Shima et al. show that there is a positive cooperativity in the binding of kinesin-1 to GDP microtubules. Binding of kinesin-1 triggers conformational changes in GDP microtubules, which then take a GMPCPP-microtubule–like conformation. These effects require microtubule to bind with kinesin in the nucleotide-free state.
Abstract
Kinesin-1, the founding member of the kinesin superfamily of proteins, is known to use only a subset of microtubules for transport in living cells. This biased use of microtubules is proposed as the guidance cue for polarized transport in neurons, but the underlying mechanisms are still poorly understood. Here, we report that kinesin-1 binding changes the microtubule lattice and promotes further kinesin-1 binding. This high-affinity state requires the binding of kinesin-1 in the nucleotide-free state. Microtubules return to the initial low-affinity state by washing out the binding kinesin-1 or by the binding of non-hydrolyzable ATP analogue AMPPNP to kinesin-1. X-ray fiber diffraction, fluorescence speckle microscopy, and second-harmonic generation microscopy, as well as cryo-EM, collectively demonstrated that the binding of nucleotide-free kinesin-1 to GDP microtubules changes the conformation of the GDP microtubule to a conformation resembling the GTP microtubule.
Introduction
Kinesin-1 (KIF5), the founding member of the kinesin superfamily proteins, plays essential roles by transporting various cargoes such as organelles and protein complexes in the cell (Hirokawa et al., 2010). For efficient intracellular transport, each cargo should be transported properly to its cognate destination in the cell. For example, axonal plasma membrane proteins such as amyloid precursor proteins and GAP43 are transported to the axon, but not to dendrites in neurons (Nakata and Hirokawa, 2003). As the molecular basis for this polarized transport, microtubules at the initial segment of the axon were found to have higher affinity to kinesin-1 than other microtubules in the neuronal cell body and dendrites. Application of Taxol interferes with the preferential binding of kinesin-1 to initial segment microtubules as well as polarized transport into the axon, suggesting that microtubules provide the directional cues for axonal transport.
Similar preferential use of subsets of microtubules was also reported in nonneuronal cells (Reed et al., 2006). Constitutively active kinesin-1 often shows asymmetric distribution in various cells when the expression level is comparable to that of authentic kinesin-1 (Fig. S1 A). Subsequent studies showed that this functional specification of cellular microtubules can be transient in immature neurons or nonneuronal cells. In juvenile hippocampal neurons, for example, kinesin-1 shows stochastic cycles of accumulation in and disappearance from the growth cones of different neurites (Jacobson et al., 2006). Similar results are obtained with nonneuronal cells (Fig. S1 B).
Posttranslational modifications such as acetylation or detyrosination of tubulin have been initially proposed as underlying mechanisms, but in vitro experiments with purified microtubules showed that these posttranslational modifications have only slight effects on the affinity or landing rate of kinesin-1 (Kaul et al., 2014). Instead, we have proposed that the conformational switching between the GTP and GDP forms of microtubules is recognized by kinesin-1 (Nakata et al., 2011; Yajima et al., 2012) based on in vitro experiments with purified microtubules and staining with an antibody that is reported to recognize the GTP form of microtubules (Dimitrov et al., 2008). However, this was inconsistent with the widely accepted GTP-cap model of microtubule dynamics (Mitchison and Kirschner, 1984). In this model, GTP is hydrolyzed in the microtubule lattice, and the body of the microtubule is composed of GDP-bound tubulins, while GTP-bound tubulins remain only on the growing ends as the GTP cap. Therefore, the GTP form of microtubules would only be found at the growing plus ends of microtubules as the GTP cap, which cannot explain the presence of GTP-form, stable microtubules in physiological conditions.
To clarify these fundamental issues, we have reexamined the interaction between kinesin-1 and GDP microtubules in vitro and found that there is a positive cooperativity in the binding of kinesin-1 to GDP microtubules. In a series of biophysical studies as well as cryo-EM reconstructions, we show that binding of kinesin-1 triggers allosteric conformational changes in GDP microtubules to a conformation resembling GTP microtubules without nucleotide exchange on tubulin. Based on these results, we propose that the binding of kinesin-1 to the GDP microtubule would increase the affinity by switching the conformation to a GTP-microtubule–like one. This positive feedback mechanism would enable the self-organization of the high-affinity subset of cellular microtubules, which serve as guidance cues for polarized transport.
Results
In vitro motility assay with a substoichiometric KIF5C dimer
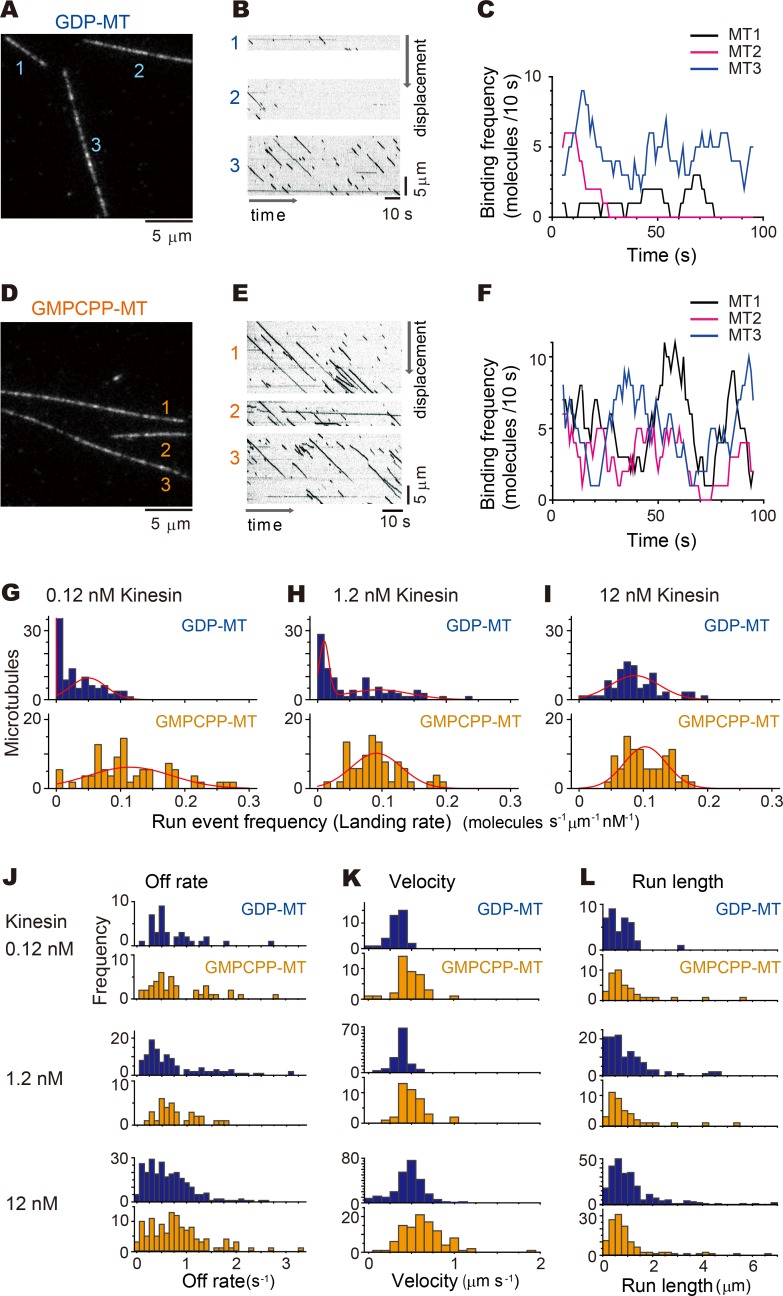
To gain insight into the stochastic and selective use of cellular microtubules by KIF5C, a neuronal isoform of kinesin-1 (Nakata and Hirokawa, 2003; Jacobson et al., 2006; Reed et al., 2006; Nakata et al., 2011; Fig. S1), we have investigated the preferential binding of KIF5C to guanylyl-(α,β)-methylene-diphosphonate (GMPCPP) microtubules over GDP microtubules (Nakata et al., 2011; Morikawa et al., 2015) in quantitative in vitro experiments. In these and the following experiments, GMPCPP (a slowly hydrolyzable GTP analogue) was used to stabilize the GTP conformation of microtubules. For the stabilization of the GDP microtubules, the buffer was supplemented with 30% (vol/vol) glycerol and kept at 37°C to prevent their depolymerization. Throughout these experiments, we noticed a tendency for KIF5C to stochastically and preferentially move along a subset of GDP microtubules when its motility was examined with a concentration of KIF5C much lower (<1 nM) than that of tubulin (∼10 nM). Fig. 1 A shows the experiment with 0.12 nM KIF5C (truncated dimer aa 1–560) and a saturating concentration (1 mM) of ATP. The GDP microtubules are immobilized to the cover glass via biotin–avidin linkage. In this field of view, one microtubule (#2) was first used as the substrate for the motility of KIF5C, but another microtubule (#3) took over. For the next several minutes, only microtubule #3 was frequently used as the track for the motility of KIF5C (Video 1 and Fig. 1, A–C). Similar heterogeneity or preferential binding of kinesin to a subset of microtubules have been previously reported by several groups (Vilfan et al., 2001; Muto et al., 2005), but these studies used Taxol-stabilized microtubules and a much higher concentration of kinesin. In our experimental conditions, however, run events were observed equally among microtubules stabilized with GMPCPP (Fig. 1, D–F).
Figure 1.
Biased use of a subset of GDP microtubules in vitro. (A–F) GDP microtubules (GDP-MT; A–C) or GMPCPP microtubules (GMPCPP-MT; D–F) were anchored by sparsely (three or four sites per micrometer microtubule protofilament) labeled biotin to the avidin-coated glass surface and the dimeric KIF5C moved along the microtubules in the presence of 1 mM ATP. (B and E) Kymographs of KIF5C movement along the microtubules in the presence of 0.12 nM KIF5C. Each kymograph corresponds to the KIF5C movement along the microtubules shown in A and D. Note that brightness of the kymographs is inversed. See also Video 1. (C and F) Time courses of the binding frequency of KIF5C to each microtubule in A and D. (G–I) Histograms of mean KIF5C binding frequency onto GDP (top) and GMPCPP microtubules (bottom) during 100-s incubation with 0.12 nM (G), 1.2 nM (H), and 12 nM (J) KIF5C. GDP microtubules showed lower binding frequency than GMPCPP microtubules under 0.12 nM KIF5C (G; P < 0.00001, Steel–Dwass test). Two-peak distribution with a sharp peak near zero and a broad peak around 0.1 molecule s−1 µm−1 nM−1 was evident with GDP microtubules at lower KIF5C concentrations (G and H; see Tables S1, S2, and S3 for statistical details). The peak around zero disappeared with 12 nM KIF5C (I), and the binding frequency distribution was similar in GDP microtubules and GMPCPP microtubules (I; P = 0.53, Steel–Dwass test). It should be noted that these three KIF5C conditions were examined with the same experimental chambers for GDP microtubules and GMPCPP microtubules, respectively. Thus, the zero peak in GDP microtubules with lower KIF5C concentrations would not be due to collapse of the GDP microtubules. (J–L) Histograms of the off rates (J), velocities (K), and run lengths (L) of the KIF5C dimer on these microtubules. There were no significant differences in these parameters, as summarized in Tables S1, S2, and S3.
This apparent heterogeneity among GDP microtubules was examined by observing more than 50 microtubules. The run events were first observed for 100 s with 0.12 nM or 1.2 nM KIF5C, concentrations much lower than the tubulin dimer concentration (∼10 nM) of the microtubules. There were multiple run events on some microtubules, but no KIF5C run events were observed for ∼40% (0.12 nM) or 10% (1.2 nM) of the microtubules during the 60-s time window of observation (Fig. 1, G and H, top). The concentration of KIF5C was then increased to 12 nM, which was comparable to the concentration of tubulin in the same experimental chamber. Approximately equal numbers of multiple run events were observed on all microtubules (Fig. 1 I, top), suggesting that the heterogeneity was not due to collapse of GDP microtubules during sample preparation or observation. Moreover, this heterogeneity was only observed with GDP microtubules and not with GMPCPP microtubules, even with 0.12 nM KIF5C (Fig. 1, G–I, bottom; and Video 1).
The run event frequency or the landing rate was further analyzed statistically. The distribution of the landing rate was fitted by expectation-maximization algorithm for Gaussian mixture model (Scrucca et al., 2016). When KIF5C was added at a concentration of 12 nM, approximately equal to the concentration of tubulin dimer (Fig. 1 I; see Tables S1, S2, and S3 for statistical details), the landing rate distributed with a single peak around 0.1 molecules per micrometer of microtubule length per second per nanomolar KIF5C (hereafter denoted as molecules s−1 µm−1 nM−1) for both GDP and GMPCPP microtubules. At much lower concentrations of KIF5C (0.12 nM), the distribution of the landing rate showed broader distributions by stochasticity due to the smaller numbers of landing events (Fig. 1 G). The distribution for GMPCPP microtubules was still fitted well to a single Gaussian distribution, but there was a distinct second peak near zero for GDP microtubules. At ∼10 times lower concentration of KIF5C (1.2 nM) than microtubules, the number of landing events was high enough that the two Gaussian distribution is clear for GDP-microtubules (Fig. 1 H) with two peaks around 0.01 and 0.1 molecules s−1 µm−1 nM−1. This two-peak distribution could not be attributed to the stochastic noise, because the landing rate to the GMPCPP-microtubules showed a single peak around 0.1 molecules s−1 µm−1 nM−1. Here, it should be noted that the observed landing rate of 0.01–0.1 molecules s−1 µm−1 nM−1 is ∼100–1,000 times slower than the diffusion-limited encounter rate of ∼10 molecules s−1 µm−1 nM−1 (Hackney, 1995). This implies that only ∼0.1–1% of diffusional collision of kinesin to microtubules leads to a run event.
Other motility-related parameters were also examined from the same dataset, but off rate, velocity, and run length showed no or only slight (within 40%) differences among the experimental conditions (Fig. 1, J–L; and Tables S1, S2, and S3).
