Local activity of the small GTPase Cdc42 is critical for cell polarization. Gallo Castro and Martin describe a new Cdc42 GTPase-activating protein (GAP) Rga3, which together with two other GAPs, constrains Cdc42-GTP zones during mitotic cycles but not during sexual reproduction.
Abstract
The formation of a local zone of Cdc42 GTPase activity, which governs cell polarization in many cell types, requires not only local activation but also switch-off mechanisms. In this study, we identify Rga3, a paralog of Rga4, as a novel Cdc42 GTPase-activating protein (GAP) in the fission yeast Schizosaccharomyces pombe. Contrary to Rga4, Rga3 localizes with Cdc42-GTP to sites of polarity. Rga3 is dispensable for cell polarization during mitotic growth, but it limits the lifetime of unstable Cdc42-GTP patches that underlie cell pairing during sexual reproduction, masking a partly compensatory patch-wandering motion. In consequence, cells lacking rga3 hyperpolarize and lose out in mating competition. Rga3 synergizes with the Cdc42 GAPs Rga4 and Rga6 to restrict Cdc42-GTP zone sizes during mitotic growth. Surprisingly, triple-mutant cells, which are almost fully round, retain pheromone-dependent dynamic polarization of Cdc42-GTP, extend a polarized projection, and mate. Thus, the requirement for Cdc42-GTP hydrolysis by GAPs is distinct during polarization by intrinsic or extrinsic cues.
Introduction
Polarization is critical for cell physiology. In most eukaryotic cell types, despite a great variety of shapes and functions, small GTPases, in particular the highly conserved Rho family GTPase Cdc42, regulate cell polarization (Etienne-Manneville, 2004). First identified for its role in the polarization of Saccharomyces cerevisiae cells for bud formation (Adams et al., 1990), the function of Cdc42 has been conserved through evolution as illustrated by cross-species complementation (Munemitsu et al., 1990; Shinjo et al., 1990; Miller and Johnson, 1994; Sasamura et al., 1997) and its requirement for polarization in numerous cell types including the fission yeast Schizosaccharomyces pombe (Miller and Johnson, 1994), Caenorhabditis elegans zygotes (Gotta et al., 2001; Kay and Hunter, 2001), Drosophila melanogaster neuroblasts (Atwood et al., 2007), or mammalian epithelia and oocytes (Wu et al., 2007; Wang et al., 2013).
Cdc42 is under complex regulation and cycles between active and inactive states (Vetter and Wittinghofer, 2001). When bound to GTP, Cdc42 activates effectors including nucleators of actin assembly such as formins, regulators of vesicle secretion such as the exocyst complex, and members of the p21-activated kinase (PAK) family (Perez and Rincón, 2010). These collectively convert a localized Cdc42 signal into effective cell polarization. Cdc42 activation relies on guanine nucleotide exchange factors (GEFs), which promote exchange of GDP for GTP. For its inactivation, Cdc42 has intrinsic GTPase activity, which is also promoted by GTPase-activating proteins (GAPs). Cdc42, which associates with membranes through a prenyl moiety, can also be sequestered in the cytosol by GDP dissociation inhibitors (GDIs; DerMardirossian and Bokoch, 2005). Importantly, cycling of Cdc42 GTP-bound active and GDP-bound inactive states is critical for its function in cell polarization. In fission yeast, both Cdc42 disruption and constitutive activation lead to cell rounding and lethality, with disruption causing small round cells and constitutive activation causing large ones (Miller and Johnson, 1994; Bendezú et al., 2015). In consequence, the local activity of Cdc42 is critical for cell polarization.
Local activity results in part from localized GEFs, of which there are two in S. pombe: Scd1 and Gef1. These are together essential and localize at sites of growth and division (Coll et al., 2003; Hirota et al., 2003). However, local activation is not sufficient to generate locally restricted Cdc42 activity: cells also need to inactivate Cdc42 to prevent the spread of the active form. For instance, we recently observed that the cell pole–restricted activity of the related small GTPase Ras1 requires inactivation by its GAP, the deletion of which leads to loss of spatial information and Ras1-GTP distribution over the whole plasma membrane (Merlini et al., 2018). In fission yeast, two Cdc42 GAPs, Rga4 and Rga6, both of which localize to cell sides, contribute to the spatial restriction of active Cdc42 to the cell tips, with rga4Δ and rga6Δ mutant cells exhibiting enlarged cell width (Tatebe et al., 2008; Revilla-Guarinos et al., 2016). However, even double-mutant cells retain polarized Cdc42-GTP zones, albeit a bit wider, suggesting that negative controls of Cdc42 activity remain in place. Cdc42 inactivation may also involve detachment from the membrane and sequestration in the cytosol by GDI. In S. cerevisiae, the GDI plays an important role in promoting Cdc42-GDP recycling to the polarity patch and accumulation of Cdc42 at its site of activity, with GAP activity also recently implicated (Slaughter et al., 2009; Woods et al., 2016). In S. pombe, Cdc42-GDP exhibits fast lateral mobility at the plasma membrane such that the slower-diffusing Cdc42-GTP form can polarize independently of the GDI (Bendezú et al., 2015). Consistently, deletion of the sole GDI yields no overt phenotype (Nakano et al., 2003; Bendezú et al., 2015).
Cell polarization can take many shapes and forms for which the same polarity components are reused. In fission yeast, Cdc42 activity can respond to intracellular microtubule-deposited landmarks that recruit it to cell poles (Tatebe et al., 2008; Kokkoris et al., 2014). There, it oscillates in an anticorrelated manner (Das et al., 2012) to drive the bipolar growth patterns of these cells and defines cellular dimensions during mitotic growth (Kelly and Nurse, 2011; Bendezú et al., 2015). Upon spore germination, Cdc42-GTP polarizes spontaneously, forming dynamic zones that stabilize upon spore outgrowth (Bonazzi et al., 2014). During sexual differentiation, Cdc42-GTP also forms unstable zones that explore the cell periphery upon pheromone exposure and become stabilized upon increased pheromone concentrations (Bendezú and Martin, 2013). In the physiological process, cells of opposite mating type (plus [P or h+] or minus [M or h–]) express and secrete distinct pheromones and cognate receptors, which activate a common Ras1-MAPK signaling cascade (Merlini et al., 2013). In potential partner cells, Cdc42 patches act as a source of pheromone such that facing patches in partner cells stabilize each other, driving cell pairing (Merlini et al., 2016). Consequently, alterations in Cdc42 patch dynamics modify partner cell choice. For instance, excessive pheromone perception compromises Cdc42 dynamics outside the cell tip region, leading to preferential mating between just-divided sister cells (Bendezú and Martin, 2013); excessive Ras1 activity promotes zone stabilization at reduced pheromone concentration, leading to defective cell pairing (Merlini et al., 2016; Khalili et al., 2018).
The oscillatory patterns described above imply the existence of positive and negative feedbacks. Positive feedbacks are well described from work in S. cerevisiae, where they underlie the spontaneous polarization of Cdc42 in the absence of landmarks (Gulli et al., 2000; Irazoqui et al., 2003; Wedlich-Soldner and Li, 2003). One prominent feedback involves the formation of a complex between a Cdc42 effector PAK kinase and a Cdc42 GEF through a scaffold protein, thought to amplify initial stochastic variations in Cdc42 activity to promote symmetry breaking (Kozubowski et al., 2008). The molecular controls of negative feedback that destabilize the patch remain largely unclear. Phosphorylation of the Cdc42 scaffold by the PAK kinase may contribute in dampening the positive feedback reaction (Das et al., 2012; Howell et al., 2012; Kuo et al., 2014; Rapali et al., 2017). Mechanical constraints from the cell wall may contribute to patch destabilization (Bonazzi et al., 2014). The role of Cdc42 GAP proteins in promoting negative feedback has not been systematically explored.
In this study, we describe a novel Cdc42 GAP Rga3. We show that Rga3 is a paralog of Rga4, which arose from gene duplication in the Schizosaccharomyces lineage. In contrast with Rga4 and Rga6, Rga3 is recruited to sites of Cdc42 activity, yet it synergizes with these two GAPs during mitotic growth to restrict Cdc42-GTP zone size and cell dimensions. During pheromone-dependent polarization, Rga3 is recruited to the Cdc42 patch, where it promotes its dynamics and modulates partner choice. Surprisingly, a triple GAP mutant, though lacking polarity during mitotic growth, retains almost complete ability to polarize during sexual differentiation and mates, indicating fundamental differences in Cdc42-GTP zone regulation in distinct contexts.
Results
Rga3 is a paralog of Rga4
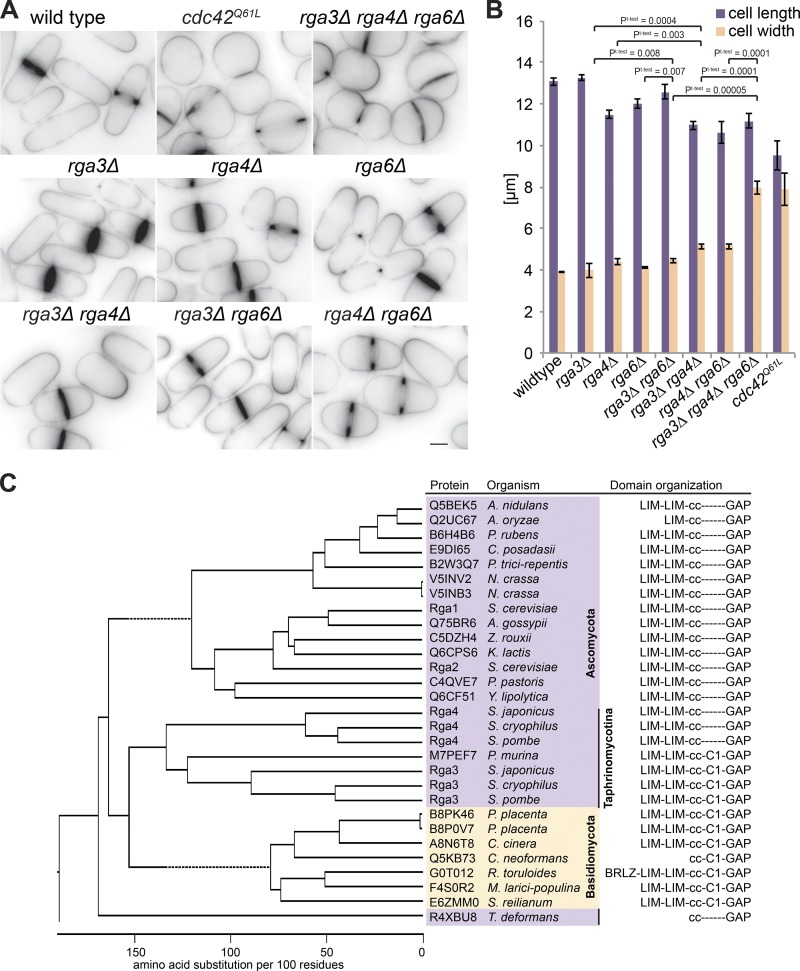
Two Cdc42 GAPs have been reported in S. pombe: Rga4 and Rga6 (Tatebe et al., 2008; Revilla-Guarinos et al., 2016). Recent work showed that Rga6 collaborates with Rga4 in the control of cell dimensions as double deletion of rga4 and rga6 leads to shorter and wider cells than either rga4Δ or rga6Δ single mutants (Revilla-Guarinos et al., 2016). However, the phenotype of this double mutant is much weaker than that caused by overexpression of a constitutively active allele of Cdc42 (Cdc42Q61L), which leads to complete polarity loss and formation of round cells with cytokinesis defects (Fig. 1, A and B; Miller and Johnson, 1994; Bendezú et al., 2015). This discrepancy suggests the existence of other GAP(s) promoting Cdc42-GTP hydrolysis.
