Abstract
Lineage relationships among the large number of heterogeneous cell types generated during development are difficult to reconstruct in a high-throughput manner. We recently established a method, scGESTALT, that combines cumulative editing of a lineage barcode array by CRISPR-Cas9 with large-scale transcriptional profiling using droplet-based single-cell RNA sequencing. The technique generates edits in the barcode array over multiple timepoints using Cas9 and pools of single-guide RNAs introduced during early and late zebrafish embryonic development, which distinguishes it from similar Cas9 lineage tracing methods. The recorded lineages are captured along with thousands of cellular transcriptomes to build lineage trees with hundreds of branches representing relationships among profiled cell types. Here we provide details for (i) generating transgenic zebrafish; (ii) performing multi-timepoint barcode editing; (iii) building single-cell RNA-seq libraries from brain tissue; and (iv) concurrently amplifying lineage barcodes from captured single cells. Generating transgenic lines takes 6 months while performing barcode editing and generating single-cell libraries involve 7 days of hands-on time. scGESTALT provides a scalable platform to map lineage relationships between cell types in any system that permits genome editing during development, regeneration or disease.
Keywords: neurobiology, neural development, developmental biology, lineage tracing, transcriptomics, single-cell sequencing, scRNA-seq, genetic engineering, CRISPR-Cas, zebrafish, transgenesis, Tol2, lineage barcodes, lineage tree
EDITORIAL SUMMARY
This protocol describes how to generate transgenic zebrafish expressing a barcode array that can be edited by CRISPR/Cas9 at multiple developmental stages. Single cell RNA sequencing of edited barcodes and cellular transcriptomes allows reconstruction of lineage relationships.
INTRODUCTION
Single-cell technologies have rapidly progressed to enable large-scale profiling of cell-to-cell heterogeneity in tissues during development, homeostasis and disease1–3. It is now feasible to characterize cell identities based on transcriptional, epigenetic and protein marker landscapes by capturing tens of thousands of single cells or nuclei in a short timeframe and at a low cost. These methods rely on droplet microfluidics, high density arrays or combinatorial indexing for massively parallel experiments4–17. Among the various single-cell assays, single-cell RNA-sequencing (scRNA-seq) is widely used to categorize cell types using gene expression signatures in many tissues and animals, including humans, without requiring extensive prior knowledge of cell composition. Concomitant with technological advances, substantial efforts have been made to develop computational approaches to correct for batch effects, enable efficient data clustering, and compare datasets between different conditions or methods18–21. The resultant cell type catalogs are revealing new insights into cellular heterogeneity such as de novo identification of rare cell types and gene markers, delineation of cellular dynamics via developmental trajectories and cell cycle states, and changes in structure of profiled cell types during disease22–38.
Large-scale reconstruction of lineage relationships at single-cell resolution
Despite the ability to define cell types on a large scale, it has been challenging to determine the lineage relationships between cell types and characterize the progressive steps in their specification39–41. Reconstruction of lineage trees that capture the patterns and timings of lineage segregation can provide information about the structural and functional organization of tissues and their dysregulation in congenital disease and cancer; this may eventually provide a means for translation into in vitro reprogramming and organoid systems. The realization of such lineage trees would require simultaneous measurements of cell type identities and lineage relationships at high throughput. Lineage tracing methods using barcoded viral libraries, Cre-loxP recombination, somatic mutations and transposon tagging have been linked with next-generation sequencing, but have not been combined with simultaneous cell type identification at single-cell resolution42–48. Coupling transcriptomic and lineage readouts with scRNA-seq would provide a powerful assay to perform unbiased lineage tree reconstruction.
Lineage recording with CRISPR-Cas9 genome editing
The CRISPR-Cas9 nuclease system has become a widely-used tool in genome engineering with applications extending beyond gene editing to regulation of gene expression, epigenetic editing, base editing, knockout screening, in situ labeling, RNA tracking, protein localization, chromatin interaction profiling, and signal recording, among others49–65. Recently, CRISPR-Cas9 has been adapted for use in several lineage tracing strategies including GESTALT, LINNAEUS, MEMOIR and ScarTrace66–71. In general, these methods rely on the introduction of Cas9-induced stochastic mutations (insertions or deletions during DNA repair following double strand breaks) to predefined target sites in transgenes, and use the edited sequences as lineage barcodes for clonal tracing and lineage tree reconstruction.
We have established an approach, scGESTALT72, which extends the CRISPR-Cas9 lineage recording capabilities of GESTALT66, with large-scale transcriptional profiling using droplet-based scRNA-seq, inDrops8, to reconstruct developmental lineage trees using zebrafish as a model system (Fig. 1). Our method makes use of combinatorial and cumulative additions of Cas9-induced mutations to a genomic CRISPR barcode array at multiple timepoints to write permanent records of lineage histories as cells divide72. The edited barcodes are expressed from a transgene, captured within cellular transcriptomes and sequenced at single-cell resolution with inDrops, which enables the indexing of >12,000 cells/h into nanoliter-size droplets73. Using tools adapted from phylogenetics, branching lineage trees are generated using patterns of shared mutations between barcodes with the tips of the trees representing profiled cell types defined using associated scRNA-seq gene expression signatures.
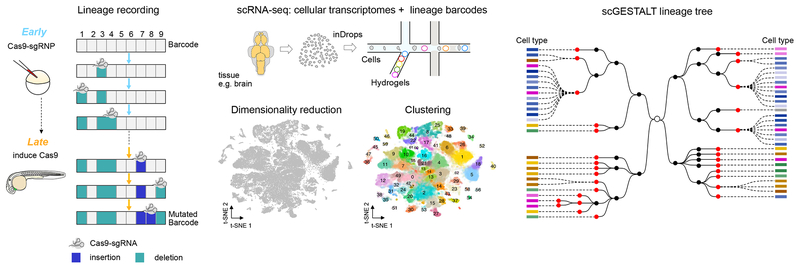
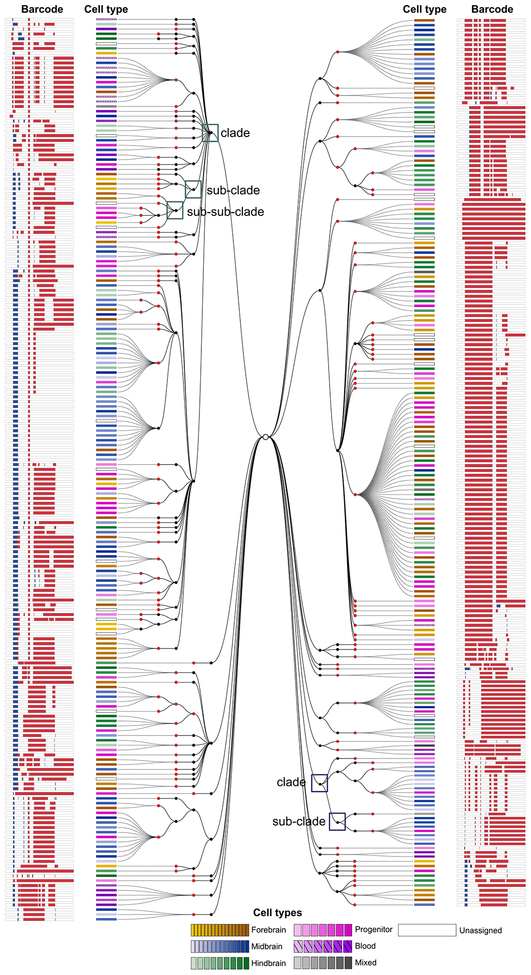
Figure 1. Simultaneous recovery of lineages and cell types at single-cell resolution using scGESTALT.
Left panels, During early and late developmental time periods, CRISPR-Cas9 induced indels record cell lineages as mutated genomic barcode sequences. Middle panels, Tissues of interest, such as brain, are dissociated into a single-cell suspension and loaded into a microfluidics device (inDrops). Single cells are encapsulated in droplets and indexed using hydrogels (color-coded to indicate different cell identifier primers) that are coated with oligodT primers. Polyadenylated cellular transcriptomes and scGESTALT lineage barcodes bind to the oligodT sequences and are simultaneously extracted from the same cells. Transcriptome libraries are sequenced to generate gene expression matrices for thousands of single cells. Gene expression profiles are used to perform dimensionality reduction using principal component analysis and visualized in two dimensions on a t-distributed stochastic neighbor embedding (t-SNE) plot. Single cells are represented as grey dots on the shown plot. A modularity-based clustering algorithm (Louvain) is used to cluster cells into discrete cell types using significant principal components. A t-SNE plot of 58,492 cells from n = 22 animals is color-coded to show 63 distinct clusters that were identified from zebrafish juvenile brains72. Right panels, scGESTALT libraries are sequenced to obtain lineage barcodes of profiled single cells. The inDrops index sequences are used to match transcriptomes and lineage barcodes for the same cells. Cell lineage trees are generated using maximum parsimony based on patterns of shared edits. Black and red nodes represent early and late barcode edits, respectively. Dashed lines connect profiled single cells to nodes on the tree. Cells connected to the same node are clonal (i.e. contain the same lineage barcode). Each cell is categorized into a discrete cell type (color coded rectangles) based on prior transcriptional clustering analysis. Brown shades represent forebrain cell types, blue shades represent midbrain cell types, green shades represent hindbrain cell types, and pink shades represent progenitor cell types. This procedure was approved by the HU/FAS Committee on the Use of Animals in Research & Teaching under Protocol No. 25–08. Figure adapted from ref72.
Here we explain in detail how to perform scGESTALT multi-timepoint barcode editing with transgenic zebrafish. We describe the generation of scRNA-seq transcriptome and lineage barcode libraries using the juvenile brain as an experimental system. Due to the inherent scalability of droplet-based scRNA-seq, scGESTALT can be used to profile thousands of cells from a single animal. Furthermore, scGESTALT can be adapted for use in any organism or in vitro model system that is compatible with genome editing and single-cell technologies to characterize lineages in development, regeneration, cellular reprogramming and disease states.
Comparison with other lineage tracing methods
To generate large-scale lineage trees or perform large-scale clonal analysis, lineage tracing methods should possess high multiplexing capacities for unique and unambiguous labeling of ancestral cells. Approaches that incorporate high-throughput sequencing of diverse DNA or RNA lineage records are advantageous over the limited spectral labeling capacity of fluorescent reporters. For example, virally-delivered DNA barcode libraries can in principle encode high sequence complexity. However, the relative representation of each barcode in the library and its successful recovery can confound data interpretation. If subsets of barcodes are overrepresented in the viral library, it will result in coalescence of independent clones marked by the same barcode into a single clone, and rare barcodes would be underrepresented and potentially missed during data analysis. Inefficiencies in viral transduction of certain cell types can also preclude its use in various tissues or organisms. Furthermore, as the barcodes are not mutable (i.e. cannot be edited during development), only clonal analysis can be performed. In this case, all descendants of an ancestrally marked founder cell can be identified (all cells in a clone inherit the same barcode sequence), but lineage relationships between the clonal descendants cannot be inferred. Another approach known as Polylox barcoding, has been recently described to potentially generate hundreds of thousands in vivo barcode sequences (via Cre-induced recombination of a synthetic DNA cassette inserted into the genome), however its compatibility with single-cell analysis and use for generating lineage trees have not been demonstrated44. Somatic mutations accumulated over an animal’s lifespan can be harnessed to generate detailed lineage trees and do not require building transgenics or introducing barcoding reagents43. However, methods for retrieving somatic mutations from single cells involve whole genome amplification and sequencing strategies that can introduce technical artefacts and are currently cost-prohibitive for analyzing large single-cell populations43,74,75. In contrast, scGESTALT barcodes can be recovered by targeted enrichment from a predefined genetic locus. Coupling scGESTALT to the inDrops scRNA-seq platform also provides several advantages such as high scalability, low library preparation cost (currently ~$0.06 per cell), high single-cell capture rates (>70% of cells), low cell input requirements, no cell capture size bias, and simultaneous transcriptome profiling for cell type or cell state identification73. Recently, inDrops has also been coupled with Tol2 transposon-based barcoding (TracerSeq) for lineage tracing in zebrafish37.
Two other methods have linked CRISPR-Cas9 lineage tracing with scRNA-seq in zebrafish68,69. These techniques generate barcode edits in a multi-copy transgenic reporter (in tandem or in distinct genomic loci) by injecting Cas9 protein or mRNA along with one sgRNA targeting a reporter-specific sequence. This restricts lineage analysis to early embryonic stages as Cas9 editing terminates upon degradation of Cas9/sgRNA or mutation of all potential sites, and the observed number of distinct lineage barcodes is limited to ~1,00068. Multi-copy integrations also require that sequences from all sites are recovered to obtain full lineage information, which can be challenging with sparse scRNA-seq data69. The scGESTALT barcode array consists of 9 distinct CRISPR sites in tandem, each of which can be targeted by a different sgRNA. It also enables multi-timepoint editing by targeting subsets of the CRISPR sites during early and late embryogenesis using a combination of injection and transgenic Cas9/sgRNA expression. Such a modular system enables longer periods of lineage recording and increases barcode diversity such that >10,000 barcodes are observed72. Furthermore, the use of single-copy barcode integrations facilitates unequivocal profiling of the complete barcode array extracted from each cell without the need for inference of dropout events (i.e. partial loss of lineage information due to distribution of lineage recording sites across multiple loci).
Limitations of scGESTALT
Several considerations have to be made when choosing scGESTALT for lineage recording. First, the edited barcode is recovered from fewer than 30% of profiled single cells, possibly due to low expression level of the barcode, inefficient capture of barcode transcript within droplets, or amplification bottlenecks during sequencing library preparation. If using this method for profiling lineages of rare cell types, an enrichment step (e.g. FACS) for populations of interest may be required to obtain sufficient lineage barcodes. Future optimizations using stronger promoter sequences to drive barcode expression or improved library preparation could address these issues. Alternatively, the barcode could be extracted from genomic DNA rather than the transcriptome68. Second, due to the tandem arrangement of CRISPR sites on the barcode array, some lineage records can be erased by large deletions that span multiple sites. Third, barcode diversity is limited to ~10,000 unique labels due to relatively quick saturation of Cas9 editing activities. To follow fate specification of progenitors over longer timeframes (e.g. days or weeks) and generate lineage trees with more branches, the timing and duration of editing will need to be further modulated, and the diversity of barcodes will need to be increased to millions of unique labels. Finally, tissue dissociation results in loss of precise spatial information of profiled cells, thus spatial distributions of clones and distantly related cells cannot be investigated.
