Abstract
Cancers are comprised of heterogeneous subpopulations with various tumor-initiating capacities, yet key stem cell genes associated with enhanced tumor initiating capacities and their regulatory mechanisms remain elusive. Here we analyzed patient-derived xenografts from melanoma, colon, and pancreatic cancer tissues and identified enrichment of tumor-initiating cells in MHC class I-hi cells, where CDK1, a master regulator of the cell cycle, was upregulated. Overexpression of CDK1, but not its kinase-dead variant, in melanoma cells increased their spheroid forming ability, tumorigenic potential, and tumor-initiating capacity; inhibition of CDK1 with pharmacological agents reduced these characteristics, which was unexplained by the role of CDK1 in regulating the cell cycle. Proteomic analysis revealed an interaction between CDK1 and the pluripotent stem cell transcription factor Sox2. Blockade or knockdown of CDK1 resulted in reduced phosphorylation, nuclear localization, and transcriptional activity of Sox2. Knockout of Sox2 in CDK1-overexpressing cells reduced CDK1-driven tumor-initiating capacity substantially. Furthermore, GSEA analysis of CDK1-hi tumor cells identified a pathway signature common in all three cancer types, including E2F, G2M, MYC, and spermatogenesis, confirming a stem-like nature of CDK1-hi tumor cells. These findings reveal a previously unrecognized role for CDK1 in regulating tumor-initiating capacity in melanoma and suggest a novel treatment strategy in cancer via interruption of CDK1 function and its protein-protein interactions.
Keywords: tumor initiation, CDK1, Sox2, phosphorylation, nuclear localization
Introduction
Over the past decade, it has become clear that cancers are comprised of heterogeneous cell populations with different tumorigenic capacities, phenotypes and functions [1]. Numerous studies have identified tumor subpopulations with enhanced tumorigenic capacities by using a variety of markers from various cancer types [2]. The frequency of tumor initiating cells (TICs) varies among reports and is influenced by tumor cell isolation methods [3–5] and many other factors including model system, source of tumor samples, and interpretation of the findings [3–9]. These studies point to a need for further understanding of the underlying mechanisms driving tumor initiating potential and the genes and transcription factors that contribute to this process.
Sex determining region Y-box 2 (Sox2) is a transcription factor containing a high-mobility-group domain, which enables sequence-specific DNA binding activity [10]. Sox2 is not only an essential embryonic stem cell transcription factor, but also a critical factor for induced pluripotent stem cells [11, 12]. Sox2 is amplified in many cancers and correlated with poor prognosis [13, 14]. Recent studies have shown the role of Sox2 in the maintenance of TICs [15, 16]; however, the regulation of Sox2 and the mechanism of Sox2-mediated tumor initiation are still largely unexplored.
Cyclin-dependent kinases (CDKs) are a family of protein kinases driving the major events of cell cycle in eukaryotic cells [17]. Over the past two decades, a vast body of literature has illustrated that the dysregulation of CDKs induces not only rapid tumor growth but also continued or spontaneous proliferation of cancer cells [18]. In addition to the role in cell cycle regulation, CDKs are involved in cell cycle-independent processes such as DNA damage repair, epigenetics, stemness, metabolism and transcriptional functions [19], indicating broader roles of CDKs. In addition, recent studies have discovered that CDKs 4 and 6 contribute to cancer stemness in breast cancer [20], lung cancer and ovarian cancer [21]. CDK4/6 are also reported to play a role in driving epithelial to mesenchymal transition and metastasis in breast cancer [22]. Hence targeting this group of proteins holds immense promise in the context of cancer.
In this study, using patient-derived xenograft (PDX) tumors of melanoma, colon cancer and pancreatic cancer, we identified CDK1 as the most significantly upregulated gene in TIC-enriched, major histocompatibility (MHC) class I (MHC I)hi tumor cells. We found CDK1 enhanced not only tumor growth but also tumor initiating capacity in human melanoma. We provide evidence that CDK1 binds to Sox2 and regulates its phosphorylation, nuclear translocation and transcriptional activity, thereby promoting tumor initiating potential. These findings reveal a new regulator of Sox2 in cancer cells, and implicate the CDK1-Sox2 interaction as a potential target in cancer therapy.
Materials and Methods
Animal use
Athymic (nu/nu) mice (NCI) or NOD/SCID/IL2Rγnull (NSG) mice (Jackson Laboratories) were used. All animal experiments were approved of by the Institutional Animal Care and Use Committee at the University of Colorado and performed under the institutional guidelines for the use of laboratory animals.
Cell lines and tissue culture
1205Lu, WM239A, A375 were obtained from Rockland Immunochemicals, Inc. HCT116 cell line was obtained from ATCC. Cell lines were cultured in RPMI media containing 2% L Glutamine (Gibco), 10% FCS (BenchmarkTm), 2% penstrep (Corning) and used with in the first 20 passages for experiments described. The cells where routinely checked for mycoplasma by PCR as previously described [23]. All the cell lines were authenticated using STR analysis by Barbara Davis Center Bioresource Core.
Enzymatic digestion of PDX tumors
Surgical specimens of human melanoma, colon cancer and pancreatic cancer were provided by patients in the University of Colorado Hospital cancer clinics after written signed consent under Institutional Review Board-approved protocols, adhering to Health Insurance Portability and Accountability Act Regulations. To establish PDX models, parts of fresh tumor tissues were subcutaneously implanted into pockets made by a small incision on the flank of 5–6-week-old female athymic (nu/nu) mice (NCI) as previously described [24].
Supplementary Table 1 summarizes the information of the PDX tumors used in this study. The PDX tumors were harvested from mice and mechanically dissociated, weighed, and placed into 8 tubes to assess the effects of different digestion methods. Single-cell suspensions were obtained using combinations of digestion with either collagenase I (1 mg/ml = 235 U/ml, Sigma-Aldrich) + hyaluronidase (1 mg/ml = 850 U/ml, Sigma-Aldrich) or collagenase IV (200 U/ml, Worthington) at 37°C, for either 20 minutes or 2 hours, followed by further treatment with or without 0.05% trypsin-EDTA for 5 minutes at 37°C. Viable cell counts were determined using trypan blue staining, and tumor cells were sorted by flow cytometry.
Flow cytometry-based negative and positive selection of PDX tumor cells
Prior to fluorescence-activated cell sorting (FACS), single cell suspensions obtained by the above methods were filtered sequentially using 70- and 40-μm cell strainers and then treated with 1 × Red Blood Cell Lysis Buffer (eBioscience) to remove red blood cells. Human tumor cells were negatively selected by excluding mouse stromal cells that were labeled with a combination of surface markers of mouse cells using APC-labeled anti-mouse MHC class I (H-2Kd, eBioscience), PE-Cy7-labeled anti-mouse CD45 (eBioscience), and PE-Cy7-labeled anti-mouse CD31 (eBioscience) antibodies. These negatively selected human tumor cells were further labeled with FITC-anti-human HLA-ABC (BD Pharmingen) and positively selected. Dead cells were eliminated by using 4’,6-diamidino-2-phenylindole (DAPI) staining, and cells were sorted using the MoFlo XDP100 cell sorter (DakoCytomation). Viable cell counts were determined by analyzing percentages of cells stained/unstained by antibodies.
In vivo tumor initiation assay and phenotypic plasticity of PDX tumor cells
After sorting, human melanoma and colon cancer PDX tumor cells were counted, resuspended in 100 μl of RPMI medium containing 25% high protein matrigel (#354248, BD Biosciences) and subcutaneously injected onto the flanks of 6–8-week-old NSG mice (Jackson Laboratories). Tumor formation was assessed by palpating tumors at the injection sites. The frequency of TICs was calculated using L-Calc Software (Stem Cell Technologies). The significance was determined by chi-square analysis. To investigate phenotypic plasticity, the tumors from MHC Ihi, MHC Ilo and negative selection groups were further digested using collagenase IV for 2 hours and analyzed for their MHC I expression after negative selection as described above.