Figure 1.
Rga3 is a paralog of Rga4 and contributes to cellular dimensions. (A) Medial-plane inverted images of indicated strains stained with calcofluor. Bar, 3 µm. (B) Average cell length and width of indicated strains grown in EMM-ALU at 25°C (n = 3 experiments; n > 145 cells each). Error bars show SD. Student’s t test P values are indicated. (C) Phylogenetic tree of ClustalW alignment of selected RhoGAP domain–containing proteins from Ascomycetes (purple) and Basidiomycetes (yellow) species. The Taphrinomycotina group to which fission yeasts belong is highlighted. Domain architecture (derived from SMART analysis) is shown on the right. Note that the C1 domain is present in Basidiomycetes and some Taphrinomycotina sequences but is absent in all other ascomycete sequences.
Basic Local Alignment Search Tool (BLAST) searches with Rga4 against the fission yeast genome revealed Rga3 as a reciprocal best hit. Further searches for RhoGAP domain–containing sequences in ascomycetes and basidiomycetes as well as phylogenetic analysis of proteins from selected species showed that Rga3 and Rga4 cluster together on the same branch as the S. cerevisiae Cdc42 GAPs Rga1 and Rga2 (Fig. 1 C). Reciprocal BLAST analysis did not reveal any clear orthologue pair with the S. cerevisiae GAPs, consistent with the notion that S. cerevisiae underwent a whole-genome duplication during its evolution (Wolfe and Shields, 1997). Furthermore, with the exception of S. cerevisiae and a few other species, all ascomycetes and basidiomycetes contain a single rga4-like gene, whereas all Schizosaccharomyces species have two (Fig. 1 C). Thus, Rga3 and Rga4 likely arose from gene duplication in the Schizosaccharomyces lineage.
All Rga4-like GAPs share the same domain structures: two N-terminal LIM domains, a central coiled-coil region, and a C-terminal RhoGAP domain (Fig. 1 C). Interestingly, Rga3 contains an additional C1 domain, predicted to bind lipids (Kaibuchi et al., 1989; Ono et al., 1989). This domain architecture is not shared by any other ascomycete protein except by the basal Taphrinomycotina group, to which the Schizosaccharomyces lineage belongs. Rga4-like GAPs also exhibit similar domain architecture, including a C1 domain, in the closely related basidiomycete fungi. BLAST searches with the Rga3 or Rga4 GAP domain through other eukaryotic phyla returned Chimaerin-family RacGAPs as closest hits, which also bear a linked C1 domain (Canagarajah et al., 2004). Thus, a likely scenario is that the Rga3/4 ancestral state contained a C1 domain, which was subsequently lost in Rga4 and other ascomycete species.
Rga3 is a Cdc42 GAP
Deletion of rga3 did not lead to any evident morphological phenotype during vegetative growth. Indeed, cells lacking rga3 showed similar cell length and width as WT cells (Fig. 1, A and B). However, deletion of rga3 in combination with rga4Δ and rga6Δ led to cells significantly wider and more rounded compared with the single or double mutants, a phenotype very similar to that of overexpression of a constitutively active allele of Cdc42 (Fig. 1, A and B). Similarly, rga3Δrga4Δ and rga3Δrga6Δ double mutants were also significantly wider than either single mutant. These phenotypes strongly suggest that Rga3 could be a Cdc42 GAP.
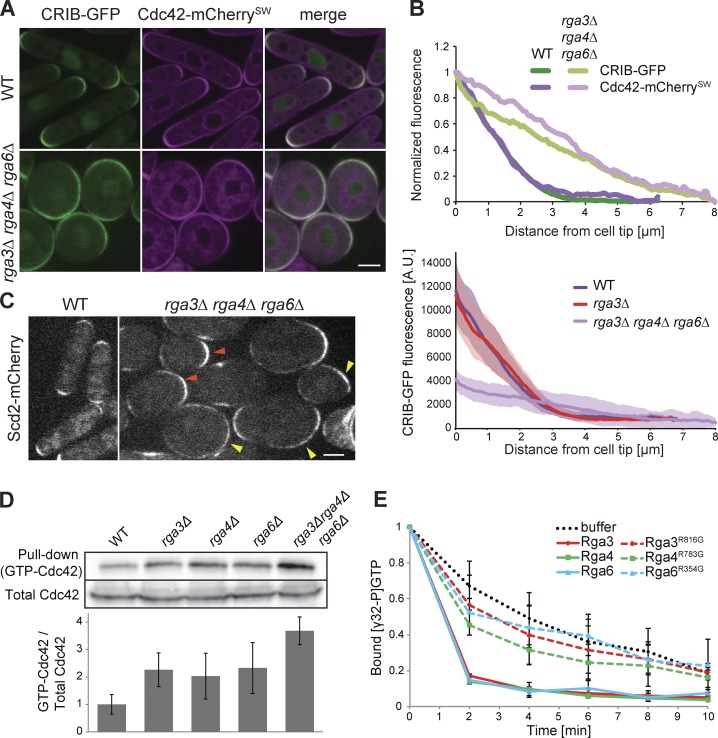
The dramatic morphological change in rga3Δrga4Δrga6Δ cells raised the question of how Cdc42 activity is distributed along the cortex. To address this question, we coimaged Cdc42-mCherrySW, a functional internally mCherry-tagged allele of Cdc42 encoded at the endogenous locus (Bendezú et al., 2015), and Cdc42/Rac-interactive binding peptide (CRIB)-GFP in WT and rga3Δrga4Δrga6Δ strains. The CRIB domain, which specifically binds Cdc42-GTP, is used as a marker for active Cdc42. In contrast with the restricted distribution of CRIB around the tips of WT cells, CRIB formed very large cortical domains occupying broad zones all over the cell cortex in the triple mutant, with Cdc42-mCherrySW showing a similar distribution (Fig. 2 A). Cortical profile measurements confirmed a much broader distribution of both Cdc42 and its active form (Fig. 2 B). Of note, the very tight overlap between the normalized Cdc42 and CRIB distribution profiles, indicative of Cdc42 protein enrichment to sites of activity in WT cells (Bendezú et al., 2015), was largely preserved in the triple-mutant condition. Thus, the correlation between Cdc42 activity and its local enrichment does not depend on GAP proteins (Fig. 2 A). The Cdc42-GTP–binding scaffold protein Scd2 (Endo et al., 2003; Wheatley and Rittinger, 2005) similarly localized to a broad cortical region (Fig. 2 C). We observed a broad variation of zone sizes for both CRIB and Scd2, ranging from zones covering an enlarged cell tip to almost complete cell surface coverage. These results indicate that Rga3 in conjunction with other GAP proteins serve to restrict the size of the Cdc42-GTP domain.
Figure 2.
Rga3 is a Cdc42 GAP. (A) Medial-plane spinning-disk confocal images of Cdc42-mCherrySW and CRIB-GFP in WT and rga3Δrga4Δrga6Δ. (B) The top graph shows average cortical profiles of Cdc42 and CRIB fluorescence intensity normalized to maximum and minimum values. For rga3Δrga4Δrga6Δ, only cells in which CRIB-GFP formed defined zones were used (n ≥ 20), so the described phenotype is underestimated in this quantification. For clarity, SDs are omitted from this graph. The bottom graph shows the nonnormalized CRIB-GFP fluorescence values for WT, rga3Δ, and triple rga3Δrga4Δrga6Δ mutants. Confidence bands show SD. n = 20 profiles per curve. (C) Medial-plane spinning-disk confocal images of Scd2-mCherry in WT and rga3Δrga4Δrga6Δ. Orange arrowheads indicate enlarged zones at cell poles; yellow arrowheads indicate zones extending over a large part of the cell cortex. (D) Cdc42-GTP pulldown assay. GST-CRIB was coupled to glutathione beads and mixed with extracts of WT, rga3Δ, rga4Δ, rga6Δ, or rga3Δrga4Δrga6Δ cells expressing Cdc42-mCherrySW. Cdc42 was revealed in input and pulldown fractions with anti-mCherry antibody. One representative experiment is shown on top, and the average of three experiments is reported on the graph below. (E) In vitro GAP assay. [γ-32P]GTP-loaded recombinant Cdc42 was incubated with recombinant full-length GAPs (WT or catalytically inactive; point mutations indicated) or with buffer, and Cdc42-associated radioactivity was determined by scintillation counting. Data are normalized to initial value. Bars, 3 µm. Error bars show SD.
To test whether Rga3 is indeed a Cdc42 GAP, protein extracts from WT, single rga3Δ, rga4Δ, and rga6Δ mutant and triple-mutant cells carrying Cdc42-mcherrySW were used to determine the total amount of GTP-bound Cdc42 by pulldown with GST-CRIB. Extracts of cells lacking rga3 showed an increase in GTP-bound Cdc42 compared with WT extracts, comparable with the levels shown upon rga4 and rga6 deletion. Moreover, the triple mutant presented a further increase in the fraction of active Cdc42 (Fig. 2 D), consistent with the additive phenotypes seen in vivo. While these results are consistent with the idea that Rga3, Rga4, and Rga6 function as Cdc42 GAPs in vivo, we note that the significant increase in Cdc42-GTP in rga3Δ is somewhat at odds with the absence of morphological phenotype. As quantification of CRIB distribution was indistinguishable in rga3Δ and WT cells (Fig. 2 B), it is possible that the detected differences in Cdc42-GTP levels may be amplified in vitro in the course of the pulldown experiment.
To further verify Rga3’s direct role as a Cdc42 GAP, we performed an in vitro GTPase assay on Cdc42. Purified recombinant Cdc42 preloaded with [γ-32P]GTP was incubated with recombinant Rga3, Rga4, or Rga6 to test the ability of these GAPs to increase the rate of Cdc42-GTP hydrolysis. All three GAPs including Rga3 increased the rate of Cdc42-GTP hydrolysis. Point mutations in the catalytic arginine finger of each GAP, predicted to affect GTP hydrolysis (Scheffzek et al., 1998), blocked their ability to increase the intrinsic GTPase activity of Cdc42 (Fig. 2 E). Taken together, these data demonstrate that Rga3 is a Cdc42 GAP.
