Sui et al. report the cryo-EM structure of the conserved luminal domain of the lipid droplet (LD)-formation protein seipin. The structure reveals key features of this domain and suggests a new model for seipin’s role in LD formation.
Abstract
Metabolic energy is stored in cells primarily as triacylglycerols in lipid droplets (LDs), and LD dysregulation leads to metabolic diseases. The formation of monolayer-bound LDs from the endoplasmic reticulum (ER) bilayer is poorly understood, but the ER protein seipin is essential to this process. In this study, we report a cryo–electron microscopy structure and functional characterization of Drosophila melanogaster seipin. The structure reveals a ring-shaped dodecamer with the luminal domain of each monomer resolved at ∼4.0 Å. Each luminal domain monomer exhibits two distinctive features: a hydrophobic helix (HH) positioned toward the ER bilayer and a β-sandwich domain with structural similarity to lipid-binding proteins. This structure and our functional testing in cells suggest a model in which seipin oligomers initially detect forming LDs in the ER via HHs and subsequently act as membrane anchors to enable lipid transfer and LD growth.
Introduction
Nearly all organisms have the capacity to buffer fluctuations in energy availability by storing highly reduced carbons as triacylglycerol (TG) in lipid droplets (LDs; Onal et al., 2017; Walther et al., 2017; Henne et al., 2018). Although LDs can be formed in most cells, the molecular mechanisms for LD biogenesis remain largely unknown. In the most widely held model (Walther et al., 2017), TGs and other neutral lipids, synthesized by enzymes in the ER membrane, phase separate to form an oil lens within the ER bilayer. Lenses subsequently grow and bud toward the cytosol, forming a monolayer-bound LD that can be targeted by specific proteins (Joshi et al., 2017; Walther et al., 2017; Henne et al., 2018). ER proteins are thought to be crucial for controlling the formation process, but little is known about the underlying mechanisms for generating homogeneously sized LDs.
The ER protein seipin is a central player in LD formation. Seipin, encoded by the gene Bernadelli-Seip congenital lipodystrophy type 2 (BSCL2; Magré et al., 2001; Gomes et al., 2004), is an integral membrane protein with short N- and C-terminal segments in the cytosol, two transmembrane (TM) helices, and an evolutionarily conserved ER luminal domain (Fig. 1 A; Lundin et al., 2006). Missense mutations of human BSCL2 leading to lipodystrophy occur mainly in the ER luminal region (e.g., L91P and A212P), suggesting this part of seipin is crucial for its function. In fly and mammalian cells, seipin forms mobile foci in the ER that are recruited to and stabilized at sites of nascent LD formation, where these foci appear to be required for LD growth (Hölttä-Vuori et al., 2013; Wang et al., 2014, 2016; Grippa et al., 2015; Salo et al., 2016). In the absence of seipin, cells form many small LDs, possibly due to premature budding, that fail to grow (Grippa et al., 2015; Salo et al., 2016; Wang et al., 2016). Cells also have so-called supersized LDs that likely form from coalescence of the smaller LDs (Szymanski et al., 2007; Fei et al., 2008; Wolinski et al., 2011; Grippa et al., 2015; Salo et al., 2016; Wang et al., 2016). Seipin deficiency also leads to the recruitment of aberrant proteins to LDs and possibly to alterations in ER Ca2+ homeostasis and lipid metabolism such as phosphatidic acid accumulation (Fei et al., 2011; Han et al., 2015; Wolinski et al., 2015). Previous studies of seipin have led to different functional models including a regulator of the ER Ca2+ pump SERCA, a molecular scaffold and regulator of lipid metabolism enzymes and a structural protein facilitating LD growth at ER–LD contact sites (Sim et al., 2012; Bi et al., 2014; Talukder et al., 2015; Pagac et al., 2016; Wang et al., 2016). However, the molecular function of seipin in LD biogenesis remains unclear.
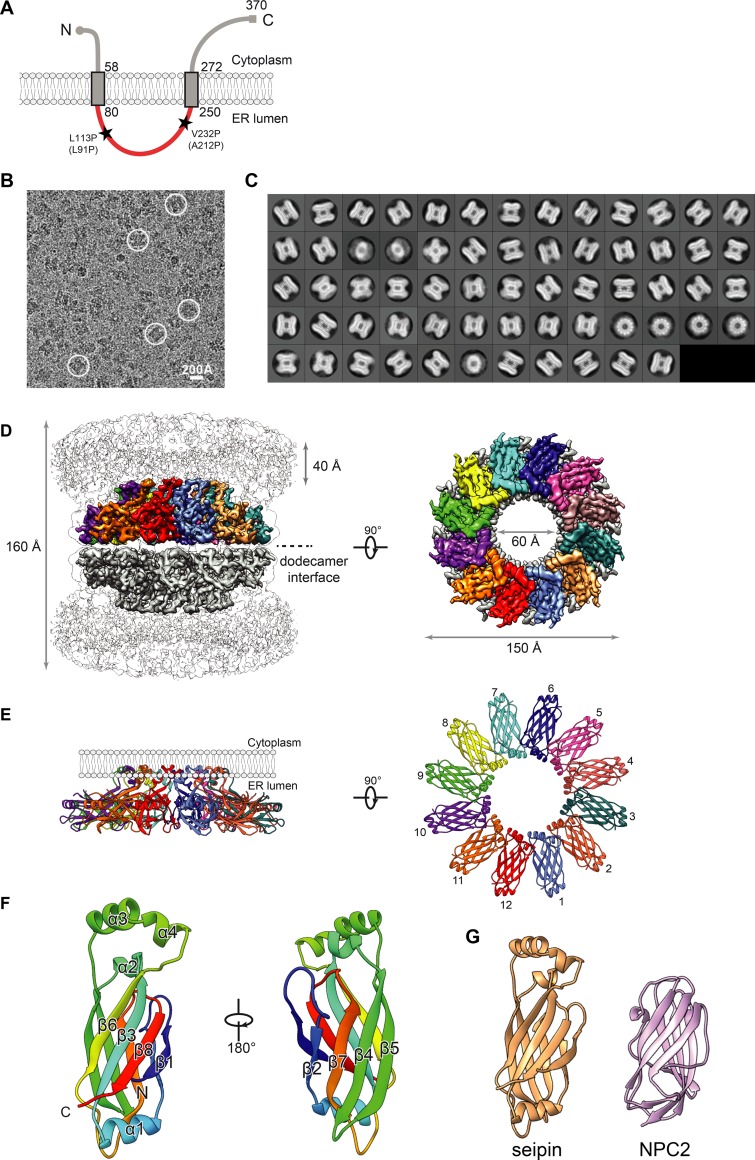
Figure 1.
Cryo-EM map and molecular model of seipin. (A) Model of D. melanogaster seipin topology in the ER membrane. The conserved ER-luminal domain is in red. Human pathogenic mutations L91P and A212P and their equivalent positions in D. melanogaster seipin are shown. (B) Representative cryo-EM image of purified seipin in digitonin. White circles indicate representative particle images. (C) 2D averages calculated with all seipin particles from combined datasets. The box dimension is 335 Å. (D) Unsharpened (transparent) and sharpened (solid colored) cryo-EM density maps of a seipin oligomer. The barrel-like structure is a head-to-head dimer of dodecamers interacting via luminal domains. The 40-Å region at the top represents poorly resolved TM regions. Each monomer in the upper dodecamer ring is shown in different colors. The en face view is shown from the perspective of the ER membrane. (E) Ribbon diagram side (left) and top (right) views of the luminal domains. (F) Model and structural elements of seipin monomers containing residues 88–240 corresponding with the ER luminal domain. Each domain contains a β sandwich of eight β strands and four α helices. Helices 3 and 4 comprise hydrophobic sequences positioned at the ER luminal leaflet. (G) Structural comparison of seipin (orange) and the cholesterol-binding protein NPC2 (pink; PDB accession ID: 2HKA); mean root mean square displacement of 4.3 Å over 106 residues.