NanoString stem cell gene array
The gene expression was analyzed by the University of Arizona Genetics Core using a nCounter® Stem Cell Panel (Human) (NanoString Technologies) consisting of 193 pre-selected stem cell genes. Supplementary Table 2 summarizes the information of the genes analyzed in this study and the final results. Briefly, total RNA from sorted cells was hybridized with capture and reporter probes on the nCounter Prep Station. The counts of gene specific fluorescent barcodes represent the expression levels of the mRNA. The data were normalized to the average counts for all housekeeping genes in each sample and analyzed in nSolver software (NanoString Technologies).
Generation of CDK1 overexpressing cells
1205Lu human melanoma cells were transfected a with pCMV-Neo-Bam, pCMV-Neo-Bam CDK1 WT or pCMV-Neo-Bam CDK1 DN (D146N, dominant negative) vectors (all from Addgene) using Lipofectamine 2000 (Thermo Fisher Scientific) according to manufacturer’s protocol. Cells were selected with Geneticin (400 μg/ml G418, Thermo Fisher Scientific) for at least 2 weeks to generate stable cell lines.
Generation of Sox2 CRISPR/Cas9 knockout cells
Sox2 guide RNA (ACCGAGGGGAGCCCGCAGGT) on a pSpCas9(BB)-2A-Puro vector [25] was purchased from GenScript. The cells were transfected using Lipofectamine 2000 (Thermo Fisher Scientific) and selected with puromycin (1 μg/ml) for 2 weeks, followed by single cell sorting using the MoFlo XDP100 cell sorter to generate stable clones from single cells.
Stem cell antibody array
The Human Pluripotent Stem Cell Antibody Array (R&D Systems) with 15 stem cell markers was performed according to the manufacturer’s instructions. In brief, cells were collected and lysed in the RIPA lysis buffer (Sigma) containing proteinase inhibitors. Immunoprecipitation was performed as previously described [26] using either control IgG or anti-CDK1 antibody (ab32384, Abcam). After incubation in blocking buffer for one hour, the array membranes were incubated with the immunoprecipitated samples overnight at 4°C. The membranes were then washed and incubated with the detection antibody cocktail for 2 hours at room temperature. Membranes were washed again and then probed with streptavidin-HRP for 30 minutes at room temperature. The signals were developed using SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher Scientific) and imaged using the Odyssey Fc system (Li-cor).
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using SimpleChIP® Plus Enzymatic Chromatin IP Kit with agarose beads (Cell Signaling Technology), according to the manufacturer’s instructions. In short, proteins were cross-linked to DNA with 1% formaldehyde (Thermo Fisher Scientific) for 10 minutes at room temperature. The formaldehyde was inactivated by glycine for 5 minutes at room temperature and cells were then collected for nuclei preparation and chromatin digestion. Chromatin was digested with a micrococcal nuclease for 20 minutes at 37°C into an average DNA fragment length of 200 to 900 bp, according to manufacturer’s instruction. Nuclei were lysed by 3 sets of 20-second pulses using a Virsonic 100 Ultrasonic Sonicator (Virtis) at setting 6 with a 1/8-inch probe. Chromatin extracts containing DNA fragments were immunoprecipitated using Sox2 polyclonal antibody (#2748, Cell Signaling Technology). The immunoprecipitated complexes were captured by protein G agarose beads (Cell Signaling Technology), cross-links were reversed, and DNA was analyzed using qPCR. ChIP primers are listed in Supplementary Table 3. Primers were designed based on the two predicted Sox2 binding sites (AGCAAGGCCAGCAG (−) strand and GCACTGCTCCTCAG (+) strand) from the search.
All additional procedures, including flow cytometry analysis of MHC I expression, qRT-PCR, immunofluorescent staining of cells and paraffin tissues, analysis of immunofluorescent images, immunoblotting and immunoprecipitation, transcription activity reporter assay for Sox2 and NF-κB, spheroid formation assay, in vivo tumor initiation assay of human melanoma cells, cell cycle analysis, siRNA knockdown, public gene expression data analysis, and statistical analysis are included as methods in Supplementary Information.
Results
Tumor initiating cells are enriched in MHC Ihi tumor cells
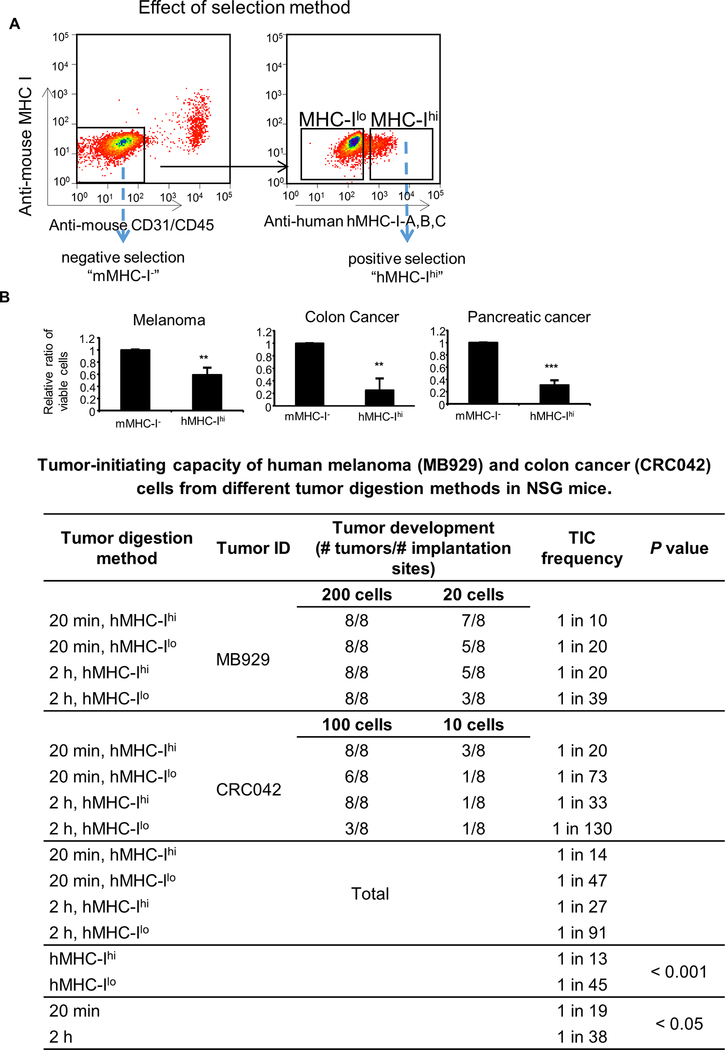
To determine the effects of tumor isolation methods on tumor cell selection, we used PDX tumors from melanoma, colon cancer, and pancreatic cancer (Supplementary Fig. S1A). We found the addition of trypsin reduced the number of viable melanoma cells (−44%), but no obvious effect was observed by the use of different collagenase in melanoma. On the other hand, the use of collagenase (either I or IV) for 20 minutes resulted in a decrease in the number of collected cells in melanoma when compared to the 2 hour digestion (−59%). This decrease from 20 minutes incubation was also observed when colon and pancreatic PDX tumors were analyzed, indicating that the shorter, 20 minutes, enzymatic treatment led to a partial collection of tumor cells.
