Abstract
Genetics plays a role, to a greater or lesser extent, in all diseases. Variations in our DNA and differences in how that DNA functions (alone or in combinations), alongside the environment (which encompasses lifestyle), contribute to disease processes. This review explores the genetic basis of human disease, including single gene disorders, chromosomal imbalances, epigenetics, cancer and complex disorders, and considers how our understanding and technological advances can be applied to provision of appropriate diagnosis, management and therapy for patients.
Keywords: cancer, genetics, genomics, molecular basis of health and disease
Introduction
When most people consider the genetic basis of disease, they might think about the rare, single gene disorders, such as cystic fibrosis (CF), phenylketonuria or haemophilia, or perhaps even cancers with a clear heritable component (for example, inherited predisposition to breast cancer). However, although genetic disorders are individually rare, they account for approximately 80% of rare disorders, of which there are several thousand. The sheer number of rare disorders means that, collectively, approximately 1 in 17 individuals are affected by them. Moreover, our genetic constitution plays a role, to a greater or lesser extent, in all disease processes, including common disorders, as a consequence of the multitude of differences in our DNA. Some of these differences, alone or in combinations, might render an individual more susceptible to one disorder (for example, a type of cancer), but could render the same individual less susceptible to develop an unrelated disorder (for example, diabetes). The environment (including lifestyle) plays a significant role in many conditions (for example, diet and exercise in relation to diabetes), but our cellular and bodily responses to the environment may differ according to our DNA. The genetics of the immune system, with enormous variation across the population, determines our response to infection by pathogens. Furthermore, most cancers result from an accumulation of genetic changes that occur through the lifetime of an individual, which may be influenced by environmental factors. Clearly, understanding genetics and the genome as a whole and its variation in the human population, are integral to understanding disease processes and this understanding provides the foundation for curative therapies, beneficial treatments and preventative measures.
With so many genetic disorders, it is impossible to include more than a few examples within this review, to illustrate the principles. For further information on specific conditions, there are a number of searchable internet resources that provide a wealth of reliable detail. These include Genetics Home Reference (https://ghr.nlm.nih.gov/), Gene Reviews (https://www.ncbi.nlm.nih.gov/books/NBK1116/), the ‘Education’ section from the National Human Genome Research Institute (https://www.genome.gov/education/) and Online Mendelian Inheritance in Man (https://www.omim.org/). In this review, an understanding and knowledge of basic principles and techniques in molecular biology, such as the structure of DNA and the PCR will be assumed, but explanations and animations of PCR (and some other processes) are available from the DNA Learning Center (https://www.dnalc.org/resources/). The focus here will be on human disease, although much of the research that defines our understanding comes from the study of animal models that share similar or related genes.
The human genome and variation
The human genome and the human genome reference sequence
The complete instructions for generating a human are encoded in the DNA present in our cells: the human genome, comprising roughly 3 billion bp of DNA. Scientists from across the world collaborated in the ‘Human Genome Project’ to generate the first DNA sequence of the entire human genome (published in 2001), with many additions and corrections made in the following years. Genome sequence information for humans and many other species is freely accessible through a number of portals, including the National Center for Biotechnology Information (NCBI; https://www.ncbi.nlm.nih.gov/) and Ensembl (http://www.ensembl.org/), which also provide a wealth of related information.
The majority of our DNA is present within the nucleus as chromosomes (the nuclear DNA or nuclear genome), but there is also a small amount of DNA in the mitochondria (the mtDNA or mitochondrial genome). Most individuals possess 23 pairs of chromosomes (Figure 2), therefore much of the DNA content is present in two copies, one from our mother and one from our father.
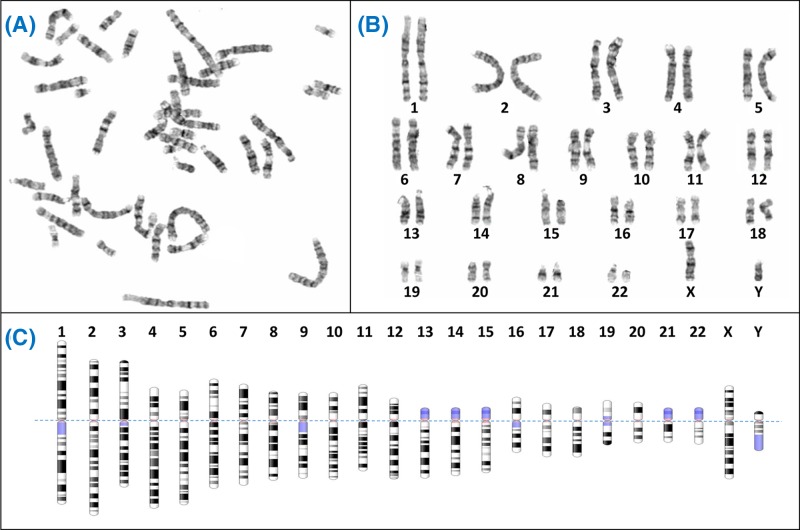
Figure 2. Giemsa banding (G-banding) to form a karyogram.
(A) Metaphase spreads like this are obtained from cultured cells arrested in metaphase using colcemid, followed by Giemsa staining to create characteristic light and dark bands. Generally the dark bands represent regions which are AT-rich and gene-poor. (B) The chromosomes from the spread are arranged in pairs to view the karyotype, often using specialist software like Cytovision. (C) Diagrammatic representations of the G-banding patterns, called ideograms, are used as a reference. The ideograms have been aligned at the centromere (dotted line); blue shaded regions are highly variable – note for example the variation between p arms of chromosome 13, 14 and 15 in (B). In fact the p arms of the acrocentric chromosomes (13, 14, 15, 21, 22) all have very similar content, which includes the nucleolar organiser regions or NORs. Each NOR contains a tandem repeat of ribosomal DNA (rDNA) which encodes the rRNAs. Between all five acrocentrics there are approximately 300–400 rDNA repeats, though the actual number varies between individuals. Chromosome ideograms from NCBI Genome Decoration Page.
The human nuclear genome encodes roughly 20000 protein-coding genes, which typically consists of both protein-coding (exon) and non-coding (intron) sequences. Our genome also contains roughly 22000 genes that encode RNA molecules only; some of these RNAs form components of the translation machinery (rRNA, tRNA) but there are many more that perform various roles within the cell, including regulation of expression of other genes. In fact it is now believed that as much as 80% of our genome has biological activity that may influence structure and function. The human genome also contains over 14000 ‘pseudogenes’; these are imperfect copies of protein-coding genes that have lost the ability to code for protein. Although originally considered as evolutionary relics, there is now evidence that some may be involved in regulating their protein-coding relatives, and in fact dysregulation of pseudogene-encoded transcripts has been reported in cancer. Additionally, sequence similarity between a pseudogene and its normal counterpart may promote recombination events which inactivate the normal copy, as seen in some cases of perinatal lethal Gaucher disease. Furthermore, some pseudogenes have the potential to be harnessed in gene therapy to generate functional genes by gene editing approaches. The distribution of genes between chromosomes is not equal: chromosome 19 is particularly gene-dense, while the autosomes for which trisomy is viable (13, 18, 21) are relatively gene-poor (Table 1).
Table 1. DNA and gene content of human chromosomes.
| Chromosome | Approximate length (bp) | Protein-coding genes | Non-protein coding genes | Pseudogenes |
|---|---|---|---|---|
| 1 | 248956422 | 2047 | 1964 | 1233 |
| 2 | 242193529 | 1303 | 1605 | 1033 |
| 3 | 198295559 | 1075 | 1160 | 768 |
| 4 | 190214555 | 753 | 984 | 732 |
| 5 | 181538259 | 881 | 1200 | 710 |
| 6 | 170805979 | 1041 | 989 | 803 |
| 7 | 159345973 | 989 | 977 | 893 |
| 8 | 145138636 | 670 | 1041 | 629 |
| 9 | 138394717 | 778 | 786 | 678 |
| 10 | 133797422 | 728 | 880 | 568 |
| 11 | 135086622 | 1312 | 1053 | 815 |
| 12 | 133275309 | 1036 | 1197 | 627 |
| 13 | 114364328 | 321 | 586 | 378 |
| 14 | 107043718 | 820 | 857 | 519 |
| 15 | 101991189 | 613 | 986 | 513 |
| 16 | 90338345 | 867 | 1033 | 467 |
| 17 | 83257441 | 1185 | 1198 | 531 |
| 18 | 80373285 | 269 | 608 | 246 |
| 19 | 58617616 | 1474 | 895 | 514 |
| 20 | 64444167 | 543 | 594 | 250 |
| 21 | 46709983 | 231 | 403 | 183 |
| 22 | 50818468 | 492 | 513 | 332 |
| X | 156040895 | 843 | 640 | 872 |
| Y | 57227415 | 63 | 108 | 392 |
| Mitochondrial | 16569 | 13 | 24 |
Note that although these numbers seem very precise they should be taken as indicative only, since (i) chromosomes of each individual will vary from the reference sequence, and (ii) the human reference genome sequence is continuously updated with corrections (the data here are from GRCh38.p12, which represents a particular ‘build’ of the human genome). Note that the data for the acrocentric chromosomes 13, 14, 15, 21, 22 does not include the shared ribosomal DNA array repeats present on the p arms (see Figure 2). Data from Ensembl, June 2018.
From the very beginning of the Human Genome Project, it was recognised that there was a huge amount of DNA sequence variation between healthy individuals, and therefore there is no such thing as a ‘normal’ human DNA sequence. However, if we are to describe changes to the DNA sequence, we need to describe these changes with respect to some baseline; this baseline is the human reference genome sequence.
Variation versus mutation
A geneticist’s definition of mutation is ‘any heritable change to the DNA sequence’, where heritable refers to both somatic cell division (the proliferation of cells in tissues) and germline inheritance (from parent to child). Such changes to the DNA may have no consequences but sometimes lead to observable differences in the individual (the ‘phenotype’). Consequently, in the past such alterations in the human population, particularly when they were associated with a disease state, were referred to as ‘mutations’. However, for many people this terminology has negative connotations, and brings to mind the ‘mutants’ seen in science fiction and zombie films! Therefore modern practice, particularly for medical genetics within the context of a health service, is to refer to differences from the reference sequence as ‘variants’. Variants may be further classified as benign (not associated with disease) or pathogenic (associated with disease), although there are increasing numbers of human DNA variants identified for which we are still not sure of the effect; these are termed ‘variants of uncertain significance’ or VUS (Table 2).
Table 2. International Agency for Research on Cancer variant classification.
| Variant class | Description | Surveillance recommendations | Predictive testing |
|---|---|---|---|
| 5 | Definitely pathogenic | Full high risk surveillance according to current guidelines | Genetic testing offered to at-risk family members |
| 4 | Likely pathogenic | Full high risk surveillance according to current guidelines | Genetic testing offered to at-risk family members |
| 3 | Uncertain | Surveillance based on family history and other known risk factors | No genetic testing offered |
| 2 | Likely not pathogenic | Treat as if ‘no mutation’ was detected | No genetic testing offered |
| 1 | Not pathogenic | Treat as if ‘no mutation’ was detected | No genetic testing offered |
Although this system was designed for classification of variants in relation to a potential role in cancer predisposition, it can also be used to classify variants in other situations.
Where two (or more) different versions of a DNA sequence exist in the population, these are referred to as ‘alleles’: each allele represents one particular version (or variant) of that sequence. By analysing many human genomes we can calculate the frequency at which a particular variant occurs in the population, often expressed as the ‘minor allele frequency’ or MAF. Where the MAF is at least 1%, a variant can be called a ‘polymorphism’, although this is a fairly arbitrary cut-off.
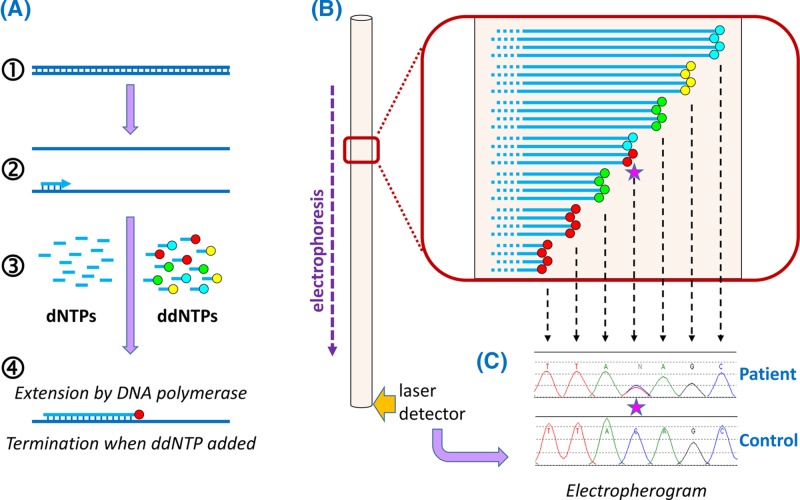
Single nucleotide variants: The most frequent variants in our genome are substitutions that affect only one base pair (bp), referred to as single nucleotide variants (SNV) or as single nucleotide polymorphisms (SNP) (Figure 1) depending upon the MAF. It has been estimated that there are at least 11 million SNPs in the human genome (averaging approximately 1 per 300 bp). It also seems likely that if we sequenced the genomes of everyone on the planet, for most positions in our genome we would discover at least one individual with an SNV, wherever such variation is compatible with life.
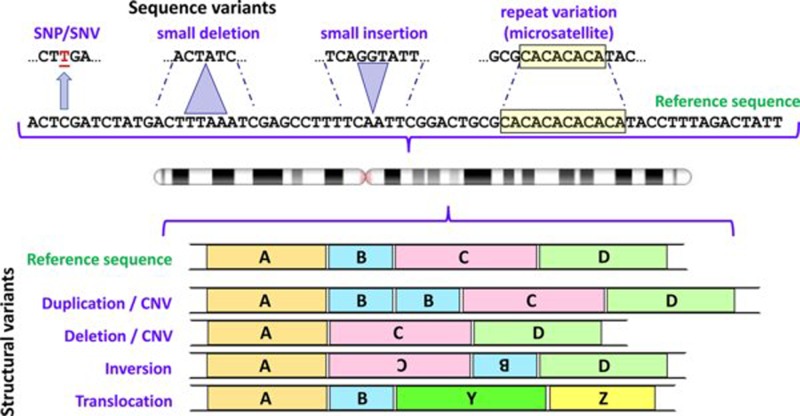
Figure 1. Some types of variants found in human genomes.
Variation involving one or a few nucleotides are shown above the chromosome icon, and structural variants below; in each case the variants are depicted in relation to the reference sequence. For depiction of structural variants A, B, C and D represent large segments of DNA; Y and Z represent segments of DNA from a different chromosome. Note that differentiation between CNVs and deletions/insertions depends upon the size of the relevant DNA segment (see text for further details). Abbreviation: CNV, copy number variant. Chromosome ideogram from NCBI Genome Decoration Page.
Insertions and deletions (indels): Insertions or deletions of less than 1000 bp are also relatively common in the human genome, with the smallest indels being the most numerous.
Structural variants: Structural variants are defined as variants affecting segments of DNA greater than 1000 bp (1 kb). They include translocations, inversions, large deletions and copy number variants (CNV). CNVs are segments of our genome that range in size from 1000 to millions of bp, and which, in healthy individuals, may vary in copy number from zero to several copies (Figure 1). By analysis of many human genomes it is apparent that CNV exists for approximately 12% of the human genome sequence. The largest CNVs may contain several entire genes. Where the population frequency of a CNV reaches 1% or more, it may be referred to as a copy number polymorphism (CNP).
Repeat variations: Human genomes contain large numbers of repetitive sequences. These include ‘interspersed repeats’ which constitute approximately 45% of our genome, and represent remnants of mobile DNA elements (transposons). There are also several classes of ‘tandem repeats’, in which the repeated units are side-by-side in a head-to-tail fashion forming arrays of repeats of the same (or very similar) sequence. The number of repeats in each array can vary, generating multiple alleles, so that these loci have high variability within the population, and can be used in identifying individuals (see below). Tandem repeats include minisatellites and microsatellites (Figure 1/Table 3). Although generally inherited stably (i.e. with the same number of repeats) from parent to child, expansions in some microsatellites are associated with disease.
Table 3. Comparison of minisatellites and microsatellites.
| Minisatellites | Microsatellites | |
|---|---|---|
| Number within the human genome | Approximately 1500 | Approximately 500000 |
| Locations within our genome | Mostly near the ends of chromosomes (telomeres) | Scattered throughout the length of all chromosomes |
| Unit repeat length1 | Approximately 10 to >100 bp | (12) 2 to approximately 6 bp |
| Number of repeat units within the array | Usually from approximately 60 to >1000 | Usually ∼6 to ∼14 |
| Used in | DNA fingerprinting | DNA profiling; genetic linkage studies |
| Also known as | Variable number tandem repeats (VNTR) | VNTR, short tandem repeats (STR), simple sequence repeats (SSR) |
Note that repeats with unit lengths of 7–9 bp may be classified as micro- or minisatellites depending on their biological behaviour.
Many (but not all) authors include mononucleotide repeats in the category of microsatellites.
Variation between healthy individuals
Given that no two individuals look exactly alike (apart from identical twins) it will come as no surprise that this is reflected in our DNA. What is surprising is the amount of variation between us. Looking at any one human genome, compared with the reference sequence, we would find approximately 3 million SNPs, and approximately 2000 structural variants. The genomes of any two unrelated individuals will differ in approximately 0.5% of their DNA (approximately 15 million bp), and most of this variation can be attributed to CNVs and large deletions. Although much of the variation in our genome lies within the non-coding DNA, we now know that, on average, each individual has several hundred variants that are either known, or predicted, to be damaging to gene function, including roughly 85 variants that lead to truncated (incomplete) protein products. Furthermore, the total number of functional genes per human genome may vary by up to 10% between individuals as a consequence of CNVs, large deletions and loss-of-function variants. Faced with this enormous level of variation you might wonder, not why some individuals are affected by disease due to inherited ‘mutations’, but rather how any of us manage to remain relatively healthy! Clearly there is no requirement for all of our genes to be functional: for many genes only one working copy is required, and in other cases there appears to be a level of redundancy or plasticity built into the system. However it is becoming increasingly apparent that some of the variations in our genomes may lead to higher susceptibility to common diseases.
Variation between populations
The greatest amount of variation is found within populations of African ancestry, which is consistent with initial migration out of Africa, with each group of migrants taking subsets of variants with them. Common variants tend to be shared between all populations, whereas rare variants are more likely to be specific to particular populations or related populations. Some of the differences will be related to environmental adaptation, for example skin pigmentation or enzymes to detoxify dietary plant toxins. These same enzymes are also responsible for the metabolism of many pharmaceutical (and recreational) drugs; genetic variants may lead to some individuals being ultrarapid metabolisers or poor metabolisers, which may translate into poor drug response or adverse side effects. For example, deficiency in dihydropyrimidine dehydrogenase, leading to a toxic response to the cancer treatment 5-fluorouracil, is two to three times more common in African-American populations than in Caucasians.
DNA profiling
In the early 1980s, with the discovery of minisatellites, which are highly variable within the population but inherited stably from parent to child, it became possible to use these in forensic analyses and paternity testing, to generate unique patterns (similar to supermarket barcodes) for each individual, a technique referred to as ‘DNA fingerprinting’. This technology needed large amounts of sample (micrograms of DNA) and tended to be time-consuming (1–2 weeks) in addition to requiring use of radioactive labels. Towards the end of the 1980s, microsatellites were first reported and since these could be analysed with simple and rapid PCR-based assays, needing only approximately 1 nanogram of sample DNA, ‘DNA profiling’ using microsatellites quickly replaced the earlier DNA fingerprinting approach. Forensic DNA profiling in the U.K. currently analyses 16 microsatellites from across the genome, together with a region from the amelogenin gene present on both X and Y chromosomes that is 4 bp different in size between them, allowing gender identification. The process is similar to QF-PCR for prenatal aneuploidy testing, which will be discussed later. Finding a perfect match between the two samples (e.g. from crime scene and suspect) strongly suggests that these came from the same individual – the likelihood of finding a perfect match between samples from two different individuals is estimated at 1 in a billion – unless of course they are identical twins. On the other hand, if the two samples do not match, it can be concluded that the crime scene sample was not from the suspect. Likewise, in paternity testing, DNA profiling can exclude a man as the father of a child, but cannot prove he is the father with absolute certainty. DNA profiling is also useful in helping to identify human remains, for example where decomposition makes physical identification difficult. The fact that certain variants (including microsatellite alleles) are more frequently found in populations of particular ancestry means that the capability already exists to make some inferences on likely ancestral origin based on only a DNA sample and research is underway to establish whether particular features (for example, eye colour, hair colour and even facial characteristics) can be predicted from DNA. Thus the DNA profiling of the future may generate an identikit image of a wanted individual.
De novo mutations and mosaicism
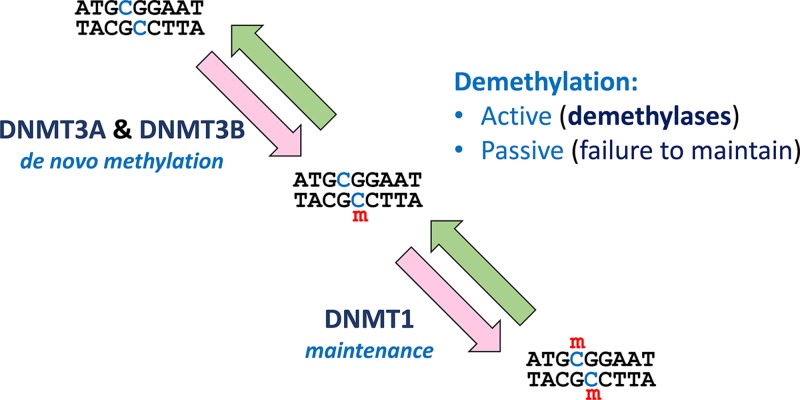
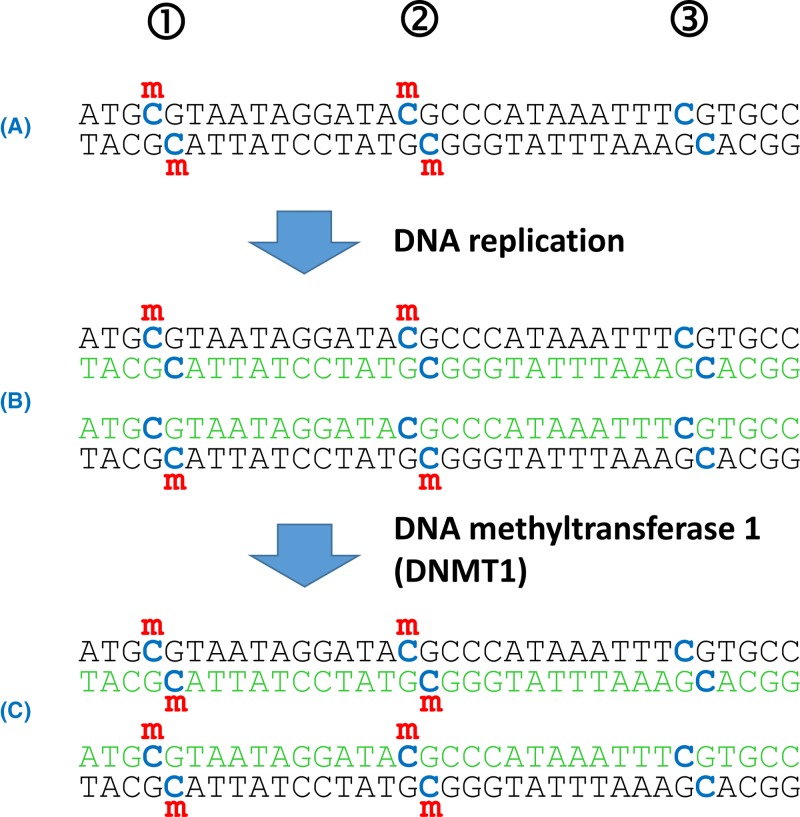
Most of the variants in our genome were inherited from one of our parents. However, our DNA is constantly bombarded with DNA damaging agents and furthermore every time a cell’s DNA is replicated prior to division there is opportunity for errors. Genomic sequencing of trios (child plus both parents) has demonstrated that on average each individual has 74 de novo SNVs that were not present in either parent, in addition to approximately three de novo insertions/deletions. Approximately 1–2% of children will have a de novo CNV greater than 100 kb in size. Microsatellites have a relatively high mutation frequency, with gain or loss of a repeat unit occurring in roughly 1 per 1000 microsatellites per gamete per generation. In contrast with aneuploidy, which is most often a consequence of meiotic error during oocyte generation, new mutations are almost four times more common in the male germline than the female germline, which is likely to relate to the high number of cell divisions during spermatogenesis. For both sexes the new mutation rate increases with age, though again, the increase is more marked in the male germline. Most new mutations will have little or no effect on health, particularly those outside coding sequences, but some are associated with disease.
If a new mutation occurs during embryogenesis or development this can lead to mosaicism, where some cells in the individual have that new variant while others do not. Mosaicism for a new mutation may also be present in the gonads (‘gonadal mosaicism’), such that a new variant may be transmitted to less than 50% of the offspring, depending upon the percentage of gonadal cells in which the new variant is present. New mutations occurring during embryogenesis and development also generate a few differences between the genomes of identical twins.
Very rarely fusion of two embryos will generate a chimera: an individual that has two genetically distinct cell lines present. Where the same sex chromosome constitution is present in both cell lines chimerism might only come to light with the observation of apparent non-maternity or non-paternity amongst offspring (where one cell line predominates in the gonads and the other predominates in blood cells). Fusion of two embryos of different sex can lead to characteristics of both genders being present, and chimerism is found in approximately 13% of cases of hermaphroditism.
Summary
The massive amount of variation between individual human genomes can make it very difficult to determine which variants are benign and which might be associated with a disease. Even where a disease-associated variant is present, this will be present within a genomic context of millions of other differences from the ‘reference’ sequence, some of which may impact upon the severity of that disease in the individual. Thus it will become increasingly common to investigate wider genomic influences when considering contribution of variants to disease. Note that several scientific conventions are used when referring to chromosomes, genes, proteins and variants affecting them; these ensure unambiguous communication between scientists and health professionals. International System for Human Cytogenetic Nomenclature (ISCN) is used for describing karyotypes and changes at the chromosomal level. Individual loci and genes, for which there are often multiple different historical names, have now been assigned specific unique names by the HUGO Gene Nomenclature Committee (HGCN) (https://www.genenames.org/). Sequence variants are described according to Human Genome Variation Society (HGVS) guidelines (http://varnomen.hgvs.org/) for both DNA and proteins. Finally, since the same names are applied to genes and the proteins they encode, italics are used to refer to the gene, with standard font used when referring to the protein.
Chromosome structure and chromosomal disorders
Introduction
Almost every human cell contains a full diploid genome, consisting of 2 metres of DNA arranged into 46 chromosomes: 22 homologous autosomal pairs, and the sex chromosomes comprising two X chromosomes in females and an X and a Y in males. The exceptions are anucleate cells like erythrocytes (red blood cells), cell fragments (platelets) and haploid germline cells (sperm and eggs) which contain 23 chromosomes. Although mechanisms have evolved which ensure that during cell division, daughter cells will inherit a complete genome, those mechanisms occasionally make mistakes. This can lead to cells with chromosomal abnormalities, which can be categorised as numerical abnormalities, i.e. the resulting daughter cell contains too many or too few chromosomes, or structural abnormalities, where more complex rearrangements of the genome have taken place.
The normal chromosome complement of a species (i.e. the number, size and shape of chromosomes) is called its karyotype. According to the ISCN, the ‘normal’ human karyotype is denoted by either 46,XX (female) or 46,XY (male). Human chromosomes consist of DNA which is wrapped around a core of histone proteins to form chromatin. Most of the time, chromatin exists in a diffuse form within a cell’s nucleus, however, during metaphase of the cell division cycle, the chromosomes condense. It is these condensed chromosomes which can be stained with a variety of chemicals, and which can then be observed under a light microscope, to reveal the characteristic banding patterns. The bands reflect regions of chromatin with different characteristics, and therefore different functional elements. A photographic representation of a person’s metaphase chromosomes, arranged by size, may be referred to as a karyogram or karyotype (Figure 2A,B) and a graphical representation is called an ideogram (Figure 2C). The available stains for chromosomes differ in their chemical properties and consequently in the resulting banding pattern. The most commonly used stain is called Giemsa after the chemist who developed it in 1904; the resulting banding pattern of chromosomes is referred to as G-banding. The microscopic analysis of stained chromosomes is termed cytogenetics. Depending on the quality of the chromosome preparation, trained cytogeneticists can identify abnormalities with a resolution of approximately 3–4 Mb (millions of bp), however, abnormalities below this resolution threshold cannot be identified using conventional cytogenetics and require alternative, molecular techniques (see section ‘Genetic testing in the diagnostic laboratory’).
When viewing condensed metaphase chromosomes under a microscope, some key features can be identified (Figure 3). All mammalian chromosomes have a centromere, which appears like a narrow waist, here proteins attach for separation of chromosomes during cell division. In humans, the centromere is located between the two arms of the chromosome, the shorter arm is called the ‘p’ arm (for ‘petite’), while the longer arm is called ‘q’ (‘queue’). Depending on the location of the centromere relative to the two arms, human chromosomes are classified as ‘metacentric’, where the centromere is more or less in the middle of the chromosome, ‘submetacentric’, where the centromere is somewhat offset from the centre or ‘acrocentric’, where the centromere is significantly offset from the centre, with only a very short p arm. In some species such as the mouse, the centromere is located at one end of the chromosome, termed as telocentric. In humans, chromosomes 1, 3, 16, 19 and 20 are metacentric, chromosomes 13, 14, 15, 21, 22 and Y are acrocentric, while the remainder are submetacentric. In eukaryotes, the structures at the ends of each linear chromosome are called telomeres and consist of 300–8000 repeats of the sequence TTAGGG, which forms a loop at the end. One function of telomeres is to protect the ends of chromosomes from being recognised as ‘damaged DNA’ and erroneously repaired by the cell’s DNA repair machinery. They also accommodate the loss of sequences during each round of replication, which occurs as a result of the so-called ‘end replication problem’. In cells without the enzyme telomerase (which extends existing telomeres), a short stretch of sequence is lost from the 5′ end of the newly replicated strand with each cell division, which ultimately can lead to cell senescence.
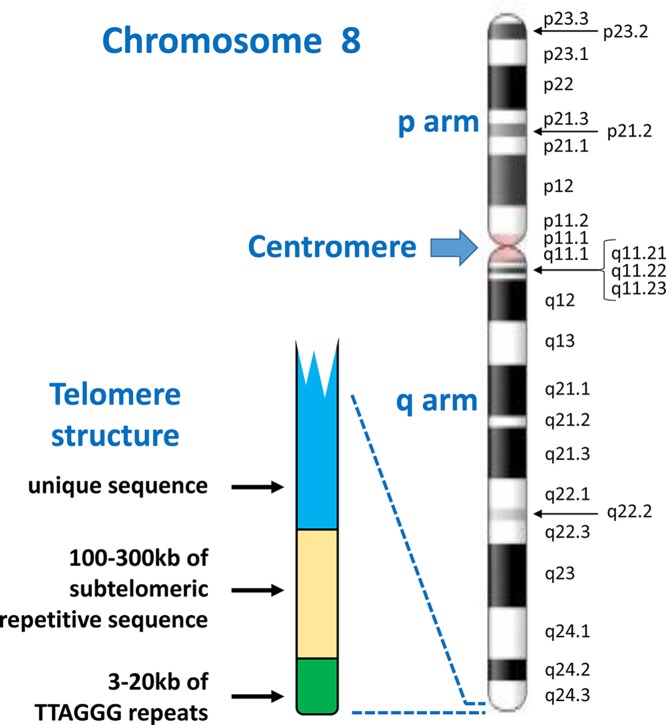
Figure 3. Chromosome structure and band nomenclature.
This ideogram of the complete chromosome 8 illustrates the general structure of all human chromosomes: short (p) and long (q) arms, joined at the centromere. Each chromosome has a characteristic G-banding pattern, with each band annotated, for example p22 or q23. In chromosomes which are less condensed, more bands are seen as separate entities, while bands may merge together in more condensed chromosomes (for example, q21.1, q21.2 and q21.3 appear as a single band [q21] in a more condensed chromosome 8). The approved way of stating the location q21.1 is q-two-one-point-one (not q-twenty-one-point-one). Telomeres, with a shared structure, are present at both ends of each chromosome. Each telomere is composed of arrays of TTAGGG repeats, followed by a subtelomere, which is formed of repetitive sequences which can be similar between several telomeres. Chromosome ideogram from NCBI Genome Decoration Page.
Numerical abnormalities
An abnormality where a cell contains more than two complete sets of the human haploid genome (69 chromosomes or more) is termed as polyploidy. Triploidy (three haploid sets of chromosomes) occurs in 1–3% of pregnancies and usually arises from fertilisation of a single egg with two sperms or sometimes from fertilisation involving a diploid gamete (egg or sperm). Viability of triploid foetuses is usually very low and leads to early spontaneous abortion during pregnancy while tetraploidy (four haploid sets of chromosomes) is even rarer and not compatible with life. However, a situation where the chromosome number is not an exact multiple of the haploid chromosome number is called aneuploidy.
