Keratin 6 (K6) isoforms are induced in wound-proximal keratinocytes after injury to skin. Paradoxically, absence of K6 isoforms leads to faster directional cell migration. Wang et al. report that K6 promotes collective keratinocyte migration by interacting with desmoplakin and myosin IIA and stabilizing cell adhesion.
Abstract
The a and b isoforms of keratin 6 (K6), a type II intermediate filament (IF) protein, are robustly induced upon injury to interfollicular epidermis. We previously showed that complete loss of K6a/K6b stimulates keratinocyte migration, correlating with enhanced Src activity. In this study, we demonstrate that this property is cell autonomous, depends on the ECM, and results from elevated speed, enhanced directionality, and an increased rate of focal adhesion disassembly. We show that myosin IIA interacts with K6a/K6b, that its levels are markedly reduced in Krt6a/Krt6b-null keratinocytes, and that inhibiting myosin ATPase activity normalizes the enhanced migration potential of Krt6a/Krt6b-null cells. Desmoplakin, which mediates attachment of IFs to desmosomes, is also expressed at reduced levels and is mislocalized to the nucleus in Krt6a/Krt6b-null cells, correlating with defects in cell adhesion. These findings reveal that K6a/K6b modulate keratinocyte migration by regulating cell–matrix and cell–cell adhesion and highlight a role for keratins in collective cell migration.
Introduction
Given its role as a vital barrier, the skin has the ability to rapidly repair wounds after injury or other forms of aggression. Wound healing is a complex process that involves several cellular and noncellular elements including immune and endothelial cells, fibroblasts, keratinocytes, and the ECM (Martin, 1997; Gurtner et al., 2008). Upon injury, surviving wound-proximal keratinocytes transiently suspend terminal differentiation and undergo dramatic changes to prepare for migration (Coulombe, 2003). These entail dramatic changes in cell size and shape, in cell–cell and cell–matrix adhesion, and in the cytoskeleton, including changes in the composition and intracellular organization of keratin intermediate filaments (IFs; Paladini et al., 1996). Normally restricted to ectoderm-derived epithelial appendages (e.g., hair follicle, nail, tooth, thymus, and various glands) and palmar/plantar epidermis in adult skin epithelia, the expression of the type II IFs K6a and K6b isoforms and their type I partners K16 and K17 is rapidly and robustly induced near the wound after injury (Paladini et al., 1996; McGowan and Coulombe, 1998a; Takahashi et al., 1998). In epidermis, this induction occurs at the expense of the K1/K10 keratin pair normally expressed during terminal differentiation (McGowan and Coulombe, 1998a; Jacinto et al., 2001; Bernot et al., 2002). Expression of K6a, K6b, K16, and K17 persists at least until the wound is closed and barrier function is restored, suggesting an important physiological role during repair. Studies making use of null mouse models have established that in addition to affecting the growth (Kim et al., 2006) and migration (Wong and Coulombe, 2003) of keratinocytes, these keratin proteins modulate the expression of several inflammatory and immune mediators in a cell-autonomous fashion (Depianto et al., 2010; Lessard et al., 2013; Chung et al., 2015; Hobbs et al., 2015).
Mice carrying a germline null mutation at the Krt6a/Krt6b locus physically appear normal at birth but die during the first week of life secondary to massive oral epithelial blistering (reflecting tissue fragility), resulting in poor nutrition and ultimately a failure to thrive (Wong et al., 2000). When skin from newborn Krt6a/Krt6b-null mice is seeded for explant culture ex vivo, keratinocytes exhibit an enhanced migration potential correlating with increased phosphorylation of specific tyrosine epitopes (Wong and Coulombe, 2003), one of which is the tyrosine kinase Src (Rotty and Coulombe, 2012). Followup research showed that K6a and/or K6b bind Src directly, dampening its enzymatic activity and decreasing its partitioning to detergent-resistant membranes in keratinocytes (Rotty and Coulombe, 2012). These findings suggest that the induction of K6a/K6b proteins in wound-proximal keratinocytes attenuates their migration potential in vivo (Rotty and Coulombe, 2012).
We hypothesized that K6a/K6b may be important for the proper coordination and regulation of collective keratinocyte migration during reepithelialization. In this study, we report that the enhanced cell migration observed in the absence of K6a/K6b proteins is both a cell-collective and cell-autonomous property and is directly related to modulation of cell–cell and cell–matrix adhesion. We uncover a new interaction between K6 and myosin IIA, an F-actin–dependent motor protein with pleiotropic roles in cytoarchitecture, adhesion, and force generation (Vicente-Manzanares et al., 2009), and a new interdependence between K6 and desmoplakin (DP), which mediates attachment of keratin IFs to desmosomes (Simpson et al., 2011). Our findings suggest that the physiological basis for K6a/K6b induction in response to injury lies in the need to optimize adhesion dynamics in a manner that fosters efficient collective cell migration and wound reepithelialization.
Results
Enhanced migration of Krt6a/Krt6b-null keratinocytes is a cell-autonomous trait
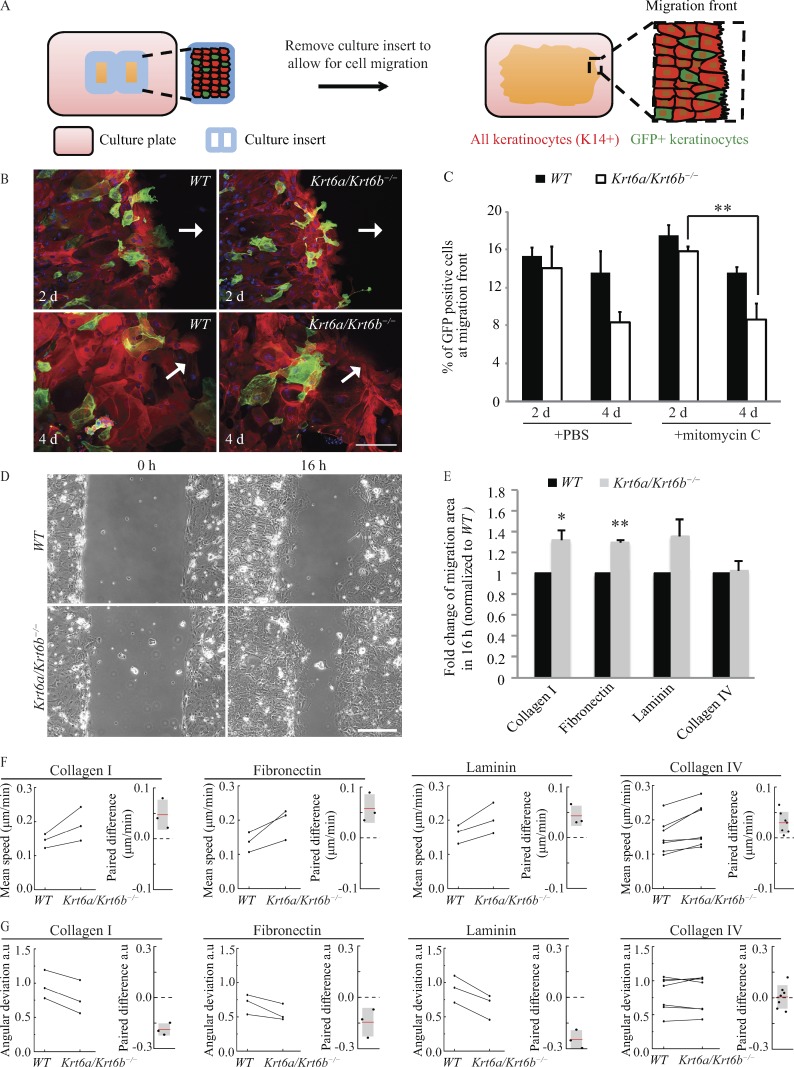
We first investigated whether the increased migration of Krt6a/Krt6b-null keratinocytes represents a cell-autonomous property or is influenced by motogenic factors present in the extracellular milieu or expressed at the cell surface. To assess this, we devised a coculture migration assay that uses keratinocytes isolated from newborn mice constitutively expressing a reporter GFP-actin fusion protein (Vaezi et al., 2002) as a tracer for cells featuring normal migratory properties. These tracer cells were mixed in a 1:4 ratio with keratinocytes derived from either newborn WT or Krt6a/Krt6b-null mice (C57Bl/6 strain) and seeded for culture on type I collagen–coated slides featuring inserts (Fig. 1 A; see Materials and methods). Upon cell confluence, the inserts were removed from the plates to allow for cell migration. 2 and 4 d later, the samples were fixed, and the percentage of GFP-positive keratinocytes at the migration front was quantified. In the event that Krt6a/Krt6b-null keratinocytes secrete or display a factor capable of enhancing migration, the tracer cells should keep pace with Krt6a/Krt6b-null cells and exhibit a similar enhancement of migratory properties with time.
Figure 1.
Increased migration of Krt6a/Krt6b-null keratinocytes is cell autonomous and a result of enhanced speed and more coordinated movement. (A) Schematic of the coculture migration assay. GFP (green) labeled tracer keratinocytes were mixed with either WT or Krt6a/Krt6b-null keratinocytes and plated within a culture insert placed on top of a coverslip coated with type I collagen. K14 (red) labeled all keratinocytes. When cells were confluent, the culture insert was removed to allow for cell migration. The cells at migration front (rectangular area with dashed lines) were imaged, and the percentage of GFP-positive cells within these areas (≥15 images) was quantified. (B) WT and tracer keratinocytes seemed to have equivalent potential for migration, whereas Krt6a/Krt6b-null keratinocytes migrated at a faster rate than tracer keratinocytes. Representative images show cells that were treated with PBS vehicle control. K14 is in red, and DAPI is in blue. GFP (green) staining represents tracer keratinocytes expressing GFP-tagged actin. Arrows point toward the direction of migration. (C) Data represent the mean + SEM for at least three biological replicates. **, P < 0.01, Student’s two-tailed t test. (D) Phase-contrast imaging was performed to monitor cell migration (Video 1). Representative images show WT and Krt6a/Krt6b-null keratinocytes migrating on type I collagen. Bars, 200 µm. (E) Krt6a/Krt6b-null keratinocytes had an increased area of migration when migrating on type I collagen, fibronectin, and laminin but not on type IV collagen. Data represent the mean + SEM of three or more biological replicates. *, P < 0.05; **, P < 0.01, Student’s two-tailed t test. (F and G) PIV analysis showed that compared with a WT cell sheet, a Krt6a/Krt6b-null cell sheet had an enhanced speed of migration on type I collagen, fibronectin, laminin, and to a lesser extent, type IV collagen. Krt6a/Krt6b-null cell sheet was more coordinated than WT cell sheet on type I collagen, fibronectin, and laminin, but not on type IV collagen (Videos 2 and 3). Experiments from the same day were paired and linked with a line. Paired difference was calculated by subtracting value of WT cells from that of Krt6a/Krt6b-null cells.
For cocultures involving GFP-actin and WT keratinocytes, the percentage of GFP-positive keratinocytes at the migration front were similar after both 2 and 4 d of comigration (15.4 ± 0.8% vs. 13.6 ± 1.1%; Fig. 1, B and C), suggesting that introducing GFP-actin into keratinocytes minimally impacted cell migration and that GFP-actin and WT keratinocytes have an equivalent potential for migration in this experimental setting. In cocultures involving GFP-actin and Krt6a/Krt6b-null keratinocytes, by contrast, the fraction of GFP-actin–labeled cells at the migration front underwent a marked decrease over the course of 2 d of comigration (14.0 ± 2.3% after 2 d vs. 8.4 ± 0.6% after 4 d; P = 0.06; Fig. 1, B and C), implying that Krt6a/Krt6b-null keratinocytes migrate faster than GFP-actin–expressing “tracer” keratinocytes. A similar outcome was obtained when this competition assay was repeated after mitomycin C treatment, which inhibits cell proliferation (Fig. 1 C; Wawersik et al., 2001). From these results, we conclude that the enhanced migration potential of Krt6a/Krt6b-null keratinocytes largely represents a cell-autonomous trait that does not require active cell proliferation.
Migrating Krt6a/Krt6b-null keratinocytes show enhanced directionality and speed depending on the ECM substratum
Activated epidermal keratinocytes use distinct integrin pairs to interact with the provisional ECM as they migrate into wounds (O’Toole, 2001). Using live microscopy (Fig. 1 D and Video 1; see Materials and methods), we next sought to determine whether the increased migration of Krt6a/Krt6b-null keratinocytes depends on the ECM. Compared with WT control cells, we found that Krt6a/Krt6b-null keratinocytes migrated faster when plated on type I collagen (1.32-fold; P = 0.026) or fibronectin (1.30-fold; P = 0.0003) and exhibited a similar trend on laminin (1.36-fold; P = 0.096; Fig. 1 E). All three ECM proteins are enriched in the provisional tissue matrix of wounds (Arnoux et al., 2005). By contrast, no difference was observed when WT and Krt6a/Krt6b-null keratinocytes migrated on type IV collagen (1.02-fold; P = 0.817; Fig. 1 E), which is prevalent in the basal lamina of intact skin. To better define the migration phenotype of migrating cells, we next used particle image velocimetry (PIV; Petitjean et al., 2010; Weiger et al., 2013; Lee et al., 2016; see Materials and methods) to generate a velocity map of the entire sheet of migrating keratinocytes from time-lapse images (see Fig. S1). Images were divided into subwindows (smaller than an individual cell), and intracellular details within these subwindows were used as tracers to calculate correlations between successive images, thus generating a map of local displacements (Petitjean et al., 2010). This PIV analysis revealed that Krt6a/Krt6b-null cells collectively displayed enhanced speed when migrating on type I collagen, fibronectin, and laminin, and less so when plated on type IV collagen (Fig. 1 F). Relative to WT cells, Krt6a/Krt6b-null keratinocytes also exhibited more coordinated movements when plated and migrating on type I collagen, fibronectin, or laminin as indicated by a smaller angular deviation in their displacement (see Materials and methods)—a behavior that was not observed on type IV collagen (Fig. 1 G). The PIV analyses also clearly showed that directionality vectors are similar across large fields of keratinocytes in each genotype, strongly suggesting that the cells largely migrate as a coordinated sheet (Videos 2 and 3). These findings show that the Krt6a/Krt6b knockout–induced enhancement of keratinocyte migration depends on ECM composition and that it leads to elevated speed and improved directionality for individual keratinocytes that make up the migrating sheet, highlighting the complexity of this migration behavior.
