Intracellular pathogens can modulate host Rabs and SNAREs to support their replication and immune evasion. Singh et al. show that the Salmonella effector SipA functionally mimics an R-SNARE and recruits host Q-SNAREs to promote membrane fusion. Thus, SNARE mimicry by this intracellular pathogen effector modulates the host trafficking machinery for Salmonella survival.
Abstract
SipA is a major effector of Salmonella, which causes gastroenteritis and enteric fever. Caspase-3 cleaves SipA into two domains: the C-terminal domain regulates actin polymerization, whereas the function of the N terminus is unknown. We show that the cleaved SipA N terminus binds and recruits host Syntaxin8 (Syn8) to Salmonella-containing vacuoles (SCVs). The SipA N terminus contains a SNARE motif with a conserved arginine residue like mammalian R-SNAREs. SipAR204Q and SipA1–435R204Q do not bind Syn8, demonstrating that SipA mimics a cognate R-SNARE for Syn8. Consequently, Salmonella lacking SipA or that express the SipA1–435R204Q SNARE mutant are unable to recruit Syn8 to SCVs. Finally, we show that SipA mimicking an R-SNARE recruits Syn8, Syn13, and Syn7 to the SCV and promotes its fusion with early endosomes to potentially arrest its maturation. Our results reveal that SipA functionally substitutes endogenous SNAREs in order to hijack the host trafficking pathway and promote Salmonella survival.
Introduction
Intracellular pathogens employ various strategies to modulate host cell trafficking pathways through their effector molecules to gain entry and subsequent survival within the host cells (Alix et al., 2011; Asrat et al., 2014). Salmonella is also shown to modulate the host cell endolysosomal system (Brumell and Grinstein, 2004; Steele-Mortimer, 2008) by translocating a repertoire of virulence effectors into host cells through two different type III secretion systems (TTSSs) on chromosomal pathogenicity islands I and II (SPI-1 and -2, respectively) to promote bacterial invasion and intracellular survival (LaRock et al., 2015; Jennings et al., 2017). Previous studies have shown that SPI-1–encoded effector proteins like SipA, SopA, SopB, SopD, SopE, and SopE2 are shown to promote bacterial invasion into host cells (Hardt et al., 1998; Hueck, 1998; Galán and Collmer, 1999; Zhou et al., 1999b; Stender et al., 2000; Raffatellu et al., 2005), whereas SPI-2 effectors are generally required for intracellular survival (Zaharik et al., 2002; Waterman and Holden, 2003). However, these boundaries between SPI-1 and SPI-2 have gradually diminished as several SPI-1 TTSS effectors are now found to contribute to the intracellular survival of the bacteria (Hardt et al., 1998; Hernandez et al., 2004; Brawn et al., 2007; Giacomodonato et al., 2007; Patel et al., 2009).
One of the well-characterized Salmonella effector proteins is Salmonella invasion protein A (SipA), which is a multifunctional protein facilitating bacterial uptake into host cells by promoting actin polymerization (Zhou et al., 1999a,b; Jepson et al., 2001; Raffatellu et al., 2005) as well as inducing intestinal inflammation (Zhang et al., 2003; Hapfelmeier et al., 2004; Silva et al., 2004). Subsequent studies have shown that host caspase-3 cleaves SipA at a specific recognition motif DEVD, leading to the formation of two functional domains (SipA1–425 and SipA426–685) required for the pathogenesis of Salmonella (Srikanth et al., 2010). The actin-binding activity of SipA is located in the C-terminal domain (SipA426–685) of the protein (Zhou et al., 1999a) and plays an important role in the entry of Salmonella into epithelial cells (Zhou et al., 1999b). However, multiple functions are associated with the N-terminal domain of SipA (SipA1–425). The 1–105 residues in the N-terminal region SipA is required for its secretion by binding with the chaperone InvB (Bronstein et al., 2000; Lilic et al., 2006). In addition, amino acid residues spanning positions 294–424 of SipA make a functional domain that induces proinflammatory responses and polymorphonuclear leukocyte transepithelial migration in the host (Lee et al., 2000; Silva et al., 2004; Wall et al., 2007). Moreover, deletion analysis of SipA has shown that the central region of SipA also harbors two distinct functional domains, F1 (SipA170–271) and F2 (SipA280–394), that are involved in SipA focus formation and SipA–SipA interactions, respectively (Schlumberger et al., 2007).
In this study, we have shown that the N-terminal domain of SipA mimics as a cognate SNARE of host Syn8 and thereby binds and recruits Syntaxin8 (Syn8) on SCVs. Subsequently, we have shown that SipA functionally substitutes as an R-SNARE and forms complexes with three host Q-SNAREs, namely Syn8, Syn13, and Syn7, on SCVs to promote fusion with early endosomes (EEs) and arrest SCV maturation toward lysosomes.
Results
SipA specifically binds and recruits host Syn8 on SCVs
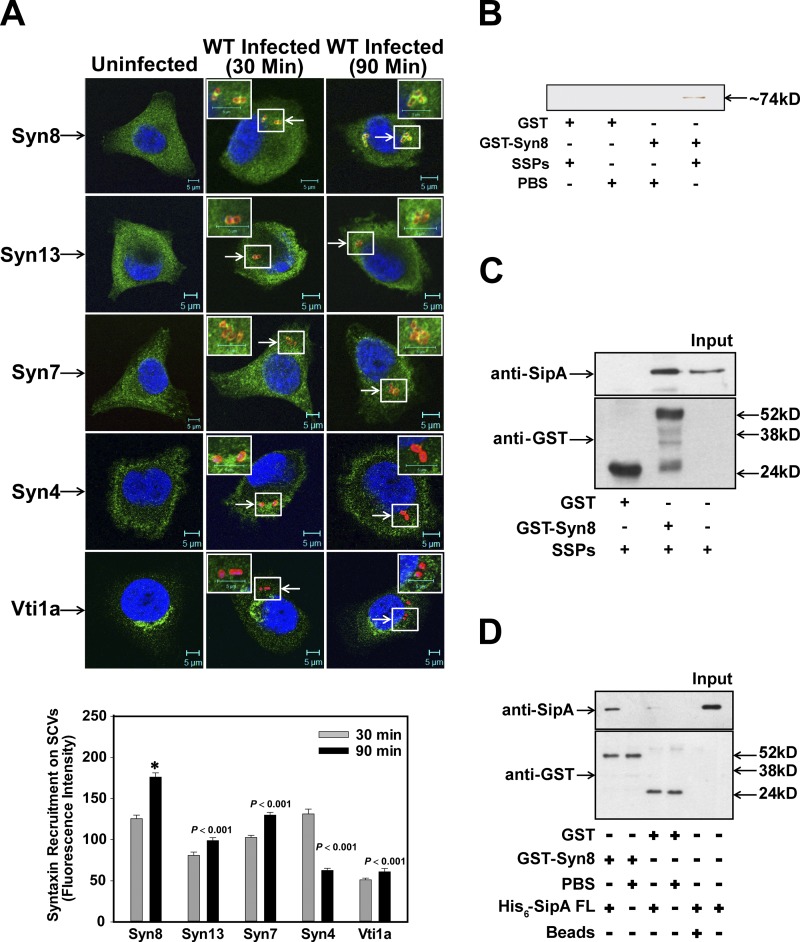
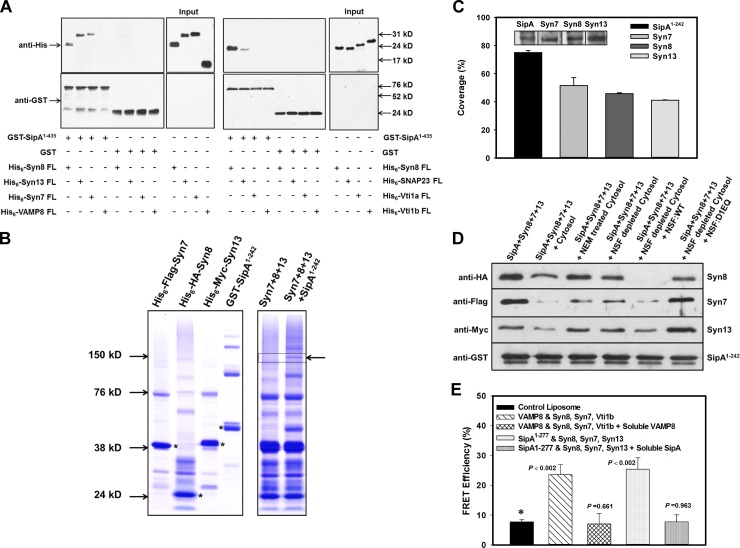
To characterize the maturation of Salmonella in HeLa cells, we analyzed the recruitment of different Syntaxins on SCVs by immunofluorescence using specific antibodies during maturation in HeLa cells and found that SCV:WT recruits significantly higher amounts of Syn8 at 90 min post infection (p.i.) than Syn13 and Syn7 (Fig. 1 A). In addition, SCVs were also found to recruit VAMP7 and VAMP8 90 min p.i. (Fig. S1). In contrast, no significant recruitment of Vti1a was detected on SCV even after 90 min p.i. (Fig. 1 A). However, Syn4 appears to be partially colocalized with SCV at 30 min p.i., which is lost at 90 min p.i. (Fig. 1 A). To identify the bacterial protein involved in the recruitment of Syn8, GST-Syn8 was immobilized on beads and incubated with Salmonella secretory proteins (SSPs). Pulldown results showed that GST-Syn8 specifically interacts with a ∼74-kD Salmonella effector protein (Fig. 1 B), which was identified as SipA by mass spectrometry (Fig. S2 A) and confirmed by using anti-SipA antibody (Fig. 1 C). Binding of SipA with Syn8 was confirmed by a protein–protein interaction wherein immobilized GST-Syn8 was incubated with His6-SipA and the binding of SipA was detected with anti-SipA antibody (Fig. 1 D).
Figure 1.
SipA specifically binds and recruits host Syn8 on SCVs. (A) Immunofluorescence studies examining the recruitment of Syn8, Syn7, Syn13, Syn4, and Vti1a on SCVs using specific antibodies; cells were analyzed by confocal microscopy. Arrows indicate respective proteins on SCVs. Bottom: Relative levels of different Syntaxins on SCVs at indicated time points. Syn (green) and Salmonella (red) are shown. Results are mean ± SEM of three independent preparations. Levels of significance are indicated by P values in comparison with Syn8 (*). (B) To identify Syn8 binding protein from Salmonella, GST-Syn8 was immobilized on beads and incubated with SSPs as described in Materials and methods. (C) Identification of Salmonella-interacting protein by Western blot analysis using anti-SipA antibody. (D) To detect the binding of Syn8 with SipA, GST-Syn8 was immobilized, and beads were incubated with His6-SipA. Binding of SipA with Syn8 was detected by Western blot analysis using anti-SipA antibody. Free GST immobilized on the beads was used as a control. All results are representative of three independent experiments.
SipA mimics as a cognate R-SNARE of Syn8
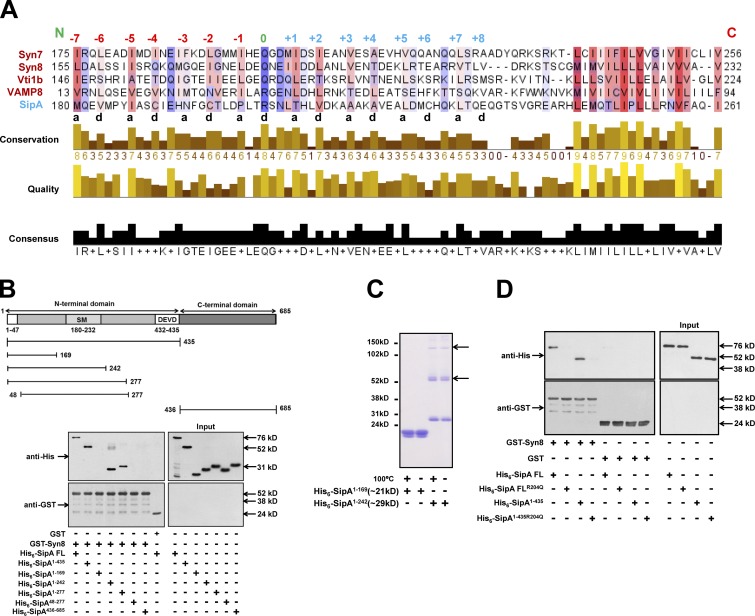
Syn8 is a SNARE protein and forms a complex with Syn7, Vti1b, and VAMP8 (Antonin et al., 2000a) to promote membrane fusion (Steegmaier et al., 1998). To understand the nature of binding between SipA and Syn8, bioinformatic analyses were performed between SipA and Syn8 along with its known cognate SNARE partners like Syn7, Vti1b, and VAMP8 using NCBI COBALT (Papadopoulos and Agarwala, 2007). The alignment results (Fig. 2 A) revealed that amino acid residues spanning between 180–232 of SipA (SipA180–232) contain a typical heptad repeat with hydrophobic residues in the “a” and “d” positions and a conserved arginine residue in the center-like “0” layer of mammalian R-SNAREs (Fasshauer et al., 1998). Moreover, we found that SipA180–232 is a helical structure (Fig. S2 B) and contains a SNARE motif (SM). As functional SNARE complexes are formed by the interaction of three Q-SNAREs with one R-SNARE (Fasshauer et al., 1998), it could be possible that SipA mimics as a cognate R-SNARE and binds with Syn8.