Rga3 colocalizes with active Cdc42 independently of its GAP domain, and its cortical localization is mainly dependent on the C1 domain
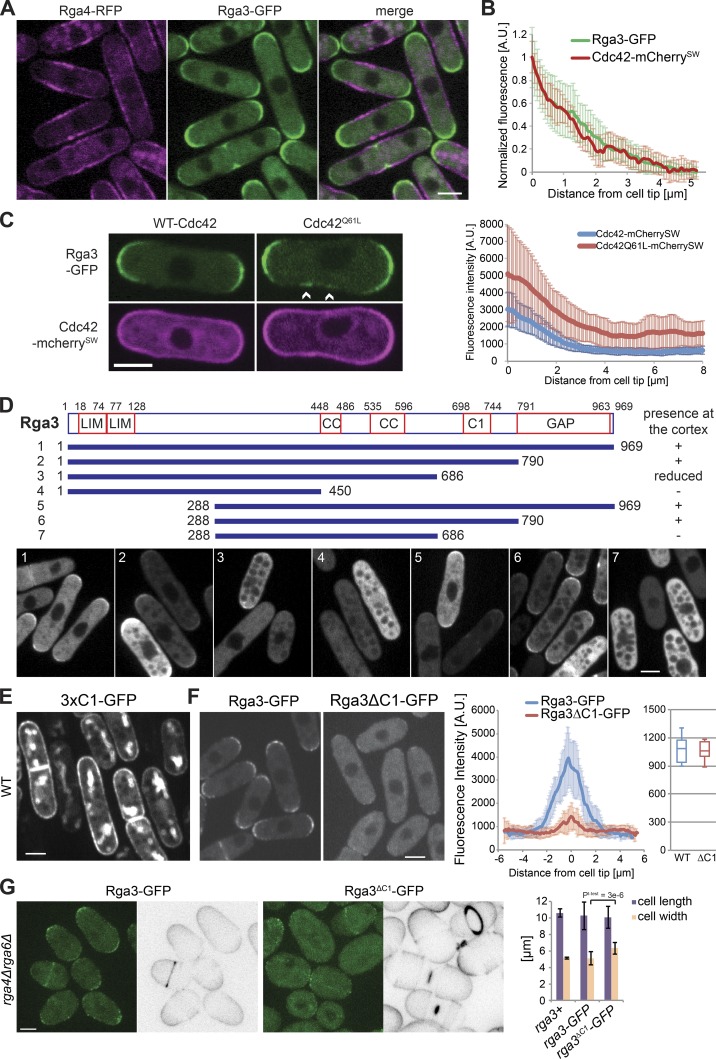
Given the observation that Rga3 and Rga4 are likely paralogs, have similar protein structure, and share function in the regulation of Cdc42, we compared the localization of the two GAP proteins. Tagging Rga3 with GFP yielded a functional protein as judged by the morphology of rga4Δrga6Δ rga3-GFP cells, which was indistinguishable from that of rga4Δrga6Δ cells (see Fig. 3 G). Strikingly, while Rga4 forms clusters at cell sides (Das et al., 2007; Tatebe et al., 2008), Rga3 localized to sites of cell division and cell tips, where its distribution was similar to that of Cdc42 (Fig. 3, A and B). Rga3 was also cytosolic. Furthermore, whereas Rga4-RFP covers the nongrowing cell pole of monopolar tea1Δ cells (Tatebe et al., 2008), Rga3-GFP was present only at the growing cell pole (Fig. S1 A). Thus, Rga3 localization to cell tips correlates with active growth.
Figure 3.
Rga3 colocalizes with active Cdc42 and binds the cortex through its C1 domain. (A) Images of Rga3-GFP and Rga4-RFP. (B) Average cortical profiles of Cdc42-mCherrySW and Rga3-GFP fluorescence intensity normalized to maximum and minimum values. n = 24 tips. (C) Images of Rga3-GFP strains expressing a pREP41-Cdc42-mCherrySW (WT or Q61L) plasmid (left). Arrowheads indicate zones of Rga3-GFP at the cell sides. Average cortical profiles of Rga3-GFP fluorescence (right). In the strain carrying the Cdc42Q61L allele, only rod-shaped cells were analyzed. n = 18. (D) Schematic representation of Rga3 fragments whose localization was tested upon GFP tagging and plasmid expression. Cortex-binding ability is summarized on the right. Representative images of plasmid-expressing rga3Δ cells are shown at the bottom. Note that Rga3 fragments are excluded from vacuoles. CC, coiled coil. (E) Image of cells expressing a triple-tandem copy of Rga3 C1 domain from plasmid. (F) Images of Rga3-GFP and Rga3ΔC1-GFP expressed from the native genomic locus (left). Average cortical profiles of Rga3-GFP and Rga3ΔC1-GFP fluorescence intensity (middle). Whole-cell intensity of Rga3-GFP and Rga3ΔC1-GFP (right). n = 11. (G) Calcofluor and GFP fluorescence images of rga4Δrga6Δ cells expressing Rga3-GFP or Rga3ΔC1-GFP (left). Average cell length and width of the same strains (n > 50; right). All micrographs are medial plane spinning-disk confocal images. Bars, 3 µm. Student’s t test P value is indicated. Error bars show SD.
Rga3 cell tip localization was largely independent of F-actin, vesicular trafficking, and microtubules: treatment with latrunculin A (an actin-depolymerizing agent), brefeldin A (a drug that blocks vesicular trafficking), or methyl benzimidazol-2-yl-carbamate (MBC; a microtubule-depolymerizing agent) did not lead to delocalization of Rga3 from the tips of the cell (Fig. S1 B). By contrast, we found that Rga3 localization was dependent on Cdc42-GTP. Indeed, cells expressing from plasmid a constitutively GTP-bound allele of Cdc42 (Cdc42Q61L), which decorates the entire cell cortex, showed Rga3 localization at the sides of the cells in addition to cell tips (Fig. 3 C). Note that Cdc42Q61L was expressed here for a shorter amount of time than in Fig. 1 A to visualize Rga3 localization in rod-shaped cells before cell rounding. This suggests that Rga3 is directly or indirectly recruited to cell poles by active Cdc42.
To further understand the determinants of Rga3 localization, we generated truncated GFP-tagged forms of Rga3, expressed from plasmids, and assessed their localization in WT and rga3Δ cells (Fig. 3 D). The full-length protein localized at cell tips as expected, and no difference was observed between WT and rga3Δ backgrounds (Fig. 3 D, fragment 1). Interestingly, truncation of the GAP domain did not impair Rga3 localization to the cortex, suggesting recruitment to cell poles does not occur through direct interaction between Cdc42-GTP and the GAP domain (Fig. 3 D, fragment 2). By contrast, further truncation of the C1 domain or the N terminus containing both LIM domains reduced and in combination abolished Rga3 cortical localization (Fig. 3 D, fragments 3 and 5–7). The central coiled-coil regions also contributed, as truncation of these in combination with the C1 domain abrogated cortical localization (Fig. 3 D, fragment 4). We conclude that multiple Rga3 domains contribute to its localization at cell tips, with the C1 domain playing a critical role. Of note, all fragments that associated with the cell cortex were enriched at the cell tips, not the cell sides, indicating that the multiple determinants of Rga3’s cell tip recruitment do not mask an Rga4-like cell side localization.
Because the C1 domain is essential for Rga3 localization in the absence of the LIM or coiled-coil domains and is unique to Rga3, we further probed its function. A construct with three tandem copies of the C1 domain decorated the entire cell cortex, consistent with the predicted role of this domain as lipid binder (Fig. 3 E). We constructed an Rga3 allele lacking only the C1 domain (Rga3ΔC1-GFP) expressed as sole copy from the native genomic locus. Rga3-GFP and Rga3ΔC1-GFP were expressed at similar levels but with distinct localization pattern: while Rga3-GFP decorated the whole tip cortex, Rga3ΔC1-GFP was mainly cytosolic, with only a few punctae of weak fluorescence intensity at the cell tips (Fig. 3 F). The C1 domain was also required for Rga3 function, as the deletion of the C1 domain in an rga4Δrga6Δ background produced rounded cells similar to rga3Δrga4Δrga6Δ triple mutants (Fig. 3 G). We conclude that the C1 domain of Rga3, though not sufficient to direct Rga3 to cell poles, is a major contributor of its localization to sites of polarity.
Because Rga3 is indirectly recruited by active Cdc42, it may form a negative feedback on Cdc42 activity. Cdc42 activity has been shown to oscillate between the two cell tips in an anticorrelated manner, a behavior that requires negative feedback (Das et al., 2012). We thus compared Cdc42 oscillations in WT and rga3Δ cells. However, analysis of CRIB-GFP signal oscillations over time showed a similar anticorrelation between the two cell poles in the two strains, with a small decrease in frequency in rga3Δ cells (Fig. S2). Thus, consistent with the lack of morphological phenotype of rga3Δ cells, these observations indicate that negative regulation by Rga3 only functions redundantly with other Cdc42 GAPs during the vegetative life stage.
Rga3 limits growth projection in response to pheromone and confers a competitive advantage during sexual reproduction
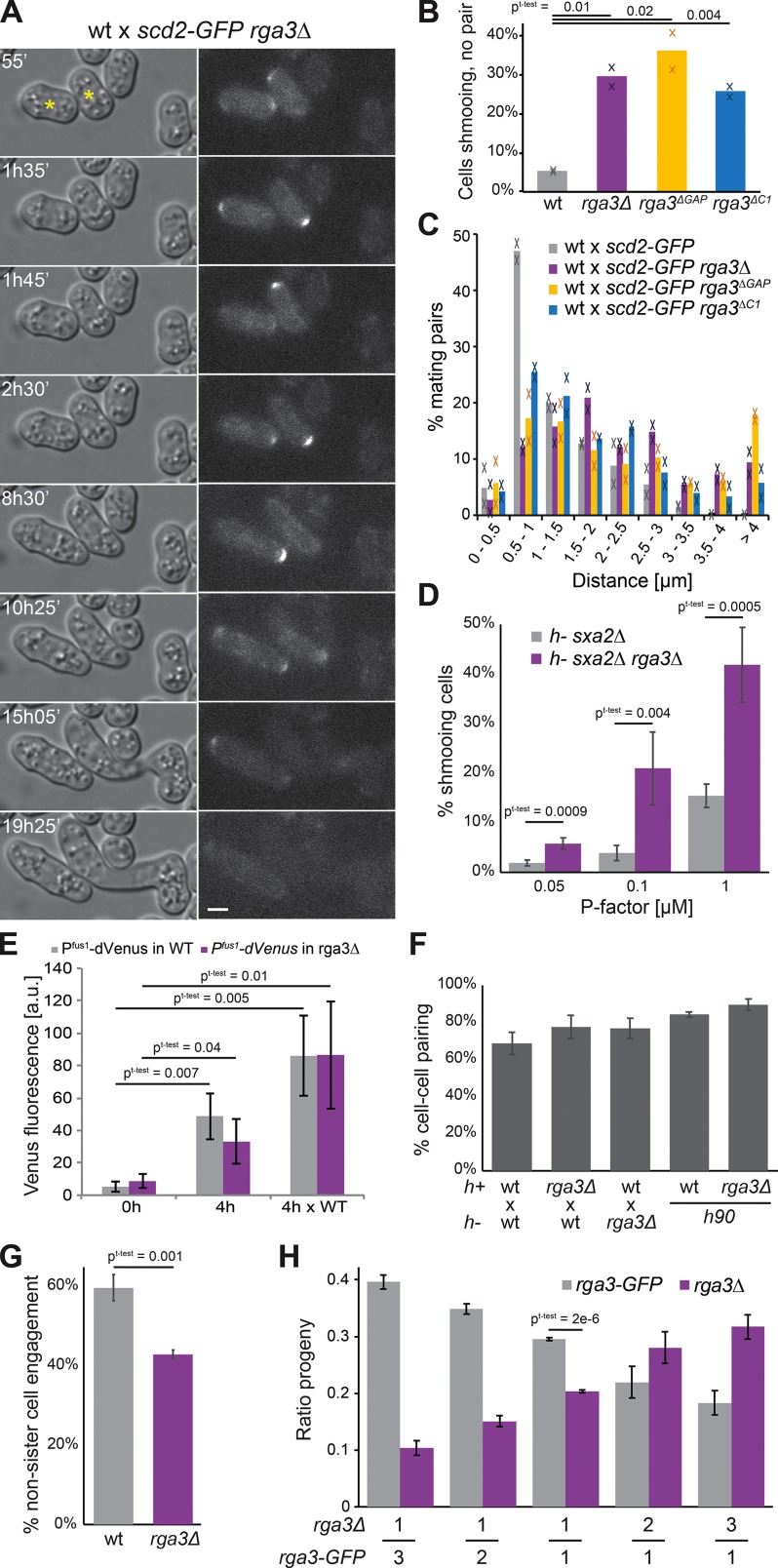
The absence of a detectable phenotype of rga3Δ cells raises the question of what selective pressures promoted the maintenance of the rga3 gene after gene duplication. We found that in contrast with the lack of morphological phenotype during vegetative growth, rga3Δ cells exhibited alterations in mating morphologies, extending long or poorly oriented shmoos. In a first series of experiments, h+ WT or rga3Δ cells expressing Scd2-GFP were mixed with WT h− cells and allowed to mate. Almost 30% of h+ rga3Δ scd2-GFP cells were found to extend a projection yet failed to find a partner after 20 h in mating conditions (Fig. 4, A and B). When observed in time-lapse experiments, these cells did not always persistently grow from a single pole as 10% alternated between the two poles (Fig. 4 A and Video 1). Similar observations were made using the rga3ΔC1 allele as well as an rga3ΔGAP allele expressed as sole copy from the native genomic locus (Fig. 4 B), indicating that Rga3 localization and GAP activity are required during mating. Pair-forming rga3Δ, rga3ΔC1, and rga3ΔGAP cells also extended longer shmoos and consequently formed pairs with more distant cells: while the majority of WT cell pairs formed between cells initially distant by <1.5 µm, most rga3Δ × WT pairs formed between cells distant by >2 µm (Fig. 4, A and C). In a second set of experiments, we treated h− cells lacking the P factor protease Sxa2 with increasing concentrations of synthetic P factor pheromone and quantified the proportion of shmooing cells after 24 h. rga3Δ cells formed shmoos at lower pheromone concentrations than rga3+ cells (Fig. 4 D), suggesting a higher sensitivity to pheromone consistent with the aberrant formation of shmoos in mating mixtures.