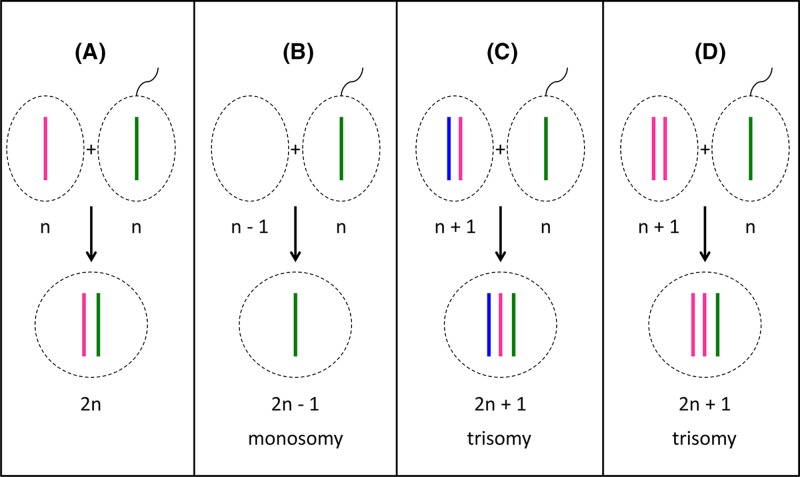
Aneuploidy usually arises because a gamete is formed that contains more or fewer chromosomes than the normal complement. This results from a phenomenon called non-disjunction, where the replicated chromosomes do not separate properly at cell division, and can happen during either meiosis I (non-disjunction of paired chromosomes) or meiosis II (non-disjunction of sister chromatids) (Figure 4). Non-disjunction generates germ cells which either contain an extra copy of one of the chromosomes or lack one chromosome. Fertilisation then leads to the formation of a zygote with an extra chromosome or a missing chromosome respectively (Figure 5). Non-disjunction most commonly occurs during meiosis II of oocyte formation, and is influenced by the mother’s age and other environmental factors. The risk of delivering a trisomic foetus increases from 1.9% in women aged 25–29 years to over 19% in women aged over 39 years. There is also evidence that folic acid deficiency, smoking, obesity and low-dose irradiation with radioactive contaminants increases the risk of non-disjunction.
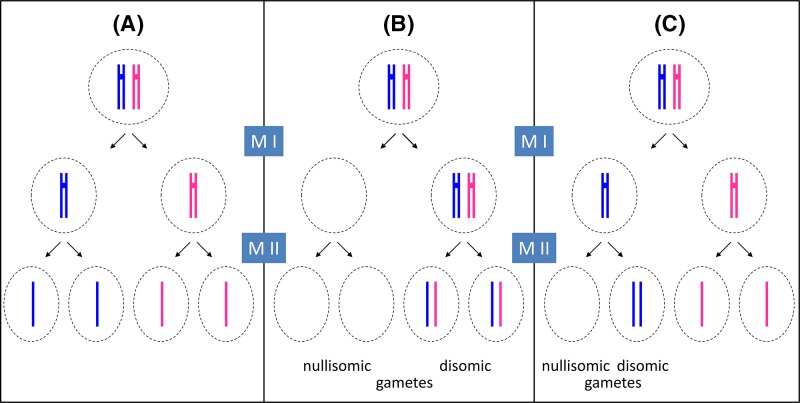
Figure 4. Principles of meiosis and non-disjunction.
For simplicity, only one pair of newly replicated autosomes is shown in two different colours to distinguish the maternal from the paternal chromosome and crossover is not considered. During spermatogenesis, all four meiotic products can form the gametes (sperm), while in oogenesis, only one of the four products will actually become the ovum (egg) as one daughter cell forms a polar body at meiosis I (MI) and another forms a polar body at meiosis II (MII). For clarity, all four potential meiotic products are shown. (A) During normal meiosis, four haploid meiotic products are formed. (B) If non-disjunction occurs during MI, two daughter cells are formed which completely lack this particular chromosome (nullisomic for this chromosome), while two others contain two copies of the chromosome (disomic). (C) If non-disjunction occurs during MII, one nullisomic and one disomic daughter cell is formed, while the remaining two form normally.
Figure 5. Fertilisation outcomes.
(A) Fertilisation of a normal oocyte with a normal sperm cell leads to the formation of a diploid (2n) zygote. (B) If a nullisomic oocyte is fertilised, the resulting zygote will be monosomic for one chromosome. (C,D) Fertilisation of a disomic oocyte results in trisomic zygotes. Note that in (C), the oocyte has resulted from non-disjunction in meiosis I, and the resulting zygote contains one chromosome (ignoring crossover) from each maternal grandparent as well as the paternal contribution. In (D), the oocyte has resulted from non-disjunction in meiosis II, and the resulting zygote contains two chromosomes (aside from crossover regions) from one grandparent.
Examples of syndromes caused by aneuploidy
Most aneuploidies are lethal. However, those that are viable are listed in Table 4, together with approximate incidence rates and common symptoms. Foetuses with trisomy 13 or 18 may survive to term, while individuals with trisomy 21 can survive beyond the age of 40. Presence of an extra autosome generally leads to severe developmental abnormalities, and only trisomies of small, gene-poor chromosomes (Table 1) appear to be tolerated. Autosomal monosomies have even more severe consequences, as they invariably lead to miscarriage during the early stages of pregnancy. The developmental consequences of such trisomies and monosomies are a result of an imbalance of the levels of critical gene products encoded on the affected chromosomes. For example, the major features of Down syndrome (DS) are associated with the presence of three copies of a 1.6-Mb region at chromosome location 21q22.2, called the Down Syndrome Critical Region.
Table 4. Viable aneuploidies.
| Aneuploidy | Common name | Estimated incidence among life-births | Symptoms can include |
|---|---|---|---|
| Trisomy 13 | Patau syndrome | Approximately 1:16000 | Severe intellectual disability, heart defects, brain or spinal cord abnormalities, small or poorly developed eyes, extra fingers or toes, cleft lip and palate, weak muscle tone |
| Trisomy 18 | Edwards syndrome | Approximately 1:5000 | Intrauterine growth retardation, low birth weight, heart defects and abnormalities of other organs, small, abnormally shaped head, small jaw and mouth, clenched fists, severe intellectual disability |
| Trisomy 21 | Down syndrome | Approximately 1:800 | Mild to moderate intellectual disability, characteristic facial appearance, weak muscle tone, heart defects, digestive abnormalities, hypothyroidism, increased risk of hearing and vision problems, leukaemia, Alzheimer’s disease |
| Trisomy X | Triple X syndrome | Approximately 1:1000 | Increased height, increased risk of learning disabilities, delayed development of speech, language and motor skills, weak muscle tone, behavioural and emotional difficulties, seizures, kidney abnormalities |
| 47,XYY | Approximately 1:1000 | Increased height, increased risk of learning disabilities, delayed development of speech, language, and motor skills, weak muscle tone, hand tremors, seizures, asthma, scoliosis, behavioural and emotional difficulties | |
| 47,XXY | Klinefelter syndrome | 1:500 to 1:1000 | Small testes, low testosterone levels, delayed and incomplete puberty, breast enlargement, reduced facial and body hair, infertility, increased height, increased risk of breast cancer, learning disabilities, delayed speech and language development |
| 48,XXXY | Approximately 1:18000 to 1:40000 | Small testes, low testosterone levels, delayed and incomplete puberty, breast enlargement, reduced facial and body hair, infertility, increased height, tremors, dental problems, peripheral vascular disease, deep vein thrombosis, asthma, type 2 diabetes, seizures, heart defects, delayed speech and language development, learning disabilities | |
| 45,X | Turner syndrome | Approximately 1:2500 | Short stature, early loss of ovarian function, infertility, absence of puberty, webbing of the neck, skeletal abnormalities, kidney problems, heart defects |
Common names are given, where available, together with estimated incidence rates and symptoms frequently associated with the condition.
Having an abnormal number of sex chromosomes generally has milder consequences than abnormal numbers of autosomes and is discussed in more detail in the section ‘The sex chromosomes, X and Y’.
Structural abnormalities
DNA damage, e.g. by radiation or mutagenic chemicals, can lead to chromosome breaks. Complex cell cycle checkpoints prevent cells with unrepaired chromosome breaks, in particular free broken ends (i.e. ends without telomeres), from entering mitosis. DNA repair mechanisms exist which recognise chromosome breaks and attempt to repair them. However, these mechanisms occasionally repair broken chromosomes incorrectly, which can then result in chromosomes with structural abnormalities. Errors during recombination, e.g. between mispaired homologues, may also result in such abnormalities.
If a single chromosome sustains breaks, incorrect repair can lead to material being lost (deletion), inverted or incorporated into a circular structure: a ring chromosome. The resulting structurally abnormal chromosomes can be stably propagated during cell division, as long as they possess a single centromere. Chromosomes without a centromere are eventually lost. Chromosomes with two centromeres are rarely found, in these cases one centromere appears to be suppressed.
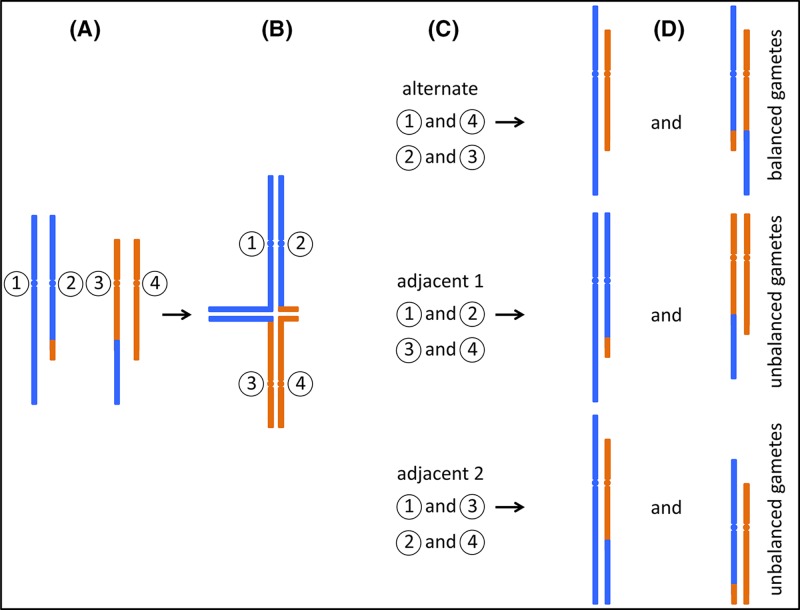
If single breaks occur in two separate chromosomes, incorrect joining of the resulting fragments may lead to the exchange of material between chromosomes (translocation). In a balanced reciprocal translocation DNA from two different chromosomes is exchanged without net loss. If both resulting hybrid (or ‘derivative’) chromosomes carry one centromere, they will be replicated and segregated stably. However, during gamete formation, it can happen that only one of the hybrid chromosomes, together with one of the unaltered chromosomes, are segregated into a gamete (Figure 6). Fertilisation of such gametes leads to the formation of a zygote with partial trisomy of genetic material from one of the chromosomes involved in the translocation, and partial monosomy of material from the other participating chromosome. Depending on the location of the breakpoints, and therefore on how much genetic material is present in trisomic or monosomic form, such embryos can be viable, but have a high risk of developmental abnormalities. Nevertheless, approximately 1 in 500 individuals carry a balanced reciprocal translocation. These carriers frequently appear asymptomatic, however, there is an increased miscarriage rate associated with either parent being a translocation carrier and offspring of carriers may present with congenital abnormalities.
Figure 6. Segregation of reciprocal translocations.
(A) A carrier of a reciprocal translocation has one unaltered copy of each chromosome that participates in the translocation, together with two hybrid chromosomes. Only the relevant chromosomes are shown, for illustration each is labelled with a circled number. (B) During meiosis, replicated sister chromatids pair up with their homologues. In the case of a translocation carrier, so-called ‘quadrivalents’ can form, in which four instead of two chromosomes pair up. (C) Three possible segregation paths are illustrated. During ‘alternate’ segregation, chromosomes 1 and 4, and chromosomes 2 and 3 are segregated into separate gametes. ‘Adjacent 1’ and ‘Adjacent 2’ segregation leads to different combinations as indicated. Note that other segregation patterns can also occur, e.g. where three chromosomes segregate into one gamete, and only one into the other. (D) Only alternate segregation leads to gametes which either carry the two unaltered, ‘normal’ chromosomes, or the two hybrid chromosomes. Zygotes formed from these gametes are expected to be phenotypically normal (unless there is a critical gene disruption at the translocation breakpoint). However, in the other two instances, all gametes carry one unaltered and one hybrid chromosome. Fertilisation of these gametes leads to zygotes carrying partial trisomy of one chromosomal segment, and partial monosomy of a different segment.
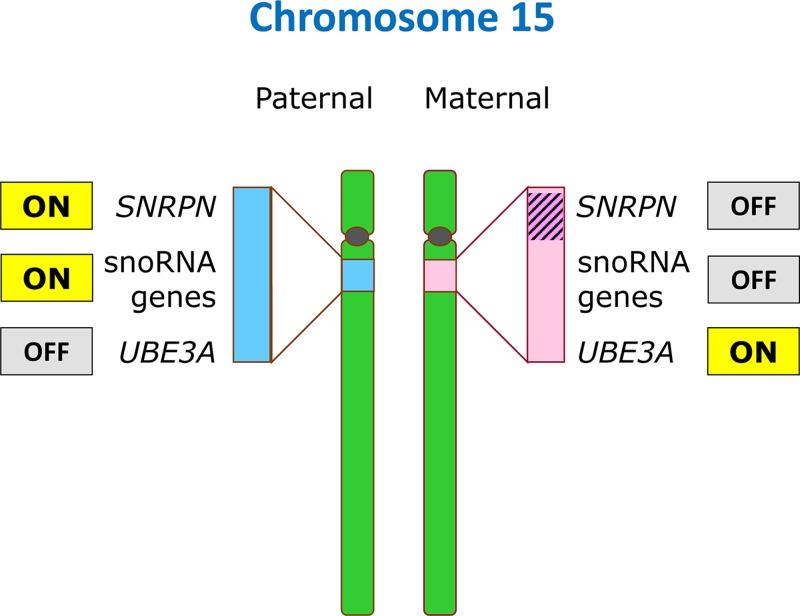
A second type of translocation is the Robertsonian translocation. Here, two acrocentric chromosomes both break at the centromere, lose their short p arms, and form a single chromosome, containing one centromere and the q arms of both original chromosomes. Carriers of Robertsonian translocations are usually phenotypically normal since only a small amount of genetic material, the nucleolar organiser region (NOR), is present in the short arms of all acrocentric chromosomes (see Figure 2). Therefore, the loss of two short arms can be compensated by the remaining acrocentric chromosomes. However, similar to reciprocal translocations, gamete formation and subsequent fertilisation can lead to the formation of zygotes with either monosomy or trisomy of one of the participating acrocentric chromosomes and therefore children with chromosomal imbalances. As is the case with meiosis in carriers of translocations, meiosis in carriers of inversions can also lead to the formation of gametes carrying an unbalanced combination of chromosomes. Therefore, such carriers may also have children with chromosomal imbalances. Carrier frequencies for Robertsonian translocations and inversions which are not considered normal variants are estimated to be 1:1000 and 1:2000 respectively.
Truly balanced translocations and inversions do not lead to the net loss of genetic material, therefore only affect the phenotype of the carrier if either a chromosome break has disrupted an important gene or a break affects the expression of a gene without disrupting its coding region, e.g. by juxtaposing the complete coding region of one gene to the control sequences of a different gene.
Microdeletions, microduplications, CNVs
Molecular genetic analysis of patients with symptoms that cannot be explained using cytogenetics can lead to the identification of the underlying causes, which in many cases are microdeletions, microduplications and other CNVs. Such variations can involve single genes or relatively few genes, which can then allow researchers to determine which particular gene is responsible for specific symptoms. Table 5 shows example of microdeletion and microduplication syndromes, together with key genes, where known, and associated symptoms. Note that in some cases, both microdeletion of a key region as well as microduplication of the same region have been identified as causative for ‘reciprocal’ syndromes. An example is a 3.6-Mb region at 17p11.2, which, when deleted, causes Smith–Magenis syndrome, but when duplicated, causes Potocki–Lupski syndrome.
Table 5. Some microdeletion and microduplication syndromes.
| Syndrome | Chromosomal location and key genes (if identified) | Typical size of deletion/duplication | Estimated incidence among live-births | Typical phenotypic features (not exhaustive, and not all these features are seen in all cases) |
|---|---|---|---|---|
| Di George syndrome/22q11 deletion syndrome |
22q11.2 TBX1, COMT |
3 Mb deletion (90% of cases) | 1/4000 | Congenital heart defects, cleft palate, developmental delay, learning difficulty, increased risk of mental illness, recurrent infections |
| Williams syndrome/Williams–Beuren syndrome |
7q11.3 CLIP2, ELN, GTF2I, GTF2IRD1, LIMK1 |
1.5–1.8 Mb deletion | 1/7500 to 1/10000 | Supravalvular aortic stenosis, joint problems and loose skin, mild to moderate intellectual disability, characteristic ‘elfin’ facial appearance |
| Smith–Magenis syndrome |
17p11.2 RAI1 |
Approximately 3.6 Mb deletion | 1/15000 to 1/25000 | Mild to moderate intellectual disability, disturbed sleep patterns, behaviour problems including aggression and self-harm |
| Cri-du-chat syndrome |
5p15.2 CTNND2 |
Approximately 5–40 Mb deletion | 1/15000 to 1/50000 | Cat-like cry, microcephaly, severe psychomotor problems and severe intellectual disability |
| Wolf–Hirschhorn syndrome |
4p16.3 NSD2, LETM1, MSX1 |
Approximately 5–18 Mb deletion | 1/50000 | Characteristic ‘Greek warrior helmet’ facial appearance, delayed growth and development, mild to severe intellectual disability |
| Potocki–Lupski syndrome |
17p11.2 RAI1 |
Approximately 3.6 Mb duplication | 1/25000 | Developmental delay, mild to moderate learning disability, behavioural problems |
| Cat eye syndrome/Schmid–Fraccaro syndrome |
22q11 ADA2, CECR2 |
2–5 Mb duplication or triplication | 1/50000 to 1/150000 | Preauricular skin tags or pits, ocular coloboma, anal atresia with fistula, heart and renal malformations |
Where specific genes have been identified as associated with particular features of the syndrome these are noted, but this does not exclude a role for additional genes in the region. Extent of the deletions/duplications often varies between patients but in general larger imbalances are associated with greater severity of symptoms.
The sex chromosomes, X and Y
Introduction
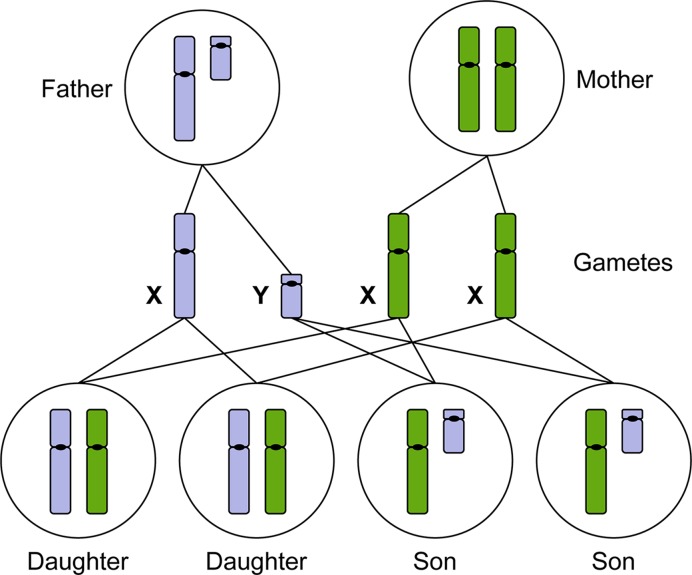
Primary sex determination in mammals is chromosomal, meaning that the development of the gonads into male (testes) or female (ovary) is determined by the sex chromosomes. The female caries two X chromosomes (46,XX) and the male has one X and one Y chromosome (46,XY). In some animals, sex determination is in part, or whole, environmentally determined (for example by temperature in most turtles), but in mammals, initiation of sexual fate is entirely driven by the chromosomes. Along with the haploid set of autosomes (22 in humans), each egg of the female has a single X chromosome, while the male can generate a sperm carrying either an X or a Y chromosome. If the egg receives an X chromosome from the sperm, the resulting XX individual will form ovaries through development and be female. If the egg receives a Y chromosome from the sperm, the resulting XY individual will form testes and be male (Figure 7). The Y chromosome is relatively small (57 Mb, with 171 genes and of these only approximately one-third are protein encoding (see Table 1)), but carries a gene that is crucial for the formation of the testes, encoding a testis-determining factor (TDF), also known as the sex-determining region Y (SRY) (Figure 8). All else being wild-type, an individual carrying a normally functioning copy of this gene will develop as a male. Thus, if the Y chromosome is missing (45,X) or if SRY is deleted, female development will ensue, although, two X chromosomes are needed for complete ovarian development. Development of primary sexual characteristics aside from the gonads, that is the reproductive structures (penis, epididymides, seminal vesicles and prostate gland in males; oviducts, vagina, cervix and uterus in females) as well as secondary sexual characteristics (mammary glands in females, along with other sex-specific features such as size, musculature, facial hair and vocal cartilage) are determined by hormones that are secreted by the gonads and this is influenced by many other genetic and environmental factors. Oestrogen, secreted by the ovaries, directs female development, while the newly formed testes secrete anti-Müllerian duct hormone and testosterone which masculinises the foetus.
Figure 7. The X and Y chromosomes determine male or female sexual development.
Males produce haploid gametes (sperm) that are either 23,X or 23,Y. Females produce haploid gametes (eggs) that are 23,X. Daughters inherit an X chromosome from their mother and an X chromosome from their father. Sons inherit an X chromosome from their mother and a Y chromosome from their father (paternal chromosomes indicated in blue, maternal chromosomes indicated in green).
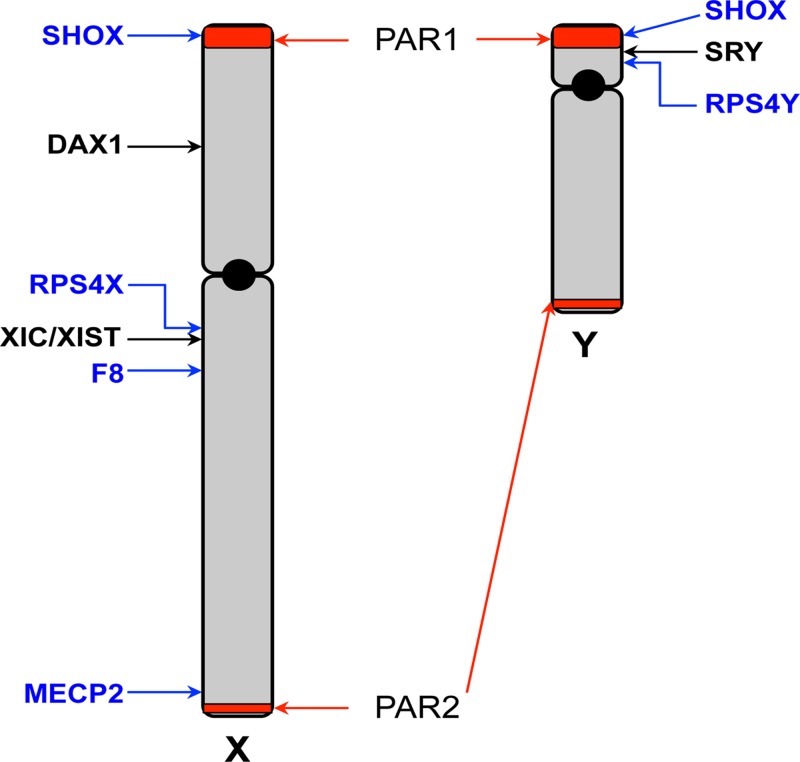
Figure 8. Schematic map of the X and Y chromosomes.
The X and Y chromosomes are depicted, showing the short (p) and long (q) arms and centromeres (black circle). The pseudoautosomal regions (PAR) 1 and 2 are highlighted in red. The sex-determining genes SRY and DAX1 are indicated. The location of the X inactivation centre (XIC) and the XIST gene is shown. Locations of other genes specifically mentioned in the text are indicated.
The X chromosome is relatively large (156 Mb) and incorporates approximately 1500 genes more than half of which are protein encoding (see Table 1), the vast majority of these have nothing to do with sex determination and are needed by both males and females. As such, the imbalance between males and females with respect to the number of X chromosomes in the genome (and therefore potential gene expression levels) needs to be rectified and this is accomplished by different mechanisms in different animal species. This balancing phenomenon, referred to as ‘dosage compensation’ is achieved in mammals by a process termed X chromosome inactivation. Early in development, in every cell in the female embryo, one of the two X chromosomes becomes inactivated, such that the majority, but not all, of the genes are not expressed from the inactive chromosome (Xi). As a consequence, the levels of expression of these genes on the active X chromosome (Xa) in female cells are equivalent to levels in male cells that only have one X chromosome. With respect to which of the two X chromosomes are inactivated, this occurs randomly from one cell to another, but then persists through subsequent cell divisions. This means that as development continues, female tissues become a patchwork, an expression-mosaic, with one of the two X chromosomes activated in some patches and the other X chromosome activated in adjacent patches. The result of this process is visible in female tortoiseshell cats, which are heterozygous for X-linked black and orange coat colour genes; the consequence of X inactivation is evident as random black and orange patches of fur in the adult. As a stochastic process, the proportion of cells that have inactivated the paternally inherited X chromosome, versus the maternally inherited X chromosome, will average 50% each. However, from one individual to another and even from one tissue to another within an individual, there can be considerable skewing from the 50% mean. Think of the tortoiseshell cat, most show roughly equivalent areas of orange and black fur, but some are more black than orange, while others are more orange than black. All female mammals are effectively expression-mosaics with respect to their X chromosomes.
One consequence for geneticists of the male-specific Y chromosome and X-inactivation in females is that the terminology of recessive and dominant allele variants becomes complicated. In addition, as described below, pathogenic allele variants on the X-chromosome can show hugely variable disease penetrance in females.
The pseudoautosomal regions
During human oogenesis, the two X chromosomes synapse in meiosis I and engage in crossover, exactly as the autosomes do. In male spermatogenesis, despite the X and Y chromosomes being of very different sizes and different genetic make-up, the chromosomes do pair (and undergo recombination) in meiosis, at short regions of homology at the ends of each chromosome. These regions are termed pseudoautosomal regions (PAR) 1 and 2 (Figure 8), because they are present on both the X and Y chromosomes; most of the genes in these regions are not subject to X inactivation in females and they behave like autosomal sequences in terms of inheritance patterns. All genes tested within the larger PAR1 escape inactivation in female cells, thus both alleles are expressed in both male and female cells. The smaller PAR2 region has been a recent acquisition in evolutionary terms, there is no equivalent in the mouse (and even some primates). PAR2 genes behave differently. The two most telomeric genes of PAR2, IL9R and CXYorf1, escape inactivation and are expressed from the Xi as well as Xa in female cells. However, two other genes of PAR2, SYBL1 and HSPRY3, do become inactivated on Xi. To compensate for this in male cells, the Y chromosome alleles of these two genes are hypermethylated and not expressed, thus in both female and male cells, only one allele is expressed.
In addition to genes within the PARs, there are several homologous gene pairs (or gametologues) present on the X and Y chromosomes, that are located in the X and Y-specific regions, that do not undergo recombination. Consequently these gene pairs have diverged from one another through evolution and often have quite different sequence from each other, although may retain a similar function. An example is the RPS4X and RPS4Y pair, which encode ribosomal proteins of essentially the same function, but differ in 19 of the 263 encoded amino acids. RPS4X escapes inactivation, therefore both alleles are expressed in female cells, while male cells express both single alleles RPS4X and RPS4Y.
SRY, DAX1 and sex determination
The SRY gene is located on the short arm of the Y chromosome, just 5 kb away from the PAR1 boundary (Figure 8). It encodes a transcription factor and is a trigger for driving male sexual development. In 46,XY individuals where the gene is dysfunctional or deleted, female development ensues. Furthermore, in approximately 80% of 46,XX male cases, the SRY gene is found translocated to an X chromosome.
In the developing embryo, a long and narrow structure called the genital ridge is the precursor to gonad formation in both sexes. The somatic cells of the genital ridge differentiate into either Sertoli cells, which promote the testicular differentiation programme or into granulosa cells, which promote ovarian differentiation. Expression of SRY in the genital ridge induces the start of Sertoli cell differentiation. While expression of SRY is brief, it initiates a cascade of events that will lead to male development. The next gene in the cascade is SOX9, an autosomal gene (located on chromosome 17) which also encodes a transcription factor and is essential for testes development. Expression of SOX9 in the genital ridge acts to induce the expression of several other genes required for testicular development, and also anti-Müllerian duct hormone which suppresses ovarian development. Thus the reverse is the case during ovarian development in XX individuals, where SOX9 is repressed. Some rare 46,XX individuals who have an extra copy of the SOX9 gene, develop as males (despite the absence of an SRY gene). Conversely, individuals who have a non-functional variant of SOX9 or gene deletion, develop a syndrome called campomelic dysplasia (which involves multiple organ systems) and 75% of 46,XY patients with this syndrome develop as phenotypic females or hermaphrodites.
It was initially thought that female development was the default state in the absence of SRY, however, this does not accurately reflect the situation that female sexual development is an active, genetically controlled process. A gene on the X chromosome, DAX1 (aka NROB1), which encodes a hormone receptor/transcriptional regulator, is required for female sexual development. DAX1 is expressed in the genital ridge shortly after SRY and antagonises the function of SRY by interfering with the induction of SOX9, in a dose-dependent manner. Normally, in 46,XY genital ridge cells, DAX1 is expressed from the X chromosome and SRY from the Y chromosome, in a ratio that leads to testes development. In 46,XX genital ridge cells, one copy of DAX1 is expressed (the other is inactivated on Xi), in the absence of SRY, to produce ovaries. However, if there are two active copies of DAX1 (for example, through gene duplication on Xa), along with SRY in 46,XY individuals, this leads to poorly formed gonads that produce neither anti-Müllerian duct hormone nor testosterone and individuals appear phenotypically female.
Following initiation of the female developmental pathway, several genes play a key role in female sexual development. The WNT4A gene (chromosome 1) encodes a secreted factor that is essential for the growth of ovarian follicle cells and is down-regulated by SOX9. The gene NR5A1/SF1 (on chromosome 9) encodes another transcriptional regulator important in both male and female pathways (as well as in the adrenal glands). Along with SRY, SF1 co-regulates SOX9 expression and is therefore critical in male sex determination. However, this multifunctional protein also plays a role later in ovarian follicular development. Thus, mutations in this gene can lead to disorders of sex development (DSD), including sex reversal, in both XX and XY individuals. Thus the SRY and DAX1 genes (present on Y and X chromosomes respectively) determine sex, as they act to flip the switch between male and female sexual fates. However, in each case, there are critical downstream regulators that promote one programme and/or inhibit the other programme and variant or mutated forms of these genes can cause DSD.
The human Y chromosome has 63 protein encoding genes, and aside from genes within the PARs and the gametologues, the majority are expressed in the testis and are involved in male fertility. The Y chromosome also has a large number (392 at last count) of pseudogenes. These have resulted from the fact that the Y chromosome (outside the PARs) has no recombination partner. As a consequence, through evolution, harmful gene mutations cannot uncouple (and thereby be selected against) from necessary genes and therefore such deleterious mutations, now in the form of pseudogenes, hitchhike along with the necessary genes.
The process of X inactivation
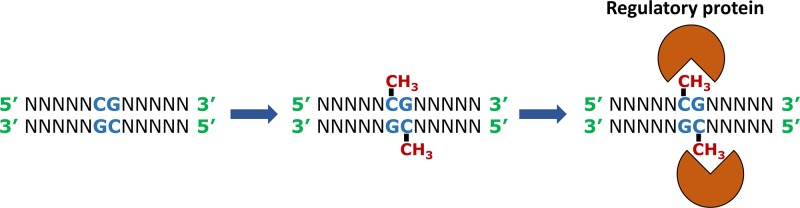
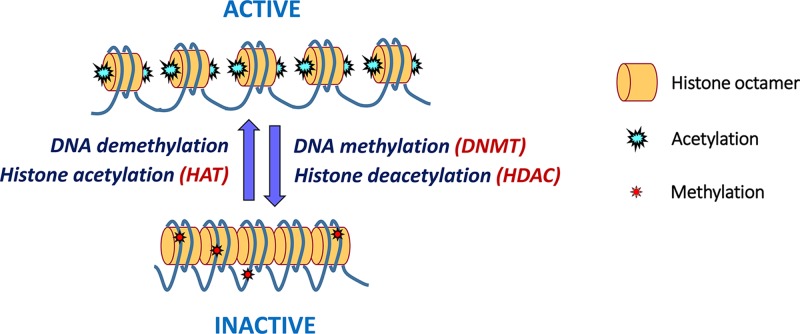
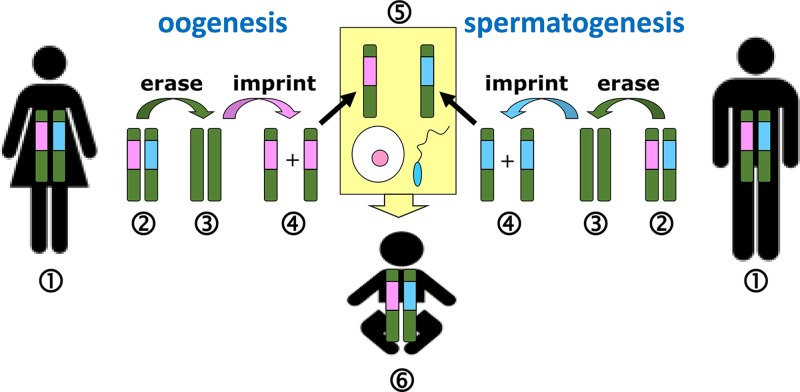
Early in female development (the early blastocyst), one X chromosome in each 46,XX cell becomes inactivated. This is initiated from a region called the X-inactivation centre (XIC) at Xq13 and in humans occurs at random, it can be either the X chromosome inherited from the mother, or the one inherited from the father. Inactivation starts with the expression of a long, non-coding RNA located within the XIC, called the X-inactive specific transcript (XIST), from the chromosome that will be silenced (Figure 9). XIST RNA coats the chromosome from which it is expressed, spreading from the XIC outwards in both directions along the entire length of the chromosome. This then leads to several epigenetic changes along the coated chromosome, including depletion of RNA polymerase II, loss of histone acetylation and an increase in histone ubiquitination and repressive methylation to silence gene expression on Xi. The Xi chromosome becomes condensed and can be seen microscopically as a dense area to the side of the nucleus referred to as the Barr body (as it was first described by the cytogeneticist Murray Barr). The inactivation is stable through subsequent cell divisions so that the same Xi is maintained in each cell lineage throughout development and adult life, with the exception of the germline. In germ cells X inactivation is reversed, so that all oocytes contain an active X.