Figure 2.
SipA mimics as a cognate R-SNARE of Syn8. (A) Bioinformatics analyses of the protein sequences of SipA, Syn8, Syn7, Syn13, and VAMP8 were performed using NCBI COBALT to determine the presence of SMs in SipA. (B) To determine the region of SipA interacting with Syn8, GST-Syn8 was immobilized, and beads were incubated with His6-SipA1–435, His6-SipA436–685, His6-SipA1–169, His6-SipA1–242, His6-SipA1–277, or His6-SipA48–277. Binding of different SipA-truncated proteins with Syn8 was detected by Western blot analysis using anti-His antibody. (C) SipA1–169 and SipA1–242 were analyzed by nonreducing SDS-PAGE as indicated to determine the formation of homodimers. Arrows indicate the multimers of indicated proteins. (D) To demonstrate that SipA binds with Syn8 through the SM, SipAR204Q and SipA1-435R204Q were generated, and binding of His6-SipAR204Q or His6-SipA1-435R204Q with GST-Syn8 was determined as described in the Bioinformatics analysis section of Materials and methods. . All results are representative of three independent experiments.
To validate these observations, we made several truncated proteins of SipA, namely SipA1–435, SipA436–685, SipA1–169, SipA1–242, SipA1–277, and SipA48–277, and we determined their binding with Syn8 (Fig. 2 B). Our results showed that SipA1–435 specifically binds with Syn8 like SipA:full length (FL), whereas no binding of SipA436–685 with Syn8 was detected (Fig. 2 B). Moreover, we found that SM-deleted SipA1–169 fails to bind with Syn8, and this binding is restored with SM containing truncated proteins like SipA1–242 and SipA1–277. We also found that the initial 48 amino acids deleted SipA48–277, which contains SM and does not bind with Syn8 (Fig. 2 B). Moreover, it has been shown that some R-SNAREs can efficiently form homodimers in vitro through the cysteine residues present in the SM (Flanagan et al., 2015). Remarkably, we also found that SipA1–242 specifically forms homodimers/multimers when the purified protein was analyzed by nonreducing SDS-PAGE (Fig. 2 C). To unequivocally prove that SipA acts as an R-SNARE, we mutated the conserved arginine residue in the SM to glutamine. Our results showed that SipAR204Q and SipA1–435R204Q do not bind with Syn8 (Fig. 2 D). Taken together, these results demonstrated that SipA mimics as an R-SNARE to bind with Syn8.
N-terminal domain of SipA recruits Syn8 on SCV
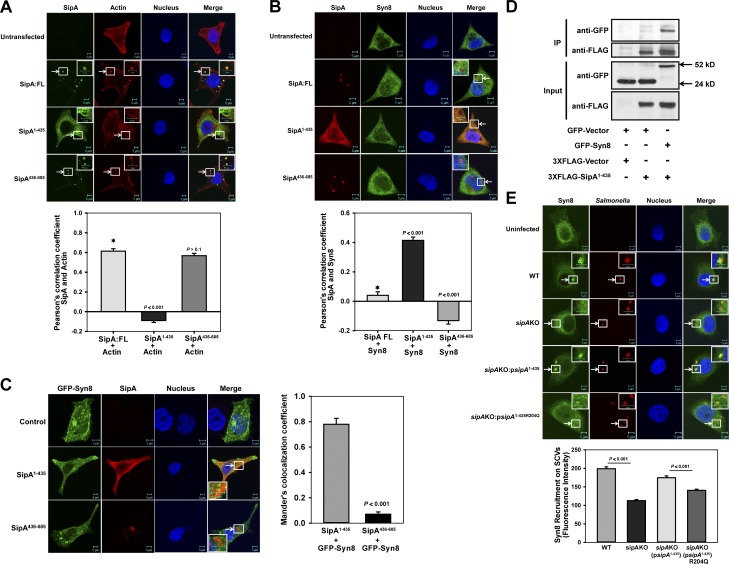
To determine the localization of N-terminal SipA with Syn8 within the cells, SipA:FL, SipA1–435 or SipA436–685 was expressed in HeLa cells as N-terminal FLAG-tagged proteins, and cells were stained with phalloidin or anti-Syn8 antibody. Our results showed that SipA:FL and SipA436–685 significantly colocalized with phalloidin-labeled actin (Fig. 3 A). In contrast, we observed that SipA1–435 does not colocalize with actin and predominantly labels intracellular vesicular membranes (Fig. 3 A) that are positive for Syn8 (Fig. 3 B). Interestingly, SipA:FL was also found to be partially colocalized with Syn8, whereas no significant association of SipA436–685 with Syn8 was detected (Fig. 3 B). Similarly, GFP-Syn8 was found to be colocalized with FLAG-tagged SipA1–435 when coexpressed in HeLa cells (Fig. 3 C). These results are further confirmed by the fact that immobilized anti-FLAG antibody pulled out the Syn8 from GFP-Syn8 and FLAG-tagged SipA1–435 coexpressing HeLa cell lysate (Fig. 3 D). To confirm the role of SipA in the recruitment of Syn8 on SCVs in HeLa cells, a sipA knockout (KO) Salmonella (Salmonella:sipAKO) was generated and characterized (Fig. S3). Our results showed that recruitment of Syn8 to SCV:sipAKO and Salmonella:sipAKO complemented with SipA1–435R204Q was reduced significantly in comparison with SCV:WT. However, Salmonella:sipAKO complemented with N-terminal domain of SipA (sipAKO:psipA1-435) restored the recruitment of Syn8 on SCVs (Fig. 3 E). Subsequently, we found that SipA1–435 is localized on both sipAKO:psipA1–435 and sipAKO:psipA1–435R204Q-Salmonella containing SCVs in HeLa cells, but only sipAKO:psipA1–435 significantly recruits Syn8 (Fig. S4, A and B). These results unambiguously proved that Syn8 is recruited on SCVs by the N-terminal domain of SipA in the host cells.
Figure 3.
N-terminal domain of SipA recruits Syn8 on SCV. (A) SipA:FL, SipA1–435, or SipA436–685 were expressed in HeLa cells as N-terminal FLAG-tagged proteins, and cells were stained with phalloidin (red) and anti-FLAG antibody (green). (B) SipA:FL, SipA1–435, or SipA436–685 was expressed in HeLa cells as above, and cells were stained with anti-FLAG antibody (red) and anti-Syn8 antibody (green). (C) HeLa cells were cotransfected with GFP-Syn8 (green) and FLAG-tagged SipA1–435 or SipA436–685 as described in Materials and methods. Cells were stained with anti-FLAG antibody (red). Colocalization of SipA1–435 and GFP-Syn8 was determined by confocal microscopy. (D) Immunoprecipitation (IP) of GFP-Syn8 from FLAG-SipA1–435 and GFP-Syn8 coexpressing HeLa cell lysate using immobilized anti-FLAG antibody. Binding of GFP-Syn8 with FLAG-SipA1–435 was determined by Western blot analysis using anti-GFP antibody. (E) HeLa cells were infected with indicated Salmonella strains, and cells were stained 90 min p.i. with anti-Salmonella antibody (red) and anti-Syn8 antibody (green). Cells were analyzed by confocal microscopy. Arrows indicate colocalization of the respective proteins (A–C) or on SCVs (D). All results are mean ± SEM of three independent experiments, and levels of significance are indicated by P values in comparison with control (*) below the respective figures.
SipA is intracellularly cleaved and recruits Syn8 on SCV by the N-terminal domain
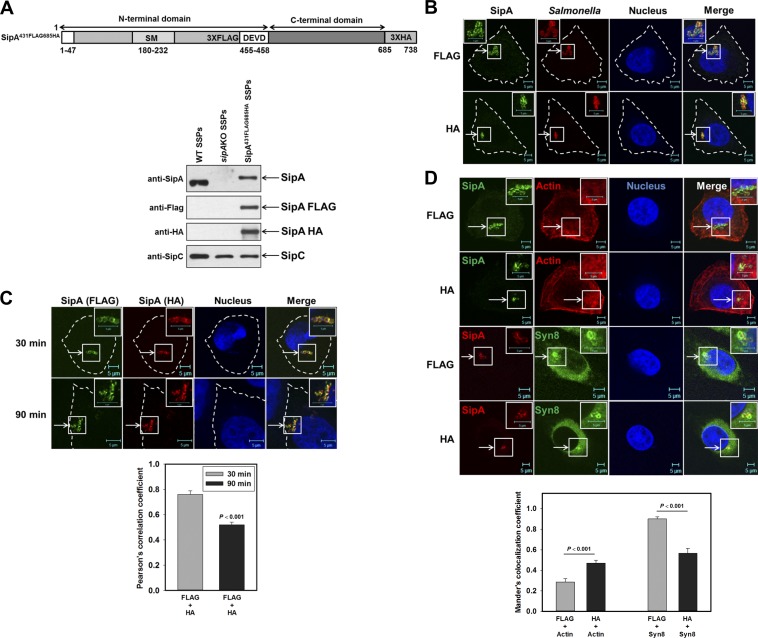
Although SipA is cleaved by host caspase-3 at the DEVD motif (Srikanth et al., 2010) into N-terminal and C-terminal fragments, the role of the N-terminal fragment in intracellular trafficking of Salmonella is not clearly depicted. To determine the role of these fragments of SipA in Salmonella-infected HeLa cells, we generated a dual-tagged SipA construct where the FLAG tag was inserted before the DEVD motif and the HA tag was introduced at the C-terminal end of the protein (Fig. S4, C–E). Subsequently, this construct was introduced into the sipA chromosomal locus of the Salmonella (Salmonella:sipA431FLAG-685HA) by homologous recombination for expressing dual-tagged SipA under the control of its native promoter. For functional characterization, we prepared the secretory proteins from Salmonella:WT and Salmonella:sipA431FLAG-685HA bacteria and found that Salmonella:sipA431FLAG-685HA efficiently secretes SipA431FLAG-685HA protein in culture medium, which is detected by both anti-FLAG and anti-HA antibodies (Fig. 4 A). In addition, 90 min p.i. of HeLa cells with Salmonella:sipA431FLAG-685HA, SipA was detected on SCVs by both anti-FLAG and anti-HA antibodies (Fig. 4 B). Most interestingly, Salmonella:sipA431FLAG-685HA-infected HeLa cells, when stained with both anti-FLAG and anti-HA antibodies 90 min p.i., showed significantly less colocalization between FLAG and HA (Fig. 4 C) in comparison with 30 min p.i., suggesting that SipA might be cleaved intracellularly into N-terminal and C-terminal fragments. Consequently, we found that SipA stained with anti-FLAG antibody does not significantly colocalize with phalloidin-labeled actin in Salmonella:sipA431FLAG-685HA-infected cells in comparison with anti-HA antibody–stained cells (Fig. 4 D). However, SipA stained with anti-FLAG antibody in Salmonella:sipA431FLAG-685HA-infected cells predominantly colocalized with Syn8 (Fig. 4 D). However, as expected, we observed significant overlap between FLAG and HA as both antibodies recognized uncleaved SipA in infected cells.
Figure 4.
Salmonella:sipA431FLAG-685HA recruits Syn8 on SCVs. (A) Top: Schematic diagram showing the position of FLAG and HA tag in the sipA431FLAG-685HA construct. Bottom: Western blot analysis showing the secretion of SipA431FLAG-685HA like SipA:WT protein in the culture supernatant (SSPs). (B) HeLa cells were infected with Salmonella:sipA431FLAG-685HA, and localization of SipA 90 min p.i. was determined using anti-FLAG or anti-HA antibody in respective cells (green). Salmonella was stained with anti-Salmonella antibody (red). (C) Similarly, cells were infected with Salmonella:sipA431FLAG-685HA for indicated times, and localization of SipA in the same cells was detected using anti-FLAG (green) and anti-HA (red) antibodies to determine intracellular cleavage of SipA. (D) To determine the colocalization of SipA with Syn8, cells were infected with Salmonella:sipA431FLAG-685HA and stained with anti-FLAG or anti-HA antibody 90 min p.i. Cells were also costained with phalloidin (actin) or anti-Syn8 antibody as indicated. Arrows indicate colocalization of the respective proteins (C) or on SCVs (B and D). All results are mean ± SEM of three independent experiments, and levels of significance are indicated by P values below the respective figures.