Experimental design
The scGESTALT protocol is divided into four experimental sections: (i) generating two transgenic zebrafish lines: one for barcode expression and the other for inducible Cas9 and ubiquitous sgRNA expression; (ii) performing multi-timepoint barcode editing using a combination of injection and induction of barcoding reagents for early and late labeling of cells, respectively; (iii) transcriptome profiling using inDrops; and (iv) concurrently extracting lineage barcodes from captured single cells. Transcriptome and scGESTALT libraries are sequenced separately and gene expression and lineage information for each cell are matched using the inDrops cell identifier index.
Generating transgenics for barcoding.
Two transgenic lines expressing different components of the scGESTALT barcoding strategy are established using the Tol2 transposon system (Fig. 2). In this system, a DNA sequence of interest flanked by minimal cis-sequences from the left and right ends of the medaka Tol2 element is randomly integrated into the genome in the presence of Tol2 transposase by a cut-and-paste mechanism76. Thus, a transgenic construct can be cloned in between the Tol2 ends and the resulting plasmid is injected along with mRNA encoding Tol2 transposase into zebrafish embryos. Translated Tol2 protein will mediate excision and stable integration of the transgenic construct into the genome of embryonic cells during early development, including some cells that give rise to germ cells. The injected fish are outcrossed and screened for successful germline transmission of the Tol2 transgenic construct into the F1 generation to establish transgenic animals. The lineage barcode array construct comprises a heat shock-inducible promoter driving ubiquitous DsRed transgene expression with the scGESTALT CRISPR array inserted in the 3′UTR upstream of the SV40 polyadenylation signal. In addition, the construct contains a heart-specific GFP reporter as a transgenesis marker (cmlc2:GFP). Upon heat shock, the wildtype or edited barcode is expressed as part of the DsRed mRNA and can be extracted from the cellular transcriptome. The barcode transgenic line is screened by qPCR to determine transgene copy number; single-copy integrants are identified and outcrossed to expand and maintain the line. A separate transgenic line is established using a Tol2 construct comprising a heat shock inducible promoter driving Cas9-t2A-GFP and 5 independent U6 promoters driving 5 distinct sgRNAs targeting the barcode array. This line does not need to be surveyed for single-copy integration. The two lines are available upon request from the Schier or Gagnon labs (see REAGENTS). Depending on the user’s goal, it is possible to make changes to both of the above constructs and generate new transgenics. For example, cell- or tissue-specific promoters can be used to drive Cas9 or barcode expression. In addition, modifications can be made to integrate the constructs into other organisms or cell lines.
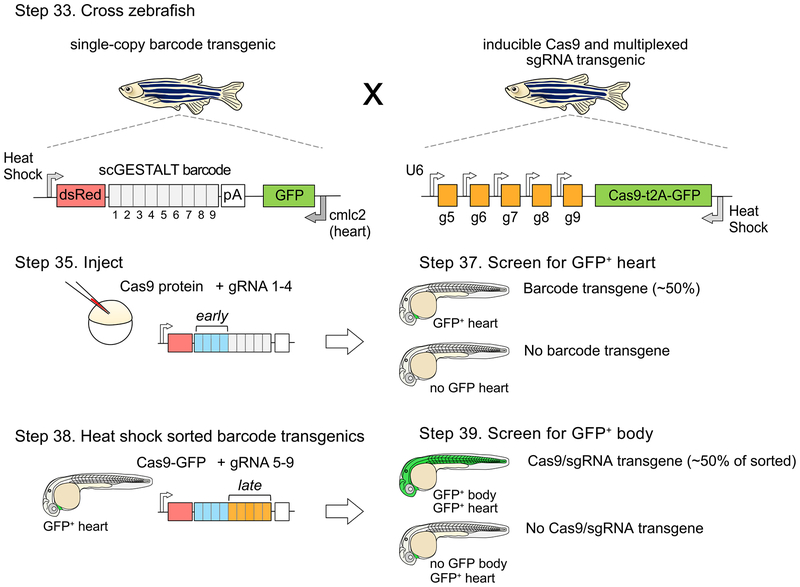
Figure 2. Strategy for a CRISPR-Cas9 system that enables early and late barcode editing.
Zebrafish with single-copy heat shock promoter-driven scGESTALT barcode (to promote ubiquitous barcode expression at stage of interest) are crossed to zebrafish that express heat shock-inducible Cas9 and U6-driven sgRNAs 5–9. The barcode is cloned downstream of the dsRed coding sequence and upstream of the SV40 polyadenylation sequence (pA). Resulting embryos are injected with Cas9 protein and sgRNAs 1–4 at the one-cell stage (blue bars; early editing). The embryos are screened for GFP positive heart transgenics (cmlc2 promoter drives heart-specific GFP expression) at 30 hpf to identify embryos containing the barcode transgene, and sorted embryos are heat shocked to induce transgenic Cas9 for a second round of editing (orange bars; late editing). The embryos are screened again for ubiquitous GFP expression (Cas9 is linked to GFP with a t2A self-cleaving peptide), which indicates successful Cas9 transgene induction. Double transgenic embryos are grown for downstream profiling, and heat shocked at time of interest (e.g. juvenile stage 23–25 dpf) to induce expression of the edited barcode array prior to scRNA-seq analysis. Protocol steps for each stage are indicated. Adapted with permission from ref72.
Early and late barcoding.
To enable recording of lineages during early and late development, the scGESTALT barcode transgenic is crossed to the Cas9/sgRNA transgenic. Single-cell embryos are injected with Cas9 protein and a pool of sgRNAs which target the first 4 sites of the CRISPR array. This strategy initiates an ‘early’ round of Cas9 editing activity that introduces mutations at target sites 1–4 of the barcode. Upon zygotic genome activation, sgRNAs targeting sites 5–9 of the array are expressed from U6 promoters. Embryos are then heat shocked at a time of choosing (e.g. 30 hours post-fertilization, hpf) to induce ubiquitous expression of transgenic Cas9, and initiate barcode editing at target sites 5–9 of the array. The sequential activation of this ‘early and late’ editing strategy enables longer lineage recording, higher barcode diversity, and encodes the relative order of mutation patterns in the two halves of the array, thereby facilitating lineage tree reconstruction72. Edited double transgenic embryos are selected by sequentially screening for strong heart-specific GFP expression to identify the barcode transgene, and ubiquitous GFP expression to identify the Cas9/sgRNA transgene, and are grown until the user is ready to perform scRNA-seq (Fig. 2). Due to the modular nature of the barcoding strategy, if the user is interested in tracing early lineage segregations, it is possible to barcode cells during early embryogenesis only by injecting Cas9 protein or mRNA together with all 9 sgRNAs targeting the CRISPR array. The sequences for the 9 sgRNA target sites are provided in Supplementary Table 1. Generally, the spectrum of barcode edits is dominated by deletions, which can be categorized into intra-site (small deletion restricted to one target site) and inter-site (larger deletions that span two or more sites). Insertions are observed at a low frequency. The combination of small indels and larger deletions in early and/or late editing generates high overall diversity of lineage barcodes (<0.5% of lineage barcodes were found to overlap between n=8 different embryos72). We direct the reader to ref72 for further information about the nature and frequency of barcode repair products.
Single-cell transcriptome profiling.
scRNA-seq is performed to simultaneously extract edited lineage barcodes and cellular transcriptomes. For droplet-based scRNA-seq, samples are first dissociated into single-cell suspensions prior to loading on the device. We processed juvenile zebrafish brains to provide a snapshot of the heterogeneity of cell types and lineages in the vertebrate brain72, and here provide details for brain dissociation and scRNA-seq using inDrops. However, the user can perform similar analyses using any tissue or developmental stage of interest for which a single-cell dissociation protocol that yields high cell viability is available. The inDrops protocol has been described in detail previously73. Briefly, it involves (i) obtaining a compatible microfluidic chip, (ii) synthesizing indexed hydrogel beads, (iii) encapsulating and indexing single cells in nanoliter-size droplets, and (iv) building transcriptome libraries for next-generation sequencing. As single cells are lysed during inDrops, polyadenylated cellular mRNA and edited scGESTALT lineage barcode mRNA are hybridized to oligodT primers on hydrogel beads (Fig. 3a). Following reverse transcription of hybridized mRNA, the cDNA is indexed with an inDrops cell identifier, subjected to second strand synthesis and linearly amplified by in vitro transcription (Fig. 3b). A fraction of the resulting amplified RNA is chemically fragmented for standard transcriptome library preparation, and converted to a DNA library by another round of reverse transcription followed by addition of Illumina-compatible sequencing adapters by PCR. The remaining unfragmented RNA is used for building scGESTALT libraries. scGESTALT can also be adapted for scRNA-seq with other droplet-based (e.g Drop-seq9, 10× Genomics10) or plate/array-based methods. However, amplification and library construction of lineage barcodes as detailed below will have to be empirically determined by the user.
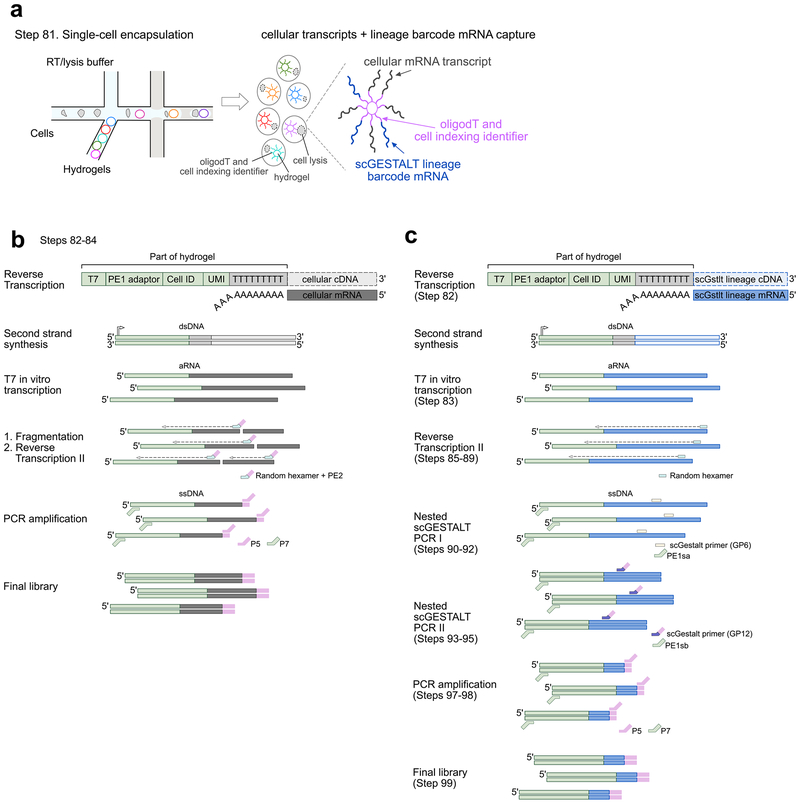
Figure 3. Transcriptome and scGESTALT library preparation overview.
a. Single cells are encapsulated and indexed in droplets using the inDrops platform (left, Step 82). Upon lysis (cells shown with dotted lines), polyadenylated cellular transcripts and scGESTALT lineage barcodes hybridize to oligodT primers on hydrogels. Hydrogels are color-coded to indicate distinct indexing identifiers. Adapted with permission from ref72.
b. Overview of transcriptome library preparation steps from single cells (Steps 82–84). Hydrogels are coated with oligonucleotides containing T7 promoter, an adaptor sequence (PE1), a cell indexing identifier (Cell ID), unique molecular identifier (UMI), and oligodT sequence. Polyadenylated cellular mRNA hybridizes to the oligodT sequence and is reverse transcribed into cellular cDNA sequence. Second strand synthesis is carried out to generate double stranded DNA (dsDNA) that is used for T7 in vitro transcription and generates linearly amplified RNA (aRNA). The aRNA is chemically fragmented and then subjected to a second round of reverse transcription using an oligonucleotide that contains a random hexamer and an adaptor sequence (PE2). The resulting single stranded DNA (ssDNA) is PCR amplified with Illumina P5 and P7 adaptor sequences containing overlap with PE2 and PE1 sequences, and a limited number of cycles to generate the final sequencing-ready transcriptome library. ssDNA fragments that do not contain the PE1 adaptor sequence will not be amplified.
c. Overview of scGESTALT lineage barcode library preparation steps from single cells. Polyadenylated lineage barcode mRNA, consisting of dsRed and edited scGESTALT CRISPR array (see Fig. 2, Step 33), are expressed from a transgene upon heat shock of the animal prior to brain dissection and dissociation. The lineage barcode mRNA hybridizes to oligodT sequence on the hydrogel and is reverse transcribed into lineage barcode cDNA. Second strand synthesis and T7 in vitro transcription is carried out similar to the transcriptome library preparation (Step 82). However, the aRNA is not fragmented in order to preserve the full lineage barcode sequence (Step 83, part of the of reverse transcribed product is not fragmented and stored at −80 °C). Instead reverse transcription is carried out using random hexamers to generate full-length ssDNA (Steps 85–89). The edited scGESTALT CRISPR array sequence is selectively enriched from the full-length ssDNA using a nested PCR approach. The first round amplifies a longer piece of dsDNA using a primer that hybridizes in the dsRed sequence (GP6) and one that overlaps the PE1 adaptor sequence (PE1sa) (Steps 90–92). The resulting dsDNA is used in a second round to target the edited lineage barcode sequence using a primer that binds upstream proximal to the start of the CRISPR lineage array sequence (GP12) and a primer that overlaps the adaptor sequence (PE1sb) (Steps 93–95). The resulting dsDNA is PCR amplified (Steps 97–98) similar to the transcriptome library preparation to generate the final sequencing-ready scGESTALT lineage barcode library (Step 99).
