These studies reveal a previously unrecognized role for Cnbp as a novel transcriptional regulator engaged downstream of innate immune receptors controlling the c-Rel–IL-12–Th1 axis, which has important implications for both host defense and inflammatory disease.
Abstract
An inducible program of inflammatory gene expression is a hallmark of antimicrobial defenses. Recently, cellular nucleic acid–binding protein (CNBP) was identified as a regulator of nuclear factor-kappaB (NF-κB)–dependent proinflammatory cytokine gene expression. Here, we generated mice lacking CNBP and found that CNBP regulates a very restricted gene signature that includes IL-12β. CNBP resides in the cytosol of macrophages and translocates to the nucleus in response to diverse microbial pathogens and pathogen-derived products. Cnbp-deficient macrophages induced canonical NF-κB/Rel signaling normally but were impaired in their ability to control the activation of c-Rel, a key driver of IL-12β gene transcription. The nuclear translocation and DNA-binding activity of c-Rel required CNBP. Lastly, Cnbp-deficient mice were more susceptible to acute toxoplasmosis associated with reduced production of IL-12β, as well as a reduced T helper type 1 (Th1) cell IFN-γ response essential to controlling parasite replication. Collectively, these findings identify CNBP as important regulator of c-Rel–dependent IL-12β gene transcription and Th1 immunity.
Introduction
The acute inflammatory response is induced as a first line of defense against pathogen invasion. Innate immune cells, including macrophages and dendritic cells (DCs), recognize pathogen-associated molecular patterns via pattern recognition receptors (PRRs) to drive these responses. Multiple PRRs have been identified, including TLRs, Nod-like receptors, Aim2-like receptors, Rig-I–like receptors, and C-type lectins (Trinchieri and Sher, 2007; Thompson et al., 2011; O’Neill et al., 2013; Cao, 2016; Chen et al., 2016). PRR–pathogen-associated molecular pattern interactions promote the activation of signal transduction pathways that converge on transcription factors, resulting in changes in expression of hundreds of genes involved in antimicrobial defense, phagocytosis, cell migration, metabolic reprogramming, tissue repair, and regulation of adaptive immunity (Trinchieri and Sher, 2007; Medzhitov and Horng, 2009; Liu et al., 2016). Signal-dependent activation of NF-κB, IFN regulatory factors (IRFs), and STATs control the timing and duration of these transcriptional programs (Ghosh and Hayden, 2008; Smale and Natoli, 2014). Inappropriate regulation of these processes leads to chronic inflammation and diseases such as autoimmunity, atherosclerosis, and cancer.
Cellular nucleic acid–binding protein (CNBP), also called zinc-finger protein 9 (ZNF9), is a highly conserved zinc-finger protein with seven tandem repeats of cysteine-cysteine-histidine-cysteine (CCHC) zinc-finger knuckles and one arginine–glycine–glycine box (Calcaterra et al., 2010). This DNA- and RNA-binding protein with broad sequence specificity has been associated with diverse cellular functions, including transcription and translation (Chen et al., 2013; Margarit et al., 2014; Benhalevy et al., 2017). Cnbp is linked to age-related sporadic inclusion body myositis, an inflammatory muscle disease characterized by progressive muscle weakness and atrophy (Niedowicz et al., 2010). Furthermore, a dominantly transmitted (CCTG)n expansion in intron 1 of the Cnbp gene leads to myotonic dystrophy type 2 (Raheem et al., 2010; Sun et al., 2011; Meola and Cardani, 2015; Thornton et al., 2017), a disease associated with a high frequency of autoantibodies and autoreactive T cells (Tieleman et al., 2009). Recently, Cnbp was linked to regulation of immune gene expression. RNA interference–based studies in Raw 264.7 macrophage cell lines indicated that knockdown of CNBP resulted in reduced expression of NF-κB–dependent inflammatory genes. Induction of proinflammatory cytokines such as IL-6 required CNBP for their sustained expression (Lee et al., 2017). Collectively, these observations suggest that CNBP functions to coordinate immune gene expression.
The IL-12 family of cytokines, particularly IL-12 and IL-23, are critical proinflammatory cytokines produced by macrophages and DCs. These cytokines act as a bridge between the innate and the adaptive immune response. IL-12 shares ligand and receptor subunits with a related cytokine, IL-23. The IL-12 heterodimer is composed of the p35 subunit (IL-12α) and the shared p40 β chain (IL-12β). IL-12β also dimerizes with IL-23p19 (IL-23α) to generate the biologically active IL-23 cytokine (Trinchieri, 1995; O’Garra and Murphy, 2009; Teng et al., 2015). Products from microorganisms, including bacteria, intracellular parasites, fungi, double-stranded RNA, bacterial DNA, and CpG-containing oligonucleotides, are potent inducers of IL-12 in macrophages, monocytes, neutrophils, and DCs. The induction of IL-12 is critical for host resistance against different categories of pathogens, including CMV (Orange et al., 1995), Mycobacterium tuberculosis (Hölscher et al., 2001; Cooper et al., 2002; Khader et al., 2006), Listeria (Tripp et al., 1993), Leishmania (Sypek et al., 1993), Toxoplasma (Gazzinelli et al., 1993, 2014; Sher et al., 2003; Dupont et al., 2015), and Salmonella (Lehmann et al., 2001; Schulz et al., 2008) through the establishment of T helper type 1 cell (Th1) and IFN-γ–mediated immune responses (Biron and Gazzinelli, 1995; Gazzinelli et al., 2014).
The inducible expression of IL-12 is regulated at the level of gene transcription (Sanjabi et al., 2000; Smale, 2012). The mouse IL-12β promoter contains several cis-elements that bind a number of transcription factors, including NF-κB/Rel family members, IRF5, and CCAAT/enhancer–binding protein (Sanjabi et al., 2000, 2005; Bradley et al., 2003; Koshiba et al., 2013). The NF-κB/Rel family of transcription factors includes the proteins RelA (p65), RelB, c-Rel, NF-κB1 (p105/p50), and NF-κB2 (p100/p52; Caamaño and Hunter, 2002; Dev et al., 2011; Hayden and Ghosh, 2011). These proteins can homo- and heterodimerize and possess unique specificities in regulating target gene expression. Rel-binding sites have been identified in the promoters of cytokine and immune effectors in macrophages. Many proinflammatory genes induced downstream of TLRs and other PRRs engage the RelA–p50 heterodimeric complex to control their expression. In the case of IL-12β, c-Rel has been defined as the critical NF-κB family member responsible. While the molecular mechanisms involved in c-Rel binding to the IL-12β promoter and c-Rel–dependent transcription of IL-12β have been studied extensively, it is still unclear what governs the highly specific effects of c-Rel for IL-12β regulation.
Here, we generated mice lacking Cnbp and found that macrophages lacking Cnbp were compromised in their ability to induce IL-12 p40 (IL-12β) in response to diverse microbial pathogens that engage multiple PRRs. Importantly, our studies reveal that CNBP is not a broad regulator of the NF-κB/Rel family of transcription factors as was suggested from earlier studies (Lee et al., 2017). Rather, CNBP controlled the activity of only one member of the NF-κB/Rel family: c-Rel. c-Rel, but not RelA (p65), nuclear translocation was dependent on CNBP. Furthermore, we provide in vivo evidence for the importance of the CNBP–c-Rel–IL-12 pathway in controlling acute toxoplasmosis in vivo. Collectively, these studies reveal a previously unrecognized role for CNBP as a novel transcriptional regulator engaged downstream of innate immune receptors controlling the c-Rel–IL-12–Th1 axis.
Results
Identification of CNBP
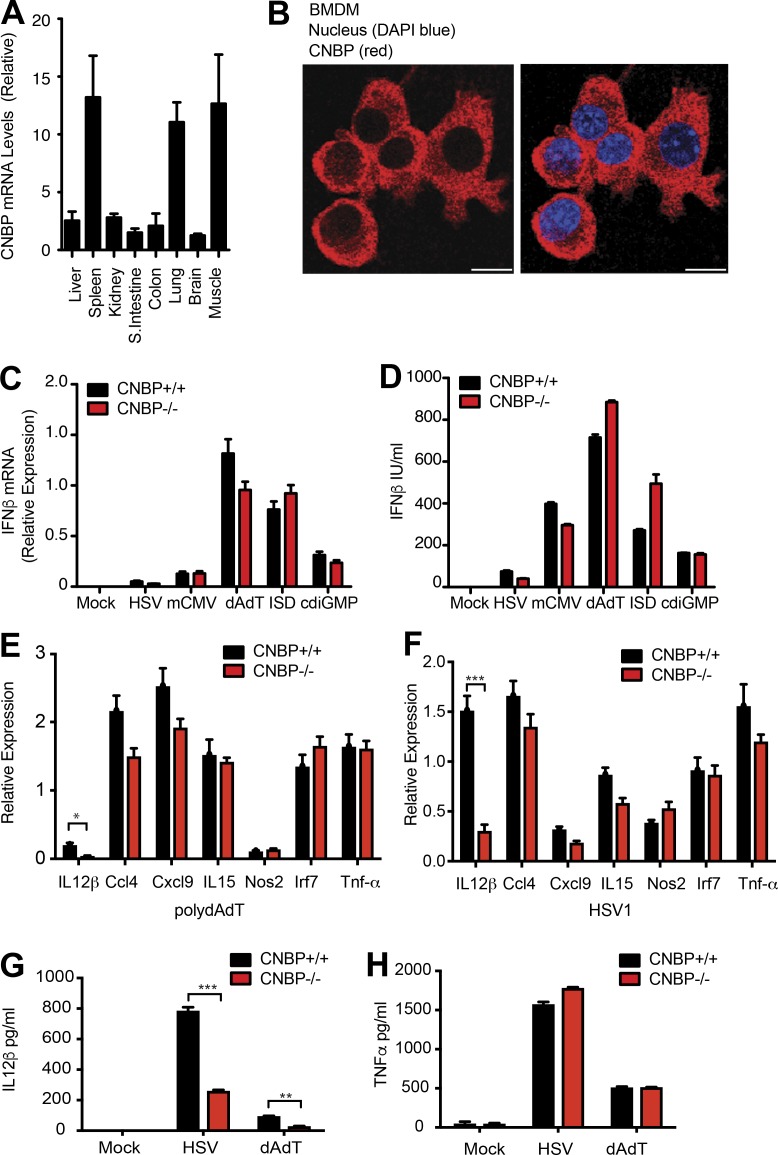
To systematically identify proteins involved in the recognition of foreign nucleic acids, we took advantage of immune stimulatory oligonucleotides containing AT-rich motif (ATr-ODN) that we had previously shown were potent stimulators of inflammatory cytokines via the STING (stimulator of IFN genes) pathway (Sharma et al., 2011; Gallego-Marin et al., 2018). In an effort to identify receptors or regulators of double-stranded DNA (dsDNA)–induced signaling pathways, ATr-ODN and a nonstimulatory ODN (ATr-AA ODN) were used in pull-down experiments to capture DNA-binding proteins from macrophage cytosolic extracts. ATr-ODN–binding proteins were eluted and separated by SDS-PAGE. Immunoblotting using antibodies to known components of the cytosolic DNA–sensing pathway identified STING, DDX3x, and TBK1 in these pull-downs (Fig. S1 A). To identify additional components of these pathways, we also performed liquid chromatography mass spectrometry (LC-MS) on these ATr-ODN pull-downs. The most enriched of the proteins identified in the ATr-ODN pull-downs, but not the nonstimulatory ATr-AA pull-downs, was CNBP (Fig. S1 B). CNBP (also called zinc-finger protein 9) is a highly conserved zinc-finger protein with seven tandem repeats of CCHC zinc-finger knuckles and one arginine–glycine–glycine box. Cnbp mRNA is constitutively expressed in numerous tissues but particularly enriched in the spleen, lung, and muscle (Fig. 1 A). Immunofluorescence staining of endogenous CNBP protein in macrophages showed that CNBP was predominantly localized in the cytoplasm at steady state (Fig. 1 B). This is consistent with our ability to isolate this protein from cytosolic extracts of macrophages.
