasite clones containing the pfhrp3 gene were favored over pfhrp3-deleted clones in a genetic cross, the same has not been demonstrated for pfhrp2 in vitro [41].
Second, our study was restricted to the DRC. While this may raise questions about generalizability, the DRC is a large and geographically diverse country that borders nine other countries in East, West, and Southern Africa, suggesting that our findings have relevance throughout the region.
Independent of concerns about evolutionary selection, our findings represent the first report of geographical clustering of pfhrp2-deleted P. falciparum parasites in Africa, where HRP2-based RDT use is most common [1]. Malaria is widespread in the DRC; at least 34.8 percent of the children in this cohort were infected with P. falciparum parasites. These and other infected African children bear the brunt of malaria’s heavy toll. Rapid and accurate diagnosis of P. falciparum infection, especially among clinically ill patients presenting for evaluation and treatment, is essential for curbing its effects. RDTs that detect antigens other than HRP2 such as lactate dehydrogenase (LDH) and aldolase are commercially available, but they are not widely utilized in sub-Saharan Africa due to concerns about heat stability and inferior P. falciparum sensitivity, higher costs, and limited global availability compared to HRP2-based RDTs [42, 43]. Multilateral donors currently invest hundreds of millions of dollars in the procurement and distribution of HRP2-based RDTs. Our findings underscore the need for surveillance of pfhrp2-deleted mutants and suggest that alternative diagnostics need to be considered in areas where these deletions are common. Use of RDTs that detect multiple antigens (e.g. HRP2 and LDH) or novel P. falciparum-specific antigens may be required.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Notes
Acknowledgements. This work would not have been possible without the administrators of the 2013–2014 DRC DHS and the thousands of subjects who participated. We thank Sheila Akinyi Okoth, Ph.D., Naomi Lucchi, Ph.D., and Venkatachalam Udhayakumar, Ph.D., at the US Centers for Disease Control for assistance with the microsatellite PCR assays.
Presentations. Preliminary findings were presented during an oral presentation at the Journées Scientifiques de Lutte contre le Paludisme 2016 conference in Kinshasa, Democratic Republic of Congo on April 27, 2016.
Changes in affiliation. N/A
Financial support. This work was supported by the National Institutes of Allergy and Infectious Diseases [5T32AI007151 to JBP and 5R01AI107949 to SRM], the Population Research Infrastructure Program through funding awarded by the Eunice Kennedy Shriver National Institute of Child Health and Human Development to the Carolina Population Center [P2C HD050924 to MJ and CK], the National Science Foundation [BCS-1339949 to ME], and the UK Medical Research Council (MRC) and Department for International Development (DfID) under the MRC/DfID concordat [to RV and ACG].
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. WHO. World Malaria Report 2015. Geneva: World Health Organization, 2015. [Google Scholar]
- 2. WHO. Malaria rapid diagnostic test performance. Results of WHO product testing of malaria RDTs: Round 6 (2015). Geneva: World Health Organization, 2015. [Google Scholar]
- 3. Iqbal J, Siddique A, Jameel M, Hira PR. Persistent histidine-rich protein 2, parasite lactate dehydrogenase, and panmalarial antigen reactivity after clearance of Plasmodium falciparum monoinfection. J Clin Microbiol 2004; 42:4237–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Abba K, Deeks JJ, Olliaro P, et al. Rapid diagnostic tests for diagnosing uncomplicated P. falciparum malaria in endemic countries. Cochrane Database Syst Rev 2011:CD008122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abdallah JF, Okoth SA, Fontecha GA, et al. Prevalence of pfhrp2 and pfhrp3 gene deletions in Puerto Lempira, Honduras. Malar J 2015; 14:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Akinyi S, Hayden T, Gamboa D, et al. Multiple genetic origins of histidine-rich protein 2 gene deletion in Plasmodium falciparum parasites from Peru. Sci Rep 2013; 3:2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baldeviano GC, Okoth SA, Arrospide N, et al. Molecular epidemiology of Plasmodium falciparum malaria outbreak, Tumbes, Peru, 2010–2012. Emerg Inf Dis 2015; 21:797–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koita OA, Doumbo OK, Ouattara A, et al. False-negative rapid diagnostic tests for malaria and deletion of the histidine-rich repeat region of the hrp2 gene. Am J Trop Med Hyg 2012; 86:194–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murillo Solano C, Akinyi Okoth S, Abdallah JF, et al. Deletion of Plasmodium falciparum Histidine-Rich Protein 2 (pfhrp2) and Histidine-Rich Protein 3 (pfhrp3) genes in Colombian parasites. PLoS One 2015; 10:e0131576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wurtz N, Fall B, Bui K, et al. Pfhrp2 and pfhrp3 polymorphisms in Plasmodium falciparum isolates from Dakar, Senegal: impact on rapid malaria diagnostic tests. Malar J 2013; 12:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kumar N, Pande V, Bhatt RM, et al. Genetic deletion of HRP2 and HRP3 in Indian Plasmodium falciparum population and false negative malaria rapid diagnostic test. Acta Trop 2013; 125:119–21. [DOI] [PubMed] [Google Scholar]
