Collins et al. use yeast genetics, biochemistry, and structure probing to dissect the role of the assembly factor Ltv1 in 40S ribosome maturation. Ribosomes from Ltv1-deficient cells have substoichiometric amounts of Rps10 and Asc1 and misfolded head rRNA, leading to defects in translational fidelity and ribosome-mediated RNA quality control, demonstrating a role for Ltv1 in chaperoning the assembly of the subunit head.
Abstract
The correct assembly of ribosomes from ribosomal RNAs (rRNAs) and ribosomal proteins (RPs) is critical, as indicated by the diseases caused by RP haploinsufficiency and loss of RP stoichiometry in cancer cells. Nevertheless, how assembly of each RP is ensured remains poorly understood. We use yeast genetics, biochemistry, and structure probing to show that the assembly factor Ltv1 facilitates the incorporation of Rps3, Rps10, and Asc1/RACK1 into the small ribosomal subunit head. Ribosomes from Ltv1-deficient yeast have substoichiometric amounts of Rps10 and Asc1 and show defects in translational fidelity and ribosome-mediated RNA quality control. These defects provide a growth advantage under some conditions but sensitize the cells to oxidative stress. Intriguingly, relative to glioma cell lines, breast cancer cells have reduced levels of LTV1 and produce ribosomes lacking RPS3, RPS10, and RACK1. These data describe a mechanism to ensure RP assembly and demonstrate how cancer cells circumvent this mechanism to generate diverse ribosome populations that can promote survival under stress.
Graphical Abstract
Introduction
Ribosomes are conserved RNA-protein machines responsible for protein synthesis in all cells. Composed of 79 ribosomal proteins (RPs) and four ribosomal RNAs (rRNAs) in eukaryotes, there are nearly infinite possibilities for incorrect ribosome assembly, but only one fully assembled ribosome. This constitutes an important challenge for cells as defects in ribosome assembly are the molecular cause for a number of human diseases (Ellis and Lipton, 2008; Burwick et al., 2011). This challenge is exacerbated by parallel assembly pathways (Davis et al., 2016; Sanghai et al., 2018), leading to the formation of diverse intermediates, which renders quality control difficult.
Ribosomes lacking Rps26 disrupt protein homeostasis because they recognize a distinct set of mRNAs (Ferretti et al., 2017), illustrating the importance of ensuring the correct incorporation of all RPs into ribosomes. Similarly, Asc1-deficient ribosomes have defects in the translation of small mRNAs (Thompson et al., 2016). Furthermore, Rps26-deficient ribosomes play physiological roles during high-salt and high-pH stress, where they promote the preferential translation of mRNAs involved in the response to these stresses (Ferretti et al., 2017). Thus, these seemingly misassembled ribosomes also play important cellular roles. Finally, tumor development also leads to changes in the stoichiometry of RP expression (Vlachos et al., 2012; Guimaraes and Zavolan, 2016; Ajore et al., 2017; Kulkarni et al., 2017). How cells ensure the correct assembly of all RPs and how this becomes a regulated process under certain physiological conditions to produce ribosomes lacking individual proteins remain unknown.
Ribosome assembly requires a large machinery comprising ∼200 assembly factors (AFs), which transiently associate with nascent ribosomes to facilitate and regulate their assembly (Kressler et al., 2017; Peña et al., 2017). Ltv1 is one of these AFs, promoting the assembly of the small (40S) ribosomal subunit. Deletion of Ltv1 leads to a cold-sensitive growth phenotype and affects export of the nascent subunits to the cytoplasm (Loar et al., 2004; Seiser et al., 2006). Ltv1 binds directly to the AF Enp1, the RPs Rps3, Rps15, and Rps20 (these proteins are also referred to as uS3, uS19, and uS10), and is located on the solvent-side of the nascent beak structure (Schäfer et al., 2006; Campbell and Karbstein, 2011; Strunk et al., 2011; Ghalei et al., 2015; Mitterer et al., 2016; Heuer et al., 2017; Johnson et al., 2017; Scaiola et al., 2018). Ltv1 release from precursor 40S (pre-40S) subunits requires phosphorylation by the casein kinase Hrr25/CK1δ (Schäfer et al., 2006; Ghalei et al., 2015). Despite the plethora of studies, the exact role of Ltv1 in 40S maturation remains unknown.
In this study, we use a combination of genetic analyses, ribosome purifications, translational fidelity assays, and RNA structure probing in yeast as well as drug sensitivity and growth assays, ribosome purifications, and translational fidelity assays in breast cancer and glioma cells to dissect the role of Ltv1 in 40S ribosome maturation. The data show that ribosomes from ΔLtv1 yeast have substoichiometric amounts of Rps10 and Asc1 and misfolded head rRNA, demonstrating a role for Ltv1 in chaperoning the correct assembly of the small subunit head. These deficiencies result in defects in translational fidelity and ribosome-mediated RNA quality control. Intriguingly, some human cancer cell lines have down-regulated LTV1, leading to the production of ribosomes deficient in RPS3, RPS10, and RACK1, which are expected to perturb protein homeostasis in ways that might support tumor development.
Results
Ribosomes from ΔLtv1 yeast have mispositioned Rps3
Ltv1 binds the solvent side of the beak structure, where it interacts with Rps3, Rps15, and Rps20 (Schäfer et al., 2006; Campbell and Karbstein, 2011; Strunk et al., 2011; Ghalei et al., 2015; Mitterer et al., 2016; Heuer et al., 2017; Johnson et al., 2017; Scaiola et al., 2018). Although Ltv1 is not essential in yeast, its deletion leads to cold-sensitive slow-growth (Loar et al., 2004), an effect linked to ribosome misassembly (Guthrie et al., 1969). Thus, to assess the structural consequences of Ltv1 deletion, we used RNA structure probing. After incubation with dimethylsulfate (DMS), which modifies A and C residues that are not protected by base pairing or protein binding, RNA was extracted, and modifications were detected using reverse transcription (Fig. 1 A). We probed for changes in residues 1,170–1,380 of the small (40S) subunit head. Only a small subset of these nucleotides are differentially modified in the ribosomes from Ltv1-containing and deficient cells as shown in Fig. 1. These center around Rps20 and the C-terminal portion of Rps3 and extend to the Rps17 (eS17) and Asc1 binding sites (Fig. 1, A–C).
Figure 1.
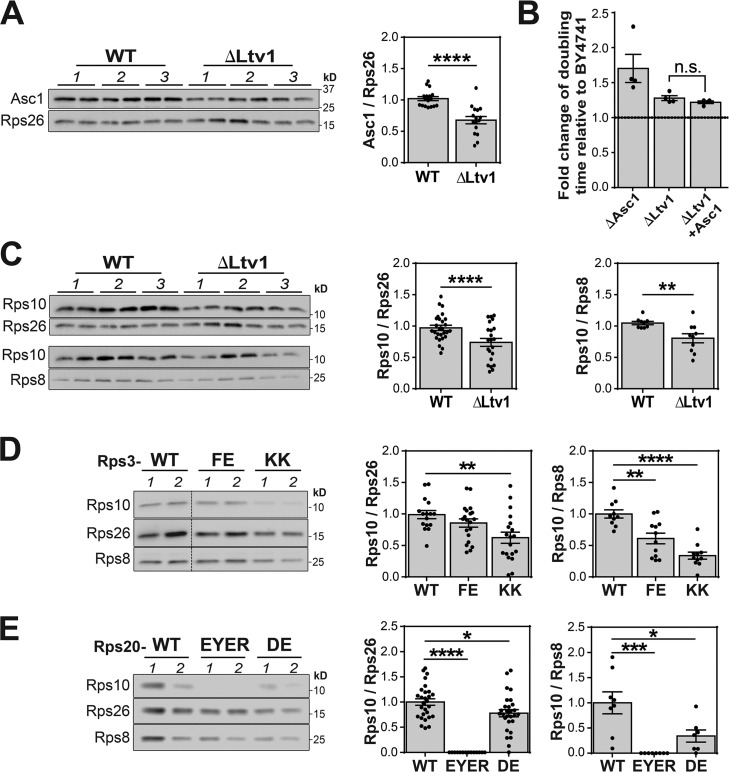
Structure probing and genetic data indicate repositioning of Rps3. (A) Primer extension pattern of ribosomes from n = 3 biological replicates of WT and ΔLtv1 cells in the presence (+) or absence (–) of DMS. Differentially accessible residues as quantified below are highlighted in red (exposed) or blue (protected). Gray bars show ΔLtv1, and black bars show WT. (B and C) Quantification was done using Image Lab 6.0 and normalized to band 1, which causes a stop in all lanes. Differentially accessible residues are shown in the secondary (B) and tertiary (C) structure of the head rRNA. Rps3 (red), Rps10 (blue), Rps20 (green), Rps29 (cyan), Asc1 (yellow), and Rps17 (brown) binding sites are indicated in the colored boxes (B) or in the structure of mature 40S ribosome (C; PDB ID: 4V88). Enp1 and a peptide of Ltv1 are shown in purple. Their position was derived from a superposition of 40S in 4V88 and pre-40S in 6FAI. (D) Genetic interactions between Ltv1 deletion and mutations at the interface between Rps3 and Rps17. (E) Genetic interactions between Ltv1 deletion and mutations in the Rps3 protein network. Fold-changes were determined by comparison of the effects from Rps3, Rps20, or Rps17 mutations in the presence or absence of Ltv1. Data in D and E are three to five technical repeats of 3–12 biological replicates. Error bars represent the SEM, and significance was determined using an unpaired t test. ****, P < 0.0001.