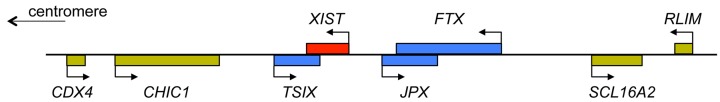
Figure 9. The XIC.
A simplified view of genes located at the XIC (not to scale), which maps at Xq13.2 on the human X chromosome and spans over 1 Mb. XIST (red) is surrounded by a number of other non-coding RNA genes (blue), which have an effect upon XIST regulation, including TSIX. Additionally, there are protein encoding genes (yellow), within the XIC region and recently RLIM has been shown to regulate XIST expression. Deletions across this region affect the process of X chromosome inactivation, but the function of all the genes and sequence regions located at XIC are yet to be fully understood.
At the XIC another non-coding transcript is expressed, partially overlapping and in the opposite (antisense) direction to XIST, the TSIX transcript is specifically expressed from the active X chromosome Xa (Figure 9). TSIX acts locally as a negative regulator of XIST expression and protects Xa against inactivation. In conditions of aneuploidy, with more than two X chromosomes, only one X remains activated, which reveals that there is a counting mechanism at play. For each autosome set, one X chromosome remains active, although how this occurs is currently poorly understood. Deletion of XIC sequences (including the XIST and TSIX genes) from one X chromosome still allows inactivation of the other, wild-type X chromosome, in 46,XX individuals. Furthermore, in transgenic mice, introduction of an XIC into an autosome, renders the autosome subject to silencing. These studies show that the XIC (and XIST) is required for initiation of chromosome inactivation in cis, but that the counting mechanism must involve factors and regions outside XIST and TSIX. It is thought that X chromosome inactivation (and the counting mechanism) must be regulated by X-encoded activators and autosomally encoded suppressors which control XIST. Recently, an X-linked gene, RLIM (or RNF12), located 500 kb upstream of XIST, has been identified as an important regulator of XIST expression, as the RLIM protein works to degrade an inhibitor of XIST transcription. However, the complexities of this process are yet far from being fully understood.
Approximately one-fifth of the genes on the X chromosome escape inactivation on Xi. These either have a Y chromosome homologue (for example, located in a PAR, as described above), or those that do not have a Y homologue tend to lie in clusters (mostly on the short arm, Xp) and apparently the 2:1 dosage (expression in female:male cells) is not problematic. It is thought that these genes are surrounded by a DNA sequence that binds a protein factor (termed CTCF) that can insulate the genes from the actions of XIST.
Aneuploidy
Due to the low gene count of the Y chromosome and the process of X inactivation, having an abnormal number of sex chromosomes has milder consequences than abnormal numbers of autosomes. Females with Triple X syndrome (47,XXX) or males with 47,XYY tend to be taller than average, but usually show few other physical differences (Table 4) and have normal fertility, thus can go undiagnosed. X inactivation in 47,XXX cells will lead to the inactivation of two X chromosomes, so despite the presence of the trisomy, the vast majority of the X-linked genes will be expressed from just the single Xa in each cell. However, overexpression of genes that escape X inactivation (as described above) gives rise to the syndrome. In 47,XYY men, there is double the Y chromosome gene dose. Therefore, there will be double the levels of Y-specific gene expression and an extra dose of products of the genes that have an X chromosome homologue (one dose from X and two doses from Y). Males with 47,XYY, as well as the above noted features, tend to have an increased risk of behavioural, emotional and social difficulties.
Men with Klinefelter syndrome (47,XXY) carry an extra X chromosome and tend to be sterile, however symptoms are frequently very subtle (Table 4) and only noticed at puberty. Again, one of the two X chromosome will be inactivated (Xi) in each cell, therefore, 47,XXY cells will only show overexpression of genes (compared with 46,XY cells) that escape X inactivation (in 47,XXX individuals, this extra expression is in the context of female development, while in 47,XXY individuals it is in the context of male development, so can have different consequences). Men with 48,XXXY display a more severe syndrome, resulting from the extra overexpression of genes that escape X inactivation, as well as the expression of Y-specific genes.
Complete loss of the X chromosome, 45,Y is early embryonic lethal. However, females with monosomy of chromosome X (45,X) or partial loss of an X chromosome, develop Turner syndrome (Table 4). In 45,X cases where an entire sex chromosome (X or Y) is lost, the remaining X chromosome does not undergo inactivation, however, this leads to half the normal expression levels of genes that do not undergo X chromosome inactivation. As such, the loss of dosage of several of these genes contributes to the syndrome. In cases where there is only partial loss of the X chromosome, such individuals will display some features of the syndrome. A gene that lies within PAR1, with alleles on both X and Y chromosomes, SHOX (Figure 8), is responsible for approximately two-thirds of the height deficit seen in Turner syndrome individuals. With loss of this region of the X chromosome, SHOX is expressed from the other allele, therefore at only half the usual levels and this is not sufficient (haploinsufficiency) to fully achieve its required growth-related function. Heterozygous loss-of-function mutations in this gene alone in both males and females causes Leri–Weill dyschondrosteosis which is characterised by skeletal dysplasia and short stature. Loss of function in both alleles causes Langer mesomelic dysplasia, which is associated with severe limb aplasia and severe height deficit. Conversely, duplication of the SHOX gene is associated with tall stature.
Pathogenic single gene variants
Haemophilia A, an example of X-linked recessive inheritance
Haemophilia A is a condition where blood clotting is defective, due to deficiency in the activity of one of the blood clotting factors, factor VIII. Symptoms can vary considerably, from mild cases, where patients only bleed excessively after major trauma or surgery, to severe cases, where patients suffer up to 30 annual episodes of spontaneous or excessive bleeding, even after minor trauma. Factor VIII is encoded by the F8 gene, located on chromosome Xq28 (Figure 8) and the gene is subject to X inactivation. There is no Y homologue, therefore if males inherit a pathogenic variant allele on the maternal X chromosome (or in 30% cases have a de novo mutation), they will be affected and disease severity will reflect the type of mutation (ranging from expression of a dysfunctional protein to complete absence of the factor). Females who are heterozygous, carrying one copy of a pathogenic variant allele, are generally asymptomatic and thus haemophilia shows a typical X-linked recessive inheritance pattern (see section on ‘Single-gene disorders’). However, as a result of X inactivation, some cells will express the wild-type F8 allele (from Xa), while other cells will express the pathogenic variant allele and the overall expression ratio between the two alleles can be 50:50 or skewed to one or the other. No cell will express both alleles. As a consequence, most female carriers produce enough factor VIII (between 30 and 70% of normal circulating levels, the variation depending both upon the nature of the variant allele and the degree to which it is Xi silenced) to appear largely unaffected. However, a significant proportion of female carriers do show some bleeding problems (for example, heavy menstrual bleeding) and in some cases (carriers who have less than 30% of normal factor VIII levels) can show mild haemophilia symptoms. In rare cases, where females inherit two variant alleles, they are more severely affected, as in males.
Rett syndrome, an example of X-linked dominant inheritance
Rett syndrome is an X-linked, dominant, neurodevelopmental disorder seen predominantly in females and becomes apparent in babies between the age of 6 and 18 months. After an initial phase of apparently normal development, individuals develop severe mental and physical disabilities, displaying coordination problems, slower growth, repetitive movements, seizures, scoliosis and other problems. The age at which symptoms first appear and the severity, varies considerably from one individual to another. Rett syndrome affects approximately 1 in 10000 females and is a single gene disorder involving the X-linked MECP2 gene. Due to the severity of symptoms, this usually arises as a de novo mutation. Boys with a similar mutation have a more severe phenotype, for example congenital encephalopathy, and die shortly after birth. MECP2 encodes a protein that binds to methylated DNA and has an important epigenetic function as a repressor of gene expression. Although the gene is normally expressed throughout the body, its function is essential in mature nerve cells and the phenotypic consequences of loss-of-function mutations (for example, loss of expression mutations or mutations that give rise to a non-functional protein) are most profound in the brain. The nature of the mutation and the extent to which the allele has lost function dictates one variable seen in disease severity. Additionally, the MECP2 gene is subject to X chromosome inactivation, this therefore also contributes to the variation seen in disease severity. If the mutant allele shows skewed inactivation (such that this allele is more frequently inactivated on Xi than the wild-type allele), this can result in considerably milder symptoms. It has been proposed that following X inactivation during development, cells which inactivated the chromosome carrying the wild-type MECP2 allele and therefore express the mutant MECP2 allele, may be selected against, resulting in a skewed X inactivation body pattern. In addition other genetic factors may exacerbate or alleviate the disease pathogenicity to contribute to the variation observed. The levels of MECP2 are critical and both too little and too much are deleterious. Therefore, mutations that result in overexpression of this gene give rise to a different syndrome (MECP2 duplication syndrome). In males MECP2 duplication leads to severe intellectual disability and epilepsy; similar duplications in females lead to a more variable condition depending upon the proportion of cells that inactivate the X chromosome containing the duplication.
In conclusion, the presentation and severity of syndromes and diseases resulting from variants of the sex chromosomes are not only influenced by the nature of the variant itself, but also by the sex-linked ploidy of these chromosomes and the consequences of X chromosome inactivation.
Single-gene disorders
Introduction
Many conditions and diseases depend on the genotype at a single locus (or gene), with inheritance following Mendel’s laws of segregation, independent assortment and dominance. Therefore, these diseases are often called ‘Mendelian’ although not all inherited disorders follow Mendel’s laws (e.g. triplet-repeat diseases and imprinting disorders). To date, over 6000 phenotypes have been identified for which the molecular basis is known, these phenotypes and the associated genes are collected in the database OMIM (‘Online Mendelian Inheritance in Man’, https://www.omim.org/). Some well-characterised examples are shown in Table 6.
Table 6. Examples of Mendelian diseases.
| Inheritance pattern | Disease | Gene/region | Nature of variants | Estimated frequency |
|---|---|---|---|---|
| Autosomal dominant | Glut1 deficiency (De Vivo disease) | SLC2A1 | Mutations reduce or eliminate function | Rare, approximately 1/90000 |
| Osteogenesis imperfecta (brittle bone disease) | COL1A1 or COL1A2 (90%) (also CRTAP or P3H1) | COL1A1/COL1A2 – usually missense mutations that lead to protein (collagen) of altered structure | 6–7/100000 | |
| Achondroplasia | FGFR3 | Activating point mutations | 1/15000 to 1/40000 | |
| Autosomal recessive | Phenylketonuria | PAH | Many different mutations, including missense, non-sense, splicing mutations | 1/10000 to 1/15000 |
| Cystic fibrosis | CFTR | Over 2000 different variants known | 1/2500 to 1/3500 in Caucasians, less common in other ethnic groups | |
| Sickle-cell anaemia | HBB | Various missense variants, gene deletions | 1/70000 to 1/80000 in the U.S.A., more common in other countries | |
| X-linked recessive | Haemophilia A | F8 | Missense and nonsense mutations | 1/4000 to 1/5000 males |
| Duchenne muscular dystrophy | DMD | Usually deletions or duplications | 1/3500 to 1/5000 (Duchenne and Becker muscular dystrophy together) | |
| X-linked dominant | Fragile X syndrome | FMR1 | CGG trinucleotide repeat expansion | 1/4000 (males), 1/8000 (females) |
| Rett syndrome | MECP2 | Missense mutations, abnormal epigenetic regulation | 1/8500 females | |
| X-linked hypophosphatemic rickets | PHEX | Deletions, insertions, missense, nonsense, splicing mutations | 1/20000 | |
| Y-linked | Nonobstructive spermatogenic failure | USP9Y | Most commonly deletions | 1/2000 to 1/3000 |
Diseases are shown together with their inheritance patterns, the affected gene, the most commonly found types of mutation, and estimated incidence rates. Note, some diseases, for example osteogenesis imperfecta (of which there are several forms), can be caused by pathogenic variants in one of a number of different genes.
Modes of inheritance and examples
Mendelian diseases can be recognised by their characteristic patterns of inheritance in family trees or pedigrees. Pedigrees can also reveal if the locus in question resides on an autosome or a sex chromosome and if a genetic variant is dominant or recessive. To understand the concepts of dominant and recessive variants, it is important to recall that each diploid human cell carries two copies (called alleles) of each autosomal gene, one inherited from the mother and one from the father. Frequently, these alleles are not identical. A person carrying two identical copies of the same allele on both autosomes is homozygous for this allele, while a person carrying two different alleles is heterozygous for the locus. A dominant allele is one which leads to a particular phenotype (e.g. a genetic disorder) no matter if the second allele is ‘normal’ or not. This is because the second, ‘normal’ allele cannot compensate for the effect of the dominant allele. A recessive allele, on the other hand, does not lead to a phenotype on its own, here, the ‘normal’ allele is sufficient or can compensate. However, if a person inherits recessive alleles of a gene from both parents, then the corresponding phenotype is displayed because no ‘normal’ allele is present to compensate. Consequently, five distinct types of Mendelian inheritance patterns can be distinguished.
Autosomal dominant
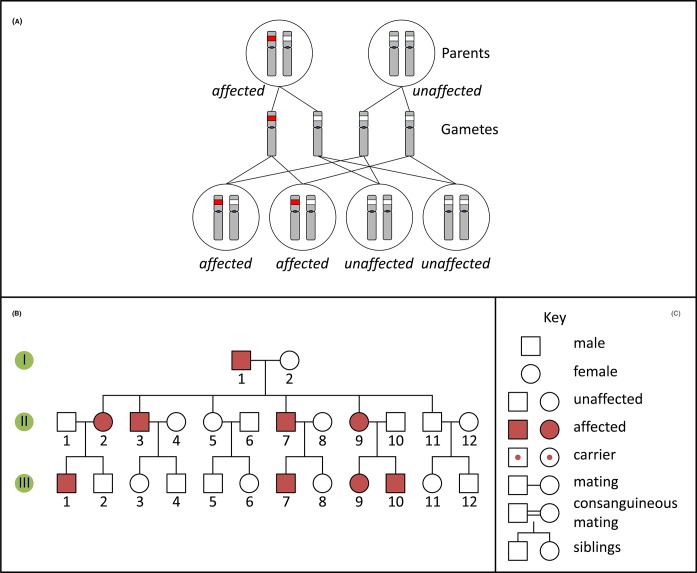
A person with an autosomal dominant disorder usually has at least one similarly affected parent and on average 50% of their children will have the disorder. Such conditions are revealed in pedigrees (Figure 10) because the disease occurs in each generation, affects both males and females, and transmission can occur from either parent to offspring of either sex. Frequently, but not always, autosomal dominant disorders are caused by genetic variants which convey a novel function to a gene product (termed gain-of-function). In this case, the presence of a ‘normal’, non-pathogenic allele of the gene on the homologous autosome cannot compensate for the altered function of the mutated gene. Note that autosomal dominant disorders can also be caused by loss-of-function alleles, if 50% of normal gene expression from the normal allele is not sufficient, a phenomenon termed haploinsufficiency.
Figure 10. Autosomal dominant inheritance and pedigree.
(A) Inheritance pattern of an autosomal dominant variant (red). Only the relevant chromosomes are shown. (B) Pedigree of a family with an autosomal dominant condition. (C) Key for pedigree symbols.
Autosomal recessive
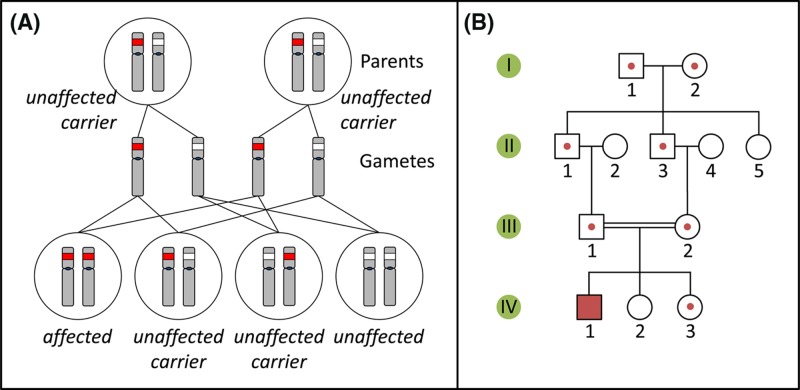
Autosomal recessive conditions are caused by loss-of-function pathogenic variants which on their own do not lead to a recognisable phenotype. Here, the presence of a second, functional allele of the gene in question on the homologous autosome is sufficient to compensate. Consequently, such conditions only manifest themselves in individuals who carry pathogenic variants at both the homologous loci (either two identical or two different recessive variants). Usually, such individuals have two unaffected parents, who are both non-symptomatic heterozygous carriers of a single pathogenic allele (Figure 11). The children of two carrier parents have a 25% chance of inheriting both pathogenic variants, while the children of affected individuals are obligate carriers of a pathogenic variant. Disease incidence is frequently increased in families where parents are consanguineous (related by descent). In families with multiple affected generations, autosomal recessive diseases often skip one or more generations.
Figure 11. Autosomal recessive inheritance and pedigree.
(A) Inheritance pattern of an autosomal recessive variant (red). Only the relevant chromosomes are shown. (B) Pedigree of a family with an autosomal recessive condition. See Figure 10C for key to symbols.
X-linked recessive
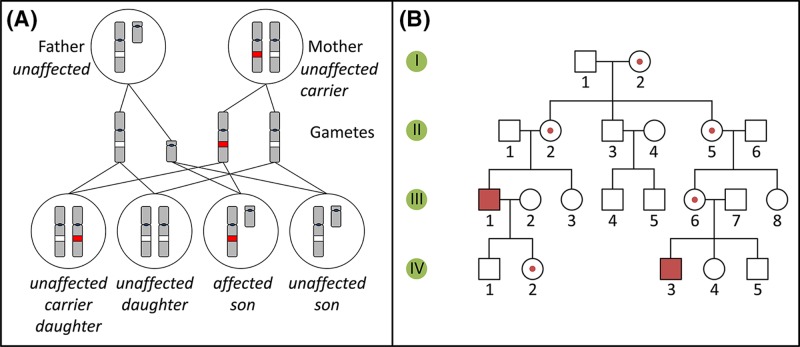
Diseases which are caused by recessive variants in loci located on the X chromosome affect females and males differently. Males have a single X chromosome, therefore, if they carry a pathogenic variant, they have no second allele to compensate for its effect, and will be affected by the disease. All their daughters will inherit their X chromosome, therefore will be carriers, while their sons will be unaffected (Figure 12). Since females carry two X chromosomes, they will typically only be affected by the disease if they inherited one pathogenic variant of the relevant gene from their (affected) father and a second pathogenic variant from their mother, who could be an unaffected carrier or a homozygous, affected individual. However, due to the phenomenon of X chromosome inactivation in females, such variants are often not completely recessive and can show some aspects of the phenotype in female carriers (as explained in the section ‘The sex chromosomes, X and Y’).
Figure 12. X-linked recessive inheritance and pedigree.
(A) Inheritance pattern of an X-linked recessive variant (red). Only the relevant chromosomes are shown. (B) Pedigree of a family with an X-linked recessive condition. See Figure 10C for key to symbols.
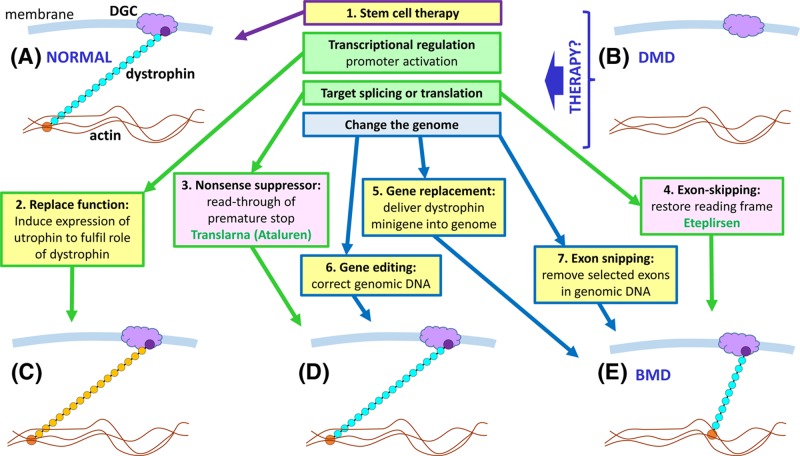
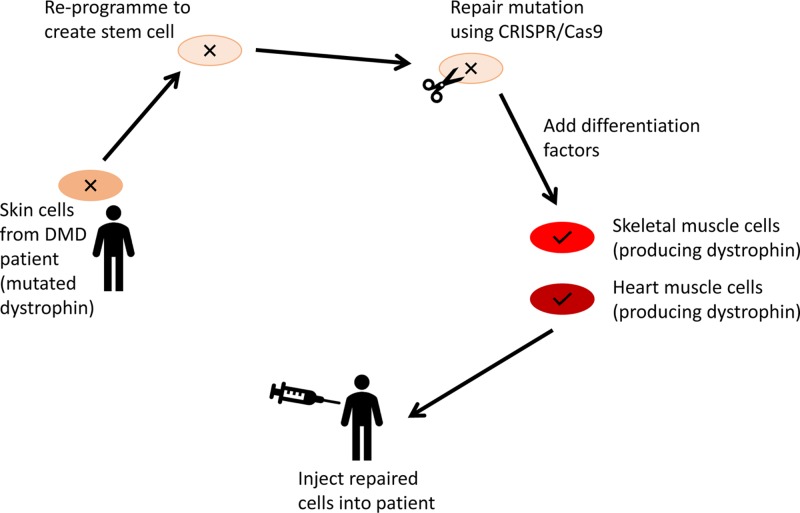
X-linked dominant
A dominant pathogenic variant on the X chromosome will typically affect both males and females (but this is also complicated by X-inactivation). All daughters of an affected male will inherit the condition, while all of his sons will be unaffected. In the case of an affected female, if she has a single pathogenic variant, her children will have a 50% chance of being affected. However, if she has inherited a pathogenic variant from each of her parents, both her parents will typically have been affected, and all her children will also be affected. The actual situation may be more complicated, for example in late-onset conditions if an apparently unaffected parent died before they developed the disorder or if one of the pathogenic alleles is non-penetrant due to non-random X-inactivation. X-linked dominant disorders are rare, but examples are shown in Table 6.
Y-linked
Since the Y chromosome is very small and only contains comparatively few genes, Y-linked single-gene disorders are even rarer than X-linked dominant ones. As much of the Y chromosome exists in a hemizygous state (with the exception of genes with homologues on the X chromosome), recessive and dominant definitions do not apply; as such, the phenotype of Y chromosome variants will be manifest. Consequently, affected males also have affected fathers, unless a de novo mutation has occurred, and all their sons will be affected. Since daughters of affected males will inherit their father’s normal X chromosome, and not the affected Y chromosome, they will be unaffected, and their offspring will also be unaffected. An example of a Y-linked condition is nonobstructive spermatogenic failure, which leads to fertility problems in males which may be addressed by assisted reproductive methods such as in vitro fertilisation (IVF).
Types of variants and their effects
Mendelian disorders are caused by pathogenic variants at single loci (in single genes), therefore, it is relevant to briefly discuss what kinds of mutations are involved and what their consequences upon gene function are. This depends on where within the sequence of a gene the change has occurred (e.g. within the coding region, in a control region or in a region involved in post-transcriptional modification).
Mutations can be categorised into those where nucleotides are exchanged against different ones where the total number of nucleotides do not change, and those where nucleotides are deleted, inserted or a combination thereof, with a concomitant change in the overall number of nucleotides. Where only a few nucleotides are involved, this is referred to as a microlesion, if only a single nucleotide is involved, this is referred to as a point mutation.
The following section will briefly describe mutations in coding regions, but microlesions outside the coding region of a gene can still have severe consequences. Mutations that change the regulation of a gene’s expression (e.g. promoter mutations) can lead to the production of too much or too little of the resulting protein, which may lead to a noticeable phenotype. Mutations can also occur in the conserved intronic sequences directly adjacent to intron–exon boundaries. Such mutations can then lead to aberrant splicing of the resulting transcript, with subsequent consequences on the encoded protein. In addition, mutations in non-coding RNAs can have profound effects, for example in one of the numerous miRNAs, which act to control the expression of other genes.
Point mutations
A point mutation is one which changes one nucleotide by substitution (one base pair is replaced by another), deletion or addition. If a substitution point mutation occurs within the coding region of a gene, various outcomes are possible: silent or synonymous mutations lead to the exchange of one codon for a different codon which still encodes the same amino acid. For example, the codons ATT, ATC and ATA all code for the amino acid isoleucine and if a mutation changed ATT to ATC, this would not lead to a change in the encoded protein sequence, therefore, such mutations are not expected to change the function of the encoded proteins. Missense mutations lead to a change in the codon such that it encodes a different amino acid. For example, a change from GAG to GTG will lead to the incorporation of valine into the resulting protein instead of glutamic acid. Such changes could lead to the complete loss-of-function of the resulting protein, or a dramatic change in function, or may have only a small effect that can be tolerated. This depends on the context of the amino acid within the mature protein, its function and on the chemical characteristics of the amino acids that are exchanged. Nonsense mutations lead to a codon for an amino acid being exchanged for one of the three stop codons. For example, a change from TAC to TAG leads to a change from the codon for tyrosine to a stop codon. Translation of the resulting mutated coding sequence leads to the formation of a prematurely truncated polypeptide, and most such truncations lead to non-functional proteins, although if they occur towards the C-terminal end of the protein they may have less effect.
Insertions, deletions and indels
The term ‘indel’ refers to mutations that change the total number of nucleotides in a genome, by insertion, deletion or a combination of both. Indels generally refer to small changes from a point mutation, up to 1 kb. Indels occurring in sequences which control levels of gene expression or transcript splicing will lead to aberrant gene expression or splicing, but the effect of indels in coding sequences depends on the actual number of nucleotides inserted or deleted.
Deletion of three (or a multiple of three) nucleotides from a coding sequence corresponds to the deletion of one (or more) codons and will lead to the expression of a protein where one (or more) amino acids are deleted, without changes to the remaining amino acid sequence (Figure 13). Such polypeptides may still be functional. However, if a number of nucleotides which is not divisible by three is deleted from a coding region, all subsequent codons will be altered, a phenomenon called a ‘frameshift’ (Figure 13). A ribosome translates mRNA molecules one triplet (codon) at a time, before moving to the next triplet. If a single nucleotide is deleted, the ribosome will still translate one triplet at a time, but from the deletion point onwards, each triplet will now differ from those originally present. This will lead to the formation of a polypeptide whose sequence will differ completely from that originally present. Frequently, such frame shifts lead to stop codons being brought into the reading frame soon after the deletion point, thereby truncating the protein.
Figure 13. Insertions and deletions.
(1) A wild-type sequence consisting of three-letter words. (2) Insertion of three letters inserts another three-letter word, and keeps the remainder of the sequence unchanged. (3,4) Deletion of a number of letters which is a multiple of three removes a number of words, but leaves the remainder of the sequence unchanged. Note that the resulting sentence still makes (more or less) sense. Deletion (5) or insertion (6) of a number of letters which is not a multiple of three changes all three-letter words after the point of deletion/insertion. The resulting sentence does not make sense any more. These two cases are examples of frameshift mutations.
Insertion of nucleotides follows the same principle. Insertion of three (or a multiple of three) nucleotides leads to the insertion of one (or more) amino acids into the translated protein, while insertion of a number of nucleotides not divisible by three will lead to a frameshift mutation.
Examples of single-gene disorders
A special case of insertion mutations are the nucleotide repeat expansion disorders, typically triplet-repeat disorders (Table 7). Trinucleotide repeats are repetitive sequences where triplets of nucleotides are repeated in tandem multiple times; in some cases these repeats are located in coding sequences and are translated as stretches of polypeptide consisting of a repeat of the same amino acid. Other trinucleotide repeats are located in non-coding sequences. It is possible that the presence of expanded repeats within a transcript is toxic for the RNA processing machinery in the nucleus. The nucleotide sequence of trinucleotide repeats is often (CAG)n or (CTG)n, where n is the number of repeat units, but other trinucleotide repeats are also known. One unusual aspect of these repeats is that they can become unstable and expand, such that the number of repeats increases dramatically from one generation to the next, a phenomenon termed ‘anticipation’. In addition, the repeats (over a certain number) can be unstable through somatic cell division, such that cells of some tissues show hugely varying numbers of repeats in the expanded allele. In all known cases of trinucleotide repeat expansion disorders, individuals carrying a number of repeats up to a threshold do not show any clinical symptoms, while individuals carrying longer repeats show progressively severe symptoms.
Table 7. Examples of nucleotide repeat expansion disorders.
| Disease | Gene | Repeat | Normal range | Pathogenic range | Disease features | Estimated incidence |
|---|---|---|---|---|---|---|
| Huntington disease | HTT | CAG (encoding glutamine) | 9–35 | 36–39 (possibly pathogenic) >39 (pathogenic) |
Uncontrolled movements, emotional problems, loss of cognitive ability | 3–7/100000 |
| Myotonic dystrophy type 1 | DMPK | CTG (3′-UTR) | 5–37 | 50–150 (mildly affected) 100–1000 (classic symptoms) >2000 (congenital onset) |
Progressive muscle wasting and weakness, muscle contractions, cataracts, cardiac abnormalities | >1/8000 |
| Fragile X-associated tremor/ataxia syndrome (FXTAS) | FMR1 | CGG (5′-UTR) | 5–40 | 55–200 (pre-mutation with respect to FXS) | Ataxia, tremors, cognitive decline, learning disabilities, blood pressure problems | 1/4000 males, 1/8000 females (milder in females) |
| Fragile X syndrome (FXS) | FMR1 | CGG (5′-UTR) | 5–40 | 200-several thousand | Developmental problems including learning disabilities and intellectual impairment, autistic spectrum disorders, attention deficit | |
| Friedreich ataxia | FXN | GAA (in intron 1) | 5–33 | 66 to >1000 | Impaired muscle coordination, loss of strength and sensation, muscle stiffness, impaired speech, hearing, and vision, heart disease | 1/40000 in people of European, Middle Eastern or North African ancestry |
Four genes subject to repeat expansions are shown, with the gene affected, repeat sequence, normal and pathogenic range of repeat, main disease features and estimated incidence.
Huntington disease
Huntington disease (HD) is one of the trinucleotide repeat expansion disorders where the CAG repeat encodes a polyglutamine tract within the coding region of the huntingtin gene HTT on chromosome 4p16. It is a progressive neurodegenerative disorder with patients suffering from progressive neural cell loss and atrophy. Symptoms start with personality and mood changes, followed by a steady deterioration of physical and mental abilities. The function of the huntingtin protein is unclear, but it is essential for development. Inheritance follows an autosomal dominant pattern, caused by a gain-of-function associated with the repeat expansion. Unaffected individuals carry between 9 and 35 CAG repeats, incomplete penetrance occurs in carriers of 36–39 repeats, while the disease is fully penetrant when 40 or more repeats are present. Alleles containing 250 and more repeats have been reported.
While repeat alleles of 9–30 are almost always transmitted without change to the next generation, larger alleles show instability, both in somatic tissues and in the germline, with a tendency towards expansion from one generation to the next. There is a correlation between the number of repeats and the severity of disease and also an inverse correlation between the number of repeats and the age of disease onset. The degree of repeat instability is also largely proportional to the number of repeats, and is also affected by the sex of the transmitting parent, with larger expansions occurring in male transmission. This leads to ‘anticipation’ where an apparently healthy individual might have a child with late onset HD and a grandchild with more severe symptoms and an earlier onset, and so on.
Achondroplasia
Achondroplasia (ACH) is the most common form of dwarfism in humans and is inherited in an autosomal dominant fashion with 100% penetrance. Individuals with ACH have shortened limbs, a large head, and a trunk of relatively normal size.
