Voltage stimulation is commonly used to study pannexin 1 (Panx1). However, whether Panx1 is a voltage-gated channel remains controversial. Michalski et al. demonstrate that Panx1 is a channel with weak voltage dependence, whose activity can be tuned by N-terminal modifications.
Abstract
Pannexins are a family of ATP release channels important for physiological and pathological processes like blood pressure regulation, epilepsy, and neuropathic pain. To study these important channels in vitro, voltage stimulation is the most common and convenient tool, particularly for pannexin 1 (Panx1). However, whether Panx1 is a voltage-gated channel remains controversial. Here, we carefully examine the effect of N-terminal modification on voltage-dependent Panx1 channel activity. Using a whole-cell patch-clamp recording technique, we demonstrate that both human and mouse Panx1, with their nativeN termini, give rise to voltage-dependent currents, but only at membrane potentials larger than +100 mV. This weak voltage-dependent channel activity profoundly increases when a glycine–serine (GS) motif is inserted immediately after the first methionine. Single-channel recordings reveal that the addition of GS increases the channel open probability as well as the number of unitary conductance classes. We also find that insertions of other amino acid(s) at the same position mimics the effect of GS. On the other hand, tagging the N terminus with GFP abolishes voltage-dependent channel activity. Our results suggest that Panx1 is a channel with weak voltage dependence whose activity can be tuned by N-terminal modifications.
Introduction
Pannexins are membrane channels important for regulated ATP release in vertebrates (Bao et al., 2004; Dahl and Keane, 2012; Dahl, 2015). Attributed to their broad distribution, pannexins are implicated in many physiological processes (Penuela et al., 2014a). For example, pannexin 1 (Panx1) in vascular smooth muscle cells regulates systemic blood pressure via α1-adrenergic receptors (Billaud et al., 2015). Also, Panx1 mutants enhance ATP release and promote metastatic progression of breast cancer (Furlow et al., 2015). More recently, Panx1 was demonstrated to mediate hyperalgesia in neuropathic pain (Weaver et al., 2017). These examples, and more, highlight pannexins as promising targets for treating diseases like hypertension, ischemia, cancer, and chronic pain (Velasquez and Eugenin, 2014; Thompson, 2015; Jiang and Penuela, 2016).
Panx1 activates through diverse mechanisms (Sandilos and Bayliss, 2012; Dahl, 2015; Chiu et al., 2018). Opening of ATP-gated P2X7 channels, for example, triggers Panx1 activation (Pelegrin and Surprenant, 2006; Qiu and Dahl, 2009; Silverman et al., 2009; Gulbransen et al., 2012). Stimulating the N-methyl-d-aspartate receptors also triggers Panx1 activation in hippocampal neurons (Thompson et al., 2008; Weilinger et al., 2012, 2016; Isakson and Thompson, 2014). In addition, activation of the α1-adrenergic receptor or the type-1 tumor necrosis factor receptor has been suggested to enhance Panx1 activity through tyrosine phosphorylation at the intracellular loop (Billaud et al., 2015; Lohman et al., 2015). In apoptotic cells, caspases cleave the Panx1 C terminus to unplug its pore, through which phagocyte-recruiting signals are released (Chekeni et al., 2010; Sandilos et al., 2012; Chiu et al., 2017). Besides these mechanisms, studies suggest that Panx1 senses intracellular Ca2+, extracellular K+, membrane stretch, and positive membrane potentials (Bao et al., 2004; Bruzzone et al., 2005; Locovei et al., 2006; Jackson et al., 2014; Wang et al., 2014). Panx1 therefore likely possesses the intriguing ability to sense, respond to, and coordinate channel opening through various mechanisms.
While the collection of activation stimuli continues to expand, voltage has been most commonly used to study Panx1. Voltage-stimulated Panx1 typically gives rise to outwardly rectifying currents, which are readily attenuated by several small molecules such as carbenoxolone (CBX; Bruzzone et al., 2005) and probenecid (Silverman et al., 2008). We and many others took advantage of this property to study mechanisms of Panx1 channel function (Michalski and Kawate, 2016; Chiu et al., 2018; Dahl, 2018). Several studies, however, questioned such voltage-dependent channel activity. Endogenous human Panx1 (hPanx1) in immune cells or ectopically expressed hPanx1 in human embryonic kidney (HEK) 293T cells failed to respond to voltages even at +80–90 mV (Chekeni et al., 2010; Sandilos et al., 2012). Another study reported that mouse Panx1 (mPanx1) expressed in HEK293T cells only infrequently (∼15% of cells) gave rise to voltage-dependent currents (Dourado et al., 2014). Where do these discrepancies come from? Do species or experimental conditions matter? More detailed studies are needed to reconcile these discrepancies.
In this study, we characterize voltage-dependent activity of Panx1 over an extended range of membrane potentials. We also reexamined our hPanx1 expression constructs and carefully investigated the effects of N-terminal modifications on the voltage-dependent channel activity. Using two mammalian expression systems, we report that both WT hPanx1 and WT mPanx1 poorly respond to voltage stimuli and that a subtle modification of the N terminus substantially enhances the voltage-dependent currents.
Materials and methods
Chemicals and reagents
All chemicals were purchased from Sigma-Aldrich unless otherwise noted.
Cell culture
HEK293 (CRL-1573) and Chinese hamster ovary (CHO; CCL-61) cell lines were purchased from the American Type Culture Collection and therefore were not further authenticated. The mycoplasma contamination test was confirmed to be negative at the American Type Culture Collection. HEK293 cells were maintained in Dulbecco's modified Eagle medium (Gibco) supplemented with 10% FBS (Atlanta Biologicals) and 10 µg/ml gentamicin (Gibco). CHO cells were maintained in F-12K media (Gibco) supplemented with 10% FBS and 10 µg/ml gentamicin. Cells were incubated at 37°C with 5% CO2 in a humidified incubator.
Molecular biology
DNA corresponding to hPanx1 (available from GenBank under accession no. NP_056183.2) and mPanx1 (available from GenBank under accession no. NM_019482) were synthesized based on protein sequence (GenScript). Panx1 constructs were cloned into the BamHI and XhoI sites of the pIE2 (Michalski and Kawate, 2016), pNGFP-EU2, or pNCGFP-EU2 vectors. This cloning strategy resulted in insertions of two amino acids (glycine–serine, GS) right after the first methionine and three amino acids (Ala–Ser–Ser, ASS) before the stop codon. To restore the native N terminus, these linkers were removed by QuikChange mutagenesis (Agilent Technologies). Insertions, deletions, and point mutations were performed using overlapping PCR and QuikChange mutagenesis. To generate GFP-tagged versions of constructs lacking the BamHI and XhoI sites, the respective constructs were digested with NdeI and PstI, gel purified, and ligated to a similarly digested pCGFP-EU2[Panx1] vector. All constructs were generated on the full-length Panx1 genes and verified by DNA sequencing.
