Introduction
The use of diagnostic imaging and image-guided interventions using low-dose ionizing radiation has increased dramatically in recent years. This is explained by dramatic advances in imaging technology which provide invaluable diagnostic information for clinical decision-making.1 Although the benefits of appropriate use of advanced imaging technology outweigh the risks, the medical use of X-ray-based imaging techniques has become a leading source of man-made radiation exposure to the general population. According to a recent report by the National Council on Radiation Protection and Measurements, the total radiation exposure from medical imaging has increased six-fold from the early 1980s to the present, and currently almost 40% of medical radiation exposure (excluding radiotherapy) comes from cardiovascular imaging and image-guided interventions.2,3 This has prompted renewed interest in the potential long-term risks of low-dose radiation exposure for patients, physicians, and technical staff members.4,5 Clinical decision-making inherently requires balancing the potential benefits of e.g. a cardiac-imaging procedure and intervention with the projected risks, including those from radiation exposure. Although risk estimates for low-dose radiation exposures and international guidelines exist,6,7 these have been developed predominantly for the purpose of radiation protection and the development of occupational dose limits for radiation exposed workers (such as some physicians). Risk estimates for medical low-dose radiation exposure are associated with substantial uncertainties—to some extent this is due to the fact that our understanding of the biological effects of low-dose radiation exposure in humans is incomplete.6,8
Radiation dose from medical imaging, commonly referred to as effective dose, is expressed in units of millisieverts, which is the weighted average of the absorbed dose in mGy multiplied by two weighting factors that depend on the type of tissue irradiated and the specific type of radiation.9 The effective dose allows for a rough estimation of the risk of a partial or whole body exposure to ionizing radiation.10 Cardiovascular imaging may involve considerable radiation exposure. For example, coronary computed tomographic angiography (CCTA) is commonly used to manage patients with suspected coronary artery disease (CAD), new-onset heart failure with reduced heart function, and for a wide range of acute indications such as acute aortic syndromes, pulmonary embolism, as well as surgical or transcatheter treatment planning of aortic diseases.11 Typical effective doses from CCTA can range from 0.06 to 18.0 mSv with a median of 12 mSv.12–14 For patients at intermediate risk for obstructive CAD, single-photon emission computed tomography myocardial perfusion imaging (SPECT MPI) with injected radioactive tracers has also been the cornerstone for diagnosis, risk stratification, and management. The median effective dose for SPECT MPI was reported to be 10 mSv (range, 10–25 mSv).15 The effective doses from these medical radiation exposure are equivalent to having hundreds of chest X-rays. Of note, patients undergoing cardiac imaging may undergo not one but a series of tests or procedures involving ionizing radiation exposure, which can result in cumulative exposure of >100 mSv, a threshold-level documented to increase potential cancer risk.8,16,17 It should be noted, however, that these effective doses are only gross estimates, especially for partial body exposures, and may suffer from inherent relative uncertainties of about ±40% due to methodological limitations.18,19
Current recommended models for assessing radiation risk
Current cancer risk models for low-radiation exposure often use the linear no-threshold (LNT) model, which assumes that the risk of cancer increases linearly with the exposure, and that the detriments (solid cancers and leukaemias) associated with high-dose and high-dose rate exposures in atomic bomb survivors and from accidental high-dose occupational exposures can be extrapolated to the low-dose range.20,21 The use of this model is reasonable for purposes of developing dose limits for occupational exposure—where erring on the side of higher risk is desirable—but whether the LNT model accurately describes the relationship between low-dose exposure and the development of cancer remains unclear and controversial. Other models, for example, assume that a dose below a certain threshold is not harmful, and the hormesis model, even posits that low-dose radiation might sometimes be beneficial. Estimating the radiation risk of low-lose radiation (≤100 mSv), thus, remains challenging due to the lack of sufficiently large and well-controlled cohorts for epidemiological studies to quantify a likely small excess cancer risk at low doses, relative to the high ‘natural’ cancer rate of 40%.8,22,23 Since the 1970s, the current risk estimates that inform health protection strategies are based on the LNT model, an approach recommended by the International Commission of Radiological Protection6 and endorsed by the Biological Effects of Ionizing Radiation VII report of the US National Academy of Sciences.7 In fact, a significantly increased cancer risk of developing both solid cancers and leukaemia is observed in epidemiologic studies of atomic bomb survivors, in those exposed to lower doses of radiation (5–150 mSv) and in a major international study of >400 000 nuclear industry radiation workers who were exposed to low-dose radiation (5–150 mSv) and an average dose of radiation of 20 mSv.20,24,25 Although some studies have shown the extent of DNA damage to cells is linearly related to dose,26,27 others show that there may be threshold effects,28 highlighting that cellular and tissue-level responses to radiation-induced damage are not always linear. In addition, recent studies have challenged the validity of the LNT model for evaluating radiation at low doses because of differences in biological responses of living cells and tissues to low- vs. high doses of radiation. Consequently, the use of biomarkers to measure the cellular effects of low-dose radiation exposure has emerged as an alternative approach to assess the potential risk of radiation.29,30
Utilizing biomarkers for estimating biological effects of low-dose radiation exposure
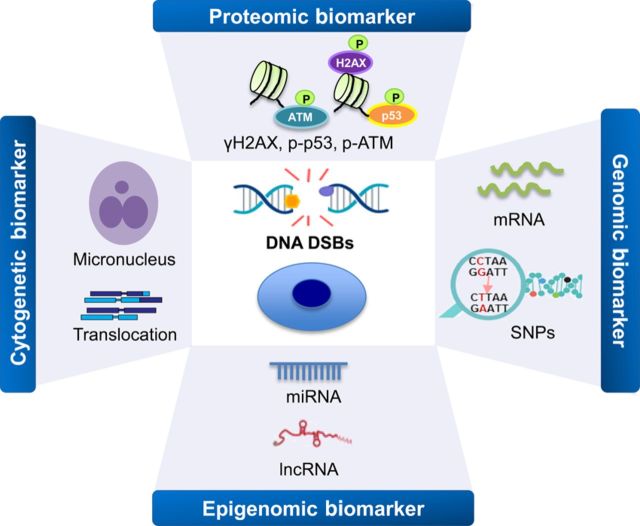
Different types of biomarkers have shown promise as predictors of radiation dose and risk, including chromosome damage (e.g. aberrations and micronuclei), post-translational modification, changes in gene expression and protein synthesis, and epigenomic modifications (Figure 1, Table 1). Exposure of cells to therapeutic doses of radiation initiates a large-scale activation of specific DNA damage signalling and repair mechanisms, a process known as the DNA damage response (DDR) pathway. This leads to the activation of a number of genes and proteins whose products trigger apoptosis, cell-cycle arrest, chromatin remodelling, and DNA repair, which minimize the risk of heritable mutations implicated in the process of carcinogenesis in human.31,32 Misrepair of these DNA double-strand breaks (DSBs) can produce many different types of chromosomal aberrations. Cytogenetic biomarkers that can be used for analysis of these aberrations (e.g. dicentrics, translocations, premature chromosome condensation, and micronuclei) in peripheral blood lymphocytes have been extensively validated as biomarkers of somatic chromosomal damage and intermediate end points in carcinogenesis after radiation exposure. For example, studies have found significantly increased chromosome abnormalities in blood lymphocytes obtained from adult and paediatric patients after CT scans.33,34 However, because of low sensitivity, long processing time, and tedious scoring methods of these chromosomal aberration biomarkers after radiation exposure, the application of these markers to doses <100 mGy is limited at the present.35,36
Figure 1.