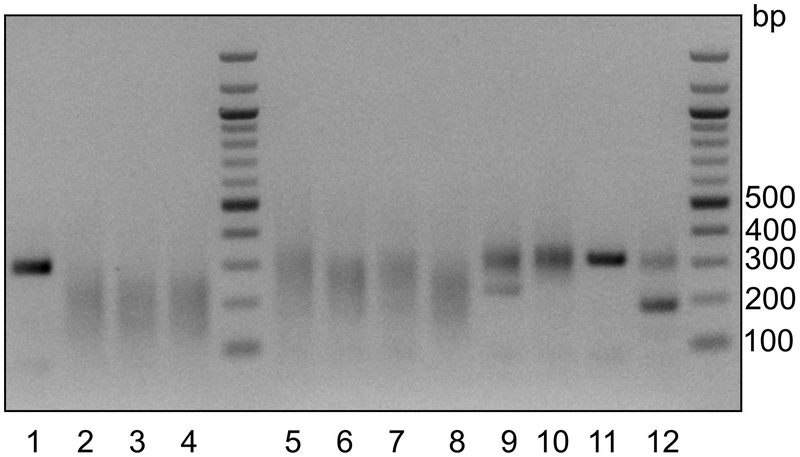
Targeted amplification of lineage barcodes.
After in vitro transcription of indexed cDNA, a fraction of the full-length amplified RNA is reverse transcribed using random hexamers. scGESTALT lineage barcodes are amplified from the resulting cDNA using a nested PCR strategy (Fig. 3c). First, a larger fragment is enriched using primers that flank the DsRed transgene and a universal inDrops adaptor sequence located downstream of the cell identifier and UMI (unique molecular identifier, for counting number of unique transcripts of a gene) sequences. Following purification, the PCR product is used to amplify a shorter lineage barcode fragment using primers that flank the barcode array and a universal inDrops adaptor sequence. Finally, Illumina-compatible DNA libraries are generated following the same strategy for transcriptome libraries. If mixing multiple libraries in a sequencing run, it is critical that the same multiplexing primers are used for both transcriptome and lineage barcode libraries to ensure that these can later be matched to each other. In this protocol, we provide details for preparing “V3” inDrops transcriptome and scGESTALT libraries, which are compatible with standard Illumina sequencing primers. If using “V2” inDrops libraries, they will need to be sequenced with custom read and index primers as described previously73.
Experimental controls.
A negative control for editing involves no injection of Cas9 protein and sgRNAs 1–4 followed by no heat shock Cas9 induction. In this case, no substantial editing of the barcode array at sites 1–9 should be observed by PCR amplification from genomic DNA or using next generation sequencing from genomic DNA and/or scRNA-seq libraries. Similarly, injection of Cas9 protein alone (no sgRNAs 1–4) followed by no heat shock Cas9 induction, should also result in little to no editing at sites 1–9. In some cases, low levels of editing may be observed at sites 5–9 due to cross-activity between injected Cas9 protein and U6-expressed sgRNAs 5–9. When no Cas9 protein and sgRNAs 1–4 are injected, but Cas9 is induced at 30 hpf by heat shock, editing is expected to be largely confined to sites 5–9. In contrast, performing two-timepoint editing by Cas9 protein and sgRNAs 1–4 injection followed by heat shock Cas9 induction, should result in edits observed throughout sites 1–9 of the barcode array. …
Data Analysis.
The data analysis consists of three major parts: (i) analysis of the transcriptome data for cell type identification, (ii) analysis of the scGESTALT lineage barcodes for identification of editing events, and (iii) generation of a single-cell resolved lineage tree with associated cell types. The inDrops.py pipeline (see Software in REAGENTS below) will generate a gene expression matrix that can be used as input for identification of cell types and gene markers using an analysis tool designed for processing sparse scRNA-seq data (e.g. Seurat18,77). The inDrops.py pipeline can also be used to obtain sequence reads for scGESTALT lineage barcodes that are sorted by the inDrops cell identifier index (i.e. obtain lineage reads for single cells). Next, the sorted scGESTALT lineage barcode reads are further formatted using the scGestaltPrepFunc.R script (Supplementary Software 2) and processed with the scGESTALT analysis pipeline (see Software in REAGENTS below, also provided as Supplementary Software 1). This will generate a table containing the lineage barcode sequence for each cell as well as its inDrops cell identifier index sequence. Next, for each cell from which a lineage barcode was successfully recovered and processed, the matching transcriptome and hence cell identity is assigned using the Transcriptome-scGESTALTMatchPipe.R and MatchPipeFunc.R scripts (Supplementary Software 3–4). At this stage, a table will be generated (GestMaster.txt, see TestData in Supplementary Software 1) containing the scGESTALT lineage barcode, cell cluster identity (obtained using Seurat for cell type classification) and the inDrops cell identifier. This can be used as input for drawing the overall multi-timepoint edited lineage tree using the scGESTALT analysis pipeline.
MATERIALS
REAGENTS
Nuclease-free water (Life Technologies, cat. no. M5310–1L)
NotI-HF (NEB, cat. no. R3189)
E.Z.N.A. Cycle Pure Kit (Omega, cat. no. D6492–02) or equivalent
E.Z.N.A. Plasmid DNA Mini Kit I (Omega, cat. no. D6942–02) or equivalent
E.Z.N.A. Total RNA Kit I (Omega, cat. no. R6834–02) or equivalent
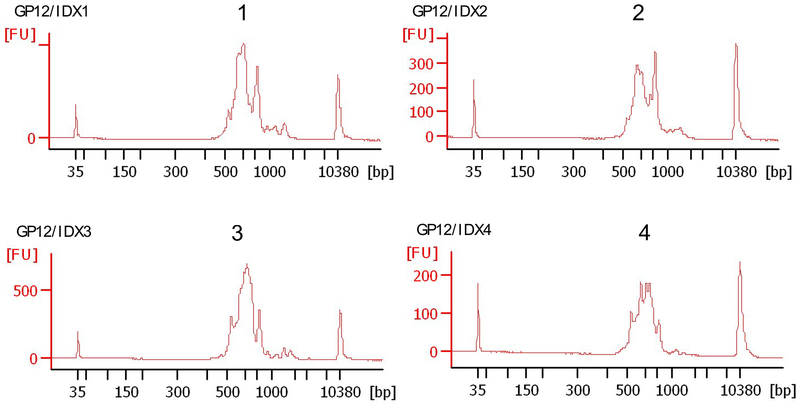
RNA Clean & Concetrator-25 (Zymo Research, cat. no. R1018)
mMESSAGE mMACHINE SP6 (Thermo Fisher Scientific, AM1340)
NaOH, 1M (Honeywell Research Chemicals, cat. no. 319511–1L)
Tris-HCl pH 7.5, 1M (VWR, cat. no. 75800–958)
Tris–HCl pH 7.0, 1 M (Thermo Fisher Scientific, cat. no. AM9851)
Tris–HCl pH 8.0, (Thermo Fisher Scientific, cat. no. 15568025)
EnGen sgRNA Synthesis Kit, S. pyogenes (NEB, cat. no. E3322)
CRITICAL sgRNA oligonucleotide sequences (sgRNA 1–4) provided (see DNA oligonucleotide sequences below) are designed for use with this kit
EnGen Cas9 NLS, S. pyogenes (NEB, cat. no. M0646)
iTaq Universal SYBR Green Supermix (Biorad, cat. no. 1725120)
Phusion High-Fidelity DNA polymerase (NEB, cat. no. M0530)
CRITICAL Amplification of the scGESTALT library has been optimized using this enzyme Q5 High-Fidelity DNA polymerase (NEB, cat. no. M0491)
CRITICAL Amplification of the scGESTALT library has been optimized using this enzyme KAPA 2× HiFi HotStart PCR mix (KAPABiosystems, cat. no. KK2601)
dNTP mix, 10 mM each (NEB, cat. no. N0447)
100 bp DNA ladder (NEB, cat. no. N3231)
Papain Dissociation System (Worthington, cat. no. LK003150)
CRITICAL Brain dissociation has been optimized using this kit Neurobasal media (Thermo Fisher Scientific, cat. no. 21103049)
CRITICAL Brain dissection and dissociation have been optimized using this media B27 supplement, serum free, 50× (Thermo Fisher Scientific, cat. no. 17504044)
CRITICAL Brain dissection has been optimized using this supplement for high cell viability DPBS, no calcium, no magnesium (Thermo Fisher Scientific, cat. no. 14190144) DBPS, calcium, magnesium, glucose, pyruvate (Thermo Fisher Scientific, cat no. 14287080)
35 μm cell strainer (e.g. VWR, cat. no. 352235)
20 μm cell strainer (e.g. Sysmex, cat. no. 04–004-2325)
Trypan blue, 0.4% (wt/vol) (Thermo Fisher Scientific, cat. no. T10282)
OptiPrep Density Gradient Medium (Sigma-Aldrich, cat. no. D1556–250ML)
Agencourt AMPure XP magnetic beads (Beckman Coulter, cat. no. A63881)
Random Hexamers, 50 μM (Fisher Scientific, cat. no. N8080127)
PrimeScript Reverse Transcriptase (Takara Clonetec, cat. no. 2680A)
RNaseOUT Recombinant Ribonuclease Inhibitor (Fisher Scientific, cat. no. 10777019)
Phenol red, 0.5% (wt/vol) (Sigma-Aldrich, cat. no. P0290)
Methylene blue, 0.1% (wt/vol) (Aqua Solutions, cat. no. 5761–500ML)
Instant Ocean salt (Instant Ocean)
Agarose (e.g. National Diagnostics, cat. no. EC-202)
Sylgard 184 (Electron Microscopy Sciences, cat. no. 24236–10)
Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, cat. no. Q33230)
Tricaine (Sigma-Aldrich, cat. no. E10521–50G)
CAUTION: Use gloves when handling (can cause skin irritation).
MiSeq v2 300 cycle kit (Illumina, cat no. MS-102–2002)
NextSeq 500/550 High Output v2 75 cycle kit (Illumina, cat. no. FC-404–2005)
PhiX Control v3 (Illumina, FC-110–3001)
Zebrafish (Danio rerio)
wildtype strains (e.g. TL/AB, TL or AB; available from ZIRC, http://zebrafish.org),
- Tg(hsp70:DsRed-barcodev7; myl7:eGFP) strain (allele number a167; scGESTALT barcode transgenic; available by request from Schier or Gagnon labs),
- Tg(hsp70:Cas9-t2a-GFP; 5×U6:sgRNA) strain (allele number a168; inducible Cas9 and U6-driven sgRNA transgenic; available by request from Schier or Gagnon labs)
CAUTION This procedure was approved by the Harvard University/Faculty of Arts & Sciences Standing (HU/FAS) Committee on the Use of Animals in Research & Teaching under Protocol No. 25–08. The HU/FAS animal care and use program maintains full AAALAC accreditation, is assured with OLAW (A3593–01), and is currently registered with the USDA.