Because both Rps17 and Asc1 interact with the C-terminal tail of Rps3 (Fig. 1 C), we hypothesized that the Rps3 tail was mispositioned in the ribosomes from ΔLtv1 cells. To test this model, we probed for synthetic genetic interactions between Rps17 and Ltv1. If the Rps3 tail was mispositioned, mutations that affect the binding of Rps17 should have smaller effects in the absence of Ltv1 than in its presence as the tail already is mispositioned, thus perturbing the relevant interactions. We therefore produced two mutants in the Rps3/Rps17 interface, Rps3_I208A-E210R-P211A (Rps3_IEP) and the equivalent Rps17_L16A-R19E-Y20A-T39A-Q42A (Rps17_LRYTQ; Fig. S1 A). These mutations give small (<1.2-fold) but reproducible effects on growth in the presence of Ltv1 (Fig. S1 B). In contrast, these mutations have no defect in the absence of Ltv1, producing the epistasis effect expected from mispositioning of the Rps3-tail (Fig. 1 D).
Asc1 recruitment requires interactions with the Rps3 tail
Like Rps17, Asc1 binds the Rps3 tail (Fig. 1 C). Thus, the structural changes observed in the upper part of the head could be due to defective Asc1 recruitment. To first test whether the Rps3-tail affected recruitment of Asc1, we produced a yeast strain encoding Rps3 truncated after residue 214 (Rps3_214), which removes the interface with Asc1 (Fig. 1 C). Asc1 is required for growth on nonfermentable carbon sources such as glycerol (Rachfall et al., 2013; Thompson et al., 2016). As expected, if the Rps3-tail were required for Asc1 recruitment, tail truncation to abolish the interaction with Asc1 produces a severe growth defect on glycerol-containing plates but has no effect on YPD (Fig. 2 A).
Figure 2.
Recruitment of Asc1 requires the Rps3 tail. (A) Asc1 and the C-terminal tail of Rps3 are required for growth on glycerol, a nonfermentable carbon source. V.O., vector only. (B) Asc1 occupancy in ribosomes from WT or C-terminally truncated Rps3 (Rps3214) cells was investigated by Coomassie-stained SDS-PAGE of purified 40S ribosomes. (C) In vitro binding of Asc1 to Asc1-deficient ribosomes containing full-length Rps3 or Rps3214. Shown is the Coomassie-stained SDS–PAGE gel of the pellet fractions. The empty lanes on the right show that Asc1 alone does not pellet.
To confirm that the Asc1-interacting residues in Rps3 are required for Asc1 recruitment to ribosomes, we purified ribosomes from yeast containing WT Rps3 or Rps3_214 and used SDS-PAGE to test for Asc1 binding. Indeed, ribosomes from Rps3_214 yeast lack both full-length Rps3 and Asc1 (Fig. 2 B). Furthermore, binding of recombinant Asc1 to Asc1-deficient ribosomes requires full-length Rps3 and is abrogated in ribosomes containing Rps3_214 (Fig. 2 C). Together, these data show that an interaction with the C-terminal tail of Rps3 is required for Asc1 binding to ribosomes.
Ribosomes from ΔLtv1 yeast have reduced levels of Asc1
Having established that the interaction between Rps3 and Asc1 is required for Asc1 recruitment to 40S subunits, we used Asc1 recruitment to confirm mispositioning of the Rps3 tail in ribosomes from ΔLtv1 yeast. If indeed the Rps3 tail was mispositioned in these ribosomes, as suggested by the DMS probing data and the synthetic rescue of mutations in the Rps3-Rps17 interface, then we would expect that ribosomes from ΔLtv1 cells are deficient in Asc1.
To test this prediction, we used Western blot analysis to measure Asc1 occupancy relative to Rps26. Indeed, ribosomes from ΔLtv1 yeast have reduced levels of Asc1 (Fig. 3 A). Furthermore, ΔLtv1 cells are deficient for growth on glycerol, akin to ΔAsc1 yeast, albeit with a reduced effect as expected, because Asc1 is reduced and not entirely absent (Fig. 3 B). Thus, Ltv1 is required for the efficient recruitment of Asc1 to 40S ribosomes. Overexpression of Asc1 in the ΔLtv1 background does not rescue Asc1 deficiency (Fig. 3 B), suggesting that it is not reduced availability but weakened affinity that plays a role. This is not unexpected as many RPs are produced in excess and then degraded (Tsay et al., 1988). Taken together, the DMS footprinting, the genetic interactions between Rps3 and Rps17, and the reduced binding of Asc1 all suggest that the Rps3 tail is mispositioned in the ribosomes from ΔLtv1 cells.
Figure 3.
Ltv1 is required for stoichiometric assembly of Rps10 and Asc1. (A) The levels of Asc1 relative to Rps26 in ribosomes purified from WT or ΔLtv1 cells were determined by Western blotting. Data are averages from two technical repeats of three biological replicates (1–3). (B) Similar to ΔAsc1 cells, ΔLtv1 cells grow slowly in glycerol. Overexpression of Asc1 does not rescue this effect (n = 4). (C–E) Changes in the RP composition of the head structure were determined by Western blotting of ribosomes purified from WT and ΔLtv1 cells (C) or cells containing mutations in the Rps3 protein network (D and E). Note that the same gel is used and shown to evaluate Asc1, Rps10, and Rps26 levels in A and C. A separate gel is run to evaluate Rps8 relative to Rps10. The data are averages from four to six technical repeats from two biological replicates. In all cases, significance was determined using an unpaired t test, and the error bars represent the SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Ribosomes from ΔLtv1 yeast have reduced levels of Rps10
Next, we tested whether the Rps3 tail was the only part of Rps3 mispositioned in ribosomes from ΔLtv1 cells or whether the two globular domains were also affected. The N-terminal domain of Rps3 interacts with Rps10, Rps20, and Rps29 (eS10, uS10, and uS14; Fig. 1 C), and Ltv1 binds Rps3 and Rps20 (Campbell and Karbstein, 2011; Ghalei et al., 2015; Mitterer et al., 2016). To decipher how Ltv1 depletion affects the interaction network between Rps3, Rps10, Rps20, and Rps29, we introduced mutations at the interfaces between these proteins and then tested whether their impact was changed in the presence or absence of Ltv1.
We mutated Rps3 residues located at the interface with Rps10 (Rps3_F24A-E28R; Rps3_FE) and Rps20 (Rps3_K7A-K10A, Rps3_KK; Mitterer et al., 2016), respectively. Additionally, mutations in Rps20 residues located at the interface with Rps3 (Rps20_D113K-E115K, Rps20_DE; Mitterer et al., 2016) and Rps29 (Rps20_E80K-Y82A-E83K-R85E, Rps20_EYER; Fig. S1 A), were introduced. We then used quantitative growth assays to measure their effects in yeast strains where the endogenous copy of Rps3 (or Rps20) was under a galactose-inducible/glucose-repressible promoter, and WT or mutant Rps3 (or Rps20) were provided on centromeric plasmids. These mutations all provided small but reproducible (1.2- to 3.5-fold) effects on yeast cellular growth (Fig. S1 D) as expected, because these mutations are located at the interfaces between essential RPs. Western blot analysis suggests that these mutations do not substantially affect their expression levels (Fig. S1 E).
Next, we asked whether these effects on growth are different in the presence or absence of Ltv1. If a mutation has the same defect in the presence or absence of Ltv1, then the ratio will be 1, signifying the absence of a genetic interaction. In contrast, larger or smaller defects in the absence of Ltv1 signify synthetic genetic interactions (a sensitivity or a resistance to Ltv1 deletion, respectively). Interestingly, the Rps3_FE and Rps3_KK mutations, which affect the interface with Rps10 and Rps20, respectively, both give small synthetic defects with Ltv1 deletion (Fig. 1 E). In addition, the Rps20_EYER mutation, which disrupts the binding of Rps29, essentially produces a synthetic lethal phenotype (Figs. 1 E and S1 F).
In contrast, the Rps20_DE mutation, which is in the Rps20_Rps3 interface opposite of Rps3_KK, is rescued by deletion of Ltv1 (Fig. 1 E). The differing effects from Rps3_KK and Rps20_DE suggest that Rps3 and/or Rps20 are positioned differently in the presence and absence of Ltv1.
To test whether ΔLtv1 yeast are generally sensitive to mutations in RPs, we produced mutants in Rps26, which disrupt its interaction with Rps14 (Fig. S2 A). Rps26_D52R, Rps26_E46R_D52R, Rps26_E46R_D52R_V58D, and Rps26_E46R_D52R_S29V show ∼1.2–1.8-fold growth defects in the presence of Ltv1 (Fig. S2 B) but grow better than WT Rps26 in its absence (Fig. S2 C). This is the opposite observation of that made with the Rps3_FE, Rps3_KK, and Rps20_EYER mutations, demonstrating that ΔLtv1 yeast are not generally sensitive to RP mutations. We do not know whether this rescue is related to the effects on Rps17 mutations (Fig. 1 D), perhaps linking Rps26 and Rps17, which are both on the platform.
The synthetic phenotypes of the Rps3_FE, Rps3_KK, and Rps20_EYER mutations and the Ltv1 deletion could indicate that Ltv1 deletion and RP mutations have the same effect on a third player, which is exacerbated by the double mutation, leading to a more catastrophic effect. Because the RPs mutations are in the interface between these proteins, the simplest interpretation was that they affected the binding of one or more of these RPs and that Ltv1 deletion was doing the same.
To determine whether Ltv1 was playing a role in recruitment of beak proteins, we purified ribosomes from WT and ΔLtv1 yeast and used Western analysis to test whether these are deficient in recruitment of Rps10, Rps20, and Rps29 relative to Rps26 and Rps8. Because antibodies against Rps20 or Rps29 were unavailable, we individually HA-tagged Rps20 or Rps29. The data in Fig. 3 C demonstrate that relative to Rps8 or Rps26, Rps10 levels are reduced in ribosomes from ΔLtv1 yeast, relative to ribosomes from Ltv1-containing yeast. In contrast, neither Rps20 nor Rps29 are decreased (Fig. S1, G and H). Nevertheless, we note that the HA-tag on Rps29 introduces sensitivity to Ltv1 deletion (Fig. S1 I), indicating that this tag may also perturb the arrangement of RPs in the head. Although Ltv1 depletion leads to reduced Rps10 in WT and Rps29-HA yeast, indicating that at least Rps10 is similarly positioned as in WT ribosomes, we cannot exclude the possibility that HA-tagging Rps29 reduces its occupancy, thus masking effects from Ltv1 deletion.
