Abstract
Multiple follicular lymphoma (FL) susceptibility single-nucleotide polymorphisms in the human leukocyte antigen (HLA) class I and II regions have been identified, including rs6457327, rs3117222, rs2647012, rs10484561, rs9268853 and rs2621416. Here we validated previous expression quantitative trait loci results with real-time reverse transcription quantitative PCR and investigated protein expression in B-lymphoblastoid cell lines and primary dendritic cells using flow cytometry, cell-based enzyme-linked immunosorbent assay and western blotting. We confirmed that FL-protective rs2647012-linked variants, in high linkage disequilibrium with the extended haplotype DRB1*15:01-DQA1*01:02-DQB1*06:02, correlate with increased HLA-DQB1 expression. This association remained significant at the protein level and was reproducible across different cell types. We also found that differences in HLA-DQB1 expression were not related to changes in activation markers or class II, major histocompatibility complex, transactivator expression, suggesting the role of an alternative regulatory mechanism. However, functional analysis using Regulome DB did not reveal any relevant regulatory candidates. Future studies should focus on the clinical relevance of increased HLA-DQB1 protein expression facilitating tumor cell removal through increased immune surveillance.
Keywords: follicular lymphoma, human leukocyte antigen, gene expression, protein expression
INTRODUCTION
Follicular lymphoma (FL) is a common type of B-cell lymphoma, although its causes remain unclear.1 Multiple non-coding FL susceptibility single-nucleotide polymorphisms (SNPs) have been identified in the human leukocyte antigen (HLA) class I and II regions including rs6457327,2 rs10484561,3 rs2647012,4 rs3117222,5 rs92688536 and rs26214166 (Supplementary Table S1). We previously investigated the role of rs6457327-, rs10484561- and rs2647012-linked SNPs as expression quantitative trait loci (eQTL) using public mRNA sequencing data on B-lymphoblastoid cell lines (B-LCLs). We found that FL-protective rs2647012-linked variants significantly correlated with HLA expression changes, particularly with increased HLA-DQB1 gene expression.7 SNPs linked to rs10484561 did not significantly alter gene expression, and correlations of rs6457327 with increased levels of HLA-B and HCG22 expression were ambiguous.7 To gain further insights into the functionality of these FL susceptibility variants, we investigated their effects on both transcript and protein expression. Given the high linkage disequilibrium (LD) in the region, with rs10484561 and rs2647012 being part of the extended haplotypes DRB1*01:01-DQA1*01:01-DQB1*05:013 and DRB1*15-DQA1*01-DQB1*06:02,8 respectively, correlations were also investigated for HLA alleles and inferred HLA amino acids. Primary lipopolysaccharide (LPS)-activated monocyte-derived dendritic cells (DCs), which are specialized antigen-presenting cells that may be more relevant for lymphoma studies,9,10 were also analyzed in this study.
