Marié et al. describe a futile cycle between constitutive histone acetyltransferases and deacetylases (HDACs) that regulates interferon-stimulated gene transcription through availability of the epigenetic reader, Brd4. Pharmacologic targeting of the HDAC/Brd4 axis resolves aberrant gene expression associated with genetic susceptibility to autoimmune interferonopathies.
Abstract
In contrast to the common role of histone deacetylases (HDACs) for gene repression, HDAC activity provides a required positive function for IFN-stimulated gene (ISG) expression. Here, we show that HDAC1/2 as components of the Sin3A complex are required for ISG transcriptional elongation but not for recruitment of RNA polymerase or transcriptional initiation. Transcriptional arrest by HDAC inhibition coincides with failure to recruit the epigenetic reader Brd4 and elongation factor P-TEFb due to sequestration of Brd4 on hyperacetylated chromatin. Brd4 availability is regulated by an equilibrium cycle between opposed acetyltransferase and deacetylase activities that maintains a steady-state pool of free Brd4 available for recruitment to inducible promoters. An ISG expression signature is a hallmark of interferonopathies and other autoimmune diseases. Combined inhibition of HDAC1/2 and Brd4 resolved the aberrant ISG expression detected in cells derived from patients with two inherited interferonopathies, ISG15 and USP18 deficiencies, defining a novel therapeutic approach to ISG-associated autoimmune diseases.
Graphical Abstract
Introduction
The type I interferons (IFN-α/β) are central mediators of innate immunity and inflammatory responses. IFN expression is induced in a wide variety of cell types in response to microbial and viral infection and cellular stress, through activation of cellular pathogen recognition receptors by pathogen-associated molecular patterns (Levy et al., 2011). Constitutive IFN production at basal levels also contributes to immune system homeostasis through tonic signaling (Gough et al., 2012). Once produced, secreted IFNs induce antiviral and antimicrobial responses in target cells through activation of a receptor-mediated JAK-STAT signal transduction system, involving tyrosine phosphorylation of STAT1 and STAT2, leading to their assembly with the DNA binding protein IFN regulatory factor 9 (IRF9) into the trimeric transcription factor complex ISGF3, followed by nuclear translocation, assembly on chromatin, and recruitment of the transcriptional apparatus (Au-Yeung et al., 2013). Induction of IFN-stimulated gene (ISG) expression by activated STAT factors is rapid and transient, correlating closely with complex assembly on chromatin (Lerner et al., 2003). The majority of the direct transcriptional capability of ISGF3 is dependent on a carboxyl-terminal transactivation domain of the STAT2 protein (Qureshi et al., 1996) that facilitates the assembly of the transcriptional apparatus (Bhattacharya et al., 1996; Park et al., 1999; Paulson et al., 1999, 2002; Lau et al., 2003). Transcriptional induction by STAT2 depends on its interaction with coactivators, modification of chromatin, and recruitment of components of the basal transcription machinery, but its detailed mechanism of action remains ill defined (Au-Yeung et al., 2013).
Several unusual characteristics of ISG transcription have been described, possibly related to the involvement of this pathway in antiviral immunity, causing it to evolve under the selective pressure of virus-encoded inhibitory proteins (Levy and García-Sastre, 2001). For instance, STAT2 recruits a distinct initiation complex that is resistant to virus-dependent proteolysis (Paulson et al., 2002), and multiple steps in ISG transcription are targeted by viral virulence factors (Fleming, 2016). ISGF3 recruits lysine acetyltransferase coactivators through interactions with the transactivation domains of both STAT1 and STAT2 (Wojciak et al., 2009), and acetyltransferase activity is required for induction of gene expression (Gnatovskiy et al., 2013). STAT2 also recruits a required chromatin remodeling complex containing the DNA helicases Rvb1 and Rvb2, which facilitate RNA polymerase recruitment to target promoters (Gnatovskiy et al., 2013), as well as components of the Mediator complex, which provides communication between chromatin-bound ISGF3 and RNA polymerase II (RNAPII; Lau et al., 2003). Interestingly, STAT2-dependent transcription also requires the lysine deacetylase activity of histone deacetylase (HDAC) enzymes as a positive regulator of transcription (Yu et al., 2002; Nusinzon and Horvath, 2003; Chang et al., 2004; Sakamoto et al., 2004), unlike the more common role of HDACs as inhibitors and silencers of gene expression (Kouzarides, 2007). Pharmacologic or genetic impairment of HDAC activity prevents ISG induction, but the mechanism underlying this HDAC requirement remains unknown.
Lysine deacetylases have been widely studied in the context of histone deacetylation, where they are commonly recruited by corepressor complexes to silenced genes (Kouzarides, 2007). It has been hypothesized that histone acetylation activates chromatin by modulating the association of nucleosomes with DNA through partial neutralization of histone-positive electrostatic charge, a process that is opposed by HDACs through reversal of histone acetylation (Cosgrove et al., 2004). More recently, however, it has become clear that HDACs play a much broader role, including the identification of nonhistone protein substrates (Glozak et al., 2005; Spange et al., 2009) and the documentation of HDACs associated with transcriptionally active as well as transcriptionally repressed chromatin (Wang et al., 2009).
The hallmark of eukaryotic transcriptional initiation of protein coding genes is the recruitment and activation of RNAPII, a process requiring DNA-bound activators, the general transcription machinery, and chromatin-modifying enzymes, leading to formation of a preinitiation complex. Preinitiation complex formation has long been recognized as an important regulatory event and possibly the rate-limiting step for transcriptional control of gene expression (Darnell, 1982). However, gene expression requires additional transcriptional, cotranscriptional, and posttranscriptional events including efficient transcriptional elongation, cotranscriptional processing, 3′ end formation, and nuclear-cytoplasmic transport, all of which contribute to the efficiency of gene expression. In fact, it has been long appreciated that the transition from transcriptional initiation to processive elongation is a discrete mechanistic step, characterized by natural pause sites, abortive transcriptional events, and polymerase backtracking (Boeger et al., 2005; Lis, 2007).
Evidence that post-initiation events can be subject to regulation has come from a variety of studies. Efficient transcriptional elongation is dependent on the cyclin-dependent kinase P-TEFb, which phosphorylates the carboxyl-terminal domain (CTD) heptad repeat of RNAPII at serine 2 and overcomes the inhibitory activity of two negative regulators, NELF and DSIF (Peterlin and Price, 2006; Kwak and Lis, 2013). The P-TEFb complex is recruited to promoters of many genes by the acetylated lysine-binding protein Brd4 (Brès et al., 2008), following the release of P-TEFb from inhibitory complexes through posttranslational modification (Cho et al., 2010). Brd4 is a large BET protein composed of a double amino-terminal bromodomain mediating recruitment to acetylated chromatin and an extra terminal domain that binds to the core complex of P-TEFb. Although there is evidence that transcription of some genes may be independent of P-TEFb (Gomes et al., 2006; Oven et al., 2007), this factor is recruited to many promoters during transcriptional induction, including ISGs (Patel et al., 2013), and it facilitates the movement of RNAPII past pause sites, activates 5′-end capping, associates with and continues to phosphorylate elongating RNAPII, and is essential to control expression from numerous promoters, such as HSP70, c-MYC, and the HIV LTR (Peterlin and Price, 2006). Additional evidence for regulation of gene expression after transcriptional initiation comes from the characterization of poised genes. Many genes, such as HSP70 and c-MYC, display constitutively recruited RNAPII even before induction and in the absence of gene expression, and transcriptional induction is accomplished by converting poised, paused polymerases into efficiently elongating enzymes (Lis, 2007). Recent genome-wide analyses of chromatin modifications have demonstrated that many inactive yet permissive genes display hallmarks for active genes, such as trimethylation of histone H3 on lysine 4 and constitutively associated RNAPII (Lee et al., 2006; Rasmussen, 2008), suggesting that post-initiation regulation may be a common regulatory strategy.
The need for stringent regulation of ISG expression is exemplified by a set of autoimmune inflammatory diseases collectively known as interferonopathies, including Aicardi–Goutières syndrome (Crow and Manel, 2015). These Mendelian disorders are associated with constitutive up-regulation of IFN or ISG expression, caused by gain or loss of function of individual positive or negative regulators of the IFN pathway. Due to the key involvement of IFN in these syndromes, modulating the abnormal ISG expression has become an attractive therapeutic target (Rodero and Crow, 2016).
We have explored the biochemical basis for the activating role of HDACs during ISG expression. Our results pinpointed HDAC action at the transition between committed transcriptional initiation and processive transcriptional elongation, mediated by the recruitment of P-TEFb by Brd4 to ISG promoters. In particular, HDAC activity was critical for the availability of Brd4 to be mobilized to newly activated promoters. Inhibition of HDAC activity or Brd4 recruitment abrogated ISG transcription and cooperatively reduced pathogenic ISG expression associated with autoimmune syndromes. These results delineate a Brd4-dependent epigenetic control point that couples transition from initiation to concerted elongation to provide precise regulation of acutely inducible gene expression, which can be targeted to modulate inflammatory syndromes.
Results
ISG transcription requires class I HDAC activity
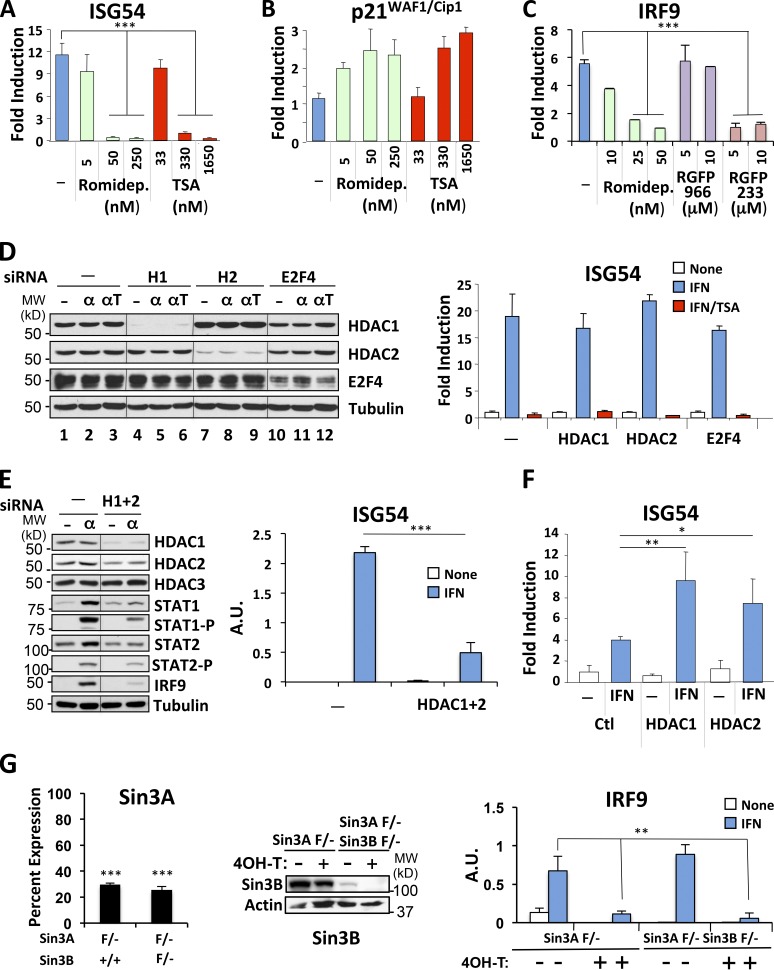
Trichostatin A (TSA) is a potent general inhibitor of class I and II HDAC enzymes, whereas valproate, which also inhibits ISG expression (Chang et al., 2004), preferentially inhibits class I enzymes (Göttlicher et al., 2001). To gain more insight into which enzymes is required for ISG transcription, we tested the efficacy of romidepsin, an inhibitor with high specificity for class I HDAC (Furumai et al., 2002; Narita et al., 2009). Cells were stimulated with IFN in the presence of increasing concentrations of romidepsin ranging from 5 to 250 nM, since its IC50 for class I HDAC is ∼40 nM and for class II is >500 nM (Furumai et al., 2002). Romidepsin inhibited ISG54 expression partially at 5 nM and completely at 50 and 250 nM, equivalent to the effect of inhibitory concentrations of TSA (Fig. 1 A). As control, both romidepsin and TSA treatments led to p21Cip1/WAF1 expression (Fig. 1 B), indicative of HDAC inhibition (Archer et al., 1998). We extended this observation to another ISG, IRF9, showing that romidepsin inhibits IRF9 expression in a dose-dependent manner with complete inhibition at ∼50 nM. To further identify what HDAC enzyme is involved in ISG transcription, we tested the effect of RGFP966, an inhibitor with selective specificity for HDAC3, and RGPF233, which preferentially inhibits HDAC1 and 2 (Park et al., 2004). Interestingly, RGFP966 had very little effect on ISG transcription, whereas RGFP233 almost completely abrogated IRF9 expression (Fig. 1 C).
Figure 1.
Class I HDAC mediates ISG expression. (A and B) HeLa cells were treated with IFN-α for 6 h in the absence or presence of increasing concentrations of Romidepsin or TSA, as indicated. RNA was quantified by real-time RT-PCR for ISG54 and p21WAF1/Cip1 expression, normalized to GAPDH, and represented as fold induction over untreated cells. (C) HeLa cells were treated with IFN-α for 6 h in the absence or presence of increasing concentrations of romidepsin, RGFP966, or RGFP233, as indicated. RNA was quantified by real-time RT-PCR for IRF9 expression, normalized to GAPDH, and represented as fold induction over untreated cells. (D and E) HEK293 cells were transfected with siRNA against HDAC1, HDAC2, or E2F4 separately (D) or with a combination of HDAC1 and HDAC2 targeting oligonucleotides (E), and cells were stimulated with IFN-α for 6 h (α) in the presence (αΤ) or absence of TSA, as indicated. Whole-cell extracts were analyzed for expression of the indicated proteins by Western blotting (left). ISG54 expression was quantified by real-time RT-PCR and represented in arbitrary units (right). MW, molecular weight. A.U., arbitrary units. (F) HEK293T cells were transfected with pcDNA3 (Ctl), HDAC1, or HDAC2 expression constructs, and ISG54 expression was quantified after stimulation with IFN-α for 10 h. Representative data from two experiments are shown. (G) Sin3A F/– and Sin3A F/– Sin3B F/– immortalized mouse embryonic fibroblasts expressing Cre-ERT2 were treated for 3 d with 4OH-tamoxifen (4OH-T) or left untreated. Nuclear extracts were analyzed for expression of Sin3B by Western blotting (middle). Expression of Sin3A, represented as percent expression relative to levels before tamoxifen treatment (left) and expression of IRF9 mRNA before and after tamoxifen (right), were quantified by real-time RT-PCR and normalized to GAPDH mRNA expression. Weak expression of Sin3B protein in Sin3A F/– Sin3B F/– fibroblasts before 4OH-T treatment is likely due to some leakiness of Cre recombinase expression. Quantitative data are representative examples of three (A and F) or two (B, C, E, and G) independent experiments, each performed in duplicate, and error bars represent +SD. *, P < 0.05; **, P < 0.005; ***, P < 0.001.
