Drugs that target insulin secretion are useful to understand β cell function and the pathogenesis of diabetes. Kalwat et al. investigate an aureolic acid that inhibits insulin secretion and reveal that it disrupts Wnt signaling, interferes with gene expression, and suppresses Ca2+ influx in β cells.
Abstract
Modulators of insulin secretion could be used to treat diabetes and as tools to investigate β cell regulatory pathways in order to increase our understanding of pancreatic islet function. Toward this goal, we previously used an insulin-linked luciferase that is cosecreted with insulin in MIN6 β cells to perform a high-throughput screen of natural products for chronic effects on glucose-stimulated insulin secretion. In this study, using multiple phenotypic analyses, we found that one of the top natural product hits, chromomycin A2 (CMA2), potently inhibited insulin secretion by at least three potential mechanisms: disruption of Wnt signaling, interference of β cell gene expression, and partial suppression of Ca2+ influx. Chronic treatment with CMA2 largely ablated glucose-stimulated insulin secretion even after washout, but it did not inhibit glucose-stimulated generation of ATP or Ca2+ influx. However, by using the KATP channel opener diazoxide, we uncovered defects in depolarization-induced Ca2+ influx that may contribute to the suppressed secretory response. Glucose-responsive ERK1/2 and S6 phosphorylation were also disrupted by chronic CMA2 treatment. By querying the FUSION bioinformatic database, we revealed that the phenotypic effects of CMA2 cluster with a number of Wnt–GSK3 pathway-related genes. Furthermore, CMA2 consistently decreased GSK3β phosphorylation and suppressed activation of a β-catenin activity reporter. CMA2 and a related compound, mithramycin, are known to have DNA interaction properties, possibly abrogating transcription factor binding to critical β cell gene promoters. We observed that CMA2 but not mithramycin suppressed expression of PDX1 and UCN3. However, neither expression of INSI/II nor insulin content was affected by chronic CMA2. The mechanisms of CMA2-induced insulin secretion defects may involve components both proximal and distal to Ca2+ influx. Therefore, CMA2 is an example of a chemical that can simultaneously disrupt β cell function through both noncytotoxic and cytotoxic mechanisms. Future therapeutic applications of CMA2 and similar aureolic acid analogues should consider their potential effects on pancreatic islet function.
Introduction
Insulin secretion from β cells is one of the most important functions of the pancreatic islet. Type 1 and type 2 diabetes combined afflict 9.4% of Americans (Centers for Disease Control and Prevention, 2017) and result from the autoimmune destruction of β cells or defects in insulin secretion and action, respectively. Diabetes is usually treated by insulin replacement or by β cell–targeted therapeutics, which are typically inducers of insulin secretion. However, insulin hypersecretion also occurs in type 2 diabetes and other disorders such as persistent hyperinsulinemic hypoglycemia of infancy and polycystic ovary syndrome (Cordain et al., 2003; Molven et al., 2004; Stanley, 2011). Accordingly, suppression of insulin secretion has been considered in the past as a therapeutic avenue (Hansen et al., 2004). Pancreatic islet β cells sense hyperglycemia and respond by metabolizing glucose, increasing their intracellular [ATP/ADP] ratio and causing the closure of ATP-sensitive potassium channels. These steps lead to the opening of voltage-dependent Ca2+ channels, Ca2+ influx, and insulin granule exocytosis denoted as “triggering” (Henquin, 2000). Glucose metabolism also mediates an amplifying pathway through the production of other signals that drive insulin secretion independent from further Ca2+ influx (Gembal et al., 1992; Sato et al., 1992). Much is known about the triggering pathway; however, less is understood about the glucose-mediated metabolic amplifying pathway, which can account for half of the insulin secretion response (Henquin, 2009; Henquin et al., 2017). In particular, the signaling pathways and transcriptional outputs that are chronically required for regulated insulin secretion are not completely understood. Further development of knowledge and tools pertaining to these aspects of β cell function are required to uncover new directions for research and to guide disease therapies.
Marine natural products are a rich resource for potential new chemical tools and therapies. A unique custom library of natural compounds assembled at University of Texas (UT) Southwestern has been used in screens to discover new leads for chemotherapies (Hu et al., 2011; Potts et al., 2013, 2015; Vaden et al., 2017); inhibitors of endocytosis (Elkin et al., 2016); antibiotics (Hu et al., 2013); and, in our recent screening efforts, modulators of pancreatic β cell function (Kalwat et al., 2016b). As a result, we discovered that the natural product chromomycin A2 (CMA2) is a potent inhibitor of insulin secretion. The CMA2 is a member of a family of glycosylated aromatic polyketides called aureolic acids. This family includes a variety of chromomycins, olivomycins, chromocyclomycin, and duhramycin A as well as the founding member mithramycin (Lombó et al., 2006). This class of compounds is known to interact with the minor groove of DNA in a nonintercalative way with a preference for G/C-rich regions (Hou et al., 2016). This mechanism is more subtle and distinct from other natural product transcription inhibitors like actinomycin D, which binds DNA at the transcription elongation complex (Sobell, 1985), or α-amanatin, which directly inhibits RNA polymerase II (Bushnell et al., 2002). Based on their known activities, aureolic acids have been pursued as anticancer chemotherapies. In addition to their anticancer properties (Lombó et al., 2006; Singh et al., 2017), these compounds have been implicated as inhibitors of Wnt signaling (Toume et al., 2014) and DNA gyrase (Simon et al., 1994) as well as inducers of autophagy (Guimarães et al., 2014; Ratovitski, 2016). Despite this array of activities, the specific genes and pathways involved in the mechanisms of action of CMA2 and mithramycin are only just being fleshed out. In this study we have determined that CMA2 alters β cell function through possibly indirect mechanisms by interfering with Ca2+ influx, suppressing β cell gene expression and disrupting GSK3/β-catenin signaling.
Materials and methods
Antibodies and reagents
CMA2 was purified from extracts of Streptomyces anulatus as described below or purchased (Santa Cruz). Mithramycin was generously provided by R. Kittler (UT Southwestern, Dallas, TX). Pyrvinium was a gift from C. Thorne (University of Arizona, Tucson, AZ). Antibodies, their dilutions for immunoblotting, and their sources are listed in Table 1. All other reagents were obtained through Thermo Fisher Scientific unless otherwise stated.
Table 1. Antibodies used for immunoblotting.
| Antibody | Host species | kD | Company | Cat. no. | Dilution |
|---|---|---|---|---|---|
| ERK1/2 | Rabbit | 42/44 | Cobb laboratory | Y691 | 1:3,000 |
| pERK1/2 | Mouse | 42/44 | Cell Signaling | 9106 | 1:2,000 |
| rpS6-p235/36 | Rabbit | 32 | Cell Signaling | 2211 | 1:1,000 |
| rpS6 | Mouse | 32 | Santa Cruz | sc-74459 | 1:3,000 |
| β-Catenin | Mouse | 92 | BD | 610154 | 1:1,000 |
| GSK3 α/β | Mouse | 51/46 | Santa Cruz | sc-7291 | 1:250 |
| pSer9/21-GSK3 | Rabbit | 51/46 | Cell Signaling | 9331 | 1:1,000 |
Secondary antibodies were donkey anti-mouse 800 and donkey anti-rabbit 700 from LI-COR Biosciences and were used at 1:10,000.
Isolation of natural product fractions and bacterial strain information
The library of microbial and invertebrate natural product fractions was subjected to liquid chromatography-mass spectrometry analysis using an Agilent Model 6130 single quadrupole mass spectrometer with an HP1200 high-performance liquid chromatographer (HPLC). A photodiode array detector provided a chemical fingerprint of all fractions on the basis of molecular weight and UV profile. Fractions were dereplicated using various compound databases, including Antibase, Reaxys, and Scifinder Scholar. The sediment was desiccated and stamped onto acidic agar plates (20 g starch, 1 g NaNO3, 0.5 g K2HPO4, 0.5 g MgSO4, 0.5 g NaCl, 0.01 g FeSO4, 10 µM cadaverine, 10 µM spermidine, 750 ml seawater, and 15 g agar, pH adjusted to 5.3 with phosphate buffer). Bacterial colonies were selected and streaked for purity using the same agar medium. Standard procedures for 16S rRNA analysis were used for phylogenetic characterization of bacterial strains. Analysis of the Streptomyces sp. strain SN-B-022-1 by 16S rRNA revealed 99% identity to S. anulatus.