Plasmids
pCS2-zT2TP (available from Koichi Kawakami lab78)
pTol2-hspDRv7_scGstlt (Addgene, plasmid ID 108870)
pTol2-hsp70l:Cas9-t2A-GFP, 5×U6:sgRNA (Addgene, plasmid ID 108871)
DNA oligonucleotide sequences (purified by standard desalting, can be ordered from IDT)
| Name | Sequence | Purpose | Step |
|---|---|---|---|
| qPCRctrl F | 5′-TCAGTCAACCATTCAGTGGCCCAT-3′ | PCR amplify ultraconserved genomic region | 23 |
| qPCRctrl R | 5′-CAGGAAAGGGAATGCAGGGTTTGT-3′ | PCR amplify ultraconserved genomic region | 23 |
| qPCRdsRed F | 5′-GAGCGCGTGATGAACTTCGAGG-3′ | PCR amplify dsRed transgenic region | 23 |
| qPCRdsRed R | 5′-CAGCCCATAGTCTTCTTCTGCATTACG-3′ | PCR amplify dsRed transgenic region | 23 |
| sgRNAl | 5′TTCTAATACGACTCACTATAGACAGCAGTATCATCGACTAGTTTTAGAGCTAGA-3′ | Synthesize sgRNA1 for CRISPR-Cas9 lineage barcoding by injection | 27 |
| sgRNA2 | 5′-TTCTAATACGACTCACTATAGAGAGCGCGCTCGTCGACTAGTTTTAGAGCTAGA-3′ | Synthesize sgRNA2 for CRISPR-Cas9 lineage barcoding by injection | 27 |
| sgRNA3 | 5′-TTCTAATACGACTCACTATAGTCAGCA GTACTACTGACGAGTTTTAGAGCTAGA-3′ | Synthesize sgRNA3 for CRISPR-Cas9 lineage barcoding by injection | 27 |
| sgRNA4 | 5′-TTCTAATACGACTCACTATAGACAGCAGTGTGTGAGTCTAGTTTTAGAGCTAGA-3′ | Synthesize sgRNA4 for CRISPR-Cas9 lineage barcoding by injection | 27 |
| scGESTALT F | 5′-TCGAGCTCAAGCTTCGG-3′ | PCR amplify scGESTALT fragment for assessing editing efficiency | 43 |
| scGESTALT R | 5′-CTGCCATTTGTCTCGAGGTC-3′ | PCR amplify scGESTALT fragment for assessing editing efficiency | 43 |
| inDrops_GP6 | 5′-GAGGACTACACCATCGTGGAG-3′ | PCR amplify long scGESTALT fragment from inDrops library | 90 |
| inDrops_PE1Sa | 5′-CTCTTTCCCTACACGACGCTGGGTGTCGGGTGCAG-3′ | PCR amplify long scGESTALT fragment from inDrops library | 90 |
| inDrops_GP12 | 5′-TCGTCGGCAGCGTCAGATGTGTATAA GAGACAG NNNNNNNNNTCGAGCTCAA GCTTCGGAC-3′, where N stands for a random base | PCR amplify short scGESTALT fragment from inDrops library | 93 |
| inDrops_PE1Sb | 5′-CTCTTTCCCTACACGACGCT-3′ | PCR amplify short scGESTALT fragment from inDrops library | 93 |
| R1-PCRix | 5′-AATGATACGGCGACCACCGAGATCTACACxrefTCGTCGGCAGCGTC-3′, where xref is an index sequence for multiplexing (see Supplementary Table 2) | PCR amplify transcriptome or scGESTALT final sequencing library | 97 |
| R2-PCR | 5′-CAAGCAGAAGACGGCATACGAGATGGGTGTCGGGTGCAG-3′ | PCR amplify transcriptome or scGESTALT final sequencing library | 97 |
EQUIPMENT
1.5 ml microcentrifuge tubes (e.g. USA Scientific, cat. no. 1415–2600)
1.5 ml microcentrifuge tubes (e.g. USA Scientific, cat. no. 1415–2500)
0.2 ml PCR tubes (e.g. VWR, cat. no. 53509–304)
50 ml tubes (e.g. VWR, cat. no. 352070)
15 ml tubes (e.g. VWR, cat. no. 352096)
5 ml sterile serological pipettes (e.g. VWR, cat. no. 89130–896)
NanoDrop (Thermo Scientific)
PCR and qPCR machines (e.g. Bio-Rad)
DNA electrophoresis system (e.g. Bio-Rad)
Benchtop centrifuge for 1.5 ml tubes (e.g. Eppendorf, cat. no. 54124)
Refrigerated centrifuge for 1.5 ml tubes (e.g. Eppendorf, cat. no. 5424 R)
Minifuge (e.g. Benchmark, cat. no C1008-C)
Water bath (e.g. VWR)
Heat block for 1.5 ml tubes (e.g. VWR)
Vortex (e.g. Scientific Industries, cat. no. SI-0236)
Magnetic rack for 0.2 ml PCR strip (e.g. Permagen, cat. no. MSR1224)
2100 Electrophoresis Bioanalyzer Instrument (Agilent, cat. no. G2939AA)
Laser-based micropipette puller (e.g. Sutter Instruments, cat. no. P-2000)
Microinjection glass capillaries (World Precision Instruments, cat. no. TW100F-4)
Fine forceps (Fine Science Tools, cat. no. 11254–20)
Fine scissors (Fine Science Tools, cat. no. 14040–10)
Microloader tips for glass capillaries (e.g. Eppendorf, cat. no. 930001007)
Injection molds (e.g. Adaptive Science Tools, model no. TU-1)
Pneumatic PicoPump (World Precision Instruments, model no. PV820) or equivalent
Piconozzle Kit v2 (World Precision Instruments, cat. no. 5430-ALL) or equivalent
Manipulator, magnetic stand and iron plate (Narishige, model. no. M-152, GJ-1, IP) or equivalent
Treadlite II Foot Switch (Lineamaster, model no. T-91-S) or equivalent
Micrometer for calibration (AmScope, cat. no MR100) or equivalent
Dissection microscope with light source (e.g. Stemi 2000 stereomicroscope, Zeiss)
Fluorescent microscope with GFP filter (e.g. Zeiss)
10-cm Petri dishes (e.g. VWR, cat. no. 25384–342)
Insect pins (Fine Science Tools, cat. no. 26002–15)
Breeding tanks and 0.5 L, 1 L and 2 L tanks (e.g. Techniplast)
95%O2: 5%CO2 gas tank (Airgas, X02OX95C2003102)
Gas regulator (e.g. VWR, 55850–392)
Tygon E-3603 tubing for bubbling gas (e.g. Component Supply)
Hemocytometer (e.g., Bulldog Bio, cat. no. DHCN420)
Qubit (Thermo Fisher Scientific, cat. no. Q33226)
Qubit Assay Tubes (Thermo Fisher Scientific, cat. no. Q32856)
REAGENT SETUP
Zebrafish
Adult fish are maintained at a density of 8–10 fish per 2 L tank with a 14-h light:10-h dark cycle at 28.5 °C. Wildtype TL/AB strain is bred in house by crossing wildtype TL strain with wildtype AB strain and is used in all experiments described in this protocol. Animals used for breeding are typically 3 to 18 months old.
CAUTION This procedure was approved by the HU/FAS Committee on the Use of Animals in Research & Teaching under Protocol No. 25–08.
Phenol red
Dilute 0.5% (wt/vol) phenol red stock 1:5 with nuclease-free water. Use this diluted stock for injections, and store at room temperature (23 °C – 26 °C) for at least one year.
Blue water
Combine 20 L of water in a carboy with 20 ml of 0.1% (wt/vol) Methylene blue and 5gof Instant Ocean sea salt, and adjust pH to 7.0 with sodium bicarbonate buffer. Store at room temperature for up to 1 month.
50 mM NaOH
Combine 19 ml of nuclease-free water with 1 ml of 1M NaOH. Store at room temperature for up to 3 months.
MESAB, 25×
Combine 489.5 ml of water with 2g tricaine powder and 10.5 ml of 1M Tris (pH 9.0), and adjust pH to 7. Make 10 ml aliquots and store at −20 °C for up to 1 year.
B27 supplement
Upon receiving the product, thaw the solution completely at room temperature. Make 500 μl aliquots and store at −20 °C for up to 1 year.
RNA elution buffer
Combine 9.9 ml of nuclease-free water, 100 μl of 1 M Tris–HCl (pH 7.0), and 2 μl of 0.5 M EDTA (pH 8.0). Store 1 ml aliquots at −20 °C for up to 1 year.
DNA elution buffer
Combine 9.9 ml of nuclease-free water, 100 μl of 1 M Tris–HCl (pH 8.0), and 2 μl of 0.5 M EDTA (pH 8.0). Store 1 ml aliquots at −20 °C for up to 1 year.
R1-PCRix/R2-PCR primer mix
For each variant of R1-PCRix, combine 25 μl of 10 μM R1-PCRix primer and 25 μl of 10 μM R2-PCR primer in DNA elution buffer. Store primer mixes at −20 °C for up to 1 year. See Supplementary Table 1 for a list of 24 R1-PCRix primer sequences.
EQUIPMENT SETUP
Preparation of injection needles
Pull glass capillaries for injection as described previously79.
Embryo microinjection dish
Boil 3 g of agarose in 100 ml blue water in a microwave until the agarose dissolves. Let it cool slightly on bench, and pour 25–30 ml per 10-cm Petri dish. Gently layer an injection mold (pre-wetted with water to avoid introducing bubbles) on top of the agarose. Let the agarose solidify and remove the injection mold. The microinjection dish can be stored at 4 °C for two weeks.
Microinjection setup
Configure and connect the Pneumatic PicoPump, piconozzle, manipulator, magnetic stand, iron plate and foot switch according to the manufacturer’s instructions. Connect the Pneumatic PicoPump to a nitrogen tank according to the manufacturer’s instructions.
Preparation of 95%O2: 5%CO2 gas tank
Fit the gas tank with the gas regulator and Tygon tubing according to the manufacturer’s instructions.
Preparation of sylgard dissection dishes
Mix 45 ml of Sylgard elastomer base with 4.5 ml of Sylgard elastomer curing agent (10:1 ratio; both reagents provided in Sylgard 184) in a 50 ml tube. Mix thoroughly by inverting several times and transferring back and forth between two 50 ml tubes. Pour ~10 ml of the mix per 10-cm Petri dish. Gently tap the dish to force air bubbles to rise to the top. Allow to cure and set overnight. The dishes can be stored at room temperature for at least one year.
inDrops microfluidics platform setup
Detailed instructions for setting up the inDrops platform have been described previously73. Note that the system should be primed and ready for cell barcoding prior to starting single-cell preparation (Step 48). Alternatively, if working in a pair, one person should get the device ready while the other is preparing cells for encapsulation.
Software
inDrops pipeline inDrops.py: https://github.com/indrops/indrops
scGESTALT pipeline: https://github.com/aaronmck/SC_GESTALT
R studio: https://www.rstudio.com
Seurat: https://satijalab.org/seurat/
PROCEDURE
Tol2 transposase mRNA synthesis TIMING 1 d
-
1
Linearize 5 μg of pCS2-zT2TP plasmid using NotI enzyme according to the manufacturer’s instructions.
-
2
Purify the linearized plasmid with a mini quick-spin DNA column (e.g. E.Z.N.A. Cycle Pure kit) according to the manufacturer’s protocol and elute in 50 μl nuclease-free water.
-
3In vitro transcribe Tol2 mRNA using mMESSAGE mMACHINE SP6 kit. Assemble the reaction below (20 μl total) and follow the manufacturer’s protocol.
Component Amount (μl) Final Concentration 2× NTP/CAP 10 1× 10× buffer 2 1× Linear, purified pCS2-zT2TP 6 ~20–30 ng/μl Enzyme mix 2 Total 20 -
4
Purify Tol2 mRNA using a mini quick-spin RNA column (e.g. E.Z.N.A. Total RNA kit I) according to the manufacturer’s protocol and elute in 50 μl nuclease-free water. Measure mRNA concentration using NanoDrop. Successful in vitro transcription reactions will yield mRNA at concentrations of 300–500 ng/μl. Dilute Tol2 mRNA to 100 ng/μl using nuclease-free water. PAUSE POINT Store the mRNA in 3 μl aliquots at −80 °C for at least one year.
-
5
Purify DNA from minipreps of pTol2-hspDRv7_scGstlt and pTol2-hsp70l:Cas9-t2A-GFP, 5×U6:sgRNA plasmids using a plasmid purification kit (e.g. E.Z.N.A. Plasmid DNA Mini Kit I). To remove residual RNase A, purify ~2 μg of miniprep DNA with a mini quick-spin DNA column (e.g. E.Z.N.A. Cycle Pure kit) according to the manufacturer’s protocol and elute in 30 μl nuclease-free water. Measure DNA concentrations using NanoDrop. Dilute each of the plasmids to 25 ng/μl using nuclease-free water.
PAUSE POINT Store diluted plasmids at −20 °C for at least one year.
Zebrafish transgene microinjection TIMING 3 h
-
6
The day prior to performing microinjections, set up 6 to 10 overnight tanks of wild type zebrafish for mating as described previously78.
CAUTION All vertebrate animal work must be performed in accordance with relevant guidelines and regulations.
-
7On the morning of microinjections, assemble reagent mixes (3 μl total) as follows for each plasmid. Vortex to mix, centrifuge briefly, and hold on ice.
Component Amount (μl) Final Concentration pTol2-hspDRv7_scGstlt OR pTol2-hsp70l:Cas9-t2A-GFP, 5×U6:sgRNA plasmid (25 ng/μl) 1.5 12.5 ng/μl Tol2 mRNA (100 ng/μl) 1.0 33.3 ng/μl Phenol red (0.1% wt/vol) 0.5 0.017% Total 3 -
8
Stagger mating of fish to enable several rounds of injections to be performed at the one-cell stage (~20–30 min after fertilization). For example, mating can be initiated in two tanks at once, followed by ~35 min-spaced intervals between each subsequent mating pairs.
-
9
While fish are mating, prepare an injection needle by breaking the tip of a pulled glass capillary with fine forceps. Load 1.5 μl of the pTol2-hspDRv7_scGstlt injection mix (Step 7) into the needle with a microloader tip and attach it to the piconozzle and manipulator. Set the pressure to 20 psi and calibrate the needle with a micrometer as described previously79.
-
10
Collect fertilized eggs into Petri dishes with blue water. Transfer ~100 one-cell stage embryos to the agarose ramp in the injection dish (distribute the embryos across 4 rows of the ramp), and add enough blue water to just cover the embryos. Pierce the chorion with the calibrated needle, move the needle into the cytoplasm, and inject 1 nl of the reagent mix. Repeat until all embryos are injected. Transfer the injected embryos with blue water into a new Petri dish and incubate at 28.5 °C. Keep some uninjected control embryos to assess the health of the clutch.
-
11
Repeat steps 8–10 for the pTol2-hsp70l:Cas9-t2A-GFP, 5×U6:sgRNA injection mix (Step 7).
-
12
At the end of the day, remove any unfertilized (embryos will appear as if still at single-cell stage rather than having undergone multiple rounds of cell division) or damaged (embryos are squashed or have become deformed from the injection procedure) embryos from the injected batches.
Sorting and rearing transgenic fish TIMING 1 d sort, 2–3 months rear
-
13
Heat shock fish injected with pTol2-hsp70l:Cas9-t2A-GFP, 5×U6:sgRNA at 24 hpf. Transfer 20–30 healthy looking embryos into a 1.5 ml microcentrifuge tube and add 500 μl of blue water. Use as many tubes as required depending on how many embryos survived injection (20–30 embryos per tube). Incubate the tubes in a heat block set at 37 °C for 30 min. Transfer heat shocked embryos back into Petri dishes with blue water and incubate at 28.5 °C. Alternatively, one can perform the heat shock in 50 ml tubes (with 5 ml blue water) in a water bath at 37 °C for 30 min.
-
14
GFP expression starts to become apparent ~2 h after heat shock and becomes bright enough for easy screening and sorting ~4 h after heat shock. Place the petri dish from Step 13 on the stage of a fluorescent microscope fitted with a GFP filter. Swirl the petri dish to bring the embryos to the center and turn on the fluorescent light. Look through the eyepiece to identify fish that exhibit mosaic GFP fluorescence throughout the body. Using a plastic or glass transfer pipette, transfer the brightest GFP positive fish into a new Petri dish containing 20 ml blue water. Incubate at 28.5 °C until 5 dpf and then transfer into 2 L tanks (8–10 larvae per tank) in a zebrafish facility to raise to adulthood. CRITICAL STEP: Seeding tanks at a low density will enable fish to grow and reach sexual maturity faster.