Krt6a/Krt6b-null cells display increased focal adhesion turnover on type I collagen
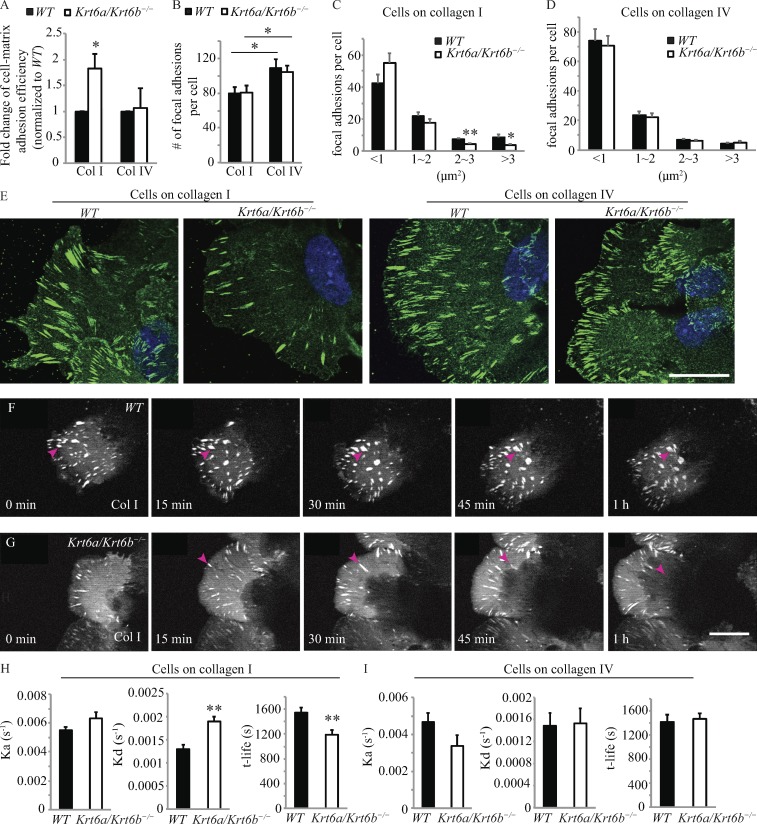
Actively migrating cells exhibit a self-sustained polarized cytoarchitecture (Kriebel et al., 2003; Ridley et al., 2003; Mayor and Etienne-Manneville, 2016). At their front end, they extend lamellipodia and filopodia, in the direction of migration, that then attach to the ECM via the formation of new integrin-based focal adhesions, and at their rear end, they dismantle previously formed focal adhesions to detach from the substratum and enable forward movement. The observation that Krt6a/Krt6b-null keratinocytes show enhanced migration on type I but not type IV collagen is intriguing in light of the fact that adhesion of keratinocytes to both ECM molecules is mediated by the same integrin receptor α2β1 (Vandenberg et al., 1991; Tuckwell et al., 1995). This prompted us to probe deeper into cell–matrix adhesion and focal adhesions in particular.
We found that skin keratinocytes freshly harvested from Krt6a/Krt6b-null and WT newborn mice differed in their adhesive properties to type I collagen, with significantly higher adhesion observed for null cells, but showed no difference when plated on type IV collagen (Fig. 2 A). Followup studies showed that both WT and Krt6a/Krt6b-null keratinocytes exhibit a higher total number of focal adhesions when plated on type IV relative to type I collagen; otherwise there was no difference between genotypes on each of these matrices (Fig. 2 B). Breaking down focal adhesions by size revealed ECM-specific differences between genotypes. Krt6a/Krt6b-null keratinocytes showed a lesser number of large focal adhesions (≥2 µm2) than WT keratinocytes on type I collagen (Fig. 2, C and E), suggesting enhanced turnover of focal adhesions in the absence of K6a/K6b. In contrast, the distribution of focal adhesion sizes was indistinguishable in Krt6a/Krt6b-null and WT keratinocytes plated on type IV collagen (Fig. 2, D and E).
Figure 2.
Increased disassembly rate results in shorter lifetime of focal adhesions in Krt6a/Krt6b-null keratinocytes. (A) Krt6a/Krt6b-null keratinocytes adhered more efficiently to type I collagen than WT keratinocytes. WT and Krt6a/Krt6b-null keratinocytes had similar adhesion efficiency to type IV collagen. Data represent the mean + SEM of six biological replicates. (B) Cells possessed increased number of focal adhesions on type IV collagen than cells on type I collagen. Data represent the mean + SEM of 30 cells from two or three biological replicates. (C) On type I collagen, Krt6a/Krt6b-null keratinocytes have less large focal adhesion than WT keratinocytes. Data represent the mean + SEM of 30 cells from three biological replicates. (D) The size of focal adhesions was not affected by the loss of K6a/K6b when keratinocytes were migrating on type IV collagen. Data represent the mean + SEM of 30 cells from two biological replicates. (E) Immunofluorescence images of endogenous paxillin (green) in WT and Krt6a/Krt6b-null keratinocytes on type I and type IV collagens. Cells were located at the migration leading edge. DAPI, blue. (F and G) Time-lapse apotome microscopy of mCherry–paxillin accumulation in focal adhesions in WT and Krt6a/Krt6b-null cells (Video 4). Arrowheads followed the same focal adhesion over time. Bars, 20 µm. (H) Compared with WT keratinocytes, Krt6a/Krt6b-null keratinocytes had an enhanced rate of disassembly and a decreased focal adhesions lifetime on type I collagen (Video 4). Data represent the mean + SEM of 10 cells. *, P < 0.05; **, P < 0.01, Student’s two-tailed t test. (I) WT and Krt6a/Krt6b-null keratinocytes have similar focal adhesion turnover rate on type IV collagen (Video 5). Data represent the mean + SEM of five cells.
Next, we investigated focal adhesion dynamics in WT and Krt6a/Krt6b-null keratinocytes undergoing migration on type I or IV collagen. mCherry-tagged paxillin, a highly reliable marker of focal adhesion (Laukaitis et al., 2001; Webb et al., 2004; Hu et al., 2007), was used as a reporter for live-imaging studies (see Materials and methods). We found that focal adhesions first appeared at the leading edge of both WT and Krt6a/Krt6b-null migrating cells and became larger as a function of time as they moved gradually toward the center of cells (Fig. 2, F and G; and Videos 4 and 5). Interestingly, the focal adhesions that formed in Krt6a/Krt6b-null keratinocytes plated on type I collagen followed a similar spatiotemporal pattern but appeared to turn over more rapidly compared with WT keratinocytes (Fig. 2, F and G). To confirm this, we used ImageJ to systematically measure the intensity of the mCherry fluorescence associated with individual focal adhesions for every time frame recorded from formation to disappearance. From these individual intensity measurements, we calculated the rates of focal adhesion assembly and disassembly and their lifetime using an established algorithm (Stehbens et al., 2014; Stehbens and Wittmann, 2014). When plated on type I collagen, mCherry-paxillin–labeled focal adhesions disassemble at markedly faster rates (by 1.47-fold; P = 0.0005) in Krt6a/Krt6b-null keratinocytes relative to WT keratinocytes. When combined with the slightly increased assembly rate occurring in Krt6a/Krt6b-null keratinocytes (by 1.15-fold; P = 0.1), this yields a significantly shorter overall focal adhesion lifetime in the mutant cells (P = 0.006; Fig. 2 H and Video 4). This behavior was specific for cells plated on type I collagen as no statistically significant differences were seen in the assembly and disassembly rates and lifetime of focal adhesions in Krt6a/Krt6b-null and WT keratinocytes migrating on type IV collagen (Fig. 2 I and Video 5). These findings suggest that the enhanced migration of Krt6a/Krt6b-null keratinocytes on type I collagen is facilitated in part by an enhanced rate of disassembly and overall turnover of focal adhesions.
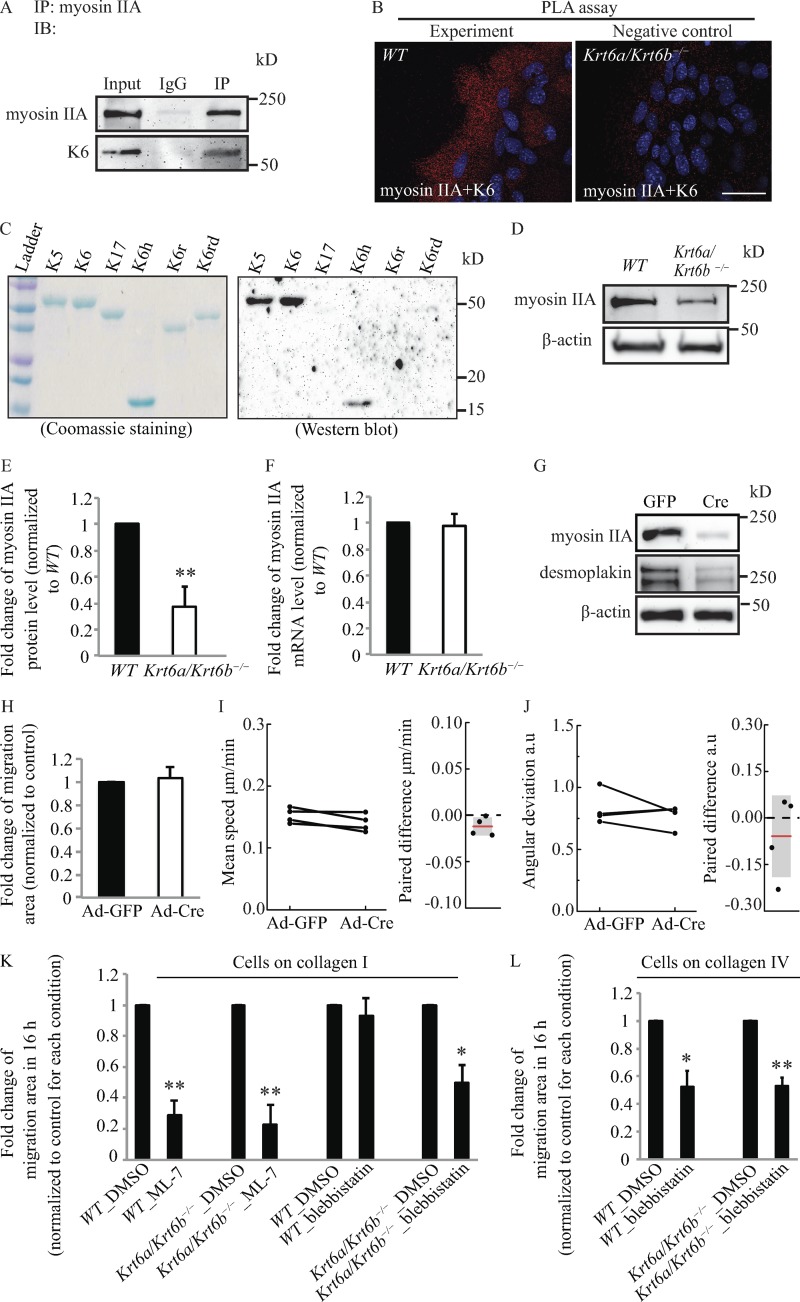
K6 interacts with and stabilizes myosin IIA
We next explored how K6a/K6b might regulate focal adhesion disassembly by searching for proteins that impact focal adhesion turnover and interact with keratins. Myosin II plays essential roles in adhesion and migration in part by regulating the rates of maturation and disassembly of focal adhesions through actin-myosin contraction (Vicente-Manzanares et al., 2009; Parsons et al., 2010). Fibroblasts deficient in a specific myosin II isoform, myosin IIA, exhibit reduced contractile properties and enhanced migration as single cells (Even-Ram et al., 2007). Myosin IIA also functions as a tumor suppressor in squamous cell carcinoma (Schramek et al., 2014). Myosin IIA was previously shown to interact with K8/K18 in A549 cells in culture and in mouse liver (Kwan et al., 2015). We therefore probed for a potential interaction between K6 and myosin IIA in mouse skin keratinocytes. Coimmunoprecipitation assays revealed that endogenous myosin IIA and K6 proteins physically interact in keratinocytes (Fig. 3 A). Proximity ligation assays (PLAs) suggested that K6 and myosin IIA are physically proximal (<40 nm) in the cytoplasm of skin keratinocytes (Fig. 3 B). Far-Western assays involving purified proteins in vitro suggested that purified myosin II and K6 can directly interact with each other (Fig. 3 C). Accordingly, we next examined the status of myosin IIA in Krt6a/Krt6b-null keratinocytes. Relative to WT cells, actively migrating Krt6a/Krt6b-null cells exhibited a marked reduction in myosin IIA protein levels (P = 0.009; Fig. 3, D and E) whereas the Myh9 mRNA remained within normal range (Fig. 3 F), suggesting that K6 plays a role in stabilizing myosin IIA protein levels in normal skin keratinocytes.