SipA forms a SNARE complex with Syn8, Syn13, and Syn7 to promote membrane fusion
Subsequently, attempts were made to identify the Q-SNARE partners of SipA as fusion requires interaction of three Q-SNAREs with one R-SNARE (Fasshauer et al., 1998). As SCVs also recruit the endosomal Q-SNAREs Syn13 and Syn7, we determined the interaction of GST-SipA1–435 with His6-Syn13 or His6-Syn7. Our results showed that SipA1–435 also interacts with Syn13, Syn7, and SNAP23. In contrast, SipA did not interact with VAMP8, Vti1a, and Vti1b (Fig. 5 A). Next, we analyzed whether SipA1–242, which has an SM, can form a SNARE complex with Syn8, Syn13, and Syn7 using a similar procedure described previously (Shi et al., 2016). Accordingly, mixtures of equimolar amounts of purified His6-HA-Syn8, His6-Myc-Syn13, and His6-FLAG-Syn7 proteins (Fig. 5 B, left) were incubated for 2 h at 24°C in the presence or absence of purified GST-SipA1–242 and then resolved by SDS-PAGE without heat denaturation under nonreducing conditions (Fig. 5 B, right). Interestingly, a band corresponding with high molecular weight (MW) was detected by Coomassie staining only when SipA1–242 was present in the reaction mixture (Fig. 5 B, right). This band was cut out and subjected to mass spectrometry analysis. The mass spectrometry data were analyzed against a local database comprising Syn8, Syn13, Syn7, and SipA proteins. Our results showed that the high-MW SDS-resistant indicated band was composed of all four proteins—Syn8, Syn13, Syn7, and SipA—with similar percent coverage (Fig. 5 C). This result was further confirmed by Western blot analysis using specific antibodies (Fig. 5 C, inset). These results clearly demonstrated that Syn8, Syn13, and Syn7 form an SDS-resistant complex with SipA.
Figure 5.
Determination of SipA-mediated formation of functional SNARE complex required for membrane fusion. (A) To identify SipA-binding SNARE partners, GST-SipA1–435 was immobilized on beads, and binding with indicated His6-SNARE proteins was determined as described in Materials and methods. His6-Syn8 was used as a positive control. (B) To determine whether SipA, Syn8, Syn7, and Syn13 form SDS-resistant complexes, GST-SipA1–242, His6-HA-Syn8, His6-Myc-Syn13, and His6-FLAG-Syn7 proteins were purified and analyzed by SDS-PAGE without heat denaturation under nonreducing conditions, shown as Coomassie-stained bands (left). Strongly stained bands (asterisks) correspond with the expected size of the monomer of the respective proteins. Purified His6-HA-Syn8, His6-Myc-Syn13, and His6-FLAG-Syn7 proteins were incubated in the presence and absence of GST-SipA1–242 and analyzed by SDS-PAGE as described in Materials and methods. Arrow indicates a band corresponding with high-MW hybrid complex only when SipA1–242 was present (right). (C) Percent coverage of all four proteins in the indicated hybrid complex as revealed by proteomic analysis. Inset shows the Western blot analysis using specific antibodies. (D) To determine formation of functional SNARE complex by SipA with Syn8, Syn13, and Syn7, we analyzed the disassembly of the SNARE complex by NSF. GST-SipA1–242 was immobilized on beads and incubated with His6-HA-Syn8, His6-Myc-Syn13, and His6-FLAG-Syn7 proteins to form a SNARE complex. Subsequently, disassembly of SNARE complex was determined in the presence and absence of untreated, NEM-treated, or NSF-depleted cytosol by Western blot analysis as described in Materials and methods. All results are representative of three independent experiments. (E) To directly demonstrate fusion between SipA and host Q-SNAREs, we determined the fusion of donor liposome containing SipA1–277 with acceptor liposome containing Syn8, Syn13, and Syn7 by fusion-induced lipid mixing using a standard FRET-based assay as described in Materials and methods. The fusion between donor liposomes containing VAMP8 with acceptor liposomes containing Syn8, Syn7, and Vti1B was used as control. All results are mean ± SD of four independent experiments and are expressed as percent FRET efficiency.
To further confirm that SipA, Syn8, Syn13, and Syn7 form a functional SNARE complex, we analyzed disassembly of the SNARE complex by NSF. NSF is an AAA+ ATPase, and it can be inactivated by N-ethylmaleimide (NEM) treatment (Whiteheart et al., 1994). It has been shown that cytosolic α-SNAP first binds with SNARE complexes, which subsequently recruit NSF. Finally, ATP hydrolysis of NSF disassembles SNARE complex (Jahn and Scheller, 2006). Thus, GST-SipA1–242 was immobilized on beads and incubated with equimolar concentrations of His6-HA-Syn8, His6-Myc-Syn13, and His6-FLAG-Syn7 proteins to form a SNARE complex. Beads were washed and incubated in the presence of untreated cytosol, NEM-treated cytosol, or NSF-depleted cytosol in PBS containing 3 mM MgCl2 and 1 mM ATP for 45 min at 24°C. Finally, proteins present on the washed beads were detected by Western blot analysis using specific antibodies. The results presented in Fig. 5 D show that a significant amount of Syn8, Syn13, and Syn7 proteins were lost from the immobilized complex in the presence of cytosol. In contrast, there was no significant loss of Syn8, Syn13, and Syn7 proteins from the immobilized complex when the complex was incubated with NEM-treated cytosol or NSF-depleted cytosol under the same conditions. In addition, we found that NSF-depleted cytosol supplemented with NSF:WT dissociated the target Q-SNAREs from the complex, whereas addition of NSF:D1EQ, a dominant-negative mutant of NSF, was unable to release Syn8, Syn13, and Syn7 from the complex (Fig. 5 D). These results demonstrated that SipA forms a functional SNARE complex with Syn8, Syn13, and Syn7.
To directly demonstrate whether such SNARE pairing is sufficient to cause membrane fusion, we reconstituted purified SipA1–277 and Syn8, Syn13, and Syn7 in liposomes and monitored fusion-induced lipid mixing using a standard FRET-based assay (Shi et al., 2016). Mixing of donor liposomes containing SipA1–277 with acceptor liposomes containing Syn8, Syn13, and Syn7 showed efficient FRET, thus indicating membrane fusion (Fig. 5 E). The extent of fusion was comparable with that seen with a canonical SNARE pair of donor liposomes with VAMP8 (R-SNARE) and acceptor liposomes with Syn8, Syn7, and Vti1b (Q-SNAREs). Importantly, respective fusion was completely inhibited by the addition of the soluble cytoplasmic domains of VAMP8 or SipA1–242. No significant fusion was observed between donor and acceptor liposomes without addition of proteins.
SipA promotes fusion of SCV with EEs
To determine the functional significance of SipA-mediated recruitment of the EE SNARE Syn8 (Subramaniam et al., 2000) on SCV, we analyzed the in vitro fusion of purified SCV:WT or SCV:sipAKO with EEs. We found ∼50% inhibition of fusion between SCV:sipAKO and EEs in comparison with the fusion of SCV:WT with EEs (Fig. 6 A), indicating that SipA-mediated recruitment of Syn8 is necessary for optimal fusion of SCVs with EEs. Residual fusion SCV:sipAKO with EEs could be due to the presence of Rab5 on SCVs (Mukherjee et al., 2000). To unequivocally demonstrate the role of Rab5 and Syn8 in this fusion process, fusion was performed in the presence of Rab5- or Syn8-specific antibodies. We observed that addition of anti-Rab5 antibodies inhibited ∼80% fusion between SCVs with EEs, whereas anti-Syn8 antibody almost completely abrogated the fusion (Fig. 6 B). Subsequently, attempts were made to determine the role of Syn13 and Syn7 in the fusion between SCVs and EEs. Therefore, fusion was performed in the presence of the cytoplasmic domain of respective SNAREs, which acts as a dominant-negative mutant (Pulido et al., 2011). Our results showed that addition of dominant-negative mutants of Syn8, Syn13, or Syn7 in the fusion reaction significantly inhibited the fusion between SCV:WT and EE (Fig. 6 C). In correlation, we also found that both purified SCV and EEs contain Syn8, Syn7, Syn13, and Rab5 (Fig. S5).
Figure 6.
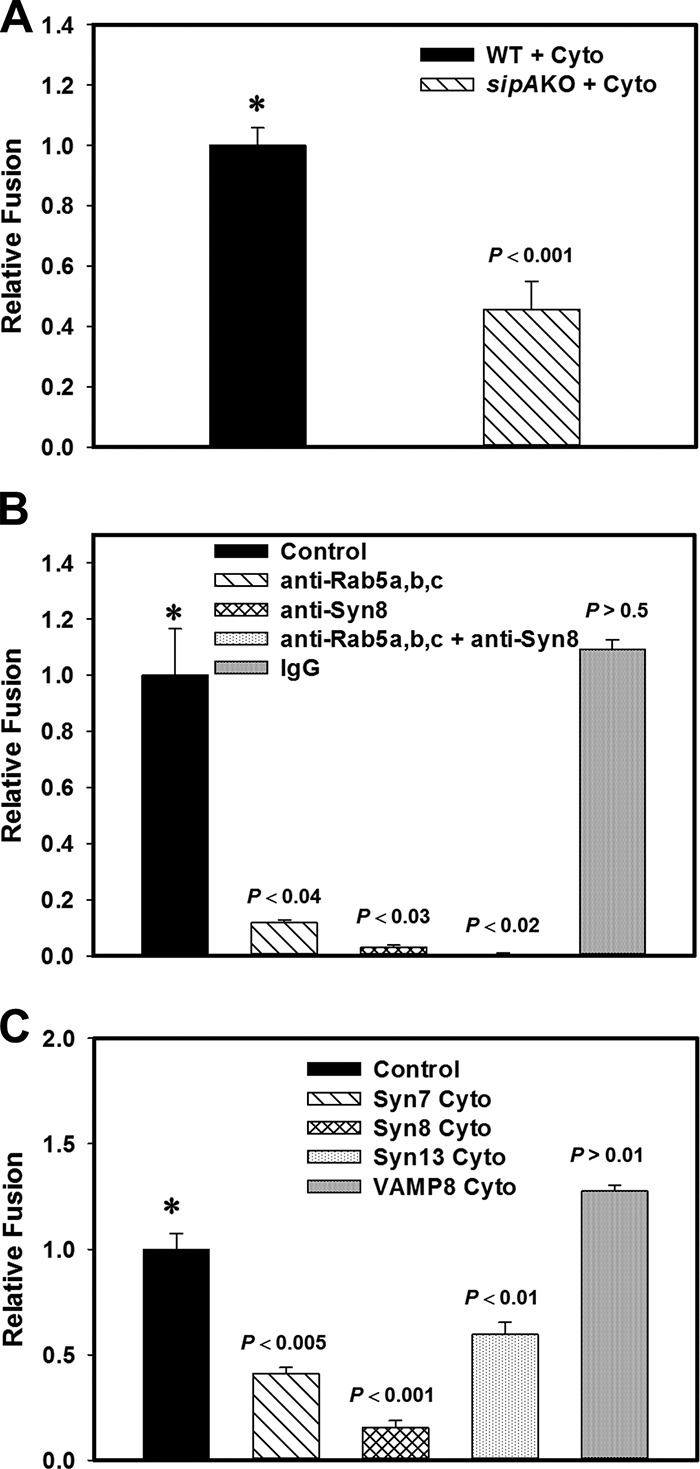
SipA acts as a cognate R-SNARE to promote fusion of SCVs with EEs. (A) To determine the role of Syn8, in vitro fusion was performed between SCVs containing biotinylated Salmonella:WT or Salmonella:sipAKO and EEs containing avidin-HRP in the presence of ATP-regenerating fusion buffer supplemented with gel-filtered cytosol for 1 h at 37°C. Fusion was measured as described in Materials and methods. (B) To determine the role of Syn8 and Rab5, a similar fusion assay was performed between SCV:WT and EE in the presence of indicated antibodies. (C) To determine the role of identified SipA cognate SNAREs, fusion was performed between SCV:WT and EE in presence of the cytoplasmic domain of indicated SNARE proteins. Fusion obtained with SCV:WT with EE was chosen as 1 U in all experiments, and the results are expressed as relative fusion of three independent experiments ± SEM. 1 U corresponds with ∼30 ng, 34 ng, or 36 ng HRP activity per mg protein in the fusion assay reported in A–C, respectively. Levels of significance are indicated by P values.