To confirm that the synthetic genetic defects between mutations in Rps3 and Rps20 and deletion of Ltv1 indeed arose because Rps10 levels were reduced in the mutant ribosomes, we also purified ribosomes from strains bearing Rps3_FE and Rps3_KK or Rps20_EYER and Rps20_DE and used Western blot analysis to determine their Rps10 levels relative to strains encoding the corresponding WT protein. As expected from our model, ribosomes from the Rps3_FE and Rps3_KK strains show reduced levels of Rps10 relative to both Rps26 and Rps8 (Fig. 3 D). Similarly, Rps20_EYER and DE mutants have reduced levels of Rps10 (Fig. 3 E).
Together, the analysis of purified ribosomes demonstrates that Ltv1 is required for the efficient recruitment of Rps10 and Asc1 but not Rps20 and Rps29. Furthermore, given the Ltv1-dependent changes in RNA accessibility near Rps3, the opposing Ltv1 genetic interactions of the Rps3_KK and Rps20_DE mutations (two residues that interact in mature ribosomes from WT yeast) and the perturbed binding of Rps10, Asc1, and Rps17, three proteins that bind directly to Rps3, we suggest that ribosomes from ΔLtv1 yeast have mispositioned Rps3. Mispositioned Rps3 also explains why Rps10 recruitment is affected by Ltv1 deficiency as Rps10 is recruited after Ltv1 is released as their binding sites overlap (Strunk et al., 2011; Heuer et al., 2017; Johnson et al., 2017; Scaiola et al., 2018).
Ribosomes from ΔLtv1 yeast are prone to mistakes in translation, resistant to stresses, and deficient in nonfunctional rRNA decay (NRD)
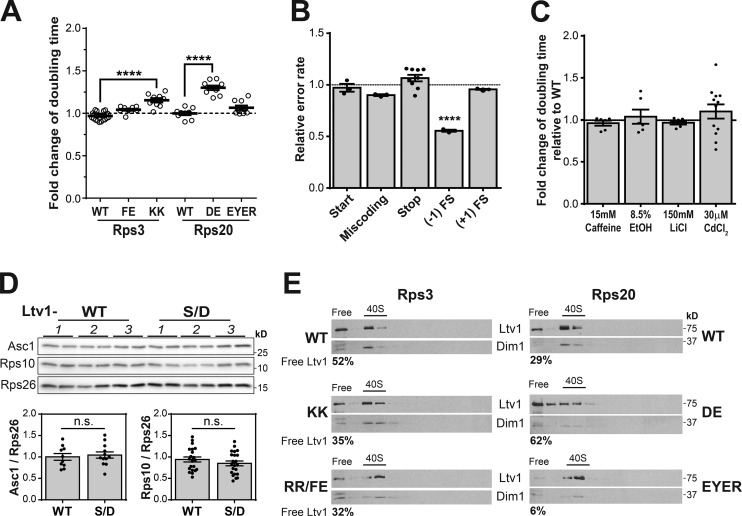
The data above show that Ltv1 chaperones the assembly of Rps3 such that its deletion leads to accumulation of ribosomes with mispositioned Rps3 and substoichiometric Rps10 and Asc1. To test whether these deficiencies contribute to mistakes by the ribosome during translation, we used reporter plasmids (Harger and Dinman, 2003; Keeling et al., 2004; Salas-Marco and Bedwell, 2005; Cheung et al., 2007) to assay ribosome fidelity during start and stop codon selection, decoding, and frameshifting. Deletion of Ltv1 leads to small but highly reproducible defects in recognition of the start and stop codons and increased miscoding (Fig. 4 A). In contrast, +1 frameshifting is reduced.
Figure 4.
Ribosomes from ΔLtv1 yeast display functional differences. (A) Effects on translational fidelity in ΔLtv1, ΔAsc1, and Rps10-reduced cells. Three to six technical repeats of three to six biological replicates were obtained. (B) Increased stress resistance in ΔLtv1, ΔAsc1, and Rps10 reduced cells (n = 6–12). The change in doubling time from addition of stress media was determined in ΔLtv1 and ΔAsc1 cells and normalized to the effect in WT cells. Similarly, Rps10-reduced cells were normalized to Rps10-replete cells. (C) ΔLtv1 cells are deficient in NRD (n = 5–6). Significance was determined by a two-tailed t test. (D) ΔLtv1 cells are sensitive to H2O2 (n = 6, with three technical repeats). In all cases, error bars represent the SEM, and unless otherwise noted, significance was determined using an unpaired student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Stochastic stop codon read-through can produce phenotypic diversity within a genetically identical colony, thereby promoting survival under stress (True and Lindquist, 2000). Our data indicate that ribosomes from ΔLtv1 yeast are more prone to making mistakes during translation, leading to defects in recognition of all codons including the stop codon. To test whether this confers stress resistance, we tested the sensitivity of WT and ΔLtv1 yeast to caffeine, ethanol, LiCl, and CdCl2. ΔLtv1 yeast were partially resistant to caffeine, CdCl2, and ethanol treatment. No resistance to LiCl was observed (Fig. 4 B).
To test whether the translational fidelity defects and stress resistance observed in ΔLtv1 yeast were due to the accumulation of Rps10 and/or Asc1-deficient ribosomes, we also investigated the effects from Asc1 or Rps10 deletion/depletion on translational fidelity and stress sensitivity (Fig. 4, A and B). These data show that the effects from deletion of Asc1 and depletion of Rps10 on ribosomal fidelity largely mirror those observed from Ltv1 deletion, as expected if they arise from the substoichiometric incorporation of these two proteins. The somewhat larger effects on start codon recognition from Asc1 deletion are likely due to deletion versus substoichiometric incorporation. Interestingly, Rps10 depletion has effects on frameshift maintenance that are not observed or are opposed by Asc1 deficiency. This might arise from the suggested interaction of structured frameshifting elements in the mRNA with the mRNA entry channel (Caliskan et al., 2014).
Rps10 depletion and Ltv1 deletion have similar effects on stress resistance (Fig. 4 B), suggesting that the substoichiometric incorporation of Rps10 underlies this effect. A notable exception to this observation is the resistance to LiCl. Although both Asc1 deletion and Rps10 depletion lead to LiCl resistance, ΔLtv1 yeast are unaffected, suggesting either that codepletion has unexpected effects or that there are additional defects in ribosomes from ΔLtv1 yeast that we have yet to discover. Asc1 deletion produces stress resistance that also overlaps that of ΔLtv1 yeast but is more mixed.
Asc1 is required for the related pathways of no-go decay (NGD) and nonfunctional rRNA decay (NRD; Ikeuchi and Inada, 2016; Limoncelli et al., 2017; Sitron et al., 2017). These pathways are induced by ribosomes stalled either on mRNAs containing stem loops, polylysine sequences, or other unfavorable codons (NGD) or due to defects in the 40S subunits (NRD; Doma and Parker, 2006; LaRiviere et al., 2006; Cole et al., 2009; Arthur et al., 2015; Joazeiro, 2017). Because Asc1 is substoichiometric in ribosomes from ΔLtv1 yeast, we hypothesized that ΔLtv1 yeast are defective in NRD and NGD. To test this hypothesis, we used an NRD reporter assay to measure the levels of defective 18S rRNA (A1755C) relative to 25S rRNA (Limoncelli et al., 2017). This is done by providing the yeast with a plasmid encoding the rRNA operon where both 18S and 25S rRNA are uniquely tagged to allow for their detection by Northern blotting. To account for differences in ribosome assembly between different strains, we normalize the mutant 18S/25S ratio against WT 18S/25S levels in each yeast strain. As previously shown (LaRiviere et al., 2006; Cole et al., 2009; Limoncelli et al., 2017), in WT cells, 18S-A1755C RNA is reduced (relative to WT 18S RNA) due to its efficient degradation via NRD. As also previously shown (LaRiviere et al., 2006; Cole et al., 2009; Limoncelli et al., 2017), NRD requires both Dom34 and Asc1 (Fig. 4 C). Importantly, the data also demonstrate that akin to ΔAsc1 yeast, ΔLtv1 yeast are deficient in NRD (Fig. 4 C). Because NRD appears to be highly related to NGD and requires the same factors (Cole et al., 2009; Passos et al., 2009; Shoemaker et al., 2010) including Asc1 (Ikeuchi and Inada, 2016; Sitron et al., 2017), we suggest that ΔLtv1 yeast are also defective in NGD.
Although NGD has been discovered using artificial stem loop–containing RNAs (Doma and Parker, 2006), its in vivo substrates likely include oxidized RNAs (Simms et al., 2017). Thus, Asc1- and Ltv1-deficient yeast should be defective in clearing ribosomes from oxidized mRNAs and thus be sensitive to oxidative stress. To test this hypothesis, we used a survival assay to measure the sensitivity of WT and ΔLtv1 yeast to H2O2. This analysis showed that ΔLtv1 yeast are more sensitive to oxidative stress (Fig. 4 D).
Hrr25 is not required for beak protein incorporation
Ltv1 promotes the proper positioning of Rps3 and efficient Asc1 and Rps10 recruitment. Ltv1 could accomplish this role either by actively supporting changes in the beak structure or because Hrr25-dependent Ltv1 release, required for assembly to continue (Ghalei et al., 2015), depends on incorporation of beak proteins. In that model, Ltv1 has a passive role as a road block to maturation.
To test whether Hrr25 was required for the chaperoning function of Ltv1 as implied by the second model, we took advantage of a phosphomimetic Ltv1 mutant (Ltv1-S/D), whose release is Hrr25 independent (Ghalei et al., 2015). If indeed Hrr25 was monitoring RP assembly and using Ltv1 to block downstream assembly, then the Ltv1-S/D strain should have essentially the same phenotypes as the Ltv1 deletion strain as both bypass the requirement for Hrr25. In contrast, if the binding of Ltv1 was facilitating proper beak protein assembly, then we predict that the phosphomimetic Ltv1-S/D should rescue the Ltv1 deletion phenotypes even though its release is not Hrr25 regulated.