Figure 3.
K6a/K6b may regulate cell migration via interacting with myosin IIA. (A) Coimmunoprecipitation assay demonstrated that myosin IIA interacted with K6. n = 3 biological repeats. IB, immunoblot; IP, immunoprecipitation. (B) PLA showed that myosin IIA associated with K6. n = 4 biological repeats. Bar, 50 µm. The white values for all signals were set the same in WT and Krt6a/Krt6b-null keratinocytes. (C) Far-Western assay indicates that K6 may directly interact with myosin II via K6 head domain. Myosin II was the bait protein. Myosin IIA antibody was used to detect the binding of myosin II to proteins on the membrane. (D and E) Myosin IIA protein level was reduced in whole-cell lysates harvested from 6-d Krt6a/Krt6b-null skin explant culture. β-Actin was used as a loading control. Each bar represents the mean + SEM of six biological replicates. (F) The absence of K6a/K6b proteins did not affect the mRNA level of myosin IIA as measured by qRT-PCR with Gapdh and Rn18s as loading controls. Each bar represents the mean + SEM of three biological replicates. (G) Western blot analysis of whole-cell lysates harvested from 6-d skin explant culture indicated that adenovirus-delivered Cre (Ad-Cre)–induced Myh9 deletion in WT keratinocytes led to a decrease in DP protein level. Data represent the results of three biological repeats. (H) Cre-recombinase–mediated knockout of floxed-Myh9 did not result in increased connective cell migration (Video 6). Data represent the mean + SEM of four biological replicates. (I and J) PIV analysis indicated that neither speed nor angular deviation was affected by Cre-mediated knockout of Myh9. Experiments from the same day were paired and linked with a line. Paired difference was calculated by subtracting the value derived from GFP-expressing control cells from that of the Cre-expressing cells. (K) On type I collagen, MLCK inhibitor, ML-7, inhibited the migration of WT and Krt6a/Krt6b-null keratinocytes. Inhibition of myosin II by blebbistatin significantly decreased migratory potential of Krt6a/Krt6b-null keratinocytes but only minimally affected that of WT keratinocytes on type I collagen. Data represent the mean + SEM of four to five biological replicates. (L) On type IV collagen, blebbistatin reduced the migration of both WT and Krt6a/Krt6b-null keratinocytes. Data represent the mean + SEM of four biological replicates. *, P < 0.05; **, P < 0.01, Student’s two-tailed t test.
We next tested for a functional role for myosin IIA in migrating skin keratinocytes by inducing a Cre recombinase–mediated deletion of the Myh9 allele. Although this strategy significantly lowered myosin IIA protein levels (Fig. 3 G), it had no impact on migration efficiency as a whole (Fig. 3 H) or on migration speed and directionality in particular (Fig. 3, I and J). However, depletion of myosin IIA had a sizable impact on cell–cell adhesion as manifested through the occurrence of unusual gaps between migrating keratinocytes (Fig. S2 A and Video 6) along with a reduction in the levels of DP (Fig. 3 G). Additional information relating K6a/K6b to cell–cell adhesion is reported below.
The lack of an impact of Myh9 silencing on keratinocyte collective migration may reflect redundancy among myosin II isoform A (Myh9), B (Myh10), and C (Myh14; see Vicente-Manzanares et al., 2009; Human Protein Atlas, http://www.proteinatlas.org), prompting us to resort to a pharmacological approach to directly inhibit myosin contractility. Phosphorylation on Ser 19 of myosin II regulatory light chain by myosin light chain kinase (MLCK) regulates actomyosin contractility (Somlyo and Somlyo, 1994) and enhances myosin II ATPase activity (Sellers et al., 1981). We treated WT and Krt6a/Krt6b-null keratinocytes with ML-7, an inhibitor for MLCK (Totsukawa et al., 2004; Sarkar et al., 2009), and blebbistatin, an inhibitor of myosin II ATPase activity (Kovács et al., 2004). Similar to previous research involving fibroblasts (Totsukawa et al., 2004), ML-7 treatment attenuated the migration of WT and Krt6a/Krt6b-null keratinocytes plated on type I collagen to a comparable degree, i.e., by ∼71% (P = 0.002) and ∼78% (P = 0.004), respectively (Fig. 3 K). Interestingly, this was accompanied by increased average size for focal adhesions (Fig. S2 B). Treatment with blebbistatin significantly decreased the migration of Krt6a/Krt6b-null keratinocytes plated on type I collagen (by ∼50%; P = 0.021) but minimally impacted that of WT keratinocytes (Fig. 3 K). On type IV collagen, blebbistatin treatment decreased the migration of both WT and Krt6a/Krt6b-null keratinocytes, i.e., by ∼48% (P = 0.025) and ∼47% (P = 0.005), respectively (Fig. 3 L). Further, blebbistatin treatment disrupted focal adhesion structure in both Krt6a/Krt6b-null and WT keratinocytes (Fig. S2, B and C). These findings establish that myosin II ATPase activity is essential for the enhanced migration potential exhibited by Krt6a/Krt6b-null keratinocytes plated on type I collagen. They also support a novel role for K6a/K6b protein in interacting with and regulating myosin II, adding to the notion that the role of myosin II in the setting of keratinocyte migration is complex (see Discussion).
Actively migrating Krt6a/Krt6b-null cells show striking alterations in DP and cell–cell adhesion
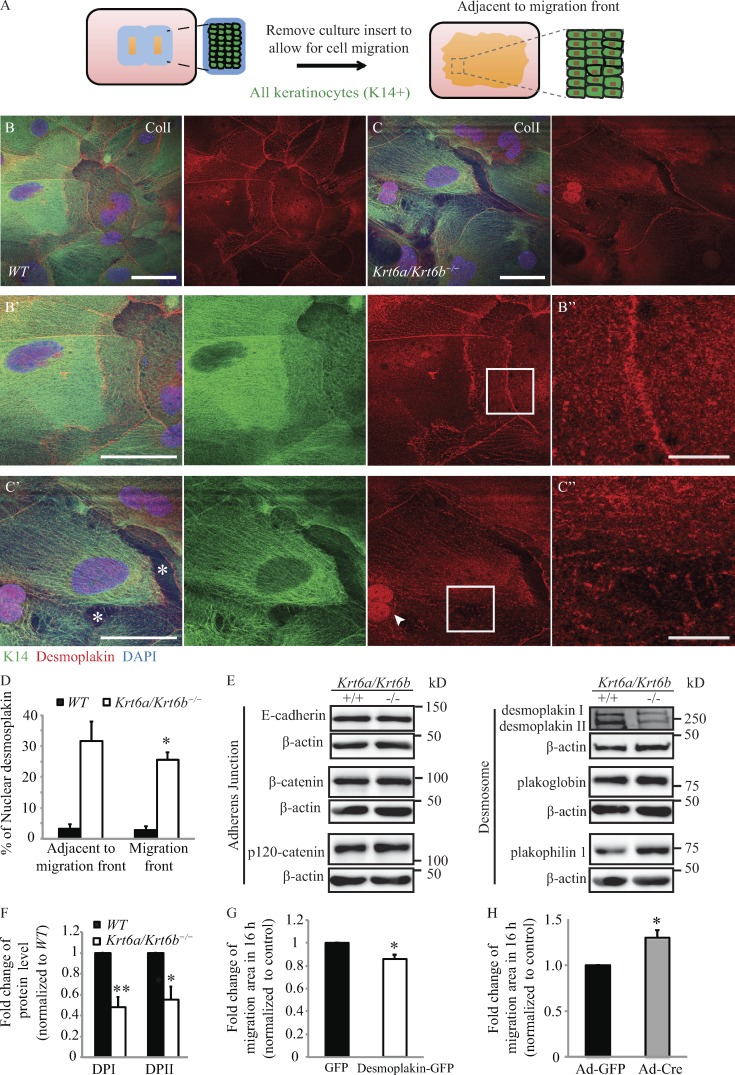
Keratin IFs are robustly tethered at sites of desmosome cell–cell adhesion in the intact epidermis (Simpson et al., 2011). Such anchorage of keratin IFs at desmosomes is key to maintaining the integrity and resilience of keratinocyte sheets and keratinocyte-rich tissue assemblies (Kröger et al., 2013). Mechanistically, the amino-terminal head region of select type II keratins (including K1, K2, K5, and K6) directly associates with the carboxy terminus of DP, a key constituent of desmosomal plaques (Kouklis et al., 1994). After wounding, however, keratinocytes proximal to the wound edge display a reduced number of desmosomes along with intercellular gaps (e.g., Paladini et al., 1996; Garrod et al., 2005; Savagner et al., 2005), suggesting that attenuated cell–cell adhesion favors reepithelialization. In support of the latter, overexpression of a deletion mutant that contains only the first 584 amino acids of DP protein (thus missing the entire C-terminal tail domain) uncouples IFs from desmosomes and leads to accelerated keratinocyte migration ex vivo (Setzer et al., 2004).
We observed that migrating Krt6a/Krt6b-null keratinocytes show clear signs of defective cell–cell adhesion. 4 d after inserts were removed from the culture chambers (see diagram in Fig. 4 A), we found that WT keratinocytes that had migrated into the wound space were tightly packed and organized as a cohesive cell sheet on type I collagen (Fig. 4 B). In contrast, for Krt6a/Krt6b-null keratinocyte sheets on type I collagen, we observed frequent gaps between cells (Fig. 4 C), pointing to defects in cell–cell adhesion and sheet formation. Similar findings were also observed for Krt6a/Krt6b-null keratinocyte cultures after 2 d migration (not depicted). These observations were extended by immunostainings for DP, E-cadherin, plakophilin 1, and β-catenin (Fig. 4, B–C″; and Fig. S3 A–D′). As expected, DP localized to the outer cellular membrane and the cytoplasm in WT keratinocytes (Fig. 4, B–B″). Although this also proved true in Krt6a/Krt6b-null keratinocytes, these cells exhibited decreased DP signal at cell–cell borders relative to WT controls and showed DP inside the nucleus (Fig. 4, C–C″; and Fig. S4). Quantitation of microscopy images showed that 25.5 ± 2.4% (migration front) and 31.5 ± 6.5% (adjacent to migration front) of Krt6a/Krt6b-null keratinocytes had prominent intranuclear staining for DP (Figs. 4 D and S4). By contrast, only 3.0 ± 1.4% and 3.0 ± 1.5% of WT keratinocytes showed distinct intranuclear DP staining, respectively, at the migration front and area behind it (Figs. 4 D and S4). Such alterations in subcellular localization were paralleled by a marked reduction (∼52% for DPI, P = 0.003; ∼45% for DPII, P = 0.016) in the steady-state levels of DP protein in whole-cell lysates prepared from 6-d skin explant cultures (Fig. 4, E and F). Among several other adhesion proteins examined, a difference was also observed in the level of plakophilin 1, given an approximately threefold increase in Krt6a/Krt6b-null keratinocytes upon Western blot analysis (P = 0.029; Figs. 4 E and S3 E). Plakophilin 1 has been shown to be critical for maintaining desmosomal adhesion and tight junction (Rietscher et al., 2016). Although they colocalized in both WT and Krt6a/Krt6b-null keratinocytes, plakophilin 1 and β-catenin were not organized in a fine line at cell–cell interfaces in Krt6a/Krt6b-null compared to WT keratinocytes (Fig. S3, C–D′). Moreover, in the absence of K6a/K6b, there was increased plakophilin 1 in the nucleus and in the cytoplasm (Fig. S3 D). Target-specific quantitative PCR assays indicated that mRNA levels for genes encoding for both adherens junction and desmosomal proteins were similar between WT and Krt6a/Krt6b-null cultures (Fig. S5 A), suggesting that the alterations reported above reflect altered posttranscriptional regulation of select adhesion proteins in the absence of K6a/K6b.
Figure 4.
Migrating Krt6a/Krt6b-null keratinocytes show disrupted cell–cell adhesion, and loss of DP promotes collective keratinocyte migration. (A) Schematic of the experiment. The rectangular area (with dashed lines) shows area adjacent to migration front. (B and C) In the absence of K6a/K6b proteins, the amount of DP at cell–cell adhesion (B′, B″, C′, and C″) was reduced, and DP localized to the nucleus of keratinocytes on type I collagen. Asterisks mark the gaps between keratinocyte sheets. Arrowheads point to nuclear DP. Bars: 50 µm (B, B′, C, and C′); 10 µm (B″ and C″). (D) The percentage of cells that showed a distinct nuclear localization of DP is presented as the mean + SEM of three biological replicates. (E) Western blot analysis of whole-cell lysates harvested from 6-d skin explant culture showed that Krt6a/Krt6b-null keratinocytes had a reduced level of DP and increased level of plakophilin 1. β-Actin was used as a loading control. n = six biological repeats. (F) Quantification of Western blot results of six biological replicates. Data represent the mean + SEM. (G) The overexpressing of DP-GFP in Krt6a/Krt6b-null keratinocytes led to a reduction in migration area. Data represent the mean + SEM of five biological replicates. (H) Adenovirus delivered Cre (Ad-Cre)–induced Dsp deletion in WT keratinocytes resulted in an increase in migration area (Video 7). Data represent the mean + SEM of three biological replicates. *, P < 0.05; **, P < 0.01, Student’s two-tailed t test.