Role of Syn8 in the survival of Salmonella in host cells
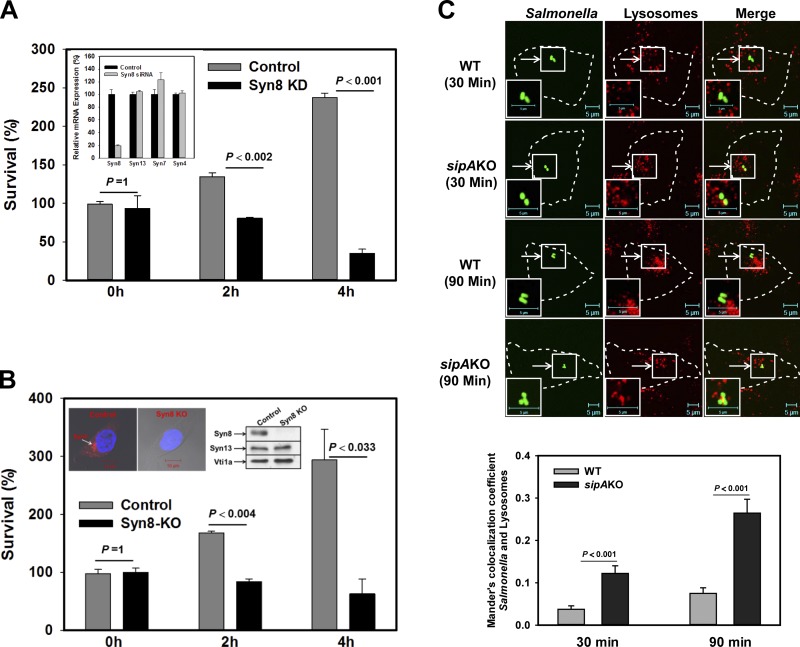
To determine the role of Syn8 in the survival of Salmonella in HeLa cells, Syn8 was specifically knocked down in HeLa cells by siRNA. Syn8 knockdown was confirmed by quantitative PCR (qPCR; Fig. 7 A, inset). Subsequently, cells were infected with Salmonella:WT, and numbers of bacteria present in the respective cells at the indicated time points were determined. We found that knockdown of Syn8 in HeLa cells inhibited ∼80% growth of Salmonella 4 h p.i. in comparison with control cells (Fig. 7 A). To unequivocally prove the role of Syn8 in the survival of Salmonella in the host cells, Syn8 was knocked out in the HeLa cells using CRISPR. Syn8 KO by CRISPR was confirmed by immunofluorescence and Western blot analysis using specific antibodies (Fig. 7 B, inset). Our results showed that KO of Syn8 in the HeLa cells inhibited the growth of Salmonella almost to the similar extent as observed with siRNA-mediated knockdown of Syn8 in HeLa cells (Fig. 7 B).
Figure 7.
Role of Syn8 in the survival of Salmonella in host cells. (A) To determine the role of Syn8 in the survival of Salmonella in HeLa cells, Syn8 was knocked down with Syn8 siRNA, and cells were infected with Salmonella:WT as described in Materials and methods. Cells were lysed at the indicated time points after infection, and the number of bacteria present in the cells was calculated by measuring colony-forming units. Results are expressed as percent survival, arbitrarily chosen as 100% survival at 0 h of infection with Salmonella in control cells. All results are mean ± SEM of three independent experiments, and levels of significance are indicated by P values. Inset shows the specific knockdown of Syn8 by siRNA in HeLa cells by qPCR. (B) A similar assay was performed in CRISPR-mediated KO of Syn8 in HeLa cells. Inset shows the specific KO of Syn8 by CRISPR in HeLa cells by immunofluorescence and Western blot analysis using specific antibodies. (C) Lysosomes of HeLa cells were labeled with Dextran–Texas Red and were infected with GFP-Salmonella:WT or GFP-Salmonella:sipAKO. Cells were analyzed by confocal microscopy at indicated time points to determine the localization of respective Salmonella in the lysosomes. Bottom: Quantitation for the same. P values are indicated.
To investigate whether recruitment of Syn8 on SCVs is required to inhibit transport to the lysosomes, lysosomes of HeLa cells were labeled with Dextran–Texas Red, and cells were infected with GFP-Salmonella:WT or GFP-Salmonella:sipAKO. Cells were analyzed at indicated time points to determine the localization of respective Salmonella in the lysosomes. Our results showed significant colocalization of Salmonella:sipAKO with lysosomes compared with Salmonella:WT after 90 min infection (Fig. 7 C). Taken together, our results demonstrate that SipA-mediated recruitment of Syn8 on SCVs promotes fusion of SCVs with EEs and prevents its transport to the lysosomes.
Discussion
Phagosome maturation depends on the sequential recruitment and removal of different Rabs and SNAREs (Brumell and Scidmore, 2007; Haraga et al., 2008; McGhie et al., 2009), which are master regulators of intracellular trafficking (Wickner and Schekman, 2008; Stenmark, 2009; Pfeffer, 2013). We and others have shown that Salmonella modulates the endolysosomal pathway by targeting these proteins to establish their niche in host cells by their effectors (Hashim et al., 2000; Mukherjee et al., 2000; McGhie et al., 2009; Agbor and McCormick, 2011; Madan et al., 2012). Thus, Salmonella, via its effector molecules, modulates membrane-fusion events in the host cells to avoid its targeting to the lysosomes. The current model of vesicle fusion suggests that the membrane fusion is regulated by Rab GTPases. These proteins act as molecular switches and activate specific SNARE proteins to drive membrane fusion between donor and acceptor compartments. The specificity of membrane fusion is further governed by the interactions of an R-SNARE with three cognate Q-SNAREs (Jahn and Scheller, 2006). It is now well evident that some of the Salmonella effectors like SopB, SopE, SptP, etc. target host GTPases (LaRock et al., 2015), but very little is known about the interaction of Salmonella effector with host SNAREs.
In this study, we have determined the recruitment of different Syntaxins on SCVs at various stages of their maturation in HeLa cells. Our results have shown that SCVs predominantly recruit higher amounts of Syn8 than Syn13 or Syn7. Subsequently, we identified that SipA is involved in the binding and recruitment of Syn8 on SCVs. SipA is shown to facilitate bacterial invasion into host cells by modulating actin polymerization (Zhou et al., 1999a,b; Jepson et al., 2001; Raffatellu et al., 2005). Subsequent research has shown that host caspase-3 cleaves SipA at the DEVD motif, leading to the formation of two functional domains (Srikanth et al., 2010). The actin-binding activity of SipA is located in the C-terminal domain, whereas the function of the N-terminal domain is associated with multiple functions. In this study, we have found that the N-terminal domain of SipA specifically recruits Syn8 on SCVs.
To understand the mechanism of interaction between SipA and Syn8, we analyzed SipA and Syn8 sequences. Our results show that SipA180–232 has a typical SM containing a conserved arginine residue in the center like mammalian R-SNAREs (Fasshauer et al., 1998). In addition, our results show that SipA1–242, which contains an SM, specifically forms homodimers/multimers, which is in correlation with the fact that R-SNAREs can efficiently form homodimers in vitro through the cysteine residues present in the SM (Flanagan et al., 2015). Interestingly, bioinformatics analysis predicted that SipA1–277 also contains a putative transmembrane domain. Therefore, it is tempting to speculate that SipA mimics as an R-SNARE and binds with Syn8. Indeed, three lines of evidence have demonstrated that SipA mimics as an R-SNARE to bind Syn8: (1) SM-deleted SipA1–169 truncated protein fails to bind with Syn8; (2) binding with Syn8 is restored with an SM containing truncated proteins SipA1–242; and (3) mutation in the conserved arginine residue in the SM of SipA, SipAR204Q, and SipA1–435R204Q fails to bind Syn8. Interestingly, our results show that SipA48–277, which contains an SM, does not interact with Syn8. It has been shown that the N-terminal region of the SNARE protein acts as a nucleation center for the interaction with cognate SNAREs (Fasshauer and Margittai, 2004; Pobbati et al., 2006; Ellena et al., 2009). Therefore, it might be possible that the initial 48 amino acids of N-terminal SipA are required for nucleation with Syn8. To determine the significance of interaction between SipA and Syn8 within cells, SipA:FL, SipA1–435, or SipA436–685 was expressed in HeLa cells. Consistent with previous findings (Zhou et al., 1999a), we have also found that SipA436–685 colocalized with actin, whereas SipA1–435 predominantly labeled Syn8-positive intracellular vesicular membranes. In addition, we found that infection with Salmonella:sipAKO significantly reduced the recruitment of Syn8 on SCVs in comparison with SCV:WT and that the recruitment of Syn8 on SCVs is restored by infection with Salmonella:sipAKO complemented with SipA1–435 (sipAKO:psipA1–435). Most interestingly, Salmonella:sipAKO complemented with SipA1–435R204Q fails to recruit Syn8 on SCVs significantly compared with SCV:WT. These results unequivocally prove that SipA mimics as a cognate R-SNARE and recruits Syn8 on SCVs, which is a Q-SNARE (Subramaniam et al., 2000).
To unambiguously prove that the N terminus of SipA recruits Syn8 on SCVs in Salmonella infection, we made a transgenic Salmonella where we introduced a dual-tag SipA construct (SipA431FLAG-685HA) into the sipA chromosomal locus of the Salmonella (Salmonella:sipA431FLAG-685HA). Interestingly, infection of HeLa cells with Salmonella:sipA431FLAG-685HA and subsequent staining with both anti-FLAG and anti-HA antibodies does not show complete colocalization between FLAG and HA. As SipA FL protein should be recognized by both antibodies, this result suggests that SipA is probably cleaved at the DEVD motif as observed previously (Srikanth et al., 2010) to generate intracellular N-terminal and C-terminal fragments. Consequently, we found that SipA stained with anti-FLAG antibody in Salmonella:sipA431FLAG-685HA-infected cells predominantly colocalized with Syn8. Thus, these results translate our in vitro finding that Salmonella infection in HeLa cells also recruits Syn8 on SCVs by the N-terminal domain of SipA.
Subsequently, we have tried to identify SNARE partners as fusion requires interaction of three Q-SNAREs with one R-SNARE (Fasshauer et al., 1998). Interestingly, it has been shown that Syn8 is preferentially associated with EEs (Subramaniam et al., 2000) and regulates endosomal trafficking (Prekeris et al., 1999). Moreover, previous studies have shown that Syn8 forms a complex with Syn7, Vti1b, and VAMP8 to promote homotypic fusion of EEs and late endosomes (Antonin et al., 2000a,b), whereas replacement of VAMP8 with VAMP7 in the same SNARE complex stimulated fusion of late endosomes with lysosomes (Pryor et al., 2004). In addition, it has been shown that Syn13, Vti1a, Syn6, and VAMP4 regulate homotypic fusion of EEs (Brandhorst et al., 2006). Among these EE SNAREs, our results show that SipA binds with Q-SNAREs like Syn8, Syn13, Syn7, and SNAP23. However, SipA does not interact with R-SNARE, VAMP8, or Vti1a and Vti1b. SipA binding with SNAP23 is an interesting observation. It could be possible that SipA, which functionally mimics as VAMP8, binds with SNAP23 as it has been shown previously that SNAP23 binds and forms complexes with several VAMPs including VAMP8 (Foster et al., 1998; Hong, 2005). As SipA binds and recruits endosomal Q-SNAREs like Syn8, Syn13, and Syn7, we have tried to determine whether SipA forms a SNARE complex with Syn8, Syn13, and Syn7. Indeed, we have found that these four proteins form an SDS-resistant SNARE complex. To determine whether SipA forms a functional SNARE complex with Syn8, Syn13, and Syn7, we analyzed disassembly of SNARE complex by NSF:WT and NSF:D1EQ, a dominant-negative mutant of NSF (Whiteheart et al., 1994). Our results show that NSF dissociates the target Q-SNAREs from the complex, demonstrating that SipA forms a functional SNARE complex with Syn8, Syn13, and Syn7. Subsequently, we have shown that such SNARE pairing is sufficient to cause membrane fusion between donor liposomes containing SipA1–277 with acceptor liposomes containing Syn8, Syn13, and Syn7. These results suggest the molecular mimicry of SipA as an R-SNARE to promote membrane fusion for modulating the endolysosomal pathway in host cells.