Measurement of binding affinity of the KIF5C monomeric motor domain on GDP microtubules
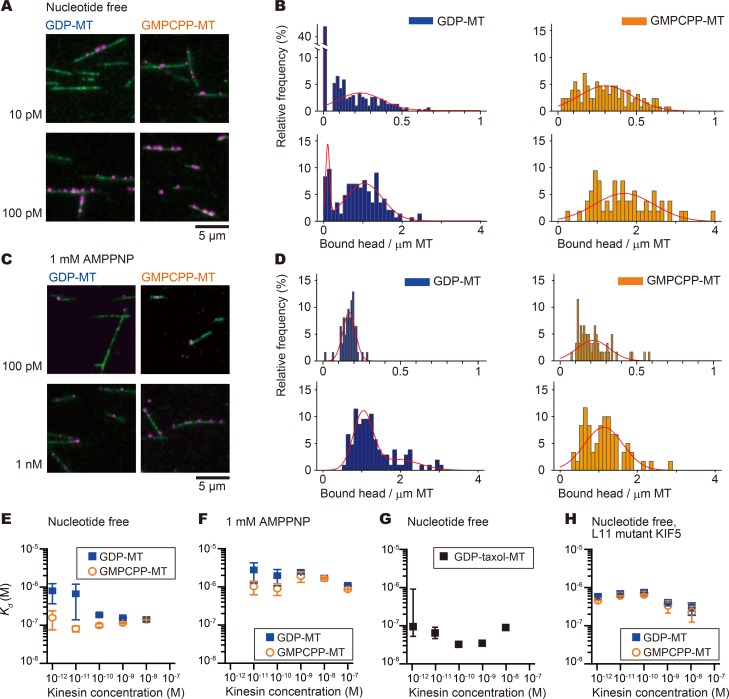
To quantify this apparently cooperative binding of KIF5C to GDP microtubules, we next measured the binding affinity of KIF5C to GDP and GMPCPP microtubules with substoichiometric concentrations of KIF5C. To measure the affinity at 1 nM or below, we used a single-molecule assay. Rhodamine-labeled microtubules were anchored to the surface of the flow cell, to which the monomeric motor domain of KIF5C (K351) fluorescently labeled with DY-647 was introduced with or without nucleotide.
During this experiment, we noticed that the GDP microtubules appeared to be heterogeneously labeled with KIF5C monomers in some conditions. This tendency was statistically tested by fitting the distribution of the density of bound KIF5C spots on each microtubule by using the expectation-maximization algorithm for Gaussian mixture model (Scrucca et al., 2016). With limited concentrations (10 and 100 pM) of KIF5C in a nucleotide-free state, there were two populations of GDP microtubules. Some were not bound with KIF5C, but others were bound with multiple KIF5C molecules. GMPCPP microtubules were more uniformly labeled in the same conditions (Fig. 2, A and B). This heterogeneity or the two-peak distribution was reversed in the presence of adenylyl-imidodiphosphate (AMPPNP), a non-hydrolysable ATP analogue (Fig. 2, C and D). These results are apparently consistent with the single motor motility assay (Fig. 1).
Figure 2.
Cooperative binding of KIF5C to GDP microtubules. (A–D) The binding affinity of the KIF5C motor domain (monomer) to GDP or GMPCPP microtubules (MTs) was examined using single-molecule assays in the absence of nucleotide (A and B) or in the presence of 1 mM AMPPNP (C and D). (A and C) Examples of the raw results. Each image shows the KIF5C motor domain (magenta) bound on GDP microtubules (left columns) or GMPCPP microtubules (right columns) in the absence of nucleotide (A) or in the presence of 1 mM AMPPNP (C). The concentration of the kinesin used for each experiment is shown on the left side of the columns. (B and D) Distribution of the density of bound KIF5C on the microtubules. The results were fitted with a mixed Gaussian model to examine the heterogeneity in binding affinity. Two-peak distribution was evident with nucleotide-free KIF5C and GDP microtubules (B, left). See Tables S4, S5, and S6 for details. (E–H) Kd Dissociation constants of KIF5C. Affinity (equilibrium Kd) plotted against the concentration of KIF5C monomeric motor domain. Error bars indicate SEM (see Materials and methods for details). (E–G) Affinity of the wild-type KIF5C motor domain to GDP, GMPCPP, or Taxol-stabilized GDP microtubules. The nucleotide conditions for kinesin and the microtubule conditions are indicated in each panel. (H) Affinity of the L11 mutant KIF5C (K11C, the L11-α4 junction was replaced with the corresponding sequence of KIF1A). This mutant showed constant affinity to both GDP and GMPCPP microtubules, which is consistent with our previous report that this mutant cannot discriminate axonal microtubules from dendritic microtubules in neurons (Nakata et al., 2011). The numbers of the observed microtubules and bound KIF5 molecule are summarized in Tables S4, S5, and S6.
Next, we converted this binding frequency data to the affinity (dissociation constant [Kd]) from the number of KIF5C spots on the microtubules and the total length of the microtubules in the view field. As we have reported previously (Nakata et al., 2011; Morikawa et al., 2015), KIF5C showed approximately fivefold higher affinity to GMPCPP microtubules than to GDP microtubules. This was only seen when the concentration of KIF5C was low (1–10 pM) and in the absence of nucleotides (Fig. 2 E, blue markers). By increasing the KIF5C concentration from 1 pM to 10 nM, the Kd value decreased steeply at ∼100 pM. The Hill constant of this change was 1.8 ± 0.1 (mean ± SEM), suggesting positive cooperativity as the underlying mechanism. Contrastingly, the Kd values were insensitive to the KIF5C concentrations with GMPCPP microtubules (Fig. 2 E, orange markers) and in the presence of a saturating concentration of AMPPNP (Fig. 2 F).
Furthermore, the application of Taxol increased the affinity even at low concentration of KIF5C, and the dependence to the KIF5C concentrations was reversed (Fig. 2 G), which is consistent with the previous in vivo results (Nakata and Hirokawa, 2003). In the following assays with neurons (Nakata et al., 2011), mutations at the L11-α4 junction reversed the preferential movement of KIF5C on axonal microtubules over dendritic microtubules. Consistently, the same mutation decreased the affinity to GMPCPP microtubules, and the dependence on the KIF5C concentrations was lost for GDP microtubules (Fig. 2 H).
Binding of KIF5C transiently converts GDP microtubules to a high-affinity state
From these results, we surmised a hypothesis that GDP microtubules can take two states: a low-affinity state and a high-affinity state. The binding of KIF5C would convert the microtubule from the low-affinity state to the high-affinity state. At limited KIF5C concentrations, only some microtubules might be converted to the high-affinity state, but all microtubules will be converted with sufficiently high concentrations of KIF5C.
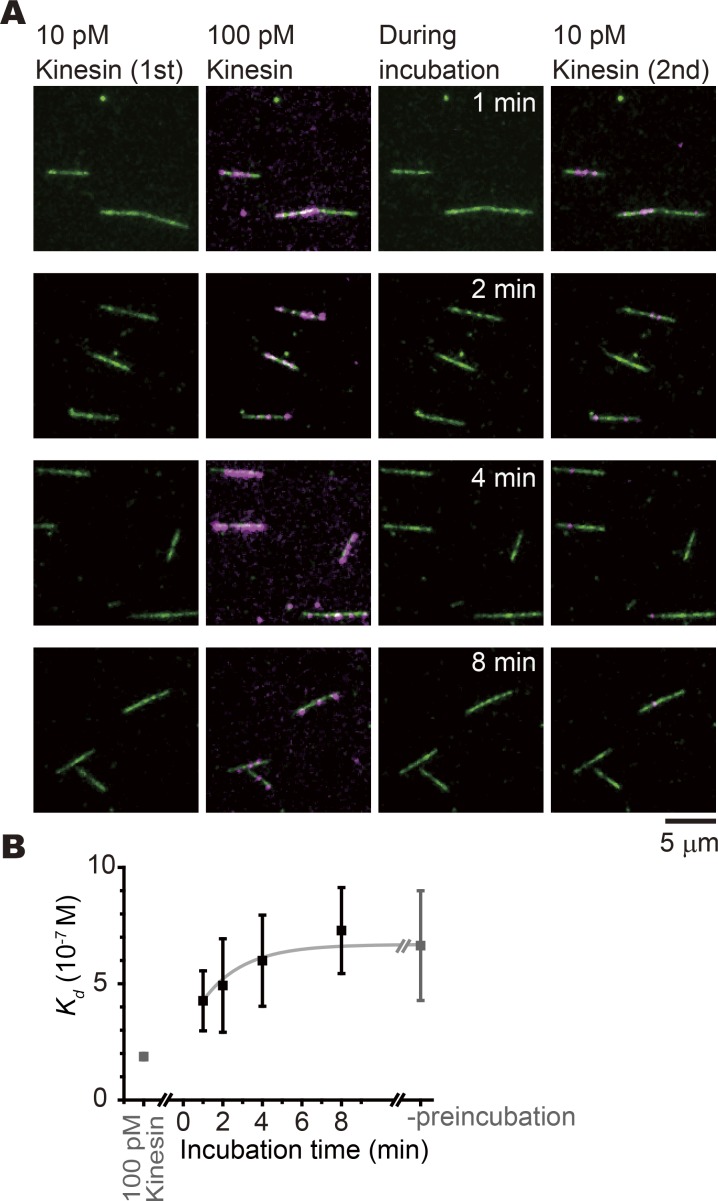
We tested this hypothesis using the following experiment (Fig. 3 A). GDP microtubules were first attached to the surface of the flow cell. Then, KIF5C (monomer, without nucleotide) was added at 10 pM. Most microtubules are not associated with KIF5C at this concentration. The concentration of the KIF5C monomer was further increased to 100 pM so that multiple KIF5C molecules were bound to most of the microtubules. The bound KIF5C molecules were then washed out by flushing with 10 mM ATP and 500 mM K-Pipes. After incubating with the observation buffer (45 mM K-Pipes) for 1–8 min, the KIF5C monomer (without a nucleotide) was reintroduced at 10 pM.
Figure 3.
Transient increase of binding affinity of GDP microtubules by pretreatment with KIF5C. (A) Images of the KIF5C (magenta) motor domain and GDP microtubules (green) in each step of the experiment. First, GDP microtubules were incubated with 10 pM KIF5C in the absence of nucleotide (left column). Following incubation with 100 pM KIF5C (second left column), KIF5C was washed out from the observation chamber using 0.5 M Pipes buffer, and microtubules were incubated in the observation solution (second right column). After the incubation time indicated left side of the images, 10 pM KIF5C was reintroduced in the chamber (right column). (B) Incubation time dependency of the Kd of KIF5C and GDP microtubule after temporal binding of 100 pM KIF5C. Data points are shown in black. Kd values before incubation with 100 pM kinesin and during the incubation with 100 pM kinesin are shown in gray for comparison. The time constant for the decay was estimated to be 2.1 ± 1.1 min (mean ± SEM) by fitting with an exponential decay (gray curve). Nmicrotubules are 35, 43, 28, and 34, and Nbound are 74, 51, 20, and 11 for the data points for 1, 2, 4, and 8 min, respectively.
When 10 pM KIF5C was reintroduced immediately after the washout, many KIF5C signals were found on GDP-microtubules, confirming that pretreatment with higher concentration of KIF5C increased the affinity of GDP microtubules. However, this effect was transient and lasted only a few minutes. After an 8-min incubation, the affinity decayed to the initial level (Fig. 3 B).
Binding of KIF5C elongates the axial pitch of the GDP-microtubules
These results collectively suggest a positive cooperativity in the binding of KIF5C to GDP microtubules, and this positive cooperativity would be mediated by the transient changes in microtubules. How can KIF5C binding convert GDP microtubules to the high-affinity state?
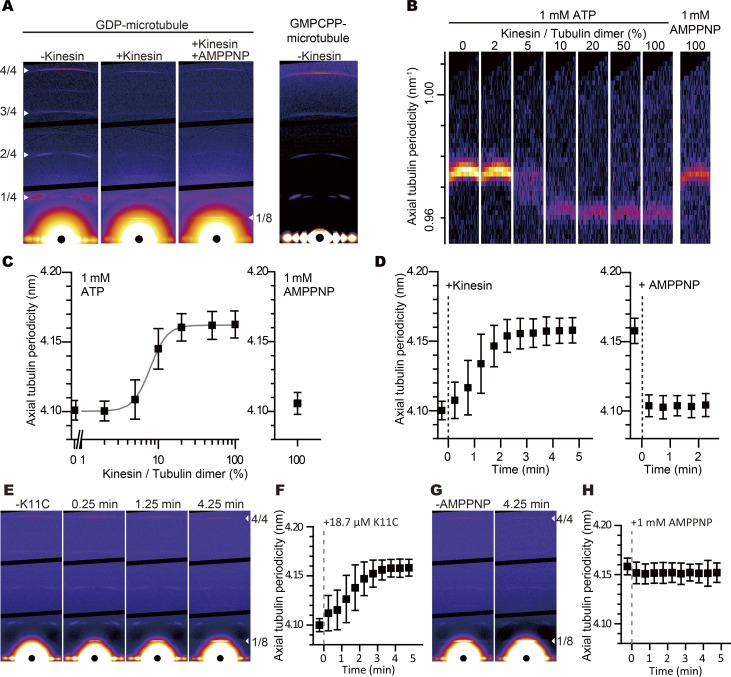
Since the high-affinity-state GDP microtubule and GMPCPP microtubules seem to have similar Kd values, we surmised that binding of KIF5C might change the microtubule lattice from the GDP conformation to a conformation similar to a GMPCPP microtubule. To examine this hypothesis, we first focused on the axial pitch of the microtubule, because it is well established that GMPCPP microtubules have a 2% longer axial pitch (4.2 nm/monomer) than GDP microtubules (4.1 nm/monomer; Hyman et al., 1995; Yajima et al., 2012; Alushin et al., 2014; Kamimura et al., 2016). We, therefore, examined the axial pitch of KIF5C-decorated GDP microtubules with x-ray fiber diffraction, since it gives accurate values for the axial pitch of microtubules in solution (<0.2% error; Kamimura et al., 2016).
