Abstract
The development of new heterologous hosts for polyketides production represents an excellent opportunity to expand the genomic, physiological, and biochemical backgrounds that better fit the sustainable production of these valuable molecules. Cyanobacteria are particularly attractive for the production of natural compounds because they have minimal nutritional demands and several strains have well established genetic tools. Using the model strain Synechococcus elongatus, a generic platform was developed for the heterologous production of polyketide synthase (PKS)-derived compounds. The versatility of this system is based on interchangeable modules harboring promiscuous enzymes for PKS activation and the production of PKS extender units, as well as inducible circuits for a regulated expression of the PKS biosynthetic gene cluster. To assess the capability of this platform, we expressed the mycobacterial PKS-based mycocerosic biosynthetic pathway to produce multimethyl-branched esters (MBE). This work is a foundational step forward for the production of high value polyketides in a photosynthetic microorganism.
Keywords: Cyanobacteria, Heterologous production, PKS-derived compounds, Synthetic Biology
1. Introduction
Polyketides are included in a large family of natural products that possess a wide variety of relevant pharmacological and biological activities. Numerous polyketides and their semisynthetic derivatives have been approved for clinical use in humans and animals, including antibiotics, antifungal agents, immunosuppressants, antiparasitic agents, and insecticides1. All these natural products share a common mechanism of biosynthesis and are produced by a class of enzymes called polyketide synthases (PKSs). These enzymes condense and reduce a small series of simple 2-, 3-, and 4-carbon acyl groups derived from coenzyme A (CoA) thioesters (for example malonyl-CoA, methylmalonyl-CoA, and ethylmalonyl-CoA) to build the polyketide backbone in a step-wise manner that resembles fatty acid biosynthesis2. Besides their essential role in the biosynthesis of a vast diversity of natural products, the versatility of PKSs can be further emphasized as they can be redesigned and repurposed to produce novel molecules that could be used as fuels, industrial chemicals, and monomers1,3–8
Most polyketide producers are slow-growing, recalcitrant to genetic manipulation, or even non-culturable2,9. Therefore, as an alternative to studying these polyketides and their encoding gene clusters from the native producers, the heterologous production of polyketide molecules has proven to be a valuable strategy. Since its early days10,11, the heterologous expression of the PKS biosynthetic gene clusters has greatly facilitated our understanding of the basic enzymatic mechanisms involved in polyketide biosynthesis and enabled the manipulation of polyketide-encoding pathways either to improve their production or in order to generate novel molecules with novel properties1,8,12–15.
Photosynthetic microorganisms are attractive platforms for the production of specialty chemicals, valuable drugs, and fuels because they can use CO2 and sunlight as sole sources of carbon and energy16. Cyanobacteria are the only prokaryotes that can perform plant-like oxygenic photosynthesis. In comparison to eukaryotic algae, several strains of cyanobacteria have facile genetics that take advantage of their natural competency to take up exogenous DNA and their capability to integrate DNA into their chromosome through homologous recombination. In this regard, Synechococcus elongatus strain PCC 7942 (thereafter S. elongatus) is an excellent host strain that grows rapidly and segregates mutations efficiently. S. elongatus’s fundamental biological processes and primary metabolism have been extensively studied17–19. In addition, genome-scale metabolic models, as well as a broad array of advanced genetic tools, facilitate the engineering and construction of complex metabolic systems20–22. S. elongatus and other cyanobacteria have been used as photosynthetic biocatalysts for the biosynthesis of several industrial compounds23–25. However, synthetic biology and metabolic engineering of cyanobacteria have only rarely been applied to natural product pathways26,27.
In this work, we demonstrate the potential of the photosynthetic microorganism S. elongatus as a versatile platform for the heterologous biosynthesis of PKS-derived molecules. To facilitate the construction of diverse recombinant cyanobacteria, we developed a modular design where each module comprises a set of enzymes that perform a specific function required for the biosynthesis of the final polyketides and polyketide-like molecules. As a proof of concept, we focus on the type I iterative Mycobacterium tuberculosis PKS MAS system (comprising mas, papA5, fadD28 genes) and show that the engineered S. elongatus strains produce (2S)-methylmalonyl-CoA PKS-derived molecules.
2. Materials and Methods
2.1. Strain construction and culture conditions
Plasmid DNA was introduced by natural transformation in a sub-strain of S. elongatus PCC 7942 (AMC2302), which was cured of its small plasmid, pANS28,29. Recombinant DNA harboring the functional modules I through III (Fig. 1) and an antibiotic resistance gene were integrated into the chromosome at neutral site 1 (NS1), 2 (NS2), and 3 (NS3) or were carried on self-replicating plasmids based on the native pANS plasmid of S. elongatus PCC 794221,29,30 Plasmids and cyanobacterial strains constructed in this study are listed in Table 1 and 2, respectively. S. elongatus strain AMC2302 was grown in BG11 medium31 as 50-ml or 100-ml cultures at 30°C with continuous shaking or on agar plates (40 ml, 1.5% agarose) and continuous illumination of 150-μmol photons m−2 s−1. Media were supplemented with appropriate antibiotics at the following final concentrations: 2 μg/ml each spectinomycin (Sp) and streptomycin (Sm), 2 μg/ml gentamycin (Gm), 5 μg/ml kanamycin (Km), and 10 μg/ml nourseothricin (Nt).
Figure 1: Modular-design of essential components for the production of PKS-derived compounds in S. elongatus.

Module I is responsible for the biosynthesis of the carboxyacyl-CoA precursors using three different routes: module IA contains matB and epi genes encoding for an acyl-CoA synthetase and epimerase, respectively; module IB contains mutA, mutB (encoding for the methylmalonyl-CoA mutase complex), and epi genes; module IC contains accA2, pccE, and pccB genes (encoding for the propionyl-CoA carboxylase complex). Module II is responsible for the post-translational modification of the PKS and the controlled orthogonal expression of the PKS gene cluster using two T7 RNAP expression devices. In module IIA, the T7 RNAP is expressed from the synthetic E. coli consensus conII promoter (PconII) with riboswitch F (rswF); in module IIB the T7 RNAP is expressed from a weakened version of the conII promoter (PconII*) with riboswitch B (rswB). In both modules, the sfp gene encoding for the phosphopantetheinyl transferase is expressed from the conII promoter with riboswitch F (rswF). Module III is responsible for the biosynthesis of the PKS-derived products, in this case, the mycobacterial PKS MAS pathway that includes papA5 (polyketide-associated protein A5), fadD28 (acyl-AMP ligase) and mas (mycocerosic acid synthase) genes. These genes were engineered in a single operon expressed from PT7.
Table 1.
Plasmids developed
| Plasmid | Description | Antibiotic resistance | Reference |
|---|---|---|---|
| pJR03 | pANS-aadA-PconII-matB | Sp Sm | This work |
| pJR04 | NS2TC-aphI-PconII-epimerase | Tc Km | This work |
| pJR05 | pAUS-aadA-PconII-mutA-mutB | Sp Sm | This work |
| pJR06 | pANS-aadA-PconII-accA2-pccE-pccB | Sp Sm | This work |
| pJR07 | NS1-nat1-PT7-papA5-fadD28-mas | Nt | This work |
| pJR08 | NS3-aacC1-PconII-rswF-sfp-PconII-rswF-77RNAP | Gm | This work |
| pJR09 | NS3-aacC1-PconII-rswF-sfp-PconII*-rswB-T7RNAP | Gm | This work |
| pAM5467 | NS2TC-aacC1-PT7-yfp | Gm | This work |
| pAM5470 | NS1-aadA-PconII*-rswB-T7RNAP | Sp Sm | This work |
| pAM5471 | NS1-aadA-PconII*-rswF-T7RNAP | Sp Sm | This work |
Table 2.