Pharmacologic inhibition by romidepsin and RGFP233 strongly suggested that HDAC1 and 2 are required for ISG transcription. We therefore depleted HDAC 1 and 2 by RNA interference. Knockdown of HDAC1 or HDAC2 individually did not affect ISG induction (Fig. 1 D, right), in spite of major reductions of the respective HDAC proteins (Fig. 1 D, left). Interestingly, knockdown of HDAC2 led to a commensurate increase in HDAC1 levels (Fig. 1 D, lanes 7–9). These two proteins exist together in several multiprotein deacetylase complexes, suggesting likely functional redundancy between them. Therefore, we simultaneously depleted HDAC1 and HDAC2. Reduction of HDAC1 and HDAC2 together led to a significant (P < 0.002) impairment in ISG54 induction in response to IFN (Fig. 1 E, right), in spite of only partial knockdown of the HDAC2 protein (Fig. 1 E, left), suggesting that these two enzymes together are critical for IFN responsiveness. As expected, depletion of the control protein E2F4 had no effect on ISG54 induction (Fig. 1 D). We also examined expression of the IFN-inducible protein IRF9, as another measure of IFN responsiveness (Levy et al., 1990). IRF9 was highly induced in IFN-stimulated cells. However, knockdown of HDAC1 and 2 impaired induction of IRF9 induction (Fig. 1 E). STAT1 and to a lesser extent STAT2 are also IFN-inducible proteins. Their induction was also impaired by HDAC knockdown (Fig. 1 E), although the ability of residual protein to be phosphorylated was largely unimpaired, as previously shown (Chang et al., 2004).
We ectopically expressed HDAC1 and HDAC2 by transfection in 293T cells, which led to a sensitized response to IFN (Fig. 1 F). Heightened expression of HDAC2 caused a 50% increase in IFN-stimulated ISG54 expression (P < 0.1), whereas HDAC1 caused a >100% increase (P < 0.02; Fig. 1 F), an effect that was not observed following expression of a catalytically impaired HDAC1 mutant or of other deacetylases (data not shown). Expression of neither enzyme affected basal expression of ISG54.
HDAC1 and 2 are members of Sin3 complexes, which in mammalian cells contain either Sin3A or Sin3B and are commonly associated with gene repression (Hayakawa and Nakayama, 2011). However, the results described thus far indicating a potentiating role for HDAC1 and 2 in ISG expression and a recent report of a positive role for Sin3A during IFN responses (Icardi et al., 2012) prompted us to investigate the requirement for Sin3A and B proteins. Deletion of Sin3A is incompatible with cell growth (Dannenberg et al., 2005). Therefore, we examined IFN responses in cells containing a conditional allele of Sin3A following acute deletion, in the presence or absence of the related Sin3B gene (David et al., 2008). Immortalized mouse fibroblasts containing a single conditional allele of Sin3A (Sin3A-F/–) and expressing a tamoxifen-regulated Cre recombinase protein, with or without deletion of Sin3B (Sin3B+/+ or F/–), were examined for ISG expression before and after treatment with 4-hydroxy-tamoxifen and/or IFN (Fig. 1 G). Tamoxifen treatment significantly (P < 0.001) reduced the expression of Sin3A mRNA (Fig. 1 G, left), whether or not Sin3B was expressed (Fig. 1 G, middle). Loss of Sin3A significantly impaired induction of mouse IRF9 in response to IFN treatment (P < 0.007; Fig. 1 G, right), whereas loss of Sin3B had no significant effect on gene induction. Interestingly, loss of either Sin3A or Sin3B lowered basal expression of IRF9, indicating a possible role for IFN priming in maintaining basal expression levels (Gough et al., 2012). Residual IFN responsiveness was detected in Sin3 mutant cells, possibly due to incomplete loss of Sin3 complexes or to redundant activity from non–Sin3-containing HDAC complexes.
HDAC inhibition does not prevent transcription in vitro
Previous results showed that treatment of cells with HDAC inhibitors blocked ISG transcription as measured by endogenous gene expression, transcriptional run-on assays, and promoter-reporter assays (Génin et al., 2003; Nusinzon and Horvath, 2003; Chang et al., 2004). To determine whether HDAC activity was required for transcriptional initiation, we adopted an in vitro transcription system using a genomic segment containing the promoter of the ISG54 gene as template (Levy et al., 1986). In vitro transcription reactions were programmed with nuclear extracts from 293T cells with or without active ISGF3.
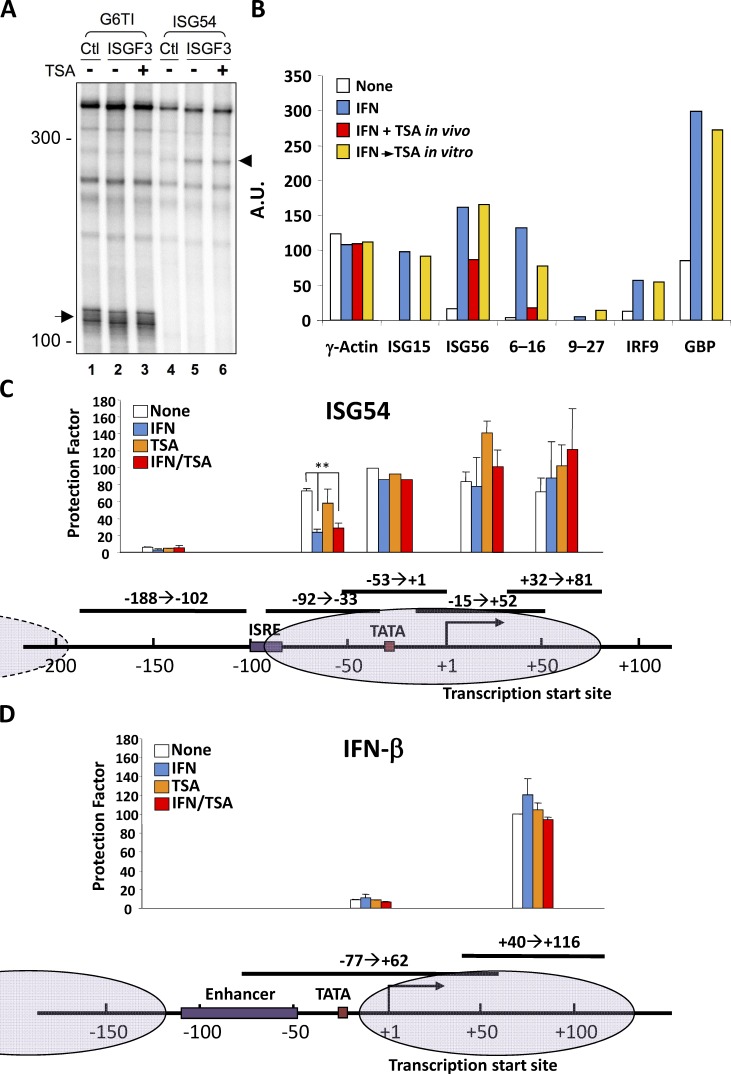
293T cell extracts supported basal transcription from a control template (G6TI) driven by Sp1 (Gill et al., 1994), as indicated by a correctly initiated runoff product (Fig. 2 A, lanes 1–3, arrow). However, transcription from the ISG54 promoter was only supported by nuclear extracts containing ISGF3 (Fig. 2 A, lanes 5 and 6, arrowhead), which resulted in a correctly initiated transcript from the ISG54 template without affecting transcription from the control. Significantly, neither basal nor ISGF3-dependent transcription was inhibited by inclusion of TSA in the reaction, indicating that HDAC activity was not required for transcription in vitro on naked DNA templates (Fig. 2 A, lanes 3 and 6).
Figure 2.
HDAC inhibition does not prevent transcription in vitro or IFN-stimulated chromatin remodeling in vivo. (A) Nuclear extracts from HEK293T cells expressing activated recombinant ISGF3 (STAT1, STAT2, IRF9, and JAK1) or transfected with vector (Ctl) were programmed with G6TI-CAT (G6TI) or p107 (ISG54) templates. Radiolabeled RNA transcripts were resolved by polyacrylamide-urea gel electrophoresis. Arrow and arrowhead indicate position of the transcripts from G6TI-CAT and p107, respectively. Molecular sizes indicated in nucleotides. (B) HeLa cells were either left untreated or treated with IFN-α in the absence or presence of TSA (TSA in vivo) before nuclei were isolated for run-on transcription. Where indicated, TSA was added to the elongation reaction in vitro (TSA in vitro). Specific signals from γ-actin, ISG15, ISG56, 6-16, 9-27, IRF9, and GBP transcription were quantified following filter hybridization and normalized to the signal for GAPDH, arbitrarily set to 100. Representative data from three independent experiments are shown. A.U., arbitrary units. (C and D) FS2 human diploid fibroblasts were starved for 72 h before being treated with IFN-α for 7.5 h in the absence or presence of TSA. Mononucleosomal DNA fraction was purified from MNase-digested nuclei and analyzed for nucleosome positions by PCR quantitation of protected fragments. All samples were quantified by real-time PCR, except the −53 to +1 ISG54 fragment, which was quantified by gel electrophoresis (C). The same mononucleosomal fractions were assayed for two fragments of the IFN-β promoter as control (D). Normalized protection factor for each sample was expressed proportionally to the signal obtained for fragment +40/+116 protected by the fixed nucleosome N2 at IFN-β promoter-proximal region, which was arbitrarily set at 100. Putative nucleosome positions are diagramed. **, P < 0.002 from representative experiment (of two) performed in duplicate, and error bars represent +SD.
Runoff transcription is largely a measure of transcriptional initiation. We asked whether HDAC activity was required for transcription in isolated nuclei, a measure of in vitro elongation of polymerase molecules previously initiated in vivo. Nuclei were isolated from cells untreated or treated with IFN-α and pulse labeled in vitro with radioactive nucleotides, and specific transcriptional elongation was quantified by filter hybridization (Fig. 2 B). Transcription of several example ISGs was stimulated in nuclei isolated from IFN-treated cells, whereas housekeeping gene transcription was equivalent regardless of how the cells were treated before nuclei isolation. Nuclei from cells stimulated with IFN in the presence of TSA displayed impaired ISG transcription, consistent with a requirement for HDAC activity for ISG expression in vivo. However, similar to the in vitro runoff result, addition of TSA in vitro failed to affect either housekeeping or ISG transcription. Therefore, HDAC activity does not appear to be required for polymerase initiation in vitro or for polymerase elongation in vitro, at least for previously initiated and committed polymerases. We note that this assay, which produces relatively short bursts of transcriptional elongation, may not be sensitive to regulators of transcriptional processivity.
IFN-stimulated chromatin remodeling
The involvement of HDAC and Sin3-containing complexes in ISG expression prompted us to examine IFN-dependent changes in chromatin architecture. To this end, we explored the nucleosome environment of the ISG54 gene by micrococcal nuclease (MNase) protection. The mononucleosomal genomic DNA fraction was recovered from untreated or IFN-stimulated human diploid fibroblasts following MNase digestion of nuclei, and segments of the ISG54 promoter were quantified by using PCR primers tiled across the promoter-proximal region. Efficient recovery of a specific DNA segment is indicative of protection from enzyme digestion by a placed nucleosome, whereas relative sensitivity to digestion is indicative of nucleosome-free or randomly placed nucleosome regions (Keene and Elgin, 1981; Levy and Noll, 1981). As control, we examined the MNase sensitivity of the IFN-β promoter proximal region, documenting the previously described nucleosome-free and fixed nucleosome elements (Agalioti et al., 2000) that were unaffected by IFN treatment.
The ISG54 promoter distal region upstream of the IFN-stimulated response element (ISRE) was relatively nucleosome free and was not affected by IFN or TSA treatments (Fig. 2 C). In contrast, genomic segments surrounding the TATA box and the transcriptional initiation site were relatively MNase resistant (Fig. 2 C), equivalent to the nuclease resistance of control loci, such as the IFN-β promoter proximal region (Fig. 2 D). Again, nuclease sensitivity was minimally affected by IFN or TSA treatments. The only ISG54 promoter region affected by IFN treatment was the −92 to −33 segment containing the ISRE. This region was resistant to digestion in unstimulated cells but became substantially more sensitive following IFN treatment (P < 0.002; Fig. 2 C), indicative of an alteration of nucleosome positioning. However, increased nuclease sensitivity following IFN treatment was not prevented by cotreatment with TSA.
These results suggest that the ISG54 ISRE and promoter-proximal region are packaged in a nucleosome, bounded by a relatively nucleosome-free region flanking the ISRE, which is altered in response to IFN. STAT2 recruits the DNA helicase proteins Rvb1 and 2, which are required for ISG expression (Gnatovskiy et al., 2013). However, STAT2 and Rvb recruitment to chromatin are not dependent on HDAC activity (Chang et al., 2004; Gnatovskiy et al., 2013), consistent with the lack of involvement of HDAC activity in nucleosome repositioning, as monitored by nuclease sensitivity.