GDP microtubules were aligned by the shear flow device (Kamimura et al., 2016), and the fiber diffraction pattern was recorded by irradiating the sample with an x-ray beam. Among the layer lines that reflect the ∼4-nm axial tubulin repeat in the microtubule, the stronger fourth-order reflection at ∼1-nm axial spacing (Kamimura et al., 2016) was used for the measurement of axial pitch (Fig. 4 A).
Figure 4.
Elongation of the axial pitch of GDP microtubule measured by x-ray fiber diffraction. (A) X-ray fiber diffraction patterns of GDP microtubules (94 µM) without KIF5C, with 94 µM KIF5C, and with 94 µM KIF5C plus 1 mM AMPPNP and those of GMPCPP microtubules without KIF5C (from left to right panels). The bottom black circle in each panel is an x-ray beam stop placed around the image center. The arrowheads on the left side of the first panel indicate the first to fourth diffraction peaks (from bottom to top of the image) corresponding the axial tubulin periodicity (∼4 nm). The arrowhead on the right side of the third panel indicates the first diffraction peak corresponding to the bound kinesin periodicity (∼8 nm). Without KIF5C, the diffraction peaks of both the GDP and GMPCPP microtubules fit well with the previously reported tubulin axial periodicity (4.1 nm for GDP microtubules and 4.2 nm for GMPCPP microtubules). (B) The images of the fourth diffraction peak of GDP microtubules. As the KIF5C concentration increased, the peak shifted toward the center; however, the peak returned to the initial position by addition of 1 mM AMPPNP. (C) The relation of KIF5C concentration and axial tubulin periodicity of GDP microtubules in the presence of 1 mM ATP and 1 mM AMPPNP. The Hill coefficient of this reaction was estimated to be 6.4 ± 1.5 by fitting the data to Hill equation (gray line). (D) Time course of the axial tubulin periodicity after adding 19 µM KIF5C to 94 µM GDP microtubule (the microtubule concentration is expressed as tubulin dimer concentration) and 1 mM AMPPNP. (C and D) Error bars indicate the diffraction peak FWHM. Among the four individual trials, the SDs of the axial tubulin periodicity of each data point were below 0.1 Å. (E and F) The time course of the layer line positions after the addition of mutant KIF5C (K11C, the L11-α4 junction was replaced with the corresponding sequence of KIF1A). K11C was added at 18.7 µM, which corresponds to the 20% (molar ratio) of tubulin in the assay chamber. The lattice of the GDP microtubule was elongated both in magnitude and duration similarly to wild-type KIF5C (Fig. 4 D). Error bars indicate the diffraction peak FWHM. SD of the three independent experiments was below 0.1 Å. (G and H) The time course after the addition of excess (1 mM) AMPPNP to the K11C–GDP microtubule complex. The microtubule pitch was not shortened. These results suggest that the L11-α4 junction would be involved in the ATP-dependent shortening of the microtubule lattice, but not in the elongation of the microtubule lattice upon binding of KIF5C. Error bars represent FWHM. SD of three individual experiments was below 0.1 Å. Note the presence of the peak at 8 nm−1 (arrowheads at 1/8 in E and G), which demonstrates the binding of K11C to the microtubules in these experiments.
As shown in Fig. 4 B, the layer line shifted inwardly by the addition of KIF5C (the monomeric motor domain). Before the addition of KIF5C, the axial pitch was 4.10 ± 0.006 nm (mean ± SEM; Fig. 4 C). The addition of KIF5C to the same sample to the concentration of 2% of tubulin (molar ratio) did not change the pitch. When the amount of KIF5C was increased to 5%, the layer line broadened. After the amount of KIF5C was increased to 10% or more, the layer line sharpened again, and the axial pitch was elongated to 4.16 ± 0.008 nm. Finally, AMPPNP was added to the same sample after increasing the KIF5C amount to 100%. Then, the layer line returned to the initial position (4.10 nm). Here, the 8 nm-layer line remains (layer lines 1/8 in Fig. 4 A). This layer line is from KIF5C on each tubulin dimer of the microtubule, indicating that KIF5C remains bound to microtubules after the addition of AMPPNP. It should be noted here that the presence of KIF5C does not affect this measurement, because reflections from KIF5C should match those from the tubulin dimer. The eighth-order reflection of the ∼8-nm tubulin dimer periodicity should overlap with the fourth-order reflection of the tubulin monomer repeat. Furthermore, the 10-fold increase in the amount of KIF5C on the microtubule (from 10% to 100%) did not change the layer line position. This result experimentally confirms that the presence of KIF5C on the microtubule did not affect the measurement.
The shorter pitch of GDP microtubules without KIF5C is in agreement with our previous results with GDP microtubules with a substoichiometric concentration of Taxol measured using the same method (Kamimura et al., 2016). The longer pitch with KIF5C is closer to the pitch of the GMPCPP microtubules measured at the same temperature (37°C). Thus, the results of x-ray fiber diffraction are consistent with the hypothesis that KIF5C-bound GDP microtubules take a conformation similar to GMPCPP microtubules.
Cooperativity in axial pitch elongation and its time course
It should also be noted that the changes in the layer line (Fig. 4, B and C) are very similar to the results of the affinity measurement in Fig. 2. The layer line broadens at ∼5%, which suggests mixed populations with different pitches. The shift of the layer line at ∼5% KIF5C is steep, with a Hill coefficient of 6.4 ± 1.5 (mean ± SEM). Furthermore, the addition of AMPPNP to the sample with 100% KIF5C returned the layer line to the initial position. These results collectively suggest that a substoichiometric amount of bound KIF5C can cooperatively change the conformation of GDP microtubules, but KIF5C in the AMPPNP-bound state would reverse the conformational changes.
We also measured the time course of the response. As shown in Fig. 4 D, it took ∼2 min for elongation by KIF5C, which could reflect the slow kinetics of the conformational changes in the microtubule, because the binding kinetics of KIF5C is much faster as judged from the 8-nm layer line. In contrast, the addition of AMPPNP to bound KIF5C returned the layer line position within 30 s. This kinetics was much faster than the reversal of the microtubule affinity after KIF5C washout experiments in Fig. 3. However, the corresponding measurements were difficult with this x-ray fiber diffraction experiment, because it requires rapid separation of KIF5C from the floating microtubules in the rotating flow chamber.
Here, the L11-α4 junction mutant KIF5C did induce microtubule elongation upon binding (Fig. 4, E and F), but the pitch was not returned by the addition of AMPPNP (Fig. 4, G and H). These results suggest that the L11-α4 junction would be involved in the ATP-dependent shortening of the microtubule lattice, but not in the elongation of the microtubule lattice upon binding of KIF5C. The structural basis for this observation is discussed in the cryo-EM section.
Single-molecule measurement of the axial elongation of microtubules
The measurements presented in Figs. 1 and 2 are in the regimes where each single microtubule was measured, while the x-ray diffraction represents the average of millions of microtubules in the x-ray spot. Therefore, we further confirmed the axial elongation of microtubules at the single microtubule level using fluorescent speckle microscopy. Each microtubule was polymerized from a small seed of surface-stabilized microtubules and sparsely labeled with DY-647 fluorescent dye. The labeling density with DY-647 was titrated so that the average distance between the nearest DY-647 speckles was ∼3 µm. Although the microtubules were attached to the cover glass only at the small seed regions near their minus ends, the high viscosity of solution containing 0.5% methylcellulose suppressed Brownian movement of microtubules, so that the positions of the DY-647–labeled speckles were precisely measured by Gaussian fitting of the signal with a standard error of ∼10 nm. Thus, the length of the microtubule was measured as the distance between the speckles with the precision of 10 nm/3 µm = 0.3%.
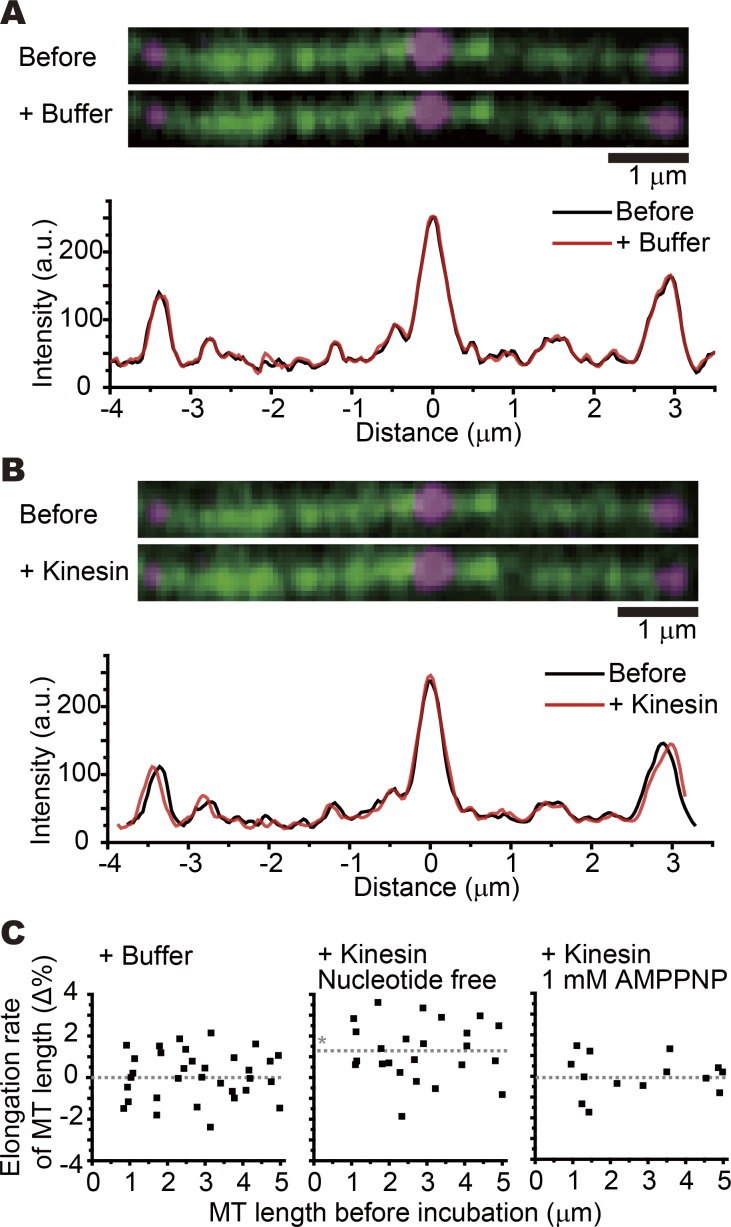
Fig. 5 A shows an example of the control experiment. The positions of the three speckles overlapped completely after buffer exchange. The microtubule was then incubated with KIF5C without nucleotide (Fig. 5 B). When the position of the central speckle was aligned, the left speckle shifted toward the left and the right speckle shifted toward the right, indicating the elongation of the microtubule. Fig. 5 C shows the summary of the experiments. To avoid biases or errors, speckle pairs were chosen so that the distances distributed equally between 1 and 5 µm. The vertical axis shows the relative elongation of the speckle distance (ΔL/L in percent). Consistent with the results from x-ray fiber diffraction, the distance between two speckles was elongated 1.3% from the initial distance by addition of the KIF5C motor domain (monomer) in the absence of a nucleotide (P < 0.01, Steel–Dwass test). On the contrary, the distance did not change significantly in the presence of 1 mM AMPPNP. These results confirm that the shift in the layer line position in the x-ray fiber diffraction experiment reflects the elongation of the axial pitch of the microtubule by the binding of KIF5C.
Figure 5.
Single-molecule measurement of the axial elongation of microtubules. Fluorescent speckle microscopy of GDP microtubules showed that KIF5C elongates the microtubule length. Tetramethylrhodamine-labeled microtubules were polymerized from the DY-647–labeled seeds, which were anchored on a glass surface via biotin-avidin linkage. The polymerized microtubules were sparsely labeled with DY-647. (A) Images of the microtubule (green) with the DY-647 speckles (magenta) before (top) and after (middle) introducing buffer in the chamber. The bottom graph shows the fluorescent intensity of DY-647 along the microtubule before (black line) and after (red line) introducing buffer in the chamber. (B) Images of the microtubule (green) with the DY-647 speckles (magenta) before (top) and after (middle) introducing KIF5C in the chamber in the absence of ATP. The bottom graph shows the fluorescent intensity of DY-647 along the microtubule before (black line) and after (red line) introducing KIF5C in the chamber. Bars, 1 µm (A and B). (C) The elongation rates of the microtubule (MT) on introducing buffer (left, Nmicrotubules = 33), KIF5C without nucleotide (middle, Nmicrotubules = 24), and KIF5C with 1 mM AMPPNP (right, Nmicrotubules = 14). The gray dotted lines indicate the mean elongation rate. The asterisk indicates significant difference (P < 0.01, Steel–Dwass test). See also Fig. S2 for the hyperstabilization of GDP microtubules by KIF5C and Fig. S3 for the reversal of the microtubule elongation after removal of KIF5C.
We next examined the shortening (recovery) of microtubule length after washing out KIF5C. The minus end of the GDP microtubule was attached to the cover glass. After incubating with KIF5C (nucleotide-free state), a constant flow of buffer containing 300 mM K-Pipes, 40% (vol/vol) glycerol, and 1 mM Mg-ATP was applied to wash out the KIF5C motor domain. KIF5C was completely removed from the microtubule within 10 s (Fig. S3 A), while the shortening of the microtubule length was delayed by 30–150 s (Fig. S3 B). This result suggests that GDP microtubules can keep the KIF5C-induced elongated form on the order of minutes after KIF5C dissociation.