-
15
Screen and sort fish injected with pTol2-hspDRv7_scGstlt at 30 hpf. Check for bright GFP fluorescence in the heart using a fluorescent microscope. Using a plastic or glass transfer pipette, transfer GFP positive fish into a new Petri dish containing 20 ml blue water. Incubate at 28.5 °C until 5 dpf and then transfer into tanks (8–10 larvae per tank) in a zebrafish facility to raise to adulthood.
Screen for germline transmission TIMING 2 d, then 2–3 months to rear
-
16
Identify adult founder fish with germline integration by outcrossing individual injected F0 animals to wildtype fish of the opposite sex. Collect embryos the next morning in blue water and incubate at 28.5 °C. Keep potential founders separate until screening of clutches is complete.
-
17
Screen clutches from pTol2-hsp70l:Cas9-t2A-GFP, 5×U6:sgRNA F0 fish by heat shocking embryos at 24 hpf as described in Steps 13–14. The screened F1 are henceforth referred to as “hsp:Cas9, U6:sgRNA” transgenic line.
-
18
Screen clutches from pTol2-hspDRv7_scGstlt F0 fish by GFP heart expression at 30 hpf as described in Step 15. The screened F1 are henceforth referred to as “scGESTALT” transgenic line.
-
19
Incubate sorted F1 embryos at 28.5 °C until 5 dpf and then transfer into 2 L tanks (8–10 larvae per tank) in a zebrafish facility to raise to adulthood.
PAUSE POINT Maintain stable lines by outcrossing. Adults can be maintained for up to 2 years.
Determine copy number of scGESTALT barcode TIMING 1 d
-
20
Fin clip adult F1 pTol2-hspDRv7_scGstlt fish. Anesthetize fish in batches of 3–4 with 100 ml of 1× MESAB and snip a small section of the tail fin using fine scissors. Transfer the tail fin into a PCR strip tube and hold on ice. Transfer each fin clipped fish to individual 0.5 L tanks to recover and number them uniquely. Repeat until all F1s have been fin clipped.
-
21
Prepare genomic DNA using a modified HotSHOT method80. Add 50 μl of 50 mM NaOH to each fin clip and incubate at 95 °C for 20 min in a thermocycler.
-
22
Add 5 μl of 1M Tris-HCl pH 7.5 and vortex to mix. Centrifuge briefly. PAUSE POINT genomic DNA can be stored at 4 °C for at least 3 months.
-
23Set up qPCR reactions in triplicate. For each genomic DNA sample, set up qPCR reactions with two sets of primers, one for DsRed (to determine copy number of scGESTALT barcode) and another for an ultraconserved control region that is known to be present in 2 copies80. Thus, each sample will have a total of 6 reactions. In addition, set up qPCR with reference standards using genomic DNA from fish with no DsRed integration, and from fish with known single and/or double copy DsRed integrations.
Component Amount (μl) Final Concentration Biorad iTaq mix 10 1× Primer (F+R mix) 10μM 0.8 0.4 μM Nuclease-free water 7.2 Genomic DNA (1:5 dilution) 2 Total 20 Primer pairs: 1. qPCRctrl F+R 2. qPCRdsRed F+R (see DNA oligonucleotide sequences)
-
24Run qPCR as follows
Cycle number Denature Anneal/Extend Final 1 95 °C, 3 min 2–36 95 °C, 10 s 55 °C, 30 s (with fluorescence readout) 37 8 °C, hold -
25Calculate copy number with the equations below
-
Perform calculations for all unknown samples and positive and negative reference standards)
-
Perform calculations for all unknown samples and positive and negative reference standards. Δ(Ct, sample) is the Δ(Ct) for each sample and positive and negative reference standards calculated using the first equation. Use the Δ(Ct) of the known single copy reference standard as Δ(Ct, reference standard). Thus, ΔΔCt for the single copy reference standard will be 0.
-
Perform calculations for all unknown samples positive and negative reference standards. The value for the known single copy reference will be 1. The values for the known negative and double copy reference standards should be around 0 and 2 respectively.
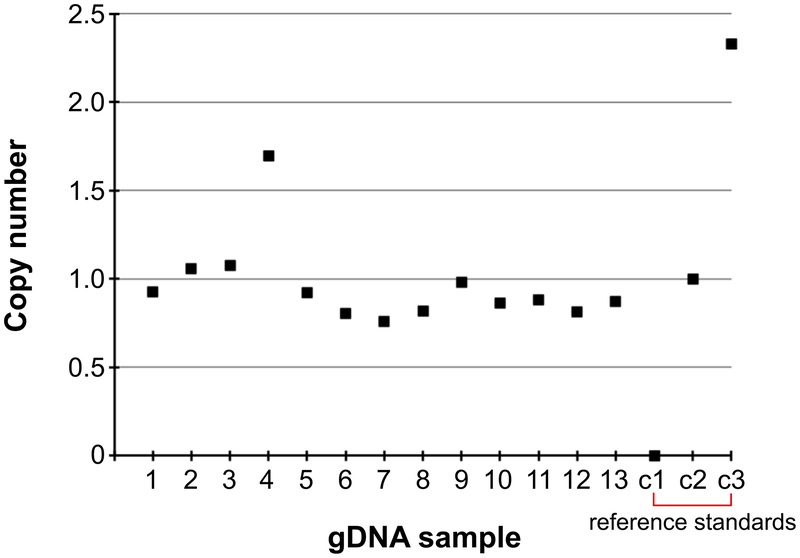
See ANTICIPATED RESULTS and Supplementary Data for examples of single-copy calculations
-
-
26
Keep single-copy scGESTALT F1 animals for all downstream experiments. Identify females and maintain them in separate 1 L tanks for barcoding experiments described below.
PAUSE POINT Maintain stable lines by outcrossing single-copy F1 animals. Adults can be maintained for up to 2 years.
scGESTALT sgRNA synthesis TIMING 2 h
-
27
Generate sgRNAs targeting sites 1–4 of the scGESTALT barcode using EnGen sgRNA synthesis kit according to the manufacturer’s guidelines. Oligonucleotide sequences are provided in the Reagents list (see DNA oligonucleotide sequences).
-
28
Purify sgRNAs with RNA Clean & Concetrator-25 kit according to the manufacturer’s guidelines and elute with 50 μl nuclease-free water.
-
29
Quantify sgRNAs using NanoDrop and dilute each sgRNA to ~350 ng/μl with nuclease-free water. Combine equal volumes (e.g. 5 μl) of sgRNAs 1–4 to make an equimolar pool. PAUSE POINT Store the sgRNA pool in 1.5 μl aliquots at −80 °C for at least one year. CRITICAL STEP Avoid freeze-thaw of sgRNA pools. Each aliquot is sufficient for 1 injection mix.
scGESTALT ‘early’ barcode editing TIMING 2 h
-
2
The day prior to performing microinjections, set up 4–6 overnight tanks with one scGESTALT F1 female and one hsp:Cas9, U6:sgRNA F1 male per tank for mating.
CRITICAL STEP Male hsp:Cas9, U6:sgRNA must be used in the ‘early and late’ barcoding experiments to avoid maternal contribution of Cas9 mRNA and sgRNAs, and enable barcode editing at later developmental timepoints. Female scGESTALT animals must be verified to have single-copy transgene integrations.
-
3On the morning of microinjections, assemble the following reagent mix (3.3 μl total). Vortex to mix and centrifuge briefly.
Component Amount (μl) Final Concentration Engen Cas9 NLS buffer 1 Phenol red (0.1%, wt/vol) 0.3 0.009% sgRNA pool (~350 ng/μl each) 1 ~106 ng/μl EnGen Cas9 NLS protein 1 Total 3.3 -
4
Incubate the mix at room temperature for 5 minutes to enable Cas9-sgRNA complexes to assemble. Hold the mix on ice until ready to start injections.
-
5
Cross scGESTALT and hsp:Cas9, U6:sgRNA fish as described in Step 8.
-
6
While fish are mating, prepare and calibrate needles for microinjection as described in Step 9. Load 1.5 μl of the Cas9-sgRNA injection mix into the needle.
-
7
Collect fertilized eggs into Petri dishes with blue water and arrange 1-cell stage embryos for injections as described in Step 10. Inject 1.5 nl of Cas9-sgRNA mix through the chorion.
-
8
Transfer injected embryos with blue water into a new Petri dish and incubate at 28.5 °C. Keep some uninjected control embryos to assess the health of the clutch. At the end of the day, remove any unfertilized or damaged embryos from the injected batches.
scGESTALT ‘late’ barcode editing TIMING 6 h
-
2
At 30 hpf, screen embryos from Step 36 for heart GFP expression as described in Step 15 to identify ones carrying the scGESTALT transgene (~50% of injected progeny).
-
3
Heat shock the sorted embryos for 30 min at 37 °C as described in Step 13.
-
4
Screen embryos 2–4 hours after heat shock for ubiquitous GFP expression as described in Step 14 to identify ones carrying the hsp:Cas9, U6:sgRNA transgene.
Rearing double transgenic fish TIMING ~25 d
-
2
Incubate sorted double transgenic embryos from Step 39 (~25% of injected progeny) at 28.5 °C until 5 dpf and then transfer into 2 L tanks (8–10 larvae per tank) in a zebrafish facility to raise to juveniles (23–25 dpf).
PAUSE POINT The rearing time is user-dependent. Animals can be used in experiments at earlier stages or grown to adults.
Identify barcode edited juveniles TIMING 1 d
-
30
The day prior to performing droplet encapsulation for scRNA-seq, identify individual juveniles from Step 40 with edited scGESTALT barcodes. Fin clip juveniles as described in Step 20. Transfer each fin clipped fish to individual 0.5 L tanks to recover and number them uniquely.
-
31
Prepare genomic DNA as described in Steps 21–22.
-
32Set up PCR reactions (10 μl) as follows
Component Amount (μl) Final Concentration 5× HF Phusion buffer 2 1× 10 mM dNTP 0.2 0.2 mM Nuclease-free water 6.2 scGESTALT primer mix (10 μM) 0.5 0.5 mM Phusion polymerase 0.1 Genomic DNA 1 Total 10 -
33Run the following program on a thermocycler
Cycle number Denature Anneal Extend Final 1 98 °C, 30 s 2–34 98 °C, 10 s 63 °C, 20 s 72 °C, 20 s 35 72 °C, 3 min 36 8 °C, hold -
34
Load PCR products on 1% agarose gel along with 100 bp DNA ladder according to standard procedures.
-
35
Run the gel and visualize bands. Successfully edited scGESTALT barcodes will appear as smears ranging from ~120–250 bp. Generally, >80% of injected embryos are efficiently edited. See ANTICIPATED RESULTS for examples of successful editing.
?Troubleshooting
-
36
Select 2–4 juveniles showing sufficient barcode editing for inDrops experiments the next day, and keep each juvenile in a separate 0.5 L tank overnight.
Prepare single-cell suspension of whole brain TIMING 2.5 h
-
9
The next day, pre-warm 40 ml zebrafish facility water in a water bath at 37 °C in a 50 ml tube.
-
10
Heat shock one 23–25 dpf juvenile selected in Step 47 to induce expression of the barcode array. Incubate the animal in a 50 ml tube with 20 ml of pre-warmed fish facility water at 37 °C for 40 min. This will promote expression of scGESTALT mRNA barcodes just prior to inDrops encapsulation. While the fish is being heat shocked, proceed with preparing dissociation solutions as described below (Steps 50–54).
-
11
Transfer 25 ml Neurobasal media to a 50 ml tube. Add 500 μl B27 supplement. Oxygenate the solution by passing 95%O2:5%CO2 gas through a tubing attached to a sterile 5 ml serological pipette for 2 min (Supplementary Fig. 1). Hold Neurobasal/B27 media on ice.
-
12
Prepare papain solution for dissociation. Add 5 ml Neurobasal media to Vial 2 (papain) of the Papain Dissociation System (Worthington). Add 0.5 ml Neurobasal media to Vial 3 (DNase). Gently resuspend the DNase and transfer 250 μl to Vial 2. Mix the papain/DNase solution by inverting the vial several times.
-
13
Oxygenate the papain/DNase mix by passing 95%O2:5%CO2 gas through a tubing attached to a sterile 5 ml serological pipette for 2 min (Supplementary Fig. 1 and Supplementary Movie). CRITICAL STEP Do not bubble gas directly into the solution as it will cause frothing and protein denaturation, and reduce papain activity. Instead, slowly move the pipette in a circular motion above the liquid’s surface being careful not to cause the liquid to spill over (adjust the gas pressure if necessary).
-
14
Prepare ovumucoid protease inhibitor (Vial 4) as described by the manufacturer.
-
15
Incubate papain/DNase mix in a 34 °C water bath while preparing the single-cell brain suspension as described below (Steps 55–61).
-
16
Anesthetize the heat shocked juvenile from Step 49 by adding 800 μl 25× MESAB.
-
17
Add 1 ml DPBS (containing calcium, magnesium, glucose, pyruvate) to a 1.5 ml microfuge tube and hold on ice.
-
18
Add 10 ml ice-cold Neurobasal/B27 media to a 10-cm sylgard dish. Add 400 μl 25× MESAB.
-
19
Transfer the anesthetized fish to the sylgard dish (Supplementary Fig. 2). Pin the fish just posterior of the head, in the middle of the trunk and near the tail using 3 insect pins.
-
20
Using a pair of fine forceps, remove the jaw, eyes, heart and gut tissues. Pierce the skin on top of the head and gently peel it back to expose the brain. Using fine forceps, gently scoop the brain out taking care not to lose part of the hindbrain in the process. Rip the brain tissue into 6–8 small pieces using two pairs of fine forceps. Typically, dissection takes ~5 min per brain.
-
21
Transfer the brain tissue pieces to the 1.5 ml microcentrifuge tube with DPBS from Step 56.
-
22
Briefly spin down the tube using a benchtop minifuge (2 second spin at room temperature). Discard DPBS and add 1 ml of ice-cold DPBS (no calcium, no magnesium). Hold the tube at room temperature.
-
23
Remove the papain/DNase mix from the water bath (Step 54). Transfer 1 ml of the papain/DNase solution to a new 1.5 ml tube and add 1 ml Neurobasal media. This dilutes the papain to 10 units/ml. Note the concentration of papain can be adjusted if needed (generally 10–20 units/ml are equally effective for dissociation).