ACH is caused by specific variants in FGFR3, the gene for fibroblast growth factor (FGF) receptor 3 (FGFR3), on chromosome 4p16. Almost all individuals with ACH are heterozygous for a variant which leads to a substitution of arginine for glycine at position 380 (p.Gly380Arg) in the mature protein. Eighty percent of ACH cases are due to spontaneous, de novo mutations, often occurring during spermatogenesis. FGFR3 is a transmembrane receptor protein which binds to FGF ligands and triggers intracellular signalling processes. One of these processes is the inhibition of chondrocyte proliferation in the growth plate of long bones. The p.Gly380Arg variant in FGFR3 generates a constitutively active version of the receptor which can be further activated by binding of FGF. Therefore, this variant acts as a gain-of-function mutation. Consequently, chondrocyte proliferation in growth plates is constitutively inhibited. While one such variant allele (in the heterozygous state) leads to ACH, homozygosity is lethal before birth or perinatally. Interestingly, loss-of-function variants in FGFR3 have also been described which cause a different condition, camptodactyly, tall stature and hearing loss (CATSHL) syndrome. This is an example where different variants of the same gene result in different phenotypes, so-called ‘allelic disorders’.
Cystic fibrosis
Cystic fibrosis (CF) mostly affects the lungs (resulting in breathing difficulty and frequent lung infections) and the pancreas (with disruption of the exocrine function), but the liver, kidney, intestines and male reproductive system are also frequently affected. It is the most common lethal genetic disease among Caucasians, and is inherited in an autosomal recessive pattern.
CF is caused by pathogenic variants in the CFTR gene, which encodes the CF transmembrane conductance regulator, a transmembrane protein which functions as a selective chloride channel. If the CFTR protein does not function properly, the chloride balance between the inside and outside of cells becomes disrupted, leading to the build-up of mucus in narrow passages in affected organs such as the lungs. Such blockages lead to damage and reduced function in these organs. The CFTR gene is located on chromosome 7q31 and encodes a protein of 1480 amino acids. The gene is approximately 250000 nts long and over 2000 pathogenic variants have been identified in its sequence. These variants fall into different classes (e.g. those where protein synthesis is defective, those where reduced amounts of normal protein is made, those where the synthesised protein is not processed properly and does not reach the membrane and others). As long as an individual carries one functional allele of CFTR, they may show no or only very mild symptoms (Figure 14), but an individual carrying two pathogenic variants will display symptoms that depend on the amount of functional protein generated. The most common pathogenic variant, representing approximately 70% of Caucasian CF alleles, is a deletion of the codon for a phenylalanine at position 508 (p.Phe508del) in the mature protein. This particular variant leads to the synthesis of a protein which does not fold properly into its 3D shape, and is degraded by the cell before it can reach the membrane, therefore representing a loss of function.
Figure 14. CF symptoms depend on residual CFTR activity.
The left-hand side shows a scale of residual CFTR protein activity, between 0 and 100%. The right-hand side shows corresponding symptoms. Note that heterozygotes, carrying one pathogenic CF allele, still retain 50% of CFTR activity, therefore may be asymptomatic or show only mild symptoms. Only patients with 5% or less residual CFTR activity show full CF symptoms. Figure adapted from Davis 2001.
Mitochondrial disorders
Mitochondria – structure and function
Mitochondria are cellular organelles containing a genome which is independent of the nuclear genome, and are thought to have arisen from a symbiotic relationship between a primitive bacterial cell and a precursor of a eukaryotic cell. Mitochondria are the ‘powerhouse’ of the cell and are responsible for generating energy in the form of ATP. In addition, these organelles are key structures in controlling the process of programmed cell death or apoptosis. As such, they embody the life and death of the cell. They are capsule-like in structure (Figure 15) with two membranes, an outer and an inner, and an intermembrane space in between the two. The inner membrane folds on itself forming cristae which increase the overall surface area and contain a series of protein complexes and transport chains involved in the production of ATP.
Figure 15. Mitochondrial structure.
The mitochondrion has two membranes; the inner membrane folds into series of cristae on which are the electron carriers and ATP synthase, responsible for the generation of ATP. The matrix of the mitochondrion contains multiple copies of the circular mtDNA (image by Mariana Ruiz Villarreal, reproduced from Wikimedia Commons: Public Domain).
The generation of energy by the mitochondria involves several stages which are collectively known as cellular respiration. Firstly, a sugar (usually glucose) taken in by a cell is broken down by glycolysis in the cytoplasm, to generate two molecules of pyruvate, NADH and ATP. The pyruvate produced in glycolysis is converted into acetyl CoA which then enters the Krebs cycle – both the reactions occur in the mitochondrial matrix. The product of the Krebs cycle is NADH, which undergoes oxidative phosphorylation in the last stage of cellular respiration. In oxidative phosphorylation, which occurs on the cristae of the inner membrane, electrons are transferred from NADH or FADH2 to O2 by a series of electron carriers (Figure 16). As a result of this process, ATP is formed.
Figure 16. The electron transport chain.
On the inner membrane of the mitochondria, electrons from NADH and FADH2 pass through the electron transport chain to oxygen which then undergoes a reduction reaction, producing water. The chain comprises five complexes and a series of electron transporters and electrons are passed from donors to acceptors down the chain, releasing energy which is utilised to generate a proton gradient across the mitochondrial membrane (image reproduced from Wikimedia Commons: Public Domain).
Control of some aspects of apoptosis or programmed cell death (so called because the survival or death of the cell is determined by a balance of cellular components) is another important mitochondrial function. In the very early stages of apoptosis, mitochondria release cytochrome c, an essential component of the electron transport chain, which initiates a pathway of procaspase activation, central to apoptosis. Key in mediating the mitochondria to release cytochrome c are proteins from the BCL family which reside in the outer mitochondrial membrane.
The mitochondrial genome
Located in the mitochondrial matrix, the mitochondrial genome exists as a circular, dsDNA molecule (Figure 17), of approximately 16500 bp encoding 37 genes, each of which is vital to the function of mitochondria. However, numerous nuclear DNA encoded proteins also contribute to the formation and function of the mitochondria. Each cell contains thousands of mitochondria and each mitochondrion contains multiple copies of the mitochondrial genome. Thirteen of the 37 mitochondrial genes encode subunits of the four respiratory complexes involved in oxidative phosphorylation, which are situated in the inner membrane, while the rest code for 22 tRNAs and 2 rRNAs. The majority of components of the respiratory complexes are expressed from the nuclear genome and transported into the mitochondria.
Figure 17. The mitochondrial genome.
Circular and double stranded, with no introns, the mitochondrial genome comprises 37 genes which code for components of the respiratory complexes involved in oxidative phosphorylation as well as for tRNA and rRNA. Many of the components of the respiratory complexes are coded for by the nuclear DNA (image reproduced from Wikimedia Commons: Public Domain).
The mitochondrial genome is unique in several ways. Almost 93% of mtDNA is coding, compared with the nuclear genome of which only 3% is coding, and mtDNA is free from introns, histones and epigenetic marks. MtDNA is subject to a mutation rate 100-times higher than that of the nuclear genome, since the mtDNA repair systems are less robust (more error prone) than nuclear DNA repair systems and because the internal environment of the organelle has more reactive molecules that can damage DNA (from the products of the respiratory transport chain). Importantly, mtDNA is only inherited maternally – a female will pass on mitochondria to all her children while a male will not normally pass on any of his mitochondria (Figure 18).
Figure 18. Mitochondrial inheritance.
In the case of a mitochondrial mutation (see following section), an affected mother (A) will pass the mutation on to all of her children since mitochondria are maternally inherited. On the other hand paternal mitochondrial mutations (B) will not be transmitted to his children.
Heteroplasmy is an important feature of the mitochondrial genome – this means that not all copies of the mitochondrial genome within a cell are the same (Figure 19). If a particular variant is present in all copies of the mitochondrial genome in a cell, the cell is said to be homoplasmic for the mutation. The situation where some mitochondria contain the mutation while others which do not are referred to as heteroplasmy. Obviously this is an important phenomenon when considering the inheritance of mitochondrial mutations (discussed below) as offspring may inherit a high or low proportion of mutant mitochondria from a carrier mother.
Figure 19. Heteroplasmy.
A primordial germ cell containing a mixture of mutant and normal mtDNA may give rise to oocytes which have high, intermediate or low mutation loads, dependent on the region of cytoplasm which contributes to it.
Mitochondria and disease
A number of conditions, from age-related hearing loss to specific forms of epilepsy and diabetes, are associated with mutations in the mitochondrial genome, with the phenotypes and severity varying widely. Often affecting tissues with high energy requirements such as muscle and nerve tissue, there is significant overlap between phenotypes caused by different mutations. As an example, two phenotypically distinct conditions caused by mitochondrial mutations are described below.
Leber hereditary optic neuropathy (LHON) is a mitochondrially inherited disorder of vision, with a reported incidence of approximately 1/30000 to 1/50000 births in European populations and symptoms which typically appear in the teens or twenties. The central area of vision tends to be most severely affected, with blurring and cloudy vision progressing to loss of sharpness and colour vision in the later stages. The pathogenesis of the condition results from cell death in the optic nerve. Mutations in a number of mitochondrial genes, including those encoding several NADH dehydrogenases (MT-ND 1, 4 and 6), are known to cause LHON.
The precise link between these mutations and the optic nerve cell death is unknown, and further complicated by the fact that up to 85% of individuals harbouring these mutations never develop visual problems. In some families, additional complications are present, including cardiac conduction and mild neurological problems. Although many different mutations have been associated with LHON, one of three missense mutations is thought to be present in 90% of affected families. Genetic diagnosis of the mutations causing mitochondrial disease such as LHON can be carried out using PCR-based techniques, such as allele-specific PCR.
Leigh syndrome, in contrast with LHON, typically appears within the first year of life. It is a severe and progressive neurological condition, with progressive loss of both mental and motor skills. Affected children usually die within 2 or 3 years of first showing symptoms, generally because of respiratory failure. With an overall similar frequency to LHON, Leigh syndrome is found in some populations (for example in Quebec, Canada) at a much higher frequency. Lesions in the basal ganglia, cerebellum and brainstem are seen on MRI scans in affected individuals. Both nuclear and mtDNA mutations have been observed in this condition, affecting complexes II, III, IV as well as ATP synthase, also known as complex V.
Therapy for mitochondrial disease
A number of traditional approaches have been used to combat the symptoms of mitochondrial disease, mainly in the form of dietary supplements, with little success. More recently, several strategies, including gene transfer using adeno-associated viral vectors have been trialled, in an attempt to replace the mutated gene.
However, as there is currently no effective strategy for treating mitochondrial disease, the approach of preventing rather than curing the conditions seems very attractive. Women who carry a mitochondrial mutation have the option of using a donor egg or adoption in order to avoid the possibility of having an affected child, but the possibility of having their own healthy biological child is more attractive to many. Thus mitochondrial replacement therapy or the ‘three parent baby’, in which the nucleus is removed from an egg containing mutated mitochondria and is placed into an enucleated egg from a healthy donor, has gained increasing interest in recent years.
There are two main approaches to achieve this – pronuclear transfer and spindle nuclear transfer (Figure 20). With pronuclear transfer, which is currently the technique approved in the U.K., the mother’s egg as well as a donor egg are both fertilised with the father’s sperm. Subsequently the nucleus from each fertilised egg is removed and the donor egg’s nucleus is then replaced with that of the mother. In the alternative technique (spindle transfer), the nucleus is removed from an unfertilised mother’s egg and this is then inserted into a donor egg which has had its nucleus removed. Fertilisation with the father’s sperm then takes place. Although the U.K. approved pronuclear transfer in 2016, the first baby to be born using this technique was a boy born to Jordanian parents at a clinic in Mexico, to avoid the transmission of a mitochondrial mutation causing Leigh syndrome. It is expected that more babies will be conceived using this technique.
Figure 20. Mitochondrial replacement therapy (‘three-parent baby’).
Eggs are harvested (1) from the mother-to-be, whose mitochondria are affected by a pathogenic DNA variant. The nucleus is extracted from the egg (2). Eggs are also harvested (3) from a donor female who has healthy mitochondria, and the nucleus is removed from the egg, leaving only the cytoplasm (4). Now the nucleus (2) is injected into the enucleated egg (4) to generate an egg (5) that has the nuclear DNA of the mother-to-be, and healthy mitochondria. This can now be fertilised in vitro using sperm from the father (6), and allowed to develop for a few days before implantation into the mother-to-be.
Epigenetics
Introduction
The nucleotide sequence of the human genome contains within it a vast amount of complex information that is required for a healthy human, and changes to the DNA sequence may lead to disease states as described in previous sections. However, there is another layer of information that is superimposed upon the nucleotide sequence information, and study of this additional information is referred to as ‘epigenetics’, a term derived from the Greek and literally meaning ‘above the genetics’. This epigenetic information takes the form of chemical groups (for example, methyl groups) attached to the DNA or attached to the histone proteins around which the DNA is wrapped in chromatin. A key concept is that epigenetic information can be inherited between cell generations.
DNA methylation
In humans (as well as other mammals) cytosines within the dinucleotide sequence CG may become methylated (Figure 21). Note the distinction between a C–G base pair (where the C and G are on opposite DNA strands) and a CG dinucleotide, which is a CG sequence along one strand of the DNA read in the 5′ to 3′direction (consequently, this is often referred to as CpG, to indicate the phosphate linkage on the DNA backbone). However, due to the complementarity of DNA there will also be a CG sequence in the 5′ to 3′ direction on the other strand of DNA (Figure 22). The methylated cytosines are recognised by specific proteins, which bind to the DNA at these locations, and often lead to the recruitment of further proteins; these protein complexes can lead to alterations in chromatin structure and thereby activity of genes, typically by affecting the modification of histones.
Figure 21. Methylation of cytosine and consequences of deamination of methyl-C.
(A) Cytosine can be methylated to generate 5-methylcytosine; this may undergo spontaneous deamination to generate thymine. The arrow indicates position of attachment to the deoxyribose ring. (B) CG dinucleotides (indicated by vertical lines) are relatively rare in the genome overall compared with the expected frequency. This is believed to be an evolutionary consequence of deamination of methylcytosine leading to conversion of many C–G base pairs within CG dinucleotides into T–A. However due to the importance of DNA methylation in transcription regulation (see Figure 25), CG dinucleotides are present at higher frequency around promoter regions (transcriptional start sites) of genes, generating so-called ‘CG islands’ (also known as CpG islands).
Figure 22. Methylation occurs on the C of the sequence CG and facilitates binding of specific regulatory proteins to the DNA.
Note that the sequence CG is palindromic, in the sense that the same sequence occurs in the 5′ to 3′ direction on both strands of the DNA. Abbreviation: N, any nucleotide.
The cell has several different enzymes which are responsible for DNA methylation, the DNA methyltransferases (DNMTs). Two of these, DNMT3A and DNMT3B, appear to specialise in de novo methylation, while the major role for DNMT1 appears to be in maintenance methylation (Figure 23). The importance of maintenance methylation is evident when DNA replication is considered (Figure 24). When methylated DNA is replicated by the action of DNA polymerase, the new strand will be generated by incorporation of standard nucleotide triphosphates, and thus no methyl groups will be present. If this DNA is replicated again, the newly synthesised DNA would have no methylation. However, hemimethylated DNA is recognised by DNMT1, which will methylate the cytosine on the complementary strand. This ensures that DNA methylation patterns are heritable between cell generations.
Figure 23. Methylation and demethylation.
The methylation process is initiated by addition of a methyl group to one strand of the DNA by DNMT3A or DNMT3B. The resultant hemimethylated DNA becomes fully methylated by the action of DNMT1. Removal of methyl groups from DNA may be active (involving DNA demethylases) or passive (in the absence of maintenance – see also Figure 24). Abbreviation: m, methyl (CH3) group.
Figure 24. DNA methylation status is heritable but requires maintenance.
(A) The Cs within CG dinucleotides (blue) represent potential methylation sites. In this piece of DNA sites 1 and 2 are fully methylated (i.e. on both strands of the DNA), but site 3 is not methylated. (B) Following replication the new strands of DNA (green) are unmethylated; further rounds of replication of this hemimethylated DNA would lead to some unmethylated DNA. However, hemimethylated DNA is a substrate for the maintenance methylase, DNMT1. (C) The daughter DNA molecules are now fully methylated at the originally methylated positions (1 and 2), but remain unmethylated at position 3, which was unmethylated in the original DNA template. Abbreviation: m, methyl (CH3) group.
Methylation patterns in DNA are dynamic and can be altered in response to stimuli or at particular stages of development. To achieve this, it is necessary to have enzymes which can remove methyl groups from DNA: the DNA demethylases.
Histone modifications
For packaging into chromatin in eukaryotes, DNA becomes wrapped around octamers of histones; each octamer includes two molecules each of H2A, H2B, H3 and H4. The histone octamer forms a disc-like shape, with the N-terminal tails of each of the individual histones protruding from this disc. The histone proteins are subject to a variety of post-translational modifications, which include acetylation, methylation, phosphorylation and ubiquitination. Like DNA methylation, these modifications may be altered in response to changing circumstances, and influence the compaction of the chromatin and its accessibility by transcription factors and other proteins (Figure 25). The best understood histone modifications are those which occur on the N-terminal tails. For example, acetylation of lysine residues in the N-terminal tails renders chromatin less compact and thus facilitates transcription. Acetyl groups can be added by the histone acetyl transferase (HAT) group of enzymes, and removed by histone deacetylases (HDACs); thus the activity of HATs will facilitate transcription, while the activity of HDACs will tend to inhibit transcription.
Figure 25. Chromatin status is influenced by DNA methylation and histone acetylation.
In active chromatin the N-terminal tails of histone proteins are acetylated (by HATs); the additional positive charges encourage a looser packing of nucleosomes that makes the DNA more accessible for other proteins and thus facilitates transcription. Addition of methyl groups to the CG dinucleotides (by DNMTs) and removal of the acetyl groups from the histones (by HDACs) provokes a more compact chromatin structure, which is not easily accessible by transcription factors, generating inactive chromatin. Reactivation of inactive chromatin is facilitated by DNA demethylases (which remove the methyl groups) and HATs.
DNA methylation status can influence histone modification and vice versa. For example, methyl-C binding proteins can recruit HDACs to sites of DNA methylation, and histone methylation may either facilitate or inhibit DNA methylation, depending upon which amino acid has been methylated within the histones. One consequence of all this modification is that, although all our cells may contain the same genome and therefore the same genes, the pattern of which genes may or may not be active in a given cell or tissue type is dependent upon the modifications present in the DNA and on the histone proteins: the epigenetics.
Chromosomal imprinting
Several regions of the human genome, involving roughly 100 genes, are rendered inactive by epigenetic mechanisms depending upon parent of origin. For some genes only the paternal allele is active, while the maternal copy is epigenetically silenced throughout the life of the individual. For other genes it is the maternal copy that is active and the paternal copy which becomes epigenetically silenced. The process of epigenetic silencing in a parent-of-origin specific manner is termed as ‘chromosomal imprinting’, and involves DNA methylation, histone modification and additional protein and RNA factors. During gametogenesis all previous imprints are removed from the DNA, and new imprints are established: female imprints during oogenesis and male imprints during spermatogenesis (Figure 26), thus each embryo should receive chromosomes with a complementary set of imprints and therefore one active copy of each imprinted gene.
Figure 26. Imprints are erased and reset during gametogenesis.
For each imprinted chromosome region, healthy adult humans (1) will have, in their somatic cells, one maternal and one paternal imprint. During gametogenesis, the imprints present (2) must first be erased (3), and then the imprints are reset (4) according to the sex of the parent. Therefore during oogenesis all relevant regions are given a female imprint, and during spermatogenesis all relevant regions are given a male imprint. Thus at fertilisation (5) the union of egg and sperm generates a baby (6) with one maternal and one paternal imprint for each imprinted region. Chromosomes are shown in green, with male and female imprints indicated by blue and pink respectively.
What is the purpose of imprinting? In general, paternal imprints lead to gene expression patterns associated with increased growth, and maternal imprints lead to gene expression patterns associated with decreased growth. Some theories regarding the origin of imprinting relate to adaptive co-evolution. Other theories are based on conflict relating to strategies for reproductive success: the mother has to make a greater investment of energy in the child than the father, and too much maternal investment in one child may be detrimental to her ability to reproduce again; however the father’s reproductive success may be facilitated by larger babies that are better equipped to survive. There is also a potential conflict between the needs of the developing foetus and the continued health of the mother. Imprinting is observed in all mammals and in some other species; there is much debate about its origins, but correct imprinting is critical for normal development.
For imprinted genes, with one copy epigenetically silenced throughout the lifetime of the individual, it is clearly vital that a functional copy of that gene is inherited from the other parent. Where this is not the case, for example due to gene mutations or errors affecting the resetting of imprints during gametogenesis, imprinting disorders will be seen.
Imprinting disorders and uniparental disomy
There are a number of genetic conditions which are related to imprinting, including Beckwith–Wiedemann syndrome, Silver–Russell syndrome, Angelman syndrome (AS) and Prader–Willi syndrome (PWS). One of the best characterised imprinted regions is located close to the centromere on the long arm of chromosome 15 (15q11.2). The genes in this region include SNRPN, two clusters of genes for small nucleolar RNAs (snoRNA), and UBE3A. The products of SNRPN and snoRNA genes appear to play roles in RNA processing within the nucleus, while the UBE3A protein functions in targeting proteins for degradation by the proteasome. Thus the products of genes in this region clearly have wide-ranging effects within the cell. SNRPN and the two snoRNA clusters are active only on the paternal chromosome 15, while UBE3A is active on the maternal copy (Figure 27). Absence of the paternally expressed genes leads to PWS (Table 8). In contrast, if there is no maternal copy of 15q11.2, and thus no active UBE3A gene, then the individual will be affected by AS. While the majority of PWS and AS cases are a consequence of microdeletions, there are other mechanisms (Table 8), including uniparental disomy (UPD).
Figure 27. Imprinting on chromosome 15.
A cluster of genes near the centromere of chromosome 15 (at band 15q11.2) is subject to imprinting in a parent-of-origin specific manner (indicated by blue and pink shading): a number of genes including SNRPN and many snoRNA genes are expressed exclusively from the paternal chromosome 15, and are silenced on the maternal 15. Conversely, the UBE3A gene is silenced on the paternal copy and active on the maternal copy. Thus loss of function of these genes has different consequences depending upon the parental origin: loss of UBE3A on the maternal 15 leads to Angelman syndrome (Table 8) whereas loss of UBE3A on the paternal 15 is without consequence since the gene is inactive anyway on the paternal copy. The mechanism involves DNA methylation (indicated by hatching) of the SNRPN region of the maternal chromosome 15.
Table 8. Summary of two disorders of imprinting, Angelman syndrome and Prader-Willi syndrome.
| Angelman syndrome | Prader-Willi syndrome | |
|---|---|---|
| Key features | Moderate to severe intellectual disability (IQ generally in the range 25–54) Jerky, puppet-like movements Happy and sociable disposition Seizures |
Mild to moderate intellectual disability (IQ generally in the range 60–70) Insatiable appetite leading to morbid obesity Behaviour problems |
| Frequency in the population | Approximately 1 per 20000 | Approximately 1 per 15000 |
| Underlying genetic abnormality (note that in some cases, the underlying cause has not been determined) | Maternal 15q11.2 deletion (approximately 70%) Paternal UPD (approximately 4%) Imprinting defect (approximately 8%) Pathogenic variant in UBE3A (∼6%) |
Paternal 15q11.2 deletion (approximately 70%) Maternal UPD (approximately 20%) Imprinting defect (approximately 5%) |
| Key genes | UBE3A encoding a ubiquitin ligase | SNORD116 gene cluster encoding snoRNAs (other genes in the imprinted region may also influence the phenotype) |
UPD is a rare phenomenon in which both homologues of a chromosome pair are derived from the same parent. If both chromosomes 15 are maternal in origin then the child will be affected by PWS, and if both 15s are paternal in origin the child will be affected by AS. There are essentially three mechanisms which can lead to UPD, each requiring two errors affecting meiosis/mitosis. Monosomy rescue is a rare, sporadic event which can allow a monosomic zygote to survive by duplication of the monosomic chromosome; the mechanism permitting this is not entirely clear, but it will always result in UPD. Trisomy rescue is, likewise, a rare, sporadic event, in which trisomic cells lose one chromosome, for example by anaphase lag, in which one chromosome does not get incorporated into a daughter nucleus during mitosis. Depending upon which of the three chromosomes is lost, trisomy rescue leads to UPD in one third of cases. A final possibility is for a nullisomic gamete to combine with a disomic gamete (in other words, meiotic errors in both parents), which appears statistically unlikely. UPD of some chromosomes is without consequence (for example 1, 5, 9, 10, 13, 21, 22), but for chromosomes that harbour imprinted genes (which include 6, 11, 14, 15, 20), the relevant imprinting disorder will ensue.
UPD can be in the form of heterodisomy (both homologues from one parent are present) or isodisomy (two copies of one of the parental homologues). In isodisomy, because the two chromosomes are identical there will be homozygosity for all alleles and therefore in this case UPD may also unmask a recessive disorder.
Although most cases of imprinting disorders occur as a consequence of de novo mutations, there are also cases in which mutations have been transmitted through families. For example, if a new UBE3A mutation is present in a spermatozoon that fertilises an egg, the resultant child would not show any phenotype, since the paternal copy of UBE3A is epigenetically silenced anyway. If the child is a male, he could also transmit this same mutation to offspring without consequence to their health. However, if the child is female, and she transmits this mutation to her offspring, they would be affected by AS.
Interestingly, imprinting of UBE3A appears to be maintained only in the brain; other tissues have biallelic expression. Furthermore, overexpression of UBE3A, for example as a consequence of gene duplication on the maternal chromosome 15, is associated with autism.
Epigenetic contributions to other disorders
During development, epigenetic modifications at many genetic loci change as part of the differentiation process, generating tissue types with specific patterns of gene expression. Epigenetic changes may also occur in response to environmental stimuli. Deficits affecting components of epigenetic mechanisms can lead to disease states. A well-characterised example involves the MECP2 gene which encodes a methyl-C binding protein. This protein recognises and binds to methylated cytosines in DNA, acting to recruit other complexes like HDAC which can alter the transcriptional status of the DNA. Pathogenic loss-of-function variants in MECP2 lead to Rett syndrome, as discussed earlier. Another example is the rare autosomal recessive disorder: immunodeficiency, centromeric instability and facial anomalies (ICF) syndrome. Roughly half of ICF cases are a consequence of biallelic pathogenic variants in the DNMT3B gene. From mouse studies, complete loss of function of DNMT3B appears to be incompatible with life, and thus the variants that lead to ICF are generally missense, with each patient having at least one missense variant which retains some DNMT3B function. Note that epigenetic changes can also involve non-coding RNAs, for which X-inactivation provides a good example.
It is becoming increasingly apparent that changes in epigenetic programming play a role in many conditions, including cancer, mental illness, autoimmune disease, diabetes and obesity. Identical twins have different patterns of DNA methylation and histone modification that may contribute to different disease susceptibility between them. Because epigenetic marks like DNA methylation can be faithfully copied between cell generations (in other words they are heritable), epigenetics can also help explain how the foetal or childhood environment can influence later susceptibility to disease. There is also evidence that epigenetic marks, including those on imprinted genes, may be perturbed during IVF, and that this may lead to health problems, for example imprinting disorders, in some children conceived by IVF.
Complex disorders
Introduction
The clear inheritance patterns associated with single gene (monogenic) disorders have facilitated the identification of the causative genetic changes for these conditions. However, increasing attention is now focused on genetic contributions to complex multifactorial disorders like diabetes, heart disease and schizophrenia, where disease is the outcome of a complex interplay of multiple genetic and environmental influences. The impact of an individual variant in one gene may be very small, but when present together with multiple variants in other genes, in the context of a particular environment, may lead to an increased risk of disease. The same is true for many traits (for example, height) and behaviours (for example, aggression or novelty seeking).
Type 2 diabetes (T2D) exemplifies complex disorders and is one that is increasing in incidence across the world. The major environmental contributions to T2D relate to diet and exercise: high-calorie diets in the context of a less active lifestyle, often involving long periods spent in front of a computer or television. This lifestyle presents a stark contrast with the environment that our ancestors had to survive, and traits that might once have been advantageous (like an energy-saving metabolism) have become a disadvantage. Identifying the genetic factors underlying T2D is challenging because of the small effects of individual contributions.
Whereas type 1 diabetes (T1D) is characterised by loss of insulin production as a consequence of autoimmune destruction of pancreatic islet cells, T2D is most commonly associated with insulin resistance – the body is no longer able to respond appropriately to insulin. T2D typically has a late onset (>35 years) and is associated with obesity, though onset at younger ages is increasing.
Family studies
Initial clues that genetics plays a role in complex disorders like T2D tend to come from family studies, in particular, the study of twins. If one of a pair of monozygotic (identical) twins is affected by a monogenic disorder like CF it is practically certain that the other twin also has CF, in other words, is concordant, because they have virtually identical DNA. However, for dizygotic (non-identical) twins, who share, on average, 50% of their DNA, the chance of being concordant for CF is 50%. For a disorder that is completely environmental in origin there would be expected to be little difference in concordance when comparing dizygotic with monozygotic twin pairs. For T2D concordance is approximately 70% for monozygotic twins, but only approximately 25% for dizygotic twins, therefore indicating significant genetic involvement. Furthermore, the risk of T2D is higher for any individual if a parent is also affected (higher still if both are affected). The relative contribution of genetic factors to a disease phenotype is known as ‘heritability’, and for T2D estimates of the heritability vary between 25 and 80%.
Identifying genetic loci associated with complex disorders
One way of identifying the loci involved is a ‘candidate gene’ approach. For T2D potential candidates might be genes involved in glucose metabolism or the response to insulin, or in predisposition to obesity. Investigation of the PPARG gene, based on its known role in adipocyte differentiation and glucose homoeostasis, identified a common polymorphism which was protective for T2D (Table 9). However, due to the complexity of regulatory and metabolic networks in the cell, the number of potential candidate genes for T2D is immense, and fully investigating all possible candidates would require massive investment of time and money. Furthermore, by looking only at genes that are expected to play a role based on current understanding some key players might be missed.
Table 9. Examples of genetic loci implicated in T2D risk by GWAS.
| Gene | Function of encoded product(s) | Comments |
|---|---|---|
| PPARG | Peroxisome proliferator-activator receptor-γ; a transcription factor of the PPAR family which have roles in regulating cell differentiation, and metabolism of glucose and lipids | This receptor is a target for thiazolidinediones, which are insulin-sensitising drugs used in T2D treatment. Variant p.Pro12Ala (representing 12% of Caucasian and East Asian alleles) is protective for T2D |
| TCF7L2 | Transcription factor, involved in stimulation of pancreatic β-cell proliferation and in production of GLP-1, which stimulates insulin secretion | The T allele of SNP rs7903146 is not only a strong risk factor for T2D, but this allele has also been associated with better response to two common T2D medications: sulphonylurea and metformin |
| HNF1A | Hepatocyte nuclear factor 1 α; transcription factor required for normal development and function of liver and pancreatic islets. | Variant p.Glu508Lys is globally extremely rare (approximately 5 per 10000 alleles), and predominantly occurs in individuals of Native American ancestry. This allele was five times more common in a Mexican cohort with T2D than Mexican controls |
| HNF1B | Hepatocyte nuclear factor 1 β; transcription factor required for normal development and function of liver and pancreatic islets | A common variant (rs4430796) which is protective for T2D is associated with increased risk of prostate cancer |
| KCNJ11 | Subunit of potassium channels, required in pancreatic β-cells for regulation of glucose-stimulated insulin secretion | These potassium channels are targeted by sulphonylurea, a treatment for T2D. Activating variants are associated with neonatal diabetes, while loss-of-function variants lead to hyperinsulinaemia in infancy |
| KCNQ1 | Voltage-gated potassium channel, required in pancreatic β-cells for regulation of glucose-stimulated insulin secretion | For two SNPs in KCNQ1 (which lies within an imprinted region of the genome), increased risk of T2D is only seen when the risk allele is maternally transmitted |
| SLC30A8 | Zinc transporter; zinc is required as a cofactor by many proteins, and as a signal ion | A common missense variant, p.Trp325Arg, is associated with increased risk for diabetes, while several rare loss-of-function variants are protective |
| CDKN2A/2B | CDKN2A encodes two proteins: cyclin-dependent kinase inhibitor p16INK4 and the p14ARF protein, both of which function in the p53/RB pathways. CDKN2B generates an antisense, non-coding transcript from the same locus | Variants within this genomic region have shown association with cardiovascular disease, cancer, periodontitis and glaucoma (but note that it is quite feasible that an individual variant might lead to increased risk for one disease while being protective for another!) |
| CDKAL1 | Methylthiotransferase that modifies tRNA for lysine to increase stability of the codon–anticodon interaction, and thereby increase fidelity of lysine incorporation during translation | Proinsulin contains two lysine residues, one of which is at the cleavage site used to generate insulin. Mistranslation of this lysine codon may generate cleavage-resistant proinsulin |
| FTO | Fat mass- and obesity-associated gene; encodes a nucleic acid demethylase | FTO is the most significant locus identified in GWASs designed to identify obesity-related genes |
To facilitate the identification of susceptibility loci, approaches based on genetic linkage or ‘association’ have been used (Figure 28). Such approaches rely upon the observation that genetic recombination does not occur randomly across our genome, but instead tends to occur at ‘hotspots’ spread throughout chromosomes. The consequence is that particular segments of the genome tend to remain together through many generations. This means that we can use ‘genetic markers’, most often SNPs, as tags for the genome segments, and then look within populations to test whether particular markers associate with the disease. This approach is typified by the ‘genome-wide association study’ or GWAS (Figure 29).