While preparing this experiment, we also noted that the stability of the GDP microtubules was strongly affected by the binding of KIF5C, which is consistent with previous observations in HeLa cells and fungi (Marceiller et al., 2005; Uchida et al., 2008). The GDP microtubule is known to depolymerize rapidly, especially when free tubulin is washed out from the solution and in the absence of glycerol or DMSO. However, we found that the binding of KIF5C slowed depolymerization (Fig. S2 and Video 2). We measured the depolymerization rate as a function of the relative concentration of KIF5C against the tubulin dimer in the microtubule. The depolymerization rate showed a steep (100-fold) decrease at ∼10% saturation with KIF5C in the absence of a nucleotide. In contrast, the depolymerization rate showed only 10-fold decrease, even with full saturation with KIF5C, in the presence of AMPPNP. Thus, the 100-fold stabilization of the GDP microtubule could not have been caused by physical interference from the presence of bound KIF5C on the microtubule surface, but it could have been caused by stabilization of the microtubule itself due to conformational changes in the microtubule lattice induced by the binding of nucleotide-free KIF5C. It should be noted here that the depolymerization rate also changes steeply with nucleotide-free KIF5C at ∼10% saturation with KIF5C in the absence of a nucleotide, which is similar to the cooperative effects observed with affinity switching and axial pitch elongation.
Conformational changes in GDP microtubules by second-harmonic generation (SHG) microscopy
The above results collectively suggest that the conformation of the GDP microtubules changes cooperatively with the binding of nucleotide KIF5C at ∼10% saturation. X-ray fiber diffraction and speckle microscopy only demonstrated the elongation of the axial pitch, and it is not clear whether this elongation is accompanied by conformational changes in the tubulin subunits. Therefore, we examined GDP microtubules using SHG microscopy.
The nonlinear susceptibility for SHG (χ) is represented by a third-rank tensor, which reflects the structure of the molecule (Psilodimitrakopoulos et al., 2013; Kaneshiro et al., 2018 Preprint). For a cylindrically symmetric polymer such as a microtubule, most elements of the SHG tensor vanish, and the nonzero elements are all expressed with two independent components, χzxx and χzzz, where z and x are defined as the directions parallel and perpendicular to the microtubule axis, respectively (Fig. 6 A). The two components are related to the mean tilt angle (φ) of tubulin subunits from the microtubule axis (z axis) as
and
where βvvv is the magnitude of the first hyperpolarizability of a tubulin subunit and K is a proportional constant that contains the net molecular number in the focal volume. Hence, the ratio of χzxx to χzzz (hereinafter denoted by ρ) reflects the mean tilt angle of tubulin subunits in the microtubule lattice as
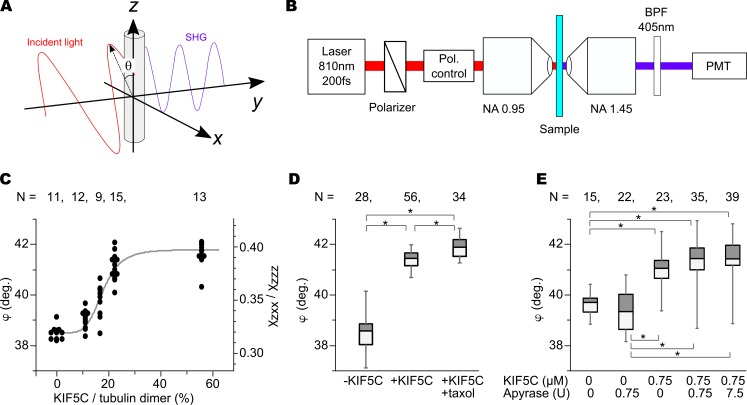
Here, this ratio is sensitive to the angular changes in the tilt angle of the tubulin subunits, but not to the axial pitch. Thus, it gives essentially different information regarding the conformational changes in the microtubule lattice. Here, the ratio ρ can be measured from the polarization dependence of SHG signal intensity by changing the polarization angle θ of incident light (Fig. 6 A) with the microscope optics shown in Fig. 6 B, as reported previously (Psilodimitrakopoulos et al., 2013).
Figure 6.
Microtubule conformational changes examined with SHG. (A) Principle of the measurement. Incident polarization θ dependence of SHG signal reflects the orientation of tubulin subunits in the microtubule. (B) Schematic drawing of the SHG microscope. BPF, bandpass filter; PMT, photomultiplier tube. (C) The ratio of the SHG polarization angle against the concentration of KIF5C motor domain. Parallel GDP microtubule bundles grown from the mitotic asters without Taxol were measured. The ratio of the polarization factor (right vertical axes) reflects the tilt angle (φ) of the dipole moment of the tubulin subunit in the microtubule lattice (left vertical axes). (D) The box-whisker plot of the SHG signal for GDP microtubules without KIF5C and Taxol, with KIF5C, and with both KIF5C and Taxol. The boxes show the first and third quadrants, and the bars in the boxes indicates the median of each dataset. The bottom and top whiskers in each plot indicate minimum and maximum values of a dataset. (E) The effect of apyrase on the SHG signal. Addition of apyrase did not affect the tubulin dipole angle of the GDP-microtubules, suggesting that shift in the SHG signals is induced by the nucleotide free KIF5C. (D and E) P values were calculated using a Steel–Dwass test. *, P < 0.001. (C–E) The numbers of tested samples are shown in the figures.
Fig. 6 C shows the dependence of ρ (right axis) and φ (left axis) on the relative molar concentration of KIF5C to the tubulin dimer. Each dot represents the experimental data value, and the solid line is the theoretical curve obtained by least-squares fitting to a modified Hill equation:
where b and c are constants corresponding to intercept and scaling factor. The parameters n and a, found to be 5.1 and 0.177, respectively, were strongly suggestive of positive cooperativity of binding of kinesin. As is shown, ρ rapidly increased at low concentrations (10–20%) of the KIF5C motor domain and no longer increased at higher concentrations (>30%). This indicates that conformational change does not linearly progress with increasing kinesin concentrations but starts to occur at a specific concentration region. Furthermore, this also indicates that the polarization changes in the SHG signal was most likely caused by the conformational changes in microtubules. KIF5C might not contribute much to the SHG signal, because the increase of KIF5C on the microtubule from 22% to 56% did not strongly affect the SHG signal (Fig. 6 C). This is the opposite of the actomyosin system, where SHG signals mostly come from the myosin head (Plotnikov et al., 2006).
We also compared the tilt angle φ of GDP–KIF5C and GDP–KIF5C–Taxol microtubules with undecorated GDP microtubules. The relative concentration of KIF5C to tubulin dimer was set to be higher than 50%. The result is shown in Fig. 6 D, where the left, middle, and right boxes are GDP, GDP–KIF5C, and GDP–KIF5C–Taxol microtubules, respectively. The mean values of φ for GDP, GDP–KIF5C, and GDP–KIF5C–Taxol were estimated to be 38.5°, 41.2°, and 41.6°, respectively. Using the Steel–Dwass test, we confirmed significance between GDP and GDP–KIF5C and between GDP–KIF5C and GDP–KIF5C–Taxol, as shown in the graph. This means that those microtubules states might have different conformations associated with the tilt of the dipole moment in a tubulin subunit. It should be noted that the change of φ may contain the tilt of the tubulin subunit not only in the direction normal to the microtubule surface but also in the tangential direction. We further assessed the possible effect of small amounts of ATP/ADP contamination in the assay system by adding apyrase, which catalytically removes nucleotide tri- or diphosphates (Fig. 6 E). The addition of apyrase did not affect tubulin dipole angles, suggesting that apo-KIF5C (KIF5C without bound nucleotide) indeed changes the tubulin dipole in GDP microtubules.
Conformational variations of microtubules by cryo-EM analysis
All the above experiments collectively support the idea that the binding of nucleotide-free KIF5C triggers cooperative conformational changes in GDP microtubules, while AMPPNP-bound KIF5C does not.
To confirm this idea directly and to elucidate the underlying atomic mechanisms, we compared the conformations of microtubules using cryo-EM. Using a seam search protocol developed by Zhang and Nogales (2015), we examined the cryo-EM structures of GDP microtubules (MT(GDP)) complexed with KIF5C in the various nucleotide states: KIF5C(ø)–MT(GDP), KIF5C(ATP)–MT(GDP), and KIF5C(PNP)–MT(GDP). The GMPCPP microtubule (MT(GMPCPP)) complexed with apo-KIF5C (KIF5C(ø)–MT(GMPCPP)) was also visualized for comparison with a previous study (Alushin et al., 2014). These reconstructions were performed at ∼6 Å overall resolution, and the average resolution in the tubulin region was better than 5 Å (Figs. 7 A and S4).
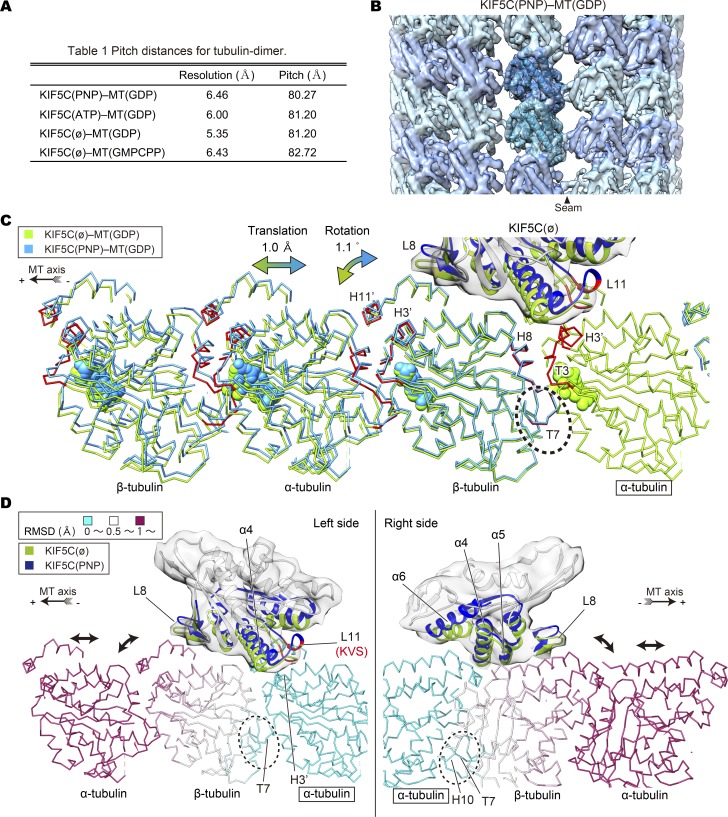
Figure 7.
Microtubule conformational changes examined with cryo-EM. (A) The estimated resolutions according to the Fourier shell correlation 0.143 criteria, and pitch distances for tubulin dimer calculated from the helical symmetry parameters. (B) Cryo-EM reconstruction of KIF5C(PNP)–MT(GDP) and the docked atomic model showing only the microtubule surface. (C) Cα chain trace models of KIF5C(PNP)–MT(GDP) and KIF5C(ø)–MT(GDP) superimposed at α-tubulin on the right end. Note the tilt angle changes (1.1°) at the intradimer interface, which leads to the extension of the pitch (1.0 Å). Tubulin subunits and KIF5C in KIF5C(PNP)–MT(GDP) are shown in cyan and blue, and those in KIF5C(ø)–MT(GDP) are shown in green. Helices and loops involved in the rearrangement of lattice are highlighted in red. (D) The Cα chain trace model of KIF5C(ø)–MT(GDP) colored by root mean square deviation values between the Cα atoms in C, viewing from the left side and the right side. The interface elements of KIF5C were highlighted as in C. See Fig. S4 for resolution and the statistics of cryo-EM maps.
According to the helical parameters iteratively refined and measured from asymmetric (C1) reconstruction, microtubules adopt at least three distinct conformations (Fig. 7 A). The axial periodicity or the tubulin-dimer length was shortest in KIF5C(PNP)–MT(GDP). It was 0.9 Å or 1% longer in both KIF5C(ø)–MT(GDP) and KIF5C(ATP)–MT(GDP), which quantitatively agrees with the results of the above experiments (Figs. 4 and 5). KIF5C(ø)–MT(GMPCPP) was two times longer (i.e., 2 Å longer) than KIF5C(PNP)–MT(GDP), as reported previously (Alushin et al., 2014). Slight discrepancies in the absolute values of the axial pitch between x-ray fiber diffraction and cryo-EM might be caused by the effect of the shear flow during x-ray fiber diffraction measurements or inaccuracies in cryo-EM magnification.
Mechanism of conformational changes in microtubule by KIF5C
To investigate the atomic mechanisms of the conformational changes by KIF5C binding, the atomic models of α- and β-tubulin monomers (Protein Data Bank accession no. 3J6F) were overlaid by rigid-body fitting (Fig. 7 B). The fitting was good for all reconstructions with high cross correlation coefficients (>0.96). We also docked KIF5C atomic models (Protein Data Bank accession nos. 4HNA and 4LNU) to our density maps by rigid-body fitting. We then superimposed KIF5C(ø)–MT(GDP) and KIF5C(PNP)–MT(GDP) by using one α-tubulin subunit as the reference point (Fig. 7, C and D; Fig. 8 A; and Video 3). This representation clearly visualized the relative movement of tubulin subunits in the microtubule lattice. β-Tubulin slightly rotates toward the plus-end side around the T7 loop, so that the helices H11′ and H3′ in the KIF5C(ø)–MT(GDP) approach the neighboring α-tubulin at the plus-end side (Fig. 7 C). As a consequence, the neighboring α-tubulin is pushed toward the plus-end side by 1.0 Å, leading to elongation of the tubulin-dimer periodicity. The conformation of KIF5C(ATP)–MT(GDP) was essentially the same as KIF5C(ø)–MT(GDP) (Fig. 8, B and C; and Video 4). For KIF5C(ø)–MT(GMPCPP), the tubulin subunits moved similarly, but two times greater in angle and two times farther in distance, than those in KIF5C(ø)–MT(GDP) (Fig. 8 D and Video 5). Thus, both KIF5C binding and GTP binding to β-tubulin induced similar conformational changes in the microtubule lattice but by twofold different magnitudes. These observations are perfectly consistent with the results of tilt-angle changes detected by SHG measurements (Fig. 6).
Figure 8.