We first tested whether Ltv1-S/D had the same genetic interactions as ΔLtv1. Indeed, Ltv1-S/D rescued the synthetic lethality of the Rps20-EYER mutation and the defect from Rps3_FE. In addition, the size of the synthetic defect from Rps3_KK was reduced, and Rps20_DE now had the same kind of effect as Rps3_KK (Fig. 5 A). Furthermore, ribosomes from Ltv1-S/D cells had no defects in translation fidelity and a slightly decreased propensity to frameshift into the −1 direction (Fig. 5 B). Consistent with the fidelity defects being responsible for promoting stress resistance, Ltv1-S/D showed the same stress resistance as WT cells (Fig. 5 C). Additionally, Western analysis indicates that Ltv1-S/D rescues the depletion of Asc1 and Rps10 observed in ΔLtv1 yeast (Fig. 5 D). Finally, Ltv1-S/D rescues the sensitivity to oxidative stress (Fig. 4 D). Because Ltv1-S/D is released independently of Hrr25 (Ghalei et al., 2015), these data are inconsistent with a role for Hrr25 in monitoring head RP incorporation.
Figure 5.
Ltv1 facilitates RP assembly. (A) Genetic interactions between Ltv1-S/D and mutations in the Rps3 protein network. Effect from Rps3 and Rps20 mutations in the Ltv1-S/D background was normalized to the effect in WT background n = 3–6 biological replicates with three to five technical repeats. (B) Translational fidelity in Ltv1-S/D cells (n = 3–9). (C) No stress resistance in Ltv1-S/D cells (n = 3–12). (D) Ribosomal protein occupancy in Ltv1-S/D cells. The data are averages from two technical repeats of three to five biological replicate experiments. In all cases, the error bars represent the SEM, and the significance was determined using an unpaired t test. ****, P < 0.0001. (E) Release of Ltv1 is promoted by the Rps3 interaction network. Sucrose-gradient and Western analysis of lysates from cells containing WT or mutant Rps3 (left) and Rps20 (right), respectively. The percentage of free Ltv1 (vs. ribosome-bound Ltv1) is given below each Western blot.
To test whether incorporation of beak RPs also affected Ltv1 release, we measured the fraction of free and ribosome-bound Ltv1 in cells with mutations in the network of RPs. For this experiment, we performed sucrose gradient fractionation of yeast lysates and then used Western analysis to monitor the sedimentation of Ltv1. When bound to 40S subunits, Ltv1 migrates into the gradient, whereas free Ltv1 will remain on top of the gradient. This analysis shows that more Ltv1 is retained on 40S subunits in the case of the Rps3_KK, Rps3_RR/FE, and the Rps20_EYER mutants, which disrupt the Rps3/Rps20, Rps3/Rps10, and Rps20/Rps29 interfaces, respectively (Fig. 5 E). The effect on Ltv1 release from these mutations, which affect Rps10 binding, are surprising, as Rps10 binds after Ltv1 release (Strunk et al., 2011; Heuer et al., 2017; Johnson et al., 2017; Scaiola et al., 2018). We reconcile this paradox by suggesting that Rps10 binding renders Ltv1 release irreversible.
In contrast, the Rps20_DE mutant has increased free Ltv1, consistent with the opposite synthetic growth phenotype of this mutant (Fig. 1 B). This observation is somewhat surprising as Rps20_DE interacts with Rps3_KK in mature ribosomes (Fig. 1 A; Rabl et al., 2011). Thus, the opposing effects from these mutants on Ltv1 release and on growth in the ΔLtv1 background suggest that before the release of Ltv1, the interaction between these residues on the Rps20/Rps3 interface is not made. Of note, the Rps20_DE results are somewhat at odds with those observed previously (Mitterer et al., 2016). We do not have any explanation for these differences.
Breast cancer cells lines have substoichiometric LTV1, RPS3, RPS10, and Asc1/RACK1
Some cancer cells display a loss of RP stoichiometry (Vlachos et al., 2012; Guimaraes and Zavolan, 2016; Ajore et al., 2017; Kulkarni et al., 2017). Because Ltv1 promotes incorporation of Rps10 and Asc1, we wondered whether cancer cells might down-regulate LTV1, thereby producing RPS10- and Asc1/RACK1-deficient ribosomes (the mammalian homologue of Asc1 is referred to as RACK1).
We tested MDA–MB-231 triple-negative breast cancer cells in which the CK1δ-dependent release of LTV1 can be targeted with the kinase inhibitor SR3029 (Ghalei et al., 2015; Rosenberg et al., 2015) as well as SF268 glioma cells. Although both cell lines have similar LTV1 levels (Fig. 6 A), they responded differentially to a heterozygous LTV1 knockout. Both cell types grow with similar doubling times with both copies of LTV1 intact, but deleting one allele of LTV1 in the breast cancer cells has a 2.5-fold effect on proliferation rates and a smaller, 1.5-fold effect in glioma cells (Fig. S3 A). In contrast, relative to glioma cells, LTV1 heterozygous breast cancer cells are less sensitive to SR3029, a kinase inhibitor that blocks LTV1 release, even though both LTV1+/+ cell lines are equally sensitive (Fig. S3, B and C). LTV1 knockdown rescues drug sensitivity (Ghalei et al., 2015; Rosenberg et al., 2015). Thus, the reduced SR3029 response in the LTV1+/− breast cancer cells is consistent with reduced LTV1 levels in breast cancer relative to glioma cells.
Figure 6.
LTV1 deficiency in human cancer cells. (A) Western blot for LTV1 from total protein in SF268 glioma and MDA–MB-231 breast cancer cell lines and their LTV1+/− heterozygous deletion derivatives. n = 3. (B and C) Northern blots and their quantification relative to U2 and SRP of total RNA from glioma and cancer cells (n = 6). (D) Analysis of ribosome content from equally loaded sucrose gradients of cancer cell lines (n = 3). (E) LTV1 distribution probed by sucrose-gradient and Western blot analysis of cell lysates from glioma (left) and breast cancer (right) cells. (F) Quantification of data in E (n = 3). (G) Occupancy of RPS3, RPS10, and RACK1 in ribosomes purified from glioma and cancer cells (n = 9–12). (H) Translational fidelity of glioma and breast cancer cells (n = 12). Error bars represent the SEM, and significance was determined using an unpaired student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (I) cBioPortal analysis of genomic alterations in LTV1, BYSL (hEnp1), and CSNK1D (hCK1δ) from two datasets of cancer cell lines. Studies cited in this figure are from cBioPortal (Cerami et al., 2012; Gao et al., 2013).
We therefore considered the possibility that breast cancer cells make more ribosomes so that the equal amounts of LTV1 would not be sufficient for all ribosomes in breast cancer cells. To test this model, we used Northern analysis, which indicates that breast cancer cells have ∼1.5-fold more 18S and 28S rRNA than glioma cells (Fig. 6, B and C). Similarly, quantification of polysome profiles shows that breast cancer cells have ∼1.5-fold more ribosomes (Figs. 6 D and S3 D). Together with the equal levels of LTV1, this observation suggests that the LTV1 occupancy on pre-40S subunits is higher in glioma cells than breast cancer cells. If this is true, then more LTV1 would cosediment with 40S precursors in breast cancer cells, while in glioma cells a larger amount of LTV1 should remain free. To test this prediction, we probed the polysomes in breast cancer and glioma cells for LTV1. Indeed, in breast cancer cells, most LTV1 is 40S-bound, whereas in glioma cells, nearly half remains unbound (Fig. 6, E and F), as predicted if LTV1 is limiting in breast cancer but not glioma cells.
Next, we tested whether the reduced LTV1 occupancy on pre40S ribosomes led to loss in stoichiometry of RPS10 and Asc1/RACK1 as observed in yeast cells. We purified ribosomes from both cell lines, and then used antibodies against RPS3, RPS10, RPS20, RPS29, and RACK1 to quantify their levels relative to RPS5. Excitingly, these analyses demonstrate that relative to glioma cells, breast cancer cells have reduced levels of RPS3, RPS10, and RACK1 in their ribosomes whereas as in yeast cells, RPS20 and RPS29 are unchanged (Figs. 6 G and S3 E).
To test whether these differences in RP binding lead to translational fidelity defects as observed in yeast, we used reporters for stop codon read-through and miscoding (Oishi et al., 2015). Although there were no differences between the cell lines in the decoding error rate (Fig. S3 F), the breast cancer cells had about twofold higher propensity to read through a stop codon than the glioma cells (Fig. 6 H). Taken together, these data indicate that in MDA–MB-231 breast cancer cell lines, the LTV1 levels are insufficient relative to their high rate of ribosome production, leading to the production of 40S ribosomal subunits with substoichiometric levels of RPS3, RPS10, and RACK1. These differences in the RP stoichiometry are likely to underlie the defects in stop codon recognition.
To test whether the LTV1 insufficiency observed in this study in MDB–MB-231 breast cancer cell lines was unique or more generally observed in cancer cells, we used the cBioPortal to investigate genomic alterations in LTV1, its binding partner BYSL (hEnp1), and its regulator casein kinase 1δ (CSNK1D). Interestingly, although deletions of LTV1 were slightly more common than amplifications, for BYSL or CSNK1D, mostly amplifications were observed (Fig. 6 H). These data indicate that LTV1 insufficiency and the resulting substoichiometric incorporation of RPS10 and RACK1 are commonly observed in tumor cell lines and presumably the cancer cells from which these lines were originally derived.
Discussion
Ltv1 chaperones head assembly
In ribosomes from ΔLtv1 yeast, incorporation of Rps10 and Asc1, two proteins that bind Rps3, into the small subunit head is reduced. In addition, genetic interactions between Ltv1 deletion and mutations in RPs from the 40S beak strongly suggest that Rps3 is mispositioned. These data are supported by RNA structure probing, which reveals changes adjacent to Rps17, Asc1, and Rps20. Thus, these data provide strong evidence for a role of Ltv1 in positioning of Rps3 and incorporating Rps10 and Asc1 into the head structure of the small ribosomal subunit.