S. elongatus strains
| Strain | Genotype | Antibiotic resistance | Reference |
|---|---|---|---|
| AMC2302 | S. elongatus PCC 7942 WT strain cured of the small pANS plasmid | - | 52 |
| JR101 | AMC2302; pANS-aadA-PconII-matB; NS2TC::aphI-PconII-epimerase; NS1::nat1-PT7-papA5-fadD28-mas; NS3::aacC1-PconII-rswF-sfp-PconII-rswF-T7RNAP | Sp Sm Km Nt Gm | This work |
| JR102 | AMC2302; pANS-aadA-PconII-matB; NS2TC::aphI-PconII-epimerase; NS1::nat1-PT7-papA5-fadD28-mas; NS3::aacC1-PconII-rswF-sfp-PconII*-rswB-T7RNAP | Sp Sm Km Nt Gm | This work |
| JR103 | AMC2302; pANS-aadA-PconII-matB; NS2TC::aphI-PconII-epimerase; NS3::aacC1-PconII-rswF-sfp-PconII-rswF-T7RNAP | Sp Sm Km Gm | This work |
| JR104 | AMC2302; pANS-aadA-PconII-matB; NS2TC::aphI-PconII-epimerase; NS3::aacC1-PconII-rswF-sfp-PconII*-rswB-T7RNAP | Sp Sm Km Gm | This work |
| JR105 | AMC2302; pANS-aadA-PconII-mutA-mutB; NS2TC::aphI-PconII-epimerase; NS1::nat1-PT7-papA5-fadD28-mas; NS3::aacC1-PconII-rswF-sfp-PconII-rswF-T7RNAP | Sp Sm Km Nt Gm | This work |
| JR106 | AMC2302; pANS-aadA-PconII-mutA-mutB; NS2TC::aphI-PconII-epimerase; NS1::nat1-PT7-papA5-fadD28-mas; NS3::aacC1-PconII-rswF-sfp-PconII*-rswB-T7RNAP | Sp Sm Km Nt Gm | This work |
| JR107 | AMC2302; pANS-aadA-PconII-accA2-pccE-pccB; NS1::nat1-PT7-papA5-fadD28-mas; NS3::aacC1-PconII-rswF-sfp-PconII-rswF-T7RNAP | Sp Sm Nt Gm | This work |
| JR108 | AMC2302; pANS-aadA-PconII-accA2-pccE-pccB; NS1::nat1-PT7-papA5-fadD28-mas; NS3::aacC1-PconII-rswF-sfp-PconII*-rswB-T7RNAP | Sp Sm Nt Gm | This work |
| JR109 | AMC2302; pANS-aadA-PconII-accA2-pccE-pccB | Sp Sm | This work |
| JR110 | AMC2302; NS1::aadA-PconII*-rswB-T7RNAP; NS2::aacC1-PT7-yfp | Sp Sm Gm | This work |
| JR111 | AMC2302; NS1::aadA-PconII*-rswF-T7RNAP; NS2::aacC1-PT7-yfp | Sp Sm Gm | This work |
E. coli strains were grown at 37°C with LB medium in tubes placed on a roller drum, or on agar plates supplemented with appropriate antibiotics at standard concentrations.
2.2. General methods for vector and plasmid assembly
Plasmid preparations were carried out with QIAprep Spin Miniprep Kit (Qiagen). Restriction digests were typically performed for 3 to 5 h, using 5U of enzyme per pg of plasmid DNA in a final volume at least 50 times greater than the volume of the enzyme added. PCR amplifications were carried out with Q5 enzyme (New England BioLabs) according to the manufacturer’s instructions. DNA purification and concentration of restriction digests and PCR products were performed with DNA Clean & Concentrator TM-5 (Zymo). Nucleic acid concentrations were measured with a UV-Vis spectrophotometer NanoDrop 2000c. Cloning reactions were performed by Gibson assembly using GeneArt Seamless Cloning and Assembly Kit (Life Technologies).
Vector backbones were constructed in silico using the CYANO-VECTOR portal http://golden.ucsd.edu/CyanoVECTOR/ then assembled from compatible devices as described previously21. Briefly, DNA devices including S. elongatus recombination or replicon sequences, E. coli origin of replication, antibiotic selection markers and expression cassettes were released from donor plasmids by restriction digests using Zral or EcoRV-HF (New England BioLabs). Then, assembly reactions were carried out using equimolar ratios of each device adjusted to a final concentration of about 8 ng/μl. To obtain final constructs, inserts were either obtained by PCR, or synthesized and then assembled into vector backbones linearized by restriction digests.
Intermediate plasmids and destination vectors, and primer sequences used in this work are listed in the Supplementary Table S1 and Table S2, respectively. Details on the construction of each plasmid developed for the platform are provided as supplementary material (Fig. S1). The vectors were designed to be easily used for the exchange of different modules. Codon optimized synthetic gene sequence is detailed in Supplementary Materials and Methods. Synthetic DNA fragments were ordered as synthetic genes (GenScript) or gene blocks (SGI-DNA) and the matB gene was codon optimized using the COOL algorithm32
2.3. Yellow Fluorescent Protein (YFP) fluorescence measurements
First, cyanobacterial strains were adjusted to an optical density at 750 nm (OD750) of 0.2 and pre-grown for 48 h in liquid culture. Then, cultures were adjusted to OD750 of 0.1 before fluorescence measurements. For induction, 2-mL cultures were grown in 24-well plates and adjusted to final concentrations of 0, 0.25, 0.5, 1, 2 and 4 mM theophylline (Sigma-Aldrich). Theophylline was dissolved in DMSO at different stock concentrations (0, 25, 50, 100 and 200 mM) to obtain a final ratio of inducer solution to culture of 1% v/v or 2% v/v (for 4 mM theophylline). The plates were set on a shaker (125 rpm) at 30°C under continuous illumination of 100 μmol photons m−2 s−1 for 24 h. This time point was selected based in our previous work22, where we described and analyzed theophylline-responsive riboswitches. The optical densities and emission intensities of YFP were measured from 200 μL of culture in 96 well plates with a Tecan Infinite(R) M200 plate reader (TECAN). Black-walled 96 well plates (Greiner) were used to measure fluorescence intensities. The excitation and emission wavelengths were set to EX490/9 and EM535/20 for YFP. Measurements were taken from 2 or 3 independent strains and biological-triplicate cultures for each strain.
2.4. Phosphopantetheinyl Transferase (PPTase) in vitro assays
Soluble protein extracts from S. elongatus AMC2302 recombinant strains with a protein concentration of approximately 3 mg/mL were added to a volume of premade 10× PPTase reaction buffer (500 mM Na-HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic sodium salt) and 100 mM MgCl2, pH 7.6) that brought the total reaction concentration to 50 mM Na-HEPES, pH 7.6; 10 mM MgCl2; 100 μM TAMRA (Carboxytetramethylrhodamine)-CoA; TCEP (Tris(2-carboxyethyl)-phosphine) 5 mM and apo-ACP 25 mM. In the positive control, 2 μM Sfp was added instead of the protein extracts of the recombinant strains. Samples were incubated at 37°C overnight and then the detection of the ACP phosphopantetheinylation was carried out on SDS-PAGE and the reporter-labeled ACP was visualized by UV fluorescence.
2.5. Bioconversion assays
The cyanobacterial strains were pre-grown in liquid cultures (with appropriate antibiotics) for 3 to 4 days prior to induction. Then, the cultures were adjusted to an OD750 of 0.4, and the theophylline inducer was added. These cultures were grown for 19 h while shaking at 30°C under continuous illumination at 150 μmol photons m−2 s−1, and then supplemented with the appropriate precursors (n-octanol and propionate or methylmalonate), and 14C-acetate (58.9 mCi/mmol, PerkinElmer) to label the products. All assays were performed in triplicate with independent strains. Samples were collected 48 h after the precursors were added, stored at −80°C and lyophilized before processing for TLC (Thin Layer Chromatography) analysis.
For the E. coli bioconversion assay, an overnight culture of the strain RQ18 was used to inoculate fresh LB medium supplemented with 20 μM biotin and the appropriate antibiotics. At mid-exponential phase, the culture was induced with 0.1 mM IPTG, and 30 mM propionate, 0.5 mM n-octanol, and 14C-acetate (58.9 mCi/mmol, PerkinElmer) label were added. The induced culture was incubated for 20 h at 22°C. Then, the sample was collected and total lipids were extracted according to the Blight and Dyer method33.
2.6. TLC analysis
Total lipid samples were extracted by the Bligh and Dyer method with minor modifications33. First, the culture was lyophilized, then the cell pellet/lyophilized sample was resuspended in water. Subsequently, a solution of 1:2 chloroform: methanol with acetic acid 1% (final v/v) was added, and the samples were vortexed. Then chloroform and NaCl 1 M were added and finally centrifugated to separate phases. Total lipid extracts were analyzed by radio-TLC on silica gel 60 F254 plates (+/− 0.2 mm, Merck) using the solvent system hexane/diethyl-ether/acetic acid (90: 7.5: 1, v/v/v) for the mobile phase. The lipids were visualized by using a Typhoon Phosphorimager (GE Healthcare) and the spots corresponding to methyl-branched esters (MBEs) from the S. elongatus recombinant strains were assigned by Rf (Ratio of front) comparison with MBE synthesized by the E. coli RQ1 producer strain used as a standard8.