Overview of the DNA damage-associated biomarkers of ionizing radiation. Multiple types of biomarkers are available for measuring radiation exposure and monitoring the DNA damage and repair, such as cytogenetic, proteomic, genomic, and epigenomic biomarkers. ATM, ataxia-telangiectasia mutated; DSBs, double-strand breaks; lncRNA, long non-coding RNA; miRNA, microRNA; p, phosphate group; p53, tumour protein p53; SNPs, single nucleotide polymorphisms.
Table 1.
Radiation biomarkers used for studying the risk after different diagnostic procedures
| Procedure | Biomarkers | References |
|---|---|---|
| [18F] FDG PET/CT (∼5 mSv) | Proteomic marker (γ-H2AX foci) | May et al.93 |
| CCTA (∼8 mSv) | Proteomic marker (γ-H2AX foci) | Grudzenski et al.94 |
| CCTA (∼36.9 mSv) | Proteomic and genomic markers | Nguyen et al.28 |
| CCTA (∼11.4 mSv) | Proteomic marker (γ-H2AX foci) | Kuefner et al.95 |
| CCTA (∼6.4 mSv) | Proteomic marker (γ-H2AX foci) | Brand et al.54 |
| CT (∼6.3 mSv) | Proteomic marker (H2AX foci) | Rothkamm et al.38 |
| Invasive angiography (∼18.2 mSv) | Proteomic and genomic markers | Lee et al.39 |
| Invasive angiography (∼12 mSv) | Cytogenetic marker (MN assay) | Andreassi et al.96 |
| SPECT MPI (∼10.0 mSv) | Proteomic and genomic markers | Lee et al.39 |
PET, positron emission tomography; CT, computed tomography; CCTA, coronary computed tomography angiography; SPECT-MPI, single-photon emission computed tomography myocardial perfusion imaging; MN, micronucleus.
The measurement of γ-H2AX foci formation has been applied as a biomarker of human low-dose radiation exposure that is more sensitive than quantification of cytogenetic biomarkers (e.g. dicentric chromosomes, micronuclei, and translocations), as foci formation can be detected at lower doses, <10–20 mGy.37–44 Although the overall γ-H2AX levels in cells and/or tissues can be obtained by using immunoblotting or the enzyme-linked immunosorbent assay,45,46 detection of individual DSBs by microscopy through foci counting is still the prevailing approach for clinical application since it is the most sensitive method.47 In response to DSB generation, histone H2AX is phosphorylated within seconds to form γ-H2AX, with γ-H2AX levels peaking at ∼30 min. Subsequent to this phosphorylation event, modifications to several other proteins, including phosphorylation of tumour protein 53 (p53) and ataxia-telangiectasia mutated (ATM), have been reported by our group and others as useful biomarkers for low-dose radiation exposure in lymphocytes and fibroblasts.28,39,42,48 Upon rapid activation by ionizing radiation, the kinase activity of ATM leads to phosphorylation and activation of a number of DNA repair and checkpoint proteins, including p53, H2AX, Chk2, and SMC1.49 For example, a high frequency and similar kinetics of co-localization of γ-H2AX and p53 with pATM foci were observed following exposure to irradiation.50–52 Although phosphorylation of H2AX may not exclusively reflect DSBs, it is still the best biomarker based on its cell cycle-independent induction, strong correlation with repair kinetics, and repair pathway independence. However, the extensive use of phosphorylated DNA damage marker proteins (e.g. γ-H2AX, p53, and ATM) alone as biomarkers of direct radiation exposure in biological samples is limited due to several factors including the transient character of foci formation, the lack of specificity for radiation, and the variation of foci frequencies between individuals.53 Despite these limitations, these biomarkers have the potential to reveal heightened sensitivity against low-dose radiation if samples can be collected at multiple time points within appropriate time windows. For examples, the biological effect of different scan modes in different CT generations was reliably compared using γ-H2AX immunofluorescence microscopy.38,40,54 Our recent studies also demonstrated the distinct levels of phosphorylation of H2AX, p53, and ATM in lymphocytes isolated from adult patients undergoing several cardiac medical imaging tests such as CCTA, SPECT MPI, and invasive X-ray angiography.28,39 Specifically, the loss of foci has been demonstrated to be correlated with DSB repair, suggesting that the kinetics of foci loss of these protein biomarkers might be also used as an indicator of individual susceptibility to low-dose radiation exposure in vivo or in vitro studies.28,38–40,43 With the demonstrated utility of γ-H2AX foci measurements in clinical application, multiple evaluation procedures such as cytometric assessment,39 automated assay and image processing,55 and image analysis algorithms56,57 have been developed for optimizing the methods of foci assessment and detection. For example, recently, a fully automated, high-throughput analysis platform, the Rapid Automated Biodosimetry Tool, was developed to screen γ-H2AX fluorescence labelling in fingerstick-derived blood samples and allows the analysis of up to 30 000 samples per day.58 To further increase the speed, throughput, and reliability of automated analysis, optimization of the protocols and regular calibration or adequate concurrent analysis of reference samples is necessary.
The transcriptional changes related to DNA damage are also central components of the DDR.59 Previous studies investigating the influence of dose and dose rate on radiation-induced gene expression profiles have found that a dose as low as 10 mGy can trigger gene expression modifications in human cells, and that low-dose transcriptional responses (25–100 mGy) may differ from those observed at high-dose (>100 mGy) radiation.60,61 For example, a linear increase in genes involved in p53-regulated pathways such as cyclin-dependent kinase inhibitor 1A (Cdkn1a), growth arrest and DNA-damage-inducible protein 45 alpha (Gadd45a), and Mdm2 p53 binding protein homolog (Mdm2) was found between 25 and 500 mGy, whereas at 25 mGy, only genes involved in the regulation of cell death processes were induced.61,62 Consistent with these findings, we recently demonstrated a concerted elevation in the gene expression of six DNA damage response genes (e.g. Bax, Ddb2, Mdm2, Tp53, Bbc3, and Atf6) in T-lymphocytes isolated from a small subset of adult patients post-SPECT MPI, most patients after radiation exposure from CCTA, and all undergoing invasive X-ray angiography.28,39 These changes were measureable as early as 2 h after radiation exposure and in some patients were extended to 48 h. Thus, changes in gene expression profiling may be potentially useful to estimate radiation exposure, providing several advantages over the more traditional cytogenetic assays that are more labour-intensive and time-consuming, and requiring relatively long-lived (>24 h) changes and γ-H2AX foci analysis that shows normally a very early and transient response of cells to DSBs, with the caveat that accurate measurements must be performed within a shorter window of exposure when using gene expression profiling as well as taking into account inter-patient variability which may potentially be resolved by having a large enough group size.