Figure 1.
Identification of CNBP, a cytosolic dsDNA–binding protein regulating the cytosolic dsDNA–induced IL-12β response. (A) Tissue-based expression of Cnbp in mouse (normalized to Gapdh); n = 3. (B) Confocal microscopy of primary BMDMs probed with anti-CNBP. Bars, 10 μm. (C and D) IFN-β mRNA and protein levels in Cnbp+/+ and Cnbp−/− BMDMs treated with DNA ligands, including HSV, mCMV, dAdT, ISD, or c-di-GMP, were detected through qRT-PCR (C) and ELISA (D). (E and F) qRT-PCR analysis of Il12b, Ccl4, Cxcl9, Il15, Nos2, Irf7, or Tnf-α mRNA in Cnbp+/+ and Cnbp−/− BMDMs left unstimulated or transfected with poly(dA:dT) (E) or HSV1 infection (F). (G and H) IL-12β (G) or TNF-α (H) protein levels in Cnbp+/+ and Cnbp−/− BMDMs treated with HSV or dA:dT were detected through ELISA. Error bars represent SD of triplicate technical replicates (C, E, and F) or SEM of triplicate biological replicates (A, D, G, and H). All data are representative of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
CNBP regulates the cytosolic dsDNA–induced IL-12β response
To investigate the contribution of CNBP to the cytosolic DNA–sensing pathway in macrophages, we generated Cnbp-deficient mice. While Cnbp-deficient mice are viable, homozygous KO animals were born at a lower than expected Mendelian frequency. Heterozygous (HET) mice were intercrossed, resulting in 29.5% WT, 55.5% heterozygous, and 15% homozygous mice. Homozygous CNBP-KO animals were runted at birth, but they reached weights equivalent to their WT littermate controls by 6 wk. Bone marrow–derived macrophages (BMDMs) were generated from Cnbp+/+ and Cnbp−/− littermate controls. Macrophage differentiation was examined by staining cells for CD11b and F4/80. Both Cnbp+/+ and Cnbp−/− macrophages had equivalent F4/80 and CD11b staining patterns indicating normal macrophage differentiation (Fig. S1 C). There were also normal numbers of macrophages and DCs in the peritoneum (Fig. S1, D and E). Quantitative PCR (qPCR) and immunoblotting confirmed loss of CNBP expression in BMDMs, bone marrow–derived DCs (BMDCs), and peritoneal macrophages (PECs) from Cbnp−/− mice as indicated (Fig. S1, F and G). Lastly, numbers of CD4+ and CD8+ T cells were equivalent in the spleens of Cnbp+/+ and Cnbp−/− littermate controls (Fig. S1, H and I).
Type I IFN induction is a hallmark of the cytosolic DNA–sensing pathways, and we first investigated if CNBP was involved in its production in macrophages. Cnbp+/+ and Cnbp−/− macrophages were infected with HSV (HSV1-ICP0–deficient mutant) and mouse CMV (mCMV) or stimulated with poly(deoxyadenylic-deoxythymidylic) (poly(dA:dT)), the immune-stimulatory dsDNA IFN stimulatory DNA (ISD), and Bis-(3′-5′)-cyclic dimeric guanosine monophosphate (c-di-GMP), all of which signal via the STING pathway and IFNβ levels measured. In all cases, IFNβ mRNA and protein levels were induced at similar levels between the genotypes (Fig. 1, C and D). Consistent with this result, replication of mCMV and murine gammaherpesvirus 68 was unaffected in Cnbp−/− MEFs and comparable to that seen in WT cells (Fig. S1, J and K). These results indicate that CNBP is not a receptor for cytosolic dsDNA controlling the type I IFN response.
We expanded this analysis to evaluate the inducible expression of a broader panel of immune response genes in cells exposed to poly(dA:dT) or HSV1. While the inducible expression of several genes, including Ccl4, Cxcl9, Il-15, Nos2, Irf7, and Tnf-α, was induced normally in Cnbp+/+ and Cnbp−/− macrophages, the only impact on the DNA-sensing pathway we observed was on the induction of the IL-12β gene (Fig. 1, E and F). The effect of HSV1 on IL-12 mRNA was observed across a time course of HSV1 infection (Fig. S1 L). Consistently, the protein level of IL-12β in Cnbp−/− BMDMs following infection with HSV1 or transfection with poly(dA:dT) was impaired, while production of TNF-α was comparable between genotypes (Fig. 1, G and H). Collectively, these results indicate that CNBP was not involved in the DNA-sensing STING–IRF3–IFN-β pathway but identified a role for CNBP in controlling IL-12β gene expression.
CNBP broadly regulates IL-12 response
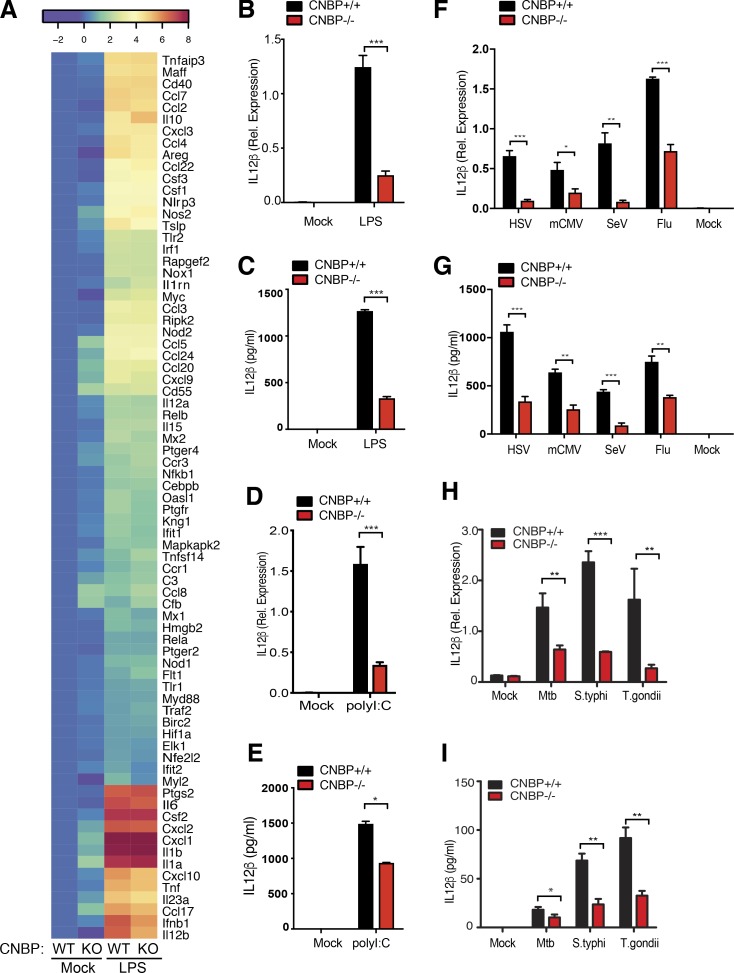
To broaden our investigation of CNBP in the control of inflammatory responses, we used multiplex gene expression analysis (NanoString) to compare the inducible expression of a panel of 254 inflammatory genes in Cnbp+/+ and Cnbp−/− macrophages stimulated with LPS. These genes represent a broad range of inflammatory cytokines, type I IFNs, IFN-stimulated genes, and other pathways that included apoptosis, epidermal growth factor, interleukin signaling, Ras, T cell receptor, and TLR signaling genes. While most of these LPS induced genes were induced at equivalent levels in Cnbp+/+ and Cnbp−/− macrophages, only a few genes, including Il12b, Ifnb, Il6, and Cxcl10, were reduced in Cnbp−/− macrophages compared with control cells, with Il12b being the most affected (Fig. 2 A). A heat map depicting ∼80 of the most highly inducible LPS-regulated genes in WT and KO cells is shown. We confirmed these effects for IL-12β using quantitative RT-PCR (qRT-PCR) and ELISA (Fig. 2, B and C). Analysis of a time course of these responses revealed that CNBP controlled the induction of IL-12β mRNA at the early phase of the LPS response (before 4 h; Fig. S2 A). In contrast to IL-12β, the impact of CNBP on IL-6 gene expression was modest at all time points examined (Fig. S2 B). The role of CNBP in the control of IL-12β was not restricted to BMDMs, as similar results were seen in DCs (BMDCs) and PECs from Cnbp−/− mice (Fig. S2, C and D). The requirement for CNBP in controlling IL-12β gene expression was also observed in cells stimulated with poly(I:C), which signals via TLR3/MDA5 (Fig. 2, D and E), and in cells infected with Sendai virus (SeV) and influenza virus, which signal via RIG-I (Fig. 2, F and G; and Fig. S2 E). We also evaluated the contribution of CNBP in controlling IL-12β gene transcription and IL-12β production in macrophages infected with bacterial (M. tuberculosis and Salmonella typhimurium), and parasitic pathogens (Toxoplasma gondii). In all cases, the inducible expression of IL-12β and production of IL-12β protein was impaired in macrophages from Cnbp−/− mice (Fig. 2, H and I).
Figure 2.
CNBP is a broad regulator of the IL-12β response in the TLR and RIG-I–like receptor pathway. (A) Heat map of gene expression in WT and KO BMDMs treated or not treated with LPS and analyzed by NanoString; n = 3. (B and C) Analysis of IL-12β by qRT-PCR (B) and ELISA (C) in WT and Cnbp−/− BMDMs left unstimulated or stimulated with LPS. (D and E) Analysis of IL-12β by qRT-PCR (D) and ELISA (E) in WT and Cnbp−/− BMDMs left unstimulated or stimulated with poly(I:C). (F and G) IL-12β mRNA and protein levels in WT and Cnbp−/− BMDMs uninfected (0 h) or infected with HSV, mCMV, SeV, or flu virus were detected through qRT-PCR (F) and ELISA (G). (H and I) IL-12β mRNA and protein levels in WT and Cnbp−/− BMDMs uninfected (0 h) or infected with M. tuberculosis, Salmonella, or T. gondii were detected through qRT-PCR (H) and ELISA (I). Error bars represent SD of triplicate technical replicates (B–I). All data are representative of three independent experiments with similar results. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Biologically active IL-12 consists of the p35 subunit (IL-12α) and the p40 subunit (IL-12β). IL-12β also dimerizes with IL-23p19 (IL-23α) to generate the biologically active IL-23 cytokine. Thus, we investigated whether CNBP also regulated IL-12α and IL-23α. As shown in Fig. S2 F, CNBP was also required for the induction of IL-12α and IL-23α in response to LPS, and this effect was not restricted to BMDMs, as similar results were seen in DCs (BMDCs) and PECs (Fig. S2, G and H). Analysis of a time course of these responses revealed that CNBP controlled the induction of IL-12 and IL-23 mRNA expression at early time points during the LPS response (before 4 h; Fig. S2, I and J).