As a step toward unraveling seipin’s function, we sought to elucidate its molecular structure. Using cryo-EM, we report in this study a structural model of Drosophila melanogaster seipin solved for the luminal domain at ∼4.0 Å resolution. This structure reveals that seipin forms a dodecamer, with each monomer positioning a hydrophobic helix (HH) near the ER bilayer and having a β-sandwich domain with similarity to lipid-binding proteins. We validated this structure and tested several of its key features in vitro and in cells. Our results suggest a new model for how seipin functions to detect forming LDs and promote their growth.
Results and discussion
Determination of a cryo-EM map of D. melanogaster seipin
We purified recombinant D. melanogaster seipin in detergents, and gel-filtration chromatography revealed it to be an oligomer (Fig. S1, A and B), similar to reports for other species (Binns et al., 2010; Sim et al., 2014). Consistently, negative-stain EM showed that seipin particles were monodisperse, and 2D averages demonstrated distinct views with round or multilayer barrel shapes (Fig. S1 C).
We next analyzed purified seipin by cryo-EM. The 2D averages of cryo-EM particle images (Fig. 1, B and C) appeared similar to those obtained with negative-stain EM. 3D classification of cryo-EM images of seipin purified in digitonin or n-dodecyl β-D-maltoside (DDM) detergents showed similar overall conformations, allowing for combined image processing of these two datasets (Fig. S1 D).
An initial round of 3D classification performed without symmetry demonstrated a multilayer barrel-shaped protein complex containing two middle layers, which correspond with the luminal domains, and top and bottom layers, which are less well resolved and represent the TM domains. Because 12 repetitive features were clearly discernable in the 2D averages, we subsequently applied D12 symmetry for further processing (Fig. S1 D). Focusing on the middle layers, where the EM density was best resolved, we selected a homogeneous subset of particles to generate a final cryo-EM map at an overall resolution of ∼4.0 Å (Figs. 1 D and S2, A–C). This map revealed that the barrel-shaped structure consists of two dodecamer rings interacting head to head via seipin luminal domains. This dimer of dodecamers is likely due to nonphysiological contacts; we also found single-ring dodecamers in protein preparations, and we did not find this superdimer structure in preparations of seipin from other species (not depicted).
The cryo-EM map for most of the seipin luminal domain is of high quality, showing large amino acid side-chain densities and well-separated β-strand density, whereas the loop regions distal from the symmetry axis appeared more flexible with lower resolution (Fig. S2, A and D). The N- and C-terminal sequences located on the cytoplasmic side of the ER membrane as well as the TM domains were poorly resolved in the density map, likely due to conformational flexibility.
A molecular model for D. melanogaster seipin
The cryo-EM map quality enabled us to build an atomic model of monomeric seipin spanning amino acid residues 88–240, corresponding with the ER luminal domain. Starting from a manually built partial model corresponding with the highest-resolution region, the combination of Rosetta de novo model building (Wang et al., 2015) and model completion with RosettaES (Frenz et al., 2017) confidently placed nearly the entire monomeric sequence. This monomeric model was subsequently refined in the context of the symmetric oligomeric assembly (Fig. 1, E and F).
The molecular model reveals three prominent features. First, seipin forms a ring-shaped dodecamer of laterally interacting subunits, with an overall diameter of ∼150 Å (Fig. 1, D and E). Second, in each seipin monomer, a helical, hydrophobic region (α3 and α4; residues 175–192) of the luminal domain was oriented toward and adjacent to the ER luminal membrane leaflet (Fig. 1 E). Third, the remainder of each luminal domain consists of two β-sheets, each containing four antiparallel β strands (Figs. 1 F and S2 D). This region of seipin formed a similar fold as the lipid-binding domain of Niemann-Pick type C2 protein (NPC2; Fig. 1 G; Xu et al., 2007). Consistently, seipin exhibits weak similarities to NPC2 in hidden Markov model searches with the HHPred algorithm (Söding et al., 2005).
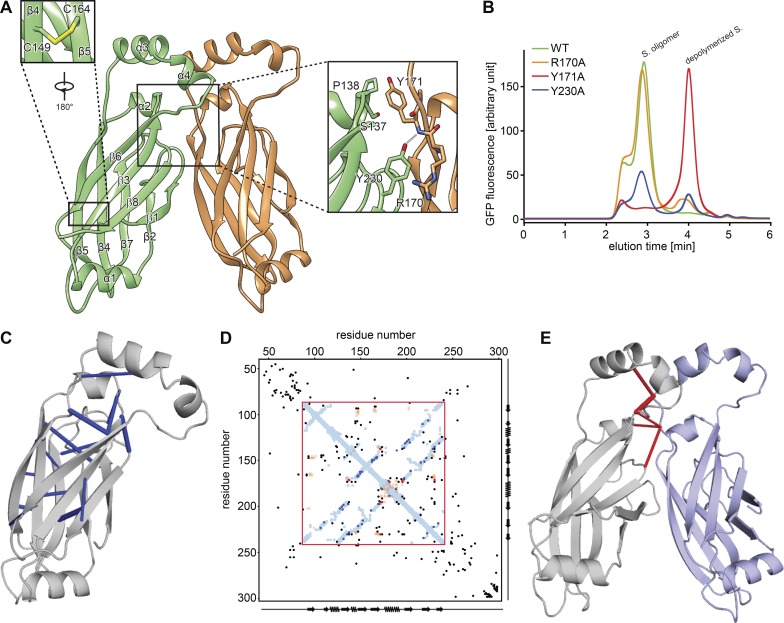
To test the structural model, we investigated the interface between monomers. Our model predicted that Tyr230 forms a π–cation interaction with Arg170 and a hydrogen bond with the peptide backbone amino group of Tyr171 (Fig. 2 A). To test the requirement of these residues for seipin oligomer formation, we expressed and analyzed mutant forms of seipin by fluorescence-detection size-exclusion chromatography (FSEC) of GFP fusion proteins expressed in cells. Tyr171Ala resulted in a complete shift of the gel filtration peak to a lower molecular weight, indicating weakened oligomerization (Fig. 2 B). Expression of either Tyr230Ala or Arg170Ala resulted in a partial oligomerization defect. The model also shows Arg165 forming a π−cation interaction with Phe94 on the neighboring monomer (Fig. S3 B). However, Arg165Ala mutation did not alter seipin oligomerization (Fig. S3 C), and either Arg165Ala or Phe94Ala only showed an effect on oligomerization when combined with a Tyr230Ala mutation, suggesting a less critical role for Arg165 and Phe94 in oligomer formation (Fig. S3 C). These results indicate that the seipin structural model correctly predicts key interactions between seipin monomers in vitro, although the precise interactions at this interface require further investigation.
Figure 2.
Analysis of a seipin oligomer’s inter- and intramolecular interactions. (A) Model of interactions between seipin monomers. Enlarged views (boxed) show the intramolecular disulfide bond (C149 and C164) and key interactions between monomers. (B) Fluorescence-based gel-filtration analyses of seipin (S.) variants expressed in HEK cells. (C) Molecular model of seipin with significant intramolecular evolutionary couplings (blue) as revealed by evolutionary coupling analysis and folding (EV fold; Marks et al., 2011, 2012). (D) Overlay of seipin’s amino acid evolutionary couplings with distances derived from the molecular model for D. melanogaster seipin. Amino acid positions as well as secondary structure elements are shown on x and y axes. The sequence in the red box represents the cryo-EM structure. Blue and orange dots are residues closer than 5 Å within or between seipin monomers, respectively. Black, blue, and red dots represent the top 125 evolutionary couplings, with the most significant ones observed in the fly seipin structures shown in C (blue) and E (red). (E) Molecular model of seipin with significant intermolecular evolutionary couplings shown in red.
As an alternative means to validate the model, we analyzed 1,591 seipin protein sequences from different species for evolutionary covariation that predicts physical constraints of protein structure (Marks et al., 2011, 2012). Remarkably, overlaying of the seipin evolutionary couplings with proximities of residues derived from our model showed extensive overlap within a monomer (Fig. 2, C, D [red box], and E), supporting the validity of our model. Additionally, the model placed two cysteines (C149 and C164) in close spatial proximity, consistent with the density in the cryo-EM map indicating a disulfide bond (Figs. 2 A and S2 D). This analysis also was consistent with our findings for residues interacting at monomer interfaces as Tyr230 is highly conserved (Fig. S3 A) and four of the five residues with most enriched evolutionary couplings are in this region (see, for example, Tyr230 and Tyr171; Fig. 2 E).
