Abstract
Background: Small cell lung carcinoma (SCLC) is an aggressive, recalcitrant cancer, often metastatic at diagnosis and unresponsive to chemotherapy upon recurrence, thus it is challenging to treat.
Methods: Sixty-three human SCLC lines and three NSCLC lines were screened for response to 103 US Food and Drug Administration–approved oncology agents and 423 investigational agents. The investigational agents library was a diverse set of small molecules that included multiple compounds targeting the same molecular entity. The compounds were screened in triplicate at nine concentrations with a 96-hour exposure time using an ATP Lite endpoint. Gene expression was assessed by exon array, and microRNA expression was derived by direct digital detection. Activity across the SCLC lines was associated with molecular characteristics using pair-wise Pearson correlations.
Results: Results are presented for inhibitors of targets: BCL2, PARP1, mTOR, IGF1R, KSP/Eg5, PLK-1, AURK, and FGFR1. A relational map identified compounds with similar patterns of response. Unsupervised microRNA clustering resulted in three distinct SCLC subgroups. Associating drug response with micro-RNA expression indicated that lines most sensitive to etoposide and topotecan expressed high miR-200c-3p and low miR-140-5p and miR-9-5p. The BCL-2/BCL-X L inhibitors produced similar response patterns. Sensitivity to ABT-737 correlated with higher ASCL1 and BCL2. Several classes of compounds targeting nuclear proteins regulating mitosis produced a response pattern distinct from the etoposide response pattern.
Conclusions: Agents targeting nuclear kinases appear to be effective in SCLC lines. Confirmation of SCLC line findings in xenografts is needed. The drug and compound response, gene expression, and microRNA expression data are publicly available at http://sclccelllines.cancer.gov .
Small cell lung cancer (SCLC) is an aggressive carcinoma that was named in the Recalcitrant Cancer Act. SCLC is a neuroendocrine lung malignancy that affects more than 200 000 people every year with high mortality. In the United States, SCLC comprises 13% to 15% of lung cancer, SCLC recurs rapidly, and less than 5% of patients survive five years. Only two drugs, etoposide and topotecan, are US Food and Drug Administration (FDA) approved for SCLC. First-line therapy for SCLC is etoposide with a platinum complex (cisplatin or carboplatin). While initial response is 60% to 80%, responses tend to be short lived and recurrent disease is often no longer responsive to chemotherapy ( 1–3 ).
SCLC cells have little cytoplasm and faint or absent nucleoli. SCLC has a high mitotic rate and often areas of necrosis ( 4 ). Treatment resistance has been attributed to the persistence of a cancer stem-like subpopulation that exhibits multiple drug resistance ( 5 , 6 ). In culture, SCLC lines grow as floating clusters or spheroids, which are often difficult to disaggregate.
SCLC has unique biology and chromosomal changes, and active early development pathways ( 7–9 ). Genetic somatic alterations in SCLC including mutations (8.88 mutations per megabyte), insertions, deletions, copy number variations, and chromosomal rearrangements are among the highest in cancer ( 10–13 ). The most frequent genetic alterations are deletion or mutation in retinoblastoma protein (RB1) and deletion or mutation of p53 (TP53). Beyond these changes, genetic alterations in SCLC are varied and nonrecurrent ( 14 ).
ASCL1 transcription factor is highly expressed in neuroendocrine lung cancers ( 15 , 16 ). Antiapoptotic regulator BCL2 is an ASCL1 target. BCL2-targeted therapy was effective in SCLC xenografts ( 17 ). However, in clinical trial, a BCL2 inhibitor failed to show therapeutic benefit ( 18 ). SCLC proteomic profiling identified DNA repair enzymes PARP1 and checkpoint kinase 1 as potential targets ( 19 , 20 ). In addition, EZH2, which epigenetically silences genes during development, may be involved in SCLC ( 21 ).
Mitotic kinesin inhibitors are being examined as anticancer agents ( 22–24 ). Kinesin Eg5 is targeted by compounds in clinical trial ( 25–27 ). IGF1R and FGFRs are overexpressed by an SCLC subset ( 28 ). FGFR1 is amplified in 6% of SCLCs ( 29 ). IGF1R expression is frequent in SCLCs ( 30 ). A subset of SCLCs harbor PIK3CA mutations ( 31 ). Compounds targeted to these proteins are in development; however, clinical results have been disappointing.
MicroRNAs are implicated in SCLC as regulators of cell viability and drug sensitivity ( 32 ). MicroRNAs are being explored as biomarkers of disease and as drug response or therapeutic targets ( 33 , 34 ). From 1977 through 1992, 126 SCLC cell lines were established ( 9 , 35 , 36 ). The current study was undertaken to explore the response of a panel of SCLC lines to FDA-approved anticancer drugs and a library of investigational agents, along with exon and microRNA arrays. These data are publicly available at: http://sclccelllines.cancer.gov .
Methods
Cell Lines
The SCLC lines used were purchased from American Type Culture Collection (ATCC) (Manassas, VA), Sigma-Aldrich, or were from the National Cancer Institute (NCI) repository ( 37 ). NCI-H28 mesothelioma, NCI-H2066 mixed SCLC/NSCLC NCI-H1650 NSCLC, and A549/ATCC NSCLC, purchased from ATCC, were included as comparators. Cells were maintained in 5% CO 2 -humidified incubators at 37 °C and maintained in the medium specified, supplemented with additives. The Supplementary Methods and Supplementary Table 1 (available online) describe the properties for each line along with expression for canonical SCLC and neuroendorcine genes. A subset of the lines is included in the cancer cell line encyclopedia along with genomic data; we note key genomic alterations for this subset of the cell lines in the Supplementary Methods and Supplementary Table 2 (available online). The SCLC lines were authenticated using the Applied Biosystems Identifiler kit. Samples were taken for analysis within passages 2-5. For 48 lines, ATCC’s STR profiles ( http://www.atcc.org ) were used for authentication. Eleven cell lines were authenticated using STR data from Dr. A. Gazdar (UT Southwestern; personal communication) and two from the SIGMA STR database ( http://www.sigmaaldrich.com/life-science/cell-culture/ecacc-cell-lines.html ). Nine SCLC lines had unique STR profiles indicating no contamination with other cell lines. All SCLC lines were verified mycoplasma and pathogen free ( Supplementary Table 1 , available online). Cells were used up to 20 passages. The A549/ATCC NSCLC line was a screen control.
Compounds
One hundred and three FDA-approved anticancer drugs (available from NCI at: http://dtp.nci.nih.gov/branches/dscb/oncology_drugset_explanation.html ) and a library of 423 investigational agents, acquired by synthesis or purchase, were screened.
Screen
Twelve lines (11 SCLC and A549/ATCC) were screened per run. Day 1, the cells were collected and suspended by exposure to Accutase (see SOPs at http://sclccelllines.cancer.gov ) in 300 mL of media, then plated (5x10 3 - 20x10 3 in 42 μL) in 384-well plates (CulturPlates, PE, Waltham, MA) using a Tecan Freedom Evo. After incubation overnight, the Tecan Evo was used for compound addition. Each compound was tested at nine concentrations (10 μM to 1.5 nM; DMSO concentration 0.25%), then the plates were incubated for 96 hrs. The screen controls were: topotecan (10 μM), doxorubicin (10 μM), tamoxifen (200 μM), and DMSO (0.25%). The incubation was terminated by adding ATP Lite (Perkin Elmer Inc., Waltham, MA), and luminescence was determined. All assays were performed in triplicate.