Followup studies provided evidence that altering DP levels impacts the migratory properties of keratinocyte sheets. On one hand, overexpression of DP in faster-migrating Krt6a/Krt6b-null keratinocytes resulted in a modest, albeit significant, reduction in migration area (by 14%; P = 0.03) relative to controls (Fig. 4 G; note that transfection efficiency in these primary cell cultures was only ∼23%). On the other hand, Cre-mediated deletion of Dsp in Dspfl/fl keratinocytes, in which the Krt6a/Krt6b locus is intact, resulted in a ∼1.30-fold increase in migration area (P = 0.02; Fig. 4 H and Video 7). These findings suggest that the increased migratory potential of Krt6a/Krt6b-null keratinocytes is related in part to a reduction in the levels of DP protein and the associated changes in cell–cell adhesion.
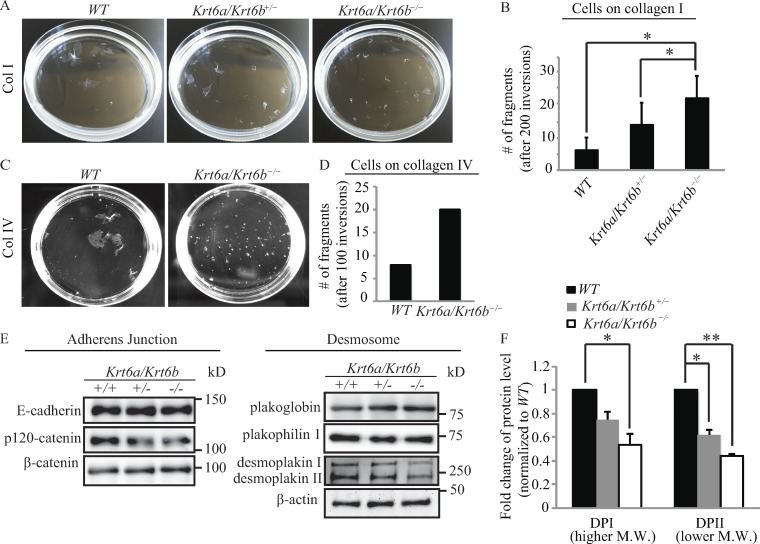
Confluent cultures of Krt6a/Krt6b-null cells exhibit weakened cell–cell adhesion
We next investigated the strength of cell–cell adhesion by subjecting keratinocyte sheets forming postconfluence to dispase lifting followed by mechanical stress–induced tumbling in solution (Yin et al., 2005; Kröger et al., 2013). We found that relative to WT keratinocytes, confluent sheets formed by Krt6a/Krt6b-null keratinocytes on type I collagen fragmented earlier (not depicted) and yielded an approximately threefold increase in the number of fragments (P = 0.022; Fig. 5, A and B). This differential outcome is likely not due to cell fragility or lysis since the activity level of soluble lactate dehydrogenase (LDH), an intracellular marker (Yin et al., 2005), was comparable in cells corresponding with the two genotypes and markedly lower than that observed in WT keratinocytes subjected to deliberate lysis as a positive control (Fig. S5 B). Relative to WT controls, cell sheets formed by culturing Krt6a/Krt6b-null keratinocytes on type IV collagen also fragmented earlier (not depicted) and yielded more fragments (Fig. 5, C and D), pointing to an interesting difference with findings focused on cell–matrix adhesion.
Figure 5.
The loss of K6a/K6b proteins impairs the mechanical integrity of keratinocyte sheets in stationary culture. (A and C) Dispase-based assay revealed that Krt6a/Krt6b-null keratinocyte sheets display compromised stress resilience. (B) Quantification of the total number of fragments generated after 200 inversions. Data represent as the mean + SEM of four biological replicates. (D) Quantification of the total number of fragments generated after 100 inversions. Data represent the mean + SEM of two biological replicates. (E) By Western blot, the protein level of DP was reduced in the absence of K6a/K6b. Whole-cell protein lysates were harvested from keratinocytes that were grown in the same condition as that for the keratinocytes used in dispase-based assay. β-Actin was used as a loading control. n ≥ three biological repeats. (F) Data shown represent the mean + SEM of three biological replicates. *, P < 0.05; **, P < 0.01, Student’s two-tailed t test. M.W., molecular weight.
Next, we used Western immunoblotting to assess the steady-state level of key proteins residing at adherens junctions and desmosomes in whole-cell protein lysates prepared from keratinocytes under the conditions used for the dispase assay. When cultured on type I collagen and relative to WT cells, Krt6a/Krt6b hemizygous null cells showed 25% (P = 0.056) and 38% (P = 0.012) decreases, respectively, in the levels of DPI and DPII. Krt6a/Krt6b homozygous null cells showed as much as 47% (P = 0.041) and 56% (P = 0.002) decreases in DPI and DPII, respectively (Fig. 5, E and F). No significant change was observed in the level of any of the other adhesion proteins tested, including E-cadherin, p120-catenin, β-catenin, plakoglobin, and plakophilin 1 (Fig. 5 E). These findings establish that keratinocytes lacking K6a/K6b exhibit weakened cell–cell adhesion strength in both stationary and migratory settings, correlating with markedly reduced levels of DP protein.
Discussion
The findings reported in this study significantly expand emerging evidence showing that IFs and associated elements exert a substantive impact on collective cell migration. For instance, the genetic loss of plectin, a pleiotropic “cytolinker” protein integrating IFs with other cytoskeletal elements at cell–matrix adhesion sites, at the surface of the nucleus, and elsewhere in the cell enhances keratinocyte migration and gives rise to enhanced Src kinase activity (Osmanagic-Myers et al., 2006; see also Seltmann et al., 2013) as is the case for Krt6a/Krt6b-null keratinocytes (Rotty and Coulombe, 2012). Keratinocytes that are deficient for all keratin proteins (global knockout) also exhibit increased migration and velocity, and in such cells, integrin β4 is misregulated and fails to interact with plectin (Seltmann et al., 2013, 2015). In addition, suppressing the expression of keratin 8, or alternatively of plakoglobin, a keratin-binding protein that occurs in the intracellular plaques of desmosomes and adherens junction, impairs the collective migration of Xenopus laevis endodermal cells ex vivo and in vivo. The latter trait entails a loss of cadherin (and tension)-mediated cell polarization (Weber et al., 2012) along with defects in the spatial regulation of traction forces resulting from altered focal adhesion dynamics, reorganization of the actin cytoskeleton, increased phosphomyosin II light chain levels, and elevated Rho-GTPase activity (Sonavane et al., 2017). Similarly, the compound loss of vimentin, glial acidic fibrillary protein, and nestin IFs restricts the collective migration of astrocytes in primary culture secondary to alterations in actin-driven treadmilling of adherens junctions and in focal adhesion dynamics and mechanical coupling to the actomyosin system (De Pascalis et al., 2018). A common thread across these studies is that altering IFs invariably results in altered regulation of key protagonists involved in cell migration including F-actin, the actomyosin system, cell–matrix adhesion (focal adhesions in particular), and relevant protein kinases.
The impact of K6a/K6b’s loss depends on the ECM since Krt6a/Krt6b-null keratinocytes are faster on type I collagen, fibronectin, and laminin but not on type IV collagen. Type I and type IV collagens interact with keratinocytes through a common integrin receptor, α2β1, but via distinct determinants on their extracellular domains (Vandenberg et al., 1991; Tuckwell et al., 1995). We find that the steady-state levels of α2 integrin are similar, but interestingly, those of β4 integrin (which interacts with plectin) are lower in migrating Krt6a/Krt6b-null keratinocytes relative to WT keratinocytes (Fig. S5 C). Further studies are needed to elucidate why the interaction of Krt6a/Krt6b-null keratinocytes with type I and IV collagens triggers distinct downstream effects and behaviors (e.g., cell attachment, cell migration, focal adhesion dynamics, response to blebbistatin) even though they may proceed through the same integrin receptors. There is evidence in the literature showing that stiffer substrate promotes more directed, faster migration along with enhanced formation of focal adhesions (Ng et al., 2012; Yeh et al., 2017). We envision that the fibrillar organization and rigidity of type IV and type I collagen along with how they each relate to mechanically sensitive signaling pathways in skin keratinocytes differ in significant ways. Atomic force microscopy has been used to assess the mechanical properties of collagen fibers (McDaniel et al., 2007; Wenger et al., 2007) and may be brought to bear to address this possibility.
Keratinocytes adhere to the ECM via actin-associated focal adhesions and keratin-associated hemidesmosomes (Simpson et al., 2011). Seltmann et al. (2015) showed that K5, a type II keratin related to K6, represses keratinocyte migration by regulating the localization and dynamics of hemidesmosome cell–matrix adhesion complexes. A recent study revealed that IFs can also interact with focal adhesions, affecting their size, number, and turnover (De Pascalis et al., 2018). In this study, we showed that the enhanced migration of Krt6a/Krt6b-null keratinocytes entails striking alterations in focal adhesion dynamics. We previously reported that several integrin-bound targets of Src kinase exhibit enhanced activity in the absence of K6a/K6b protein (Rotty and Coulombe, 2012), and thus it appears likely that K6a/K6b impact focal adhesion dynamics through regulating Src kinase as well as myosin II. Src activity is a key driver of cell migration, and one of the specific mechanisms involved is enhancement of focal adhesion turnover, possibly coupled to increased actomyosin contractility (Webb et al., 2004). We also uncovered an interaction and interdependence between K6a/K6b and myosin IIA, along with a role for myosin II motor activity during keratinocyte migration. We propose that the impact of K6a/K6b loss on focal adhesion size and rate of turnover is related to the markedly reduced levels of myosin IIA protein. Although not required for the assembly and stability of nascent focal adhesions initially forming in lamellipodia, myosin II plays a role during their subsequent maturation, in part via an impact on F-actin bundling and contractility (Choi et al., 2008; Vicente-Manzanares et al., 2009; Pasapera et al., 2010). Likewise, low myosin IIA levels may contribute to the failure of focal adhesions to mature long enough to reach larger sizes in Krt6a/Krt6b-null keratinocytes plated on type I collagen. Additionally, our finding that loss of myosin IIA reduces DP protein level in WT keratinocytes is consistent with a recent study showing that myosin IIA is essential for assembly of junctional complexes (Ozawa, 2018).
The functional importance of keratin for cell–cell adhesion between nonmigrating keratinocytes is already established. Mouse skin keratinocytes deficient for all keratins exhibit reduced levels of DP, desmoglein, and plakophilin 1 at the plasma membrane along with cell–cell adhesion defects (Kröger et al., 2013). In this study, we have shown that cell–cell adhesion is significantly weakened in the absence of K6a/K6b in both stationary and migrating keratinocytes. The notion that Krt6a/Krt6b-null keratinocyte sheets display enhanced directionality and speed while having partially disrupted cell–cell adhesion is intriguing. It may be that wound-proximal keratinocytes display enhanced desmosome disassembly and dynamics (Paladini et al., 1996; Garrod et al., 2005; Savagner et al., 2005) to promote directionality and migration speed during reepithelialization. Our data show that in both stationary and migration settings, reduced expression of DP occurs at the protein, not the mRNA, level, and otherwise that DP is redistributed away from the cell surface and into the nucleus. Others have shown that deletion of other key components of desmosomes including plakophilin 1 (South et al., 2003) and plakoglobin (Yin et al., 2005) each results in enhanced migration of skin keratinocytes akin to what is observed in Krt6a/Krt6b-null cells. Additionally, loss of Perp, a component of tessellate junctions and other junctional and nonjunctional plasma membrane regions, also leads to enhanced migration in vitro but delays wound healing in vivo, suggesting that efficient wound reepithelialization requires proper regulation of desmosomes and cell–cell adhesion (Beaudry et al., 2010; Franke et al., 2013).
Experiments in which DP was overexpressed in Krt6a/Krt6b-null keratinocytes or silenced in WT keratinocytes converge in establishing that this desmosome constituent plays a significant role during keratinocyte migration. Although one might reasonably assume that DP’s impact on migration entails an impact on desmosome regulation or function, our results raise the prospect that mislocalization of DP to the nucleus may play a role as well. This observation builds on our recent demonstration of the presence of keratin proteins inside the nucleus of skin keratinocytes (Hobbs et al., 2015, 2016). Vasioukhin et al. (2001) reported that a transfected DP mutant lacking the keratin-interacting domain at the C terminus localizes to the nucleus when expressed in Dsp-null keratinocytes, but the associated functional consequences have not been determined. Other key components of desmosomes, including plakophilins 1–3 and plakoglobin, have been shown to localize to the nucleus where they carry out specific functions (Mertens et al., 1996; Schmidt et al., 1997; Bonné et al., 1999; Maeda et al., 2004). Thus, plakophilin 1 binds to DNA and influences cell survival after DNA damage (Sobolik-Delmaire et al., 2010), whereas plakoglobin acts as a transcriptional activator (Maeda et al., 2004). Nuclear localization of DP in skin keratinocytes thus represents another observation that merits further investigation.
How the alterations in cell–matrix and cell–cell adhesion are interrelated in the absence of K6a/K6b proteins in collectively migrating keratinocytes is unclear at present. Key activities with an already identified role given our studies, such as Src kinase, PKC, and myosin IIA, could be part of this integration. Whether other proteins, such as the receptor for activated kinase 1 (RACK1), may also play a key role as part of a broader interdependent network involving plectin, keratin, adhesion proteins, PKC (and possibly other proteins) now emerges as an issue of interest. RACK1 is a scaffolding protein that interacts with plectin and is involved in the trafficking of key signaling effectors, including PKCδ and Src, to specific subcellular locations (Ron et al., 1994; Chang et al., 2002; Osmanagic-Myers and Wiche, 2004). RACK1 has been reported to regulate focal adhesion and migration through its Src binding site (Cox et al., 2003). RACK1 and PKCα have been implicated in the regulation of desmosome cell–cell adhesion (Kröger et al., 2013). Of interest, we recently reported that expression of K16, a key type I partner protein for K6a/K6b, is associated with the release of RACK1 from the keratin filament cytoskeleton, along with misregulation of PKCδ in skin keratinocytes (Kerns et al., 2016). Owing to their roles as multifaceted scaffolds, RACK1, plectin, and IFs are examples of proteins and assemblies that are poised to coordinate and integrate the regulation of cell–matrix and cell–cell adhesion events and facilitate collective cell migration in vivo.