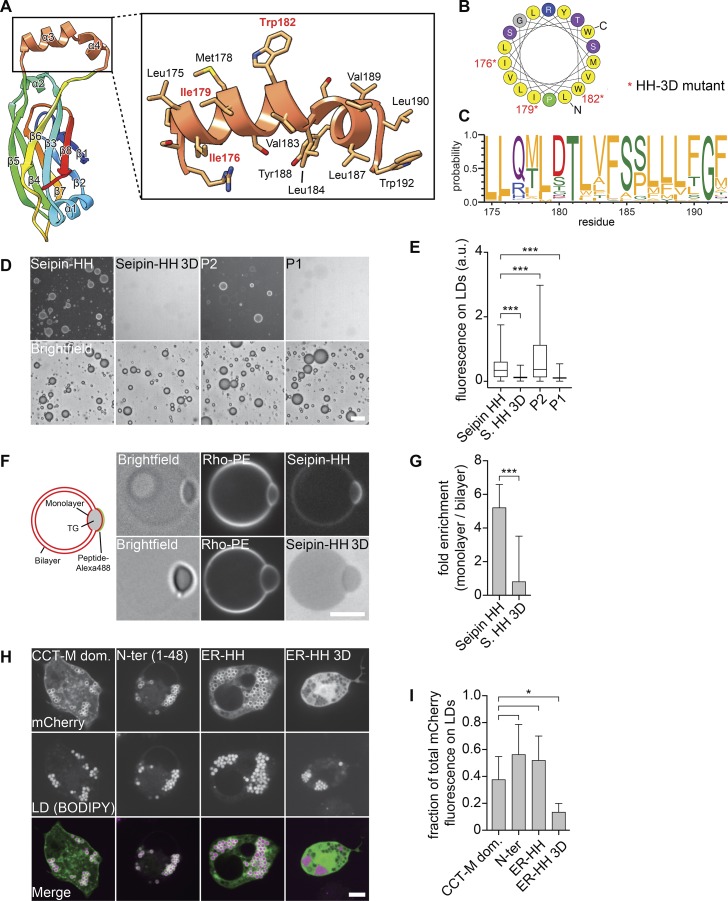
The HHs of the seipin luminal domain bind monolayers of LDs in vitro and in cells
The positioning of multiple HHs close to the ER bilayer is a distinctive feature of the seipin structure. This helical region of each monomer (Fig. 3 A) exhibited a high degree of evolutionary conservation with respect to hydrophobicity (Fig. 3, B and C). Similar helices with large hydrophobic residues from other LD proteins bind phospholipid packing defects of the LD surface monolayer (Prévost et al., 2018). We thus hypothesized that these HHs function to detect packing defects of the ER membrane due to neutral lipid accumulation and lens formation. To test this hypothesis, we incubated a peptide from the seipin-HH with artificial LDs and found it bound to monolayer surfaces, similar to previous results with the LD-targeting amphipathic helix derived from the LD protein CCT1 (Fig. 3, D and E; Prévost et al., 2018). In contrast, a seipin-HH peptide with three Asp residues (seipin-HH 3D) failed to bind LDs. The seipin-HH peptide also bound predominantly to monolayers of LDs contained in giant unilamellar vesicles (GUVs), whereas seipin-HH 3D showed no binding to either the bilayer or monolayer (Fig. 3, F and G). Consistent with these results, seipin-HH expressed as an mCherry fusion protein targeted to LDs in D. melanogaster cells similar to the M domain of CCT1 (Kory et al., 2015), but seipin-HH 3D did not (Fig. 3, H and I). These results support the hypothesis that the HHs serve to recognize and target the protein to phospholipid-packing defects of lipid lenses within the ER bilayer.
Figure 3.
The HH of the seipin ER luminal domain targets to LDs. (A) Molecular structure of the HH highlighting residues 172–192 in orange. (B) Helical plot of residues Leu175–Trp192. Nonpolar residues are shown in yellow (Gautier et al., 2008). Asterisks indicate residues mutated to Asp in the seipin 3D mutant. (C) The helical region residue distribution for the top 200 seipin sequences (retrieved from the Pfam database; corresponding with residues 175–192 of D. melanogaster seipin) shows evolutionary conservation of hydrophobicity (Crooks et al., 2004). Residues are colored according to their physicochemical properties, with hydrophobic residues in orange. (D) The seipin HH binds artificial LDs in vitro. An Alexa Fluor 488–labeled peptide comprising residues 174–193 but not a version with the 3D mutation (replacing Ile176, Ile179, and Trp182 with Asp) binds to artificial LDs. (E) Quantification of fluorescent signals from >2,000 artificial LDs per peptide as in C as a boxplot representation. (F) The seipin HH binds to the phospholipid monolayer in vitro. Seipin helix peptide but not the mutated 3D version preferentially binds to the monolayer of TG lenses incorporated into GUVs. Graphical representation and representative confocal images show the peptide and phospholipid signals, respectively. (G) Quantification of enrichment on a monolayer versus a bilayer of ≥18 GUVs per peptide as mean ± SD. (H) Binding of seipin HH to LDs in cells. The mCherry-tagged N-terminal amphipathic helical sequence (1–48), the luminal HH (174–193) of seipin, and a seipin-HH 3D mutant of the luminal helix were expressed in D. melanogaster S2 cells and analyzed by confocal imaging for LD binding. As a control, the CTP-phosphocholine cytidylyltransferase (CCT) M domain was expressed in S2 cells from the same vector. Bars: 20 µm (D); 5 µm (F and H). (I) Quantification of mCherry fluorescence on LDs versus total signal per cell as shown in H as mean ± SD from ≥9 cells per construct. *, P < 0.01; ***, P < 0.0001.
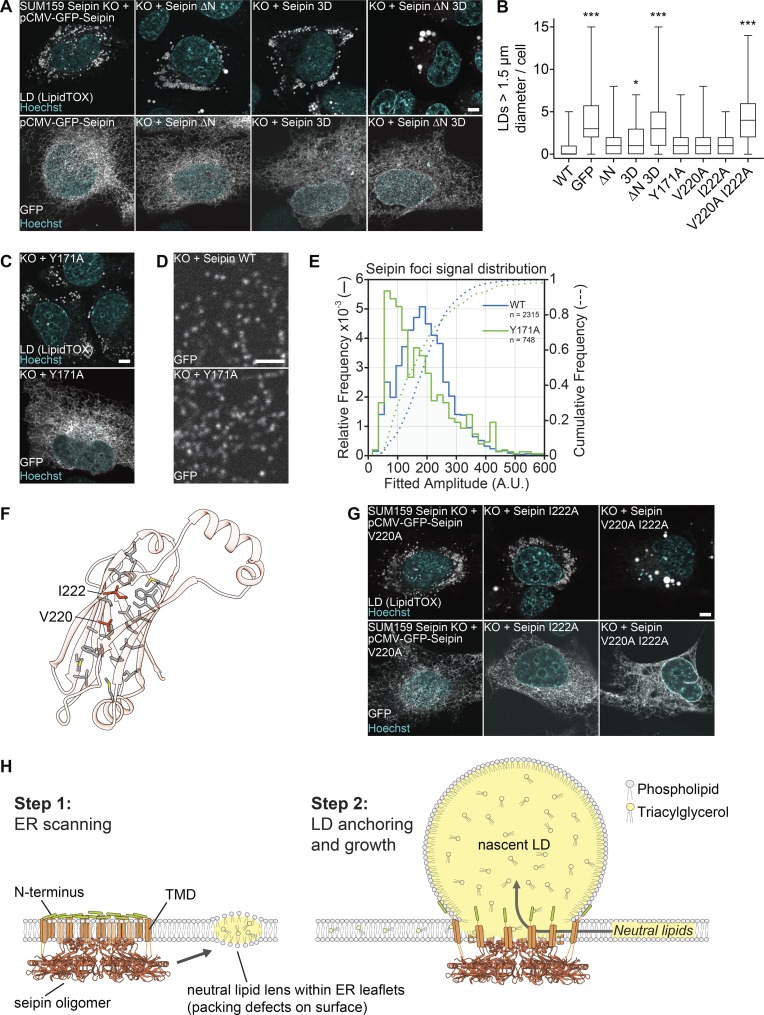
Luminal or N-terminal helical sequences and the luminal domain are required for LD formation
We next used the seipin structure to identify key features of the protein required for its function in cells. First, we tested the requirement for the HHs by assaying seipin function in LD formation after expression of either WT seipin or a variant containing negatively charged residues in the seipin-HH 3D in SUM159 cells lacking seipin. After oleate addition to induce LD formation, WT cells formed many small LDs, and cells lacking seipin formed numerous smaller LDs and some supersized LDs (>1.5 µm). Reexpression of WT seipin as an N-terminal GFP-fusion protein rescued the seipin knockout phenotype, whereas GFP alone did not (Figs. 4 A and S3 D). Surprisingly, expression of seipin-HH 3D also rescued the seipin deletion phenotype (Fig. 4, A and B), indicating that this helical region was not required for LD formation. We previously found that the N-terminal sequence of seipin was also sufficient to bind LDs but not required for LD formation (Fig. 3 H; Wang et al., 2016). Therefore, we reasoned that these different LD-binding helices may function in a complementary manner during LD formation. Indeed, expression of a double mutant of the N terminus deletion and seipin-HH 3D was not able to rescue the LD phenotype of seipin knockout cells (Fig. 4, A and B) despite proper folding and localization to the ER (Figs. 3 A and S3 G).