RNAPII recruitment and activation do not require HDAC activity
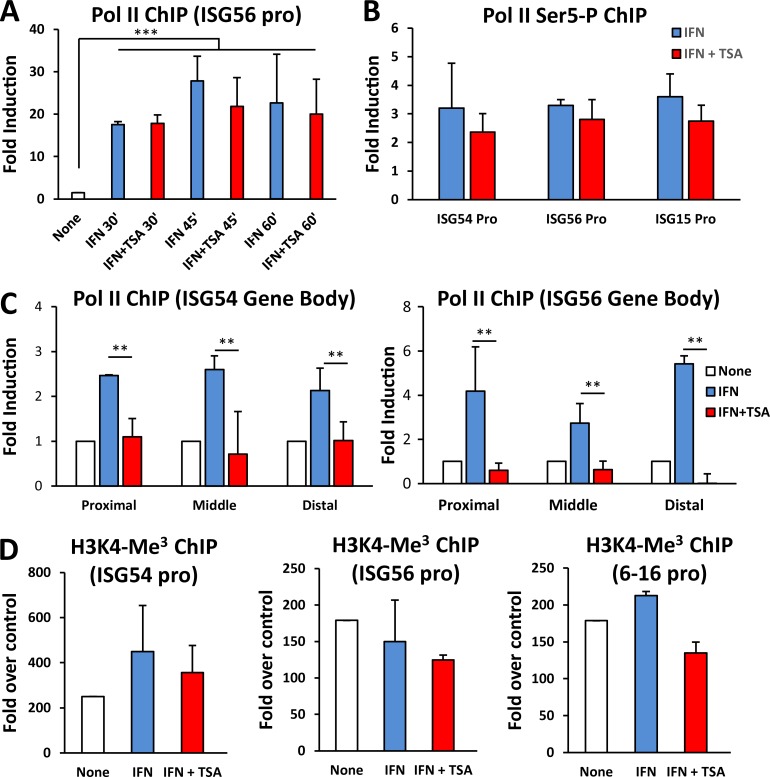
The carboxyl-terminal transactivation domain of STAT2 functions to recruit coactivator components, the Mediator complex, and presumably RNAPII (Bhattacharya et al., 1996; Qureshi et al., 1996; Paulson et al., 1999, 2002; Lau et al., 2003). Since RNAPII recruitment to chromatin is a prerequisite for transcription, we asked whether this step required HDAC activity. Chromatin immunoprecipitation (ChIP) analysis of RNAPII on the ISG56 promoter revealed recruitment in response to IFN stimulation that was rapid and robust (P < 0.001; Fig. 3 A). RNAPII was readily detectable on the promoter after 30 min of IFN treatment and was retained at 45 and 60 min after treatment. Strikingly, promoter recruitment of RNAPII was not blocked by cotreatment with TSA (Fig. 3 A). The presence of RNAPII at ISG promoters in the absence of transcription suggests that HDAC inhibits a post-initiation step. A hallmark of transcriptional initiation is carboxyl-terminal phosphorylation of RNAPII (Meinhart et al., 2005), with Ser 5 phosphorylation by transcription factor IIH (TFIIH) considered indicative of promoter clearance following preinitiation complex formation (Thomas and Chiang, 2006). RNAPII recruited to ISG promoters in response to IFN treatment was phosphorylated on Ser 5 regardless of HDAC inhibition (Fig. 3 B). RNAPII was also recruited to the body of transcription units in response to IFN (Fig. 3 C), although the density of RNAPII within the gene body was lower than that at the promoter. Significantly, RNAPII recruitment within the ISG56 and ISG54 transcription units was largely abrogated in the absence of HDAC activity (P < 0.05; Fig. 3 C), consistent with the observed inhibition of gene expression.
Figure 3.
RNAPII recruitment and activation do not require HDAC activity. (A) 2fTGH cells were either left untreated or treated with IFN-α in the absence or presence of TSA for the indicated time (minutes). ChIP assays were performed with antibodies against Pol II, and recovered ISG56 promoter sequences were quantified by real-time PCR relative to input and represented as fold induction relative to untreated cells. (B) As in A, except that antibodies against phosphorylated Pol II at serine-5 in the CTD were used for immunoprecipitation, following 60-min IFN-α treatments. Recovered ISG54, ISG56, and ISG15 promoter sequences were quantified by real-time PCR relative to input and represented as fold induction relative to untreated cells, which were arbitrarily set to 1. (C) As in A, except that cells were treated with IFN-α for 60 min and proximal, middle, and distal fragments along the ISG54 gene (left) and ISG56 gene (right) were assayed. Regions of ISG54 analyzed were centered around +2380 (proximal), +4540 (middle), and +7590 (distal); regions of ISG56 were +2940 (proximal), 7120 (middle), and +11040 (distal), relative to the TSS. (D) As in A, except that antibodies against trimethylated lysine-4 on histone H3 were used. Promoter regions of ISG54, ISG56, and 6-16 genes were quantified by real-time PCR and reported as fold over the signal detected with a nonspecific antibody. **, P < 0.05; ***, P < 0.001 from representative experiment (of two) performed in duplicate, and error bars represent +SD.
A common hallmark of active or potentially active genes is the presence of trimethylation on lysine 4 of histone H3 (H3K4me3) at promoter or transcriptional start regions (Vermeulen and Timmers, 2010). This modification usually occurs subsequent to the assembly of the general transcription machinery on the promoter. Unexpectedly, we detected high levels of H3K4me3 on ISG promoters (Fig. 3 D), even before IFN stimulation. However, neither IFN stimulation nor TSA cotreatment significantly affected the level of this histone mark. Taken together, these results demonstrate that while RNAPII can be efficiently recruited by STAT2, incorporated into a preinitiation complex at ISG promoters, and serve as a substrate for modification by TFIIH, it fails to successfully transit the transcription unit in the absence of HDAC activity, resulting in impaired transcription.
HDAC activity is required for P-TEFb recruitment
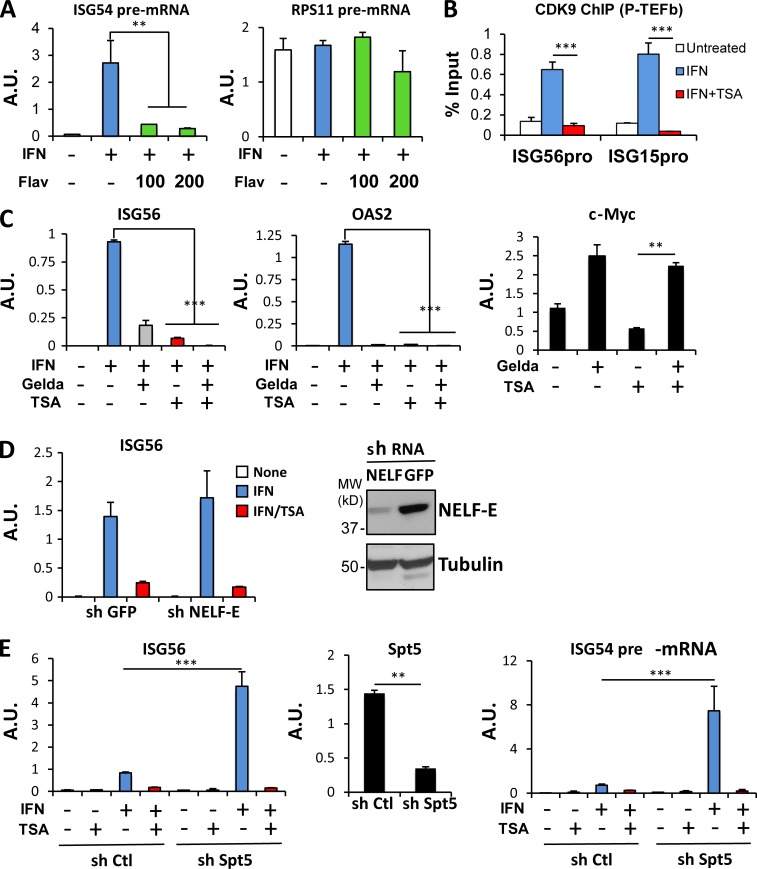
A crucial step driving the transition from promoter clearance to productive elongation is the recruitment of the CDK9/cyclin T1-containing P-TEFb complex. To test the importance of P-TEFb for ISG transcription, we used flavopiridol, a CDK9-selective inhibitor (Chao and Price, 2001). To examine effects on transcription, we scored the abundance of unspliced pre-mRNA transcripts in nuclei. Flavopiridol inhibited ISG54 induction (P < 0.03) at concentrations that did not prevent transcription of constitutively expressed RPS11 (Fig. 4 A, right). Higher concentrations of flavopiridol inhibited expression of all genes tested (data not shown), consistent with previous reports (Chao and Price, 2001). These results suggest that ISG54 transcription is acutely sensitive to the action of P-TEFb relative to constitutively expressed genes.
Figure 4.
HDAC activity is required for P-TEFb recruitment. (A) HeLa cells were either left untreated or treated with IFN-α for 1 h in the presence or absence of 100 or 200 nM of the CDK9 inhibitor flavopiridol (Flav). Using primers spanning intron-exon junctions, nuclear pre-mRNA abundance for ISG54 and RPS11 was quantified using real-time RT-PCR and normalized to GAPDH mRNA. (B) 2fTGH cells were either left untreated or treated with IFN-α in the absence or presence of TSA for 60 min. ChIP assays were performed with antibodies against CDK9, and recovered ISG56 and ISG15 promoter sequences were quantified by real-time PCR relative to input and represented as percentage of the input signal. (C) BT474 cells were either left untreated or treated with IFN-α in the absence or presence of TSA after exposure to geldanamycin (Gelda) for 24 h, as indicated. mRNAs for ISG56, OAS2, and c-Myc were quantified by real-time RT-PCR and normalized to GAPDH. (D) Control and NELF-E KD cells were either left untreated or treated with IFN-α in the absence or presence of TSA. ISG56 mRNA was quantified using real-time RT-PCR and normalized to GAPDH mRNA abundance (left). Knockdown was verified by Western blotting using anti–NELF-E antibodies (right). (E) Control and Spt5 KD cells were either left untreated or treated with IFN-α in the absence or presence of TSA. ISG56, Spt5 mRNA, and ISG54 pre-mRNA were quantified using real-time RT-PCR and normalized to GAPDH mRNA abundance. **, P < 0.03; ***, P < 0.005 from representative experiment (of two) performed in duplicate, and error bars represent +SD. A.U., arbitrary units.
To directly probe the involvement of P-TEFb in ISG expression, we assessed its recruitment to chromatin in response to IFN treatment. P-TEFb was absent from ISG promoters before IFN treatment (Fig. 4 B) but was rapidly recruited to the promoters of both ISG56 and ISG15, as measured by ChIP for CDK9. Notably, chromatin-bound CDK9 was not observed in cells treated with IFN in the presence of TSA (P < 0.005), suggesting that P-TEFb recruitment required HDAC activity.
In addition to the RNAPII carboxyl-terminal tail, another CDK9 substrate is the negative elongation factor (NELF; Kwak and Lis, 2013). It has been recently reported that transcriptional inhibition by HDAC inhibitors in a different biological context was dependent on HSP90 function, whose ability to stabilize the NELF complex and therefore block elongation was dependent on HDAC activity (Greer et al., 2015). Greer et al. (2015) showed that repression of ERBB2 and MYC expression by HDAC inhibitors is antagonized by geldanamycin treatment, a potent HSP90 inhibitor that led to loss of NELF and rescue of elongation. However, geldanamycin destabilization of NELF failed to rescue ISG transcription from HDAC inhibition; instead, ISG expression was partially inhibited by geldanamycin alone. However, repressed c-Myc expression was normalized by concomitant TSA and geldanamycin treatment (Fig. 4 C), consistent with previous observations (Greer et al., 2015).
We also tested more directly whether NELF dismissal could circumvent the HDAC requirement for ISG transcription. The NELF complex was depleted by RNA interference targeting NELF-E, since depletion of any of the four NELF subunits leads to functional loss of the entire complex (Narita et al., 2007). Again, destabilization of the NELF complex through reduced NELF-E did not significantly alter ISG induction in response to IFN and failed to rescue gene expression in absence of HDAC activity (Fig. 4 D).
The transcription elongation factor DRB sensitivity-inducing factor (DSIF) is another target of CDK9 that has been implicated in promoter proximal pausing of Pol II (Yamaguchi et al., 2013). To test its role, we targeted Spt5, the major subunit of DSIF, by RNA interference. Interestingly, down-regulation of Spt5 substantially increased ISG induction in response to IFN, but, similar to NELF depletion, failed to rescue gene expression when HDAC activity was inhibited (Fig. 4 E, left). DSIF also plays a role in coordinating elongation with mRNA splicing and nuclear export (Diamant et al., 2012), which could complicate interpretation of changes in mRNA abundance in its absence. To ascertain whether the role of DSIF in ISG regulation was transcriptional, we scored the abundance of ISG54 pre-mRNA transcripts as a measure of ongoing transcription after IFN stimulation. The effect of DSIF depletion was even greater on primary transcripts, suggesting that DSIF plays a major role as a negative regulator of ISG transcription (Fig. 4 E, right). However, ISG transcription remained sensitive to HDAC inhibition even in absence of the negative regulation imposed by DSIF. These results demonstrate that displacement of NELF or inactivation of DSIF are insufficient, at least individually, to allow transition from transcription initiation to transcriptional elongation in absence of HDAC activity, suggesting that P-TEFb likely targets additional substrates to enhance elongation (Ramanathan et al., 2001; Glover-Cutter et al., 2009; Hsin et al., 2011).