Exposure to radiation is also known to lead to epigenomic alteration, which will affect gene regulation after DNA damage induction. MicroRNAs (miRNAs) have recently emerged as promising biomarkers for the detection of various pathological conditions, including post-exposure to radiation.63,64 After DNA damage induction, post-transcriptional regulation by miRNAs occurs between transient post-translational protein modifications (seconds/minutes) such as phosphorylation and ubiquitination, and gene transcriptional events (hours/days).65 Serum miRNAs that fall under the ‘omics’ biodosimetry approach provide simple and attractive biomarkers that may effectively determine individual radiation exposure because of their inherent stability.66–68 Previous studies have shown modulated expression profiles in miRNA expression following exposure to low- and high-dose radiation.69,70 For example, miR-150 demonstrated a dose and time-dependent depletion in serum from mice irradiated at a range of 1–8 Gy.71 A significant modification of expression upon radiation exposure was also observed for miR-34-a-5p and miR-182-5p in human T lymphocytes, which exhibit strong pro-apoptotic and anti-proliferative properties,72 and dual properties as both an oncogene and tumour suppressor depending on the cellular model, respectively.73 In addition, the expression of miR-20 and miR-21 was significantly decreased in low dose (50 mGy) irradiated human B lymphoblast cell lines,74 thus indicating potential key roles of miRNAs in estimation of the dose and the regulation of cellular response to which the individual was exposed. In addition to miRNAs, long non-coding RNAs (lncRNAs) are a less investigated class of mRNA-like transcripts and their expression has been shown to be associated with cellular response to radiation-induced DNA damage. So far, only a few radiation-responsive lncRNAs have been found. For example, the expression of several lncRNAs such as lncRNACCND1, gadd7, ANRIL, and PANDA were found to be induced by DNA damage, and lncRNA-RoR, loc285194, and lncRNA-p21 were shown to be regulated by the p53 pathway, which is involved in the DDR.75 The two other lncRNAs (e.g. TP53TG1 and FAS-AS1), direct target of TP53, were also up-regulated by radiation exposure in human T lymphocytes.76 Although the deregulation and biological functions of radiation-responsive miRNAs and lncRNAs remain largely unknown considerable evidence suggests that miRNAs and lncRNAs may serve as a potentially rich source of biomarkers for studying radiation exposure, predisposition, and individual susceptibility.68,77
The identification of radiation exposure-related biomarkers will enable us to better understand how humans react to radiation exposure, and may provide a model to estimate individual sensitivity to radiation in the future. The main features of DNA damage-associated biomarkers are summarized in Table 2. As the formation of DNA damage is not unique to radiation, studies should take into account how the utility of these biomarkers can be affected by various factors that may affect individual sensitivity, such as age, gender, genetic susceptibility, and exposure to other environmental carcinogens such as tobacco smoke.
Table 2.
Principal of features of DNA damage-associated biomarkers
| Biomarkers | Advantages | Limitations | Readout/time of onset | Cell types |
|---|---|---|---|---|
| Cytogenetic (e.g. micronuclei, translocations, dicentrics) | Standardized protocol and relatively low costs High specificity to IR and low background in non-exposed population (dicentrics) Easy identification (micronuclei) Can be used in cases of long-term IR (translocations) |
Laborious, time-consuming, sophisticated, variability in scoring cells Limited sensitivity at dose <0.1 Gy High background frequency (translocation and micronuclei) |
Days to weeks Retrospective (translocations) |
WB PBMC |
| Proteomic (e.g. γ-H2AX, pATM, pP53) | Highly sensitive and linear with radiation dose : 0.01–8 Gy Can detect radiosensitive individuals Potentially high-throughput analysis |
Not specific to IR (also formed in response to UV and other genotoxins) Fast decline of the signal Variation of foci frequency between individuals |
Minutes to days | PBMC Fibroblasts |
| Genomic (e.g. mRNA, SNPs) | High-throughput analysis Linearly dose dependent to IR |
Bioinformatic challenge and high cost (RNA-Seq) | 1–3 days | PBMC WB Cell lines |
| Epigenomic (e.g. miRNA, lncRNA) | Relatively stable Potentially high-throughput analysis Cell- or tissue-type-specific expression |
Lack of data on specificity and sensitivity | Hours to days | Serum PBMC Cell lines |
WB, whole blood; PBMC, peripheral blood mononuclear cell; IR, ionizing radiation; UV, ultraviolet; ATM, ataxia-telangiectasia mutated; lncRNA, long non-coding RNA; miRNA, microRNA; p, phosphate group; p53, tumour protein p53; SNPs, single nucleotide polymorphisms.
Strategies for assessing individual radiation sensitivity using biomarkers and cellular models
The induction or suppression of DDR pathways are important determinants of how patients respond to radiation exposure.78 To maintain the benefits of cardiac medical imaging tests while minimizing the radiation risk, a better understanding of individual differences in radiation sensitivity and molecular events involved in cellular response to low-dose radiation is needed. The occurrence of individual variability in response to radiation sensitivity has been extensively reviewed in previously published reports.79,80 In recent years, cell-based and genetic studies have provided the molecular and genetic basis of cellular effects of radiation by identifying the genes and pathway involved.61,81,82 For example, genome-wide transcriptomic analysis of a small area of human tissue exposed in vivo to low-dose radiation yielded considerable individual variability of radiation response.83 Our recently published prospective cohort study of 63 patients undergoing SPECT MPI investigated the biological effects of low-dose radiation using proteomic and genomic biomarkers and found marked variation in individual response to low-dose radiation.39 However, individual radiosensitivity can arise from both genetic predisposition and/or other factors (e.g. diet, tobacco use, or prescribed medications). Controlling such confounding factors is difficult, compromising assessments of in vivo radiation responses between individuals.
Alternatively, patient-derived primary cells can serve as a predictor of individual variability in response to low-dose radiation, providing better control of the confounding factors via standardization of cell culture conditions. Peripheral blood lymphocytes have been commonly used for identifying biomarkers and studying individual response to low-dose radiation from cardiac medical imaging, because these primary cells are easily accessible and represent one of the most radiosensitive cells types in the body.39,62 However, such cells have limitations due to their low proliferation potential, and their use is further complicated by the fact that different cell types (e.g. proliferative vs. non-proliferative) within the same individual may show varying responses to low-dose radiation. Therefore, it is highly desirable to have an in vitro platform in which different cell types from the same individual can be exposed to the same in vitro low-dose radiation for measurement of cellular responses with sensitive biomarkers that focus on the DNA damage response, alterations in chromatin structure, gene expression, and proteomics.
In this context, human induced pluripotent stem cells (hiPSCs) are an attractive option as they are easily accessible and can be derived from fibroblasts or peripheral blood mononuclear cells. In fact, hiPSCs have greatly expanded the realm of possibilities for both basic research and potential clinical applications, including development of personalized cell-based assays and well-defined in vitro platforms utilizing specific types of cells derived from patients, which may help elucidate the molecular basis of diseases and lead to the discovery of clinically relevant biomarkers and potential therapeutic targets (Figure 2). For example, patient-specific iPSCs have been widely used as an in vitro platform for disease modelling, drug screening, drug discovery and toxicity assays, and precision medicine.84,85 These iPSCs are capable of differentiating into various cell types, including cardiomyocytes and endothelial cells, providing an effective system for studying individual variability in response to low-dose radiation across various cell types. By using genetic and molecular approaches, iPSCs-based and patient-specific platforms will allow us to identify potential candidate genes that may contribute to individual variation in response to radiation. It is important to note, however, that the iPSC-based platform is in itself limited by the lack of differentiation protocols into certain cell types, as well as challenges in manufacturing scale and long-term culture. While limitations remain that prevent the full application of iPSCs at the present, such as the absence of well-defined controls, genetic aberrations caused by reprogramming factors, and lack of large numbers of iPSC lines, these hurdles are expected to be overcome in the near future with the ongoing development of more effective reprogramming methods and creation of large iPSC biobanks worldwide.
