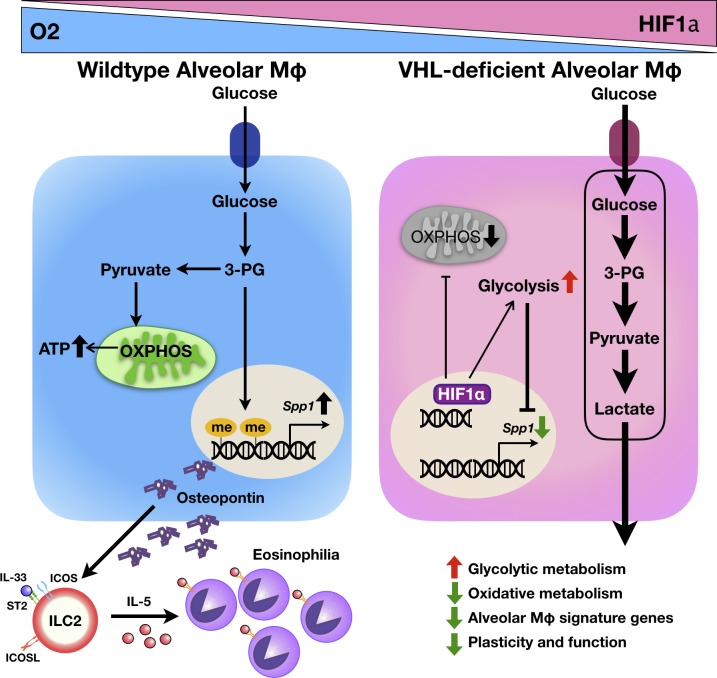
Zhang et al. report an essential role of the E3 ligase VHL in regulating the metabolic fitness and effector function of alveolar macrophages to prime ILC2 activation through osteopontin during pulmonary type 2 inflammation and fibrosis.
Abstract
Metabolic pathways such as glycolysis or oxidative phosphorylation play a key role in regulating macrophage function during inflammation and tissue repair. However, how exactly the VHL–HIF–glycolysis axis is involved in the function of tissue-resident macrophages remains unclear. Here we demonstrate that loss of VHL in myeloid cells resulted in attenuated pulmonary type 2 and fibrotic responses, accompanied by reduced eosinophil infiltration, decreased IL-5 and IL-13 concentrations, and ameliorated fiber deposition upon challenge. VHL deficiency uplifted glycolytic metabolism, decreased respiratory capacity, and reduced osteopontin expression in alveolar macrophages, which impaired the function of type 2 innate lymphoid cells but was significantly reversed by HIF1α inhibition or ablation. The up-regulated glycolysis altered the epigenetic modification of osteopontin gene, with the metabolic intermediate 3-phosphoglyceric acid as a key checkpoint controller. Thus, our results indicate that VHL acts as a crucial regulatory factor in lung inflammation and fibrosis by regulating alveolar macrophages.
Graphical Abstract
Introduction
Type 2 immune responses are critical in driving lung inflammation, which involves both innate and adaptive cell types including the early cytokine production of IL-33, IL-25, and thymic stromal lymphopoietin by lung epithelial cells; the induction and activation of type 2 innate lymphoid cells (ILC2s) and antigen-presenting cells; and the subsequent T helper type 2 (Th2) cell differentiation and the production of more Th2 cytokines, which further causes the recruitment, infiltration, proliferation, and activation of other effector cells and subsequent lung inflammation (Hammad and Lambrecht, 2015). Chronic type 2 cytokine production is also linked to the development of tissue remodeling, regeneration, and even pathological fibrosis (Eming et al., 2017). Macrophages are critically involved in these processes to maintain tissue integrity (Wynn and Vannella, 2016). In the lung, alveolar macrophages (AMs) are uniquely positioned in the lumens of airway spaces in close contact with lung epithelium and are involved in cellular debris clearance and pathological situations such as pathogen invasion or tissue damage (Hussell and Bell, 2014). However, the detailed mechanisms of their function and regulation are far from clear.
Pulmonary inflammation and injury result in hypoxic responses and the accumulation of hypoxia inducible factors (HIFs) including HIF1α and HIF2α, which in turn drives chronic inflammatory and profibrotic responses (Darby and Hewitson, 2016). Under normoxic conditions, HIF1α is kept at a low level via the hydroxylation by prolyl hydroxylase domain enzymes and the recognition and ubiquitination by the E3 ligase von Hippel–Lindau protein (VHL), followed by proteasomal degradation (Semenza, 2011). Hypoxia reduces the activity of prolyl hydroxylase domain enzymes, which leads to the stabilization of HIFs and the initiation of HIF-dependent transcriptional program. Previous studies have documented the critical roles of HIFs in the immune regulation (Palazon et al., 2014): for example, HIF1α is essential for myeloid cell–mediated inflammation (Cramer et al., 2003); HIF1α and its pharmaceutical inducers positively regulate innate antibacterial defense responses (Peyssonnaux et al., 2005). On the other hand, hypoxic signaling pathway induces pathogenic fibrosis by promoting vascular remodeling, extracellular matrix turnover, and epithelial to mesenchymal transition through HIF-dependent mechanisms (Higgins et al., 2007; Tzouvelekis et al., 2007; Goodwin et al., 2018). However, how VHL–HIF pathway is involved in the regulation of lung inflammation remains elusive.
Emerging evidence suggests that inflammation-inflicted metabolic changes impact cellular signaling pathways or epigenetic modifications, in addition to the classical roles of metabolism in ATP production and biosynthesis (Phan et al., 2017). In macrophages, LPS-induced conversion to glycolytic metabolism is linked to myeloid cell infiltration and proinflammatory responses (Cramer et al., 2003; Palsson-McDermott et al., 2015), whereas type 2 cytokines IL-4 and IL-13 induce the transition toward oxidative phosphorylation, which is involved in the production of tissue repair factors (Jha et al., 2015; Huang et al., 2016). Several metabolic enzymes or intermediates are recently shown to act as key regulators in macrophages. For example, succinate and its dehydrogenase are critical for LPS-stimulated IL-1β production via promoting ROS generation and stabilizing HIF1α (Tannahill et al., 2013; Mills et al., 2016), whereas the glycolytic product lactate or its derivative induces anti-inflammatory repair/fibrotic responses dependent on IL-4 receptor signaling and HIF1α (Colegio et al., 2014; Minutti et al., 2017). Apparently, the exact roles of VHL–HIF–glycolysis axis in macrophage-driven inflammation and fibrosis in a tissue-specific context need to be investigated.
To address these questions, we generated mice with myeloid cell–restricted deletion of VHL and examined pulmonary type 2 and fibrotic responses upon papain and bleomycin challenge. Unexpectedly, loss of VHL ameliorated lung inflammation and fibrosis, accompanied by significant reduction in ILC2 activation and eosinophil infiltration. The HIF1α-driven glycolysis in AMs inhibited the transcription of osteopontin through epigenetic modification. Our work thus sheds light on the VHL–HIF1α–glycolysis axis in controlling the function of AMs and the development of type 2 inflammation.
Results
VHL deficiency dampens protease-elicited airway inflammation
To explore the role of VHL in tissue-resident macrophages, we generated Vhlfl/fl LysM-Cre mice and detected comparable frequencies and patterns of F4/80+ CD11b+ populations in the spleen, colon, and bone marrow between control and knockout mice, whereas mature AMs, characterized as CD11blo F4/80+ cells in WT mice, showed foamy CD11bhi F4/80hi phenotype in the bronchoalveolar lavage (BAL) fluid and lungs of Vhlfl/fl LysM-Cre mice (Fig. S1, A–C). There was no spontaneous pulmonary inflammation, and cell numbers of myeloid subsets were equivalent between control and knockout mice at steady state (Fig. S1, C and D). Cre-mediated recombination in the Vhl locus indeed resulted in much less Vhl mRNA expression in AMs of Vhlfl/fl LysM-Cre mice (Fig. S1 E).