Figure 4.
Testing key features of the seipin luminal domain in cells. (A) The hydrophobic and N-terminal helices are required for seipin function. SUM159 seipin-knockout (KO) cells were transfected with seipin constructs with N-terminal GFP and analyzed for LD phenotype after 24 h oleate treatment. Top: LipidTOX staining. Bottom: Localization to the ER using GFP fluorescence. Representative images are shown. (B) LD size of ≥47 transfected cells per construct from experiments in A, C, and G represented as boxplots. *, P < 0.01; ***, P < 0.0001 compared with WT sample. (C) Seipin Y171A mutant rescues seipin deficiency. Transfection and cell treatment as in A. Top: LD phenotype. Bottom: Localization to the ER. (D) Seipin WT and Y171A form fluorescent foci in the ER. Cells were imaged without oleate addition. To monitor puncta, low transfected cells were monitored. (E) Seipin puncta of 38 cells expressing seipin WT and 31 cells expressing seipin Y171A as in D were tracked and quantified over time. The comparative foci signal distributions are shown for seipin WT (blue lines) and Y171A (green lines). (F) Hydrophobic residues form a putative pocket in the luminal domain of seipin. Residues mutated in B and G are indicated. (G) Analysis of dmSeipin luminal domain mutants as in A. Bars: 5 µm (A, C, and G); 2 µm (D). (H) Model for the molecular function of seipin during LD formation. TMD, TM domain.
Next, we tested whether interactions of the luminal domains are required for seipin function in LD formation. Each of the interface mutant proteins rescued the LD phenotype of seipin-deletion cells, including those that destabilized the oligomer in in vitro analyses (Fig. 4, B and C; and Fig. S3, D–F). Even the Y171A mutant, which most compromised oligomerization in vitro, rescued the LD phenotype and maintained the ability to form foci in the ER, likely indicating the assembly of oligomers or higher-order complexes (Fig. 4 D). Tracking of seipin foci of WT or the Y171A mutant over time revealed that the Y171A mutant was present in foci with similar intensity as WT seipin but also in another population with less GFP intensity (Fig. 4 E), suggesting seipin foci with fewer seipin molecules. Indeed, negative-stain EM analyses of purified Y171A protein revealed significantly smaller particles (Fig. S3 H). These results suggest that interactions of the luminal domains likely contribute to but are not strictly required for seipin oligomerization in vivo.
We also tested the requirement of the luminal domain for seipin function by mutating two residues (V220 and I222) at the center of a hydrophobic cavity within the β-sandwich domain to alanines (Fig. 4 F). Although expression of either single mutant rescued the seipin-deletion phenotype, expression of a V220A/I222A mutant was unable to rescue the phenotype, highlighting the importance of the luminal domain to seipin function (Fig. 4 G). Whether the double mutant fails to rescue because it impairs folding or lipid binding is unknown.
Collectively, our data indicate that D. melanogaster seipin forms a ring-shaped dodecamer with N- and C-terminal segments oriented toward the cytoplasm and an assembly of folded domains that are localized in the ER lumen. The dimensions of this complex are consistent with a barrel-shaped ring that connects the ER membrane with nascent LDs during their formation. The molecular structure of seipin argues strongly that seipin performs a structural and possibly a lipid transfer role in organizing LD formation and suggests that other effects of seipin deficiency such as changes in ER calcium homeostasis found with seipin deficiency are indirect.
Many avenues of evidence indicate that the seipin luminal domain is crucial for its function. This domain is highly conserved and is the location of numerous lipodystrophy mutations (Magré et al., 2001), and we show that mutating the luminal domain impairs seipin function in LD biogenesis (Fig. 4, F and G) The structure of the D. melanogaster luminal domain defines features of this domain that shed light on how seipin functions. For example, the structure suggests that seipin oligomers position multiple HHs on either side of the ER membrane, possibly to detect phospholipid packing defects due to forming neutral lipid lenses. This is consistent with previous studies that indicate that seipin foci and nascent LD foci are initially separate but interact and subsequently colocalize during LD formation (Grippa et al., 2015; Salo et al., 2016; Wang et al., 2016). An oligomeric structure of multiple LD-binding helices may increase the ability of helices to detect lipid lenses through increased avidity, a property that is relevant to the binding of amphipathic helices to LD surfaces (Prévost et al., 2018). The requirement for a strictly dodecameric oligomer seems unlikely. Data from other species suggest the number of monomers can vary between species (unpublished data), suggesting that there is flexibility in the numbers of seipin molecules in a macromolecular structure. Also, a mutation at the luminal interface that weakened oligomerization in vitro still formed foci and rescued the seipin-deficiency phenotype in cells (Fig. 4, C and D). This suggests that other regions of the protein, such as the TM domains, contribute to oligomerization in cells and may be essential for seipin function.
The luminal domain may not only provide an anchor for the complex on the luminal side of the ER membrane but may also function to mediate lipid transfer to growing LDs. The β-sandwich fold of the luminal domain is structurally similar to lysosomal NPC2, which binds and solubilizes cholesterol in the lumen of the lysosome to deliver it to membrane-embedded NPC1 for export (Wang et al., 2010). Other related NPC2-type proteins (e.g., in Camponotus japonicus) have a similar β-sandwich structure that allows binding of semiochemicals including fatty acids used for chemical communication (Ishida et al., 2014). Our structure and sequence alignments suggest that like NPC2, seipin has a binding pocket of sufficient size for accommodating hydrophobic molecules (Fig. 4 F). We speculate that the NPC2-like luminal domain of seipin participates in transferring lipids from the ER luminal leaflet to the nascent LDs to maintain the proper balance or composition of phospholipids or neutral lipids. The identity of a lipid that binds to seipin is unknown and under investigation.
Based on the seipin structure, we suggest a new model for LD formation (shown in Fig. 4 H). In this model, seipin forms an oligomeric complex in the ER that moves throughout the reticular network in the absence of LDs. Once neutral lipids are synthesized and their concentration in the ER exceeds a critical concentration, lipid lenses form and disrupt phospholipid bilayer packing in the ER membrane, resulting in localized surface defects (Prévost et al., 2018). Seipin complexes may recognize the phospholipid packing defects at lipid lenses by binding via their many amphipathic and HHs located at the cytoplasmic N terminus and in the ER lumen, respectively. Subsequently, a seipin oligomer becomes localized to a neutral lipid lens (Wang et al., 2016). As nascent LDs grow toward the cytosol, seipin may anchor the nascent LD to the ER (via N-terminal helix binding) and allow for maintenance of the ER–LD connection to enable LD growth and prevent nascent premature severing as found with seipin deficiency (Grippa et al., 2015; Salo et al., 2016; Wang et al., 2016). In this model, oligomerization could also serve to restrict the diameter of the neck of the budding LDs. Finally, seipin may facilitate lipid transfer to nascent LDs. Our structural model thus provides a new framework for the further molecular dissection of the LD formation pathway.