3. Results
3.1. Modules for the production of PKS-derived compounds
For a heterologous host to be suitable for the biosynthesis of PKS-derived compounds, it must be able to: 1) produce the appropriate PKS carboxyacyl-CoA substrates (extender units); 2) carry out the post-translational modification of the heterologous PKS ACP domain(s); and 3) harbor a robust and preferably regulated expression system for the often large and polycistronic PKS gene clusters. Therefore, we designed and constructed for S. elongatus three independent interchangeable and combinable modules (Fig. 1). Each module comprises a set of enzymes to perform the specific functions listed above. Module I is dedicated to the production of different carboxyacyl-CoAs used as PKS substrates. Module II includes T7 gene 1 (hereafter T7RNAP), which encodes the T7 RNA polymerase (T7 RNAP), for the robust and controlled expression of a PKS gene cluster driven by a T7 promoter (PT7), and a promiscuous phosphopantetheinyl transferase (PPTase) for the post-translational modification of the PKS34. Finally, module III harbors the PKS gene cluster driven by PT7.
Considering that the biosynthesis of many industrially relevant polyketides needs malonyl-CoA, methylmalonyl-CoA and/or ethylmalonyl-CoA as substrates35, three different pathways capable of generating these carboxyacyl-CoAs, but mainly focusing in methylmalonyl-CoA production, were designed as Module I. The first pathway involves matB from Streptomyces coelicolor and the epimerase (epi) gene from Propionibacterium shermanii. MatB is an acyl-CoA synthetase36 with a relaxed substrate specificity that allows the in vitro synthesis of different PKS extender units such as malonyl-CoA, methylmalonyl-CoA, ethylmalonyl-CoA, hydroxymalonyl-CoA, and methoxymalonyl-CoA from the corresponding carboxylic acid substrates36. The broad substrate specificity of this enzyme would contribute to the versatility of the platform strain because it would enable the intracellular production of more than one PKS extender unit, depending on the exogenous supply of the carboxylic acid. Because PKSs recognize only the 2S isomer of the α-substituted carboxyacyl-CoAs, and considering that MatB produces a racemic mixture of them, the epimerization of (2R)-α-substituted extender units formed by MatB would increase the availability of the correct 2S isomer before it can be accepted by the PKS acyltransferase domains. In this sense, we decided to co-express the methylmalonyl-CoA epimerase (epi) from P. shermanii37 to shift the equilibrium of the reaction towards the correct 2S isomer and therefore improve the production of the final polyketide. Hereafter this module will be named module IA (Fig. 1).
The second pathway for methylmalonyl-CoA production involves the sequential action of two enzymes, methylmalonyl-CoA mutase complex (mutAB) and methylmalonyl-CoA epimerase from P. shermanii, which convert succinyl-CoA to (2R)-methylmalonyl-CoA and then to (2S)-methylmalonyl-CoA37. This biosynthetic route was designed as module IB (Fig. 1).
The third pathway involves the S. coelicolor propionyl-CoA carboxylase (PCC) complex, which comprises the AccA2, PccB, and PccE subunits38. This enzyme complex has also been demonstrated to have a relaxed substrate specificity in vitro, being able to produce (2S)-methylmalonyl-CoA and (2S)-ethylmalonyl-CoA from their corresponding substrates, propionyl-CoA and butyryl-CoA, respectively38. This module was named IC (Fig. 1). Because the PCC substrate, propionyl-CoA, is not readily available in most bacteria, the PCC biosynthetic route requires an exogenous supply of propionate, which has to be further converted into propionyl-CoA. We hypothesized that this latter step could be performed by the S. elongatus native acetyl-CoA synthetase/acetyl-CoA ligase (AcsA, SynPCC7942_1352) because its orthologous protein from Synechococcus sp. PCC 7002 showed in vitro CoA ligase activity towards various organic acids, including acetate and propionate39.
The main genes of module I, namely MatB, MutAB complex, and the PCC complex, were cloned in a plasmid vector derived from S. elongatus small plasmid pANS (formerly identified as pUH24)40 downstream of a synthetic E. coli promoter (PconII)21,41 (Table 1). We expected that placing these genes in a replicative vector and downstream of a constitutive, medium strength promoter42 should ensure high levels of gene expression and the availability of the precursor molecules. What is more, pANS vector can also replicate in Anabaena sp PCC 7120 that could be used as an alternative cyanobacterial host26,29. The epi gene was also cloned downstream of PconII but in an integrative vector that recombines into the neutral site NS2 on the S. elongatus chromosome (Table 1)30.
Module II was designed to enable the orthogonal expression of the PKS biosynthetic gene cluster and the post-translational modification of the acyl carrier protein domain(s) (ACP) of the PKS. A variety of constitutive and inducible expression systems have been used in S. elongatus21,22,43–46, but compared to many other applications, the expression of PKS gene clusters presents additional challenges. PKSs are often arranged as large polycistronic transcriptional units, and although low levels of expression may be essential in some cases, several studies have suggested that the biosynthesis of polyketides and other bioactive specialized metabolites (at detectable levels) may require the use of strong promoters14. Therefore, we hypothesized that a controlled T7 expression system would be well suited for the expression of PKS gene clusters and could be engineered in S. elongatus. Typically, the gene encoding T7 RNAP is under the control of a derivative of the lac promoter, which is tightly regulated by the Lacl repressor47. IPTG inducible promoter systems have been used in cyanobacteria and in S. elongatus in particular; however, they usually exhibit high basal activity levels in the absence of inducer, and poor induction ratios22,44,48. In comparison, theophylline-inducible riboswitches that control translation initiation provide tighter control of gene expression with higher induction ratios22. Therefore, for better control of the T7 RNAP expression, we constructed expression devices in which the transcription of the T7 RNAP was driven by two different variants of the conll promoter, while the translation of the T7 RNAP was controlled by theophylline-dependent riboswitches (see below).
For the post-translational modification of ACP domains within PKSs, a 4’-phosphopantetheinyl transferase (PPTase) activity is essential to catalyze the addition of the 4’-phosphopantetheine (PPant) moiety onto this domain to generate its active holo form34,49. The S. elongatus genome does not encode PKS or non-ribosomal peptide synthase (NRPS) clusters50,51. Although a Sfp-type PPTase was identified in its genome52, further characterization of this PPTase showed that it exhibits very low or no activity towards ACPs of different PKSs52. Therefore, as part of module II, we constructed a device in which the promiscuous Sfp-PPTase from Bacillus subtilis49 was expressed from PconII and placed under the translational control of riboswitch F (Fig. 1).
Module III is specifically dedicated to the enzymes involved in the biosynthesis of a final product. Flere, we focused on the production of multimethyl-branched wax esters (MBE) using the PKS-based mycocerosic biosynthetic pathway (MAS) from Mycobacterium tuberculosis. The MAS pathway is naturally involved in the biosynthesis of the multimethyl-branched fatty acids that form part of the complex lipid phthiocerol dimycocerosate53. The PKS MAS system has been previously repurposed for the production of multimethyl-branched wax esters (MBEs) in E. coli8. MBEs have been shown to have exceptional physicochemical properties as bio-lubricants12. This system relies on the expression of FadD28 (acyl-AMP ligase), the PKS Mas (mycocerosic acid synthase), and PapA5 (polyketide-associated protein A5)8. The three coding sequences for these enzymes were cloned in an operon configuration downstream of PT7 (Fig. 1).
Modules II and III, specifically target neutral sites for chromosomal integration by homologous recombination in S. elongatus genome. Therefore, these modules are specific for S. elongatus. However, the neutral site could be replaced to be compatible with another strain of cyanobacteria21.