We then investigated the effects of cell sorting on the collection of viable tumor cells. We compared two cell sorting methods. The first method was negative selection where unstained human tumor cells were sorted away from mouse stromal cells stained with anti-mouse CD31, anti-mouse CD45, and anti-mouse MHC I antibodies [7, 8]. The second method was positive selection where human tumor cells were further positively stained with anti-human HLA-ABC (human MHC I) antibody [5] (Fig. 1A). We found that the numbers of tumor cells sorted by the positive selection method were lower than those sorted by the negative selection method in all three tumor types (47%, 8%, and 28% in melanoma, colon, and pancreatic tumors, respectively) (Fig. 1B, upper panel), indicating that the positive selection protocol led to a partial collection of tumor cells. Altogether, these results demonstrate that the shorter enzymatic treatment and the positive selection of MHC Ihi tumor cells both resulted in a partial selection of tumor cells.
Figure 1. Effect of tumor isolation methods on viable cell number and tumor initiating capacity.
A, FACS sorting of PDX tumor cells that are negative for mouse CD31, mouse CD45 and mouse MHC I expression (negative selection, “mMHC-I-”) and those that are further positive for human MHC I expression (positive selection, “hMHC-Ihi”). B, Viable cell numbers and TIC frequency of PDX tumors. Upper panel, The number of viable cells after positive (“hMHC-Ihi”) selections were compared to those with negative (“mMHC-I-”). Mean ± SE, n = 3–4, **, P < 0.01; ***, P < 0.001. Lower table, Table depicting TIC frequency of PDX tumors. Tumor tissues were digested by collagenase IV for either 20 minutes (20 min) or 2 hours (2 h), excluded of mouse stromal cells (CD31/CD45/MHC-I), and sorted using human-MHC I staining to provide hMHC-Ihi cells and hMHC-Ilo tumor cells. Twenty or 200 cells of MB929 PDX tumor and 10 or 100 cells of CRC042 PDX tumor were subcutaneously injected into NSG mice with high-protein matrigel, and monitored for tumor formation for 24 weeks. L-Calc Software was used to calculate the TIC frequency (with 95% confidence interval).
To determine whether partially selected tumor cells were enriched for TICs, we implanted cells from the melanoma PDX tumor, MB929, or colon cancer PDX tumor, CRC042, into NSG mice using high-protein matrigel. We found that the TIC frequency was higher in MHC Ihi cells (1 in 13) when compared to MHC Ilo cells (1 in 45) (Fig. 1B, lower table). Similarly, the frequency was higher in cells isolated from a 20 minute digestion (1 in 19) compared to those isolated from a 2 hour digestion (1 in 38), demonstrating that both MHC Ihi cells and cells isolated using a shorter digestion time were enriched for TICs.
Because the selection of cells by MHC I expression led to more significant differences in tumor initiating capacity, we decided to investigate the underlying mechanisms that contribute to higher TIC frequency in MHC Ihi cancer cells.
CDK1 is upregulated in MHC Ihi tumor cells and controls MHC I expression
We first examined whether MHC Ihi or MHC Ilo cancer cells were phenotypically static or plastic. Despite the differences in tumor size and frequency, developing tumors from MHC Ihi and MHC Ilo cells from melanoma PDX (Supplementary Fig. S1B) reconstituted the heterogeneity in MHC I expression after reinjection into NSG mice, suggesting dynamic phenotypic switching between these populations.
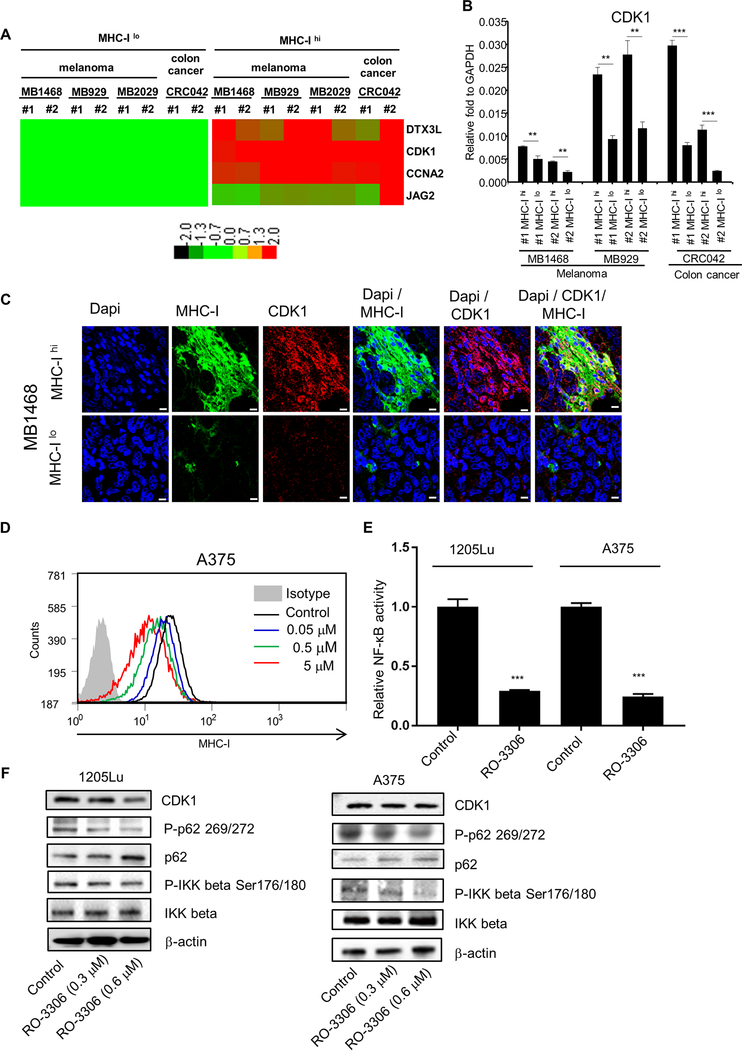
We speculated that MHC Ihi expression was related with unique gene signatures and profiles that enhanced tumor-initiating capacity. We performed NanoString® nCounter Analysis to identify stem cell genes differentially expressed between MHC Ihi and MHC Ilo tumor cells. Three melanoma (MB1468, MB929, and MB2029) and one colon cancer (CRC042) PDX tumors were used for this analysis. DTX3L, CDK1, CCNA2, and JAG2 were identified as some of the genes that were differentially expressed between human MHC Ihi and MHC Ilo cells (P < 0.05, fold change > 1.5) (Fig. 2A, Supplementary Table 2). Among them, CDK1 was consistently upregulated across all MHC Ihi samples. This finding was further validated by qRT-PCR (Fig. 2B). The ratio of CDK1 mRNA levels between MHC Ilo and MHC Ihi cells was 1.6- to 2.1-fold, 2.2- to 5.7-fold, and 3.7- to 4.6-fold in MB1468, MB929, and CRC042 samples, respectively. Immunohistochemical staining of melanoma PDX tumors (Fig. 2C) and the original patient samples (Supplementary Fig. S2A) showed the heterogeneous and clustered expression of CDK1 and MHC I which co-stained well in tumor tissues. To understand the relation between CDK1 and MHC I, we treated melanoma cells with a CDK1 inhibitor RO-3306 [27] or knocked out CDK1 using CRISPR. MHC I expression was downregulated by the CDK1 inhibitor in A375 (Fig. 2D) and 1205Lu (Supplementary Fig. S2B) cell lines as well as in A375 cells where CDK1 was knocked out (Supplementary Fig. S2C), indicating that CDK1 upregulates MHC I expression. Previous reports have suggested that CDK1 affects p62 phosphorylation [28] which in turn regulates NF-κB pathway through IKKβ phosphorylation [28]. The NF-κB pathway has been shown to play a vital role in positively regulating MHC I expression [29, 30]. Consistent with these reports, CDK1 inhibitor downregulated NF-κB transcriptional activity in luciferase assays (Fig. 2E), which correlated with the reduction in phospho-p62 and phospho-IKKβ (Fig. 2F). In line with these findings, NF-κB inhibition downregulated MHC I expression in A375 cells (Supplementary Fig. S2D). Together, these data demonstrate that CDK1 upregulates MHC I expression through the NF-κB pathway.