-
24
Discard DPBS from the brain tissue pieces (step 61) and add 1 ml of the diluted papain/DNase mix from Step 62. Transfer the entire contents of the 1.5 ml microcentrifuge tube to a 15 ml tube, and oxygenate the solution as described in Step 52.
CRITICAL STEP Re-oxygenation maintains cell viability during dissociation.
-
25
Incubate the brain tissue pieces in a 34 °C water bath for 20 min.
-
26
Gently triturate the tissue 10 times with a p1000 pipette set at 800 μl.
CRITICAL STEP Do not triturate too vigorously and avoid introducing bubbles. Generally, pipetting at a rate of 1 sec per up and down motion of the pipette is desirable.
-
27
Re-oxygenate the solution as described in Step 52 for 1 min. Incubate in a 34 °C water bath for an additional 5–6 min.
-
28
Gently triturate the tissue 15–20 times with a p1000 pipette as described in Step 65. CRITICAL STEP Typically, this range of trituration is sufficient for efficient tissue dissociation. However depending on the strength of the papain, if large pieces of tissues are still visible, it may be necessary to perform additional pipetting or increase incubation time by 5–10 min. If longer incubation time at 34 °C is necessary, repeat Step 66 just prior to re-incubation.
-
29
Transfer the entire contents of the 15 ml tube to a 1.5 microcentrifuge tube. Spin at 500 g for 4 min at 4 °C.
-
30
While tube is spinning, prepare inhibitor solution as follows. Transfer 0.9 ml EBSS (Vial 1) solution to a 15 ml tube. Add 100 μl of resuspended ovumucoid protease inhibitor from Step 53. Add 50 μl of resuspended DNase (Vial 3) from Step 51. Oxygenate the mix as described in Step 52 and hold on ice.
-
31
Following centrifugation in Step 68, discard the papain supernatant. A clear cell pellet should be visible. Resuspend the pellet with 1 ml of the prepared inhibitor solution.
-
32
Spin at 500 g for 4 min at 4 °C.
-
33
Discard the supernatant and gently resuspend the pellet with 1 ml ice-cold DPBS (no calcium, no magnesium).
-
34
Sequentially filter the resuspension first through a 35 μm cell strainer, followed by a 20 μm cell strainer. Perform all steps on ice with pre-chilled tubes.
-
35
Spin at 500 g for 4 min at 4 °C.
-
36
Discard the supernatant and add 500 μl ice-cold DBPS (no calcium, no magnesium). Do not pipette the pellet. This step is to remove any residual debris.
-
37
Spin at 500 g for 4 min at 4 °C.
-
38
Discard the supernatant and resuspend the pellet with 400 μl ice-cold DPBS (no calcium, no magnesium). Hold on ice.
-
39
Determine cell concentration and viability. Add 10 μl of the cell suspension to 10 μl of Trypan blue in a 1.5 ml microcentrifuge tube. Mix by gentle pipetting and load 10 μl onto a hemocytometer (e.g. Bulldog Bio). Count cells and determine the concentration as described by the manufacturer. Cell viability should be >70% (ideally >80%).
CRITICAL STEP High cell viability is required to proceed with scRNA-seq experiments. Poor viability will result in fewer cells passing data quality filtering steps and can introduce more ambient RNA (i.e. non-cell associated RNA, such as free floating RNA from dead or dying cells) into cell suspension, compromising single-cell measurements.
?Troubleshooting
-
40
Dilute cells to ~200,000 cells/ml with DPBS (no calcium, no magnesium).
-
41
Add 300 μl 36% (vol/vol) OptiPrep to 300 μl diluted cells and gently resuspend the cell suspension. Final cell concentration is 100,000 cells/ml in 18% optiprep/DPBS (vol/vol). Proceed immediately to inDrops cell encapsulation.
inDrops single-cell encapsulation TIMING 30–60 min
-
5
Run the inDrops device as described previously73. Depending on the desired number of cells to be profiled, cells can be collected at a rate of 10,000–20,000 cells per hour. Generally, single-cell transcriptomes are obtained for ~70% of cells introduced into the device.
Reverse transcription in droplets TIMING 3 h
-
3
Proceed to reverse transcription as described previously73. If more than 3,000 cells were captured, split the emulsion into fractions containing ~3,000 cells.
PAUSE POINT: Libraries post-reverse transcription can be stored at −80 °C for at least 3 months
inDrops transcriptome library preparation TIMING 2 d
-
37
Generate single-cell transcriptome libraries as described previously73 with the following modification. After in vitro transcription (IVT, for linear amplification) of the second-strand synthesis product, purify the reaction with 1.3× volume of AMPure beads according to the manufacturer’s instructions, and elute with 25 μl RNA elution buffer.
-
38
Use 9 μl of the purified post-IVT product to proceed with fragmentation of amplified RNA and complete library preparation as described previously73.
PAUSE POINT Store remaining unfragmented RNA at −80 °C for at least 3 months.
scGESTALT library preparation: reverse transcription TIMING 2.5 h
-
42Assemble the following reaction (11 μl total) on ice using unfragmented RNA from Step 84.
Component Amount (μl) Final Concentration Unfragmented RNA 5 50 μM random hexamer 1.5 3.75 μM 10 mM dNTP 1 0.5 mM Nuclease-free water 3.5 Total 11 -
43
Vortex the mixture and briefly centrifuge. Incubate the reaction at 70° C (lid 105 °C) for 3 min and immediately cool on ice.
-
44Add the following reverse transcription reagents on ice and mix by pipetting (20 μl total). Briefly spin down.
Component Amount (μl) Final Concentration Mixture from Step 86 11 5× PrimeScript buffer 4 1× Nuclease-free water 3.5 RNaseOut (40 U/μl) 1 2 U/μl PrimeScript RT enzyme (200 U/μl) 0.5 5 U/μl Total 20 -
45
Incubate the reaction mix in a thermocycler at 30 °C for 10 min, followed by 42 °C for 1 h, and then 70 °C for 15 min.
-
46
Purify the reverse transcription product with 1.2× volume AMPure beads (24 μl) and elute with 20 μl DNA elution buffer.
PAUSE POINT Purified cDNA can be stored at −20 °C for at least 3 months.
scGESTALT library preparation: PCR round 1 TIMING 1.5 h
-
6Set up a PCR reaction (25 μl) for targeted amplification of scGESTALT barcodes as follows
Component Amount (μl) Final Concentration 5× Q5 reaction buffer 5 1× 10 mM dNTP 0.5 0.2 mM 10 μM inDrops_GP6 1.25 0.5 μM 10 μM inDrops_PE1Sa 1.25 0.5 μM cDNA from Step 89 7.5 Q5 DNA polymerase 0.25 Nuclease-free water 4.25 Q5 enhancer 5 Total 25 CRITICAL STEP This PCR amplifies a longer scGESTALT fragment using a forward primer that binds in the DsRed transgene upstream of the scGESTALT barcode. We have found that using a nested PCR strategy improves specificity of amplification.
-
7Run the following program on a thermocycler
Cycle number Denature Anneal Extend Final 1 98 °C, 30 s 2–16 98 °C, 10 s 61 °C, 25 s 72 °C, 30 s 17 72 °C, 2 min 18 8 °C, hold -
8
Add 25 μl nuclease-free water to the PCR product and purify with 0.6× volume AMPure beads (30 μl). Elute the product with 20 μl DNA elution buffer.
PAUSE POINT Purified PCR product can be stored at −20 °C for at least one week.
scGESTALT library preparation: PCR round 2 TIMING 1.5 h
-
4Set up a second PCR reaction (25 μl) for targeted amplification of scGESTALT barcodes as follows
Component Amount (μl) Final Concentration 5× HF Phusion reaction buffer 5 1× 10 mM dNTP 0.5 0.2 mM 10 μM inDrops_GP12 1.25 0.5 μM 10 μM inDrops_PE1Sb 1.25 0.5 μM PCR product from Step 92 8 Phusion DNA polymerase 0.25 Nuclease-free water 8.75 Total 25 CRITICAL STEP This PCR amplifies the desired scGESTALT fragment using a forward primer that binds 5′ of the scGESTALT barcode sequence. The primer contains 9×N sequences, where N is a random base, to introduce sequence variety.
-
5Run the following program on a thermocycler
Cycle number Denature Anneal Extend Final 1 98 °C, 30 s 2–9 98 °C, 10 s 60 °C, 25 s 72 °C, 30 s 16 72 °C, 2 min 17 8 °C, hold -
6
Add 25 μl nuclease-free water to the PCR product and purify with 0.6× volume AMPure beads (30 μl). Elute the product with 15 μl DNA elution buffer.
PAUSE POINT Purified PCR product can be stored at −20 °C for at least one week.
-
7
Check concentration of 1 μl of eluate using Qubit according to the manufacturer’s guidelines. Generally, concentrations range from 1–2.5 ng/μl. If concentrations are substantially higher, repeat steps 93–94 with lower DNA input and/or fewer PCR cycles.
scGESTALT library preparation: final PCR TIMING 1.5 h
-
39Set up a PCR reaction (25 μl) for final library amplification as follows
Component Amount (μl) Final Concentration 2× KAPA HiFi HotStart PCR mix 12.5 1× Eluate from step 95 (1:10 dilution) 5 R1-PCRix/ R2-PCR primer mix (5 μM) 2.5 0.5 μM Nuclease-free water 5 Total 25 CRITICAL STEP If multiplexing libraries, use different R1-PCRix/R2-PCR primer mix variants. For each scGESTALT library, ensure that the same primer pairs as those used for building the transcriptome libraries are used. For example, if multiplexing two libraries (e.g. A and B) in the same sequencing run, use R1-PCRix1/R2-PCR (for library A) and R1-PCRix2/R2-PCR (for library B) for both transcriptome and scGESTALT sequencing.
-
40Run the following program on a thermocycler
Cycle number Denature Anneal Extend Final 1 98 °C, 30 s 2–3 98 °C, 20 s 55 °C, 30 s 72 °C, 30 s 4–9/11a 98 °C, 20 s 65 °C, 30 s 72 °C, 30 s 12 72 °C, 2 min 13 8 °C, hold aThe number of cycles performed at annealing temperature of 65 °C typically ranges from 6–8. This number can also be determined empirically by qPCR as described previously70. -
41
Add 25 μl nuclease-free water to the PCR product and purify with 0.6× volume AMPure beads (30 μl). Elute the product with 25 μl RNA elution buffer. The eluate is the final sequencing-ready library.
PAUSE POINT Libraries can be stored at −20 °C for at least a month.
-
42
Check concentration of 1 μl of eluate using Qubit Generally, concentrations range from 2.5–6 ng/μl.
scGESTALT library preparation: library quality check TIMING 2 h
-
47
Run a BioAnalyzer DNA HS assay to determine library size with 1 μl of eluate from Step 99. See ANTICIPATED RESULTS for examples of successful libraries.
-
48
Determine the molar concentration of the library. Dilute the library to 10 nM. If sequencing multiple libraries together, combine equal volumes of libraries (each at 10 nM) to be multiplexed in 1.5 ml low adhesion microcentrifuge tubes.
PAUSE POINT Libraries can be stored at −20 °C for at least a month or at −80 °C for a longer period.
Sequencing transcriptome and scGESTALT libraries TIMING 1 d
-
9
Sequence transcriptome libraries using NextSeq 75 cycle high-output kit with standard Illumina primers.
Read 1 (transcript read) is 61 cycles
Index 1 (part 1 of inDrops cell barcode) is 8 cycles
Index 2 (library index) is 8 cycles
Read 2 (part 2 of inDrops cell barcode [8bp,] followed by 6 bp UMI) is 14 cycles
-
10
Sequence scGESTALT libraries using MiSeq 300 cycle kits and 20% PhiX spike-in with standard Illumina primers.
Read 1 (scGESTALT lineage barcode) is 250 cycles
Index 1 (part 1 of inDrops cell barcode) is 8 cycles
Index 2 (library index) is 8 cycles
Read 2 (part 2 of inDrops cell barcode [8bp,] followed by 6 bp UMI) is 14 cycles
Sequencing data processing TIMING 1 d
-
8
Process raw transcriptome reads as described previously73. The inDrops bioinformatics pipeline (inDrops.py) is available at https://github.com/indrops/indrops. Bowtie version1.1.1 is used with parameter −e 200; UMI quantification is used with parameter −u 2.
-
9
Process scGESTALT reads in parallel using the inDrops.py script and stop just prior to the transcriptome mapping step. Modify the Trimmomatic settings to LEADING: “10”; SLIDINGWINDOW: “4:5”; MINLEN: “16”. This will generate files with inDrops cell identifiers and the scGESTALT lineage barcode sequence. The data can be further processed using the scGestaltPrepFunc.R script in Supplementary Software 2 or a similar custom script.
-
10
Process the resulting files using the pipeline available at https://github.com/aaronmck/SC_GESTALT (Supplementary Software 1). This will generate tables containing the lineage barcode sequence for each cell.
Downstream data processing TIMING 3 d
-
43
Using Seurat (or related scRNA-seq analysis tool), identify cell clusters (i.e. different cell types) using the transcriptome scRNA-seq data (for tutorial refer to: https://satijalab.org/seurat/). In brief, gene expression profiles are used to perform dimensionality reduction using principal component analysis with highly variable genes. A modularity-based clustering algorithm (Louvain) is used to cluster cells into discrete cell types using significant principal components. The result is visualized in two dimensions on a t-distributed stochastic neighbor embedding (t-SNE) plot (Fig. 1). For further details of how clustering is performed for the zebrafish juvenile brain dataset, we refer the reader to the Methods section of ref72. See ANTICIPATED RESULTS for examples of clustering analyses from zebrafish brain scRNA-seq.
-
44
Match transcriptome (from Step 108) and lineage barcodes (from Step 107) for each cell using the inDrops cell identifier index sequence (Supplementary Software 3–4) and generate lineage trees (based on maximum parsimony) using the pipeline available at https://github.com/aaronmck/SC_GESTALT. See ANTICIPATED RESULTS for an example lineage tree.