RESULTS AND DISCUSSION
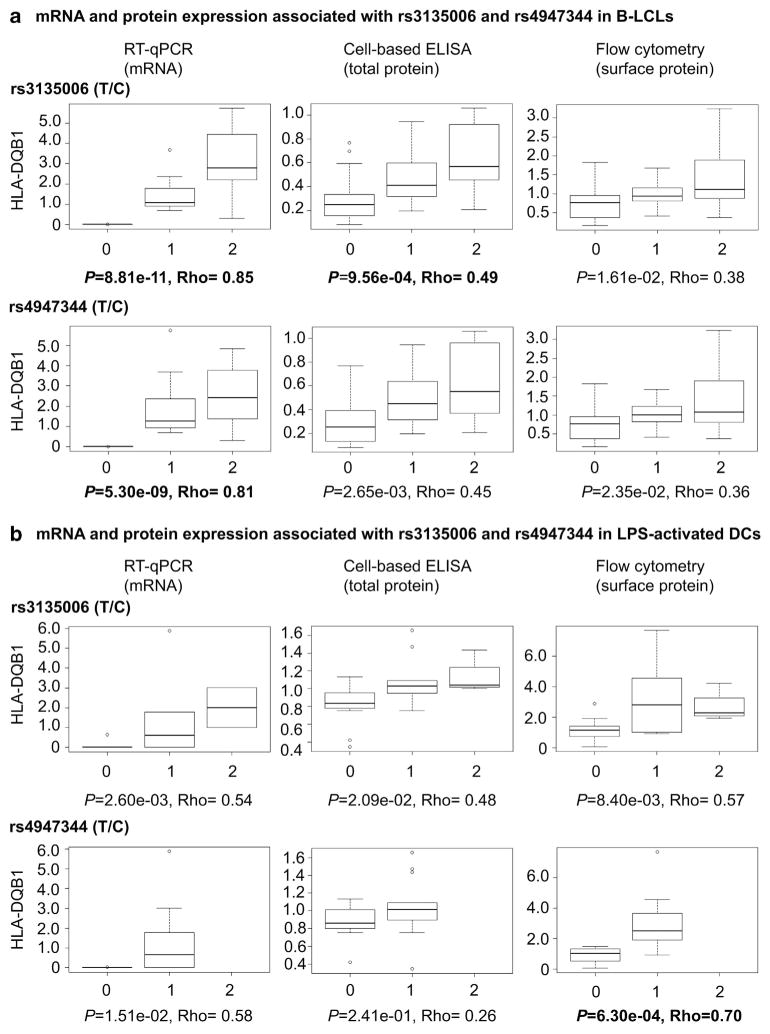
In agreement with our previous eQTL analysis,7 the strongest associations were observed for rs2647012-linked variants and increased HLA-DQB1 transcript levels in B-LCLs (8.81 × 10−11 ≤ P ≤3.25 × 10−4; Supplementary Table S2), with rs3135006 and HLA-DQB1*06 displaying the most significant association (Figure 1; Supplementary Table S2). Amino acids −5L, 87F and 125G corresponding to DQB1*06:02 also correlated with increased HLA-DQB1 expression (Supplementary Table S4). Two top eQTLs, rs4947344 and rs3135006, as well as the HLA-DQB1*06 and DRB1*15 alleles, also modestly correlated with increased HLA-DQB1 transcript levels in LPS-activated DCs (approaching significance at Bonferroni-corrected P-values P =1.51 × 10−2, P =2.60 × 10−2, P =8.87 × 10−3 and P =9.91 × 10−3, respectively; Supplementary Table S3).
Figure 1.
HLA mRNA and protein expression in B-LCLs and dendritic cells. Box-and-whisker plots display HLA mRNA and protein expression associated with rs3135006 (T/C) and rs4947344 (T/C) in B-LCLs (a) and LPS-activated DCs (b). mRNA was quantified by RT–qPCR using Taqman gene expression assays. Cell-based ELISA was used to measure total protein levels, whereas surface protein expression was determined by flow cytometry. The number of variant alleles per carrier are 0, 1 and 2. P-values are deemed significant at P≤2.1 × 10 −3 (highlighted in bold).
In line with the transcription data, the top rs2647012-linked eQTLs, predominantly rs3135006 (P =9.56 × 10 −4) and rs9273448 (P =7.82 × 10 −4), the HLA alleles DQB1*06 (P =9.56 × 10−4) and DRB1*15 (P =1.32 × 10−3), and most HLA-DQB1 amino acids (2.21 × 10 −5 ≤ P≤1.88 × 10−2) also correlated with increased levels of HLA-DQB1 protein expression, as observed by cell-based enzyme-linked immunosorbent assay (ELISA; Supplementary Table S4) and flow cytometry (approaching significance at 1.29 × 10−2 ≤ P≤3.41 × 10−2; Supplementary Table S5). Western blot analysis using polyclonal HLA-DQA1 antibodies (Supplementary Figure S1A), rather than monoclonal antibodies against monomeric HLA-DQB1 (Supplementary Figure S1B), confirmed that total levels of heterodimeric HLA-DQA1/DQB1 were increased in B-LCLs carrying rs2647012-linked protective alleles. Importantly, similar protein expression patterns were observed by flow cytometry in LPS-activated DCs, for which rs4947344 (P =6.30 × 10−4), rs2647012 (P =1.45 × 10 −3), rs3135006 (approaching significance at P =8.40 × 10−3) and DQB1*06 (P =1.46 × 10−4) showed positive associations with HLA-DQB1 expression (Supplementary Table S7). This trend was also observed for rs3135006 by cell-based ELISA (approaching significance at P =2.09 × 10 −2; Supplementary Table S6). Because of high LD in the region and the limited number of haplotypes in our data set, stepwise regression analysis failed to detect evidence for combinatory effects of alleles, SNPs and amino acids on HLA-DQB1 expression (data not shown).
