Ransohoff discusses why animal models have uniformly failed to predict success in neurodegenerative clinical trials.
Abstract
Richard M. Ransohoff, Entrepreneur-in-Residence at Third Rock Ventures and Visiting Scientist at Harvard Medical School, provides his personal opinion on using animal models to address current challenges and opportunities in drug development for neurodegeneration.
“All models are wrong, but some are useful.” (Box, 1979)
Why animal models?
The thesis proposed in this article is that it’s not helpful for neurodegeneration drug development to perform preclinical efficacy experiments in animal models. George Box’s epigram quoted above is commonly projected at the beginning of discussions of this topic, suggesting a “don’t let the perfect be the enemy of the good” stance. It isn’t perhaps widely known among audience members, and possibly speakers as well, that Box was addressing participants at a statistical workshop and that the example he offered was Boyle’s Ideal Gas Law, which is useful but not true for any real gas.
By contrast, results from using animal models to predict success in neurodegenerative disease clinical experiments have been uniformly disappointing and, in that sense, not useful. Neurological disease clinical experiments are monstrously expensive, all the while consuming time and effort on the part of research subjects, trialists, and the innumerable functions that pharmaceutical companies deploy in these studies. Therefore, it’s potentially worth considering where the problem lies: with the models used; the protocols for applying the models in preclinical efficacy studies; the underlying therapeutic hypothesis; the conduct of the clinical experiments; elsewhere; or all of the above.
The motivation for using animal models of neurodegenerative disease to predict clinical success is evident and even praiseworthy. Despite longstanding, well-founded, and well-conducted research and development efforts, not a single treatment that modifies the overall natural history for sporadic neurodegenerative disease has proven its value in registration-sized clinical trials. Stubborn searching for efficacious treatments against this bleak background should be saluted and supported. Because neurodegenerative syndromes such as dementia (including Alzheimer’s disease [AD] and frontotemporal dementia [FTD]) unfold over decades, trial design is extremely challenging. This difficulty is multiplied by the absence of registration end points other than clinical rating scales, which are difficult to implement, imprecise, and noisy, raising the specter of both type I and II errors.
Discovery and characterization of Mendelian forms of dementia such as familial AD (FAD) caused by dominantly acting mutations in amyloid precursor protein (APP) or presenilin 1 (PSEN1) genes transformed the understanding of AD pathogenesis. Genetically engineered mice that express FAD-associated mutant forms of both APP and PSEN1 (here collectively termed APP/PS1 mice) have been reported by several groups and yield invaluable insights into the mechanisms and consequences of amyloid deposition in the intact brain. APP/PS1 animals overproduce amyloidogenic peptides (Aβ peptides) derived from APP and demonstrate excess Aβ oligomers in brain interstitial fluid and parenchymal amyloid plaques, as well as cognitive deficits, all in an age-dependent manner. It’s tantalizing to consider that these mice represent a model of the clinical disease AD. Models that use FAD transgenes or knockin genes to cause central nervous system (CNS) amyloid deposition are often referenced using the shorthand term “AD mice.”
Unfortunately, AD animal models, to date, lack the ability to forecast success in the clinic. One example is provided by Tg2576, a model strain based on a single FAD transgene, and therefore relatively uncomplicated with regard to background strain. Further, in Tg2576, abundant data confirm the relationship between amyloid pathology and impaired performance on cognitive tests. In preclinical experiments, Tg2576 mice have been improved or cured no less than 300 times (Zahs and Ashe, 2010). Yet none of these remedies has transitioned through clinical experiments to approval and benefit for patients.
How large is the problem?
Mind-numbing facts are used to communicate the magnitude of the societal challenge represented by age-related dementia: ~135 million people are estimated to be living with dementia by 2050 (by which time a baby born in 2019 will be only 31 yr old). Annual cost of care for one person with dementia is estimated to be $30,000 (2018), with projected worldwide costs of care totaling slightly more than $4 trillion per year.
Biomedical research to address this and other medical conditions takes place at substantial cost as well. It’s estimated that development of a drug product to modify the natural history of AD will take, on average, 13 yr and cost more than $5.5 billion (in 2018 US dollars; Cummings et al., 2018a). As of early 2018, https://clinicaltrials.gov/ listed 112 intervention studies for AD, of which 71 (63%) represented attempts to modify the natural history of disease. Even moderate success would constitute a monumental achievement, as the introduction in 2025 of an agent that delays AD onset by 5 yr would halve disease prevalence by 2050 (Cummings et al., 2018a,b).