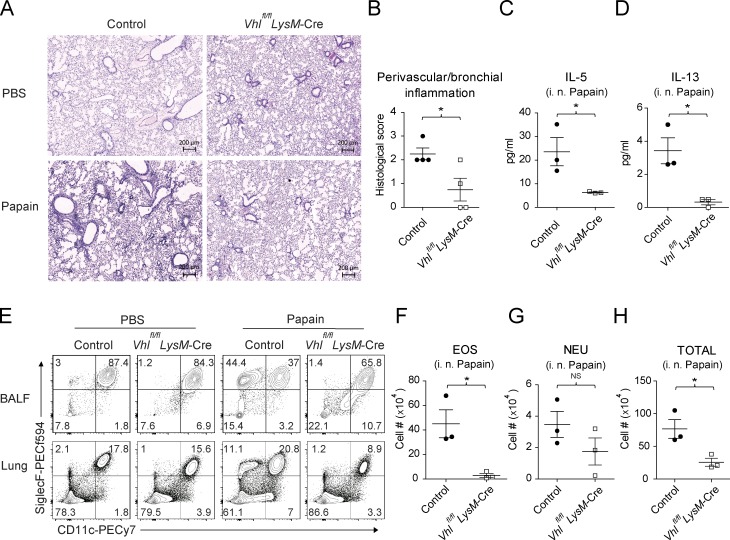
To determine whether VHL deletion in myeloid cells affected lung inflammation, we adopted a papain-induced airway inflammation model. Compared with control mice, Vhlfl/fl LysM-Cre mice developed much less perivascular and bronchial inflammation upon papain challenge (Fig. 1, A and B). Inhalation of papain resulted in dose-dependent development of airway inflammation accompanied by increased concentrations of IL-5, IL-13, and TGFβ1 in the BAL fluid of control mice, which was not observed in Vhlfl/fl LysM-Cre mice (Fig. 1, C and D; and Fig. S1 F). Additionally, VHL depletion led to significantly impaired airway eosinophilia (Fig. 1, E and F), with comparable neutrophils but much less total number of leukocytes (Fig. 1, G and H), suggesting a positive role that VHL played in the eosinophilic airway inflammation. Notably, the concentrations of other proinflammatory cytokines including IL-33 and IL-6 were comparable between VHL-deficient mice and littermate controls (Fig. S1, G and H). While VHL-sufficient mice developed lung eosinophilia upon papain challenge, VHL-deficient mice failed to mount type 2 immune responses in the pulmonary (Fig. S1 D). Overall, these data indicated that myeloid VHL positively regulated papain-induced airway inflammation.
Figure 1.
VHL deficiency attenuates papain-induced lung inflammation. (A) Representative H&E staining of lung sections of control and Vhlfl/fl LysM-Cre mice challenged with PBS or papain. Bar, 200 µm. (B) Perivascular and peribronchial inflammation scored from stained lung sections. (C and D) Concentrations of IL-5 and IL-13 in the BAL fluid. (E) Flow cytometry of CD45+ cells in BAL fluid and lungs. BALF, BAL fluid. (F–H) Absolute numbers of CD11b+ CD11c− SiglecF+ eosinophils (EOS), CD11b+ Ly6C− Ly6G+ neutrophils (NEU), and total CD45+ leukocytes in the BAL fluid. *, P < 0.05 (Student’s t test). Data are from one experiment representative of three independent experiments (mean and SEM of three to six mice per group).
VHL deficiency curtails bleomycin-induced pulmonary fibrosis
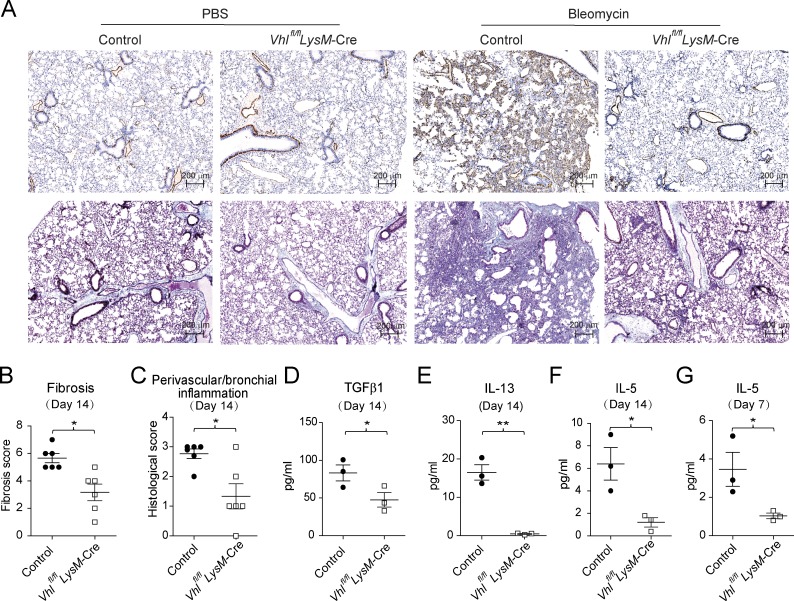
To investigate a potential role of VHL in tissue remodeling, we adopted a bleomycin-induced pulmonary fibrosis model. VHL deficiency protected mice from weight loss during the inflammation phase (data not shown) and led to reduced α-smooth muscle actin and collagen deposition, as well as fibrotic masses during the fibrotic phase (Fig. 2, A and B). In addition, perivascular or bronchial inflammation was attenuated in Vhlfl/fl LysM-Cre mice compared with littermate controls, together with decreased production of profibrotic cytokines TGFβ1 and IL-13 (Fig. 2, C–E). We also found that IL-5 levels and eosinophil infiltration were significantly lower in VHL-deficient mice compared with the control mice (Fig. 2, F and G; and Fig. S2, A and B). Furthermore, VHL-deficient AMs failed to exhibit enhanced proliferation during papain-induced lung inflammation and bleomycin-induced fibrosis progression, resulting in fewer cell numbers relative to control mice (Fig. S1 D and Fig. S2, B and C). Notably, Vhlfl/fl LysM-Cre mice showed normal T cell activation (Fig. S2, D and E) and T cell subset differentiation including FoxP3+ Treg, IFNγ+ Th1, and IL-4+ Th2 cells (Fig. S2 F). Altogether, these data demonstrated that VHL expression in myeloid cells was required for bleomycin-induced pulmonary inflammation and fibrosis, probably via regulation of type 2 innate immune responses.
Figure 2.
Vhlfl/fl LysM-Cre mice are protected from bleomycin-induced pulmonary fibrosis. (A) Lung sections of control and Vhlfl/fl LysM-Cre mice challenged with PBS or bleomycin (5 mg/kg), stained with α-SMA (top) or Masson's blue (bottom). Bar, 200 µm. (B and C) Fibrosis and perivascular and peribronchial inflammation scored from stained lung sections. (D and E) Concentrations of TGFβ1 and IL-13 in the BAL fluid. (F and G) Concentration of IL-5 in the BAL fluid on days 14 and 7. *, P < 0.05; **, P < 0.01 (Student’s t test). Data are from one experiment representative of three independent experiments (mean and SEM of three to six mice per group).
Loss of myeloid VHL affects ILC2 accumulation
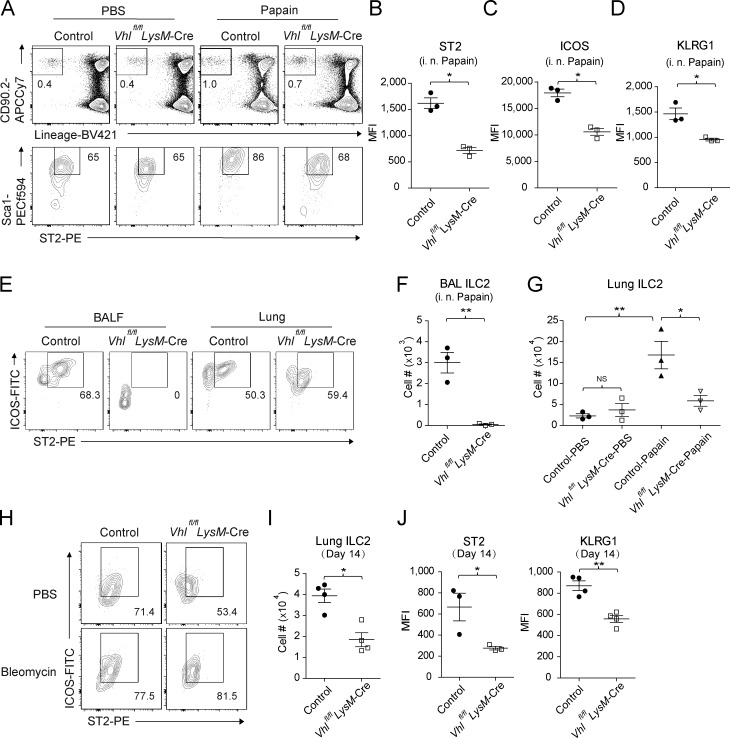
ILC2s are considered as important initiators for type 2 immunity and wound healing (Zaiss et al., 2015). Therefore, we analyzed ILC2s by measuring the expression of surface markers including CD90.2 (Thy1.2), the IL-33 receptor chain T1/ST2, ICOS, and KLRG1. Papain exposure resulted in increased frequencies of lung ILC2s in the control mice, which was severely diminished in VHL-deficient mice (Fig. 3 A), with weakened expression of activation markers ST2, ICOS, and KLRG1 (Fig. 3, B–D); reduced BAL ILC2 infiltration and total lung ILC2 numbers (Fig. 3, E–G); and compromised IL-5 and IL-13 cytokine productions in the lung of Vhlfl/fl LysM-Cre mice (Fig. S3, A and B). Consistently, decreased ILC2 cell numbers were also detected during the early fibrotic phase after bleomycin challenge in Vhlfl/fl LysM-Cre mice, along with reduced ST2 and KLRG1 expression compared with littermate controls (Fig. 3, H–J). To rule out that other myeloid cells contributed to the resistance of VHL-deficient mice to papain, we selectively depleted AMs in WT mice before papain challenge using clodronate liposomes (clophosomes). AM depletion resulted in impaired airway eosinophilia and defective ILC2 activation, resembling the phenotype observed in Vhlfl/fl LysM-Cre mice (Fig. S3, C–G). These findings supported an essential role that AMs played in promoting inflammatory and fibrotic responses, probably by priming lung ILC2 activation and effector function.
