Abstract
Saliva contains many proteins that have an important role in biological process of the oral cavity and is closely associated with many diseases. Although the dog is a common companion animal, the composition of salivary proteome and its relationship with that of human are unclear. In this study, shotgun proteomics was used to compare the salivary proteomes of 7 Thai village dogs and 7 human subjects. Salivary proteomes revealed 2,532 differentially expressed proteins in dogs and humans, representing various functions including cellular component organization or biogenesis, cellular process, localization, biological regulation, response to stimulus, developmental process, multicellular organismal process, metabolic process, immune system process, apoptosis and biological adhesion. The oral proteomes of dogs and humans were appreciably different. Proteins related to apoptosis processes and biological adhesion were predominated in dog saliva. Drug-target network predictions by STITCH Version 5.0 showed that dog salivary proteins were found to have potential roles in tumorigenesis, anti-inflammation and antimicrobial processes. In addition, proteins related to regeneration and healing processes such as fibroblast growth factor and epidermal growth factor were also up-regulated in dogs. These findings provide new information on dog saliva composition and will be beneficial for the study of dog saliva in diseased and health conditions in the future.
Introduction
Saliva is an important fluid that maintains homeostasis in the oral cavity. It contains many kinds of proteins and peptides including immunoglobulins, enzymes and cytokines [1]. Saliva has numerous functions such as moistening food and bolus formation, lubrication of the oral mucosa, maintaining the mineralization of teeth, tissue defense and buffering system of the oral cavity [2]. Human saliva has been well studied and in terms of human medicine has been employed in diagnostic tests for oral diseases, cancer, and systemic diseases, since saliva constituents provide information on health status [3,4]. In the veterinary field, a variety of techniques such as immunofluorometric assay, enzyme-linked immunosorbent assay and radioimmunoassay, have been utilized in developing salivary protein detection [5–8].
Dogs are a major reservoir for zoonotic infections. The resident pathogenic oral bacteria or viruses can be transmitted to humans mainly by infected saliva. Therefore, saliva becomes more important for public health considerations. However, the composition of the dog salivary proteome, which may also be associated with human pathogenic organisms, and its relationship with that of their owners remain unclear. Alteration of dog or human saliva protein composition by age, food consumption, environmental changes, and health condition may increase the risk of dog-associated zoonotic infection. Major proteins identified by a proteomics approach in dog saliva were involved in metabolism, however, major proteins in human saliva were cytoskeletal and inflammation-related [9]. Dog saliva has a more basic pH and higher buffering capacity than human saliva, and has different electrolyte composition in calcium, potassium and sodium [10]. The aim of this study was to identify proteins present in dog or human salivary samples utilizing shotgun proteomics. Better understanding of salivary functions as a result of proteome information will further studies of pathophysiological mechanisms of diseases in dogs.
Materials and methods
Dogs
Saliva was obtained from 7 healthy Thai village dogs (1–3 years old) at the Veterinary Teaching Hospital of Mahidol University, Thailand. The study was approved by the Faculty of Veterinary Science-Animal Care and Use Committee (FVS-ACUC) (Protocol No. MUVS-2015-19). Written informed consent forms were obtained from all dog owners. Health status of the dogs was determined by veterinarians. The dogs were included in this study according to the following criteria: none of the dogs had received antibiotics within the 3-month period before sample collection, no signs of oral diseases (clinically healthy, probing depth < 3 mm and no gingival inflammation) or systemic diseases. Dog saliva was allowed to drip from the mouth into a collecting vessel, or was collected using a syringe at the buccal area from healthy dogs under anesthesia.
Human subjects
Seven subjects were recruited from the Dental Hospital, Faculty of Dentistry, Mahidol University, Thailand. Whole, unstimulated saliva was collected following informed consent from healthy volunteers by Navazesh’s method [11] between 07:00 and 10:00. All subjects showed no sign of periodontitis (probing depth < 3 mm and no attachment loss). The study protocol was approved by the Ethics Committee of the Faculty of Dentistry/Faculty of Pharmacy, Mahidol University Institutional Review Board (COA.No.MU-DT/PY-IRB 2011/012.3103). Written informed consent forms were obtained from all subjects.
Saliva preparation
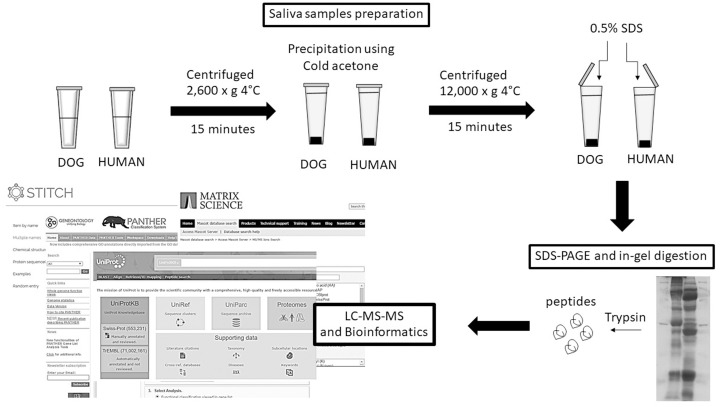
Protease inhibitor cocktail (Roche, Mannheim, Germany) was added to the saliva samples immediately after collection and they were stored at -80 °C until use. Saliva was centrifuged at 2,600 × g at 4 °C for 15 min and the supernatant was collected. Protein concentrations of samples were estimated using Bradford Protein assay [12]. Portions of saliva containing 10 μg protein from each dog were pooled and proteins were precipitated with 3 volumes of ice-cold acetone at -20 °C for 16 h. The precipitant was collected by centrifugation at 12,000 × g at 4 °C for 15 min. The supernatant was discarded and protein pellet was allowed to air dry. Human salivary proteins were prepared using same procedure. The workflow of shotgun proteomics analysis is shown in Fig 1.
Fig 1. Workflow of shotgun proteomics analysis.
Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) for shotgun proteomics
Protein pellets from dog or human samples were suspended in 0.5% SDS and heated at 60 °C for 30 min. Samples containing 50 μg protein were mixed with SDS-sample buffer containing 0.5 M dithiothreitol (DTT), 10% w/v SDS, 0.4 M Tris-HCl pH 6.8 and 50% v/v glycerol and proteins were separated by SDS-PAGE (12.5% acrylamide gel) at 90 volts. Proteins were stained with Coomassie Brilliant Blue R-250.