Figure 4.
Rga3 limits growth projection in response to pheromone and confers a competitive advantage during sexual reproduction. (A) Differential interference contrast and Scd2-GFP time-lapse images of h+ rga3Δ scd2-GFP cells mated to h− WT scd2-mCherry (not depicted). Asterisks indicate rga3Δ scd2-GFP cells. Bar, 3 µm. Time is in minutes from start of imaging. (B) Percentages of cells growing a cell projection that does not result in cell pairing in crosses of h+ WT or rga3Δ-expressing Scd2-GFP with h− WT after 20 h on MSL-N pads. Average and individual data points from two experiments with n > 1,000 cells each are shown. (C) Quantification of the distance between partner cells before shmoo formation from mating mixtures as in A. n > 60 in each of two experiments. (D) Percentage of h− sxa2Δ and h− sxa2Δ rga3Δ cells extending a growth projection 24 h after 0.05, 0.1, or 1 µg/ml P factor addition on MSL-N pads. n = 4 experiments with >1,400 cells each. (E) Quantification of fluorescence of a double-Venus transgene expressed under Pfus1 promoter in cells before (0 h) or after (4 h) nitrogen starvation in absence or presence (× WT) of opposite mating type partners. n = 3 experiments. (F) Percentage of cell pair formation of heterothallic and homothallic WT and rga3Δ cells. n > 1,700 in three experiments. (G) Percentage of zygotes formed by two nonsister cells in homothallic WT and rga3Δ cells. n = 3 experiments. (H) Ratio of rga3-GFP and rga3Δ progenies resulting from competitive mating, where an h− WT strain was mixed 1:1 with two h+ rga3Δ and h+ WT strains present in indicated ratios. n ≥ 3 experiments with n > 170 colonies. Error bars show SD. Student’s t test P values are indicated.
The long shmoos of rga3Δ cells are reminiscent of pheromone hypersignaling mutants (Bendezú and Martin, 2013). To test whether rga3Δ cells hyperactivate the pheromone-signaling pathway, we measured the fluorescence produced by Venus under control of the Pfus1 pheromone-responsive promoter (Petersen et al., 1995). Consistent with previous work, Pfus1-driven Venus expression was induced upon sexual differentiation in nitrogen-starved cells in the presence of mating partners. We also found that it was expressed, though at somewhat lower levels, upon nitrogen starvation in absence of cells of opposite mating type. Expression levels were highly variable across the cell population, likely due to heterogeneity in the timing of sexual differentiation. However, in both cases, Pfus1-Venus levels were indistinguishable in rga3Δ and WT cells, suggesting that the absence of Rga3 does not significantly increase the transcriptional output of the pheromone-signaling cascade, at least in this assay (Fig. 4 E). Similar observations were made using a Pste6-Venus reporter (not depicted).
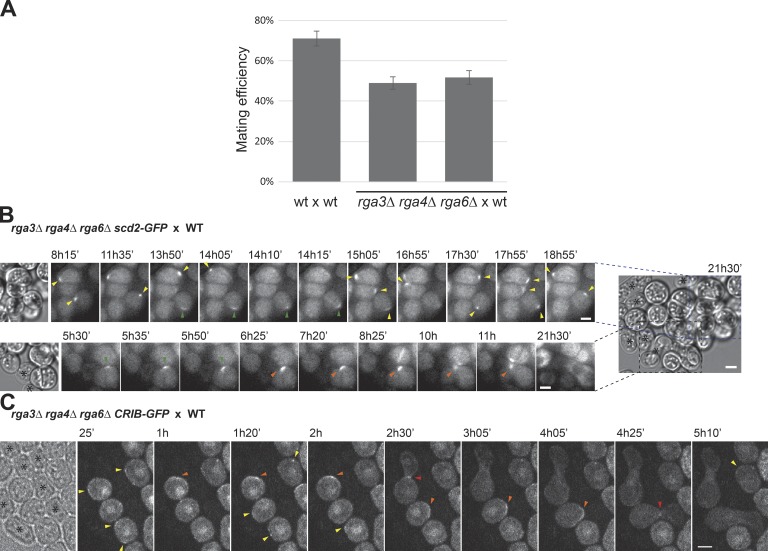
We tested whether the formation of growth projections at lower pheromone concentrations in rga3Δ has physiological consequences on mating outcome. The overall mating efficiency, measured as the percentage of cells engaged with a partner cell, was similar for rga3Δ and WT cells (Fig. 4 F). However, we found that mate preference was significantly altered. Homothallic (self-fertile) WT cells can switch mating type such that two sister cells are potential mating partners (Klar, 2007) but preferentially mate with nonsister cells (Fig. 4 G; Bendezú and Martin, 2013). Conversely, homothallic rga3Δ cells displayed a small preference for sister cells (Fig. 4 G). In competitive mating assays, rga3+ cells also outperformed rga3Δ cells. We mixed different ratios of rga3Δ and rga3-GFP h+ cells labeled with distinct antibiotic markers (hygromycin-B and G418 resistance, respectively), with WT h− cells marked with myo52-tomato (with nourseothricin resistance) and quantified the proportion of hygromycin-B– and G418-resistant spore progeny. rga3Δ cells were significantly depleted from the progeny of 1:1 mixes relative to rga3-GFP (Fig. 4 H). This competitive disadvantage was also observed with 1:2 and 1:3 ratios of parental cells (Fig. 4 H). We conclude that the more sensitive morphological response to pheromone in rga3Δ mutants decreases the ability of cells to efficiently pair with nonsister cells, producing a significant competitive disadvantage during sexual reproduction.
Rga3 localizes to Cdc42 dynamic patches and promotes their exploratory dynamics
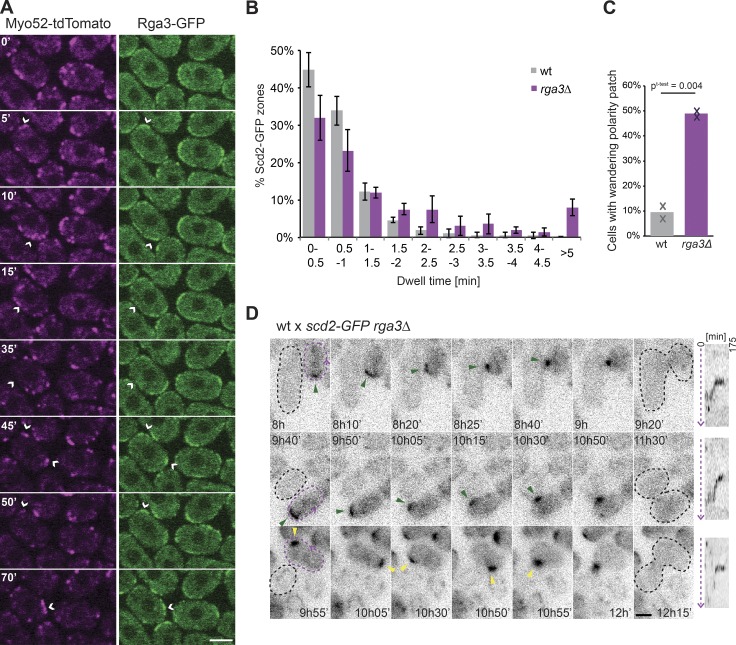
Similar to its localization during vegetative growth, Rga3 localized to sites of polarity during sexual reproduction. We labeled sites of polarity with the type V myosin Myo52. During early stages of the mating process, when Cdc42 undergoes dynamic polarization cycles, Rga3-GFP colocalized with Myo52 (Fig. 5 A), which was previously shown to label sites of Cdc42 activation (Bendezú and Martin, 2013). In shmooing cells, Rga3 localized to the shmoo tip, coincident with Myo52. At the site of cell fusion, Rga3 colocalized with the focus of Myo52 (Fig. S1 C). In these assays, we were unable to detect any specific localization of Rga3ΔC1-GFP.
Figure 5.
Rga3 localizes to Cdc42 dynamic patches and promotes their exploratory dynamics. (A) Medial spinning-disk time-lapse confocal images of Rga3-GFP and Myo52-tdTomato in mating mixes during early stages of mating. Time is in minutes. Arrowheads highlight the coincidence of Rga3-GFP and Myo52-tdTomato. (B) Distribution of Scd2-GFP zone dwell time in h− WT and rga3Δ cells treated with 0.02 µg/ml P factor on MSL-N pads. n = 4 experiments with n = 10 cells each. (C) Frequency of wandering behavior for Scd2-GFP patches in h+ WT and rga3Δ cells mated with h− WT. n ≥ 69 in each of two experiments. Student’s t test P value is indicated. (D) Time-lapse images of Scd2-GFP in h+ rga3Δ cells (dashed purple) mated with h− WT (dashed black) showing two examples of Scd2-GFP patch wandering from the cell tip (top and middle) and one example of Scd2-GFP patch assembly–disassembly behavior before stabilization at the cell side (bottom). Green arrowheads indicate wandering patch movement. Yellow arrowheads highlight assembly–disassembly patch dynamics. To the right of the time lapse are kymographs of the rga3Δ cell cortex along the dashed purple arrow. All micrographs are medial-plane spinning-disk confocal images. Error bars show SD. Time is in minutes from start of imaging. Bars, 3 µm.
To relate the mating behaviors of rga3Δ to the activity state of Cdc42, we used Scd2-GFP as a marker for Cdc42-GTP and measured its dynamic properties upon pheromone exposure. In heterothallic h− sxa2Δ cells treated with 0.02 µM P factor, 90% WT cells displayed an exploratory Scd2 patch with a dwell time of <1.5 min (Bendezú and Martin, 2013; Merlini et al., 2016). Similarly treated cells lacking rga3 showed a more stable Scd2-GFP patch, with only 65% cells displaying a dwell time <1.5 min and nearly 10% cells showing a nondynamic patch, leading to cell shmooing (Fig. 5 B), consistent with the higher pheromone sensitivity shown above (Fig. 4 D). In rga3Δ mating mixtures, consistent with the more persistent growth projections shown above (Fig. 4, B and C), the Scd2-GFP signal was often stable at the cell cortex for extended periods of time, with fewer cells exhibiting dynamic polarization. Notably, although rga3Δ cells like WT could engage partners from cell sides (26 ± 7% and 29 ± 5%, respectively), in 49% of these, the polarity patch first stabilized at the cell tip before exhibiting wandering motion to the cell side, rather than displaying the assembly–disassembly process typical of WT cells (Fig. 5, C and D; and Videos 2 and 3). Similar patch wandering was observed in <10% of scd2-GFP WT cells engaging partners from cell sides. In sum, these data indicate that Rga3 modulates the dynamic behavior of the Cdc42 patch. Because Rga3 is recruited to polarity sites in response to Cdc42 activity, these data further suggest Rga3 may form a negative feedback promoting Cdc42-GTP zone turnover.