How does Ltv1 chaperone the assembly of the 40S head? Our data show that the phosphomimetic Ltv1-S/D, which is released independently of Hrr25 (Ghalei et al., 2015), rescues nearly all phenotypes from Ltv1 deletion including the positioning of Rps3 and the recruitment of Rps10 and Asc1. These data are inconsistent with a model in which beak protein incorporation is monitored by Hrr25, and they suggest that instead beak assembly is facilitated by the presence of Ltv1. How does Ltv1 facilitate RP assembly? The observation that Rps3_KK and Rps20_DE, mutations of residues that interact with each other in mature ribosomes, have opposing effects on Ltv1 deletion and Ltv1 release indicates that Ltv1 does not simply prealign these RPs for their interactions. Perhaps Ltv1 instead plays an opposite role, preventing premature formation of protein–protein interactions within the Rps3 network, which could stabilize misfolded rRNA structures.
We also considered the possibility that the effects from Ltv1 deletion are partially or fully mediated by partial Enp1 depletion (Strunk et al., 2011). If this were the case, then we would predict that the defects from Ltv1 deletion would be rescued by overexpression of Enp1. To test this model, we cloned Enp1 into yeast expression vectors, under the constitutive TEF and Cyc1 promoters, respectively. As expected, the TEF promoter produces more Enp1 (Fig. S5 A), and Gal:Enp1 cells supplemented with TEF-Enp1–encoding plasmids grow faster than the same cells supplemented with Cyc1-Enp1–encoding plasmids (Fig. S5 B). Nevertheless, in the absence of Ltv1, Enp1 overexpression via the TEF promoter inhibits yeast growth, in contrast with what would be expected if the effects on cell growth from Ltv1 deletion arose from partial depletion of Enp1. We thus consider it unlikely that the effects on subunit assembly from Ltv1 deletion are mediated by Enp1 depletion.
Assembly of the small ribosomal subunit head
Based on these data as well as others in the literature, we suggest the following model for assembly of the small subunit beak structure (Fig. 7 A). Enp1 binds early to pre40S subunits and is present in nuclear precursors (Schäfer et al., 2003). Because Rps20 and Rps29 bind under Rps3 (Fig. 1 C), we assume that these are recruited before Rps3. Thus, we suggest that after initial Enp1 recruitment and nucleolar rRNA maturation, Ltv1, Rps29, and Rps20 are recruited next. Previous data show that Ltv1 binds Rps20 (Mitterer et al., 2016), and we have shown that Yar1 delivers Rps3 to Ltv1 (Ghalei et al., 2015). Thus, Rps3 recruitment follows Ltv1 binding, and we have shown in this study that its proper positioning in mature ribosomes depends on the presence of Ltv1. Intriguingly, Ltv1 release repositions both Rps3 subdomains (Strunk et al., 2011; Mitterer et al., 2016; Johnson et al., 2017). This last observation is broadly consistent with structure probing data by Hector et al. (2014), who have demonstrated structural changes in the Rps3 and Rps17 binding sites during the latest stages of 40S maturation, when Ltv1 is released. This finding indicates that the Ltv1-dependent changes in 40S structure monitored in this study are also relevant to rearrangements during 40S maturation.
Figure 7.
Model of Ltv1-dependent head structure assembly. (A) Model for the Ltv1-dependent assembly of the small subunit head structure. Ribosomal proteins are indicated by their corresponding number, and assembly factors are defined as: L, Ltv1; E, Enp1; Y, Yar1; and H, Hrr25/Ck1δ. (B) Model of small subunit head assembly in ΔLtv1 cells.
ΔLtv1 yeast (Fig. 7 B) correctly incorporate Rps10 and Asc1 in a fraction of their ribosomes, whereas others lack these proteins. It is unknown whether in the ΔLtv1 yeast some ribosomes are completely assembled while others lack both proteins, or whether there are partially assembled subpopulations.
Ltv1 functions analogously to bacterial RimM
As we have previously pointed out (Ghalei et al., 2015), there is functional homology between Ltv1 and bacterial RimM. Assembly intermediates in RimM deficient bacteria lack S3, S10, and S14 (Clatterbuck Soper et al., 2013), the bacterial homologues for Rps3, Rps20, and Rps29, all of which are missing in 40S assembly intermediates from ΔLtv1 yeast (Strunk et al., 2011; Johnson et al., 2017). Strikingly, the changes in head accessibility produced by Ltv1 deletion are also highly similar to those observed upon RimM deletion (Fig. S4). The head is the last substructure of the small subunit to assemble in vivo, and kinetic research of its folding in vitro indicates that in particular, the beak folds very slowly, perhaps rearranging from misfolded structures (Adilakshmi et al., 2008). This rationalizes the need for a beak chaperone such as RimM and Ltv1. The observation that Ltv1 deficiency leads to mispositioned Rps3 and substoichiometric levels of Rps10 and Asc1 indicates that bacterial ribosomes lacking RimM might similarly misposition Rps3.
Recruitment of Asc1 via interactions with the Rps3 tail
Our data demonstrate that interactions between the C-terminal tail of Rps3 and Asc1 are required for Asc1 recruitment. Previous data from Limoncelli et al. (2017) are consistent with these findings as they demonstrate that yeast strains containing Rps3 truncated at residue 211 are deficient in NRD. We rationalize this finding in this study as we show that these ribosomes lack Asc1. The even larger stability of mutant 18S rRNA observed in the Rps3 truncation relative to the Asc1 deletion might reflect an additional functional role for Rps3 as suggested by Limoncelli et al. (2017), or alternatively, be related to the deletion of the snoRNA in the Asc1 intron that was apparently not complemented by Limoncelli et al. (2017).
Interestingly, Rps3 levels appear to be unchanged in ribosomes from ΔLtv1 yeast but reduced in breast cancer cells. We believe that this most likely reflects quantitative differences and not qualitative differences between yeast and human ribosomes. Our data strongly suggest that Rps3 is mispositioned in yeast ribosomes from ΔLtv1 yeast, thus blocking Asc1 recruitment and leading to genetic interactions with Rps17. Furthermore, the Rps3 position affects salt-dependent binding strength (Schäfer et al., 2006). Thus, we suggest that the affinity of Rps3 in this mispositioned assembly state is slightly different between yeast and human ribosomes, leading to dissociation of Rps3 during purification of human ribosomes, but retention in yeast ribosomes.
LTV1 is substoichiometric in breast cancer cells, producing ribosomes with reduced Rps10 and RACK1
Haploinsufficiency of several RPs can lead to a diverse set of diseases (ribosomopathies), which share an increased propensity to develop specific forms of cancer (Ellis and Lipton, 2008; Burwick et al., 2011; Vlachos, 2017). Partial depletion of Rps26 leads to the accumulation of ribosomes lacking Rps26 (Ferretti et al., 2017). These ribosomes have a different translational program than “normal” ribosomes, translating a set of mRNAs that are poorly translated in WT cells. The disrupted protein homeostasis resulting from translation by Rps26-deficient ribosomes might be mirrored in diseases arising from haploinsufficiency of other RPs. Interestingly, Rps26-deficient ribosomes are generated under specific stress conditions, demonstrating the existence of cellular pathways for their production (Ferretti et al., 2017).
Ribosomal protein gene deletions resulting in unequal RP stoichiometries also arise spontaneously in cancer cells, where they are associated with poor outcomes (Guimaraes and Zavolan, 2016; Ajore et al., 2017; Kulkarni et al., 2017), suggesting that preexisting pathways leading to the formation of different ribosome populations are exploited by tumor cells to drive an oncogenic translational program.
In this study, we show that cancer cells have different levels of LTV1 relative to 40S ribosomes, leading to the formation of Rps3- and RACK1-deficient ribosomes in MDA–MB-231 triple-negative breast cancer cells. Although additional experiments on the specific translational program imparted by deficiency of Rps10, Rps3, and RACK1 await further study, data in the literature indicate a role for Asc1 in promoting the translation of mRNAs with small open reading frames (Thompson et al., 2016). These include many efficiently translated mRNAs, and Asc1 deficiency has relatively larger effects on the translation of well-translated mRNAs. Similarly, Rps3 promotes translation of mRNAs with optimal Kozak contexts (Dong et al., 2017). Thus, deficiency of Rps3 and Asc1 is likely to affect well-translated mRNAs more strongly than poorly translated mRNAs, perhaps changing protein homeostasis toward otherwise highly regulated genes.
Ribosomes from Ltv1-deficient yeast and mammalian cells are more prone to stop codon read-through. Work with the yeast Sup35 prion, a translation termination factor, has shown that under stress conditions, the increased read-through can provide an advantage as it provides a fertile ground for evolution and thus adaptation to stress (True and Lindquist, 2000). In cancer cells, this could lead to further adaptations to the unique stresses that cancer cells are exposed to. In addition, we show that yeast cells lacking Ltv1 are uniquely vulnerable to oxidative stress, possibly as a result of the defects in NRG and NGD, which lead to the accumulation of oxidized mRNAs and rRNAs. In cancer cells, this might present a unique vulnerability that could be exploited therapeutically.
Materials and methods
Yeast strains and cloning
Yeast strains (Table S1) were obtained from Open Biosystems or produced by PCR-based homologous recombination (Longtine et al., 1998), verified by PCR and Western blotting when antibodies were available. Plasmids used in this study are listed in Table S2.
Translational fidelity by luciferase assay
Translational fidelity in yeast was measured using dual-luciferase reporter plasmids (Harger and Dinman, 2003; Keeling et al., 2004; Salas-Marco and Bedwell, 2005; Cheung et al., 2007) and the Dual-Luciferase Reporter assay system (Promega) according to the manufacturer’s protocol. Briefly, cells were grown to mid-log phase in sterile Nunc MicroWell 96W Microplates (VWR), pelleted, and resuspended in passive lysis buffer. Firefly and renilla signals were measured sequentially by addition of 15 µl Luciferase Assay reagent II and Stop & Glo Reagent to 5–10 µl sample lysate in black Fluotrac 600 96W Microplates (Greiner Bio-One). Luciferase activity was measured immediately in a 2104 EnVision Multilabel plate reader (PerkinElmer). We calculated the firefly-to-renilla ratio for each sample and then normalized the values observed for each cell type relative to its WT control. Significance was determined using an unpaired t test.
