Maria Bohnert discusses the potential functional insight into lipid droplet formation provided by Sui et al.’s new cryo-EM structure of Drosophila seipin.
Abstract
The lipid droplet (LD) biogenesis protein seipin is crucial for formation of normal LDs, but its exact functional role has been enigmatic. In this issue, Sui et al. (2018. J. Cell Biol. https://doi.org/10.1083/jcb.201809067) report the cryo–electron microscopy structure of seipin, which provides novel insights into how seipin might mediate LD formation.
Seipin is a key player in lipid storage. On a cellular level, loss of seipin results in aberrant lipid droplet (LD) biogenesis, leading to formation of various types of altered LDs. Loss of functional seipin on a systemic level in patients suffering from Berardinelli-Seip congenital lipodystrophy type 2, a disease caused by seipin mutations, results in a severe defect in adipogenesis (1). Although the importance of seipin for LD biology and human health is clear, the exact molecular function of this mysterious protein has been debated. Seipin resides in the ER membrane and comprises two transmembrane segments linked by a large, conserved luminal domain and two cytosolic termini (1). Seipin forms oligomers (2) that are ring shaped according to negative-stain EM (3). Furthermore, a number of seipin interaction partners has been identified, among them several key lipid metabolism enzymes (4).
Different hypotheses about the molecular role of seipin have been proposed (4). LD biogenesis starts at the ER, where neutral lipid synthesizing enzymes are located. LDs likely develop through accumulation of neutral lipid lenses between the ER membrane leaflets, which grow and bud toward the cytosol, leading to formation of balls of neutral lipids that are covered by a monolayer of phospholipids and proteins (1). One prominent hypothesis on seipin function states that the oligomeric complex has a structural role at the LD–ER interface for biogenesis and/or maintenance of normal LDs, and indeed, seipin has been detected at LD–ER contact sites (5, 6). A different hypothesis proposes a role of seipin in phospholipid and/or neutral lipid metabolism, possibly by acting as a scaffold that spatially controls these multistep processes. Furthermore, regulatory roles of seipin in ER calcium flux, cytoskeleton dynamics, or lipolysis have been suggested, as well as primary roles in adipogenesis. Detailed structural information on seipin promises more clarity on the mechanistic role of seipin, and has been long awaited.
In this issue, Sui et al. (7) report on a cryo-EM structure of seipin from Drosophila melanogaster that is similar to the structure of human seipin recently reported by Yan et al. (8). The cryo-EM map of fly seipin reveals a symmetrical dodecameric ring structure, while the human protein forms an undecameric ring, indicating that formation of ring-shaped oligomers is a conserved feature, while the exact size of the oligomer seems to be species specific. Both maps yielded high resolution atomic models of the conserved luminal domain of seipin, which is of particular interest, as it houses most of the missense mutations occurring in seipin-related lipodystrophy. In both models, a large part of this domain folds into two adjacent four-stranded β-sheets that together form a hollow fold with a hydrophobic pocket with similarities to lipid-binding domains (LBDs), for example the LBD of Niemann-Pick type C2 protein. The β-sheets are antiparallel and intertwined, one sheet consisting of β-strands 1, 8, 3, and 6 and opposed by strands 2, 7, 4, and 5 in the second sheet. The loop connecting strands 5 and 6 forms an α-helix located at the inner rim of the seipin ring and faces the surface of the ER membrane. This α-helix is enriched in conserved, large hydrophobic amino acids, a feature previously identified as a targeting determinant of a class of LD proteins. Unlike typical phospholipid bilayers, the monolayer that covers LDs uniquely exposes large, persistent membrane packing defects that are believed to be recognized by these large hydrophobic amino acids (9). Indeed, while full-length seipin is an ER protein, the α-helix alone preferentially binds to LDs. A similar preference for LDs has previously been described for a further α-helical subdomain of seipin located in its cytosolic N terminus (10), and simultaneous mutation of both of these helices makes seipin nonfunctional (7).