Why don’t AD animal model studies translate to the clinic?
Not an easy problem
Seeking to answer this question, one confronts an embarrassment of riches. First, the diseases themselves are unimaginably complex. Confining oneself to AD, the diagnosis is made with complete certainty only at the time of autopsy, with the finding of extracellular amyloid deposits and intracellular aggregates of hyperphosphorylated tau protein (encoded by the microtubule-associated protein tau gene, MAPT) in a person who demonstrated a clinical syndrome of dementia compatible with AD. Each of these phenomena (amyloid plaques, neurofibrillary tau tangles, and cognitive and behavioral manifestations of AD) comprises a substantial discipline in itself. Amyloid deposits consist of much more than β sheets composed of Aβ peptides, with a substantial fraction of cases showing deposition (for example) of α-synuclein, the protein which aggregates into Lewy bodies in the substantia nigra of individuals with Parkinson’s disease. Turning to the tau tangles, there are at least 80 phosphorylation sites in the protein, which are subjected to dynamic posttranslational modification by multiple kinases and phosphatases. It’s clearly a formidable problem to decipher which of these events, and in what order, leads to the sequential loss of tau’s physiological function and its toxic aggregation into tangles. Mathematical models have recently been deployed to address the problem (Stepanov et al., 2018).
Large-scale, intensive genetic studies over decades have uncovered alleles associated with risk for sporadic AD, the most consequential of which is APOE ε4, by virtue of its effect size and allele frequency (Karch and Goate, 2015). As a broad and simplifying statement (thereby leaving out much of importance), these risk genomic variants mainly map to genes associated with increased amyloid pathology and altered microglial reaction. The genetic architecture of both dominant and sporadic AD points to amyloid as the essential initiating factor in disease pathogenesis. Yet, frustratingly, the relationship between amyloid deposition and cognitive loss is quite imperfect, and tau pathology along with synapse loss appear to associate much more closely with clinical impairment (Nelson et al., 2012). Only one well-established relationship between a risk variant for sporadic AD, that for bridging integrator 1 (BIN1) and tau pathology, unambiguously violates the appropriate emphasis on amyloid for using genetics to understand AD pathogenesis. Notably, however, although genetic variants in MAPT account for a variety of neurodegenerative conditions collectively termed tauopathies and which includes FTD, there are only very sparse genetic links between MAPT and AD. Finally, the relentless neuropathological progression of tau pathology in AD brain may be mediated in part by a prionoid intercellular spreading process (Brettschneider et al., 2015), which raises the degree-of-difficulty for animal modeling considerably. Summarizing this paragraph, one can surely place part of the blame for ineffective translation from animals to humans on the incontestable complexity of neurodegeneration as exemplified by AD.
Lessons from failed clinical trials
Drug trials to treat AD have been notoriously difficult, with only one approved drug (not disease modifying) between 2002 and 2014 (Cummings et al., 2018b). Negative outcome in a clinical experiment, in which there is no difference between active drug and placebo for the primary outcome measure, arises axiomatically either because the “drug fails the trial” (for reasons including therapeutic hypothesis being wrong, excess toxicity, or insufficient tolerability) or the “trial fails the drug” (for reasons including inappropriate patient selection or stratification or because the dose used doesn’t cover the target over the treatment interval). Negative results can also emerge due to bad luck; for example, if there is considerably less than expected worsening among the subjects receiving placebo. Of many lessons recovered from the post-failure debris of the last 15 years, several are particularly applicable to AD. First, clinical diagnosis is insufficient to ensure that amyloid pathology underlies a patient’s dementia, and biomarker assurance of diagnosis should be performed unless there are overwhelming and persuasive reasons to defer that step (Cummings et al., 2018b). As one example, several early studies of amyloid-removal agents proved to be significantly underpowered when it was appreciated that as many as 30% of trial subjects lacked cerebral amyloidosis. A second hard-won lesson points to a virtually absolute requirement for CNS pharmacodynamic biomarkers, which indicate that the clinical trial dose of the drug engaged the CNS target and induced a predictable biological effect. Another lesson is well known from first principles but difficult to follow in practice: phase 3 experiments based on unplanned post-hoc subgroup analysis from phase 2 are unlikely to be successful (Cummings et al., 2018b).