TIMING
Zebrafish transgenesis
Steps 1–5, Tol2 mRNA synthesis: 1 d
Steps 6–12, microinjection: 1 d
Steps 13–15, screening and rearing fish: 2–3 months
Steps 16–19, germline transmission: 2 d, wait for adults 2–3 months
Steps 20–26, copy number determination: 1d
scGESTALT barcode editing
Steps 27–29, sgRNA synthesis: 2 h
Steps 30–37, early timepoint Cas9 editing: 2 h
Steps 38–39, late timepoint Cas9 editing: 6 h
Step 40, growing to juveniles: 25 d
Steps 41–47, identifying edited fish: 1d
inDrops transcriptome and lineage profiling
Steps 48–80, single-cell processing of brain tissue: 2.5 h
Step 81, cell encapsulation: 30–60 minStep
82, reverse transcription: 3 h
Steps 83–84, transcriptome library preparation: 2 d
Steps 85–89, scGESTALT reverse transcription: 2.5 h
Steps 90–92, scGESTALT PCR round 1: 1.5 h
Steps 93–96, scGESTALT PCR round 2: 1.5 h
Steps 97–100, scGESTALT final library PCR: 1.5 h
Steps 101–102, check library with BioAnalyzer: 2 h
Sequencing and raw data processing
Steps 103–104, Miseq and NextSeq runs: 1 d
Steps 105–107, raw data processing: 1d
Downstream data processing
Steps 108–109, clustering analysis: 3 d (preliminary analysis)
TROUBLESHOOTING
Extensive troubleshooting pointers for running the inDrops device and preparing inDrops transcriptome libraries have been described previously73.
There are two main parts in the scGESTALT protocol where issues may arise.
No or inefficient editing of the barcode (Step 46).
This usually indicates problems with the editing reagents. Ensure that the Cas9 protein used for injection is active by testing it for successful editing of endogenous genes e.g. tyr gene encoding tyrosinase results in loss of pigmentation by 48 hpf when mutated (see ref81 for details and tyr sgRNA sequences). If Cas9 is active, then the sgRNA pool is likely compromised. Remake the sgRNAs, aliquot in single-use amounts and store at −80 °C. Do not freeze/thaw sgRNAs and discard each aliquot after use. After heat shock of the embryos at 30 hpf, GFP expression is used as a proxy for successful Cas9 transgene induction. If weak or no GFP is observed, it indicates that heat shock was not successful. Test the heat shock conditions empirically and if necessary increase time of heat shock by 10 min. Do not heat shock for >45 min as it can result in large deletions in the lineage barcode array. Alternatively, screen F1 transgenic animals to identify ones with robust GFP expression following heat shock.
Low cell viability (Step 78).
High quality single-cell preparations are critical for obtaining successful single-cell transcriptome and scGESTALT libraries. If cell viability is <70%, repeat dissociation with another sample. Ensure that all media is properly oxygenated (this is the most critical part of the protocol), especially during the papain incubation at 34 °C (steps 63–64). Do not incubate tissue for >30 min at 34 °C and do not triturate the sample vigorously after papain digestion. Gently pipette cells during DBPS washes and final resuspension.
ANTICIPATED RESULTS
The protocol described herein enables the user to apply CRISPR-Cas9 for cumulative, combinatorial and heritable genomic barcode editing at multiple timepoints to generate permanent records of cell lineages during development. As the mutated barcodes are also expressed as mRNA, both lineage information and cell type identity can be simultaneously extracted from single cells at high throughput using droplet-based scRNA-seq. Cell relationships at the level of gene expression and lineages can then be represented on lineage trees, which can be explored to identify timing and patterns of lineage segregation within or between tissue regions and cellular subtypes.
Below, we describe results obtained at various stages of the protocol that can be used to gauge the experiment’s success.
Barcode copy number determination by qPCR (Step 25).
The number of integrations of the scGESTALT lineage barcode in transgenic F1 can be determined by qPCR. Figure 4 shows representative qPCR results from this assay. Animals with a calculated copy number of less than or equal to 1 (below 0.6 are not considered integrants) are considered to be single integrants, and can be used for downstream experiments and further expanded to maintain single-copy transgenic lines. Raw data for the calculations are included in Supplementary Data.
Figure 4. Barcode copy number determination.
Copy number of scGESTALT barcode integration was determined using qPCR. Samples 1–13 represent different genomic DNA samples (n = 13 animals). Reference standards: c1 is negative control (n = 1 animal), c2 is known to contain 1 copy (n = 1 animal), c3 is known to contain 2 copies (n = 1 animal). This procedure was approved by the HU/FAS Committee on the Use of Animals in Research & Teaching under Protocol No. 25–08.
Identification of successful barcode editing (Step 46).
Genomic DNA from edited fish is extracted, and lineage barcodes are PCR amplified and visualized on an agarose gel. As shown in Figure 5, band patterns that appear as smears ranging from 120–250 bp are indicative of efficiently edited barcodes. Depending on whether barcodes were edited at only ‘early’ timepoints by injections, only ‘late’ timepoints by Cas9 induction, or both ‘early and late’ timepoints by combination of Cas9 injection and induction, the gel band patterns have slight distinctions. If the pattern is dominated by short bands at ~100–150bp, it is advisable to avoid using the corresponding animal in scRNA-seq experiments, since it is likely that this individual’s barcode edits will be predominantly large deletions, reducing the diversity of edits and the resolution of lineage trees. In addition, since Cas9-induced mutations are stochastic, gel band patterns between animals that were edited in a similar manner will be variable.
Figure 5. scGESTALT barcode editing.
scGESTALT barcode zebrafish were crossed to zebrafish that express heat shock-inducible Cas9 and U6-driven sgRNAs 5–9. Resulting embryos were injected with Cas9 protein and sgRNAs 1–4 at the one-cell stage. Embryos were heat shocked at 30 hpf to induce transgenic Cas9 for a late round of editing. Double transgenic (scGESTALT+, hsp:Cas9+; lanes 2–8, n = 7 embryos) and single transgenic (scGESTALT+, hsp:Cas9−; lanes 9–12, n = 4 embryos) were identified by screening for GFP expression. The gel shows PCR results of amplifying the scGESTALT barcode (unedited = ~300 bp). Large smear patterns (120–250bp) are observed in early and late edited embryos (lanes 2–8), whereas embryos that were only mutated at sites 1–4 display less editing (lanes 9–12. The band at ~200 bp in lane 12 likely represents large deletion(s) between sites 1–4 that occurred early in development and was inherited by most cells. Note that samples with such dominant large deletions should not be used for downstream experiments and analyses as they are likely to have low barcode diversity). Sample in lane 11 was likely not efficiently injected. Lane 1 represents a control embryo, which was injected with Cas9 protein only (no sgRNAs 1–4, n = 1 embryo) and was not heat shocked. As expected, the barcode is not edited in this case. This procedure was approved by the HU/FAS Committee on the Use of Animals in Research & Teaching under Protocol No. 25–08.
BioAnalyzer electropherograms of successful scGESTALT libraries (Step 101).
The profiles of amplified lineage barcode libraries are shown in Figure 6. A successful library typically ranges in size between 500–800bp with an average of 600–650 bp. Some larger fragments may be observed, but are generally not problematic for sequencing. Alternatively, the ratio of AMPure beads at steps 95 and 99 can be increased (e.g. to 0.8× or 1×) to remove those fragments. If fragments are considerably smaller than this range, it may indicate that the barcode contains large deletions. Refer to the previous section to determine how to avoid using animals with undesirable barcode mutation patterns.
Figure 6. BioAnalyzer electropherograms of scGESTALT sequencing libraries.
Traces were obtained from 23–25 dpf juvenile zebrafish brains (n = 1 animal). The single-cell emulsion (step 82) from one inDrops collection was split into 4 fractions of ~3,000 cells each, and the resulting libraries were indexed uniquely. The average size of the libraries is 600–650bp. Peaks at 35 and 10380 bp represent gel migration markers. This procedure was approved by the HU/FAS Committee on the Use of Animals in Research & Teaching under Protocol No. 25–08.
Detailed discussion of results from inDrops transcriptome libraries have been described before73. It is worth noting that zebrafish neural cells have low RNA content, with estimates in the 1–2 picogram range23. This is still sufficient for scRNA-seq profiling and BioAnalyzer electropherograms display similar profiles as previously described for inDrops libraries from other cell types73.
Clustering analyses and lineage trees (Steps 108–109).
The digital gene expression matrix output (rows contain gene counts and columns represent single cells) from the inDrops processing pipeline is used to identify cell types using any relevant scRNA-seq computational tool. We will use Seurat18 as an example for the rest of this section. First, the matrix is column-normalized and log-transformed (see tutorial at https://satijalab.org/seurat/). Next, low quality single-cell libraries are filtered out from the dataset. The metrics for what constitutes ‘low quality’ will vary between cell types, tissues, dissociation protocols and sequencing depth. For zebrafish brain data, we filtered cells with fewer than 500 expressed genes or greater than 9% mitochondrial content72. Potential cell doublets or multiplets should also be filtered out (e.g. by identifying cells with high numbers of gene counts and unique molecular identifiers that are outliers of a normal distribution). Next, highly variable genes are identified and used for principal component analysis. The top significant principal components are used for dimensionality reduction and for grouping all cells into distinct clusters (using a Louvain modularity algorithm). Clusters are expected to be supported by cells from multiple biological replicates, otherwise it may indicate technical artefacts in the data. The resulting clusters can be compared to each other to identify differentially expressed genes, which may serve as gene markers for individual cell types. Clusters can be classified using the identified gene markers and validated with in situ hybridizations.
To generate scGESTALT lineage trees, the inDrops index sequences are used to match transcriptomes and lineage barcodes for the same cells. Cell lineage trees are generated using maximum parsimony (adapted from phylogenetics) based on patterns of shared edits (for more details see refs66,72). To represent early and late editing, the tree is anchored with edits that occur at sites 1–4 of the barcode array (early editing from injections at the 1-cell stage), and is then extended with additional edits accumulated at sites 5–9 (late editing from heat shock-induced Cas9 activity). A clade on the tree represents all lineage barcodes that share at least one common edit, and sub-clades that branch from the original clade contain increasingly restricted subsets of barcodes that contain the previous edit(s) as well as additional shared edits (Fig. 1, Fig. 7). Individual cells (identified by their inDrops index) containing each of the recovered barcodes is connected to the tips of the tree (i.e. the terminal lineage barcode sequence). Cells connected to the same terminal node are clonal (i.e. contain the same lineage barcode). Cell annotations (e.g. cell type and spatial/regional origin) can be color coded and added to the tree to explore various lineage relationships, such as to determine the relationship between cells belonging to the same cluster or cells from different regions of the tissue.
Figure 7. Zebrafish brain lineage tree generated using scGESTALT.
An example of a reconstructed lineage tree from a single juvenile zebrafish brain. 376 edited barcodes were recovered from single cells using inDrops. A cell lineage tree was generated from the barcodes based on shared edits using a maximum parsimony approach. Black nodes represent early barcode edits (Cas9 and sgRNA injection at 1-cell stage, Step 35); red nodes represent late edits (heat shock-induced Cas9 transgene expression, Step 38). Dashed lines join single cells to terminal nodes (represent the final edited barcode sequence) on the tree. Distinct cell types (identified from simultaneous transcriptome capture and cell clustering analyses) are color coded as indicated in the legend. The edited barcode for each cell is shown as a white bar with deletions (red) and insertions (blue). Examples of clades and subclades are indicated on the tree. A clade on the tree represents all lineage barcodes that share at least one common edit, and sub-clades that branch from the original clade contain increasingly restricted subsets of barcodes that contain the previous edit(s) as well as additional shared edits. Adapted with permission from ref72. This procedure was approved by the HU/FAS Committee on the Use of Animals in Research & Teaching under Protocol No. 25–08.
Supplementary Material
Supplementary Data. Raw data used for calculation of scGESTALT lineage barcode copy by qPCR. The number of integrations of the scGESTALT barcode in transgenic F1 is determined by qPCR. Genomic DNA from 13 animals (gDNA 33–50) was used in the assay. Genomic DNA from an animal with no barcode integration (control zero), an animal with known single copy integration (control single), and an animal with known double copy integration (control double) are included as standards. For each genomic DNA sample, qPCR reactions are set up in triplicate using primers for an ultraconserved control region that is known to be 2 copies, and for the DsRed region to determine copy number of the scGESTALT lineage barcode. The Ct number for each reaction is shown. The average Ct for each region is calculated (Ctrl Ct and DsRed Ct). The delta Ct and delta delta Ct are calculated and used to determine copy number (refer to Procedure Step 25 for details). Animals with a calculated copy number of less than or equal to 1 (but greater than or equal to 0.6) are considered to be single integrants.
Supplementary Figure 1. Papain oxygenation setup. Left and middle panels, 95%O2: 5%CO2 gas tank fitted with a gas regulator, Tygon E-3603 tubing and a 5 ml serological pipette. Right panel, Oxygenation of papain/DNase mix in Neurobasal media (small vial to the right) is performed by bubbling 95%O2: 5%CO2 gas through tubing attached to a sterile 5 ml serological pipette for 2 min (Procedure Step 52). EBSS buffer (large vial to the left, used for resuspending ovumucoid, Procedure Step 53) and Neurobasal media (Procedure Step 50) are oxygenated in a similar manner.
Supplementary Figure 2. Zebrafish brain dissection. Top panels, Anesthetized fish is transferred to a sylgard dish covered with Neurobasal media and MESAB (left). The fish is pinned just posterior of the head, in the middle of the trunk and near the tail using 3 insect pins (right, asterisks mark pin positions). Bottom panels, The jaw, eyes, heart and gut tissues are removed. The skin on top of the head is pierced and peeled back to expose the brain (left, circle marks the exposed brain). Gently scoop the brain out taking care not to lose part of the hindbrain in the process (right, whole brain is encircled).