Materials and methods
Seipin expression and purification
The sequence of D. melanogaster seipin (FlyBase ID: FBpp0070426) was codon optimized for bacterial expression and synthesized and cloned into the pET28a+ expression vector with the enzyme restriction sites of NcoI and NotI to produce C-terminally 6×His-tagged seipin. The integrity of the plasmid was confirmed by sequencing. The plasmid was transformed into the BL21(DE3) Escherichia coli strain (New England Biolabs) for protein expression. 1-liter cultures of Luria–Bertani medium containing 50 µg/ml kanamycin were grown at 37°C to an OD600 of 1.5–1.7, and then incubated at 4°C for ∼15–20 min. Protein expression was induced by adding isopropyl β-D-1-thiogalactopyranoside (Roche) to a final concentration of 0.5 mM and an additional 1 ml kanamycin at 50 mg/ml into 1 liter culture. After overnight growth (∼14 h) at 190 rpm and 16°C, cells were harvested by centrifugation, suspended in TMSG buffer (50 mM Tris-HCl, pH 8.0, 5 mM MgCl2, 400 mM NaCl, and 10% vol/vol glycerol), and either stored at −80°C or used immediately. Typically, a cell pellet from 2-liter cultures was resuspended in 40 ml TMSG buffer.
All purification procedures were performed at 4°C. For each 50-ml cell suspension, one tablet of protease inhibitor cocktail (Roche) was added, and the cells were lysed by sonication. The lysate was cleared by centrifugation at 11,594 g for 30 min. The membrane-containing supernatant was then centrifuged at 185,511 g for 1 h. The membrane pellet was collected and homogenized with a Dounce homogenizer in equilibration buffer (50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 5 mM MgCl2, 5% vol/vol glycerol, and 50 mM imidazole). Protein was extracted by adding DDM to a 1% wt/vol final concentration with gentle rocking for 1 h. The insoluble fraction was removed by centrifugation at 184,000 g for 30 min. The recombinant protein in the supernatant was affinity purified using Ni-NTA affinity resin (QIAGEN). Briefly, for DDM-solubilized membrane suspension from 6-liter cultures, 1 ml prewashed Ni-NTA resin by TMSG buffer was added into the suspension. After gentle stirring for 1 h, the resin was collected and washed with 10 bed volumes of wash buffer (50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 5 mM MgCl2, 5% vol/vol glycerol, 50 mM imidazole, and 0.1% wt/vol DDM) containing 5 mM ATP. The protein was then eluted by 10 column volumes elution buffer (50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 5 mM MgCl2, 5% vol/vol glycerol, 500 mM imidazole, and 0.1% wt/vol DDM). The eluted protein was collected and concentrated to 500 µl in a 100-kD cutoff Amicon protein concentrator (EMD Millipore) and loaded onto a Superose 6 10/300 GL size-exclusion column (GE Healthcare) equilibrated with gel-filtration buffer (50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 5 mM MgCl2, and 0.05% wt/vol DDM or 0.05% wt/vol digitonin). The peak fractions containing seipin were pooled, flash frozen, and stored at −80°C or placed on ice for immediate use.
EM sample preparation and data acquisition
Negatively stained specimens were prepared by an established protocol with minor modifications (Booth et al., 2011). Specifically, 2.5 µl purified seipin in DDM or digitonin at 0.2–0.3 mg/ml were applied to glow-discharged copper EM grids covered with a thin layer of continuous carbon film, and the grids were stained with 1.5% (wt/vol) uranyl formate for 30 s. These grids were imaged on a Tecnai T12 microscope (Thermo Fisher Scientific) operated at 120 kV and equipped with a 4,000 × 4,000 charge-coupled device camera (UltraScan 4000; Gatan). A nominal magnification of 52,000× corresponding with a pixel size of 2.13 Å on the specimen and a defocus of ∼1.5 µm were used to record the images.
For cryo-EM, 2.5–3.5 µl purified seipin was applied to Quantifoil holey carbon grids (Cu R1.2/1.3; 400 mesh) glow discharged for 30 s. Our initial trials showed that very few particles appeared in the vitreous ice even with a high protein concentration of ∼5 mg/ml. Attempts to further increase protein concentration led to severe protein aggregation as revealed by cryo-EM analysis. To overcome this problem, the grids were overlaid with graphene oxide according to a published protocol (Bokori-Brown et al., 2016; Martin et al., 2016). This treatment substantially increased the number of particles embedded in vitreous ice. Optimal particle distribution was obtained with a protein concentration of 0.5–1.5 mg/ml. After applying the protein, the grids were blotted with a Whatman filter paper (grade 595) for 3 s with 90% humidity and plunge frozen in liquid ethane cooled by liquid nitrogen using a Vitrobot (Thermo Fisher Scientific) or Cryoplunge 3 System (Gatan). Cryo-EM data were collected on a Titan Krios electron microscope (Thermo Fisher Scientific) at the Howard Hughes Medical Institute cryo-EM facility at the Janelia Research Campus. Image were recorded using SerialEM (Mastronarde, 2005) and a K2 Summit direct electron detector (Gatan) in superresolution counting mode. Refer to Table S1 for more information about data collection.
EM data processing
For negative-stain EM data, the images were binned over 2 × 2 pixels, yielding a pixel size of 2.13 Å, for further processing using Simplified Application Managing Utilities for EM Labs (SAMUEL) scripts (Liao et al., 2014). For cryo-EM data, drift correction was performed using MotionCor2 (Zheng et al., 2017), and images were binned 2 × 2 by Fourier cropping to a pixel size of 2.62 Å. The defocus values were determined using CTFFIND4 (Rohou and Grigorieff, 2015) and motion-corrected sums without dose-weighting. Motion-corrected sums with dose-weighting were used for all other image processing. Particle picking was performed using a semiautomated procedure (Ru et al., 2015). 2D classification of selected particle images were performed by samclasscas.py, which uses SPIDER operations to run 10 cycles of correspondence analysis, K-means classification, and multireference alignment, or RELION 2D classification (Scheres, 2012a,b). Initial 3D models were generated with 2D averages using SPIDER 3D projection matching refinement (samrefine.py) starting from a cylindrical density that mimics the general shape and size of seipin. 3D classification and refinement were performed using relion_refine_mpi in RELION. One round of 3D classification without applying symmetry was performed on the total 270,716 particles to remove bad particles and to select particles with homogenous signal for the middle stacks. Subsequently, particles from classes 3 and 5 were combined for one round of 2D classification followed by 3D refinement with D12 symmetry applied. The resulting model showed high signal-to-noise ratio in the middle stack region, whereas the distal region exhibited weak density. The next round of 3D classification focused on the middle stack region (red mesh) and yielded a total of six classes. Among them, particles belonging to class 4 were used to produce the final seipin cryo-EM map with an overall resolution of 4 Å. All refinements followed the gold-standard procedure in which two half datasets are refined independently. The overall resolutions were estimated based on the gold-standard Fourier shell correlation (FSC) = 0.143 criterion. Local resolution variation of cryo-EM maps was calculated using relion_postprocess_mpi with the –locres option. The amplitude information of the final maps was corrected by applying a negative B factor using relion_postprocessing with the –auto_bfac option. The number of particles in each dataset and other details related to data processing are summarized in Table S1.
Model building and refinement
The seipin monomer and dimer maps were segmented and extracted in UCSF Chimera by using the integrated program Segger (Pintilie et al., 2010). The seipin monomer map was used to build the seipin model. Ab initio model building was performed in COOT (Emsley and Cowtan, 2004). The regions with high resolution in the monomer map, including the first two layers of all four layers of seipin, which correspond with residues from Ala125 to Glu210, could be manually built with confidence. For the low-resolution regions, the Rosetta package was used build the whole-seipin model (see below). After building the model, the monomer structure was docked into dimer map, and the dimer model was manually adjusted and refined in Phenix real-space refinement package (Adams et al., 2010). The refined model was visually inspected and adjusted in COOT, and the resulting map was further put back through the real-space refinement procedure to undergo further refinement. This iterative process was repeated until the dimer model reached optimal geometric statistics as evaluated by MorProbility (Chen et al., 2010). Finally, the seipin dodecamer structure was obtained by docking the dimer model into the full dodecamer map in UCSF Chimera (Pettersen et al., 2004).