Exon and MicroRNA Arrays
Total RNA was extracted using Qiagen miRNeasy Mini Kit (Qiagen, Valencia, CA) according to manufacturer's instructions. An agilent RNA Integrity Number (RIN) greater than 8.5 indicated good-quality RNA. Sense strand cDNA from 100 ng total RNA was fragmented and labeled using Affymetrix WT terminal labeling kit. Samples were hybridized with Human Exon 1.0 ST Arrays (Affymetrix) at 45°C, 60 rpm, for 16 hrs. Arrays were washed and stained using Affymetrix Fluidics Station 450 and scanned on an Affymetrix GeneChip scanner 3000 7G. mRNA expression data was normalized using Robust Multi-Array Average (RMA) and summarized at the gene level using AROMA ( 38 ).
For microRNA profiling, total RNA (100 ng) was ligated to unique oligonucleotide tags without amplification using the NanoString kit. Samples were hybridized for 16 hours to NanoString human miRNA probeset. Each consists of a Reporter Probe, with the fluorescent signal on its 5' end, and a Capture Probe with biotin on the 3' end. Purification of bound probes was performed with a magnetic bead-based wash on the nCounter Prep Station, followed by immobilization in the cartridge. The miRNA data was normalized and log2 plus 1 transformed.
Statistical Analysis
Screen statistical validity was verified using a Z’ value of greater than 0.5 as representative of a high-quality assay ( 39 ). Concentration response data were fit with a four-parameter curve fit and IC 50 s determined as the mean of three replicates. Pair-wise Pearson correlations between the negative log 10 IC 50 and log 2 gene or miRNA expression are presented. The constellation relationship map was created using the pair-wise correlations and a threshold for an edge connecting two compounds determined by a network topology best fitting a scale-free topology. The threshold was selected by finding the value maximizing the R-squared for the regression of the log of the frequency distribution vs the log of the number of edges. Unsupervised clustering of SCLC lines based on microRNA expression was performed using partitioning around medoids algorithm. The clusters were selected by maximum cluster silhouette width. Response differences between subgroups were tested using analysis of variance. Agents with the smallest P values were evaluated. All statistical tests were two-sided, and P values of lses than .05 were considered statistically significant. Analysis was performed using R ( http://www.R-project.org ). The data are publicly available at: http://sclccelllines.cancer.gov . The Affymetrix mRNA and NanoString microRNA raw data files are available for direct download from the Gene Expression Omnibus ( http://www.ncbi.nlm.nih.gov/geo/ ). The accession number for the overall project is GSE73162 ( http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73162 ), Affymetrix: GSE73160 ( http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73160 ), and Nanostring: GSE73161 ( http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE73161 ).
Results
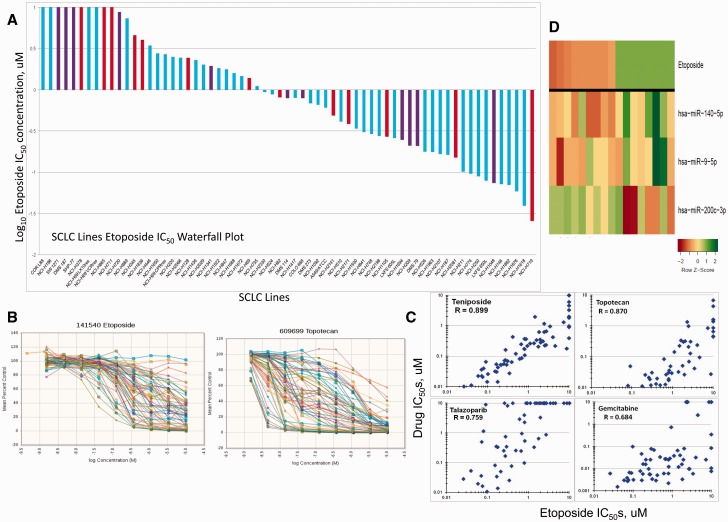
SCLC lines generally grow in suspension as loose or tight clusters/spheroids. Up to 20 000 cells were required in a 384-well format to obtain a reliable ATP baseline. Etoposide and topotecan are the only drugs FDA-approved for SCLC; therefore, these drugs were used as comparators for other agents. The IC 50 s for etoposide span 2.5 logs ( Figure 1A ). The heterogeneity of response to etoposide and topotecan is evident in Figure 1B . SCLC lines derived from treatment-naive patients were distributed throughout the response range. The sensitivity to etoposide, and teniposide, topotecan, talazoparib and gemcitabine, was highly related (R = 0.89, 0.87, 0.76, and 0.68, respectively) ( Figure 1C ). Associating drug response with microRNA expression indicated that SCLC lines most sensitive to etoposide expressed high miR-200c-3p and low miR-140-5p and miR-9-5p ( Figure 1D ). The mean log 10 IC 50 across all compounds was used as a global metric for sensitivity and was associated with etoposide sensitivity (R = 0.64).
Figure 1.
Analysis of etoposide response. A) Waterfall plot showing the range (25 nM to > 10 uM) of IC50s for small cell lung cancer (SCLC) lines exposed to etoposide for 96 hourrs. Blue bars are SCLC lines developed from previously treated tumors; red bars are SCLC lines developed from treatment-naive tumors; purple bars are SCLC lines developed from tumors of unknown treatment status. B) Concentration response curves for the 63 SCLC lines exposed to etoposide or topotecan for 96 hours. C) Etoposide IC50 plotted vs the teniposide, topotecan, talazoparib, and gemcitabine IC50s showing a correlation of R = 0.89, 0.87, 0.76, and 0.68, respectively, for the SCLC lines. D) Heat map of the etoposide eight most sensitive and eight most resistant SCLC lines along with microRNA expression from counts for miR-140-5p, miR-200c-3p, and miR-9-5p. The heat map is shown as Z score to allow comparison across scales, where yellow is the mean, greeen is high, and red is low. IC50 = the inhibitory concentration producing 50% growth inhibition.