Figure 2. Identification of CDK1 from stem cell gene array.
A, nCounter stem cell gene array analysis between MHC-Ilo and MHC-Ihi cells. RNAs were isolated from 4 PDX tumors (MB1496, MB929, MB2029 and CRC042). #1 and #2 are biological replicates. Data were normalized by nSolver (NanoString), and displayed as a comparison to the expression levels from MHC-Ilo cells. Genes with significant changes were selected based on P < 0.05, fold change > 1.5, and an original readout > 50. The heat map was generated with TreeView. Red represents upregulated genes; Green represents downregulated genes. B, qRT-PCR validation of CDK1. #1 and #2 are biological replicates. Mean ± SE, n = 3. **, P < 0.01; ***, P < 0.001. C, Immunofluorescent images representing the localization of MHC I expression (Alexa flour 488, green) and CDK1 expression (Alexa flour 594, red) in MB1468 PDX tumor. Upper and lower panels represent regions where tumor cells highly expressed MHC I (MHC-Ihi) and poorly expressed MHC I (MHC-Ilo), respectively. Dapi (blue) was used as the nuclear stain. Scale bar = 10 μm. D, Flow cytometry data depicting the MHC I expression after CDK1 inhibitor RO-3306 treatment in A375 cells. E, Luciferase assays depicting the transcriptional activity of NF-κB after CDK1 inhibitor RO-3306 treatment (2 μM) in 1205Lu and A375 cells. Mean ± SE, n = 3. ***, P < 0.001. F, Immunoblots showing the phosphorylation of p62 and IKKβ after RO-3306 treatment (0.3 or 0.6 μM) in 1205Lu and A375 cells.
CDK1 promotes tumor initiating capacity
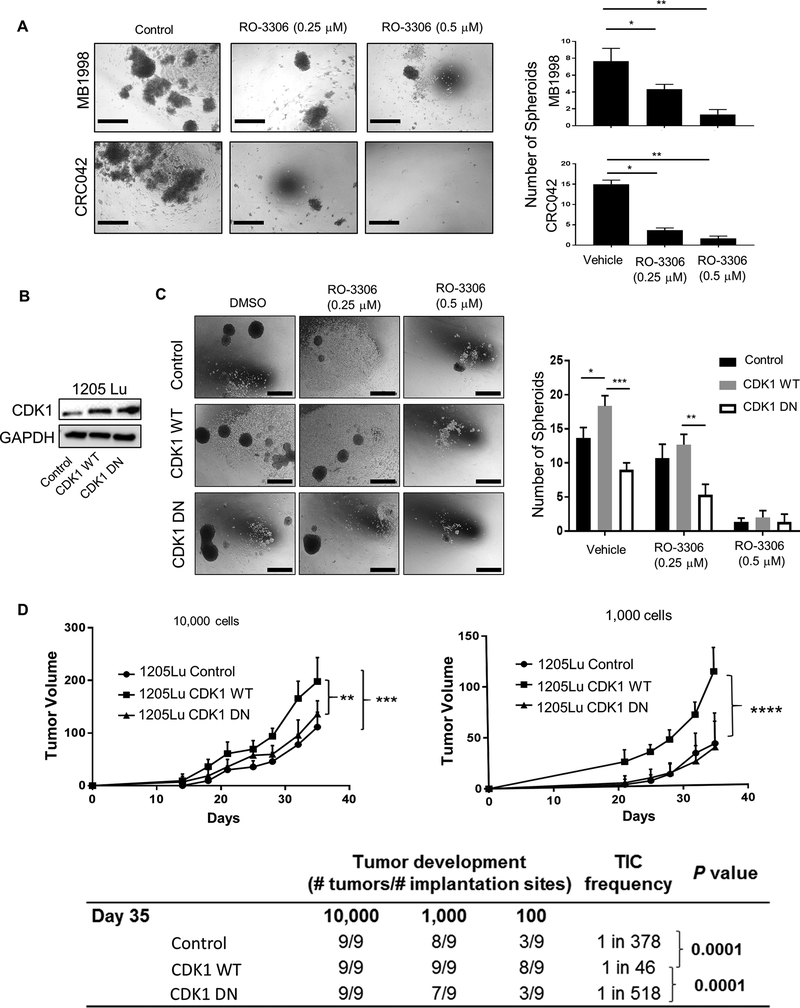
Because CDK1 was upregulated in TIC-enriched MHC Ihi tumor cells, we hypothesized that CDK1 might be involved in driving tumor initiation. Consistent with this hypothesis, MHC Ihi cells from both melanoma (MB1998) and colon cancer (CRC042) PDX tumors displayed higher spheroid forming capacity in vitro when compared to MHC Ilo cells (Supplementary Fig. S3A). When tumor cells were treated with RO-3306, the treatment resulted in a dramatic decrease in spheroid formation in MB1998 and CRC042 cells (Fig. 3A) as well as A375 and WM239A cells (Supplementary Fig. S3B). To confirm the effect of CDK1 on increased spheroid forming capacity, we overexpressed wild-type CDK1 (CDK1-WT) and a kinase-dead variant (CDK1-DN) in 1205Lu cells (Fig. 3B). Overexpression of CDK1-WT but not CDK1-DN led to the increased spheroid formation, which was reduced by the CDK1 inhibitor (Fig. 3C).
Figure 3. Effect of CDK1 inhibitor on spheroid formation, tumor growth and initiation.
A, in vitro spheroid formation of PDX tumor cells from MB1998 and CRC042 (negatively selected away from mouse stromal cells), treated with CDK1 inhibitor RO-3306 for 10 days. Left panel, Representative picture of spheroids. Right panel, Number of spheroids per well. Scale bar = 100 μm. B, Immunoblots confirming the overexpression of CDK1 wild-type (WT) and D146N (DN) in 1205Lu cells compared to control vector (Neo-Bam)-transfected cells. C, in vitro spheroid formation of 1205Lu control, CDK1 WT and CDK1 DN cells treated with RO-3306 for 10 days. Left panel, Representative picture of spheroids. Right panel, Number of spheroids per well. Scale bar = 100 μm. D, in vivo TIC frequency and tumor growth experiment. Titrating numbers of 1205Lu control, CDK1 WT and CDK1 DN cells were injected into NSG mice subcutaneously. Upper panel, Tumor growth curve of 10,000 and 1,000 cell injection (Tumors from 100 cell injection were palpable but small, and hence were not plotted in the graph). Lower table, TIC frequency was determined at day 35 using 10,000, 1,000 and 100 cell injection data. Mean ± SE, n = 3 (A, C) or 9 (D). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We then investigated the role of CDK1 in vivo. To determine its effect on tumor initiating capacity, limiting dilutions (10,000, 1,000, and 100 cells) of 1205Lu cells overexpressing CDK1-WT or CDK1-DN were injected into NSG mice, and their tumor formation and TIC frequency was compared to control-vector-transfected 1205Lu cells. We found the tumor volume (Fig. 3D, upper panel) and TIC frequency (Fig. 3D, lower table) were significantly increased in the CDK1-WT overexpressing cells in comparison to vector-control or CDK1-DN cells. Mice harboring 1205Lu tumors treated orally with AZD5438, another CDK1 inhibitor used in xenograft models [31], yielded similar results with a reduction in tumor volume and a 10-fold lower TIC frequency in the treated group (Supplementary Fig. S3C). Consistent with the in vitro data shown in Fig. 2D and Supplementary Fig. S2B and C, CDK1 inhibitor reduced MHC I expression in tumor cells in vivo (Supplementary Fig. S3D).