CNBP negatively regulates the IL-10 response
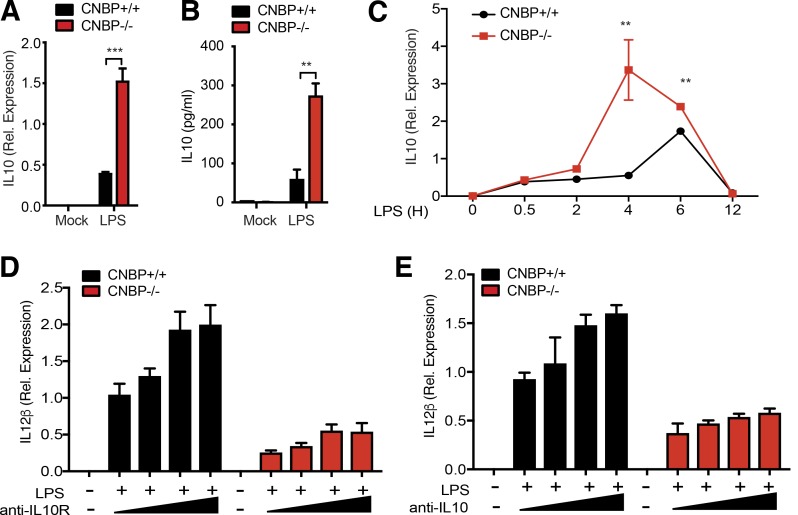
In addition to the marked effects observed on IL-12β, NanoString analysis also revealed that the levels of IL-10 were considerably higher in Cnbp−/− macrophages relative to their WT counterparts (Fig. 2 A). We confirmed these effects at the mRNA and protein level by qRT-PCR and ELISA, respectively (Fig. 3, A and B). IL-10 mRNA levels peaked at earlier time points in Cnbp−/− macrophages after LPS stimulation (Fig. 3 C). Since IL-10 is known to repress IL-12β expression (D’Andrea et al., 1993; Aste-Amezaga et al., 1998), we wanted to determine if the elevated IL-10 contributed to the reduction in IL-12β observed in Cnbp−/− macrophages. To test this directly, we incubated both WT and Cnbp−/− macrophages with anti–IL-10R blocking antibody before LPS stimulation and measured IL-12β production. As previously, shown the levels of IL-12β were reduced in Cnbp−/− relative to their WT counterparts. Anti–IL-10R blockade did not rescue this defect (Fig. 3 D). Similar observations were made using a cytokine blocking antibody. An anti–IL-10 cytokine antibody showed similar results (Fig. 3 E). We also examined whether the elevated IL-10 was an indirect consequence of CNBP’s effect on IL-12β expression. We incubated WT macrophages with anti–IL-12 neutralizing antibody before LPS stimulation and measured IL-10 production. The induction of IL-10 was not affected by anti–IL-12 neutralizing antibody, suggesting that IL-12 does not regulate IL-10 expression in this context (Fig. S3). Collectively, these observations indicate that CNBP-mediated IL-12 production occurs independently of the elevated production of IL-10.
Figure 3.
CNBP negatively regulates IL-10. (A and B) Analysis of IL-10 by qRT-PCR (A) and ELISA (B) in Cnbp WT or KO BMDMs left unstimulated or stimulated with LPS. (C) qRT-PCR analysis of IL-10 mRNA in Cnbp WT or KO BMDMs left unstimulated or stimulated with LPS at different time courses. (D and E) WT and Cnbp−/− macrophages were incubated with anti–IL-10R blocking antibody (D) or anti–IL-10 neutralizing antibody (E) before LPS stimulation, and IL-12β mRNA expression levels were determined by qPCR. Error bars represent SD of triplicate technical replicates. All data are representative of three independent experiments with similar results. **, P < 0.01; ***, P < 0.001.
CNBP translocates to the nucleus depending on the TLR–MyD88–IRAK–TAK1 signaling pathway
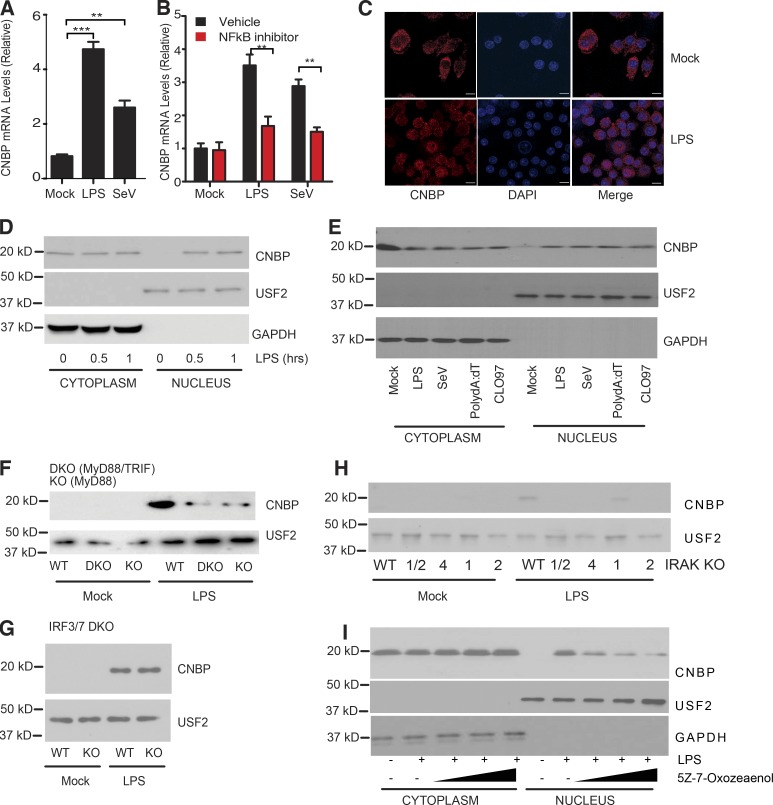
We next wanted to understand how CNBP impacted IL-12β gene expression and the molecular basis for the specificity of this response. CNBP is a relatively abundant protein in myeloid cells (Fig. 1 A), but CNBP expression could be further up-regulated in cells activated with LPS or infected with SeV (Fig. 4 A). To further explore the mechanisms by which CNBP was induced under these conditions, BMDMs were pretreated with DMSO (control) or BAY11-7082, an irreversible inhibitor of the IκB kinase, followed by LPS stimulation or SeV infection. The up-regulation of CNBP was abolished in BAY11-7082–treated BMDMs relative to the control cells (Fig. 4 B), indicating that the inducible expression of CNBP was dependent on the NF-κB signaling pathway.
Figure 4.
CNBP translocates to the nucleus depending on TLR–MyD88–IRAK–TAK1 signaling. (A) qRT-PCR analysis of CNBP expression in BMDMs stimulated with LPS or SeV. (B) WT BMDMs were pretreated with the NF-κB inhibitor BAY11-7082 (10 μM) or DMSO, followed by LPS stimulation or SeV infection. (C) Confocal analysis of CNBP distribution in primary macrophage stimulated with LPS. Bars, 10 μm. (D) The cytosolic and nuclear extracts were analyzed for CNBP by Western blotting in WT BMDMs treated with LPS. Anti-GAPDH or anti-USF2 was used for the cytosolic or nuclear loading control, respectively. (E) The cytosolic and nuclear extracts were analyzed for CNBP by Western blotting in WT BMDMs treated with LPS, SeV, poly(dA:dT), or CL097. (F–H) Nuclear extracts were analyzed for CNBP by Western blotting in MyD88−/− and MyD88−/−TRIF−/− (F), IRF3/7−/− (G), or IRAK1/2−/−, IRAK4−/−, IRAK1−/−, and IRAK2−/− (H) BMDMs treated with LPS. (I) The cytosolic and nuclear extracts were analyzed for CNBP by Western blotting in WT BMDMs treated with the TAK1 kinase inhibitor 5Z-7-Oxozeaenol in the presence of LPS. Error bars represent SD of triplicate technical replicates (A and B). All data are representative of three independent experiments with similar results. **, P < 0.01; ***, P < 0.001.
CNBP is predominantly localized in the cytosol. In Raw 264.7 macrophages, CNBP relocalizes from the macrophage to the cytosol in response to LPS (Lee et al., 2017). To test this in primary cells, we used confocal microscopy to monitor CNBP localization both at baseline and following exposure to LPS. Confocal microscopy revealed that CNBP translocated from the cytosol to the nucleus in response to LPS (Fig. 4 C). We confirmed this by purifying nuclear extracts and monitoring CNBP nuclear accumulation by immunoblotting. In resting cells, CNBP was exclusively cytosolic excluded from the nuclear compartment. Upon LPS stimulation, CNBP translocated to the nucleus within 30 min (Fig. 4 D). CNBP also translocated to the nucleus in cells exposed to SeV, poly(dA:dT), or CL097, a TLR7 ligand, indicating that multiple TLRs and other PRRs trigger CNBP nuclear translocation (Fig. 4 E). These experiments were controlled by detecting USF2 and GAPDH, proteins that are found exclusively in the nuclei and cytosol, respectively.
To evaluate if nuclear translocation of CNBP proceeded via Toll/IL-1 receptor (TIR) domain signaling, we monitored CNBP nuclear translocation in macrophages from mice lacking MyD88 and TRIF. These double-KO mice fail to induce downstream TLR signaling (Yamamoto et al., 2003). The nuclear translocation of CNBP was significantly reduced in macrophages from these double-KO mice (Fig. 4 F). This effect was largely MyD88 dependent, since macrophages from MyD88 single-KO mice were as compromised as those from double-KO mice (Fig. 4 F). We also evaluated the role of the TRIF-IRF3 pathway in controlling CNBP nuclear translocation by monitoring nuclear translocation of CNBP in macrophages from IRF3/7 double-KO mice. CNBP nuclear translocation proceeded normally in these cells (Fig. 4 G). Downstream of MyD88, the IL-1 receptor–associated kinases (IRAKs) IRAK1, 2, and 4 are engaged to control NF-κB signaling. Similar to what we observed in MyD88-deficient macrophages, the LPS-induced nuclear accumulation of CNBP was reduced in cells lacking these IRAK kinases (Fig. 4 H). IRAK2 and IRAK4 appeared to be the predominant drivers of this response, since IRAK1 KO had residual CNBP nuclear translocation. Consistent with a role for IRAKs as regulators of CNBP nuclear translocation, the inducible expression of IL-12β was compromised in cells lacking these IRAKs (Fig. S4 A). Expression of IL-6, a known target gene of IRAK signaling, but not IFN-β, served as positive and negative controls in these assays, respectively (Fig. S4, B and C). Finally, downstream of the IRAK kinases, the kinase TAK1 is an important driver of TLR-MyD88–dependent signaling. We tested the effect of 5Z-7-Oxozeaenol, an inhibitor of TAK1 kinase. Cells treated with 5Z-7-Oxozeaenol had a dose-dependent decrease in CNBP nuclear translocation (Fig. 4 I). These observations suggest that CNBP nuclear translocation proceeds via a TLR–MyD88–IRAK–TAK1 pathway in cells exposed to LPS and that CNBP nuclear translocation is a common signal downstream of multiple PRRs.
CNBP regulates IL12β via the NF-κB/Rel family member c-Rel
Multiple DNA elements in the promoter of the IL-12β gene contribute to the inducible expression of IL-12β. These include DNA-binding sites for the NF-κB/Rel family of transcription factors, an IFN-sensitive response element (ISRE) that binds IRFs and activator protein 1 (AP1) elements that respond to MAPK (Fig. 5 A). To pinpoint which if any of these pathways might be regulated by CNBP, we took advantage of reporter genes with multimerized DNA elements that bind each of these factors. In Hek293 cells, ectopic expression of CNBP alone did not induce any of these reporter genes, although expression of STING induced both the NF-κB and ISRE reporters (Fig. 5 B). We next tested the ability of CNBP to turn on expression of a reporter gene driven by the IL-12β proximal promoter. CNBP alone weakly induced the reporter but in the presence of either c-Rel or p65 elicited a robust induction of the reporter gene (Fig. 5 C). These observations indicate that CNBP has the capacity to synergize with c-Rel or p65 to turn on IL-12β gene transcription. As described earlier, Cnbp-deficient macrophages had normal levels of TNF-α, a well-characterized NF-κB target gene, in response to multiple ligands, including LPS (Fig. 1). The phosphorylation and degradation of IκBα were intact in cells lacking CNBP (Fig. S4 D). Similarly, the phosphorylation and nuclear translocation of the NF-κB family member p65 were intact between both genotypes (Fig. S4 E). We also evaluated the activation of the TBK1–IRF3 signaling pathway by monitoring TBK1 phosphorylation. Again, TBK1 phosphorylation was equivalent in Cnbp+/+ and Cnbp−/− macrophages (Fig. S4 F). These results indicate that NF-κB and TBK1-IRF3 activation were intact in cells lacking CNBP.