Protein identification by LC-MS/MS
Protein bands on SDS-PAGE gels were excised manually according to molecular size using a scalpel and subjected to in-gel digestion followed by High-performance liquid chromatography-Tandem mass spectrometry (LC/MS/MS) using an in-house method as described previously [13]. Briefly, gel plugs were dehydrated with 100% acetonitrile (ACN), reduced with 10 mM DTT and alkylated in 100 mM IAA solution. After that, proteins in gel plugs were digested by trypsin at 37 °C overnight. The peptides were extracted by 50% ACN in 0.1% trifluoroacetic acid followed by drying at 40 °C overnight. The peptides were analyzed by nanoLC-MS/MS (nanoACQUITY UPLC/ SYNAPT HDMS, Waters, Milford, USA) coupled to mass spectrometry (Electrospray ionization/Quadrupole-Time of flight (ESIQ-TOF), Waters, Milford, USA). Finally, the raw MS/MS data were subjected to DecyderMS [14,15] for quantitation, and the analyzed MS/MS data were sent to identify by Mascot MS/MS Ions Search [16] using NCBI protein database. Gene ontology annotations included biological process, cellular component and molecular function were performed using Panther database [17]. Protein-chemical interactions were analyzed according to STITCH 5.0 database [18].
Immunoblotting analysis
Immunoblotting analysis were performed using 10 μg protein from each group. Samples were separated by 7.5% or 10% SDS-PAGE and transferred to nitrocellulose membranes and subsequently probed with antibodies. Antibodies to Amylase, Baculoviral IAP repeat-containing protein 2 (BIRC2) and Carbonic anhydrase 6 were from Abcam, Cambridge, MA, USA., MyBioSource, Inc., San Diego, CA, USA. and Santa Cruz Biotechnology, Inc., USA. Immunoreactive protein bands were visualized using an ECL detection reagent (PerkinElmer, Inc., Waltham, MA, USA).
Statistical analysis
Data were analyzed using an Analysis of Variance (ANOVA) to determine differences between groups at a significance level of 95%.
Results
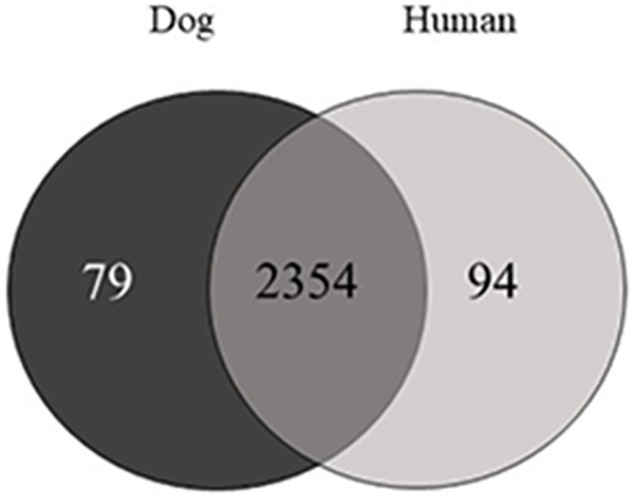
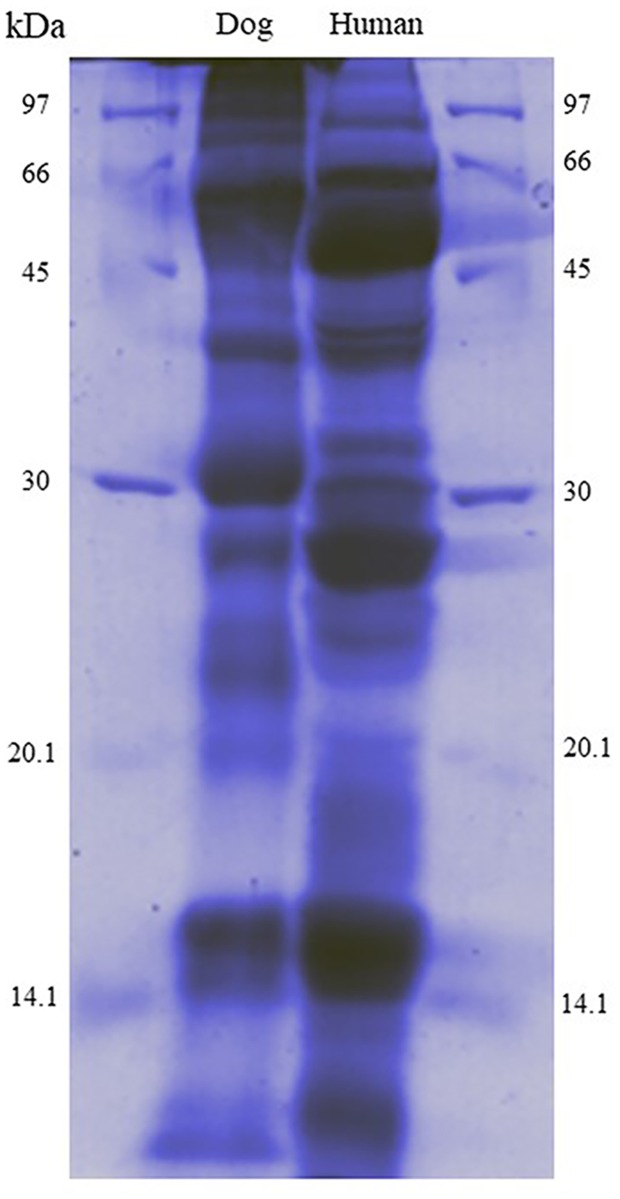
A total of 2,532 proteins (S1, S2 and S3 Tables) showed significantly different expression in dogs when compared to humans, with 79 proteins being found only in dog saliva (Fig 2). The salivary protein profiles from SDS-PAGE showed different banding patterns between dog and human saliva (Fig 3). Predominant bands at high molecular weight were observed in dog saliva.
Fig 2. Venn diagram identifying the overlapped and unique proteins of dog and human saliva.
Fig 3. Representative SDS-PAGE of 50 μg protein from dog saliva pool and human saliva pool.