Whole-cell recording
Two days before recording, HEK293 cells (passage no. 10–40) were plated at low density onto 12-mm glass coverslips (VWR) in wells of a six-well plate (Greiner). Cells were transfected after 24 h with 300–800 ng plasmid DNA using FuGENE6 (Promega) according to the manufacturer’s instructions, and used for whole-cell electrophysiological recordings 16–24 h later. For CHO cell recordings, cells (passage no. 10–40) were plated to high density into wells of a six-well plate and transfected the next day with 2 µg plasmid DNA using FuGENE6 according to the manufacturer’s instructions. After 24 h, cells were washed once with PBS, trypsinized, and plated onto glass coverslips at low density. Recordings were obtained within 2 h of plating. Borosilicate glass pipettes (Harvard Apparatus) were pulled and heat polished to a final resistance of 1–6 MΩ and backfilled with (in mM) 147 NaCl, 10 EGTA, and 10 HEPES (adjusted to pH 7.0 with NaOH). Patches were obtained in an external buffer containing (in mM) 147 NaCl, 2 KCl, 2 CaCl2, 1 MgCl2, 13 glucose, and 10 HEPES (adjusted to pH 7.3 with NaOH). A rapid solution exchange system (RSC-200; Bio-Logic) was used for recordings in which patches were perfused with drugs. Currents were recorded using an Axopatch 200B patch-clamp amplifier (Axon Instruments), filtered at 2 kHz (Frequency Devices), digitized with a Digidata 1440A (Axon Instruments) with a sampling frequency of 10 kHz, and analyzed using the pCLAMP 10.5 software (Axon Instruments). Recordings of Panx1 were obtained by performing voltage steps, in which cells were held at −60 mV and stepped between various voltages in 20-mV increments for 1 s.
Single-channel recording
CHO cells were plated onto glass coverslips and cotransfected with 100 ng pIE2 hPanx1 construct and 500 ng empty pIE2 vector (to help visualize transfected cells). Recordings were performed 24–48 h after transfection. Pipette tips were coated with Sylguard 184 (Dow Corning) and heat polished to a final resistance of 4–8 MΩ. The bath solution was the same as the whole-cell recordings, and the pipette solution contained (in mM) 110 CsCl, 37 tetraethylammonium chloride, 10 EGTA, and 10 HEPES adjusted to pH 7.0 with CsOH. After the outside-out configuration was obtained, the holding potential was increased to +60 or +120 mV. For single-channel recordings of WT hPanx1 at +60 mV, the presence of a channel was confirmed by stepping the holding potential to +120 mV. To confirm the channel identity, 100 µM CBX was perfused for 10 s using the rapid solution exchange system. Total recording lengths ranged from 30 s to 6 min. Recordings were sampled at 10 kHz and filtered at 1 kHz.
Single-channel data analysis
For WT hPanx1, raw traces were analyzed to obtain open probability (Po) and unitary G using the idealization function of the QuB software suite (https://milesculabs.biology.missouri.edu/QuB.html). In patches that contained more than one-step currents, Po was calculated under the assumption that each step correlated with gating of an independent channel. Patches with more than three steps were not used to calculate Po. For hPanx1 with the N-terminal GS insertion (hPanx1+GS), numerous subtransitions between closed and open states or various open states made it difficult to resolve distinct conductance classes at +60 mV and +120 mV.
Cell surface biotinylation
Constructs used in this assay were cloned into the pIE2 vector modified with a C-terminal FLAG tag or the N- or C-GFP EU2 vector. HEK293 cells were plated onto six-well plates and transfected at 100% confluency with 2 µg DNA using JetPrime (Polyplus) following the manufacturer’s instructions. After 24 h, cells were suspended, transferred to a 2-ml centrifuge tube, and washed twice with 1 ml PBS (Thermo Fisher Scientific). Surface membrane proteins were biotin labeled by resuspending cells in 2 ml PBS supplemented with 0.5 mg/ml sulfo-NHS-SS-biotin (Thermo Fisher Scientific) and rotated for 40 min at 4°C. The reaction was quenched by washing cells twice with 2 ml PBS supplemented with 50 mM NH4Cl, followed by a final wash with 1 ml PBS. Cells were lysed in 200 µl radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 3 mM MgCl2, 1% NP-40, 0.5% deoxycholate; Anatrace), 0.1% SDS, and 20 mM HEPES, pH adjusted to 7.4 with NaOH) supplemented with 1x protease inhibitor cocktail (Thermo Fisher Scientific) and rotated for 30 min. The lysate was clarified by centrifugation at 21,000 g for 15 min, and the supernatant was recovered. The “Input” samples were generated by mixing 30 µl of lysate with 15 µl 3x SDS sample buffer supplemented with 60 mM dithiothreitol (Thermo Fisher Scientific). Streptactin sepharose high-performance resin (GE Healthcare) was equilibrated in RIPA buffer, and 35 µl of 50% slurry was added to the remaining lysates and rotated for 2 h 30 min. Samples were washed six times by pelleting resin at 21,000 g for 2 min and resuspending in 700 µl RIPA buffer. Biotinylated proteins were eluted by incubating resin with 50 µl of 1.5x SDS sample buffer supplemented with 75 mM dithiothreitol for 30 min at 55°C with intermittent vortexing. The supernatants were recovered after a final spin at 21,000 g for 2 min and used as the “pull-down” samples. Samples were resolved on 9% SDS-PAGE gels and blotted onto nitrocellulose (Bio-Rad). Membranes were blocked with Tris-buffered saline/0.1% Tween (Anatrace) supplemented with 5% milk (Bio-Rad) and 1% BSA overnight; probed first with anti-GFP (1:2,000; Clontech), anti-FLAG (1:2,000; clone M2), or anti-actin monoclonal antibodies (1:2,000; line AC-40), then with goat anti–mouse alkaline phosphatase conjugate secondary antibody (1:2,000; Bio-Rad); and developed with colorimetric alkaline phosphatase substrate (Bio-Rad).
Fluorescence detection size exclusion chromatography (FSEC)
All constructs used for this experiment were cloned into the pNGFP-EU2 or pCGFP-EU2 vectors. FSEC experiments were performed as described previously (Kawate and Gouaux, 2006). Briefly, HEK293 cells were plated onto six-well plates and transfected at 100% confluency (usually next day) using 2 µg DNA and JetPrime according to the instructions. After 24 h, cells were suspended and transferred to a 2-ml centrifuge tube and centrifuged at 5,000 g for 5 min. Cells were washed with 1 ml PBS and centrifuged again at 5,000 g for 5 min. Cells were suspended in 150 µl solubilization buffer (1x PBS supplemented with 1% C12E8 and 1x protease inhibitor cocktail) and rocked for 30 min at 4°C. Samples were centrifuged at 21,000 g for 5 min at 4°C. The supernatants were transferred to 1.5-ml ultracentrifuge tubes and centrifuged again at ∼200,000 g for 20 min. 50 µl of each supernatant was injected into a Superose 6 Increase 10/300 GL column preequilibrated with running buffer (1x PBS supplemented with 0.5 mM C12E8) using a flow rate of 0.5 ml/min. The eluate was monitored using a fluorescence detector (Shimadzu RF-20Axs; excitation: 480, emission: 508).