Cells lacking all Cdc42 GAPs retain a pheromone-dependent polarization ability
The effect of rga3Δ during mating incited us to investigate more globally whether Cdc42 GAPs contribute to the pheromone-dependent Cdc42 dynamic polarization. Unexpectedly, triple rga3Δrga4Δrga6Δ mutants retained an ability to mate and fuse with WT cells (Fig. 6 A). In addition, both Scd2-mCherry and CRIB-GFP still formed discrete zones that formed and disappeared around the cell periphery. Observation of Scd2 zones over long time periods revealed both instances of patch assembly–disassembly and wandering as described above for rga3Δ (Fig. 6 B and Video 4). CRIB zones were often less well defined than in WT cells, but they also formed smaller zones than in vegetative cells (compare Fig. 6 C with Fig. 2 A). Upon cell pairing, both Scd2 and CRIB became clearly polarized near the mating partner toward which the triple-mutant cells grew (Fig. 6, B and C). We conclude that Cdc42 GAPs modulate the dynamic behavior of Cdc42-GTP patches but are dispensable for their assembly and disassembly. Thus, the formation of polarized zones of Cdc42-GTP relies on partly distinct regulatory mechanisms during the vegetative and sexual life cycles of the fission yeast.
Figure 6.
Cells lacking all three Cdc42 GAPs retain pheromone-dependent polarization ability. (A) Percentage of cell pair formation of heterothallic h− WT and rga3Δrga4Δrga6Δ cells (two independent strains) with h+ WT. n > 1,400 in three experiments. Error bars indicate SD. (B) Time-lapse images of Scd2-mCherry in h− rga3Δrga4Δrga6Δ cells mating with unlabeled h+ WT. Two examples extracted from the same large group of cells illustrate dynamic polarization (top) and shmoo formation (bottom). (C) Medial spinning-disk confocal time-lapse CRIB-GFP images in h+ rga3Δrga4Δrga6Δ cells mating with unlabeled h− WT. In B and C, the first image is a transmitted light image on which WT cells are marked with asterisks. Yellow arrowheads highlight assembly–disassembly patch dynamics. Green arrowheads indicate wandering patch movement. Orange arrowheads show the patch at the site of polarized growth. Red arrowheads point to fusion with partner cells. Time is in minutes from start of imaging. Bars, 3 µm.
Discussion
Spatial regulation of Cdc42 GTPase activity is critical for cell polarization. In this study, we identified and studied the function of Rga3, a novel Cdc42 GAP. Bioinformatics analysis revealed that Rga3 is a paralog of the previously characterized Rga4, with strong sequence homology throughout their sequences except for the unique presence in Rga3 of a C1 domain, a lipid-binding domain that typically binds phorbol esther and diacyl glycerol (Das and Rahman, 2014). The Rga3 domain architecture is found in related GAPs in basidiomycetes as well as in the Taphrinomycotina group, a basal ascomycete lineage that includes the fission yeast clade and Pneumocystis pathogens. The association of C1 and RhoGAP domain is also present in other eukaryotic lineages, including metazoans, in the Chimaerin family of RacGAPs (Yang and Kazanietz, 2007), which contains the GAP domain most closely related to that of Rga3 and Rga4. By contrast, GAP proteins in other ascomycete species lack C1 domains altogether. These observations suggest that the association of C1 and RhoGAP domains represents the ancestral state of this gene family. A possible evolutionary scenario is that during early ascomycete evolution, an ancestral C1-containing GAP gene underwent duplication with one gene copy subsequently losing its C1 domain. The fission yeast clade retained both gene copies, yielding Rga3 and Rga4, whereas one copy was lost in all other ascomycetes.
Duplication of the ancestral gene must have been accompanied by evolution of alternate membrane localization determinants in the two resulting genes. Indeed, Rga3 and Rga4 decorate distinct, complementary cortical regions. In Rga3, the C1 domain is necessary and sufficient for plasma membrane localization, consistent with its predicted lipid-binding activity, but it does not confer on its own specificity for cell poles. Localization to sites of polarity also requires the LIM domains and coiled-coil regions of Rga3, likely through protein–protein interaction, but interestingly not the GAP domain. Of note, none of the Rga3 fragments tested displayed Rga4-like cell side localization, suggesting that localization to sites of Cdc42 activity may represent the ancestral state. Consistent with this idea, the related S. cerevisiae GAPs also localize to sites of polarity (Caviston et al., 2003). Whether the extant cell side localization of Rga4 masks an ancestral polar localization remains to be tested. Interestingly, the observation that Rga3 is recruited by GTP-locked Cdc42, yet its localization at sites of polarity is independent from its GAP domain suggests this recruitment does not simply occur through direct binding of the GAP domain to Cdc42-GTP but indirectly, for instance through protein–protein interaction with a Cdc42-GTP effector. In this way, Rga3 may form a negative feedback on Cdc42 activity.
Rga3 promotes Cdc42-GTP zone instability during mating
The maintenance of duplicated gene copies in the fission yeast genome and the unique localization of Rga3 at the polarity patch suggest Rga3 (or Rga4) acquired a specific function distinct from that of the two other GAPs. During mitotic growth, however, though Rga3 participates in Cdc42 inactivation with Rga4 and Rga6, it does not appear to play any unique function. Analysis of Cdc42-GTP distribution and oscillatory patterns did not show crucial changes in rga3Δ mutants, with only a reduction in the oscillation frequency observed, difficult to reconcile with any direct role for a negative regulator. By contrast, during mating, the absence of Rga3 led to noticeable changes in Cdc42 patch dynamics: the Cdc42 patch exhibited a longer dwell time, leading to exacerbated outgrowth, and showed reduction in assembly–disassembly behavior. One question is whether Rga3 directly promotes patch destabilization or acts more indirectly by lowering downstream signaling. Previous work indeed showed that patch stabilization occurs upon increased signaling in strains lacking negative pheromone-GPCR control (Bendezú and Martin, 2013), which signals through a MAPK cascade proposed to be partly under Cdc42 regulation (Marcus et al., 1995; Tu et al., 1997; Weston et al., 2013). However, we could not detect a significant increase in transcriptional reporters of MAPK activity in rga3Δ cells. This suggests that MAPK output is not significantly altered in the absence of Rga3, though we note that small changes are unlikely to be detected with this assay due to high cell-to-cell variability. Thus, we favor the view that by being recruited to the polarity patch, Rga3 directly promotes patch destabilization by reducing Cdc42 activity.
Remarkably, the cell is largely able to compensate the reduction in assembly–disassembly behavior upon loss of Rga3 through an alternate patch-wandering strategy. Indeed, although rga3Δ cells exhibit a measurable disadvantage when placed in competitive mating situations, they remain able to extend a growth projection from their side and display unaltered mating efficiency when unchallenged. Patch wandering is normally rarely observed in S. pombe, but it is the principal mode of gradient tracking in S. cerevisiae. In this case, movement of the patch up-gradient is proposed to rely on dilution effects from vesicle delivery and on feedback regulation through local signal of the pheromone GPCR-MAPK (Dyer et al., 2012; Hegemann et al., 2015; McClure et al., 2015). The patch-wandering behavior observed in rga3Δ cells may rely on similar mechanisms, which may exist also in WT fission yeast cells to refine the position of the patch. It is interesting that the presence or absence of a single Cdc42 GAP partly converts patch dynamic behavior from assembly–disassembly to wandering, suggesting that cells may naturally also be able to switch between these two modes of cell pairing.
Different GAP requirement for Cdc42-GTP polarization in distinct life stages
One surprising observation is the very distinct cell polarization outcome in the absence of the three Cdc42 GAPs Rga3, Rga4, and Rga6 during the mitotic and sexual life cycles. Mitotic cells lacking the three GAPs formed very large Cdc42-GTP domains that could extend over more than half of the cell surface, and therefore, they acquired a round cell shape. This phenotype, similar to expression of GTP-locked Cdc42, is consistent with the idea that spatial restriction of Cdc42 activity is a dynamic process that relies not only on local activation at cell tips but also on GTP hydrolysis to counteract Cdc42 diffusion. An essentially identical outcome is observed for Ras1 GTPase normally selectively active at cell poles: upon deletion of the only Ras1 GAP, the GTPase is now active around the entire cell cortex (Merlini et al., 2018). By contrast, cells lacking all three Cdc42 GAPs were still almost fully mating competent and retained the ability to polarize active Cdc42 as labeled by the CRIB reporter or the Cdc42-GTP–binding Scd2 scaffold. The ability of the triple-GAP mutant to exhibit assembly–disassembly Cdc42 dynamics is remarkable given the very poor ability of the rga3Δ single mutant to do so outside cell poles. One possibility is that the shape of the cell constrains the sites of polarity. Indeed, rod-shaped WT cells show preference for pheromone-dependent polarization at cell tips (Bendezú and Martin, 2013). Rga3-dependent patch destabilization may thus serve to overcome this preference and promote polarization along cell sides. Spherical cells such as the triple-GAP mutant may lack cortical region preference, perhaps due to unrestricted distribution of other cortical landmarks, and thus show a more random choice of polarity site. In any case, we conclude that the three Cdc42 GAPs studied here, though modulating patch dynamic properties, are not essential for Cdc42-GTP polarization during mating and that the polarization mechanisms of Cdc42-GTP can be distinct in different life stages of the same cell. This finding represents an interesting paradigm of the emerging theme that polarity mechanisms show important context-dependent variation (St Johnston, 2018).
A key question is what accounts for this difference. During mitotic growth, cells polarize in response to cell-intrinsic landmarks deposited at cell poles by microtubules (Martin, 2009). As microtubules orient along cell length by sliding along cell sides (Martin and Arkowitz, 2014), the loss of cell shape in triple-GAP mutants likely does not permit landmark positioning, in turn compounding the effect of the GAP deletions. By contrast, during mating, cells polarize in response to extrinsic pheromone gradients (Merlini et al., 2013). Thus, one possibility is that the positional cue provided by external pheromone gradients is sufficient for polarization even from a round cell shape. However, this is probably a too simplistic view as GAP-deleted cells without close WT partners, unlikely to experience any sharp pheromone gradients, are also seen to efficiently polarize often in a direction unrelated to that of candidate partner cells (see Fig. 6 B). It is possible that there exists yet another Cdc42 GAP specific for the sexual life cycle. However, none of the other encoded GAP genes appear to increase levels upon pheromone exposure (Mata and Bähler, 2006), though this does not exclude other posttranscriptional regulations. Alternatively, Cdc42-GTP polarization may depend on other modes of negative regulation during mating. Possibilities include GDI-mediated membrane extraction, which plays little role during mitotic growth (Nakano et al., 2003; Bendezú et al., 2015), endocytosis-dependent Cdc42-GTP retrieval from the membrane (Slaughter et al., 2009; Orlando et al., 2011; Watson et al., 2014), GEF inhibition (Kuo et al., 2014), or a GAP-independent boost of the intrinsic Cdc42 GTPase activity promoting the return of Cdc42 to the basal state.