Figure 5.
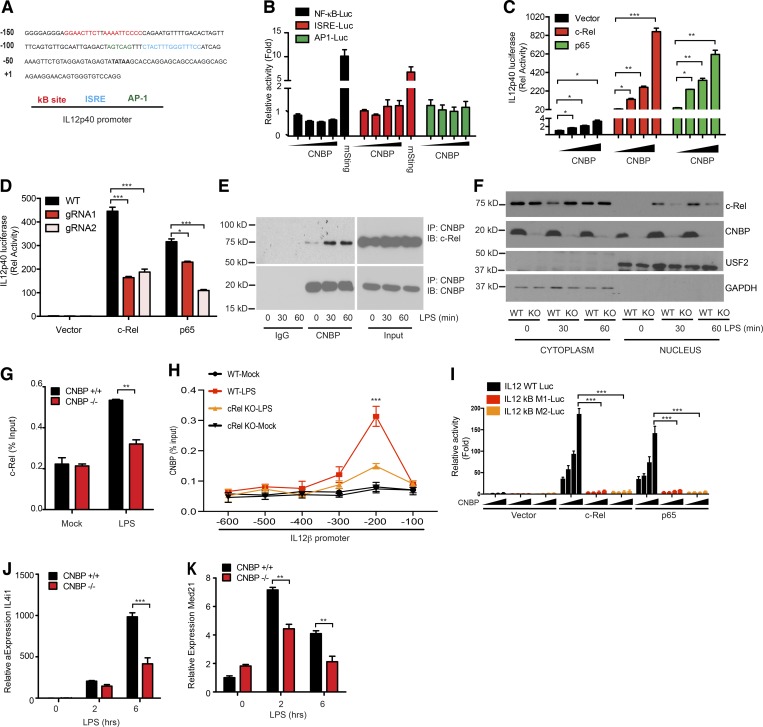
CNBP regulates IL-12β via the NF-κB/Rel family member c-Rel. (A) IL-12β promoter analysis. (B) Luciferase activity of NF-κB, ISRE, or AP1 in Hek293 cells after 36-h transfection with increasing amounts of plasmids encoding Cnbp or mSting. (C) Luciferase activity of IL12p40 in Hek293 cells after 36-h cotransfection of c-Rel or p65 with increasing amounts of CNBP. (D) Luciferase activity of IL12p40 in Hek293 Cnbp WT or KO cells after 36-h transfection of c-Rel or p65. Plasmids containing different guide RNAs targeting CNBP or scramble control guide RNAs were cotransfected with Cas9 plasmid into Hek293 cells using Lipofectamine 2000, and cells were selected with puromycin resistance after 36 h. Cells were passaged for 1–2 wk before luciferase experimental use. (E) Coimmunoprecipitation of CNBP and c-Rel in LPS-stimulated BMDMs. (F) c-Rel nuclear translocation analysis in CNBP WT and KO BMDMs stimulated with LPS. (G) ChIP followed by qPCR (ChIP-qPCR) of c-Rel at the Il12p40 promoter in CNBP WT and KO BMDMs stimulated with LPS. (H) ChIP-qPCR of CNBP at the Il12p40 promoter in c-Rel WT and KO BMDMs stimulated with LPS. Il-12β regions (horizontal axis) are as follows: −600, positions −600 to −489; −500, positions −500 to −394; −400, positions −406 to −297; −300, positions −300 to −192; −200, positions −200 to −78; and −100, positions −100 to +10. (I) Luciferase activity of IL12p40 or IL12p40 containing different κB site mutations in Hek293 cells after 36-h cotransfection of c-Rel or p65 with increasing amounts of CNBP. (J and K) qRT-PCR analysis of Il4i1 (J) and Med21 (K) in CNBP WT or KO BMDMs unstimulated or stimulated with LPS. Error bars represent SEM of triplicate biological replicates. All data are representative of three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Extensive work has revealed the importance of a c-Rel–containing NF-κB complex for activation of the endogenous IL-12β gene. Given the relatively specific effect of Cnbp deficiency for IL-12β gene expression and the normal activation of NF-κB/p65 signaling in Cnbp-deficient macrophages, we hypothesized that CNBP might regulate c-Rel. To address this directly, we took three different approaches. First, we used CRISPR/Cas9 to generate Hek293 cells lacking CNBP and evaluated the ability of c-Rel to turn on the IL-12β promoter–driven reporter gene in these cells. Ectopic expression of c-Rel induced the reporter gene in Hek293 cells, and this effect was impaired in Hek293 cells lacking CNBP (Fig. 5 D). We next tested if CNBP could interact with c-Rel in macrophages. An endogenous coimmunoprecipitation experiment was performed, and the result showed that CNBP associated with c-Rel at steady state and that this interaction was enhanced following LPS treatment of BMDMs (Fig. 5 E). Third, because c-Rel nuclear translocation is required for IL-12β gene induction, we examined c-Rel nuclear translocation in Cnbp+/+ and Cnbp−/− macrophages. As shown in Fig. 5 F, c-Rel translocated to the nucleus following LPS stimulation in Cnbp+/+ macrophages, but this was severely compromised in Cnbp−/− macrophages. In line with this finding, a chromatin immunoprecipitation (ChIP) assay demonstrated that the LPS-induced binding of c-Rel to the IL-12β promoter was greatly reduced in Cnbp-deficient BMDMs (Fig. 5 G). We also evaluated the ability of CNBP to bind the IL-12β promoter using ChIP. We used primers that amplified ∼100-bp regions from 0 to −600 bp. ChIP analysis using an antibody to CNBP in primary BMDMs revealed a very strong binding of CNBP to the region of the IL-12β promoter spanning from −100 to −200 relative to the transcription start site following LPS treatment (Fig. 5 H). Furthermore, CNBP binding to this site was reduced in c-Rel KO BMDMs, suggesting that CNBP could bind to the IL-12β promoter in a manner that was dependent on c-Rel. c-Rel binds the κB sites of the IL-12β promoter located in this region. To further demonstrate that CNBP binds the IL-12β promoter through κB sites with c-Rel, luciferase reporter gene experiments were performed using two different reporter gene constructs with mutations in the IL-12β promoter κB-binding sites. As shown in Fig. 5 I, in the presence of c-Rel, CNBP induced robust activation of the IL-12β reporter gene. This effect was abolished when the IL-12β promoter with two distinct mutations in κB-binding sites was used. Collectively, these results indicate that CNBP collaborates with c-Rel to turn on IL-12β expression, and this effect was dependent on functional κB DNA-binding elements within the IL-12β promoter. Genome-wide transcriptomics studies have defined the genes induced in macrophages following LPS that are dependent on c-Rel (unpublished data). These include Il12b as well as Il4i1 and Med21. Consistent with a specific effect of CNBP on c-Rel activity, the induction of Il4i1 and Med21 were also compromised in cells from Cnbp-deficient mice (Fig. 5, J and K). Together, these data indicate that CNBP regulates c-Rel nuclear translocation and binding to the IL-12β promoter to turn on c-Rel target genes.
CNBP protects mice against infection with T. gondii
To assess the biological significance of CNBP-mediated regulation of IL-12, we used the T. gondii infection model. We first tested if T. gondii induced the nuclear translocation of CNBP. CNBP translocated to the nucleus of DCs after T. gondii infection in vitro (Fig. 6 A). We found similar findings in vivo. Mice were infected with T. gondii (strain ME49), and splenocytes were collected at days 0, 3, 5, and 7 after infection. CNBP translocated to the nucleus in splenocytes by 5 d after infection (Fig. 6 B). This time point coincided with the peak of IL12p40 in these animals (Fig. S5 A). The nuclear translocation of CNBP following T. gondii infection was impaired in MyD88−/− mice (Fig. S5 B).
Figure 6.
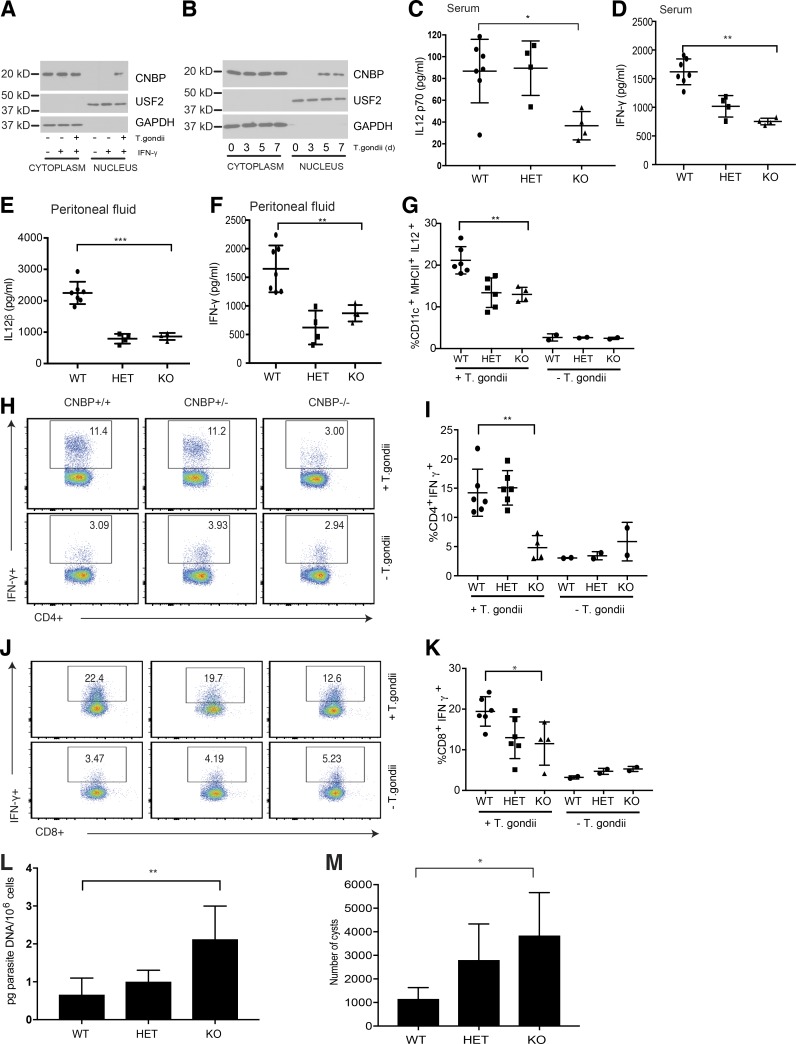
CNBP protects mice against infection with T. gondii. (A) CD11c+ splenocytes isolated by magnetic purification from mice injected with Flt3L were infected with T. gondii. The cytosolic and nuclear extracts were analyzed for CNBP by Western blotting. (B) Splenocytes from WT mice infected with T. gondii at different times after infection (days 0, 3, 5, and 7) were analyzed for CNBP translocation by Western blotting. (C and D) ELISA quantification of IL-12p70 (C) and IFN-γ (D) levels in the serum 7 d after T. gondii infection in WT, Cnbp+/− (HET), and Cnbp−/− (KO) mice. (E and F) ELISA quantification of IL-12β (E) and IFN-γ (F) levels in the peritoneal fluid 7 d after T. gondii infection in WT, Cnbp+/− (HET), and Cnbp−/− (KO) mice. (G) Frequency of IL-12p40–producing DCs (CD11c+MHC II+) in CNBP WT, HET, and KO mice. (H–K) Splenocytes were stimulated for 4 h with PMA/ionomycin in the presence of brefeldin A. The frequency of IFN-γ–producing CD4+ (H and I) and CD8+ (J and K) T cells was measured by flow cytometry. Flow plots are gated on CD3+CD4+ (H) or CD3+CD8+ (J) T cells. (L) Parasite burden in PECs at day 7 after infection was determined by qRT-PCR. (M) Brain cysts in long-term infection with T. gondii. WT, HET, and KO mice were infected with T. gondii, and brains were collected at day 44 after infection. Each symbol represents an individual mouse; small horizontal lines indicate the mean. All data are representative of at least two to three independent experiments with similar results. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
The ability of IL-12 to control T-lymphocyte responses leading to the production of IFN-γ in T. gondii–infected mice is well established. Previous studies have shown that IFN-γ produced by both CD4+ and CD8+ T cells plays an essential role in the protective response to T. gondii in vivo. To evaluate the impact of Cnbp deficiency on IL-12 production, IFN-γ production and protective CD4 and CD8 T cell responses in vivo, Cnbp+/+, Cnbp+/−, and Cnbp−/− mice (generated from intercrossing heterozygous parents) were infected with T. gondii. Spleen size and weight increased as infection progressed in all mice, as expected (Fig. S5, C and D). The levels of biologically active IL12p70 in the serum was greatly reduced in Cnbp−/− mice relative to Cnbp+/+ mice (Fig. 6 C). Consistent with these findings, the levels of IFN-γ were also reduced in Cnbp−/− mice relative to their WT counterparts (Fig. 6 D). Levels of IL-12β and IFN-γ in the peritoneal fluid of mice challenged with T. gondii were also reduced in Cnbp+/− and Cnbp−/− mice relative to their WT counterparts (Fig. 6, E and F). IL12-KO mice infected with T. gondii were used as positive controls, highlighting the importance of IL-12 in controlling IFN-γ production in this model (Fig. S5 E). TNF-α protein levels were comparable between Cnbp+/+ and Cnbp−/− mice (Fig. S5 F). DCs are thought to be important producers of IL-12 in vivo. IL-12–producing CD11c+ DCs were also reduced in Cnbp+/− and Cnbp−/− mice relative to their WT littermate controls (Fig. 6 G). We also analyzed IL-12 production from purified DC subsets in the spleen. All DC subsets from Cnbp-deficient mice (namely CD8+ DCs, CD8−CD11b– DCs, and CD8–CD11b+DCs (moDCs)) produced less IL-12 in vivo (Fig. S5 G).