Results
Panx1 gives rise to voltage-dependent currents at extremely high voltages
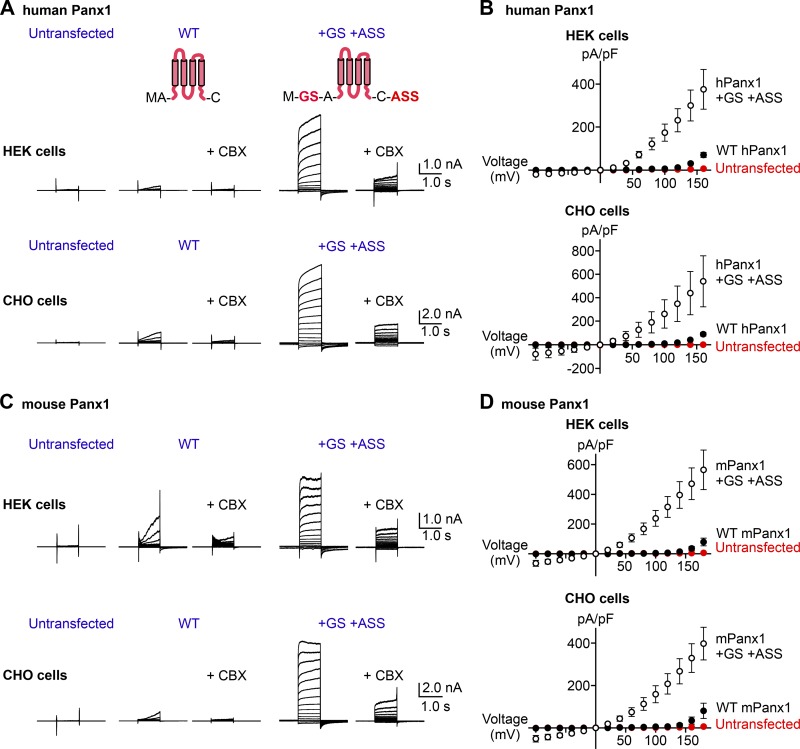
We and others previously demonstrated that the full-length hPanx1 presents robust CBX-sensitive channel activity at membrane potentials >+20 mV (Pelegrin and Surprenant, 2006; Michalski and Kawate, 2016). Other studies, however, report that hPanx1 does not respond to voltage stimuli at all or inconsistently give rise to voltage-dependent currents (Ma et al., 2009; Chekeni et al., 2010; Sandilos et al., 2012). To investigate a potential reason for these discrepancies, we reexamined our hPanx1 expression constructs we used in our previous studies. Due to our cloning strategy, our hPanx1 constructs contained insertions of GS right after the first methionine and ASS before the stop codon. We had overlooked these artificial insertions, as we assumed the addition of a few small amino acids at either terminus would not disrupt the structure and function of hPanx1. To assess whether these seemingly innocuous insertions affect the voltage-dependent channel activity, we restored the native termini of the hPanx1 and performed whole-cell patch-clamp experiments using HEK cells. To our surprise, hPanx1 with its native N and C termini (“WT hPanx1”) only weakly responded to voltage stimuli such that we did not observe detectable currents at voltages below +120 mV (Fig. 1, A and B). This was in contrast to the GS/ASS-inserted hPanx1 (hPanx1+GS/+ASS), which gave rise to strong currents at membrane potentials as low as +20 mV (Fig. 1, A and B). Importantly, we obtained consistent results using more than 30 cells with different passage numbers and from different transfections. We confirmed that these currents were mediated by hPanx1, as untransfected cells did not present detectable currents even at +160 mV, and the currents recorded from hPanx1-transfected HEK cells were CBX-sensitive (Fig. 1, A and B). We also confirmed that the weak voltage-dependent currents by WT hPanx1 and the stronger currents by hPanx1+GS/+ASS were not specific to HEK cells, as we obtained essentially the same results from these constructs expressed in CHO cells (Fig. 1, A and B). These results indicate that WT hPanx1 give rises to voltage-dependent currents, but only at extremely high voltages (>+100 mV). The large hPanx1 currents at lower voltages we reported previously (Michalski and Kawate, 2016) were due to the addition of a few amino acids in the expression constructs.
Figure 1.
Adding a few amino acids at the Panx1 termini augments voltage-dependent channel activity. (A) Whole-cell current recordings of voltage-clamped HEK cells (top) or CHO cells (bottom) expressing hPanx1. Shown are representative recordings of at least 10 different cells untransfected (left), transfected with WT hPanx1 (middle), or transfected with hPanx1+GS/+ASS (right). Cells were held at −60 mV and stepped between −100 mV and +160 mV for 1.0 s in 20-mV increments. CBX (50 µM) was applied using a rapid solution exchanger. Cartoons represent Panx1 topology and construct modifications. (B) Current density–voltage plots of hPanx1 constructs expressed in HEK cells (top) or CHO cells (bottom). (C) Whole-cell current recordings of HEK cells (top) or CHO cells (bottom) expressing mPanx1. Cells were either untransfected (left), transfected with WT mPanx1 (middle), or transfected with mPanx1+GS/+ASS (right). Shown are representative recordings from at least five different cells. (D) Current density–voltage plots of mPanx1 constructs expressed in HEK cells (top) or CHO cells (bottom). Each point represents the mean current density at each voltage, and bars represent SEM.
mPanx1 has also been widely used in the pannexin field. For example, it has been used to study single-channel conductance (Ma et al., 2012; Romanov et al., 2012; Poon et al., 2014; Wang et al., 2014), ion selectivity (Ma et al., 2012; Romanov et al., 2012), and voltage-gated channel opening (Romanov et al., 2012; Nomura et al., 2017). Like hPanx1, however, voltage-dependent activation of mPanx1 has been questioned; several studies suggest that mPanx1 is constitutively active (Sandilos et al., 2012; Chiu et al., 2018), but others failed to observe consistent currents (Ma et al., 2009; Dourado et al., 2014). To assess whether mPanx1 behaves similarly to hPanx1, we performed whole-cell patch-clamp recordings using HEK cells transfected with either WT mPanx1 or mPanx1+GS/+ASS. We found that WT mPanx1 also weakly responded to voltage stimuli and showed currents only above +100 mV (Fig. 1, C and D). Likewise, mPanx1+GS/+ASS gave rise to strong currents at voltages as low as +20 mV. We also found the same trend using mPanx1 expressed in CHO cells (Fig. 1, C and D). These data indicate that both hPanx1 and mPanx1 channels hardly open at a voltage range typically used in pannexin studies and respond to only extreme membrane potentials (>+100 mV). Because addition of only a few amino acids could significantly alter Panx1 channel activity, it is possible that small construct differences may contribute to discrepancies between laboratories in the field.
N-terminal insertion of a single amino acid could augment Panx1 channel activity
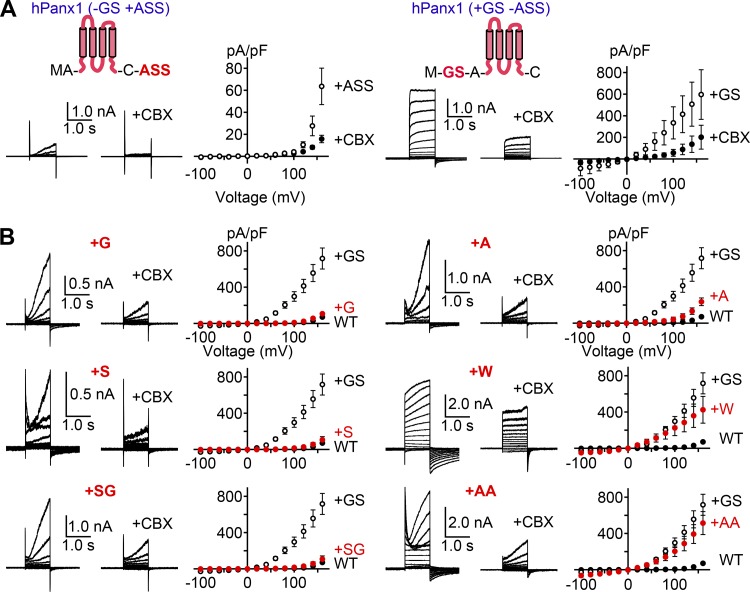
Strong voltage-dependent currents from the +GS/ASS construct suggest that even subtle modifications at the beginning and/or the end of Panx1 could profoundly enhance channel activity. To determine which insertion causes this unexpected channel activity enhancement, we tested hPanx1 constructs that have either N-terminal GS or C-terminal ASS. As shown in Fig. 2 A, addition or removal of the C-terminal ASS had no effect on voltage-dependent channel activity. In contrast, hPanx1 with N-terminal GS alone (“hPanx1+GS”) responded to voltages as low as +20 mV. These data indicate that the enhancement of hPanx1 channel activity is solely due to the N-terminal modification.