Figure 2.
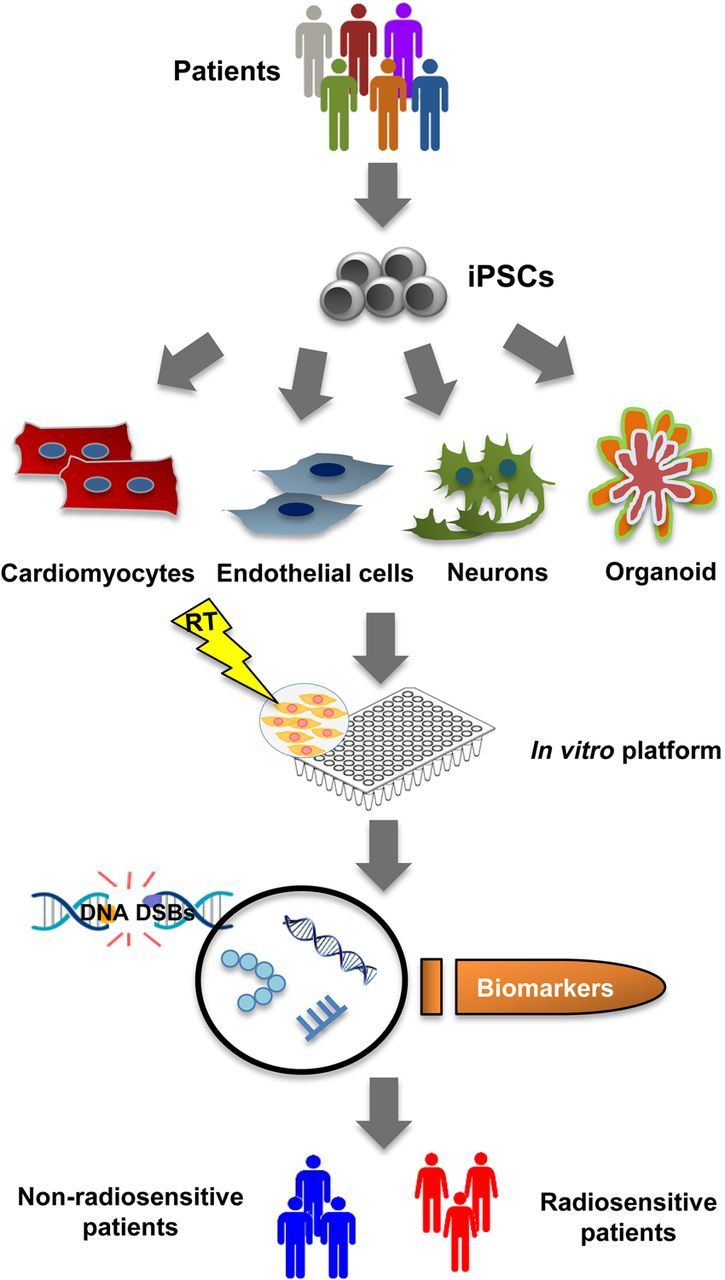
Schematic of a donor-specific cell-based platform for predicting individual radiosensitivity. Induced pluripotent stem cells obtained through the reprogramming of somatic cells from an individual can be differentiated into various cell types. Different cell types from the same individual can then be exposed to radiation for measurement of cellular responses in vitro using multiple biomarkers. iPSCs, induced pluripotent stem cells; RT, radiation; DNA DSBs, DNA double-strand breaks.
Recent advances in three-dimensional (3D) culture techniques that can independently manipulate genetics and microenvironmental factors also may be used as a platform to better understand the fundamental biological response to normal and disease processes, and to test novel therapeutic strategies, often using patient-derived cells or tissues.86 Although no approach is currently ready for routine clinical practice, 3D culture techniques may provide an integrative tool to generate individualized predictive or prognostic information for preclinical therapeutic testing, which is the ultimate objective of precision medicine and targeted therapy. There are a number of studies evaluating the effects of radiation in 3D culture models in terms of DNA damage and apoptosis. For example, treatment of organotypic slice cultures derived from human glioblastoma with the chemotherapeutic drug after irradiation-induced variable DNA damage and strongly affected proliferation and cell death rates, making this a unique model to explore susceptibility of individual tumours for specific therapies.87 In addition, foci formation of DNA damage marker proteins, such as p53-binding protein 1(53BP1), phosphorylated ATM, and γH2AX, was detected in a 3D tissue model after radiation, and foci diameter growth was shown to be correlated with chromatin remodelling to facilitate DNA repair.88 Explanting the living tissue or cell of a patient into a 3D culture model will require a high degree of standardization and reproducibility across experiments. There are still important requirements to be met for drug screening application, such as the ability to replicate complex heterogeneous cell mixtures from patients and the degree of adaptability using a high-throughput screening platform.
Conclusion
Identification of biomarkers capable of providing an accurate estimation of radiation risk caused by low-dose radiation and predicting individual radiation sensitivity may improve our understanding of the biology pertaining to low-dose radiation. Using a multi-parametric approach that includes mass spectrometry, second-generation sequencing, and high-throughput evaluation of single nucleotide polymorphisms (SNPs), we may be able to identify the underlying factors that modulate radiation sensitivity. The use of various ‘omics’ technologies together with the emergence of public data repositories may be highly useful to reduce study bias, increase statistical power, and improve overall biological understanding of underlying factors that modulate radiation sensitivity. However, care should be taken during horizontal data integration (frequently used in meta-analysis involving the combination and multi-faceted analysis of different data sets measuring the same molecular events) or vertical data integration (combining data collected at different levels in the ‘omics-cascade’) in the context of (i) data management due to the sheer size of raw data generated and (ii) the complexities of existing analytical approaches especially in dealing with high-throughput studies with high dimensionality but of relatively small sample size.89–92 Ultimately, development of cellular models that are donor-specific and obtainable non-invasively, along with use of a panel of multiple biomarkers, will provide crucial information elucidating the interplay of genes, proteins, and possible pathways responsible for individual responses to low-dose radiation. This information may provide us with a better understanding of how low-dose radiation affects living tissues so that we may develop novel strategies to minimize individual risk.
It is important to note that changes in these radiation biomarkers does not necessarily equate to increased cancer risk and interpretation of all findings using biomarkers should be limited to the cellular response to low-dose radiation-induced damage in the short-term. Measuring the potential risk of low-dose radiation-induced cancer is particularly difficult, because it is complicated by much higher potential risk of inherent risk of cancer and the omnipresent background radiation, making accurate estimates infeasible using any existing strategies.