In summary, the data reported in this study support the hypothesis that the wound-inducible K6a/K6b proteins foster collective keratinocyte migration in part by stabilizing integrin-mediated cell–matrix adhesion and desmosome-mediated cell–cell adhesion (Fig. S5 D). Upon injury to the skin, keratinocytes at the wound edge tend to down-regulate the surface display of adhesion molecules as part of a strategy to facilitate mobilization and increase their speed of migration. Induction of K6a/K6b and other proteins in this setting may represent a countermeasure that promotes cell–matrix and cell–cell adhesion to a degree that promotes cohesive collective cell migration and efficient wound closure.
Materials and methods
Mouse lines
All experiments involving mice were reviewed and approved by the Institutional Animal Care and Use Committee at Johns Hopkins University and the University of Michigan. Krt6a/Krt6b hemizygous null mice (Wong et al., 2000) are viable and were maintained in the inbred C57Bl/6 background (Rotty and Coulombe, 2012). K14-GFP-actin mice (Vaezi et al., 2002), Dspfl/fl mice (Vasioukhin et al., 2001), and Myh9fl/fl mice (Ma et al., 2009; Jacobelli et al., 2010; Nguyen-Ngoc et al., 2017) were gifts from A. Ewald (Johns Hopkins University, Baltimore, MD).
Coculture migration assay
Skin keratinocytes were isolated from P0–P2 pups as described (Rotty and Coulombe, 2012). GFP-actin expressing keratinocytes (Vaezi et al., 2002) were mixed (1:4 ratio) with WT or Krt6a/Krt6b-null keratinocytes (C57Bl/6 strain). A coverslip was placed within each well of a six-well plate, and type I collagen (354236; Thermo Fisher Scientific) was used to precoat the coverslip for 30 min at 37°C. A culture insert (80209; Ibidi) was firmly placed on top of each coverslip. Cells (3.2 × 105) were plated within the insert. Keratinocytes were cultured to confluence in mKER medium (DMEM [3 parts], Ham’s F-12 [1 part], 10% FBS, 1 nM cholera toxin, 5 µg/ml insulin, 10 ng/ml EGF, 25 µg/ml gentamicin, 400 ng/ml hydrocortisone, 5 µg/ml transferrin, 2 nM 3,3′,5-triiodo-l-thyronine, and 60 µg/ml penicillin; see Wang et al., 2016) for 2 d. In some experiments, cells were treated with 10 µg/ml mitomycin C (Sigma-Aldrich) or PBS (vehicle control) immediately after removing the culture insert for 2 h on day 3. After migrating for the indicated time period, cells were fixed and stained for GFP (green); K14 (red), which stains all keratinocytes; and DAPI (blue), which stains DNA. Images of cells at the migration front were taken with a 10× objective, and the percentage of GFP-positive cells within these areas was calculated by dividing the number of GFP-positive cells by the number of K14-positive cells (the total cell number).
Immunofluorescence staining and imaging
Keratinocytes were fixed in 4% PFA for 10 min, permeabilized with 0.5% Triton X-100/PBS for 5 min, and blocked with 5% normal goat serum (NGS)/1× PBS for 30 min at room temperature. Primary antibodies were diluted in 2.5% NGS/1× PBS and incubated with samples overnight at 4°C. Secondary antibodies were diluted in 2.5% NGS/1× PBS and incubated with samples for 1 h at room temperature. Primary antibodies used include chicken anti-K14 (1:500; 906001; BioLegend), rabbit anti-DP (1:200; NW6; K. Green, Northwestern University, Evanston, IL), rabbit anti-GFP (1:200; 29A1; M. Matunis, Johns Hopkins University, Baltimore, MD), mouse anti-paxillin (1:200; 612405; BD), rabbit anti–E-cadherin (1:100; 3195S; Cell Signaling Technology), rabbit anti–plakophilin 1 (1:100; ab183512; Abcam), and mouse anti–β-catenin (1:50; 610153; BD). Secondary antibodies used were goat anti–chicken IgY (1:1,000), goat anti–mouse IgG (1:1,000), or goat anti–rabbit IgG (1:1,000), all labeled with Alexa Fluor (Thermo Fisher Scientific). Immunofluorescence microscopy was performed using a Zeiss AxioObserver Z1 microscope equipped with a Plan Apochromat 10×/0.45 M27 objective or a Plan Apochromat 20×/0.8 M27 objective, an Apotome attachment, and Zen software (Zeiss) or a Zeiss LSM 800 confocal laser scanning microscope equipped with a Plan Apochromat 63×/1.40 oil differential interference contrast (DIC) M27 objective and Zen software. All images in a given experiment were taken under identical imaging parameters.
Phase-contrast microscopy of collective cell migration and analysis
Skin keratinocytes were isolated from P0–P2 pups as described (Rotty and Coulombe, 2012). 3.2 × 105 cells were plated within a culture insert in a chamber slide (155380; Thermo Fisher Scientific) or in a 12-well glass-bottom plate (P12-1.5H-N; Cellvis). 50 µg/ml type I collagen (354236; Thermo Fisher Scientific), type IV collagen (356233; Thermo Fisher Scientific), fibronectin (354008; Corning), or laminin (354232; Thermo Fisher Scientific) was used to precoat the chamber slide for 30 min at 37°C. Keratinocytes were cultured in mKER medium for 2 d and upon reaching confluence, culture inserts were removed to create a “wound” space allowing for migration. For inhibitor treatment migration assay, 20 µM ML-7 (ab120848; Abcam) or 30 µM blebbistatin (ab120425; Abcam) was added to cells immediately after the removal of inserts. Because it took 1 h to set up imaging, the videos and quantifications represented the effect of 1-h pretreatment with each drug and 16-h migration in the presence of each drug. Phase-contrast imaging (McGinnis et al., 2015) was performed using an AxioObserver Z1 microscope equipped with an incubation chamber (37°C and 5% CO2), a Zeiss EC Plan Neofluar 10×/0.3 Ph1 objective, and AxioVision software at 10-min intervals for 16 h. The migration area was calculated by subtracting the area measured at time 0 from the area measured after 16 h. PIV analysis of live-cell phase-contrast time-lapse videos was performed using MatLab as described (Weiger et al., 2013; Lee et al., 2016). Two interrogation window sizes (64 × 64 and 32 × 32 pixels) were used to segment the images. Then, multiple iterations, with 50% overlap for each interrogation step, were performed. Because the area of an interrogation window is smaller than that of an individual cell, the resulting flow field shows the collective migration dynamic on a subcellular level. After the images were processed to emphasize the edges of each cell sheet, they were binarized, and the edges were identified using Dijkstra’s algorithm (Dijkstra, 1959). We filtered out the flow vectors that were outside the cell edges to collect information on actual motion within the cell sheets. Based on this motion information, the mean flow speed and angular deviation across each entire cell sheet can be calculated. We defined the angular deviation as follows (Lee et al., 2016):
θi denotes the angle of the ith velocity vector. N is the number of velocity vectors in the flow fields. According to this definition, the value of angular deviation ranges from 0 (coordinated motion) to (chaotic motion; Lee et al., 2016). We then averaged the mean speed and angular deviation derived from all cell edges in each experiment.
Adhesion assay
96-well microplates (CLS3603; Sigma-Aldrich) were coated with 50 µg/ml type I collagen (354236; Thermo Fisher Scientific) or type IV collagen (356233; Thermo Fisher Scientific) at 37°C for 30 min. After washing with PBS, wells were blocked by incubation with 0.5 mg/ml heat-treated BSA in PBS-ABC (containing 1 mM CaCl2 and 1 mM MgCl2) for 1 h at 37°C (Levy et al., 2000). Freshly isolated keratinocytes (2 × 104) were plated in each well in mKER medium and incubated at 37°C for 2 h. The nonadherent cells were washed off with PBS-ABC. The adherent cells were lysed with medium containing 1% Triton X-100. The relative efficiency of cells to matrix was determined by measuring the level of LDH, which is released upon cell lysis, using the using the Cytotoxicity Detection Kit (4744926001; Sigma-Aldrich).
Quantification of focal adhesion number and size
Keratinocytes isolated from P0–P2 pups (3.2 × 105 cells) were plated within a culture insert in each well in a 12-well glass-bottom plate (P12-1.5H-N; Cellvis). 50 µg/ml type I collagen or type IV collagen was used to precoat each well for 30 min at 37°C. Keratinocytes were cultured in mKER medium for 2 d, and upon reaching confluence, culture inserts were removed to create a wound space allowing for migration. After 1 d, cells were fixed and stained for paxillin. Imaging was performed using a Zeiss LSM 800 confocal laser scanning microscope equipped with a Plan Apochromat 63×/1.40 oil DIC M27 objective and Zen software. All images in a given experiment were taken under identical imaging parameters. 30 WT and 30 Krt6a/Krt6b-null keratinocytes on each collagen matrix were quantified. Counting the number of focal adhesions per cell was done manually, and the size of individual focal adhesions was measured using ImageJ (National Institutes of Health).
Time-lapse microscopy of focal adhesion turnover
Freshly isolated keratinocytes were transfected with a plasmid encoding mCherry-tagged paxillin (50526; Addgene) using the nucleofection method (Lonza) before plating in chamber slides with culture inserts. The chamber slides were coated with either 50 µg/ml type I collagen or type IV collagen. After removing the culture inserts, keratinocytes were allowed to migrate for at least 8 h before imaging using an AxioObserver Z1 fluorescence microscope equipped with an incubation chamber (37°C and 5% CO2), a EC Plan Neofluar 40×/1.30 oil DIC M27 objective or a Plan Apochromat 63×/1.40 oil DIC M27 objective, and AxioVision software or MetaMorph microscopy automation and image analysis software at 3-min intervals for 3 h. Oil was used as the imaging medium. At each time point, five slices of z stack images were captured for every cell, spanning a height of 1.48 µm in the z axis.
Quantification of focal adhesion dynamics
An established algorithm (Stehbens and Wittmann, 2014) was used to calculate the rates of assembly, disassembly, and lifetime of focal adhesions. The focal adhesions (FAs) that had their entire lifespan within the video-recording interval, did not split or merge, and showed very similar sizes between WT and Krt6a/Krt6b-null cells were selected for analysis. ImageJ was used to measure the intensity of focal adhesion–associated mCherry-paxillin, designated as I(FA). A region near the focal adhesion was chosen as background, designated as I(BKG). The signal derived from one adhesion plaque at any given time was calculated by subtracting I(BKG) from I(FA). A three-frame running average of these intensity values was then applied to smooth variations between frames (Stehbens and Wittmann, 2014):
The data I(t) were fitted to a model that assumes that the focal adhesion assembly phase and disassembly phase follow a sigmoid, logistic function and a single exponential decay, respectively. Doing so enabled us to then calculate, for each focal adhesion, the rate of assembly (ka), rate of disassembly (kd), and lifetime (tlife; Stehbens and Wittmann, 2014):
10 focal adhesions per cell, 10 cells per genotype on type I collagen, and five cells per genotype on type IV collagen were selected for the analysis of focal adhesion turnover.
Harvesting protein lysates from skin explant culture
Ex vivo explant cultures derived from P0–P2 mouse skin were seeded as described (Mazzalupo et al., 2002; Wong and Coulombe, 2003; Rotty and Coulombe, 2012). In brief, skin punches (4.0 mm; Acuderm) were used to make circular skin biopsies. After the skin punches had been cultured for 6 d in mKER medium, cellular outgrowths from skin explants of the same genotype were pooled together and solubilized with 6.5 M urea buffer (50 mM Tris, pH 7.5, 1 mM EGTA, 6.5 M urea, 2 mM DTT, 1 mM PMSF, 50 mM NaF, 1 mM Na3VO4, and proteinase inhibitors including 2 µg/ml antipain, 10 µg/ml aprotinin, 10 µg/ml benzamidine, 1 µg/ml leupeptin, 1 µg/ml chymostatin, and 1 µg/ml pepstatin A) to obtain whole-cell lysates. Protein concentration was determined using a commercial kit (Bio-Rad), with BSA (Thermo Fisher Scientific) as a standard. Cell lysates were prepared in Laemmli SDS-PAGE sample buffer (with 5% β-mercaptoethanol) and stored at −20°C until use.