Problems with design of preclinical experiments
Just as clinical researchers have made mis-steps in conduct of experimental treatment trials in AD, preclinical scientists using animal models have stumbled, pari passu. The assessment of potential AD treatments requires numerous far-from-obvious safeguards against avoidable error. An investigation into animal studies of neurological disease, in which more than 4,400 datasets subjected to 160 meta-analyses (one for each candidate treatment) ranging from acute (stroke; intracerebral hemorrhage) to chronic (AD) and phasic (experimental autoimmune encephalomyelitis) conditions, concluded, in part, that “there are too many animal studies with statistically significant results in the literature of neurological disorders” (Tsilidis et al., 2013). In particular, there were twice as many significant positive results (nearly 50%) as expected (less than 25%) from evaluation of the most precise and objective study within each meta-analysis. There were no differences among neurological conditions being modeled. Common problems included that studies were too small and that blinding was not conducted (or not mentioned and therefore unlikely to have been a high priority). However, the success rate was excessive regardless of attention to the present checklist of best practices (randomization, blinding, and regard for animal welfare). The data strongly suggested that reporting bias (i.e., the tendency to publish only positive results, as these alone are deemed interesting) underlay the excess of success. Conversely, the successes within studies of adequate size (≥500 animals) with other quality indicators present represented a minuscule portion of all experiments (~5%; Tsilidis et al., 2013). The solution for this torrent of false-positive preclinical interventional studies of neurological disease isn’t obvious, but the authors referred the reader to “caveat emptor” assistance in the form of a database termed Collaborative Approach to Meta-Analysis and Review of Animal Data in Experimental Studies (CAMARADES), which is publicly available (http://www.dcn.ed.ac.uk/camarades/). Other concerns with preclinical AD studies are also inherent in present practice: although age is the most consequential risk factor for disease, many experiments are conducted in young mice with aggressive amyloid deposition phenotypes. Additionally, although sex is a readily recorded demographic trait and affects the susceptibility to and course of AD (Deming et al., 2018), most animal studies examine only a single sex.
The models themselves do not accurately represent disease
Once the difficulty of the problem and the concerns related to preclinical and clinical experimental protocols are considered, there remain incontrovertible limitations to the available animal models for neurodegeneration and AD in particular. The models readily available for AD preclinical research, based on FAD transgenes, overproduce amyloidogenic peptides and exhibit amyloid plaques in cortex as well as hippocampus. However, this pathway to CNS amyloidosis doesn’t model the pathogenic process ongoing in sporadic AD, where the accumulation of amyloid pathology is associated to impaired clearance, not excess production (Mawuenyega et al., 2010). This distinction between the “FAD mice” and the typical human disease also extends to the genetics of sporadic AD. In particular, the APOE ε4 risk allele, associated in cognitively normal subjects with increased amyloid pathology as monitored by imaging and fluid biomarkers, also caused decreased Aβ peptide clearance in mice carrying two FAD transgenes and overproducing amyloid (Castellano et al., 2011).
The next most salient concern regards tau pathology. Mice that deposit amyloid can exhibit aggregated hyperphosphorylated tau, but intracellular neurofibrillary tangles and neuronal cell death have not been reported. To overcome this disconnect between mouse model and human disease, scientists have made ingenious, well-considered, and valiant attempts. Mice carrying transgenes that encode familial FTD have been used to mimic AD tauopathy, but the lack of neuropathological and phenotypic similarity has been evident (Sasaguri et al., 2017). Breeding FTD with FAD mice yields an aggressive phenotype with both plaques and tangles and diverges from sporadic AD both in the mechanism of the amyloidosis and in the character of the tauopathy (Sasaguri et al., 2017). Over the 25 years of experimentation using FAD genes to model AD, numerous improvements in sophistication have been made (Sasaguri et al., 2017). Mice expressing wild-type human MAPT gene in the absence of mouse tau show AD-like tau pathology and may be considered to model this aspect of disease, albeit with mild behavioral phenotype and lack of neuron loss (Andorfer et al., 2003). It has also been demonstrated that material selectively extracted from AD brain and purified appropriately can “seed” intraneuronal tau inclusions in a fashion that mimics AD disease progression through functionally and anatomically connected brain regions (Guo et al., 2016). Despite this progress, it hasn’t been feasible to date to capture the neuropathological and behavioral features of AD in a preclinical model.
What then can we do?