Figure 28. The principles of genetic association.
SNP1 (with alleles C and T) and SNP2 (with alleles A and G) are two polymorphic sites present on one chromosome. Within one of the group of ancestors, a new mutation (yellow star) has occurred very close to the position of SNP1; this is a new pathogenic variant which contributes to a particular disease condition (yellow individual). Because SNP1 is so close to the pathogenic variant, there will be little or no recombination between these sites down the generations, whereas recombination is likely to occur between SNP2 and the pathogenic variant. Thus when the descendants are genotyped, SNP2 has identical allele distribution in both healthy and affected individuals (no association of SNP2 with the disease). However, for SNP1 there is an excess of the T allele (and a corresponding deficit in the C allele) in the affected population, in other words SNP1 shows association with the disease. In reality, for complex conditions where there may be many different predisposing variants in several different genes, the scale of association would be less extreme, requiring analysis of thousands of individuals.
Figure 29. Genome wide association study for T2D-related loci.
Appropriate study groups are selected from the general population (1). For example, a group of individuals who have T2D (2) and a group of individuals who are healthy to act as controls (3). These groups must be matched as far as possible in terms of their constitution to avoid confounding effects – for example, matched for gender, ethnicity, smoking, socioeconomic status, education and so on. For each individual, thousands of SNPs across the genome are genotyped (4), and then the overall allele frequencies are compared between the two groups for SNPs across each chromosome (5). Each spot on the graph represents one SNP at a known location on a particular chromosome. The expectation is that, for the vast majority of SNPs, the MAF will be similar between the two groups. However, where there is a difference, either a significantly higher (6) or significantly lower (7) MAF in the affected group, this identifies a specific chromosome location which may play a role in T2D. Note that the actual variants which either predispose to obesity or protect from T2D may be close to the relevant SNPs (i.e. genetic linkage) rather than the SNPs themselves being causative or protective.
One problem with GWASs is that even with high statistical stringency, reproducibility is not guaranteed and results from different studies can appear contradictory, although this may reflect that there are too many other variables between the study groups. Large meta-analyses attempt to pull results from multiple GWAS together to determine significance on a large scale. It is important to note that association of a SNP with a particular trait or condition does not necessarily indicate causation, instead it is quite likely that the SNP, through close linkage, has been inherited along with the change which contributes to the trait (Figure 28). Once a locus has been identified by GWAS the next step is to look at the genomic region to see which genes in that region are likely to be relevant to the observed phenotype, and to see if the association can be confirmed by other studies, which will need to include functional studies in cells and/or animal models. Over 120 genetic loci have been identified by GWAS for T2D; some of these are shown in Table 9.
GWAS can be an effective way of linking common variants with associated diseases, but rare variants may also play a significant role within some families and subpopulations, for example the p.Glu508Lys allele of HNF1A in Native Americans (Table 9). Identification of additional rare variants is likely to come from approaches involving genome sequencing. It should be noted that variants might either increase or decrease disease risk, depending upon their functional effect (see KCNJ11 and SLC30A8 in Table 9). The mechanism by which variants might contribute to complex disorders is often not obvious; for some loci implicated in T2D, there is a clear link to pancreatic function/glucose homoeostasis or obesity, but the significance of other loci identified by GWAS is less clear. At first glance, the function of the CDKAL1 product (tRNA modification) has no relationship to diabetes, but a potential impact emerges when the effect of lysine mistranslation on proinsulin cleavage is considered. Another locus, CDKN2A/2B, encodes products involved in cell cycle/cell proliferation, and it is feasible that there might be a link to the overall number of islet cells in the mature pancreas (having more islet cells may equate to better ability to sustain insulin production).
Epigenetics
Despite the large number of T2D-associated loci that have been identified by GWAS, these are not able to explain all of the heritability of T2D. However, it is becoming increasingly clear that epigenetics also plays a significant role in complex diseases like T2D. Imprinting may be involved: the risk that offspring will be affected by T2D is greater when the mother is affected than when the father is affected. Interestingly, this is the reverse of the situation for T1D, for which the risks for a child are higher if the father is affected, than if the mother is affected. A parent-of-origin effect has been observed for some alleles of KCNQ1 which only increase T2D risk when transmitted maternally.
The impact of epigenetics appears to begin preconceptionally: mouse experiments have demonstrated effects on health and also in DNA methylation patterns for offspring following paternal exposure to high-fat diet or low nutrition. Early foetal life is also a critical time. Key observations have come from the study of individuals who were exposed to an adverse intrauterine environment, particularly during first trimester, as a consequence of severe famine during the ‘Dutch hunger winter’ of 1944/45. These individuals had normal birth weight (since the famine had ended by the later stages of the pregnancy), but had significantly increased rates of obesity and T2D as adults, suggesting that a form of foetal programming had occurred. This is supported by the observation of altered methylation patterns 60 years later when comparing the DNA of these individuals with that of their siblings.
Lifestyle choices and environmental exposures also lead to epigenetic change. Clearly, sedentary lifestyles combined with high calorie obesogenic diets contribute directly to T2D risk, but there are also many studies that demonstrate epigenetic changes associated with different foods. Tobacco smoking leads to decreased methylation in several genes associated with T2D, including KCNQ1, and exercise has been shown to promote methylation changes in T2D-associated genes as well as altering histone deacetylase expression. Many epigenetic marks in the genome can change throughout the life of an individual as a result of changing lifestyle (Figure 30), and may provide a useful target for management of T2D risk. The epigenetic marks present in the DNA of monozygotic twins have been shown to diverge as they age, and, when comparing individuals with/without particular complex disorders, differential epigenetic marks can be demonstrated in key genes. Epigenome-wide association studies (EWAS), which operate on similar principles to GWAS, may help to elucidate the epigenetic basis of complex diseases.
Figure 30. Many factors, environmental, genetic and epigenetic interact together in the overall risk for T2D.
Epigenetic factors can alter throughout the life of an individual, and may be affected by parental environment preconception, and maternal environment during pregnancy. A multitude of inherited variants (some of which may be protective) combine with epigenetic status to generate an overall genetic risk for T2D, but the disease state will generally only be manifested in the presence of environmental triggers. Although inherited genetic variants are hard to change, it is apparent that environmental change (improved diet and more exercise) may impact on disease severity not only directly, but also indirectly by epigenetic changes. Pictograms from PictArts.
Summary
There is still a long way to go in understanding genetic contributions in complex diseases. T2D has only reached epidemic proportions over the last few decades, but the majority of genetic variants implicated as risk factors for T2D have been around for much longer in the human gene pool. Therefore, it is the environmental context of high calorie intake plus sedentary lifestyle which potentiates the effect of the genetic risk variants in T2D. Furthermore, it is important to recognise that variants which appear to be ‘bad’ as risk factors for one disease may in fact be protective against another (see HNF1B in Table 9), which underlines the fact that there is no such thing as a ‘perfect’ human genome!
Cancer: mutation and epigenetics
Introduction
Cancer affects approximately 1 in 4 people worldwide, with an estimated 14.1 million new cases in 2012 and a prediction that there will be 23.6 million new cases per year by 2030. In the U.K. there are nearly 990 new cases diagnosed every day, which equates to a new diagnosis approximately every 2 min. In the U.S.A., there are over 4600 new cases every day (a new case every 19 s). These rates are rising, due both to the increase in population size and increasing longevity. More than one-third of cancer cases in the U.K. are diagnosed in people aged 75 and older.
There are many different types of cancer and these types reflect the tissue and cell type from which the cancer originated. The most common types in the U.K. are cancer of the breast, prostate, lung and bowel, which together comprise over 53% of all cancer cases. Overall, half of the people diagnosed with cancer survive their disease for more than 10 years and this is improving, from only 24%, 40 years ago. However, each type of cancer shows a very different mortality rate, for example 98% of people diagnosed with testicular cancer survive for more than 10 years after diagnosis, while the figure is only 1% for pancreatic cancer.
Although the different types of cancer show different disease patterns and survival rates, the commonality of all cancers is that the cells have lost the normal controls over growth and movement. Cancer cells proliferate in an uncontrolled way and this can lead to the formation of a lump, or tumour. When the growth of the cells within the mass is limited and the cells retain certain normal features, do not invade adjacent tissues, nor spread to other parts of the body, the tumour is called ‘benign’. However, if the growth becomes more uncontrolled, such that the cells divide indefinitely and spread to other parts of the body, the cancer is termed malignant and the tumours that form at secondary sites are called metastases and it is metastatic disease that is the primary cause of cancer mortality.
There are several contributing factors that enable cancer cells to grow in this uncontrolled way and Hanahan and Weinberg described these in a seminal review in 2011 as the ‘hallmarks of cancer’. These features of cancer cells can be summarised as follows: (i) they have an inherent ability to divide; (ii) the cells do not respond to factors in the body that might inhibit their growth; (iii) they develop ways to avoid being destroyed by the immune system; (iv) they overcome the in-built clock that limits normal somatic cell division; (v) the cells release factors that in turn promote the surrounding normal cells to release other factors that will support the growth of the cancer cells, in other words, cancer cells can promote a permissive environment for their growth; (vi) they acquire the ability to move and invade other tissues; (vii) they promote the growth of a blood supply to the tumours to provide the oxygen and nutrients the cancer cells need to grow; (viii) the cancer cell genome becomes more prone to mutation; (ix) they become resistant to the normal mechanisms of cell death and (x) the cells adjust their metabolic pathways to better support rapid cell proliferation. All these changes from a normal cell are brought about by different somatic mutations in the cancer cell genome, and/or by epigenetic modifications. As such, cancer is essentially a disease of mutation.
The genome of a cancer cell is full of somatic mutations. Indeed, in some cancers, such as lung cancer and melanoma, there may be hundreds of thousands of mutations (Figure 31). Many such mutations are observable at the level of the karyotype, including whole and part chromosome duplications, deletions, inversions and translocations. In addition, cancer cells typically carry large numbers of point and micromutations.
Figure 31. Different cancer types typically display different numbers of mutations in the cell genome.
These can range from less than one hundred observed in some retinoblastomas to hundreds of thousands in lung cancer.
Many of these mutations will have been involved in the disease process to some degree (called causative or driver mutations), however, the cell also accumulates mutations that are not causative, i.e. ‘passenger’ mutations (as many as 99.9% of the mutations present) and one challenge is to distinguish between the two (see next section on ‘Genomics’). While there are a number of genes known to be powerful oncogenes when mutated or critical tumour suppressor genes, cancer cells never have only one causative mutation. This is because for the cancer cell to acquire all of the changes (as described above), mutations in numerous genes are needed to overcome normal growth regulatory processes. This is also reflected by the incidence of cancer. The statistics show that the biggest risk factor for cancer is age. The older an individual is, the more likely they are to develop cancer, indeed most cancer types are quite rare in young people (Figure 32). The very shape of the incidence curve reveals that cancer is caused by multiple changes to the cellular genome. Mutations occur and accumulate in the cells of the body throughout life. The vast majority of these will be harmless and may not affect the phenotype of the cell at all. However, over time, as mutations accrue, there is a risk, a statistical chance, that eventually a cell undergoes enough causative mutations that it begins to develop cancerous properties and with time, this may progress to a cancerous state. Agents that speed up the mutation rate in cells will also increase the risk of cancer, so for example, overexposure to sunlight (a component of which is mutagenic UV rays), will increase the risk of melanoma, a type of skin cancer. Similarly, cigarette smoke contains multiple mutagens and smoking tobacco increases the risk of lung cancer.
Figure 32. Cancer increases with age.
Theoretical cancer incidence curves are shown reflecting a set mutation rate. If only one mutation could cause a normal cell to become cancerous, the cancer incidence rate would be linear (blue line). The actual incidence increase with age (red curve) reflecting the accumulation of cancer causing changes that act together over time.
Oncogenes
Oncogenes are genes whose activation contributes to the development of cancer. Oncogenes are usually mutated versions or pathogenic variants of normal cellular genes (the unmutated genes are often called proto-oncogenes for this reason), and this reflects that the normal function of the gene is involved in the control of cell growth in some way. Thus genes that promote or are involved in cell division (mitosis) or inhibit programmed cell death (apoptosis), differentiation, quiescence or senescence are genes that when mutated, could become oncogenic. In addition, some pathogens carry oncogenes, for example a small number of viruses can lead to a higher risk of specific cancers, because they encode genes that promote cell proliferation or survival. It is estimated that approximately 15% of cancers have developed with the involvement of an infectious agent, for example some human papillomaviruses can increase the risk of developing cervical cancer.
Many proteins expressed by potential oncogenes act in a process called signal transduction, or are transcription factors that are activated by this mechanism. Signal transduction is the method by which a cell converts a signal, typically received from the outside of the cell, into a change in gene expression which will lead to a response (Figure 33). For example, if a growth factor acts upon a cell, this triggers a signal that passes through the cytoplasm, to induce the expression of genes required to initiate cell division. This is usually (but not exclusively) achieved by the growth factor interacting with a receptor at the cell surface. The interaction activates the receptor, which leads to a cascade of changes in the state of other factors that are located in the cytoplasm. Transcription factors are often activated by this process through phosphorylation upon specific serine or threonine residues, resulting in their translocation to the nucleus to regulate gene expression (Figure 34). Importantly, once the signal has concluded, all components are deactivated. Mutations in genes involved in this process that lead to overactivation of the protein, or just too much of it, can result in excess signalling, instructing the cell to divide continuously.
Figure 33. Signal transduction.
The cell receives signals from contact with other cells, from the extracellular matrix and from soluble molecules, including secreted proteins. The information received is integrated and transduced to the nucleus. The cytoplasmic signalling pathways and networks that are activated will depend upon the cell’s status and which genes are currently expressed. The combined signalling input can result in an altered programme of gene expression to achieve one of several possible responses.
Figure 34. A simplified, typical signal transduction pathway.
Many growth factors (secreted proteins) interact with a receptor at the cell surface and both ligand and receptor can act in the form of a monomer or in a complex. The interaction causes the receptor to become active, and this will lead to a signalling cascade (depicted by the dashed red lines), passed from one molecule to the next. The signal may culminate in the activation of a transcription factor with a consequent change in gene expression, or influence mitochondrial membrane integrity and cell survival, or have an alternative destination to elicit a cellular response. The activation signal may be transmitted in several ways, the most common method is through the action of kinases, enzymes which phosphorylate their substrates (depicted by P), thereby activating the substrate for the next step. While a simple pathway is depicted, the reality is that a vast network of complex interactions occur, integrating numerous signals allowing for an extensive array of subtly different responses.
Essentially all types of mutation have been observed in the conversion of a proto-oncogene into an oncogene, including gene duplication, point mutation, partial deletion or rearrangement and chromosomal translocation. Oncogenic mutations tend to be gain-of-function and thus are usually dominant. Such mutations generally lead to overexpression of the gene or overactivation.
Oncogene activation by overexpression
There are three main mutational mechanisms by which gene overexpression is achieved (Figure 35). These are: amplification of the gene, with increased expression due to the increased copy number, novel juxtaposition of sequences that enhance expression (for example, through chromosomal translocation) and mutations in the gene expression control sequences that either prevent gene silencing or directly enhance expression. In addition, epigenetic modification of the gene promoter sequences can act to increase or suppress expression.
Figure 35. Oncogenic mutations.
Examples of oncogene activating mutations are depicted. (A) Gene amplification leading to increased expression of the product. (B) Reciprocal chromosomal translocation leading to enhanced expression of a gene at the breakpoint, as observed in follicular lymphoma and the 18:14 translocation involving the BCL2 gene and the IGH locus (above); or as observed in chronic myeloid leukaemia and the 9:22 translocation leading to a BCR-ABL fusion protein (fused in frame with respect to the ORF). (C) Loss of a protein regulatory region by small deletion. (D) Activating change in the coding sequence brought about by a point mutation (exemplified by RAS genes). In each case, the gene region is depicted by a coloured box and expression indicated by a bent arrow.
An example of mutation leading to overexpression is seen with the gene encoding the receptor protein HER2 (aka ERBB2, a member of the epidermal growth factor receptor (EGFR) family of tyrosine kinases), which is frequently mutated in breast cancer. One type of HER2 mutation found in cancer cells is amplification. The whole gene is duplicated resulting in more than one copy, sometimes several copies. This leads to excess production of the protein within the cell. As a consequence, the cell sends more signals to the nucleus to initiate mitosis, which contributes to the cancerous state by increasing cell proliferation. This knowledge led to the development of a drug called Herceptin, which blocks the action of HER2 and is an effective co-treatment for those breast cancers that overexpress HER2.
Another example of mutation causing gene overexpression is exemplified by the BCL2 gene in follicular lymphoma, a type of B-cell cancer. The BCL2 protein sits at the surface of the mitochondria and inhibits a form of programmed cell death called apoptosis. If a cell is destined to die (and there are several instances where this is normal), overexpression of BCL2 can inhibit cell death. Follicular lymphoma cells display a typical chromosomal translocation, juxtaposing part of chromosome 18, the location of the BCL2 gene, to chromosome 14, the location of an antibody gene (the immunoglobulin heavy chain (IGH) gene) (Figure 35). The result is that BCL2 comes under the control of an enhancer sequence which would normally drive the expression of the IGH gene in B cells, but in the mutant cells, leads to overexpression of BCL2. Thus the B cells are resistant to apoptosis and this contributes to the development of B-cell lymphoma.
Oncogene activation through increased activity
There are several mutational mechanisms by which the activity of an encoded protein can be increased or rendered constitutive. These include point (or small) mutations at critical regulatory residues, for example loss of negative regulatory phosphorylation sites, deletion of negative regulatory domains (which can occur by simple deletion or result from larger rearrangements such as chromosomal translocation), or activating mutations in catalytic domains or interaction domains.
Proteins that relay growth signals in cells have to be exquisitely regulated. These proteins generally exist in an inactive state, are briefly activated by signal transduction and then return to an inactive state, thereby tightly controlling cell proliferation. Mutations that lead to increased or constitutive activity can be oncogenic. A classic example of this occurs with the RAS genes (H-RAS, N-RAS and K-RAS). These three related genes encode small proteins that are pivotal in multiple cell signalling pathways and in the development of numerous cancers, and therefore RAS has been referred to as a ‘molecular switch’. The RAS proteins bind to either GDP or GTP (guanosine di- or tri-phosphate). When bound to GDP, the protein is inactive. As a consequence of signalling from receptors, RAS switches to bind to GTP and in doing so, changes conformation and becomes active and thereby can interact with the next protein in the chain to pass on the activation signal (Figure 36). In the absence of an activating signal, RAS proteins are rapidly deactivated via an intrinsic GTPase activity (which hydrolyses the bound GTP to GDP). At just a few key points along the gene, mutation changing a single amino acid (typically codons 12, 13 or 61), can prevent GTP hydrolysis, thus locking RAS into the active, GTP-bound state and lead to constitutive signalling.
Figure 36. RAS activation.
(A) RAS is bound to GDP in the inactive state. Signal transduction can lead to the activation of RAS, via a GEF (GDP/GTP exchange factor), which displaces GDP from RAS, allowing the binding of GTP. This causes a change in conformation of RAS that enables interaction with effector proteins, thereby passing the activation signal onwards. Deactivation of RAS occurs by hydrolysis of GTP to GDP, assisted by GTPase-activating proteins (GAP). (B) Proteins of the RAF family are among several effectors of RAS. RAF proteins are serine/threonine kinases. Activated RAS binds to RAF, leading to a change in conformation of the latter and activating its kinase activity. RAF then phosphorylates its substrate MEK (which is also a kinase) thus activating it, and so the signal proceeds. This describes part of well-known signal transduction pathway, the mitogen activated protein kinase (MAPK) pathway.
Another example of protein activation is observed with the C-SRC gene, which encodes a tyrosine protein kinase. The C-SRC protein locates to the inside surface of the plasma membrane and passes on the mitogenic signals from several growth factor receptors. The kinase activity of the protein is controlled by a regulatory site at the C-terminal end, the location of a critical tyrosine residue (Tyr527). The phosphorylation status of Tyr527 is determined by other protein tyrosine kinases and phosphatases and when Tyr527 is phosphorylated, this inhibits the kinase activity of C-SRC. Mutations that delete this residue result in a protein that is constitutively active and constantly relaying growth signals to the nucleus. The mutation can be as small as a point mutation, such that the tyrosine residue is lost or replaced by an alternative amino acid. Such mutations are frequently found in cancer of the colon, lung, liver, breast and pancreas. A further example of mutation leading to loss of a regulatory domain is seen with a chromosomal translocation that leads to the expression of a fusion protein (derived from two genes), called the BCR-ABL fusion. This particular chromosomal translocation (between chromosomes 9 and 22), called the Philadelphia chromosome, is a characteristic of chronic myeloid leukaemia and leads to the loss of a regulatory domain from the ABL1 gene, which encodes a tyrosine kinase. The fusion protein has constitutive kinase activity which promotes cell division.
Tumour suppressor genes
Tumour suppressor genes (TSG) are genes whose action inhibits the growth of tumour cells, therefore their inactivation is advantageous to a cancer cell. Consequently, the function of several TSGs is lost from all forms of cancer. The loss of function can be achieved by mutation affecting a critical region of the protein or by loss of expression, the latter is frequently brought about by deletion mutations (deleting part or all of the gene) or alternatively, by epigenetic modification of the gene regulatory sequences to suppress expression. Several TSGs function to inhibit the progression of the cell cycle, each acting at different points of the cycle, or in different tissues, or under different circumstances. The cell cycle is directed by a complex of proteins, including cyclins and their partners, the cyclin-dependent kinases (CDK). The CDKs, in turn through the cycle, phosphorylate and activate a plethora of proteins that orchestrate the cycle moving forward, from the initial growth phase (G1), through DNA synthesis (S), the completion of DNA synthesis and continued growth (G2) and mitosis (M) (Figure 37). One of the central players in the cell cycle is a TSG called the retinoblastoma (RB) gene. In the unphosphorylated state, the RB protein inhibits both entry into S phase and the progress of the cell cycle by obstructing crucial cell cycle progression factors. When a cell receives signals to undergo division, this activates the cyclin/CDK complexes and one of the substrates is RB. As RB becomes phosphorylated, it releases the cell cycle progression factors to allow the cell cycle to move forward. If RB is lost from the cell, this critical inhibitory mechanism is lost, rendering the cell subject to uncontrolled proliferation. Many cancer cells show complete loss of RB expression. This means that both alleles must be affected, either by mutation (typically deletion) or epigenetic suppression. At the level of the cell phenotype, loss of RB is recessive, with one functional copy sufficient to control the cell cycle, however, as discussed below under heritable cancers, the dominant/recessive issue is more complex.
Figure 37. The cell cycle and RB.
The cell cycle is depicted, showing the phases (divided by chevrons): growth or gap 1 (G1), DNA synthesis (S), growth or gap 2 (G2) and mitosis (M) in a circle, with exit from cycle represented as G0. Cell cycle check points are depicted as bars, G1/S check point (red: a DNA damage check), S-phase check point (blue: a DNA damage and replication fork check), G2/M check point (green: a DNA damage and completion of replication check) and spindle check point (yellow: ensuring correct alignment of the chromosomes upon the spindle, ready for division). The cyclin and CDK complexes relevant to each phase are shown. Central to cell cycle control is the TSG RB. RB becomes increasingly phosphorylated by the activated CDKs through G1 (as depicted by increasing P). As the cycle progresses, it becomes hyperphosphorylated and this allows entry into S phase and further progression. Un-phosphorylated RB blocks cell cycle progression. During G1, the cycle can be initiated via mitogenic signalling (purple arrows). Once past ‘start’ the cell is committed to cycle.
Several other TSG products also act to inhibit cell cycle components to tightly control proliferation, such that this proceeds only when required and when all conditions are optimal, acting directly to inhibit the kinase activity of CDKs. A number of these (when first characterised), were given the unimaginative name that simply refers to the apparent size of the protein (in kiloDaltons), including p21, p16, p27 etc., with a superscript to distinguish between it and other proteins of the same size. The TSG encoded protein p21Waf1 (expressed from the CDKN1A gene), inhibits CDK2 and therefore blocks entry into S phase and progression through G2, while p16Ink4a (expressed from the CDKN2A gene) inactivates the CDK4 or CDK6 complex bound to cyclin D and therefore plays a key role in the passage of cells into G0 (exiting cell cycle) and entering quiescence (reversible) or senescence (irreversible).
Numerous other TSG products function in processes that are not directly related to the cell cycle, instead affecting the growth conditions of the tumour and its environment. For example, if a solid tumour does not acquire a blood supply to provide sufficient oxygen and glucose to the dividing cells, it can grow little more than a few millimetres in diameter. The TSG termed von Hippel–Lindau (VHL) encodes an enzyme required in the processes of protein degradation; a ubiquitin ligase, its primary target, HIF (hypoxia-inducible factor), is a key protein that controls the growth of blood vessels from existing vessels (angiogenesis). Normally angiogenesis occurs when metabolic activity is high and oxygen availability low (like in a growing muscle in response to exercise), but otherwise is kept repressed. Mutations in VHL that result in the loss of the ability to cause HIF degradation, permit the persistent activation of HIF, thence the unrestrained angiogenesis necessary for tumour growth.
Genome integrity
As somatic mutation is a driving force in cancer, loss or aberrant function of genes involved in the processes of repair of damaged DNA can be tumorigenic. DNA damage can be caused by exogenous mutagenic agents, but the vast majority of mutations that are present in a cancer cell have occurred as a result of errors during DNA replication or have been caused by endogenous biochemical processes. As such, replication proofreading and repair proteins work continuously to maintain the integrity of the genome. Not all DNA repair genes are TSGs or proto-oncogenes, many are essential and therefore their mutation or loss will lead to cell death. However, if mutation of such a gene permits cell survival, but increases the chance of incorrect repair (i.e. mutation), this will increase the cancer risk.
The most commonly mutated TSG in human cancer is a gene termed TP53. The encoded TP53 (or simply p53) protein is a DNA-binding transcription factor. Normally present at low levels in the cell, it becomes stabilised and activated when the cellular genome is at risk, particularly following DNA damage (Figure 38). When DNA damage is detected by a complex of proteins, the information is transduced by either the ATM kinase or the ATR kinase to activate effectors, CHK1, CHK2 and p53 (Figure 38). P53 then acts to induce the expression of other genes that will halt the cell cycle, including p21Waf1. As such, there is an effective ‘check point’ at the G1/S boundary of the cell cycle (Figure 37). If DNA is damaged, activated p53 induces p21Waf1 expression, CDK2 is then inhibited and the cycle does not proceed. P53 is also involved in DNA repair processes, therefore, following DNA damage, the repair machinery can work while the cell cycle is delayed. Additionally, p53 can induce the expression of genes that will lead to cell death (counteracting the survival function of BCL2) if the cell is beyond repair, thus the cell can be neatly removed. The most common mutations in TP53 in cancer cells occur in the DNA-binding region of the protein and prevent p53 from binding to DNA and therefore disable its function as a transcription factor. As a consequence, cells with damaged DNA can progress through the cell cycle check points with a high risk of mutation. For this reason, p53 has been referred to as ‘the guardian of the genome’.
Figure 38. Sensing and responding to damaged DNA.
The proteins that initiate DNA repair after damage or replication blockage can be divided into sensors, transducers and effectors. The sensors: a complex of proteins recognise the broken ends of DNA double strand breaks (DSB) and complexes of different composition recognise stalled DNA replication forks (pink and yellow bubbles). These complexes attract two key kinases, ATM and ATR, the transducers, which function to phosphorylate, and thereby activate, two further kinases, CHK1 and CHK2. TP53 is stabilised both directly via phosphorylation by ATM and by phosphorylation by CHK1 and CHK2. Following on, the effectors lead to one of several responses, as indicated.
BRCA1 and BRCA2 (breast cancer 1 and 2) are also TSGs involved in DNA repair. The encoded proteins act in a complex to repair DNA double strand breaks (DSB). Mutations that lead to loss of function of the proteins or loss of expression result in a high chance of inaccurate repair of DNA DSB and thereby markedly increase the mutation rate of the cells and therefore also the cancer risk.
Some oncogene products act to down-regulate DNA repair, either as one of several functions, or in some cases, their primary function. The mitogenic protein HER2 (described above), when activated, down-regulates several DNA repair factors and this contributes to its oncogenic properties. In the last few decades, the importance of small non-coding RNAs in controlling gene expression has become apparent. Numerous tiny RNAs (called miRNAs) are expressed by cells and these function by down-regulating the expression of specific target genes. The miRNA-182 (miR-182) specifically down-regulates the expression of BRCA1 and a few other TSGs and miR-182 has been found overexpressed in several cancer cells (including breast cancer), often as a result of gene duplication. It can therefore be viewed as a distinct type of oncogene.
Cancer types show characteristic mutation profiles
Multiple powerful oncogenes have been known for several decades and historically, many were identified by virtue of the action of retroviruses in animal tumour model systems. Many of these are mutated in particular types of cancer and with different frequencies, for example RAS gene mutations are found in roughly 60% of pancreatic cancers, 50% of colon cancers, 20% of lung cancers, but rarely (1%) in kidney cancer. Several critical TSGs were discovered as a result of determining the pathogenic variant in familial cancer syndromes, such as heritable retinoblastoma. Furthermore, it has long been known that certain types of mutations are typical of particular cancer types, for example follicular lymphoma cells carry a chromosomal translocation involving the BCL2 locus and Burkitt’s lymphoma cells always harbour a translocation involving the C-MYC gene. However, the vast majority of oncogenes and TSGs make just a small contribution to the development of disease and are therefore harder to discern. Tackling this problem has been revolutionised by the huge advances in genome sequencing technologies over the last few decades. As will be described in the next section on ‘Genomics’, large sequencing projects comparing the mutated genome of a tumour with that derived from the normal tissue of the same individual has enabled the identification of causative mutations in that cancer. Taking this approach, not only have many more mutations (and therefore genes) which contribute to the cancer process been identified, but it has also been found that cancer types (and even subtypes) show characteristic somatic mutation profiles and this can then inform treatment options.
Epigenetic modifiers
In addition to mutation, overexpression of oncogenes and loss or reduced expression of TSGs are found in cancer cells through epigenetic modification of the expression control sequences. The genes whose products are epigenetic modifiers, are themselves frequently mutated in cancer cells. The mutation of these genes, (or their abnormal expression as a consequence of epigenetic modification of their control sequences) leads to aberrant chromatin modelling and the misexpression (under or over) of multiple genes. Epigenetic silencing of TSGs has been found to play a significant role in the genesis of cancer. Moreover, mutation in epigenetic modifier genes can have widespread effects and in some cases, they can be viewed as TSGs. For example, loss-of-function mutation in the DNA methyltransferase 3A (DNMT3A) gene is frequently found in blood cell tumours (lymphoid and myeloid malignancies). The loss of DNMT3A appears to increase the proliferative capacity of the cells and inhibit differentiation. Conversely, other genetic modifier genes have gain-of-function mutations in cancer cells. The histone-methyltransferase encoding gene EZH2 has been found to be activated by point mutation or is overexpressed through gene amplification in a variety of tumours. However, this gene cannot be simply classified as an oncogene, as its loss is observed in other tumours indicative of a TSG role in some cell types. Thus, altered epigenetic modification of genes is widespread in cancer cells, however its control is highly complex.
Heritable cancer and predisposition
Three factors essentially determine whether a cell becomes cancerous: the environment (including lifestyle), chance and the genotype. As described above, environmental mutagens (such as sunlight and tobacco smoke) clearly increase the risk of cancer and, although some are highly controversial, dietary factors (such as alcohol) may also influence risk. Statistical chance reflects that only a tiny proportion of all possible somatic mutations will be causative in cancer processes and this is why carcinogens are described as increasing the ‘risk’ of cancer. The genotype is all-important. Heritable loss-of-function variants in some TSGs increase the risk of cancer so substantially, that these present as dominant pathogenic variants, however, they contribute to a relatively small proportion of cancers overall (Table 10). This is exemplified by heritable loss of function variants in TP53, RB, BRCA1 and BRCA2. Breast cancer is a common cancer, and approximately 1 in 8 women in the U.K. will develop it during their lifetime, while only 3% of these cases are caused by inherited pathogenic variants in BRCA1, BRCA2 or TP53. Conversely, retinoblastoma is a very rare childhood cancer, with approximately 45 children diagnosed per year in the U.K. Of these, approximately 40% carry inherited pathogenic variants in the RB gene. Germline loss of function variants in TP53 cause Li–Fraumeni syndrome (affecting approximately 1–4 individuals per 20000), which is characterised by early-onset cancer of several different types. The risk of developing cancer in such an individual is approximately 50% by the age of 30, rising to 90% by the age 70. The apparent paradox, that loss of function of these TSGs is recessive in the cellular phenotype, yet dominant in the individual, is explained by Knudson’s ‘two hit hypothesis’, first put forward to account for this phenomenon with respect to the RB gene and familial retinoblastoma (from which the gene was discovered). The function of one allele is heritably inactivated and the other allele is inactivated through somatic mutation or epigenetic silencing, such that tumours arising in these individuals show loss of function of both RB alleles. The same applies to other TSGs and tumour types, including heritable loss of TP53 function and the development of Li–Fraumeni syndrome tumours and heritable loss-of-function variants of BRCA1 and BRCA2 in breast cancer.