Conformational changes among KIF5C–microtubule complexes. (A–D) Comparisons of positions of the tubulin monomers superimposed at α-tubulin on the right end. The translation vectors between Cα coordinate pairs are shown in black. The Cα chain trace model in each panel is KIF5C(PNP)–MT(GDP) (A, blue), KIF5C(ATP)–MT(GDP) (B, pink), KIF5C(ø)–MT(GDP) (C, green), and KIF5C(ø)–MT(GMPCPP) (D, orange). (C) KIF5C(ø)–MT(GDP) and KIF5C(ATP)–MT(GDP) adopt similar lattices. Movements of C domains are measured. (E) A GTP-like, high-affinity tubulin dimer will clash with the surrounding GDP-conformation tubulins. This shows that small amounts of KIF5C below the threshold cannot induce a conformational rearrangement of the GTP-like high-affinity microtubule. Blue, KIF5C(PNP)–MT(GDP); magenta, KIF5C(ø)–MT(GDP). Interatomic clashes and contacts were detected and colored in red using Chimera.
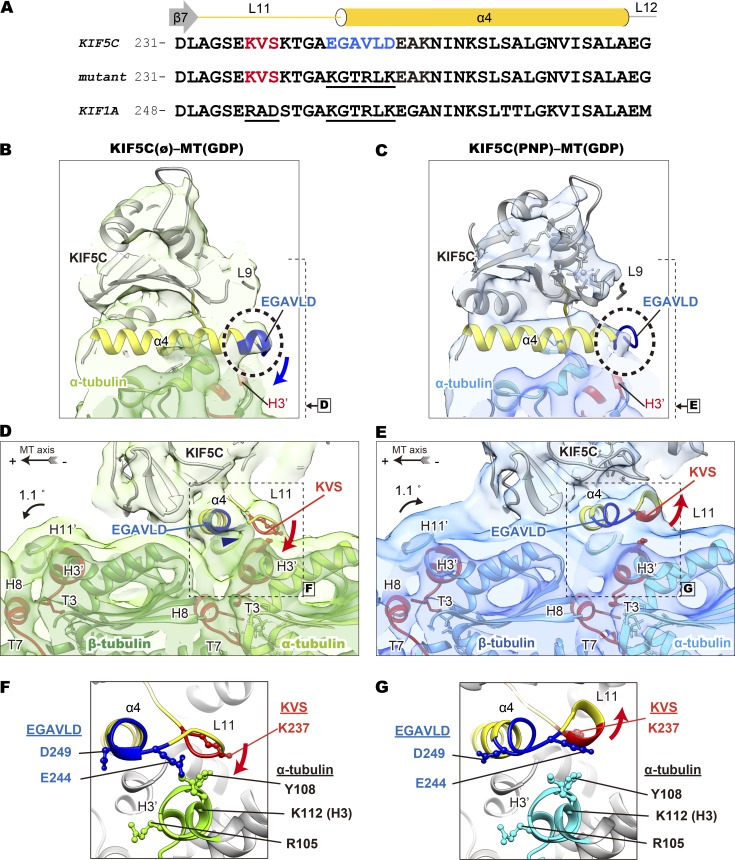
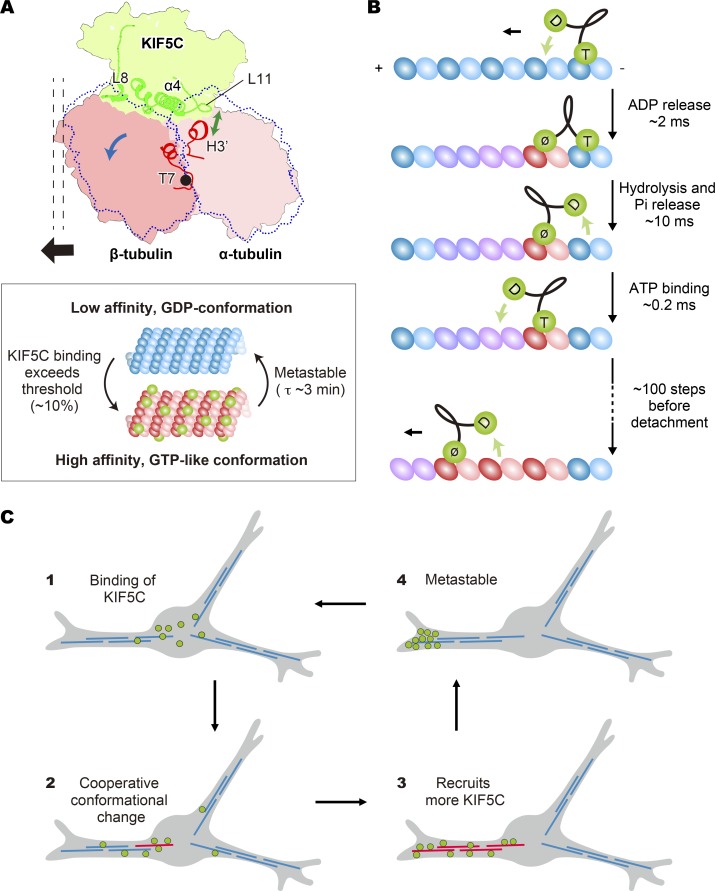
To elucidate the underlying structural basis for the tubulin rotation by KIF5C, the interfaces between the microtubule and KIF5C were examined (Figs. 7 D and 9). The atomic models of KIF5Cs in KIF5C(PNP)–MT(GDP) and KIF5C(ø)–MT(GDP) were compared after rigid-body fitting to the cryo-EM density map (Fig. 7 D). Several structural elements of KIF5C were identified that change their relative positions on the tubulin surface (hereafter, L and α are used for the loop and helix in kinesin and T and H, correspondingly, for tubulin.): L11 and α6, which interact with the α-tubulin on the left side and right side, respectively; α4 and α5, which interact with α- and β-tubulin near the intradimer interface; and L8, which interacts with β-tubulin at the plus-end side (Fig. 7 D). In the KIF5C(ø)–MT(GDP) structure, as shown in the green ribbon, the class-specific lysine-valine-serine (KVS) sequence of L11 approaches to the H3′ of α-tubulin. These interactions would be mediated by the hydrogen bond between K237 of L11 and Y108 of H3′ (Fig. 9 F). The following V238-S239 would also make hydrophobic interactions with H3′. The L8 and α5 also interact and push the C-domain region of β-tubulin in the KIF5C(ø)–MT(GDP), inducing the rotation of β-tubulin around the T7 loop toward the plus-end side (Fig. 7 D). This rotation consequently elongates the periodicity of the tubulin dimer. In contrast, these KIF5C elements were more distant from the microtubule surface in KIF5C(PNP)–MT(GDP) than in KIF5C(ø)-MT(GDP) (blue versus green ribbons in Fig. 7 D). This would weaken the rotation of tubulins and restore the curved tubulin-dimer position, triggering the contraction of the axial periodicity. Thus, KIF5C would change the axial pitch of microtubule in a nucleotide-dependent manner.
Figure 9.
Ability of the L11-α4 junction to discriminate microtubules examined with cryo-EM. (A) Sequence alignment of L11 and α4 region in KIF5C, the L11-α4 junction mutant, and KIF1A. (B–E) Comparison of cryo-EM density maps around the L11-α4 junction between KIF5C(ø)–MT(GDP) and KIF5C(PNP)–MT(GDP), viewed from the plus end (B and C) and left side (D and E). (F and G) The candidate amino acid residues proposed to be involved in the interaction between KIF5C and α-tubulin in KIF5C(ø)–MT(GDP) (F) and in KIF5C(PNP)–MT(GDP) (G). See Fig. S5 for the detailed structures of the L11-α4 junction of KIF1A and KIF5C.
It should be noted here that the conformational changes or rotation of tubulin described above would not take place independently in the microtubule lattice. If a single tubulin dimer were changed to the GTP-like conformation among GDP-conformation tubulins in the microtubule lattice, then many clashes would occur at the interfaces with the surrounding tubulin dimers (Fig. 8 E). This would suggest that the conformation would change cooperatively. Binding of a small number of KIF5Cs will not be able to trigger conformational changes, but the microtubule lattice will be strained. With more KIF5C binding, the strain will exceed a threshold, and conformational rearrangement will be triggered. Similarly, returning to the GDP conformation from the GTP-like lattice will also be cooperative, which might explain the metastability observed in the above experiments (Figs. 3 and S3) and in a previously study (Yajima et al., 2012).
Sensor region of KIF5C to discriminate microtubules
As shown above, the mutation of the L11-α4 junction (EGAVLD) to the corresponding sequence of KIF1A (KGTRLK) abolished the high-affinity binding to the longer-pitch microtubules, although it elongated GDP microtubules just as wild-type KIF5C did (Figs. 9 A and 2 F; and Fig. 4, E and F). This indicates that the L11-α4 junction would not be essential to trigger conformational changes in microtubules but would be required to discriminate conformational differences in microtubules.
Consistently, there were clear differences at this L11-α4 junction between KIF5C(ø)–MT(GDP) and KIF5C(PNP)–MT(GDP) cryo-EM density maps (Fig. 9, B–E). The density corresponding to the L11-α4 junction in KIF5C(ø)–MT(GDP) is stronger. The L11-α4 junction is closer to the H3′ of α-tubulin (Fig. 9 B), and its density appears to be connected to the microtubule surface (arrowhead in Fig. 9 D). However, the corresponding density was weaker in KIF5C(PNP)–MT(GDP) (Fig. 9 C). Quite suggestively, the L11-α4 junction densities of KIF1A are clearly separated and not connected to the H3′ in both the nucleotide-free and AMPPNP-bound conditions (arrowheads in Fig. S5, A and B). The L11-α4 junction of KIF5C in the nucleotide-free state and AMPPNP-bound state complexed with GDP-Taxol microtubules also adopt similar conformations (arrowheads in Fig. S5, C and D). These observations collectively suggest that the loop L11-α4 junction region of KIF5C takes a specific conformation in the nucleotide-free state on the GDP microtubule surface, and it would increase the binding affinity to the longer pitch of microtubules. Namely, this junction region, rather than loop L11 itself, would be the sensor to discriminate microtubule conformations. This model also explains why KIF1A does not discriminate GMPCPP microtubules from GDP microtubules, as well as the previously reported results of swap mutants between KIF5C and KIF1A (Nakata et al., 2011).
In x-ray diffraction and speckle microscopy experiments, the long-pitch microtubule returns to a short-pitch one when KIF5C binds to AMPPNP (Fig. 4, C and D; and Fig. 5 C). The observations of cryo-EM structures also suggest the structural basis for the microtubule contraction upon binding of AMPPNP to KIF5C. As shown in the previous section, AMPPNP binding to KIF5C induces the upward movement of L11 (KVS residues), and that restores the conformation of the intratubulin–dimer interface to the short-pitch microtubule (Fig. 7 D). The residues E244 and D249 of the L11-α4 junction would also be responsible for this pitch contraction movement. E244 and D249 presumably interact with K112 of H3 and R105 of H3′, respectively (Fig. 9, F and G). Since the residues corresponding to E244 and D249 are changed to lysines in KIF1A, the swap mutant would not be able to shorten the elongated the microtubule by the application of AMPPNP (Fig. 4, G and H).
Discussion
Recent structural studies have revealed conformational changes in tubulin subunits in the microtubule as the basis for the regulation of polymerization and depolymerization of microtubules or dynamic instability (Yajima et al., 2012; Alushin et al., 2014). However, tubulin subunits in the microtubule have been regarded as a static “rail” for the movement of kinesin, though many cell biological studies have reported that a small subset of microtubules are preferentially used by kinesin-1 and that those microtubules themselves serve as a “road sign” to guide transport to cognate destinations, such as the axon (Nakata and Hirokawa, 2003; Jacobson et al., 2006; Reed et al., 2006; Nakata et al., 2011). These studies consistently suggested that the rail microtubules for kinesin-1 are more stable than other nonrail microtubules, but the underlying mechanisms have been controversial or unclear.
In this study, we report that the binding of kinesin itself can induce conformational changes in GDP microtubules, the dominant nucleotide state in cells. The binding of kinesin-1 to the intradimer interface of tubulin subunits in the microtubule lattice pushes and rotates β-tubulin, leading to an increase in the axial pitch (Fig. 10 A, top, blue and black arrows). This conformation is similar to the GMPCPP microtubule, a stable microtubule lattice. KIF5C can recognize these conformational differences through interactions between the L11-α4 junction of kinesin and the H3′ helix of α-tubulin (Fig. 10 A, top, green arrow) and preferentially binds to longer-pitch microtubules, as reported here and in a previous study (Nakata et al., 2011). This positive feedback loop would explain the cooperative binding of kinesin to microtubules, as examined in detail in this study (Figs. 1 and 2) and in previous studies (Vilfan et al., 2001; Muto et al., 2005), and the preferential use of the subset of microtubules (Figs. 1 and 2; Nakata and Hirokawa, 2003; Reed et al., 2006; Nakata et al., 2011).
Figure 10.
Schematic representation of the model. (A) Summary of the results. The binding of KIF5C to β-tubulin will push this subunit to rotate around the T7 loop, leading to the elongation of the axial pitch (top, blue and black arrows). The lattice structure of the microtubule will restrict the conformation and the arrangement of tubulin subunits so that the conformational changes of the microtubule will take place in a cooperative manner (bottom). The conformational changes of the microtubule will be recognized by the interaction between kinesin’s L11-α4 junction and the H3′ helix of α-tubulin (top, green arrow). (B) A hypothetical model of the propagation of the conformational changes by the processive run of a kinesin dimer. A single protofilament of microtubule is shown with a kinesin dimer (a pair of green balls). The nucleotide states are shown in each head (ø, nucleotide-free; D, ADP; T, ATP). Bluish color shows the tubulin subunits in GDP conformation. Red to purple color shows the strain of the conformational changes propagating from the bound nucleotide-free head. (C) A model for the stochastic oscillation of kinesin accumulation in juvenile neuron. A gray Y-shaped neuron with three neurites is shown with GDP- and GTP-like conformation microtubules in blue and red, respectively. Green small circles represent kinesin molecules. See the main text for details.