Figure 3.
Defective ILC2 function in Vhlfl/fl LysM-Cre mice. (A) Flow cytometry of CD45+ cells in lungs of control and Vhlfl/fl LysM-Cre mice. Numbers adjacent to outlined areas indicate percentages of Lineage− CD90.2+ or Lineage− CD90.2+ ST2+ Sca1+ subsets. (B–D) Mean fluorescence intensity (MFI) of ST2, ICOS, and KLRG1 of ILC2s. (E) Flow cytometry of BAL and lung ILC2s, gated on CD45+ Lineage− CD90.2+. BALF, BAL fluid. (F) Absolute numbers of BAL ILC2s. (G) Absolute numbers of lung ILC2s. (H and I) Flow cytometry and absolute numbers of lung ILC2s from mice challenged with PBS and bleomycin on day 14. (J) MFI of ST2 and KLRG1 of ILC2s. *, P < 0.05; **, P < 0.01 (Student’s t test). Data are from one experiment representative of three independent experiments (mean and SEM of three to six mice per group).
Macrophages prime ILC2 through an osteopontin-dependent mechanism
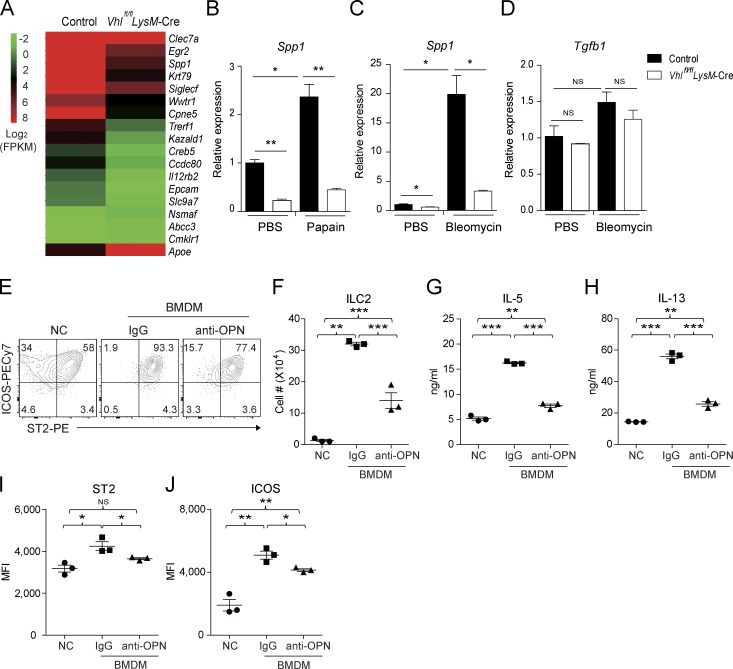
We performed mRNA sequencing analysis which revealed >100 immune-related genes that altered significantly upon deletion of VHL in AMs (Fig. 4 A) with a few genes that were previously reported as AM signature genes (Gautier et al., 2012). Among these genes, we noticed that osteopontin-encoding Spp1 expression was strongly induced in AMs of control mice both upon papain and bleomycin challenge, whereas VHL deficiency effectively blocked Spp1 but not Tgfb1 transcription (Fig. 4, B–D). Since osteopontin plays a positive role in ST2+ Th2 cell–mediated responses in allergic diseases (Xanthou et al., 2007), we hypothesized that loss of AM-derived osteopontin resulted in impaired ILC2 function in Vhlfl/fl LysM-Cre mice. We assessed the role of macrophage-derived osteopontin in regulating ILC2 by adopting an in vitro macrophage and ILC2 coculture system. Notably, macrophages significantly promoted ILC2 proliferation, activation, and cytokine production in an osteopontin-dependent manner in vitro (Fig. 4, E–J). In addition, intranasal challenge of Rag1−/− mice with recombinant osteopontin boosted lung ILC2 expansion in vivo (Fig. S4 A). Mechanistically, osteopontin promoted bone marrow ILC2 proliferation in the presence of IL-33 and IL-7 in vitro (Fig. S4 B). Osteopontin treatment resulted in increased AKT phosphorylation in ILC2s (Fig. S4 C), consistent with a previous observation in Ba/F3 cells (Lin and Yang-Yen, 2001). Collectively, these data demonstrated that osteopontin potentiated ILC2 proliferation and activation in vivo and in vitro.
Figure 4.
Macrophages potentiate ILC2s through osteopontin. (A) Expression of selected signature genes in AMs sorted from control and Vhlfl/fl LysM-Cre mice. FPKM, fragments per kilobase of exon per million reads mapped. (B) Expression of Spp1 mRNA in AMs sorted from mice challenged with PBS or papain. (C and D) Expression of Spp1 and Tgfb1 mRNA in AMs sorted from mice challenged with PBS or bleomycin at day 7. (E and F) Flow cytometry and absolute numbers of sorted bone marrow ILC2s cultured without (NC) or with BMDM ± osteopontin (OPN) antibody for 4 d. (G and H) Concentrations of IL-5 and IL-13 in supernatants of cells cultured as in E. (I and J) MFI of ST2 and ICOS of ILC2 cells cultured as in E. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). Data are from one experiment representative of three independent experiments (mean and SEM of cells pooled from at least three mice per group).
VHL regulates glycolytic metabolism in AMs via HIF1α
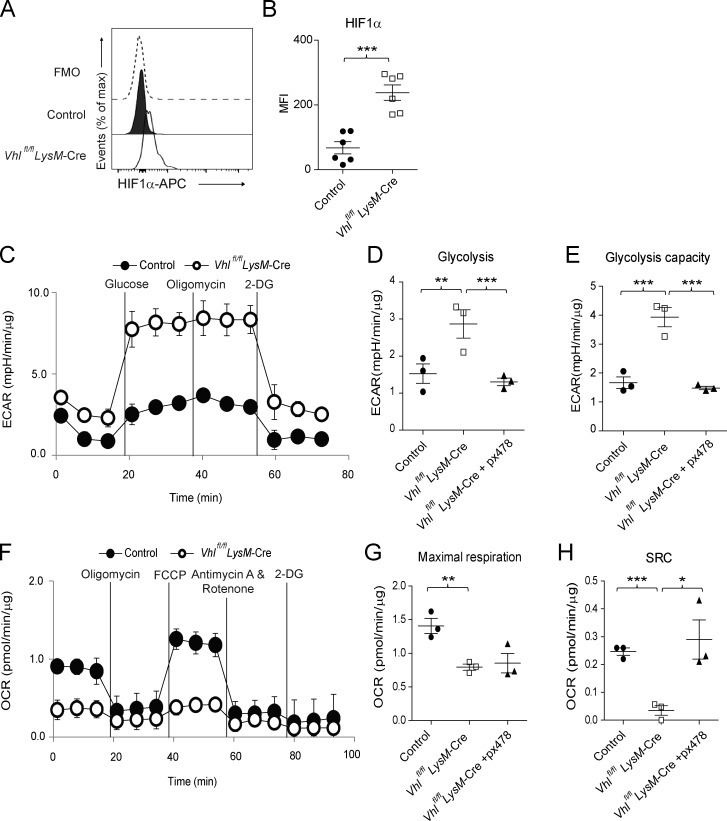
Since VHL is a well-established E3 ubiquitin ligase for HIFs (Semenza, 2011), we detected HIF1α stabilization in VHL-deficient AMs (Fig. 5, A and B). VHL-deficient AMs also exhibited increased glycolytic enzyme transcription and abnormal cell morphology, which was reversed by the genetic deletion of HIF1α (Fig. S5, A and B). We then analyzed glycolytic and mitochondrial function in AMs. Depletion of VHL in AMs led to elevated glycolysis and glycolytic capacity (Fig. 5, C–E) but reduced oxidative phosphorylation and spare respiratory capacity (SRC) compared with WT controls (Fig. 5, F–H). Importantly, pharmaceutical inhibition of HIF1α by px478 rectified the excessive glycolysis and diminished SRC (Fig. 5, D, E, and H). These results indicated that the E3 ligase VHL controlled AM metabolism through the regulation of HIF1α-driven glycolysis.
Figure 5.