To determine the functional role of Syn8 in Salmonella maturation in host cells, we reconstituted in vitro fusion between SCVs containing WT or sipAKO Salmonella with EEs. We found ∼50% inhibition of fusion between SCV:sipAKO and EEs in comparison with fusion of SCV:WT, with EEs indicating that SipA-mediated recruitment of Syn8 is necessary for optimal fusion of SCVs with EEs. However, it has been shown that SCVs also recruit Rab5 (Steele-Mortimer, 2008) and promotes fusion of phagosomes with EEs (Mukherjee et al., 2000). Similarly, we also found that addition of anti-Rab5 antibodies inhibited ∼80% of fusion between SCVs with EEs. Interestingly, addition of anti-Syn8 antibody almost completely inhibited the fusion between SCVs and EEs, indicating that the function of Syn8 is downstream of Rab5. We also found that addition of dominant-negative mutants of Syn8, Syn13, or Syn7 in the fusion reaction significantly inhibited the fusion between SCV:WT and EEs. Taken together, our results demonstrated that SipA mimics as an R-SNARE and binds with three Q-SNAREs, Syn8, Syn13, and Syn7, to promote fusion of SCVs with EEs. In correlation with our results, it has been shown recently that some of the pathogen effector molecules bind with host SNARE molecules by simulating as a SNARE partner. For example, Legionella pneumophila effector DrrA binds with Syn3 (Arasaki et al., 2012), IncA effector of Chlamydia trachomatis binds with VAMP7 and VAMP8 (Delevoye et al., 2008), Salmonella effector SipC binds with Syn6 (Madan et al., 2012), and Legionella effectors LegC2, LegC3, and LegC7 form a SNARE complex with VAMP4 (Shi et al., 2016).
It is well documented that Salmonella avoids transport to the lysosomes, but the nature of the compartment where Salmonella resides is not fully characterized. Previous studies have shown that the SCV initially recruits EE markers like EEA1, Rab5, and the transferrin receptor (Steele-Mortimer et al., 1999) followed by acquisition of Rab7 (Méresse et al., 1999). In contrast, they exclude the late endosomal marker mannose-6-phosphate receptor (Garcia-del Portillo and Finlay, 1995; Hashim et al., 2000), demonstrating that Salmonella is not targeted to late endosome. Consequently, it has been shown that the Salmonella effector SopD2 impairs Rab7 activity and thereby inhibits the recruitment of the Rab7 effectors RILP and FYCO1 on SCVs to prevent the transport of SCVs to lysosomes (D’Costa et al., 2015). In addition, we and others have shown that SCVs recruit Rab5 (Mukherjee et al., 2000; Mallo et al., 2008) but exclude the transferrin receptor (Hashim et al., 2000). Taken together, these results suggest that Salmonella resides in a unique endocytic niche and interacts with various intracellular compartments by recruiting appropriate Rab GTPases and SNAREs via their effector proteins.
It is well evident that constitutive fusion of phagosomes with EEs blocks the phagosome maturation toward lysosomes and promotes pathogen survival (Hashim et al., 2000; Mukherjee et al., 2000). Consequently, we have found that siRNA-mediated knockdown of Syn8 and KO of Syn8 by CRISPR in HeLa cells significantly inhibits the growth of Salmonella in the host cells. This is due to induced transport of Salmonella to the lysosomes as we have found that sipAKO Salmonella, which does not recruit Syn8, is significantly targeted to lysosomes in comparison with Salmonella:WT. These observations are supported by the fact that survival of sipAKO Salmonella is shown to be compromised in the host cells (Brawn et al., 2007).
This is the first demonstration that SipA mimics as an R-SNARE and thereby forms a functional SNARE complex with three host Q-SNAREs, namely Syn8, Syn13, and Syn7, to promote fusion with EEs. Constitutive fusion of SCVs with EEs inhibits SCV maturation toward lysosomes and thereby helps Salmonella survive in the host cells (Fig. 8). Our results provide mechanistic insight into how effector molecules from pathogens manipulate host cellular processes by functionally substituting endogenous SNAREs. It could be possible that pathogens have selected SMs during evolution for easy manipulation of membrane fusion for their benefit. Thus, the disruption of bacterial SNARE-like effector proteins could be a viable target for the development of a therapeutic strategy.
Figure 8.
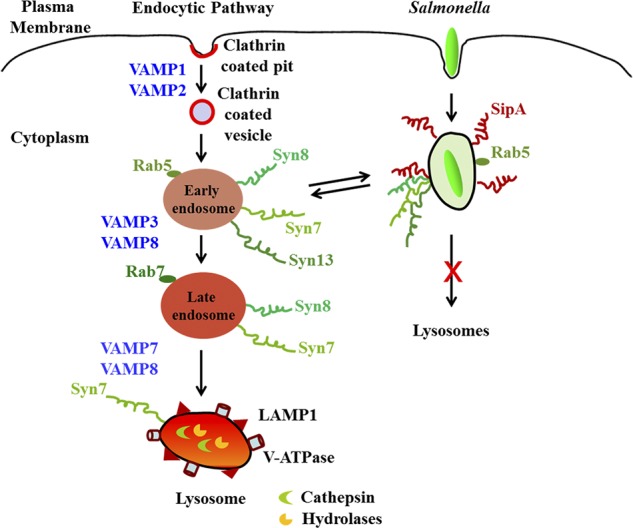
Model showing the mechanism of recruitment of Syn8 on SCV. SipA mimics as an R-SNARE and thereby recruits the host Q-SNAREs Syn8, Syn13, and Syn7 on SCVs to promote fusion with EEs and inhibits transport to lysosome.
Materials and methods
Reagents and antibodies
Unless otherwise stated, all reagents were obtained from Sigma-Aldrich. Tissue culture supplies were obtained from Greiner Bio-One and Biological Industries. Dextran–Texas Red (70,000 MW), Hoechst, Lipofectamine 2000, Lipofectamine RNAiMAX, Prolong Gold antifade mounting reagent, rabbit polyclonal anti-GFP, Alexa Fluor 546 phalloidin, and Alexa Fluor–tagged secondary antibodies were obtained from Thermo Fisher Scientific. Biotin (long arm)–N-hydroxysuccinimide, avidin D–HRP, and avidin D were purchased from Vector Laboratories; mouse monoclonal anti-GST and rabbit polyclonal anti-Rab5b were from Santa Cruz Biotechnology, Inc.; rabbit polyclonal anti-Rab5a was from Abcam; mouse monoclonal anti-His antibody was from GE Healthcare; mouse monoclonal anti-FLAG M2 antibody, EZview Red ANTI-FLAG M2 affinity gel, rabbit polyclonal anti-HA antibody, and rabbit polyclonal anti-Rab5c were from Sigma-Aldrich; mouse monoclonal anti-Myc and rabbit polyclonal IgG antibody were from Cell Signaling Technology; rabbit polyclonal anti-Syn7, rabbit polyclonal anti-Syn13, and rabbit polyclonal anti-NSF were from Synaptic Systems; mouse monoclonal anti-Syn4, anti-Vti1a, and anti-GM130 were from BD; and goat polyclonal anti-Salmonella CSA-1 antibody was from KPL. Antibodies against Syn8 and SipA were generated in rabbit and mouse, respectively, using standard methods. All secondary antibodies labeled with HRP were purchased from Jackson ImmunoResearch Laboratories, Inc. Other reagents used were of analytical grade. Gel-filtered cytosol used in the fusion assay was prepared from HeLa cells as described previously (Madan et al., 2008). NSF was depleted from cytosol using immobilized anti-NSF antibody as described previously (Mukherjee et al., 2000).
Plasmids
pFPV25.1 plasmid for constitutive expression of GFP in Salmonella (Brandhorst et al., 2006) was provided by R. Valdivia (Duke Center for Microbial Pathogenesis, Durham, NC). pBAD24, an arabinose-inducible expression vector (Guzman et al., 1995), was provided by A. Surolia (National Institute of Immunology, New Delhi, India). Syn13-EGFP was provided by J. Brumell (University of Toronto, Toronto, Canada). His6-tagged FL Syn7, Syn8, Syn13, Vti1b, and VAMP8 along with their cytoplasmic domain expression constructs were provided by R. Jahn and G. Mieskes (Max Planck Institute for Biophysical Chemistry, Göttingen, Germany). GFP-VAMP7 and GFP-VAMP8 were provided by T. Galli (Institut Jacques Monod, Paris, France). NSF:WT and NSF:D1EQ were gifts from S.W. Whiteheart (University of Kentucky, Lexington, KY). pSpCas9(BB)-2A-GFP plasmid (PX458) for generating Syn8-KO cells was provided by F. Zhang (Massachusetts Institute of Technology, Cambridge, MA). pET-28a (Novagen; Merck KGaA) and pGEX-4T2 (GE Healthcare) vectors were used for the expression of all N-terminal His6-tagged proteins and GST-tagged proteins, respectively. p3×FLAG-Myc-CMV-26 vector (Sigma-Aldrich) was used for expressing SipA as N-terminal 3×FLAG-tagged proteins in HeLa cells.
Bacterial strains and growth conditions
Salmonella (SL1344 strain) was provided by D.W. Holden (Imperial College, London, UK). All other Salmonella strains were derived from Salmonella (SL1344 strain). Escherichia coli strains SM10λpir, SY327λpir, and the suicide vector pRE112, used for the generation of sipAKO and Salmonella:sipA431FLAG-685HA strains, were gifts from O.S. Mortimer (National Institutes of Allergy and Infectious Diseases, Hamilton, MT). Bacteria were routinely grown overnight in Luria-Bertani broth supplemented with streptomycin (100 µg/ml), ampicillin (100 µg/ml), kanamycin (50 µg/ml), or chloramphenicol (30 µg/ml) as appropriate with constant shaking (250 rpm) at 37°C. The late log phase Salmonella (OD600 of 0.8–0.9) was harvested by centrifugation and used for infection experiments. SSPs were prepared from the spent culture medium as described previously (Mukherjee et al., 2000).
Cells
HeLa cells (human epithelial carcinoma cell line) were obtained from ATCC. HeLa cells were cultured in complete medium (DMEM containing 10% FCS and 50 µg/ml gentamicin) at 37°C in a humidified incubator with 5% CO2.
Identification of Syn8 binding protein(s) from SSPs
To identify Salmonella effector protein(s) interacting with Syn8, 5 µg GST-Syn8 was immobilized on glutathione beads and incubated with 5 mg SSP for 2 h at RT. Subsequently, beads were washed (Madan et al., 2012), and proteins bound to the beads were resolved in 12% SDS-PAGE. The gel was silver stained to detect effector protein(s) specifically interacting with GST-Syn8. Finally, the identified band was excised from the gel, trypsin digested, and analyzed by mass spectrometry at a commercial facility (The Center for Genomic Application, New Delhi, India). Salmonella protein was also confirmed by Western blot analysis using anti-SipA antibody.
Bioinformatics analysis
Bioinformatics analysis of SipA was done in order to check for the presence of characteristic SMs or domains. The protein sequence of SipA was aligned with Syn8 and its cognate SNARE partner (Syn7, Vti1b, and VAMP8) protein sequences using NCBI COBALT (Papadopoulos and Agarwala, 2007). The obtained multiple sequence alignment result was further analyzed through Jalview program (Waterhouse et al., 2009).
Interaction between Syn8 and SipA or its truncations
SipA and its truncations were cloned into a pET28a expression vector and purified as a His6-tagged protein. Similarly, GST-tagged fusion proteins were cloned in pGEX-4T-2 and purified. To determine the direct interaction of Syn8 with SipA, an in vitro protein–protein interaction assay was performed using 2.5 µg immobilized GST-Syn8 on glutathione beads and incubated with 0.5 µg His6-SipA or an equimolar concentration of SipA-truncated proteins for 1 h at 4°C. Beads were washed extensively (Madan et al., 2012), and binding of SipA or its truncated proteins with Syn8 was detected by Western blot analysis using anti-SipA or anti-His antibodies. GST immobilized on the beads was used as a control. A similar assay was performed to determine the binding between GST-SipA1–435 with different His6-tag SNARE proteins.
Salmonella infection in HeLa cells
HeLa cells (0.3 × 106) were seeded on coverslips placed in six-well plates and grown for 12 h at 37°C in a humidified incubator with 5% CO2 in complete medium. Cells were washed and incubated with the indicated Salmonella strain at 50 MOI for 10 min in 1 ml FCS-free medium at 37°C. After infection, cells were washed to remove uninternalized bacteria and incubated with complete DMEM containing 100 µg/ml gentamicin (to kill extracellular bacteria) for indicated periods of time at 37°C.