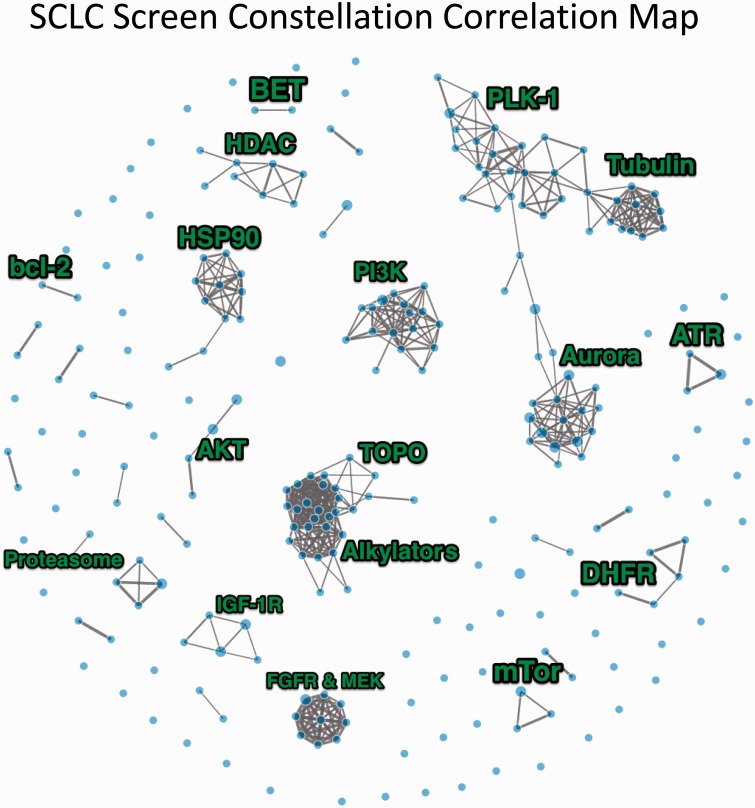
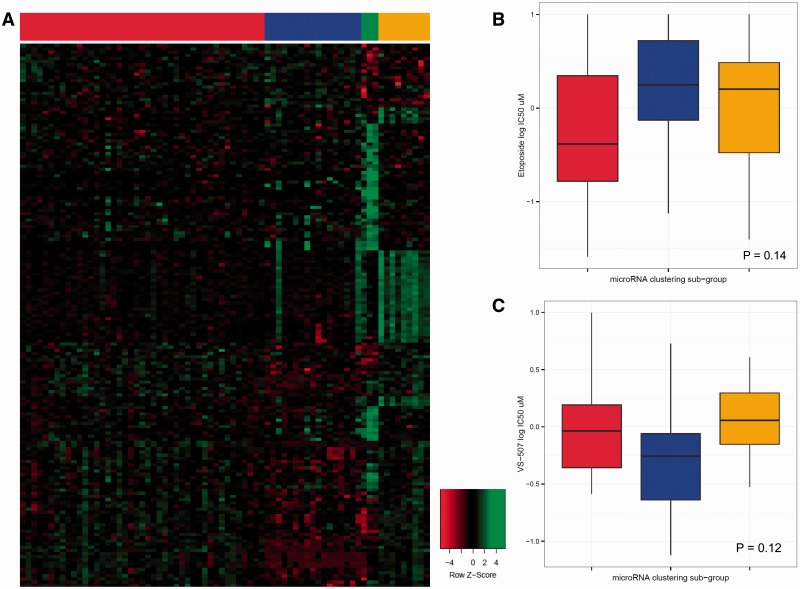
There were about 150 compounds that were inactive against the SCLC lines, including SMO inhibitors, MDM2 inhibitors, VEGFR, ALK, and C-MET inhibitors; Raf and B-Raf inhibitors; and a similar number of compounds with greaeter than 2 log concentration response across the SCLC lines ( Supplementary Table 3 , available online). After filtering agents with a log 10 IC 50 range of less than 0.5, 345 compounds (out of 523 tested) remained. A constellation relational map was developed. The drugs were connected if the pair-wise correlation of the log 10 IC 50 was greater than 0.77 ( Figure 2 ). The threshold for the correlation was estimated so that the topology best fit a scale-free network (R 2 = 0.98). The relational map identified compound clusters with similar patterns of sensitivity. Many clusters were based on molecular target or pathway and others based on similar cellular effects such as DNA or microtubule damaging. Supplementary Figure 1 (available online) is an unsupervised clustering of the data. Unsupervised microRNA clustering resulted in three distinct subgroups ( Figure 3A ). The red subgroup was most sensitive to etoposide ( P = .14) ( Figure 3B ). The blue subgroup was most sensitive to the stem-like cell-directed agent VS-507 (salinomycin) ( P = .12) ( Figure 3C ) ( 40 ). SCLC lines that were sensitive to VS-507 expressed high C20orf27, GEMIN4, HOMER2, LRP3, PCOLCE2, PFAS, RPL18, and RRP1B ( Supplementary Figure 2 , available online).
Figure 2.
Constellation relational map showing response similarity connections among the approved and investigational anticancer agents tested in the small cell lung cancer (SCLC) lines at a stringency of 0.77. The line thickness is directly proportional to the pair-wise correlation.
Figure 3.
MicroRNA small cell lung cancer (SCLC) line clustering. A) SCLC subgroups based upon unsupervised microRNA clustering. Left to right are the red , blue , and goldenrod clusters, with the green cluster being normal cell comparators. B) Box plot showing the median and association of etoposide response of the SCLC lines, with the SCLC microRNA subgroups ( P = .14). C) Box plot showing the median and association of VS-507 (salinomycin) response of the SCLC lines, with the SCLC microRNA subgroups ( P = .12). Unsupervised clustering of the SCLC lines based on microRNA expression was performed using partitioning around medoids algorithm. The number of clusters was selected by maximum width of cluster silhouette. Differences in drug response between subgroups were tested using analysis of variance, and agents with the smallest P values were evaluated. Analysis was performed using R ( http://www.R-project.org ).
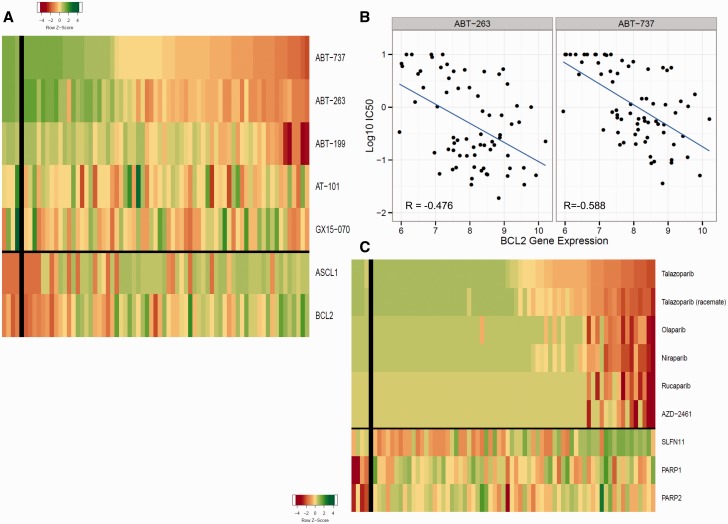
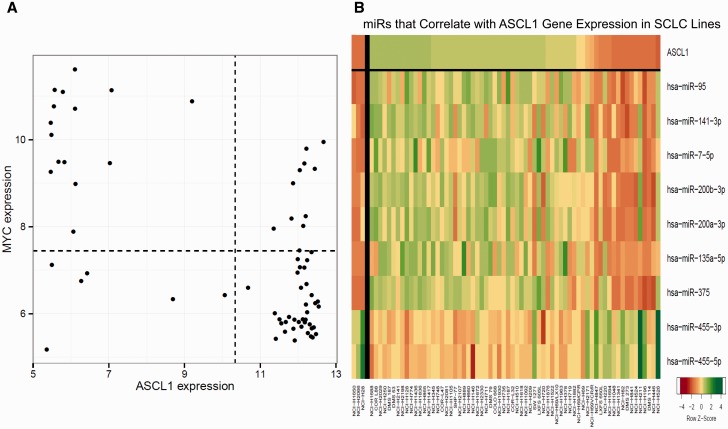
Many SCLC lines express high BCL-2. ABT-737, ABT-263, and ABT-199 primarily target BCL-2 and BCL-X L , while AT-101 and GX15-07 target BCL-2, BCL-X L , and MCL-1 ( 41 ). The three BCL-2/BCL-X L inhibitors produced similar patterns of response ( Figure 4A ). Sensitivity to ABT-737 correlated well with higher ASCL1 (R = –0.50) and BCL2 (R = –0.59) ( Figure 4A ). There was a negative correlation between log10 IC 50 for ABT-263 and ABT-737 and log 2 expression of BCL2 (R = –0.48 and R = –0.59, respectively) ( Figure 4B ). While most lines express high ASCL1, those lines expressing little ASCL1 tended to express high MYC ( Figure 5A ). Seven miRs positively correlated with ASCL1 (miR-95, miR-141-3p, miR-7-5p, miR-200b-3p, miR-200a-3p, and miR-375), and two miRs negatively correlated with ASCL1 (miR-455-3p and miR-455-5p) ( Figure 5B ). The pattern of response to each PARP-1 inhibitor was similar ( Figure 4C ). SLFN11 expression correlated with sensitivity to the PARP-1 inhibitors (R = –0.42). There was no association of PARP-1 or -2 expression with PARP-1 inhibitor response.
Figure 4.