Figure 2.
N-terminal insertion alters voltage-dependent Panx1 channel activity. (A) Whole-cell recordings of HEK cells expressing hPanx1+ASS (left) or hPanx1+GS (right). Cells were held at −60 mV and stepped between −100 and +160 mV for 1.0 s in 20-mV increments. CBX (50 µM) was applied using a rapid solution exchange system. Shown are representative recordings from at least three different cells. Each point represents the mean current density at each voltage, and bars represent SEM. (B) Whole-cell recordings of hPanx1 constructs featuring variable amino acid insertions immediately following the start methionine. Recordings were obtained from transfected HEK cells held at −60 mV and stepped between −100 and +160 mV for 1.0 s. Each point represents the mean current density at each voltage, and bars represent SEM.
Is the enhancement of Panx1 channel activity specific to the GS motif? To assess whether different insertions mimic the GS-mediated gain-of-function effect, we varied the inserted residues at the N terminus and examined voltage-dependent channel activity using a whole-cell patch-clamp method. We found that insertion of glycine (G) or serine (S) alone did not affect hPanx1 channel activity and showed slowly activating currents only above +100 mV (Fig. 2 B). Likewise, insertion of alanine (A) at this position had little effect on hPanx1 channel activity. Interestingly, insertion of the SG motif also failed to enhance hPanx1 channel activity. Insertion of tryptophan (W) or AA, on the other hand, enhanced the channel activity like insertion of the GS motif (Fig. 2 B). These results suggest some specificity in the motifs that can augment Panx1 channel activity. We also noticed that channel activation and inactivation kinetics varied substantially depending on the amino acids inserted at this position. Altogether, these data suggest that the hPanx1 N terminus is sensitive to modifications, and even a single amino acid insertion can substantially enhance channel activity.
N-terminal GFP fusion abolishes Panx1 channel activity
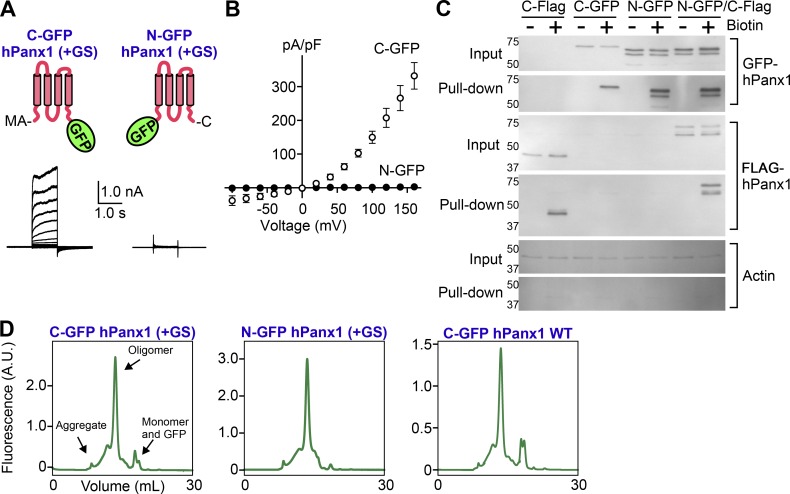
Fluorescent proteins such as GFP are commonly attached to Panx1 for studying subcellular localization and trafficking (Bhalla-Gehi et al., 2010; Gehi et al., 2011; Bao et al., 2012). Because N-terminal modification could enhance Panx1 channel activity, we wondered whether a GFP tag might affect its activity. To facilitate whole-cell recordings, we used hPanx1+GS constructs for these experiments. When GFP was attached to the C terminus, hPanx1 gave rise to outwardly rectifying voltage-dependent currents, just like the untagged parent construct (Fig. 3, A and B). In contrast, when GFP was attached to the N terminus, we did not detect voltage-dependent currents even at +160 mV (Fig. 3, A and B). These imply that that an N-terminal GFP tag abolishes hPanx1 channel activity, whereas the C-terminal GFP has little effect.
Figure 3.
N-terminal GFP abolishes Panx1 channel activity. (A) Whole-cell recordings of WT hPanx1 or GFP-tagged Panx1 expressed in HEK cells. Cells were held at a resting membrane potential of −60 mV and stepped between −100 and +160 mV in 20-mV increments for 1.0 s. Shown are representative recordings from 8–10 cells. Cartoons represent positions of the tagged GFPs. (B) Current density–voltage plots. Each point in the current density–voltage plots represents the mean conductance at each voltage, and error bars represent SEM. (C) Surface biotinylation assay of hPanx1, GFP-tagged Panx1, or N-GFP C-FLAG–tagged Panx1. Panx1 constructs were transfected into HEK cells and treated with sulfo-NHS-SS-biotin. Biotin-labeled proteins were analyzed by SDS-PAGE and Western blotting. Membranes were probed with GFP, FLAG, or actin antibodies. Representative blots from three independent experiments are shown. (D) FSEC elution profiles of N- and C-terminally GFP-tagged hPanx1. Constructs were transfected into HEK cells, solubilized in detergent, and analyzed by FSEC. A.U., arbitrary unit.
An alternative explanation for this result could simply be that the N-terminal GFP tag interferes with trafficking of the channel to the cell membrane. We performed cell-surface biotinylation to test this possibility (Fig. 3 C). Using a membrane-impermeable, amine-reactive, biotin-conjugated molecule, we labeled and enriched for membrane proteins located on the surface of transfected HEK cells and used Western blotting to identify Panx1. Both N-GFP and C-GFP Panx1 proteins could be identified in the pull-down samples, suggesting both fusion proteins traffic to the cell surface. Actin could be identified in the protein lysates but not in the pull-down samples, suggesting this assay specifically pulls down membrane proteins. We noticed, however, that the N-GFP Panx1 construct migrates faster than the C-GFP counterpart when resolved by SDS-PAGE, and additionally, that N-GFP Panx1 appears as a doublet compared with the single band observed for C-GFP Panx1. One possibility for this difference could be that the C terminus of N-GFP Panx1 gets cleaved by proteases before or during sample preparation. To test this possibility, we added a C-terminal FLAG tag to the N-GFP Panx1 construct (N-GFP C-FLAG Panx1). We were able to pull down this doubly tagged Panx1 and identify both the N-terminal GFP tag and the C-terminal FLAG tag at similar molecular weights. This suggests that N-terminally tagged Panx1 is not susceptible to proteases and has intact termini.
To analyze whether modifications of the N terminus affect hPanx1 channel assembly, we took advantage of the FSEC. This protocol enables one to characterize assembly, stability, and monodispersity of a detergent-solubilized GFP-fusion protein based on the size exclusion chromatography elution profiles (Kawate and Gouaux, 2006). The FSEC traces of both C-GFP and N-GFP hPanx1 presented a major peak at a molecular weight corresponding to the size of a mature oligomer (6–7 mers), indicating that these proteins remained properly folded and assembled after solubilization in a detergent-containing buffer (Fig. 3 D). There was little aggregation (i.e., void peak) or dissociation (i.e., monomeric peak), suggesting that the majority of WT hPanx1 was properly folded in HEK cells. The effect of N-terminal GS on the assembly was minimal, as removal of these two residues did not affect the FSEC profile (Fig. 3 D, right panel). Therefore, we conclude that N-terminally attached GFP renders hPanx1 insensitive to voltage stimuli, and this is not caused by poor membrane localization or unwanted proteolysis of the channel.