Materials and methods
Strain construction, media, and growth conditions
Strains used in this study are listed in Table S1. All plasmids used are listed in Table S2. Standard genetic manipulation methods for S. pombe transformation and tetrad dissection were used. For microscopy of fission yeast cells during exponential growth, cells were grown in Edinburgh minimal medium (EMM) supplemented with amino acids as required. For biochemistry experiments, cells were grown in rich yeast extract (YE) medium. For assessing cells during the mating process, liquid or agar minimal sporulation medium without nitrogen (MSL-N) was used (Egel et al., 1994; Vjestica et al., 2016). All live-cell imaging during sexual lifecycle was performed on MSL-N agarose pads (Vjestica et al., 2016). Mating assays were performed as by Vjestica et al. (2016).
All cells were grown at 25°C unless differently specified. For cell length measurements, all strains had identical auxotrophies and were grown in EMM-ALU (adenine, leucine, and uracil) to mid–log phase before calcofluor (Sigma-Aldrich) addition to a final concentration of 5 µg/ml from a 200× stock solution. For plasmid expression, all strains were grown in EMM-AU for 20 h before imaging unless differently specified.
Construction of YSM3149 in Fig. 3 F was obtained by integration of the C-terminal part of the gene lacking the C1 domain at its genomic locus. First, an expression plasmid was generated: Rga3 GAP domain was amplified from pSM1337 (see Table S2) with primers osm4400 (5′-CGggatccGGGTCGGTTTTGCCTCAAGTAATTG-3′) and osm4401 (5′-CCCcccgggGCCAGATTATGAAGAATCTTATCCA-3′) digested with XmaI and BamHI (lowercase sites in the sequences above) and ligated to similarly treated pSM1339 (see Table S2) to generate plasmid pREP41-rga3(-C1)-EGFPc. Second, 819 bp from the C-terminal part of this plasmid was amplified with osm4770 (5′-CGACGGATCCCCGGGTTAATTAAATTCAGCATAGAGTACC-3′)-osm4769 (5′-TTCTTCTCCTTTACTGTTAATTAACAGATTATGAAGAATC-3′) and cloned with the In-Fusion cloning kit (Takara Bio Inc.) into pSM206 (pFA6a-GFP(S65T)-kanMX6) previously digested with PacI, resulting in pSM2197. Finally, standard S. pombe transformation was performed on YSM3130 by amplification of pSM2197 fragment with primers osm774 (5′-GATTTACAGCATGCTACCAAAAAGAGTACGGCTCTTCAGTTTATGCTTGATAACGTGGATAAGATTCTTCATAATCTGCGGATCCCCGGGTTAATTAA-3′)-osm773 (5′-TTATTATTATCGTAAAGTTCCTTCTTCCCTAGAAAAATACAAGTTCCCCGATATTTTAATCATTATTCATAAAACAAGGAATTCGAGCTCGTTTAAAC-3′).
Constructions of YSM3251 and YSM3252 in Fig. 4 (B and C) were obtained by transformation of strain YSM3152 with a PCR product obtained from the amplification of pSM646 (pFA6a-NatMX6) with primers osm5235 (5′-CCAACGAGGATGCCTCCTCCTGGGCCTTCACCAACGATGTTTGGACGATCGTTAGAAAATCAATTGAAAATTGAAGGGCGGATCCCCGGGTTAATTAA-3′) and osm773 (5′-TTATTATTATCGTAAAGTTCCTTCTTCCCTAGAAAAATACAAGTTCCCCGATATTTTAATCATTATTCATAAAACAAGGAATTCGAGCTCGTTTAAAC-3′). YSM3156 and YSM3157 were obtained by cross with a strain previously transformed with pSM2187 (see Table S2).
The C1 × 3 GBlock sequence is as follows: 5′-TACCAGATCTCTCGAGTGTTTCAcGTTAATGCaATATTCAAGCCCTCAAGGTGTTATATtTGCTCGGAGAGCGTATGGGGATCTGAACTCCGCTGCTTcCATTGCTCAATcTCATGCCATTCGCGGTGTTTAAAAAGGCTGTTTGCcGAGTCGGTtTTcCATGTcAATGCCATcTTCAAaCCtTCccgcTGcTATATATGCTCtGAaAGtGTtTGGGGtTCtGAgttaCGtTGtTTcCAcTGtTCtATTTCgTGtCATTCcaGaTGccttAAgcGtttgTTcGCtGAGTCcGTcTTTCAcGTtAAcGCtATtTTCAAGCCCTCtcGaTGTTAcATtTGtTCcGAatcgGTATGGGGcTCtGAACTtaggTGCTTcCATTGtTCcATTTCtTGCCAcTCtCGcTGcTTgAAgAGattaTTTGCcGAGTCtgGATCCCCGGGTATGA-3′ was used. Manual codon degeneration (lowercase letters) was required to avoid sequence repetitions. Restriction enzyme sites (XhoI and BamHI) are underlined.
Mating assays
Mating assays were performed as by Vjestica et al. (2016). Briefly, precultures of cells were grown over night in MSL-N at 25°C to reach an OD600 of 0.5–1. Cultures were then diluted to an OD600 of 0.025 in MSL-N and grown for 16–20 h to an OD600 of 0.5–1 at 30°C. Cells were washed three times with MSL-N, diluted to an OD600 of 1.5 in 1–3 ml MSL-N, and incubated at 30°C for 1–4 h (depending on the mating stage to be visualized). Cells were mounted onto MSL-N agarose pads (2% agarose) before imaging in overnight videos at room temperature (∼22°C) or incubated at 18°C overnight before imaging. Mating efficiency was measured as by Dudin et al. (2015, 2016). Sister cell engagement and zone dwell time were measured as by Bendezú and Martin (2013).
For pheromone treatments, P factor pheromone was purchased from Pepnome and used from a stock solution of 1 mg/ml in methanol. Indicated concentrations of pheromones were directly added to the melted MSL-N agarose before mounting cells on the pads. Cells were then imaged overnight or incubated at 18°C overnight before imaging. For Fig. 4 D, the pheromone treatment was performed on pads and left at 25°C for 24 h.
Competition assay
Mating assays were performed by mixing YSM1232 (h+), YSM3160 (h+), and YSM952 (h−; see Table S1) at different ratios. Cell mixes were plated on MSL-N plates for 24 h at 25°C. The cells were then transferred into an Eppendorf tube and treated with glusulase (1:200 from stock solution; PerkinElmer) in H2O overnight at 25°C. Only spores survived the treatment as confirmed by microscopic inspection. Spores were then plated on YE and grown for 3 d at 30°C. The plates were then replicated on selective media in order to determine the genotype of each colony.
Microscopy
Microscopy was performed on live cells either on a spinning-disk confocal microscope or a DeltaVision epifluorescence system (Applied Precision Ltd.). Spinning-disk microscopy was performed using a Leica DMI4000B inverted microscope equipped with HCX Plan Apochromat 100×/1.46 NA oil objective and a PerkinElmer UltraView confocal system (including a Yokogawa CSU22 real-time confocal scanning head, solid-state laser, and a cooled 14-bit frame-transfer electron-multiplying charge-coupled device C9100-50 camera).
For cell length measurements, images of calcofluor-stained cells were taken on a DeltaVision platform composed of a customized Olympus IX-71 inverted microscope and a UPlan Apochromat 100× 1.4 NA oil objective, a CoolSNAP HQ2 camera (Photometrics), and an Insight SSI 7 color combined-unit illuminator. All microscopy was performed at room temperature.
Image analysis
For cell length and width measurements on calcofluor-stained cells, a line was drawn manually across the length and width of septated cells from the middle of one tip to the other, and the length measured using the ImageJ Measure tool (National Institutes of Health).
Fluorescence image quantification
Quantification of Cdc42-mCherrySW and CRIB-GFP distribution in Fig. 2 A was done on the sum projection of five consecutive images. The intensity profile along a 3-pixel-wide segmented line along half of the cell cortex was measured for at least 20 tips for both red and green channels. Profiles were centered on the maximum pixel value for the CRIB channel and subsequently split into two half tips.
Quantification of Cdc42-mCherrySW and Rga3-GFP distribution in Fig. 3 B was done on the sum projection of five consecutive images. The intensity of a 3-pixel-wide segment was collected from 24 half-cell tips for both red and green channels. The values were normalized to the maximum and minimum pixel values.
Quantification of Cdc42-mCherrySW (WT and Q61L expressed from plasmid) and Rga3-GFP distribution in Fig. 3 C was done on the sum projection of 10 images obtained collected every 30 s for 5 min. The intensity of a 3-pixel-wide segment was collected from 18 half-cell tips for both red and green channels. Only rod-shaped cells were taken into analysis.
Quantification of Rga3-GFP (WT and ΔC1) distribution in Fig. 3 F was done on the sum projection of five consecutive images. The intensity of a 3-pixel-wide segment was collected from at least 20 tips for the green channel. Total intensity quantification was performed by drawing a line along the cell cortex with ImageJ polygon tool and subtracting the background.
For Fig. 4 E, WT or rga3Δ h+ cells expressing a Pfus1-double-Venus reporter were treated as for mating assays. After shift on MSL-N, cells were mixed with WT cells expressing Scd2-mCherry that were either h+ (for analysis in the absence of mating partners) or h− (for analysis in presence of mating partners). Scd2-mCherry cells were used to measure background fluorescence in the Venus channel. Samples were imaged just after mixing (time 0) or after 4 h. Cell fluorescence was measured in ImageJ by placing a square in the middle of each cell and calculated as (Pfus1-Venus signal – local background)t = x – average of (Venus signal in Scd2-mCherry cells – local background)t = x. The average of at least 20 cells per each time point was calculated. Kymographs in Fig. 5 D were constructed in ImageJ version 1.51 by drawing a 3-pixel-wide line around the cell cortex and using the ImageJ Reslice tool.
CRIB oscillations
The analysis on CRIB-GFP oscillatory behavior was done as by Das et al. (2012). Briefly, time-lapse videos of WT and rga3Δ cells expressing CRIB-GFP were obtained. Cells were imaged on EMM-ALU 2% agarose pads every 1 min for 30 min by spinning-disk confocal microscopy. 20 cells were used for the analysis. Only bipolar cells were taken into consideration. CRIB-GFP intensity was obtained by drawing a 3-pixel-wide segment at each cell tip. The traces were normalized to a linear regression of another cell intensity over time to compensate for fluorescence loss due to imaging, normalized to the maximum value, and plotted against time. The frequency was calculated by identifying individual peaks on the plot and calculating the average time interval between them. Cross-correlation was calculated using the Excel Correl function.
Biochemistry methods
Recombinant protein production
Expression of GST proteins or GST was induced in BL21 bacteria from pGEX-4T-1–derived plasmids. In brief, cells were grown overnight in Luria–Bertani (LB) medium supplemented with 100 µg/ml ampicillin at 37°C. 250 ml LB-ampicillin was inoculated with 6.25 ml saturated culture and grown for 3 h at 37°C. Protein expression was induced by the addition of 100 µM IPTG for 5 h at 18°C. Bacterial pellets were resuspended in 5 ml PBS (137 mM NaCl, 2.7 mM KCl, 1.4 mM KH2PO4, and 4.3 mM NaH2PO4, pH 7.4), digested with 1 mg/ml lysozyme, treated with 1 µg/ml DNase I, sonicated 6× for 30 s (40% amplitude), incubated with 1% Triton X-100 in PBS buffer at 4°C for 30 min, and centrifuged for 15 min at 4°C at 10,000 g. Soluble extract was incubated with 200 µl glutathione–Sepharose beads at 50% slurry for 2 h at 4°C. Finally, beads were washed 3× with cold PBS and eluted in four steps in 100 µl elution buffer (15 mM reduced glutathione and 50 mM Tris-HCl, pH 8). Protein concentration was determined from SDS-PAGE gels by using BSA as a standard protein at different concentrations (4, 2, 0.8, 0.4, and 0.2 µg/µl).