Future directions
Despite these inherent limitations, radiation biomarkers can better inform us about the mechanisms modulating individual radiation risk, which can lead to the development and adherence to measures to minimize risk. In the past years, radiation dose reduction has been successfully achieved by several remarkable technical refinements. Future studies should focus on identifying highly sensitive cell injury biomarkers for very low-dose radiation (<3.0 mSv) and finding a rapid, practical, and quantifiable measure of biological response to low-dose radiation amiable to high-throughput population testing that can be used to rank individuals in their radiosensitivity. In addition, the expanded scale on the automated platform maintaining reduced variation will provide adequate statistical power to detect a modest effect of underlying traits of individual radiation sensitivity. Given the necessity of cardiovascular imaging, this information will be invaluable for clinicians and patients who rely on these tests to guide the diagnosis and management of complex cardiovascular disease.
Authors’ contributions
J.C.W. handled funding and supervision. J.C.W., W.H.L. acquired the data. J.C.W., W.H.L. drafted the manuscript. J.C.W., W.H.L., P.K.N., D.F. made critical revision of the manuscript for key intellectual content.
Acknowledgements
We thank Blake Wu for his assistance with manuscript preparation. Due to space limitation, we are unable to include all of the important papers relevant to radiation biomarkers and apologize to those investigators who have otherwise contributed significantly to this field.
Conflict of interest: none declared.
Funding
This work is supported by research grants from American Heart Association Beginning Grant in Aid 16BGIA27760016 (W.H.L.) and Burroughs Wellcome Foundation Innovation in Regulatory Science Award, NIH R24 HL117756, NIH R01 HL123968, and NIH R01 HL126527 (J.C.W.).
References
- 1. Chen J, Einstein AJ, Fazel R, Krumholz HM, Wang Y, Ross JS, Ting HH, Shah ND, Nasir K, Nallamothu BK. Cumulative exposure to ionizing radiation from diagnostic and therapeutic cardiac imaging procedures: a population-based analysis. J Am Coll Cardiol 2010;56:702–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schauer DA, Linton OW. NCRP Report No. 160, Ionizing Radiation Exposure of the Population of the United States, medical exposure – are we doing less with more, and is there a role for health physicists? Health Phys 2009;97:1–5. [DOI] [PubMed] [Google Scholar]
- 3. Fazel R, Gerber TC, Balter S, Brenner DJ, Carr JJ, Cerqueira MD, Chen J, Einstein AJ, Krumholz HM, Mahesh M, McCollough CH, Min JK, Morin RL, Nallamothu BK, Nasir K, Redberg RF, Shaw LJ. Approaches to enhancing radiation safety in cardiovascular imaging: a scientific statement from the American Heart Association. Circulation 2014;130:1730–1748. [DOI] [PubMed] [Google Scholar]
- 4. Lauer MS. Elements of danger – the case of medical imaging. N Engl J Med 2009;361:841–843. [DOI] [PubMed] [Google Scholar]
- 5. Smith-Bindman R, Lipson J, Marcus R, Kim KP, Mahesh M, Gould R, Berrington de Gonzalez A, Miglioretti DL. Radiation dose associated with common computed tomography examinations and the associated lifetime attributable risk of cancer. Arch Intern Med 2009;169:2078–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. The 2007 Recommendations of the International Commission on Radiological Protection. ICRP publication 103. Ann ICRP 2007;37:1–332. [DOI] [PubMed] [Google Scholar]
- 7. NRC. Health Risks from Exposure to Low Levels of Ionizing Radiation: BEIR VII, Phase II. Washington, DC, USA: National Academic Press, 1999. [PubMed] [Google Scholar]
- 8. Brenner DJ, Doll R, Goodhead DT, Hall EJ, Land CE, Little JB, Lubin JH, Preston DL, Preston RJ, Puskin JS, Ron E, Sachs RK, Samet JM, Setlow RB, Zaider M. Cancer risks attributable to low doses of ionizing radiation: assessing what we really know. Proc Natl Acad Sci USA 2003;100:13761–13766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gerber TC, Carr JJ, Arai AE, Dixon RL, Ferrari VA, Gomes AS, Heller GV, McCollough CH, McNitt-Gray MF, Mettler FA, Mieres JH, Morin RL, Yester MV. Ionizing radiation in cardiac imaging: a science advisory from the American Heart Association Committee on Cardiac Imaging of the Council on Clinical Cardiology and Committee on Cardiovascular Imaging and Intervention of the Council on Cardiovascular Radiology and Intervention. Circulation 2009;119:1056–1065. [DOI] [PubMed] [Google Scholar]
- 10. Lin EC. Radiation risk from medical imaging. Mayo Clinic Proc 2010;85:1142–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. American College of Cardiology Foundation Task Force on Expert Consensus D Mark DB, Berman DS, Budoff MJ, Carr JJ, Gerber TC, Hecht HS, Hlatky MA, Hodgson JM, Lauer MS, Miller JM, Morin RL, Mukherjee D, Poon M, Rubin GD, Schwartz RS. ACCF/ACR/AHA/NASCI/SAIP/SCAI/SCCT 2010 expert consensus document on coronary computed tomographic angiography: a report of the American College of Cardiology Foundation Task Force on Expert Consensus Documents. J Am Coll Cardiol 2010;55:2663–2699. [DOI] [PubMed] [Google Scholar]
- 12. Dey D, Slomka PJ, Berman DS. Achieving very-low-dose radiation exposure in cardiac computed tomography, single-photon emission computed tomography, and positron emission tomography. Circ Cardiovasc Imaging 2014;7:723–734. [DOI] [PubMed] [Google Scholar]
- 13. Hausleiter J, Meyer T, Hermann F, Hadamitzky M, Krebs M, Gerber TC, McCollough C, Martinoff S, Kastrati A, Schomig A, Achenbach S. Estimated radiation dose associated with cardiac CT angiography. J Am Med Assoc 2009;301:500–507. [DOI] [PubMed] [Google Scholar]
- 14. Stehli J, Fuchs TA, Bull S, Clerc OF, Possner M, Buechel RR, Gaemperli O, Kaufmann PA. Accuracy of coronary CT angiography using a submillisievert fraction of radiation exposure: comparison with invasive coronary angiography. J Am Coll Cardiol 2014;64:772–780. [DOI] [PubMed] [Google Scholar]
- 15. Einstein AJ, Pascual TN, Mercuri M, Karthikeyan G, Vitola JV, Mahmarian JJ, Better N, Bouyoucef SE, Hee-Seung Bom H, Lele V, Magboo VP, Alexanderson E, Allam AH, Al-Mallah MH, Flotats A, Jerome S, Kaufmann PA, Luxenburg O, Shaw LJ, Underwood SR, Rehani MM, Kashyap R, Paez D, Dondi M, Group II. Current worldwide nuclear cardiology practices and radiation exposure: results from the 65 country IAEA Nuclear Cardiology Protocols Cross-Sectional Study (INCAPS). Eur Heart J 2015;36:1689–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pierce DA, Shimizu Y, Preston DL, Vaeth M, Mabuchi K. Studies of the mortality of atomic bomb survivors. Report 12, Part I. Cancer: 1950-1990. Radiat Res 1996;146:1–27. [PubMed] [Google Scholar]