Cultivation and extraction of SN-B-022-1
Bacterium SN-B-022 was cultured in 5 × 2.8–liter Fernbach flasks each containing 1 liter of a seawater-based medium (10 g starch, 4 g yeast extract, 2 g peptone, 1 g CaCO3, 40 mg Fe2(SO4)3 · 4H2O, and 100 mg KBr) and shaken at 200 rpm at 27°C. After 7 d cultivation, sterilized XAD-7-HP resin (20 g/liter) was added to adsorb organic chemicals produced by the bacterium, and the culture and resin were shaken at 200 rpm for 2 h. The resin was extracted with acetone and filtered through cheesecloth. The acetone-soluble portion was dried in vacuo to yield 2.0 g extract.
Purification of CMA2 from SN-B-022-1
The extract (300 mg out of 2.0 g) was subjected to reversed-phase medium-pressure liquid chromatography with 5% CH3OH/H2O for 1 min, followed by a linear gradient of CH3OH/H2O from 5% to 100% over 15 min, 4 min of column washing with pure CH3OH, and substitution of the column with 5% CH3OH/H2O for 2 min to provide 20 library fractions (SNB-022-1 to SNB-022-20). CMA2 (1.2 mg; tR = 32.1 min) was obtained from the active fraction SN-B-022-1 (88 mg) by reversed-phase HPLC (5 µm; 10.0 mm × 250 mm) using 25% CH3OH/H2O (0.1% formic acid) for 5 min, followed by a linear gradient of CH3OH/H2O from 25% to 85% over 30 min and 5 min of column washing with pure CH3CN. Application of the same HPLC purification method to SN-B-022-3 (14 mg) enabled isolation of additional samples of CMA2 (2.6 mg). The compound was identified by comparison of NMR data with literature values (Toume et al., 2014), and the structure of CMA2 was confirmed independently through analysis of two-dimensional NMR (heteronuclear single quantum coherence and heteronuclear multiple bond correlation) data.
Cell culture and treatments
MIN6 β cells (originally obtained from J. Hutton, University of Colorado, Aurora, CO) were cultured in DMEM (which contains 25 mM glucose) supplemented with 15% FBS, 100 U/ml penicillin, 100 µg/ml streptomycin, 292 µg/ml L-glutamine, and 50 µM β-mercaptoethanol as described previously (Kalwat et al., 2016a). For signaling and RNA collection experiments, MIN6 cells were seeded in 12-well dishes at ∼400,000 cells per well and grown 4–7 d with medium changed every 2–3 d before use in experiments. For CMA2 or mithramycin treatments for RNA isolation, cells were treated overnight and harvested after 24 h. Media pH was not altered by the presence of 100 nM CMA2. Prior to glucose stimulation experiments, MIN6 cells were washed twice with and incubated for 2 h in freshly prepared glucose-free modified KRBH (5 mM KCl, 120 mM NaCl, 15 mM Hepes, pH 7.4, 24 mM NaHCO3, 1 mM MgCl2, 2 mM CaCl2, and 1 mg/ml radioimmunoassay-grade BSA). Cells were lysed in 25 mM Hepes, pH 7.4, 1% NP-40, 10% glycerol, 50 mM sodium fluoride, 10 mM sodium pyrophosphate, 137 mM NaCl, 1 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride, 10 µg/ml aprotinin, 1 µg/ml pepstatin, and 5 µg/ml leupeptin and cleared of insoluble material by centrifugation at 10,000 g for 10 min at 4°C for subsequent use. The Ca2+ influx measurements were completed in MIN6 cells plated in black-walled clear-bottom 96-well tissue culture imaging plates. MIN6 cells were preincubated for 1 h in glucose-free KRBH, followed by loading with Fura-PE3 (5 µM) for 25 min and a 5-min washout. Using a Synergy2 H1 plate reader (BioTek), a 5-min baseline was recorded by monitoring excitation (340/380 nm)/emission (520 nm) followed by 30 min of recording after addition of 20 mM glucose. Area-under-the-curve calculations were performed in GraphPad Prism 7. For β-catenin reporter assays, stable Super TOPFlash HEK293 cells were plated 1e4 per well and cultured for 48 h before treatment with indicated doses of CMA2 or CHIR99021 for 24 h. Medium was decanted, and 20 µl passive lysis buffer was added. After a 10-min incubation, 80 µl Promega LARII firefly luciferase substrate working solution was added with an injector on a Synergy2 H1 plate reader (BioTek). Protein concentration of HEK293 cell lysates was measured by Bradford assay, and SuperTOPFlash signal was normalized to the protein concentration.
Human islet culture and treatments
Cadaveric human islets were obtained through the Integrated Islet Distribution Program. Islets were isolated by the affiliated islet isolation center and cultured in PIM medium (PIM-R001GMP; Prodo Labs) supplemented with glutamine/glutathione (PIM-G001GMP; Prodo Labs), 5% Human AB serum (100512; Gemini Bio Products), and ciprofloxacin (61-277RG; Cellgro, Inc.) at 37°C and 5% CO2 until shipping at 4°C overnight. Details of donor information are provided in Table 2. Upon receipt, human islets were cultured in Connaught Medical Research Laboratories (CMRL) medium (CMRL-1066) containing 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. For static culture insulin secretion experiments, human islets were hand-picked under a dissection microscope equipped with a green Kodak Wratten #58 filter (Finke et al., 1979) and placed into low-binding 1.5-ml tubes with ∼50 islets per tube. Islets were washed twice with KRBH (134 mM NaCl, 4.8 mM KCl, 1 mM CaCl2, 1.2 mM MgSO4, 1.2 mM KH2PO4, 5 mM NaHCO3, 10 mM Hepes, pH 7.4, and 0.1% BSA) and preincubated in KRBH supplemented with 2 mM glucose for 1 h before switching to KRBH containing either 2 or 16 mM glucose for 1 h. Supernatants were collected, centrifuged at 10,000 g for 5 min, and transferred to fresh tubes for storage at −80°C. Total insulin content was extracted by acid-ethanol overnight at −80°C and was neutralized with an equal volume of 1 M Tris, pH 7.4, before assay. Secreted insulin and insulin content were measured using ELISA (Mercodia) and HTRF (Cisbio) assays.
Table 2. Human islet donor characteristics.
| RRID (IIDP) | Culture time prior to shipping | Age | Sex | Race | BMI | HbA1c | GSIS SI (IIDP) | Viability | Purity |
|---|---|---|---|---|---|---|---|---|---|
| SAMN08768703 | 4 d | 44 | Male | White | 34.6 | 5.4% | 2.4 | 95 | 95 |
| SAMN08768735 | 2 d 17 h | 68 | Male | White | 29.7 | 5.2% | 2.6 | 95 | 90 |
| SAMN09432880 | 3 d 6 h | 51 | Male | Hispanic | 35.7 | 5.4% | 96 | 80 |
BMI, body mass index; GSIS SI, glucose-stimulated insulin secretion stimulation index; HbA1c, hemoglobin A1c; IIDP, Integrated Islet Distribution Program.
Secreted GLuc assays
The InsGLuc-MIN6 cells were generated as previously described (Kalwat et al., 2016b). Cells were plated in 96-well dishes at 100,000 cells per well and incubated for 2–3 d before overnight (24 h) treatment with natural products. For washout experiments, drug media was replaced with culture media for an additional 24 h as indicated in the figure legends. Cells were then washed twice with KRBH and preincubated in 100 µl of KRBH (250 µM diazoxide included where indicated in figure legends) for 1 h. The solution was then removed and cells were washed one time with 100 µl KRBH and then incubated in KRBH with or without the indicated glucose concentration (or 250 µM diazoxide, 35 mM KCl, and 20 mM glucose where indicated) for 1 h. 50 µl KRBH was collected from each well and pipetted into a white opaque 96-well plate Optiplate-96 (PerkinElmer). Fresh GLuc assay working solution was then prepared by adding coelenterazine stock solution into assay buffer to a final concentration of 10 µM. 50 µl working solution was then rapidly added to the wells using a Matrix 1,250-µl electric multichannel pipette for a final concentration of 5 µM coelenterazine. After adding reagent to the plate, plates were spun briefly and luminescence was measured on a Synergy2 H1 plate reader (BioTek). The protocol was set to shake the plate orbitally for 3 s and then read the luminescence of each well with a 100-ms integration time and gain set to 150.