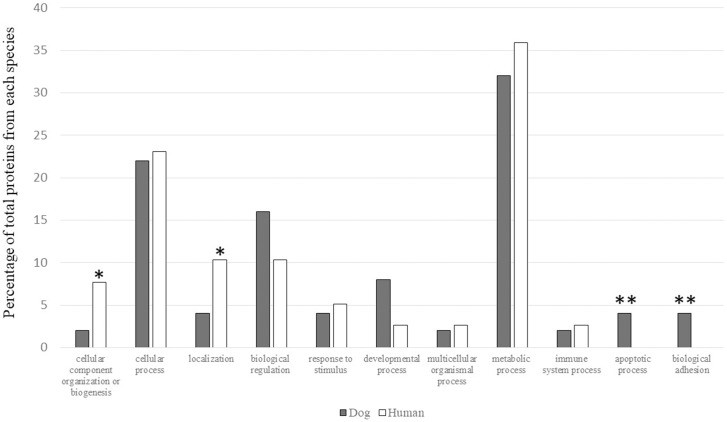
According to the functional annotation of salivary proteins using Gene Ontology Annotation Database, the identified proteins were classified as cellular component organization or biogenesis, cellular process, localization, biological regulation, response to stimulus, developmental process, multicellular organismal process, metabolic process, immune system process, apoptosis and biological adhesion as shown in Fig 4.
Fig 4. Comparison of functional characteristic of proteins found only in dogs and human.
*: Dominant functions found in human. **: Dominant functions found only in dogs.
The most abundant proteins found only in dog saliva were proteins with metabolic process (e.g. mitochondrial 10-formyltetrahydrofolate dehydrogenase precursor) (32%) followed by proteins with cellular process (e.g. 60S ribosomal protein L13) (22%) and biological regulation (e.g. histone H2A type 2-A) (16%). Unique proteins with dominant functions found in each species were presented in Table 1.
Table 1. Unique proteins from dominant functions found only in dog or human.
| Dogs: Apoptotic process and Biological adhesion | Human: Cellular component organization or biogenesis and Localization | ||
|---|---|---|---|
| Protein name | Known functions | Protein name | Known functions |
| Baculoviral IAP repeat-containing protein 2 (BIRC2) | Apoptosis inhibitor and inflammatory signaling and immunity, cell proliferation, cell invasion and metastasis [19,20] | 5-methylcytosine rRNA methyltransferase NSUN4 isoform a (NSUN4) | Cell proliferation and differentiation, protein biosynthesis and cancer [29] |
| Dexamethasone-induced Ras-related protein 1 isoform X1 (RASD1) | Intracellular signaling process related to cell morphology, cell growth and cell-extracellular matrix interaction in variety of cell types including heart, liver, brain and immune cells [21–24] | Actin-related protein 8 isoform X1 (ACTR8) | Processes of vesicle motility, mitosis, actin filament dynamics, and modulation of chromatin structure [30] |
| E3 ubiquitin-protein ligase XIAP isoform X1 (XIAP) | Apoptosis inhibition process inflammatory signaling pathway, cell proliferation and metastasis and cell invasion [25–27] | DNA repair protein RAD50 (RAD50) | DNA repairing process [31] |
| Notch-related protein or Neurogenic locus notch homolog protein 4 (NOTCH4) | Acts as a receptor involved in developmental process such as differentiation, proliferation and program cell death [28] | Dynein heavy chain 6-axonemal isoform X1 (DNAH6) | Cellular movement [32] |
| H chain H-crystal structure of the Dbl and pleckstrin homology domains of Dbs in complex with RhoA (RHOA) | Actin cytoskeleton organization, cell cycle progression, cell transformation and invasion [33,34] | ||
| Rabenosyn-5 isoform X1 (ZFYVE20) | Vesicle transportation [35] | ||
When compared with dog saliva, proteins involved in cellular component organization or biogenesis and localization were more prevalent in human saliva as shown in Fig 4. These proteins included 5-methylcytosine rRNA methyltransferase NSUN4 isoform a (NSUN4), actin-related protein 8 isoform X1 (ACTR8), DNA repair protein RAD50 (RAD50), dynein heavy chain 6-axonemal isoform X1 (DNAH6), H chain H-crystal structure of the Dbl and pleckstrin homology domains of Dbs in complex with RhoA (RHOA) and rabenosyn-5 isoform X1 (ZFYVE20) (Table 1).
Interestingly, proteins involved in apoptosis processes and in biological adhesion function were detected only in dog saliva (Fig 4). Apoptosis-related proteins included E3 ubiquitin-protein ligase XIAP isoform X1 (XIAP) and Baculoviral IAP repeat-containing protein 2 (BIRC2), while biological adhesion-related proteins included notch-related protein (Neurogenic locus notch homolog protein 4 or NOTCH4) and dexamethasone-induced Ras-related protein 1 isoform X1 (RASD1) (Table 1). Other proteins that were only present in dog saliva are sirtuin and suppressin. Sirtuin is an important protein related to cellular homeostasis such as carbohydrate, protein and lipid metabolism, aging and may play a role in type II diabetes pathogenesis [36,37]. Suppressin is the other protein that was only present in dog saliva. This protein plays a role in immunological regulation by controlling the functions of lymphocytes [38]. Moreover, proteins related to angiogenesis, cell regeneration and wound healing such as fibroblast growth factor 12 isoform X3, fibroblast growth factor receptor substrate 3 isoform X1, epidermal growth factor receptor kinase substrate 8-like protein 1 isoform X4, and growth hormone-inducible transmembrane protein, were found to up-regulate in dog saliva (Table 2).
Table 2. Expression level of proteins in dog compared to human.