VHL regulates glycolytic metabolism in AMs. (A and B) HIF1α expression in CD45+ CD11c+ SiglecF+ AMs from control and Vhlfl/fl LysM-Cre mice. FMO, fluorescence minus one. (C) Real-time ECAR of sorted AMs, followed by sequential treatment (black vertical lines) with glucose, oligomycin, and 2-deoxy-glucose (2-DG). (D and E) Maximal ECAR in AMs stimulated without or with px478 for 24 h. (F) Real-time OCR of sorted AMs, followed by sequential treatment (black vertical lines) with oligomycin, FCCP, antimycin A and rotenone, and 2-DG. (G and H) Maximal OCR and SRC in AMs stimulated without or with px478 for 24 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). Data are from one experiment representative of three independent experiments (A and B, mean and SEM of three to six mice per group; C–H, mean and SEM of cells pooled from at least two mice per group).
VHL promotes osteopontin expression by epigenetic modification
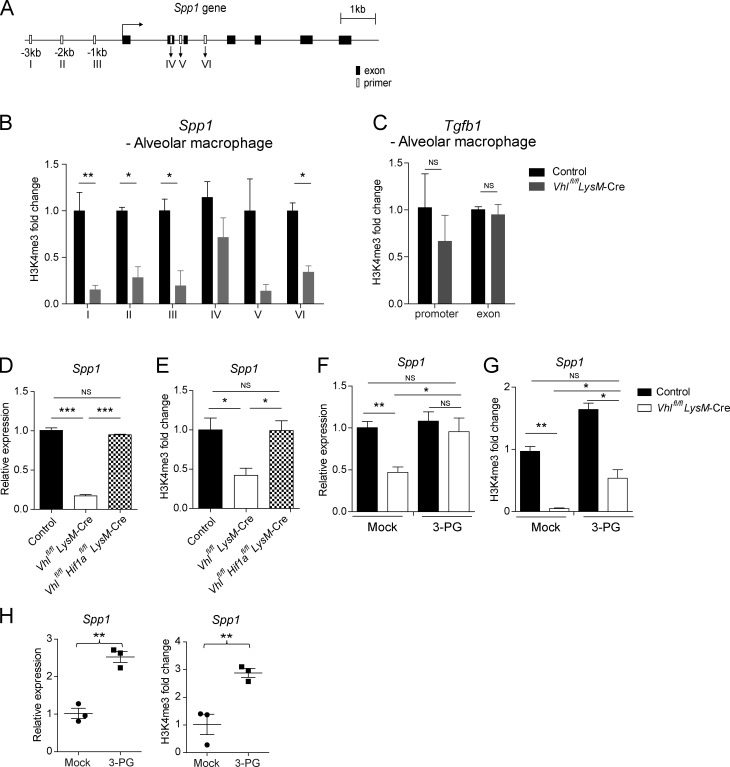
To investigate the molecular mechanisms by which elevated glycolysis altered Spp1 gene expression, we performed chromatin immunoprecipitation assay to determine transcriptional activity across Spp1 gene locus. Notably, we detected significantly decreased histone H3 trimethylation at lysine 4 residue (H3K4me3) at the promoter and putative H3K4me3 sites of Spp1 gene in VHL-deficient AMs compared with WT controls, with no difference being observed on the Tgfb1 gene locus (Fig. 6, A–C). Genetic depletion of HIF1α rescued the suppressed Spp1 expression by the absence of VHL (Fig. 6, D and E).
Figure 6.
Epigenetic regulation of osteopontin expression by VHL. (A) Schematics of Spp1 gene with putative histone H3K4me3 binding sites (empty boxes) and exons (solid boxes). (B and C) Binding of H3K4me3 to the Spp1 gene and the promoter and exon regions of Tgfb1 gene in sorted AMs from control or Vhlfl/fl LysM-Cre mice. Relative binding activity was normalized by binding in control cells. (D and E) Spp1 mRNA expression and H3K4me3 modification in BMDMs. (F and G) Spp1 mRNA expression and H3K4me3 modification in BMDMs treated without or with 3-PG for 24 h. (H) Spp1 expression and bindings of H3K4me3 on the Spp1 gene promoter in IL-4–stimulated BMDMs in the absence or presence of 3-PG for 24 h. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). Data represent the mean and SEM from three independent experiments (B–H, mean and SEM of cells pooled from at least two mice per group).
Next, we used a collection of glycolytic enzyme inhibitors and glucose metabolites to screen for potential regulators of Spp1 expression in macrophages in vitro (Fig. S5, C–E). Of note, the pyruvate kinase isoform 2 (PKM2) inhibitor oxalate significantly up-regulated Spp1 expression in both control and VHL-deficient macrophages (Fig. S5, F and G). Moreover, knocking down the glycolytic enzyme PKM2 but not TCA cycle–related enzymes in bone marrow–derived macrophages (BMDMs) resulted in elevated Spp1 expression (Fig. S5 H). Markedly, treatment with the glycolytic metabolite 3-phosphoglyceric acid (3-PG) enhanced Spp1 transcription in VHL-deficient macrophages, and the reduced H3K4me3 modification was also reversed by 3-PG treatment (Fig. 6, F and G). Furthermore, 3-PG significantly augmented gene expression as well as H3K4me3 deposition of Spp1 in IL-4–stimulated macrophages (Fig. 6 H). These results suggested that the glycolytic intermediate 3-PG functioned as a key metabolite involved in AM epigenetic regulation.
HIF1α deficiency rectifies defective ILC2 responses in VHL-deficient mice
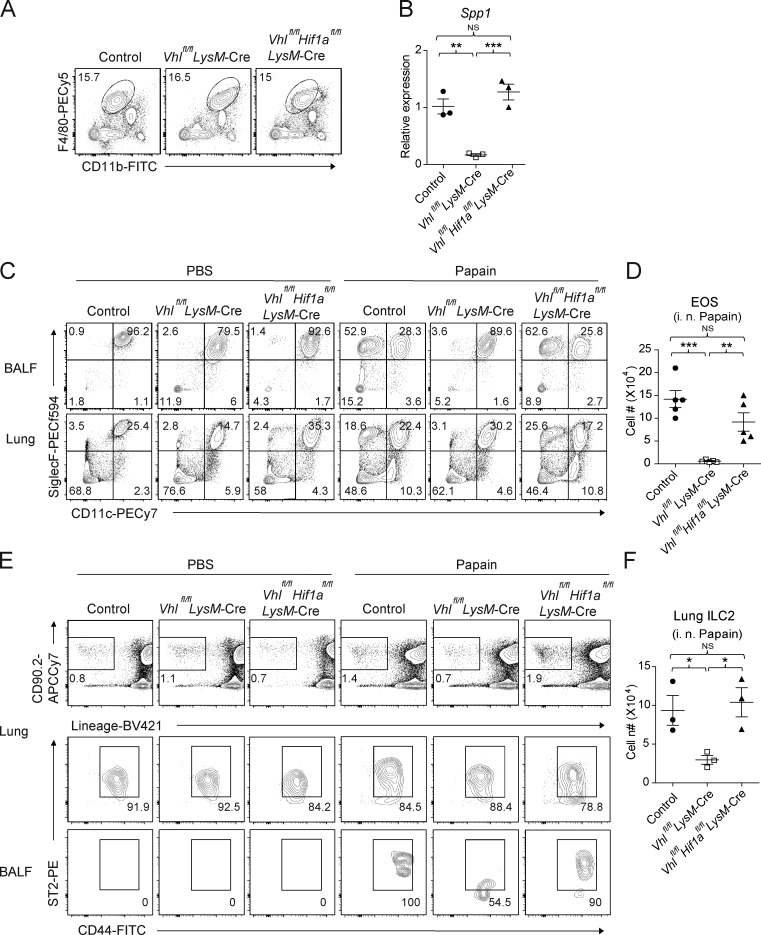
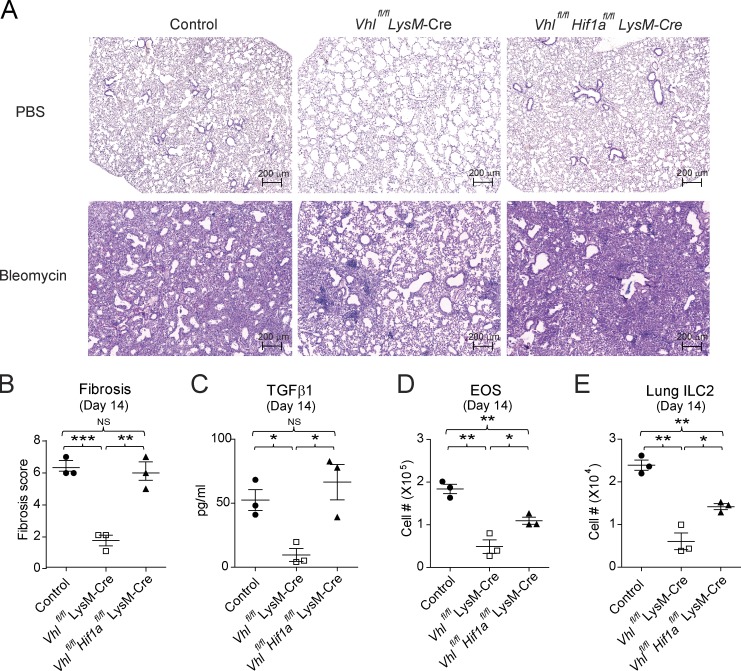
Since HIF1α was up-regulated in VHL-deficient AMs, we next investigated whether the defective ILC2-mediated immune responses in Vhlfl/fl LysM-Cre mice could be restored by genetic ablation of HIF1α. Analysis of Vhlfl/fl Hif1afl/fl LysM-Cre double knockout (DKO) mice revealed that HIF1α and VHL double deficiency restored the abnormalities and Spp1 expression in AMs from VHL-deficient mice (Fig. 7, A and B). The pulmonary eosinophilia in DKO mice was increased to that of their WT control following intranasal challenge of papain (Fig. 7, C and D). In addition, papain inhalation resulted in BAL ILC2 infiltration and lung ILC2 proliferation in Vhlfl/fl Hif1afl/fl LysM-Cre mice but not Vhlfl/fl LysM-Cre mice (Fig. 7, E and F). Moreover, in contrast to VHL-deficient mice, bleomycin-injured DKO mice developed similar pulmonary fibrotic phenotype to WT control mice, with partial but significant rescue of eosinophil and ILC2 accumulation (Fig. 8). These data further demonstrated a critical role of VHL–HIF1α axis in the regulation of AM function in pulmonary type 2 and fibrotic responses.