Immunofluorescence
HeLa cells grown on coverslips were fixed with 4% paraformaldehyde, pH 7.4, in PBS at RT for 20 min. After fixation, cells were washed three times with PBS and blocked in blocking buffer (PBS containing 3% BSA and 0.2% saponin) for 1 h at RT. After blocking, cells were incubated with appropriate primary antibody in blocking buffer for 12 h at 4°C, washed, and probed with Alexa Fluor–labeled secondary antibodies for 1 h at RT. Cells were washed and stained with Hoechst to label the nucleus for 5 min at RT. Coverslips were air dried and mounted on glass slides using Prolong Gold antifade mounting reagent for 12 h at RT. Cells were observed under a Zeiss LSM 510 META or LSM 700 confocal laser scanning microscope using an oil-immersion 63× objective.
Quantification of fluorescence intensity for Syntaxin recruitment was performed using Zeiss 510 META software. Each dataset was analyzed using at least 50 infected cells containing >100 Salmonella. Results are expressed as fluorescence intensity in AU after subtracting background fluorescence intensity. To determine the colocalization between markers, Pearson’s correlation coefficient or Manders’ colocalization coefficient of the whole cell or region of interest was determined using Zeiss 510 META software. At least 50 transfected or infected cells were imaged for each condition, and results are representative of three independent experiments.
Localization of SipA with Syn8 in HeLa cells
To determine the localization of SipA in HeLa cells, FLAG-tagged SipA:FL, SipA1–435, or SipA436–685 protein was overexpressed in cells. Briefly, HeLa (0.2 × 106) cells were plated in complete medium, washed, and transfected with FLAG-tagged construct (2 µg) of SipA:FL, SipA1–435, or SipA436–685 using Lipofectamine 2000 according to the manufacturer’s protocol. Cells were washed and fixed with 4% paraformaldehyde for 20 min at RT. Cells were immunostained with anti-FLAG and anti-Syn8 antibodies. SipA-overexpressed cells were also stained with phalloidin to determine its localization with actin. Finally, cells were viewed with a Zeiss LSM 510 META laser scanning confocal microscope.
Immunoprecipitation
HeLa cells were cotransfected with FLAG-SipA1–435 and GFP-Syn8 using Lipofectamine 2000 according to the manufacturer’s protocol. Cells were washed three times and lysed in 1 ml radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, and complete protease inhibitor cocktail). Lysates were centrifuged at 12,000 rpm for 30 min at 4°C, and supernatants were collected. Subsequently, 40 µl anti-FLAG M2 affinity gel was equilibrated with RIPA buffer and incubated with 1 mg lysate for 12 h at 4°C on a rotatory shaker. Beads were washed three times with RIPA buffer followed by three times with chilled PBS. Finally, proteins bound to the beads were resolved in 12% SDS-PAGE and analyzed by Western blotting using anti-FLAG or anti-GFP antibodies. FLAG-SipA1–435– and GFP-Syn8–coexpressing cells were also immunostained with anti-FLAG antibody and visualized with a confocal microscope.
Determination of the formation of the SNARE complex by SipA
To determine the formation of SDS-resistant SNARE complex by SipA with Syn8, Syn13, and Syn7, equimolar amounts of 2.5 µM each of His6-HA-Syn8, His6-Myc-Syn13, and His6-FLAG-Syn7 purified proteins were incubated in 50 µl PBS for 2 h at 24°C on a rotary mixer in the presence or absence of purified GST-SipA1–242. Both samples were resolved by SDS-PAGE (4–12% [wt/vol] NuPAGE Bis · Tris gel; Invitrogen) without heat denaturation under nonreducing conditions, followed by Coomassie blue staining. A band corresponding with high MW was detected by Coomassie staining only in the presence of SipA1–242, which was cut out and analyzed by nano–liquid chromatography–tandem mass spectrometry as described previously (Rastogi et al., 2016). Briefly, the indicated protein band was sliced out as gel pieces and destained with 50% acetonitrile in 25 mM ammonium bicarbonate solution. Subsequently, gel pieces were dehydrated using 100% acetonitrile followed by rehydration with 50 mM ammonium bicarbonate solution containing 10 mM DTT. Finally, gel pieces were resuspended in 100 µl digestion buffer (50 mM ammonium bicarbonate solution containing 1 mg/ml trypsin gold) and incubated for 20 h at 37°C. The mixture was centrifuged (2 min at 1,000 g), and the supernatant containing digested peptides was acidified using trifluoracetic acid. The acidified digested peptides were concentrated to 50 µl and desalted using C-18 Zip-Tip by standard protocol. The digested peptides were vacuum dried, dissolved in solvent A (5% acetonitrile containing 0.1% formic acid), and loaded for reverse-phase chromatography using a C-18 Picofrit analytical column in a Thermo Fisher Scientific Proxeon Nano liquid chromatographer. Samples were run at a flow rate of 300 nl/min using a linear gradient of solvent B (95% acetonitrile containing 0.1% formic acid); 70 min in 5–40% solvent B, 10 min in 40–80% solvent B, 10 min in 80% solvent B, 5 min in 80–5% solvent B, and 25 min in 5% solvent B. Mass spectrometry was performed in an Orbitrap Velos mass spectrometer, and data were analyzed against a local database comprising of Syn8, Syn13, Syn7, and SipA proteins using Proteome Discoverer Software (1.3.0.339 DBV version; Thermo Fisher Scientific).
Disassembly of SNARE complex by NSF
To determine whether SipA forms a functional SNARE complex with Syn8, Syn13, and Syn7, we analyzed disassembly of SNARE complex by NSF, which can be inactivated by NEM treatment. Accordingly, 6 µg GST-SipA1–242 was immobilized on beads. Beads were washed and incubated with 3 µg His6-HA-Syn8, His6-Myc-Syn13, and His6-FLAG-Syn7 proteins each in PBS for 1 h at 24°C to form a SNARE complex. Subsequently, beads containing SNARE complex were washed and resuspended in 200 µl PBS containing 3 mM MgCl2 and 1 mM ATP in the presence of 100 µg untreated cytosol, NEM-treated cytosol, or NSF-depleted cytosol for 45 min at 24°C. Finally, proteins present on the washed beads were separated by SDS-PAGE and identified by Western blot analysis using anti-GST, anti-HA, anti-Myc, and anti-FLAG antibodies. To unequivocally determine the role of NSF, beads containing SNARE complex were incubated in the presence of NSF-depleted cytosol containing 3 µg NSF:WT or NSF:D1EQ, a dominant-negative mutant of NSF, under same conditions and analyzed using same procedure.
Preparation of proteoliposomes and membrane-fusion assays
To determine membrane fusion, liposomes containing purified proteins were prepared by detergent removal as described previously (Shi et al., 2016). In brief, a stock solution of DOPC:DOPE:DOPS:cholesterol (Avanti Polar Lipids) in a 56:22:11:11 molar ratio was prepared in 20 mM Hepes buffer saline (HBS; Hepes buffer containing 150 mM NaCl and 5% cholate, pH 7.4). Proteins in HBS and 1% CHAPS were mixed with the lipid solution to a final protein/lipid molar ratio of 1:500. The donor set of liposomes contained the indicated R-SNARE and NBD-PE (2 mol%; Invitrogen), and the acceptor set contained cognate Q-SNAREs and Texas Red DHPE (1 mol%; Invitrogen). Proteoliposomes were formed by dialysis against HBS for 2 d at 4°C in 3.5-kD cutoff tubing (Invitrogen). Donor and acceptor liposomes were each diluted to a final concentration of 10 µM in HBS and incubated for 1 h at RT in the absence or presence of 10 µM of the cytosolic domains of respective R-SNAREs. Liposomes prepared without the addition of proteins served as controls. Mixtures were transferred to 96-well plates, and fluorescence emission of samples was acquired from 500–800 nm at an excitation wavelength of 450 nm using a Tecan M-200 Infinite Pro plate reader. FRET efficiency was calculated from the formula 100 × (1 − FD+A/FD), where FD is the fluorescence emission at 540 nm of donor (D) liposomes alone, and FD+A is the fluorescence emission at 540 nm of donor liposomes with acceptor (A) liposomes. Results are expressed as percent FRET efficiency.
Generation and characterization of sipAKO Salmonella
Salmonella:sipAKO (sipAKO) strain was generated by a suicide vector (pRE112)–based allelic exchange method of homologous recombination using a deletion construct containing 1 kb upstream and downstream regions of sipA as described previously (Edwards et al., 1998). In addition, SipA1–435 and SipA1–435R204Q were complemented into Salmonella:sipAKO (sipAKO:psipA1–435 and sipAKO:psipA1–435R204Q) using an arabinose-inducible Salmonella expression vector, pBAD24 (Guzman et al., 1995).
Preparation of transgenic Salmonella:sipA431FLAG-685HA strain
To generate Salmonella:sipA431FLAG-685HA strain, first, the sipA1–431 fragment was PCR amplified using a specific forward primer with a SapI restriction site (forward, 5/-GTGTGTGCTCTTCTATGGTTACAAGTGTAAGGACTCAGCCC-3/) and a reverse primer containing 3×FLAG sequence with a SapI restriction site (reverse, 5/-GTGTGTGCTCTTCTATCCTTGTCATCGTCATCCTTGTAATCGATGTCATGATCTTTATAATCACCGTCATGGTCTTTGTAGTCCATAAAAGAGGTTGTTTCACCCGTAGTGCC-3/). Similarly, a sipA432–685 fragment was PCR amplified with a specific forward primer with a SapI restriction site (forward, 5/-GTGTGTGCTCTTCTGATGAAGTCGATGGCGTAACCAGCAAG-3/) and a reverse primer containing a 3×HA sequence with a SapI restriction site (reverse, 5/-GTGTGTGCTCTTCTTTAAGCGTAATCTGGAACGTCGTATGGATACGATCCTGCATAGTCCGGGACGTCATAGGGATAGCCCGCATAGTCAGGAACATCGTATGGGTAACGCTGCATGTGCAAGCCATCAACGG-3/). Fragments were digested with SapI and ligated to generate sipA431FLAG-685HA construct. Similarly, sipA upstream fragment (SipA U) was amplified with XbaI forward (forward, 5/-GTTCTAGACCAGCAGCCTGAATGCGCTGG-3/) and SapI reverse primers (reverse, 5/-GTGTGTGCTCTTCTCATTATTAATATCCTCTTCTGTTATCCTTGCAGGAAG-3/). Similarly, sipA downstream fragments (SipA D) were amplified with SapI forward (forward, 5/-GTGTGTGCTCTTCTTAATTAACCGGGAAAGATGCGATGAATATGG-3/) and SacI reverse primers (reverse, 5/-GTGAGCTCATAATCTGCCGCCAGATAGAATCGCC-3/). All underlined sequences denote restriction sites. Finally, SipA U, sipA431FLAG-685HA, and SipA D were digested in SapI and ligated to generate a sipA431FLAG-685HA construct containing upstream and downstream sequences of sipA. This construct was introduced into a sipA chromosomal locus of the Salmonella:sipAKO through Pre112 suicide vector to generate the Salmonella:sipA431FLAG-685HA strain.
Determination of Salmonella trafficking to the lysosome
To compare the trafficking of WT and sipAKO Salmonella toward lysosomes, HeLa cells were incubated with 250 µg/ml Dextran–Texas Red (70,000 MW) for 4 h in complete medium, washed, and chased for 20 h to label the lysosomes. Subsequently, cells were infected with either GFP:WT or GFP:sipAKO Salmonella and chased for indicated periods of time. Cells were washed, fixed, and analyzed by confocal microscopy.
Preparation of biotinylated SCVs
To determine the fusion between WT and sipAKO Salmonella with EE, Salmonella was biotinylated using N-hydroxysuccinimide–biotin as described previously (Madan et al., 2008). HeLa cells were incubated with biotinylated Salmonella for 30 min (MOI 1:50) at 37°C, washed, and chased for an additional 60 min. Subsequently, SCVs were purified using a method described previously (Lührmann and Haas, 2000). Briefly, infected cells were resuspended in homogenization buffer (HB; 250 mM sucrose, 0.5 mM EGTA, and 20 mM Hepes-KOH, pH 7.2) and homogenized, and then postnuclear supernatant (PNS) was prepared. To purify the SCV, PNS was diluted to 39% (wt/vol) sucrose and loaded on top of 65% sucrose and 55% sucrose solutions. On top of the PNS, 32.5% and 10% sucrose solutions were loaded and centrifuged (100,000 g) at 4°C for 1 h. SCVs were collected from 55–65% sucrose space and further diluted to a final sucrose concentration of 11% with HB and without sucrose. Finally, an SCV fraction was placed on a 15% Ficoll cushion and centrifuged (18,000 g) at 4°C for 20 min. Purified SCV was resuspended in HB.