Bcl-2 and PARP inhibitors. A) Heat map showing the IC 50 response of the small cell lung cancer (SCLC) lines, arranged by response to ABT-737, for five Bcl-2 inhibitors. The most sensitive SCLC lines are: NCI-H2107, NCI-H889, NCI-H1963, NCI-H1105, and NCI-H748; the least sensitive SCLC lines are: NCI-H378, NCI-841, NCI-H196, DMS273, and DMS114. The heat map is shown as Z score to allow comparison across scales, where yellow is the mean, green is high, and red is low. The gene expression values were: ASCL1 mean = 9.96 (range = 4.97–12.67), and the correlation with response to ABT737 was R = -0.497; BCL2 mean = 7.93 (range = 5.95–10.2), and the correlation with response to ABT737 was R = -0.588. B) ABT-263 and ABT-737 log 10 IC 50 plotted vs log 2 BCL2 gene expression showing a correlation of R = - 0.476 with ABT-263 response and R = -0.588 with ABT-737 response for the SCLC lines. C) Heat map showing the IC 50 response of the SCLC lines, arranged by response to talazoparib, for six PARP inhibitors. The most sensitive SCLC lines are: NCI-H211, NCI-H209, NCI-H774, NCI-H526, and NCI-H1048; the least sensitive SCLC lines are: SW1271, NCI-2029, NCI-H196, NCI-H1688, and COR L88. The heat map is shown as Z score to allow comparison across scales, where yellow is the mean, green is high, and red is low. The gene expression values were: SLFN11 mean = 7.15 (range = 4.43–10.04), and the correlation with response to talazoparib was R = -0.513. IC 50 = the inhibitory concentration producing 50% growth inhibition.
Figure 5.
ASCL-1 and MYC analysis. A) Plot of the relative gene expression of ASCL-1 vs the relative gene expression of c-Myc. Each point is a small cell lung cancer (SCLC) line. In general, there was an inverse relationship between the expression of ASCL-1 and c-Myc. The correlation is R = -0.59. B) Heat map organized by expression of ASCL-1 and showing the miRs that are positively and negatively correlated with expression of ASCL-1. The heat map is shown as Z score to allow comparison across scales, where yellow is the mean, green is high, and red is low. The correlation values are: miR-95 (R = 0.66), miR-141-3p (R = 0.61), miR-7-5p (R = 0.54), miR-200b-3p (R = 0.66), miR-200a-3p (R = 0.62), miR-200c-3p (R = 0.59), miR-135a-5p (R = 0.59), miR-375 (R = 0.69), miR-455-3p (R = -0.58), and miR-455-5p (R = -0.62).
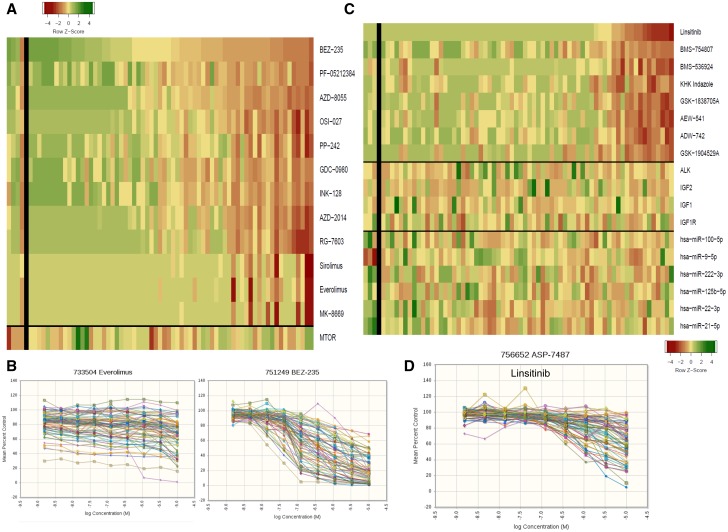
A subset of lines was sensitive to mTOR inhibitors ( Figure 6A ). The concentration response curves showed the characteristic flat concentration response to everolimus, a rapalog, in contrast to BEZ-235, an ATP-binding site inhibitor ( Figure 6B ). There was no association between MTOR expression and response to mTOR inhibitors. There was no association between response to IGF-1R inhibitors and expression of ALK, IGF-1R, IGF-1, IGF-2, or to six microRNAs ( Figure 6C ). The linsitinib concentration response curves shown in Figure 6D indicate that sensitive lines have IC 50 s in the 0.5 to 1 μM range.
Figure 6.
mTOR and IGF1R inhibitors. A) Heat map showing the IC 50 response of the small cell lung cancer (SCLC) lines, arranged by response to BEZ-235 for 12 mTOR inhibitors. The most sensitive SCLC lines are: LXFS 650L, DMS 114, NCI-H1882, NCI-H719, and NCI-H1048; the least sensitive SCLC lines are: NCI-H711, NCI-1417, NCI-H378, DMS187, and NCI-H660. The heat map is shown as Z score to allow comparison across scales, where yellow is the mean, green is high, and red is low. The gene expression values were: MTOR mean = 9.04 (range = 7.84–10.69), and the correlation with response to sirolimus was R = -0.23. B) Concentration response curves for the 63 SCLC lines exposed to everolimus or BEZ-235 for 96 hours. C) Heat map showing the IC 50 response of the SCLC lines, arranged by response to linsitinib for eight IGF1R inhibitors. The most sensitive SCLC lines are: LXFS 605L, NCI-H187, NCI-H526, COLO 668, and NCI-H1876; the least sensitive SCLC lines are: NCI-H1048, DMS 53, DMS273, DMS114, and COR L88. The heat map is shown as Z score to allow comparison across scales, where yellow is the mean, green is high, and red is low. There is no association with the expression of the gene shown or the microRNAs shown. D) Concentration response curves for the 63 SCLC lines exposed to linsitinib for 96 hours.
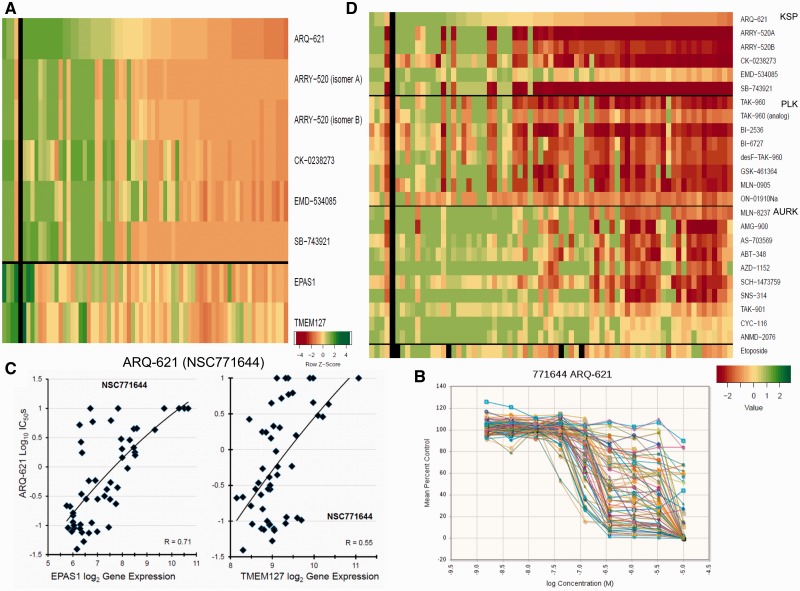
Approximately half of the 63 SCLC lines were sensitive to KSP/Eg5 inhibitors ( Figure 7A ). The ARQ-621 concentration response curves indicate marked heterogeneity of response, with responsive lines having IC 50 s of 0.1 to 0.3 μM ( Figure 7B ). Sensitivity to ARQ-621 was associated with low EPAS1/Hif2A and TMEM127 ( Figure 7C ). The polo-like kinase inhibitors and aurora kinase inhibitors had response patterns similar to the KSP/Eg5 inhibitors ( Figure 7D ). The response pattern for nuclear protein targeting agents was distinct from the etoposide response pattern.