- 17. Einstein AJ, Weiner SD, Bernheim A, Kulon M, Bokhari S, Johnson LL, Moses JW, Balter S. Multiple testing, cumulative radiation dose, and clinical indications in patients undergoing myocardial perfusion imaging. J Am Med Assoc 2010;304:2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Martin CJ. Effective dose: how should it be applied to medical exposures? Br J Radiol 2007;80:639–647. [DOI] [PubMed] [Google Scholar]
- 19. Geleijns J, Joemai RM, Cros M, Hernandez-Giron I, Calzado A, Dewey M, Salvado M. A Monte Carlo simulation for the estimation of patient dose in rest and stress cardiac computed tomography with a 320-detector row CT scanner. Phys Med 2015;31:1029–1034. [DOI] [PubMed] [Google Scholar]
- 20. Preston DL, Ron E, Tokuoka S, Funamoto S, Nishi N, Soda M, Mabuchi K, Kodama K. Solid cancer incidence in atomic bomb survivors: 1958-1998. Radiat Res 2007;168:1–64. [DOI] [PubMed] [Google Scholar]
- 21. Preston DL, Pierce DA, Shimizu Y, Cullings HM, Fujita S, Funamoto S, Kodama K. Effect of recent changes in atomic bomb survivor dosimetry on cancer mortality risk estimates. Radiat Res 2004;162:377–389. [DOI] [PubMed] [Google Scholar]
- 22. Tubiana M, Feinendegen LE, Yang C, Kaminski JM. The linear no-threshold relationship is inconsistent with radiation biologic and experimental data. Radiology 2009;251:13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Brenner DJ. Extrapolating radiation-induced cancer risks from low doses to very low doses. Health Phys 2009;97:505–509. [DOI] [PubMed] [Google Scholar]
- 24. Pierce DA, Preston DL. Radiation-related cancer risks at low doses among atomic bomb survivors. Radiat Res 2000;154:178–186. [DOI] [PubMed] [Google Scholar]
- 25. Cardis E, Vrijheid M, Blettner M, Gilbert E, Hakama M, Hill C, Howe G, Kaldor J, Muirhead CR, Schubauer-Berigan M, Yoshimura T, Bermann F, Cowper G, Fix J, Hacker C, Heinmiller B, Marshall M, Thierry-Chef I, Utterback D, Ahn YO, Amoros E, Ashmore P, Auvinen A, Bae JM, Bernar J, Biau A, Combalot E, Deboodt P, Diez Sacristan A, Eklof M, Engels H, Engholm G, Gulis G, Habib RR, Holan K, Hyvonen H, Kerekes A, Kurtinaitis J, Malker H, Martuzzi M, Mastauskas A, Monnet A, Moser M, Pearce MS, Richardson DB, Rodriguez-Artalejo F, Rogel A, Tardy H, Telle-Lamberton M, Turai I, Usel M, Veress K. The 15-country collaborative study of cancer risk among radiation workers in the nuclear industry: estimates of radiation-related cancer risks. Radiat Res 2007;167:396–416. [DOI] [PubMed] [Google Scholar]
- 26. Amundson SA, Do KT, Vinikoor LC, Lee RA, Koch-Paiz CA, Ahn J, Reimers M, Chen Y, Scudiero DA, Weinstein JN, Trent JM, Bittner ML, Meltzer PS, Fornace AJ Jr. Integrating global gene expression and radiation survival parameters across the 60 cell lines of the National Cancer Institute Anticancer Drug Screen. Cancer Res 2008;68:415–424. [DOI] [PubMed] [Google Scholar]
- 27. Suzuki K, Okada H, Yamauchi M, Oka Y, Kodama S, Watanabe M. Qualitative and quantitative analysis of phosphorylated ATM foci induced by low-dose ionizing radiation. Radiat Res 2006;165:499–504. [DOI] [PubMed] [Google Scholar]
- 28. Nguyen PK, Lee WH, Li YF, Hong WX, Hu S, Chan C, Liang G, Nguyen I, Ong SG, Churko J, Wang J, Altman RB, Fleischmann D, Wu JC. Assessment of the radiation effects of cardiac CT angiography using protein and genetic biomarkers. JACC Cardiovasc Imaging 2015;8:873–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Averbeck D. Does scientific evidence support a change from the LNT model for low-dose radiation risk extrapolation? Health Phys 2009;97:493–504. [DOI] [PubMed] [Google Scholar]
- 30. Tubiana M, Aurengo A, Averbeck D, Masse R. Recent reports on the effect of low doses of ionizing radiation and its dose-effect relationship. Radiat Environ Biophys 2006;44:245–251. [DOI] [PubMed] [Google Scholar]
- 31. Gulston M, de Lara C, Jenner T, Davis E, O'Neill P. Processing of clustered DNA damage generates additional double-strand breaks in mammalian cells post-irradiation. Nucleic Acids Res 2004;32:1602–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mitelman F. Recurrent chromosome aberrations in cancer. Mutat Res 2000;462:247–253. [DOI] [PubMed] [Google Scholar]
- 33. M'Kacher R, Violot D, Aubert B, Girinsky T, Dossou J, Beron-Gaillard N, Carde P, Parmentier C. Premature chromosome condensation associated with fluorescence in situ hybridisation detects cytogenetic abnormalities after a CT scan: evaluation of the low-dose effect. Radiat Prot Dosimetry 2003;103:35–40. [DOI] [PubMed] [Google Scholar]
- 34. Stephan G, Schneider K, Panzer W, Walsh L, Oestreicher U. Enhanced yield of chromosome aberrations after CT examinations in paediatric patients. Int J Radiat Biol 2007;83:281–287. [DOI] [PubMed] [Google Scholar]
- 35. Bauchinger M. Quantification of low-level radiation exposure by conventional chromosome aberration analysis. Mutat Res 1995;339:177–189. [DOI] [PubMed] [Google Scholar]
- 36. Rana S, Kumar R, Sultana S, Sharma RK. Radiation-induced biomarkers for the detection and assessment of absorbed radiation doses. J Pharm Bioallied Sci 2010;2:189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beels L, Bacher K, De Wolf D, Werbrouck J, Thierens H. Gamma-H2AX foci as a biomarker for patient X-ray exposure in pediatric cardiac catheterization: are we underestimating radiation risks? Circulation 2009;120:1903–1909. [DOI] [PubMed] [Google Scholar]
- 38. Rothkamm K, Balroop S, Shekhdar J, Fernie P, Goh V. Leukocyte DNA damage after multi-detector row CT: a quantitative biomarker of low-level radiation exposure. Radiology 2007;242:244–251. [DOI] [PubMed] [Google Scholar]
- 39. Lee WH, Nguyen P, Hu S, Liang G, Ong SG, Han L, Sanchez-Freire V, Lee AS, Vasanawala M, Segall G, Wu JC. Variable activation of the DNA damage response pathways in patients undergoing single-photon emission computed tomography myocardial perfusion imaging. Circ Cardiovasc Imaging 2015;8:e002851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lobrich M, Rief N, Kuhne M, Heckmann M, Fleckenstein J, Rube C, Uder M. In vivo formation and repair of DNA double-strand breaks after computed tomography examinations. Proc Natl Acad Sci USA 2005;102:8984–8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Golfier S, Jost G, Pietsch H, Lengsfeld P, Eckardt-Schupp F, Schmid E, Voth M. Dicentric chromosomes and gamma-H2AX foci formation in lymphocytes of human blood samples exposed to a CT scanner: a direct comparison of dose response relationships. Radiat Prot Dosimetry 2009;134:55–61. [DOI] [PubMed] [Google Scholar]