Preparation of EEs
EEs were prepared as described previously (Mukherjee et al., 2000). Briefly, HeLa cells were incubated with 1 mg/ml avidin-HRP for 1 h at 4°C, washed, and chased for 5 min at 37°C to label the EE compartment. Cells were washed with HB, and PNS was prepared. Subsequently, PNS was diluted with HB (1:3) and centrifuged at 60,000 g for 1 min at 4°C. Finally, the supernatant was again centrifuged at 100,000 g for 5 min at 4°C to prepare enriched EEs.
In vitro fusion between biotinylated SCVs and avidin-HRP–labeled EEs
To determine the fusion between biotinylated SCVs and EEs, 5 µg of each respective SCV-containing biotinylated Salmonella was mixed with 5 µg of avidin-HRP–loaded EEs in fusion buffer (250 mM sucrose, 0.5 mM EGTA, 20 mM Hepes-KOH, pH 7.2, 1 mM DTT, 1.5 mM MgCl2, and 100 mM KCl) containing an ATP-regenerating system (1 mM ATP, 8 mM creatine phosphate, 31 U/ml creatine phosphokinase, and 0.25 mg/ml avidin D as scavenger) supplemented with 25 µg gel-filtered cytosol with a total reaction volume of 50 µl and incubated for 60 min at 37°C as described previously (Mukherjee et al., 2000). The fusion reaction was stopped by chilling on ice. The avidin-HRP–biotin bacterial complex was recovered by centrifugation (10,000 g for 5 min) after solubilization of the membrane in solubilization buffer (PBS containing 0.5% Triton X-100 with 0.25 mg/ml avidin D as a scavenger). The enzymatic activity of avidin-HRP associated with the biotinylated bacteria was measured as a fusion unit. Specific fusion value was determined by subtracting the values corresponding with HRP activity obtained when the endosomes and SCVs were mixed in fusion buffer without cytosol. Results are expressed as relative fusion in comparison with control.
KO of Syn8 by CRISPR/Cas9
To knock out Syn8 in HeLa cells by CRISPR/Cas9, the Syn8 genomic DNA sequence was analyzed using the Broad Institute’s single-guide (sgRNA) designer tool for designing appropriate sgRNA. Two different sgRNAs were used to obtain Syn8-KO cells: sgSYN81, 5′-CCCTGGTGAGTCCCGGGTGA-3′; and sgSYN82, 5′-TGCAGACTCCGCCCGCCGCT-3′. Subsequently, sgRNAs were cloned into the pSpCas9(BB)-2A-GFP plasmid (PX458), and HeLa cells were transfected with both Cas9/sgRNA expression vectors as described previously (Ran et al., 2013). GFP-positive cells were sorted after 24 h transfection using the BD FACSAria III sorter. Subsequently, cells were appropriately diluted to seed single cells per well in a 96-well plate and allowed to grow in normal culture conditions. Finally, individual colonies were screened by Western blot and immunofluorescence analyses using specific antibodies.
Determination of the role of Syn8 in the survival of Salmonella in HeLa cells
To understand the role of Syn8 in the survival of Salmonella in HeLa cells, Syn8 siRNA (5′-CCTCTTGGATGATCTTGTA-3′) was transfected into 30–40% confluent HeLa cells using Lipofectamine RNAiMAX. Knockdown of Syn8 in cells was checked by real-time PCR (qPCR) using Syn8 gene-specific forward and reverse primers. The relative Syn8 gene expression was normalized using 18s rRNA as an internal control. Subsequently, cells were washed and infected with Salmonella:WT at 50 MOI at 37°C for 30 min. Cells were washed and incubated with complete medium containing 100 µg/ml gentamicin for 30 min at 37°C to kill extracellular bacteria. Cells were then incubated with complete medium containing 10 µg/ml gentamicin for the indicated periods of time at 37°C. At respective times, cells were washed and lysed in 1% Triton X-100 containing 0.1% SDS for 5 min (Yu et al., 2014). Serial dilutions of the lysate were plated on Luria–Bertani agar plates containing streptomycin (100 µg/ml) at 37°C for 12 h, and numbers of viable bacteria were measured by colony-forming units. Similar experiments were performed in CRISPR-mediated KO of Syn8 in HeLa cells.
Statistical analysis
Statistical analysis was performed using SigmaPlot version 12. Statistical differences were determined by one-way ANOVA or Kruskal-Wallis test followed by post hoc pairwise multiple comparison analysis using Bonferroni’s t test, Dunn's Method, Tukey’s test, or paired t test with 95% confidence intervals. P values of <0.05 were considered significant for all analyses.
Online supplemental material
Fig. S1 shows the recruitment of VAMP7 and VAMP8 on SCVs. Fig. S2 shows the identification of Syn8 binding protein as SipA by mass spectrometry. Fig. S3 shows the generation and characterization of sipAKO Salmonella. Fig. S4 shows the localization of SipA on SCVs containing sipAKO:SipA1–435 Salmonella and generation of SipA431FLAG-685HA construct. Fig. S5 shows the presence of different proteins on the SCV and endosomes.
Supplementary Material
Acknowledgments
We thank Dr. Ayub Qadri and Dr. Rajesh S. Gokhale of National Institute of Immunology for critically reviewing the manuscript. We also acknowledge the help of Dr. Shanta Sen of the mass spectrometer facility at the National Institute of Immunology (New Delhi, India).
This work is supported by grants from Department of Biotechnology [BT/PR13081/BRB/10/1379/2015] and a Government of India Science and Engineering Research Board J.C. Bose Fellowship (SR/S2/JCB-24/2009) to A. Mukhopadhyay. P.K. Singh and R.M. Lomash are supported by fellowships from the Council of Scientific and Industrial Research, Government of India (09/485(0206)/2010-EMR-I to P.K. Singh and 09/485(0136)/2005-EMR-I to R.M. Lomash). A. Kapoor and K. Kumar are supported by Department of Biotechnology, Government of India, fellowships from the project to A. Mukhopadhyay (BT/PR13081/BRB/10/1379/2015).
The authors declare no competing financial interests.
Author contributions: A. Mukhopadhyay conceived and coordinated the study and wrote the paper. P.K. Singh, A. Kapoor, R.M. Lomash, and K. Kumar performed experiments and analyzed results. S.C. Kamerkar and T.J. Pucadyil did liposome experiments. All authors reviewed the results and approved the final version of the manuscript.
References
- Agbor T.A., and McCormick B.A.. 2011. Salmonella effectors: important players modulating host cell function during infection. Cell. Microbiol. 13:1858–1869. 10.1111/j.1462-5822.2011.01701.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alix E., Mukherjee S., and Roy C.R.. 2011. Subversion of membrane transport pathways by vacuolar pathogens. J. Cell Biol. 195:943–952. 10.1083/jcb.201105019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W., Holroyd C., Fasshauer D., Pabst S., Von Mollard G.F., and Jahn R.. 2000a A SNARE complex mediating fusion of late endosomes defines conserved properties of SNARE structure and function. EMBO J. 19:6453–6464. 10.1093/emboj/19.23.6453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonin W., Holroyd C., Tikkanen R., Höning S., and Jahn R.. 2000b The R-SNARE endobrevin/VAMP-8 mediates homotypic fusion of early endosomes and late endosomes. Mol. Biol. Cell. 11:3289–3298. 10.1091/mbc.11.10.3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasaki K., Toomre D.K., and Roy C.R.. 2012. The Legionella pneumophila effector DrrA is sufficient to stimulate SNARE-dependent membrane fusion. Cell Host Microbe. 11:46–57. 10.1016/j.chom.2011.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asrat S., de Jesús D.A., Hempstead A.D., Ramabhadran V., and Isberg R.R.. 2014. Bacterial pathogen manipulation of host membrane trafficking. Annu. Rev. Cell Dev. Biol. 30:79–109. 10.1146/annurev-cellbio-100913-013439 [DOI] [PubMed] [Google Scholar]
- Brandhorst D., Zwilling D., Rizzoli S.O., Lippert U., Lang T., and Jahn R.. 2006. Homotypic fusion of early endosomes: SNAREs do not determine fusion specificity. Proc. Natl. Acad. Sci. USA. 103:2701–2706. 10.1073/pnas.0511138103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawn L.C., Hayward R.D., and Koronakis V.. 2007. Salmonella SPI1 effector SipA persists after entry and cooperates with a SPI2 effector to regulate phagosome maturation and intracellular replication. Cell Host Microbe. 1:63–75. 10.1016/j.chom.2007.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronstein P.A., Miao E.A., and Miller S.I.. 2000. InvB is a type III secretion chaperone specific for SspA. J. Bacteriol. 182:6638–6644. 10.1128/JB.182.23.6638-6644.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumell J.H., and Grinstein S.. 2004. Salmonella redirects phagosomal maturation. Curr. Opin. Microbiol. 7:78–84. 10.1016/j.mib.2003.12.005 [DOI] [PubMed] [Google Scholar]
- Brumell J.H., and Scidmore M.A.. 2007. Manipulation of rab GTPase function by intracellular bacterial pathogens. Microbiol. Mol. Biol. Rev. 71:636–652. 10.1128/MMBR.00023-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Costa V.M., Braun V., Landekic M., Shi R., Proteau A., McDonald L., Cygler M., Grinstein S., and Brumell J.H.. 2015. Salmonella Disrupts Host Endocytic Trafficking by SopD2-Mediated Inhibition of Rab7. Cell Reports. 12:1508–1518. 10.1016/j.celrep.2015.07.063 [DOI] [PubMed] [Google Scholar]
- Delevoye C., Nilges M., Dehoux P., Paumet F., Perrinet S., Dautry-Varsat A., and Subtil A.. 2008. SNARE protein mimicry by an intracellular bacterium. PLoS Pathog. 4:e1000022 10.1371/journal.ppat.1000022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards R.A., Keller L.H., and Schifferli D.M.. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene. 207:149–157. 10.1016/S0378-1119(97)00619-7 [DOI] [PubMed] [Google Scholar]
- Ellena J.F., Liang B., Wiktor M., Stein A., Cafiso D.S., Jahn R., and Tamm L.K.. 2009. Dynamic structure of lipid-bound synaptobrevin suggests a nucleation-propagation mechanism for trans-SNARE complex formation. Proc. Natl. Acad. Sci. USA. 106:20306–20311. 10.1073/pnas.0908317106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D., and Margittai M.. 2004. A transient N-terminal interaction of SNAP-25 and syntaxin nucleates SNARE assembly. J. Biol. Chem. 279:7613–7621. 10.1074/jbc.M312064200 [DOI] [PubMed] [Google Scholar]
- Fasshauer D., Sutton R.B., Brunger A.T., and Jahn R.. 1998. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA. 95:15781–15786. 10.1073/pnas.95.26.15781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan J.J., Mukherjee I., and Barlowe C.. 2015. Examination of Sec22 Homodimer Formation and Role in SNARE-dependent Membrane Fusion. J. Biol. Chem. 290:10657–10666. 10.1074/jbc.M114.626911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster L.J., Yeung B., Mohtashami M., Ross K., Trimble W.S., and Klip A.. 1998. Binary interactions of the SNARE proteins syntaxin-4, SNAP23, and VAMP-2 and their regulation by phosphorylation. Biochemistry. 37:11089–11096. 10.1021/bi980253t [DOI] [PubMed] [Google Scholar]
- Galán J.E., and Collmer A.. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science. 284:1322–1328. 10.1126/science.284.5418.1322 [DOI] [PubMed] [Google Scholar]
- Garcia-del Portillo F., and Finlay B.B.. 1995. Targeting of Salmonella typhimurium to vesicles containing lysosomal membrane glycoproteins bypasses compartments with mannose 6-phosphate receptors. J. Cell Biol. 129:81–97. 10.1083/jcb.129.1.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomodonato M.N., Uzzau S., Bacciu D., Caccuri R., Sarnacki S.H., Rubino S., and Cerquetti M.C.. 2007. SipA, SopA, SopB, SopD and SopE2 effector proteins of Salmonella enterica serovar Typhimurium are synthesized at late stages of infection in mice. Microbiology. 153:1221–1228. 10.1099/mic.0.2006/002758-0 [DOI] [PubMed] [Google Scholar]
- Guzman L.M., Belin D., Carson M.J., and Beckwith J.. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121–4130. 10.1128/jb.177.14.4121-4130.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S., Ehrbar K., Stecher B., Barthel M., Kremer M., and Hardt W.D.. 2004. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72:795–809. 10.1128/IAI.72.2.795-809.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraga A., Ohlson M.B., and Miller S.I.. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53–66. 10.1038/nrmicro1788 [DOI] [PubMed] [Google Scholar]