Model building with Rosetta
A polyalanine model was initially built into the density. While secondary structure elements were clearly identified, ambiguity in loop density made topology determination and, consequently, sequence registration of the model difficult. Using the polyalanine model as an input to guide placement, we ran de novo model building with Rosetta (Wang et al., 2015). Initially, Rosetta was able to place a sequence corresponding with 72 residues (i.e., a stretch spanning residues 127–200), and this model was consistent with abovementioned manually built model within the region. By using this model as an input for another two rounds of de novo modeling, the entire C terminus was built (residues 201–239), and several strands of the N terminus were built (residues 93–97 and 109–114).
This monomeric model was then completed using RosettaES (Frenz et al., 2017). RosettaES showed very good convergence for all missing regions except over a stretch around residues 99–103. This corresponds with a region of relatively poorly resolved density. By using the monomeric structure energy and visual inspection, one conformation of this loop was selected and refined (Wang et al., 2015) in the context of the C12 complex with the half of entire density map. In total, 1500 refined models were generated. Inspection of the five lowest-energy models yielded low every good convergence (<1 Å root mean square displacement) except over the aforementioned loop, indicating confidence in the assigned model.
Data availability
The cryo-EM density map of seipin has been deposited in the Electron Microscopy Data Bank under accession no. EMD-9146. Atomic coordinates for the atomic models has been deposited in the Protein Data Bank under accession no. 6MLU.
FSEC assay
The seipin coding sequence was cloned into the in-house–generated pFasBacMam vector with the cytomegalovirus promoter for overexpression of target protein in mammalian cells where seipin was tagged by an EGFP at the N terminus. The plasmid was transfected into HEK293F cells by polyethylenimine (PEI) as described previously (Kawate and Gouaux, 2006; Goehring et al., 2014) in a six-well plate format. In brief, 40,000 HEK293F cells were seeded into each well of six-well plate, and PEI transfection were done when cells reached ∼50% confluence (usually after 24 h). For transfection, 100 µl FreeStyle 293 expression medium (Thermo Fisher Scientific) containing 1 µg DNA plasmid and 100 µl medium containing 3 µg PEI were mixed. After incubation at room temperature for 30 min, the total 200-µl mixture was added into one well. Cells were harvested after ∼48 h transfection and washed once by PBS, and then the pellet was either stored at −80°C or used immediately. To lyse the cells, the pellet from one well of a six-well plate was resuspended in 500 µl buffer containing 50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 5 mM MgCl2, and 1% wt/vol DDM and supplemented with protease inhibitor cocktail (Roche). The mixture was placed on a shaker with gentle shaking in a cold room for 1 h and then centrifuged at 17,000 g on a precooled bench-top centrifuge for 20 min at 4°C. Then, the detergent-solubilized supernatant was collected and injected into a high-performance liquid chromatography (HPLC) system (Waters) equipped with a fluorescent detector with excitation and detecting wavelengths of 488 and 520 nm, respectively. The total 20-µl protein sample was injected into the HPLC system coupled with a reverse-phase gel-filtration column (5 µm; 7.8 × 50 mm) with a pore size of 500 Å (Sepax Technologies). Gel-filtration analysis was performed with running buffer containing 50 mM Tris-HCl, pH 8.0, 400 mM NaCl, 5 mM MgCl2, and 0.05% wt/vol DDM. To generate different seipin mutants, the QuikChange site-directed mutagenesis kit (Agilent) was used with the protocol provided by the manufacturer.
Cell culture and transfection
For D. melanogaster S2 cells, transfection and oleic acid treatment were performed as described previously (Prévost et al., 2018) with the following modifications: the cells were treated with oleic acid overnight, starting ∼6–24 h after transfection with plasmids encoding mCherry-tagged constructs. Oleic acid (complexed to BSA at a 3:1 molar ratio) was used at a concentration of 1 mM. For experiments with SUM159 cells, the culture was grown as described previously (Jayson et al., 2018). For seipin rescue experiments, cells were transfected using FuGENE HD transfection reagent (Promega) 1 d before addition of 0.5 mM oleate to induce LD formation. Before imaging at 37°C, cells were washed in medium containing DMEM/F12 without phenol red (Thermo Fisher Scientific) and stained with LipidTox deep red and Hoechst dyes (Thermo Fisher Scientific).
Microscopy
Imaging experiments were performed on Nikon Eclipse Ti inverted microscopes equipped with CSU-X1 or W1 spinning-disk confocal scan heads (Yokogawa), 405-, 488-, and 639-nm laser lines, 20× Plan Apochromat 0.7 NA, 60× Plan Apochromat 1.40 NA, or 100× Apochromat total internal reflection fluorescence 1.4 NA objectives (Nikon), Zyla 4.2 Plus sCMOS, or iXon 897 electron-multiplying charge-coupled device cameras (Andor) and NIS Elements AR software (Nikon) to image EGFP, Alexa Fluor 488, mCherry, LTOX deep red, and Hoechst fluorophores.
In vitro assays
GUVs and in vitro droplets were generated as described previously (Prévost et al., 2018). The phospholipid composition was 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC):1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE):L-a-phosphatidylinositol from bovine liver (liver PI) 65:27:8. For GUV experiments, 0.1 mol 1,2-dioleoylsn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (rhodamine PE) was added to the phospholipid mixture. GUVs were incubated with 5% triolin in buffer (20 mM Tris, pH 7.5, 100 mM NaCl, and 400 mM glucose) for 10 min. All Alexa Fluor 488–labeled peptides used in this study were purchased from Bio-Synthesis and dissolved in DMSO. For binding assays, 1 µM peptide was added to GUVs or LDs and incubated at least 5 min before imaging at 23°C.
Image analysis
Representative microscopy images were adjusted for contrast and converted to 8-bit using FIJI software (ImageJ; National Institutes of Health; Schindelin et al., 2012). Binding of peptides to in vitro–generated LDs was quantified using CellProfiler software (Carpenter et al., 2006). Fluorescence intensity on each LD (ring structure segmented by brightfield images) was quantified measuring integrated fluorescence intensity (illumination corrected and background subtracted) normalized by LD area.
GUV-binding assays were quantified manually using FIJI software. Average maximum-fluorescence intensities on monolayer and bilayer areas of GUVs were quantified (background subtracted) to calculate enrichment on the monolayer.
For seipin rescue experiments, transfected cells were automatically detected in the GFP channel, and LD size and number per cell were measured in LipidTOX channel with a CellProfiler workflow. For untransfected SUM159 WT cells, cell area was segmented using LipidTOX signal.
Localization of mCherry-tagged constructs to LDs in D. melanogaster S2 cells was quantified in CellProfiler by segmenting LDs in the BODIPY channel and measurement of background-subtracted mCherry signal in this region versus total cellular fluorescence.
For tracking of seipin foci, cells were optically sectioned using a spinning-disk microscope to capture most of the thin periphery of the cell, where the ER is organized in a relatively planar network. The images were acquired continuously using 57-ms exposures for 4.9 s total using an electron-multiplying charge-coupled device camera. Prior to the analysis of the seipin foci intensities, the cells were cropped such that the region of interest was limited to the planar ER network. Fluorescence intensities of the diffraction-limited seipin foci were detected and quantified by fitting a 2D Gaussian function using theoretically approximated sigma values for imaging conditions (Aguet et al., 2013). The detected puncta were then tracked over time and subsequently filtered to extract only those events that were tracked for >1.7 s and that did not merge or split with other foci. Using a custom MATLAB script, the maximum fitted amplitude was extracted for each trajectory from datasets derived from seipin-knockout cells expressing WT seipin or seipin Y171A and plotted as the relative and cumulative frequency distributions.