- 42. Lassmann M, Hanscheid H, Gassen D, Biko J, Meineke V, Reiners C, Scherthan H. In vivo formation of gamma-H2AX and 53BP1 DNA repair foci in blood cells after radioiodine therapy of differentiated thyroid cancer. J Nucl Med 2010;51:1318–1325. [DOI] [PubMed] [Google Scholar]
- 43. Kuefner MA, Grudzenski S, Schwab SA, Wiederseiner M, Heckmann M, Bautz W, Lobrich M, Uder M. DNA double-strand breaks and their repair in blood lymphocytes of patients undergoing angiographic procedures. Invest Radiol 2009;44:440–446. [DOI] [PubMed] [Google Scholar]
- 44. Lobrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, Barton O, Jeggo PA. GammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell Cycle 2010;9:662–669. [DOI] [PubMed] [Google Scholar]
- 45. Matsuzaki K, Harada A, Takeiri A, Tanaka K, Mishima M. Whole cell-ELISA to measure the gammaH2AX response of six aneugens and eight DNA-damaging chemicals. Mutat Res 2010;700:71–79. [DOI] [PubMed] [Google Scholar]
- 46. Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 2003;421:961–966. [DOI] [PubMed] [Google Scholar]
- 47. Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proc Natl Acad Sci USA 2003;100:5057–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Grudzenski S, Raths A, Conrad S, Rube CE, Lobrich M. Inducible response required for repair of low-dose radiation damage in human fibroblasts. Proc Natl Acad Sci USA 2010;107:14205–14210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 2007;26:7741–7748. [DOI] [PubMed] [Google Scholar]
- 50. Osipov AN, Pustovalova M, Grekhova A, Eremin P, Vorobyova N, Pulin A, Zhavoronkov A, Roumiantsev S, Klokov DY, Eremin I. Low doses of X-rays induce prolonged and ATM-independent persistence of gammaH2AX foci in human gingival mesenchymal stem cells. Oncotarget 2015;6:27275–27287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Limoli CL, Giedzinski E, Bonner WM, Cleaver JE. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma-H2AX formation, and Mre11 relocalization. Proc Natl Acad Sci USA 2002;99:233–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Momcilovic O, Knobloch L, Fornsaglio J, Varum S, Easley C, Schatten G. DNA damage responses in human induced pluripotent stem cells and embryonic stem cells. PloS ONE 2010;5:e13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ainsbury EA, Bakhanova E, Barquinero JF, Brai M, Chumak V, Correcher V, Darroudi F, Fattibene P, Gruel G, Guclu I, Horn S, Jaworska A, Kulka U, Lindholm C, Lloyd D, Longo A, Marrale M, Monteiro Gil O, Oestreicher U, Pajic J, Rakic B, Romm H, Trompier F, Veronese I, Voisin P, Vral A, Whitehouse CA, Wieser A, Woda C, Wojcik A, Rothkamm K. Review of retrospective dosimetry techniques for external ionising radiation exposures. Radiat Prot Dosimetry 2011;147:573–592. [DOI] [PubMed] [Google Scholar]
- 54. Brand M, Sommer M, Achenbach S, Anders K, Lell M, Lobrich M, Uder M, Kuefner MA. X-ray induced DNA double-strand breaks in coronary CT angiography: comparison of sequential, low-pitch helical and high-pitch helical data acquisition. Eur J Radiol 2012;81:e357–e362. [DOI] [PubMed] [Google Scholar]
- 55. Ivashkevich AN, Martin OA, Smith AJ, Redon CE, Bonner WM, Martin RF, Lobachevsky PN. gammaH2AX foci as a measure of DNA damage: a computational approach to automatic analysis. Mutat Res 2011;711:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Qvarnstrom OF, Simonsson M, Johansson KA, Nyman J, Turesson I. DNA double strand break quantification in skin biopsies. Radiother Oncol 2004;72:311–317. [DOI] [PubMed] [Google Scholar]
- 57. Roch-Lefevre S, Mandina T, Voisin P, Gaetan G, Mesa JE, Valente M, Bonnesoeur P, Garcia O, Voisin P, Roy L. Quantification of gamma-H2AX foci in human lymphocytes: a method for biological dosimetry after ionizing radiation exposure. Radiat Res 2010;174:185–194. [DOI] [PubMed] [Google Scholar]
- 58. Garty G, Chen Y, Turner HC, Zhang J, Lyulko OV, Bertucci A, Xu Y, Wang H, Simaan N, Randers-Pehrson G, Lawrence Yao Y, Brenner DJ. The RABiT: a rapid automated biodosimetry tool for radiological triage. II. Technological developments. Int J Radiat Biol 2011;87:776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Svensson JP, Stalpers LJ, Esveldt-van Lange RE, Franken NA, Haveman J, Klein B, Turesson I, Vrieling H, Giphart-Gassler M. Analysis of gene expression using gene sets discriminates cancer patients with and without late radiation toxicity. PLoS Med 2006;3:e422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Amundson SA. Functional genomics in radiation biology: a gateway to cellular systems-level studies. Radiat Environ Biophys 2008;47:25–31. [DOI] [PubMed] [Google Scholar]
- 61. Amundson SA, Lee RA, Koch-Paiz CA, Bittner ML, Meltzer P, Trent JM, Fornace AJ Jr. Differential responses of stress genes to low dose-rate gamma irradiation. Mol Cancer Res 2003;1:445–452. [PubMed] [Google Scholar]
- 62. Nosel I, Vaurijoux A, Barquinero JF, Gruel G. Characterization of gene expression profiles at low and very low doses of ionizing radiation. DNA Repair (Amst) 2013;12:508–517. [DOI] [PubMed] [Google Scholar]
- 63. Cui W, Ma J, Wang Y, Biswal S. Plasma miRNA as biomarkers for assessment of total-body radiation exposure dosimetry. PloS ONE 2011;6:e22988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Templin T, Amundson SA, Brenner DJ, Smilenov LB. Whole mouse blood microRNA as biomarkers for exposure to gamma-rays and 56Fe ion. Int J Radiat Biol 2011;87:653–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wouters MD, van Gent DC, Hoeijmakers JH, Pothof J. MicroRNAs, the DNA damage response and cancer. Mutat Res 2011;717:54–66. [DOI] [PubMed] [Google Scholar]
- 66. Halimi M, Parsian H, Asghari SM, Sariri R, Moslemi D, Yeganeh F, Zabihi E. Clinical translation of human microRNA 21 as a potential biomarker for exposure to ionizing radiation. Transl Res 2014;163:578–584. [DOI] [PubMed] [Google Scholar]
- 67. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 2008;105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Acharya SS, Fendler W, Watson J, Hamilton A, Pan Y, Gaudiano E, Moskwa P, Bhanja P, Saha S, Guha C, Parmar K, Chowdhury D. Serum microRNAs are early indicators of survival after radiation-induced hematopoietic injury. Sci Transl Med 2015;7:287ra69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Aypar U, Morgan WF, Baulch JE. Radiation-induced genomic instability: are epigenetic mechanisms the missing link? Int J Radiat Biol 2011;87:179–191. [DOI] [PubMed] [Google Scholar]