Western blotting
Protein lysates were subjected to electrophoresis on SDS-PAGE gels (4–15% precast polyacrylamide gels [Bio-Rad] were used for DP and myosin IIA; 10% polyacrylamide gels were used for the remaining proteins in this study) and transferred onto 0.45-µm nitrocellulose membranes (Bio-Rad). Analyses of DP and myosin IIA required a transfer buffer containing a high concentration of both Tris and glycine (50 mM Tris, 380 mM glycine, 0.1% SDS, and 20% methanol) for ∼10–16 h at 20 V at 4°C; for other proteins, a regular buffer (25 mM Tris, 190 mM glycine, 0.1% SDS, and 20% methanol) was used for 1 h and 20 min at 100 V at 4°C. Membranes were blocked with 5% milk in TBST for 30 min at room temperature. Primary antibodies were diluted in 5% BSA in TBST and applied to membranes overnight at 4°C. The primary antibodies used include rabbit anti-DP (1:1,000), rabbit anti–myosin IIA (1:1,000; 909801; BioLegend), rabbit anti-K6 (1:5,000; McGowan and Coulombe, 1998b), rabbit anti–E-cadherin (1:1,000), mouse anti–β-catenin (1:1,000), rabbit anti–p120-catenin (1:1,000; 4989; Cell Signaling Technology), chicken anti-plakoglobin (1:5,000, 1,407, or 1,408; K. Green), rabbit anti–plakophilin 1 (1:1,000; ab183512; Abcam), rabbit anti–integrin α2 (1:1,000; ab181548; Abcam), mouse anti–integrin β4 (1:100; sc-514252; Santa Cruz), and mouse anti–β-actin (1:2,000; A5441; Sigma-Aldrich). Membranes were washed three times (5 min each time) with TBST before incubation with HRP-conjugated secondary antibodies (1:5,000) for 1 h at room temperature. Proteins on membranes were detected using SuperSignal West Pico Chemiluminescent Substrate (34080; Thermo Fisher Scientific) or ECL Select Western Blotting Detection Reagent (45-000-999; Thermo Fisher Scientific). The FluorChem Q imaging system (ProteinSimple) was used to detect signals, and ImageJ was used for quantification.
Analysis of mRNA from skin explant culture
RNA was extracted from keratinocytes after 6 d of skin explant culture using the NucleoSpin RNA kit (740955; Macherey-Nagel) according to the manufacturer’s instructions. cDNA Synthesis was performed with the iScript cDNA Synthesis Kit (1708891; Bio-Rad). Quantitative RT-PCR (qRT-PCR) was performed with the iTAQ Universal SYBR master mix (1725121; Bio-Rad) on a Bio-Rad CFX96 machine. Oligonucleotide primers for the target genes analyzed were as follows: Myh9 (forward, 5′-GGCCCTGCTAGATGAGGAGT-3′; reverse, 5′-CTTGGGCTTCTGGAACTTGG-3′), Cdh1 (forward, 5′-CAGGTCTCCTCATGGCTTTGC-3′; reverse, 5′-CTTCCGAAAAGAAGGCTGTCC-3′), Ctnna1 (forward, 5′-AAGTCTGGAGATTAGGACTCTGG-3′; reverse, 5′-ACGGCCTCTCTTTTTATTAGACG-3′), Ctnnb1 (forward, 5′-ATGGAGCCGGACAGAAAAGC-3′; reverse, 5′-CTTGCCACTCAGGGAAGGA-3′), Ctnnd1(forward, 5′-GTGGAAACCTACACCGAGGAG-3′; reverse, 5′-CGTCTAGTGGTCCCATCATCTG-3′), Dsp (forward, 5′-GGATTCTTCTAGGGAGACTCAGT-3′; reverse, 5′-TCCACTCGTATTCCGTCTGGG-3′), Jup (forward, 5′-TGGCAACAGACATACACCTACG-3′; reverse, 5′-GGTGGTAGTCTTCTTGAGTGTG-3′), Pkp1 (forward, 5′-AACCACTCTCCGCTCAAGAC-3′; reverse, 5′-CTTCTGCCGTTTGACGGTCAT-3′), Dsg1a (forward, 5′-ACTGTGTTAAATGTCATCGAGGG-3′; reverse, 5′-TGCCTGTTCTTGAGTCAACAAC-3′), Dsc1 (forward, 5′-GGTCAAGGAATCAAAACACAGC-3′; reverse, 5′-CCAAGCCGAGGTTGAGTGAAA-3′), Gapdh (forward, 5′-AAATGGTGAAGGTCGGTGT-3′; reverse, 5′-ACTCCACGACATACTCAGCAC-3′), and Rn18s (forward, 5′-CCTGTGCCTTCCTTGGA-3′; reverse, 5′-CATTCGAACGTCTGCCCTATC-3′).
Immunoprecipitation
Confluent WT keratinocyte primary cultures were lysed in 1% Triton X-100 buffer (40 mM Hepes, pH 7.5, 120 mM NaCl, 1 mM EDTA, 1% Triton X-100, 10 mM sodium pyrophosphate, 50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, and protease inhibitors including 2 µg/ml antipain, 10 µg/ml aprotinin, 10 µg/ml benzamidine, 1 µg/ml leupeptin, 1 µg/ml chymostatin, and 1 µg/ml pepstatin A), incubated for 15 min on a shaker at 4°C, and spun down at maximum speed (16.1 × 1,000 g) for 10 min at 4°C. Cell lysates were precleared with 30 µl protein G–coupled Sepharose beads (17-0618-01; GE Healthcare) for 30 min at 4°C. The Bradford assay was used to determine the concentration of protein lysates. Mouse anti–myosin IIA antibodies (ab55456; Abcam) or mouse IgG antibodies (sc-2025; Santa Cruz) were added to lysates (1 µg antibody per 1 mg lysates) and incubated for 3 h on a shaker at 4°C. Protein G–coupled Sepharose beads (30 µl) were then added for an additional 45-min incubation with the protein lysates/antibody mixture. Bead-bound proteins were eluted by adding 2× SDS sample buffer containing 5% β-mercaptoethanol and analyzed via Western blotting.
PLA
Glass coverslips were placed in each well of 12-well plates and coated with type I collagen, and a culture insert was firmly placed on the coverslip. Keratinocytes were isolated from P0–P2 pups, plated within a culture insert at 3.2 × 105 cells per insert and cultured in mKER medium for 2 d. Upon reaching confluence, culture inserts were removed to trigger cell migration. After 1 d, cells were fixed with 4% PFA at room temperature for 15 min, permeabilized with 0.1% Triton at room temperature for 10 min, and blocked with 2.5% donkey serum in PBS at room temperature for 1 h. Fixed cells were incubated overnight at 4°C with mouse anti–myosin IIA and rabbit anti-K6a primary antibodies (see above). Donkey anti-mouse secondary antibody conjugated to oligonucleotide (minus; DUO92004; Sigma-Aldrich) and donkey anti-rabbit secondary antibody conjugated to oligonucleotide (plus; DUO92002; Sigma-Aldrich) were added to the cells and incubated for 1 h at 37°C. The ligation and amplification steps were performed according to the manufacturer’s instructions (Duolink PLA; Sigma-Aldrich). Fluorescence was detected using an AxioObserver Z1 microscope equipped with an EC Plan Neofluar 40×/1.30 oil DIC M27 objective, an Apotome attachment, and Zen software. All images in a given experiment were taken under identical imaging parameters.
Cloning of mouse K6b fragments
The regions encoding head domain, rod domain, and rod and tail domains in the mouse K6b cDNA cloned in a pET-8c vector (Chung et al., 2012; Rotty and Coulombe, 2012) were obtained via PCR and subcloned into a pT7-HMT vector, which contains a His-tag for protein purification (Feng and Coulombe, 2015). The oligonucleotide primers used were as follows: K6b head domain (1–471 bp; forward, 5′-CCCCATATGGCTAGCAAAACCACCATCAA-3′; reverse, 5′-CGGGATCCTTACCTGACCCGCTGGAT-3′); K6b rod domain (472–1,419 bp; forward, 5′-CCCCATATGACTGAGGAGAGGGAGCAGAT-3′; reverse, 5′-CGGGATCCTTACAACCTGCACTCCTCTCC-3′); and K6b rod and tail domains (472–1,689 bp; forward, 5′-CCCCATATGACTGAGGAGAGGGAGCAGAT-3′; reverse, 5′-CGGGATCCTCAGTGCCTGTAGCTCTT-3′).
Protein purification
Plasmids encoding K5 (pET-K5; Lee and Coulombe, 2009), K6b (pET-K6b; Rotty and Coulombe, 2012), K17 (pET-K17; Chung et al., 2012), and different subdomains of K6b were transformed into Escherichia coli strains BL21 (DE3) or BL21 (DE3) plysS to produce the corresponding recombinant proteins (Coulombe and Fuchs, 1990). Inclusion bodies were recovered, dissolved in 6.5 M urea buffer (Wang et al., 2016), and applied to HiTrapQ column followed by a MonoQ column for purification (Feng and Coulombe, 2015; Wang et al., 2016).
Far-Western assay
5 µg of each bait protein (purified recombinant keratin proteins: e.g., K5, K6, and K17) was electrophoresed on duplicate 10% SDS-PAGE gels. One gel was stained with Coomassie dye to assess protein loading. The other gel was electroblotted to a nitrocellulose membrane (Bio-Rad) with 0.45-µm pore size. The membrane was blocked in 5% milk in TBST for 30 min. After washing briefly with TBST, the membrane was incubated with the target protein, myosin protein (1.5 µg/ml; MY02-A; Cytoskeleton) in 5% BSA in TBST on a shaker at room temperature for 4 h. After several washes in TBST, the membrane was blocked in in 5% milk in TBST for 30 min at room temperature. Primary antibody (mouse anti–myosin IIA; ab55456; Abcam) was diluted (1:1,000) in 5% BSA in TBST and applied to the membrane overnight at 4°C. Membranes were washed three times (5 min each) with TBST before incubation with HRP-conjugated secondary antibody (1:2,000) for 1 h at room temperature. After three TBST washes (5 min each), membrane bound proteins of interest were detected using Amersham ECL Select Western Blotting Detection Reagent and the FluorChem Q imaging system (ProteinSimple).
Nucleofection
Plasmids (e.g., DP-GFP, 32227, Addgene; or pEGFP-N1 control, a gift from C. Machamer, Johns Hopkins University, Baltimore, MD) were transfected into freshly isolated keratinocytes using the nucleofection method (Lonza). Briefly, 16.4 µl nucleofection solution and 3.6 µl supplement solution from the P1 Primary Cell 4D-Nucleofector X Kit (V4XP-1032; Lonza) was mixed with plasmid DNA (0.64 µg). Keratinocytes (6.4 × 105) were pelleted by centrifugation (500 g for 5 min at 4°C) and resuspended in the nucleofection mixture. The mixture was transferred into one well in a 16-well strip and placed into the 4D-nucleofector system (Lonza). At 5 min after an electrical pulse (generated using the proprietary “Primary” program provided by the manufacturer), medium was added into the strip to suspend cells. Finally, the cell suspension was plated and then cultured for ∼48 h before analysis.
Dispase and LDH release assays
Keratinocytes were isolated from newborn mice (P0–P1), plated onto a type I or type IV collagen (50 µg/ml)-coated six-well plate at 1.5–2 million cells per well and incubated in mKER medium for 24 h. Cells were switched to CnT57 medium (CELLnTEC), cultured until confluence (∼8 d), and finally, switched to CnT57 medium supplemented with 1.2 mM calcium for an additional 2 d. Cell sheets formed were lifted off the plate using 3 ml dispase II/PBS solution (2.4 U/ml; 04942078001; Roche) and transferred into a 15-ml conical tube in a total volume of 10 ml PBS buffer. The tubes were then inverted using a SCILOGEX MS-RD-Pro LCD Digital Tube Rotator at 60 rpm for 20 rounds (for cells on type I collagen) or 10 rounds (for cells on type IV collagen; 1 round = 10 inversions). After each round, cell sheets were inspected for fragmentation. To obtain whole-cell protein lysates from such cultures, the cell sheets were lysed in urea buffer (50 mM Tris, pH 7.5, 1 mM EDTA, 6.5 M urea, and 2 mM DTT) supplemented with protease and phosphatase inhibitors (50 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 2 µg/ml antipain, 10 µg/ml aprotinin, 10 µg/ml benzamidine, 1 µg/ml leupeptin, 1 µg/ml chymostatin, and 1 µg/ml pepstatin A) instead of being lifted with dispase.
To access the occurrence of cell lysis during the sheet fragmentation assay, LDH activity was measured using the Cytotoxicity Detection Kit (4744926001; Sigma-Aldrich; Huen et al., 2002) according to the manufacturer’s instructions. Briefly, background control was obtained from the mixture of 7 ml PBS and 3 ml dispase solution. After transferring the detached cell sheet along with dispase solution into PBS, the mixture was used as a low-activity control. A high-activity control was measured by lysing cells with 100 µl lysis buffer, rocking the lysate at room temperature for 15 min, and adding 100 µl lysate into the mixture of 7 ml PBS and 3 ml dispase solution. 50 µl from each sample was used for LDH detection. The absorbance of each sample was read at 490 nm using Spectra Max M5 (Molecular Devices).
Adenovirus infection
Newborn skin keratinocytes (2 × 105) in primary culture were infected with 2 µl adenovirus (1 × 1010 PFU/ml) delivering either GFP (1060; Vector Biolabs) or Cre recombinase (1045; Vector Biolabs) for 1 h (Di Cunto et al., 1998) in DMEM (11885092; Life Technologies). Cells were then cultured in CnT57 medium (CELLnTEC) for 2 d before experimentation.
Statistical analysis
The Student’s two-tailed t test was applied to assess statistical significance.