| Protein name | Accession number | Peptide sequence | Score | Fold change in dog |
|---|---|---|---|---|
| Amylase | gi|578798954 | TGSGDIENYNDATQVR | 91.58 | -4.6 |
| Carbonic anhydrase 6 | gi|28812184 | SYDIAQHEPDGLAVLAALVK | 62.64 | 4.76 |
| Cystatin S | gi|4503109 | IIPGGIYDADLNDEWVQR | 86.73 | -7.6 |
| Cystatin SA | gi|4503105 | IIEGGIYDADLNDER | 32.31 | -3.88 |
| Cystatin SN | gi|19882251 | QLCSFEIYEVPWENRR | 19.36 | -9.54 |
| Epidermal growth factor receptor kinase substrate 8-like protein 1 isoform X4 | gi|545487378 | QRDVLEVLDDRR | 14.84 | 3.78 |
| Fibroblast growth factor 12 isoform X3 | gi|578807164 | SSGTPTMNGGK | 15.71 | 1.78 |
| Fibroblast growth factor receptor substrate 3 isoform X1 | gi|578811367 | RHGGGTR | 15.56 | 0.04 |
| Growth hormone-inducible transmembrane protein | gi|118200356 | TRIGIR | 16.39 | 0.86 |
| Histatin | gi|4504529 | EFPFYGDYGSNYLYDN | 18.43 | -16.74 |
| Ig H-chain, partial | gi|185315 | GITGTT | 17.42 | 11.27 |
| IgGFc-binding protein-like | gi|545488815 | VAGLCGNFNRDPADDVDGPDPR | 10.28 | 8.02 |
| Immunoglobulin alpha-2 heavy chain, partial | gi|184761 | QEPSQGTTTFAVTSILR | 93.14 | -9.1 |
| Immunoglobulin epsilon variable region, partial | gi|288189027 | DDSKNMLYLHMNR | 20.57 | -7.44 |
| Immunoglobulin heavy chain constant region CH2, partial | gi|124390009 | QISVSWFR | 42.53 | 6.2 |
| Immunoglobulin heavy chain variable region, partial | gi|112694973 | VQCEVQVLASGGGLAQPGGSLR | 19.26 | 6.18 |
| Immunoglobulin heavy chain VDJC region, partial | gi|553426 | STXGGTAALGCLVK | 23.24 | -6.28 |
| Immunoglobulin heavy chain, partial | gi|219566253 | SPSLESRLTINK | 9.84 | -10.76 |
| Immunoglobulin heavy variable 3–49*03, partial | gi|371570901 | GLIQPGRSLR | 4.67 | -8.6 |
| Immunoglobulin kappa light chain, partial | gi|3169770 | DSTYSLSSTLTXSK | 55.61 | 9.32 |
| Immunoglobulin L/VH3-9/N13/DH2-15/N5/JH4 heavy chain variable region, partial | gi|209165484 | SYVVTAEYYFD | 14.22 | -11.08 |
| Immunoglobulin variable region, partial | gi|323432812 | TVAXPSVFIFPPSDEQLK | 19.22 | -9.54 |
| Immunoglobulin VH_3c kappa chain, partial | gi|339272251 | SGTASVLCLLNNFYPR | 99.69 | -6.56 |
| Interferon alpha-1/2-like | gi|345806671 | RDPPGSPR | 27.01 | 5.88 |
| Interferon alpha-8 precursor | gi|42476083 | ALILLAQMRR | 21.42 | 11.42 |
| Interferon regulatory factor 6 | gi|343131263 | HATRHSPQQEEENTIFK | 9.88 | 6.88 |
| Interferon-induced protein with tetratricopeptide repeats 3 | gi|545494542 | AKSTEEGK | 11.18 | 3.44 |
| Interleukin 12 receptor, beta 1, isoform CRA_c, partial | gi|119605061 | SGDGVAEPR | 34.84 | -6.32 |
| Interleukin 17 receptor, isoform CRA_c | gi|119578144 | GAEGVCGIQNGSLRWAEVKR | 24.61 | -12.26 |
| Interleukin-18 receptor accessory protein isoform X1 | gi|530368833 | TETTGR | 7.63 | -6.16 |
| Lactoperoxidase | gi|345805633 | FWWENPGVFTEK | 44.89 | -2.62 |
| Lactoperoxidase isoform 3 preproprotein | gi|231569458 | NGFPLPLAR | 43.32 | 3.32 |
| Mucin-5AC | gi|545531528 | ALSGVVEGTAAAFANTWK | 65.56 | 1.1 |
| Mucin-7-like | gi|545560548 | NNVQQYTVDR | 47.9 | 2.66 |
| Mucin-16 | gi|545535651 | VALGLAGISMDPK | 10.52 | -3.54 |
| Mucin-19-like | gi|545547107 | DCLCTIFGNYVK | 19.42 | 5.9 |
| Olfactory marker protein | gi|545536254 | MAEDGPKQPQLSMPLVLDPDLTK | 10.05 | 9.34 |
| Olfactory receptor 10T2 | gi|52218846 | VLGMPVATK | 9.43 | -16.52 |
| Olfactory receptor 5AP2 | gi|50979290 | DVKKALK | 6.95 | -6.58 |
| Olfactory receptor 5B12 | gi|52317108 | MRSPEGR | 6.31 | 8.08 |
| Otolin | gi|359323777 | TGLKGEAGDMGIPGPPGVVGPQGPK | 3.27 | 4.72 |
| Prolactin-induced protein, partial | gi|116642859 | TYLISSIPLXGAFNYK | 51.56 | -6.44 |
| Sirtuin | gi|119581643 | EAGAGR | 7.81 | 22.6 |
| Suppressin | gi|3293442 | IHADAKR | 7.51 | 11.18 |
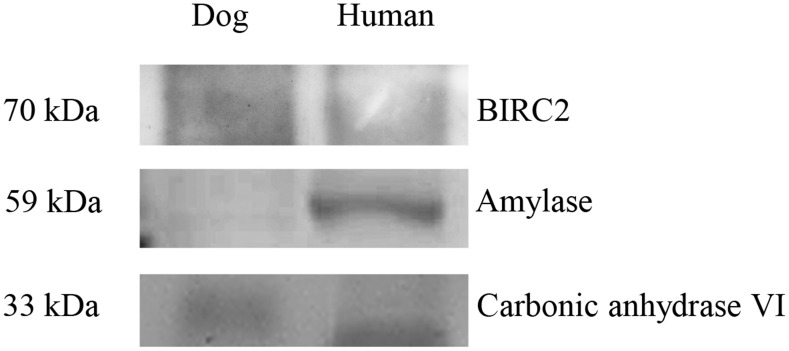
Besides these, proteins with antimicrobial properties were also found in dog saliva such as cystatin S, SA and SN, histatin, interferon alpha-1/2-like, lactoperoxidase and prolactin-inducible protein. Interestingly, levels of these proteins in dog saliva were much lower than in human saliva (Table 2). However, immunoglobulins, interferon and interleukin detected may play more of a role in the oral defense mechanism. Increased levels of carbonic anhydrase 6 with high buffering capacity were demonstrated in dog saliva. Various mucins were identified in both species, such as mucin-5AC, mucin-7-like, mucin-16 and mucin-19-like, but only mucin-16 was present in dogs at lower amounts than in human saliva. In addition, a much lower level of amylase was present in dog saliva. Validation of proteins expression were performed by immunoblotting analysis (Fig 5). The expression of BIRC2 and carbonic anhydrase 6 were detected in dog saliva whereas amylase protein was detected in human saliva. These results were consistent with shotgun proteomics.
Fig 5. Immunoblotting analysis of salivary proteins (BIRC2, amylase and carbonic anhydrase 6) was performed in dog and human samples.