Immunoblotting and microscopy
40–50 µg cleared cell lysates were separated on 10% gels by SDS-PAGE and transferred to nitrocellulose for immunoblotting. All membranes were blocked in Odyssey blocking buffer (LI-COR Biosciences) for 1 h before overnight incubation with primary antibodies diluted in blocking buffer. After three 10-min washes in 20 mM Tris-HCl, pH 7.6, 150 mM NaCl, and 0.1% Tween-20 (TBS-T), membranes were incubated with fluorescent secondary antibodies for 1 h at room temperature. After three 10-min washes in TBS-T, membranes were imaged on an Odyssey scanner (LI-COR Biosciences). For experiments in which cells were fixed and stained with DAPI, MIN6 cells were plated in 96-well plates at 50,000 cells per well 24 h before treatment. After treatment, cells were fixed and permeabilized in 4% paraformaldehyde and 0.18% Triton X-100 in PBS, pH 7.4, for 10 min, washed three times for 5 min each, and then stained with DAPI (300 nM) in PBS for 10 min. The cells were washed again twice with PBS and taken immediately for imaging. Plates were imaged in the UT Southwestern High-throughput Screening core facility using a GE INCell 6000 automated microscope (GE Healthcare) and a 10× objective lens at room temperature using a DAPI filter. Nuclei counting was performed using a CellProfiler workflow (Carpenter et al., 2006).
Quantitative PCR (RT-qPCR)
The RNA was isolated from MIN6 cells using the PureLink RNA Mini Kit (Life Technologies). 500 ng MIN6 RNA was converted into cDNA using the iScript kit (Bio-Rad), and the resulting cDNA was diluted 10-fold with water. 1 μl diluted cDNA was used in 10 µl qPCR reactions using 2× SYBR Bio-Rad master mix and 250 nM of each primer. Reactions were measured using a QuantStudio 7 Flex (Applied Biosystems), and 18S RNA was used as the reference gene. Relative expression was calculated by the 2^(-ΔΔCt) method. All primer sequences are listed in Table 3.
Table 3. Primers used for RT-qPCR.
| Name | Gene name | Species | Sequence (5′–3′) | Amplicon | PrimerBank ID |
|---|---|---|---|---|---|
| mUcn3.fwd | Urocortin 3 | Mouse | AAGCCTCTCCCACAAGTTCTA | 175 | 21492632a1 Wang et al. (2012) |
| mUcn3.rev | GAGGTGCGTTTGGTTGTCATC | ||||
| mh18S.fwd | 18S | Human/mouse | GTAACCCGTTGAACCCCATT | 151 | |
| mh18S.rev | CCATCCAATCGGTAGTAGCG | ||||
| mIns12.fwd | Insulin I/II | Mouse | TGAAGTGGAGGACCCACAAGT | 125/131 | Chuang et al. (2008) |
| mIns12.rev | AGATGCTGGTGCAGCACTGAT | ||||
| mPdx1.fwd | Pdx1 | Mouse | CCCCAGTTTACAAGCTCGCT | 177 | 6679269a1 Wang et al. (2012) |
| mPdx1.rev | CTCGGTTCCATTCGGGAAAGG |
Statistical analysis
Quantitated data are expressed as mean ± SEM. Data were evaluated using Student’s t test or ANOVA with appropriate multiple comparisons test as indicated and considered significant if P < 0.05. Graphs were made in GraphPad Prism 7.
Results
Identification of CMA2 as an insulin secretion inhibitor
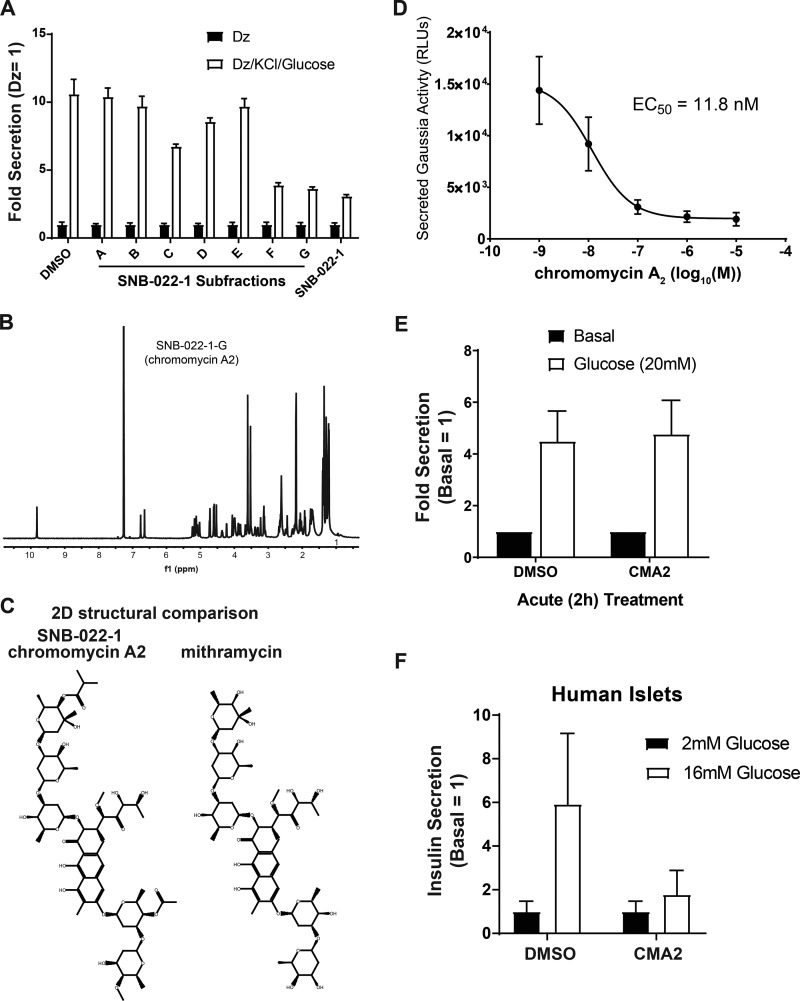
We previously performed a natural product screen using specialized MIN6 β cells containing an insulin promoter–driven Gaussia Luciferase (GLuc)-linked insulin transgene, InsGLuc-MIN6 cells (Kalwat et al., 2016b). These cells were treated chronically (24 h) with compounds, preincubated in Krebs-Ringer Bicarbonate Hepes (KRBH) in the absence of compounds, and then stimulated. This strategy allowed us to probe a specific pharmacological space to enrich for hit compounds with long-lasting effects. In the screen, we stimulated the cells using the diazoxide paradigm to drive both the triggering and metabolic amplifying pathways of insulin secretion (Henquin, 2000; Kalwat and Cobb, 2017). In this paradigm, diazoxide locks the ATP-sensitive potassium channel open, preventing Ca2+ influx and insulin secretion until extracellular KCl is provided to depolarize the membrane and elicit the triggering pathway. Addition of glucose under these conditions results in the metabolic amplification of insulin secretion without any further increase in Ca2+ influx. This approach provided a platform to later assign the actions of hit compounds to triggering and/or amplification and gave maximal dynamic range in the assay. From the screen, we selected natural product fractions that appeared nontoxic after 24 h but still potently inhibited glucose-stimulated insulin secretion. Among the top hits was fraction SNB-022-1 derived from S. anulatus. In rescreening, we found this fraction retained the expected inhibitory activity (Fig. 1 A). Further fractionation, high-resolution mass spectrometry analyses, and NMR analyses showed the major component of the fraction to be the aureolic acid CMA2 in subfractions F and G (Fig. 1 B). The structure of CMA2 is shown next to an aureolic acid family member, mithramycin, for comparison (Fig. 1 C). We determined the EC50 of CMA2 in our reporter assay to be 11.8 nM (Fig. 1 D). Acute treatment (2 h) with CMA2 was insufficient to inhibit glucose-stimulated Ins-GLuc secretion in our reporter β cells (Fig. 1 E), arguing against a rapid mechanism of action. We confirmed the inhibitory effects of 24 h treatment with CMA2 on glucose-stimulated insulin secretion from human islets (Fig. 1 F).
Figure 1.
Activity-guided fractionation identifies CMA2 as an insulin secretion inhibitor in human islets. (A) Natural product subfractions from SNB-022-1 were tested in quadruplicate for inhibitory activity in Ins-GLuc MIN6 β cells stimulated with the diazoxide (Dz) paradigm. Cells were treated 24 h with the original SNB-022-1 fraction, subfractions (10 µg/ml), or DMSO (0.1%). Cells were preincubated in glucose-free KRBH for 1 h followed by 1 h in the presence of either Dz (250 µM) or Dz + 40 mM KCl and 20 mM glucose. Secreted luciferase activity relative to Dz control is shown as the mean ± SEM. (B) Nuclear magnetic resonance spectra of the purified compound from fraction SNB-022-1 identified as CMA2. (C) The spectra in B matched CMA2, and the structure is shown. The structure of the closely related mithramycin is shown for comparison. (D) The Ins-GLuc MIN6 cells were treated 24 h with different doses of CMA2 followed by a glucose-stimulated secretion assay, and the resulting relative luciferase units (RLUs) were used to determine the effective concentration EC50. Data are the mean ± SEM (n = 4). (E) The Ins-GLuc MIN6 cells were treated with CMA2 (100 nM) starting at the beginning of the 1-h KRBH preincubation and during the 1-h glucose (20 mM) stimulation. Secreted luciferase activity was measured at the end of the assay. Data represent the mean ± SEM of three independent experiments performed in triplicate. (F) Human islets were hand-picked into groups of 50 and treated 24 h with CMA2 at the indicated doses in 500 µl complete CMRL medium. The islets were then subjected to static glucose-stimulated insulin secretion assays, and the secreted and total insulin concentrations were measured. Data are the mean ± SEM from three different batches of donor islets.