Online supplemental material
Fig. S1 shows a representative example of PIV analysis of WT keratinocytes migrating on laminin. Fig. S2 shows the impact of disrupting myosin II function by silencing Myh9 or with ML-7 and blebbistatin treatments on cell–cell adhesion and the size and organization of focal adhesions. Fig. S3 illustrates the status of E-cadherin, plakophilin 1, and β-catenin in migrating Krt6a/Krt6b-null keratinocytes. Fig. S4 shows that DP accumulates in nucleus in Krt6a/Krt6b-null keratinocytes adjacent to or at the migration front edge. Fig. S5 shows mRNA levels for genes encoding protein components of adherens junctions and desmosomes, shows the degree of cytolysis and integrin α2 and β4 levels in WT control and Krt6a/Krt6b-null keratinocytes, and illustrates the possible role of K6a/K6b in wound healing. Video 1 shows phase-contrast time-lapse imaging of WT and Krt6a/Krt6b-null keratinocytes migrating on type I collagen. Video 2 shows time-lapse videos assessing the directionality and speed of WT control and Krt6a/Krt6b-null keratinocytes migrating on type I collagen over the course of ∼16 h. Video 3 shows time-lapse videos of directionality and speed for WT and Krt6a/Krt6b-null keratinocytes migrating on type IV collagen over the course of ∼16 h. Video 4 shows time-lapse imaging of mCherry–paxillin dynamics comparing WT with Krt6a/Krt6b-null keratinocytes migrating on type I collagen. Video 5 shows time-lapse imaging of mCherry–paxillin dynamics comparing WT with Krt6a/Krt6b-null keratinocytes migrating on type IV collagen. Video 6 shows phase-contrast time-lapse imaging comparing Myh9+/+ with Myh9−/− keratinocytes migrating on type I collagen. Video 7 shows phase-contrast time-lapse imaging comparing Dsp+/+ with Dsp−/− keratinocytes migrating on type I collagen.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Ewald for providing K14-GFP-actin mice, Dspfl/fl mice, and Myh9fl/fl mice; Dr. Carolyn Machamer for pEGFP-N1 plasmid; Dr. Beau Su for PLA assay protocol; Drs. Janice Evans and Roman Giger for sharing microscope set-ups for live cell imaging; and Drs. Torsten Wittmann and Samantha Stehbens for help with focal adhesion turnover calculations. We are grateful to members of the Coulombe laboratory and to Drs. Andrew Ewald, Peter Devreotes, Michael Matunis, and Wan-yee Tang for help and advice.
This work was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (AR044232 to P.A. Coulombe).
The authors declare no competing financial interests.
Author contributions: F. Wang, S. Chen, and P.A. Coulombe designed the experiments; F. Wang, S. Chen, and H.B. Liu performed the experiments and analyzed the data; S. Chen and C.A. Parent conducted the PIV analyses; F. Wang and P.A. Coulombe collated the data and wrote the manuscript. All authors contributed to the manuscript.
References
- Arnoux V., Côme C., Kusewitt D.F., Hudson L.G., and Savagner P.. 2005. Cutaneous wound reepithelialization. In Rise and Fall of Epithelial Phenotype . Molecular Biology Intelligence Unit. Springer, Boston, MA: 111–134. doi:. 10.1007/0-387-28671-3_8 [DOI] [Google Scholar]
- Beaudry V.G., Ihrie R.A., Jacobs S.B., Nguyen B., Pathak N., Park E., and Attardi L.D.. 2010. Loss of the desmosomal component perp impairs wound healing in vivo. Dermatol. Res. Pract. 2010:759731 10.1155/2010/759731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernot K.M., Coulombe P.A., and McGowan K.M.. 2002. Keratin 16 expression defines a subset of epithelial cells during skin morphogenesis and the hair cycle. J. Invest. Dermatol. 119:1137–1149. 10.1046/j.1523-1747.2002.19518.x [DOI] [PubMed] [Google Scholar]
- Bonné S., van Hengel J., Nollet F., Kools P., and van Roy F.. 1999. Plakophilin-3, a novel armadillo-like protein present in nuclei and desmosomes of epithelial cells. J. Cell Sci. 112:2265–2276. [DOI] [PubMed] [Google Scholar]
- Chang B.Y., Harte R.A., and Cartwright C.A.. 2002. RACK1: a novel substrate for the Src protein-tyrosine kinase. Oncogene. 21:7619–7629. 10.1038/sj.onc.1206002 [DOI] [PubMed] [Google Scholar]
- Choi C.K., Vicente-Manzanares M., Zareno J., Whitmore L.A., Mogilner A., and Horwitz A.R.. 2008. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 10:1039–1050. 10.1038/ncb1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B.M., Murray C.I., Van Eyk J.E., and Coulombe P.A.. 2012. Identification of novel interaction between annexin A2 and keratin 17: evidence for reciprocal regulation. J. Biol. Chem. 287:7573–7581. 10.1074/jbc.M111.301549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B.M., Arutyunov A., Ilagan E., Yao N., Wills-Karp M., and Coulombe P.A.. 2015. Regulation of C-X-C chemokine gene expression by keratin 17 and hnRNP K in skin tumor keratinocytes. J. Cell Biol. 208:613–627. 10.1083/jcb.201408026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe P.A. 2003. Wound epithelialization: accelerating the pace of discovery. J. Invest. Dermatol. 121:219–230. 10.1046/j.1523-1747.2003.12387.x [DOI] [PubMed] [Google Scholar]
- Coulombe P.A., and Fuchs E.. 1990. Elucidating the early stages of keratin filament assembly. J. Cell Biol. 111:153–169. 10.1083/jcb.111.1.153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox E.A., Bennin D., Doan A.T., O’Toole T., and Huttenlocher A.. 2003. RACK1 regulates integrin-mediated adhesion, protrusion, and chemotactic cell migration via its Src-binding site. Mol. Biol. Cell. 14:658–669. 10.1091/mbc.e02-03-0142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pascalis C., Pérez-González C., Seetharaman S., Boëda B., Vianay B., Burute M., Leduc C., Borghi N., Trepat X., and Etienne-Manneville S.. 2018. Intermediate filaments control collective migration by restricting traction forces and sustaining cell-cell contacts. J. Cell Biol. 217:3031–3044; Advance online publication 10.1083/jcb.201801162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depianto D., Kerns M.L., Dlugosz A.A., and Coulombe P.A.. 2010. Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat. Genet. 42:910–914. 10.1038/ng.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cunto F., Topley G., Calautti E., Hsiao J., Ong L., Seth P.K., and Dotto G.P.. 1998. Inhibitory function of p21Cip1/WAF1 in differentiation of primary mouse keratinocytes independent of cell cycle control. Science. 280:1069–1072. 10.1126/science.280.5366.1069 [DOI] [PubMed] [Google Scholar]
- Dijkstra E.W. 1959. A note on two problems in connexion with graphs. Numer. Math. 1:269–271. 10.1007/BF01386390 [DOI] [Google Scholar]
- Even-Ram S., Doyle A.D., Conti M.A., Matsumoto K., Adelstein R.S., and Yamada K.M.. 2007. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk. Nat. Cell Biol. 9:299–309. 10.1038/ncb1540 [DOI] [PubMed] [Google Scholar]
- Feng X., and Coulombe P.A.. 2015. A role for disulfide bonding in keratin intermediate filament organization and dynamics in skin keratinocytes. J. Cell Biol. 209:59–72. 10.1083/jcb.201408079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke W.W., Heid H., Zimbelmann R., Kuhn C., Winter-Simanowski S., Dörflinger Y., Grund C., and Rickelt S.. 2013. Transmembrane protein PERP is a component of tessellate junctions and of other junctional and non-junctional plasma membrane regions in diverse epithelial and epithelium-derived cells. Cell Tissue Res. 353:99–115. 10.1007/s00441-013-1645-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod D.R., Berika M.Y., Bardsley W.F., Holmes D., and Tabernero L.. 2005. Hyper-adhesion in desmosomes: its regulation in wound healing and possible relationship to cadherin crystal structure. J. Cell Sci. 118:5743–5754. 10.1242/jcs.02700 [DOI] [PubMed] [Google Scholar]
- Gurtner G.C., Werner S., Barrandon Y., and Longaker M.T.. 2008. Wound repair and regeneration. Nature. 453:314–321. 10.1038/nature07039 [DOI] [PubMed] [Google Scholar]
- Hobbs R.P., DePianto D.J., Jacob J.T., Han M.C., Chung B.M., Batazzi A.S., Poll B.G., Guo Y., Han J., Ong S., et al. . 2015. Keratin-dependent regulation of Aire and gene expression in skin tumor keratinocytes. Nat. Genet. 47:933–938. . 10.1038/ng.3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs R.P., Jacob J.T., and Coulombe P.A.. 2016. Keratins are going nuclear. Dev. Cell. 38:227–233. 10.1016/j.devcel.2016.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu K., Ji L., Applegate K.T., Danuser G., and Waterman-Storer C.M.. 2007. Differential transmission of actin motion within focal adhesions. Science. 315:111–115. 10.1126/science.1135085 [DOI] [PubMed] [Google Scholar]
- Huen A.C., Park J.K., Godsel L.M., Chen X., Bannon L.J., Amargo E.V., Hudson T.Y., Mongiu A.K., Leigh I.M., Kelsell D.P., et al. . 2002. Intermediate filament-membrane attachments function synergistically with actin-dependent contacts to regulate intercellular adhesive strength. J. Cell Biol. 159:1005–1017. 10.1083/jcb.200206098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto A., Martinez-Arias A., and Martin P.. 2001. Mechanisms of epithelial fusion and repair. Nat. Cell Biol. 3:E117–E123. 10.1038/35074643 [DOI] [PubMed] [Google Scholar]
- Jacobelli J., Friedman R.S., Conti M.A., Lennon-Dumenil A.M., Piel M., Sorensen C.M., Adelstein R.S., and Krummel M.F.. 2010. Confinement-optimized three-dimensional T cell amoeboid motility is modulated via myosin IIA-regulated adhesions. Nat. Immunol. 11:953–961. 10.1038/ni.1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns M.L., Hakim J.M., Lu R.G., Guo Y., Berroth A., Kaspar R.L., and Coulombe P.A.. 2016. Oxidative stress and dysfunctional NRF2 underlie pachyonychia congenita phenotypes. J. Clin. Invest. 126:2356–2366. 10.1172/JCI84870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Wong P., and Coulombe P.A.. 2006. A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature. 441:362–365. 10.1038/nature04659 [DOI] [PubMed] [Google Scholar]
- Kouklis P., Hutton E., and Fuchs E.. 1994. Making a connection: direct binding between keratin intermediate filaments and desmosomal proteins. J. Cell Biol. 127:1049–1060. 10.1083/jcb.127.4.1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács M., Tóth J., Hetényi C., Málnási-Csizmadia A., and Sellers J.R.. 2004. Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem. 279:35557–35563. 10.1074/jbc.M405319200 [DOI] [PubMed] [Google Scholar]
- Kriebel P.W., Barr V.A., and Parent C.A.. 2003. Adenylyl cyclase localization regulates streaming during chemotaxis. Cell. 112:549–560. 10.1016/S0092-8674(03)00081-3 [DOI] [PubMed] [Google Scholar]
- Kröger C., Loschke F., Schwarz N., Windoffer R., Leube R.E., and Magin T.M.. 2013. Keratins control intercellular adhesion involving PKC-α-mediated desmoplakin phosphorylation. J. Cell Biol. 201:681–692. 10.1083/jcb.201208162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan R., Chen L., Looi K., Tao G.Z., Weerasinghe S.V., Snider N.T., Conti M.A., Adelstein R.S., Xie Q., and Omary M.B.. 2015. PKC412 normalizes mutation-related keratin filament disruption and hepatic injury in mice by promoting keratin-myosin binding. Hepatology. 62:1858–1869. 10.1002/hep.27965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laukaitis C.M., Webb D.J., Donais K., and Horwitz A.F.. 2001. Differential dynamics of α 5 integrin, paxillin, and α-actinin during formation and disassembly of adhesions in migrating cells. J. Cell Biol. 153:1427–1440. 10.1083/jcb.153.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.H., and Coulombe P.A.. 2009. Self-organization of keratin intermediate filaments into cross-linked networks. J. Cell Biol. 186:409–421. 10.1083/jcb.200810196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.M., Stuelten C.H., Parent C.A., and Losert W.. 2016. Collective cell migration over long time scales reveals distinct phenotypes. Converg. Sci. Phys. Oncol. 2:025001 10.1088/2057-1739/2/2/025001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J.C., Piña-Paz S., Rotty J.D., Hickerson R.P., Kaspar R.L., Balmain A., and Coulombe P.A.. 2013. Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proc. Natl. Acad. Sci. USA. 110:19537–19542. 10.1073/pnas.1309576110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy L., Broad S., Diekmann D., Evans R.D., and Watt F.M.. 2000. β1 integrins regulate keratinocyte adhesion and differentiation by distinct mechanisms. Mol. Biol. Cell. 11:453–466. 10.1091/mbc.11.2.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Takeda K., Singh A., Yu Z.X., Zerfas P., Blount A., Liu C., Towbin J.A., Schneider M.D., Adelstein R.S., and Wei Q.. 2009. Conditional ablation of nonmuscle myosin II-B delineates heart defects in adult mice. Circ. Res. 105:1102–1109. 10.1161/CIRCRESAHA.109.200303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda O., Usami N., Kondo M., Takahashi M., Goto H., Shimokata K., Kusugami K., and Sekido Y.. 2004. Plakoglobin (γ-catenin) has TCF/LEF family-dependent transcriptional activity in β-catenin-deficient cell line. Oncogene. 23:964–972. 10.1038/sj.onc.1207254 [DOI] [PubMed] [Google Scholar]
- Martin P. 1997. Wound healing--aiming for perfect skin regeneration. Science. 276:75–81. 10.1126/science.276.5309.75 [DOI] [PubMed] [Google Scholar]
- Mayor R., and Etienne-Manneville S.. 2016. The front and rear of collective cell migration. Nat. Rev. Mol. Cell Biol. 17:97–109. 10.1038/nrm.2015.14 [DOI] [PubMed] [Google Scholar]