Discussion
From protein profiles on SDS-PAGE analysis, dominant bands in dogs were found at high molecular weight while dominant bands in human saliva were found at both high and low molecular weight. The difference between the saliva proteins of dog and human may indicate the possibility of zoonotic pathogen reservoir and/or transfer. Shotgun proteome analysis showed that levels of proteins involved in metabolic processes were highest in dog saliva including apolipoprotein, histone, PHD finger protein, RNA-binding motif protein, transcription elongation factor A protein and Zinc finger protein. The level of apolipoprotein A-I in saliva was found to correlate with the levels of high-density lipoprotein (HDL) in plasma [39]. This protein has been used as a biomarker in coronary heart disease and Parkinson disease in human [40]. The presence of higher amount of apolipoprotein in dog saliva compared to human may be related to low incident of atherosclerosis in dog [41]. The different amount of amylase in each species may explain the difference between the food consumption behaviour [42]. From our results, dog saliva had lower amounts of alpha-amylase than human saliva which is possibly due to the natural selection associated with digestion capability and food appreciation. The study of diet and evolution of amylase gene revealed that high salivary amylase expression was observed in human populations with high-starch diets which may from natural selection and evolution [42]. Since the dog is carnivorous animal, amylase activity in this species is not as dominant as omnivorous species as humans. However, dogs in Thailand are often fed with rice and starch so amylase may present in their saliva.
Our study showed no lactoferrin in dog saliva which is consistent with a report by Sousa‐Pereira et al. [9]. Cystatin A, B, C, D, E/M and S were previously identified in dog saliva [9,43], but only cystatin S, SA and SN were detected in our study. The different results between the present study and other studies may due to the different methods for sample collection or preparation, and to different breeds of dogs since there are reports showing inter-individual variations and differences between breeds [9,43,44]. Numerous studies of cystatins in dogs have been reported, however, only serum cystatin C is evaluated as a biomarker in dog’s glomerular disease [45]. In humans, cystatin S, SA and SN in both serum and saliva were reported for their potential uses as biomarkers for cancer diseases and autoimmune disorders, and antibacterial properties against periodontal pathogens [46–48]. A future study of these proteins in dog saliva would be of interest in biomarkers and antimicrobial research. Interestingly, fibroblast growth factor and growth hormone were present in higher amounts in dog saliva. These might indicate that these proteins are protective factors in defense and repair after fighting. Other wild animals may also have similar salivary composition, however, salivary protein compositions in most wild animals have not yet been reported. Keratins were down-regulated in dog saliva, which may be from the difference in turnover rate of the oral epithelium between dogs and human [49–51].
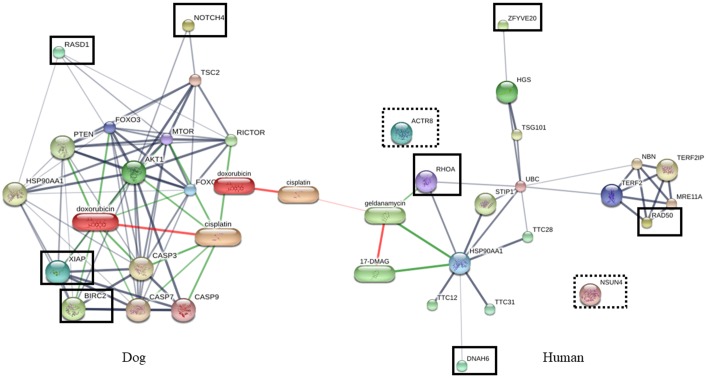
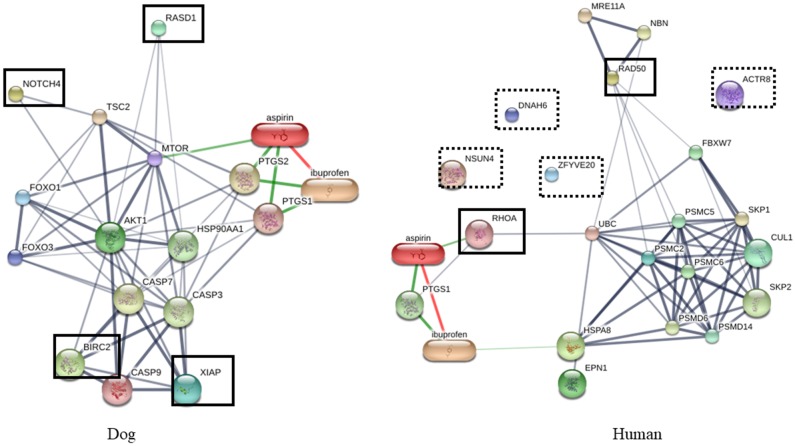
To determine the roles of XIAP, BIRC2, NOTCH4 and RASD1 in dogs and the proteins dominating in humans, which are NSUN4, ACTR8, RAD50, DNAH6, RHOA and ZFYVE20, interactions of the proteins were predicted by STITCH Version 5.0 (http://stitch.embl.de) (Figs 6–8). Proteins playing roles in apoptosis processes and biological adhesion were dominant in dog saliva. These functions are mainly associated with cancer. Therefore, these proteins were filtered and constructed with interaction networks using anti-cancer and anti-inflammatory agents, since inflammation always occurs with tumorigenesis. The proteins in dog were found to be more associated with cancer pathways (Fig 6) and inflammatory pathways than in human (Fig 7). Pathogenesis of cancer is related to dysregulation of apoptosis and the main problem in cancer treatment is chemotherapeutic resistance. Inhibitor of apoptosis proteins or IAPs (XIAP and BIRC2), and NOTCH signaling proteins are known to be involved in anti-apoptosis process and are elevated in many cancer cells, especially breast cancer [52–54]. The popular anti-cancer agents, doxorubicin and cisplatin, are associated with XIAP via protein kinase B (Akt) (Fig 6) [55]. Moreover, IAPs and NOTCH signaling proteins were also related to inflammatory pathway via Akt as shown in Fig 7. These proteins are related to inflammatory pathways by controlling homeostasis of many cells such as endothelial cells, smooth muscle cells, fibroblasts, lymphocytes and dendritic cells [56]. Since apoptosis resistance is one of the most important facets of tumorigenesis, alteration of these pathways might improve efficiency of chemotherapeutic drugs. Mammary gland tumor is the most common tumor in dogs, so higher level of these proteins in dog saliva may be related to the higher incidence rate of breast cancer in dogs than in human [52,57].
Fig 6. Protein-drug interaction network of proteins dominate in human and dogs.
Interaction network was predicted by STITCH version 5.0 (http://stitch.embl.de). Proteins network related to anticancer agents. Bold box: proteins related to the drugs, Dashed box: proteins non-related to the drugs.