Next, we examined whether the absence of CNBP during T. gondii infection altered the CD4 and CD8 T cell response. T cells from Cnbp+/+, Cnbp+/−, and Cnbp−/− infected mice were stimulated ex vivo with PMA and ionomycin and IFN-γ production from CD4+ and CD8+ T cells measured. The percentage of IFN-γ–positive CD4 T cells was significantly reduced in Cnbp−/− mice (Fig. 6, H and I). Similarly, the percentage of IFN-γ–positive CD8 T cells was also lower in these animals (Fig. 6, J and K). Lastly, we measured parasite loads in these animals. Cnbp-deficient mice had an increased parasite burden during the acute infection (Fig. 6 L). Chronic infection is associated with the development of toxoplasmic encephalitis. To evaluate chronic infection, the brains of animals were harvested at day 44 after infection and parasite loads measured (Fig. 6 M). CNBP-KO mice had higher numbers of cysts than their WT counterparts, suggesting that CNBP functions during both the acute infection and in long-term immunity to T. gondii. Altogether, these results indicate that Cnbp-deficient mice fail to mount protective IL-12 and IFN-γ responses in vivo, resulting in a reduced Th1 cell immune response and an inability to control parasite replication.
Discussion
The NF-κB/Rel family of transcription factors are major regulators of the innate and adaptive immune response (Silverman and Maniatis, 2001; Zhang and Ghosh, 2001). Active p65/p50 dimers are found in the nucleus of macrophages following ligation of multiple classes of PRRs, including TLRs, C-type lectin receptors, RIG-I like receptors, and DNA sensors. NF-κB DNA-binding elements in the promoters of immune genes are bound by these complexes, leading to the inducible expression of hundreds of cytokines and antimicrobial molecules in temporally distinct manners (Caamaño and Hunter, 2002; Dev et al., 2011). The c-Rel/NF-κB family member appears to be unique in its ability to activate transcription of the IL-12β gene. The NF-κB–binding sites in the promoter of the IL-12β gene display comparable binding affinity for both p65–p50 and c-Rel–p50 heterodimeric complexes (Sanjabi et al., 2000). Despite this, however, only c-Rel has been shown to be essential for IL-12β production in macrophages stimulated through multiple TLRs and in cells infected with intracellular pathogens. These findings therefore implicate c-Rel as an essential and specific regulator of IL-12β gene expression, distinguishing c-Rel from the more broadly acting RelA (Sanjabi et al., 2000; Mason et al., 2002). While the molecular mechanisms involved in c-Rel binding to the IL-12β promoter and c-Rel–dependent transcription of IL-12β have been studied extensively, it is still unclear what governs the highly specific effects of c-Rel for IL-12β regulation.
In this study, we define CNBP as a new regulator of this c-Rel–dependent IL-12β response. We identified CNBP in dsDNA pull-down experiments that were designed to identify cytosolic nucleic acid sensors. RNA sensors such as RIG-I and DNA sensors such as cGAS and AIM2 are activated when nucleic acids gain access to the cytosol during infection, leading to protective antimicrobial defense mechanisms (Thompson et al., 2011). In an effort to broaden our understanding of the DNA-sensing pathway in particular, we used MS to identify cytosolic DNA–binding proteins and leveraged loss-of-function approaches to link candidate proteins to innate immunity in macrophages. We generated mice lacking Cnbp and investigated type I IFN induction in response to dsDNA viruses, poly(dA:dT) and ISD (synthetic dsDNAs), and cyclic dinucleotides. The hallmark response of this pathway, namely the type IFN response, was intact in cells lacking Cnbp, as was transcription of the NF-κB–regulated cytokine TNF-α. However, we found that cells lacking Cnbp were compromised in their ability to turn on transcription of IL-12β. The importance of CNBP for IL-12β gene transcription was also observed with LPS, one of the most potent inducers of IL-12β expression as well as poly(I:C) and SeV, which signal via MDA-5 and RIG-I, respectively.
In response to multiple ligands, CNBP translocated to the nucleus, and in the case of LPS, this proceeded in a manner dependent on MyD88, IRAK4, IRAK1, and TAK1 kinase. It is likely that for different PRRs (e.g., SeV signaling via RIG-I), the pathway components involved would be different. Multiple genetic elements have been defined in the IL-12β promoter, including those that bind NF-κB, IRFs (ISRE), or AP1. Overexpression of CNBP did not induce reporters driven by κB, ISRE, or AP1; however, CNBP could induce expression of a reporter gene consisting of the IL-12 promoter, an effect that was greatly enhanced by coexpression of either c-Rel of RelA (p65). Recently, Lee et al. (2017) identified CNBP as a broad regulator of NF-κB activation controlling the expression of a broad panel of inflammatory cytokines in macrophage cell lines. In our study, using a clean genetic approach, we found that the activation of NF-κB assessed by measuring IκBα phosphorylation and degradation as well as RelA (p65) nuclear translocation proceeded normally in cells lacking CNBP. In contrast, we found that c-Rel was the major target of CNBP. First, c-Rel–dependent activation of the IL-12 promoter–driven reporter gene was impaired in HEK293 cells generated to lack CNBP. Second, we found that CNBP interacted with c-Rel and that c-Rel nuclear translocation and DNA binding to the IL-12β promoter using ChIP after LPS treatment were all dependent on CNBP. ChIP analysis in primary BMDMs also revealed binding of CNBP to the IL-12β promoter. We observed very strong binding of CNBP to the −100 to −200 site within the IL-12β promoter, where the κB binding sites reside after LPS treatment. CNBP binding to this region of the IL-12β promoter was reduced in c-Rel KO macrophages. Furthermore, the κB-binding elements were essential for this effect, since mutations at this site abrogated this response. Our data support a model whereby CNBP controls c-Rel nuclear translocation. It is possible that a stable c-Rel–CNBP complex is formed in the cytoplasm and that upon LPS stimulation, this complex translocates to the nucleus. Another possibility is that the c-Rel–CNBP complexes are essential for retention of c-Rel in the nucleus after inducible translocation and DNA binding. A third possibility is that CNBP influences the nuclear translocation of c-Rel by indirect mechanisms. Consistent with this latter possibility, although CNBP and c-Rel interact, only a modest fraction of c-Rel is stably associated with CNBP. Extensive work from Smale and colleagues have defined the importance of c-Rel in controlling IL-12β gene transcription (Sanjabi et al., 2000). Surprisingly, c-Rel regulates only a very restricted set of target genes. RNA-sequencing experiments comparing WT and c-Rel–deficient macrophages stimulated with lipid A show that IL-12β and a limited set of genes, including Il4i1 and Med21, represent bona fide c-Rel targets. Consistently, in our experiments, both Il4i1 and Med21 expression were also decreased in Cnbp-deficient macrophages stimulated with LPS. Together, all of these findings implicate CNBP as a highly specific regulator of c-Rel and define a novel TLR–CNBP–c-Rel signaling axis controlling IL-12β expression. Our studies also reveal that CNBP nuclear translocation and regulation of IL-12β are new signaling events downstream of multiple PRRs.
Classically, IL-12 is induced by TLRs in response to microbial products (Seki et al., 2002; Bafica et al., 2005; Andrade et al., 2013; Schamber-Reis et al., 2013). CNBP was essential for IL-12β production in macrophages stimulated with any ligand tested or microbial infection. This included viruses, M. tuberculosis, S. typhimurium, and T. gondii, suggesting that IL-12β induction, regardless of the stimuli, required CNBP. Moreover, we provide important in vivo evidence for the importance of CNBP in the immune system. T. gondii is an intracellular protozoan parasite whose elimination requires production of IFN-γ that activates various cell-intrinsic antiparasitic defense pathways within infected cells (Yap and Sher, 1999). The importance of IL-12 in the induction T-lymphocyte production of IFN-γ and resistance to T. gondii and other intracellular pathogens in vivo is well established (Jordan et al., 2010). Consistent with the important role of CNBP in controlling the innate production of IL-12 in vitro, Cnbp-deficient mice have reduced IL-12β levels in vivo. This leads to a reduction in IFN-γ produced by both CD4+ and CD8+ T cells and an inability to control T. gondii growth in vivo. CD8a+ DCs and monocyte-derived DCs are the important producers of IL-12 in vivo during T. gondii infection (Mashayekhi et al., 2011; Goldszmid et al., 2012; Dupont et al., 2015). In our studies, we found that DCs from Cnbp-deficient mice had reduced IL-12 levels in vivo. Both CD8α+ DCs and CD8α− DCs produced IL-12 in a CNBP-dependent manner. Consistently, IFN-γ production by CD4 T and CD8 T cells was impaired in Cnbp-KO mice infected with T. gondii. Our in vitro and vivo experiments using the T. gondii infection model further demonstrated that the kinetics of CNBP nuclear translocation correlated with the peak of IL-12 production. These observations indicate that both macrophages and DCs produce IL-12 in a CNBP-dependent manner, with the latter being more relevant in vivo for development of Th1 immunity and control of T. gondii in vivo. Interestingly, previous studies with c-Rel–KO mice describe a c-Rel–independent pathway to IL-12 with c-Rel playing an important role in T cell responses to T. gondi infection in vivo (Mason et al., 2004). Deciphering the cell-type–specific contributions in vivo of CNBP-dependent control of Th1 immunity to T. gondii will led to new insights into c-Rel–dependent and independent control of T. gondii and related pathogens.