CMA2 and mithramycin have differential effects on triggering and amplifying insulin secretion and gene expression in β cells
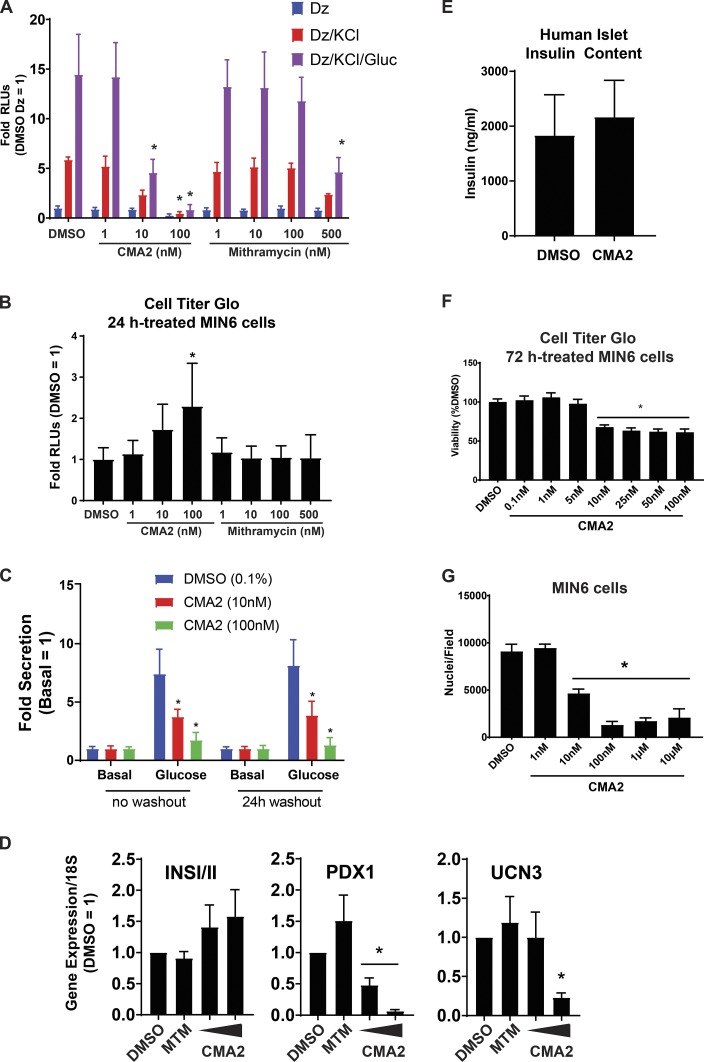
Given what is known about mithramycin–DNA binding activity (Hou et al., 2016), the effect of mithramycin on gene expression in glioblastoma models (Singh et al., 2017) and the structural similarities between CMA2 and mithramycin (Fig. 1 C), we tested CMA2 and mithramycin in our insulin secretion reporter assays. CMA2 was ∼50 times more potent than mithramycin at inhibiting glucose-stimulated secretion after a 24-h incubation (Fig. 2 A). We assessed cell viability after secretion experiments using the Cell Titer Glo assay, which indirectly measures relative ATP concentrations. The CMA2-treated cells had an elevated Cell Titer Glo signal, while mithramycin-treated cells did not (Fig. 2 B). To determine whether the effects of CMA2 were reversible in our assay, we performed washout experiments with CMA2 in InsGLuc-MIN6 cells. In agreement with our previous results (Fig. 1 D and 2 A), cells treated for 24 h followed by our standard 2-h starvation/stimulation washout in KRBH had suppressed glucose-induced secretion (Fig. 2 C). The inhibitory effects of CMA2 remained even after a 24-h washout before the starvation/stimulation protocol (Fig. 2 C).
Figure 2.
CMA2 has distinct effects on β cells compared with the analogue mithramycin. (A) Ins-GLuc MIN6 cells were treated 24 h with either CMA2 or mithramycin (MTM) at the indicated doses, and then secretion was stimulated using the diazoxide (Dz; 250 µM) paradigm as previously in the presence or absence of 40 mM KCl and 20 mM glucose (Gluc). Secreted luciferase activity was measured, and the data are the mean ± SEM from three independent experiments. *, P < 0.05 versus respective DMSO by two-way ANOVA. (B) Lysates of the diazoxide-treated samples in A were subjected to Cell Titer Glo assays to measure viability. The luciferase activity is shown with respect to DMSO control, and the data are the mean ± SEM (n = 3). (C) The Ins-GLuc MIN6 cells were incubated in culture media for 24 h with indicated treatments and then assayed for glucose-stimulated secretion in KRBH in the absence of treatment (no washout; left). In parallel, cells were also incubated for 24 h with indicated treatments followed by a 24-h washout in culture media before the secretion assay (24-h washout; right). Data are the mean ± SEM (n = 3). *, P < 0.05 versus respective DMSO by two-way ANOVA. (D) The MIN6 cells were treated with CMA2 for 24 h, and RNA was isolated for RT-qPCR against INSI/II, PDX1, and UCN3. Data were normalized to 18S rRNA by the 2^-ΔΔCt method and are the mean ± SEM (n = 3). *, P < 0.05 versus DMSO control. (E) Insulin content was measured in lysates from human islets that were treated 24 h with CMA2 (100 nM) or DMSO (0.1%). Data are the mean ± SEM (n = 3). (F) The MIN6 cells were treated with the indicated doses of CMA2 for 72 h and subjected to a Cell Titer Glo assay to measure viability. Data are the mean ± SEM normalized to the percent of DMSO (n = 3). *, P < 0.05 versus DMSO by Student’s t test. (G) The data in F were confirmed by treating MIN6 cells with CMA2 for 48 h followed by fixation and staining with DAPI. Nuclei counts per well are shown as the mean ± SEM (n = 3). *, P < 0.05 versus DMSO by Student’s t test.
Aureolic acid compounds are known to influence transcription; therefore, we hypothesized that CMA2 may exert some of its effects on β cells by suppressing the expression of critical genes. To assess the effects on gene expression, we treated MIN6 cells with CMA2 or mithramycin for 24 h and extracted RNA for reverse transcription–quantitative PCR (qPCR; RT-qPCR) analysis. The CMA2, but not mithramycin, suppressed expression of an important β cell transcription factor, PDX1, at as low as 10 nM (Fig. 2 D). Expression of Urocortin 3, a marker of β cell maturation and paracrine regulation of somatostatin secretion (van der Meulen et al., 2012, 2015), was also suppressed, but only at 100 nM CMA2, while insulin gene (INSI/II) expression was unaltered (Fig. 2 D). CMA2 (100 nM for 24 h) also had no effect on insulin content in human islets (Fig. 2 E). While treatment with CMA2 for 24 h increased the Cell Titer Glo signal in MIN6 cells (Fig. 2 B), a longer-term 72-h treatment resulted in reduced viability as determined by the same assay (Fig. 2 F) and reduced cell number determined by counting nuclei (Fig. 2 G).
Chronic CMA2 treatment interferes with triggering Ca2+ influx in MIN6 β cells
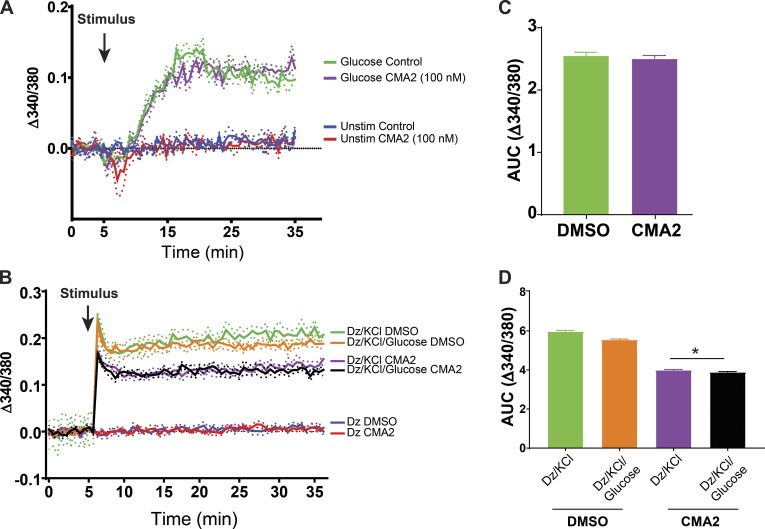
We next measured Ca2+ influx in MIN6 cells pretreated for 24 h with 100 nM CMA2. In response to glucose, Ca2+ influx was not different between vehicle- and CMA2-treated cells (Fig. 3, A and C), suggesting the dramatic ablation of glucose-stimulated insulin secretion was not primarily due to a defect in glucose-stimulated Ca2+ influx. However, we also tested Ca2+ influx under the diazoxide paradigm. Diazoxide and KCl clamped the Ca2+ levels as expected as glucose stimulation did not cause additional changes in intracellular calcium levels (Fig. 3, B and D, green vs. orange and purple vs. black). With a 24-h CMA2 pretreatment, we did observe a suppression of the triggering Ca2+ influx (Fig. 3, B and D).