Expression of soluble HLA-DQB1, generated upon allele-specific alternative splicing,11–13 could potentially be influenced by FL-protective rs2647012-linked SNPs. However, no transmembrane exclusions (Supplementary Figure S2) or soluble HLA-DQB1 were found for B-LCLs, and no significant differences in soluble HLA-DQB1 expression were observed for LPS-activated DCs (data not shown).
Previous associations of rs2647012-linked variants with increased HLA-DRB1 expression level7 were validated here by real-time reverse transcription quantitative PCR (RT-qPCR; Supplementary Tables S2 and S3). However, these findings were not confirmed at the protein level (Figure 1; Supplementary Figure S1D; Supplementary Tables S4–S7), except for rs9267955 (P =8.45 × 10−5, Supplementary Table S4). The HLA-DRB1*15 allele correlated with increased HLA-DRB1 protein levels in B-LCLs (3.40 × 10−5≤ P≤9.72 × 10−4, Supplementary Tables S4 and S5). rs2647012-linked variants did not influence HLA-DQA1, HLA-DQA2 or HLA-DPB1 protein levels (Supplementary Tables S2–S7). Despite observed variations at the mRNA level (Supplementary Table S2), rs10484561-linked variants and rs6457327 also did not alter protein expression (Supplementary Tables S4–S7). Moreover, no significant differences in HLA transcript or protein levels were observed for the recently reported FL susceptibility loci, rs3117222,5 rs26214166 and rs92688536 (Supplementary Figure S1E and Supplementary Tables S2–S5). More sensitive assays in future studies should confirm these discordances between transcript and protein levels, which could be due to numerous factors including differences in posttranslational events and stability of both mRNA and protein.14–16
Increases in HLA expression level are generally observed in activated antigen-presenting cells and concur with upregulation of activation and co-stimulatory markers.17 However, no significant differences were observed between rs2647012-linked SNPs and CD40, CD80, CD86 or CD20 protein levels in B-LCLs or LPS-activated DCs (data not shown), suggesting that the observed differences in HLA-DQB1 expression are not due to activation. Class II, major histocompatibility complex, transactivator (CIITA), located on chromosome 16, is the major regulator of HLA class II transcription,18 and defective CIITA expression was previously reported in a B-cell lymphoma cell line.19 Although SNPs can affect gene expression in trans,20 no significant changes in CIITA transcript or protein expression were observed (data not shown), suggesting that other regulatory mechanisms may be responsible for the changes observed in HLA class II expression.
To further investigate potential underlying regulatory mechanisms, the rs2647012-linked eQTLs were analyzed using RegulomeDB21 (Supplementary Figure S3, Supplementary Table S8). Several SNPs (rs2647003, rs2647046, rs2858310 and rs9469220) were annotated as likely functional on the basis of significant binding evidence with regulatory elements (RegulomeDB score 1). In our analysis, these SNPs correlated with significant changes in HLA transcript but not protein levels (Supplementary Tables S2, S4–S7). In contrast, the top eQTLs (rs3135006, rs9273448, rs2647012 and rs4947344) that did associate with significantly increased HLA-DQB1 protein levels (Supplementary Tables S4, S7) were not predicted as likely functional on the basis of RegulomeDB (score 6). It is possible that other unknown variants in the region in high LD with the investigated loci may be altering the regulation of HLA-DQB1 protein expression, or influencing protein translation and degradation.22,23 Similar to mechanisms proposed for HLA eQTLs, gene expression and risk of multiple sclerosis,24 it is possible that a combination of regulatory mechanisms that increase HLA-DQB1 expression and structural characteristics of specific HLA-DR/DQ receptors is responsible for the reduced FL risk observed in large population studies. HLA haplotype–eQTL interaction studies in diverse populations will help to further clarify these associations.