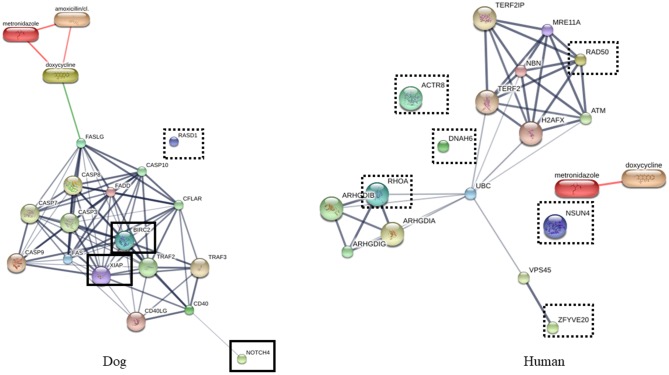
Fig 8. Protein-drug interaction network of proteins dominate in human and dogs.
Interaction network was predicted by STITCH version 5.0 (http://stitch.embl.de). Proteins network related to antimicrobial agents. Bold box: proteins related to the drugs, Dashed box: proteins non-related to the drugs.
Fig 7. Protein-drug interaction network of proteins dominate in human and dogs.
Interaction network was predicted by STITCH version 5.0 (http://stitch.embl.de). Proteins network related to anti-inflammatory agents. Bold box: proteins related to the drugs, Dashed box: proteins non-related to the drugs.
In addition, the antimicrobial properties of saliva were reported [58–61], and the interaction networks of some proteins obtained from shotgun proteomics and anti-bacterial agents including doxycycline, amoxicillin-clavulanic acid and metronidazole were analyzed by STITCH Version 5.0 (http://stitch.embl.de). The salivary proteins in the dog were found to be more associated with antimicrobial drugs than human as shown in Fig 8. The association between antimicrobial agents and the proteins was predicted via FAS/FAS ligand (FASLG) and Casp7. Anti-inflammatory and antimicrobial pathway of FASLG and Casp7 were reported [61,62]. Moreover, Torres et al. also reported that the proteins serve immune functions and show relations to antimicrobial mechanisms were the most abundant proteins in dog saliva [43]. Higher antimicrobial activities of dog salivary proteins than human was proposed. The ability of zoonotic pathogens in dog oral cavity to grow and transfer to humans may be regulated by these proteins.
Many methods have been reported for collecting dog saliva, however, no standard operating procedure has been set up [63]. Different salivary compositions have been obtained but these may be influenced by material collection methods such as cotton, polyester and polyethylene [64,65]. Since our study collected saliva directly from rom the dog’s mouth, from dripping or using a syringe to gently draw saliva out of the buccal cavity, there was no interference from collecting devices. Saliva volume and protein concentration differed considerably between dogs even though they had similar weights.
This study is one of the first to identify dog salivary proteins by using a shotgun proteomic technique. The main advantages of this proteomic technique are that low levels of a specific protein can be detected, and salivary proteins can be collected easily and noninvasively. However, the importance of saliva preparation has to be considered for downstream protein analysis. The inclusion of protease inhibitor and application of strict temperature control are recommended during saliva preparation, and separation of tissue material and debris is effected by centrifugation techniques. Proteins in samples were separated by molecular weight on one-dimensional gel electrophoresis, then bands were digested by trypsin and analyzed by tandem mass spectrometry. Overall, this technique can produce abundant protein information from small amounts of samples and has high sensitivity.
Proteome information from dog saliva has been limited, resulting in difficulties in applying to veterinary medicine. Here we have established novel dog saliva proteome data for Thai village dogs. The use of saliva as a non-invasive technique for developing disease diagnostics, therapeutics and prevention in the veterinary field has a promising future. Our findings are not only useful for animal health science but also have potential of utilizing dogs as an animal model. Since this study used only Thai village dogs, it should be of interest to extend the study to more varieties of breed to be able to better understand about behavior and salivary composition.
Supporting information
(PDF)
(PDF)
(PDF)
Acknowledgments
The authors thank Dr. Sineepat Talungjit and Dr. Sujiwan Seubbuk for helping with sample collection.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
P. Sanguansermsri and R. Surarit are rescipients of Thailand Research Fund through the Royal Golden Jubilee Ph.D. Program [Grant Number PHD/0057/2554] http://rgj.trf.or.th/main/th/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Zhang L, Henson BS, Camargo PM, Wong DT. The clinical value of salivary biomarkers for periodontal disease. Periodontol. 2000. 2009;51:25–37. 10.1111/j.1600-0757.2009.00315.x [DOI] [PubMed] [Google Scholar]
- 2.Chiappin S, Antonelli G, Gatti R, Elio F. Saliva specimen: a new laboratory tool for diagnostic and basic investigation. Clin Chim Acta. 2007;383:30–40. 10.1016/j.cca.2007.04.011 [DOI] [PubMed] [Google Scholar]
- 3.Lawrence HP. Salivary markers of systemic disease: noninvasive diagnosis of disease and monitoring of general health. J Can Dent Assoc. 2002;68:170–5. [PubMed] [Google Scholar]
- 4.Kawas SA, Rahim ZHA, Ferguson DB. Potential uses of human salivary protein and peptide analysis in the diagnosis of disease. Arch Oral Biol. 2012;57:1–9. 10.1016/j.archoralbio.2011.06.013 [DOI] [PubMed] [Google Scholar]
- 5.Vincent I, Michell A. Comparison of cortisol concentrations in saliva and plasma of dogs. Res Vet Sci. 1992;53:342–5. [DOI] [PubMed] [Google Scholar]
- 6.German A, Hall E, Day M. Measurement of IgG, IgM and IgA concentrations in canine serum, saliva, tears and bile. Vet Immunol Immunopathol. 1998;64:107–21. [DOI] [PubMed] [Google Scholar]
- 7.Parra MD, Tecles F, Subiela SM, Cerón JJ. C-reactive protein measurement in canine saliva. J Vet Diagn Invest. 2005;17:139–44. 10.1177/104063870501700207 [DOI] [PubMed] [Google Scholar]
- 8.Parra MD, Väisänen V, Cerón JJ. Development of a time-resolved fluorometry based immunoassay for the determination of canine haptoglobin in various body fluids. Vet Res. 2005;36:117–29. 10.1051/vetres:2004054 [DOI] [PubMed] [Google Scholar]
- 9.Sousa‐Pereira P, Cova M, Abrantes J, Ferreira R, Trindade F, Barros A, et al. Cross‐species comparison of mammalian saliva using an LC—MALDI based proteomic approach. Proteomics. 2015;15:1598–607. 10.1002/pmic.201400083 [DOI] [PubMed] [Google Scholar]