Brd4 is required for ISG transcription
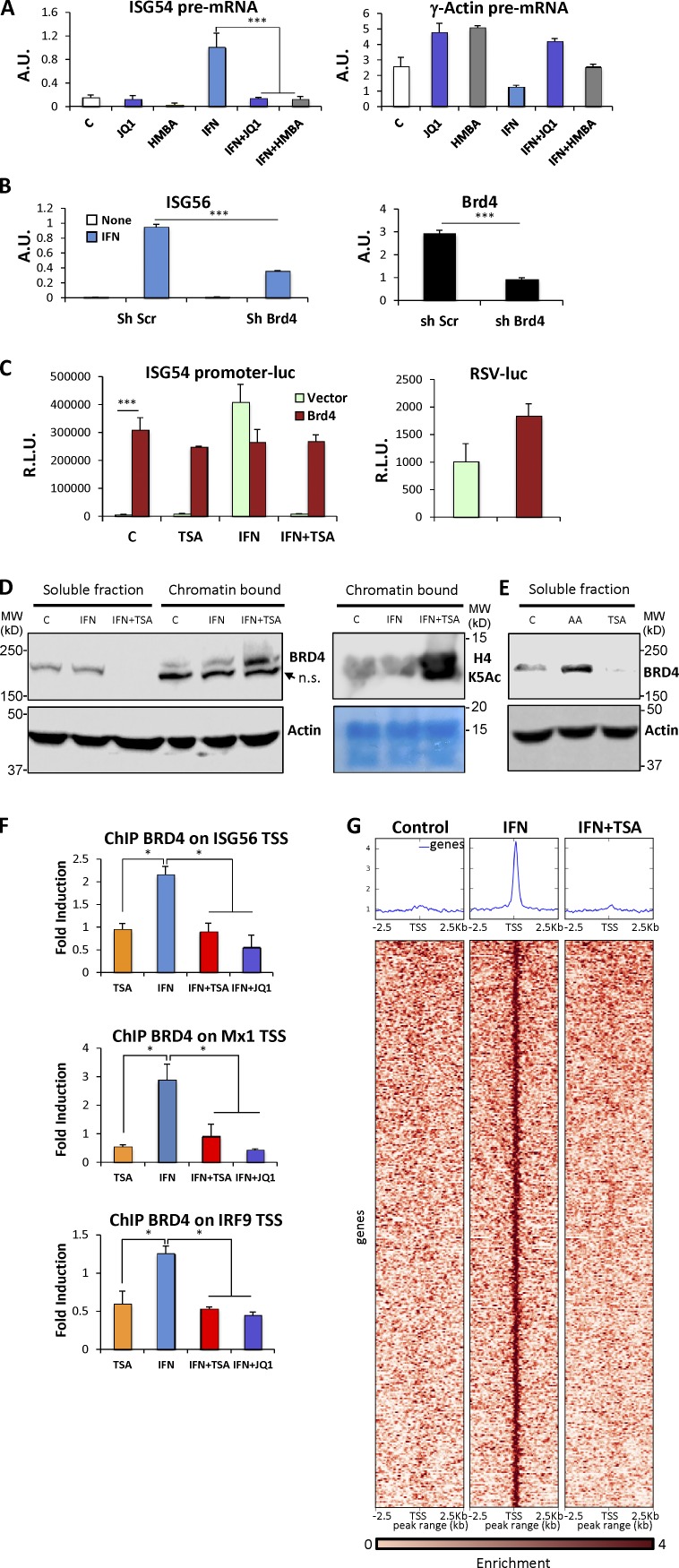
It has been shown that Brd4, an acetylated lysine binding protein, coordinates the recruitment of P-TEFb to regulate transcription of target genes, including ISGs (Jang et al., 2005; Yang et al., 2005; Hargreaves et al., 2009; Patel et al., 2013). We hypothesized that perturbing the nuclear acetylation state with a potent HDAC inhibitor could impair proper recruitment of Brd4. To examine this possibility, we tested the effect of Brd4 inhibition on ISG expression. To focus on transcriptional events, we again monitored induction of ISG unspliced pre-mRNA. JQ-1, a BET protein domain selective inhibitor that impairs Brd4 recruitment (Filippakopoulos et al., 2010), blocked induction of ISG54 nascent transcripts in IFN-treated cells (Fig. 5 A, left), without affecting the nuclear abundance of constitutively expressed γ-actin nascent transcripts (Fig. 5 A, right). Similarly, hexamethylene bisacetamide (HMBA), another inhibitor of BET bromo-domain proteins (Nilsson et al., 2016), blocked ISG transcription without affecting γ-actin expression (Fig. 5 A). Because these agents inhibit multiple BET proteins (Filippakopoulos et al., 2010; Nilsson et al., 2016), we examined the specific role of Brd4 by RNA interference. Brd4 was depleted by using a lentiviral-transduced hairpin, reducing Brd4 mRNA levels by >80% (P < 0.001; Fig. 5 B). Depletion of Brd4 significantly impaired ISG induction in response to IFN (P < 0.001; Fig. 5 B). Conversely, we tested the effect of Brd4 overexpression on ISG transcription by monitoring the activity of ISG54-luciferase. Overexpression of Brd4 significantly increased ISG54 promoter activity in the absence of IFN treatment (P < 0.001; Fig. 5 C, left), but showed only a modest stimulatory effect on the constitutive promoter from Rous sarcoma virus (Fig. 5 C, right). Surprisingly, Brd4-driven ISG54 promoter activity was largely resistant to HDAC inhibition, with no significant differences in Brd4-driven expression with or without TSA and IFN. In contrast, in the absence of ectopic Brd4 expression, IFN-driven ISG54 promoter activity was completely suppressed by TSA. To examine the role of acetylated lysine binding in the observed action of Brd4, we expressed mutant versions of Brd4 lacking bromodomain 1 (ΔBD1) or bromodomain 2 (ΔBD2) or containing point mutations in both bromodomains (Brd4 YY) that abrogate acetylated lysine binding (Patel et al., 2013). None of these constructs rescued ISG expression (Fig. S1). Interestingly, expression of either ΔBD2 or Brd4 YY construct impaired IFN-stimulated promoter activity, suggesting a dominant-negative action. This result suggests that high amounts of available Brd4 protein led to its facilitated recruitment to ISG promoters in an acetylated lysine binding–dependent manner, allowing unregulated transcription at least from transiently transfected DNA templates.
Figure 5.
Brd4 is required for ISG transcription. (A) HeLa cells were either left untreated or treated with IFN-α for 1 h in the presence or absence of JQ-1 or HMBA. Using primers spanning intron-exon junctions, nuclear pre-mRNA for ISG54 and γ-actin was quantified by real-time RT-PCR and normalized to GAPDH mRNA. (B) Control and Brd4 KD cells were either left untreated or treated with IFN-α. ISG56 and Brd4 mRNA were quantified using real-time RT-PCR and normalized to GAPDH mRNA abundance. A.U., arbitrary units. (C) HEK293T cells were transfected with pcDNA3 (Ctl) or Brd4 expression constructs along with a luciferase reporter driven by the ISG54 promoter. 24 h after transfection, cells were either left untreated or treated with IFN-α in the absence or presence of TSA before being assayed for luciferase activity (left). HEK293T cells were transfected with pcDNA3 (Ctl) or Brd4 expression constructs along with a luciferase reporter driven by the constitutive Rous sarcoma virus promoter and assayed for luciferase activity after 36 h (right). R.L.U., relative light units. (D) HeLa cells were either left untreated or treated with IFN-α for 2 h in the presence or absence of TSA. Nuclear proteins were extracted as a soluble fraction and a chromatin-bound fraction and analyzed by Western blotting using anti-Brd4, H4-AcK5, and actin antibodies, as indicated. (E) Soluble nuclear proteins from HeLa cells stimulated with IFN-α in the absence or presence of TSA or anacardic acid (AA) for 60 min were analyzed by Western blotting. (F) HeLa cells were treated for 60 min as shown, and ChIP assays were performed with antibodies against Brd4 and analyzed for recovered ISG56, Mx1, and IRF9 TSS spanning sequences by real-time PCR, normalized to input, and represented as fold enrichment over an untreated sample. (G) Profile and heat map of the enrichment of Brd4 peaks ±2.5 kb relative to TSS for genes showing IFN-dependent enrichment of Brd4. Each row represents a unique gene segment. *, P < 0.05; ***, P < 0.001 for representative experiment (of two) performed in duplicate, and error bars represent +SD.
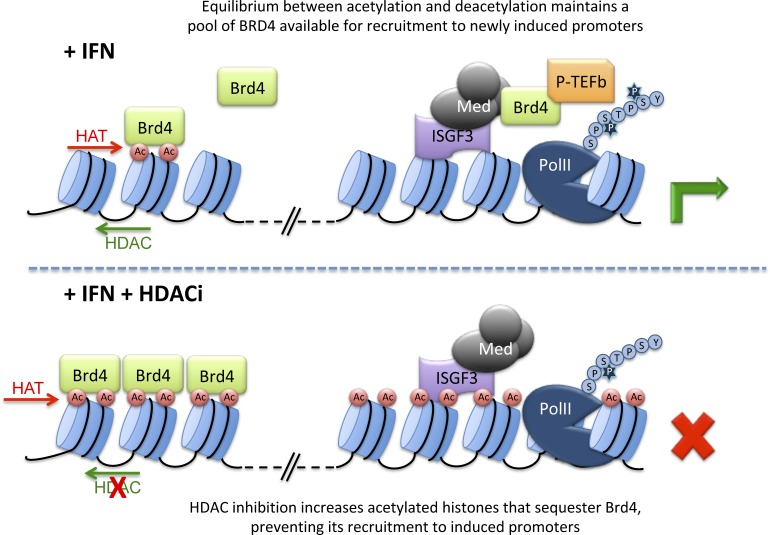
Brd4 associates with elongating polymerase to affect transcriptional elongation (Kanno et al., 2014). We reasoned that HDAC inhibition would lead to a global increase in acetylation-dependent bromodomain binding sites in chromatin which could trap Brd4, thus limiting its accessibility to ISG promoters. To test this notion, we examined global Brd4 distribution in cells treated with IFN in the presence or absence of TSA, quantifying nuclear Brd4 in soluble versus chromatin-bound fractions (Fig. 5 D). Most Brd4 was detected in the soluble nucleoplasm fraction in untreated or IFN-treated cells (Fig. 5 D, compare lanes 1 and 2 with 5 and 6). In contrast, soluble Brd4 was undetectable in nuclei of TSA-treated cells (Fig. 5 D, lane 3). Instead, Brd4 was exclusively chromatin-bound (Fig. 5 D, lane 6). As expected, treatment with TSA led to a large increase in histone acetylation (Fig. 5 D, middle), including increased acetylated H4K5, a selective binding site for Brd protein bromodomains (Marchand and Caflisch, 2015).
A corollary to a requirement for continued HDAC activity to maintain soluble Brd4 would be that continued histone acetyltransferase (HAT) activity is needed to drive chromatin acetylation, which is opposed by constitutive HDAC activity. To test this notion, we analyzed Brd4 solubility in nuclei from cells treated with the general HAT inhibitor, anacardic acid (Balasubramanyam et al., 2003). Soluble Brd4 was enriched by this treatment (Fig. 5 E, lane 2), opposite to its depletion by TSA (Fig. 5 E, lane 3). The abundance of total nuclear BRD4 was unchanged by either treatment (Fig. S1 D). These results indicate that blocking ongoing HDAC activity allows Brd4 to accumulate on acetylated nucleosomes, removing it from a more soluble available pool, whereas blocking constitutive HAT activity does the opposite, consistent with the notion that the steady-state levels of available Brd4 are maintained by the balance between constitutive HAT and HDAC activities.
To directly assess the effect of IFN and HDAC inhibition on Brd4 recruitment, we assayed the presence of Brd4 at ISG promoters by ChIP. Brd4 was recruited to three typical ISG promoters after 1 h of IFN stimulation, as previously described (Patel et al., 2013). Importantly, this recruitment was abrogated by TSA, and, as expected, by JQ-1 (Fig. 5 F). We also performed global ChIP-seq for Brd4 recruitment. IFN-dependent Brd4 recruitment was observed at 366 TSS-proximal regions, and this recruitment was abrogated by TSA treatment (Fig. 5 G, Table S1). In contrast, Brd4 recruitment at other genomic loci was not sensitive to TSA (Fig. S2). Enrichr pathway analysis (Kuleshov et al., 2016) of genes associated with IFN-dependent Brd4 recruitment (Tables S2 and S3) showed significant enrichment of genes regulated in virus-infected cells (P < 0.0015) and genes regulated by IFN (Table S4; P < 0.00015; Rusinova et al., 2013).
Combined HDAC and Brd4 inhibition as a potential therapy for type I interferonopathies
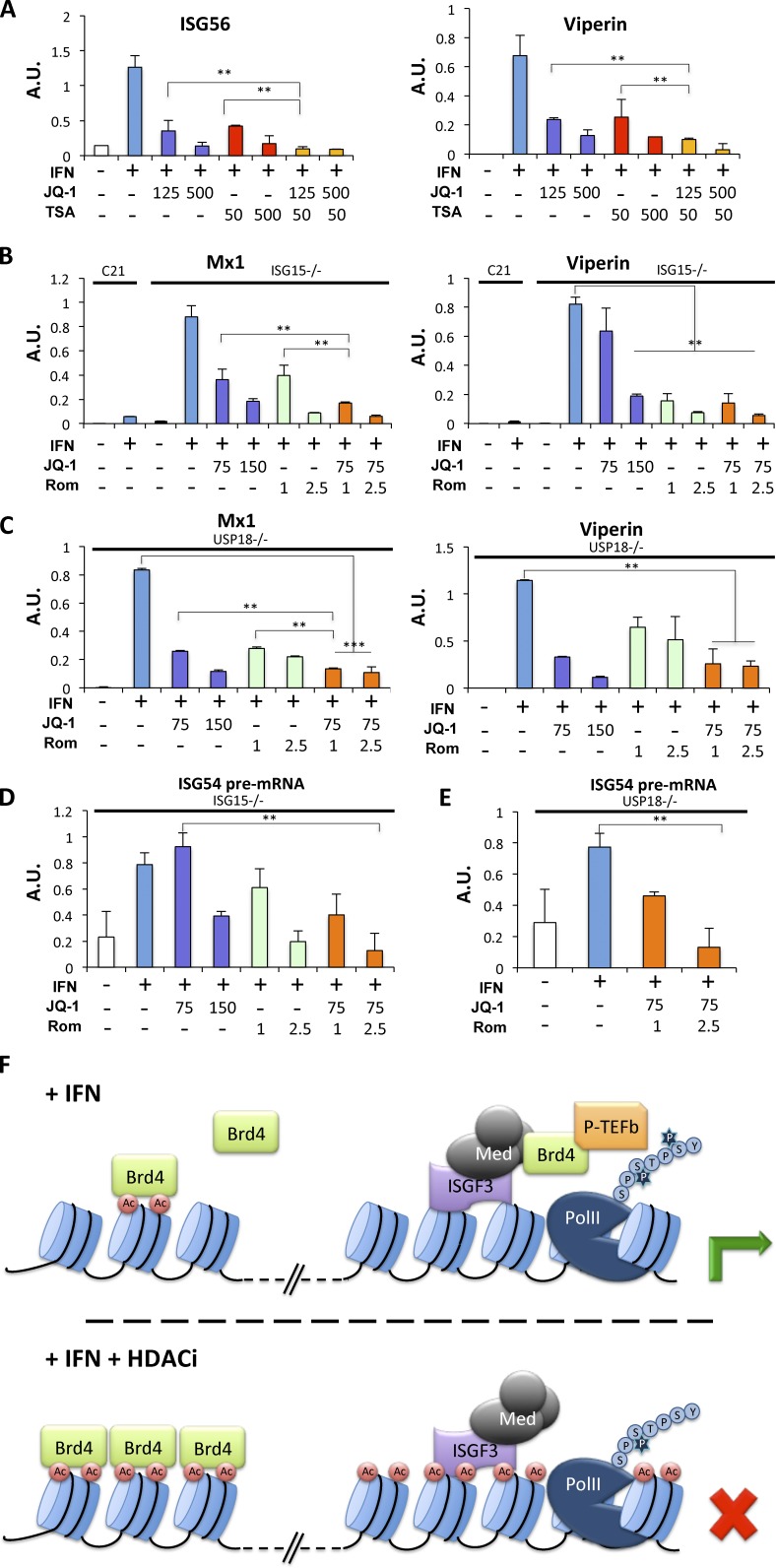
Type I interferonopathy refers to a group of monogenic autoinflammatory diseases in which a constitutive up-regulation of type I IFN production or signaling is associated with pathogenesis (Crow, 2011). Similarly, a common feature of most systemic lupus erythematosus patients is elevated serum levels of type I IFN and an ISG signature (Crow, 2014), and impeding this pathway can be beneficial (Kirou and Gkrouzman, 2013). We reasoned that combined HDAC and Brd4 inhibitor treatment should efficiently thwart ISG expression and could be therapeutically beneficial. As a proof of principle, we treated IFN-stimulated HEK 293T cells with TSA, JQ-1, or a combination of both drugs. As predicted, a combination of JQ-1 and TSA even at low concentrations (as low as 125 nM for JQ-1 and 50 nM for TSA) almost fully inhibited expression of ISG (Fig. 6 A). We extended this study to hTERT immortalized fibroblasts from patients harboring symptoms of type I interferonopathy due to inborn mutations of the negative regulators ISG15 or USP18 (Malakhova et al., 2006; Zhang and Zhang, 2011; Zhang et al., 2015). Mutant fibroblasts failed to properly down-regulate IFN signaling and therefore exhibited heightened expression of a subset of ISGs, including Mx1 and Viperin (Zhang et al., 2015). As shown in Fig. 6 B, high-level Mx 1 mRNA present in ISG15-deficient fibroblasts was significantly inhibited (P < 0.009) after combined treatment with romidepsin and JQ-1 and normalized to levels comparable to those observed in control fibroblasts isolated from healthy individuals. Similarly, increased levels of viperin in IFN-treated cells were significantly reduced by the combined inhibitor regimen (P < 0.007). Likewise, the high ISG expression in USP18-deficient fibroblasts was significantly inhibited (P < 0.0002) by cotreatment with JQ-1 and romidepsin (Fig. 6 C), and inhibition by cotreatment was significantly reduced compared with either agent alone (P < 0.005). Inhibition of heightened ISG expression by combined inhibitor treatment was also observed in cells from two additional ISG15 mutant patients (Fig. S3, A and D), while constitutive gene expression was unperturbed (Fig. S3, B, C, E, and F). ISG expression in primary cells ex vivo was also impaired by HDAC and Brd4 inhibitors (Fig. S4), suggesting that this approach could be a suitable therapy for suppression of IFN responses. To ascertain that the target of this drug combination was primary transcription, we confirmed that the abundance of ISG54 primary transcripts was effectively inhibited by combined JQ-1 and romidepsin in both ISG15- and USP18-deficient fibroblasts (Fig. 6, D and E).