Despite these intimidating hurdles, it’s not an option to declare defeat in our efforts to address age-related dementia because the human burden is too large to ignore. The principle problem facing drug development for neurodegeneration is how to achieve an adequate probability of success to justify the extremely large investments in financial and human resources involved in that effort. If the present animal models are not able to predict clinical success, that fact must be accepted and should change practice. It is well established that drugging genetically validated targets approximately doubles the probability of success (Nelson et al., 2015). In that regard, there has been no paucity of work to identify gene variants that modify AD risk, and some of them introduce consequential amino acid substitutions or affect the abundance of alternatively spliced variants, thereby changing protein function. In some cases, noncoding single nucleotide polymorphisms have been firmly linked to nearby genes by the detection of rare coding-region variants which also affect AD risk. The various genes implicated by these means have been integrated into systems biology hypothesis–generating exercises. Other extensions of genetic architecture have included clarifying through human brain transcriptomics which genes harboring risk alleles are expressed in individual CNS cell types such as microglia (Gosselin et al., 2017).
Current and prospective animal models remain extraordinarily valuable for investigating biological processes that occur in vivo in the context of amyloid or tau pathology and will unequivocally suggest potential drug targets. In the drug development process, these models can also serve as the preferred testing platform for prospective pharmacodynamic biomarkers. In the war against age-related dementia, we’re in “all hands on deck” status, and animal models of neurodegeneration are among our most valuable allies when they are asked to carry out tasks of which they are capable.
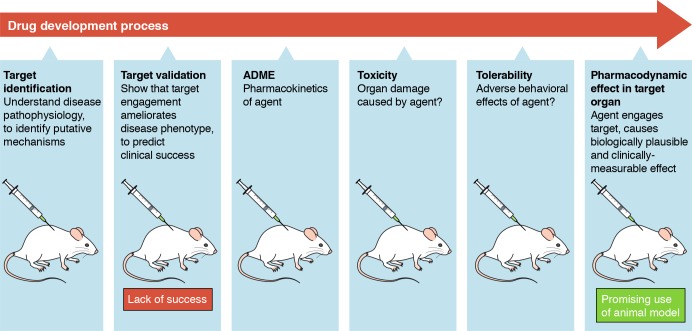
Use of animal models across the process of drug development. The cartoon indicates selected stages of drug development for which animal models may be applied. ADME: Absorption, Distribution, Metabolism, and Excretion.
References
- Andorfer C., et al. . 2003. J. Neurochem. 86:582–590. 10.1046/j.1471-4159.2003.01879.x [DOI] [PubMed] [Google Scholar]
- Box G.E.P. 1979. Robustness in the strategy of scientific model building. In Robustness in Statistics. 10.1016/B978-0-12-438150-6.50018-2 [DOI] [Google Scholar]
- Brettschneider J., et al. . 2015. Nat. Rev. Neurosci. 16:109–120. 10.1038/nrn3887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano J.M., et al. . 2011. Sci. Transl. Med. 3:89ra57 10.1126/scitranslmed.3002156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings J., et al. . 2018a Alzheimers Dement. (N. Y.). 4:195–214. 10.1016/j.trci.2018.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings J., et al. . 2018b J. Alzheimers Dis. 64(s1):S3–S22. 10.3233/JAD-179901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deming Y., et al. Acta Neuropathol. 2018 doi: 10.1007/s00401-018-1881-4. [DOI] [Google Scholar]
- Gosselin D., et al. . 2017. Science. 356:eaal3222 10.1126/science.aal3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J.L., et al. . 2016. J. Exp. Med. 213:2635–2654. 10.1084/jem.20160833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch C.M., and Goate A.M.. 2015. Biol. Psychiatry. 77:43–51. 10.1016/j.biopsych.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mawuenyega K.G., et al. . 2010. Science. 330:1774 10.1126/science.1197623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson M.R., et al. . 2015. Nat. Genet. 47:856–860. 10.1038/ng.3314 [DOI] [PubMed] [Google Scholar]
- Nelson P.T., et al. . 2012. J. Neuropathol. Exp. Neurol. 71:362–381. 10.1097/NEN.0b013e31825018f7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaguri H., et al. . 2017. EMBO J. 36:2473–2487. 10.15252/embj.201797397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanov A., et al. . 2018. PLoS One. 13:e0192519 10.1371/journal.pone.0192519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsilidis K.K., et al. . 2013. PLoS Biol. 11:e1001609 10.1371/journal.pbio.1001609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahs K.R., and Ashe K.H.. 2010. Trends Neurosci. 33:381–389. 10.1016/j.tins.2010.05.004 [DOI] [PubMed] [Google Scholar]