Here, it should be noted that conformational changes in microtubules are highly cooperative. As reproduced by different assays (Figs. 4 C and 6 A), there is a switch-like change (the Hill coefficient for this reaction was around 5) with a threshold around 5–10% saturation. In most of these experiments, monomeric KIF5C-motor domain was used in the nucleotide-free state and was unlikely to hop around multiple tubulin dimers. This high cooperativity would reflect the two-dimensional lattice alignment of tubulin subunits in the microtubule (Fig. 10 A, bottom), which would be similar to the well-studied cooperative binding to oxygen by tetrameric hemoglobin. A single subunit cannot move alone independently from the surrounding subunits (Fig. 8 E). More kinesin binding above threshold (∼10% saturation) would be required to induce the conformational changes globally including tubulin subunits not bound to kinesin to avoid breaking of the lattice (Fig. 10 A, bottom).
In more physiological-like conditions, namely with kinesin-1 dimers and ATP, much smaller number of kinesin-1 dimers might be able to trigger the conformational changes. During the processive movement of a kinesin-1 dimer, the leading head bound to the microtubule is mostly in the nucleotide free state (Fig. 10 B). The ATP-bound state lasts a much shorter time (∼2 ms) and always in the trailing head. The AMPPNP-bound conformation of kinesin would reflect the isomerization intermediate state, a transient state between the ATP binding and hydrolysis (Nitta et al., 2004; Chang et al., 2013). Its duration would be even shorter than 1 ms, and would be too short to reverse the microtubule conformation. In fact, the microtubules remained in the high affinity state for several minutes after removal of KIF5C monomer by ATP in Fig. 3. Here, KIF5C monomers will bind and hydrolyze ATP before detachment from the microtubule.
Then, the processive movement of a kinesin-1 dimer will repetitively bind to the β-tubulin on the plus-end side in the nucleotide-free state, ∼100 steps before detachment (Fig. 10 B). Thus, a single kinesin-1 dimer would be able to induce conformational changes to ∼100 tubulin dimers along a protofilament, and a few run events would suffice to trigger the conformational changes as observed in the experiments in Fig. 1.
Generally, self-organization or spontaneous symmetry breaking is driven by such highly cooperative positive feedback loops. In neurons and other cells, tubulin is one of the most abundant proteins. The tubulin concentration in living cells is very high (∼10% of the total protein [Eipper, 1975] or ∼20 µM concentration in the cytosol [Gard and Kirschner, 1987], which is more than 1,000 times higher than kinesin-1 [Kuznetsov and Gelfand, 2001]). If some microtubules are converted to the higher-affinity state in this limited kinesin-1 environment, then they will obtain most kinesin-1 via a winner-take-all mechanism. This mechanism might explain the asymmetric distribution of constitutively active kinesin-1 in various cell types (Fig. S1 A). For example, stochastic accumulation of KIF5C to a single neurite in an immature neuron (Jacobson et al., 2006; Nakata et al., 2011) can be explained as follows (Fig. 10 C). Assume all microtubules in the neuron are in the low-affinity, short-pitch conformation (GDP structure; Fig. 10 C1). KIF5C will first distribute randomly among multiple neurites, and the microtubules in one neurite with more KIF5C will be triggered to change to the high-affinity, longer-pitch conformation (Fig. 10 C2). More KIF5C will then bind to those microtubules, and many microtubules in that neurite will be in the high-affinity conformation. The increase of local concentration of KIF5C in this neurite will facilitate the conversion of other microtubules in this neurite into the high-affinity state, which will recruit even more KIF5C. Then, KIF5C will be transported to the end of that neurite, resulting in the stochastic accumulation to one of the neurites (Fig. 10 C3). When most KIF5C molecules in the cell are accumulating at the end of the neurite, the frequency of the run events will decrease, and the microtubules will return to the low-affinity state (Fig. 10 C4). Newly synthesized KIF5C molecules or those diffusively (or retrogradely) transported back to the cell body will restart the process (Fig. 10 C1), so that the process appears like a stochastic oscillation in juvenile neurons (Jacobson et al., 2006; Nakata et al., 2011) or nonneuronal cells (Fig. S1 B). In neurons, KIF5C conveys cargos containing plasma membrane precursors (Okada et al., 1995; Deng et al., 2014) as well as the signaling molecules that determine the axonal polarity (Tahirovic and Bradke, 2009). The biased transport triggered by this cooperative positive feedback loop would specify that neurite into a future axon, especially when external polarization cues are not available, such as in low-density dissociation cultures, as observed previously (Bradke and Dotti, 1997).
Similar cooperative conformational changes have recently been reported for F-actin (Ngo et al., 2015, 2016; Umeki et al., 2016). Myosin-2 binds cooperatively to F-actin by extending the pitch, while cofilin binds cooperatively by shortening the pitch. This allosteric regulation by cooperative conformational changes in actin is proposed to contribute to the establishment of the front–rear polarity of locomoting cells (cofilin at the leading edge and myosin-2 at the trailing tail). Thus, cooperative conformational changes in cytoskeletal polymers might be a general strategy used by the cell to establish and maintain polarity.
Materials and methods
Snapshot analyses of KIF5 localization in various cells
Cell lines (Vero, HeLa, and MDCK) were obtained from RIKEN cell bank (RCB0001, RCB0007, and RCB0995, respectively) and maintained in high-glucose DMEM (Wako) supplemented with 10% FBS (Thermo Fisher) and antibiotics (penicillin-streptomycin; Thermo Fisher). The expression plasmid was constructed by inserting the coding sequence for aa 1–560 of mouse KIF5C using the EcoRI and SalI sites in the multiple cloning site of pEGFP-N1 (Clontech). The cells were transfected with Transfectin (Bio-Rad) or X-treme gene (Roche) and plated to the glass bottom dish (MatTek) coated with Matrigel (BD) for HeLa and MDCK or collagen (Cellmatrix type-IA, Nitta Gelatin) for Vero. The transfected cells were observed within 12–16 h after transfection, and only cells with low expression levels were selected for the observation, because asymmetric distribution of KIF5 tended to be less clear in cells with higher expression levels, particularly more than 24 h after transfection. The cells were observed under an atmospheric CO2 environment in L15 medium (Gibco) supplemented with 10% FBS. For HeLa and MDCK cells, an inverted microscope IX71 (Olympus) was used for observation with a 100× objective lens (UPlan SApo 100×/1.40; Olympus) and an EMCCD camera Luca (Andor) using μManager (Edelstein et al., 2010) as the controlling software. Images of Vero cells were taken with the same objective lens on an inverted microscope (IX81; Olympus). MetaMorph software (Molecular Devices) was used to control the microscope and camera (sCMOS camera ORCA FLASH 4.0; Hamamatsu). Images were pseudocolored using ImageJ.
Time-course analysis of KIF5 localization in Vero cells
Vero cells were transfected and plated to a collagen-coated glass-bottom dish as described above. The cells were observed with a total internal reflection fluorescence microscope with a Plan-Apo 60×/NA 1.42 oil objective (Olympus) and an sCMOS camera (Zyla4.2; Andor) on an IX83 microscope (Olympus). The system was controlled with MetaMorph. Each cell was examined every 30 min for 6 h. For each time point of observation, 300 frames of images were recorded at 10 Hz for 30 s The trajectories of KIF5C movement were shown as the SD map (Reed et al., 2006) of each 300-frame image sequence using ImageJ.
Protein sample preparation
Tubulin was purified from porcine brain through four cycles of polymerization and depolymerization using 1 M Pipes buffer (1 M Pipes-KOH, 1 mM EGTA, and 1 mM MgCl2, pH 6.8) for effective removal of microtubule-associated proteins (Castoldi and Popov, 2003). The labeled tubulin was prepared by incubating polymerized microtubule with tetramethylrhodamine succinimidyl ester (C-1171; Life Technologies), Alexa Fluor 647 NHS Ester (A-20106; Life Technologies), or NHS-LC-biotin (21336; Life Technologies) for 30 min at 37°C (Desai and Mitchison, 1998). Then, we purified functional labeled tubulin using two cycles of polymerization and depolymerization. The K351 KIF5C motor domain and K560 KIF5C dimer recombinants were expressed in BL21-CodonPlus(DE3)-RIL (Agilent) and Sf9 cells, respectively (Okada and Hirokawa, 1999). Both expressed KIF5C proteins were then purified using their His6 tag by TALON metal affinity resin (Clontech). For fluorescent imaging, K351 was labeled with DY-647-maleimide (Dyomics; Okada and Hirokawa, 1999). Excess DY-647 was removed using the Micro Bio-Spin 30 size exclusion column (Bio-Rad). GMPCPP was prepared enzymatically from GMPCP (M3170; Sigma-Aldrich) together with nucleotide diphosphate kinase (N0379; Sigma-Aldrich) and then purified using a Mono Q 5/50 GL column (GE Healthcare; Hyman et al., 1992).
Fluorescent microscopy
The nonlabeled tubulin, tetramethylrhodamine-labeled tubulin, and biotin-labeled tubulin were copolymerized to yield microtubules with a rhodamine and biotin labeling ratio of 2% and 3%, respectively. For GMPCPP microtubules, we repeated three cycles of GMPCPP uptake in the presence of 0.2 mM GMPCPP to obtain microtubules with over 90% occupancy of GMPCPP in β-tubulins (Yajima et al., 2012). To prepare GDP microtubules, tubulins were copolymerized in the presence of 1 mM GTP. Then, the buffer was exchanged for GPEM buffer (100 mM Pipes-KOH, 30% [vol/vol] glycerol, 2 mM MgSO4, and 1 mM EGTA, pH 6.9) by centrifugation. To ensure GTP hydrolysis in the microtubule, GDP microtubules were used for the experiments after more than 3 h of incubation at room temperature. The glass chambers for observation were prepared as follows. After sonication in 1 N KOH and plasma treatment (Diener), the surface of the cover glasses (C022221S; Matsunami Glass) was silanized with N-2-(aminoethyl)-3-aminopropyl-triethoxysilane (KBE-603; Shin-Etsu Chemical) and then incubated with 200 mg/ml NHS-PEG (ME-050-TS; NOF) with or without 1 mg/ml NHS-PEG-biotin (BI-050-TS; NOF) for 3 h at room temperature to make PEG-biotin–coated or PEG-coated glasses (Yokota et al., 2009). The PEG-biotin–coated glass and the PEG-coated glass were separated using a 30-µm layer of double-sided tape (5603; Nitto-Denko) to make the flow chamber. Microtubules were immobilized on the PEG-biotin–coated glass surface via Neutravidin (31000; Thermo Fisher), and the glass surface was then blocked with imaging solution (45 mM Pipes-KOH, 2 mM MgSO4, 1 mM EGTA, 1% [wt/vol] Pluronic F-127, 1 mg/ml casein, 1 mM d-biotin, 2 mM dithiothreitol, 0.2 mg/ml glucose oxidase, 40 µg/ml catalase, 1 mM glucose, and 30% [vol/vol] glycerol, pH 7.2). Finally, K351-DY-647 or K560-GFP was added to the imaging solution and introduced into the chamber. The images of microtubules were observed under total internal reflection fluorescence microscopy (IX81) with a 100× objective lens (UPlanSApo, NA 1.40, oil; Olympus) and recorded at the frame rate of 10 frames per second using an iXon3 EM CCD camera (Andor) and MetaMorph software. During observation, the temperature of the chamber was kept at 37°C. The images were processed for background subtraction and further analysis using ImageJ.
Statistical analyses
The distribution of the landing rates (Fig. 1, G–I) and the bound head density (Fig. 2, B and D) was analyzed using a expectation-maximization algorithm for Gaussian mixture model using mclust package (Scrucca et al., 2016) in R (Ihaka and Gentleman, 1996). P values for comparison of motility parameters (Fig. 1, G–L), microtubule elongation rates (Fig. 5 C), and SHG signals (Fig. 6, D and E) were calculated by a nonparametric multiple comparison test (Steel–Dwass) using the pSDCFlig function in the NSM3 package (Schneider et al., 2018) in R.
Fluorescent speckle microscopy
GDP microtubules labeled with DY-647 and biotin were prepared as described above and then shortened to be less than 2 µm long by pipetting. These microtubule seeds were immobilized on a PEG-biotin–coated glass surface via Neutravidin. After washing with the imaging solution, we introduced the solution containing 6 µM tubulin, 120 nM tetramethylrhodamine-labeled tubulin, 1 mM GTP, and 1 mM MgSO4 into the chamber to polymerize the tubulins at the ends of the immobilized microtubules. Following incubation at 37°C for 2 min, the solution in the chamber was changed to 6 µM tubulin, 120 nM DY-647–labeled tubulin, 1 mM GTP, and 1 mM MgSO4 and then washed out with the imaging solution immediately after the change. To make multiple DY-647–labeled speckles in the microtubule, we repeated this tubulin polymerizing procedure four times. Then, the imaging solution containing 0.5% (wt/vol) methylcellulose (Methocel MC; Sigma-Aldrich) was introduced in the chamber to suppress fluctuation of the microtubules. The microtubules were observed using the aforementioned microscopy system with 0.5-s exposure. The central positions of the DY-647 speckles were determined by 2D Gaussian fitting of the image using Mark2 software (provided by K. Furuta, National Institute of Information and Communications Technology, Kobe, Japan; Furuta and Toyoshima, 2008). The SD of the determined position was 10 nm. To measure the compaction of the elongated GDP microtubule, KIF5C was washed out by 0.2-µl/s constant flow of buffer containing 300 mM K-Pipes, 40% (vol/vol) glycerol, 1 mM EGTA, 1 mM ATP, 2 mM MgSO4, 2 mM dithiothreitol, 0.2 mg/ml glucose oxidase, 40 µg/ml catalase, 1 mM glucose, and 0.1% Pluronic F-127, pH 7.2, using a Cavro syringe pump (30062929; Tecan). To measure the microtubule depolymerization rate, the solution in the assay chamber was replaced by imaging solution without glycerol, and microtubules were then observed under microscopy.