Cdc42-GTP pulldown
The Cdc42-GTP pulldown assay was performed as by Tatebe et al. (2008). Cdc42-mCherrySW strains grown overnight at 30°C in 150 ml YE media were used to produce protein extracts as in Revilla-Guarinos et al. (2016). Briefly, cell extracts from WT, rga3Δ, rga4Δ, rga6Δ, and rga3Δrga4Δrga6Δ cells expressing Cdc42-mCherrySW were obtained by mechanical breakage of the cells using glass beads and FastPrep. Cells were resuspended in 500 µl lysis buffer (50 mM Tris-HCl, pH 7.5, 20 mM NaCl, 0.5% NP-40, 10% glycerol, 0.1 mM DTT, and 2 mM MgCl2). Protein extracts were then incubated with bacterially expressed GST-CRIB (see the Recombinant protein production section) and Sepharose slurry for 1 h at 4°C in binding buffer (25 mM Tris-HCl, pH 7.5, 1 mM DTT, 30 mM MgCl2, 40 mM NaCl, and 0.5% NP-40). The bead pellet was then washed three times with 25 mM Tris-HCl, pH 7.5, 1 mM DTT, 30 mM MgCl2, 40 mM NaCl, and 1% NP-40 and two times with the same buffer without NP-40.
Samples were loaded on 12% protein gel and resolved by SDS-PAGE for Western blot analysis. Standard protocols were used for SDS-PAGE and Western blot analysis. Antibodies used on Western blots were mouse monoclonal (6G6) to RFP (ChromoTek).
GAP assay
1 µg purified Cdc42 (see the Recombinant protein production section) was incubated in 70 µl loading buffer (20 mM Tris-HCl, pH 7.5, 25 mM NaCl, 5 mM MgCl2, and 0.1 mM DTT) containing 1.5 µl [γ-32P]GTP for 10 min at 30°C. The mixture was then put on ice, and 10 µl was added to 50 µl reaction buffer (20 mM Tris, pH 7.5, 2 mM GTP, and 0.6 µg/µl BSA) with 5 µg GAPs (see the Recombinant protein production section) or 50 mM Tris, pH 8. The tube was transferred at 30°C, and every 2 min, 10 µl reaction was diluted in 990 µl cold washing buffer (20 mM Tris-HCl, pH 7.5, 50 mM NaCl, and 5 mM MgCl2). The samples were filtered through prewetted nitrocellulose (EMD Millipore) filters, washed three times with 4 ml cold washing buffer, and air dried. The amount of filter-bound radioactive nucleotide was determined by scintillation counting.
Sequence analysis
All RhoGAP-containing proteins on the Pfam site (Pfam 31.0 on 12th June 2017; http://pfam.xfam.org/) from the following species were selected: Ascomycetes: Aspergillus nidulans, Penicillium rubens, Aspergillus oryzae, Coccidioides posadasii, Pyrenophora tritici-repentis, Neurospora crassa, Taphrina deformans, S. pombe, Schizosaccharomyces japonicus, Schizosaccharomyces cryophilus, Pneumocystis murina, Pneumocystis jirovecii, Pichia pastoris, Kluyveromyces lactis, S. cerevisiae, Aphis gossypii, Zygosaccharomyces rouxii, and Yarrowia lipolytica; and Basidiomycetes: Ustilago maydis, Sphacelotheca reilianum, Rhodosporidium toruloides, Melampsora larici-populina, Tremella mesenterica, Cryptococcus neoformans, and sequences of the RhoGAP domain aligned using the ClustalW method in MegAlign (Lasergene). This identified Rga3 as an Rga4 paralog. An alignment using whole-protein sequences from entries on the same phylogenetic branch as Rga3 and Rga4 was subsequently performed and is shown in Fig. 1 C. Domain architecture was derived from simple modular architecture research tool (SMART) analysis (http://smart.embl-heidelberg.de/).
Statistics
All comparison were performed using Student’s t test with two-tailed distribution assuming normal distribution. Figures were prepared with ImageJ 64 and assembled using Adobe Illustrator CS5. All error bars are SD. All experiments were done at least three independent times except those described in Figs. 4 (B and C) and 5 C.
Online supplemental material
Fig. S1 shows localization of Rga3 to sites of polarity independent of the cytoskeleton. Fig. S2 shows minor changes in Cdc42-GTP oscillations during mitotic growth in rga3Δ cells. Table S1 shows strains used in this study. Table S2 shows plasmids used in this study. Video 1 shows exacerbated growth phenotypes of rga3Δ cells during mating. Video 2 shows examples of rga3Δ cells with patch wandering behavior. Video 3 shows an example of an rga3Δ cell with patch assembly–disassembly behavior. Video 4 shows how cells lacking Rga3, Rga4, and Rga6 retain polarization ability during mating.
Supplementary Material
Acknowledgments
We thank Laura Merlini (University of Lausanne, Lausanne, Switzerland) for the Pfus1-Venus and Pste6-Venus constructs and members of the laboratory for critical comments on the manuscript.
This work was funded by a Faculty of Biology and Medicine fellowship (University of Lausanne) to D. Gallo Castro as well as a Swiss National Science Foundation research grant (310030B_176396) and a European Research Council Consolidator grant (CellFusion) to S.G. Martin.
The authors declare no competing financial interests.
Author contributions: D. Gallo Castro: conceptualization, investigation, methodology, visualization, writing – review and editing, and funding acquisition; S.G. Martin: conceptualization, investigation, methodology, visualization, writing – original draft and editing, funding acquisition, and supervision.
References
- Adams A.E., Johnson D.I., Longnecker R.M., Sloat B.F., and Pringle J.R.. 1990. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J. Cell Biol. 111:131–142. 10.1083/jcb.111.1.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwood S.X., Chabu C., Penkert R.R., Doe C.Q., and Prehoda K.E.. 2007. Cdc42 acts downstream of Bazooka to regulate neuroblast polarity through Par-6 aPKC. J. Cell Sci. 120:3200–3206. 10.1242/jcs.014902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú F.O., and Martin S.G.. 2013. Cdc42 explores the cell periphery for mate selection in fission yeast. Curr. Biol. 23:42–47. 10.1016/j.cub.2012.10.042 [DOI] [PubMed] [Google Scholar]
- Bendezú F.O., Vincenzetti V., Vavylonis D., Wyss R., Vogel H., and Martin S.G.. 2015. Spontaneous Cdc42 polarization independent of GDI-mediated extraction and actin-based trafficking. PLoS Biol. 13:e1002097 10.1371/journal.pbio.1002097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonazzi D., Julien J.D., Romao M., Seddiki R., Piel M., Boudaoud A., and Minc N.. 2014. Symmetry breaking in spore germination relies on an interplay between polar cap stability and spore wall mechanics. Dev. Cell. 28:534–546. 10.1016/j.devcel.2014.01.023 [DOI] [PubMed] [Google Scholar]
- Canagarajah B., Leskow F.C., Ho J.Y., Mischak H., Saidi L.F., Kazanietz M.G., and Hurley J.H.. 2004. Structural mechanism for lipid activation of the Rac-specific GAP, beta2-chimaerin. Cell. 119:407–418. 10.1016/j.cell.2004.10.012 [DOI] [PubMed] [Google Scholar]
- Caviston J.P., Longtine M., Pringle J.R., and Bi E.. 2003. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell. 14:4051–4066. 10.1091/mbc.e03-04-0247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll P.M., Trillo Y., Ametzazurra A., and Perez P.. 2003. Gef1p, a new guanine nucleotide exchange factor for Cdc42p, regulates polarity in Schizosaccharomyces pombe. Mol. Biol. Cell. 14:313–323. 10.1091/mbc.e02-07-0400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J., and Rahman G.M.. 2014. C1 domains: structure and ligand-binding properties. Chem. Rev. 114:12108–12131. 10.1021/cr300481j [DOI] [PubMed] [Google Scholar]
- Das M., Wiley D.J., Medina S., Vincent H.A., Larrea M., Oriolo A., and Verde F.. 2007. Regulation of cell diameter, For3p localization, and cell symmetry by fission yeast Rho-GAP Rga4p. Mol. Biol. Cell. 18:2090–2101. 10.1091/mbc.e06-09-0883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M., Drake T., Wiley D.J., Buchwald P., Vavylonis D., and Verde F.. 2012. Oscillatory Dynamics of Cdc42 GTPase in the Control of Polarized Growth. Science. 337:239–243. 10.1126/science.1218377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerMardirossian C., and Bokoch G.M.. 2005. GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol. 15:356–363. 10.1016/j.tcb.2005.05.001 [DOI] [PubMed] [Google Scholar]
- Dudin O., Bendezú F.O., Groux R., Laroche T., Seitz A., and Martin S.G.. 2015. A formin-nucleated actin aster concentrates cell wall hydrolases for cell fusion in fission yeast. J. Cell Biol. 208:897–911. 10.1083/jcb.201411124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudin O., Merlini L., and Martin S.G.. 2016. Spatial focalization of pheromone/MAPK signaling triggers commitment to cell-cell fusion. Genes Dev. 30:2226–2239. 10.1101/gad.286922.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer J.M., Savage N.S., Jin M., Zyla T.R., Elston T.C., and Lew D.J.. 2012. Tracking Shallow Chemical Gradients by Actin-Driven Wandering of the Polarization Site. Curr. Biol. PubMed 10.1016/j.cub.2012.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egel R., Willer M., Kjaerulff S., Davey J., and Nielsen O.. 1994. Assessment of pheromone production and response in fission yeast by a halo test of induced sporulation. Yeast. 10:1347–1354. 10.1002/yea.320101012 [DOI] [PubMed] [Google Scholar]
- Endo M., Shirouzu M., and Yokoyama S.. 2003. The Cdc42 binding and scaffolding activities of the fission yeast adaptor protein Scd2. J. Biol. Chem. 278:843–852. 10.1074/jbc.M209714200 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S. 2004. Cdc42--the centre of polarity. J. Cell Sci. 117:1291–1300. 10.1242/jcs.01115 [DOI] [PubMed] [Google Scholar]
- Gotta M., Abraham M.C., and Ahringer J.. 2001. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Curr. Biol. 11:482–488. 10.1016/S0960-9822(01)00142-7 [DOI] [PubMed] [Google Scholar]
- Gulli M.P., Jaquenoud M., Shimada Y., Niederhäuser G., Wiget P., and Peter M.. 2000. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell. 6:1155–1167. 10.1016/S1097-2765(00)00113-1 [DOI] [PubMed] [Google Scholar]
- Hegemann B., Unger M., Lee S.S., Stoffel-Studer I., van den Heuvel J., Pelet S., Koeppl H., and Peter M.. 2015. A Cellular System for Spatial Signal Decoding in Chemical Gradients. Dev. Cell. 35:458–470. 10.1016/j.devcel.2015.10.013 [DOI] [PubMed] [Google Scholar]
- Hirota K., Tanaka K., Ohta K., and Yamamoto M.. 2003. Gef1p and Scd1p, the Two GDP-GTP exchange factors for Cdc42p, form a ring structure that shrinks during cytokinesis in Schizosaccharomyces pombe. Mol. Biol. Cell. 14:3617–3627. 10.1091/mbc.e02-10-0665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell A.S., Jin M., Wu C.F., Zyla T.R., Elston T.C., and Lew D.J.. 2012. Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell. 149:322–333. 10.1016/j.cell.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irazoqui J.E., Gladfelter A.S., and Lew D.J.. 2003. Scaffold-mediated symmetry breaking by Cdc42p. Nat. Cell Biol. 5:1062–1070. 10.1038/ncb1068 [DOI] [PubMed] [Google Scholar]