Figure 6.
Combined HDAC and Brd4 inhibition is a potential therapy for type I interferonopathies. (A) HEK293T cells were either left untreated or treated with IFN-α for 6 h in the presence or absence of JQ-1, TSA, or a combination of the two inhibitors, as indicated (nM). ISG56 and Viperin mRNA were quantified using real-time RT-PCR and normalized to GAPDH mRNA abundance. (B) hTert-immortalized human fibroblasts from an ISG15-deficient patient and a healthy donor (C21) were treated with IFN-α for 8 h, washed with PBS, and incubated in the absence of IFN for 3 d. Where indicated, cells were treated with JQ-1, romidepsin, or a combination of the two drugs (nM) for the final 24 h before RNA extraction. Mx1 and Viperin mRNA were quantified using real-time RT-PCR and normalized to GAPDH mRNA abundance. (C) As in B, except that hTert-immortalized fibroblasts from a USP18-deficient patient were used. (D) As in B, except that nuclear pre-mRNA for ISG54 from ISG15-deficient cells was scored. (E) As in D, except that pre-mRNA for ISG54 from hTert-immortalized USP18-deficient fibroblasts was quantified. A.U., arbitrary units. (F) Proposed model of Brd4 sequestration in the absence of constitutive HDAC function, as described in the text. *, P < 0.02; **, P < 0.01; ***, P < 0.005; ****, P < 0.002 for representative experiments (of three) performed in duplicate, and error bars represent +SD.
Discussion
The data described in this report are consistent with the model that ISG transcription depends on HDAC activity to facilitate the transition from committed initiation to processive elongation through, at least in part, the targeted recruitment of P-TEFb to ISG promoter-proximal regions mediated by the Brd4 protein, and that sequestration of Brd4 by acetylated chromatin impairs ISG expression (Fig. 6 F). Previous reports have substantiated a requirement of HDAC activity for IFN-stimulated transcription (Génin et al., 2003; Nusinzon and Horvath, 2003; Chang et al., 2004; Sakamoto et al., 2004), and this observation has been extended to additional inducible expression systems, such as in response to IFN-γ (Zupkovitz et al., 2006), glucocorticoids (Tichonicky et al., 1981; Plesko et al., 1983; Bresnick et al., 1990; Mulholland et al., 2003; Kadiyala et al., 2013) or in STAT5-dependent transcription (Rascle et al., 2003; Xu et al., 2003). Broad requirement for HDAC activity during acute induction of gene expression from stimulus-dependent promoters suggests a common positive role for HDAC enzymes for this subset of genes, in addition to its well-appreciated repressive role of maintaining histone deacetylation on silent chromatin.
We found that HDAC activity was not required to reorganize promoter chromatin in response to IFN, allow chromatin binding of the activated ISGF3 transcription factor complex, recruit RNAPII to ISG promoters and promoter-proximal regions, or allow transcriptional initiation, as judged by Ser 5 phosphorylation of RNAPII. However, accumulation of RNAPII within gene bodies was impaired in the absence of HDAC activity, indicative of failed elongation. These findings suggest that HDAC activity is required for the transition to processive elongation, possibly to overcome promoter-proximal pausing. Indeed, we found evidence for significant promoter-proximal pausing at ISG promoters by the large accumulation of RNAPII at promoter-proximal sites relative to gene bodies (Fig. 3), a common observation for paused genes (Mayer et al., 2017). However, unlike other instances of promoter pausing, e.g., poised promoters regulated through a pause-release mechanism (Young, 2011), ISG promoters only displayed signs of polymerase pausing following IFN stimulation. Thus, these promoters appear to combine two distinct mechanisms of transcriptional control, inducible recruitment of polymerase followed by an HDAC-dependent elongation barrier.
Promoter-proximal pausing is a well-documented barrier to successful transcription, commonly associated with insufficient inactivation of negative regulatory factors such as DSIF and NELF and impaired activation of polymerase elongation by P-TEFb (Sehgal et al., 1979; Wada et al., 1998). Indeed, transcription from ISG promoters was exquisitely sensitive to the P-TEFb inhibitor flavopiridol (Fig. 4 A), and P-TEFb recruitment required HDAC activity (Fig. 4 B). However, the action of neither DSIF nor NELF could individually account for the requirement of HDAC activity (Fig. 4). Therefore, the recruitment and activity of P-TEFb likely provides essential functions in addition to dismissal of NELF and modulation of DSIF.
A parsimonious interpretation of our data would posit a continuous cycle of histone acetylation and deacetylation that regulates the pool of Brd4 protein available for recruitment to ISGF3-bound mediator at IFN inducible promoters (Fig. 6 F). Under steady-state conditions, Brd4 binds chromatin as a consequence of interactions between its paired bromodomains and acetylated histones and exists in an equilibrium between bound and transiently free due to deacetylation of histones that releases a portion of bound Brd4. Deacetylation, mediated by HDAC1/2/Sin3A, is countered by continued reacetylation by HAT complexes, resulting in rebinding of Brd4 to chromatin. This constant release-and-rebind scenario maintains a small pool of Brd4 that can be mobilized for binding to newly available recruitment sites, for instance, at activated inducible promoters, where it recruits P-TEFb. In the absence of constitutive HDAC activity, Brd4 is trapped on chromatin, due to the absence of the transient removal of the sequestering acetylation marks throughout the genome that greatly outnumber the abundance of Brd4. In contrast, the absence of continuous HAT activity causes Brd4 release due to unopposed HDAC-mediated histone deacetylation. However, while this free Brd4 would be available for remobilization to newly inducible promoters, transcription is not supported in the absence of active acetyltransferases, due to their essential role in other aspects of transcriptional activation (Utley et al., 1998). Dependence on the availability of mobile Brd4 would only apply to stimulus-dependent promoters, since constitutively transcribed genes would have preassociated Brd4 regardless of the pool of free protein (Zhao et al., 2011). We note, however, that we cannot rule out the possibility that there are additional requirements for HDAC activity for ISG induction beyond regulation of BRD4 availability.
There is precedent for this notion from other gene induction mechanisms. Greer et al. (2015) also noted that HDAC activity enhanced transcriptional elongation and that HDACi led to a global redistribution of Brd4. Similarly, Wang et al. (2009) described an HAT/HDAC cycle at active genes in T lymphocytes, but they concluded that HDAC activity was more involved in resetting chromatin following gene induction and in maintaining poised promoters in an inactive state. Cellular stress activates a gene expression program that is dependent on HDAC activity and has been shown to be dependent on transient release of Brd4 from chromatin, following dephosphorylation of H3S10 (Ai et al., 2011; Hu et al., 2014). Distinct from ISG induction, cellular stress induces global changes in histone modifications and Brd4 availability, whereas IFN treatment appears to cause only gene-specific changes at inducible promoters. Moreover, ISG induction appears to depend on a constitutive cycle of global acetylation and deacetylation to modulate Brd4 availability, a cycle that is not itself stimulus dependent. Why cellular stress requires global changes in histone acetylation to mobilize Brd4 while the constitutive cycle is sufficient to allow expression of ISGs may relate to the magnitude of the respective responses or the number of activated genes.
Aberrant ISG regulation results in interferonopathies and other autoimmune diseases (Crow and Manel, 2015; Rodero and Crow, 2016). There is strong evidence that an ISG expression signature is at least in part causative in such diseases, rather than being simply correlative. Therefore, inhibition of enhanced ISG expression has become an attractive anti-inflammatory therapeutic target, with the caveat that ISG inhibition could increase the risk for viral infection (Kalunian, 2016). An attractive alternative to abrogating IFN receptor signaling in autoimmunity would be to modulate aberrant constitutive signaling back toward physiological levels. Treatment of patient cell lines from two distinct interferonopathies with HDAC and Brd4 inhibitors reduced aberrant ISG expression comparable to the JAK inhibitors ruxolitinib and tofacitinib (Fig. S3) with little apparent toxicity (Fig. S5). Our results from combined low-dose HDAC and Brd inhibitors, drugs available for clinical use in humans and well tolerated by patients, may provide a viable approach to reducing inflammation without abrogating beneficial IFN signaling.
Materials and methods
Cell culture
HeLa S3, HEK293, HEK293T, 2fTGH, FS2 human diploid fibroblasts, BT474 (a gift from B. Neel, New York University [NYU] School of Medicine, New York, NY), Sin3A and Sin3B conditionally-deficient immortalized mouse fibroblasts (a gift from G. David, NYU School of Medicine), and patient-derived hTERT immortalized human fibroblast cells (provided by D. Bogunovic, Mount Sinai Icahn School of Medicine, New York, NY) were maintained in DMEM supplemented with 10% calf serum and antibiotics. Sin3A and Sin3B conditional cells (Dannenberg et al., 2005) expressed tamoxifen-inducible Cre recombinase (McDonel et al., 2012), which was activated by incubation of cells for 3 d with 100 nM 4-hydroxytamoxifen (Sigma-Aldrich). Transfections of cells were performed by using the calcium phosphate method, and cellular extracts were collected for protein and RNA analysis, as previously described (Chang et al., 2004). Where indicated, cells were also treated with IFN-α2a (Hoffman-La Roche) at 1,000 U/ml, TSA (Reagents Direct) at 500 ng/ml unless indicated otherwise, romidepsin (depsipeptide; a gift from Fujisawa Pharmaceuticals) as indicated, flavopiridol (Selleckchem) at 100 or 200 nM, JQ-1(+; a gift from J. Bradner, Harvard University, Cambridge, MA) as indicated, RGFP233 (a gift from J. Buxbaum, The Scripps Research Institute, La Jolla, CA), RGFP966 (Selleckchem) at 5 or 10 µM, geldanamycin (Sigma-Aldrich) at 20 µM, HMBA (Sigma-Aldrich) at 10 mM, or anacardic acid (EMD Millipore) at 200 µM. Unless otherwise indicated, IFN-α treatments were for 6 h, and inhibitors were added 15 min before IFN stimulation. Expression constructs for Brd4 and HDAC1 and 2 were gifts of E. Hernando (NYU School of Medicine) and C. Callebaut and E. Verdin (University of California, San Francisco, San Francisco CA), respectively. Cell viability was determined for triplicate samples by using the tetrazolium salt WST-8 method (Bimake).
Primary cells analyzed ex vivo
Human peripheral blood mononuclear cells (PBMCs), provided by D. Bogunovic, were incubated in RPMI 1640 medium supplemented with 10% heat-inactivated FBS for 5 h in the absence or presence of IFN and inhibitors before isolation of RNA by the Trizol method. Primary murine embryo fibroblasts (EMD Millipore) were incubated for 6 h in DMEM supplemented with 10% FBS in the presence or absence of IFN and inhibitors before RNA isolation.
In vitro transcription measurements
In vitro transcription runoff assays were performed essentially as described (Dignam et al., 1983). HEK293 cells were used as a source of nuclear extract, supplemented with recombinant ISGF3 produced by transfection. Recombinant ISGF3 was produced by transfecting HEK293 cells with expression plasmids for STAT1, STAT2, IRF9, and JAK1 (Bluyssen and Levy, 1997). Transcription extracts were programmed with HindIII-digested ISG54 p107, containing promoter sequences from −547 to +283 of the human ISG54 gene (Levy et al., 1986), or PvuII-digested G6TI-CAT control DNA template containing six SP1 sites derived from the SV40 early promoter (Gill et al., 1994; Carey et al., 2010).