Statistical analysis
Statistical significance of data from in vitro LD binding experiments and in vivo protein localization and rescue experiments using GFP-tagged seipin constructs was determined by a Kruskal-Wallis test for nonnormally distributed data followed by Dunn’s multiple comparisons test. GUV-binding experiments were analyzed by a Mann-Whitney test in Prism 7 (GraphPad Software). For all analyses, P values <0.01 were considered significant.
Online supplemental material
Fig. S1 (A and B) shows seipin expression, purification, and negative-stain EM analyses of purified seipin. Fig. S1 D shows a 2D average of seipin by cryo-EM in digitonin and DDM and demonstrates cryo-EM data processing. Fig. S2 (A–D) shows local resolution of the cryo-EM map, the FSC curve, angular distribution of cryo-EM particles, and selected cryo-EM density superimposed with an atomic model, respectively. Fig. S2 E shows seipin protein sequence alignment from different species. Fig. S3 shows both in vitro and in vivo structure–function tests of seipin. Fig. S3 A shows the structural conservation of seipin. Fig. S3 B displays the connecting density mediating seipin oligomerization, with emphasis on the interaction of R165 with F94 from the adjacent monomer. Fig. S3 (C and G) shows gel-filtration analyses of different seipin mutant forms. Fig. S3 (D–F) shows the cell-based in vivo analysis of selected seipin mutants. Fig. S3 H shows negative staining and 2D average of Y171A mutant. Table S1 summarizes cryo-EM data collection, refinement, and validation statistics.
Supplementary Material
Acknowledgments
We thank Z. Yu and H.-T. Chou for cryo-EM at Howard Hughes Medical Institute Janelia and C. Xu and K. Song for cryo-EM data collection at the University of Massachusetts, S. Upadhyayula for help with image analyses, members of the Liao and Farese Jr./Walther laboratories for discussions, T. Rapoport for reading the manuscript, and G. Howard for editorial assistance.
This work was supported by National Institutes of Health grants 1R01GM123089 (to F. DiMaio), 1R01GM124348-01 (to R.V. Farese Jr.), and 1R01GM097194 (to T.C. Walther). T.C. Walther is an investigator of the Howard Hughes Medical Institute. X. Sui was supported by the American Heart Association postdoctoral fellowship (18POST34030308). H. Arlt was supported by a Deutsche Forschungsgemeinschaft research fellowship (AR1164/1-1).
The authors declare no competing financial interests.
Author contributions: X. Sui, H. Arlt, M. Liao, R.V. Farese Jr., and T.C. Walther conceived the project. X. Sui, H. Arlt, M. Liao, R.V. Farese Jr., and T.C. Walther designed experiments, and X. Sui and H. Arlt performed experiments. F. DiMaio helped with protein de novo structure model building. K.P. Brock and D.S. Marks performed the evolutionary analysis of protein structure. Z.W. Lai performed mass spectrometry analyses of protein samples. X. Sui, H. Arlt, R.V. Farese Jr., and T.C. Walther wrote the manuscript. All authors analyzed and discussed the results and contributed to the manuscript.
References
- Adams P.D., Afonine P.V., Bunkóczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. . 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguet F., Antonescu C.N., Mettlen M., Schmid S.L., and Danuser G.. 2013. Advances in analysis of low signal-to-noise images link dynamin and AP2 to the functions of an endocytic checkpoint. Dev. Cell. 26:279–291. 10.1016/j.devcel.2013.06.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi J., Wang W., Liu Z., Huang X., Jiang Q., Liu G., Wang Y., and Huang X.. 2014. Seipin promotes adipose tissue fat storage through the ER Ca2+-ATPase SERCA. Cell Metab. 19:861–871. 10.1016/j.cmet.2014.03.028 [DOI] [PubMed] [Google Scholar]
- Binns D., Lee S., Hilton C.L., Jiang Q.X., and Goodman J.M.. 2010. Seipin is a discrete homooligomer. Biochemistry. 49:10747–10755. 10.1021/bi1013003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokori-Brown M., Martin T.G., Naylor C.E., Basak A.K., Titball R.W., and Savva C.G.. 2016. Cryo-EM structure of lysenin pore elucidates membrane insertion by an aerolysin family protein. Nat. Commun. 7:11293 10.1038/ncomms11293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth D.S., Avila-Sakar A., and Cheng Y.. 2011. Visualizing proteins and macromolecular complexes by negative stain EM: from grid preparation to image acquisition. J. Vis. Exp. 58:3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. . 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.B., Arendall W.B. III, Headd J.J., Keedy D.A., Immormino R.M., Kapral G.J., Murray L.W., Richardson J.S., and Richardson D.C.. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66:12–21. 10.1107/S0907444909042073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G.E., Hon G., Chandonia J.M., and Brenner S.E.. 2004. WebLogo: a sequence logo generator. Genome Res. 14:1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Fei W., Shui G., Gaeta B., Du X., Kuerschner L., Li P., Brown A.J., Wenk M.R., Parton R.G., and Yang H.. 2008. Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol. 180:473–482. 10.1083/jcb.200711136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W., Shui G., Zhang Y., Krahmer N., Ferguson C., Kapterian T.S., Lin R.C., Dawes I.W., Brown A.J., Li P., et al. . 2011. A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7:e1002201 10.1371/journal.pgen.1002201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenz B., Walls A.C., Egelman E.H., Veesler D., and DiMaio F.. 2017. RosettaES: a sampling strategy enabling automated interpretation of difficult cryo-EM maps. Nat. Methods. 14:797–800. 10.1038/nmeth.4340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier R., Douguet D., Antonny B., and Drin G.. 2008. HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics. 24:2101–2102. 10.1093/bioinformatics/btn392 [DOI] [PubMed] [Google Scholar]
- Goehring A., Lee C.H., Wang K.H., Michel J.C., Claxton D.P., Baconguis I., Althoff T., Fischer S., Garcia K.C., and Gouaux E.. 2014. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 9:2574–2585. 10.1038/nprot.2014.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes K.B., Fernandes A.P., Ferreira A.C., Pardini H., Garg A., Magré J., and Pardini V.C.. 2004. Mutations in the seipin and AGPAT2 genes clustering in consanguineous families with Berardinelli-Seip congenital lipodystrophy from two separate geographical regions of Brazil. J. Clin. Endocrinol. Metab. 89:357–361. 10.1210/jc.2003-030415 [DOI] [PubMed] [Google Scholar]
- Grippa A., Buxó L., Mora G., Funaya C., Idrissi F.Z., Mancuso F., Gomez R., Muntanyà J., Sabidó E., and Carvalho P.. 2015. The seipin complex Fld1/Ldb16 stabilizes ER-lipid droplet contact sites. J. Cell Biol. 211:829–844. 10.1083/jcb.201502070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Binns D.D., Chang Y.F., and Goodman J.M.. 2015. Dissecting seipin function: the localized accumulation of phosphatidic acid at ER/LD junctions in the absence of seipin is suppressed by Sei1p(ΔNterm) only in combination with Ldb16p. BMC Cell Biol. 16:29 10.1186/s12860-015-0075-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W.M., Reese M.L., and Goodman J.M.. 2018. The assembly of lipid droplets and their roles in challenged cells. EMBO J. 37:e98947 10.15252/embj.201898947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölttä-Vuori M., Salo V.T., Ohsaki Y., Suster M.L., and Ikonen E.. 2013. Alleviation of seipinopathy-related ER stress by triglyceride storage. Hum. Mol. Genet. 22:1157–1166. 10.1093/hmg/dds523 [DOI] [PubMed] [Google Scholar]
- Ishida Y., Tsuchiya W., Fujii T., Fujimoto Z., Miyazawa M., Ishibashi J., Matsuyama S., Ishikawa Y., and Yamazaki T.. 2014. Niemann-Pick type C2 protein mediating chemical communication in the worker ant. Proc. Natl. Acad. Sci. USA. 111:3847–3852. 10.1073/pnas.1323928111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayson C.B.K., Arlt H., Fischer A.W., Lai Z.W., Farese R.V., and Walther T.C.. 2018. Rab18 is not necessary for lipid droplet biogenesis or turnover in human mammary carcinoma cells. Mol. Biol. Cell. 29:2045–2054. 10.1091/mbc.E18-05-0282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi A.S., Zhang H., and Prinz W.A.. 2017. Organelle biogenesis in the endoplasmic reticulum. Nat. Cell Biol. 19:876–882. 10.1038/ncb3579 [DOI] [PubMed] [Google Scholar]