X-ray fiber diffraction
187 µM tubulin dimer was polymerized in polymerization buffer (20 mM Pipes-KOH, 1 mM EGTA, and 2 mM MgSO4, pH 6.6) with 2 mM GTP or 0.5 mM GMPCPP and 2 mM MgSO4. After incubation at 37°C, 10 mM dithiothreitol, 2 mM ATP, 2 mM MgSO4 and 1% (wt/vol) methylcellulose (Methocel MC) were added to the microtubule specimen. After centrifugation at 14,000 g for 30 s to remove air bubbles, the specimen was placed in the space between a quartz disc and a ring-shaped coverslip. Microtubules were oriented under shear flow by rotating the quartz disc at 10–16 rotations per second. A synchrotron radiation x-ray beam at BL45XU beamline (Fujisawa et al., 2000) of SPring-8 (Japan Synchrotron Radiation Research Institute) irradiated the oriented microtubules (Kamimura et al., 2016). The diffraction images were recorded with a Pilatus 300K-W detector system (Dectris) with 30-s exposure. KIF5C and 1 mM AMPPNP were added to the space between the disc and the coverslip 20–40 s before taking the images. We used a 4.97-nm signal obtained from lead stearate to calibrate the spacing of diffraction peaks. Images were processed for background subtraction using ImageJ. The tubulin axial periodicity was determined by fitting the intensity of the fourth-diffraction peak with a Lorentzian function. The error bars in Fig. 4 indicate the full width at half maximum (FWHM) of the fitted function.
Sample preparations for the SHG microscopy
Xenopus laevis egg and sperm were gifts from H. Inomata (RIKEN, Hyogo, Japan). Permeabilized Xenopus sperm nuclei (Desai et al., 1999) were immobilized on a surface of a KOH-washed quartz coverslip. Following wash of the surface by 7 mg/ml casein, aster-like microtubule formation was initiated by incubating the nuclei with the M-phase egg extract (Field et al., 2014) for 5 min at 37°C. Excess egg extract was washed out with 500 mM Pipes-KOH, 10 mM MgCl2, 5 mM EGTA, and 30% (vol/vol) glycerol, pH 6.9. After exchanging buffer with GPEM50 (50 mM Pipes-KOH, 1 mM EGTA, 2 mM MgSO4, and 30% [vol/vol] glycerol, pH 6.7), 15 µM tubulin (including 2% of Alexa Fluor 647–labeled tubulin) with 1 mM GTP and 0.7 mg/ml casein was introduced into the chamber. The chamber was incubated at 37°C for 20 min to polymerize microtubules. Finally, the chamber was washed with GPEM50, and K351 with or without 1 mM AMPPNP was introduced into the chamber.
SHG microscopy
The SHG data were acquired using a custom-built microscope based on a Nikon TE2000-U (Fig. 6 B; Kaneshiro et al., 2018 Preprint). The sample was illuminated by a mode-locked Ti/sapphire laser (Chameleon Vision II; Coherent, Inc.) with an 810-nm wavelength, 80-MHz repetition rate, and 200-fs pulse duration through a Glan-laser polarizing prism (GL10-B; Thorlabs), high-speed polarization controller (350-160 and 350-80; Conoptics, Inc.), and a 40× dry objective lens (NA 0.95, CFI Apo; Nikon). The emitted light was detected with a photon-counting photomultiplier tube module (H10680-210; Hamamatsu Photonics) through a 100× objective lens (NA 1.45, CFI Apo, oil; Nikon) and filters (FF01-680/SP and FF01-405/10-25; Semrock). The SHG intensity data on the incident polarization angle (θ) were fitted with the following theoretical function to obtain three fitting parameters α, χzzz, and χzxx:
where α denotes the angle of average orientation of a microtubule bundle and χzzz and χzxx are two components of SHG susceptibility tensor (Psilodimitrakopoulos et al., 2013; Kaneshiro et al., 2018 Preprint).
Grid preparation and cryo-EM data collection
The KIF5C motor domain (mouse KIF5C residues 1–345 and a His7 tag) was purified by immobilized metal affinity chromatography and cation exchange chromatography (AKTA Explorer 10S, RESOURCE S column; GE Healthcare). The purified KIF5C was dialyzed against and concentrated in a buffer (10 mM Hepes-NaOH, pH 7.4, 100 mM NaCl, 1 mM MgCl2, and 20% [wt/vol] sucrose), frozen in liquid N2, and stored at −80°C. For GDP microtubule complexed with KIF5C (KIF5C–MT(GDP)), tubulin was diluted to 80 µM by PEM buffer (20 mM Pipes-KOH, pH 6.6, 1 mM EGTA, and 2 mM MgSO4), incubated on ice for 5 min in PEM buffer with 1 mM GTP and 1 mM MgSO4, and polymerized at 37°C for 30 min. At the end of polymerization, 0.1 mM MgSO4, 10 mM DTT, and 0.1 mM ATP for KIF5C(ATP)–MT(GDP) or 0.1 mM AMPPNP for KIF5C(PNP)–MT(GDP) were mixed into the microtubule solution. After 5 min, K351 diluted to 500 µM by a dilution buffer (10 mM Hepes-NaOH, pH 7.4, 50 mM NaCl, and 1 mM MgCl2) was added to microtubule solution to a final 20% molar ratio to tubulin. A 4-µl drop of KIF5C–MT(GDP) (40 µM tubulin) was placed onto a glow-discharging holey carbon grid (R2/2; Quantifoil) and applied in a semiautomated vitrification device (Vitrobot Mark IV; FEI) with 100% humidity at 27°C. After 30 s, a 9-µl drop of 40 µM KIF5C solution containing 5 mM ATP or AMPPNP, or 10 U/ml apyrase for KIF5C(ø)–MT(GDP), was added on a drop of microtubule. After 60 s, the remaining solution on the grid was wicked away with a piece of Whatman no. 1 filter paper and the grid was plunge-frozen into liquid ethane. GMPCPP microtubule complexed with nucleotide-free KIF5C (KIF5C(ø)–MT(GMPCPP)) was prepared as previously described (Morikawa et al., 2015) using the Vitrobot. Data acquisition was performed using a 200-kV field emission cryo-EM (Tecnai Arctica; FEI) at 78,000-fold nominal magnification with an FEI Falcon II direct detection camera under low-dose conditions using the data-acquisition software Serial EM (Mastronarde, 2005). All data were collected as a movie with seven subframes with a total electron dose of 50 e−1/Å2 at a pixel size of 1.28 Å/pixel. The defocus range of the dataset was set to −1.5 to −2.5 µm.
Image processing and 3D reconstruction of cryo-EM images
The movie data were processed for motion correction with the software MotionCorr (Li et al., 2013). Motion-corrected and summed images were analyzed its defocus and astigmatism by using CTFFIND3 (Mindell and Grigorieff, 2003) and images without significant drift and astigmatism were used for further analysis. Images of a 14-protofilament KIF5C-MT complex were selected and semiautomatically straightened using the “unbend” program of Ruby-Helix (Metlagel et al., 2007). Segments were extracted at a spacing of 80 Å using a box size of 768 × 768 pixels. The stack file combined by the addtostack command of IMOD (Kremer et al., 1996) was then applied to the seam-search scripts (Zhang and Nogales, 2015). We used EMD-6353, the kinesin-decorated GDP microtubule, as the 3D reference model to precisely separate α-tubulin and β-tubulin (Zhang et al., 2015). Local refinement was performed twice using Frealign version 9 (Grigorieff, 2007), followed by determination of seam position. Using refined alignment particles with the correct seam, 3D structures were reconstructed assuming pseudohelical symmetry (HP) and no symmetry (C1). The helical parameters for three-start tubulin monomers (twist and rise) were measured using the Relion helix toolbox program (Scheres, 2012) from the C1 reconstruction and applied to the next HP reconstruction. Two rounds of seam search were performed to obtain the final reconstruction. The resolution was analyzed by Fourier shell correlation (Van Heel, 1987). Local resolutions were estimated with ResMap program (Kucukelbir et al., 2014). A negative B-factor (−120 and −200) was applied to sharpen the final map using Bfactor program (included with the FREALIGN distribution; Grigorieff, 2016).
Atomic model fitting
For microtubules, the atomic structure of tubulin (Protein Data Bank accession no. 3J6F; Alushin et al., 2014) was fitted rigidly by monomer into the cryo-EM densities using the Fit in Map tool in UCSF Chimera (Pettersen et al., 2004; KIF5C(PNP)–MT(GDP): ccc = 0.96, KIF5C(ø)–MT(GDP): ccc = 0.96, KIF5C(ATP)–MT(GDP): ccc = 0.95, KIF5C(ø)–MT(GMPCPP): ccc = 0.94).
For KIF5C, the atomic structures of the KIF5 motor domain (Protein Data Bank accession nos. 4HNA [ATP form] and 4LNU [nucleotide-free form]; Gigant et al., 2013; Cao et al., 2014) were rigid-body fitted (KIF5C(PNP)–MT(GDP): ccc = 0.92 [4HNA]; KIF5C(ø)–MT(GDP): ccc = 0.94 [4LNU]).
Accession numbers
Cryo-EM maps and the fitted atomic coordinates have been deposited in the Electron Microscopy Data Bank and Protein Data Bank under the accession codes EMD-6779 and 5XXT (KIF5C(ø)–MT(GDP)), EMD-6781 and 5XXV (KIF5C(PNP)–MT(GDP)), EMD-6782 and 5XXW (KIF5C(ATP)–MT(GDP)), and EMD-6783 and 5XXX (KIF5C(ø)–MT(GMPCPP)), respectively.
Online supplemental material
Fig. S1 shows the stochastic and transient asymmetric distribution of kinesin in nonneuronal cells. Fig. S2 shows hyperstabilization of GDP microtubules by KIF5C. Fig. S3 shows compaction of the elongated GDP microtubule after kinesin removal. Fig. S4 shows resolution and statistics for the cryo-EM reconstructions. Fig. S5 shows the position of the elements of a kinesin–microtubule surface. Table S1 lists numbers of samples and events for Fig. 1 (G–L). Table S2 lists statistics for the mixed Gaussian fitting in Fig. 1 (G–I). Table S3 lists multiple comparison statistics by Steel–Dwass test. Table S4 lists numbers of microtubules and kinesin molecules for Fig. 2. Table S5 shows statistics for the mixed Gaussian fitting in Fig. 2 B. Table S6 shows statistics for the mixed Gaussian fitting in Fig. 2 D. Video 1 shows KIF5C dimer molecules (magenta) moving along GDP and GMPCPP microtubules (green). Video 2 shows depolymerization kinetics of GDP microtubules in the presence of KIF5C monomer (K351). Video 3 shows a comparison of the microtubule lattices between KIF5C(PNP)–MT(GDP) and KIF5C(ø)–MT(GDP). Video 4 shows a comparison of the microtubule lattices between KIF5C(PNP)–MT(GDP) and KIF5C(ATP)–MT(GDP). Video 5 shows a comparison of the microtubule lattices between KIF5C(PNP)–MT(GDP) and KIF5C(ø)–MT(GMPCPP).
Supplementary Material
Acknowledgments
We thank T. Hikima and M. Yamamoto for the use of SPring-8 RIKEN beamline BL45XU (proposal number 20140094 and 20150029); R. Zhang, T. Imasaki, and T. Osanai for cryo-EM image processing and 3D reconstruction; and Y. Oguchi for setting up the microfluidic system. We also thank S. Xu, J. Asada, M. Komeno, M. Kakiuchi, and K. Ito for their technical and secretarial assistance; W. Kylius for editing the manuscript; and T. Ogawa, H. Yajima, H. Sato, T. Akamatsu, H. Fukuda, and all members of the Hirokawa Lab for technical help and valuable discussions.
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology through a Grant-in-Aid for Specially Promoted Research (grant 23000013 to N. Hirokawa) and Grant-in-Aid for Scientific Research (KAKENHI grant 16H06372 to N. Hirokawa; grants 16H05119, 15H01334, 26115721, 26650069, and 25293046 to Y. Okada; grants 15H01656 and 17H05897 to H. Shigematsu; and grant 15K08168 to R. Nitta), the Uehara Memorial Foundation (Y. Okada), the Takeda Science Foundation (Y. Okada and R. Nitta), the Mochida Memorial Foundation for Medical and Pharmaceutical Research (R. Nitta), the RIKEN Special Postdoctoral Researchers Program (T. Shima), the All RIKEN Research Project on Single Cell (Y. Okada), and the RIKEN Pioneering Project on Dynamic Structural Biology (Y. Okada, H. Shigematsu, and M. Shirouzu).
The authors declare no competing financial interests.
Author contributions: Y. Okada and N. Hirokawa conceived the idea and designed the project. T. Shima, S. Uemura, and Y. Okada performed the in vitro experiments with fluorescent microscopes, while T. Kambara and Y. Okada did the experiments in living cells. Cryo-EM experiments were done by M. Morikawa, H. Shigematsu, M. Shirouzu, and R. Nitta. J. Kaneshiro, T. Ichimura, and T. Watanabe built the SHG microscope and analyzed the data through discussion with T. Shima and Y. Okada. T. Shima, M. Morikawa, Y. Okada, S. Kamimura, T. Yagi, and H. Iwamoto did the x-ray fiber diffraction experiments at SPring-8. All the authors discussed the results, and Y. Okada, T. Shima, M. Morikawa, R. Nitta, and N. Hirokawa wrote the manuscript.