N-terminal GS increases Panx1 Po
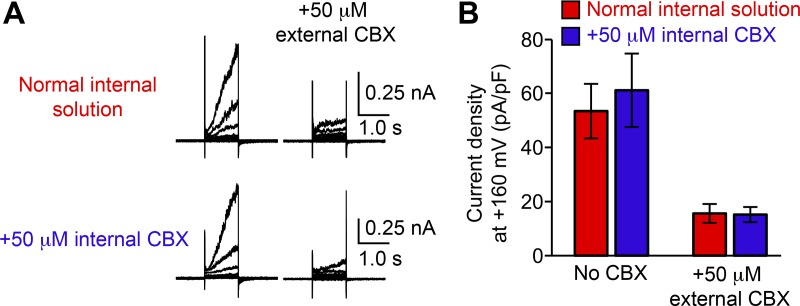
Our data imply that Panx1 is sensitive to N-terminal modifications. To further explore how such a minor manipulation—like insertion of the GS motif—enhances Panx1 channel activity, we performed single-channel recordings. Because single-channel properties of Panx1 are not fully established (Chiu et al., 2018), we used both untransfected cells and CBX to verify the recorded currents were actually mediated by Panx1. From our whole-cell recordings, it was clear that externally applied CBX effectively inhibits hPanx1 (Fig. 1). To assess whether CBX can also inhibit hPanx1 from inside, we included CBX in the pipette solution and measured voltage-dependent channel activity. Under these conditions, WT hPanx1 expressed in HEK cells gave rise to voltage-dependent currents indistinguishable from those recorded in the absence of CBX (Fig. 4, A and B). Importantly, extracellularly applied CBX attenuated WT hPanx1–mediated currents with internal CBX, excluding potential contamination of unknown channel activity (Fig. 4 A). These results indicate that CBX needs to be externally applied for inhibiting voltage-dependent Panx1 currents. We therefore used the outside-out patch configuration for our single-channel recordings.
Figure 4.
CBX inhibits Panx1 only from the extracellular side. (A) Representative recordings from WT hPanx1–transfected HEK cells with normal internal solution (top) or internal solution containing 50 µM CBX (bottom). Cells were held at −60 mV and equilibrated for ∼3 min before stepping between −100 and +160 mV for 1.0 s in 20-mV increments. External CBX (50 µM) was applied using a rapid solution exchanger. Shown are representative recordings for four different cells. (B) Current density of recordings shown in A. Bars represent the mean current density of patches obtained with either internal solution with and without external CBX application, and error bars represent SEM. Student’s t test revealed no statistically significant difference with and without internal CBX.
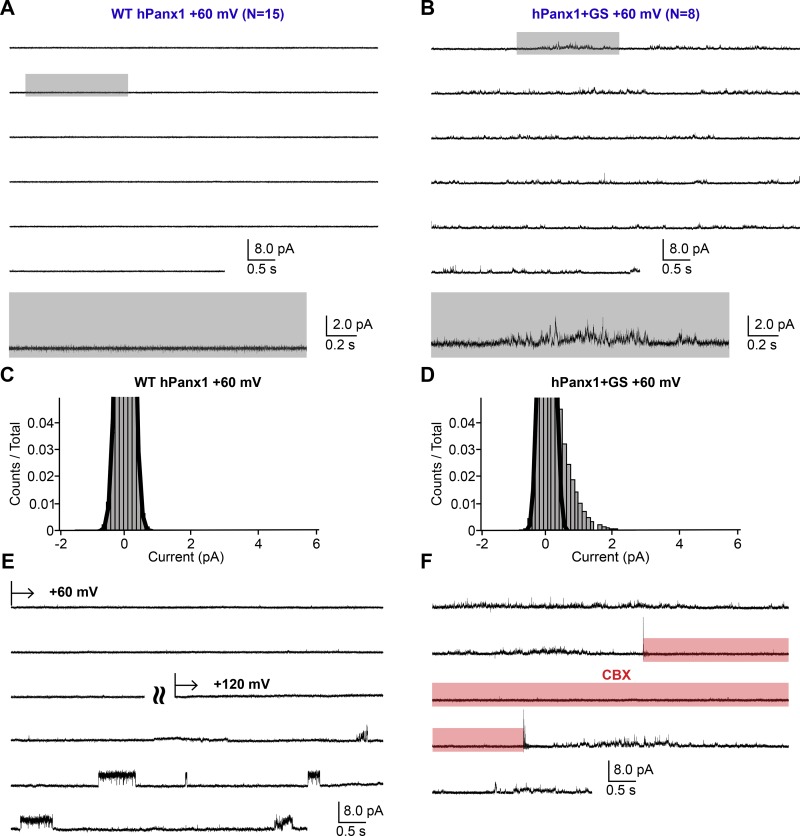
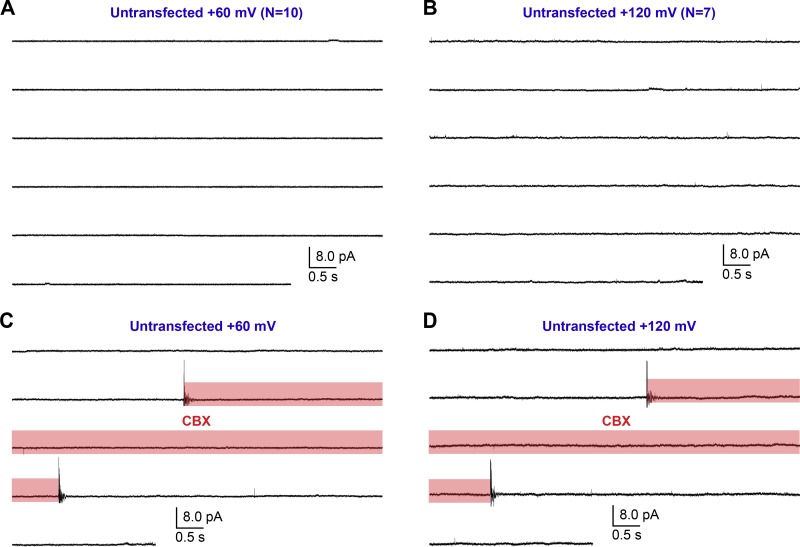
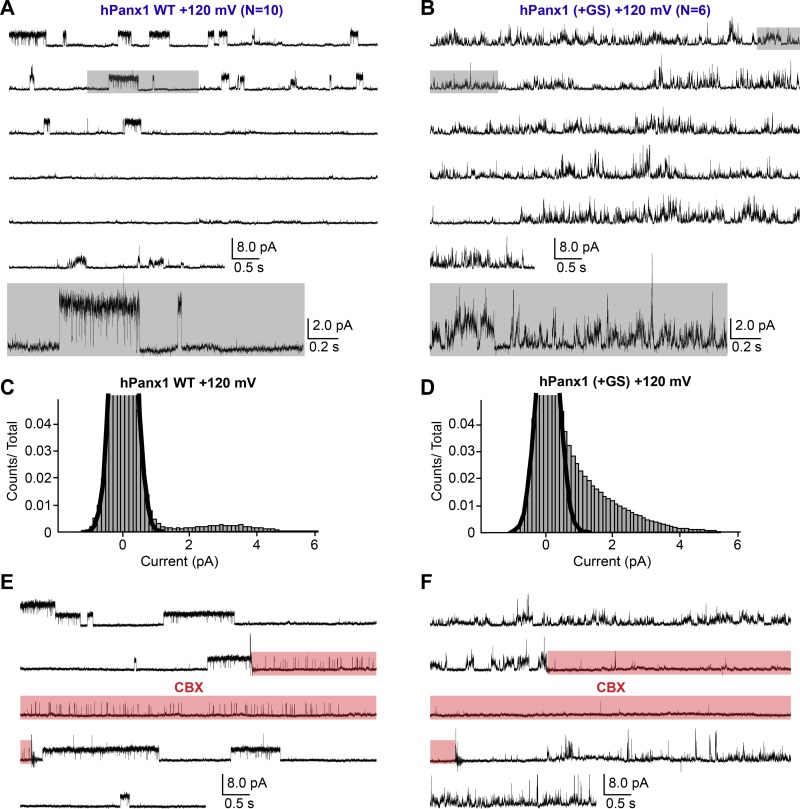
Consistent with the macroscopic observations (Fig. 1, A and B), WT hPanx1 presented no channel activity at +60 mV (Fig. 5, A and C). We confirmed the presence of a WT hPanx1 channel by measuring channel activity at +120 mV from the same patch (Fig. 5 E). We also confirmed that untransfected cells do not give rise to apparent channel activity at either +60 or +120 mV (Fig. 6, A and B). On the other hand, hPanx1+GS presented conspicuous channel activity at +60 mV, which was reversibly suppressed by externally applied CBX (Fig. 5, B, D, and F). While we were able to observe some clear steps representing discrete unitary currents between 1 and 3 pA, it was difficult to assign specific unitary conductance to those steps, as many transient currents also existed and obscured the overall current histogram (Fig. 5 D). These results suggest that the N-terminal insertion of GS increases the Po of hPanx1 at this membrane potential. At +120 mV, WT hPanx1 showed a discrete unitary current at 3.3 ± 0.4 pA with the Po of 7.3 ± 2.7% (Fig. 7, A and C). These currents were CBX-sensitive, though we observed clear residual channel activity (Fig. 7 E). CBX did not seem to induce these residual currents, as patches from untransfected cells did not show any channel activity in the presence of CBX (Fig. 6, C and D). In contrast, hPanx1+GS presented nondiscrete unitary currents ranging from 1 to 6 pA at +120 mV that were strongly attenuated by CBX (Fig. 7, B, D, and F). Like at +60 mV, it was difficult to assign particular unitary conductance to these currents. Nevertheless, overall Po of hPanx1+GS was much higher than that of WT hPanx1. Altogether, these single-channel recordings suggest that the insertion of the GS motif at the N terminus generates extra channel conductance classes and substantially increases hPanx1 channel Po.
Figure 5.
Single-channel recordings of hPanx1 at +60 mV. (A and B) Representative single-channel traces of WT hPanx1 (A; n = 15) and hPanx1+GS (B; n = 8). Outside-out patches were held at +60 mV. Highlighted gray box indicates the location of the inset underneath each respective recording. (C and D) Pooled all-amplitudes histogram generated from WT hPanx1 (C) and hPanx1+GS (D) at +60 mV. Histograms were generated from four separate recordings for each channel. Black lines represent the closed population estimated by fitting a Gaussian function. (E) Representative trace of WT hPanx1 recording confirming the existence of the channel. The patch was held at +60 mV for ∼2 min, then stepped to +120 mV. Curved lines indicate separate recording segments. (F) Representative trace of hPanx1+GS showing reversible channel activity at +60 mV. CBX (100 µM) was applied using a rapid solution exchanger for the indicated period (red box).