Prior to this work linking CNBP to the IL-12β and Th1 immune response, CNBP was shown to regulate forebrain development by controlling cell proliferation and apoptosis (Calcaterra et al., 2010). It is worth noting here that Cnbp-deficient mice were viable and, in terms of their immune systems, were comparable to WT mice under steady state conditions. In addition to these effects, CNBP has been implicated in the transcriptional and posttranscriptional control of gene expression. Early studies identified CNBP as a regulator of the transcription of c-myc, wnt, and skeletal muscle chloride channel 1 (Chen et al., 2013; Margarit et al., 2014). Recently, a photoactivatable ribonucleoside-enhanced crosslinking and immunoprecipitation (PAR-CLIP) study indicated that CNBP preferentially bound G-rich elements in mRNA coding sequences, increasing their translational efficiency (Benhalevy et al., 2017). There is little evidence linking CNBP to the immune system. CNBP has been implicated in myotonic dystrophy type 2 and sporadic inclusion body myositis, both of which are associated with increased autoantibodies, suggesting some links to the immune response (Tieleman et al., 2009; Niedowicz et al., 2010). In macrophages, the ability of Cnbp to regulate NF-κB and sustained expression of IL-6 was the first evidence of an immune role for this factor (Lee et al., 2017). Consistent with the effects we describe herein, this study also demonstrated using a zebrafish model that Cnbp-deficient zebrafish were more susceptible to infection with Shigella flexneri infection, suggesting a conservation of the immune regulatory effects we describe.
In conclusion, our findings provide critical new insights into the regulation of the innate immune response in macrophages and DCs, describing a previously unknown CNBP–c-Rel signaling axis important in controlling IL-12β expression downstream of multiple PRRs. This pathway also has important effects in vivo controlling Th1 immunity and resistance to the parasitic pathogen T. gondii. Given the importance of IL-12 in the control of Th1 immunity and the potential for dysregulation of the IL-12/Th1 response in inflammatory diseases such as inflammatory bowel disease, the identification of CNBP as a specific regulator of c-Rel–dependent IL-12 induction has enormous implications for both host defense and inflammatory disease. IL-12 is a prominent target for clinical intervention in the treatment of inflammatory diseases and cancer. Our work unveils a new target CNBP that may provide a unique handle to specifically curtail this IL-12 response.
Materials and methods
Generation of CNBP-KO mice
CNBP-deficient mice were generated using embryonic stem (ES) cells obtained from the Knockout Mouse Repository (University of California, Davis). ES cells (Cnbptm1a(KOMP)Wtsi) were generated by replacing the CNBP genomic locus with a neomycin cassette under control of a Pgk1 promoter. ES cells were injected into blastocytes to generate chimeric mice at the University of Massachusetts Medical School Transgenic Core. CNBP heterozygous mice were obtained by gamete line transmission from mating the chimeric mice with WT C57BL/6 mice. CNBP heterozygous mice were intercrossed to generate WT and KO alleles for experimentation. Littermate controls were used throughout. Myd88−/−; Myd88−/−Trif−/− mice were from S. Akira (Osaka University, Osaka, Japan). Irf3−/−-Irf7−/− mice were obtained from T. Taniguchi (The University of Tokyo, Tokyo, Japan). c-Rel−/− mice were obtained from S. Smale (University of California, Los Angeles, Los Angeles, CA) and I. Brodsky (University of Pennsylvania, Philadelphia, PA). All mouse strains were bred and maintained under specific pathogen–free conditions in the animal facilities at the University of Massachusetts Medical School in accordance with the Institutional Animal Care and Use Committee.
Cell culture and stimulation
BMDMs were generated by differentiating bone marrow cells in DMEM supplemented with 10% FCS, 1% penicillin/streptomycin cocktail and 20% L929 supernatant for 7 d. BMDCs were generated by differentiating bone marrow cells in RPMI 1640 medium supplemented with 10% FCS, 1% penicillin/streptomycin cocktail, and recombinant GM-CSF (20 ng/ml) for 10 d. For all experiments, cells were plated one day before stimulation. Cells were stimulated at the following concentrations (unless mentioned otherwise): LPS (100 ng/ml), poly(I:C) (25 μg/ml), and BAY 11–7082 (10 μM). Transfection of BMDMs with poly(dA:dT) and ISD was performed using Lipofectamine 2000 (Invitrogen). BMDMs were infected with HSV (HSV-1-ICP0–deficient mutant; 10 MOI [multiplicity of infection]) from D. Knipe (Harvard Medical School, Boston, MA), mCMV (10 MOI), IAV (5 MOI), SeV (200 IU/ml), M. tuberculosis (10 MOI), Salmonella (10 MOI), and T. gondii (5 MOI) for the indicated hours for mRNA and protein analysis.
Parasites
The T. gondii strain ME49 was maintained in C57BL/6 mice by serial inoculation of brain homogenate–containing cysts. ME49 tachyzoites were maintained in human foreskin fibroblast cells (Hs27). Parasite suspensions were prepared by collection of infected cells and disruption by repeated passage through a 27-G needle followed by centrifugation at 900 g for 10 min. For soluble tachyzoite antigen preparation, the parasites were submitted to three cycles of freezing at −70°C and thawing at 37°C followed by four 20-s rounds of sonication. The resulting homogenate was then centrifuged at 600 g for 10 min, and the supernatant was collected and its protein concentration determined by the Bradford method (Invitrogen).
In vivo experimental infections
Mice were inoculated i.p. with 25 cysts obtained from brain homogenates of 6- to 8-wk-infected mice. Mice were monitored for survival or sacrificed 7 d after infection in order to collect peritoneal exudate cells and fluid, blood, liver, and spleens. Samples from each mouse were individually processed and analyzed. Quantification of T. gondii DNA was performed by amplification of B1 gene using primers (forward: 5′-CTGGCAAATACAGGTGAAATG-3′; reverse: 5′-GTGTACTGCGAAAATGAATCC-3′) and analyzed based on a standard curve of purified parasite DNA. PCR reactions were setup in a final 20 ml volume using 5 ng of total tissue DNA, 200 ng of each primer and 1× of the iQ SYBR Green Supermix (Bio-Rad).
In vitro infection
Macrophages cultures were induced or not overnight with IFN-γ (40 ng/ml) before infection. Cells were infected with ME49 tachyzoites (5 MOI), and after 2 h, unbound parasites were washed off. The cultures were incubated for 24 h to obtain supernatant and for 4 h for RNA extraction.
Affinity purification with biotinylated oligonucleotides
Cytosolic cell extracts were generated from 10 × 106 murine BMDMs or 20 × 106 THP1 cells using low-salt lysis buffers. The lysates were precleared with Streptavidin ultralink agarose protein G resin for 2 h at 4°C and the resin washed out. 3 μg of Biotinylated oligonucleotides were then added to precleared lysates for binding overnight (∼18 h) at 4°C. Streptavidin ultralink agarose G beads were added to bind biotinylated complexes for 2 h at 4°C. The bound complexes were pelleted by spinning at 6,500 RPM for 5 min at 4°C. Several rounds of increasingly stringent washes were performed to reduce nonspecific binding partners. The majority of the sample was run under nondenaturing conditions on a stacking gel composition PAGE. A shotgun approach was used to run the sample (i.e., the sample was run so that all of it had migrated into the gel). The gel was subsequently stained with MS-compatible Biosafe Coomassie blue (Bio-Rad) to identify the band of proteins in the gel, and the band was excised. A small aliquot was subjected to normal SDS-PAGE gel and Western blotting.
LC-MS/MS
Isolated proteins in the complex were identified and analyzed by LC/MS at the Mass Spectrometry Core Facility at the University of Massachusetts. Briefly, DNA-bound protein (≥2 pmol protein) was centrifuged and the subsequent pellet was vacuum dried. The pellet was then dissolved in 20 μl of 0.1% Rapigest detergent in 0.1 M tetramethylammonium bicarbonate. Disulfides were reduced with dithiothreitol for 30 min at 60°C. Sulfhydrol was alkylated with 20 nmol iodoacetamide for 3 h in the dark at 22°C. Trypsin digestion and capillary LC-MS/MS were performed on the ThermoFinnigan LTQ quadrupole ion trap system. Proteins were identified from the product ion scan spectra using SEQUEST search engine with species subset of the International Protein Index (IPI) protein database. Peptide and protein data were assembled in Scaffold software and analyzed against proteins bound to control DNA ODN. Proteins were subsequently identified by their Gene Ontology annotations and sorted through the domain database at Pfam (Wellcome Trust, Sanger Institute) for initial bioinformatics predictions.
RNA extraction and qRT-PCR
Total RNA from BMDMs, BMDCs, and peritoneal exudate cells was extracted with an RNeasy RNA extraction kit (Qiagen) according to the manufacturer’s instructions. Genomic DNA in RNA purifications was eliminated using on-column DNase I digestion (Qiagen). Equal amounts of RNA (1,000 ng) were reverse transcribed using the iScript cDNA synthesis kit (Bio-Rad). Diluted cDNAs (1:100 final) were subjected to qPCR analysis using iQ SYBR Green Supermix reagent (Bio-Rad). Gene expression levels were normalized to Gapdh as housekeeping genes. Relative mRNA expressions were calculated by the change-in-cycling-threshold method as 2−ddC(t). The specificity of amplification was assessed for each sample by melting curve analysis. Primer sequences are provided in Table S1.
Cytokine analysis
Cell culture supernatant was assayed for cytokine levels using commercially available sandwich ELISA kits for IL12p40, IL12p70, IL10, and IFN-γ (R&D Systems), IL6 (BD Biosciences), and TNF-α (eBioscience). The murine IFN-β kit has been described previously (Roberts et al., 2007). All experiments for cytokine analysis by ELISA were performed in biological triplicates.
NanoString analysis
Cell stimulation and RNA isolation were performed as described above. The nCounter analysis system was used for multiplex mRNA measurements using the custom gene expression code set against 200 immune genes. Total RNA (100 ng) was hybridized overnight with the gene expression code set and analyzed on an nCounter Digital Analyzer (NanoString Technologies). RNA hybridization, data acquisition, and analysis were performed per the manufacturer’s specifications. RNA counts were processed to account for hybridization efficiency, and mRNA expression across experimental groups was normalized to the geometric mean of six housekeeping genes
ChIP
ChIP experiments were performed essentially as described previously (Atianand et al., 2016). Briefly, 1 × 107 primary BMDMs were used to perform immunoprecipitation with mouse monoclonal anti-CNBP. qPCR was performed on immunoprecipitated and input fractions from the immunoprecipitation. Primer sequences used for qRT-PCR are shown in Table S1.
Purification of CD11c+ DCs
Mouse CD11c+ DCs were purified from spleens of mice injected with BL6-FLT3L–producing cells. Briefly, mice were injected subcutaneously with 2 × 106 cells in 300 μl PBS. Spleens were harvested within 2 wk after injection and CD11c+ DCs were isolated using the CD11c MicroBeads UltraPure kit (Miltenyi Biotec). B16-FLT3L–producing cells were from G. Dranoff (Harvard Medical School, Boston, MA).
Flow cytometry
Mononuclear cells were incubated with 50 ng/ml PMA (Sigma) and 500 ng/ml ionomycin (Sigma) in the presence of GolgiStop (BD) in complete T cell media at 37°C for 4 h. Cells were washed with FACS buffer, stained with Live/Dead fixable aqua dead cell marker (Invitrogen), and incubated with Fc block before staining. The following antibodies were used for staining: PerCP-Cy5-CD8, APC-Cy7-CD8, APC-F4/80, Alexa Fluor 700–CD11c, FITC-MHCIIb, PE-Cy7-CD11b, eFluor450-Ly6C, APC-Cy7-CD4, and FITC-CD3. Intracellular staining for APC-IL4, PE-Cy7-IL17a, eFluor450-IFN-γ, and PE-IL12. Intracellular cytokine staining was performed by fixing cells in 2% paraformaldehyde, followed by permeabilization and staining (BD Biosciences). Flow cytometric analyses were performed on an LSRFortessa (BD Biosciences) and analyzed using FlowJo Software (Tree Star).