Run-on nuclear transcription experiments were conducted essentially as described (Decker et al., 1989; Chang et al., 2004). In brief, 10 million HeLa S3 cells were used per point. Nuclei were isolated from cells in RSBG40 lysis buffer (10 mM Tris, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 10% glycerol, 0.25% NP-40, 0.5 mM DTT, 0.5 mM PMSF, and 1× protease inhibitor cocktail). Polymerase elongation was released in the presence of radiolabeled UTP, with or without addition of 150 ng/ml TSA. Radiolabeled nuclear RNA was isolated by TRIzol reagent (Invitrogen). 6 µg plasmid DNA was spotted onto nitrocellulose for slotblot hybridization, and radioactive signals were quantified by phosphorimaging (Bio-Rad). DNA probes used were the following: Vector (pGEM1; Promega), γ-actin cDNA (Cleveland et al., 1980), GAPDH (Dani et al., 1984), ISG15 TaqI fragment from exon 2 (Reich et al., 1987), ISG56 (Larner et al., 1984), IRF9 (Veals et al., 1992), GBP (Decker et al., 1989), 6-16, and 9-27 (Friedman et al., 1984).
Protein assays
Nuclear extracts were prepared and analyzed by immunoblotting, as described (Paulson et al., 1999). In brief, cells were lysed in RSBG40, soluble nuclear proteins were extracted from pelleted nuclei in RSB supplemented with 150 mM NaCl, and chromatin-bound proteins were subsequently extracted in 400 mM NaCl buffer. Alternatively, whole-cell lysates were prepared as described (Paulson et al., 1999). Antibodies used were anti-IRF9 (Veals et al., 1993), anti-STAT1, anti-STAT2, anti-phospho-STAT1 and anti-phospho-STAT2 (Invitrogen), anti–α-tubulin (T9026; Sigma), anti-HDAC1 (clone 2E10, 05-614; Millipore Sigma), anti-HDAC2 (clone 3F3, 05-814; Millipore Sigma), anti-E2F4 (A-20, sc-1082; Santa Cruz Biotechnology), anti-Sin3B (AK-12; Santa Cruz Biotechnology), anti–NELF-E (H140; Santa Cruz Biotechnology), anti-Brd4 (A-301-985A100; Bethyl Laboratories), anti-actin (Clone C4, MAB1501; Millipore Sigma). Luciferase assays were performed using standard methods as previously described (Marié et al., 2000).
Real-time RT-PCR
Cytoplasmic and nuclear RNA was isolated and converted to cDNA as described (Marié et al., 1998). Relative abundance of specific mRNA sequences was determined by real-time fluorescent PCR, as described (Wang et al., 2006), by using SYBR Green (Molecular Probes), with comparison to a standard curve generated by serial dilution of a cDNA sample containing abundant target sequences and normalization to the expression of GAPDH. All PCR reactions were performed in triplicate, PCR efficiencies were >85%, linearity of standard curves was determined by least-squares linear regression, and standard errors were typically <10% of mean values. Sequences of primers used are available on request.
ChIP-qPCR and ChIP-seq
ChIP was performed as described (Chang et al., 2004). In brief, cells from two 15-cm plates (∼40 million) were treated with 1% formaldehyde for 30 min at 4°C, and fixation was quenched by addition of 125 mM glycine. Fixed cells were lysed in 10 ml cold buffer A (50 mM Hepes, pH 7.4, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 10% glycerol, 0.5% NP-40, and 0.25% Triton X-100), and nuclei were collected by centrifugation and washed with 10 ml cold buffer B (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.5 mM EGTA, and 200 mM NaCl). Nuclei were extracted in 2 ml cold buffer C (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 5% glycerol, 0.1% sodium deoxycholate, 0.1% SDS, and 1% Triton X-100). Genomic DNA was sheared by sonication, either by using Sonic Dismembrator Model 500 (Fisher Scientific) in a dry ice ethanol bath, or by using BiorupterPlus (Diagenode) in a 4°C water bath, to achieve chromatin fragments between 100 and 500 bp. Sheared chromatin from 4–10 million cells was used for each immunoprecipitation. Immunoprecipitates were collected on protein A Sepharose or protein A magnetic beads and digested with proteinase K in 100 µl buffer D (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.5% SDS, 25 mM EDTA, 100 µg/ml proteinase K) at 55°C for 3 h. Cross-links were reversed by incubating the digested chromatin at 65°C overnight. Immunoprecipitated DNA sequences were recovered by alcohol precipitation and quantified by real-time PCR compared with input genomic DNA.
ChIP-Seq libraries were made using the KAPA Hyper Prep kit (KK8504; Roche) using 25 ng DNA per sample, according to the manufacturer’s protocol for end repair and A-tailing. For adapter ligation, 8-nt multiplexing IDT (Integrated DNA Technologies) adapters (5 µM) were used. 0.8X post-ligation cleanup was performed with AMPure XP beads (Beckman Coulter) on adapter-ligated libraries. Libraries were size selected before PCR amplification with a 0.6X right-side selection and 1.2X rebind and cleanup. Six PCR cycles were used for library amplification. Libraries were sequenced on an Illumina HiSeq 4000 instrument, using a single read 50 protocol; eight samples were pooled in one lane of a high-output single read flow cell. Sequencing results were demultiplexed and converted to FASTQ format using Illumina Bcl2Fastq software.
ChIP-Seq data analysis was performed using the HiC-Bench pipeline (Lazaris et al., 2017). GenomicTools (Tsirigos et al., 2012), SAMtools (Li et al., 2009), DeepTools (Ramírez et al., 2016), BEDTools (Quinlan and Hall, 2010), BigBed Tools (Kent et al., 2010), Picard Tools (https://broadinstitute.github.io/picard/), and R (https://www.R-project.org/) were used to analyze the sequenced reads and check the quality of alignment. Bowtie2 (Langmead, 2010) was used to align the raw reads to the hg19 reference genome. After alignment, MACS2 peak caller (Feng et al., 2012) was used with parameters --nomodel (for bypassing the shifting model), --extsize:200, and P value 0.01 to identify enriched peaks, and these peaks were visualized in the UCSC Genome Browser (Kent et al., 2002). DiffBind (Ross-Innes et al., 2012) package was used to determine the differentially bound peaks. Samples and their peaks were loaded using samplesheet information with AnalysisMethod: DBA_DESEQ2 and th:0.05. A matrix was computed based on the reads with score: DBA_SCORE_RPKM, and a contrast was set up with category:DBA_condition, minMembers = 2. Differential binding affinity analysis was then performed by considering the full library size and annotated using bioMart. The Intervene tool (Khan and Mathelier, 2017) was used to compare peak overlaps between treatment groups. Biological pathways enriched for genes associated with IFN-induced peaks were identified by using Enrichr (Kuleshov et al., 2016) Disease/drugs module with default parameters. Genes associated with IFN-regulated peaks were queried for regulation by IFN by comparison to Interferome v2 (Rusinova et al., 2013).
Antibodies that were used for ChIP included 2 µg anti-RNA Pol-II Ab (N-20, sc-899; Santa Cruz Biotechnology), 5 µl anti-RNA Pol-II H14 (serine-5; MMS-134R; Covance), 2 µg anti H3K4 (Me3; ab85580-25; Abcam), 2 µg anti-CDK9 (C20, sc-484; Santa Cruz Biotechnology), and 5 µg anti-Brd4 (A-301-985A100; Bethyl Laboratories).
MNase-PCR nucleosome mapping
FS2 cells were starved for 72 h in DMEM supplemented with 0.02% FBS before treatment with IFN-α2a (1,000 U/ml) with or without TSA (1 µg/ml). Cells were fixed with 1% formaldehyde for 30 min at 4°C, lysed in 10 mM Tris-HCl, pH 7.5, 10 mM CaCl2, 3 mM MgCl2, 0.4% NP-40, 10 mM sodium butyrate, 0.1 µM bezamidine, 0.5 mM PMSF, 1× protease inhibitor cocktail, and nuclei were collected and resuspended in the same buffer at a final genomic DNA concentration of 1.25 µg/µl. Nuclei were digested with 4 U/ml micrococcal nuclease for 10–15 min at 37°C, and mononucleosomal DNA was purified by agarose electrophoresis. Nuclease protection of specific DNA segments was determined by PCR and normalized to a region from the ε-globin promoter protected by the N1 nucleosome (Gong et al., 1996; Gui and Dean, 2001).
RNA interference and shRNA knockdown
HEK293 cells were transfected twice sequentially with 20 nM siRNA oligonucleotides using the calcium phosphate precipitation method and treated with IFN-α and TSA for 6 h. The siRNA oligonucleotides targeted HDAC1 and 2 (Dharmacon SMARTpool M-003494, L-003495) or E2F4 (Balciunaite et al., 2005), a gift from B. Dynlacht (NYU School of Medicine). shRNA against NELF-E (clone NM_002904.4-1195s1c1) and Brd4 (clone NM_058243.1-1707s1c1) were Mission shRNA from Sigma-Aldrich, and shRNA against Spt5 was a gift from R. Dikstein (Weizmann Institute of Science, Rehovot, Israel).
Statistical analysis
Quantitative gene expression and ChIP-PCR data reflect the analysis of a minimum of duplicate samples. Luciferase data are averages of triplicate samples. Data are represented as means with error bars reflecting SD. Statistical significance was assessed by using Student’s t test. Representative experiments of a minimum of three replicates are shown, with the exception of the ChIP-Seq, which was performed once on duplicate samples.
Human subjects
The experiments described in this report involving patient-derived cells were determined to be exempt human research as defined by DHHS regulations 45 CFR 46.101(b) (4) by the Program for the Protection of Human Subjects of Icahn School of Medicine (IRB-16-01589).
Online supplemental material
Fig. S1 presents characterization of the requirement for the acetyl-lysine–binding bromodomains of BRD4 for its function in the IFN pathway. Fig. S1 also demonstrates that total nuclear BRD4 abundance is unchanged following treatment of cells with anacardic acid or TSA. Fig. S2 presents ChIP-seq analysis of control genes not affected by IFN or IFN + TSA treatment. Fig. S3 provides characterization of IFN and inhibitor responses in fibroblast cells from independent interferonopathy patients P2 and P3, along with additional control fibroblasts from healthy donors. Fig. S4 presents data on the effectiveness of HDACi on primary cells (human PBMCs from a healthy donor and mouse primary fibroblasts) analyzed ex vivo. Fig. S5 presents data showing that inhibitors used had minimal toxicity in vitro. Table S1 lists genes associated with IFN-induced ChIP-seq peaks that were abrogated by HDAC inhibition. Table S2 lists gene sets associated with “Virus perturbations from GEO up” pathways enriched for IFN-induced ChIP-Seq peaks from Enrichr analysis. Table S3 lists gene sets associated with “Virus perturbations from GEO up” pathways enriched for IFN-induced ChIP-Seq peaks from Enrichr analysis. Table S4 lists genes whose expression is regulated by IFN that were enriched for IFN-induced ChIP-Seq peaks.
Supplementary Material
Acknowledgments
We thank Karina Ray, Varshini Vasudevaraja, Aristotelis Tsirigos, and Adriana Heguy (NYU Division of Advanced Research Technologies) for assistance with DNA sequencing and data analysis. Special thanks to Dusan Bogunovic (Icahn School of Medicine) for the gift of human fibroblasts from interferonopathy patients. We thank Gregory David, Ekaterini Platanitis, Ben Neel, Rivka Dikstein, Zhijian Chen, Brian Dynlacht, Joel Buxbaum, James Bradner, Eva Hernando, Christian Callebaut, Eric Verdin, Keiko Ozato, and Laszlo Tora for gifts of reagents, helpful discussions, and encouragement. We are indebted to Fujisawa Pharmaceuticals and the National Cancer Institute Division of Cancer Treatment and Diagnosis for the gift of romidepsin (depsipeptide), and we thank Celeste Greer and Tae Hoon Kim (Yale University and University of Texas) for sharing ChIP-Seq datasets.
This work was supported in part by National Institutes of Health grants R01AI2890 to D.E. Levy and U54AI057158 to D.E. Levy and I.J. Marié. DNA sequencing at the NYU Genome Technology Center was supported in part by Cancer Center Support Grant P30CA016087 to the Laura and Isaac Perlmutter Cancer Center.
The authors declare no competing financial interests.
Author contributions: I.J. Marié designed and conducted experiments, analyzed data, and wrote the manuscript; H.-M. Chang conducted experiments; and D.E. Levy designed experiments, analyzed data, and wrote the manuscript.