3.2. Development of theophylline-responsive T7 expression circuits
To establish a functional, inducible, and tightly regulated T7 RNAP-based expression circuit for S. elongatus, we initially constructed devices in which the transcription of the T7 RNAP was driven by PconII and its translation was controlled by either the theophylline-dependent riboswitches B or F22,46 The induction ratios of these riboswitches were previously evaluated in devices in which PconII drove the expression of the yfp gene (PconII-yfp)22. While riboswitch F enabled a higher level of expression after induction, riboswitch B provided tighter control with a lower uninduced level of expression22. Based on these studies we evaluated these regulatory parts to develop the regulated T7 RNAP expression circuits in S. elongatus.
To characterize the expression circuits, we constructed a reporter device in which the yfp gene was driven by PT7 (PT7-yfp) and two devices in which the T7RNAP gene was placed under the control of the conII promoter and riboswitch F (PconII-rswF-T7RNAP) or riboswitch B (PconII-rswB-T7RNAP). The reporter device was integrated at NS2 on the S. elongatus chromosome30. The T7RNAP expression device, either PconII-rswF-T7RNAP or PconII-rswB-T7RNAP, was integrated at NS1 on the S. elongatus chromosome30. Both expression circuits (PconII-rswF-T7RNAP/PT7-yfp and PconII-rswB-T7RNAP/PT7-yfp) were then characterized in the S. elongatus strains by measuring YFP-fluorescence one day after addition of 2 mM theophylline or 1% DMSO vehicle control. In absence of the theophylline inducer, both circuits produced relatively high levels of the YFP reporter (high background). With the inducer, only a small increase or no increase of the YFP-fluorescence level was measured. These results indicated that neither of these two circuits was satisfactory for controlling gene expression in S. elongatus, but it is worth noting that the highest levels of YFP-fluorescence were produced by the strains in which the T7RNAP gene was placed under the control of PconII-rswF (data not shown).
In order to reduce the background levels of the T7 RNAP, we engineered a weakened version of PconII to drive the T7RNAP expression. This targeted mutation in the −10 sequence of PconII (indicated as PconII*) led to significantly lower basal levels of uninduced YFP fluorescence in strains containing either of the two riboswitches. As shown in Fig. 2, the two new T7RNAP expression devices controlled by PconII*-rswB or PconII*-rswF produce a clear dose-dependent response to the theophylline inducer (Fig. 2). Although the highest levels of YFP expression were similar with both riboswitches F and B, lower concentrations of theophylline (1 mM compared to 2 mM) were needed for the riboswitch F to reach maximal expression. On the other hand, riboswitch B showed a tighter control in the absence of the inducer (Fig. 2). Note that under the low-density growth conditions of these strains for this experiment, a concentration of 4 mM theophylline produced lower YFP expression presumably due to some theophylline toxicity22.
Figure 2: Characterization of T7 RNAP expression devices in S. elongatus.

(a) Scheme of the theophylline-regulated T7 RNAP expression circuits. The expression of the T7 RNAP was driven by a mutated PconII (PconII*) with lower activity and was translationally controlled by (b) riboswitch F (PconII*-rswF-T7RNAP/PT7-yfp) or (c) riboswitch B (Pconn*-rswB-T7RNAP/PT7-yfp). YFP-reporter fluorescence served as an easily readable output. Three independent clones were analyzed for each construct (represented with different shades of grey) and error bars indicate standard deviations of three replicates of the experiment.
We chose to continue our studies with two of the T7 RNAP expression devices: 1) the original PconII and riboswitch F device, which did not show theophylline regulation but produced the highest levels of expression and 2) the mutated PconII (PconII*) and riboswitch B device, which enabled a tighter control of the T7 RNAP and allows testing of different expression levels. We chose the first expression device because we assumed that in some cases the production of polyketides may be limited by the levels of their biosynthetic enzymes. Therefore, having higher expression levels of the MAS pathway would give us a better chance of detecting MBE production in cyanobacteria. These expression devices were used in the construction of modules IIA and IIB (Fig. 1).
3.3. Analysis of protein expression in recombinants S. elongatus
Using our synthetic biology tools for cyanobacteria21, we assembled the functional modules (Fig. 1) into appropriate plasmid vectors. Details on each plasmid developed for the platform are presented in Supplementary Fig. 1 and their construction is described in Supplementary Methods. The recombinant cyanobacterial strains described in Table 2 were obtained by sequential transformation with the indicated recombinant vectors. Each of the final six recombinant strains that were constructed carried a different combination of the functional modules I, II, and III.
The heterologous proteins that are involved in the biosynthesis of methylmalonyl-CoA (Module I) including MatB, the MutA subunit, and the epimerase were FLAG-tagged for western blot analyses. These assays indicated that MutA was not expressed with the correct size in some S. elongatus strains and in others strains it was not expressed at detectable levels (Supplementary Fig. 2), consequently we did not pursue any further development of these strains. On the other hand, MatB and the epimerase were expressed with the expected molecular weights on western blots (Supplementary Fig. 2). The PccB subunit of the PCC complex was analyzed with a specific anti-PccB antibody38 and showed correct expression (Supplementary Fig. 3). The three enzymes of the PKS pathway, PapA5, FadD28, and Mas, were His-tagged and were detected as soluble proteins in the cyanobacterial strains by western blot analyses (Supplementary Fig. 4). These results showed that S. elongatus, which does not have PKS and NRPS genes in its genome50,51, is capable of expressing the proteins for a complex heterologous pathway including the Mas protein, which is a large multidomain PKS enzyme. In addition, the fact that the MAS pathway was expressed in all of the recombinant cyanobacterial strains analyzed also indicates that both T7 RNAP expression devices constructed and tested here can successfully drive the expression of the genes cloned in module III.
3.4. Functional analysis of phosphopantetheinyl transferase
To determine Sfp expression and functional activity in S. elongatus, we employed an assay for the addition of the fluorescently labeled pantetheine analogue (TAMRA-CoA) to the type II E. coli fatty acid synthase ACP (AcpP)54 Soluble protein extract from each recombinant cyanobacterium containing the heterologous Sfp enzyme or from S. elongatus wild-type strain AMC2302 was added to recombinant apo-AcpP in the presence of TAMRA-CoA. The detection of the ACP phosphopantetheinylation was carried out on SDS-PAGE; and the reporter-labeled ACP was visualized by UV fluorescence (Fig. 3 and Supplementary Fig. 5). As shown in Fig. 3 and Supplementary Fig. 5, soluble protein extracts from the recombinant cyanobacterial strains pursued in this study allowed the conversion of E. coli ACP to its fluorescently labeled derivative. These results indicate that S. elongatus wild-type strain AMC2302 does not exhibit promiscuous native PPTase activity, as was previously reported52, and that the heterologous Sfp is expressed in its active form in all the S. elongatus recombinant strains analyzed. Therefore, considering that we had previously shown that the Mas synthase is a suitable substrate for Sfp8,55, we were confident that the Mas enzyme would also be adequately phosphopantetheinylated in the recombinant strains here developed.
Figure 3: PPTase in vitro assays.

Sfp functional in vitro assay by detection of E. coli ACP phosphopantetheinylation with fluorescently labeled CoA (TAMRA-CoA). Proteins were separated by SDS-PAGE and visualized by UV fluorescence. Lanes: 1, purified E. coli ACP plus TAMRA-CoA and purified Sfp (positive control); 2, purified E. coli ACP (negative control); 3, 4, 5, purified E. coli ACP plus TAMRA-CoA and soluble protein extracts from three independent clones from strain JR102; 6, purified E. coli ACP plus TAMRA-CoA and protein extract from strain JR104; 7, purified E. coli ACP plus TAMRA-CoA and wild-type S. elongatus protein extract (negative control).
3.5. Production of PKS-derived MBE by S. elongatus
After the expression of the key proteins encoded by functional modules I, II, and III was confirmed, we further analyzed these strains for the production of MBE. Bioconversion assays were performed on four different strains: JR101 (modules IA + IIA + III), JR102 (modules IA + IIB + III), JR107 (modules IC + IIA + III), and JR108 (modules IC + IIB + III).
While module I was constitutively expressed from PconII, the expression and activation of module III depended on the expression of the T7 RNAP and Sfp placed downstream of PconII (or PconII*) and under the translational control of a theophylline-inducible riboswitch (F or B). The biosynthesis of (2S)-methylmalonyl-CoA required methylmalonate (module IA) or propionate (module IC) as substrates, and the transesterification of the PKS-ACP-bound multimethyl-branched fatty acid (MBFA) required an alcohol substrate n-octanol to produce MBE as the final product. Cell toxicity analyses with methylmalonate, propionate, and n-octanol were carried out to determine the optimal concentration of these precursors in the cyanobacterial cultures (Supplementary Fig. 6).