In summary, here we report a significant correlation between rs2647012-linked SNPs and HLA-DQB1 protein expression in cells pertinent to lymphoma. These FL-protective SNPs are in LD with the extended DRB1*15:01-DQA1*01:02-DQB1*06:02 haplotype8,25 and thus, the HLA-DRB1*15 and HLA-DQB1*06 alleles and several of the inferred HLA-DQB1 amino acids were also correlated with increased HLA-DQB1 expression. Owing to the high LD among the SNPs and HLA alleles, the effects on gene expression from the individual SNPs, HLA alleles and amino acids will require further investigation. Although the actual mechanisms remain unclear, our results could be clinically relevant as increased HLA-DQB1 expression may enhance antigen presentation, immune surveillance and the elimination of FL tumor cells. Future efforts should focus on the identification of regulatory mechanisms that alter DQB1 protein expression, the determination of FL-relevant antigens and the influence of HLA FL susceptibility loci on antigen presentation.
MATERIALS AND METHODS
Variants selected for study
Thirty-three rs2647012-linked SNPs that correlated with HLA expression changes in our previous eQTL study7 were selected for validation and follow-up, as well as rs6457327, rs10484561 and two surrogates in high LD (r2>0.8) with the latter. Three recently reported SNPs (rs3117222,5 rs92688536 and rs26214166) associated with FL were also analyzed (Supplementary Table S1).
Amino acids at HLA-DQB1 were determined for each cell line by using alignments from the IMGT/HLA database (http://www.ebi.ac.uk/ipd/imgt/hla/) release 3.14.0 (accessed 12 October 2013). Polymorphic aminoacid sites were limited to sites with frequency from 0.05 to 0.50 in our sample. Combinations of alleles at multiallelic sites were tested in each unique combination as described previously.26
Cell cultures
Peripheral blood mononuclear cells from 24 healthy Caucasians (Astarte Biologics, Redmond, WA and Cellular Technology Limited, Cleveland, OH, USA) and 40 B-LCLs of European descent (CEPH collection and SNP500 Human Variation Panel; Coriell Institute, Camden, NJ, USA) were used in this study. Genotypes of interest were either available or obtained by TaqMan genotyping. Monocytes, isolated from peripheral blood mononuclear cells using Monocyte Isolation Kit II (Miltenyi Biotec, Auburn, CA, USA) were differentiated into mature DCs as previously described.27
Transcript and protein expression analyses
mRNA transcript levels were determined by duplex RT-qPCR using TaqMan gene expression assays (Applied Biosystems, Grand Island, NY, USA). HLA-DQB1 mRNA splicing transcripts were evaluated by RT–PCR using intron-spanning primers (Supplementary Materials and methods). Using HLA-specific antibodies, total HLA protein expression was measured by cell-based ELISA (R&D Systems, Minneapolis, MN, USA) and visualized by chemiluminescent western blotting. Surface protein levels were detected by flow cytometry. Secreted HLA-DQB1 was measured in cell culture supernatants using conventional ELISA (USCN Life Science, Wuhan, China).
Statistics and bioinformatics
Single outliers were eliminated on the basis of the Grubbs’ outlier test.28 Pooled normalized averages were analyzed using a Spearman’s rank correlation test with t-distribution approximation. Correlations between genotype and gene expression were estimated with respect to the minor allele. P-values were deemed significant at P≤2.1 × 10−3 based on Bonferroni correction for six independent loci and four main genes (HLA-DQB1, HLA-DQA1, HLA-DRB1, HLA-DQA2) at α =0.05. To investigate potential overlap with known and predicted regulatory elements, the SNPs were analyzed using RegulomeDB (http://regulome.stanford.edu).21
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants CA154643 and CA104682 (CFS).