Figure 3.
Diazoxide paradigm unmasks effect of CMA2 on triggering Ca2+ influx. (A–D) The MIN6 cells were treated 24 h with CMA2 (100 nM) and then prepared for Ca2+ influx measurements using Fura-PE3 (5 µM). (A) Cells were preincubated in KRBH without glucose for 1 h and loaded with Fura-PE3 for 30 min followed by a baseline Ca2+ measurement for 5 min. 20 mM glucose was added, and Fura fluorescence was measured for 30 min. (B) Diazoxide (250 µM) was included in the KRBH during the baseline reading and throughout the stimulation protocol. Data are shown as traces representing the mean ± SEM (shown as dots; n = 3). (C and D) Area under the curve (AUC) analyses for their respective traces. C shows data in A; D shows data in B. *, P < 0.05 CMA2 versus DMSO by one-way ANOVA.
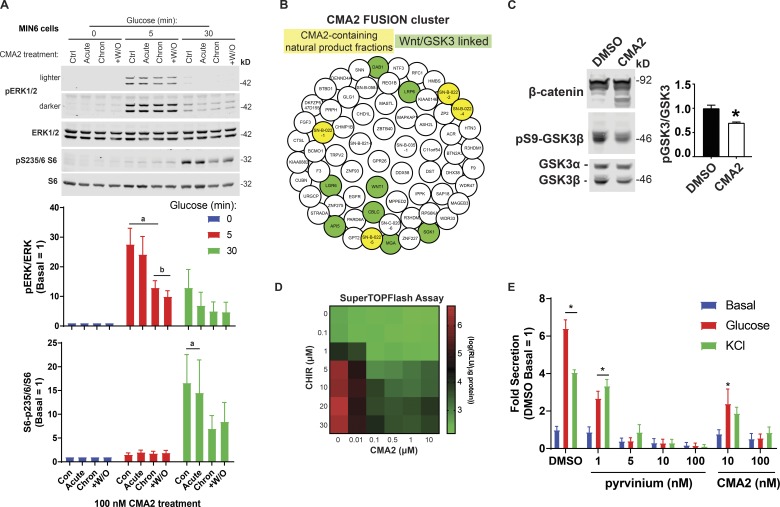
To address whether well-known glucose-mediated β cell signaling pathways are disrupted by CMA2, we tested glucose-induced phosphorylation of ERK1/2 MAP kinase (Khoo et al., 2003) and ribosomal protein S6 (Moore et al., 2009; Guerra et al., 2017). MIN6 cells were treated with DMSO or CMA2 for 24 h, and the following day, they were preincubated in KRBH containing either DMSO or CMA2 to compare effects on cells treated acutely (2 h in CMA2-KRBH), chronically (24 h in CMA2-medium + 2 h in CMA2-KRBH), or chronically with a washout (24 h in CMA2-medium + 2 h in DMSO-KRBH). After the 2-h KRBH incubation, the cells were stimulated with glucose, which increased ERK1/2 phosphorylation by 5 min (Fig. 4 A). Chronic CMA2 treatment suppressed the fold increase in ERK1/2 activation at the 5-min time point. By 30 min, all CMA2 treatments had suppressed glucose-stimulated ERK1/2 phosphorylation relative to the 0-min time point (Fig. 4 A). With respect to S6 phosphorylation, only in control and acute CMA2-treated cells was pS235/6-S6 induced by 30 min, whereas both chronic CMA2 treatments blunted this event (Fig. 4 A).
Figure 4.
CMA2 dysregulates β cell signaling through ERK, S6, and the Wnt–GSK3β pathways. (A) The MIN6 cells were treated 24 h with DMSO (0.1%) or CMA2 (100 nM; chronic) and the next day incubated 2 h in KRBH without glucose but containing either DMSO or CMA2. Acutely treated cells received CMA2 only during the 2-h KRBH incubation. Washout (+W/O) were cells treated 24 h with CMA2, which received only DMSO during the 2 h KRBH incubation. Cells were then either unstimulated or stimulated with 20 mM glucose for 5 and 30 min and lysed for immunoblotting against pERK1/2, ERK1/2 and pS235/6-S6, and S6. Bar graphs of quantified immunoblots are shown as the mean ± SEM (n = 3). a, P < 0.05 versus respective 0 min glucose; b, P < 0.05 versus control glucose 5 min by two-way ANOVA. (B) The FUSION cluster of CMA2 and implicated genes. The CMA2-containing natural product fractions are highlighted in yellow, and genes linked to the Wnt pathway are highlighted in green. (C) The MIN6 cells treated 24 h with CMA2 (100 nM) or DMSO (0.1%) showed increased β-catenin degradation bands as well as decreased GSK3β Ser9 phosphorylation, indicating increased GSKβ activity. Bar graph shows GSK3β Ser9 phosphorylation as fold of total GSK3β normalized to DMSO control. Data are the mean ± SEM (n = 3). *, P < 0.05. (D) The Super TOPFlash assay in stable HEK293 cells treated 24 h with different dose combinations of CHIR99021 and CMA2. Displayed data are in log10 format (n = 3). (E) Ins-GLuc MIN6 cells were treated overnight in culture media with the indicated doses of pyrvinium, CMA2, or DMSO followed by a 1-h incubation in glucose-free KRBH and a 1-h stimulation with either 20 mM glucose or 35 mM KCl. Data are the mean ± SEM (n = 3). *, P < 0.05 versus DMSO basal by two-way ANOVA.
CMA2 disrupts GSK3–β-catenin pathway signaling in MIN6 β cells
To discern the signaling pathways affected by CMA2, we used a database developed on campus called Functional Signature Ontology (FUSION; Potts et al., 2015). The FUSION database works by measuring genetic perturbation signatures in response to whole-genome siRNA and synthetic and natural product screening (Potts et al., 2013, 2015; Vaden et al., 2017; Das et al., 2018). This approach allows small molecules and natural products to be linked to genes that may be implicated in their mechanisms of action. We applied FUSION to CMA2 and found that suppression of genes connected to Wnt signaling and glycogen synthase kinase 3β (GSK3β) cluster with CMA2. The resulting network allowed us to query what genes may be involved in the actions of CMA2. Through FUSION, we found that CMA2-containing natural product fractions clustered with a small set of genes. Using the online tool Enrichr (Chen et al., 2013; Kuleshov et al., 2016), we determined this cluster contained Wnt–GSK3β-linked genes (DAB1, LRP6, WNT1, LGR6, API5, CBLC, MGA, and SGK1; Fig. 4 B). In agreement with this finding, chromomycin compounds were previously linked to Wnt pathway inhibitory activity (Toume et al., 2014). When GSK3β is not phosphorylated at serine 9, it is active and can phosphorylate β-catenin, targeting it for proteasomal degradation (Xu et al., 2009). Chronic CMA2 treatment of MIN6 cells also resulted in suppressed GSK3β serine 9 phosphorylation and increased degradation of β-catenin (Fig. 4 C). Wnt pathway activation or small-molecule inhibitors of GSK3 (CHIR99021) lead to accumulation of β-catenin, thereby increasing expression of Wnt-responsive genes (Liu and Habener, 2010). We observed similar CMA2-mediated inhibition in the β-catenin reporter Super TOPFlash assay (Veeman et al., 2003) in HEK293 cells, even in the face of high doses of CHIR99021, which activates reporter expression (Fig. 4 D). Conversely, a potent Wnt pathway inhibitor pyrvinium, which induces the degradation of β-catenin (Thorne et al., 2010), also dramatically suppressed glucose-stimulated Ins-GLuc secretion (Fig. 4 E), in agreement with published findings (Sorrenson et al., 2016).