We first analyzed the two strains containing module IA (MatB and epimerase) for (2S)-methylmalonyl-CoA biosynthesis, modules IIA or MB for T7 RNAP and Sfp expression, and module III for the Mas and accessory enzymes expression. In addition, strains without module III and a wild-type strain were included as negative controls. For bioconversion assays the strains were induced with 2 mM theophylline and after 24 h supplemented with 10 mM methylmalonate, 0.25 mM n-octanol, and 2 μCi 14C-acetate (58.9 mCi/mmol, PerkinElmer) to label the products. We used 14C-acetate as an easy, sensitive and quick method for analyzing de novo lipids biosynthesis. S. elongatus is an obligate autotrophic microorganism and it strictly depends upon the generation of photosynthetically derived energy for growth, therefore 14C-acetate was not used as an energy source for the microorganism. After addition of the precursors, the cultures were grown for 48 h, and the in vivo synthesis of MBE was analyzed in total lipid extracts from each culture by TLC fractionation and autoradiography (Fig. 4b and 4c). Additional bioconversion assays were carried out as controls where one or both precursors (methylmalonate and n-octanol) were absent. Labeled MBE obtained through a bioconversion assay using the E. coli strain RQ1 was used as a standard8.
Figure 4. Functional analysis of modules IA, II (A and B), and III.

(a) Scheme of the full pathways for MBE biosynthesis using module IA for (2S)-methylmalonyl-CoA production. (b) Radio-TLC analysis of total lipid fraction from S. elongatus strain JR101: lanes 1, 2, 3, 4, 5, 6; JR103 (without module III): lane 7; and from E. coli RQ1 MBE producer strain: lane 8. The lipid fractions on lanes 1, 2, and 3, were each extracted from one of three cultures grown from three independent clones as biological replicates. These cultures were supplemented with methylmalonate and n-octanol, while E. coli RQ1 was supplemented with propionate and n-octanol for the bioconversion assay. The cultures used for lanes 4, 5, and 6 were grown from the same clone as for lane 1 but in the absence of one or both precursors, n-octanol and methylmalonate, as indicated below each lane. (c) Radio-TLC analysis of total lipid fraction from S. elongatus strain JR102: lanes 1, 2, 3, 4, 5, 6; JR104 (without module III): lane 7; wild-type strain AMC2302: lane 8, and from E. coli RQ1 MBE producer strain: lane 9. The lipid fractions on lanes 1, 2, and 3 were each extracted from one of three cultures grown from three independent clones as biological replicates. These cultures were supplemented with methylmalonate and n-octanol, while E. coli RQ1 culture was supplemented with propionate and n-octanol for the bioconversion assay. The cultures used for lanes 4, 5, and 6 were grown from the same clone as for lane 1 but in the absence of either or both precursors, n-octanol and methylmalonate, as indicated below each lane. Modules analyzed are indicated on the left of each TLC and each color of the boxes makes reference to the colors used in Fig. 1 for each module. MBE, multimethyl-branched wax esters. Spots that are not annotated were not analyzed by LC/MS. A comparison of the pattern shows that their presence is independent of the modules. The second spot, from top to down, appears only when n-octanol was supplemented to the media.
The results of Fig. 4 clearly indicate that S. elongatus was successfully engineered to synthesize MBE. The production of MBE was indeed detected only when both precursors and all the enzymes of the MAS pathway (module III) were present. To confirm the identity of the putative MBE compounds, analytical characterization of purified MBE was carried out by LC-MS/MS experiments as detailed in Supplementary Methods (Supplementary Fig. 7). From this analysis, we conclude that MBE synthesized by S. elongatus recombinant strains are formed by the esterification of the MBFA derived from four iterative MAS-catalyzed extension steps of a C16:1 fatty acid and the n-octanol as acceptor alcohol. These results also indicate that (2S)-methylmalonyl-CoA is synthesized through the MatB-epimerase pathway (module IA) using methylmalonate as precursor. These assays further confirmed that both modules II (A and B) produce functional T7 RNAP and PPTase, which enable the expression and activation of module III enzymes. Finally, these findings indicate that the MAS biosynthesis system (PapA5-FadD28-Mas) cloned as module III is properly expressed and functional in S. elongatus. Next, we sought to test module 1C for the biosynthesis of (2S)-methylmalonyl-CoA from propionate by the PCC pathway. This biosynthetic route was considered more challenging because it required the expression of a complex of three different subunits (AccA2, PccB, and PccE), and an exogenous supply of propionate, previously reported as toxic for S. elongatus at low concentrations39, for conversion into propionyl-CoA. As mentioned earlier, we hypothesized that this enzymatic step could be performed by the S. elongatus native acetyl-CoA synthetase/acetate-CoA ligase (AcsA, SynPCC7942_1352). Preliminary analyses indicated that at a final concentration of 1 mM, propionate was not toxic for S. elongatus (Supplementary Fig. 6a). The bioconversion assays and MBE production analyses for JR107 and JR108 strains (as well as a wild-type strain as negative control) were performed as described above, but the cultures were adjusted to final concentrations of 1 mM propionate, 0.25 mM n-octanol, and 2 μCi 14C-acetate label (58.9 mCi/mmol, PerkinElmer). The results shown in Fig. 5 indicate that module IC for (2S)-methylmalonyl-CoA biosynthesis in cyanobacteria is also functional and enables MBE production.
Figure 5. Functional analysis of module IC.

(a) Scheme of full pathways for MBE biosynthesis using module IC for (2S)-methylmalonyl-CoA production. (b) Radio-TLC analysis of total lipid fraction from S. elongatus strain JR107: lanes 1, 2, 3, 4, 5 and E. coli RQ1 MBE producer strain: lane 6. The lipid fractions on lanes 1,4, and 5 were each extracted from one of three cultures grown from three independent clones as biological replicates. These cultures and the E. coli RQ1 culture were supplemented with propionate and n-octanol for the bioconversion assay. The cultures used for lanes 2 and 3 were grown from the same clone as for lane 1 but in the absence of one or both precursors, n-octanol and propionate, as indicated below each lane. (c) Radio-TLC analysis of total lipid fraction from S. elongatus strain JR108: lanes 1, 2, 3, 5, 6, wild-type strain AMC2302: lane 4 and E. coli RQ1 MBE producer strain: lane 7. The lipid fractions on lanes 1, 5, and 6 were each extracted from one of three cultures grown from three independent clones as biological replicates. These cultures and the E. coli RQ1 culture were supplemented with propionate and n-octanol for the bioconversion assay. The cultures used for lanes 2 and 3 were grown from the same clone as for lane 1 but in the absence of either or both precursors, n-octanol and propionate, as indicated below each lane. Modules analyzed are indicated on the left of each TLC and each color of the boxes makes reference to the colors used in Fig. 1 for each module. MBE, multimethyl-branched wax esters.
The analysis of TLCs shown in Figures 4 and 5 and LC-MS/MS experiments (Supplementary Fig. 7) demonstrate that strains JR101 (modules IA + IIA + III), JR102 (modules IA + IIB + III), JR107 (modules IC + IIA + III), and JR108 (modules IC + IIB + III) produced (2S)-methylmalonyl-CoA PKS-derived MBE. These results provide evidence that (2S)-methylmalonyl-CoA is synthesized in vivo through both pathways (MatB-epimerase and PCC) and that (2S)-methylmalonyl-CoA is available as a substrate for the multidomain Mas PKS enzyme.
3.6. Effect of inducer concentration and MBE production
In order to study the response of the two selected regulatory modules IIA (PconII-rswF-sfp-PconII-rswF-T7RNAP) and IIB (PconII-rswF-sfp-PconII*-rswB-T7RNAP) at different concentrations of theophylline and determine their effect on MBE production, we carried out bioconversion assays with the strains JR101 and JR102 induced with a range of theophylline concentrations (0, 0.5, 1, 2, 4, and 8 mM). The TLC analyses showed that the production of MBE responded to the addition of the inducer in a concentration-dependent manner in both strains analyzed (Fig. 6). Similar to the results found with the T7 RNAP driving the YFP reporter, module IIB showed a tight inducer-dependent response in the strain JR102, and MBE levels were dependent on the concentration of the theophylline inducer up to 8 mM. Surprisingly, MBE production in the strain harboring module IIA also showed a dose-dependent response, however this regulation showed less MBE production at lower concentrations of theophylline and maximum MBE production at 4 mM theophylline with a decrease in MBE at 8 mM. This result suggests that the highest levels of expression of module III was detrimental for MBE production for unknown reasons. MBE production depends on the expression levels of the MAS pathway and other factors such as the Mas/Sfp ratio, the availability of the MBE precursors, and the general fitness of the strains. Obtaining maximum MBE production will require balancing all of these factors, which will be facilitated by the modular design utilized in this work.