Figure 7.
Nuclear kinase inhibitors. A) Heat map showing the IC 50 response of the small cell lung cancer (SCLC) lines, arranged by response to ARQ-621 for six KSP/Eg5 inhibitors. The most sensitive SCLC lines are: NCI-H69, NCI-H1105, NCI-H211, NCI-H2107, and NCI-H1048; the least sensitive SCLC lines are: NCI-H69/LX10, DMS187, NCI-H2029, NCI-H196, and DMS53. The heat map is shown as Z score to allow comparison across scales, where yellow is the mean, green is high, and red is low. B) Concentration response curves for the 63 SCLC lines exposed to ARQ-621 for 96 hours. C) ARQ-621 log 10 IC 50 plotted vs log 2 EPAS1 and TMEM127 gene expression showing a correlation of R = 0.71 for EPAS1 with ARQ-621 response and R = 0.55 for TMEM127 with ARQ-621 response for the SCLC lines. The expression values were: EPAS1 mean = 7.58 (range = 5.72–12.72), and TMEM127 mean = 9.09 (range = 8.15–11.07). D) Heat map showing the log 10 IC 50 response of the SCLC lines, arranged by response to ARQ-621 for six KSP/Eg5 inhibitors, eight Polo-like kinase inhibitors, 10 aurora kinase inhibitors, and showing response to etoposide. Green indicates IC 50 ≥ 10 -5 ; yellow indicates IC 50 ≥ 10 -7 ; red indicates IC 50 ≥ 10 -9 . IC 50 = the inhibitory concentration producing 50% growth inhibition.
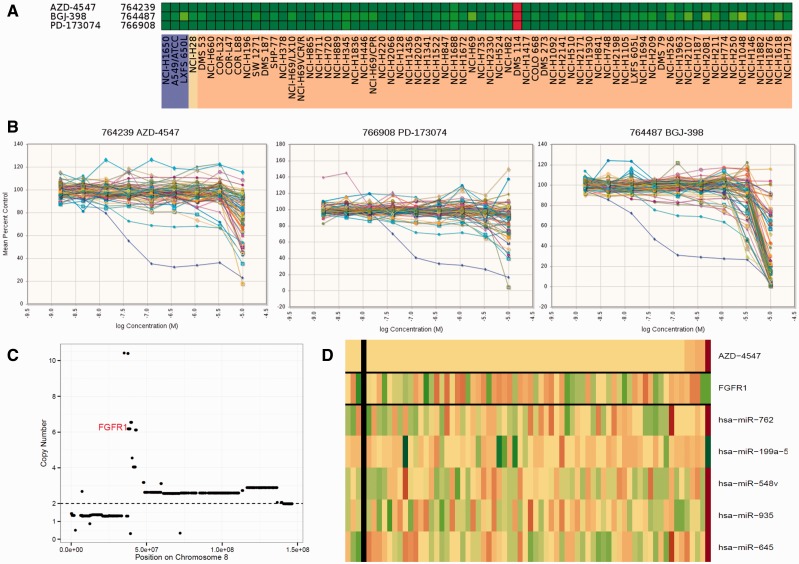
The concentration response curves indicated that the lines were generally unresponsive to three FGFR1 inhibitors ( Figure 8A ). However, DMS114 was exceptionally sensitive to all three FGFR1 inhibitors ( Figure 8B ). The FGFR1 gene is located on chromosome 8, and CCLE copy number data describe an amplification event encompassing FGFR1 with an estimated copy number of 6 for DMS114 ( Figure 8C ). Response to the FGFR1 inhibitor AZD-4547 along with FGFR1 expression is shown in Figure 8D . The DMS114 response to the FGFR1 inhibitors was associated with low microRNAs miR-762, miR-548v, miR-935, and miR-645 and moderately associated with high miR-199a-5p.
Figure 8.
An exceptional responder. A) Heat map showing the IC 50 response of the small cell lung cancer (SCLC) lines response to three FGFR1 inhibitors. Green indicates log 10 ≥ 10 -5 ; yellow indicates IC 50 ≥ 10 -7 ; red indicates IC 50 ≥ 10 -9 . B) Concentration response curves for the 63 SCLC lines exposed to the FGFR1 inhibitors AZD-4547 (NSC764239), PD-173074 (NSC766908), and BGJ-398 (NSC764487) for 96 hours. C) DMS114 chromosome 8 map showing copy number on the y-axis . The data were derived from the CCLE database. D) Heat map showing the IC 50 response of the SCLC lines, arranged by response to AZD-4547, gene expression of FGFR1, and microRNAs miR-548v, miR-935, miR-762, miR-645, and miR-199a-5p by the SCLC lines. The most sensitive SCLC lines are: DMS114, NCI-H1688, NCI-H526, NCI-H2107, and NCI-H211.The heat map is shown as Z score to allow comparison across scales, where yellow is the mean, green is high, and red is low. IC 50 = the inhibitory concentration producing 50% growth inhibition.
Thirteen MEK inhibitors were tested. The DMS114 line, a cell line with mixed SCLC and NSCLC properties, NCI-H2066, and the NSCLC line A549/ATCC responded to four MEK inhibitors ( Supplementary Figure 3 , A and B, available online). There was no association between MAP2K1/MEK expression and response to MEK inhibitors ( Supplementary Figure 3 A, available online).
The NCI-H378 line was an exceptional responder to three kinase inhibitors ( Supplementary Figure 4 A, available online). The three compounds were: Motesanib (AMG-706, NSC760843), an inhibitor of VEGFR1, 2, and 3, as well as c-Kit; VE-821, an ATR kinase inhibitor; and ITRI-260, a Flt3 inhibitor. The 'all-cells' concentration response curves showed that most lines had little response ( Supplementary Figure 4 B, available online). There was no correlation between ATR expression and response to VE-821. MiR-1306-3p was uniquely poorly expressed by the NCI-H378 line ( Supplementary Figure 4 C, available online). Two lines (DMS114 and NCI-H211) were responsive to the CDK inhibitor palbociclib (NSC758247) ( Supplementary Figure 5 , available online).
Discussion
SCLC is a therapeutic puzzle, initially responsive to etoposide plus platinum, the established treatment, and, upon recurrence, resistant to chemotherapeutics. Recently, immunotherapy, anti-PD1, and anti-CTLA4 demonstrated activity in recurrent SCLC ( 42 , 43 ). The SCLC lines that were sensitive to etoposide were usually sensitive to teniposide, topotecan, talazoparib, and gemcitabine. This pattern of sensitivity and resistance persisted for the more than 500 agents tested, indicating that the mechanism(s) of resistance were unrelated to the drug mechanism. Preventing resistance in responsive SCLC and overcoming resistance (de novo or post-treatment) requires a better understanding of SCLC biology.
Unsupervised clustering of gene and miR expression showed clear discrimination of the SCLC and NSCLC cell lines. The unsupervised miR clustering provided distinct subgrouping. There were trends toward miR subgroups aligning with compound response. One microRNA subgroup was more sensitive to VS-507 (salinomycin), a compound directed toward drug-resistant cancer stem-like cells ( 40 ). The expression of some miRs was characteristic of particular lines such as DMS114 and NCI-H378, which showed exceptional response to specific targeted compounds. The data clustered by compound pattern were visualized on a constellation relational map.