- 10.Larmas M, Scheinin A. Studies on dog saliva: I. Some physico-chemical characteristics. Acta Odontol Scandi. 1971;29:205–14. [DOI] [PubMed] [Google Scholar]
- 11.Navazesh M. Methods for collecting saliva. Ann NY Acad Sci. 1993;694:72–7. [DOI] [PubMed] [Google Scholar]
- 12.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. [DOI] [PubMed] [Google Scholar]
- 13.Paemanee A, Wikan N, Roytrakul S, Smith DR. Application of GelC-MS/MS to proteomic profiling of Chikungunya virus infection: preparation of peptides for analysis. Methods Mol Biol. 2016;1426:179–93. 10.1007/978-1-4939-3618-2_16 [DOI] [PubMed] [Google Scholar]
- 14.Johansson C, Samskog J, Sundström L, Wadensten H, Björkesten L, Flensburg J. Differential expression analysis of Escherichia coli proteins using a novel software for relative quantitation of LC‐MS/MS data. Proteomics. 2006;6:4475–85. 10.1002/pmic.200500921 [DOI] [PubMed] [Google Scholar]
- 15.Thorsell A, Portelius E, Blennow K, Westman‐Brinkmalm A. Evaluation of sample fractionation using micro‐scale liquid‐phase isoelectric focusing on mass spectrometric identification and quantitation of proteins in a SILAC experiment. Rapid Commun Mass Sp. 2007;21:771–8. [DOI] [PubMed] [Google Scholar]
- 16.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–67. 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2 [DOI] [PubMed] [Google Scholar]
- 17.Mi H, Lazareva-Ulitsky B, Loo R, Kejariwal A, Vandergriff J, Rabkin S, et al. The Panther database of protein families, subfamilies, functions and pathways. Nucleic Acids Res. 2005;33(Suppl 1):D284–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szklarczyk D, Santos A, Von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH5: augmenting protein—chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2015;44(D1):D380–4. 10.1093/nar/gkv1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cartier J, Berthelet J, Marivin A, Gemble S, Edmond V, Plenchette S, et al. Cellular inhibitor of apoptosis protein-1 (cIAP1) can regulate E2F1 transcription factor-mediated control of cyclin transcription. J Biol Chem. 2011;286:26406–17. 10.1074/jbc.M110.191239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou AY, Shen RR, Kim E, Lock YJ, Xu M, Chen ZJ, et al. IKKε-mediated tumorigenesis requires K63-linked polyubiquitination by a cIAP1/cIAP2/TRAF2 E3 ubiquitin ligase complex. Cell Rep. 2013;3:724–33. 10.1016/j.celrep.2013.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCormick F. Ras‐related proteins in signal transduction and growth control. Mol Reprod Dev. 1995;42:500–6. 10.1002/mrd.1080420419 [DOI] [PubMed] [Google Scholar]
- 22.Kemppainen RJ, Behrend EN. Dexamethasone rapidly induces a novel ras superfamily member-related gene in AtT-20 cells. J Biol Chem. 1998;273:3129–31. [DOI] [PubMed] [Google Scholar]
- 23.Tu Y, Wu C. Cloning, expression and characterization of a novel human Ras-related protein that is regulated by glucocorticoid hormone. BBA-GENE Struct Expr. 1999;1489:452–6. [DOI] [PubMed] [Google Scholar]
- 24.Lindsey J. Dexamethasone-induced Ras-related protein 1 is a potential regulatory protein in B lymphocytes. Int Immunol. 2007;19:583–90. 10.1093/intimm/dxm023 [DOI] [PubMed] [Google Scholar]
- 25.Kluger HM, McCarthy MM, Alvero AB, Sznol M, Ariyan S, Camp RL. et al. The X-linked inhibitor of apoptosis protein (XIAP) is up-regulated in metastatic melanoma, and XIAP cleavage by phenoxodiol is associated with carboplatin sensitization. J Transl Med. 2007; 10.1186/1479-5876-5-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang R, Li B, Wang X, Lin F, Gao P, Cheng SY, et al. Inhibiting XIAP expression by RNAi to inhibit proliferation and enhance radiosensitivity in laryngeal cancer cell line. Auris Nasus Larynx. 2009;36:332–9. 10.1016/j.anl.2008.08.006 [DOI] [PubMed] [Google Scholar]
- 27.Damgaard RB, Nachbur U, Yabal M, Wong WW, Fiil BK, Kastirr M, et al. The ubiquitin ligase XIAP recruits LUBAC for NOD2 signaling in inflammation and innate immunity. Mol Cell 2012;46:746–58. 10.1016/j.molcel.2012.04.014 [DOI] [PubMed] [Google Scholar]
- 28.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6. [DOI] [PubMed] [Google Scholar]
- 29.Chi L, Delgado-Olguín P. Expression of NOL1/NOP2/sun domain (Nsun) RNA methyltransferase family genes in early mouse embryogenesis. Gene Expr Patterns. 2013;13:319–27. 10.1016/j.gep.2013.06.003 [DOI] [PubMed] [Google Scholar]
- 30.Schafer D, Schroer T. Actin-related proteins. Annu Rev Cell Dev Bi. 1999;15:341–63. [DOI] [PubMed] [Google Scholar]
- 31.Williams RS, Williams JS, Tainer JA. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem Cell Biol. 2007;85:509–20. 10.1139/O07-069 [DOI] [PubMed] [Google Scholar]
- 32.Asai DJ, Brokaw C.J. Dynein heavy chain isoforms and axonemal motility. Trends Cell Biol. 1993;3:398–402. [DOI] [PubMed] [Google Scholar]
- 33.Del Peso L, Hernández-Alcoceba R, Embade N, Carnero A, Esteve P, Paje C, et al. Rho proteins induce metastatic properties in vivo. Oncogene. 1997;15:3047–57. 10.1038/sj.onc.1201499 [DOI] [PubMed] [Google Scholar]
- 34.Snyder JT, Worthylake DK, Rossman KL, Betts L, Pruitt WM, Siderovski DP, et al. Structural basis for the selective activation of Rho GTPases by Dbl exchange factors. Nat Struct Mol Biol. 2002;9:468–75. [DOI] [PubMed] [Google Scholar]
- 35.Nielsen E, Christoforidis S, Uttenweiler-Joseph S, Miaczynska M, Dewitte F, Wilm M, et al. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J Cell Biol. 2000;151:601–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, et al. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–6. 10.1038/nature06261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guarente L. Sirtuins, aging, and medicine. New Engl J Med. 2011;364:2235–44. 10.1056/NEJMra1100831 [DOI] [PubMed] [Google Scholar]
- 38.Ban EM, Leboeuf RD. Suppressin: an endogenous negative regulator of immune cell activation. Immunol Res. 1994;13:1–9. [DOI] [PubMed] [Google Scholar]
- 39.Fitzpatrick J, Lenda RB, Jones CL. Method and device for detection of Apo A, Apo B and the ratio thereof in saliva. 2004. www.google.ch/patents/US6808889 (accessed 18 January 2017).