Figure 7.
HIF1α mediates VHL-regulated type 2 responses. (A) Expression of F4/80 and CD11b among CD45+ cells in the lungs of control, Vhlfl/fl LysM-Cre, and Vhlfl/fl Hif1afl/fl LysM-Cre mice. (B) Expression of Spp1 mRNA in AMs. (C) Flow cytometry of CD45+ cells in BAL fluid and lungs of the indicated mouse strains challenged with PBS or papain. (D) Absolute numbers of CD11b+ CD11c− SiglecF+ eosinophils (EOS) in BAL fluid. (E) Flow cytometry of CD45+ cells in lungs and BAL fluid. Numbers adjacent to outlined areas indicate percentages of Lineage− CD90.2+ or Lineage− CD90.2+ ST2+ CD44+ subsets. BALF, BAL fluid. (F) Absolute numbers of lung ILC2s. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). Data are from one experiment representative of three independent experiments (mean and SEM of three to six mice per group).
Figure 8.
HIF1α deficiency restores pulmonary fibrosis in Vhlfl/fl LysM-Cre mice. (A) Representative H&E staining of lung sections of control, Vhlfl/fl LysM-Cre, and Vhlfl/fl Hif1afl/fl LysM-Cre mice challenged with PBS or 5 mg/kg bleomycin. Bar, 200 µm. (B) Fibrosis scored from stained lung sections. (C) Concentrations of TGFβ1 in the BAL fluid. (D and E) Absolute numbers of eosinophils and lung ILC2s. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (Student’s t test). Data are from one experiment representative of three independent experiments (mean and SEM of three to six mice per group).
Discussion
The degree and duration of tissue injury–elicited inflammatory responses dictate the final outcome of effective tissue repair, dysregulation of which can lead to excessive tissue damage or pathological fibrosis. Hypoxia has been linked to pulmonary fibrosis progression through induction of profibrotic lung macrophages (Ueno et al., 2011; Philip et al., 2017). In addition, HIF1α-mediated glycolytic reprogramming promotes myofibroblast differentiation and fibrotic progression (Goodwin et al., 2018). In contrast to previous observations, our study demonstrated that loss of VHL in myeloid lineages resulted in attenuated inflammatory and fibrotic responses via controlling AMs, which are master regulators of lung immune homeostasis (Hussell and Bell, 2014). We have shown that absence of VHL caused glycolytic upshift, impaired mitochondrial respiration capacity, and altered signature gene expression of AMs in a HIF1α-dependent manner. Interestingly, one recent study has shown that VHL plays a critical role in AM terminal differentiation, self-renews, and functions at steady state by using the Vhlfl/fl CD11c-Cre mice (Izquierdo et al., 2018). Consistently, we detected foamy AM morphology, abnormal terminal differentiation markers, and impaired proliferation during inflammation upon loss of VHL. Moreover, we have also shown that VHL was required for macrophage and ILC2 crosstalk through regulating osteopontin expression in AMs via epigenetic modification. Our study thus offers a new perspective for how type 2 responses to inhaled irritants are regulated, which may have implications to use AMs as potential target cells in intervening pulmonary inflammatory and fibrotic diseases.
During allergic inflammation or helminth infection, ILC2s are the initial source of IL-5, IL-13, and amphiregulin, which serve as the dominant mediators of tissue inflammation, repair, and fibrosis (Zaiss et al., 2015). Upon allergen exposure, lung ILC2s drive Arginase 1+ macrophage accumulation to promote wound healing and tissue fibrosis (Van Dyken et al., 2014; Wynn and Vannella, 2016). These studies, among others, highlight macrophages as effector cells downstream of ILC2s and Th2 cells via type 2 cytokines. Here we instead identified ILC2s as effector cells for AMs, probably through osteopontin production. Osteopontin is involved in the pathogenesis of asthma and pulmonary fibrosis, and its expression positively correlates with disease severity in asthmatic patients (Pardo et al., 2005; Konno et al., 2011). Our data suggested that osteopontin synergized with IL-33 to activate PI3K/Akt signaling and further promoted the proliferation and cytokine production of ILC2s. Hence, our results highlight that osteopontin mediates the crosstalk between AMs and ILC2s as a tissue-specific modulator for the progression of type 2 inflammation and fibrosis in the injured lung.
Metabolic reprogramming plays an essential role in macrophage activation and function (O’Neill and Pearce, 2016), and oxidative phosphorylation is linked to type 2 responses and IL-4–induced alternative activation of macrophages during wound healing and resistance to helminth (Eming et al., 2017). At hypoxic inflammation sites, changes in macrophage metabolism affect their cellular functions such as phagocytosis, motility and aggregation, as well as proinflammatory cytokine production (Palazon et al., 2014). Here we found that the E3 ligase VHL regulated the transcription of Spp1 through repression of HIF1α–mediated glycolytic metabolism in AMs. Spp1 has been recognized as a hypoxia responsive gene in tumor cells and an ICOS-inducible gene in follicular Th cells (Raja et al., 2014; Leavenworth et al., 2015), yet its cell type–specific transcriptional regulation is incompletely understood. Our data demonstrated that AMs mainly adopted oxidative phosphorylation at steady state and that VHL inactivation resulted in metabolic shift toward glycolysis, which was reversed by pharmaceutical inhibition or deletion of HIF1α. In addition, knockdown of the pyruvate kinase isoform PKM2, a critical glycolytic enzyme, up-regulated Spp1 transcription in macrophages. Therefore, we speculate that the accelerated glycolysis leads to transcriptional repression of the Spp1 gene in macrophages, most likely via epigenetic modification.
Importantly, we identified the glycolytic metabolite 3-PG as a checkpoint controller for osteopontin expression in macrophages. Recently, 3-PG was identified as an M2 (IL-4) related metabolite during macrophage polarization (Jha et al., 2015). Here we showed that the decreased H3K4me3 modification of Spp1 in VHL-deficient macrophages could be restored, to a significant degree, by the addition of 3-PG. In yeast, 3-PG acts as a metabolic checkpoint for the formation of a multicomponent enzyme complex including serine metabolic enzymes and PKM2 homologue, which senses both serine metabolism and glycolysis and regulates H3K4 methylation and histone phosphorylation (Li et al., 2015). Indeed, we observed that the PKM2 inhibitor oxalate could also reverse the decreased Spp1 H3K4me3 modification in VHL-deficient macrophages. We thus speculate that 3-PG controls the epigenetic modification of Spp1 via both serine biosynthetic and glycolytic pathways, the bifurcation of 3-PG metabolism. Alternatively, 3-PG modifies Spp1 methylation via serving as a source of serine-mediated one-carbon metabolism, which has diverse roles from biosynthesis to methylation reactions (Locasale, 2013), whereas the effect of PKM2 on osteopontin expression could be due to the role of PKM2 as a coactivator of HIF1α in a positive-feedback loop (Luo et al., 2011). Obviously, further investigation of the detailed biochemical processes by 3-PG is needed to understand the underlying molecular mechanisms, which may reveal new therapeutic targets for treating pulmonary distress such as allergy and fibrosis via metabolic manipulation.