- Kawate T., and Gouaux E.. 2006. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 14:673–681. 10.1016/j.str.2006.01.013 [DOI] [PubMed] [Google Scholar]
- Kory N., Thiam A.R., Farese R.V. Jr., and Walther T.C.. 2015. Protein Crowding Is a Determinant of Lipid Droplet Protein Composition. Dev. Cell. 34:351–363. 10.1016/j.devcel.2015.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao M., Cao E., Julius D., and Cheng Y.. 2014. Single particle electron cryo-microscopy of a mammalian ion channel. Curr. Opin. Struct. Biol. 27:1–7. 10.1016/j.sbi.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C., Nordström R., Wagner K., Windpassinger C., Andersson H., von Heijne G., and Nilsson I.. 2006. Membrane topology of the human seipin protein. FEBS Lett. 580:2281–2284. 10.1016/j.febslet.2006.03.040 [DOI] [PubMed] [Google Scholar]
- Magré J., Delépine M., Khallouf E., Gedde-Dahl T. Jr, Van Maldergem L., Sobel E., Papp J., Meier M., Mégarbané A., Bachy A.; BSCL Working Group, et al. . 2001. Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 28:365–370. 10.1038/ng585 [DOI] [PubMed] [Google Scholar]
- Marks D.S., Colwell L.J., Sheridan R., Hopf T.A., Pagnani A., Zecchina R., and Sander C.. 2011. Protein 3D structure computed from evolutionary sequence variation. PLoS One. 6:e28766 10.1371/journal.pone.0028766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks D.S., Hopf T.A., and Sander C.. 2012. Protein structure prediction from sequence variation. Nat. Biotechnol. 30:1072–1080. 10.1038/nbt.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin T.G., Boland A., Fitzpatrick A.W.P., and Scheres S.H.W.. 2016. Graphene Oxide Grid Preparation. 10.6084/m9.figshare.3178669.v1 [DOI]
- Mastronarde D.N. 2005. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152:36–51. 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- Onal G., Kutlu O., Gozuacik D., and Dokmeci Emre S.. 2017. Lipid Droplets in Health and Disease. Lipids Health Dis. 16:128 10.1186/s12944-017-0521-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagac M., Cooper D.E., Qi Y., Lukmantara I.E., Mak H.Y., Wu Z., Tian Y., Liu Z., Lei M., Du X., et al. . 2016. SEIPIN Regulates Lipid Droplet Expansion and Adipocyte Development by Modulating the Activity of Glycerol-3-phosphate Acyltransferase. Cell Reports. 17:1546–1559. 10.1016/j.celrep.2016.10.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pintilie G.D., Zhang J., Goddard T.D., Chiu W., and Gossard D.C.. 2010. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 170:427–438. https://doi.org/ 10.1016/j.jsb.2010.03.007 [DOI] [PMC free article] [PubMed]
- Prévost C., Sharp M.E., Kory N., Lin Q., Voth G.A., Farese R.V. Jr., and Walther T.C.. 2018. Mechanism and Determinants of Amphipathic Helix-Containing Protein Targeting to Lipid Droplets. Dev. Cell. 44:73–86. 10.1016/j.devcel.2017.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohou A., and Grigorieff N.. 2015. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192:216–221. 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ru H., Chambers M.G., Fu T.M., Tong A.B., Liao M., and Wu H.. 2015. Molecular Mechanism of V(D)J Recombination from Synaptic RAG1-RAG2 Complex Structures. Cell. 163:1138–1152. 10.1016/j.cell.2015.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo V.T., Belevich I., Li S., Karhinen L., Vihinen H., Vigouroux C., Magré J., Thiele C., Hölttä-Vuori M., Jokitalo E., and Ikonen E.. 2016. Seipin regulates ER-lipid droplet contacts and cargo delivery. EMBO J. 35:2699–2716. 10.15252/embj.201695170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. 2012a A Bayesian view on cryo-EM structure determination. J. Mol. Biol. 415:406–418. 10.1016/j.jmb.2011.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H. 2012b RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180:519–530. 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim M.F., Dennis R.J., Aubry E.M., Ramanathan N., Sembongi H., Saudek V., Ito D., O’Rahilly S., Siniossoglou S., and Rochford J.J.. 2012. The human lipodystrophy protein seipin is an ER membrane adaptor for the adipogenic PA phosphatase lipin 1. Mol. Metab. 2:38–46. 10.1016/j.molmet.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim M.F., Talukder M.U., Dennis R.J., Edwardson J.M., and Rochford J.J.. 2014. Analyzing the functions and structure of the human lipodystrophy protein seipin. Methods Enzymol. 537:161–175. 10.1016/B978-0-12-411619-1.00009-4 [DOI] [PubMed] [Google Scholar]
- Söding J., Biegert A., and Lupas A.N.. 2005. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33:W244–W248. 10.1093/nar/gki408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski K.M., Binns D., Bartz R., Grishin N.V., Li W.P., Agarwal A.K., Garg A., Anderson R.G., and Goodman J.M.. 2007. The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. USA. 104:20890–20895. 10.1073/pnas.0704154104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukder M.M., Sim M.F., O’Rahilly S., Edwardson J.M., and Rochford J.J.. 2015. Seipin oligomers can interact directly with AGPAT2 and lipin 1, physically scaffolding critical regulators of adipogenesis. Mol. Metab. 4:199–209. 10.1016/j.molmet.2014.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.C., Chung J., and Farese R.V. Jr. 2017. Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Biol. 33:491–510. 10.1146/annurev-cellbio-100616-060608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.W., Miao Y.H., and Chang Y.S.. 2014. Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J. Cell Sci. 127:1214–1228. 10.1242/jcs.137737 [DOI] [PubMed] [Google Scholar]
- Wang H., Becuwe M., Housden B.E., Chitraju C., Porras A.J., Graham M.M., Liu X.N., Thiam A.R., Savage D.B., Agarwal A.K., et al. . 2016. Seipin is required for converting nascent to mature lipid droplets. eLife. 5:e16582 10.7554/eLife.16582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.L., Motamed M., Infante R.E., Abi-Mosleh L., Kwon H.J., Brown M.S., and Goldstein J.L.. 2010. Identification of surface residues on Niemann-Pick C2 essential for hydrophobic handoff of cholesterol to NPC1 in lysosomes. Cell Metab. 12:166–173. 10.1016/j.cmet.2010.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R.Y., Kudryashev M., Li X., Egelman E.H., Basler M., Cheng Y., Baker D., and DiMaio F.. 2015. De novo protein structure determination from near-atomic-resolution cryo-EM maps. Nat. Methods. 12:335–338. 10.1038/nmeth.3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolinski H., Kolb D., Hermann S., Koning R.I., and Kohlwein S.D.. 2011. A role for seipin in lipid droplet dynamics and inheritance in yeast. J. Cell Sci. 124:3894–3904. 10.1242/jcs.091454 [DOI] [PubMed] [Google Scholar]
- Wolinski H., Hofbauer H.F., Hellauer K., Cristobal-Sarramian A., Kolb D., Radulovic M., Knittelfelder O.L., Rechberger G.N., and Kohlwein S.D.. 2015. Seipin is involved in the regulation of phosphatidic acid metabolism at a subdomain of the nuclear envelope in yeast. Biochim. Biophys. Acta. 1851:1450–1464. 10.1016/j.bbalip.2015.08.003 [DOI] [PubMed] [Google Scholar]
- Xu S., Benoff B., Liou H.L., Lobel P., and Stock A.M.. 2007. Structural basis of sterol binding by NPC2, a lysosomal protein deficient in Niemann-Pick type C2 disease. J. Biol. Chem. 282:23525–23531. 10.1074/jbc.M703848200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S.Q., Palovcak E., Armache J.P., Verba K.A., Cheng Y., and Agard D.A.. 2017. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 14:331–332. 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density map of seipin has been deposited in the Electron Microscopy Data Bank under accession no. EMD-9146. Atomic coordinates for the atomic models has been deposited in the Protein Data Bank under accession no. 6MLU.