Table 10. Hereditary cancer and the associated genes for which high risk variants have been identified (the list is not exhaustive).
| Primary organ affected | Syndrome, further information | Gene symbol | Estimated frequency within the cancer type |
|---|---|---|---|
| Bowel (fourth most common in U.K., 41804 in 2015) | Familial adenomatous polypsis | APC | 1% of bowel cancer |
| MYH associated polyposis | MYH | Rare | |
| Lynch syndrome (also increased risk of other cancers, see below) | MLH1, MSH2, MSH6, PMS2 | 3% of bowel cancer | |
| Peutz Jeghers syndrome (also increased risk of other cancers) | STK11 | Very rare | |
| Juvenile Polyposis Syndrome | BMPR1A, SMAD4 | Unknown | |
| Breast (most common cancer in women, approximately 12.5%, 55122 in 2015) | BRCA1, BRCA1, TP53, PTEN, PALB2 | 5–10% of breast cancers are associated with the inheritance of high risk variants | |
| Kidney (seventh most common in U.K.) | VHL syndrome | VHL | 2–4% of kidney cancer |
| Tuberous sclerosis | TSC1, TSC2 | ||
| Birt Hogg Dube syndrome | FLCN | ||
| Isolated hereditary papillary renal cell cancer | MET | ||
| Hereditary leiomyomatosis and renal carcinoma | FH | ||
| Melanocyte/skin (melanoma: approximately 15400/year) | Familial melanoma | CDKN2A and unknown | Approximately 10% of melanoma |
| Ovary (approximately 2% women) | BRCA1, BRCA2 | 5–15% of ovarian cancer | |
| Lynch syndrome | (as above) | ||
| Pancreas (1.4% people) | Unknown or as part of several other syndromes | Approximately 10% of pancreatic cancer | |
| Prostate (approximately 12.5% men, 47151 in 2015) | BRCA2 (MLH1, MSH2, MSH6) | ||
| Retina (approximately 45 children/year) | Familial retinoblastoma | RB | Approximately 40% of retinoblastoma |
| Thyroid (approximately 3400/year) | Medullary thyroid cancer (3–10% of thyroid cancer) | Unknown | Approximately 25% of medullary thyroid cancer |
| Uterus (approximately 2% women) | Unknown | ||
| Lynch syndrome | (as above) | ||
| Cowden syndrome | PTEN |
The population frequency % given under organ affected, indicates the proportion of individuals in the U.K. that are likely to develop this cancer at some point in their life. Numbers indicate new cases diagnosed in the U.K. Statistics were derived from the Cancer Research UK website.
Some heritable pathogenic allele variants increase the cancer risk by a small but significant degree, for example patients with a hyperactivated variant of the PIK3CD gene (a proto-oncogene which functions in signal transduction) suffer from the dominant disorder activated PI3Kδ syndrome (APDS) but also have an increased risk of B-cell lymphoma.
Aside from the clear cancer-associated, high risk, inherited gene variants, the genetic profile of an individual has a profound effect upon the predisposition to cancer. Thousands of allele polymorphisms or variants may increase or decrease the cancer risk (relative to each other) by a tiny degree, or under specific circumstances. Combinations of many such medium or low risk alleles together may have a compound effect upon risk. These low, medium and condition-specific risk alleles are being identified and explored using GWAS approaches and mass sequencing endeavours like the 100,000 Genomes Project (see next section). As such, although the statistics currently give this impression, there is really no clear distinction between ‘heritable’ and ‘spontaneous’ cancers, it is better described as a sliding scale of inherited risk. The large proportion of cancers currently not classified as heritable, nevertheless arose against a genomic background with a certain risk value. The massive GWAS and sequencing studies currently underway hold enormous promise for the future; there will come a time when the genotype of an individual can be used to determine the lifetime risk of certain diseases, particularly cancer and this can then be used to suggest preventative lifestyle measures or treatments.
Genomics
Introduction
Whereas genetic studies have traditionally focused on the effects of variants in individual genes, there is a shift towards consideration of the impact of the whole genome in health and medicine. Many conditions with a strong genetic basis, like T2D, epilepsy, hypertrophic cardiomyopathy and intellectual disability are associated, not with pathogenic variants in single genes, but rather, with variants in any one of a growing number of genes. There are (in 2018) 84 genes in which variants are reported to be associated with epilepsy as a core symptom, and several hundred other genes in which variants lead to conditions with epilepsy appearing as part of a wider spectrum of symptoms. There are over 600 genes in which pathogenic variants have been reported to be associated with intellectual disability, and the list is expanding. This diversity underlies the fact that for many individuals and families affected by genetic disease, there has been a ‘diagnostic odyssey’, with a series of misdiagnoses over many years, and perhaps a series of genetic tests, which may or may not have culminated in a definitive outcome.
As explained earlier, cancer results from an accumulation of somatic mutations which lead to disruption of the pathways which normally regulate processes including cell proliferation, cell death and cell motility. These pathways collectively involve input from the products of hundreds of genes (over 1% of our genome), and it is a challenge to identify all the genes in which mutation can contribute to cancer (the causative mutations). In addition to this, the inherited genome of all individuals influences the risk of developing certain types of cancers, upon which the somatic mutation profile builds.
Whole genome approaches, both GWAS and full sequencing are now being used by collaborative groups of scientists and consortiums to investigate the genetic predisposition to disease and the contribution of somatic mutation. To help address the gaps in our understanding of genes involved in rare diseases and complex disorders and genes associated with cancer, as well as to enhance understanding of our genome as a whole, the 100,000 Genomes Project was initiated in 2012 by Genomics England (owned and funded by the U.K. Department of Health).
The 100,000 Genomes Project and Scottish Genomes Partnership
The 100,000 Genomes Project set out with two targets. Firstly, to fully sequence entire ‘normal’ and ‘tumour’ genomes from approximately 50000 cancer patients, in order to be able to identify all the alterations in the cancer genomes; and secondly to fully sequence the genomes of approximately 17000 rare disease patients together with two very close relatives (ideally mother and father) (Table 11). Analysis of parent-child trios facilitates identification of de novo mutations that may contribute to disease, as well as recognition of variants that may be benign (present in a healthy parent). The Scottish Genomes Partnership (SGP) plans to sequence genomes of at least 3000 individuals, with goals that are similar to those of the 100,000 Genomes Project.
Table 11. Breakdown of genome targets of the 100,000 Genomes Project.
| Cases (affected patients) | Genomes sequenced per case | Total genomes sequenced | |
|---|---|---|---|
| Cancer | Approximately 25000 | Tumour | Approximately 50000 |
| Blood | |||
| Rare disease | Approximately 17000 | Patient | Approximately 50000 |
| Mother* | |||
| Father* | |||
| Total | Approximately 42000 | Approximately 100000 |
In a few cases, it may be another close relative.
Each genome sequenced generates roughly 200 GB of data, which represents huge challenges for data storage and analysis, as well as data security. Nevertheless, the benefits should include not only a molecular diagnosis for thousands of rare disease patients, but also a huge amount of data that will contribute to our understanding of the role of genomic variations and mutations in disease, and to the development of better diagnostic approaches and improved therapies that are targeted to key alterations.
Genetic modifiers
The complexity of, and interactions between, biochemical pathways and physiological processes within the body mean that it is important to consider not only pathogenic variants in a single gene, but to also take into account the potential effects of other genetic variants elsewhere in the genome. This is exemplified by long QT syndrome (LQTS; named for an alteration seen on electrocardiogram traces of the heart’s rhythm, where the ‘QT interval’ is very prolonged) which presents as a defect in the electrical activity of the heart affecting approximately 1 in 2000 individuals, and can result in sudden death. The commonest cause of autosomal dominant LQTS is a loss-of-function variant in one copy of the KCNQ1 gene. Some common variants in another gene, NOS1AP, have been demonstrated to have a small but significant effect on QT interval, even in healthy individuals. These variants in NOS1AP can cause further prolongation of the QT interval that is associated with an increased risk of sudden cardiac death in carriers of a pathogenic KCNQ1 variant.
Variants in several genes have been identified as potential modifiers of lung disease phenotypes in CF, and it is likely that multiple genetic modifiers exist for a major proportion of genetic diseases. Identification and understanding of genetic modifiers and the mechanisms by which they operate will extend the range of potential therapeutic targets, and also allow better prognostic information and risk stratification. It is hoped that initiatives like the 100,000 Genomes Project and SGP will contribute to the identification of such genetic modifiers.
Investigating cancer predisposition
There are many examples in which the identical pathogenic variant leads to different severity of disease in different individuals. For many variants the penetrance (the proportion of individuals with the variant who exhibit the phenotype) is less than 100%, and the expression (pattern of effects observed in individuals with the variant) can also vary. For example, inherited pathogenic BRCA1 variants are associated with predisposition to breast and ovarian cancer. However, some women inherit a clearly pathogenic BRCA1 variant but are not affected during their lifetime. Women who inherited the same pathogenic BRCA1 variant may be affected by breast cancer only (unilateral or bilateral), by ovarian cancer, or by both breast and ovarian cancer, and in fact other cancer types may also be seen as a consequence of the inherited BRCA1 variant. Differences in penetrance and expression are likely to depend to a great extent upon a combination of environmental and genetic modifiers. Thus, while such individuals carry a high risk pathogenic variant, multiple other genes contribute to the overall risk of disease.
The majority of cancers arise in individuals who do not carry a high risk pathogenic variant (such as the BRCA1/2 variants). Nevertheless, the genetic make-up of the individual influences the chance of cancer occurring, affording protection against risk, or increasing the predisposition. By comparing the genotypes of individuals who have suffered from one type of cancer with those who have not, it is possible to begin to build information about the relative risk that certain gene variants might hold, also, to relate the risk to particular conditions. Such studies typically involve GWAS approaches (see Figures 28 and 29), but can also be revealed by full genome sequence analysis. For example, a specific SNP might be linked with some protection for the individual from the damaging effects of cigarette smoke in the lung, but might have little effect on lung cancer incidence in a non-smoker. Similarly, another variant might increase the cancer risk, but only in a smoker. Risk of lung cancer attributed to such variants therefore would be conditional upon the smoking status. Similarly, risk alleles might reflect diet or other environmental factors, or may show a disease association that is not dependent upon any known environmental condition. In relation to cancer, such studies hold great promise for future cancer prevention, as building a risk profile for individuals would facilitate informed lifestyle choices and preventative treatment measures (as is currently the case with prophylactic surgery for BRCA1 and BRCA2 pathogenic variant carriers).
Identifying causative or driver mutations in cancer
Cancer cells harbour hundreds, even thousands of somatic mutations, but only a small proportion of these are causative in the cancer process; the vast majority are generally ‘passenger’ mutations. By comparing the genomic sequence of the cancer cells with that of the unaffected genome from the same individual (from non-cancer tissue, usually the blood), all of the somatic mutations that have occurred in the cancer cell can be identified. Then by comparing the genes that have been subject to mutation, between cancers arising in hundreds of different people, it is possible to establish which genes are commonly mutated in a given cancer type. On the basis that mutation occurs largely at random, the chance that a passenger mutation in a given gene will be observed in multiple cancer samples is low. Conversely, if a causative mutation occurs, this will give the cell a growth advantage and it will be selected during the ‘evolution’ of the cancer. Therefore, genes that are found to be mutated in multiple different cancer samples constitute candidate driver mutations. The gene’s properties and function can then be researched to assess if it can indeed contribute to cancerous processes.
Understanding the contribution of risk alleles and the role of all the cancer-associated mutations is crucial to enable the design and successful delivery of novel therapies which target particular mutations present in a given cancer. Furthermore, it appears likely that future genetic diagnostics will be aiming to identify not only the primary causative variants in rare and complex diseases, but also the genetic modifier variants that may influence disease severity and progression: a genomic rather than genetic approach.
Genetic testing in the diagnostic laboratory
Introduction
Advances in technology, particularly in DNA sequencing technologies, have led to identification of even more genes for which loss or changes in function can lead to disease. This in turn increases the need for genetic testing, to confirm potential diagnoses, to provide information on recurrence risk (for example, in future pregnancies) and to facilitate testing in relatives where appropriate. The range of tests available in genetic diagnostic laboratories has changed dramatically in recent years, with time-consuming tests like Southern blot being replaced by approaches with much faster turn-around times.
Karyotyping
Genetic testing has its roots in being able to visualise whole chromosomes in a process known as karyotyping (see Figure 2). This process usually requires culture of cells (can take 1–2 weeks) and arrests cells in metaphase, the stage of the cell cycle in which the chromosomes are in their most condensed form and thus easiest to visualise. In early karyotyping analyses, the imaged chromosomes were simply arranged, in order of their size and position of the centromere. Advances in this procedure came in the 1970s when various banding techniques were developed, including G-banding, currently the most widely used technique in the U.K. and U.S.A. Following trypsin digestion, Giemsa dye is used to stain the chromosomes, with AT-rich, gene-poor regions staining more readily than the gene-rich regions. The resultant pattern allows the chromosomes to be distinguished and thus permits the presence of large abnormalities such as deletions, duplications, inversions and translocations to be assessed. One drawback of karyotyping is the lack of resolution; in general changes of less than 3–4 Mb are virtually impossible to detect.
Fluorescence in situ hybridisation
A later development, in the 1980s, was fluorescence in situ hybridisation (FISH), which uses labelled DNA probes to assess the presence, copy number and location of the complementary sequence of DNA in an individual’s genome by hybridisation. The sample under investigation can comprise interphase cells or cultured cells arrested in metaphase, and is immobilised on a surface, usually a glass slide. After incubation with the fluorescently tagged probe followed by washing to remove unbound probe, binding of the probe is viewed using a fluorescence microscope (Figure 39). An example of a condition where FISH might be useful is DiGeorge syndrome (DGS), a genetic condition usually caused by microdeletions of 3 kb or less, including the TBX1 gene, at 22q11.2. Conventional karyotyping does not have sufficient resolution to identify such deletions. However, using FISH probes for the TBX1 gene, the deletion can be easily identified. Various types of FISH probes can be used, including single locus probes (like the TBX1 probe), centromeric probes (which recognise centromeres of specific chromosomes), subtelomeric probes (which target unique sequences close to the telomere) and chromosome paints (which can target the non-repetitive content of an entire chromosome). Chromosome paints can facilitate the identification of the chromosomal origin of constituent parts of rearranged chromosomes. A drawback of FISH is the cost of the probes (£ 50–100) plus the time taken for the analysis, so that there is a limit to the number of loci that it would be economically feasible to test for in one patient. Thus the subsequent development of microarrays has superseded the use of FISH, particularly where the clinical features do not narrow the region of the genome that is likely involved.
Figure 39. FISH.
(A) One or more fluorescently labelled probes are required for the targets that are to be detected. The patient sample containing chromosomal DNA (which may be cultured cells for metaphase FISH or uncultured cells for interphase FISH) is immobilised on a microscope slide. Probes and chromosomes are denatured and allowed to hybridise together, thus localising the fluorescent probes to the regions where complementary chromosomal DNA is present. (B) Probes for detection of DiGeorge syndrome. One probe (red) targets the DiGeorge locus at 22q11.2 (including the TBX1 gene), and a control probe (green) targeting genes at 22q13 (ARSA and SHANK3) helps to identify both copies of chromosome 22. Note that FISH probes are very long, typically hundreds of kb, in order to render the target detectable; this means that a small deletion may be missed. (C) A fluorescence microscope is used to visualise the results; the image here includes an interphase nucleus as well as a metaphase spread. The chromosomal material has been counterstained blue by DAPI. The normal chromosome 22 is indicated by ‘n’ in the metaphase spread and in the inset; both red and green probes hybridise (the presence of two separate spots for each probe is due to the presence of sister chromatids each possessing a copy of the relevant locus). The chromosome 22 with deletion of the DiGeorge locus (indicated by ‘d’ in the metaphase and inset) shows hybridisation only to the control probe. Note that the interphase nucleus shows only the number of loci present (two green control loci and one red DiGeorge locus), not their location with respect to each other. Note also that in interphase nuclei the chromosomes are very extended so that even loci on the same chromosome become widely separated. FISH image courtesy of West of Scotland Genetics Service. Chromosome ideogram from NCBI Genome Decoration Page.
Mutation specific assays
Although karyotyping and FISH offer a large scale view of chromosomes, there are often times where resolution at the nucleotide level is required. Conditions where the underlying genetic change is known allows the design of specific assays which can be used in a clinical context. Using CF as an example, it is known that by testing for a defined set of 31 pathogenic variants, it is possible to detect almost 90% of CF cases in a Caucasian population. One of the approaches used is known as allele-specific PCR (or amplification refractory mutation system, ARMS).
This allele-specific PCR is based on the principle that complementarity at the 3′ binding site of the primer is crucial for amplification to occur. Although base mismatches at other points in the primer may be tolerated, mismatches at the 3′ end prevent amplification in the extension phase of the PCR. Thus it is possible to design two sets of primers – one which matches the ‘normal’ allele and one set where the 3′ end of one primer is complementary to the pathogenic allele (Figure 40). By determining which set of primers generate a PCR product, it can be determined whether normal, pathogenic or both alleles are present in the individual. This process is repeated for multiple variants, often carried out simultaneously in the same reaction.
Figure 40. Allele-specific PCR by positioning the variant at the 3′ end of one primer.
(A) As with all PCRs, both forward and reverse primers are required, one of which will be a common primer (here the forward primer) and one of which will have specific versions for each allele (here the reverse primer). The specificity is generated by the sequence at the 3′ end of the primer. For assay of a SNP with two alleles, two PCR amplifications are set up, both containing the common primer, but containing the alternate versions of the allele-specific primer. (B) An assay for a T/C SNP. (1) One version of the reverse primer has an A at the 3′ end; this matches the T allele, and extension can occur from the primer when the T allele is present, so PCR products are obtained from homozygotes or heterozygotes for T (TT or TC). (2) The reverse primer with A at the 3′ end does not allow extension, so PCR would fail if only the C allele were present (CC homozygotes). (3) Reverse primer ending in G does not allow PCR amplification when the template contains only the T allele (TT homozygotes). (4) Reverse primer ending in G allows extension if the C allele is present (CC or TC).
Several other techniques, including reverse dot blot and the oligonucleotide ligation assay are alternatives to consider when detecting pathogenic variants that affect one or a few nucleotides. For conditions where pathogenic change is more likely to be a deletion or duplication, for example DGS or Duchenne muscular dystrophy (DMD), a technique such as multiplex ligation-dependent probe amplification (MLPA) may be used (Figure 41). Although FISH (Figure 39) can also detect deletions such as those found in DGS, MLPA, utilising multiple smaller probes across a larger genomic region, can provide a higher resolution view of the extent of any deletion. However FISH is able to provide information on chromosomal position of the target sequence(s), whereas MLPA only informs on copy number.
Figure 41. The MLPA assay and application to diagnosis of DGS.
(A) Short regions of DNA, roughly 50–70 bp, are selected as targets, indicated by S and T (1). Single-stranded oligonucleotide probe pairs are designed for each target (2), with half of the target sequence present in the ‘left’ probe and the other half in the ‘right’ probe. Each left probe contains an additional sequence ‘F’ and each right probe contains an additional sequence ‘R’. The right probes also contain a ‘stuffer’ sequence which is a different length for each target to be detected. Genomic DNA is denatured and probes allowed to anneal (3). Annealed probe pairs will lie precisely adjacent to each other on the target DNA (4) allowing DNA ligase to join left and right probes together into a single molecule (5). The ligated probes are denatured away from the target (6), and then PCR amplification is carried out (7) using the same primer pair for all ligated probes (fluorescently-labelled F and the complement of R). The final quantity of each amplified product is dependent upon the copy number of that target sequence within the sample genome. (B) For each target the amount of fluorescent product from the test sample is compared with the amount of product from a control genome, and the ratio is plotted. This plot depicts typical MLPA results for 29 target sequences across the 22q11 region for a patient suspected of having DGS. The gene targeted by each probe is indicated below the plot; distance along the X-axis indicates distance along the chromosome. A ratio of approximately 1.0 indicates normal copy number (blue squares). However the results for 14 targets (including two for the TBX1 gene) give a ratio of only 0.5, indicating a halving of the copy number (one copy instead of the two expected from two complete copies of chromosome 22). This result confirms a diagnosis of DGS. (C) The region of chromosome 22 which is targeted by MLPA probes from the ‘P250-B2 DiGeorge’ kit from MRC-Holland. Cat eye syndrome (CES) is caused by duplication of the region indicated by the green bar; duplications can be identified by amplification ratios of 1.5 compared with control. Roughly 90% of DGS cases result from a 3-Mb deletion, indicated by the blue bar, and including approximately 60 genes, while approximately 8% of cases have a 1.5-Mb deletion, indicated by the purple bar, involving 28 genes. A small number of cases have atypical deletions which may be larger than 3 Mb. While FISH using the TBX1 probe (red) can identify a deletion in the region, MLPA can provide a more accurate picture of the extent of any imbalance due to the greater number of probes used. The P250-B2 DiGeorge kit also contains probes that target relevant regions on chromosomes 4q, 8p, 9q, 10p and 17p, in which copy imbalance generates phenotypes which overlap with DGS; thus a single MLPA assay can assess multiple target regions. Chromosome ideogram from NCBI Genome Decoration Page.
Sanger sequencing
The techniques described above are useful in detecting conditions where there is already some knowledge of where in the gene the pathogenic variant is likely to be, for example it is known that p.Phe508del accounts for approximately 70% of CF alleles worldwide, which can be easily detected using allele-specific PCR. However, many conditions do not have a subset of common mutations, a good example being the inherited predisposition to breast cancer, where pathogenic variants are spread throughout the relevant genes, which include BRCA1 and BRCA2. In order to detect these types of changes it is often necessary to determine the sequence of the whole gene(s) in a patient and to then compare this with a reference genome to identify changes in the patient. For many years this has been done using Sanger sequencing, a technique which was first developed in the 1970s, but which has undergone many developments to generate the currently used automated method (Figure 42).
Figure 42. Automated Sanger sequencing.
(A) PCR products (1) are generated from the region to be sequenced so that there are billions of template molecules for the sequencing reaction. A sequencing primer is annealed to the denatured PCR products (2). A mixture of deoxynucleotide triphosphates (dNTPs) and dideoxynucleotide triphosphates (ddNTPs) is added (3), which are used by DNA polymerase (4) to synthesise a new strand. The four ddNTPs are each labelled with a different fluorescent dye: ddATP green, ddCTP blue, ddGTP yellow and ddTTP red. When a dNTP has been added, DNA synthesis can continue in the normal way. However, ddNTPs are chain terminators, so that once a ddNTP is added, synthesis will terminate; the colour of fluorescence of the terminated chain will indicate which nucleotide is present at that position (here the red fluorescence indicates a T). Because there are billions of templates, and because ddNTPs are added randomly, terminating different chains at different positions, billions of terminated chains are generated, with many chains terminated at each nucleotide position. (B) The products of the sequencing reaction are denatured away from the template and electrophoresed to separate them by size; the shortest chains will move fastest. A laser-based detector registers the colours of fluorescence emitted as each of the sequencing products travels past. (C) The data from the detector is processed to generate an ‘electropherogram’, in which successive peaks represent products which are each one nucleotide longer than the previous one, allowing the sequence to be read by using the fluorescent colour to identify the nucleotide(s) at that position in the DNA. Here the control sequence is TTACAGC, while the patient sample shows a heterozygous substitution of T in the middle of this sequence: since two different alleles are present in the patient, both are represented in the sequence trace at this position, indicated by the pink star.
Aneuploidy testing by quantificative fluorescence PCR
Karyotyping was the traditional approach to diagnose aneuploidy, but the complete process took approximately 1–2 weeks due to the time taken to culture cells. To provide a rapid result, for example for testing during pregnancy, interphase FISH has also been used, but more recently quantitative fluorescence PCR (QF-PCR) has become the technique of choice for initial aneuploidy testing. This involves the analysis of microsatellites with high variability in the population. The copy number of each chromosome tested is deduced from a combination of the number of different microsatellite alleles present, and their ratios (Figure 43). For each chromosome to be tested (usually 13, 18, 21 and sometimes including sex chromosomes) four or five loci are analysed along the length of the chromosome. This approach is generally effective in rapidly (next day) identifying trisomy, and a positive finding can be confirmed by karyotyping or another test if desired. Sometimes one or more of the loci used are homozygous and therefore non-informative, but results from the other loci on that chromosome are generally informative, allowing a result to be reported. Consanguinity is likely to lead to multiple uninformative microsatellites, in which case it is possible to use additional loci, or if that also fails, to use FISH/karyotyping.
Figure 43. QF-PCR.
Several microsatellite loci (here R, S and T) on the relevant chromosome(s) are analysed by PCR using an appropriate pair of flanking primers, one of which is fluorescently labelled, so that all products will be fluorescent, and quantity of product is measurable by amount of fluorescent product generated. The particular loci are selected on the basis of high variability (many different alleles) in the population. (A) Where there are two copies of the chromosome, there should be two copies of each microsatellite, which ideally have different repeat numbers. Thus at locus R one chromosome has 12 repeats, whilst the other has 14 repeats. Following PCR and electrophoretic analysis (see Figure 42), two product peaks are generated, one for each allele, at a 1:1 ratio. The same pattern is observed for loci S and T. (B) Trisomy should result in three copies of each microsatellite. Where there are three different alleles present (as for locus R) three peaks, at a ratio of 1:1:1, will be generated by the analysis. If two of the chromosomes share the same allele while te other is different (as for loci S and T) there will be two peaks with a ratio of 1:2 or 2:1. Thus disomy can be differentiated from trisomy by number of peaks generated and the ratios between them. Note that if all chromosomes present share the same allele at a particular locus, then only one peak is generated and that locus is therefore uninformative.
New approaches to diagnostics
Where there is clear suspicion relating to a particular gene or group of genes, it may be that specific assays, as discussed above, are the most cost-effective diagnostic approach (Figure 44). But it is not always possible to pinpoint the relevant genes or genomic region from the patient symptoms. In such circumstances whole genome approaches are frequently used as the first-line strategy. Developmental delay and learning difficulties, for example can be associated with not only full aneuploidy, but also with gains and/or losses involving large segments of DNA which might be the consequence of microdeletions, microduplications or unbalanced rearrangements of parental translocations/inversions or specific gene variants.
Figure 44. Selecting an appropriate test.
Where the pathogenic variant in a family is known then testing will generally use a specific assay for that variant. For new diagnoses the most appropriate and cost-effective test must be selected according to technology and expertise available to the laboratory as well as the clinical features of the patient. Definitive findings (blue, underlined) can be reported in cases where a specific family variant was being tested for, or where a clearly pathogenic variant that is causative of the patient phenotype is detected. Note that use of some techniques, such as karyotyping, is declining with the advent of NGS- and array-based approaches. *It is possible NGS gene panels may be used in future even where the disease gene is known. Abbreviations: ASP, allele-specific PCR; NGS, next generation sequencing; SS, Sanger sequencing; WES, whole exome sequencing; WGS, whole genome sequencing.
Microarrays
Microarrays allow simultaneous analysis of hundreds of thousands of individual targets across the genome. For whole genome analysis, SNP genotyping by array has largely replaced the earlier array comparative genomic hybridisation (aCGH) approach, so only SNP arrays will be described here. There are several different approaches (or ‘platforms’) for SNP array, but all are based on oligonucleotide ‘probes’ for each of the targets to be assayed, that are immobilised on a solid surface (Figure 45).
Figure 45. Basic microarray.
The microarray (1) is a grid of hundreds of thousands of microscopic ‘spots’; each spot (2) contains billions of copies of an oligonucleotide ‘probe’ that represents a specific target. These single-stranded probes will be able to hybridise to, and therefore capture (3), complementary ssDNA from a denatured sample – for example DNA from a patient (purple strands). Fluorescent label (yellow starbursts) can be attached to the patient DNA prior to hybridisation to facilitate detection – the presence and amount of label at each spot on the array is determined by scanning of the entire array.
DNA from patient samples can be fragmented, denatured and hybridised to the microarray; each spot on the microarray will capture its complementary sequence from the patient sample, provided that the particular sequence is present in the patient’s genome. The quantity of patient DNA that is captured on each spot will be proportional to the amount of that sequence present in the sample. The hybridisation process is optimised such that only perfectly complementary matches can occur. Thus the hybridisation can be sensitive to a single nucleotide difference, allowing genotyping to determine which allele of a particular SNP is present, for example by having one spot on the array for each of the two alleles. A typical SNP array can be used to genotype hundreds of thousands of SNPs from across the genome of a patient, and the interpretation of allele ratios plus total fluorescence (Figure 46) can reveal multiple chromosomal imbalances (Figure 47).
Figure 46. SNP array data can reveal copy number imbalance and homozygosity associated with consanguinity and UPD.
For ease of display and interpretation in SNP arrays the two alleles of each SNP are conventionally designated A and B, thus, for a T/C SNP, T would become the ‘A’ allele and C would become the ‘B’ allele. Each SNP is genotyped and the result plotted (as a green spot) according to position along the chromosome in terms of ‘B allele ratio’, which will be 0% for AA, 50% for AB and 100% for BB. Trisomy is revealed by B allele frequencies of 33 and 66%, and by an overall gain in fluorescence. In monosomy there is only one allele for each SNP so there will be no heterozygosity, and the total fluorescence will be only half the expected value. Long runs of homozygosity resulting from UPD or consanguinity will generate normal fluorescence levels. Other states can also be diagnosed, for example mosaicism (mixtures of two or more cell lines) will lead to skewed B allele ratios.
Figure 47. SNP array demonstrates areas of loss and gain across the genome at high resolution.
(A) ‘LogR’ plot for all 22 autosomes and the sex chromosomes demonstrates balanced copy number for most chromosomes. The pink star indicates a duplication (upward shift of LogR) affecting the chromosome 2p terminus, which is expanded in (i). The blue star indicates a deletion affecting the 15q terminus, which is expanded in (ii). The green star indicates apparent loss affecting the X chromosome but this reflects the presence of only one X chromosome in a male. (B) The B allele frequency plot confirms the 2p duplication (i) and 15q deletion (ii); each spot represents the result for a single SNP – there are 843551 SNPs represented in this array. (C,D) The sample in this case came from the child of a balanced reciprocal translocation carrier. The chromosomes 2 and 15 of the parent are depicted in (C), with arrows indicating the breakpoints. The array result indicates that the child received an unbalanced arrangement from this parent: a normal chromosome 2 together with the translocation 15 (D). Note that the array result (A,B) also demonstrates a duplication of chromosome 15 material (yellow arrow) associated with the translocation breakpoint. This would have been below the resolution of a standard karyotype, but is clear from the array result. Small gains and losses can be seen in other chromosomes on close inspection; these may represent CNVs. Array images courtesy of West of Scotland Genetics Service using Illumina CytoSNP 850K Beadchip; chromosome images generated using CyDAS (www.cydas.org).
SNP arrays are more effective than karyotyping at revealing chromosomal abnormalities, since the resolution is higher: typically 50 kb, with a resolution of 10 kb in critical regions for diagnostic SNP arrays, compared with approximately 3–4 Mb for karyotyping.
Next-generation sequencing
While microarrays provide whole genome coverage at high resolution, many genetic conditions result from much smaller alterations, typically SNVs or small indels. Sanger sequencing provides accurate data but, even when automated, only one or two genes can generally be analysed at a time. A number of newer technologies, collectively referred to as next-generation sequencing (NGS), overcome the limitations of Sanger sequencing and can provide an entire human genome sequence by ‘massively parallel sequencing’. DNA from a patient sample can be fragmented and then the fragments can be sequenced as part of massive arrays generating millions or even billions of short sequence ‘reads’ per run. Bioinformatics is then used to map all these reads to the reference genome, which can then be assembled into a whole genome sequence for that individual (Figure 48). Differences from the reference sequence are identified by the software, and this is where the limitations of whole genome approaches start to become apparent. Because of the sheer amount of variation present in each human genome, the task of filtering the information to identify relevant variants is enormous. In fact, in 2010 Elaine Mardis described the whole genome approach as ‘the $1,000 genome, the $100,000 analysis’ in recognition of the fact that while obtaining the full genome sequence is now relatively cheap, the subsequent analysis requires huge input of time and effort. Development of improved bioinformatics approaches will undoubtedly decrease the cost of analysis, however whole-genome sequencing (WGS) is currently beyond the scope of routine diagnostics in a healthcare context.
Figure 48. NGS data analysis.
The screenshots cover one exon from the analysis of a patient sample with an epilepsy gene panel that examines exon sequences and flanking regions from 104 genes. (A) The top of the screenshot provides chromosomal context (in this case 14q32). The ‘read depth’ window provides an indication of the number of times each nucleotide was represented among all the reads. Individual reads are shown as blue (forward strand read) or green (reverse strand read) bars in the ‘pile-up’. Each read is approximately 100 nucleotides in length, and represents the output from one of the millions of massively parallel sequencing reactions. The reads are aligned to the matching segment of the genome reference sequence, to generate the ‘pile-up’ view. Where the sequence of a read differs from the reference sequence that nucleotide is highlighted in a different colour. Differences seen in only one or two of the reads are likely sequencing errors (for example, those indicated by orange arrows), while the pink arrow indicates a position at which approximately 50% of the reads differ from the reference, which indicates a heterozygous variant. At the bottom is shown the intron/exon structure: the data are for exon 65 plus flanking sequence of the DYNC1N1 gene. (B) The view is zoomed in to the heterozygous variant detected in the first nucleotide of exon 65 (the second base of a codon); this is a characterised variant known as rs138428684, present in 1 per 1000 European alleles, changing the coded amino acid from threonine to arginine at position 3981 in the encoded protein. Readers can explore this variant in databases like Ensembl genome browser (www.ensembl.org) by inserting the variant name (rs138428684) into the search box. Images courtesy of West of Scotland Genetics Service using SeqVar software.