- Hardt W.D., Chen L.M., Schuebel K.E., Bustelo X.R., and Galán J.E.. 1998. S. typhimurium encodes an activator of Rho GTPases that induces membrane ruffling and nuclear responses in host cells. Cell. 93:815–826. 10.1016/S0092-8674(00)81442-7 [DOI] [PubMed] [Google Scholar]
- Hashim S., Mukherjee K., Raje M., Basu S.K., and Mukhopadhyay A.. 2000. Live Salmonella modulate expression of Rab proteins to persist in a specialized compartment and escape transport to lysosomes. J. Biol. Chem. 275:16281–16288. 10.1074/jbc.275.21.16281 [DOI] [PubMed] [Google Scholar]
- Hernandez L.D., Hueffer K., Wenk M.R., and Galán J.E.. 2004. Salmonella modulates vesicular traffic by altering phosphoinositide metabolism. Science. 304:1805–1807. 10.1126/science.1098188 [DOI] [PubMed] [Google Scholar]
- Hong W. 2005. SNAREs and traffic. Biochim. Biophys. Acta. 1744:120–144. 10.1016/j.bbamcr.2005.03.014 [DOI] [PubMed] [Google Scholar]
- Hueck C.J. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R., and Scheller R.H.. 2006. SNAREs--engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7:631–643. 10.1038/nrm2002 [DOI] [PubMed] [Google Scholar]
- Jennings E., Thurston T.L.M., and Holden D.W.. 2017. Salmonella SPI-2 Type III Secretion System Effectors: Molecular Mechanisms And Physiological Consequences. Cell Host Microbe. 22:217–231. 10.1016/j.chom.2017.07.009 [DOI] [PubMed] [Google Scholar]
- Jepson M.A., Kenny B., and Leard A.D.. 2001. Role of sipA in the early stages of Salmonella typhimurium entry into epithelial cells. Cell. Microbiol. 3:417–426. 10.1046/j.1462-5822.2001.00124.x [DOI] [PubMed] [Google Scholar]
- LaRock D.L., Chaudhary A., and Miller S.I.. 2015. Salmonellae interactions with host processes. Nat. Rev. Microbiol. 13:191–205. 10.1038/nrmicro3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.A., Silva M., Siber A.M., Kelly A.J., Galyov E., and McCormick B.A.. 2000. A secreted Salmonella protein induces a proinflammatory response in epithelial cells, which promotes neutrophil migration. Proc. Natl. Acad. Sci. USA. 97:12283–12288. 10.1073/pnas.97.22.12283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilic M., Vujanac M., and Stebbins C.E.. 2006. A common structural motif in the binding of virulence factors to bacterial secretion chaperones. Mol. Cell. 21:653–664. 10.1016/j.molcel.2006.01.026 [DOI] [PubMed] [Google Scholar]
- Lührmann A., and Haas A.. 2000. A method to purify bacteria-containing phagosomes from infected macrophages. Methods Cell Sci. 22:329–341. 10.1023/A:1017963401560 [DOI] [PubMed] [Google Scholar]
- Madan R., Krishnamurthy G., and Mukhopadhyay A.. 2008. SopE-mediated recruitment of host Rab5 on phagosomes inhibits Salmonella transport to lysosomes. Methods Mol. Biol. 445:417–437. 10.1007/978-1-59745-157-4_27 [DOI] [PubMed] [Google Scholar]
- Madan R., Rastogi R., Parashuraman S., and Mukhopadhyay A.. 2012. Salmonella acquires lysosome-associated membrane protein 1 (LAMP1) on phagosomes from Golgi via SipC protein-mediated recruitment of host Syntaxin6. J. Biol. Chem. 287:5574–5587. 10.1074/jbc.M111.286120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallo G.V., Espina M., Smith A.C., Terebiznik M.R., Alemán A., Finlay B.B., Rameh L.E., Grinstein S., and Brumell J.H.. 2008. SopB promotes phosphatidylinositol 3-phosphate formation on Salmonella vacuoles by recruiting Rab5 and Vps34. J. Cell Biol. 182:741–752. 10.1083/jcb.200804131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhie E.J., Brawn L.C., Hume P.J., Humphreys D., and Koronakis V.. 2009. Salmonella takes control: effector-driven manipulation of the host. Curr. Opin. Microbiol. 12:117–124. 10.1016/j.mib.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méresse S., Steele-Mortimer O., Finlay B.B., and Gorvel J.P.. 1999. The rab7 GTPase controls the maturation of Salmonella typhimurium-containing vacuoles in HeLa cells. EMBO J. 18:4394–4403. 10.1093/emboj/18.16.4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee K., Siddiqi S.A., Hashim S., Raje M., Basu S.K., and Mukhopadhyay A.. 2000. Live Salmonella recruits N-ethylmaleimide-sensitive fusion protein on phagosomal membrane and promotes fusion with early endosome. J. Cell Biol. 148:741–753. 10.1083/jcb.148.4.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos J.S., and Agarwala R.. 2007. COBALT: constraint-based alignment tool for multiple protein sequences. Bioinformatics. 23:1073–1079. 10.1093/bioinformatics/btm076 [DOI] [PubMed] [Google Scholar]
- Patel J.C., Hueffer K., Lam T.T., and Galán J.E.. 2009. Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell. 137:283–294. 10.1016/j.cell.2009.01.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S.R. 2013. Rab GTPase regulation of membrane identity. Curr. Opin. Cell Biol. 25:414–419. 10.1016/j.ceb.2013.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pobbati A.V., Stein A., and Fasshauer D.. 2006. N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science. 313:673–676. 10.1126/science.1129486 [DOI] [PubMed] [Google Scholar]
- Prekeris R., Yang B., Oorschot V., Klumperman J., and Scheller R.H.. 1999. Differential roles of syntaxin 7 and syntaxin 8 in endosomal trafficking. Mol. Biol. Cell. 10:3891–3908. 10.1091/mbc.10.11.3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryor P.R., Mullock B.M., Bright N.A., Lindsay M.R., Gray S.R., Richardson S.C., Stewart A., James D.E., Piper R.C., and Luzio J.P.. 2004. Combinatorial SNARE complexes with VAMP7 or VAMP8 define different late endocytic fusion events. EMBO Rep. 5:590–595. 10.1038/sj.embor.7400150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido I.R., Jahn R., and Gerke V.. 2011. VAMP3 is associated with endothelial weibel-palade bodies and participates in their Ca(2+)-dependent exocytosis. Biochim. Biophys. Acta. 1813:1038–1044. 10.1016/j.bbamcr.2010.11.007 [DOI] [PubMed] [Google Scholar]
- Raffatellu M., Wilson R.P., Chessa D., Andrews-Polymenis H., Tran Q.T., Lawhon S., Khare S., Adams L.G., and Bäumler A.J.. 2005. SipA, SopA, SopB, SopD, and SopE2 contribute to Salmonella enterica serotype typhimurium invasion of epithelial cells. Infect. Immun. 73:146–154. 10.1128/IAI.73.1.146-154.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rastogi R., Verma J.K., Kapoor A., Langsley G., and Mukhopadhyay A.. 2016. Rab5 Isoforms Specifically Regulate Different Modes of Endocytosis in Leishmania. J. Biol. Chem. 291:14732–14746. 10.1074/jbc.M116.716514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlumberger M.C., Käppeli R., Wetter M., Müller A.J., Misselwitz B., Dilling S., Kremer M., and Hardt W.D.. 2007. Two newly identified SipA domains (F1, F2) steer effector protein localization and contribute to Salmonella host cell manipulation. Mol. Microbiol. 65:741–760. 10.1111/j.1365-2958.2007.05823.x [DOI] [PubMed] [Google Scholar]
- Shi X., Halder P., Yavuz H., Jahn R., and Shuman H.A.. 2016. Direct targeting of membrane fusion by SNARE mimicry: Convergent evolution of Legionella effectors. Proc. Natl. Acad. Sci. USA. 113:8807–8812. 10.1073/pnas.1608755113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva M., Song C., Nadeau W.J., Matthews J.B., and McCormick B.A.. 2004. Salmonella typhimurium SipA-induced neutrophil transepithelial migration: involvement of a PKC-alpha-dependent signal transduction pathway. Am. J. Physiol. Gastrointest. Liver Physiol. 286:G1024–G1031. 10.1152/ajpgi.00299.2003 [DOI] [PubMed] [Google Scholar]
- Srikanth C.V., Wall D.M., Maldonado-Contreras A., Shi H., Zhou D., Demma Z., Mumy K.L., and McCormick B.A.. 2010. Salmonella pathogenesis and processing of secreted effectors by caspase-3. Science. 330:390–393. 10.1126/science.1194598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steegmaier M., Yang B., Yoo J.S., Huang B., Shen M., Yu S., Luo Y., and Scheller R.H.. 1998. Three novel proteins of the syntaxin/SNAP-25 family. J. Biol. Chem. 273:34171–34179. 10.1074/jbc.273.51.34171 [DOI] [PubMed] [Google Scholar]
- Steele-Mortimer O. 2008. The Salmonella-containing vacuole: moving with the times. Curr. Opin. Microbiol. 11:38–45. 10.1016/j.mib.2008.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele-Mortimer O., Méresse S., Gorvel J.P., Toh B.H., and Finlay B.B.. 1999. Biogenesis of Salmonella typhimurium-containing vacuoles in epithelial cells involves interactions with the early endocytic pathway. Cell. Microbiol. 1:33–49. 10.1046/j.1462-5822.1999.00003.x [DOI] [PubMed] [Google Scholar]
- Stender S., Friebel A., Linder S., Rohde M., Mirold S., and Hardt W.D.. 2000. Identification of SopE2 from Salmonella typhimurium, a conserved guanine nucleotide exchange factor for Cdc42 of the host cell. Mol. Microbiol. 36:1206–1221. 10.1046/j.1365-2958.2000.01933.x [DOI] [PubMed] [Google Scholar]
- Stenmark H. 2009. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10:513–525. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- Subramaniam V.N., Loh E., Horstmann H., Habermann A., Xu Y., Coe J., Griffiths G., and Hong W.. 2000. Preferential association of syntaxin 8 with the early endosome. J. Cell Sci. 113:997–1008. [DOI] [PubMed] [Google Scholar]
- Wall D.M., Nadeau W.J., Pazos M.A., Shi H.N., Galyov E.E., and McCormick B.A.. 2007. Identification of the Salmonella enterica serotype typhimurium SipA domain responsible for inducing neutrophil recruitment across the intestinal epithelium. Cell. Microbiol. 9:2299–2313. 10.1111/j.1462-5822.2007.00960.x [DOI] [PubMed] [Google Scholar]
- Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., and Barton G.J.. 2009. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 25:1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman S.R., and Holden D.W.. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell. Microbiol. 5:501–511. 10.1046/j.1462-5822.2003.00294.x [DOI] [PubMed] [Google Scholar]
- Whiteheart S.W., Rossnagel K., Buhrow S.A., Brunner M., Jaenicke R., and Rothman J.E.. 1994. N-ethylmaleimide-sensitive fusion protein: a trimeric ATPase whose hydrolysis of ATP is required for membrane fusion. J. Cell Biol. 126:945–954. 10.1083/jcb.126.4.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W., and Schekman R.. 2008. Membrane fusion. Nat. Struct. Mol. Biol. 15:658–664. 10.1038/nsmb.1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H.B., Croxen M.A., Marchiando A.M., Ferreira R.B., Cadwell K., Foster L.J., and Finlay B.B.. 2014. Autophagy facilitates Salmonella replication in HeLa cells. MBio. 5:e00865 10.1128/mBio.00865-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharik M.L., Gruenheid S., Perrin A.J., and Finlay B.B.. 2002. Delivery of dangerous goods: type III secretion in enteric pathogens. Int. J. Med. Microbiol. 291:593–603. 10.1078/1438-4221-00179 [DOI] [PubMed] [Google Scholar]
- Zhang S., Adams L.G., Nunes J., Khare S., Tsolis R.M., and Bäumler A.J.. 2003. Secreted effector proteins of Salmonella enterica serotype typhimurium elicit host-specific chemokine profiles in animal models of typhoid fever and enterocolitis. Infect. Immun. 71:4795–4803. 10.1128/IAI.71.8.4795-4803.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Mooseker M.S., and Galán J.E.. 1999a An invasion-associated Salmonella protein modulates the actin-bundling activity of plastin. Proc. Natl. Acad. Sci. USA. 96:10176–10181. 10.1073/pnas.96.18.10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Mooseker M.S., and Galán J.E.. 1999b Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science. 283:2092–2095. 10.1126/science.283.5410.2092 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.