- 40.Qiang JK, Wong YC, Siderowf A, Hurtig HI, Xie SX, Lee VM, et al. Plasma apolipoprotein A1 as a biomarker for Parkinson disease. Ann Neurol. 2013;74:119–27. 10.1002/ana.23872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xenoulis PG, Steiner JM. Lipid metabolism and hyperlipidemia in dogs. Vet J. 2010;183:12–21. 10.1016/j.tvjl.2008.10.011 [DOI] [PubMed] [Google Scholar]
- 42.Perry G, Dominy N, G Claw K, S Lee A, Fiegler H, Redon R, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–60. 10.1038/ng2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Torres SM, Furrow E, Souza CP, Granick JL, de Jong EP, Griffin TJ et al. Salivary proteomics of healthy dogs: An in depth catalog. PloS One. 2018;13:e0191307 10.1371/journal.pone.0191307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pasha S, Inui T, Chapple I, Harris S, Holcombe L, Grant MM. The saliva proteome of dogs: variations within and between breeds and between species. Proteomics. 2018;18: 10.1002/pmic.201700293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Almy FS, Christopher MM, King DP, Brown SA. Evaluation of cystatin C as an endogenous marker of glomerular filtration rate in dogs. J Vet Intern Med. 2002;16:45–51. [DOI] [PubMed] [Google Scholar]
- 46.Hu S, Wang J, Meijer J, Ieong S, Xie Y, Yu T, et al. Salivary proteomic and genomic biomarkers for primary Sjögren’s syndrome. Arthritis Rheum. 2007;56:3588–600. 10.1002/art.22954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoneda K, Iida H, Endo H, Hosono K, Akiyama T, Takahashi H, et al. Identification of Cystatin SN as a novel tumor marker for colorectal cancer. Int J Oncol. 2009;35:33–40. [PubMed] [Google Scholar]
- 48.Ganeshnarayan K, Velliyagounder K, Furgang D, Fine DH. Human salivary cystatin SA exhibits antimicrobial effect against Aggregatibacter actinomycetemcomitans. J Periodontal Res, 2012;47:661–73. 10.1111/j.1600-0765.2012.01481.x [DOI] [PubMed] [Google Scholar]
- 49.Meyer J, Marwah AS, Weinmann JP. Mitotic rate of gingival epithelium in two age groups. J Invest Dermatol. 1956;27:237–47. [DOI] [PubMed] [Google Scholar]
- 50.Skougaard M. Turnover of the gingival epithelium in marmosets. Acta Odontol Scand. 1965;23:623–43. [DOI] [PubMed] [Google Scholar]
- 51.Löe H. Physiological aspects of the gingival pocket an experimental study. Acta Odontol Scand. 1961;19:387–95. [DOI] [PubMed] [Google Scholar]
- 52.Merlo D, Rossi L, Pellegrino C, Ceppi M, Cardellino U, Capurro C, et al. Cancer incidence in pet dogs: findings of the animal tumor registry of Genoa, Italy. J Vet Intern Med. 2008;22:976–84. 10.1111/j.1939-1676.2008.0133.x [DOI] [PubMed] [Google Scholar]
- 53.Grudzien P. Notch Signaling is important in the survival, proliferation, and self-renewal of the putative breast cancer stem cell population [dissertation]. Loyola University Chicago; 2010.
- 54.Gyrd-Hansen M, Meier P. IAPs: from caspase inhibitors to modulators of NF-κB, inflammation and cancer. Nat Rev Cancer. 2010;10:561–74. 10.1038/nrc2889 [DOI] [PubMed] [Google Scholar]
- 55.Gagnon V, Van Themsche C, Turner S, Leblanc V, Asselin E. Akt and XIAP regulate the sensitivity of human uterine cancer cells to cisplatin, doxorubicin and taxol. Apoptosis. 2008;13:259–71. 10.1007/s10495-007-0165-6 [DOI] [PubMed] [Google Scholar]
- 56.Quillard T, Charreau B. Impact of notch signaling on inflammatory responses in cardiovascular disorders. Int J Mol Sci. 2013;14:6863–88. 10.3390/ijms14046863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider R. Comparison of age, sex, and incidence rates in human and canine breast cancer. Cancer. 1970;26:419–26. [DOI] [PubMed] [Google Scholar]
- 58.Hart BL, Powell KL. Antibacterial properties of saliva: role in maternal periparturient grooming and in licking wounds. Physiol Behav. 1990;48:383–6. [DOI] [PubMed] [Google Scholar]
- 59.Dale BA, Fredericks LP. Antimicrobial peptides in the oral environment: expression and function in health and disease. Curr Issues Mol Biol. 2005;7:119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Akpomie OO, Ukoha P, Nwafor OE, Umukoro G. Saliva of different dog breeds as antimicrobial agents against microorganisms isolated from wound infections. Anim Sci J. 2011;2:18–22. [Google Scholar]
- 61.Liu J, Kuszynski CA, Baxter BT. Doxycycline induces Fas/Fas ligand-mediated apoptosis in Jurkat T lymphocytes. Biochem Bioph Res Co. 1999;260:562–7. [DOI] [PubMed] [Google Scholar]
- 62.Brinkman BM, Hildebrand F, Kubica M, Goosens D, Del Favero J, Declercq W et al. Caspase deficiency alters the murine gut microbiome. Cell Death Dis. 2011;2:e220 10.1038/cddis.2011.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lensen CM, Moons CP, Diederich C. Saliva sampling in dogs: how to select the most appropriate procedure for your study. J Vet Behav. 2015;10:504–12. [Google Scholar]
- 64.Gröschl M, Köhler H, Topf HG, Rupprecht T, Rauh M. 2008. Evaluation of saliva collection devices for the analysis of steroids, peptides and therapeutic drugs. J Pharm Biomed Anal. 2008;47:478–86. 10.1016/j.jpba.2008.01.033 [DOI] [PubMed] [Google Scholar]
- 65.Takagi K, Ishikura Y, Hiramatsu M, Nakamura K, Degawa M. Development of a saliva collection device for use in the field. Clin Chim Acta. 2013;425:181–5. 10.1016/j.cca.2013.08.008 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.