Figure 6.
Single-channel recordings of untransfected CHO cells. (A and B) Outside-out patches pulled from untransfected CHO cells held at +60 mV (A; n = 10) and +120 mV (B; n = 7). (C and D) Outside-out patches held at +60 mV (C) +120 mV (D) were treated with 100 µM CBX using a rapid solution exchange system during the indicated period (red box).
Figure 7.
Single-channel recordings of hPanx1 at +120 mV. (A and B) Representative single-channel traces of WT hPanx1 (A; n = 10) and hPanx1+GS (B; n = 6). Outside-out patches were held at +120 mV. Highlighted gray box indicates the location of the inset underneath each respective recording. (C and D) Pooled all-amplitudes histogram generated from WT hPanx1 (C) and hPanx1+GS (D) at +120mV. Histograms were generated from four separate recordings for each channel. Black lines represent the closed population estimated by fitting a Gaussian function. (E and F) Representative traces of WT hPanx1 (E) and hPanx1+GS (F) showing reversible channel activity at +120 mV. CBX (100 µM) was applied using a rapid solution exchanger for the indicated period (red box).
Discussion
In this study, we demonstrate that Panx1 is a weak voltage-gated channel, and a modification of the N terminus can alter its voltage-dependent channel activity. By carefully reevaluating our full-length construct, we find that WT Panx1 with its native N terminus only activates at voltages above +100 mV. We also demonstrate that addition of one or two specific amino acid(s), GS in particular, immediately after the first methionine, substantially enhances Panx1 channel activity. In contrast, attachment of a large protein like GFP abolishes Panx1 activity. Importantly, such N-terminal manipulations do not seem to affect Panx1 channel folding or assembly. These results are consistent with the idea that the Panx1 N terminus is not merely hanging in the cytoplasm, but rather forms a structured domain and plays a role in regulating channel gating or ion permeation.
We previously reported that hPanx1 channels give rise to conspicuous voltage-dependent currents (Michalski and Kawate, 2016). We found in this study, however, that such strong voltage-dependent currents were facilitated by the GS motif at the N terminus. Indeed, WT hPanx1 with its native N terminus actually responds to voltages only beyond +100 mV. This unusually weak voltage-dependent activity might be a reason why some groups failed to record voltage-activated hPanx1 currents (Chiu et al., 2018). Notably, we found that mPanx1 also responds to voltage very weakly, and its activity is remarkably enhanced by the addition of GS. This is not limited to the channel expressed in HEK cells, as we also observed similar activities from CHO cells. These results are inconsistent with previous studies, which showed robust and perhaps constitutive channel activity from mPanx1 (Romanov et al., 2012; Chiu et al., 2018). Though the actual reason for this discrepancy is unclear, it is possible that mPanx1 with an unintentionally altered N terminus might be used in those studies. Alternatively, different degrees of posttranslational modifications, which could be due to various passage numbers or different culture conditions, may result in inconsistent mPanx1 channel activity. Indeed, phosphorylation and S-nitrosylation have been shown to modulate Panx1 channel activity (Lohman et al., 2012; Weilinger et al., 2012; Penuela et al., 2014b).
GS insertion is an excellent tool for studying Panx1, as voltage stimulation is convenient and easily controlled. But how does it enhance Panx1 channel activity? Our single-channel recordings revealed that +GS elevates the apparent Po of the hPanx1 channel, which normally opens only at an extremely high membrane potential (i.e., >+120 mV). This N-terminal manipulation also gave rise to a wide range of unitary currents, which do not seem to exist in WT hPanx1. One potential mechanism is that the natural N terminus contributes to keep the channel closed under normal conditions, rendering it insensitive to voltages up to +120 mV. Addition of GS may dislocate the N terminus, which could lead to a wobbly pore that opens more frequently and less uniformly at positive membrane potentials. Because a glycine residue at the second position could get cotranslationally myristoylated (Boutin, 1997), the N terminus of hPanx1+GS may be rearranged due to lipidation. This idea could explain why insertion of SG had no effect on hPanx1 and insertions of a hydrophobic residue such as tryptophan mimicked the effect of GS.