For mammalian fidelity assays, 1 µg reporter plasmid (Oishi et al., 2015) was transfected into 2 × 105 cells cultured in six-well dishes using Lipofectamine LTX Plus (Invitrogen). 24 h after transfection, cells from one well were trypsinized and plated onto four wells of a 96-well white plate (Costar) and incubated at 37°C for 24 h. Firefly luciferase (Fluc) and renilla luciferase (Rluc) were detected as in yeast assays.
Yeast growth assays
Growth measurements were performed using a Synergy 2 multimode microplate reader (BioTek). Cells were pregrown to mid-log phase in selective glucose-containing media, diluted to OD600 0.05 in Nunc MicroWell 96W microplates, and grown at 30°C with rapid shaking. Doubling times were extracted and normalized to cells expressing the WT protein on plasmids. For stress assays, normalization was done against growth in rich medium. Rps10 was depleted by growth of YKK722 in 100 ng/ml doxycycline.
Hydrogen peroxide survival assay
Survival after acute H2O2 exposure was determined as previously described (Lee et al., 2002), with slight modifications. Mid-log phase cells were pelleted and resuspended into PBS at a cell density of 2 × 106 cells/ml. Cells were exposed for 30 min to either PBS or PBS with 10 mM H2O2 (Sigma-Aldrich) with shaking at 30°C. 200 cells were plated on YPD plates in technical triplicates. The number of surviving colonies was counted using OpenCFU v3.9.0 (Geissmann, 2013).
NRD assays
A plasmid-based reporter system was used to measure the stability of WT or a decoding site mutant of 18S rRNA (A1755C in yeast equivalent to A1492C in Escherichia coli) encoded by the entire 35S rDNA (Limoncelli et al., 2017). WT or A1755C plasmids were transformed into BY4741, ΔLtv1, ΔAsc1, and ΔDom34 yeast strains and grown to mid-log phase in minimal galactose/DO ura medium to induce the reporter rRNAs. Total RNA was extracted using the hot phenol method and analyzed by Northern blotting using published oligonucleotides directed against the 18S and 25S reporter tags (FL125 and FL126; Limoncelli et al., 2017). Band intensity was detected by phosphoimaging, and the ratio of 18S-tag/25S-tag for each sample was measured. To control for differences in 18S rRNA levels in the different yeast strains, we normalized the mutant 18S/25S ratio with the WT 18S/25S ratio from the same yeast stain. The experiment was repeated three times using two clones for each reporter in each of the four yeast strains.
Yeast ribosome purification
Ribosomes were purified as described (Acker et al., 2007). In brief, seeded 1-liter cultures were harvested at OD600 0.4–0.8 and flash-frozen in ribosome buffer (20 mM Hepes/KOH, pH 7.4, 100 mM KOAc, and 2.5 mM Mg(OAc)2) supplemented with 1 mg/ml heparin, 1 mM benzamidine, protease inhibitor cocktail (Sigma-Aldrich), and 2 mM DTT. Frozen cells were lysed by grinding to powder in a mortar and pestle. Yeast lysates were clarified and layered over a 500-µl sucrose cushion and centrifuged in a Beckman TLA 110.1 at 70,000 rpm for 1 h. Pelleted ribosomes were resuspended in high-salt buffer (ribosome buffer, 500 mM KCl, 1 mg/ml heparin, and 2 mM DTT) and again layered over a 500-µl sucrose cushion and centrifuged at 100,000 rpm for 70 min. The ribosome pellet was resuspended in subunit separation buffer (50 mM Hepes/KOH, pH 7.4, 500 mM KCl, 2 mM MgCl2, and 2 mM DTT), and 1 mM puromycin (Sigma-Aldrich) was added. Subunits were isolated by loading onto 5–20% sucrose gradients (50 mM Hepes/KOH, pH 7.4, 500 mM KCl, 5 mM MgCl2, 2 mM DTT, and 0.1 mM EDTA) and centrifuged at 32,000 rpm for 6 h. Finally, subunits were concentrated, and buffer was exchanged into ribosome storage buffer (ribosome buffer, 250 mM sucrose and 2 mM DTT) and stored at −80°C.
Human ribosome purification
Ribosomes were purified as described (Khatter et al., 2014). Briefly, 2 × 106 cells were trypsinized and washed in 10 ml Dulbecco’s PBS (Corning), and the cell pellet was lysed on ice for 30 min in 400 µl freshly prepared lysis buffer containing 15 mM Tris, pH 7.5, 0.5% NP-40 (Sigma-Aldrich), 6 mM MgCl2, 300 mM NaCl, and RNase inhibitor (Promega). The lysate was cleared by centrifuging at 12,000 g for 10 min, and the supernatant was loaded onto a 1-M sucrose cushion containing 20 mM Tris, pH 7.5, 2 mM Mg(OAc)2, 150 mM KCl, and 2 mM DTT and centrifuged for 3 h at 100,000 rpm (TLA100.1 rotor). The ribosome pellet was suspended in 100 µl buffer containing 20 mM Tris, pH 7.5, 6 mM Mg(OAc)2, 150 mM KCl, 1 mM DTT, 200 mM sucrose, and RNase inhibitor and denatured with SDS-PAGE buffer.
Protein purification
BL21 cells containing an MBP-Asc1 expression plasmid were grown to OD600 0.8 in 2× YT medium at 37°C before induction with 0.5 mM IPTG for 3 h at 20°C. Cells were lysed by sonication in lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole, and 5 mM β-mercaptoethanol), and the lysate was applied to Ni-NTA resin (Qiagen) equilibrated with lysis buffer. Resin was washed with 20 volumes of lysis buffer containing 20 mM imidazole. Untagged Asc1 was eluted from the resin overnight with on-column treatment with tobacco etch virus (TEV) protease. Asc1 was further purified over a Mono Q column (GE Healthcare) equilibrated with 20 mM Tris, pH 7.5, 50 mM NaCl, and 2 mM DTT and eluted with a linear gradient of 100–600 mM NaCl over 30 column volumes before size-exclusion chromatography (Superdex 75; GE Healthcare) in 20 mM Tris, pH 7.5, 50 mM NaCl, and 2 mM DTT. Fractions with pure protein were pooled and concentrated using Amicon concentrators (EMD Millipore).
Ribosome binding assay by cosedimentation
6 pmol 40S ribosomal subunits purified from ΔAsc1 or Rps3-214 cells was incubated without Asc1 or increasing concentrations (120, 240, and 480 nM) of recombinant Asc1 in 50 µl RB buffer (200 mM KOAc, 20 mM Hepes/KOH, pH 7.4, 2.5 mM MgOAc, 2 mM DTT, 0.1 mg/ml heparin, and 0.5 µl RNAsin [Promega]). The mixture was incubated for 5 min on ice followed by an additional 5 min at room temperature, placed on 400 µl of 20% sucrose cushions in RB buffer, and centrifuged for 4 h at 400,000 g in a TLA 100.1 rotor. After centrifugation, the supernatants were removed, and the pellets were resuspended in SDS-loading dye and analyzed by SDS-PAGE followed by Coomassie staining.
In vivo DMS probing
In vivo DMS probing was performed essentially as described (Wells et al., 2000). In brief, cells with or without Ltv1 were grown in YPD to OD600 0.8, when 10 ml culture was mixed with either DMS (Sigma-Aldrich) or ethanol (controls). The mixtures were incubated at 30°C for 2 min. Reactions were stopped by adding 5 ml of 0.6 M 2-mercaptoethanol and 5 ml of water-saturated isoamyl alcohol on ice. Cells were pelleted and washed with 5 ml of 0.6 M 2-mercaptoethanol. RNAs were purified with the High Pure RNA isolation kit (Roche) before reverse transcription was performed using Superscript III (Thermo Fisher Scientific) according to the manufacturer’s protocol.
Generation of LTV1 knockout cell lines
The CRISPR/Cas9 nickase system (Ran et al., 2013) was used to target exon 2 of the LTV1 gene in SF268 human glioma and MDA–MB-231 human breast cancer cell lines. The two guide RNAs targeting opposite strands of LTV1 were designed and cloned individually into the gRNA scaffold of pSpCas9n(BB)-2A-Puro (PX462) V2.0 vector following the Zhang laboratory protocol (see Table S2, PK523 row). 250 ng of each PX462 construct (containing guide 1 and guide 2) was transfected into cells (5 × 104 cells/well, 24-well plate) using Lipofectamine LTX Plus (Invitrogen). 1 µg/ml puromycin selection medium was added to the cells 24 h after transfection and maintained for 5–7 d. The pool of selected cells was expanded without puromycin, genomic DNA was isolated from half the pool (Quick Extract), and the remaining cells were sorted by flow cytometry as individual cells/well onto a 96-well plate.
Mismatch repair cleavage assays were performed on genomic DNA from cell pools to ensure that cells contained insertion/deletions in the LTV1 gene. Briefly, a 630-bp fragment from exon 2 of LTV1 was amplified from genomic DNA and gel purified. 200 ng PCR product was melted and reannealed in a thermocycler before being digested with T7 endonuclease (New England Biolabs) and separated by agarose gel electrophoresis. Mismatched, T7-cleaved products were detected as bands above and below the WT amplicon. Pools that contained a high percentage of mismatch products were expanded as single-cell clones from the 96-well plate and screened for Ltv1 protein level by Western blotting.
To confirm targeting of LTV1, exon 2 was PCR amplified from genomic DNA and TA cloned into pCR2.1 vector (Invitrogen). At least six pCR2.1 clones were sequenced in both directions and aligned with WT LTV1 exon 2. A dominant indel was detected from each target, and these cell lines were characterized further.