Materials and methods
Mice
Mice used in this study were on a C57BL/6 background. Vhlfl/fl, LysM-Cre, and Rag1−/− mice were purchased from The Jackson Laboratory. Hif1afl/fl mice were a kind gift from Dr. Wenwen Zeng (Tsinghua University, Beijing, China). Vhlfl/fl mice were crossed with LysM-Cre mice to generate Vhlfl/fl LysM-Cre mice. Hif1afl/fl mice were crossed with Vhlfl/fl LysM-Cre mice to generate Vhlfl/fl Hif1afl/fl LysM-Cre mice. Animals were cohoused after weaning, and littermate controls at age 6–8 wk were used for all studies. In individual experiments, all animals of both male and female were age-matched, and exact numbers of animals used per experiment were indicated in figure legends. All mice were maintained in the animal facility under an animal protocol approved by the Institutional Animal Care and Use Committee at Tsinghua University.
Models of lung inflammation and pulmonary fibrosis
For induction of acute airway inflammation, mice were anesthetized with 3% isoflurane, treated then with 25 µg of papain (Sigma) or PBS i.n. in a volume of 40 µl every day for 3 d. Mice were killed on day 4 for analysis. For induction of bleomycin-induced pulmonary fibrosis, mice were anesthetized with 3% isoflurane, treated then with 5 mg/kg of bleomycin (Selleck) or PBS intranasally (i.n.) in a volume of 40 µl. Weight change, pulmonary inflammation, and fibrosis were assessed daily up to 21 d.
Giemsa and Oil Red O staining
Cytospins of BAL fluid cells were fixed for 10 min with methanol and stained for 30 min with a solution of 0.02% Giemsa Stain (Sigma). For Oil Red O staining, cytospins of BAL fluid cells were fixed and stained for 10 min with a solution of 0.3% Oil Red O (Solarbio), followed by 2 min of destaining in 60% isopropanol and two washes with PBS. Subsequently, nuclei were stained with hematoxylin.
Lung histology
Lungs were removed, fixed in 4% phosphate-buffered formalin, and embedded in paraffin for sectioning. H&E staining and Masson’s blue staining were performed for analysis of airway inflammation and pathological changes before microscopic analysis (Zeiss Axio Scan). The intensity of perivascular inflammatory cell infiltration or peribronchial infiltration (perivascular and bronchial inflammation) was evaluated by the observation of inflammatory cell infiltration density and alveolar wall thickening in affected parenchyma and scored on an arbitrary 1–3+ basis in a blind fashion. Fibrosis score was evaluated on collagen deposition and alveolar wall thickness and scored on an arbitrary 1–8+ basis in a blind fashion. All grading was based on the most severely inflamed or fibrotic section on each slide.
ELISA assay
The concentrations of IL-5, IL-6, IL-13, IL-33, and TGFβ1 were quantified by using mouse ELISA Kits (eBioscience) according to the manufacturer’s instructions.
Cell isolation from mouse tissues
Cells from BAL fluid were collected by flushing the lung twice with 1-ml aliquots of PBS through the trachea. The aliquots were combined and centrifuged at 300 g for 5 min. For isolation of lung tissue cells, the lungs were minced and digested with 2 mg/ml type IV collagenase (Roche) and 50 µg/ml DNase I (Roche) for 1 h at 37°C on a rotator. Large intestines were opened longitudinally, cut into 5-mm pieces, and washed with HBSS containing 5 mM EDTA five times to remove epithelial cells. Then, tissues were digested with 1 mg/ml type III collagenase (Worthington) for 1 h at 37°C on a rotator. To isolate leukocytes from the lungs and intestines, digested tissues were filtered through 70-µm cell strainers and enriched with 40% Percoll (GE Healthcare) gradient. The spleen was minced and passed through a 70-µm cell strainer without prior digestion. Bone marrow cells were flushed from femurs and tibias using a syringe with RPMI-1640 medium. Red blood cells were lysed with ACK buffer.
Flow cytometry and cell sorting
Isolated cells from tissues were kept at 4°C, and nonspecific binding was blocked with 5 µg/ml of anti-CD16/32 (93; eBioscience) before cell surfaces were stained with fluorochrome-conjugated or biotinylated monoclonal antibodies specific to mouse CD11c (N418), CD11b (M1/70), F4/80 (BM8; eBioscience), SiglecF (E50-2440; BD Biosciences), CD45 (30-F11), CD4 (GK1.5), CD8α (53–6.7), MHC class II (M5/114.15.2), Ly6G (1A8), CD25 (PC61.5; eBioscience), CD44 (IM7; eBioscience), CD62L (MEL-14; eBioscience), and IL-7α (A7R34; eBioscience). Dead cells were outgated with 7-AAD (eBioscience). For ILC2 staining, cells were first stained with biotinylated monoclonal antibodies specific for CD3ε (145−2C11), CD4, CD8α, CD11c, CD11b, CD19 (6D5), TCRβ (H57-597), TCRγδ (GL3), NK1.1 (PK136), Gr1 (RB6-8C5), Ter119 (TER-119), and B220 (RA3-6B2) and then stained with fluorochrome-conjugated secondary antibodies against streptavidin, CD90.2 (53–2.1; eBioscience), Sca1 (D7; BD Biosciences), ST2 (RMST2-2; eBioscience), CD25, CD44, CD45, IL-7α, ICOS (C398.4A; eBioscience), and KLRG1 (2F1; eBiosciences). Antibodies used are listed in Table S4.
Lung cells were used for isolation of CD11c+ SiglecF+ AMs. For isolation of Lineage− IL-7α+ ST2+ bone marrow ILC2s, bone marrow cells were first enriched by mouse Lineage Cell Depletion Kit (Miltenyi Biotec) following the manufacturer’s instructions. Enriched cells were then sorted on FACSAria (BD Biosciences). Purity of sorted cells was >99%.
Intracellular staining
For measurement of transcription factors, cells were fixed and permeabilized with the Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturer’s instructions and then stained with monoclonal antibodies specific for FoxP3 (FJK-16s; eBioscience), Ki67 (SolA15; eBioscience), and HIF1α (241812; R&D Systems). For intracellular staining for cytokines, cells were stimulated with 50 ng/ml phorbol 12-myristate 13-acetate (Sigma) and 500 ng/ml ionomycin (Sigma) in the presence of GolgiStop (BD Biosciences) in complete media for 4 h at 37°C. After surface marker staining, cells were fixed and permeabilized with the Cytofix/Cytoperm kit (BD Biosciences) and then stained with anti-IFNγ (XMG1.2; eBioscience), anti–IL-4 (11B11; eBioscience), anti–IL-5 (TRFK5), and anti–IL-13 (eBio13A; eBioscience) antibodies. Antibodies were from BioLegend unless otherwise stated. An LSR Fortessa (BD Biosciences) was used for multiparameter analysis, and data were analyzed with FlowJo software (TreeStar).
Generation of murine BMDMs
Bone marrow cells were differentiated for 7 d in DMEM containing 10% FBS, 20% L929 supernatant, and 1% penicillin–streptomycin at 37°C in a humidified incubator. Macrophages at day 7 were rested in DMEM containing 10% FBS and 1% penicillin–streptomycin overnight and stimulated for 24 h with or without 20 ng/ml mouse recombinant IL-4 (PeproTech) in the presence or absence of chemical inhibitors or metabolites including 1 mM 2-deoxyglucose (2-DG), 100 µM sodium iodoacetate, 500 µM sodium oxalate, 200 µM sodium oxamate, 5 µM px478 (Selleck), 200 µM D-fructose 1,6-biphosphate trisodium salt hydrate (F1,6BP), 400 µM D-3-PG disodium salt (3-PG), 5 mM sodium pyruvate (Gibco), or 1 mM L-lactate. Reagents were from Sigma unless otherwise stated.
In vitro ILC2 culture
Bone marrow ILC2s were sorted and cocultured with precoated BMDMs at a density of 5 × 103 cells per well in MEM-α complete media containing 10% FBS, 1% penicillin-streptomycin, 10 ng/ml IL-7 (Peprotech), and 5 ng/ml IL-33 (R&D Systems) in the absence or presence of 5 µg/ml anti-Osteopontin (R&D Systems). ILC2 proliferation and cytokine production were evaluated by FACS and ELISA on day 4.
Bone marrow ILC2s were sorted and plated at a density of 5 × 103 cells per well in the presence of 10 ng/ml IL-7 and 5 ng/ml IL-33. Indicated concentrations of mouse recombinant osteopontin (BioLegend) were added on day 1. Treated ILC2s were analyzed by FACS on day 6.
Retroviral transduction
shRNA-containing oligonucleotides (Table S1) were cloned into the LMP vector according to the manufacturer’s protocol (Open Biosystems). Plat-E packaging cells were transfected with 3 µg of retroviral vector along with 9 µl of TransIT-LT1 transfection reagent (Mirus). The culture supernatant containing retrovirus was collected 48 h after transfection and used to transduce freshly isolated bone marrow cells at day 1 in L929 supernatant-containing BMDM culture media. Transduced BMDMs were harvested, sorted, and analyzed for gene expression at day 7.