References
- Agalioti T., Lomvardas S., Parekh B., Yie J., Maniatis T., and Thanos D.. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-beta promoter. Cell. 103:667–678. 10.1016/S0092-8674(00)00169-0 [DOI] [PubMed] [Google Scholar]
- Ai N., Hu X., Ding F., Yu B., Wang H., Lu X., Zhang K., Li Y., Han A., Lin W., et al. 2011. Signal-induced Brd4 release from chromatin is essential for its role transition from chromatin targeting to transcriptional regulation. Nucleic Acids Res. 39:9592–9604. 10.1093/nar/gkr698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer S.Y., Meng S., Shei A., and Hodin R.A.. 1998. p21(WAF1) is required for butyrate-mediated growth inhibition of human colon cancer cells. Proc. Natl. Acad. Sci. USA. 95:6791–6796. 10.1073/pnas.95.12.6791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au-Yeung N., Mandhana R., and Horvath C.M.. 2013. Transcriptional regulation by STAT1 and STAT2 in the interferon JAK-STAT pathway. JAK-STAT. 2:e23931 10.4161/jkst.23931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanyam K., Swaminathan V., Ranganathan A., and Kundu T.K.. 2003. Small molecule modulators of histone acetyltransferase p300. J. Biol. Chem. 278:19134–19140. 10.1074/jbc.M301580200 [DOI] [PubMed] [Google Scholar]
- Balciunaite E., Spektor A., Lents N.H., Cam H., Te Riele H., Scime A., Rudnicki M.A., Young R., and Dynlacht B.D.. 2005. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol. Cell. Biol. 25:8166–8178. 10.1128/MCB.25.18.8166-8178.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S., Eckner R., Grossman S., Oldread E., Arany Z., D’Andrea A., and Livingston D.M.. 1996. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 383:344–347. 10.1038/383344a0 [DOI] [PubMed] [Google Scholar]
- Bluyssen H.A.R., and Levy D.E.. 1997. Stat2 is a transcriptional activator that requires sequence-specific contacts provided by stat1 and p48 for stable interaction with DNA. J. Biol. Chem. 272:4600–4605. 10.1074/jbc.272.7.4600 [DOI] [PubMed] [Google Scholar]
- Boeger H., Bushnell D.A., Davis R., Griesenbeck J., Lorch Y., Strattan J.S., Westover K.D., and Kornberg R.D.. 2005. Structural basis of eukaryotic gene transcription. FEBS Lett. 579:899–903. 10.1016/j.febslet.2004.11.027 [DOI] [PubMed] [Google Scholar]
- Brès V., Yoh S.M., and Jones K.A.. 2008. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 20:334–340. 10.1016/j.ceb.2008.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresnick E.H., John S., Berard D.S., LeFebvre P., and Hager G.L.. 1990. Glucocorticoid receptor-dependent disruption of a specific nucleosome on the mouse mammary tumor virus promoter is prevented by sodium butyrate. Proc. Natl. Acad. Sci. USA. 87:3977–3981. 10.1073/pnas.87.10.3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M.F., Peterson C.L., and Smale S.T.. 2010. G-less cassette in vitro transcription using HeLa cell nuclear extracts. Cold Spring Harb. Protoc. 2010:pdb.prot5387 10.1101/pdb.prot5387 [DOI] [PubMed] [Google Scholar]
- Chang H.M., Paulson M., Holko M., Rice C.M., Williams B.R., Marié I., and Levy D.E.. 2004. Induction of interferon-stimulated gene expression and antiviral responses require protein deacetylase activity. Proc. Natl. Acad. Sci. USA. 101:9578–9583. 10.1073/pnas.0400567101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao S.H., and Price D.H.. 2001. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J. Biol. Chem. 276:31793–31799. 10.1074/jbc.M102306200 [DOI] [PubMed] [Google Scholar]
- Cho S., Schroeder S., and Ott M.. 2010. CYCLINg through transcription: posttranslational modifications of P-TEFb regulate transcription elongation. Cell Cycle. 9:1697–1705. 10.4161/cc.9.9.11346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleveland D.W., Lopata M.A., MacDonald R.J., Cowan N.J., Rutter W.J., and Kirschner M.W.. 1980. Number and evolutionary conservation of alpha- and beta-tubulin and cytoplasmic beta- and gamma-actin genes using specific cloned cDNA probes. Cell. 20:95–105. 10.1016/0092-8674(80)90238-X [DOI] [PubMed] [Google Scholar]
- Cosgrove M.S., Boeke J.D., and Wolberger C.. 2004. Regulated nucleosome mobility and the histone code. Nat. Struct. Mol. Biol. 11:1037–1043. 10.1038/nsmb851 [DOI] [PubMed] [Google Scholar]
- Crow M.K. 2014. Type I interferon in the pathogenesis of lupus. J. Immunol. 192:5459–5468. 10.4049/jimmunol.1002795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow Y.J. 2011. Type I interferonopathies: a novel set of inborn errors of immunity. Ann. N. Y. Acad. Sci. 1238:91–98. 10.1111/j.1749-6632.2011.06220.x [DOI] [PubMed] [Google Scholar]
- Crow Y.J., and Manel N.. 2015. Aicardi-Goutières syndrome and the type I interferonopathies. Nat. Rev. Immunol. 15:429–440. 10.1038/nri3850 [DOI] [PubMed] [Google Scholar]
- Dani C., Piechaczyk M., Audigier Y., El Sabouty S., Cathala G., Marty L., Fort P., Blanchard J.M., and Jeanteur P.. 1984. Characterization of the transcription products of glyceraldehyde 3-phosphate-dehydrogenase gene in HeLa cells. Eur. J. Biochem. 145:299–304. 10.1111/j.1432-1033.1984.tb08552.x [DOI] [PubMed] [Google Scholar]
- Dannenberg J.H., David G., Zhong S., van der Torre J., Wong W.H., and Depinho R.A.. 2005. mSin3A corepressor regulates diverse transcriptional networks governing normal and neoplastic growth and survival. Genes Dev. 19:1581–1595. 10.1101/gad.1286905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell J.E., Jr 1982. Variety in the level of gene control in eukaryotic cells. Nature. 297:365–371. 10.1038/297365a0 [DOI] [PubMed] [Google Scholar]
- David G., Grandinetti K.B., Finnerty P.M., Simpson N., Chu G.C., and Depinho R.A.. 2008. Specific requirement of the chromatin modifier mSin3B in cell cycle exit and cellular differentiation. Proc. Natl. Acad. Sci. USA. 105:4168–4172. 10.1073/pnas.0710285105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T., Lew D.J., Cheng Y.S., Levy D.E., and Darnell J.E. Jr. 1989. Interactions of alpha- and gamma-interferon in the transcriptional regulation of the gene encoding a guanylate-binding protein. EMBO J. 8:2009–2014. 10.1002/j.1460-2075.1989.tb03608.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamant G., Amir-Zilberstein L., Yamaguchi Y., Handa H., and Dikstein R.. 2012. DSIF restricts NF-κB signaling by coordinating elongation with mRNA processing of negative feedback genes. Cell Reports. 2:722–731. 10.1016/j.celrep.2012.08.041 [DOI] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz R.M., and Roeder R.G.. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475–1489. 10.1093/nar/11.5.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Liu T., Qin B., Zhang Y., and Liu X.S.. 2012. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 7:1728–1740. 10.1038/nprot.2012.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P., Qi J., Picaud S., Shen Y., Smith W.B., Fedorov O., Morse E.M., Keates T., Hickman T.T., Felletar I., et al. 2010. Selective inhibition of BET bromodomains. Nature. 468:1067–1073. 10.1038/nature09504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming S.B. 2016. Viral inhibition of the IFN-induced JAK/STAT signalling pathway: Development of live attenuated vaccines by mutation of viral-encoded IFN-antagonists. Vaccines (Basel). 4:E23 10.3390/vaccines4030023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman R.L., Manly S.P., McMahon M., Kerr I.M., and Stark G.R.. 1984. Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell. 38:745–755. 10.1016/0092-8674(84)90270-8 [DOI] [PubMed] [Google Scholar]
- Furumai R., Matsuyama A., Kobashi N., Lee K.H., Nishiyama M., Nakajima H., Tanaka A., Komatsu Y., Nishino N., Yoshida M., and Horinouchi S.. 2002. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 62:4916–4921. [PubMed] [Google Scholar]
- Génin P., Morin P., and Civas A.. 2003. Impairment of interferon-induced IRF-7 gene expression due to inhibition of ISGF3 formation by trichostatin A. J. Virol. 77:7113–7119. 10.1128/JVI.77.12.7113-7119.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G., Pascal E., Tseng Z.H., and Tjian R.. 1994. A glutamine-rich hydrophobic patch in transcription factor Sp1 contacts the dTAFII110 component of the Drosophila TFIID complex and mediates transcriptional activation. Proc. Natl. Acad. Sci. USA. 91:192–196. 10.1073/pnas.91.1.192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover-Cutter K., Larochelle S., Erickson B., Zhang C., Shokat K., Fisher R.P., and Bentley D.L.. 2009. TFIIH-associated Cdk7 kinase functions in phosphorylation of C-terminal domain Ser7 residues, promoter-proximal pausing, and termination by RNA polymerase II. Mol. Cell. Biol. 29:5455–5464. 10.1128/MCB.00637-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glozak M.A., Sengupta N., Zhang X., and Seto E.. 2005. Acetylation and deacetylation of non-histone proteins. Gene. 363:15–23. 10.1016/j.gene.2005.09.010 [DOI] [PubMed] [Google Scholar]
- Gnatovskiy L., Mita P., and Levy D.E.. 2013. The human RVB complex is required for efficient transcription of type I interferon-stimulated genes. Mol. Cell. Biol. 33:3817–3825. 10.1128/MCB.01562-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes N.P., Bjerke G., Llorente B., Szostek S.A., Emerson B.M., and Espinosa J.M.. 2006. Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20:601–612. 10.1101/gad.1398206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q.H., McDowell J.C., and Dean A.. 1996. Essential role of NF-E2 in remodeling of chromatin structure and transcriptional activation of the epsilon-globin gene in vivo by 5′ hypersensitive site 2 of the beta-globin locus control region. Mol. Cell. Biol. 16:6055–6064. 10.1128/MCB.16.11.6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttlicher M., Minucci S., Zhu P., Krämer O.H., Schimpf A., Giavara S., Sleeman J.P., Lo Coco F., Nervi C., Pelicci P.G., and Heinzel T.. 2001. Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 20:6969–6978. 10.1093/emboj/20.24.6969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gough D.J., Messina N.L., Clarke C.J., Johnstone R.W., and Levy D.E.. 2012. Constitutive type I interferon modulates homeostatic balance through tonic signaling. Immunity. 36:166–174. 10.1016/j.immuni.2012.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer C.B., Tanaka Y., Kim Y.J., Xie P., Zhang M.Q., Park I.H., and Kim T.H.. 2015. Histone Deacetylases Positively Regulate Transcription through the Elongation Machinery. Cell Reports. 13:1444–1455. 10.1016/j.celrep.2015.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui C.Y., and Dean A.. 2001. Acetylation of a specific promoter nucleosome accompanies activation of the epsilon-globin gene by beta-globin locus control region HS2. Mol. Cell. Biol. 21:1155–1163. 10.1128/MCB.21.4.1155-1163.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves D.C., Horng T., and Medzhitov R.. 2009. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 138:129–145. 10.1016/j.cell.2009.05.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa T., and Nakayama J.. 2011. Physiological roles of class I HDAC complex and histone demethylase. J. Biomed. Biotechnol. 2011:129383 10.1155/2011/129383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin J.P., Sheth A., and Manley J.L.. 2011. RNAP II CTD phosphorylated on threonine-4 is required for histone mRNA 3′ end processing. Science. 334:683–686. 10.1126/science.1206034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Lu X., Liu R., Ai N., Cao Z., Li Y., Liu J., Yu B., Liu K., Wang H., et al. 2014. Histone cross-talk connects protein phosphatase 1α (PP1α) and histone deacetylase (HDAC) pathways to regulate the functional transition of bromodomain-containing 4 (BRD4) for inducible gene expression. J. Biol. Chem. 289:23154–23167. 10.1074/jbc.M114.570812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Icardi L., Mori R., Gesellchen V., Eyckerman S., De Cauwer L., Verhelst J., Vercauteren K., Saelens X., Meuleman P., Leroux-Roels G., et al. 2012. The Sin3a repressor complex is a master regulator of STAT transcriptional activity. Proc. Natl. Acad. Sci. USA. 109:12058–12063. 10.1073/pnas.1206458109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang M.K., Mochizuki K., Zhou M., Jeong H.S., Brady J.N., and Ozato K.. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol. Cell. 19:523–534. 10.1016/j.molcel.2005.06.027 [DOI] [PubMed] [Google Scholar]
- Kadiyala V., Patrick N.M., Mathieu W., Jaime-Frias R., Pookhao N., An L., and Smith C.L.. 2013. Class I lysine deacetylases facilitate glucocorticoid-induced transcription. J. Biol. Chem. 288:28900–28912. 10.1074/jbc.M113.505115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalunian K.C. 2016. Interferon-targeted therapy in systemic lupus erythematosus: Is this an alternative to targeting B and T cells? Lupus. 25:1097–1101. 10.1177/0961203316652495 [DOI] [PubMed] [Google Scholar]
- Kanno T., Kanno Y., LeRoy G., Campos E., Sun H.W., Brooks S.R., Vahedi G., Heightman T.D., Garcia B.A., Reinberg D., et al. 2014. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat. Struct. Mol. Biol. 21:1047–1057. 10.1038/nsmb.2912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene M.A., and Elgin S.C.. 1981. Micrococcal nuclease as a probe of DNA sequence organization and chromatin structure. Cell. 27:57–64. 10.1016/0092-8674(81)90360-3 [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., and Haussler D.. 2002. The human genome browser at UCSC. Genome Res. 12:996–1006. 10.1101/gr.229102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., and Karolchik D.. 2010. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 26:2204–2207. 10.1093/bioinformatics/btq351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A., and Mathelier A.. 2017. Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics. 18:287 10.1186/s12859-017-1708-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirou K.A., and Gkrouzman E.. 2013. Anti-interferon alpha treatment in SLE. Clin. Immunol. 148:303–312. 10.1016/j.clim.2013.02.013 [DOI] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell. 128:693–705. 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44(W1):W90-7 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H., and Lis J.T.. 2013. Control of transcriptional elongation. Annu. Rev. Genet. 47:483–508. 10.1146/annurev-genet-110711-155440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. 2010. Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics. Chapter 11:Unit 11 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larner A.C., Jonak G., Cheng Y.S., Korant B., Knight E., and Darnell J.E. Jr. 1984. Transcriptional induction of two genes in human cells by beta interferon. Proc. Natl. Acad. Sci. USA. 81:6733–6737. 10.1073/pnas.81.21.6733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau J.F., Nusinzon I., Burakov D., Freedman L.P., and Horvath C.M.. 2003. Role of metazoan mediator proteins in interferon-responsive transcription. Mol. Cell. Biol. 23:620–628. 10.1128/MCB.23.2.620-628.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaris C., Kelly S., Ntziachristos P., Aifantis I., and Tsirigos A.. 2017. HiC-bench: comprehensive and reproducible Hi-C data analysis designed for parameter exploration and benchmarking. BMC Genomics. 18:22 10.1186/s12864-016-3387-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.I., Jenner R.G., Boyer L.A., Guenther M.G., Levine S.S., Kumar R.M., Chevalier B., Johnstone S.E., Cole M.F., Isono K., et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 125:301–313. 10.1016/j.cell.2006.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner L., Henriksen M.A., Zhang X., and Darnell J.E. Jr. 2003. STAT3-dependent enhanceosome assembly and disassembly: synergy with GR for full transcriptional increase of the alpha 2-macroglobulin gene. Genes Dev. 17:2564–2577. 10.1101/gad.1135003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy A., and Noll M.. 1981. Chromatin fine structure of active and repressed genes. Nature. 289:198–203. 10.1038/289198a0 [DOI] [PubMed] [Google Scholar]