Immunofluorescence staining
Primary BMDMs were fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100/PBS before incubation with primary antibodies (anti-CNBP) for 2 h at room temperature. Cells were washed in PBS, followed by incubation with TRITC-conjugated anti-mouse Ig secondary antibodies. Nuclei were stained with DAPI.
CRISPR/Cas9 KO
Hek293 cells were seeded on 24-well plates; after 16 h, plasmids expressing Cas9 and single guide RNA (sgRNA) were cotransfected into Hek293 cells. At 36 h after transfection, cells were selected for puromycin resistance, then serial dilution of living cells was done to obtain a cell density of 1 cell per well on a 96-well plate. Immunoblotting was performed to ensure gene KO results after the clones were grown. sgRNA sequences are shown in Table S1.
Luciferase reporter assay
Hek293 cells were seeded on 96-well plates (4 × 104 cells per well) and then transfected with 50 ng IL12p40, IL6, or TNF luciferase reporter vector and 5 ng Renilla-luciferase reporter vector with increasing amounts of expression vector for Cnbp or plus 10 ng c-Rel or p65 expression vector. Empty control vector was added so that a total of 200 ng vector DNA was transfected into each well. Cells were collected at 36 h after transfection, and luciferase activity was measured with a Dual-Luciferase Reporter Assay System according to the manufacturer’s instructions (Promega). Firefly luciferase activities were normalized to Renilla luciferase activities. All reporter assays were repeated at least three times. Data shown are average values and SEM from one representative experiment.
Coimmunoprecipitation and Western blot analysis
Cells were lysed 36–48 h after transfection of expression plasmids using the lysis buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl,1% Triton X-100, and protease inhibitor cocktails. For immunoprecipitation, lysates were incubated with the appropriate antibodies for 2 h on ice, followed by precipitation with protein G Sepharose. Samples were separated by SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes. After blocking in PBS containing 0.1% Tween-20 and 5% skim milk, the blots were probed with indicated antibodies. Western blot visualization was done with enhanced chemiluminescence.
Statistics
GraphPad Prism 5 software (GraphPad Software) was used for data analysis using a two-tailed unpaired t test. A P value <0.05 was considered statistically significant (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Online supplemental material
Fig. S1 shows that Cnbp-deficient mice have normal immune cell phenotyping and provides additional details on DNA virus infection in CNBP-deficient BMDMs. Fig. S2 shows cytokine analysis in CNBP-KO cells after stimulation with various ligands at different time points. Fig. S3 shows that IL10 induction was not affected by IL-12 through anti–IL-12 neutralizing antibody treatment. Fig. S4 shows the cytokine inductions in IRAK1/2−/− or IRAK4−/− BMDMs and analyzes NF-κB signaling activity by detecting the translocation of p65 and phosphorylation of IκBα. Fig. S5 includes further characterization of CNBP-deficient mice after T. gondii infection. Table S1 contains primers for qPCR and ChIP-PCR and the sgRNAs used in this study.
Supplementary Material
Acknowledgments
We are grateful to Dr. Steve Smale for providing the reporter plasmid Rel Mut p40-Luc and c-Rel–deficient mice. We would like to thank Dr. Igor E. Brodsky for providing c-Rel–deficient mice, Natalia Martinez Araujo for help with T. gondii infections, and all members of the K.A. Fitzgerald laboratory for their helpful comments.
This study was supported by National Institutes of Health grants R37-AI067497 and R01-AI079293 (to K.A. Fitzgerald), R01-AI116577 and R01-NS098747 (to R.T. Gazzinelli), R01-AI106934 (to D.M. Knipe), and AI132130 (to C.M. Sassetti) and by the Charles A. King Trust Postdoctoral Research Fellowship Program, Bank of America, N.A., Co-Trustees (to Y. Chen).
The authors declare no competing financial interests.
Author contributions: K.A. Fitzgerald supervised the work. Y. Chen and K.A. Fitzgerald designed the research, analyzed results, and wrote the manuscript. Y. Chen performed the majority of the experiments, with contributions from S. Sharma, P.A. Assis, Z. Jiang, R. Elling, A.J. Olive, S. Hang, J. Bernier, J.R. Huh, C.M. Sassetti, and R.T. Gazzinelli. D.M. Knipe provided critical reagents and suggestions. All authors read and provided suggestions during the preparation of the manuscript.
References
- Andrade W.A., Souza M.C., Ramos-Martinez E., Nagpal K., Dutra M.S., Melo M.B., Bartholomeu D.C., Ghosh S., Golenbock D.T., and Gazzinelli R.T.. 2013. Combined action of nucleic acid-sensing Toll-like receptors and TLR11/TLR12 heterodimers imparts resistance to Toxoplasma gondii in mice. Cell Host Microbe. 13:42–53. 10.1016/j.chom.2012.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aste-Amezaga M., Ma X., Sartori A., and Trinchieri G.. 1998. Molecular mechanisms of the induction of IL-12 and its inhibition by IL-10. J. Immunol. 160:5936–5944. [PubMed] [Google Scholar]
- Atianand M.K., Hu W., Satpathy A.T., Shen Y., Ricci E.P., Alvarez-Dominguez J.R., Bhatta A., Schattgen S.A., McGowan J.D., Blin J., et al. 2016. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell. 165:1672–1685. 10.1016/j.cell.2016.05.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafica A., Scanga C.A., Feng C.G., Leifer C., Cheever A., and Sher A.. 2005. TLR9 regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J. Exp. Med. 202:1715–1724. 10.1084/jem.20051782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhalevy D., Gupta S.K., Danan C.H., Ghosal S., Sun H.W., Kazemier H.G., Paeschke K., Hafner M., and Juranek S.A.. 2017. The Human CCHC-type Zinc Finger Nucleic Acid-Binding Protein Binds G-Rich Elements in Target mRNA Coding Sequences and Promotes Translation. Cell Reports. 18:2979–2990. 10.1016/j.celrep.2017.02.080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron C.A., and Gazzinelli R.T.. 1995. Effects of IL-12 on immune responses to microbial infections: a key mediator in regulating disease outcome. Curr. Opin. Immunol. 7:485–496. 10.1016/0952-7915(95)80093-X [DOI] [PubMed] [Google Scholar]
- Bradley M.N., Zhou L., and Smale S.T.. 2003. C/EBPbeta regulation in lipopolysaccharide-stimulated macrophages. Mol. Cell. Biol. 23:4841–4858. 10.1128/MCB.23.14.4841-4858.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caamaño J., and Hunter C.A.. 2002. NF-kappaB family of transcription factors: central regulators of innate and adaptive immune functions. Clin. Microbiol. Rev. 15:414–429. 10.1128/CMR.15.3.414-429.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calcaterra N.B., Armas P., Weiner A.M., and Borgognone M.. 2010. CNBP: a multifunctional nucleic acid chaperone involved in cell death and proliferation control. IUBMB Life. 62:707–714. 10.1002/iub.379 [DOI] [PubMed] [Google Scholar]
- Cao X. 2016. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 16:35–50. 10.1038/nri.2015.8 [DOI] [PubMed] [Google Scholar]
- Chen Q., Sun L., and Chen Z.J.. 2016. Regulation and function of the cGAS-STING pathway of cytosolic DNA sensing. Nat. Immunol. 17:1142–1149. 10.1038/ni.3558 [DOI] [PubMed] [Google Scholar]
- Chen S., Su L., Qiu J., Xiao N., Lin J., Tan J.H., Ou T.M., Gu L.Q., Huang Z.S., and Li D.. 2013. Mechanistic studies for the role of cellular nucleic-acid-binding protein (CNBP) in regulation of c-myc transcription. Biochim. Biophys. Acta. 1830:4769–4777. 10.1016/j.bbagen.2013.06.007 [DOI] [PubMed] [Google Scholar]
- Cooper A.M., Kipnis A., Turner J., Magram J., Ferrante J., and Orme I.M.. 2002. Mice lacking bioactive IL-12 can generate protective, antigen-specific cellular responses to mycobacterial infection only if the IL-12 p40 subunit is present. J. Immunol. 168:1322–1327. 10.4049/jimmunol.168.3.1322 [DOI] [PubMed] [Google Scholar]
- D’Andrea A., Aste-Amezaga M., Valiante N.M., Ma X., Kubin M., and Trinchieri G.. 1993. Interleukin 10 (IL-10) inhibits human lymphocyte interferon gamma-production by suppressing natural killer cell stimulatory factor/IL-12 synthesis in accessory cells. J. Exp. Med. 178:1041–1048. 10.1084/jem.178.3.1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dev A., Iyer S., Razani B., and Cheng G.. 2011. NF-κB and innate immunity. Curr. Top. Microbiol. Immunol. 349:115–143. [DOI] [PubMed] [Google Scholar]
- Dupont C.D., Harms Pritchard G., Hidano S., Christian D.A., Wagage S., Muallem G., Tait Wojno E.D., and Hunter C.A.. 2015. Flt3 Ligand Is Essential for Survival and Protective Immune Responses during Toxoplasmosis. J. Immunol. 195:4369–4377. 10.4049/jimmunol.1500690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Marin C., Schrum J.E., Andrade W.A., Shaffer S.A., Giraldo L.F., Lasso A.M., Kurt-Jones E.A., Fitzgerald K.A., and Golenbock D.T.. 2018. Cyclic GMP-AMP Synthase Is the Cytosolic Sensor of Plasmodium falciparum Genomic DNA and Activates Type I IFN in Malaria. J. Immunol. 200:768–774. 10.4049/jimmunol.1701048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzinelli R.T., Hieny S., Wynn T.A., Wolf S., and Sher A.. 1993. Interleukin 12 is required for the T-lymphocyte-independent induction of interferon gamma by an intracellular parasite and induces resistance in T-cell-deficient hosts. Proc. Natl. Acad. Sci. USA. 90:6115–6119. 10.1073/pnas.90.13.6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzinelli R.T., Mendonça-Neto R., Lilue J., Howard J., and Sher A.. 2014. Innate resistance against Toxoplasma gondii: an evolutionary tale of mice, cats, and men. Cell Host Microbe. 15:132–138. 10.1016/j.chom.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., and Hayden M.S.. 2008. New regulators of NF-kappaB in inflammation. Nat. Rev. Immunol. 8:837–848. 10.1038/nri2423 [DOI] [PubMed] [Google Scholar]
- Goldszmid R.S., Caspar P., Rivollier A., White S., Dzutsev A., Hieny S., Kelsall B., Trinchieri G., and Sher A.. 2012. NK cell-derived interferon-γ orchestrates cellular dynamics and the differentiation of monocytes into dendritic cells at the site of infection. Immunity. 36:1047–1059. 10.1016/j.immuni.2012.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M.S., and Ghosh S.. 2011. NF-κB in immunobiology. Cell Res. 21:223–244. 10.1038/cr.2011.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölscher C., Atkinson R.A., Arendse B., Brown N., Myburgh E., Alber G., and Brombacher F.. 2001. A protective and agonistic function of IL-12p40 in mycobacterial infection. J. Immunol. 167:6957–6966. 10.4049/jimmunol.167.12.6957 [DOI] [PubMed] [Google Scholar]