RNA isolation and qRT-PCR
Total RNA was extracted with TRIzol (Invitrogen), and cDNA was synthesized with reverse transcription (RevertAid First Strand cDNA Synthesis Kit; ThermoFisher). Real-time PCR was performed using SYBR Green qPCR Master Mix (Bimake) with a CFX96 real-time system (Bio-Rad). The primers used in the present study are listed in Table S2. Samples were normalized to the expression of gene encoding β-actin.
Chromatin immunoprecipitation assay
For each sample, 4 × 105 cells were collected and lysed in 190 µl lysis buffer (0.5% NP-40, 0.5% Tween-20, 0.1% SDS, and proteinase inhibitor) for 5 min on ice and then subjected to enzymatic digestion by incubating with 190 µl MNase working buffer (100 mM Tris-HCl, pH 8.0, 2 mM CaCl2) and 20 µl diluted MNase (0.01 unit/µl; N3755; Sigma) at 37°C for 5 min. The reaction was terminated by adding 50 µl stop buffer (110 mM Tris-HCl, pH 8.0, 55 mM EDTA) and then 450 µl cold 2 × RIPA buffer (1% Triton X-100, 280 mM NaCl, 0.1% SDS, 0.2% sodium deoxycholate, 5 mM EGTA, and proteinase inhibitor). After centrifuge at maximum speed at 4°C for 15 min, the supernatant was transferred to a new tube, and 5% was kept as input for later purification. Each chromatin sample was supplemented with 400 µl RIPA buffer (10 mM Tris-HCl, pH 8.0, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.1% DOC, and 1 mM EDTA) and then incubated with 1 µg H3K4me3 (ab8580; Abcam) or rabbit isotype control IgG (ab171870; Abcam) overnight with rotation at 4°C. The next day, the sample was incubated with 100 µg protein A/G dynabeads (Life Technologies) for another 2 h. The beads were washed five times with 1 ml RIPA buffer and once with 1 ml LiCl buffer (250 mM LiCl, 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.5% NP-40, and 0.5% DOC). After washing, the beads were resuspended with 95 µl TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA) supplemented with 5 µl proteinase K (10 mg/ml; Roche) and incubated at 55°C with shaking for 90 min to digest proteins. The supernatant was collected, and DNA was purified by phenol-chloroform extraction and ethanol precipitation. Enrichment of histone modifications were analyzed by qPCR using validated primers described in Table S3. The ratio of precipitated DNA to input DNA was calculated for comparison of histone modification between different samples.
Seahorse cellular metabolic profiling
For real-time analysis of the extracellular acidification rate (ECAR) and the oxygen consumption rate (OCR), macrophages were analyzed with an XF-96 Extracellular Flux Analyzer (Seahorse Bioscience). In brief, 5–8 × 104 macrophages were plated in buffer-free, glucose-free media (Seahorse Bioscience) with 10 mM glucose, 2 mM glutamine, and 1 mM sodium pyruvate in a 37°C incubator without CO2 for 1 h. Three consecutive measurements of ECAR (mpH/min) and OCR (pMoles O2/min) were obtained under basal conditions and after the additions of 1 µM oligomycin, to inhibit mitochondrial ATP synthesis; 4 µM fluoro-carbonyl cyanide phenylhydrazone, a protonophore that uncouples ATP synthesis from oxygen consumption by the electron-transport chain; and 100 nM rotenone plus 1 µM antimycin A, which inhibit the electron transport chain, and 100 mM 2-deoxyglucose, which inhibits glycolysis through competitive binding to glucose hexokinase. All compounds are from Sigma-Aldrich. Basal ECAR and maximal ECAR were calculated from average of three measurements following addition of glucose and oligomycin, respectively. Basal OCR and maximal OCR were calculated from average of three measurements before addition of oligomycin and following addition of fluoro-carbonyl cyanide phenylhydrazone, respectively. The SRC is calculated as the difference between basal OCR and maximal OCR.
RNA sequencing (RNA-seq) analysis
Total RNA of sorted AMs was prepared by RNeasy mini kit (Qiagen) with RNase free DNase set (Qiagen). Sample preparation, library construction, and sequencing on BGISEQ-500 were performed at Beijing Genomics Institute at 50-bp read length. Clean tags were mapped to the mouse genome database (UCSC version mm10) by HISAT (hierarchical indexing for splicing alignment for transcripts) software. For gene expression analysis, the matched reads were calculated and then normalized to reads per kilobase per million using RNA-seq by expectation maximization software. The significance of the differential expression of genes was defined by the bioinformatics service of Beijing Genomics Institute according to the combination of the absolute value of log2-Ratio ≥ 1 and false discovery rate ≤ 0.001. Based on the reads per kilobase per million value of each signature gene in AMs, a heat map was generated in Mev4.9.0. RNA-seq data have been deposited in the National Center for Biotechnology Information Sequence Read Archive database with accession nos. SRR6407103 and SRR8121125.
Statistical analysis
Experiments were repeated at least three times (n = 3–6 each), and data were representative of three independent experiments. All data were analyzed by an unpaired two-tailed Student’s t test with GraphPad Prism 5.0. P values of <0.05 were considered statistically significant.
Online supplemental material
Fig. S1 shows analysis of steady state AMs and quantification of lung cell numbers upon papain challenge. Fig. S2 shows flow cytometric analysis and quantification of lung cell numbers after bleomycin challenge. Fig. S3 demonstrates numbers of cytokine-producing ILC2s upon papain challenge and flow cytometric analysis of lung inflammation after AM depletion with clophosomes. Fig. S4 demonstrates that osteopontin treatment enhances ILC2 activation and proliferation. Fig. S5 shows glycolytic enzyme expression in AMs and Spp1 expression in BMDMs upon glucose metabolism inhibitors treatment. Table S1 lists shRNA probes. Table S2 lists quantitative real-time PCR primers. Table S3 lists chromatin immunoprecipitation (ChIP)–qPCR primers. Table S4 lists FACS, ChIP, and neutralization antibodies.
Supplementary Material
Acknowledgments
We thank the Center for Biomedical Analysis, Tsinghua University, for technical support and members of Liu laboratory for help and suggestions.
This work is supported by funds from the National Natural Science Foundation of China (grant NSFC81630041), Ministry of Science and Technology of China (grants YFC0903900 and YFA0505802), Tsinghua-Peking Center for Life Sciences, and the National Institutes of Health (grants RO1AI23398 and R56AI37017).
The authors declare no competing financial interests.
Author contributions: W. Zhang designed and performed the experiments and analyzed the data; Q. Li conducted the epigenetic experiments; D. Li and J. Li performed the in vitro retroviral infection studies; D. Aki helped the experimental design and data analysis; and W. Zhang and Y.-C. Liu conceived the project, interpreted the data, and wrote the paper.