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supplementary Information accompanies this paper on Genes and Immunity website (http://www.nature.com/gene)
Author contributions: CFS obtained financial support for the study; FCMS, LC and CFS designed the study; FCMS, JZ, NKA, SS and JM, performed experiments; FCMS and LC analyzed data; FCMS, LC and CFS wrote the manuscript; All authors contributed to the final manuscript and approved its content.
References
- 1.Cerhan JR. Host genetics in follicular lymphoma. Best Pract Res Clin Haematol. 2011;24:121–134. doi: 10.1016/j.beha.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Skibola CF, Bracci PM, Halperin E, Conde L, Craig DW, Agana L, et al. Genetic variants at 6p21. 33 are associated with susceptibility to follicular lymphoma. Nat Genet. 2009;41:873–875. doi: 10.1038/ng.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conde L, Halperin E, Akers NK, Brown KM, Smedby KE, Rothman N, et al. Genome-wide association study of follicular lymphoma identifies a risk locus at 6p21. 32. Nat Genet. 2010;42:661–664. doi: 10.1038/ng.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smedby KE, Foo JN, Skibola CF, Darabi H, Conde L, Hjalgrim H, et al. GWAS of follicular lymphoma reveals allelic heterogeneity at 6p21. 32 and suggests shared genetic susceptibility with diffuse large B-cell lymphoma. PLoS Genet. 2011;7:e1001378. doi: 10.1371/journal.pgen.1001378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Skibola CF, Conde L, Foo JN, Riby J, Humphreys K, Sille FC, et al. A meta-analysis of genome-wide association studies of follicular lymphoma. BMC Genomics. 2012;13:516. doi: 10.1186/1471-2164-13-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vijai J, Kirchhoff T, Schrader KA, Brown J, Dutra-Clarke AV, Manschreck C, et al. Susceptibility loci associated with specific and shared subtypes of lymphoid malignancies. PLoS Genet. 2013;9:e1003220. doi: 10.1371/journal.pgen.1003220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conde L, Bracci PM, Richardson R, Montgomery SB, Skibola CF. Integrating GWAS and expression data for functional characterization of disease-associated SNPs: an application to follicular lymphoma. Am J Hum Genet. 2013;92:126–130. doi: 10.1016/j.ajhg.2012.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skibola CF, Akers NK, Conde L, Ladner M, Hawbecker SK, Cohen F, et al. Multi-locus HLA class I and II allele and haplotype associations with follicular lymphoma. Tissue Antigens. 2012;79:279–286. doi: 10.1111/j.1399-0039.2012.01845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang KC, Huang X, Medeiros LJ, Jones D. Germinal centre-like versus undifferentiated stromal immunophenotypes in follicular lymphoma. J Pathol. 2003;201:404–412. doi: 10.1002/path.1478. [DOI] [PubMed] [Google Scholar]
- 10.Jin MK, Hoster E, Dreyling M, Unterhalt M, Hiddemann W, Klapper W. Follicular dendritic cells in follicular lymphoma and types of non-Hodgkin lymphoma show reduced expression of CD23, CD35 and CD54 but no association with clinical outcome. Histopathology. 2011;58:586–592. doi: 10.1111/j.1365-2559.2011.03779.x. [DOI] [PubMed] [Google Scholar]
- 11.Briata P, Radka SF, Sartoris S, Lee JS. Alternative splicing of HLA-DQB transcripts and secretion of HLA-DQ beta-chain proteins: allelic polymorphism in splicing and polyadenylylation sites. Proc Natl Acad Sci USA. 1989;86:1003–1007. doi: 10.1073/pnas.86.3.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Královicová J, Vorechovsky I. Position-dependent repression and promotion of DQB1 intron 3 splicing by GGGG motifs. J Immunol. 2006;176:2381–2388. doi: 10.4049/jimmunol.176.4.2381. [DOI] [PubMed] [Google Scholar]