- 12. Gamboa D, Ho MF, Bendezu J, et al. A large proportion of P. falciparum isolates in the Amazon region of Peru lack pfhrp2 and pfhrp3: implications for malaria rapid diagnostic tests. PLoS One 2010; 5:e8091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li P, Xing H, Zhao Z, et al. Genetic diversity of Plasmodium falciparum histidine-rich protein 2 in the China-Myanmar border area. Acta Trop 2015; 152:26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Amoah LE, Abankwa J, Oppong A. Plasmodium falciparum histidine rich protein-2 diversity and the implications for PfHRP 2: based malaria rapid diagnostic tests in Ghana. Malar J 2016; 15:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bharti PK, Chandel HS, Ahmad A, Krishna S, Udhayakumar V, Singh N. Prevalence of pfhrp2 and/or pfhrp3 gene deletion in Plasmodium falciparum population in eight highly endemic states in India. PLoS One 2016; 11:e0157949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. WHO. False-negative RDT results and implications of new P. falciparum histidine-rich protein 2/3 gene deletions. Geneva: World Health Organization, 2016. [Google Scholar]
- 17. WHO. P. falciparum hrp2/3 gene deletions: Conclusions and recommendations of a Technical Consultation. Geneva: World Health Organization, 2016. [Google Scholar]
- 18. Meshnick SR, Janko M, Doctor S, et al. Demographic and Health Survey (DRC-DHS II) 2013–2014: Supplemental Malaria Report. Rockville, MD: DHS, 2015. [Google Scholar]
- 19. Doctor SM, Liu Y, Whitesell A, et al. Malaria surveillance in the Democratic Republic of the Congo: comparison of microscopy, PCR, and rapid diagnostic test. Diagn Microbiol Infect Dis 2016; 85:16–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Plowe CV, Djimde A, Bouare M, Doumbo O, Wellems TE. Pyrimethamine and proguanil resistance-conferring mutations in Plasmodium falciparum dihydrofolate reductase: polymerase chain reaction methods for surveillance in Africa. Am J Trop Med Hyg 1995; 52:565–8. [DOI] [PubMed] [Google Scholar]
- 21. Su XZ, Wu Y, Sifri CD, Wellems TE. Reduced extension temperatures required for PCR amplification of extremely A+T-rich DNA. Nucleic Acids Res 1996; 24:1574–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wakefield J, Kim A. A Bayesian model for cluster detection. Biostatistics 2013;14:752–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bahl A, Brunk B, Crabtree J, et al. PlasmoDB: the Plasmodium genome resource. a database integrating experimental and computational data. Nucleic Acids Res 2003; 31:212–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peakall ROD, Smouse PE. genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 2006; 6:288–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peakall R, Smouse PE. GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 2012; 28:2537–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics 2004; 20:289–90. [DOI] [PubMed] [Google Scholar]
- 27. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci U S A 1973; 70:3321–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Babiker HA, Lines J, Hill WG, Walliker D. Population structure of Plasmodium falciparum in villages with different malaria endemicity in east Africa. Am J Trop Med Hyg 1997; 56:141–7. [DOI] [PubMed] [Google Scholar]
- 30. Kateera F, Nsobya SL, Tukwasibwe S, et al. Malaria case clinical profiles and Plasmodium falciparum parasite genetic diversity: a cross sectional survey at two sites of different malaria transmission intensities in Rwanda. Malar J 2016; 15:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Swarthout TD, Counihan H, Senga RK, van den Broek I. Paracheck-Pf accuracy and recently treated Plasmodium falciparum infections: is there a risk of over-diagnosis? Malar J 2007; 6:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hawkes M, Katsuva JP, Masumbuko CK. Use and limitations of malaria rapid diagnostic testing by community health workers in war-torn Democratic Republic of Congo. Malar J 2009; 8:308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Piriou E.(Medicins Sans Frontieres). Personal communication. [Google Scholar]
- 34. ACTwatch and ASF. The DRC Outlet Survey 2013. Washington DC: PSI, 2014. [Google Scholar]
- 35. Laban NM, Kobayashi T, Hamapumbu H, et al. ; Southern Africa International Centers of Excellence for Malaria Research. Comparison of a PfHRP2-based rapid diagnostic test and PCR for malaria in a low prevalence setting in rural southern Zambia: implications for elimination. Malar J 2015; 14:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cheng Q, Gatton ML, Barnwell J, et al. Plasmodium falciparum parasites lacking histidine-rich protein 2 and 3: a review and recommendations for accurate reporting. Malar J 2014; 13:283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vinayak S, Alam MT, Sem R, et al. Multiple genetic backgrounds of the amplified Plasmodium falciparum multidrug resistance (pfmdr1) gene and selective sweep of 184F mutation in Cambodia. J Infect Dis 2010; 201:1551–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cheeseman IH, Miller B, Tan JC, et al. Population Structure shapes copy number variation in malaria parasites. Mol Biol Evol 2016; 33:603–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Spring MD, Lin JT, Manning JE, et al. Dihydroartemisinin-piperaquine failure associated with a triple mutant including kelch13 C580Y in Cambodia: an observational cohort study. Lancet Infect Dis 2015; 15:683–91. [DOI] [PubMed] [Google Scholar]
- 40. Sullivan DJ, Jr, Gluzman IY, Goldberg DE. Plasmodium hemozoin formation mediated by histidine-rich proteins. Science 1996; 271:219–22. [DOI] [PubMed] [Google Scholar]
- 41. Walker-Jonah A, Dolan SA, Gwadz RW, Panton LJ, Wellems TE. An RFLP map of the Plasmodium falciparum genome, recombination rates and favored linkage groups in a genetic cross. Mol Biochem Parasitol 1992; 51:313–20. [DOI] [PubMed] [Google Scholar]
- 42. Ali IM, Bigoga JD, Forsah DA, et al. Field evaluation of the 22 rapid diagnostic tests for community management of malaria with artemisinin combination therapy in Cameroon. Malar J 2016; 15:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. WHO. Good practices for selecting and procuring rapid diagnostic tests for malaria. Geneva, Switzerland: World Health Organization, 2011. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.