How does hPanx1+GS respond to positive membrane potentials? One possibility is that the N-terminal domain acts as a voltage sensor. Given that (a) Panx1 does not possess charged residues in the transmembrane helices and (b) the connexin N terminus harbors a voltage sensor (Harris and Contreras, 2014), this idea sounds reasonable. If the Panx1 N terminus forms a helix within the membrane pore, this helix dipole likely repositions when an unusually high voltage is applied. Another possibility is that the N terminus is not directly involved in voltage sensing but governs other channel properties such as ion selectivity or conductance. If this is the case, voltage is somehow sensed by the transmembrane or other unfound domains in the voltage field. Movement of the voltage sensor then triggers rearrangement of the N terminus, which could alter the electrostatic surface potential or physical diameter of the permeation pore, allowing more ions to flow. Notably, manipulation of a connexin N terminus, which lies within the pore lumen, also alters ion permeation properties (Maeda et al., 2009; Harris and Contreras, 2014). It is possible that the N terminus of Panx1 may also fold up into the channel cavity and forms part of the permeation pathway. This can explain why an N-terminal manipulation leads to loss of channel activity, as the permeation pathway could easily be disrupted by these modifications.
Our single-channel recordings revealed that hPanx1 opens a channel at +120 mV with an estimated unitary conductance of ∼20–40 pS. This is less than half of the channel activated by C-terminal cleavage or by α1-adrenoceptor (Chiu et al., 2017). We suspect that perhaps only a subset of the C termini move out from the hPanx1 pore at +120 mV. Unfortunately, we could not test whether unitary conductance becomes larger at higher voltages, as single-channel patches did not survive at those membrane potentials. An alternative explanation is that the mode of channel opening at such a high voltage substantially differs from other modes, which are normally tested at membrane potentials up to +80 mV.
In conclusion, we demonstrate that Panx1 hardly opens at a positive membrane potential, but its activity redoubles when a couple of specific amino acids (GS in particular) are inserted at the N terminus. While this is an artificial way to promote Panx1 activity, a natural facilitation mechanism may exist. For example, posttranslational modification or interaction with the other cytoplasmic domains at the N terminus may enhance the channel activity. Involvement of the N terminus in Panx1 channel gating/ion permeation supports that pannexins may be structural analogues of connexins and innexins. Therefore, care must be taken when manipulating the N terminus, which could be a powerful tool for studying Panx1 function.
Acknowledgments
We thank the Kawate laboratory members for discussion.
This work was supported by the National Institutes of Health (GM008267, GM114379, and NS101390).
The authors declare no competing financial interests.
Author contributions: K. Michalski, E. Henze, and T. Kawate designed the project. K. Michalski performed the whole-cell patch-clamp recordings and biochemical experiments with the help of P. Nguyen and P. Lynch. E. Henze performed single-channel recordings. K. Michalski and T. Kawate wrote the manuscript.
Sharona E. Gordon served as editor.
References
- Bao B.A., Lai C.P., Naus C.C., and Morgan J.R.. 2012. Pannexin1 drives multicellular aggregate compaction via a signaling cascade that remodels the actin cytoskeleton. J. Biol. Chem. 287:8407–8416. 10.1074/jbc.M111.306522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao L., Locovei S., and Dahl G.. 2004. Pannexin membrane channels are mechanosensitive conduits for ATP. FEBS Lett. 572:65–68. 10.1016/j.febslet.2004.07.009 [DOI] [PubMed] [Google Scholar]
- Bhalla-Gehi R., Penuela S., Churko J.M., Shao Q., and Laird D.W.. 2010. Pannexin1 and pannexin3 delivery, cell surface dynamics, and cytoskeletal interactions. J. Biol. Chem. 285:9147–9160. 10.1074/jbc.M109.082008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billaud M., Chiu Y.H., Lohman A.W., Parpaite T., Butcher J.T., Mutchler S.M., DeLalio L.J., Artamonov M.V., Sandilos J.K., Best A.K., et al. . 2015. A molecular signature in the pannexin1 intracellular loop confers channel activation by the α1 adrenoreceptor in smooth muscle cells. Sci. Signal. 8:ra17 10.1126/scisignal.2005824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutin J.A. 1997. Myristoylation. Cell. Signal. 9:15–35. 10.1016/S0898-6568(96)00100-3 [DOI] [PubMed] [Google Scholar]
- Bruzzone R., Barbe M.T., Jakob N.J., and Monyer H.. 2005. Pharmacological properties of homomeric and heteromeric pannexin hemichannels expressed in Xenopus oocytes. J. Neurochem. 92:1033–1043. 10.1111/j.1471-4159.2004.02947.x [DOI] [PubMed] [Google Scholar]
- Chekeni F.B., Elliott M.R., Sandilos J.K., Walk S.F., Kinchen J.M., Lazarowski E.R., Armstrong A.J., Penuela S., Laird D.W., Salvesen G.S., et al. . 2010. Pannexin 1 channels mediate ‘find-me’ signal release and membrane permeability during apoptosis. Nature. 467:863–867. 10.1038/nature09413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y.H., Jin X., Medina C.B., Leonhardt S.A., Kiessling V., Bennett B.C., Shu S., Tamm L.K., Yeager M., Ravichandran K.S., and Bayliss D.A.. 2017. A quantized mechanism for activation of pannexin channels. Nat. Commun. 8:14324 10.1038/ncomms14324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu Y.H., Schappe M.S., Desai B.N., and Bayliss D.A.. 2018. Revisiting multimodal activation and channel properties of Pannexin 1. J. Gen. Physiol. 150:19–39. 10.1085/jgp.201711888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl G. 2015. ATP release through pannexon channels. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370:20140191 10.1098/rstb.2014.0191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl G. 2018. The Pannexin1 membrane channel: distinct conformations and functions. FEBS Lett. 592:3201–3209. 10.1002/1873-3468.13115 [DOI] [PubMed] [Google Scholar]
- Dahl G., and Keane R.W.. 2012. Pannexin: from discovery to bedside in 11±4 years? Brain Res. 1487:150–159. 10.1016/j.brainres.2012.04.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dourado M., Wong E., and Hackos D.H.. 2014. Pannexin-1 is blocked by its C-terminus through a delocalized non-specific interaction surface. PLoS One. 9:e99596 10.1371/journal.pone.0099596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furlow P.W., Zhang S., Soong T.D., Halberg N., Goodarzi H., Mangrum C., Wu Y.G., Elemento O., and Tavazoie S.F.. 2015. Mechanosensitive pannexin-1 channels mediate microvascular metastatic cell survival. Nat. Cell Biol. 17:943–952. 10.1038/ncb3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehi R., Shao Q., and Laird D.W.. 2011. Pathways regulating the trafficking and turnover of pannexin1 protein and the role of the C-terminal domain. J. Biol. Chem. 286:27639–27653. 10.1074/jbc.M111.260711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulbransen B.D., Bashashati M., Hirota S.A., Gui X., Roberts J.A., MacDonald J.A., Muruve D.A., McKay D.M., Beck P.L., Mawe G.M., et al. . 2012. Activation of neuronal P2X7 receptor-pannexin-1 mediates death of enteric neurons during colitis. Nat. Med. 18:600–604. 10.1038/nm.2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A.L., and Contreras J.E.. 2014. Motifs in the permeation pathway of connexin channels mediate voltage and Ca (2+) sensing. Front. Physiol. 5:113 10.3389/fphys.2014.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isakson B.E., and Thompson R.J.. 2014. Pannexin-1 as a potentiator of ligand-gated receptor signaling. Channels (Austin). 8:118–123. 10.4161/chan.27978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D.G., Wang J., Keane R.W., Scemes E., and Dahl G.. 2014. ATP and potassium ions: a deadly combination for astrocytes. Sci. Rep. 4:4576 10.1038/srep04576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J.X., and Penuela S.. 2016. Connexin and pannexin channels in cancer. BMC Cell Biol. 17(suppl 1):12 10.1186/s12860-016-0094-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawate T., and Gouaux E.. 2006. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 14:673–681. 10.1016/j.str.2006.01.013 [DOI] [PubMed] [Google Scholar]