Cell culture, proliferation, and MTT assays
Cells were cultured in RPMI 1640 GlutaMax-1 medium (Gibco) supplemented with 10% FBS equivalent (Atlas Biologicals) and penicillin/streptomycin (Gibco). Cell cultures were maintained at 37°C in a water-jacketed incubator and buffered with 5% CO2.
Growth assays were performed by plating cells (1.5 × 104 cells/well, 24-well plate) in medium with or without 40 nM SR3029 and counted daily for 6–8 d. Cells from three wells were trypsinized and mixed with Trypan blue, and live cells counted using a hemocytometer. To extract doubling times, growth curves were fitted to an exponential growth equation using Prism (GraphPad Software). The average doubling time and SD from at least three replicate experiments were calculated.
The sensitivity to SR3029 (IC50) was measured by culturing cells for 72 h with a dose titration of SR3029 (10 µM to 0.16 nM at threefold dilution steps) and measuring live cells by MTT assay (CT01; EMD Millipore). Cells were plated at 5 × 104 cells/well on 96-well plates, and the next day, SR3029 in DMSO was diluted 200-fold into media and added to wells (four wells/concentration). MTT reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrasodium bromide) was added to wells and developed at 37°C for 4 h. Medium was removed, and 100 µl isopropanol/0.04 N HCl was added to each well and allowed to sit for 15 min at room temperature. 570- and 630-nm absorbance of plates was read using Synergy2 plate reader (BioTek), and IC50 was calculated using Prism software.
Northern blotting from mammalian cells
RNA was isolated from subconfluent cultures with TRIzol reagent (Ambion), denatured in formaldehyde, and then separated on an agarose gel, transferred, and probed as previously described for yeast (Ghalei et al., 2017). Band intensities were quantitated using Quantity One software.
Polysome profiling from mammalian cells
Cells were cultured in 15-cm culture dishes as above until 70–80% confluence was reached. 100 µg/ml cycloheximide (CHX) was added for 10 min followed by a wash with Dulbecco’s PBS (Corning) supplemented with 100 µg/ml CHX. Cells were scraped from plates and resuspended in 400 µl freshly prepared cold lysis buffer containing 10 mM Hepes/KOH, pH 7.4, 2% NP-40 (Sigma-Aldrich), 100 µg/ml CHX, 10 mM MgCl2, 150 mM KCl, 1 mM DTT, protease inhibitor (EDTA-free), and RNAsin inhibitor (Promega). To split polysomes, Dulbecco’s PBSS and lysis buffer were supplemented with 15 mM EDTA. Cells were lysed on ice for 15 min and clarified at 13,000 rpm at 4°C for 10 min, and equal amounts of lysate were layered onto a 10–50% sucrose gradient and spun at 40,000 rpm for 2 h. Gradients were fractionated, and the monosome and polysome content was determined using ImageJ (National Institutes of Health) to measure the total peak areas.
Antibodies
Antibodies against yRps26 and yRps10 were raised in rabbits by New England Peptide. Anti-HA is from Covance. The HRP-conjugated anti-rabbit and anti-mouse secondary antibodies were from Rockland Immunochemicals. Mammalian antibodies were from Sigma-Aldrich (hLtv1), Abgent (hRps10, hRps20), Invitrogen (hRps29), ProteinTech (hRps5), Santa Cruz (RACK1, hRps3), Abcam (GCLC), and Thermo Fisher Scientific (β-actin) and were detected with secondary antibodies (goat anti–mouse and anti–rabbit IgG) conjugated to HRP or near-infrared dyes (LI-COR Biosciences). Blots were visualized using either the Odyssey (LI-COR Biosciences) or the ChemiDoc XRS (Bio-Rad) imaging systems, and band intensity was quantitated using Odyssey or Quantity One software, respectively.
Online supplemental material
Fig. S1 shows growth defects of Rps3 network mutations. Fig. S2 shows the effect of loss of Ltv1 on mutations in Rps26. Fig. S3 shows that glioma and breast cancer cells are differentially affected by Lt1v deletion. Fig. S4 shows that RimM and Ltv1 deletion produces similar changes in the small subunit head. Fig. S5 shows the effect of Enp1 expression on the loss of Ltv1. Table S1 lists yeast strains, Table S2 lists plasmids, and Table S3 lists oligonucleotides used in this study.
Supplementary Material
Acknowledgments
We thank E. Böttger (Universitat Zurich, Zurich, Switzerland) for the mammalian translation reporter plasmids, F. LaRiviere (Washington and Lee University, Lexington, VA) for yeast NRD reporter plasmids, W.R. Roush (Scripps Florida, Jupiter, FL) for the gift of SR3029, and U. Kutay (hLtv1; ETH Zurich, Zurich, Switzerland), M. Seedorf (Rps3; Universitat Heidelberg, Heidelberg, Germany), G. Dieci (Rps8; Universita Degli Studi di Parma, Parma, italy), and A. Link (Asc1; Vanderbilt University, Nashville, TN) for antibodies. We also thank members of the Karbstein laboratory for comments on the manuscript.
This work was supported by National Institutes of Health grants R01-GM117093 (to K. Karbstein) and R01-GM086451-S (to K. Karbstein for J.C. Collins), Department of Defense grant W81XWH-16-1-0009 (to K. Karbstein), University of Florida Brain Cancer grant UFDSP00010793 (to J.R. Doherty), and Howard Hughes Medical Institute Faculty Scholar grant 55108536 (to K. Karbstein). H. Ghalei was supported in part by a Professional Golfer’s Association National Women’s Cancer Awareness Postdoctoral Fellowship.
The authors declare no competing financial interests.
Author contributions: Experiments were designed by H. Ghalei, J.C. Collins, J.R. Doherty, and K. Karbstein. H. Ghalei, J.C. Collins, J.R. Doherty, H. Huang, and R.N. Culver performed and analyzed the experiments. The manuscript was written and edited by J.C. Collins, H. Ghalei, J.R. Doherty, and K. Karbstein, and all authors support its conclusions.
References
- Acker M.G., Kolitz S.E., Mitchell S.F., Nanda J.S., and Lorsch J.R.. 2007. Reconstitution of yeast translation initiation. Methods Enzymol. 430:111–145. 10.1016/S0076-6879(07)30006-2 [DOI] [PubMed] [Google Scholar]
- Adilakshmi T., Bellur D.L., and Woodson S.A.. 2008. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 455:1268–1272. 10.1038/nature07298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajore R., Raiser D., McConkey M., Jöud M., Boidol B., Mar B., Saksena G., Weinstock D.M., Armstrong S., Ellis S.R., et al. . 2017. Deletion of ribosomal protein genes is a common vulnerability in human cancer, especially in concert with TP53 mutations. EMBO Mol. Med. 9:498–507. 10.15252/emmm.201606660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur L., Pavlovic-Djuranovic S., Smith-Koutmou K., Green R., Szczesny P., and Djuranovic S.. 2015. Translational control by lysine-encoding A-rich sequences. Sci. Adv. 1:e15000154 10.1126/sciadv.1500154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burwick N., Shimamura A., and Liu J.M.. 2011. Non-Diamond Blackfan anemia disorders of ribosome function: Shwachman Diamond syndrome and 5q- syndrome. Semin. Hematol. 48:136–143. 10.1053/j.seminhematol.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caliskan N., Katunin V.I., Belardinelli R., Peske F., and Rodnina M.V.. 2014. Programmed -1 frameshifting by kinetic partitioning during impeded translocation. Cell. 157:1619–1631. 10.1016/j.cell.2014.04.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell M.G., and Karbstein K.. 2011. Protein-protein interactions within late pre-40S ribosomes. PLoS One. 6:e16194 10.1371/journal.pone.0016194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. . 2012. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2:401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung Y.N., Maag D., Mitchell S.F., Fekete C.A., Algire M.A., Takacs J.E., Shirokikh N., Pestova T., Lorsch J.R., and Hinnebusch A.G.. 2007. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 21:1217–1230. 10.1101/gad.1528307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clatterbuck Soper S.F., Dator R.P., Limbach P.A., and Woodson S.A.. 2013. In vivo X-ray footprinting of pre-30S ribosomes reveals chaperone-dependent remodeling of late assembly intermediates. Mol. Cell. 52:506–516. 10.1016/j.molcel.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole S.E., LaRiviere F.J., Merrikh C.N., and Moore M.J.. 2009. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol. Cell. 34:440–450. 10.1016/j.molcel.2009.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J.H., Tan Y.Z., Carragher B., Potter C.S., Lyumkis D., and Williamson J.R.. 2016. Modular assembly of the bacterial large ribosomal subunit. Cell. 167:1610–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma M.K., and Parker R.. 2006. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 440:561–564. 10.1038/nature04530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Aitken C.E., Thakur A., Shin B.S., Lorsch J.R., and Hinnebusch A.G.. 2017. Rps3/uS3 promotes mRNA binding at the 40S ribosome entry channel and stabilizes preinitiation complexes at start codons. Proc. Natl. Acad. Sci. USA. 114:E2126–E2135. 10.1073/pnas.1620569114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis S.R., and Lipton J.M.. 2008. Diamond Blackfan anemia: a disorder of red blood cell development. Curr. Top. Dev. Biol. 82:217–241. 10.1016/S0070-2153(07)00008-7 [DOI] [PubMed] [Google Scholar]