Since CDK1 drives G2/M cell cycle progression [32], the reduction in spheroid forming ability and tumor initiation capacity could be explained by CDK1’s role on cell cycle arrest. To delineate CDK1’s effects on cell cycle and cell growth, melanoma cells were cultured in 2D and treated with RO-3306. Consistent with CDK1’s role in the G2/M transition, the doses of RO-3306 (0.2 – 2 μM) resulted in an increase of melanoma cells in G2/M phase (Supplementary Fig. S3E, left); however, these doses rarely induced growth arrest in 2D culture (Supplementary Fig. S3E, right). This finding could be explained by a partially redundant role of CDK1 and CDK2 in cell cycle progression of cancer cells [33] but also suggests a cell cycle-independent role for CDK1 in tumorigenesis.
Altogether, these data demonstrate that CDK1 promotes not only tumor growth but also tumor initiating capacity of human melanoma cells, which cannot be explained by CDK1’s role as a master regulator of cell cycle.
CDK1 interacts with Sox2 and phosphorylates Sox2
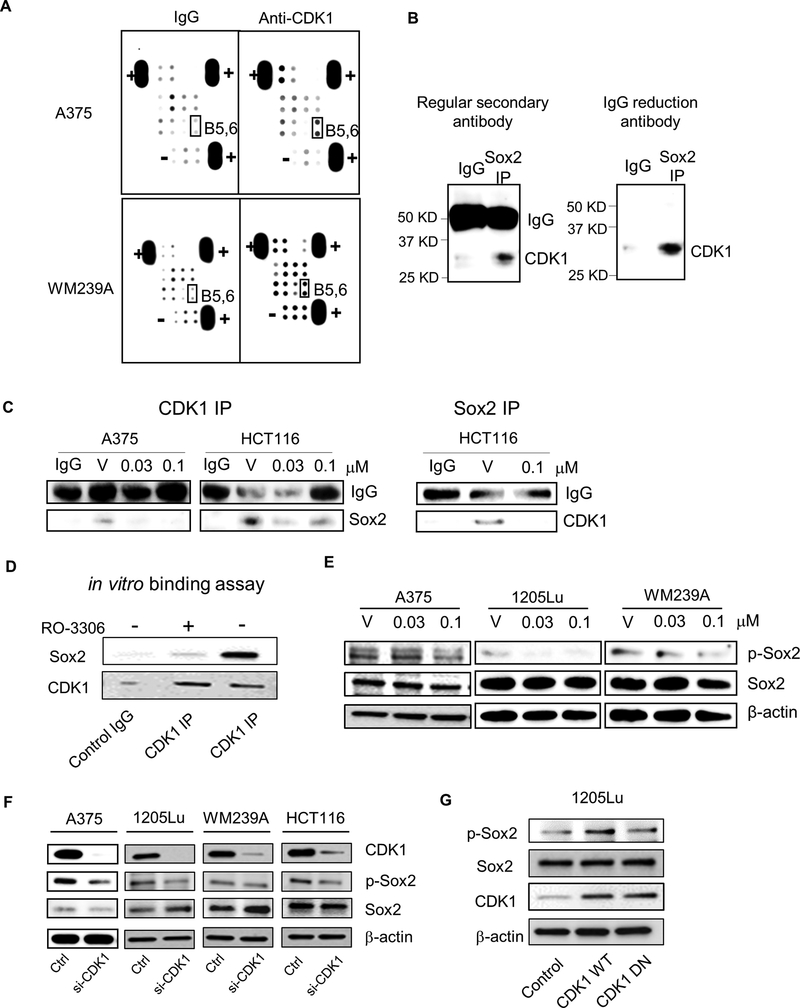
We speculated that CDK1 could interact with stem cell-related proteins, thereby promoting tumor-initiating capacity. To test this hypothesis, we used a stem cell antibody array to identify proteins that immunoprecipitated with the anti-CDK1 antibody. In both A375 and WM239A melanoma cell lines, CDK1 displayed a binding affinity to Sox2 (Fig. 4A). This result was validated in a co-immunoprecipatation (Co-IP) assay (Fig. 4B). When cells were treated with RO-3306, Co-IP of CDK1 with Sox2 (detection of Sox2 with CDK1 IP) was decreased in A375 melanoma and HCT116 colon cancer cell lines (Fig. 4C, Left panels). Co-IP of Sox2 with CDK1 (CDK1 immunoblot bands with Sox2 IP) was also decreased in HCT 116 cells (Fig. 4C, Right panel). In line with these findings, in vitro binding assays confirmed a direct binding of CDK1/Cyclin B complex to Sox2, which was inhibited by RO-3306 treatment (Fig. 4D).
Figure 4. Identification of stem cell genes binding to CDK1.
A, Stem cell gene antibody array. Lysates of A375 and WM239A cells were IP pulled-down by IgG (control) or anti-CDK1 antibody, and subjected to the antibody array. B5,6, highlighted in black box, are Sox2. “+” and “-” represent positive and negative control, respectively. B, Sox2 IP immunoblotted by anti-CDK1 antibody in colon cancer cell line HCT116. Left panel, immunoblot with anti-rabbit secondary antibody, Right panel, immunoblot with rabbit IgG reduction TrueBlot® secondary antibody. C, CDK1 and Sox2 IP immunoblot of cells treated with vehicle (V) or RO-3306. Left panel, Cell lysates were immunoprecipitated with anti-CDK1 antibody and immunoblotted with anti-Sox2 antibody. Right panel, Cell lysates were immunoprecipitated with anti-Sox2 antibody and immunoblotted with anti-CDK1 antibody. IgG served as internal control. D, Immunoblots depicting the binding of recombinant Sox2 to CDK1/Cyclin B Complex after CDK1 IP. RO3306 was used to show CDK1-dependent interaction. E, Immunoblots depicting the Sox2 phosphorylation (S249-S250-S251, p-Sox2) and Sox2 after treatment with vehicle (V) or RO-3306 (0.03 or 0.1 μM) in A375, 1205Lu and WM239A cells. β-actin used as loading control. F, Immunoblots depicting Sox2 phosphorylation (p-Sox2) and Sox2 after CDK1 siRNA knockdown in A375, 1205Lu, WM239A and HCT116 cells. G, Immunoblots depicting phosphorylated Sox2 (p-Sox2), Sox2 and CDK1 in 1205Lu cells transfected with Control, CDK1 WT and CDK1 DN vectors.
Because CDK1 is a kinase that catalyzes the transfer of phosphate groups from ATP to target substrate proteins [34], we hypothesized that CDK1 may phosphorylate Sox2 while interacting with this protein. Sox2 has been demonstrated to be phosphorylated at three consecutive serine residues, namely S249, S250, and S251 [35]. Consistent with our hypothesis, the treatment with RO-3306 led to a decrease in Sox2 phosphorylation (S249-S250-S251) in A375, 1205Lu, and WM239A melanoma cells (Fig. 4E). While we observed differences in the phosphorylation of Sox2, we did not see the effects of RO-3306 on Sox2 protein expression. Similar to these findings, knocking down CDK1 using CDK1 siRNA showed a reduction in Sox2 phosphorylation in A375, 1205Lu, WM239A, and HCT116 cells without affecting total Sox2 levels (Fig. 4F). In addition, CDK1-WT overexpression in 1205Lu cells led to increased Sox2 phosphorylation whereas this effect was not observed in cells overexpressing CDK1-DN (Fig. 4G). Together, these results demonstrate a CDK1-Sox2 interaction and subsequent phosphorylation of Sox2 in melanoma cells.