- Locovei S., Wang J., and Dahl G.. 2006. Activation of pannexin 1 channels by ATP through P2Y receptors and by cytoplasmic calcium. FEBS Lett. 580:239–244. 10.1016/j.febslet.2005.12.004 [DOI] [PubMed] [Google Scholar]
- Lohman A.W., Weaver J.L., Billaud M., Sandilos J.K., Griffiths R., Straub A.C., Penuela S., Leitinger N., Laird D.W., Bayliss D.A., and Isakson B.E.. 2012. S-nitrosylation inhibits pannexin 1 channel function. J. Biol. Chem. 287:39602–39612. 10.1074/jbc.M112.397976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman A.W., Leskov I.L., Butcher J.T., Johnstone S.R., Stokes T.A., Begandt D., DeLalio L.J., Best A.K., Penuela S., Leitinger N., et al. . 2015. Pannexin 1 channels regulate leukocyte emigration through the venous endothelium during acute inflammation. Nat. Commun. 6:7965 10.1038/ncomms8965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Hui H., Pelegrin P., and Surprenant A.. 2009. Pharmacological characterization of pannexin-1 currents expressed in mammalian cells. J. Pharmacol. Exp. Ther. 328:409–418. 10.1124/jpet.108.146365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W., Compan V., Zheng W., Martin E., North R.A., Verkhratsky A., and Surprenant A.. 2012. Pannexin 1 forms an anion-selective channel. Pflugers Arch. 463:585–592. 10.1007/s00424-012-1077-z [DOI] [PubMed] [Google Scholar]
- Maeda S., Nakagawa S., Suga M., Yamashita E., Oshima A., Fujiyoshi Y., and Tsukihara T.. 2009. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature. 458:597–602. 10.1038/nature07869 [DOI] [PubMed] [Google Scholar]
- Michalski K., and Kawate T.. 2016. Carbenoxolone inhibits Pannexin1 channels through interactions in the first extracellular loop. J. Gen. Physiol. 147:165–174. 10.1085/jgp.201511505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milescu L. 2018. QuB – The MLab Edition. Available at: https://milesculabs.biology.missouri.edu/QuB.html (accessed October 27, 2018).
- Nomura T., Taruno A., Shiraishi M., Nakahari T., Inui T., Sokabe M., Eaton D.C., and Marunaka Y.. 2017. Current-direction/amplitude-dependent single channel gating kinetics of mouse pannexin 1 channel: a new concept for gating kinetics. Sci. Rep. 7:10512 10.1038/s41598-017-10921-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelegrin P., and Surprenant A.. 2006. Pannexin-1 mediates large pore formation and interleukin-1beta release by the ATP-gated P2X7 receptor. EMBO J. 25:5071–5082. 10.1038/sj.emboj.7601378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penuela S., Harland L., Simek J., and Laird D.W.. 2014a Pannexin channels and their links to human disease. Biochem. J. 461:371–381. 10.1042/BJ20140447 [DOI] [PubMed] [Google Scholar]
- Penuela S., Simek J., and Thompson R.J.. 2014b Regulation of pannexin channels by post-translational modifications. FEBS Lett. 588:1411–1415. 10.1016/j.febslet.2014.01.028 [DOI] [PubMed] [Google Scholar]
- Poon I.K., Chiu Y.H., Armstrong A.J., Kinchen J.M., Juncadella I.J., Bayliss D.A., and Ravichandran K.S.. 2014. Unexpected link between an antibiotic, pannexin channels and apoptosis. Nature. 507:329–334. 10.1038/nature13147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu F., and Dahl G.. 2009. A permeant regulating its permeation pore: inhibition of pannexin 1 channels by ATP. Am. J. Physiol. Cell Physiol. 296:C250–C255. 10.1152/ajpcell.00433.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov R.A., Bystrova M.F., Rogachevskaya O.A., Sadovnikov V.B., Shestopalov V.I., and Kolesnikov S.S.. 2012. The ATP permeability of pannexin 1 channels in a heterologous system and in mammalian taste cells is dispensable. J. Cell Sci. 125:5514–5523. 10.1242/jcs.111062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilos J.K., and Bayliss D.A.. 2012. Physiological mechanisms for the modulation of pannexin 1 channel activity. J. Physiol. 590:6257–6266. 10.1113/jphysiol.2012.240911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandilos J.K., Chiu Y.H., Chekeni F.B., Armstrong A.J., Walk S.F., Ravichandran K.S., and Bayliss D.A.. 2012. Pannexin 1, an ATP release channel, is activated by caspase cleavage of its pore-associated C-terminal autoinhibitory region. J. Biol. Chem. 287:11303–11311. 10.1074/jbc.M111.323378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman W., Locovei S., and Dahl G.. 2008. Probenecid, a gout remedy, inhibits pannexin 1 channels. Am. J. Physiol. Cell Physiol. 295:C761–C767. 10.1152/ajpcell.00227.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman W.R., de Rivero Vaccari J.P., Locovei S., Qiu F., Carlsson S.K., Scemes E., Keane R.W., and Dahl G.. 2009. The pannexin 1 channel activates the inflammasome in neurons and astrocytes. J. Biol. Chem. 284:18143–18151. 10.1074/jbc.M109.004804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R.J. 2015. Pannexin channels and ischaemia. J. Physiol. 593:3463–3470. 10.1113/jphysiol.2014.282426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R.J., Jackson M.F., Olah M.E., Rungta R.L., Hines D.J., Beazely M.A., MacDonald J.F., and MacVicar B.A.. 2008. Activation of pannexin-1 hemichannels augments aberrant bursting in the hippocampus. Science. 322:1555–1559. 10.1126/science.1165209 [DOI] [PubMed] [Google Scholar]
- Velasquez S., and Eugenin E.A.. 2014. Role of Pannexin-1 hemichannels and purinergic receptors in the pathogenesis of human diseases. Front. Physiol. 5:96 10.3389/fphys.2014.00096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Ambrosi C., Qiu F., Jackson D.G., Sosinsky G., and Dahl G.. 2014. The membrane protein Pannexin1 forms two open-channel conformations depending on the mode of activation. Sci. Signal. 7:ra69 10.1126/scisignal.2005431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver J.L., Arandjelovic S., Brown G., K Mendu S., S Schappe M., Buckley M.W., Chiu Y.H., Shu S., Kim J.K., Chung J., et al. . 2017. Hematopoietic pannexin 1 function is critical for neuropathic pain. Sci. Rep. 7:42550 10.1038/srep42550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilinger N.L., Tang P.L., and Thompson R.J.. 2012. Anoxia-induced NMDA receptor activation opens pannexin channels via Src family kinases. J. Neurosci. 32:12579–12588. 10.1523/JNEUROSCI.1267-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weilinger N.L., Lohman A.W., Rakai B.D., Ma E.M., Bialecki J., Maslieieva V., Rilea T., Bandet M.V., Ikuta N.T., Scott L., et al. . 2016. Metabotropic NMDA receptor signaling couples Src family kinases to pannexin-1 during excitotoxicity. Nat. Neurosci. 19:432–442. 10.1038/nn.4236 [DOI] [PubMed] [Google Scholar]