Figure 6. MBE biosynthesis by JR101 and JR102 cyanobacterial strains induced with different theophylline concentrations.
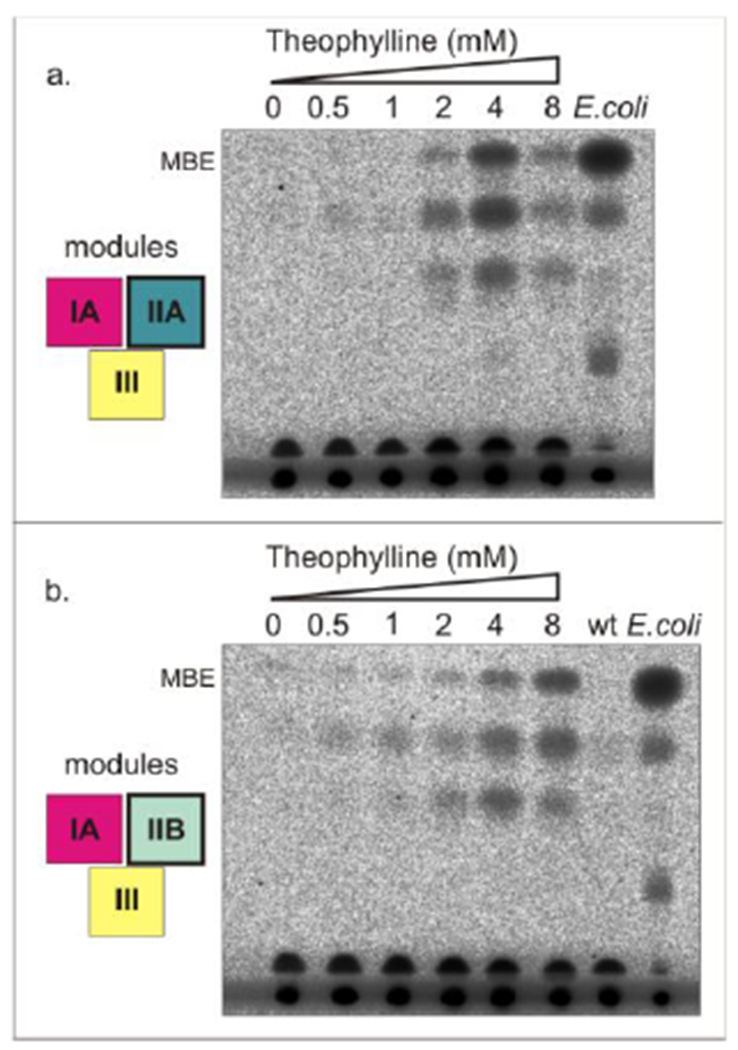
(a) Radio-TLC analysis of the total lipid fraction from S. elongatus strain JR101 induced with theophylline (0, 0.5, 1, 2, 4, 8 mM) and supplemented with methylmalonate (10 mM) and n-octanol (0.25 mM) for the bioconversion assay; and of the total lipid fraction from the E. coli RQ1 MBE producer strain, used as MBE standard, supplemented with propionate (30 mM) and n-octanol (0.5 mM). (b) Radio-TLC analysis of the total lipid fraction from S. elongatus strain JR102 induced with theophylline (0, 0.5, 1, 2, 4, 8 mM) and supplemented with methylmalonate (10 mM) and n-octanol (0.25 mM); of the total lipid fraction from wild-type S. elongatus strain induced with theophylline (2 mM), and supplemented as above with n-octanol and methylmalonate; and of the total lipid fraction from E. coli RQ1 with propionate and n-octanol supplied as above. Modules analyzed are indicated on the left of each TLC and each color of the boxes makes reference to the colors used in Fig. 1 for each module. MBE, multimethyl-branched wax esters.
4. Discussion
A wide variety of polyketide compounds are synthesized from a small subset of starter units such as acetyl-CoA and propionyl-CoA, and elongated with a specific subset of a-carboxyacyl-CoAs, which mainly include malonyl-CoA, (2S)-methylmalonyl-CoA, and (2S)-ethylmalonyl-CoA. Therefore, a robust heterologous platform for polyketide production must be endowed with the ability to synthesize these substrates. Because S. elongatus carries the metabolic pathways for the biosynthesis of acetyl-CoA and malonyl-CoA, but not for (2S)-methylmalonyl-CoA, which is the major extender unit used by PKSs after malonyl-CoA35, we focused on the heterologous production of (2S)-methylmalonyl-CoA. For this, we selected enzymes that are able to synthesize (2S)-methylmalonyl-CoA, but that due to their substrate tolerance would also enable the orthogonal production of malonyl-CoA or of other PKS extender units, such as (2S)-ethylmalonyl-CoA, providing that the cells are fed with the proper substrate. Different pathways are responsible for the biosynthesis of (2S)-methylmalonyl-CoA in bacteria2,35. Considering that the expression of recombinant genes depends upon multiple factors, in particular the heterologous host9,14,56; and the fact that not all (2S)-methylmalonyl-CoA biosynthetic pathways are energetically or kinetically equivalent, we evaluated three alternative modules (IA, IB, and IC) to produce the PKS α-carboxyacyl-CoA substrates from three different heterologous pathways: MatB-epimerase, MutAB-epimerase, and PCC. The in vivo biosynthesis of (2S)-methylmalonyl-CoA by the PCC and MatB routes in the engineered strains of S. elongatus was confirmed by the production of the final MBE product (Fig. 4, 5, and Supplementary Fig. 7). Additional experimentation will be required to determine the full potential and versatility of our in vivo heterologous production platform by determining if the PCC and MatB routes produce all the expected carboxyacyl-CoA precursors that were previously produced in vitro36,38. In addition, future work to enable the production of (2S)-methylmalonyl-CoA using the MutAB pathway in S. elongatus could prove to be useful, because we expect that its substrate could be endogenously produced in S. elongatus. However, recent genome scale metabolic predictions together with data on gene essentiality20 showed the presence of a linear, noncyclic TCA pathway in S. elongatus, with no evidence of succinyl-CoA as intermediate. Therefore, because MutAB’s substrate is succinyl-CoA, a succinyl-CoA synthetase would likely need to be co-expressed in the production strain. We also note that pANS-based shuttle vectors, where the genes of module I were cloned, also replicate stably in the filamentous cyanobacterium Anabaena sp. strain PCC 712029. Therefore, the biosynthesis of (2S)-methylmalonyl-CoA could be tested in Anabaena PCC 7120, which has a larger genome and several specialized metabolic pathways50,51, and could be a more appropriate heterologous host for the production of some polyketides.
In order to develop a strong and regulated system of expression to drive the transcription of the polyketide biosynthetic genes, we developed T7 RNAP expression circuits for S. elongatus. We constructed four different expression devices where the T7RNAP was transcribed from two promoters with distinct strengths, PconII or a weaker PconII* variant, and translated under the control of one of two theophylline-inducible riboswitches, each with different regulatory properties. Each of the four devices was characterized in S. elongatus strains carrying a reporter device where a yfp reporter gene was expressed from a T7 promoter and YFP fluorescence served as the output (Fig. 2). For MBE production, two of the T7 RNAP expression devices were selected based on the highest levels of YFP expression (module IA) and tighter regulation of YFP expression (module MB). Characterization of the two different regulatory modules IIA and MB in S. elongatus (also containing modules I and III) showed that both IIA and MB were able to regulate the expression of the PKS genes, and therefore MBE production, in a dose dependent fashion (Fig. 6).