Discussion
In this study, we identified CMA2 as a modulator of β cell function that acts through at least three mechanisms: suppression of gene expression, interference of GSK3–β-catenin signaling, and partial inhibition of Ca2+ influx. Although we uncovered certain facets of CMA2 actions, we do not yet know the extent to which each of these mechanisms contribute to the inhibition of insulin secretion or if there are additional affected mechanisms. Given the potential for aureolic acids to modulate the expression of a wide swath of the transcriptome, future studies will require in-depth gene expression analysis to determine the β cell–specific genes, which are disrupted in response to CMA2. Additionally, protein targets of CMA2 or mithramycin have yet to be identified. Elucidation of these targets may explain the differential activity of CMA2, and discovery of the genes and pathways involved in its mechanisms of action could provide inroads to pharmacologically stabilizing β cell function in a variety of disease settings.
Inhibitors of β cell function are useful for understanding disease and pathway discovery
It is common to focus on activators of insulin secretion because of their therapeutic potential; however, while secretion inhibitors are not intended to be diabetes therapeutics themselves, they are a relatively untapped resource for understanding β cell function. Determining the target genes and pathways of inhibitors of β cell function including CMA2 has the potential to elucidate factors that are important for insulin expression, processing, and secretion. We identified CMA2 as a Wnt pathway inhibitor in β cells, and the requirements for the Wnt pathway in β cell development and function are well known (Welters and Kulkarni, 2008; Liu and Habener, 2010; Bader et al., 2016; Sorrenson et al., 2016). Therefore, new chemical tools to interrogate this pathway are of immediate relevance to the field of diabetes and islet biology. The uses of aureolic acid natural products in applications such as chemotherapy warrant consideration of side effects on β cells or other sensitive tissues. At the same time, this class of compounds may help uncover pharmacologically targetable pathways to suppress insulin secretion for novel therapeutic approaches for diseases such as polycystic ovary syndrome, insulinomas, and persistent hyperinsulinemic hypoglycemia of infancy. In some cases these disorders are treatable with diazoxide, but there are individuals who are unresponsive to therapy (Snider et al., 2013). Insulin secretion inhibitors such as diazoxide, in conjunction with exogenous insulin therapy, have also been used to rest β cells, thereby prolonging endogenous functional β cell mass (Brown and Rother, 2008; van Boekel et al., 2008) as well as preserving isolated human islet function before transplant (Nijhoff et al., 2018). Therefore, identifying and segregating CMA2 target pathways will be useful for future development of chromomycin analogues.
Cytoxicity-dependent and cytoxicity-independent effects of CMA2 on β cell function
We observed that CMA2 at high doses and longer durations (48–72 h) of treatment also exhibited cytotoxic or cytostatic effects. This is likely due in part to the suppression of gene expression and blunting of prosurvival β-catenin activity. Correlative, but consistent with the effect of CMA2 on GSK3–β-catenin signaling in β cells, the small-molecule GSK3 activator pyrvinium led to dramatic suppression of glucose-stimulated insulin secretion (Fig. 4 E; Sorrenson et al., 2016). Additionally, CMA2 was identified to inhibit DNA gyrase in vitro at concentrations >50 nM (Simon et al., 1994); however, its Wnt pathway inhibitory activity is observed in cells even at doses below 5 nM, suggesting dose-dependent target selectivity. This is in line with our observations of an 11.8-nM EC50 for inhibition of glucose-stimulated insulin secretion by CMA2 and the ability of 10 nM CMA2 to suppress CHIR-induced β-catenin activity. Additionally, GSK3 was shown to exhibit genetic/signaling interactions with a substantial fraction of the kinome (Thorne et al., 2015) including the Raf-MAPK and mTOR pathways. This suggests that CMA2-mediated modulation of GSK3 function may directly or indirectly alter many pathway outputs, including MAPK and S6 phosphorylation, and contribute to β cell dysfunction in this context. While ERK1/2 were recently confirmed to be required for the first phase of insulin secretion (Leduc et al., 2017), we predict that the modest reduction in ERK activation by CMA2 treatment is likely due to indirect effects downstream of disrupted gene expression by CMA2. Indeed, while multiday treatment with CMA2 blunts cell growth, its suppression of insulin secretion after only 24 h may have independent mechanisms. For example, another natural product with growth-suppressive activities, everolimus, was recently shown to suppress insulin secretion independent of its growth inhibition effects (Suzuki et al., 2018). In many of the experiments in which cells were treated chronically (24 h) with CMA2, the medium and compound were washed off with KRBH before the 1–2 h preincubation period and stimulation. This suggests a lasting impact of CMA2 on β cells, perhaps through effects on transcription or protein stability, or that CMA2 remains inside cells even after a washout. Because 100 nM CMA2 pretreatment almost completely blocked secretion in both triggering and amplifying conditions (Fig. 2 A), we surmise that the relative reduction in KCl-stimulated Ca2+ influx likely contributes to but does not entirely account for the inhibition of secretion (Fig. 3 B). While suppression of Ca2+ influx and inhibition of insulin secretion are correlated, previous work showed that blocking Ca2+ influx by half resulted in a relatively minor decrease in insulin secretion (Boyd et al., 1986). Our findings that (1) CMA2 treatment did not suppress insulin expression or insulin content and (2) CMA2 did not blunt Ca2+ influx in response glucose stimulation alone suggest that CMA2, possibly indirectly, disrupts factors required downstream of Ca2+ for insulin secretory granule transport, docking, or exocytosis. Further experiments focused on granule trafficking and transcriptome analysis will be required to determine the specific components and steps affected by CMA2.
CMA2 and mithramycin elicit divergent phenotypes in β cells
We investigated whether the structurally similar CMA2 relative mithramycin had a similar effect on secretion, but to our surprise, as much as 50 times more mithramycin was required to have the same impact on secretion as 10 nM CMA2. While 100 nM CMA2 increased relative ATP concentrations after 24 h, 500 nM mithramycin failed to have an impact, indicating other mechanistic differences between these two products. Mithramycin was shown to have tumor-selective properties, inhibiting the function of a transcriptional trifecta (Sox2, Zeb1, and Olig2) to inhibit growth of glioblastoma while apparently sparing normal tissue (Singh et al., 2017). Given the recent work on mithramycin and less toxic isoforms (Osgood et al., 2016; Singh et al., 2017) and the high similarity between the mithramycin and CMA2 structures, modified forms of CMA2 may prove useful as therapeutics for cancer or chemical probes for research, for example. Over 30 yr ago, the differences in potency of the aureolic acid family compounds, including mithramycin and CMA2, were suggested to be a result of differential uptake of the compounds into mammalian cells (Gupta, 1982). CMA2 and mithramycin were also shown to recognize slightly different C/G-rich DNA tracts (Mansilla et al., 2010). This may be accounted for by small differences in the side chains of CMA2 and mithramycin (Barceló et al., 2010). While the mechanistic explanation for the differences between CMA2 and mithramycin in β cells are still unknown, it remains possible that in our assays, β cells are more sensitive to CMA2 compared with mithramycin due to differential uptake. However, in glioblastoma models, 50 nM mithramycin showed substantial effects on gene expression (Singh et al., 2017), suggesting 50–100 nM mithramycin is more than sufficient to elicit effects in other mammalian cells. The greater potency of CMA2 suggests nuanced mechanisms that are likely to involve signaling and gene expression critical for insulin secretion. Further clarification of the underlying mechanisms may uncover new targets for diabetes treatment and increase understanding of β cell function.
Acknowledgments
We thank current and former members of the Cobb laboratory for helpful discussions. We are grateful to Elizabeth A. McMillan for providing the FUSION Cytoscape network. Mithramycin was a generous gift from R. Kittler. The M50 Super 8x TOPFlash was a gift from R. Moon (University of Washington, Seattle, WA; Addgene plasmid 12456).
Early phases of this work were supported by a National Institutes of Health Ruth L. Kirschstein National Research Service Award DK100113 (M.A. Kalwat) and R01 DK55310 (M.H. Cobb). Other support was provided through National Institutes of Health grants U41 AT008718 (J.B. MacMillan) and R37 DK34128 (M.H. Cobb). We are grateful to the Welch Foundation for funding portions of this project (I1689 to J.B. MacMillan and I1243 to M.H. Cobb). M.G. Grzemska was supported by a training grant from the Cancer Prevention and Research Institute of Texas (RP160157).
The authors declare no competing financial interests.