References
- Colegio O.R., Chu N.Q., Szabo A.L., Chu T., Rhebergen A.M., Jairam V., Cyrus N., Brokowski C.E., Eisenbarth S.C., Phillips G.M., et al. . 2014. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 513:559–563. 10.1038/nature13490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramer T., Yamanishi Y., Clausen B.E., Förster I., Pawlinski R., Mackman N., Haase V.H., Jaenisch R., Corr M., Nizet V., et al. . 2003. HIF-1alpha is essential for myeloid cell-mediated inflammation. Cell. 112:645–657. 10.1016/S0092-8674(03)00154-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby I.A., and Hewitson T.D.. 2016. Hypoxia in tissue repair and fibrosis. Cell Tissue Res. 365:553–562. 10.1007/s00441-016-2461-3 [DOI] [PubMed] [Google Scholar]
- Eming S.A., Wynn T.A., and Martin P.. 2017. Inflammation and metabolism in tissue repair and regeneration. Science. 356:1026–1030. 10.1126/science.aam7928 [DOI] [PubMed] [Google Scholar]
- Gautier E.L., Shay T., Miller J., Greter M., Jakubzick C., Ivanov S., Helft J., Chow A., Elpek K.G., Gordonov S., et al. Immunological Genome Consortium . 2012. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol. 13:1118–1128. 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodwin J., Choi H., Hsieh M.H., Neugent M.L., Ahn J.M., Hayenga H.N., Singh P.K., Shackelford D.B., Lee I.K., Shulaev V., et al. . 2018. Targeting Hypoxia-Inducible Factor-1α/Pyruvate Dehydrogenase Kinase 1 Axis by Dichloroacetate Suppresses Bleomycin-induced Pulmonary Fibrosis. Am. J. Respir. Cell Mol. Biol. 58:216–231. 10.1165/rcmb.2016-0186OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammad H., and Lambrecht B.N.. 2015. Barrier Epithelial Cells and the Control of Type 2 Immunity. Immunity. 43:29–40. 10.1016/j.immuni.2015.07.007 [DOI] [PubMed] [Google Scholar]
- Higgins D.F., Kimura K., Bernhardt W.M., Shrimanker N., Akai Y., Hohenstein B., Saito Y., Johnson R.S., Kretzler M., Cohen C.D., et al. . 2007. Hypoxia promotes fibrogenesis in vivo via HIF-1 stimulation of epithelial-to-mesenchymal transition. J. Clin. Invest. 117:3810–3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.C., Smith A.M., Everts B., Colonna M., Pearce E.L., Schilling J.D., and Pearce E.J.. 2016. Metabolic Reprogramming Mediated by the mTORC2-IRF4 Signaling Axis Is Essential for Macrophage Alternative Activation. Immunity. 45:817–830. 10.1016/j.immuni.2016.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussell T., and Bell T.J.. 2014. Alveolar macrophages: plasticity in a tissue-specific context. Nat. Rev. Immunol. 14:81–93. 10.1038/nri3600 [DOI] [PubMed] [Google Scholar]
- Izquierdo H.M., Brandi P., Gómez M.J., Conde-Garrosa R., Priego E., Enamorado M., Martínez-Cano S., Sánchez I., Conejero L., Jimenez-Carretero D., et al. . 2018. Von Hippel-Lindau Protein Is Required for Optimal Alveolar Macrophage Terminal Differentiation, Self-Renewal, and Function. Cell Reports. 24:1738–1746. 10.1016/j.celrep.2018.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha A.K., Huang S.C., Sergushichev A., Lampropoulou V., Ivanova Y., Loginicheva E., Chmielewski K., Stewart K.M., Ashall J., Everts B., et al. . 2015. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity. 42:419–430. 10.1016/j.immuni.2015.02.005 [DOI] [PubMed] [Google Scholar]
- Konno S., Kurokawa M., Uede T., Nishimura M., and Huang S.K.. 2011. Role of osteopontin, a multifunctional protein, in allergy and asthma. Clin. Exp. Allergy. 41:1360–1366. 10.1111/j.1365-2222.2011.03775.x [DOI] [PubMed] [Google Scholar]
- Leavenworth J.W., Verbinnen B., Yin J., Huang H., and Cantor H.. 2015. A p85α-osteopontin axis couples the receptor ICOS to sustained Bcl-6 expression by follicular helper and regulatory T cells. Nat. Immunol. 16:96–106. 10.1038/ni.3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Swanson S.K., Gogol M., Florens L., Washburn M.P., Workman J.L., and Suganuma T.. 2015. Serine and SAM Responsive Complex SESAME Regulates Histone Modification Crosstalk by Sensing Cellular Metabolism. Mol. Cell. 60:408–421. 10.1016/j.molcel.2015.09.024 [DOI] [PubMed] [Google Scholar]
- Lin Y.H., and Yang-Yen H.F.. 2001. The osteopontin-CD44 survival signal involves activation of the phosphatidylinositol 3-kinase/Akt signaling pathway. J. Biol. Chem. 276:46024–46030. 10.1074/jbc.M105132200 [DOI] [PubMed] [Google Scholar]
- Locasale J.W. 2013. Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer. 13:572–583. 10.1038/nrc3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W., Hu H., Chang R., Zhong J., Knabel M., O’Meally R., Cole R.N., Pandey A., and Semenza G.L.. 2011. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 145:732–744. 10.1016/j.cell.2011.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills E.L., Kelly B., Logan A., Costa A.S.H., Varma M., Bryant C.E., Tourlomousis P., Däbritz J.H.M., Gottlieb E., Latorre I., et al. . 2016. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell. 167:457–470.e13. 10.1016/j.cell.2016.08.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minutti C.M., Jackson-Jones L.H., García-Fojeda B., Knipper J.A., Sutherland T.E., Logan N., Ringqvist E., Guillamat-Prats R., Ferenbach D.A., Artigas A., et al. . 2017. Local amplifiers of IL-4Rα-mediated macrophage activation promote repair in lung and liver. Science. 356:1076–1080. 10.1126/science.aaj2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill L.A., and Pearce E.J.. 2016. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 213:15–23. 10.1084/jem.20151570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazon A., Goldrath A.W., Nizet V., and Johnson R.S.. 2014. HIF transcription factors, inflammation, and immunity. Immunity. 41:518–528. 10.1016/j.immuni.2014.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palsson-McDermott E.M., Curtis A.M., Goel G., Lauterbach M.A.R., Sheedy F.J., Gleeson L.E., van den Bosch M.W.M., Quinn S.R., Domingo-Fernandez R., Johnston D.G.W., et al. . 2015. Pyruvate Kinase M2 Regulates Hif-1α Activity and IL-1β Induction and Is a Critical Determinant of the Warburg Effect in LPS-Activated Macrophages. Cell Metab. 21:347 10.1016/j.cmet.2015.01.017 [DOI] [PubMed] [Google Scholar]
- Pardo A., Gibson K., Cisneros J., Richards T.J., Yang Y., Becerril C., Yousem S., Herrera I., Ruiz V., Selman M., and Kaminski N.. 2005. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med. 2:e251 10.1371/journal.pmed.0020251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyssonnaux C., Datta V., Cramer T., Doedens A., Theodorakis E.A., Gallo R.L., Hurtado-Ziola N., Nizet V., and Johnson R.S.. 2005. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J. Clin. Invest. 115:1806–1815. 10.1172/JCI23865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan A.T., Goldrath A.W., and Glass C.K.. 2017. Metabolic and Epigenetic Coordination of T Cell and Macrophage Immunity. Immunity. 46:714–729. 10.1016/j.immuni.2017.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip K., Mills T.W., Davies J., Chen N.Y., Karmouty-Quintana H., Luo F., Molina J.G., Amione-Guerra J., Sinha N., Guha A., et al. . 2017. HIF1A up-regulates the ADORA2B receptor on alternatively activated macrophages and contributes to pulmonary fibrosis. FASEB J. 31:4745–4758. 10.1096/fj.201700219R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raja R., Kale S., Thorat D., Soundararajan G., Lohite K., Mane A., Karnik S., and Kundu G.C.. 2014. Hypoxia-driven osteopontin contributes to breast tumor growth through modulation of HIF1α-mediated VEGF-dependent angiogenesis. Oncogene. 33:2053–2064. 10.1038/onc.2013.171 [DOI] [PubMed] [Google Scholar]
- Semenza G.L. 2011. Oxygen sensing, homeostasis, and disease. N. Engl. J. Med. 365:537–547. 10.1056/NEJMra1011165 [DOI] [PubMed] [Google Scholar]
- Tannahill G.M., Curtis A.M., Adamik J., Palsson-McDermott E.M., McGettrick A.F., Goel G., Frezza C., Bernard N.J., Kelly B., Foley N.H., et al. . 2013. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 496:238–242. 10.1038/nature11986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzouvelekis A., Harokopos V., Paparountas T., Oikonomou N., Chatziioannou A., Vilaras G., Tsiambas E., Karameris A., Bouros D., and Aidinis V.. 2007. Comparative expression profiling in pulmonary fibrosis suggests a role of hypoxia-inducible factor-1alpha in disease pathogenesis. Am. J. Respir. Crit. Care Med. 176:1108–1119. 10.1164/rccm.200705-683OC [DOI] [PubMed] [Google Scholar]
- Ueno M., Maeno T., Nomura M., Aoyagi-Ikeda K., Matsui H., Hara K., Tanaka T., Iso T., Suga T., and Kurabayashi M.. 2011. Hypoxia-inducible factor-1α mediates TGF-β-induced PAI-1 production in alveolar macrophages in pulmonary fibrosis. Am. J. Physiol. Lung Cell. Mol. Physiol. 300:L740–L752. 10.1152/ajplung.00146.2010 [DOI] [PubMed] [Google Scholar]
- Van Dyken S.J., Mohapatra A., Nussbaum J.C., Molofsky A.B., Thornton E.E., Ziegler S.F., McKenzie A.N., Krummel M.F., Liang H.E., and Locksley R.M.. 2014. Chitin activates parallel immune modules that direct distinct inflammatory responses via innate lymphoid type 2 and γδ T cells. Immunity. 40:414–424. 10.1016/j.immuni.2014.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn T.A., and Vannella K.M.. 2016. Macrophages in Tissue Repair, Regeneration, and Fibrosis. Immunity. 44:450–462. 10.1016/j.immuni.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xanthou G., Alissafi T., Semitekolou M., Simoes D.C., Economidou E., Gaga M., Lambrecht B.N., Lloyd C.M., and Panoutsakopoulou V.. 2007. Osteopontin has a crucial role in allergic airway disease through regulation of dendritic cell subsets. Nat. Med. 13:570–578. 10.1038/nm1580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiss D.M.W., Gause W.C., Osborne L.C., and Artis D.. 2015. Emerging functions of amphiregulin in orchestrating immunity, inflammation, and tissue repair. Immunity. 42:216–226. 10.1016/j.immuni.2015.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.