- Mazzalupo S., Wawersik M.J., and Coulombe P.A.. 2002. An ex vivo assay to assess the potential of skin keratinocytes for wound epithelialization. J. Invest. Dermatol. 118:866–870. 10.1046/j.1523-1747.2002.01736.x [DOI] [PubMed] [Google Scholar]
- McDaniel D.P., Shaw G.A., Elliott J.T., Bhadriraju K., Meuse C., Chung K.H., and Plant A.L.. 2007. The stiffness of collagen fibrils influences vascular smooth muscle cell phenotype. Biophys. J. 92:1759–1769. 10.1529/biophysj.106.089003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinnis L.A., Lee H.J., Robinson D.N., and Evans J.P.. 2015. MAPK3/1 (ERK1/2) and myosin light chain kinase in mammalian eggs affect myosin-II function and regulate the metaphase II state in a calcium- and zinc- dependent manner. Biol. Reprod. 92:146 10.1095/biolreprod.114.127027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan K.M., and Coulombe P.A.. 1998a The wound repair associated keratins 6, 16, and 17: insights into the role of intermediate filaments in specifying cytoarchitecture. In Subcellular Biochemistry: Intermediate Filaments. Harris J.R., and Herrmann H., editors. Plenum Publishing Co., London, UK: 141–165. [PubMed] [Google Scholar]
- McGowan K.M., and Coulombe P.A.. 1998b Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol. 143:469–486. 10.1083/jcb.143.2.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens C., Kuhn C., and Franke W.W.. 1996. Plakophilins 2a and 2b: constitutive proteins of dual location in the karyoplasm and the desmosomal plaque. J. Cell Biol. 135:1009–1025. 10.1083/jcb.135.4.1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M.R., Besser A., Danuser G., and Brugge J.S.. 2012. Substrate stiffness regulates cadherin-dependent collective migration through myosin-II contractility. J. Cell Biol. 199:545–563. 10.1083/jcb.201207148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen-Ngoc K.V., Silvestri V.L., Georgess D., Fairchild A.N., and Ewald A.J.. 2017. Mosaic loss of non-muscle myosin IIA and IIB is sufficient to induce mammary epithelial proliferation. J. Cell Sci. 130:3213–3221. 10.1242/jcs.208546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole E.A. 2001. Extracellular matrix and keratinocyte migration. Clin. Exp. Dermatol. 26:525–530. 10.1046/j.1365-2230.2001.00891.x [DOI] [PubMed] [Google Scholar]
- Osmanagic-Myers S., and Wiche G.. 2004. Plectin-RACK1 (receptor for activated C kinase 1) scaffolding: a novel mechanism to regulate protein kinase C activity. J. Biol. Chem. 279:18701–18710. 10.1074/jbc.M312382200 [DOI] [PubMed] [Google Scholar]
- Osmanagic-Myers S., Gregor M., Walko G., Burgstaller G., Reipert S., and Wiche G.. 2006. Plectin-controlled keratin cytoarchitecture affects MAP kinases involved in cellular stress response and migration. J. Cell Biol. 174:557–568. 10.1083/jcb.200605172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa M. 2018. Nonmuscle myosin IIA is involved in recruitment of apical junction components through activation of α-catenin. Biol. Open. 7:bio-031369 10.1242/bio.031369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladini R.D., Takahashi K., Bravo N.S., and Coulombe P.A.. 1996. Onset of re-epithelialization after skin injury correlates with a reorganization of keratin filaments in wound edge keratinocytes: defining a potential role for keratin 16. J. Cell Biol. 132:381–397. 10.1083/jcb.132.3.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons J.T., Horwitz A.R., and Schwartz M.A.. 2010. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nat. Rev. Mol. Cell Biol. 11:633–643. 10.1038/nrm2957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasapera A.M., Schneider I.C., Rericha E., Schlaepfer D.D., and Waterman C.M.. 2010. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J. Cell Biol. 188:877–890. 10.1083/jcb.200906012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean L., Reffay M., Grasland-Mongrain E., Poujade M., Ladoux B., Buguin A., and Silberzan P.. 2010. Velocity fields in a collectively migrating epithelium. Biophys. J. 98:1790–1800. 10.1016/j.bpj.2010.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., and Horwitz A.R.. 2003. Cell migration: integrating signals from front to back. Science. 302:1704–1709. 10.1126/science.1092053 [DOI] [PubMed] [Google Scholar]
- Rietscher K., Wolf A., Hause G., Rother A., Keil R., Magin T.M., Glass M., Niessen C.M., and Hatzfeld M.. 2016. Growth retardation, loss of desmosomal adhesion, and impaired tight junction function identify a unique role of plakophilin 1 in vivo. J. Invest. Dermatol. 136:1471–1478. 10.1016/j.jid.2016.03.021 [DOI] [PubMed] [Google Scholar]
- Ron D., Chen C.H., Caldwell J., Jamieson L., Orr E., and Mochly-Rosen D.. 1994. Cloning of an intracellular receptor for protein kinase C: a homolog of the beta subunit of G proteins. Proc. Natl. Acad. Sci. USA. 91:839–843. 10.1073/pnas.91.3.839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty J.D., and Coulombe P.A.. 2012. A wound-induced keratin inhibits Src activity during keratinocyte migration and tissue repair. J. Cell Biol. 197:381–389. 10.1083/jcb.201107078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Egelhoff T., and Baskaran H.. 2009. Insights into the roles of non-muscle myosin IIA in human keratinocyte migration. Cell. Mol. Bioeng. 2:486–494. 10.1007/s12195-009-0094-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savagner P., Kusewitt D.F., Carver E.A., Magnino F., Choi C., Gridley T., and Hudson L.G.. 2005. Developmental transcription factor slug is required for effective re-epithelialization by adult keratinocytes. J. Cell. Physiol. 202:858–866. 10.1002/jcp.20188 [DOI] [PubMed] [Google Scholar]
- Schmidt A., Langbein L., Rode M., Prätzel S., Zimbelmann R., and Franke W.W.. 1997. Plakophilins 1a and 1b: widespread nuclear proteins recruited in specific epithelial cells as desmosomal plaque components. Cell Tissue Res. 290:481–499. 10.1007/s004410050956 [DOI] [PubMed] [Google Scholar]
- Schramek D., Sendoel A., Segal J.P., Beronja S., Heller E., Oristian D., Reva B., and Fuchs E.. 2014. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science. 343:309–313. 10.1126/science.1248627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers J.R., Pato M.D., and Adelstein R.S.. 1981. Reversible phosphorylation of smooth muscle myosin, heavy meromyosin, and platelet myosin. J. Biol. Chem. 256:13137–13142. [PubMed] [Google Scholar]
- Seltmann K., Roth W., Kröger C., Loschke F., Lederer M., Hüttelmaier S., and Magin T.M.. 2013. Keratins mediate localization of hemidesmosomes and repress cell motility. J. Invest. Dermatol. 133:181–190. 10.1038/jid.2012.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seltmann K., Cheng F., Wiche G., Eriksson J.E., and Magin T.M.. 2015. Keratins stabilize hemidesmosomes through regulation of β4-integrin turnover. J. Invest. Dermatol. 135:1609–1620. 10.1038/jid.2015.46 [DOI] [PubMed] [Google Scholar]
- Setzer S.V., Calkins C.C., Garner J., Summers S., Green K.J., and Kowalczyk A.P.. 2004. Comparative analysis of armadillo family proteins in the regulation of a431 epithelial cell junction assembly, adhesion and migration. J. Invest. Dermatol. 123:426–433. 10.1111/j.0022-202X.2004.23319.x [DOI] [PubMed] [Google Scholar]
- Simpson C.L., Patel D.M., and Green K.J.. 2011. Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nat. Rev. Mol. Cell Biol. 12:565–580. 10.1038/nrm3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolik-Delmaire T., Reddy R., Pashaj A., Roberts B.J., and Wahl J.K. III. 2010. Plakophilin-1 localizes to the nucleus and interacts with single-stranded DNA. J. Invest. Dermatol. 130:2638–2646. 10.1038/jid.2010.191 [DOI] [PubMed] [Google Scholar]
- Somlyo A.P., and Somlyo A.V.. 1994. Signal transduction and regulation in smooth muscle. Nature. 372:231–236. 10.1038/372231a0 [DOI] [PubMed] [Google Scholar]
- Sonavane P.R., Wang C., Dzamba B., Weber G.F., Periasamy A., and DeSimone D.W.. 2017. Mechanical and signaling roles for keratin intermediate filaments in the assembly and morphogenesis of Xenopus mesendoderm tissue at gastrulation. Development. 144:4363–4376. 10.1242/dev.155200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- South A.P., Wan H., Stone M.G., Dopping-Hepenstal P.J., Purkis P.E., Marshall J.F., Leigh I.M., Eady R.A., Hart I.R., and McGrath J.A.. 2003. Lack of plakophilin 1 increases keratinocyte migration and reduces desmosome stability. J. Cell Sci. 116:3303–3314. 10.1242/jcs.00636 [DOI] [PubMed] [Google Scholar]
- Stehbens S.J., and Wittmann T.. 2014. Analysis of focal adhesion turnover: a quantitative live-cell imaging example. Methods Cell Biol. 123:335–346. 10.1016/B978-0-12-420138-5.00018-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens S.J., Paszek M., Pemble H., Ettinger A., Gierke S., and Wittmann T.. 2014. CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16:561–573. 10.1038/ncb2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yan B., Yamanishi K., Imamura S., and Coulombe P.A.. 1998. The two functional keratin 6 genes of mouse are differentially regulated and evolved independently from their human orthologs. Genomics. 53:170–183. 10.1006/geno.1998.5476 [DOI] [PubMed] [Google Scholar]
- Totsukawa G., Wu Y., Sasaki Y., Hartshorne D.J., Yamakita Y., Yamashiro S., and Matsumura F.. 2004. Distinct roles of MLCK and ROCK in the regulation of membrane protrusions and focal adhesion dynamics during cell migration of fibroblasts. J. Cell Biol. 164:427–439. 10.1083/jcb.200306172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckwell D., Calderwood D.A., Green L.J., and Humphries M.J.. 1995. Integrin alpha 2 I-domain is a binding site for collagens. J. Cell Sci. 108:1629–1637. [DOI] [PubMed] [Google Scholar]
- Vaezi A., Bauer C., Vasioukhin V., and Fuchs E.. 2002. Actin cable dynamics and Rho/Rock orchestrate a polarized cytoskeletal architecture in the early steps of assembling a stratified epithelium. Dev. Cell. 3:367–381. 10.1016/S1534-5807(02)00259-9 [DOI] [PubMed] [Google Scholar]
- Vandenberg P., Kern A., Ries A., Luckenbill-Edds L., Mann K., and Kühn K.. 1991. Characterization of a type IV collagen major cell binding site with affinity to the alpha 1 beta 1 and the alpha 2 beta 1 integrins. J. Cell Biol. 113:1475–1483. 10.1083/jcb.113.6.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasioukhin V., Bowers E., Bauer C., Degenstein L., and Fuchs E.. 2001. Desmoplakin is essential in epidermal sheet formation. Nat. Cell Biol. 3:1076–1085. 10.1038/ncb1201-1076 [DOI] [PubMed] [Google Scholar]
- Vicente-Manzanares M., Ma X., Adelstein R.S., and Horwitz A.R.. 2009. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat. Rev. Mol. Cell Biol. 10:778–790. 10.1038/nrm2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Zieman A., and Coulombe P.A.. 2016. Skin Keratins. Methods Enzymol. 568:303–350. 10.1016/bs.mie.2015.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawersik M.J., Mazzalupo S., Nguyen D., and Coulombe P.A.. 2001. Increased levels of keratin 16 alter epithelialization potential of mouse skin keratinocytes in vivo and ex vivo. Mol. Biol. Cell. 12:3439–3450. 10.1091/mbc.12.11.3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb D.J., Donais K., Whitmore L.A., Thomas S.M., Turner C.E., Parsons J.T., and Horwitz A.F.. 2004. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat. Cell Biol. 6:154–161. 10.1038/ncb1094 [DOI] [PubMed] [Google Scholar]
- Weber G.F., Bjerke M.A., and DeSimone D.W.. 2012. A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev. Cell. 22:104–115. 10.1016/j.devcel.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiger M.C., Vedham V., Stuelten C.H., Shou K., Herrera M., Sato M., Losert W., and Parent C.A.. 2013. Real-time motion analysis reveals cell directionality as an indicator of breast cancer progression. PLoS One. 8:e58859 10.1371/journal.pone.0058859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenger M.P., Bozec L., Horton M.A., and Mesquida P.. 2007. Mechanical properties of collagen fibrils. Biophys. J. 93:1255–1263. 10.1529/biophysj.106.103192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P., and Coulombe P.A.. 2003. Loss of keratin 6 (K6) proteins reveals a function for intermediate filaments during wound repair. J. Cell Biol. 163:327–337. 10.1083/jcb.200305032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P., Colucci-Guyon E., Takahashi K., Gu C., Babinet C., and Coulombe P.A.. 2000. Introducing a null mutation in the mouse K6α and K6β genes reveals their essential structural role in the oral mucosa. J. Cell Biol. 150:921–928. 10.1083/jcb.150.4.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh Y.C., Ling J.Y., Chen W.C., Lin H.H., and Tang M.J.. 2017. Mechanotransduction of matrix stiffness in regulation of focal adhesion size and number: reciprocal regulation of caveolin-1 and β1 integrin. Sci. Rep. 7:15008 10.1038/s41598-017-14932-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin T., Getsios S., Caldelari R., Kowalczyk A.P., Müller E.J., Jones J.C., and Green K.J.. 2005. Plakoglobin suppresses keratinocyte motility through both cell-cell adhesion-dependent and -independent mechanisms. Proc. Natl. Acad. Sci. USA. 102:5420–5425. 10.1073/pnas.0501676102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.