Author contributions: Conceptualization: M.A. Kalwat. Data curation: M.A. Kalwat, I.H. Hwang, M.G. Grzemska, and J.Z. Yang. Formal analysis: M.A. Kalwat and I.H. Hwang. Funding acquisition: M.A. Kalwat, J.B. Macmillan, and M.H. Cobb. Investigation: M.A. Kalwat, M.G. Grzemska, and J.Z. Yang. Methodology: M.A. Kalwat. Project administration: M.A. Kalwat. Resources: I.H. Hwang, J. Macho, and K. McGlynn. Supervision: M.A. Kalwat, J.B. MacMillan, and M.H. Cobb. Validation: M.A. Kalwat, M.G. Grzemska, and J.Z. Yang. Visualization: M.A. Kalwat. Writing, original draft: M.A. Kalwat. Writing, review and editing: M.A. Kalwat, I.H. Hwang, M.G. Grzemska, and M.H. Cobb.
Richard W. Aldrich served as editor.
References
- Bader E., Migliorini A., Gegg M., Moruzzi N., Gerdes J., Roscioni S.S., Bakhti M., Brandl E., Irmler M., Beckers J., et al. 2016. Identification of proliferative and mature β-cells in the islets of Langerhans. Nature. 535:430–434. 10.1038/nature18624 [DOI] [PubMed] [Google Scholar]
- Barceló F., Ortiz-Lombardía M., Martorell M., Oliver M., Méndez C., Salas J.A., and Portugal J.. 2010. DNA binding characteristics of mithramycin and chromomycin analogues obtained by combinatorial biosynthesis. Biochemistry. 49:10543–10552. 10.1021/bi101398s [DOI] [PubMed] [Google Scholar]
- Boyd A.E. III, Hill R.S., Oberwetter J.M., and Berg M.. 1986. Calcium dependency and free calcium concentrations during insulin secretion in a hamster beta cell line. J. Clin. Invest. 77:774–781. 10.1172/JCI112374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R.J., and Rother K.I.. 2008. Effects of beta-cell rest on beta-cell function: A review of clinical and preclinical data. Pediatr. Diabetes. 9:14–22. 10.1111/j.1399-5448.2007.00272.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell D.A., Cramer P., and Kornberg R.D.. 2002. Structural basis of transcription: α-Amanitin-RNA polymerase II cocrystal at 2.8 A resolution. Proc. Natl. Acad. Sci. USA. 99:1218–1222. 10.1073/pnas.251664698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. 2006. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention 2017. National Diabetes Statistics Report, 2017. Centers for Disease Control and Prevention, US Department of Health and Human Services, Atlanta, GA. [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., and Ma’ayan A.. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14:128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang J.-C., Cha J.-Y., Garmey J.C., Mirmira R.G., and Repa J.J.. 2008. Research resource: nuclear hormone receptor expression in the endocrine pancreas. Mol. Endocrinol. 22:2353–2363. 10.1210/me.2007-0568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordain L., Eades M.R., and Eades M.D.. 2003. Hyperinsulinemic diseases of civilization: More than just Syndrome X. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 136:95–112. 10.1016/S1095-6433(03)00011-4 [DOI] [PubMed] [Google Scholar]
- Das B., Neilsen B.K., Fisher K.W., Gehring D., Hu Y., Volle D.J., Kim H.S., McCall J.L., Kelly D.L., MacMillan J.B., et al. 2018. A Functional Signature Ontology (FUSION) screen detects an AMPK inhibitor with selective toxicity toward human colon tumor cells. Sci. Rep. 8:3770 10.1038/s41598-018-22090-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkin S.R., Oswald N.W., Reed D.K., Mettlen M., MacMillan J.B., and Schmid S.L.. 2016. Ikarugamycin: A natural product inhibitor of clathrin-mediated endocytosis. Traffic. 17:1139–1149. 10.1111/tra.12425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finke E.H., Lacy P.E., and Ono J.. 1979. Use of reflected green light for specific identification of islets in vitro after collagenase isolation. Diabetes. 28:612–613. 10.2337/diab.28.6.612 [DOI] [PubMed] [Google Scholar]
- Gembal M., Gilon P., and Henquin J.C.. 1992. Evidence that glucose can control insulin release independently from its action on ATP-sensitive K+ channels in mouse B cells. J. Clin. Invest. 89:1288–1295. 10.1172/JCI115714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra M.L., Kalwat M.A., McGlynn K., and Cobb M.H.. 2017. Sucralose activates an ERK1/2-ribosomal protein S6 signaling axis. FEBS Open Bio. 7:174–186. 10.1002/2211-5463.12172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimarães L.A., Jimenez P.C., Sousa T.S., Freitas H.P., Rocha D.D., Wilke D.V., Martín J., Reyes F., Deusdênia Loiola Pessoa O., and Costa-Lotufo L.V.. 2014. Chromomycin A2 induces autophagy in melanoma cells. Mar. Drugs. 12:5839–5855. 10.3390/md12125839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R.S. 1982. Species specific differences in the toxicity of mithramycin, chromomycin A3, and olivomycin towards cultured mammalian cells. J. Cell. Physiol. 113:11–16. 10.1002/jcp.1041130104 [DOI] [PubMed] [Google Scholar]
- Hansen J.B., Arkhammar P.O., Bodvarsdottir T.B., and Wahl P.. 2004. Inhibition of insulin secretion as a new drug target in the treatment of metabolic disorders. Curr. Med. Chem. 11:1595–1615. 10.2174/0929867043365026 [DOI] [PubMed] [Google Scholar]
- Henquin J.C. 2000. Triggering and amplifying pathways of regulation of insulin secretion by glucose. Diabetes. 49:1751–1760. 10.2337/diabetes.49.11.1751 [DOI] [PubMed] [Google Scholar]
- Henquin J.C. 2009. Regulation of insulin secretion: A matter of phase control and amplitude modulation. Diabetologia. 52:739–751. 10.1007/s00125-009-1314-y [DOI] [PubMed] [Google Scholar]
- Henquin J.C., Dufrane D., Gmyr V., Kerr-Conte J., and Nenquin M.. 2017. Pharmacological approach to understanding the control of insulin secretion in human islets. Diabetes Obes. Metab. 19:1061–1070. 10.1111/dom.12887 [DOI] [PubMed] [Google Scholar]
- Hou C., Weidenbach S., Cano K.E., Wang Z., Mitra P., Ivanov D.N., Rohr J., and Tsodikov O.V.. 2016. Structures of mithramycin analogues bound to DNA and implications for targeting transcription factor FLI1. Nucleic Acids Res. 44:8990–9004. 10.1093/nar/gkw761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Espindola A.P., Stewart N.A., Wei S., Posner B.A., and MacMillan J.B.. 2011. Chromomycin SA analogs from a marine-derived Streptomyces sp. Bioorg. Med. Chem. 19:5183–5189. 10.1016/j.bmc.2011.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Wang K., and MacMillan J.B.. 2013. Hunanamycin A, an antibiotic from a marine-derived Bacillus hunanensis. Org. Lett. 15:390–393. 10.1021/ol303376c [DOI] [PubMed] [Google Scholar]
- Kalwat M.A., and Cobb M.H.. 2017. Mechanisms of the amplifying pathway of insulin secretion in the β cell. Pharmacol. Ther. 179:17–30. 10.1016/j.pharmthera.2017.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalwat M.A., Huang Z., Wichaidit C., McGlynn K., Earnest S., Savoia C., Dioum E.M., Schneider J.W., Hutchison M.R., and Cobb M.H.. 2016a Isoxazole alters metabolites and gene expression, decreasing proliferation and promoting a neuroendocrine phenotype in β-cells. ACS Chem. Biol. 11:1128–1136. 10.1021/acschembio.5b00993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalwat M.A., Wichaidit C., Nava Garcia A.Y., McCoy M.K., McGlynn K., Hwang I.H., MacMillan J.B., Posner B.A., and Cobb M.H.. 2016b Insulin promoter-driven Gaussia luciferase-based insulin secretion biosensor assay for discovery of β-cell glucose-sensing pathways. ACS Sens. 1:1208–1212. 10.1021/acssensors.6b00433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo S., Griffen S.C., Xia Y., Baer R.J., German M.S., and Cobb M.H.. 2003. Regulation of insulin gene transcription by ERK1 and ERK2 in pancreatic beta cells. J. Biol. Chem. 278:32969–32977. 10.1074/jbc.M301198200 [DOI] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. 2016. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44(W1):W90-7 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leduc M., Richard J., Costes S., Muller D., Varrault A., Compan V., Mathieu J., Tanti J.F., Pagès G., Pouyssegur J., et al. 2017. ERK1 is dispensable for mouse pancreatic beta cell function but is necessary for glucose-induced full activation of MSK1 and CREB. Diabetologia. 60:1999–2010. 10.1007/s00125-017-4356-6 [DOI] [PubMed] [Google Scholar]