Considering that different polyketide biosynthetic pathways and/or their intermediates or final products may adversely affect the cells, and in order to cope with the variability and complexity of natural products and their biosynthetic pathways, it is important to create tunable expression circuits. The versatility of the two modules tested in this work indicates that the four T7RNAP expression devices would be well suited for heterologous expression of PKS gene clusters in S. elongatus. Moreover, other combinations of new PconII mutants and different riboswitch variants22 could be generated and replaced in module II in order to adjust the expression of different PKS systems. In addition, the T7 promoter itself could be modified in order to fine tune the expression of the PKS cluster in module III57.
Compared to heterotrophic microorganisms, the use of photosynthetic cyanobacteria as production platforms for chemicals is still in its infancy, and with the growing interest of using cyanobacteria for producing a wide variety of compounds, new genetic tools are essential. The T7RNAP-based expression circuits developed here are a valuable addition to the collection of genetic tools available for S. elongatus21,22,44,46.
Specialized natural product pathways, in particular PKS and NRPS, invariably rely on the post-translational modification of the synthases to produce active enzymes. Because S. elongatus lacks a PPTase with activity towards the carrier proteins of different PKSs or NRPSs52, a promiscuous PPTase enzyme49 was included in our platform strain, and in vitro assays confirmed that the enzyme was active (Fig. 3 and Supplementary Fig. 5). Because S. elongatus lacks any PKS or NRPS biosynthetic pathways50,51, the engineering of a promiscuous PPTase activity into this strain provides a suitable platform with a clean background where heterologous production of PKS or NRPS derived molecules are less likely to face crosstalk, contamination, or competition from native pathways.
The heterologous expression of biosynthetic gene clusters in a photosynthetic microbial host that has minimal growth requirements and is amenable to genetic manipulations provides a powerful approach for exploring natural products from uncultivable organisms and metagenomic sequences. These heterologous production platforms can also be used to genetically modify biosynthetic pathways to produce novel products and for renewable large-scale production of a wide variety of compounds. These include medicines, biofuels, and polymer precursors1. In this work, we demonstrated that the expression of a heterologous PKS pathway as well as its precursor biosynthetic pathways can result in functional enzymes and the production of the expected final compound in a photosynthetic microorganism. While the production of MBE by S. elongatus stands as a proof of concept, the platform strain, along with the synthetic biology tools that we have developed, provide a starting point for the production of a wide array of polyketide compounds. The modular design of the platform described in this work will facilitate the optimization of polyketide production. Using the platform strains currently available, other (2S)-methylmalonyl-CoA derived polyketides could be produced in a sustainable manner by changing the third module with the desired PKS. Furthermore, this work suggests that production of other classes of PKSs that require malonyl-CoA, (2S)-ethylmalonyl-CoA, or other CoA precursors should also be achievable by following the same strategy.
5. Conclusion
Traditionally, the idea of heterologous production has stemmed from the desire to produce a known natural compound from elucidated pathways. However, with the advances of genome sequencing and bioinformatics, many new natural product pathways have been discovered58–61. The overwhelming number of enigmatic biosynthetic gene clusters, coupled with the limitations associated to their study in native hosts, strongly supports the development of novel heterologous production platforms. This work provides a sustainable organism with an optimized genetic background for the interrogation of the wealth of biosynthetic gene clusters for specialized metabolites with known or unknown functions, and for synthetic biology approaches to manipulate or modify those pathways.
Supplementary Material
Acknowledgements
This work was supported by Bec.AR-Fulbright fellowship to J.R., Y-TEC/ANPCyT grant PID-2013-0042 to H.G. and the National Institutes of Health NIH grant 5R01GM118815 to J.W.G. We thank Susan S. Golden for providing additional lab support, Jeffrey T. Mindrebo, Eunice Kim, Ryan Simkovsky, Brian P. Tieu, Cigdem Sancar, and Nathan Moss for research materials and technical support.
Footnotes
Competing financial interests
J.R., A.T., J.W.G., A.A., M.D.B., and H.G. are authors on a patent application entitled “Engineering polyketide synthase machinery in cyanobacteria”; U.S. Provisional Application Serial No. 62/643,370 filed on March 15, 2018.
References
- 1.Yuzawa S, Keasling JD & Katz L Bio-based production of fuels and industrial chemicals by repurposing antibiotic-producing type I modular polyketide synthases: Opportunities and challenges. J. Antibiot. (Tokyo) 70, 378–385 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Pfeifer BA & Khosla C. Biosynthesis of Polyketides in Heterologous Hosts. Microbiol. Mol. Biol. Rev 65, 106–118 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xue Q, Ashley G, Hutchinson CR & Santi DV A multiplasmid approach to preparing large libraries of polyketides. Proc. Natl. Acad. Sci. U. S. A 96, 11740–11745 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gokhale RS, Sankaranarayanan R & Mohanty D Versatility of polyketide synthases in generating metabolic diversity. Curr. Opin. Struct. Biol 17, 736–743 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Dunn BJ & Khosla C Engineering the acyltransferase substrate specificity of assembly line polyketide synthases. J. R. Soc. Interface 10, 20130297 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Q, Wu K, Cheng Y, Lu L, Xiao E, Zhang Y, Deng Z & Liu T Engineering an iterative polyketide pathway in Escherichia coli results in singleform alkene and alkane overproduction. Metab. Eng 28, 82–90 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Cai W & Zhang W Engineering modular polyketide synthases for production of biofuels and industrial chemicals. Curr. Opin. Biotechnol 50, 32–38 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menendez-Bravo S, Comba S, Sabatini M, Arabolaza A & Gramajo H Expanding the chemical diversity of natural esters by engineering a polyketide-derived pathway into Escherichia coli. Metab. Eng 24, 97–106 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Rodriguez E, Menzella HG & Gramajo H Chapter 15 Heterologous Production of Polyketides in Bacteria. Methods in Enzymology 459, (Elsevier Inc., 2009). [DOI] [PubMed] [Google Scholar]
- 10.Kealey JT, Liu L, Santi DV, Betlach MC & Barr PJ Production of a polyketide natural product in nonpolyketide-producing prokaryotic and eukaryotic hosts. Proc. Natl. Acad. Sci. U. S. A 95, 505–509 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfeifer BA, Admiraal SJ, Gramajo H, Cane DE & Khosla C Biosynthesis of complex polyketides in a metabolically engineered strain of E. coli. Science 291, 1790–1792 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Menendez-Bravo S, Roulet J, Sabatini M, Comba S, Dunn R, Gramajo H & Arabolaza A High cell density production of multimethyl-branched long-chain esters in Escherichia coli and determination of their physicochemical properties. Biotechnol. Biofuels 9, 215 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smanski MJ, Zhou H, Claesen J, Shen B Fischbach M & Voigt CA Synthetic biology to access and expand nature’s chemical diversity. Nat Rev Microbiol 14, 135–149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stevens DC, Hari TPA & Boddy CN The role of transcription in heterologous expression of polyketides in bacterial hosts. Nat. Prod. Rep 30, 1391–1411 (2013). [DOI] [PubMed] [Google Scholar]
- 15.Fujii I Heterologous expression systems for polyketide synthases. Nat. Prod. Rep 26, 155–169 (2009). [DOI] [PubMed] [Google Scholar]
- 16.Ducat DC, Way JC & Silver PA Engineering cyanobacteria to generate high-value products. Trends Biotechnol. 29, 95–103 (2011). [DOI] [PubMed] [Google Scholar]