An alternative to WGS is whole-exome sequencing (WES), in which either exon sequences are specifically captured/amplified from the sample for sequencing, or bioinformatics tools remove non-coding sequences from the subsequent analysis. A typical WGS will generate 3–4 million variants per genome, whereas WES will generate only 30000–60000. In cases where a particular variant leads to loss-of-function (e.g. nonsense, frameshift) in a gene that has been previously associated with the patient’s condition, or where the particular variant has been reported as causative for that condition, then an unambiguous diagnosis can be provided. However, large numbers of VUS are identified during NGS, in particular missense variants, or variants that might affect splicing. These can be analysed in silico, for example based on properties of the new amino acid compared with the original, or conservation of that amino acid between species, or potential to create or destroy a splice recognition site in the RNA.
The scale and difficulties associated with variant analysis mean that a preferred approach for many genetic conditions is to use a gene panel representing the specific set of genes that are known to be associated with the particular condition, for example epilepsy, cardiomyopathy, inherited cancer predisposition or even broader panels, such as for recessive paediatric-onset conditions. Even with a reduced sequencing target the problem of VUS is still significant, and many patients will still not receive a diagnosis. As knowledge improves, for example with functional studies by research laboratories, some VUS will be reclassified as either benign or pathogenic. However, the current position is that NGS technology surpasses our ability to effectively utilise the information generated for the benefit of patients.
Variants which are expected to have a severe effect on an encoded protein (for example nonsense or frameshift) are easy to classify as pathogenic. However, variants which might affect the quantity or sequence of the mRNA (for example, variants affecting splicing or promoter activity) are harder to interpret using the DNA sequence alone. Although in silico analysis can help, the optimal approach is to investigate the transcripts generated, by isolating RNA from a patient sample and conversion into complementary DNA (cDNA) for analysis. Of course, transcription and splicing patterns of many genes are tissue-specific, so there may be a requirement to use a tissue biopsy rather than a blood sample. RNA-based analysis is currently uncommon in diagnostic laboratories, with the notable exception of cancer diagnostics (see below). Nevertheless, NGS technology can also be applied to the analysis of cDNA, and this ‘RNA-seq’ approach, currently used extensively in research, has enormous potential in clinical diagnostics.
The role of molecular pathology in cancer diagnostics and management
Cancer cells accumulate large numbers of mutations, many of which are passengers, but some of which are drivers of the cancer phenotype. Identification of the driver mutations present in the cancer of a particular patient can help direct therapy. A relatively new application of genetic diagnostics is in molecular pathology, which is fast becoming a core discipline within cancer management. Immunohistochemistry is frequently used to detect levels of particular proteins in tissue sections from cancers, for example the detection of HER2 overexpression to inform decisions about use of the drug Herceptin. However, many of the genetic diagnostic approaches used for inherited disorders can also be applied to investigation of cancers. The presence of genome rearrangements that are associated with particular diagnoses and/or success of a particular therapy (Figure 35) can be identified by use of specific combinations of FISH probes applied to interphase cells or tissue sections. Likewise, techniques including allele-specific PCR and DNA sequencing can be used to identify mutations of diagnostic or therapeutic significance, and MLPA can be used to identify gene copy number changes. The presence of cancer-associated transcripts, particularly fusion transcripts such as those from BCR-ABL gene fusions, can be assessed by PCR amplification of cDNA. Such PCR-based approaches have much higher sensitivity than FISH; for example interphase FISH can detect one leukaemia cell per 200 normal cells, whereas PCR using cDNA can detect one leukaemia cell per million cells. This level of sensitivity is critical in monitoring response to therapy, and in early detection of relapse (by reappearance of fusion transcripts in blood samples). In fact, the sensitivity of PCR is being harnessed in the development of ‘liquid biopsy’, which targets circulating cancer DNA present in the blood, and has the potential to replace invasive tissue biopsies.
As with all genetic diagnostics, NGS-based approaches are starting to play a role in molecular pathology, which is not surprising, given the large numbers of mutations in each individual cancer. Gene panels can be applied to the analysis of hundreds of targets within cancer DNA for alterations that provide information relevant to diagnosis, prognosis and response to therapy. Whereas the PCR-based approaches mentioned above are effective in identifying fusion transcripts, the number of targets that can be assessed in one assay is very limited, and therefore some fusions will be missed. RNA-seq represents a powerful approach to screen for hundreds of potential fusion targets in a single assay, and will be particularly useful in initial identification of relevant gene fusions in individual patients. Following identification of the fusion, standard PCR-based approaches can be used to monitor that patient in relation to the identified fusion.
Summary
Clearly health service genetics laboratories have many different approaches available for the detection of pathogenic variants, either inherited or in relation to cancer tissue. One of the key roles of a clinical scientist is therefore to ensure that the most appropriate and most cost-effective approach is used for each patient sample. This can also require that the clinician who is referring the patient for genetic analysis provides a sufficiently clear and thorough view of the clinical features, to guide appropriate genetic analysis. However, the increasing use of NGS-based approaches will facilitate a more comprehensive genetic analysis that will improve diagnostic success rates.
Diagnosis, management and therapy of genetic disease
When considering genetic diseases, it is always vital to remember the impact they can have on the life of an individual, family and society. Diagnosis, management and therapy are all important aspects of genetic disease and these issues are discussed briefly in this section. Obtaining a clear diagnosis is often important for several reasons, potentially directing therapeutic intervention, management or informing reproductive decisions. A diagnosis can also be useful in terms of gaining access to relevant support, whether this be financial, social or practical. In addition, many individuals for whom obtaining a diagnosis has been problematic, report great relief when a diagnosis is finally made. Genetic testing can be carried out at any time during the life of an individual who is symptomatic or has a high risk. However, in some situations it is desirable to be proactive in testing whole populations in order for timely intervention to occur. This is typified by genetic screening programmes.
Newborn screening
Newborn screening for genetic conditions has been used for several decades in order to detect and treat genetic conditions and potentially avert serious outcomes. Diagnosis and treatment of the autosomal recessive condition phenylketonuria (PKU) (see Table 6), is one such success story. In this situation both parents are carriers and phenotypically unaffected but with a one-fourth chance of having an affected child. Affected individuals lack the enzyme phenylalanine hydroxylase which converts phenylalanine into tyrosine and this results in the toxic build-up of phenylalanine in the body. This has a devastating effect on the developing brain in particular, and causes irreversible damage.
Newborn screening for PKU in the U.K. began in 1969 and identifies affected babies by measuring the phenylalanine levels in a bloodspot taken from the heel between 5 and 8 days after birth. Once PKU is diagnosed, a strict phenylalanine-free diet is implemented which ideally should be continued throughout life (although adherence is less essential during adulthood with the exception of pregnancy). If this diet is implemented prior to day 23 of life, the individual will not suffer from brain damage and is likely to lead a normal life. Testing for other conditions such as CF and sickle cell disease is now also routinely offered and new conditions are added to the programme as part of regular review processes. Mass Spectrometry with its ability to give insight into the metabolome has already proven significant in these developments and will likely contribute to further expansion.
Pre-marital genetic testing for thalassemia
While newborn screening attempts to identify affected infants very early in life, other strategies are aimed at preventing the conception of affected individuals. Specifically, pre-marital or pre-conception testing identifies couples where both are carriers of the same condition so that they can make informed choices. Examples of conditions in which pre-marital testing has been successfully used are the thalassemias and sickle cell anaemia as well as Tay–Sachs disease. The widely reported pre-marital screening programme for thalassaemia in Cyprus began in 1973, with couples being required to produce a certificate confirming they had been tested for thalassemia carrier status before marriage could legally take place. In the majority of countries where this is practiced, marriage between two carrier individuals is not forbidden, however the social structure in many communities means that it is discouraged. In other settings, religious leaders, for example in some Jewish communities, have used the genetic information (with full consent from the individuals) as part of their marriage arrangement considerations. This was deemed to be both acceptable and indeed welcomed by the community, who for many years had suffered the effects of a high incidence of Tay–Sachs disease, a fatal inborn error of metabolism.
Prenatal diagnosis
Prenatal diagnosis (PND), the testing of an unborn foetus for a specific condition, is also offered by genetics services. An initial requirement of any PND is that the sample is obtained from the foetus. This can be achieved in a number of ways, primarily chorionic villus sampling (CVS) between 10 and 14 weeks of gestation, and amniotic fluid sampling, usually between 14 and 20 weeks of gestation (Figure 49). As both have an intrinsic risk of miscarriage (1–2% and 0.5–1% respectively), they tend to only be offered where there is a substantial risk of disease. Later in pregnancy, taking a cord blood sample is also a possibility, with a similar risk of miscarriage to CVS. Amniocentesis and cordocentesis have the advantage that they represent foetal rather than placental tissue – although both are derived from the embryo, the possibility exists that mosaicism may be present, resulting in the placenta and the foetus having different genotypes and thus a small risk of misdiagnosis using a CVS.
Figure 49. Amniocentesis procedure.
Under ultrasound guidance, a thin needle is passed through the abdominal wall into the amniotic sac. A small amount of amniotic fluid is then removed and subjected to analysis. The amniotic fluid contains a mixture of cells, a proportion of which will be foetal. By BruceBlaus, CC BY-SA 4.0, from Wikimedia Commons.
Once a sample has been obtained through CVS or amniocentesis, testing can be carried out. The genetic test used depends upon the reason for the PND; QF-PCR (Figure 43) for aneuploidies is currently a routine step, and additional testing can be either directed (for a particular condition or chromosomal imbalance that the foetus is at-risk for) or a whole genome approach like SNP array (Figure 47), where abnormalities of unknown aetiology have been detected on ultrasound. Individuals undergoing these procedures most often opt for termination of pregnancy if a genetic problem is found, while others use the knowledge to enable them to prepare for the birth of an affected child.
Pregnancy screening and non-invasive prenatal diagnosis
Traditionally, women have been offered maternal blood screening tests in early pregnancy, the results of which place them in either high or low risk categories for DS other trisomies. In the maternal blood sample, the level of a number of proteins is quantified and these results are combined with ultrasound measurements and factors such as age to assess risk. Women in the high risk category are then offered PND using CVS or amniocentesis to obtain a sample.
However, the relatively low sensitivity and specificity of the traditional maternal blood screening tests result in both false positives (unaffected pregnancies placed in the high risk category and therefore undergoing invasive diagnostic tests, with inherent miscarriage risk) as well as false negatives (affected pregnancies where the mother is falsely reassured by being placed in the low risk category).
Therefore, more recently, non-invasive prenatal diagnosis (NIPD) has been offered in pilot schemes in the U.K., in an attempt to reduce the number of healthy pregnancies lost due to diagnostic testing and to offer women greater choice. In NIPD, a sample of maternal blood is taken from approximately 7 weeks of gestation onwards and analysed directly, using cell-free foetal DNA in the maternal circulation (Figure 50). The amount of DNA present can be quantified and a risk for aneuploidies given with a very high sensitivity and specificity (approximately 99% for DS). This screening can be followed up by diagnostic testing if a high risk result is obtained. NIPD can also be applied to some inherited disorders, for example by testing for foetal sex by presence of Y chromosome (for X-linked recessive disorders) or looking for paternal pathogenic variants.
Figure 50. NIPD for trisomy 21.
(A) Foetal cell-free DNA (cfDNA) from the foetal circulation crosses the placenta into the maternal circulation, which thus contains both maternal and foetal cfDNA. (B) cfDNA is collected from maternal blood samples. The maternal cfDNA tends to be longer fragments than foetal cfDNA so that separation is possible but not straightforward. In general, however, there is no separation stage. In cases where the foetus is affected by trisomy 21, there will be more foetal cfDNA fragments derived from chromosome 21 (coloured red and indicated by asterisks) in comparison with a case where the foetus is unaffected. (C) The total cfDNA is analysed by DNA sequencing using NGS, allowing a count of how many reads have been obtained from each chromosome. If there was an over-representation of chromosome 21 fragments in the cfDNA sample then there will be increased representation of NGS sequence reads that match chromosome 21 (asterisked). The analysis is often quantified by calculating ratios of (for example) chromosome 21 reads to chromosome 1 reads. If only foetal cfDNA was present the chr 21:chr 1ratio would be expected to be 1:1 from an unaffected foetus, and 1.5:1 from an affected foetus, but the additional presence of disomic maternal cfDNA in the sample means that the ratio will be lower.
Preimplantation genetic diagnosis and three parent babies
As an alternative to prenatal diagnosis and possible termination of an affected pregnancy, some couples prefer to prevent the implantation of an affected embryo. Pre-implantation genetic diagnosis (PGD) is an approach which combines IVF and genetic technology to ensure only embryos unaffected by a specific genetic condition are implanted in the uterus (Figure 51). After IVF, and growth to approximately the 8-cell stage, 1 or 2 cells are removed from the developing blastocyst and, depending on the condition being tested for, undergo FISH or PCR-based analysis, looking for specific variants or imbalances. Only unaffected embryos are then implanted. Follow-up PND is recommended during the pregnancy as occasionally technical limitations may lead to false results at the genetic analysis stage. PGD has been successfully used for a number of conditions, including both single gene disorders and chromosomal disorders.
Figure 51. Pre-implantation genetic diagnosis by biopsy of early embryos.
Traditional IVF procedures (1) are used to generate fertilised embryos (2), which are allowed to develop to the 8-celled stage (3). One or two cells are then removed for genetic analysis (4) and only embryos without the condition tested for are implanted into the mother’s uterus (5).
Furthermore, as discussed earlier, in a family with a mitochondrial disorder, transmission to the next generation can be prevented using the new approach known as mitochondrial replacement therapy in which a maternal and donor egg are combined, either prior or subsequent to fertilisation.
Personalised medicine
In addition to provision of diagnostic information for genetic disease, treatment is also an important area to address. While, general therapies have been used for centuries, the concept of personalised medicine has long been seen as the ‘holy grail’ of genetics. Encompassed in the idea of giving the right treatment to the right patient at the right time, personalised medicine relies on the ability to specifically diagnose the underlying molecular and genetic aspects of the condition.
With some cancers, a personalised approach is, to some extent, mainstream. In the treatment of breast cancer, only patients whose cancer expresses specific hormone receptors on their cell surface will be offered hormonal treatments such as tamoxifen for those whose tumours overexpress the oestrogen receptor. Similar approaches in leukaemia (using the drug imatinib to target cancer cells with BCR-ABL translocations) have been used for several years and are now beginning to be used in lung cancers (using erlotonib to target cancer cells which have EGFR mutations) and a variety of others (Figures 52 and 53).
Figure 52. EGFR mutation status determines the outcome of erlotinib as a therapy.
(A) EGFR is a transmembrane receptor that has a tyrosine kinase (TK) domain. In the absence of EGF the normal receptor is in an inactive state. (B) When EGF binds, the TK domain undergoes a conformational change and becomes activated, generating signals for cell proliferation. (C) Cancer associated mutations in EGFR lead to hyperactivation of EGFR that represents a key driver of tumorigenesis. (D) The TK inhibitor erlotinib binds to EGFR and prevents downstream signalling. Therefore in cases where EGFR mutations are driving tumorigenesis, erlotinib can block this process. (E) The acquisition of a second mutation that prevents erlotinib binding leads to resistance to this inhibitor.
Figure 53. EGFR mutation status must be determined in order to ensure that erlotinib is only used in those cases where it will provide benefit.
The EGFR activating and TKI resistance mutations are clustered in the tyrosine kinase domain (see Figure 52) between amino acids 688 and 875 of the EGFR protein, so that mutation analysis can be focussed on the coding sequences for this region. Abbreviation: NSCLC, non-small cell lung cancer.
More recently, personalised approaches to single gene disorders, for example CF are beginning to be used. For CF, drugs have been developed which target the specific protein defects caused by particular mutations, for example some mutations which cause the protein to form channels that cannot open and close properly. A drug called KALYDECO (ivacaftor) can be used in these patients to help the channel to stay open allowing normal passage of ions through the open channel.
Gene therapy/gene editing
While personalised treatments in cancer and in CF target the affected protein, therapies which address the underlying genetic aspects, to ensure that a functional protein can be generated, also hold great appeal. Broadly speaking, gene therapy aims to replace the defective gene by delivering a new working copy to the cell, while gene editing aims to correct the defective gene.
One condition in which these approaches have been studied is DMD, an X-linked recessive disorder which causes progressive muscle degeneration and weakness. DMD is mainly caused by large deletions and duplications (and less commonly nonsense mutations) in the dystrophin gene and affected individuals produce virtually no dystrophin, with resultant muscle cell damage. Males with the condition commonly use a wheelchair by 12 years old and the average lifespan for an affected individual is approximately 30 years.
In a condition such as DMD, where the underlying mutation is known, a variety of approaches have been trialled (Figure 54). It is hoped that to induce exon skipping, whereby the RNA processing machinery excludes the affected exon, can result in a milder form of the condition, similar to Becker muscular dstrophy (BMD). Replacing the dystrophin gene through delivery by adenovirus particles has also been attempted, although not without difficulty due to both the size of the gene and the immune reaction to the viral particles.
Figure 54. Potential therapeutic approaches for DMD.
(A) In healthy muscle the dystrophin protein functions as a link between the actin fibres in the cell and the dystroglycan complex (DGC) in the cell membrane, preventing damage to the cell during muscle contraction. (B) In the absence of dystrophin, the muscle cell sustains damage during contractions, eventually leading to muscle cell death. A number of therapeutic approaches are possible (those in pink boxes have been trialled in DMD patients, with some success reported; the relevant drug names are in green font). (1) Stem cell therapy, by injecting either healthy, tissue matched muscle stem cells from a donor, or stem cells from the patient which have been isolated, cultured in vitro, and then subjected to genome modification (see 6,7) to correct the defect prior to injection back into the patient. (2) The utrophin protein has a similar structure and function to dystrophin, but is not normally expressed in sufficient quantity to substitute for dystrophin; by up-regulating utrophin gene expression the function of dystrophin can be replaced (C); this approach has been effective in mouse models. (3) A significant proportion (approximately 15%) of DMD is due to nonsense mutations which lead to premature translation termination, and would therefore generate non-functional protein fragments. By use of a nonsense suppressor drug, which influences the ribosome to read through nonsense codons by incorporating an amino acid and continuing translation, full length dystrophin protein can be generated (D). (4) Roughly 70% of DMD is due to deletion or duplication of exons, leading to frameshift; a proportion of microlesions also lead to frameshift. By use of molecules that target the splicing process, and cause selected exons to be skipped (removed during splicing), the reading frame can be restored, generating a shorter version of the dystrophin protein, which is, nevertheless, still able to form a link between actin and DGC (E). Although not the same as normal dystrophin, these shorter dystrophin proteins are associated with much milder symptoms, i.e. Becker muscular dystrophy (BMD). Exon skipping could also be used to skip exons harbouring nonsense mutations. (5) The full-length dystrophin gene is too large to be accommodated in current gene therapy vectors, but because shorter versions of dystrophin are effective in restoring function, gene therapy with minigenes is a possibility. (6) Genome editing using strategies like CRISPR-Cas may be utilised to correct the pathogenic change within the genome, either by in vitro targeting of stem cells removed from the patient prior to injection back into the patient, or by delivering the CRISPR-Cas system directly to the muscle cells. (7) Genome editing (exon snipping) to remove particular exons from the genome is an alternative approach to generating an in-frame gene that is free of pathogenic variants.
Clinical trials using gene therapy have had a notoriously difficult path, with death of a patient affected by ornithine transcarbamoylase (OTC) deficiency (Jesse Gelsinger) in one trial and development of leukaemia in others. Even in conditions which seem naturally amenable to gene therapy (e.g. CF with its well-understood pathophysiology and relative ease of access to affected tissues), achieving stable and sustained expression of replacement genes has proven difficult.
Recent success in treating the neuromuscular disorder spinal muscular atrophy (SMA) using a treatment known as antisense oligonucleotide therapy has aroused much interest. With the condition being caused by loss of a gene called SMN1, research has focused on attempting to restore the function of a homologue, the SMN2 gene. Under normal conditions SMN2 is largely non-functional due to a point mutation which results in exon 7 being spliced out. Antisense oligonucleotide therapy employs an oligonucleotide which binds to the SMN2 mRNA and changes how it is spliced, allowing the SMN2 protein to substitute for the absent SMN1. Trials have so far been very successful, with every indication that this is a remarkable breakthrough in treating this severe and life-limiting condition.
Great excitement has also surrounded the advent of gene editing technology, in particular the use of the CRISPR Cas9 (clustered regularly interspaced short palindromic repeats (CRISPR) associated nuclease 9) system, a genome editing tool which acts as a pair of ‘molecular scissors’ to cut a specific piece of DNA. Made from two components and originating as part of bacterial immune defences, Cas9 is a nuclease, guided to its target by a single guide RNA which binds to its cDNA sequence. The Cas9–RNA complex creates specific DSB non-homologous end joining or by homology directed repair if a suitable donor DNA is present. This allows precise sequence modification to edit out the genetic defect and replace it with a desired sequence. Recent studies, for example in DMD mouse models show promise and there are hopes that the technique may also be applicable in humans (Figure 55), although difficulties such as specificity and efficiency of the repair still have to be fully addressed, as does the issue of immunogenicity. Nevertheless this is an exciting area, holding great promise for the future.
Figure 55. The use of CRISPR/Cas9 in DMD.
In stem cells from the patient CRISPR/Cas9 can be used to remove the section of the dystrophin gene which harbours the mutation. The cells will then repair the DNA, creating a gene which, when expressed, will result in a shorter but functional form of the protein. When the cells are then grown and caused to differentiate into skeletal and heart muscle cells which can then be transplanted into the patient resulting in a milder phenotype. This has already been achieved in mice and the same treatment could potentially be applicable to humans.
Challenges in delivering a genetics service
Introduction
As options available to patients, healthcare workers and scientists continue to expand, we face an array of issues and dilemmas in genetics.
The first full human genome was sequenced at a cost of $1 billion over a period of 13 years. Today, a full genome can be sequenced in 1 h costing approximately $ 1000 (Figure 56). As we understand the genetic basis of increasing numbers of conditions, and as full genome sequencing becomes ever more available, it seems inevitable that there will be a clash between cost effectiveness and the patient’s ‘right to know’. Additionally, although it is relatively straightforward to sequence the human genome, the interpretation of these data are an altogether more complex issue, highlighted by the issues surrounding VUS.
Figure 56. Cost per genome over time.
As a comparison, expected fall in cost as predicted by Moore’s law, a commonly used model for tracking technological development, was surpassed beginning approximately 2008. Image courtesy of National Human Genome Research Institute https://www.genome.gov
VUS
VUS pose increasing dilemmas for the healthcare sector because, while the capability exists to detect virtually all variants in the human genome, the ability to understand this information has yet to catch up. Simply put, although it is possible to identify that there has been, for example a single base change in a particular gene, it is much more difficult to determine what the resultant effect of that change is at a protein or even cellular level. Variants are investigated using a range of computer algorithms, a lengthy process which examines aspects such as conservation across species, amino acid similarity, protein domain structure etc., but which will not always yield a definitive result (see Table 2). As explained earlier, each individual harbours approximately 3 million variations in their genome, the vast majority of which are harmless. Deciding which, if any, of all the variants found is responsible for, or may predispose to a specific condition is a potential minefield. Similarly challenging is the issue of which of these variants should be disclosed to patients, and with what explanation. As our ability to catalogue and share information on variants expands, their significance will likely become clearer. Ensuring that patients are kept abreast of significant discoveries relevant to their healthcare is likely to be challenging. Finally, if interpreting VUS in post-natal, child and adult genetic services is complex, the challenge is even greater in prenatal genetics, where the lack of phenotype information compounds uncertainty. Additionally, the issue of data storage is an increasingly important one, with the concern that health service systems may soon struggle to keep pace with the increasing quantity of data being generated.
Direct to consumer testing
While individuals affected with genetic conditions have typically been seen by genetics hospital services, in the past decade the availability of ‘Direct to Consumer’ (DTC) genetic testing has risen exponentially. DTC testing is ‘sold’ directly to the customer with no healthcare service/provider involvement and through companies such as ‘23andMe’ and ‘Living DNA’, individuals can be tested for both ancestry and health information. As an example, ‘23andMe’ (at the time of writing) offers to screen customers for their carrier status for 40 diseases and their susceptibility to an additional ten conditions (including Alzheimer’s, Parkinson’s and Celiac disease), as well as offering insights into ‘traits’ such as earlobe type, sweet taste preference and hair curliness. Samples, generally saliva or cheek swaps are sent through the post with results being communicated online after a period of approximately 3 months.
The testing carried out by these companies is generally SNP array-based and ranges from testing for established mutations (e.g. CF carrier screening tests for 29 mutations in the CFTR gene) to the highly controversial susceptibility testing. For example in the test used to determine an individual’s risk of developing Parkinson’s disease, two SNPs in the LRRK2 and GBA genes are used to provide an estimate of the individual’s risk of developing the condition. Evidence surrounding these SNPs is reasonably convincing, however they are only two of several such SNPs that have been reported. The U.S. Food and Drug Administration withdrew ‘23andMe’s’ licence to provide heath information in 2013 due to concerns regarding ‘medical guidance reasons and the accuracy of the data gathered’, however this has since been reinstated for a limited number of conditions. Uncoupling of genetic testing from professional genetic counselling has raised concerns and several studies have suggested that many consumers are not able to fully understand the implications of their test results. This in turn may lead to increased pressure on GPs, with whom patients are likely to consult for help interpreting this information.
Whose information is it?
As the amount of genetic information generated grows, the question of data ownership becomes increasingly pertinent. Both within and out-with individual families, it is often not clear who has a right to know certain genetic information. Consider the situation where a child has been diagnosed with DS, and more specifically with DS resulting from a translocation present in one of the parents (accounting for approximately 4% of cases). Other members of the family, (for instance, the carrier parent’s siblings) may well be at risk of having similarly affected children. The question of communicating the results to other family members is generally left to the discretion of the family themselves, however this may not be information they want to share. Emotions such as guilt and blame, as well as existing family dynamics all come into play when such personal information is disclosed. Thus at risk relatives may be left uninformed. Similarly, a young adult receiving an ‘affected’ predictive test result for a condition such as HD has simultaneously uncovered a parent’s result because it is almost certain that they inherited this affected allele from one of their parents. This is irrespective of whether the parent wished to be tested or not. In such cases, patients are advised during pre-test counselling that they may wish not to disclose to family members that they are considering testing, on the premise that once a result is known it is very difficult to conceal the news.
Perhaps arousing more debate is the question of who, if anyone, outside a family has the right to access genetic test results. Clearly such results may be of considerable use to certain public services, for example the police, however, the ethics of release of information remains unclear. Regulatory bodies such as the U.K. Driver and Vehicle Licensing Agency also have a vested interest in genetic test information – for example from patients whose genetic condition means they may not be safe to continue holding a driving licence.
Insurance is also an important issue with the moratorium on the use of genetic test results by insurers in the U.K. until 2019. Some would argue that the current health questionnaires required to gain insurance already screen for some of the information, e.g. a family history of HD, and that by including genetic testing results those not at risk would not be unfairly penalised. However, requiring genetic results to be disclosed on application could prevent some from having useful genetic testing carried out, or may prevent other vulnerable people from obtaining relevant insurance. Thus while the understanding and interpretation of genetic data are challenges for the scientific community and healthcare providers, the encompassing issues require considerable ethical debate and supporting legislation.
Concluding remarks
During recent decades the role of genetics in medicine has changed dramatically, beginning as a speciality dealing with conditions that were relatively rare in the population, and becoming a discipline that underpins developments in wider areas of patient care. Better understanding of the contributions of genetic predisposition and epigenetic changes to common diseases, and of the role of genetic alterations in response to therapy, means that genetics will be relevant to everyone in the population. Education, both for health professionals and for their patients, will play a key part in ensuring that new developments in genetics and genomics continue to be successfully translated and implemented into clinical practice. There will be an increasing need for clinical scientists, genetic technologists and bioinformaticians to provide the laboratory services, as well as an increasing need for clinicians, genetic counsellors and nurses who are conversant with the new genetic and genomic technologies. Additionally, there will be a great need for further generations of research scientists to address the huge remaining gaps in our understanding and to generate innovative approaches in the application of our knowledge to the delivery of cost-effective healthcare. For anyone exploring career options, genetics and genomics should provide many interesting and rewarding possibilities for the future!
Acknowledgments
We thank the West of Scotland Genetics Service for providing images of diagnostic results.
Abbreviations
- ACH
achondroplasia
- AS
Angelman syndrome
- BMD
Becker muscular dystrophy
- BRCA
breast cancer susceptibility
- CDK
cyclin-dependent kinase
- cDNA
complementary DNA
- CF
cystic fibrosis
- cfDNA
cell-free DNA
- CFTR
CF transmembrane conductance regulator
- CNP
copy number polymorphism
- CNV
copy number variant
- CVS
chorionic villus sampling
- DMD
Duchenne muscular dystrophy
- DNMT
DNA methyl transferase
- DS
Down syndrome
- DSB
double strand break
- DSD
disorder of sex development
- DTC
direct to consumer (genetic testing)
- FISH
fluorescence in situ hybridisation
- GWAS
genome-wide association study
- HAT
histone acetyl transferase
- HD
Huntington disease
- HDAC
histone deacetylase
- HIF
hypoxia-inducible factor
- ICF
Immunodeficiency, centromeric instability, and facial anomalies syndrome
- ISCN
International System for Human Cytogenetic Nomenclature
- IVF
in vitro fertilisation
- kb
kilobase pair (1000 bp)
- LHON
Leber hereditary optic neuropathy
- LQTS
long QT syndrome
- MAF
minor allele frequency
- Mb
million bp
- MLPA
multiplex ligation dependent probe amplification
- NGS
next-generation sequencing
- NIPD
non-invasive prenatal diagnosis
- NOR
nucleolar organiser region
- PAR
pseudoautosomal region
- PCD
programmed cell death
- PGD
pre-implantation genetic diagnosis
- PND
pre-natal diagnosis
- PWS
Prader–Willi syndrome
- QF-PCR
quantificative fluorescence PCR
- rDNA
ribosomal DNA
- SGP
Scottish Genomes Partnership
- SMA
spinal muscular atrophy
- SNP
single nucleotide polymorphism
- SNV
single nucleotide variant
- SRY
sex-determining region Y
- SS
Sanger sequencing
- SSR
simple sequence repeat
- STR
short tandem repeat
- T1D
type 1 diabetes
- T2D
type 2 diabetes
- TDF
testis determining factor
- TK
tyrosine kinase
- TKI
tyrosine kinase inhibitor
- TSG
tumour suppressor gene
- UPD
uniparental disomy
- VNTR
variable number tandem repeat
- VUS
variant of unknown significance
- WES
whole exome sequencing
- WGS
whole genome sequencing
- Xa
active X chromosome
- Xi
inactive X chromosome
- XIC
X inactivation centre
Appendix. Glossary of terms
Acetylation/acetyltransferase The process of acetylation involves the addition of acetyl group (O=C–CH3) to the target molecule, for example proteins, by the action of the appropriate acetyltransferase enzymes.
Allele A particular form of a given gene, usually one of several versions that differ in sequence and may differ in phenotype. Multiple different alleles (gene versions that differ in sequence) can exist within a population (some quite common), giving rise to phenotypic variation. Germ line differences from the sequence database for the human reference genome are called polymorphisms, or those that are more rare, called variants. Polymorphisms and variants arose in the germ line gene pool by mutation. Therefore the distinction between what is called a polymorphic or variant allele and what is called a mutant allele by research geneticists, can be blurred. In general, the frequency of the allele in the population and the extent to which it is disease causing or not, determines what it is called. Thus, the currently accepted definitions used by medical geneticists, in relation to the human reference genome are: pathogenic variant, likely pathogenic variant, VUS, likely benign variant and benign variant.
Allele-specific PCR A PCR-based method in which particular allele(s) are amplified, facilitating genotyping of the locus, typically by placing the variant nucleotide at the 3′ end of either the forward or the reverse PCR primer.
Aneuploidy An abnormal number of chromosomes in a cell, with one or more extra chromosomes or chromosomes missing.
Angiogenesis The process of ‘growing’ new blood vessels from pre-existing vessels.
Anticipation Anticipation can occur in some genetic conditions as a pathogenic variant is passed from one generation to the next. In those cases, the age of onset of the symptoms decreases from one generation to the next, and often the severity of symptoms increases as well.
Antisense oligonucleotideShort deoxynucleotide which is complementary to a ‘sense’ sequence of DNA.
Apoptosis This is a form of ‘programmed cell death’. The phenomenon results when a cell is genetically determined to die or receives internal and/or external signals to self-destruct. The cell dismantles into component parts that are neatly disposed of or recycled and this does not cause inflammation. A classic example of normal apoptosis occurs during the formation of the fingers and toes in development. During mammalian embryogenesis, as an ‘evolutionary throwback’, the digits form, linked by webbing. The cells that make up the webbing are programmed to die and as development proceeds, they die by apoptosis, separating the digits.
AssociationThe occurrence of a specific polymorphism together with a particular trait more often than would be expected by change.