In SCLC patients, 55% to 90% of tumors reportedly overexpress BCL-2 ( 44 ). High BCL-2 mRNA and positivity for ASCL1 correlated well with response to ABT-737 (obatoclax), ABT-263 (navitoclax), and ABT-199 (venetoclax). AT-101 and GX15-070 had different patterns of response, reflecting lack of BCL-2 selectivity ( 45 , 46 ). SCLC sensitivity was observed during ABT-737 preclinical studies ( 44 , 47 , 48 ). ABT-737 treatment slowed the growth of LX22 patient-derived xenograft, which had the highest BCL-2 and BCL-X L protein expression among the patient-derived xenografts tested, and the combination of ABT-737 and etoposide resulted in decreased tumor growth compared with controls. In a phase I trial of SCLC and other solid tumor patients, navitoclax produced encouraging preliminary efficacy in SCLCs with thrombocytopenia being the major adverse effect ( 49 ). However, in the phase II trial in relapsed, advanced SCLC patients, navitoclax (ABT-263) had limited single-agent activity with grade 3/4 thrombocytopenia noted in more than 40% of patients ( 11 , 18 ). Navitoclax is in clinical trial in combination with trametinib for treatment of patients with advanced or metastatic solid tumors including SCLC (NCT02079740). Venetoclax, a specific BCL-2 inhibitor, with decrease thrombocytopenia is in multiple clinical trials ( 46 ).
The observation that PARP1 and PARP2 are highly expressed in SCLC led to numerous SCLC PARP inhibitor clinical trials ( 1 , 19 , 20 ). In SCLC, veliparib is in a randomized first-line phase I/II trial (NCT01642251) with or without cisplatin and etoposide in patients with extensive-stage SCLC or metastatic large cell neuroendocrine NSCLC. Veliparib is in a phase II clinical trial (NCT01638546) with or without temozolomide in patients with relapsed or refractory SCLC. Olaparib and temozolomide are undergoing phase I/II clinical trials (NCT02446704) in recurrent SCLC. Olaparib and cediranib are in a phase II clinical trial (NCT02498613) in late-stage solid tumor patients including recurrent SCLC. In a phase I clinical trial (NCT01286987), talazoparib treatment produced an 18% (2/11 pts) RECIST-confirmed response rate in metastatic SCLC ( 50 ). Correlation of PARP inhibitors with specific genetic alterations and or protein expression in SCLC is being explored. Our data showed higher PARP1 and PARP2 expression in SCLC; however, there was no association between PARP inhibitor response and PARP1 or PARP2 gene expression. An association was noted between PARP inhibitor response and SLFN11 gene expression. SLFN11 has been associated with response to DNA-damaging agents and PARP inhibitors ( 51 ). Correlation of SLFN11 expression to responses with PARP inhibitors may be incorporated in the next PARP inhibitor trial to be conducted via ETCTN.
The ATP-binding site mTOR inhibitors were more effective against SCLC lines than were rapalogs. In an SCLC line panel, inhibiting mTOR signaling with everolimus disrupted survival ( 52 ). A single-agent phase II trial was conducted with temsirolimus in extensive-stage SCLC, and a phase II study with everolimus in relapsed SCLC showed limited activity ( 53 , 54 ). A phase I/II trial is underway testing the combination of sirolimus and auranofin in patients with advanced or recurrent SCLC or NSCLC (NCT01737502).
IGF-1R inhibition decreased cell proliferation, sensitized SCLC cells to cisplatin and etoposide, and increased survival of mice bearing SCLC xenografts ( 2 ). Cixutumumab (IMC-A12) blocked IGF1 signaling in SCLC lines and increased cell killing in combination with chemotherapy ( 55 ). A phase II trial of cisplatin and etoposide with or without vismodegib or cixutumumab did not show a difference in progression-free survival or overall survival in combination with standard chemotherapy in extensive-stage SCLC (NCT00887159). Linsitinib, which inhibits both IGF-1R and the insulin receptor, blocked the growth of 6/19 SCLC lines ( 56 ). A phase II trial of linsitinib vs topotecan in relapsed SCLC has completed accrual (NCT01533181). In our study, 18/63 SCLC lines had linsitinib IC50s of less than1 uM. Sensitivity to linsitinib did not correlate with IGF-1R, IR, IGF-1, IGF-2, IGFBP3, or IGFBP6 expression ( 56 ).
KSP/Eg5 inhibitors have undergone early clinical trial. ARRY-520 had clinical activity in multiple myeloma ( 22 , 23 , 57 , 58 ). Combination regimen trials are underway with KSP/Eg5 inhibitors. Among early PLK1 inhibitor trials was a phase II trial of BI2536 in relapsed SCLC ( 59 ). A phase II study of the aurora kinase inhibitor alisertib in combination with paclitaxel is underway as second-line therapy for SCLC (NCT02038647) ( 60 ). The pattern of response to KSP/Eg5 inhibitors and PLK inhibitors was similar while a smaller group of SCLC lines responded to AURK inhibitors.
This study was limited to well-established SCLC lines growing in culture; thus, the general applicability of the findings from this study to in vivo models, PDX models, and patients may be questioned. Extending the results of this screen to in vivo models to confirm that new agents may be worthy of clinical trial in SCLC is critical. The compounds (>500) were tested over a broad concentration range; however, no potentially therapeutic biologicals such as antibodies or antibody drug conjugates were studied. Despite these limitations, the study provides added value to the SCLC research community.
Exploration of the roles and potential therapeutic utility of miRs in SCLC is at an early stage ( 32 , 34 ). Literature indicates that miRs act by regulating gene expression and can be associated with response or lack of response to specific drugs ( 61–63 ). miRs are negative regulators of gene expression, each with a specific gene set. Through gene regulatory activity, miRs determine response to drugs. Rukov et al. developed a website associating miRs with genes and drugs ( 63 ). Two miRs associated with ASCL1 expression in the SCLC lines, miR-375 and miR-455-5p, were identified as regulating about 20 genes and more than 20 drugs. MiRs correlated with SCLC line sensitivity to drugs both positively and negatively. In searching for the ‘key' to SCLC, beyond mutations in TP53 and deletion or mutations in RB1, SCLC may be a collection of genomic changes that vary widely and comprise small fractions of the total SCLC population. Circulating miRs may have potential as biomarkers to aid in treatment selection in SCLC. The SCLC screen data including the drug and compound data, and gene and miRNA expression data, are publically available at http://sclccelllines.cancer.gov .
Funding
This work was supported in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. This research was supported (in part) by the Developmental Therapeutics Program in the Division of Cancer Treatment and Diagnosis of the National Cancer Institute.
Notes
The study funder had no role in the design or conduct of the study; the collection, management, analysis, or interpretation of the data; the preparation, review, or approval of the manuscript; or the decision to submit the manuscript for publication.