- 13.Kralovicova J, Houngninou-Molango S, Kramer A, Vorechovsky I. Branch site haplotypes that control alternative splicing. Hum Mol Genet. 2004;13:3189–3202. doi: 10.1093/hmg/ddh334. [DOI] [PubMed] [Google Scholar]
- 14.Britten AC, Mijovic CH, Barnett AH, Kelly MA. Differential expression of HLA-DQ alleles in peripheral blood mononuclear cells: alleles associated with susceptibility to and protection from autoimmune type 1 diabetes. Int J Immunogenet. 2009;36:47–57. doi: 10.1111/j.1744-313X.2008.00823.x. [DOI] [PubMed] [Google Scholar]
- 15.Wang H, Wang Q, Pape UJ, Shen B, Huang J, Wu B, et al. Systematic investigation of global coordination among mRNA and protein in cellular society. BMC Genomics. 2010;11:364. doi: 10.1186/1471-2164-11-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Gool SW, Vandenberghe P, de Boer M, Ceuppens JL. CD80, CD86 and CD40 provide accessory signals in a multiple-step T-cell activation model. Immunol Rev. 1996;153:47–83. doi: 10.1111/j.1600-065x.1996.tb00920.x. [DOI] [PubMed] [Google Scholar]
- 18.Krawczyk M, Peyraud N, Rybtsova N, Masternak K, Bucher P, Barras E, et al. Long distance control of MHC class II expression by multiple distal enhancers regulated by regulatory factor X complex and CIITA. J Immunol. 2004;173:6200–6210. doi: 10.4049/jimmunol.173.10.6200. [DOI] [PubMed] [Google Scholar]
- 19.Prod’homme T, Drenou B, De Ruyffelaere C, Barbieri G, Wiszniewski W, Bastard C, et al. Defective class II transactivator expression in a B lymphoma cell line. Leukemia. 2004;18:832–840. doi: 10.1038/sj.leu.2403315. [DOI] [PubMed] [Google Scholar]
- 20.Fairfax BP, Makino S, Radhakrishnan J, Plant K, Leslie S, Dilthey A, et al. Genetics of gene expression in primary immune cells identifies cell type-specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–510. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22:1790–1797. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doran G. The short and the long of UTRs. J RNAi Gene Silencing. 2008;4:264–265. [PMC free article] [PubMed] [Google Scholar]
- 23.Halvorsen M, Martin JS, Broadaway S, Laederach A. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genetics. 2010;6:e1001074. doi: 10.1371/journal.pgen.1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alcina A, del Abad-Grau MM, Fedetz M, Izquierdo G, Lucas M, Fernandez O, et al. Multiple sclerosis risk variant HLA-DRB1*1501 associates with high expression of DRB1 gene in different human populations. PLoS One. 2012;7:e29819. doi: 10.1371/journal.pone.0029819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akers NK, Curry JD, Conde L, Bracci PM, Smith MT, Skibola CF. Association of HLA-DQB1 alleles with risk of follicular lymphoma. Leuk Lymphoma. 2011;52:53–58. doi: 10.3109/10428194.2010.532888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foo JN, Smedby KE, Akers NK, Berglund M, Irwan ID, Jia X, et al. Coding variants at hexa-allelic amino acid 13 of HLA-DRB1 explain independent SNP associations with follicular lymphoma risk. Am J Hum Genet. 2013;93:167–172. doi: 10.1016/j.ajhg.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Skibola CF, Nieters A, Bracci PM, Curry JD, Agana L, Skibola DR, et al. A functional TNFRSF5 gene variant is associated with risk of lymphoma. Blood. 2008;111:4348–4354. doi: 10.1182/blood-2007-09-112144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grubbs FE. Procedures for detecting outlying observations in samples. Technometrics. 1969;11:1–21. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.