- 70. Ma S, Liu X, Jiao B, Yang Y, Liu X. Low-dose radiation-induced responses: focusing on epigenetic regulation. Int J Radiat Biol 2010;86:517–528. [DOI] [PubMed] [Google Scholar]
- 71. Jacob NK, Cooley JV, Yee TN, Jacob J, Alder H, Wickramasinghe P, Maclean KH, Chakravarti A. Identification of sensitive serum microRNA biomarkers for radiation biodosimetry. PLoS ONE 2013;8:e57603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature 2007;447:1130–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Krishnan K, Steptoe AL, Martin HC, Wani S, Nones K, Waddell N, Mariasegaram M, Simpson PT, Lakhani SR, Gabrielli B, Vlassov A, Cloonan N, Grimmond SM. MicroRNA-182-5p targets a network of genes involved in DNA repair. RNA 2013;19:230–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Cha HJ, Seong KM, Bae S, Jung JH, Kim CS, Yang KH, Jin YW, An S. Identification of specific microRNAs responding to low and high dose gamma-irradiation in the human lymphoblast line IM9. Oncol Rep 2009;22:863–868. [PubMed] [Google Scholar]
- 75. Nie J, Peng C, Pei W, Zhu W, Zhang S, Cao H, Qi X, Tong J, Jiao Y. A novel role of long non-coding RNAs in response to X-ray irradiation. Toxicol In Vitro 2015;30(1 Pt B):536–544. [DOI] [PubMed] [Google Scholar]
- 76. Kabacik S, Manning G, Raffy C, Bouffler S, Badie C. Time, dose and ataxia telangiectasia mutated (ATM) status dependency of coding and noncoding RNA expression after ionizing radiation exposure. Radiat Res 2015;183:325–337. [DOI] [PubMed] [Google Scholar]
- 77. Jiao Y, Liu C, Cui FM, Xu JY, Tong J, Qi XF, Wang LL, Zhu W. Long intergenic non-coding RNA induced by X-ray irradiation regulates DNA damage response signaling in the human bronchial epithelial BEAS-2B cell line. Oncol Lett 2015;9:169–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Barnett GC, West CM, Dunning AM, Elliott RM, Coles CE, Pharoah PD, Burnet NG. Normal tissue reactions to radiotherapy: towards tailoring treatment dose by genotype. Nat Rev Cancer 2009;9:134–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kato TA, Wilson PF, Nagasaw H, Peng Y, Weil MM, Little JB, Bedford JS. Variations in radiosensitivity among individuals: a potential impact on risk assessment? Health Phys 2009;97:470–480. [DOI] [PubMed] [Google Scholar]
- 80. Greve B, Bolling T, Amler S, Rossler U, Gomolka M, Mayer C, Popanda O, Dreffke K, Rickinger A, Fritz E, Eckardt-Schupp F, Sauerland C, Braselmann H, Sauter W, Illig T, Riesenbeck D, Konemann S, Willich N, Mortl S, Eich HT, Schmezer P. Evaluation of different biomarkers to predict individual radiosensitivity in an inter-laboratory comparison – lessons for future studies. PLoS ONE 2012;7:e47185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Smirnov DA, Brady L, Halasa K, Morley M, Solomon S, Cheung VG. Genetic variation in radiation-induced cell death. Genome Res 2012;22:332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG. Genetic analysis of radiation-induced changes in human gene expression. Nature 2009;459:587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Goldberg Z, Rocke DM, Schwietert C, Berglund SR, Santana A, Jones A, Lehmann J, Stern R, Lu R, Hartmann Siantar C. Human in vivo dose-response to controlled, low-dose low linear energy transfer ionizing radiation exposure. Clin Cancer Res 2006;12:3723–3729. [DOI] [PubMed] [Google Scholar]
- 84. Liang P, Lan F, Lee AS, Gong T, Sanchez-Freire V, Wang Y, Diecke S, Sallam K, Knowles JW, Wang PJ, Nguyen PK, Bers DM, Robbins RC, Wu JC. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation 2013;127:1677–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Navarrete EG, Liang P, Lan F, Sanchez-Freire V, Simmons C, Gong T, Sharma A, Burridge PW, Patlolla B, Lee AS, Wu H, Beygui RE, Wu SM, Robbins RC, Bers DM, Wu JC. Screening drug-induced arrhythmia [corrected] using human induced pluripotent stem cell-derived cardiomyocytes and low-impedance microelectrode arrays. Circulation 2013;128(11 Suppl. 1):S3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shamir ER, Ewald AJ. Three-dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol 2014;15:647–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Merz F, Gaunitz F, Dehghani F, Renner C, Meixensberger J, Gutenberg A, Giese A, Schopow K, Hellwig C, Schafer M, Bauer M, Stocker H, Taucher-Scholz G, Durante M, Bechmann I. Organotypic slice cultures of human glioblastoma reveal different susceptibilities to treatments. Neuro Oncol 2013;15:670–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Suzuki K, Nakashima M, Yamashita S. Dynamics of ionizing radiation-induced DNA damage response in reconstituted three-dimensional human skin tissue. Radiat Res 2010;174:415–423. [DOI] [PubMed] [Google Scholar]
- 89. Xia J, Fjell CD, Mayer ML, Pena OM, Wishart DS, Hancock RE. INMEX--a web-based tool for integrative meta-analysis of expression data. Nucleic Acids Res 2013;41:W63–W70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Tseng GC, Ghosh D, Feingold E. Comprehensive literature review and statistical considerations for microarray meta-analysis. Nucleic Acids Res 2012;40:3785–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kim S, Herazo-Maya JD, Kang DD, Juan-Guardela BM, Tedrow J, Martinez FJ, Sciurba FC, Tseng GC, Kaminski N. Integrative phenotyping framework (iPF): integrative clustering of multiple omics data identifies novel lung disease subphenotypes. BMC Genomics 2015;16:924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ghosh D, Poisson LM. “Omics” data and levels of evidence for biomarker discovery. Genomics 2009;93:13–16. [DOI] [PubMed] [Google Scholar]
- 93. May MS, Brand M, Wuest W, Anders K, Kuwert T, Prante O, Schmidt D, Maschauer S, Semelka RC, Uder M, Kuefner MA. Induction and repair of DNA double-strand breaks in blood lymphocytes of patients undergoing 18F-FDG PET/CT examinations. Eur J Nucl Med Mol Imaging 2012;39:1712–1719. [DOI] [PubMed] [Google Scholar]
- 94. Grudzenski S, Kuefner MA, Heckmann MB, Uder M, Lobrich M. Contrast medium-enhanced radiation damage caused by CT examinations. Radiology 2009;253:706–714. [DOI] [PubMed] [Google Scholar]
- 95. Kuefner MA, Grudzenski S, Hamann J, Achenbach S, Lell M, Anders K, Schwab SA, Haberle L, Lobrich M, Uder M. Effect of CT scan protocols on x-ray-induced DNA double-strand breaks in blood lymphocytes of patients undergoing coronary CT angiography. Eur Radiol 2010;20:2917–2924. [DOI] [PubMed] [Google Scholar]
- 96. Andreassi MG, Cioppa A, Manfredi S, Palmieri C, Botto N, Picano E. Acute chromosomal DNA damage in human lymphocytes after radiation exposure in invasive cardiovascular procedures. Eur Heart J 2007;28:2195–2199. [DOI] [PubMed] [Google Scholar]