- 17.Rubin BE, Wetmore KM, Price MN, Diamond S, Shultzaberger RK, Lowe LC, Curtin G, Arkin AP, Deutschbauer A & Golden SS The essential gene set of a photosynthetic organism. Proc. Natl. Acad. Sci. U. S. A 112, E6634–E6643 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diamond S, Jun D, Rubin BE & Golden SS The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proc. Natl. Acad. Sci. U. S. A. 112, E1916–E1925 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vijayan V, Jain IH & O’Shea EK A high resolution map of a cyanobacterial transcriptome. Genome Biol. 12, R47 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Broddrick JT, Rubin BE, Welkiec DG, Dud N, Mihf N, Diamond S, Lee JJ, Golden SS & Palsson B0 Unique attributes of cyanobacterial metabolism revealed by improved genome-scale metabolic modeling and essential gene analysis. Proc. Natl. Acad. Sci. U. S. A 113, E8344–E8353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taton A, Unglaub F, Wright NE, Zeng WY, Paz-Yepes J, Brahamsha B, Palenik B,. Peterson TC, Haerizadeh F, Golden SS & Golden JW Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res. 42, e136 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma AT, Schmidt CM & Golden JW Regulation of gene expression in diverse cyanobacterial species by using theophylline-responsive riboswitches. Appl. Environ. Microbiol 80, 6704–6713 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ruffing AM Engineered cyanobacteria: Teaching an old bug new tricks. Bioeng. Bugs 2, 136–149 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Atsumi S, Higashide W & Liao JC Direct photosynthetic recycling of carbon dioxide to isobutyraldehyde. Nat. Biotechnol 27, 1177–1180 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Ducat DC, Avelar-Rivas JA, Way JC & Silvera PA Rerouting carbon flux to enhance photosynthetic productivity. Appl. Environ. Microbiol 78, 2660–2668 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Videau P, Wells KN, Singh AJ, Gerwick WH & Philmus B Assessment of Anabaena sp. strain PCC 7120 as a heterologous expression host for cyanobacterial natural products: production of lyngbyatoxin A. ACS Synth. Biol 5, 978–988 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Knoot CJ, Ungerer JL, Wangikar PP & Pakrasi HB Cyanobacteria: promising biocatalysts for sustainable chemical production. J. Biol. Chem jbc.R117.815886 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Golden SS & Sherman LA A hybrid plasmid is a stable cloning vector for the cyanobacterium Anacystis nidulans R2. J. Bacteriol 155, 966–972 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Y, Taton A, Go M, London RE, Pieper LM, Golden SS & Golden JW. Self-replicating shuttle vectors based on pANS, a small endogenous plasmid of the unicellular cyanobacterium Synechococcus elongatus PCC 7942. Microbiol. (United Kingdom) 162, 2029–2041 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Clerico EM, Ditty JL & Golden SS Specialized techniques for site-directed mutagenesis in cyanobacteria. Methods Mol. Biol 362, 155–171 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Rippka R , Deruelles J, Waterbury JB, Herdman M & Stanier RY Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Microbiology 111, 1–61 (1979). [Google Scholar]
- 32.Chin JX, Chung BKS & Lee DY Codon Optimization OnLine (COOL): A web-based multi-objective optimization platform for synthetic gene design. Bioinformatics 30, 2210–2212 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Bligh EG & Dyer WJ A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
- 34.Beld J, Sonnenschein EC, Vickery CR, Noel JP & Burkart MD The phosphopantetheinyl transferases: catalysis of a post-translational modification crucial for life. Nat. Prod. Rep 31, 61–108 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan YA, Podevels AM, Kevany BM & Thomas MG Biosynthesis of polyketide synthase extender units. Nat. Prod. Rep 26, 90–114 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes AJ & Keatinge-Clay A Enzymatic extender unit generation for in vitro polyketide synthase reactions: Structural and functional showcasing of Streptomyces coelicolor MatB. Chem. Biol 18, 165–176 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Dayem LC, Carney JR, Santi DV, Pfeifer BA, Khosla C & Kealey JT Metabolic engineering of a methylmalonyl-CoA mutase - epimerase pathway for complex polyketide biosynthesis in Escherichia coli. Biochemistry 41, 5193–5201 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Diacovich L, Peirú S, Kurth D, Rodríguez E, Podestá F, Khosla C & Gramajo H Kinetic and structural analysis of a new group of acyl-CoA carboxylases found in Streptomyces coelicolor A3(2). J. Biol. Chem 277, 31228–31236 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Begemann MB Begemann MB, Zess EK, Walters EM, Schmitt EF, Markley AL & Pfleger BF An organic acid based counter selection system for cyanobacteria. PLoS One 8, e76594 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van der Plas J, Oosterhoff-Teertstra R, Borrias M & Weisbeek P Identification of replication and stability functions in the complete nucleotide sequence of plasmid pUFI24 from the cyanobacterium Synechococcus sp. PCC 7942. Mol. Microbiol 6, 653–664 (1992). [DOI] [PubMed] [Google Scholar]
- 41.Elledge SJ & Davis RW Position and density effects on repression by stationay and mobile DNA-binding proteins. Genes Dev 3, 185–197 (1989). [DOI] [PubMed] [Google Scholar]
- 42.Li R & Golden SS Enhancer activity of light-responsive regulatory elements in the untranslated leader regions of cyanobacterial psbA genes. Proc. Natl. Acad. Sci. U. S. A 90, 11678–11682 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang B, Wang J & Meldrum DR Application of synthetic biology in cyanobacteria and algae. Front. Microbiol 3, 344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim WJ, Lee S-M, Um Y, Sim SJ & Woo HM Development of SyneBrick vectors as a synthetic biology platform for gene expression in Synechococcus elongatus PCC 7942. Front. Plant Sci 8, 293 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao YQ, Li Q, Xia PF, Wei LJ, Guo N, Li JW & Wang SG AraBAD based toolkit for gene expression and metabolic robustness improvement in Synechococcus elongatus. Sci. Rep 7, 18059 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Taton A, Ma AT, Ota M, Golden SS & Golden JW NOT gate genetic circuits to control gene expression in cyanobacteria. ACS Synth. Biol 6, 2175–2182 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dubendorf JW & Studier FW Controlling basal expression in an inducible T7 expression system by blocking the target T7 promoter with lac repressor. J. Mol. Biol 219, 45–59 (1991). [DOI] [PubMed] [Google Scholar]
- 48.Huang HH, Camsund D, Lindblad P & Heidorn T Design and characterization of molecular tools for a synthetic biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res 38, 2577–2593 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quadri LEN, Weinreb PH, Lei M, Nakano MM, Zuber P & Walsh CT Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carder protein domains in peptide synthetases. Biochemistry 37, 1585–1595 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Shih PM, Wu D, Latifi A, Axen SD, Fewer DP, Talla E, Calteau A, Cai F, Tandeau de Marsac N, Rippka R, Herdman M, Sivonen K, Coursin T, Laurent T, Goodwin L, Nolan M, Davenport KW, Han CS, Rubin EM, Eisen JA, Woyke T, Gugger M & Kerfeld CA Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. U. S. A 110, 1053–1058 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Calteau A, Fewer DP, Latifi A, Coursin T, Laurent T, Jokela J, Kerfeld CA, Sivonen K, Piel J & Gugger M Phylum-wide comparative genomics unravel the diversity of secondary metabolism in Cyanobacteria. BMC Genomics 15, 977 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang G, Zhang Y, Lee NK, Cozad MA, Kearney SE, Luesch H & Ding Y Cyanobacterial Sfp-type phosphopantetheinyl transferases functionalize carrier proteins of diverse biosynthetic pathways. Sci. Rep 7, 11888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trivedi OA, Arora P, Vats A, Ansari MZ, Tickoo R, Sridharan V, Mohanty D & Gokhale RS Dissecting the mechanism and assembly of a complex virulence mycobacterial lipid. Mol. Cell 17, 631–643 (2005). [DOI] [PubMed] [Google Scholar]
- 54.La Clair JJ, Foley TL, Schegg TR, Regan CM & Burkart MD Manipulation of carrier proteins in antibiotic biosynthesis. Chem. Biol 11, 195–201 (2004). [DOI] [PubMed] [Google Scholar]
- 55.Ishikawa F, Haushalter RW & Burkart MD Dehydratase-specific probes for fatty acid and polyketide synthases. J. Am. Chem. Soc 134, 769–772 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cardinale S & Arkin AP Contextualizing context for synthetic biology - identifying causes of failure of synthetic biological systems. Biotechnol. J 7, 856–866 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Temme K, Hill R, Segall-Shapiro TH, Moser F & Voigt CA Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res 40, 8773–8781 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kehr JC, Picchi DG & Dittmann E Natural product biosyntheses in cyanobacteria: A treasure trove of unique enzymes. Beilstein J. Org. Chem 7, 1622–1635 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gomes ES, Schuch V & Lemos EG de M Biotechnology of polyketides: New breath of life for the novel antibiotic genetic pathways discovery through metagenomics. Brazilian J. Microbiol 44, 1007–1034 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Micallef ML, D’Agostino PM, Al-Sinawi B, Neilan B. a. & Moffitt MC Exploring cyanobacterial genomes for natural product biosynthesis pathways. Mar. Genomics 21, 1–12 (2015). [DOI] [PubMed] [Google Scholar]
- 61.Helfrich EJN, Reiter S & Piel J Recent advances in genome-based polyketide discovery. Curr. Opin. Biotechnol 29, 107–115 (2014). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