Supplementary Movie. Papain/DNase mix oxygenation. Demonstration of the proper method for oxygenation of the papain/DNase mix.
Supplementary Software 1. scGESTALT analysis pipeline scripts. This is the master pipeline for processing scGESTALT reads and generating lineage trees (Fig. 1 and Fig. 7, relevant for Step 107).
Supplementary Software 2. R script (scGestaltPrepFunc) for pre-processing of inDrops scGestalt data. The script will format the data for input to the scGestalt analysis pipeline (relevant for Step 106).
Supplementary Software 3. R script pipeline (Transcriptome-scGestaltMatchPipe) for matching transcriptome and scGESTALT lineage barcodes for profiled single cells (relevant for Step 109).
Supplementary Software 4. R script (MatchPipeFunc) containing the code of the R functions called by the scGestaltMatchPipe pipeline (relevant for Step 109).
Supplementary Table 1. Sequences of scGESTALT CRISPR target sites.
Supplementary Table 2. InDrops library multiplexing primer sequences.
ACKNOWLEDGEMENTS
We thank Daniel E. Wagner, Aaron McKenna and Shristi Pandey for discussion and advice. This work was supported by a postdoctoral fellowship from the Canadian Institutes of Health Research to B.R., NIH grants U01MH109560, R01HD85905 and DP1 HD094764 to A.F.S., and an Allen Discovery Center grant to A.F.S.
Footnotes
TWEET #CRISPRCas9 lineage recording and #scRNAseq to build single-cell resolved vertebrate lineage trees @bushranraj @schierlab
COVER TEASER Single-cell lineage tracing during development
Please indicate up to four primary research articles where the protocol has been used and/or developed.
1. Raj et al., Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. 2018. Nature Biotechnology. 10.1038/nbt.4103
2. McKenna et al., Whole-organism lineage tracing by combinatorial and cumulative genome editing, 2016. Science. 10.1126/science.aaf7907
COMPETING INTERESTS
The authors declare no competing interests.
CODE AVAILABILITY
scGESTALT computational scripts and analysis pipeline are available at https://github.com/aaronmck/SC_GESTALT and are included as Supplementary Software 1 with this protocol.
DATA AVAILABILITY
Figure 4 has associated raw data (Supplementary Data), There is no restriction on data availability.
REFERENCES
- 1.Tanay A & Regev A Scaling single-cell genomics from phenomenology to mechanism. Nature 541, 331–338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wagner A, Regev A & Yosef N Revealing the vectors of cellular identity with single-cell genomics. Nat Biotechnol 34, 1145–1160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelsey G, Stegle O & Reik W Single-cell epigenomics: Recording the past and predicting the future. Science 358, 69–75 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Han X et al. Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 172, 1091–1107.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Gierahn TM et al. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 14, 395–398 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao J et al. Comprehensive single-cell transcriptional profiling of a multicellular organism. Science 357, 661–667 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg AB et al. Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein AM et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macosko EZ et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng GXY et al. Massively parallel digital transcriptional profiling of single cells. Nat Comms 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Habib N et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cusanovich DA et al. Multiplex single-cell profiling of chromatin accessibility by combinatorial cellular indexing. Science 348, 910–914 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramani V et al. Massively multiplex single-cell Hi-C. Nat. Methods 14, 263–266 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lake BB et al. Integrative single-cell analysis of transcriptional and epigenetic states in the human adult brain. Nat Biotechnol 36, 70–80 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Preissl S et al. Single-nucleus analysis of accessible chromatin in developing mouse forebrain reveals cell-type-specific transcriptional regulation. Nat. Neurosci 21, 432–439 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mulqueen RM et al. Highly scalable generation of DNA methylation profiles in single cells. Nat Biotechnol 36, 428–431 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Butler A, Hoffman P, Smibert P, Papalexi E & Satija R Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haghverdi L, Lun ATL, Morgan MD & Marioni JC Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol 36, 421–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiselev VY, Yiu A & Hemberg M scmap: projection of single-cell RNA-seq data across data sets. Nat. Methods 15, 359–362 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Wolf FA, Angerer P & Theis FJ SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shekhar K et al. Comprehensive Classification of Retinal Bipolar Neurons by Single-Cell Transcriptomics. Cell 166, 1308–1323.e30 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pandey S, Shekhar K, Regev A & Schier AF Comprehensive Identification and Spatial Mapping of Habenular Neuronal Types Using Single-Cell RNA-Seq. Curr. Biol 28, 1052–1065.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cusanovich DA et al. The cis-regulatory dynamics of embryonic development at single-cell resolution. Nature 555, 538–542 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keren-Shaul H et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Halpern KB et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 542, 352–356 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.La Manno G et al. Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580.e19 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mi Da et al. Early emergence of cortical interneuron diversity in the mouse embryo. Science 360, 81–85 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tusi BK et al. Population snapshots predict early haematopoietic and erythroid hierarchies. Nature 555, 54–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hrvatin S et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat. Neurosci 21, 120–129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baron M et al. A Single-Cell Transcriptomic Map of the Human and Mouse Pancreas Reveals Inter- and Intra-cell Population Structure. Cell Syst 3, 346–360.e4 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haber AL et al. A single-cell survey of the small intestinal epithelium. Nature 551, 333–339 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park J et al. Single-cell transcriptomics of the mouse kidney reveals potential cellular targets of kidney disease. Science 360, 758–763 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hochgerner H, Zeisel A, Lönnerberg P & Linnarsson S Conserved properties of dentate gyrus neurogenesis across postnatal development revealed by single-cell RNA sequencing. Nat. Neurosci 21, 290–299 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Nowakowski TJ et al. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farrell JA et al. Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science 360, eaar3131 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner DE et al. Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Briggs JA et al. The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science 360, eaar5780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spanjaard B & Junker JP Methods for lineage tracing on the organism-wide level. Current Opinion in Cell Biology 49, 16–21 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Woodworth MB, Girskis KM & Walsh CA Building a lineage from single cells: genetic techniques for cell lineage tracking. Nat. Rev. Genet 18, 230–244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma J, Shen Z, Yu Y-C & Shi S-H Neural lineage tracing in the mammalian brain. Curr. Opin. Neurobiol 50, 7–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun J et al. Clonal dynamics of native haematopoiesis. Nature 514, 322–327 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lodato MA et al. Somatic mutation in single human neurons tracks developmental and transcriptional history. Science 350, 94–98 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pei W et al. Polylox barcoding reveals haematopoietic stem cell fates realized in vivo. Nature 548, 456–460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuentealba LC et al. Embryonic Origin of Postnatal Neural Stem Cells. Cell 161, 1644–1655 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harwell CC et al. Wide Dispersion and Diversity of Clonally Related Inhibitory Interneurons. Neuron 87, 999–1007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayer C et al. Clonally Related Forebrain Interneurons Disperse Broadly across Both Functional Areas and Structural Boundaries. Neuron 87, 989–998 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez-Fraticelli AE et al. Clonal analysis of lineage fate in native haematopoiesis. Nature 553, 212–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilbert LA et al. CRISPR-Mediated Modular RNA-Guided Regulation of Transcription in Eukaryotes. Cell 154, 442–451 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Perez-Pinera P et al. RNA-guided gene activation by CRISPR-Cas9–based transcription factors. Nat. Methods 10, 973–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maeder ML et al. CRISPR RNA–guided activation of endogenous human genes. Nat. Methods 10, 977–979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng AW et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Research 2013 23:10 23, 1163–1171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deng W, Shi X, Tjian R, Lionnet T & Singer RH CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. Proc. Natl. Acad. Sci. U.S.A 112, 11870–11875 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X et al. In Situ Capture of Chromatin Interactions by Biotinylated dCas9. Cell 170, 1028–1043.e19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao H-K et al. In Vivo Target Gene Activation via CRISPR/Cas9-Mediated Trans-epigenetic Modulation. Cell 171, 1495–1507.e15 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Datlinger P et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dixit A et al. Perturb-Seq: Dissecting Molecular Circuits with Scalable Single-Cell RNA Profiling of Pooled Genetic Screens. Cell 167, 1853–1866.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang W & Liu DR Rewritable multi-event analog recording in bacterial and mammalian cells. Science 360, eaap8992 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Komor AC, Kim YB, Packer MS, Zuris JA & Liu DR Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim YB et al. Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol 35, 371–376 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gaudelli NM et al. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nelles DA et al. Programmable RNA Tracking in Live Cells with CRISPR/Cas9. Cell 165, 488–496 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mikuni T, Nishiyama J, Sun Y, Kamasawa N & Yasuda R High-Throughput, High-Resolution Mapping of Protein Localization in Mammalian Brain by In Vivo Genome Editing. Cell 165, 1803–1817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Perli SD, Cui CH & Lu TK Continuous genetic recording with self-targeting CRISPR-Cas in human cells. Science 353, aag0511–aag0511 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Chow RD et al. AAV-mediated direct in vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat. Neurosci 20, 1329–1341 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKenna A et al. Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frieda KL et al. Synthetic recording and in situ readout of lineage information in single cells. Nature 541, 107–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alemany A, Florescu M, Baron CS, Peterson-Maduro J & van Oudenaarden A Whole-organism clone tracing using single-cell sequencing. Nature 556, 108 (2018). [DOI] [PubMed] [Google Scholar]
- 69.Spanjaard B et al. Simultaneous lineage tracing and cell-type identification using CRISPR-Cas9-induced genetic scars. Nat Biotechnol 36, 469–473 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kalhor R, Mali P & Church GM Rapidly evolving homing CRISPR barcodes. Nat. Methods 14, 195–200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmidt ST, Zimmerman SM, Wang J, Kim SK & Quake SR Quantitative Analysis of Synthetic Cell Lineage Tracing Using Nuclease Barcoding. ACS Synth. Biol 6, 936–942 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raj B et al. Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol 36, 442–450 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zilionis R et al. Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc 12, 44–73 (2017). [DOI] [PubMed] [Google Scholar]
- 74.Lodato MA et al. Aging and neurodegeneration are associated with increased mutations in single human neurons. Science 359, 555–559 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bae T et al. Different mutational rates and mechanisms in human cells at pregastrulation and neurogenesis. Science 359, 550–555 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kawakami K Tol2: a versatile gene transfer vector in vertebrates. Genome Biol 8 Suppl 1, S7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Satija R, Farrell JA, Gennert D, Schier AF & Regev A Spatial reconstruction of single-cell gene expression data. Nat Biotechnol 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suster ML, Abe G, Schouw A & Kawakami K Transposon-mediated BAC transgenesis in zebrafish. Nat Protoc 6, 1998–2021 (2011). [DOI] [PubMed] [Google Scholar]
- 79.Fisher S et al. Evaluating the biological relevance of putative enhancers using Tol2 transposon-mediated transgenesis in zebrafish. Nat Protoc 1, 1297–1305 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Pan YA et al. Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development 140, 2835–2846 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yin L et al. Multiplex Conditional Mutagenesis Using Transgenic Expression of Cas9 and sgRNAs. Genetics 200, 431–441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data. Raw data used for calculation of scGESTALT lineage barcode copy by qPCR. The number of integrations of the scGESTALT barcode in transgenic F1 is determined by qPCR. Genomic DNA from 13 animals (gDNA 33–50) was used in the assay. Genomic DNA from an animal with no barcode integration (control zero), an animal with known single copy integration (control single), and an animal with known double copy integration (control double) are included as standards. For each genomic DNA sample, qPCR reactions are set up in triplicate using primers for an ultraconserved control region that is known to be 2 copies, and for the DsRed region to determine copy number of the scGESTALT lineage barcode. The Ct number for each reaction is shown. The average Ct for each region is calculated (Ctrl Ct and DsRed Ct). The delta Ct and delta delta Ct are calculated and used to determine copy number (refer to Procedure Step 25 for details). Animals with a calculated copy number of less than or equal to 1 (but greater than or equal to 0.6) are considered to be single integrants.
Supplementary Figure 1. Papain oxygenation setup. Left and middle panels, 95%O2: 5%CO2 gas tank fitted with a gas regulator, Tygon E-3603 tubing and a 5 ml serological pipette. Right panel, Oxygenation of papain/DNase mix in Neurobasal media (small vial to the right) is performed by bubbling 95%O2: 5%CO2 gas through tubing attached to a sterile 5 ml serological pipette for 2 min (Procedure Step 52). EBSS buffer (large vial to the left, used for resuspending ovumucoid, Procedure Step 53) and Neurobasal media (Procedure Step 50) are oxygenated in a similar manner.
Supplementary Figure 2. Zebrafish brain dissection. Top panels, Anesthetized fish is transferred to a sylgard dish covered with Neurobasal media and MESAB (left). The fish is pinned just posterior of the head, in the middle of the trunk and near the tail using 3 insect pins (right, asterisks mark pin positions). Bottom panels, The jaw, eyes, heart and gut tissues are removed. The skin on top of the head is pierced and peeled back to expose the brain (left, circle marks the exposed brain). Gently scoop the brain out taking care not to lose part of the hindbrain in the process (right, whole brain is encircled).
Supplementary Movie. Papain/DNase mix oxygenation. Demonstration of the proper method for oxygenation of the papain/DNase mix.
Supplementary Software 1. scGESTALT analysis pipeline scripts. This is the master pipeline for processing scGESTALT reads and generating lineage trees (Fig. 1 and Fig. 7, relevant for Step 107).
Supplementary Software 2. R script (scGestaltPrepFunc) for pre-processing of inDrops scGestalt data. The script will format the data for input to the scGestalt analysis pipeline (relevant for Step 106).
Supplementary Software 3. R script pipeline (Transcriptome-scGestaltMatchPipe) for matching transcriptome and scGESTALT lineage barcodes for profiled single cells (relevant for Step 109).
Supplementary Software 4. R script (MatchPipeFunc) containing the code of the R functions called by the scGestaltMatchPipe pipeline (relevant for Step 109).
Supplementary Table 1. Sequences of scGESTALT CRISPR target sites.
Supplementary Table 2. InDrops library multiplexing primer sequences.