- Levy D.E., and García-Sastre A.. 2001. The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12:143–156. 10.1016/S1359-6101(00)00027-7 [DOI] [PubMed] [Google Scholar]
- Levy D., Larner A., Chaudhuri A., Babiss L.E., and Darnell J.E. Jr. 1986. Interferon-stimulated transcription: isolation of an inducible gene and identification of its regulatory region. Proc. Natl. Acad. Sci. USA. 83:8929–8933. 10.1073/pnas.83.23.8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.E., Lew D.J., Decker T., Kessler D.S., and Darnell J.E. Jr. 1990. Synergistic interaction between interferon-alpha and interferon-gamma through induced synthesis of one subunit of the transcription factor ISGF3. EMBO J. 9:1105–1111. 10.1002/j.1460-2075.1990.tb08216.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D.E., Marié I.J., and Durbin J.E.. 2011. Induction and function of type I and III interferon in response to viral infection. Curr. Opin. Virol. 1:476–486. 10.1016/j.coviro.2011.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.. 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J.T. 2007. Imaging Drosophila gene activation and polymerase pausing in vivo. Nature. 450:198–202. 10.1038/nature06324 [DOI] [PubMed] [Google Scholar]
- Malakhova O.A., Kim K.I., Luo J.K., Zou W., Kumar K.G., Fuchs S.Y., Shuai K., and Zhang D.E.. 2006. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 25:2358–2367. 10.1038/sj.emboj.7601149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand J.R., and Caflisch A.. 2015. Binding Mode of Acetylated Histones to Bromodomains: Variations on a Common Motif. ChemMedChem. 10:1327–1333. 10.1002/cmdc.201500141 [DOI] [PubMed] [Google Scholar]
- Marié I., Durbin J.E., and Levy D.E.. 1998. Differential viral induction of distinct interferon-alpha genes by positive feedback through interferon regulatory factor-7. EMBO J. 17:6660–6669. 10.1093/emboj/17.22.6660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marié I., Smith E., Prakash A., and Levy D.E.. 2000. Phosphorylation-induced dimerization of interferon regulatory factor 7 unmasks DNA binding and a bipartite transactivation domain. Mol. Cell. Biol. 20:8803–8814. 10.1128/MCB.20.23.8803-8814.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A., Landry H.M., and Churchman L.S.. 2017. Pause & go: from the discovery of RNA polymerase pausing to its functional implications. Curr. Opin. Cell Biol. 46:72–80. 10.1016/j.ceb.2017.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonel P., Demmers J., Tan D.W., Watt F., and Hendrich B.D.. 2012. Sin3a is essential for the genome integrity and viability of pluripotent cells. Dev. Biol. 363:62–73. 10.1016/j.ydbio.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinhart A., Kamenski T., Hoeppner S., Baumli S., and Cramer P.. 2005. A structural perspective of CTD function. Genes Dev. 19:1401–1415. 10.1101/gad.1318105 [DOI] [PubMed] [Google Scholar]
- Mulholland N.M., Soeth E., and Smith C.L.. 2003. Inhibition of MMTV transcription by HDAC inhibitors occurs independent of changes in chromatin remodeling and increased histone acetylation. Oncogene. 22:4807–4818. 10.1038/sj.onc.1206722 [DOI] [PubMed] [Google Scholar]
- Narita K., Kikuchi T., Watanabe K., Takizawa T., Oguchi T., Kudo K., Matsuhara K., Abe H., Yamori T., Yoshida M., and Katoh T.. 2009. Total synthesis of the bicyclic depsipeptide HDAC inhibitors spiruchostatins A and B, 5′'-epi-spiruchostatin B, FK228 (FR901228) and preliminary evaluation of their biological activity. Chemistry. 15:11174–11186. 10.1002/chem.200901552 [DOI] [PubMed] [Google Scholar]
- Narita T., Yung T.M., Yamamoto J., Tsuboi Y., Tanabe H., Tanaka K., Yamaguchi Y., and Handa H.. 2007. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol. Cell. 26:349–365. 10.1016/j.molcel.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Nilsson L.M., Green L.C., Muralidharan S.V., Demir D., Welin M., Bhadury J., Logan D.T., Walse B., and Nilsson J.A.. 2016. Cancer Differentiating Agent Hexamethylene Bisacetamide Inhibits BET Bromodomain Proteins. Cancer Res. 76:2376–2383. 10.1158/0008-5472.CAN-15-2721 [DOI] [PubMed] [Google Scholar]
- Nusinzon I., and Horvath C.M.. 2003. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc. Natl. Acad. Sci. USA. 100:14742–14747. 10.1073/pnas.2433987100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oven I., Brdicková N., Kohoutek J., Vaupotic T., Narat M., and Peterlin B.M.. 2007. AIRE recruits P-TEFb for transcriptional elongation of target genes in medullary thymic epithelial cells. Mol. Cell. Biol. 27:8815–8823. 10.1128/MCB.01085-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C., Lecomte M.-J., and Schindler C.. 1999. Murine Stat2 is uncharacteristically divergent. Nucleic Acids Res. 27:4191–4199. 10.1093/nar/27.21.4191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Jung Y., Kim T.Y., Kim S.G., Jong H.S., Lee J.W., Kim D.K., Lee J.S., Kim N.K., Kim T.Y., and Bang Y.J.. 2004. Class I histone deacetylase-selective novel synthetic inhibitors potently inhibit human tumor proliferation. Clin. Cancer Res. 10:5271–5281. 10.1158/1078-0432.CCR-03-0709 [DOI] [PubMed] [Google Scholar]
- Patel M.C., Debrosse M., Smith M., Dey A., Huynh W., Sarai N., Heightman T.D., Tamura T., and Ozato K.. 2013. BRD4 coordinates recruitment of pause release factor P-TEFb and the pausing complex NELF/DSIF to regulate transcription elongation of interferon-stimulated genes. Mol. Cell. Biol. 33:2497–2507. 10.1128/MCB.01180-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson M., Pisharody S., Pan L., Guadagno S., Mui A.L., and Levy D.E.. 1999. Stat protein transactivation domains recruit p300/CBP through widely divergent sequences. J. Biol. Chem. 274:25343–25349. 10.1074/jbc.274.36.25343 [DOI] [PubMed] [Google Scholar]
- Paulson M., Press C., Smith E., Tanese N., and Levy D.E.. 2002. IFN-Stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat. Cell Biol. 4:140–147. 10.1038/ncb747 [DOI] [PubMed] [Google Scholar]
- Peterlin B.M., and Price D.H.. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 23:297–305. 10.1016/j.molcel.2006.06.014 [DOI] [PubMed] [Google Scholar]
- Plesko M.M., Hargrove J.L., Granner D.K., and Chalkley R.. 1983. Inhibition by sodium butyrate of enzyme induction by glucocorticoids and dibutyryl cyclic AMP. A role for the rapid form of histone acetylation. J. Biol. Chem. 258:13738–13744. [PubMed] [Google Scholar]
- Quinlan A.R., and Hall I.M.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 26:841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi S.A., Leung S., Kerr I.M., Stark G.R., and Darnell J.E. Jr. 1996. Function of Stat2 protein in transcriptional activation by alpha interferon. Mol. Cell. Biol. 16:288–293. 10.1128/MCB.16.1.288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan Y., Rajpara S.M., Reza S.M., Lees E., Shuman S., Mathews M.B., and Pe’ery T.. 2001. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J. Biol. Chem. 276:10913–10920. 10.1074/jbc.M010975200 [DOI] [PubMed] [Google Scholar]
- Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., and Manke T.. 2016. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44(W1):W160-5 10.1093/nar/gkw257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascle A., Johnston J.A., and Amati B.. 2003. Deacetylase activity is required for recruitment of the basal transcription machinery and transactivation by STAT5. Mol. Cell. Biol. 23:4162–4173. 10.1128/MCB.23.12.4162-4173.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen T.P. 2008. Developmentally-poised chromatin of embryonic stem cells. Front. Biosci. 13:1568–1577. 10.2741/2781 [DOI] [PubMed] [Google Scholar]
- Reich N., Evans B., Levy D., Fahey D., Knight E. Jr., and Darnell J.E. Jr. 1987. Interferon-induced transcription of a gene encoding a 15-kDa protein depends on an upstream enhancer element. Proc. Natl. Acad. Sci. USA. 84:6394–6398. 10.1073/pnas.84.18.6394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodero M.P., and Crow Y.J.. 2016. Type I interferon-mediated monogenic autoinflammation: The type I interferonopathies, a conceptual overview. J. Exp. Med. 213:2527–2538. 10.1084/jem.20161596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes C.S., Stark R., Teschendorff A.E., Holmes K.A., Ali H.R., Dunning M.J., Brown G.D., Gojis O., Ellis I.O., Green A.R., et al. 2012. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 481:389–393. 10.1038/nature10730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusinova I., Forster S., Yu S., Kannan A., Masse M., Cumming H., Chapman R., and Hertzog P.J.. 2013. Interferome v2.0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41(D1):D1040–D1046. 10.1093/nar/gks1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto S., Potla R., and Larner A.C.. 2004. Histone deacetylase activity is required to recruit RNA polymerase II to the promoters of selected interferon-stimulated early response genes. J. Biol. Chem. 279:40362–40367. 10.1074/jbc.M406400200 [DOI] [PubMed] [Google Scholar]
- Sehgal P.B., Fraser N.W., and Darnell J.E. Jr. 1979. Early Ad-2 transcription units: only promoter-proximal RNA continues to be made in the presence of DRB. Virology. 94:185–191. 10.1016/0042-6822(79)90448-3 [DOI] [PubMed] [Google Scholar]
- Spange S., Wagner T., Heinzel T., and Krämer O.H.. 2009. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int. J. Biochem. Cell Biol. 41:185–198. 10.1016/j.biocel.2008.08.027 [DOI] [PubMed] [Google Scholar]
- Thomas M.C., and Chiang C.M.. 2006. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 41:105–178. 10.1080/10409230600648736 [DOI] [PubMed] [Google Scholar]
- Tichonicky L., Santana-Calderon M.A., Defer N., Giesen E.M., Beck G., and Kruh J.. 1981. Selective inhibition by sodium butyrate of glucocorticoid-induced tyrosine aminotransferase synthesis in hepatoma tissue-cultured cells. Eur. J. Biochem. 120:427–433. 10.1111/j.1432-1033.1981.tb05720.x [DOI] [PubMed] [Google Scholar]
- Tsirigos A., Haiminen N., Bilal E., and Utro F.. 2012. GenomicTools: a computational platform for developing high-throughput analytics in genomics. Bioinformatics. 28:282–283. 10.1093/bioinformatics/btr646 [DOI] [PubMed] [Google Scholar]
- Utley R.T., Ikeda K., Grant P.A., Côté J., Steger D.J., Eberharter A., John S., and Workman J.L.. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature. 394:498–502. 10.1038/28886 [DOI] [PubMed] [Google Scholar]
- Veals S.A., Schindler C., Leonard D., Fu X.Y., Aebersold R., Darnell J.E. Jr., and Levy D.E.. 1992. Subunit of an alpha-interferon-responsive transcription factor is related to interferon regulatory factor and Myb families of DNA-binding proteins. Mol. Cell. Biol. 12:3315–3324. 10.1128/MCB.12.8.3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veals S.A., Santa Maria T., and Levy D.E.. 1993. Two domains of ISGF3 gamma that mediate protein-DNA and protein-protein interactions during transcription factor assembly contribute to DNA-binding specificity. Mol. Cell. Biol. 13:196–206. 10.1128/MCB.13.1.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M., and Timmers H.T.. 2010. Grasping trimethylation of histone H3 at lysine 4. Epigenomics. 2:395–406. 10.2217/epi.10.11 [DOI] [PubMed] [Google Scholar]
- Wada T., Takagi T., Yamaguchi Y., Ferdous A., Imai T., Hirose S., Sugimoto S., Yano K., Hartzog G.A., Winston F., et al. 1998. DSIF, a novel transcription elongation factor that regulates RNA polymerase II processivity, is composed of human Spt4 and Spt5 homologs. Genes Dev. 12:343–356. 10.1101/gad.12.3.343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhu W., and Levy D.E.. 2006. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods. 39:356–362. 10.1016/j.ymeth.2006.06.010 [DOI] [PubMed] [Google Scholar]
- Wang Z., Zang C., Cui K., Schones D.E., Barski A., Peng W., and Zhao K.. 2009. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 138:1019–1031. 10.1016/j.cell.2009.06.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciak J.M., Martinez-Yamout M.A., Dyson H.J., and Wright P.E.. 2009. Structural basis for recruitment of CBP/p300 coactivators by STAT1 and STAT2 transactivation domains. EMBO J. 28:948–958. 10.1038/emboj.2009.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Nie L., Kim S.-H., and Sun X.-H.. 2003. STAT5-induced Id-1 transcription involves recruitment of HDAC1 and deacetylation of C/EBPbeta. EMBO J. 22:893–904. 10.1093/emboj/cdg094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi Y., Shibata H., and Handa H.. 2013. Transcription elongation factors DSIF and NELF: promoter-proximal pausing and beyond. Biochim. Biophys. Acta. 1829:98–104. 10.1016/j.bbagrm.2012.11.007 [DOI] [PubMed] [Google Scholar]
- Yang Z., Yik J.H., Chen R., He N., Jang M.K., Ozato K., and Zhou Q.. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol. Cell. 19:535–545. 10.1016/j.molcel.2005.06.029 [DOI] [PubMed] [Google Scholar]
- Young R.A. 2011. Control of the embryonic stem cell state. Cell. 144:940–954. 10.1016/j.cell.2011.01.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Zhang W., and Kone B.C.. 2002. Histone deacetylases augment cytokine induction of the iNOS gene. J. Am. Soc. Nephrol. 13:2009–2017. 10.1097/01.ASN.0000024253.59665.F1 [DOI] [PubMed] [Google Scholar]
- Zhang D., and Zhang D.E.. 2011. Interferon-stimulated gene 15 and the protein ISGylation system. J. Interferon Cytokine Res. 31:119–130. 10.1089/jir.2010.0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bogunovic D., Payelle-Brogard B., Francois-Newton V., Speer S.D., Yuan C., Volpi S., Li Z., Sanal O., Mansouri D., et al. 2015. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto-inflammation. Nature. 517:89–93. 10.1038/nature13801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R., Nakamura T., Fu Y., Lazar Z., and Spector D.L.. 2011. Gene bookmarking accelerates the kinetics of post-mitotic transcriptional re-activation. Nat. Cell Biol. 13:1295–1304. 10.1038/ncb2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zupkovitz G., Tischler J., Posch M., Sadzak I., Ramsauer K., Egger G., Grausenburger R., Schweifer N., Chiocca S., Decker T., and Seiser C.. 2006. Negative and positive regulation of gene expression by mouse histone deacetylase 1. Mol. Cell. Biol. 26:7913–7928. 10.1128/MCB.01220-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.