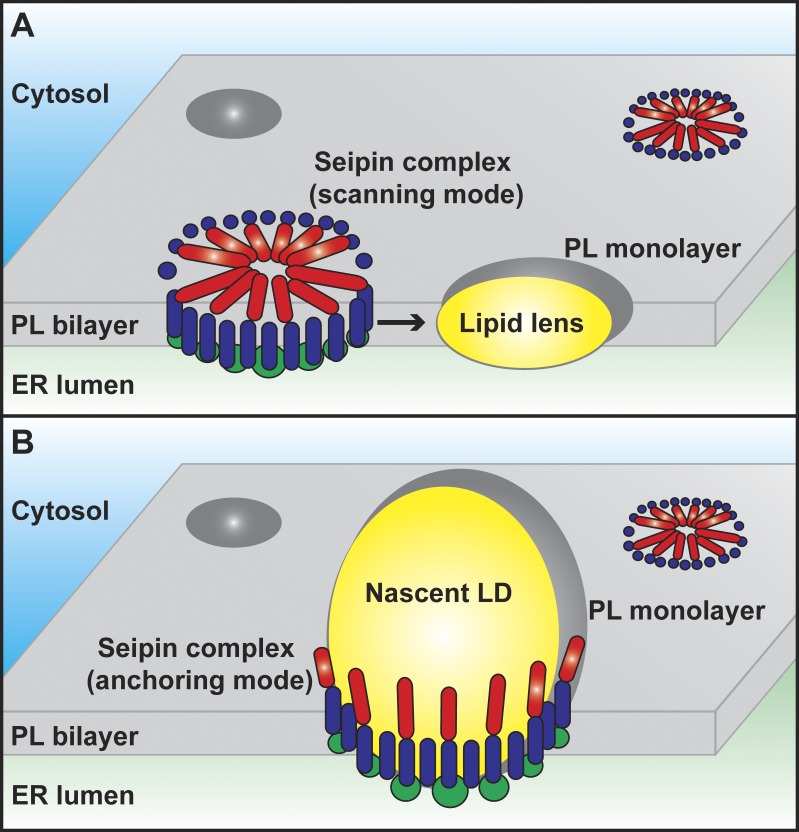
The structural information thus indicates that each seipin monomer combines, on one hand, hydrophobic features favoring the ER bilayer membrane, namely the two transmembrane domains, and on the other hand, features favoring the monolayer covering LDs, namely the luminal and N-terminal hydrophobic α-helices. Oligomerization might potentiate both affinities and thus give the seipin complex the characteristics required for a molecular mediator of the unique bilayer-to-monolayer connection at the site of LD biogenesis from the ER. Based on these features, Sui et al. (7) propose a novel model for the mechanistic role of seipin in LD biogenesis (Fig. 1). In this model, seipin oligomers move within the ER membrane and scan its surface from both sides with the hydrophobic α-helices for membrane packing defects induced by small neutral lipid lenses (Fig. 1 A). Once the oligomer detects such a lens, it stops scanning and becomes immobilized, now acting as a structural anchor that maintains the connection between the ER membrane and the surface of the developing LD (Fig. 1 B). This model is supported by earlier studies that described highly mobile seipin foci that move within the ER membrane until they eventually merge with nascent LDs (10). The N-terminal cytosolic α-helix might mediate stable docking of the LD onto the ER membrane and prevent aberrant formation of tiny LDs through premature physical separation from the parent organelle, an LD phenotype that indeed occurs in the absence of functional seipin (1).
Figure 1.
Hypothetical model of the molecular function of seipin. (A) Seipin oligomers anchored within the phospholipid (PL) bilayer of the ER via their transmembrane domains (dark blue) move within the plane of the membrane and scan its surface from both sides with hydrophobic cytosolic (red) and luminal (not depicted) helices that bind preferentially to lipid packing defects in the PL monolayer of lipid lenses and LDs. (B) Upon encountering a lipid lens, the seipin complex binds to the monolayer surface via its hydrophobic helices (red) and supports growth of the emerging LD by anchoring it to the ER bilayer. The putative LBD (green) may have a role in lipid transfer, signaling, or regulation of PL metabolism.
The seipin ring might have an additional structural role at the LD–ER connection by dictating the dimension of this unique lipidic structure. It is currently debated whether under physiological conditions, the lipidic LD–ER bridge is severed at some point in LD formation, or whether it is maintained throughout the lifetime of an LD. A seipin ring located at the LD–ER contact site could thus serve a role in maintenance of mature LDs, either though physical stabilization of the lipidic connection or through a gatekeeper function that contributes to establishing and maintaining identity of the LD surface as compared with the ER surface. Indeed, LDs in seipin mutants lose typical LD surface proteins (5, 6).
While this model is very intuitive with respect to the formation of an oligomeric ring arrangement and the requirement for bilayer-specific transmembrane domains and monolayer-specific hydrophobic α-helices, the additional presence of an LBD-like domain within the luminal part of seipin is intriguing and points toward functional implications beyond a mere structural role. Mutations within this domain are detrimental to seipin function, underlining its importance. A lipid binding role of this domain is supported by data in Yan et al. (8), who observe binding to anionic phospholipids and suggest phosphatidic acid as a potential ligand of this domain. Importantly, LBDs can fulfill diverse functions, including transfer of lipids from one place to another, lipid scavenging, lipid sensing, and presentation of lipids to other proteins. Therefore, potential roles of the putative seipin LBD include lipid sorting at the LD–ER interface, but also involvement in localized lipid metabolism reactions as well as regulatory or signaling roles; for example a possible lipid-mediated conformational switch of the seipin complex at some point during LD biogenesis, or the lipid-based integration of an external signal.
In conclusion, the seipin structures by Sui et al. (7) and Yan et al. (8) represent a milestone in the endeavor of unraveling the function of this key player in LD biogenesis and will likely tremendously advance the field by guiding future directed mechanistic investigations. Understanding the functional role of the putative seipin LBD might represent a future crucial step in this adventurous enterprise.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft, Cells-in-Motion Cluster of Excellence (EXC 1003 – CiM), University of Münster.
The author declares no competing financial interests.
References
- 1.Walther T.C., et al. . 2017. Annu. Rev. Cell Dev. Biol. 33:491–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sim M.F., et al. . 2014. Methods Enzymol. 537:161–175. [DOI] [PubMed] [Google Scholar]
- 3.Binns D., et al. . 2010. Biochemistry. 49:10747–10755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen X., and Goodman J.M.. 2017. Biochim Biophys Acta Mol Cell Biol Lipids. 1862:1205–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salo V.T., et al. . 2016. EMBO J. 35:2699–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grippa A., et al. . 2015. J. Cell Biol. 211:829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sui X., et al. J. Cell Biol. 2018 doi: 10.1083/jcb.201809067. [DOI] [Google Scholar]
- 8.Yan R., et al. . 2018. Dev. Cell. 47:248–256.e4. [DOI] [PubMed] [Google Scholar]
- 9.Prévost C., et al. . 2018. Dev. Cell. 44:73–86.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang H., et al. . 2016. eLife. 5:e16582.27564575 [Google Scholar]