- Kaibuchi K., Fukumoto Y., Oku N., Takai Y., Arai K., and Muramatsu M.. 1989. Molecular genetic analysis of the regulatory and catalytic domains of protein kinase C. J. Biol. Chem. 264:13489–13496. [PubMed] [Google Scholar]
- Kay A.J., and Hunter C.P.. 2001. CDC-42 regulates PAR protein localization and function to control cellular and embryonic polarity in C. elegans. Curr. Biol. 11:474–481. 10.1016/S0960-9822(01)00141-5 [DOI] [PubMed] [Google Scholar]
- Kelly F.D., and Nurse P.. 2011. Spatial control of Cdc42 activation determines cell width in fission yeast. Mol. Biol. Cell. 22:3801–3811. 10.1091/mbc.e11-01-0057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalili B., Merlini L., Vincenzetti V., Martin S.G., and Vavylonis D.. 2018. Exploration and stabilization of Ras1 mating zone: A mechanism with positive and negative feedbacks. PLOS Comput. Biol. 14:e1006317 10.1371/journal.pcbi.1006317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klar A.J. 2007. Lessons learned from studies of fission yeast mating-type switching and silencing. Annu. Rev. Genet. 41:213–236. 10.1146/annurev.genet.39.073103.094316 [DOI] [PubMed] [Google Scholar]
- Kokkoris K., Castro D.G., and Martin S.G.. 2014. Tea4-phosphatase I landmark promotes local growth by dual Cdc42 GEF recruitment and GAP exclusion. J. Cell Sci. 127:2005–2016. 10.1242/jcs.142174 [DOI] [PubMed] [Google Scholar]
- Kozubowski L., Saito K., Johnson J.M., Howell A.S., Zyla T.R., and Lew D.J.. 2008. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr. Biol. 18:1719–1726. 10.1016/j.cub.2008.09.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo C.C., Savage N.S., Chen H., Wu C.F., Zyla T.R., and Lew D.J.. 2014. Inhibitory GEF phosphorylation provides negative feedback in the yeast polarity circuit. Curr. Biol. 24:753–759. 10.1016/j.cub.2014.02.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus S., Polverino A., Chang E., Robbins D., Cobb M.H., and Wigler M.H.. 1995. Shk1, a homolog of the Saccharomyces cerevisiae Ste20 and mammalian p65PAK protein kinases, is a component of a Ras/Cdc42 signaling module in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA. 92:6180–6184. 10.1073/pnas.92.13.6180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.G. 2009. Microtubule-dependent cell morphogenesis in the fission yeast. Trends Cell Biol. 19:447–454. 10.1016/j.tcb.2009.06.003 [DOI] [PubMed] [Google Scholar]
- Martin S.G., and Arkowitz R.A.. 2014. Cell polarization in budding and fission yeasts. FEMS Microbiol. Rev. 38:228–253. 10.1111/1574-6976.12055 [DOI] [PubMed] [Google Scholar]
- Mata J., and Bähler J.. 2006. Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc. Natl. Acad. Sci. USA. 103:15517–15522. 10.1073/pnas.0603403103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure A.W., Minakova M., Dyer J.M., Zyla T.R., Elston T.C., and Lew D.J.. 2015. Role of Polarized G Protein Signaling in Tracking Pheromone Gradients. Dev. Cell. 35:471–482. 10.1016/j.devcel.2015.10.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L., Dudin O., and Martin S.G.. 2013. Mate and fuse: how yeast cells do it. Open Biol. 3:130008 10.1098/rsob.130008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L., Khalili B., Bendezú F.O., Hurwitz D., Vincenzetti V., Vavylonis D., and Martin S.G.. 2016. Local Pheromone Release from Dynamic Polarity Sites Underlies Cell-Cell Pairing during Yeast Mating. Curr. Biol. 26:1117–1125. 10.1016/j.cub.2016.02.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L., Khalili B., Dudin O., Michon L., Vincenzetti V., and Martin S.G.. 2018. Inhibition of Ras activity coordinates cell fusion with cell-cell contact during yeast mating. J. Cell Biol. 217:1467–1483. 10.1083/jcb.201708195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P.J., and Johnson D.I.. 1994. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol. Cell. Biol. 14:1075–1083. 10.1128/MCB.14.2.1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munemitsu S., Innis M.A., Clark R., McCormick F., Ullrich A., and Polakis P.. 1990. Molecular cloning and expression of a G25K cDNA, the human homolog of the yeast cell cycle gene CDC42. Mol. Cell. Biol. 10:5977–5982. 10.1128/MCB.10.11.5977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., Mutoh T., Arai R., and Mabuchi I.. 2003. The small GTPase Rho4 is involved in controlling cell morphology and septation in fission yeast. Genes Cells. 8:357–370. 10.1046/j.1365-2443.2003.00639.x [DOI] [PubMed] [Google Scholar]
- Ono Y., Fujii T., Igarashi K., Kuno T., Tanaka C., Kikkawa U., and Nishizuka Y.. 1989. Phorbol ester binding to protein kinase C requires a cysteine-rich zinc-finger-like sequence. Proc. Natl. Acad. Sci. USA. 86:4868–4871. 10.1073/pnas.86.13.4868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando K., Sun X., Zhang J., Lu T., Yokomizo L., Wang P., and Guo W.. 2011. Exo-endocytic trafficking and the septin-based diffusion barrier are required for the maintenance of Cdc42p polarization during budding yeast asymmetric growth. Mol. Biol. Cell. 22:624–633. 10.1091/mbc.e10-06-0484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez P., and Rincón S.A.. 2010. Rho GTPases: regulation of cell polarity and growth in yeasts. Biochem. J. 426:243–253. 10.1042/BJ20091823 [DOI] [PubMed] [Google Scholar]
- Petersen J., Weilguny D., Egel R., and Nielsen O.. 1995. Characterization of fus1 of Schizosaccharomyces pombe: a developmentally controlled function needed for conjugation. Mol. Cell. Biol. 15:3697–3707. 10.1128/MCB.15.7.3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapali P., Mitteau R., Braun C., Massoni-Laporte A., Ünlü C., Bataille L., Arramon F.S., Gygi S.P., and McCusker D.. 2017. Scaffold-mediated gating of Cdc42 signalling flux. eLife. 6:e25257 10.7554/eLife.25257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revilla-Guarinos M.T., Martin-Garcia R., Villar-Tajadura M.A., Estravis M., Coll P.M., and Perez P.. 2016. Rga6 is a Fission Yeast Rho GAP Involved in Cdc42 Regulation of Polarized Growth. Mol. Biol. Cell. 27:1524–1539. 10.1091/mbc.E15-120818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasamura T., Kobayashi T., Kojima S., Qadota H., Ohya Y., Masai I., and Hotta Y.. 1997. Molecular cloning and characterization of Drosophila genes encoding small GTPases of the rab and rho families. Mol. Gen. Genet. 254:486–494. 10.1007/s004380050443 [DOI] [PubMed] [Google Scholar]
- Scheffzek K., Ahmadian M.R., and Wittinghofer A.. 1998. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem. Sci. 23:257–262. 10.1016/S0968-0004(98)01224-9 [DOI] [PubMed] [Google Scholar]
- Shinjo K., Koland J.G., Hart M.J., Narasimhan V., Johnson D.I., Evans T., and Cerione R.A.. 1990. Molecular cloning of the gene for the human placental GTP-binding protein Gp (G25K): identification of this GTP-binding protein as the human homolog of the yeast cell-division-cycle protein CDC42. Proc. Natl. Acad. Sci. USA. 87:9853–9857. 10.1073/pnas.87.24.9853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaughter B.D., Das A., Schwartz J.W., Rubinstein B., and Li R.. 2009. Dual modes of cdc42 recycling fine-tune polarized morphogenesis. Dev. Cell. 17:823–835. 10.1016/j.devcel.2009.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston D. 2018. Establishing and transducing cell polarity: common themes and variations. Curr. Opin. Cell Biol. 51:33–41. 10.1016/j.ceb.2017.10.007 [DOI] [PubMed] [Google Scholar]
- Tatebe H., Nakano K., Maximo R., and Shiozaki K.. 2008. Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr. Biol. 18:322–330. 10.1016/j.cub.2008.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu H., Barr M., Dong D.L., and Wigler M.. 1997. Multiple regulatory domains on the Byr2 protein kinase. Mol. Cell. Biol. 17:5876–5887. 10.1128/MCB.17.10.5876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter I.R., and Wittinghofer A.. 2001. The guanine nucleotide-binding switch in three dimensions. Science. 294:1299–1304. 10.1126/science.1062023 [DOI] [PubMed] [Google Scholar]
- Vjestica A., Merlini L., Dudin O., Bendezu F.O., and Martin S.G.. 2016. Microscopy of Fission Yeast Sexual Lifecycle. J. Vis. Exp. 109:e53801 10.3791/53801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.B., Jiang Z.Z., Zhang Q.H., Hu M.W., Huang L., Ou X.H., Guo L., Ouyang Y.C., Hou Y., Brakebusch C., et al. 2013. Specific deletion of Cdc42 does not affect meiotic spindle organization/migration and homologous chromosome segregation but disrupts polarity establishment and cytokinesis in mouse oocytes. Mol. Biol. Cell. 24:3832–3841. 10.1091/mbc.e13-03-0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson L.J., Rossi G., and Brennwald P.. 2014. Quantitative analysis of membrane trafficking in regulation of Cdc42 polarity. Traffic. 15:1330–1343. 10.1111/tra.12211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Soldner R., and Li R.. 2003. Spontaneous cell polarization: undermining determinism. Nat. Cell Biol. 5:267–270. 10.1038/ncb0403-267 [DOI] [PubMed] [Google Scholar]
- Weston C., Bond M., Croft W., and Ladds G.. 2013. The coordination of cell growth during fission yeast mating requires Ras1-GTP hydrolysis. PLoS One. 8:e77487 10.1371/journal.pone.0077487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley E., and Rittinger K.. 2005. Interactions between Cdc42 and the scaffold protein Scd2: requirement of SH3 domains for GTPase binding. Biochem. J. 388:177–184. 10.1042/BJ20041838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe K.H., and Shields D.C.. 1997. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 387:708–713. 10.1038/42711 [DOI] [PubMed] [Google Scholar]
- Woods B., Lai H., Wu C.F., Zyla T.R., Savage N.S., and Lew D.J.. 2016. Parallel Actin-Independent Recycling Pathways Polarize Cdc42 in Budding Yeast. Curr. Biol. 26:2114–2126. 10.1016/j.cub.2016.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Li S., Chrostek-Grashoff A., Czuchra A., Meyer H., Yurchenco P.D., and Brakebusch C.. 2007. Cdc42 is crucial for the establishment of epithelial polarity during early mammalian development. Dev. Dyn. 236:2767–2778. 10.1002/dvdy.21309 [DOI] [PubMed] [Google Scholar]
- Yang C., and Kazanietz M.G.. 2007. Chimaerins: GAPs that bridge diacylglycerol signalling and the small G-protein Rac. Biochem. J. 403:1–12. 10.1042/BJ20061750 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.