CDK1 is critical for Sox2 nuclear localization
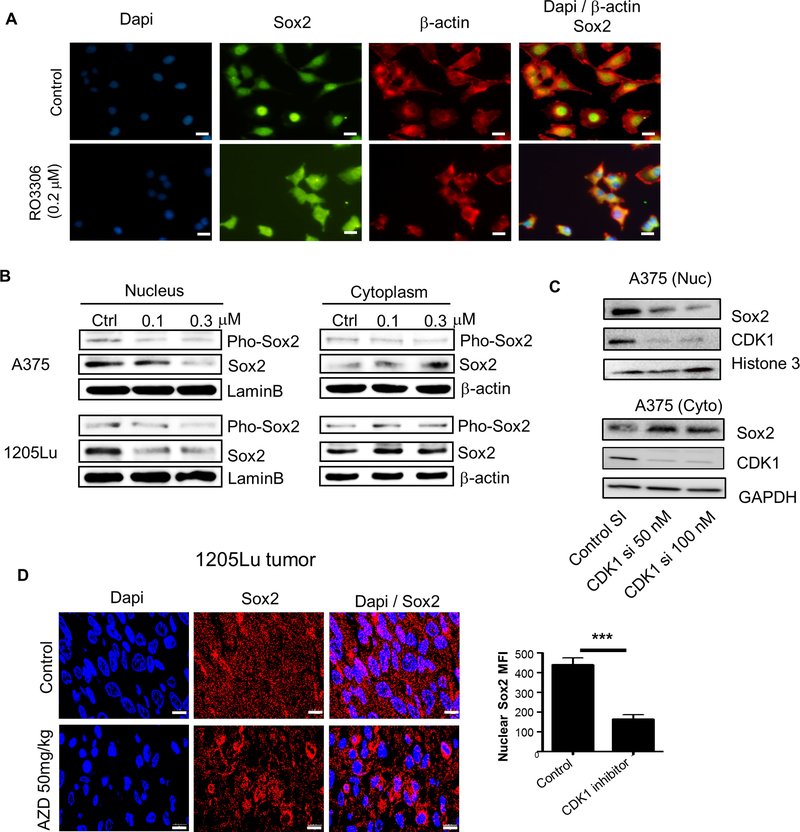
Phosphorylation of transcription factors affects their structure, cellular localization, stability, protein-protein interactions and DNA binding ability. Therefore, we analyzed the subcellular localization of Sox2 after CDK1 inhibition or knockdown. When visualized by immunofluorescence, Sox2 was predominantly located in the nucleus of A375 cells prior to treatment with RO-3306, but this nuclear localization was impaired after treatment with the inhibitor (Fig. 5A). These findings were confirmed by immunoblotting of A375 and 1205Lu cells (Fig. 5B): Sox2 was predominantly located in the nucleus without treatment, whereas RO-3306 treatment led to an increase in cytoplasmic Sox2 and a corresponding decrease in nuclear Sox2. Specific knockdown of CDK1 by siRNA showed a similar reduction of Sox2 in the nucleus with a corresponding increase in the cytoplasm (Fig. 5C). In line with these findings, immunohistochemical staining of 1205Lu tumor tissues treated with the CDK1 inhibitor, AZD, showed a reduction of Sox2 nuclear localization (Fig. 5D). Of note, the effect of CDK1 on Sox2 localization was independent of CDK1’s effect on NF-κB activity, since NF-κB inhibition did not affect Sox2 nuclear localization in A375 cells (Supplementary Fig. S4). Collectively, these results demonstrate a role for CDK1 in maintaining the nuclear localization of Sox2.
Figure 5. Effect of CDK1 inhibitor on Sox2 subcellular localization.
A, Immunofluorescent images representing the nuclear/cytoplasmic localization of Sox2 (Alexa flour 488, green) and β-actin (Alexa flour 594, red) after RO-3306 treatment in A375 cells. Dapi (blue) was used for the nuclear stain. Scale bar = 10 μm. B, Immunoblots of phosphorylated Sox2 and total Sox2 in the nucleus and cytoplasm after RO-3306 treatment in A375 and 1205Lu cells. LaminB and β-actin served as internal controls for nuclear and cytoplasmic protein, respectively. C, Immunoblots depicting the nuclear (Nuc) and cytoplasmic (Cyto) localization of Sox2 and CDK1 after CDK1 siRNA knockdown in A375 cells. D, Left panel, Immunofluorescent images representing the nuclear/cytoplasmic localization of Sox2 (Alexa flour 594, red) in 1205Lu xenograft tumors, shown in Supplementary Fig. S3C, treated with AZD5438 (50 mg/kg) by daily oral gavage for 14 days in comparison to untreated tumors. Right panel, MFI quantification of nuclear Sox2 in cells from untreated and AZD5438-treated tumors. Mean ± SE, n = 30. ***, P < 0.001. Scale bar = 10 μm.
CDK1 is necessary for Sox2 transcriptional activity
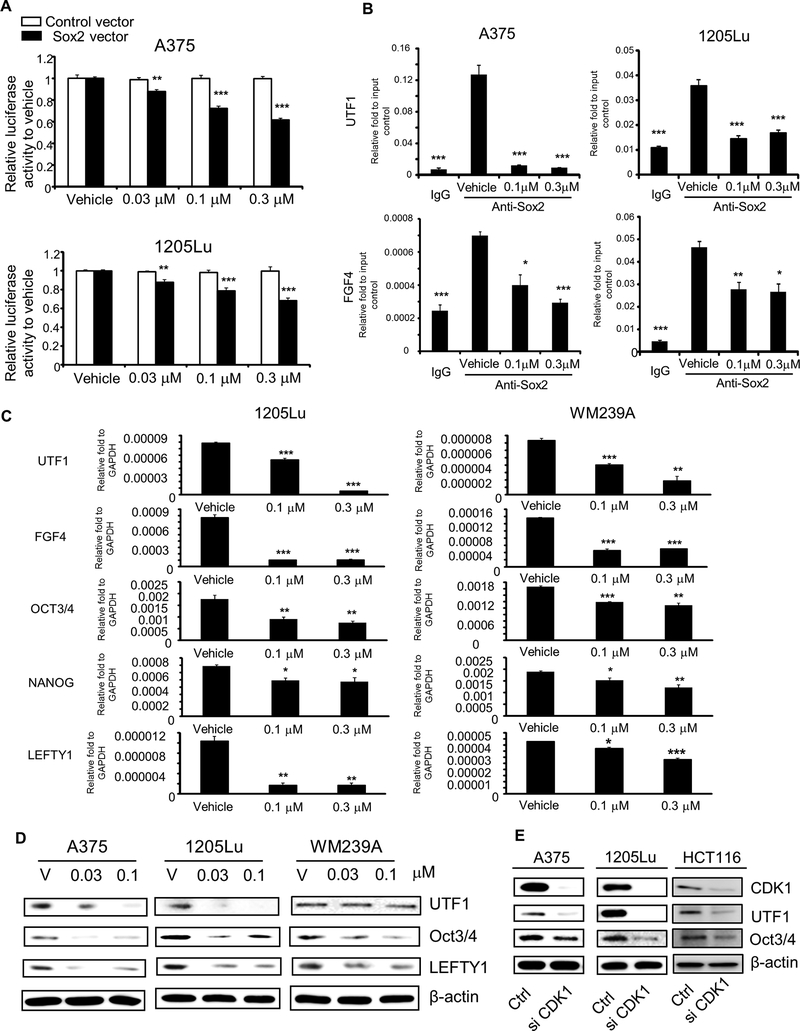
Because nuclear localization and phosphorylation are important for the function of transcription factors, it is reasonable to speculate that CDK1 regulates Sox2’s transcriptional activity. To test this hypothesis, we first transfected melanoma cells with a luciferase vector driven by a promoter containing Sox2 binding motif. When A375 and 1205Lu cells were treated with RO-3306, we observed a dose-dependent reduction in the luciferase activity in the cells transfected with Sox2 binding motif (Fig. 6A). It should be noted that A375 and 1205Lu cells transfected with control vector did not show any reduction in luciferase activity. We observed comparable results using HCT116 and WM239A cells, treated with AZD5438, another CDK1 inhibitor used for in vivo study (Supplementary Fig. S5A). Collectively, these data demonstrate that CDK1 upregulates the transcriptional activity of Sox2.