- Jordan K.A., Dupont C.D., Tait E.D., Liou H.C., and Hunter C.A.. 2010. Role of the NF-κB transcription factor c-Rel in the generation of CD8+ T-cell responses to Toxoplasma gondii. Int. Immunol. 22:851–861. 10.1093/intimm/dxq439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khader S.A., Partida-Sanchez S., Bell G., Jelley-Gibbs D.M., Swain S., Pearl J.E., Ghilardi N., Desauvage F.J., Lund F.E., and Cooper A.M.. 2006. Interleukin 12p40 is required for dendritic cell migration and T cell priming after Mycobacterium tuberculosis infection. J. Exp. Med. 203:1805–1815. 10.1084/jem.20052545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiba R., Yanai H., Matsuda A., Goto A., Nakajima A., Negishi H., Nishio J., Smale S.T., and Taniguchi T.. 2013. Regulation of cooperative function of the Il12b enhancer and promoter by the interferon regulatory factors 3 and 5. Biochem. Biophys. Res. Commun. 430:95–100. 10.1016/j.bbrc.2012.11.006 [DOI] [PubMed] [Google Scholar]
- Lee E., Lee T.A., Kim J.H., Park A., Ra E.A., Kang S., Choi H.J., Choi J.L., Huh H.D., Lee J.E., et al. 2017. CNBP acts as a key transcriptional regulator of sustained expression of interleukin-6. Nucleic Acids Res. 45:3280–3296. 10.1093/nar/gkx071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann J., Bellmann S., Werner C., Schröder R., Schütze N., and Alber G.. 2001. IL-12p40-dependent agonistic effects on the development of protective innate and adaptive immunity against Salmonella enteritidis. J. Immunol. 167:5304–5315. 10.4049/jimmunol.167.9.5304 [DOI] [PubMed] [Google Scholar]
- Liu J., Qian C., and Cao X.. 2016. Post-Translational Modification Control of Innate Immunity. Immunity. 45:15–30. 10.1016/j.immuni.2016.06.020 [DOI] [PubMed] [Google Scholar]
- Margarit E., Armas P., García Siburu N., and Calcaterra N.B.. 2014. CNBP modulates the transcription of Wnt signaling pathway components. Biochim. Biophys. Acta. 1839:1151–1160. 10.1016/j.bbagrm.2014.08.009 [DOI] [PubMed] [Google Scholar]
- Mashayekhi M., Sandau M.M., Dunay I.R., Frickel E.M., Khan A., Goldszmid R.S., Sher A., Ploegh H.L., Murphy T.L., Sibley L.D., and Murphy K.M.. 2011. CD8α(+) dendritic cells are the critical source of interleukin-12 that controls acute infection by Toxoplasma gondii tachyzoites. Immunity. 35:249–259. 10.1016/j.immuni.2011.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason N., Aliberti J., Caamano J.C., Liou H.C., and Hunter C.A.. 2002. Cutting edge: identification of c-Rel-dependent and -independent pathways of IL-12 production during infectious and inflammatory stimuli. J. Immunol. 168:2590–2594. 10.4049/jimmunol.168.6.2590 [DOI] [PubMed] [Google Scholar]
- Mason N.J., Liou H.C., and Hunter C.A.. 2004. T cell-intrinsic expression of c-Rel regulates Th1 cell responses essential for resistance to Toxoplasma gondii. J. Immunol. 172:3704–3711. 10.4049/jimmunol.172.6.3704 [DOI] [PubMed] [Google Scholar]
- Medzhitov R., and Horng T.. 2009. Transcriptional control of the inflammatory response. Nat. Rev. Immunol. 9:692–703. 10.1038/nri2634 [DOI] [PubMed] [Google Scholar]
- Meola G., and Cardani R.. 2015. Myotonic dystrophies: An update on clinical aspects, genetic, pathology, and molecular pathomechanisms. Biochim. Biophys. Acta. 1852:594–606. 10.1016/j.bbadis.2014.05.019 [DOI] [PubMed] [Google Scholar]
- Niedowicz D.M., Beckett T.L., Holler C.J., Weidner A.M., and Murphy M.P.. 2010. APP(DeltaNL695) expression in murine tissue downregulates CNBP expression. Neurosci. Lett. 482:57–61. 10.1016/j.neulet.2010.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Garra A., and Murphy K.M.. 2009. From IL-10 to IL-12: how pathogens and their products stimulate APCs to induce T(H)1 development. Nat. Immunol. 10:929–932. 10.1038/ni0909-929 [DOI] [PubMed] [Google Scholar]
- O’Neill L.A., Golenbock D., and Bowie A.G.. 2013. The history of Toll-like receptors - redefining innate immunity. Nat. Rev. Immunol. 13:453–460. 10.1038/nri3446 [DOI] [PubMed] [Google Scholar]
- Orange J.S., Wang B., Terhorst C., and Biron C.A.. 1995. Requirement for natural killer cell-produced interferon gamma in defense against murine cytomegalovirus infection and enhancement of this defense pathway by interleukin 12 administration. J. Exp. Med. 182:1045–1056. 10.1084/jem.182.4.1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raheem O., Olufemi S.E., Bachinski L.L., Vihola A., Sirito M., Holmlund-Hampf J., Haapasalo H., Li Y.P., Udd B., and Krahe R.. 2010. Mutant (CCTG)n expansion causes abnormal expression of zinc finger protein 9 (ZNF9) in myotonic dystrophy type 2. Am. J. Pathol. 177:3025–3036. 10.2353/ajpath.2010.100179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts Z.J., Goutagny N., Perera P.Y., Kato H., Kumar H., Kawai T., Akira S., Savan R., van Echo D., Fitzgerald K.A., et al. 2007. The chemotherapeutic agent DMXAA potently and specifically activates the TBK1-IRF-3 signaling axis. J. Exp. Med. 204:1559–1569. 10.1084/jem.20061845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjabi S., Hoffmann A., Liou H.C., Baltimore D., and Smale S.T.. 2000. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proc. Natl. Acad. Sci. USA. 97:12705–12710. 10.1073/pnas.230436397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjabi S., Williams K.J., Saccani S., Zhou L., Hoffmann A., Ghosh G., Gerondakis S., Natoli G., and Smale S.T.. 2005. A c-Rel subdomain responsible for enhanced DNA-binding affinity and selective gene activation. Genes Dev. 19:2138–2151. 10.1101/gad.1329805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schamber-Reis B.L., Petritus P.M., Caetano B.C., Martinez E.R., Okuda K., Golenbock D., Scott P., and Gazzinelli R.T.. 2013. UNC93B1 and nucleic acid-sensing Toll-like receptors mediate host resistance to infection with Leishmania major. J. Biol. Chem. 288:7127–7136. 10.1074/jbc.M112.407684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz S.M., Köhler G., Schütze N., Knauer J., Straubinger R.K., Chackerian A.A., Witte E., Wolk K., Sabat R., Iwakura Y., et al. 2008. Protective immunity to systemic infection with attenuated Salmonella enterica serovar enteritidis in the absence of IL-12 is associated with IL-23-dependent IL-22, but not IL-17. J. Immunol. 181:7891–7901. 10.4049/jimmunol.181.11.7891 [DOI] [PubMed] [Google Scholar]
- Seki E., Tsutsui H., Tsuji N.M., Hayashi N., Adachi K., Nakano H., Futatsugi-Yumikura S., Takeuchi O., Hoshino K., Akira S., et al. 2002. Critical roles of myeloid differentiation factor 88-dependent proinflammatory cytokine release in early phase clearance of Listeria monocytogenes in mice. J. Immunol. 169:3863–3868. 10.4049/jimmunol.169.7.3863 [DOI] [PubMed] [Google Scholar]
- Sharma S., DeOliveira R.B., Kalantari P., Parroche P., Goutagny N., Jiang Z., Chan J., Bartholomeu D.C., Lauw F., Hall J.P., et al. 2011. Innate immune recognition of an AT-rich stem-loop DNA motif in the Plasmodium falciparum genome. Immunity. 35:194–207. 10.1016/j.immuni.2011.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher A., Collazzo C., Scanga C., Jankovic D., Yap G., and Aliberti J.. 2003. Induction and regulation of IL-12-dependent host resistance to Toxoplasma gondii. Immunol. Res. 27:521–528. 10.1385/IR:27:2-3:521 [DOI] [PubMed] [Google Scholar]
- Silverman N., and Maniatis T.. 2001. NF-kappaB signaling pathways in mammalian and insect innate immunity. Genes Dev. 15:2321–2342. 10.1101/gad.909001 [DOI] [PubMed] [Google Scholar]
- Smale S.T. 2012. Transcriptional regulation in the innate immune system. Curr. Opin. Immunol. 24:51–57. 10.1016/j.coi.2011.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale S.T., and Natoli G.. 2014. Transcriptional control of inflammatory responses. Cold Spring Harb. Perspect. Biol. 6:a016261 10.1101/cshperspect.a016261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Van Ghelue M., Tranebjærg L., Thyssen F., Nilssen Ø., and Torbergsen T.. 2011. Myotonia congenita and myotonic dystrophy in the same family: coexistence of a CLCN1 mutation and expansion in the CNBP (ZNF9) gene. Clin. Genet. 80:574–580. 10.1111/j.1399-0004.2010.01616.x [DOI] [PubMed] [Google Scholar]
- Sypek J.P., Chung C.L., Mayor S.E., Subramanyam J.M., Goldman S.J., Sieburth D.S., Wolf S.F., and Schaub R.G.. 1993. Resolution of cutaneous leishmaniasis: interleukin 12 initiates a protective T helper type 1 immune response. J. Exp. Med. 177:1797–1802. 10.1084/jem.177.6.1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng M.W.L., Bowman E.P., McElwee J.J., Smyth M.J., Casanova J.L., Cooper A.M., and Cua D.J.. 2015. IL-12 and IL-23 cytokines: from discovery to targeted therapies for immune-mediated inflammatory diseases. Nat. Med. 21:719–729. 10.1038/nm.3895 [DOI] [PubMed] [Google Scholar]
- Thompson M.R., Kaminski J.J., Kurt-Jones E.A., and Fitzgerald K.A.. 2011. Pattern recognition receptors and the innate immune response to viral infection. Viruses. 3:920–940. 10.3390/v3060920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton C.A., Wang E., and Carrell E.M.. 2017. Myotonic dystrophy: approach to therapy. Curr. Opin. Genet. Dev. 44:135–140. 10.1016/j.gde.2017.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tieleman A.A., den Broeder A.A., van de Logt A.E., and van Engelen B.G.M.. 2009. Strong association between myotonic dystrophy type 2 and autoimmune diseases. J. Neurol. Neurosurg. Psychiatry. 80:1293–1295. 10.1136/jnnp.2008.156562 [DOI] [PubMed] [Google Scholar]
- Trinchieri G. 1995. Interleukin-12: a proinflammatory cytokine with immunoregulatory functions that bridge innate resistance and antigen-specific adaptive immunity. Annu. Rev. Immunol. 13:251–276. 10.1146/annurev.iy.13.040195.001343 [DOI] [PubMed] [Google Scholar]
- Trinchieri G., and Sher A.. 2007. Cooperation of Toll-like receptor signals in innate immune defence. Nat. Rev. Immunol. 7:179–190. 10.1038/nri2038 [DOI] [PubMed] [Google Scholar]
- Tripp C.S., Wolf S.F., and Unanue E.R.. 1993. Interleukin 12 and tumor necrosis factor alpha are costimulators of interferon gamma production by natural killer cells in severe combined immunodeficiency mice with listeriosis, and interleukin 10 is a physiologic antagonist. Proc. Natl. Acad. Sci. USA. 90:3725–3729. 10.1073/pnas.90.8.3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., and Akira S.. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 301:640–643. 10.1126/science.1087262 [DOI] [PubMed] [Google Scholar]
- Yap G.S., and Sher A.. 1999. Cell-mediated immunity to Toxoplasma gondii: initiation, regulation and effector function. Immunobiology. 201:240–247. 10.1016/S0171-2985(99)80064-3 [DOI] [PubMed] [Google Scholar]
- Zhang G., and Ghosh S.. 2001. Toll-like receptor-mediated NF-kappaB activation: a phylogenetically conserved paradigm in innate immunity. J. Clin. Invest. 107:13–19. 10.1172/JCI11837 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.