- Ferretti M.B., Ghalei H., Ward E.A., Potts E.L., and Karbstein K.. 2017. Rps26 directs mRNA-specific translation by recognition of Kozak sequence elements. Nat. Struct. Mol. Biol. 24:700–707. 10.1038/nsmb.3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. . 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6:pl1 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann Q. 2013. OpenCFU, a new free and open-source software to count cell colonies and other circular objects. PLoS One. 8:e54072 10.1371/journal.pone.0054072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghalei H., Schaub F.X., Doherty J.R., Noguchi Y., Roush W.R., Cleveland J.L., Stroupe M.E., and Karbstein K.. 2015. Hrr25/CK1δ-directed release of Ltv1 from pre-40S ribosomes is necessary for ribosome assembly and cell growth. J. Cell Biol. 208:745–759. 10.1083/jcb.201409056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghalei H., Trepreau J., Collins J.C., Bhaskaran H., Strunk B.S., and Karbstein K.. 2017. The ATPase Fap7 tests the ability to carry out translocation-like conformational changes and releases dim1 during 40S ribosome maturation. Mol. Cell. 67:990–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guimaraes J.C., and Zavolan M.. 2016. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 17:236 10.1186/s13059-016-1104-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C., Nashimoto H., and Nomura M.. 1969. Structure and function of E. coli ribosomes. 8. Cold-sensitive mutants defective in ribosome assembly. Proc. Natl. Acad. Sci. USA. 63:384–391. 10.1073/pnas.63.2.384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harger J.W., and Dinman J.D.. 2003. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 9:1019–1024. 10.1261/rna.5930803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector R.D., Burlacu E., Aitken S., Le Bihan T., Tuijtel M., Zaplatina A., Cook A.G., and Granneman S.. 2014. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res. 42:12138–12154. 10.1093/nar/gku815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer A., Thomson E., Schmidt C., Berninghausen O., Becker T., Hurt E., and Beckmann R.. 2017. Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae. eLife. 6:e30189 10.7554/eLife.30189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeuchi K., and Inada T.. 2016. Ribosome-associated Asc1/RACK1 is required for endonucleolytic cleavage induced by stalled ribosome at the 3′ end of nonstop mRNA. Sci. Rep. 6:28234 10.1038/srep28234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joazeiro C.A.P. 2017. Ribosomal Stalling During Translation: Providing Substrates for Ribosome-Associated Protein Quality Control. Annu. Rev. Cell Dev. Biol. 33:343–368. 10.1146/annurev-cellbio-111315-125249 [DOI] [PubMed] [Google Scholar]
- Johnson M.C., Ghalei H., Doxtader K.A., Karbstein K., and Stroupe M.E.. 2017. Structural Heterogeneity in Pre-40S Ribosomes. Structure. 25:329–340. 10.1016/j.str.2016.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeling K.M., Lanier J., Du M., Salas-Marco J., Gao L., Kaenjak-Angeletti A., and Bedwell D.M.. 2004. Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA. 10:691–703. 10.1261/rna.5147804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter H., Myasnikov A.G., Mastio L., Billas I.M., Birck C., Stella S., and Klaholz B.P.. 2014. Purification, characterization and crystallization of the human 80S ribosome. Nucleic Acids Res. 42:e49 10.1093/nar/gkt1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressler D., Hurt E., and Baßler J.. 2017. A Puzzle of Life: Crafting Ribosomal Subunits. Trends Biochem. Sci. 42:640–654. 10.1016/j.tibs.2017.05.005 [DOI] [PubMed] [Google Scholar]
- Kulkarni S., Dolezal J.M., Wang H., Jackson L., Lu J., Frodey B.P., Dosunmu-Ogunbi A., Li Y., Fromherz M., Kang A., et al. . 2017. Ribosomopathy-like properties of murine and human cancers. PLoS One. 12:e0182705 10.1371/journal.pone.0182705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRiviere F.J., Cole S.E., Ferullo D.J., and Moore M.J.. 2006. A late-acting quality control process for mature eukaryotic rRNAs. Mol. Cell. 24:619–626. 10.1016/j.molcel.2006.10.008 [DOI] [PubMed] [Google Scholar]
- Lee S.K., Yu S.L., Prakash L., and Prakash S.. 2002. Yeast RAD26, a homolog of the human CSB gene, functions independently of nucleotide excision repair and base excision repair in promoting transcription through damaged bases. Mol. Cell. Biol. 22:4383–4389. 10.1128/MCB.22.12.4383-4389.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limoncelli K.A., Merrikh C.N., and Moore M.J.. 2017. ASC1 and RPS3: new actors in 18S nonfunctional rRNA decay. RNA. 23:1946–1960. 10.1261/rna.061671.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loar J.W., Seiser R.M., Sundberg A.E., Sagerson H.J., Ilias N., Zobel-Thropp P., Craig E.A., and Lycan D.E.. 2004. Genetic and biochemical interactions among Yar1, Ltv1 and Rps3 define novel links between environmental stress and ribosome biogenesis in Saccharomyces cerevisiae. Genetics. 168:1877–1889. 10.1534/genetics.104.032656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Mitterer V., Murat G., Réty S., Blaud M., Delbos L., Stanborough T., Bergler H., Leulliot N., Kressler D., and Pertschy B.. 2016. Sequential domain assembly of ribosomal protein S3 drives 40S subunit maturation. Nat. Commun. 7:10336 10.1038/ncomms10336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi N., Duscha S., Boukari H., Meyer M., Xie J., Wei G., Schrepfer T., Roschitzki B., Boettger E.C., and Schacht J.. 2015. XBP1 mitigates aminoglycoside-induced endoplasmic reticulum stress and neuronal cell death. Cell Death Dis. 6:e1763 10.1038/cddis.2015.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passos D.O., Doma M.K., Shoemaker C.J., Muhlrad D., Green R., Weissman J., Hollien J., and Parker R.. 2009. Analysis of Dom34 and its function in no-go decay. Mol. Biol. Cell. 20:3025–3032. 10.1091/mbc.e09-01-0028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña C., Hurt E., and Panse V.G.. 2017. Eukaryotic ribosome assembly, transport and quality control. Nat. Struct. Mol. Biol. 24:689–699. 10.1038/nsmb.3454 [DOI] [PubMed] [Google Scholar]
- Rabl J., Leibundgut M., Ataide S.F., Haag A., and Ban N.. 2011. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 331:730–736. 10.1126/science.1198308 [DOI] [PubMed] [Google Scholar]
- Rachfall N., Schmitt K., Bandau S., Smolinski N., Ehrenreich A., Valerius O., and Braus G.H.. 2013. RACK1/Asc1p, a ribosomal node in cellular signaling. Mol. Cell. Proteomics. 12:87–105. 10.1074/mcp.M112.017277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., and Zhang F.. 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 154:1380–1389. 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg L.H., Lafitte M., Quereda V., Grant W., Chen W., Bibian M., Noguchi Y., Fallahi M., Yang C., Chang J.C., et al. . 2015. Therapeutic targeting of casein kinase 1δ in breast cancer. Sci. Transl. Med. 7:318ra202 10.1126/scitranslmed.aac8773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas-Marco J., and Bedwell D.M.. 2005. Discrimination between defects in elongation fidelity and termination efficiency provides mechanistic insights into translational readthrough. J. Mol. Biol. 348:801–815. 10.1016/j.jmb.2005.03.025 [DOI] [PubMed] [Google Scholar]
- Sanghai Z.A., Miller L., Molloy K.R., Barandun J., Hunziker M., Chaker-Margot M., Wang J., Chait B.T., and Klinge S.. 2018. Modular assembly of the nucleolar pre-60S ribosomal subunit. Nature. 556:126–129. 10.1038/nature26156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaiola A., Peña C., Weisser M., Böhringer D., Leibundgut M., Klingauf-Nerurkar P., Gerhardy S., Panse V.G., and Ban N.. 2018. Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit. EMBO J. 37:e98499 10.15252/embj.201798499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T., Strauss D., Petfalski E., Tollervey D., and Hurt E.. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 22:1370–1380. 10.1093/emboj/cdg121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T., Maco B., Petfalski E., Tollervey D., Böttcher B., Aebi U., and Hurt E.. 2006. Hrr25-dependent phosphorylation state regulates organization of the pre-40S subunit. Nature. 441:651–655. 10.1038/nature04840 [DOI] [PubMed] [Google Scholar]
- Seiser R.M., Sundberg A.E., Wollam B.J., Zobel-Thropp P., Baldwin K., Spector M.D., and Lycan D.E.. 2006. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics. 174:679–691. 10.1534/genetics.106.062117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker C.J., Eyler D.E., and Green R.. 2010. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 330:369–372. 10.1126/science.1192430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms C.L., Yan L.L., and Zaher H.S.. 2017. Ribosome collision is critical for quality control during no-go decay. Mol. Cell. 68:361–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitron C.S., Park J.H., and Brandman O.. 2017. Asc1, Hel2, and Slh1 couple translation arrest to nascent chain degradation. RNA. 23:798–810. 10.1261/rna.060897.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunk B.S., Loucks C.R., Su M., Vashisth H., Cheng S., Schilling J., Brooks C.L. III, Karbstein K., and Skiniotis G.. 2011. Ribosome assembly factors prevent premature translation initiation by 40S assembly intermediates. Science. 333:1449–1453. 10.1126/science.1208245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M.K., Rojas-Duran M.F., Gangaramani P., and Gilbert W.V.. 2016. The ribosomal protein Asc1/RACK1 is required for efficient translation of short mRNAs. eLife. 5:e11154 10.7554/eLife.11154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- True H.L., and Lindquist S.L.. 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 407:477–483. 10.1038/35035005 [DOI] [PubMed] [Google Scholar]
- Tsay Y.F., Thompson J.R., Rotenberg M.O., Larkin J.C., and Woolford J.L. Jr. 1988. Ribosomal protein synthesis is not regulated at the translational level in Saccharomyces cerevisiae: balanced accumulation of ribosomal proteins L16 and rp59 is mediated by turnover of excess protein. Genes Dev. 2:664–676. 10.1101/gad.2.6.664 [DOI] [PubMed] [Google Scholar]
- Vlachos A. 2017. Acquired ribosomopathies in leukemia and solid tumors. Hematology (Am. Soc. Hematol. Educ. Program). 2017:716–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos A., Rosenberg P.S., Atsidaftos E., Alter B.P., and Lipton J.M.. 2012. Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Blood. 119:3815–3819. 10.1182/blood-2011-08-375972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells S.E., Hughes J.M., Igel A.H., and Ares M. Jr. 2000. Use of dimethyl sulfate to probe RNA structure in vivo. Methods Enzymol. 318:479–493. 10.1016/S0076-6879(00)18071-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.