The authors declare that EP, MK, DS, GK, SM JM, and BAT were employed by the National Cancer Institute during the planning, conduct, and analysis of the study. The authors declare that there are no additional conflicts of interest regarding the present study. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Supplementary Material
References
- 1. Pietanza CM, Byers LA, Minna JD, Rudin CM. Small cell lung cancer: will recent progress lead to improved outcomes? Clin Cancer Res. 2015. ; 21 ( 10 ): 2244 - 2255 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. William WN, Glisson BS. Novel strategies for the treatment of small-cell lung carcinoma . Nat Rev Clin Oncol. 2011. ; 8 ( 8 ): 611 - 619 . [DOI] [PubMed] [Google Scholar]
- 3. Klijn C, Durinck S, Stawiski EW , et al. . A comprehensive transcriptional portrait of human cancer cell lines . Nat Biotechnol. 2015. ; 33 ( 3 ): 306 - 312 . [DOI] [PubMed] [Google Scholar]
- 4. Travis WD. Update on small cell carcinoma and its differentiation from squamous cell carcinoma and other non-small cell carcinomas . Mod Pathol. 2012. ; 25(Suppl 1) : S18 - S30 . [DOI] [PubMed] [Google Scholar]
- 5. Sarvi S, Mackinnon AC, Avlonitis N , et al. . CD133+ cancer stem-like cells in small cell lung cancer are highly tumorigenic and chemoresistant but sensitive to a novel neuropeptide antagonist . Cancer Res. 2014. ; 74 ( 5 ): 1554 - 1565 . [DOI] [PubMed] [Google Scholar]
- 6. Meder L, Konig K, Ozretic L , et al. . NOTCH, ASCL1, p53 and RB alterations define an alternative pathway driving neuroendocrine and small cell lung carcinomas . Int J Cancer . 2016. ; 138 ( 4 ): 927 - 938 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodriguez E, Lilenbaum RC. Small cell lung cancer: past, present and future . Curr Oncol Rep. 2010. ; 12 ( 5 ): 327 - 334 . [DOI] [PubMed] [Google Scholar]
- 8. D’Angelo SP, Pietanza MC. The molecular pathogenesis of small cell lung cancer . Cancer Biol Ther . 2010. ; 10 ( 1 ): 1 - 10 . [DOI] [PubMed] [Google Scholar]
- 9. Johnson BE, Russell E, Simmons AM , et al. . MYC family DNA amplification in 126 tumor cell lines from patients with small cell lung cancer . J Cell Biochem Suppl . 1996. ; 24 : 210 - 217 . [DOI] [PubMed] [Google Scholar]
- 10. Pleasance ED, Stephens PJ, O'Meara S , et al. . A small-cell lung cancer genome with complex signatures of tobacco exposure . Nature. 2010. ; 463 ( 7278 ): 184 - 192 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rudin CM, Durinck S, Stawiski EW , et al. . Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer . Nat Genet. 2012. ; 44 ( 10 ): 1111 - 1116 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peifer M, Fernandez-Cuesta L, Sos ML , et al. . Integrative genome analyses identify key somatic driver mutations of small cell lung cancer . Nat Genet. 2012. ; 44 ( 10 ): 1104 - 1110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. George J, Lim JS, Jang SJ , et al. . Comprehensive genomic profiles of small cell lung cancer . Nature . 2015. ; 524 ( 7563 ): 47 - 53 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lopez-Chavez A, Thomas A, Rajan A , et al. . Molecular profiling and targeted therapy for advanced thoracic malignancies: a biomarker-derived, multiarm, multihistology phase II basket trial . J Clin Oncol . 2015. ; 33 ( 9 ): 1000 - 1007 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Augustyn A, Borromeo M, Wang T , et al. . ASCL1 is a lineage oncogene providing therapeutic targets for high-grade neuroendocrine lung cancers . Proc Natl Acad Sci U S A . 2014. ; 111 ( 41 ): 14788 - 14793 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Castro DS, Martynoga B, Parras C , et al. . A novel function of the proneural factor Ascl1 in progenitor proliferation identified by genome-wide characterization of its targets . Genes Dev . 2011. ; 25 ( 9 ): 930 - 945 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shoemaker AR, Mitten MJ, Adickes J , et al. . Activity of the Bcl-2 family inhibitor ABT-263 in a panel of small cell lung cancer xenograft models . Clin Cancer Res. 2008. ; 14 ( 11 ): 3268 - 3277 . [DOI] [PubMed] [Google Scholar]
- 18. Rudin CM, Hann CL, Garon EB , et al. . Phase II study of single-agent navitoclax (ABT-263) and biomarker correlates in patients with relapsed small cell lung cancer . Clin Cancer Res . 2012. ; 18 ( 11 ): 3163 - 3169 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cardnell RJ, Feng Y, Diao L , et al. . Proteomic markers of DNA repair and PI3K pathway activation predict response to the PARP inhibitor BMN673 in small cell lung cancer . Clin Cancer Res . 2013. ; 19 ( 22 ): 6322 - 6328 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Byers LA, Wang J, Nilsson MB , et al. . Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov . 2012. ; 2 ( 9 ): 798 - 811 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Coe BP, Thu KL, Aviel-Ronen S , et al. . Genomic deregulation of the E2F/Rb pathway leads to activation of the oncogene EZH2 in small cell lung cancer . Plos One. 2013. ; 8 ( 8 ): e71670 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lonial S, Delforge M, Einsele H. et al. . The AFFIRM study: a mutlicenter phase 2 study of single-agent filanesib (ARRY-520) in patients with advanced multiple myeloma . J Clin Oncol . 2015. ; 33(Suppl) :Abstr TPS8613. [Google Scholar]
- 23. Zonder JA, Raje NS, Scott EC. et al. . A multicenter, randomized, open-label, phase 2 study of carfilzomib with or without ARRY-520 (filanesib) in patients with advanced multiple myeloma . J Clin Oncol . 2015. ; 33(Suppl) :Abstr TPS8612. [Google Scholar]
- 24. Mross KB, Scharr D, Richly H. et al. . First-in-human study of 4SC-205 (AEGIS), a novel oral inhibitor of Eg5 kinesin spindle protein . J Clin Oncol . 2014. ; 32(Suppl) :Abstr 2564. [Google Scholar]
- 25. Rath O, Kozielski F. Kinesins and cancer . Nat Rev Cancer . 2012. ; 12 ( 8 ): 527 - 539 . [DOI] [PubMed] [Google Scholar]
- 26. Vicente JJ, Wordeman L. Mitosis, microtubule dynamics and the evolution of kinesins . Exp Cell Res. 2015. ; 334 ( 1 ): 61 - 69 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Holen K, DiPaola R, Liu G , et al. . A phase I trial of MK-0731, a kinesin spindle protein (KSP) inhibitor, in patients with solid tumors . Invest New Drugs. 2012. ; 30 ( 3 ): 1088 - 1095 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matsumura Y, Umemura S, Ishii G , et al. . Expression profiling of receptor tyrosine kinases in high-grade neuroendocrine carcinoma of the lung: a comparative analysis with adenocarcinoma and squamous cell carcinoma . J Cancer Res Clin Oncol . 2015. ; 141 ( 12 ): 2159 - 2170 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sos ML, Dietlein F, Peifer M , et al. . A framework for identification of actionable cancer genome dependencies in small cell lung cancer . Proc Natl Acad Sci U S A . 2012. ; 109 ( 42 ): 17034 - 17039 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Badzio A, Wynes MW, Dziadziuszko R , et al. . Increased insulin-like growth factor 1 receptor (IGF1R) protein expression and gene copy number in small cell lung cancer . J Thorac Oncol . 2010. ; 5 ( 12 ): 1905 - 1911 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Walls M, Baxi SM, Mehta Liu KKC , et al. . Targeting small cell lung cancer harboring PIK3CA mutation with a selective oral PI3K inhibitor PF-4989216 . Clin Cancer Res. 2014. ; 20 ( 3 ): 631 - 643 . [DOI] [PubMed] [Google Scholar]
- 32. Lee JH, Voortman J, Dingemans AMC , et al. . MicroRNA expression and clinical outcome of small cell lung cancer . Plos One. 2011. ; 6 ( 6 ): e21300 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Du L, Pertsemlidis A. microRNA regulation of cell viability and drug sensitivity in lung cancer . Expert Opin Biol Ther . 2012. ; 12 ( 9 ): 1221 - 1239 . [DOI] [PubMed] [Google Scholar]
- 34. Zhang W, Dolan ME. Emerging role of microRNAs in drug response . Curr Opin Mol Ther . 2010. ; 12 ( 6 ): 695 - 702 . [PMC free article] [PubMed] [Google Scholar]
- 35. Gazdar AF, Gao B, Minna JD. Lung cancer cell lines: useless artifacts or invaluable tools for medical science? Lung Cancer . 2010. ; 68 ( 3 ): 309 - 318 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hodgkinson CL, Morrow CJ, Li Y , et al. . Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer . Nat Med. 2014. ; 20 ( 8 ): 897 - 903 . [DOI] [PubMed] [Google Scholar]
- 37. Phelps RM, Johnson BE, Ihde DC , et al. . NCI-Navy Medical Oncology Branch cell line data base . J Cell Biochem Suppl . 1996. ; 24 : 32 - 91 . [DOI] [PubMed] [Google Scholar]
- 38. Bengtsson H, Simpson K, Bullard J, Hansen K. aroma.affymetrix: A generic framework in R for analyzing small to very large Affymetrix data sets in bounded memory, Tech Report #745, Department of Statistics, University of California, Berkeley, February 2008. . http://statistics.berkeley.edu/tech-reports/745 .