References
- Alushin G.M., Lander G.C., Kellogg E.H., Zhang R., Baker D., and Nogales E.. 2014. High-resolution microtubule structures reveal the structural transitions in αβ-tubulin upon GTP hydrolysis. Cell. 157:1117–1129. 10.1016/j.cell.2014.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradke F., and Dotti C.G.. 1997. Neuronal polarity: vectorial cytoplasmic flow precedes axon formation. Neuron. 19:1175–1186. 10.1016/S0896-6273(00)80410-9 [DOI] [PubMed] [Google Scholar]
- Cao L., Wang W., Jiang Q., Wang C., Knossow M., and Gigant B.. 2014. The structure of apo-kinesin bound to tubulin links the nucleotide cycle to movement. Nat. Commun. 5:5364 10.1038/ncomms6364 [DOI] [PubMed] [Google Scholar]
- Castoldi M., and Popov A.V.. 2003. Purification of brain tubulin through two cycles of polymerization-depolymerization in a high-molarity buffer. Protein Expr. Purif. 32:83–88. 10.1016/S1046-5928(03)00218-3 [DOI] [PubMed] [Google Scholar]
- Chang Q., Nitta R., Inoue S., and Hirokawa N.. 2013. Structural basis for the ATP-induced isomerization of kinesin. J. Mol. Biol. 425:1869–1880. 10.1016/j.jmb.2013.03.004 [DOI] [PubMed] [Google Scholar]
- Deng C.-Y., Lei W.-L., Xu X.-H., Ju X.-C., Liu Y., and Luo Z.-G.. 2014. JIP1 mediates anterograde transport of Rab10 cargos during neuronal polarization. J. Neurosci. 34:1710–1723. 10.1523/JNEUROSCI.4496-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A., and Mitchison T.J.. 1998. Preparation and characterization of caged fluorescein tubulin. Methods Enzymol. 298:125–132. 10.1016/S0076-6879(98)98014-4 [DOI] [PubMed] [Google Scholar]
- Desai A., Murray A., Mitchison T.J., and Walczak C.E.. 1999. The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61:385–412. 10.1016/S0091-679X(08)61991-3 [DOI] [PubMed] [Google Scholar]
- Dimitrov A., Quesnoit M., Moutel S., Cantaloube I., Poüs C., and Perez F.. 2008. Detection of GTP-tubulin conformation in vivo reveals a role for GTP remnants in microtubule rescues. Science. 322:1353–1356. 10.1126/science.1165401 [DOI] [PubMed] [Google Scholar]
- Edelstein A., Amodaj N., Hoover K., Vale R., and Stuurman N.. 2010. Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol. 14:14–14. 10.1002/0471142727.mb1420s92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eipper B.A. 1975. Purification of rat brain tubulin. Ann. N. Y. Acad. Sci. 253(1 The Biology o):239–246. 10.1111/j.1749-6632.1975.tb19203.x [DOI] [PubMed] [Google Scholar]
- Field C.M., Nguyen P.A., Ishihara K., Groen A.C., and Mitchison T.J.. 2014. Xenopus Egg Cytoplasm with Intact Actin. Methods Enzymol. 540:399–415. 10.1016/B978-0-12-397924-7.00022-4 [DOI] [PubMed] [Google Scholar]
- Fujisawa T., Inoue K., Oka T., Iwamoto H., Uruga T., Kumasaka T., Inoko Y., Yagi N., Yamamoto M., and Ueki T.. 2000. Small-angle x-ray scattering station at the SPring-8 RIKEN beamline. J. Appl. Cryst. 33:797–800. 10.1107/S002188980000131X [DOI] [Google Scholar]
- Furuta K., and Toyoshima Y.Y.. 2008. Minus-end-directed motor Ncd exhibits processive movement that is enhanced by microtubule bundling in vitro. Curr. Biol. 18:152–157. 10.1016/j.cub.2007.12.056 [DOI] [PubMed] [Google Scholar]
- Gard D.L., and Kirschner M.W.. 1987. Microtubule assembly in cytoplasmic extracts of Xenopus oocytes and eggs. J. Cell Biol. 105:2191–2201. 10.1083/jcb.105.5.2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gigant B., Wang W., Dreier B., Jiang Q., Pecqueur L., Plückthun A., Wang C., and Knossow M.. 2013. Structure of a kinesin-tubulin complex and implications for kinesin motility. Nat. Struct. Mol. Biol. 20:1001–1007. 10.1038/nsmb.2624 [DOI] [PubMed] [Google Scholar]
- Grigorieff N. 2007. FREALIGN: high-resolution refinement of single particle structures. J. Struct. Biol. 157:117–125. 10.1016/j.jsb.2006.05.004 [DOI] [PubMed] [Google Scholar]
- Grigorieff N. 2016. Frealign: An Exploratory Tool for Single-Particle Cryo-EM. Methods Enzymol. 579:191–226. 10.1016/bs.mie.2016.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney D.D. 1995. Implications of diffusion-controlled limit for processivity of dimeric kinesin head domains. Biophys. J. 68:267S–269S. [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Niwa S., and Tanaka Y.. 2010. Molecular motors in neurons: transport mechanisms and roles in brain function, development, and disease. Neuron. 68:610–638. 10.1016/j.neuron.2010.09.039 [DOI] [PubMed] [Google Scholar]
- Hyman A.A., Salser S., Drechsel D.N., Unwin N., and Mitchison T.J.. 1992. Role of GTP hydrolysis in microtubule dynamics: information from a slowly hydrolyzable analogue, GMPCPP. Mol. Biol. Cell. 3:1155–1167. 10.1091/mbc.3.10.1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman A.A., Chrétien D., Arnal I., and Wade R.H.. 1995. Structural changes accompanying GTP hydrolysis in microtubules: information from a slowly hydrolyzable analogue guanylyl-(alpha,beta)-methylene-diphosphonate. J. Cell Biol. 128:117–125. 10.1083/jcb.128.1.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R., and Gentleman R.. 1996. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Stat. 5:299–314. 10.1080/10618600.1996.10474713 [DOI] [Google Scholar]
- Jacobson C., Schnapp B., and Banker G.A.. 2006. A change in the selective translocation of the Kinesin-1 motor domain marks the initial specification of the axon. Neuron. 49:797–804. 10.1016/j.neuron.2006.02.005 [DOI] [PubMed] [Google Scholar]
- Kamimura S., Fujita Y., Wada Y., Yagi T., and Iwamoto H.. 2016. X-ray fiber diffraction analysis shows dynamic changes in axial tubulin repeats in native microtubules depending on paclitaxel content, temperature and GTP-hydrolysis. Cytoskeleton (Hoboken). 73:131–144. 10.1002/cm.21283 [DOI] [PubMed] [Google Scholar]
- Kaneshiro J., Okada Y., Shima T., Tsujii M., Imada K., Ichimura T., and Watanabe T.M.. 2018. Second harmonic generation polarization microscopy as a tool for protein structure analysis. bioRxiv (Preprint posted June 4, 2018). 10.1101/338137 [DOI] [PMC free article] [PubMed]
- Kaul N., Soppina V., and Verhey K.J.. 2014. Effects of α-tubulin K40 acetylation and detyrosination on kinesin-1 motility in a purified system. Biophys. J. 106:2636–2643. 10.1016/j.bpj.2014.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Kucukelbir A., Sigworth F.J., and Tagare H.D.. 2014. Quantifying the local resolution of cryo-EM density maps. Nat. Methods. 11:63–65. 10.1038/nmeth.2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov S.A., and Gelfand V.I.. 2001. Purification of kinesin from the brain. Methods Mol. Biol. 164:1–7. 10.1385/1-59259-069-1:1 [DOI] [PubMed] [Google Scholar]
- Li X., Mooney P., Zheng S., Booth C.R., Braunfeld M.B., Gubbens S., Agard D.A., and Cheng Y.. 2013. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods. 10:584–590. 10.1038/nmeth.2472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marceiller J., Drechou A., Durand G., Perez F., and Poüs C.. 2005. Kinesin is involved in protecting nascent microtubules from disassembly after recovery from nocodazole treatment. Exp. Cell Res. 304:483–492. 10.1016/j.yexcr.2004.12.007 [DOI] [PubMed] [Google Scholar]
- Mastronarde D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Metlagel Z., Kikkawa Y.S., and Kikkawa M.. 2007. Ruby-Helix: an implementation of helical image processing based on object-oriented scripting language. J. Struct. Biol. 157:95–105. 10.1016/j.jsb.2006.07.015 [DOI] [PubMed] [Google Scholar]
- Mindell J.A., and Grigorieff N.. 2003. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142:334–347. 10.1016/S1047-8477(03)00069-8 [DOI] [PubMed] [Google Scholar]
- Mitchison T., and Kirschner M.. 1984. Dynamic instability of microtubule growth. Nature. 312:237–242. 10.1038/312237a0 [DOI] [PubMed] [Google Scholar]
- Morikawa M., Yajima H., Nitta R., Inoue S., Ogura T., Sato C., and Hirokawa N.. 2015. X-ray and Cryo-EM structures reveal mutual conformational changes of Kinesin and GTP-state microtubules upon binding. EMBO J. 34:1270–1286. 10.15252/embj.201490588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto E., Sakai H., and Kaseda K.. 2005. Long-range cooperative binding of kinesin to a microtubule in the presence of ATP. J. Cell Biol. 168:691–696. 10.1083/jcb.200409035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T., and Hirokawa N.. 2003. Microtubules provide directional cues for polarized axonal transport through interaction with kinesin motor head. J. Cell Biol. 162:1045–1055. 10.1083/jcb.200302175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata T., Niwa S., Okada Y., Perez F., and Hirokawa N.. 2011. Preferential binding of a kinesin-1 motor to GTP-tubulin-rich microtubules underlies polarized vesicle transport. J. Cell Biol. 194:245–255. 10.1083/jcb.201104034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo K.X., Kodera N., Katayama E., Ando T., and Uyeda T.Q.. 2015. Cofilin-induced unidirectional cooperative conformational changes in actin filaments revealed by high-speed atomic force microscopy. eLife. 4:e04806 10.7554/eLife.04806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo K.X., Umeki N., Kijima S.T., Kodera N., Ueno H., Furutani-Umezu N., Nakajima J., Noguchi T.Q.P., Nagasaki A., Tokuraku K., and Uyeda T.Q.P.. 2016. Allosteric regulation by cooperative conformational changes of actin filaments drives mutually exclusive binding with cofilin and myosin. Sci. Rep. 6:35449 10.1038/srep35449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta R., Kikkawa M., Okada Y., and Hirokawa N.. 2004. KIF1A alternately uses two loops to bind microtubules. Science. 305:678–683. 10.1126/science.1096621 [DOI] [PubMed] [Google Scholar]
- Okada Y., and Hirokawa N.. 1999. A processive single-headed motor: kinesin superfamily protein KIF1A. Science. 283:1152–1157. 10.1126/science.283.5405.1152 [DOI] [PubMed] [Google Scholar]
- Okada Y., Yamazaki H., Sekine-Aizawa Y., and Hirokawa N.. 1995. The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell. 81:769–780. 10.1016/0092-8674(95)90538-3 [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Plotnikov S.V., Millard A.C., Campagnola P.J., and Mohler W.A.. 2006. Characterization of the myosin-based source for second-harmonic generation from muscle sarcomeres. Biophys. J. 90:693–703. 10.1529/biophysj.105.071555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psilodimitrakopoulos S., Petegnief V., de Vera N., Hernandez O., Artigas D., Planas A.M., and Loza-Alvarez P.. 2013. Quantitative imaging of microtubule alteration as an early marker of axonal degeneration after ischemia in neurons. Biophys. J. 104:968–975. 10.1016/j.bpj.2013.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed N.A., Cai D., Blasius T.L., Jih G.T., Meyhofer E., Gaertig J., and Verhey K.J.. 2006. Microtubule acetylation promotes kinesin-1 binding and transport. Curr. Biol. 16:2166–2172. 10.1016/j.cub.2006.09.014 [DOI] [PubMed] [Google Scholar]
- Scheres S.H.W. 2012. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180:519–530. 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider G., Chicken E., and Becvarik R.. 2018. NSM3: Functions and Datasets to Accompany Hollander, Wolfe, and Chicken. Nonparametric Statistical Methods, Third edition, R package version 1.12. https://CRAN.R-roject.org/package=NSM3 (accessed May 7, 2018).
- Scrucca L., Fop M., Murphy T.B., and Raftery A.E.. 2016. mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. R J. 8:289–317. [PMC free article] [PubMed] [Google Scholar]
- Tahirovic S., and Bradke F.. 2009. Neuronal polarity. Cold Spring Harb. Perspect. Biol. 1:a001644 10.1101/cshperspect.a001644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida M., Mouriño-Pérez R.R., Freitag M., Bartnicki-García S., and Roberson R.W.. 2008. Microtubule dynamics and the role of molecular motors in Neurospora crassa. Fungal Genet. Biol. 45:683–692. 10.1016/j.fgb.2007.10.013 [DOI] [PubMed] [Google Scholar]
- Umeki N., Hirose K., and Uyeda T.Q.P.. 2016. Cofilin-induced cooperative conformational changes of actin subunits revealed using cofilin-actin fusion protein. Sci. Rep. 6:20406 10.1038/srep20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Heel M. 1987. Similarity measures Between Images. Ultramicroscopy. 21:95–100. 10.1016/0304-3991(87)90010-6 [DOI] [Google Scholar]
- Vilfan A., Frey E., Schwabl F., Thormählen M., Song Y.H., and Mandelkow E.. 2001. Dynamics and cooperativity of microtubule decoration by the motor protein kinesin. J. Mol. Biol. 312:1011–1026. 10.1006/jmbi.2001.5020 [DOI] [PubMed] [Google Scholar]
- Yajima H., Ogura T., Nitta R., Okada Y., Sato C., and Hirokawa N.. 2012. Conformational changes in tubulin in GMPCPP and GDP-taxol microtubules observed by cryoelectron microscopy. J. Cell Biol. 198:315–322. 10.1083/jcb.201201161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokota H., Han Y.-W., Allemand J.-F., Xi X.G., Bensimon D., Croquette V., Ito Y., and Harada Y.. 2009. Single-molecule Visualization of Binding Modes of Helicase to DNA on PEGylated Surfaces. Chem. Lett. 38:308–309. 10.1246/cl.2009.308 [DOI] [Google Scholar]
- Zhang R., and Nogales E.. 2015. A new protocol to accurately determine microtubule lattice seam location. J. Struct. Biol. 192:245–254. 10.1016/j.jsb.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Alushin G.M., Brown A., and Nogales E.. 2015. Mechanistic origin of microtubule dynamic instability and its modulation by EB proteins. Cell. 162:849–859. 10.1016/j.cell.2015.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.