Figure 6. Effect of CDK1 inhibitor on Sox2 functions.
A, Luciferase reporter assay using Sox2 response elements. White and black bars represent activities from cells transfected with control vector and Sox2-binding luciferase reporter vector, respectively. Luciferase activities after RO-3306 treatment were normalized to those with corresponding control vector or Sox2-binding luciferase reporter vector. Mean ± SE, n = 6. **, P < 0.01 compared with vehicle control; ***, P < 0.001. B, ChIP PCR assay, using rabbit anti-Sox2 antibody with DNA extracts prepared from cells treated with vehicle and RO-3306 (0.1 or 0.3 μM). Normal rabbit IgG served as binding control. Mean ± SE, n = 3. *, P < 0.05 compared with vehicle control; **, P < 0.01; ***, P < 0.001. C, qRT-PCR analysis of Sox2 downstream genes (OCT3/4, NANOG, UTF1, FGF4, and LEFTY1) after RO-3306 (0.1 or 0.3 μM) treatment in 1205Lu and WM239A cells. GAPDH served as internal control. Mean ± SE, n = 3. *, P < 0.05; **, P < 0.01; ***, P < 0.001. D, Immunoblot analysis for Sox2 downstream proteins (OCT3/4, UTF1, and LEFTY1) after RO-3306 treatment (0.03 or 0.1 μM) in A375, 1205Lu and WM239A cells. β-actin served as internal control. E, Immunoblots depicting expression of CDK1, Oct3/4, and UTF1 after CDK1 siRNA knockdown in A375, 1205Lu and HCT116 cells. The experiments were done with the samples mentioned in Figure 4F and hence the same CDK1 and β-actin blots are used in this figure.
We then used a chromatin immunoprecipitation (ChIP) assay to investigate the effect of CDK1 on the binding of Sox2 to the downstream genes, UTF1 and FGF4. We found that the DNA regions within the Sox2 enhancer of UTF1 and FGF4 were enriched with anti-Sox2 antibody compared with IgG control in both A375 and 1205Lu cells, confirming the binding of Sox2 to these regions (Fig. 6B). When the cells were treated with RO-3306, Sox2’s binding signals in UTF1 and FGF4 promoters were reduced compared with their corresponding vehicle control. These results confirm that CDK1 positively regulates Sox2’s binding to the downstream genes.
To further understand the effect of CDK1 on Sox2’s function, we treated melanoma cells with the CDK1 inhibitor and examined the expression of downstream genes. Upon treatment with the inhibitor, the expression of UTF1 and FGF4 mRNA was reduced in 1205Lu and WM239A cells, compared with vehicle treatment (Fig. 6C). Likewise, the expression of other downstream genes such as OCT3/4, NANOG, and LEFTY1 that are transcriptionally controlled by Sox2 [36] was reduced in both cells. The reduction of the gene expression was further confirmed by immunoblotting results which showed decreased protein levels of UTF1, Oct3/4, and LEFTY1 with RO-3306 treatment (Fig. 6D). Similar to the effect of the CDK1 inhibitor, knockdown of CDK1 by siRNA led to decreased transcript levels of UTF1 and FGF4 (Supplementary Fig. S5B). UTF1 and Oct3/4 protein levels in CDK1 siRNA treated cells confirmed the mRNA results (Fig. 6E). Together, the data indicate the important role of CDK1 on Sox2’s transcriptional activity.
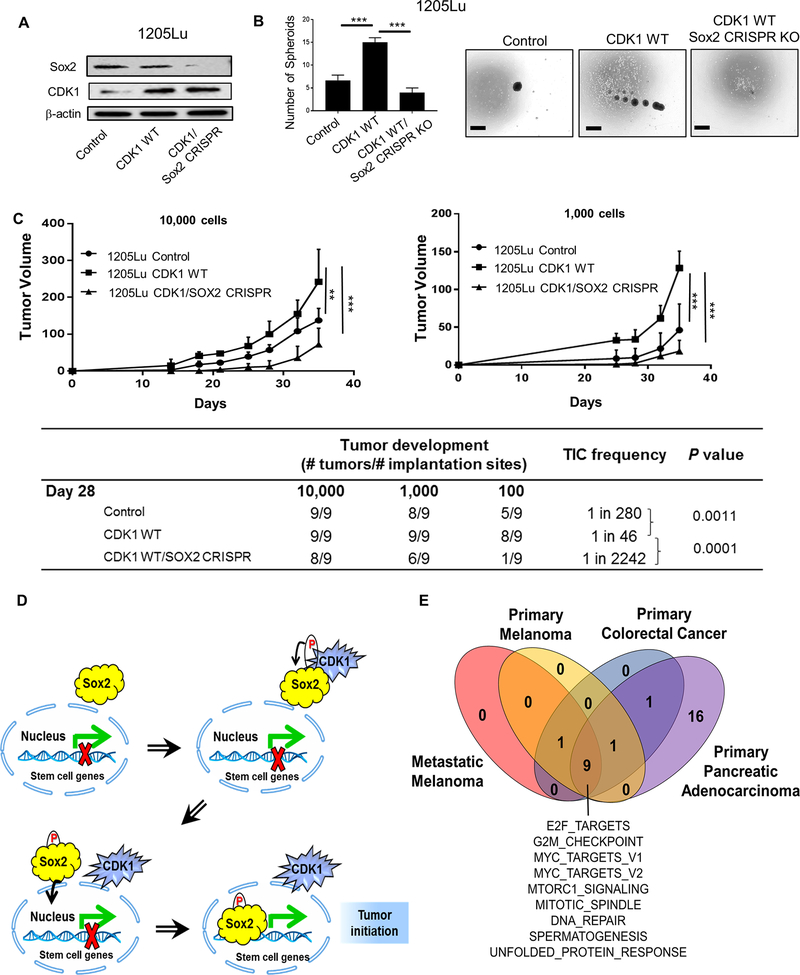
To define the biological implications of CDK1’s role on Sox2, we knocked out Sox2 using a CRISPR/Cas9 system in 1205Lu cells overexpressing CDK1-WT (Fig. 7A). Consistent with the data shown in Fig. 3C, CDK1-WT overexpression enhanced the spheroid forming capacity of 1205Lu cells compared to control, whereas this increase was abrogated by knocking out Sox2 (Fig. 7B). Indeed, Sox2 knockout not only abrogated CDK1’s effects but also further decreased the spheroid forming capacity even compared to control 1205Lu cells. Similarly, TIC frequency and tumor volume of CDK1-WT overexpressing 1205Lu cells were lower than those of control cells when Sox2 was knocked out (Fig. 7C). All together, the data indicate that CDK1-driven tumor initiating capacity substantially depends on its binding partner Sox2.