- 39. Zhang JH, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays . J Biomolec Screen . 1999. ; 4 ( 2 ): 67 - 73 . [DOI] [PubMed] [Google Scholar]
- 40. Kai M, Kanaya N, Wu SV , et al. . Targeting breast cancer stem cells in triple negative breast cancer using a combination of LBH589 and salinomycin . Breast Cancer Res Treat . 2015. ; 151 ( 2 ): 281 - 294 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lessene G, Czabotar PE, Colman PM. BCL-2 family antagonists for cancer therapy . Nature Rev Drug Discov. 2008. ; 7 ( 12 ): 989 - 1000 . [DOI] [PubMed] [Google Scholar]
- 42. Antonia SJ, Bendell JC, Taylor MH , et al. . Phase I/II study of nivolumab with or without ipilimumab for treatment of recurrent small cell lung cancer (SCLC): CA209-032 . J Clin Oncol . 2015. ; 33(Suppl) :Abstr 7503. [Google Scholar]
- 43. Ott PA, Fernandez MEE, Hiret S. et al. . Pembrolizumab (MK-3475) in patients (pts) with extensive-stage small cell lung cancer (SCLC): Preliminary safety and efficacy results from KEYNOTE-028 . J Clin Oncol . 2015. ; 33(Suppl) :Abstr 7502. [Google Scholar]
- 44. Tahir SK, Yang X, Anderson MG , et al. . Influence of Bcl-2 family members on the cellular response of small-cell lung cancer cell lines to ABT-737 . Cancer Res . 2007. ; 67 ( 3 ): 1176 - 1183 . [DOI] [PubMed] [Google Scholar]
- 45. Vogler M, Weber K, Dinsdale D , et al. . Different forms of cell death induced by putative BCL2 inhibitors . Cell Death Differ . 2009. ; 16 ( 7 ): 1030 - 1039 . [DOI] [PubMed] [Google Scholar]
- 46. Vogler M. Targeting BCL2-proteins for the treatment of solid tumors . Adv Med . 2014. ;2014:943648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oltersdorf T, Elmore SW, Shoemaker AR , et al. . An inhibitor of Bcl-2 family proteins induces regression of solid tumors . Nature. 2005. ; 435 ( 7042 ): 677 - 681 . [DOI] [PubMed] [Google Scholar]
- 48. Hann CL, Daniel VC, Sugar EA , et al. . Therapeutic efficacy of ABT-737, a selective inhibitor of BCL-2, in small cell lung cancer . Cancer Res . 2008. ; 68 ( 7 ): 2321 - 2328 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gandhi L, Camidge DR, Ribeiro de Oliveira M , et al. . Phase 1 study of navitoclax (ABT-263), a novel BCL-2 family inhibitor, in patients with small-cell lung cancer and other solid tumors . J Clin Oncol . 2011. ; 29 ( 7 ): 909 - 916 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wainberg ZA, Rafii S, Ramanathan RK. et al. . Safety and antitumor activity of the PARP inhibitor BMN673 in a phase 1 trial recruiting metastatic small-cell lung cancer (SCLC) and germline BRCA-mutation carrier cancer patients . J Clin Oncol . 2014. ; 32(Suppl) :Abstr 7522. [Google Scholar]
- 51. Tang SW, Bilke S, Cao L , et al. . SLFN11 is a transcriptional target of EWS-FLI1 and a determinant of drug response in Ewing's sarcoma . Clin Cancer Res . 2015. ; 21 ( 18 ): 4184 - 4193 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marinov M, Ziogas A, Pardo OE , et al. . AKt/mTOR pathway activation and BCL-2 family proteins modulate the sensitivity of human small cell lung cancer cells to RAD001 . Clin Cancer Res . 2009. ; 15 ( 4 ): 1277 - 1287 [DOI] [PubMed] [Google Scholar]
- 53. Pandya KJ, Dahlberg S, Hidalgo M , et al. . A randomized phase II trial of two levels of temsirolimus (CCI-779) in patients with extensive-stage small-cell lung cancer who have responding or stable disease after induction chemotherapy: a trial of the Eastern Cooperative Oncology Group (E1500) . J Thorac Oncol . 2007. ; 2 ( 11 ): 1036 - 1041 . [DOI] [PubMed] [Google Scholar]
- 54. Tarhini A, Kotsakis A, Gooding W , et al. , Phase II study of everolimus (RAD001) in previously treated small cell lung cancer . Clin Cancer Res. 2010. ; 16 ( 23 ): 5900 - 5907 . [DOI] [PubMed] [Google Scholar]
- 55. Yeh J, Litz J, Hauck P, Ludwig DL, Krystal GW. Selective inhibition of SCLC growth by the A12 anti-IGF-1R monoclonal antibody correlates with inhibition of Akt . Lung Cancer. 2008. ; 60 ( 2 ): 166 - 174 . [DOI] [PubMed] [Google Scholar]
- 56. Zinn RL, Gardner EE, Marchionni L , et al. . ERK phosphorylation is predictive of resistance to IGF-1R inhibition in small cell lung cancer . Molec Cancer Therap. 2013. ; 12 ( 6 ): 1131 - 1139 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Shah JJ, Cohen AD,, Zonder JA. et al. . Phase I trial of ARRY-520 in relapsed/refractory multiple myeloma (RR MM) . J Clin Oncol . 2010. ; 28(Suppl) :Abstr 8132. [Google Scholar]
- 58. Chandrasekaran G, Tatrai P, Gergely F. Hitting the brakes: targeting microtubule motors in cancer . Brit J Cancer . 2015. ; 113 ( 5 ): 693 - 698 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gandhi L,, Chu QS,, Stephenson J , et al. . An open label phase II trial of the Plk1 inhibitor BI2536, in patients with sensitive relapse small cell lung cancer (SCLC) . J Clin Oncol . 2009. ; 27(Suppl) :Abstr 8108. [Google Scholar]
- 60. Melichar B,, Adenis A, havel L , et al. . Phase (Ph) I/II study of investigational Aurora A kinase (AAK) inhibitor MLN8237 (alisertib): updated ph II results in patients (pts) with small cell lung cancer (SCLC), non-SCLC (NSCLC), breast cancer (BrC), head and neck squamous cell carcinoma (HNSCC), and gastroesophageal cancer (GE) . J Clin Oncol . 2013. ; 31(Suppl) :Abstr 605. [Google Scholar]
- 61. Sokilde R, Kaczkowski B, Podolska A , et al. . Global microRNA analysis of the NCI-60 cancer cell panel . Molec Cancer Therap. 2011. ; 10 ( 3 ): 375 - 384 . [DOI] [PubMed] [Google Scholar]
- 62. Liu H, D'Andrade P, Fulmer-Smentek S , et al. . mRNA amd microRNA expression profiles of the NCI-60 integrated with drug activities . Molec Cancer Therap. 2010. ; 9 ( 5 ): 1080 - 1091 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rukov JL, Wilentzik R, Jaffe I, Vinther J, Shomron N. Pharmaco-miR: linking microRNAs and drug effects . Brief Bioinform. 2014. ; 15 ( 4 ): 648 - 659 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.