- Liu Z., and Habener J.F.. 2010. Wnt signaling in pancreatic islets. Adv. Exp. Med. Biol. 654:391–419. 10.1007/978-90-481-3271-3_17 [DOI] [PubMed] [Google Scholar]
- Lombó F., Menéndez N., Salas J.A., and Méndez C.. 2006. The aureolic acid family of antitumor compounds: Structure, mode of action, biosynthesis, and novel derivatives. Appl. Microbiol. Biotechnol. 73:1–14. 10.1007/s00253-006-0511-6 [DOI] [PubMed] [Google Scholar]
- Mansilla S., Garcia-Ferrer I., Méndez C., Salas J.A., and Portugal J.. 2010. Differential inhibition of restriction enzyme cleavage by chromophore-modified analogues of the antitumour antibiotics mithramycin and chromomycin reveals structure-activity relationships. Biochem. Pharmacol. 79:1418–1427. 10.1016/j.bcp.2010.01.005 [DOI] [PubMed] [Google Scholar]
- Molven A., Matre G.E., Duran M., Wanders R.J., Rishaug U., Njølstad P.R., Jellum E., and Søvik O.. 2004. Familial hyperinsulinemic hypoglycemia caused by a defect in the SCHAD enzyme of mitochondrial fatty acid oxidation. Diabetes. 53:221–227. 10.2337/diabetes.53.1.221 [DOI] [PubMed] [Google Scholar]
- Moore C.E., Xie J., Gomez E., and Herbert T.P.. 2009. Identification of cAMP-dependent kinase as a third in vivo ribosomal protein S6 kinase in pancreatic beta-cells. J. Mol. Biol. 389:480–494. 10.1016/j.jmb.2009.04.020 [DOI] [PubMed] [Google Scholar]
- Nijhoff M., Carlotti F., Engelse M., and De K.E.. 2018. Diazoxide pretreatment improves pancreatic islet survival in vitro and functionality in vivo. Endocrine Abstracts. 56:GP113. [Google Scholar]
- Osgood C.L., Maloney N., Kidd C.G., Kitchen-Goosen S., Segars L., Gebregiorgis M., Woldemichael G.M., He M., Sankar S., Lessnick S.L., et al. 2016. Identification of mithramycin analogues with improved targeting of the EWS-FLI1 transcription factor. Clin. Cancer Res. 22:4105–4118. 10.1158/1078-0432.CCR-15-2624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts M.B., Kim H.S., Fisher K.W., Hu Y., Carrasco Y.P., Bulut G.B., Ou Y.H., Herrera-Herrera M.L., Cubillos F., Mendiratta S., et al. 2013. Using functional signature ontology (FUSION) to identify mechanisms of action for natural products. Sci. Signal. 6:ra90 10.1126/scisignal.2004657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts M.B., McMillan E.A., Rosales T.I., Kim H.S., Ou Y.H., Toombs J.E., Brekken R.A., Minden M.D., MacMillan J.B., and White M.A.. 2015. Mode of action and pharmacogenomic biomarkers for exceptional responders to didemnin B. Nat. Chem. Biol. 11:401–408. 10.1038/nchembio.1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratovitski E.A. 2016. Tumor protein (TP)-p53 members as regulators of autophagy in tumor cells upon marine drug exposure. Mar. Drugs. 14:154 10.3390/md14080154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato Y., Aizawa T., Komatsu M., Okada N., and Yamada T.. 1992. Dual functional role of membrane depolarization/Ca2+ influx in rat pancreatic B-cell. Diabetes. 41:438–443. 10.2337/diab.41.4.438 [DOI] [PubMed] [Google Scholar]
- Simon H., Wittig B., and Zimmer C.. 1994. Effect of netropsin, distamycin A and chromomycin A3 on the binding and cleavage reaction of DNA gyrase. FEBS Lett. 353:79–83. 10.1016/0014-5793(94)00998-8 [DOI] [PubMed] [Google Scholar]
- Singh D.K., Kollipara R.K., Vemireddy V., Yang X.L., Sun Y., Regmi N., Klingler S., Hatanpaa K.J., Raisanen J., Cho S.K., et al. 2017. Oncogenes activate an autonomous transcriptional regulatory circuit that drives glioblastoma. Cell Reports. 18:961–976. 10.1016/j.celrep.2016.12.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider K.E., Becker S., Boyajian L., Shyng S.L., MacMullen C., Hughes N., Ganapathy K., Bhatti T., Stanley C.A., and Ganguly A.. 2013. Genotype and phenotype correlations in 417 children with congenital hyperinsulinism. J. Clin. Endocrinol. Metab. 98:E355–E363. 10.1210/jc.2012-2169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobell H.M. 1985. Actinomycin and DNA transcription. Proc. Natl. Acad. Sci. USA. 82:5328–5331. 10.1073/pnas.82.16.5328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorrenson B., Cognard E., Lee K.L., Dissanayake W.C., Fu Y., Han W., Hughes W.E., and Shepherd P.R.. 2016. A critical role for β-catenin in modulating levels of insulin secretion from β-cells by regulating actin cytoskeleton and insulin vesicle localization. J. Biol. Chem. 291:25888–25900. 10.1074/jbc.M116.758516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley C.A. 2011. Two genetic forms of hyperinsulinemic hypoglycemia caused by dysregulation of glutamate dehydrogenase. Neurochem. Int. 59:465–472. 10.1016/j.neuint.2010.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki L., Miyatsuka T., Himuro M., Nishio R., Goto H., Uchida T., Nishida Y., Kanazawa A., and Watada H.. 2018. Everolimus directly suppresses insulin secretion independently of cell growth inhibition. J. Endocr. Soc. 2:589–596. 10.1210/js.2017-00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne C.A., Hanson A.J., Schneider J., Tahinci E., Orton D., Cselenyi C.S., Jernigan K.K., Meyers K.C., Hang B.I., Waterson A.G., et al. 2010. Small-molecule inhibition of Wnt signaling through activation of casein kinase 1α. Nat. Chem. Biol. 6:829–836. 10.1038/nchembio.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne C.A., Wichaidit C., Coster A.D., Posner B.A., Wu L.F., and Altschuler S.J.. 2015. GSK-3 modulates cellular responses to a broad spectrum of kinase inhibitors. Nat. Chem. Biol. 11:58–63. 10.1038/nchembio.1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toume K., Tsukahara K., Ito H., Arai M.A., and Ishibashi M.. 2014. Chromomycins A2 and A3 from marine actinomycetes with TRAIL resistance-overcoming and Wnt signal inhibitory activities. Mar. Drugs. 12:3466–3476. 10.3390/md12063466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaden R.M., Oswald N.W., Potts M.B., MacMillan J.B., and White M.A.. 2017. FUSION-guided hypothesis development leads to the identification of N6,N6-dimethyladenosine, a marine-derived AKT pathway inhibitor. Mar. Drugs. 15:75 10.3390/md15030075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Boekel G., Loves S., van Sorge A., Ruinemans-Koerts J., Rijnders T., and de Boer H.. 2008. Weight loss in obese men by caloric restriction and high-dose diazoxide-mediated insulin suppression. Diabetes Obes. Metab. 10:1195–1203. [DOI] [PubMed] [Google Scholar]
- van der Meulen T., Xie R., Kelly O.G., Vale W.W., Sander M., and Huising M.O.. 2012. Urocortin 3 marks mature human primary and embryonic stem cell-derived pancreatic alpha and beta cells. PLoS One. 7:e52181 10.1371/journal.pone.0052181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meulen T., Donaldson C.J., Cáceres E., Hunter A.E., Cowing-Zitron C., Pound L.D., Adams M.W., Zembrzycki A., Grove K.L., and Huising M.O.. 2015. Urocortin3 mediates somatostatin-dependent negative feedback control of insulin secretion. Nat. Med. 21:769–776. 10.1038/nm.3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veeman M.T., Slusarski D.C., Kaykas A., Louie S.H., and Moon R.T.. 2003. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr. Biol. 13:680–685. 10.1016/S0960-9822(03)00240-9 [DOI] [PubMed] [Google Scholar]
- Wang X., Spandidos A., Wang H., and Seed B.. 2012. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 40(D1):D1144–D1149. 10.1093/nar/gkr1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welters H.J., and Kulkarni R.N.. 2008. Wnt signaling: Relevance to beta-cell biology and diabetes. Trends Endocrinol. Metab. 19:349–355. 10.1016/j.tem.2008.08.004 [DOI] [PubMed] [Google Scholar]
- Xu C., Kim N.G., and Gumbiner B.M.. 2009. Regulation of protein stability by GSK3 mediated phosphorylation. Cell Cycle. 8:4032–4039. 10.4161/cc.8.24.10111 [DOI] [PMC free article] [PubMed] [Google Scholar]