Autosome A chromosome that is not a sex chromosome.
Benign growth/tumour This results from limited new growth of a cell such that a small (occasionally large) lump forms within a tissue, but does not progress on to become invasive. A classic example of this is a mole or nevus on the skin. Benign tumours are mostly harmless, but some may acquire further mutations to become cancerous.
Benign variant A variant allele which is believed to have no effect on health either in the heterozygous or homozygous state.
Biallelic Relating to both alleles of a gene; for example biallelic expression means that products are generated from both copies of the gene.
Carcinogen Any agent that acts to increase the risk of cancer. Not all carcinogens are mutagens, but many are.
Candidate gene A gene thought to have a high chance of being involved in a particular phenotype, often due to pathways it is known to be involved in.
Cell cycle The process by which a cell divides into two cells. The cycle usually follows the four stages: G1 (gap or growth 1), S (synthesis of DNA), G2 (gap or growth 2), finally mitosis (note in meiosis, the cell cycle follows a different pattern, as described below). G1, S and G2 together make up ‘interphase’.
Cell-free DNA (cfDNA)DNA which is not contained within a cell and is found in small amounts in the circulation or other fluids, e.g. urine.
Cell proliferation The term used when cells divide by mitosis, resulting in an increase in cell number.
Chromatin Describes the way the human genome is organised/packaged within a cell. Usually, DNA is wrapped around a core of histone proteins, forming nucleosomes.
Centromere The waist-like constriction of a chromosome which separates the short from the long arm. During cell division, spindle fibres attach at the centromere to pull replicated chromatids apart.
Chimera An organism consisting of cells which are genetically different, which can be generated by fusion of early embryos.
Codon Three consecutive nucleotides which instruct a ribosome to incorporate a specific amino acid into a growing polypeptide, or to stop translation. Note that strictly it makes only sense to speak of codons in mRNA sequences, but codons are also usually referred to when describing nucleotide triplets in the coding sequences of genomic DNA.
Complementary DNA (cDNA) This is generated by use of the enzyme reverse transcriptase to make a DNA copy (a ‘complementary’ copy) from RNA that has been isolated from a sample (for example, blood or tissue). This cDNA can be used to analyse splicing patterns and relative transcription levels.
Compound heterozygote An individual with (usually) pathogenic variants in both copies of a gene, where the variants are different from each other, for example a CF patient with p.Phe508del in one copy of the CFTR gene and p.Gly542X affecting the other copy.
Consanguineous Refers to families where both parents share at least one recent common ancestor.
Copy number variant (CNV)/copy number polymorphism (CNP) Segments of our genome that range in size from 1000 to millions of bp, and which, in healthy individuals, may vary in copy number from zero to several copies. Where the population frequency reaches 1% or more it may be referred to as a copy number polymorphism.
Cytogenetics The study of chromosomes.
De novo Latin for ‘anew; starting from the beginning’. Used to describe newly arisen mutations, as opposed to variants which have been inherited from a parent.
Dideoxynucleotide Used in Sanger DNA sequencing, the deoxyribose moiety of these nucleotides lacks the 3′ hydroxy group so that, while dideoxynucleotides can be incorporated into a growing DNA chain, they do not allow the addition of further nucleotides, so that the chain is terminated.
Differentiation The process by which cells and tissues acquire specialized characteristics, for example during embryonic development.
Diploid Having two copies of each autosome, and two sex chromosomes. This is the normal state of most human somatic cells.
Disorders of sex development (DSD) A diverse group of conditions that affect the development of the gonads and/or sexual differentiation and include partial or complete sex reversal in relation to the XX or XY genotype.
Dominant An allele or mutant gene version that leads to a phenotype when in a heterozygous state (for example the other allele is wild-type) is referred to as dominant, is also often used to describe a condition.
Dosage compensation The mechanism by which an imbalance in gene dose (gene copy number) is compensated for by differential gene expression. This is particularly relevant to genes residing on the X chromosome that have no Y chromosome homologue. In mammals, the process of X chromosome inactivation results in only one copy of the two alleles in female cells being available for expression, balancing with the fact that male cells only have one allele.
Dysgenesis Defective or abnormal development of an organ, for example of the gonads.
Electropherogram This is a visualisation of the results of electrophoretic separation of molecules; in the case of genetic analysis, DNA molecules can be separated based on size, and detected by the use of fluorescent labels previously attached to the DNA.
EnhancerA specific DNA sequence often adjacent to the coding region of a gene, which functions in gene regulation, for example by binding transcription factors.
Epigenetic modificationThis refers to modification marks that do not change a DNA sequence, but can effect gene expression and includes methylation of DNA bases (usually cytosine in mammals), and methylation, phosphorylation and acetylation of the proteins that DNA is wrapped around, the histones.
Epigenetics The study of changes in gene function that are heritable mitotically and/or meiotically and that are not a consequence of change in the DNA sequence.
Exome The portion of the genome that codes for proteins – the complete collection of all the exons.
Exon A section of a protein-coding gene that encodes part of the protein sequence; within the gene exons are separated by intervening sequences (introns) and in order to generate a functional mRNA the relevant exons must be spliced together to generate an uninterrupted coding sequence.
Expression
Gene expression represents the processes including transcription and translation which lead to the production of products (for example, proteins) from genes
Expression is also used to describe physical traits (or phenotypes) generated as a consequence of variants; the term expressivity is the degree to which the traits observed differ between individuals who have the same genotype.
Frameshift Ribosomes translate mRNA molecules one triplet codon at a time, in a continuous ‘reading frame’. Any mutation that leads to the insertion or deletion of a number of nucleotides into the mRNA, which is not a multiple of three, leads to a shift in this reading frame. This usually leads to premature truncation of the resulting polypeptide.
Gametologue A gene that has homologues on both X and Y chromosomes that are not subject to crossover in meiosis. These are not termed alleles due to the fact that they do not recombine and therefore evolve independently on the two chromosomes.
Gene amplification The duplication of a gene, often at the site of the original gene, leading to multiple copies. The duplicated gene may be wild-type or mutant and duplication usually results in its overexpression.
Genome The complete set of genetic information (usually DNA) of an organism, including all genes plus all other sequences, and in humans includes both nuclear and mtDNA.
Genomics In contrast with genetics, which often focuses on single genes, genomics represents the study of large groups of genes, often the entire genome of one or more organisms.
Genotype The genetic make-up of a cell or organism, relating to the sequence of the genes and genome as a whole. Often the genotype(s) at one or a few loci only are considered. Genotyping is the process of determining which alleles are present at one or more loci.
Germ cells Cells that will form the gametes, to become the haploid oocytes or sperm cells upon differentiation.
Gonadal mosaic Having cells of different genotypes within one or both gonads, often as a consequence of somatic mutation, with the consequence that an apparently de novo mutation, not present in the parent, may be transmitted to more than one child.
Haploid Having only a single copy of each autosome, and one sex chromosome. The usual state of gametes.
HaploinsufficiencyThis occurs when one allele of the homologous pair of genes in a diploid organism is lost or not expressed and this results in an abnormal phenotype. The remaining allele is expressed, but can only provide half the normal level of gene product and this is not sufficient to fully conduct the required function. Such loss-of-function mutations are dominant, as they give rise to a phenotype.
Hemizygous Having only one locus/allele within the cell.
Heteroplasmy The presence of differing mitochondrial genomes within a cell.
Heterozygous Having two different alleles at one locus.
Histone acetyl transferase (HAT)/histone deacetylase (HDAC) These enzymes add or remove acetyl groups (O=C–CH3) from histone proteins leading to changes in chromatin structure that affect function of the DNA.
Homologue
This is a genetical term referring to genes that are related by evolutionary descent, that is, homologues have evolved from the same gene in an ancient organism. There are also subdivisions of the term homologue. Paralogue: indicating descent within a single species, for example the human RAS genes (HRAS, NRAS and KRAS) are paralogues of each other. In the ancestral organism, the RAS gene underwent duplications and three of these remain in humans (and many other mammals) today that have evolved to take on slightly different roles. Orthologue: indicating the same gene in different species, for example human HRAS and mouse HRas are orthologues.
In a diploid organism, each autosome and the X chromosome in females (and therefore also each gene on these chromosomes), has a homologue, thus comprising the homologous pairs of chromosomes present in the nucleus of the diploid cell.
Homoplasmy The presence of only identical mitochondrial genomes within a cell.
Homozygous Having two identical alleles at one locus.
Ideogram A graphical representation of a cell’s or organism’s karyogram.
Imprinting (chromosomal or genomic) The process by which epigenetic marks are attached to particular loci in a parent-of-origin specific manner, leading to differential expression of maternally and paternally derived genes.
Indel A term describing any variant which represents the insertion or deletion (or a combination of both) of nucleotides at a specific position, compared with a reference genome.
Inflammation This describes an immune response, usually wounding or infection, but can be of unknown cause. Immune cells enter the damaged or infected tissue and release factors that are designed to repair the wound and combat infection. Short-lived inflammation, for example in response to a small wound, is called acute inflammation. Prolonged inflammation, sometimes of unknown cause, is called chronic inflammation and can be damaging to tissues if it continues unabated.
In silico Performed using a computer; for example the application of software or algorithms that use existing information to predict the effect of DNA variants.
Intron A segment of a gene, between two coding segments (exons), in other words an intervening sequence, which is transcribed into RNA and then removed by splicing during generation of the final mRNA.
Karyogram A (usually photographic) representation of the chromosomes of a cell, arranged in pairs.
Karyotype The number and appearance of the chromosomes in the nucleus.
KinaseKinases are enzymes that add a phosphate group onto their substrates. Protein kinases phosphorylate (by transfer of the phosphate group onto the oxygen atom of the amino acid side chain) their protein substrates upon serine, threonine or tyrosine residues.
Locus Genetical term referring to a specific location in a genome, usually defining the position of a gene or DNA sequence of interest. Plural: loci.
Metaphase One of the phases of the mitotic cell division cycle, during which chromosomes become condensed and visible under a light microscope.
Metastasis (plural metastases)/metastatic disease Cancer cells that are dividing out of control and have spread (from the primary tumour site) to other tissues or organ sites around the body.
Meiosis The final stages of germ cell division to produce four haploid gametes, each of which is genetically distinct. A germ cell undergoes DNA replication and then, before separation of the replicated ‘sister chromatids’, homologous chromosomes pair and undergo recombination, such that DNA is swapped or ‘crossed over’. Then two rounds of cell division occur: Meiosis I and Meiosis II. In Meiosis I, the chromosome pairs separate into daughter cells, in Meiosis II, the sister chromatids separate into daughter cells. In some organisms and in the production of mammalian sperm, all four meiotic products form gametes. In female mammals, only one cell develops into the ovum or oocyte, with asymmetric division of the cytoplasm, the other three meiotic nuclei are extruded as polar bodies.
Methylation/methyltransferase The process of methylation involves the addition of methyl (CH3) groups to target molecules, for example DNA or proteins, by the action of the appropriate methyltransferase enzymes.
Microarray A set of targets, most often DNA probes, arranged in a grid to facilitate testing. DNA microarrays, including SNP arrays, usually contain hundreds of thousands of probes to which sample DNA or RNA can be hybridised.
Microdeletion/microduplication A deletion/duplication that is generally defined as below the resolution of karyotyping and therefore less than 4–5 Mb, but larger than 1 kb. However, precise definitions may vary between authors.
Microsatellite A variable number tandem repeat in which the repeating unit is generally between 2 and 6 bp in length, and under some definitions includes mononucleotide repeats.
Minisatellite A variable number tandem repeat in which the repeating unit is generally between 10 and 100 bp in length.
Minor allele frequency (MAF) The frequency at which the second most common allele occurs in a population.
Missense An alteration (mutation), often affecting a single nucleotide, which leads to the change of a codon for one specific amino acid into one for a different amino acid.
Mitosis The process of somatic cell division, as part of the cell cycle after DNA replication, whereby a diploid cell goes through the stages: prophase, metaphase, anaphase and telophase; to separate the chromosomes into two nuclei. This is followed by cytokinesis, dividing the cytoplasm and organelles to produce two diploid daughter cells.
Mitochondrial replacement therapyA modification of IVF in which the mitochondria of the embryo are obtained from someone other than the mother or father of the child.
Modifier gene A gene in which variation can alter the severity or phenotype of disease caused by a pathogenic variant at another locus.
Molecular pathology The study and diagnosis of disease by analysis of molecules such as nucleic acids and proteins within tissues or body fluids.
Mosaic A condition in which cells of distinct genotypes or distinct karyotypes are present in one individual, often as a consequence of somatic mutation or mitotic non-disjunction.
Mutagen Any agent that can lead to DNA mutation. Therefore, by definition, all mutagens are potential carcinogens.
Mutant see Wild-type.
Mutation Any heritable (through somatic cell division or germline) change in the DNA sequence. It does not have to result in a phenotypic change or a change to an encoded protein sequence (which would be a silent mutation). See also the definition of wild-type (and polymorphic, variant and mutant alleles).
Non-invasive prenatal diagnosis (NIPD)A form of PND in which foetal DNA is obtained from the maternal blood rather than from a CVS or amniotic fluid sample.
Nonsense An alteration (mutation) affecting a single nucleotide which leads to the change of a codon for one specific amino acid into one of the three stop codons.
Oncogene A gene whose product can positively contribute to the cancerous process. Often an oncogene is a mutated form of a normal cellular gene.
Pathogenic variant A variant which is associated with disease.
Penetrance The extent to which a pathogenic variant leads to observable clinical symptoms, that is, the proportion of individuals with the variant who exhibit the disease phenotype. A variant with 100% penetrance would affect all carriers, whereas for a variant with 80% penetrance the particular phenotype would not be seen in 20% of carriers.
Phenotype The appearance, properties and behaviour of a cell or organism that are a direct consequence of its genotype.
Point mutation: The mutational change of one base pair, either to any of the other three possibilities, or deletion of the base pair, or addition of a new base pair in the sequence.
Polygenic The situation where a trait or phenotype is controlled by multiple genes.
Polymorphism Any variant allele that is present in the population at a frequency of at least 1% of all alleles.
Polyploidy State of a cell or organism which contains more than two complete sets of all chromosomes. Triploidy indicates the presence of three sets of chromosomes, tetraploidy of four.
PrimerA short sequence of DNA or RNA which can act as a starting place for a new DNA strand to be synthesised.
Programmed cell death see Apoptosis.
Pronuclear transfer A technique in which the mother’s egg as well as a donor egg are both fertilised with the father’s sperm. Subsequently the nucleus from each fertilised egg is removed and the donor egg’s nucleus is then replaced with that of the mother.
Pseudogene A locus that resembles a protein-coding gene, and likely arose via an ancient gene-duplication event, but which, as a consequence of mutation, is not able to generate the original protein. Pseudogenes may, however, have regulatory roles in expression of other genes.
Quiescence Describes the state when cells are not in cell cycle (reversible G0).
Recessive An allele or mutant gene version that leads only to a phenotype when in a homozygous state (or heterozygous with another pathogenic allele) is referred to as recessive. Also used to describe a condition.
Reference sequence A genomic sequence that represents a baseline from which to compare individual human sequences; the reference sequence is intended not as a ‘norm’, but only as a reference point for describing human variation.
Senescence Describes the state when cells can no longer divide, they are irreversibly in G0. They may still be able to function.
Sex chromosome Chromosomes determining the genetic gender of an organism. Human sex chromosomes are X and Y, male somatic cells carry one of each, female somatic cells carry two X chromosomes.
Signal transductionThe process by which a cell converts an external signal into a responding action. For example, when a growth factor binds to its receptor on the cell surface, this sends a cascade of signals inside the cell, to the nucleus (or other target, such as mitochondria), which can result in a change in gene expression resulting in the cell following a new action (such as initiating the cell cycle).
Silent mutation A mutation that results in no observable phenotypic effect.
Somatic cells Comprise all cells of the body, other than those that contribute to the germ line.
Spindle transfer A technique in which the nucleus is removed from an unfertilised mother’s egg and this is then inserted into a donor egg which has had its nucleus removed. Fertilisation with the father’s sperm then takes place.
Synonymous mutation A mutation in a coding region which, despite changing the DNA sequence does not change the sequence of the resulting polypeptide (due to the redundancy of the genetic code).
Tetraploidy Having four complete sets of chromosomes – i.e. four times the haploid number.
Telomere Specific, repetitive, sequences present at the ends of linear chromosomes that protect the chromosome ends and play a key role in chromosome maintenance and stability.
Translocation This describes a large rearrangement, often visible by karyotypic analysis, where a large chunk of one chromosome has moved and joined another chromosome. Reciprocal translocation is when two chromosomes have effectively exchanged large portions. In a Robertsonian translocation, two acrocentric chromosomes lose their short arms and fuse at the centromere.
Transcription factorA protein that, typically, binds to DNA (often as part of a complex of proteins) and regulates (up or down) the expression of a gene that is usually located at, or near, the site of binding.
Triploidy Having three complete sets of chromosomes – i.e. three times the haploid number.
Trisomy Three copies of a specific chromosome, e.g. three copies of chromosome 21 in Down syndrome.
Tumour suppressor gene (TSG) A gene whose product can inhibit tumour cell growth, often through inhibiting cell cycle progression. One or more TSGs are frequently silenced in cancer cells.
Ubiquitin A small protein of 76 amino acids, that can be covalently attached to other proteins, by ubiquitin ligases, resulting in a variety of potential regulatory effects that include targeting for degradation, and changes in localisation or activity of the ubiquitinated protein.
Uniparental disomy (UPD) A situation in which both homologues of a chromosome are from the same parent.
Variant Any DNA sequence that differs from the reference genome sequence.
Variant of uncertain significance (VUS) A classification applied to variants for which the effect on the phenotype is not clear, and there is insufficient evidence to support either benign or pathogenic classification.
Wild-type allele A term used by research geneticists to indicate the allele that is most commonly found in the population. Different alleles commonly found in the population are referred to as polymorphisms, or more rare alleles, variants. Medical geneticists tend not to use the term wild-type and instead refer to the human reference genome (see also Allele). Mutant alleles are those that have undergone a sequence change as a result of somatic mutation. Constitutional pathogenic variants are often referred to by research geneticists as ‘mutant’ alleles (and this term will be found in the scientific literature), but this terminology is out of favour with medical geneticists.
X chromosome inactivation The almost complete expressional silencing of one of the two X chromosomes in every mammalian female cell, with the exception of the gametes. The process is initiated in early embryonic development and persists throughout adult life.
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Author contribution
The main sections of this article were authored as follows:
- The human genome and variation by Maria Jackson
- Chromosome structure and chromosomal disorders by Gerhard H.W. May
- Single‐gene disorders by Gerhard H.W. May
- The sex chromosomes, X and Y by Joanna B. Wilson
- Mitochondrial disorders by Leah Marks
- Epigenetics by Maria Jackson
- Complex disorders by Maria Jackson and Leah Marks
- Cancer: mutation and epigenetics by Joanna B. Wilson
- Genomics by Maria Jackson and Joanna B. Wilson
- Genetic testing in the diagnostic laboratory by Maria Jackson and Leah Marks
- Diagnosis, Management and therapy of genetic disease by Leah Marks
- Challenges in delivering a genetics service by Leah Marks
Further reading: The human genome and variation
- Gonzaga-Jauregui C., Lupski J.R. and Gibbs R.A. (2012) Human genome sequencing in health and disease. Annu. Rev. Med. 63, 35–61 10.1146/annurev-med-051010-162644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E. (2018) Forensic DNA typing. Annu. Rev. Criminol. 1, 497–515 10.1146/annurev-criminol-032317-092127 [DOI] [Google Scholar]
- Richards S., Aziz N., Bale S., Bick D., Das S., Gastier-Foster J.. et al. (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels M.E. and Friedman J.M. (2015) Genetic mosaics and the germline lineage. Genes (Basel) 6, 216–237 10.3390/genes6020216 [DOI] [PMC free article] [PubMed] [Google Scholar]
Chromosome structure and chromosomal disorders
- To learn more about general genetics, consult one of many available text books. The following is freely available online.
- Griffiths A.J.F, Miller J.H., Suzuki D.T., Lewontin R.C. and Gelbart W.M. (2000) An Introduction to Genetic Analysis, 7th edn, W.H. Freeman, New York, https://www.ncbi.nlm.nih.gov/books/NBK21766/ [Google Scholar]
The sex chromosomes, X and Y
- Bonora G. and Disteche C.M. (2017) Structural aspects of the inactive X chromosome. Phil. Trans. R. Soc. B Biol. Sci. 372, 20160357, (and others in this volume of 12 contributions to a discussion meeting issue ‘X-chromsome inactivation: a tribute to Mary Lyon’) 10.1098/rstb.2016.0357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiot E., Zenaty D., Boizeau P., Haignere J., Dos Santos S. and Leger J (2016) X-chromosome gene dosage as a determinant of impaired pre and postnatal growth and adult height in Turner syndrome. Eur. J. Endocrinol. 174, 281–288 10.1530/EJE-15-1000 [DOI] [PubMed] [Google Scholar]
- Gamble T. and Zarkower D. (2012) Sex determination. Current Biol. 22:, 257–262 10.1016/j.cub.2012.02.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert S.F. (2000) Developmental Biology, 6th edn, Chromosomal Sex Determination in Mammals Sinauer Associates [Google Scholar]
- Lombardi L.M., Baker S.A. and Zoghbi H.Y. (2015) MECP2 disorders: from the clinic to mice and back. J. Clin. Invest. 125, 2914–2923 10.1172/JCI78167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro I. and Heard E. (2017) X chromosome inactivation: new players in the initiation of gene silencing. F1000 Res., 6, 344–354 10.12688/f1000research.10707.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevant I., Papaioannour M.D. and Nef S. (2018) A brief history of sex determination. Mol. Cell. Endocrinol. 10.1016/j.mce.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Tanaka S.S. and Nishinakamura R. (2014) Regulation of male sex determination: genital ridge formation and Sry activation in mice. Cell. Mol. Life Sci. 71, 4781–4802 10.1007/s00018-014-1703-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Single-gene disorders
- Chial H. (2008) Mendelian genetics: patterns of inheritance and single-gene disorders. Nat. Education 1, 63 [Google Scholar]
- Davis P.B. (2001) Cystic fibrosis. Pediatr. Rev. 22, 257–264 10.1542/pir.22-8-257 [DOI] [PubMed] [Google Scholar]
- Martiniano S.L., Sagel S.D. and Zemanick E.T. (2016) Cystic fibrosis: a model system for precision medicine. Curr. Opin. Pediatr. 28:, 312–317 10.1097/MOP.0000000000000351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nopoulos P.C. (2016) Huntington disease: a single-gene degenerative disorder of the striatum. Dialogues Clin. Neurosci. 18, 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz D.M. and Legeai-Mallet L. (2017) Achondroplasia: development, pathogenesis, and therapy. Dev. Dyn. 246, 291–309 10.1002/dvdy.24479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B.Z., Haaf J.B., Leal T. and Noel S. (2016) Cystic fibrosis transmembrane conductance regulator modulators in cystic fibrosis: current perspectives. Clin. Pharmacol. 8:, 127–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
Mitochondrial disorders
- Chinnery P. (2000) Mitochondrial Disorders Overview. SourceGeneReviews® (Adam M.P., Ardinger H.H., Pagon R.A., Wallace S.E., Bean L.J.H., Stephens K. and Amemiya A., eds), pp. 1993–2018, University of Washington, Seattle, Seattle (WA) [Google Scholar]
- Chong J.X., Buckingham K.J., Jhangiani S.N., Boehm C., Sobreira N., Smith J.D.. et al. (2015) The genetic basis of mendelian phenotypes: discoveries, challenges, and opportunities. Am. J. Hum. Genet. 97, 199–215 10.1016/j.ajhg.2015.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale H., Pfeffer G., Bargiela D., Horvath R. and Chinnery P. F. (2016) Emerging therapies for mitochondrial disorders. Brain 139, 1633–1648 10.1093/brain/aww081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznichenko A., Huyser C. and Pepper M. (2016) Mitochondrial transfer: implications for assisted reproductive technologies. Appl. Transl. Genom. 11, 40–47 10.1016/j.atg.2016.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Epigenetics
- Barlow D.P. and Bartolomei M.S. (2014) Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 6, a018382 10.1101/cshperspect.a018382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo I. (2008) Genomic imprinting and patterns of disease inheritance. Nat. Education 1, 66 [Google Scholar]
- Schuebel K., Gitil M., Domschke K. and Goldman D. (2016) Making sense of epigenetics. Int. J. Neuropsychopharmacol. 19, 1–10 10.1093/ijnp/pyw058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soshnev A.A., Josefowicz S.Z. and Allis C.D. (2016) Greater than the sum of parts: complexity of the dynamic epigenome. Mol. Cell 62, 681–694 10.1016/j.molcel.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturá-Junca P., Irarrázaval I., Rolle A.J., Gutiérrez J.I., Moreno R.D. and Santos M.J. (2015) In vitro fertilization (IVF) in mammals: epigenetic and developmental alterations. Scientific and bioethical implications for IVF in humans. Biol. Res. 48:, 68 10.1186/s40659-015-0059-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi H.Y. and Beaudet A.L. (2016) Epigenetics and human disease. Cold Spring Harb. Perspect. Biol. 8:, a019497 10.1101/cshperspect.a019497 [DOI] [PMC free article] [PubMed] [Google Scholar]
Complex disorders
- Chial H. and Craig J. (2008) Genome-wide association studies (GWAS) and obesity. Nat. Education 1, 80 [Google Scholar]
- Prasad R.B. and Groop L. (2015) Genetics of type 2 diabetes–pitfalls and possibilities. Genes 6, 87–123 10.3390/genes6010087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz L.C. (2010) The Dutch Hunger Winter and the developmental origins of health and disease. Proc. Natl. Acad. Sci. U.S.A. 107, 16757–16758 10.1073/pnas.1012911107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Cancer: mutation and epigenetics
- Aunan J.R., Cho W.C. and Søreide K. (2017) The biology of aging and cancer: a brief overview of shared and divergent molecular hallmarks. Aging Dis. 8, 628–642 10.14336/AD.2017.0103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burotto M., Chiou V.L., Lee J-M. and Kohn E.C. (2014) The MAPK pathway across different malignancies: A new perspective. Cancer 120, 3446–3456 10.1002/cncr.28864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbridge A.R., Valente L.J. and Strasser A. (2012) The role of the apoptotic machinery in tumor suppression. Cold Spring Harb. Perspect. Biol. 4, a008789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A.P., Koldobskiy M.A. and Gondor A. (2016) Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 17, 284–299 10.1038/nrg.2016.13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M. and Müller G.A. (2017) Cell cycle transcription control: DREAM/MuvB and RB-E2F complexes. Crit. Rev. Biochem. Mol. Biol. 52, 638–662 10.1080/10409238.2017.1360836 [DOI] [PubMed] [Google Scholar]
- Hanahan D. and Weinberg R.A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Pereira B. (2016) The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat. Commun. 7, 11479 10.1038/ncomms11479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior I., Lewis P.D. and Mattos C. (2012) A comprehensive survey of Ras mutations in cancer. Cancer Res. 72, 2457–2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryana B.M. and Faupel-Badgerb J.M. (2016) The hallmarks of premalignant conditions: a molecular basis for cancer prevention. Semin. Oncol. 43, 22–35 10.1053/j.seminoncol.2015.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C.J. (2004) Principles of tumor suppression. Cell 116, 235–246 10.1016/S0092-8674(03)01075-4 [DOI] [PubMed] [Google Scholar]
- Weinberg R.A. (2014) The Biology of Cancer, 2nd edn, Garland Science, ISBN: 978-0-8153-4219-9/978-0-8153-4220-5 [Google Scholar]
- Cancer Research UK, http://www.cancerresearchuk.org/health-professional/data-and-statistics
Genomics
- Feero W.G. and Gutmacher A.E. (2014) Genomics, personalized medicine, and paediatrics. Acad. Pediatr. 14, 14–22 10.1016/j.acap.2013.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomics England, 100,000 genomes project, 2018. https://www.genomicsengland.co.uk/
- Naidoo N., Pawitan Y., Soong R., Cooper D.N. and Ku C.-S. (2011) Human genetics and genomics a decade after the release of the draft sequence of the human genome. Hum. Genomics 5, 577–622 10.1186/1479-7364-5-6-577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehm H.L. (2017) Evolving healthcare through personal genomics. Nat. Rev. Genet. 18, 259–267 10.1038/nrg.2016.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scottish Genomes Partnership (2018), https://www.scottishgenomespartnership.org/
Genetic testing in the diagnostic laboratory
- Bishop R. (2010) Applications of fluorescence in situ hybridization (FISH) in detecting genetic aberrations of medical significance. Biosci. Horiz. 3, 85–95, 10.1093/biohorizons/hzq009 [DOI] [Google Scholar]
- Ferrie R.M., Schwarz M.J., Robertson N.H., Vaudin S., Super M., Malone G.. et al. (1992) Development, multiplexing, and applications of ARMS tests for common mutations in the CFTR gene. Am. J. Hum. Genet. 51, 251–262 [PMC free article] [PubMed] [Google Scholar]
- Frese K.S., Katus H.A. and Meder B. (2013) Next-generation sequencing: from understanding biology to personalized medicine. Biology (Basel) 2, 378–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis S.H. and Katsanis N. (2013) Molecular genetic testing and the future of clinical genomics. Nat. Rev. Genet. 14, 415–426 10.1038/nrg3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kchouk M., Gibrat J.-F. and Elloumi M. (2017) Generations of sequencing technologies: from first to next generation. Biol. Med. (Aligarh) 9, 395 10.4172/0974-8369.1000395 [DOI] [Google Scholar]
- Norbury G. and Norbury C.J. (2006) DNA analysis: what and when to request? Arch. Dis. Child. 91, 357–360 10.1136/adc.2005.089219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor C. (2008) Karyotyping for chromosomal abnormalities. Nat. Education 1, 27 [Google Scholar]
- O’Connor C. (2008) Fluorescence in situ hybridization (FISH). Nat. Education 1, 171 [Google Scholar]
- Stuppia L., Antonucci I., Palka G. and Gatta V. (2012) Use of the MLPA assay in the molecular diagnosis of gene copy alterations in human genetic diseases. Int. J. Mol. Sci. 13, 3245–3276 10.3390/ijms13033245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiszniewska J., Bi W., Shaw C., Stankiewicz P., Kang S.H., Pursley A.N.. et al. (2014) Combined array CGH plus SNP genome analyses in a single assay for optimized clinical testing. Eur. J. Hum. Genet. 22, 79–87 10.1038/ejhg.2013.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xuan J., Yu Y., Qing T., Guo L. and Shi L. (2013) Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 340, 284–295 10.1016/j.canlet.2012.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
Diagnosis, management and therapy of genetic disease
- Aronson S.J. and Rehm H.L. (2015) Building the foundation for genomics in precision medicine. Nature 526, 336–342 10.1038/nature15816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaterji S., Ahn E.H. and Kim D.-H. (2017) CRISPR genome engineering for human pluripotent stem cell research. Theranostics 7, 4445–4469 10.7150/thno.18456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid A. and Mahfouz M.M. (2016) Genome editing: the road of CRISPR/Cas9 from bench to clinic. Exp. Mol. Med. 48, e265 10.1038/emm.2016.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockyer E. (2016) The potential of CRISPR-Cas9 for treating genetic disorders. Biosci. Horiz. 9, 10.1093/biohorizons/hzw012 [DOI] [Google Scholar]
- Martiniano S.L., Sagel S.D. and Zemanick E.T. (2016) Cystic fibrosis: a model system for precision medicine. Curr. Opin. Pediatr. 28, 312–317 10.1097/MOP.0000000000000351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plönes T., Engel-Riedel W., Stoelben E., Limmroth C., Schildgen O. and Schildgen V. (2016) Molecular pathology and personalized medicine: the dawn of a new era in companion diagnostics–practical considerations about companion diagnostics for non-small-cell-lung-Cancer. J. Personal. Med. 6, 3 10.3390/jpm6010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt B.Z., Haaf J.B., Leal T. and Noel S. (2016) Cystic fibrosis transmembrane conductance regulator modulators in cystic fibrosis: current perspectives. Clin. Pharmacol. 8, 127–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneller J.L., Lee C.M., Bao G. and Venditti C.P. (2017) Genome editing for inborn errors of metabolism: advancing towards the clinic. BMC Medicine 15, 43 10.1186/s12916-017-0798-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumaily K.M. and Mujamammi A.H. (2017) Phenylketonuria: a new look at an old topic, advances in laboratory diagnosis, and therapeutic strategies. Int. J. Health Sci. 11, 63–70 [PMC free article] [PubMed] [Google Scholar]
Challenges in delivering a genetics service
- Blashki G., Metcalfe S. and Emery J. (2014) Genetics in general practice. Aust. Fam. Phys. 43, 428–431 [PubMed] [Google Scholar]
- Harris A., Kelly S. E. and Wyatt S. (2013) Counseling customers: emerging roles for genetic counselors in the direct-to-consumer genetic testing market. J. Genet. Couns. 22, 277–288 10.1007/s10897-012-9548-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J.S., Dolinoy D. and Tarini B. (2014) Emerging issues in public health genomics. Annu. Rev. Genomics Hum. Genet. 15, 461–480 10.1146/annurev-genom-090413-025514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su P. (2013) Direct-to-consumer genetic testing: a comprehensive view. Yale J. Biol. Med. 86, 359–365 [PMC free article] [PubMed] [Google Scholar]