Figure 7. Biological implications of CDK1’s role in Sox2 function and tumorigenesis.
A, Immunoblots of Sox2 and CDK1 in 1205Lu cells transfected with Control, CDK1 WT and CDK1 WT + Sox2 CRISPR knockout (KO). B, in vitro spheroid formation of 1205Lu cells transfected with Control, CDK1 WT and CDK1 WT/Sox2 CRISPR KO, cultured for 10 days. Left panel, Number of spheroid per well. Right panel, Representative picture of spheroids. Scale bar = 100 μm. C in vivo TIC frequency and tumor growth experiment. Titrating numbers of 1205Lu control, CDK1 WT and CDK1 WT/Sox2 CRISPR cells were injected into NSG mice subcutaneously. Upper panel, Tumor growth curve of 10,000 and 1,000 cell injection (Tumors from 100 cell injection were palpable but small, and hence were not plotted in the graph). Lower table, TIC frequency was determined at day 35 using 10,000, 1,000 and 100 cell injection data. Mean ± SE, n = 3 (B) or 9 (C). **, P < 0.01; ***, P < 0.001. D, CDK1 binds to Sox2 and regulates its phosphorylation, nuclear translocation and transcriptional activity, thereby promoting tumor initiating potential. E, GSEA results for melanoma, colorectal and pancreatic cancers enriched in CDK1hi patients. Melanoma data are from RNA-seq data of the Cancer Genome Atlas (TCGA) from cBioPortal (PMID: 26091043). Colon cancer data are from the accession number GSE14333 from NCBI Gene Expression Omnibus (PMID: 19996206). Pancreatic cancer data are from RNA-seq data of the Queensland Center for Medical Genomics from cBioPortal (PMID: 26909576). Nine Hallmark gene sets were found to be commonly enriched in the CDK1hi patients (P < 0.05).
In summary, we found CDK1 enhanced tumor growth as well as tumor initiating capacity in human melanoma. CDK1 bound to Sox2 and regulated its phosphorylation, nuclear translocation, and transcriptional activity, thereby promoting tumor initiating potential (Fig. 7D).
Discussion
A growing body of evidence suggests broader roles of CDKs [18]. The present findings indicate that CDK1 is not only a cell cycle regulator but also a kinase that catalyzes the transfer of phosphate groups from ATP to target substrate proteins [34]. CDK1 has been shown to interact and phosphorylate both its canonical cell cycle regulators and non-canonical partners including mitochondrial P53 [37], Filamin A [38], and HUR [39]. The functions of these non-canonical partners change through phosphorylation. Accordingly, CDK1 appears to be involved in protein translation, chromatin structure, cell morphogenesis, protein secretion, and nuclear transport, in addition to its role in cell cycle regulation [17, 34]. Our studies demonstrate that the effect of CDK1 in tumorigenesis and tumor initiation is from both its canonical function as a cell cycle regulator and its non-canonical function in mediating phosphorylation and nuclear localization of the pluripotent stem cell gene Sox2. Sox2 interacts with other pluripotent stem cell gene OCT3/4 to govern the transcription of NANOG and other pluripotency-associated genes including FGF4, UTF1 and LEFTY1 [46, 51, 52]. It is therefore reasonable to consider that CDK1 might interact with other stem cell factors such as OCT3/4 to facilitate tumorigenesis. Indeed, CDK1 was shown to interplay with OCT3/4 in a complex to maintain the undifferentiated status of embryonic stem cells [53]. Because the interaction between CDK1 and OCT3/4 does not seem to depend on the kinase activity of CDK1 [53], their interaction may not be direct and may need an intermediate partner such as Sox2.
Post-translational modulation of transcription factors by phosphorylation is related with either enhanced or suppressed transcriptional activity [40]. For example, phosphorylation of Sox2 at Thr118 by Akt was reported to be correlated with enhanced transcriptional activity of Sox2 in mouse embryonic stem cells [41]. On the other hand, it was shown in human embryonic stem cells that Sox2 is phosphorylated at three consecutive serine residues (S249-S250-S251) [35]; however, the biological function and regulation of this phosphorylation was undetermined in this study. Here, by using a CDK1 specific inhibitor, we have provided evidence that the phosphorylation of Sox2 at S249-S250-S251 enhances its transcriptional activity in melanoma and that this phosphorylation is regulated by CDK1. CDKs can compensate each other’s function in regulating cell cycle progression [18], and CDK2 has also been reported to regulate Sox2 phosphorylation in embryonic stem cells at different motifs (S39-S253), suggesting multiple modes of regulating Sox2 by CDKs [42]. While CDK1 is required for the maintenance of embryonic stem cells [43, 44], CDK2-mediated Sox2 regulation is dispensable for embryonic stem cell maintenance [42], therefore, CDK1 might possess a unique function in regulating stem cell function. In line with these reports, gene set enrichment analysis (GSEA) of publicly available gene expression data of CDK1hi tumors from human melanoma, colorectal cancer and pancreatic cancer showed a unique signature where pathways regulated by MYC, mTOR, DNA repair, spermatogenesis, unfolded protein response, and E2F target genes were consistently upregulated across three tumor types (Fig. 7E). Survival analysis of CDK1hi vs CDK1lo tumors also attributed worse prognosis for CDK1hi tumors in primary melanoma and colorectal cancer (Supplementary Fig. S6), with metastatic melanoma and pancreatic cancer showing a similar trend. In addition, it is important to note that MYC targets share an 85 % overlap with Sox2, which could also result in the upregulation of Sox2 target genes in CDK1hi tumors [45].
In agreement with our observation, Sox2 has been shown to play a vital role in determining self-renewal capacity and tumorigenic potential in multiple cancers [15, 46–48]. However, this particular function of Sox2 in tumorigenesis has been disputed by recent reports in melanoma [49, 50]. While Sox2 regulated self-renewal and tumorigenicity of human melanoma-initiating cells [15], Sox2 was not required for melanomagenesis and metastasis formation in genetically engineered mouse models of melanoma [49, 50]. Several factors could contribute to this discrepancy, including human vs. mouse tumors, immunocompromised vs. immunocompetent mice, transplanted vs. spontaneous tumors, and tumor initiation vs. tumor growth. Furthermore, Sox2’s effect could be more profound when limiting dilutions are used to evaluate tumor initiation from small numbers of tumor cells rather than larger cell numbers.
We also demonstrate, in this study, that phenotypic heterogeneity in tumors plays a vital role in determining TIC frequency of human cancers and that tumor cell isolation methods account for variations in TIC frequency. Previous studies have shown variations in TIC frequency, particularly in melanoma. While some studies reported an extremely high TIC frequency (1 in 4) [4, 5], other groups showed a relatively low TIC frequency (less than 1 in 500) in melanoma [8, 9]. The former used a digestion method of trypsin-EDTA, collagenase IV, and a short incubation time, combined with a tumor cell sorting method of MHC I positive selection, whereas the latter isolated tumor cells with collagenase I or IV without trypsin-EDTA, using a longer incubation time, and MHC I negative selection. We show here that the use of a shorter enzymatic treatment and MHC I positive selection leads to a partial collection of tumor cells thereby enriching TICs. We also show that the use of trypsin-EDTA leads to a partial collection of tumor cells, although its effect on TIC frequency hasn’t been examined. Hence, our findings are in line with some of the previous observations that high TIC frequency could be partially explained by their tumor cell isolation methods to enrich a highly tumorigenic subpopulation of cancer cells [3–5]. Of note, we observed this enrichment effect in colon and pancreatic cancers in addition to melanoma. Our study therefore provides important insights on how tumor cell isolation methods affect TIC frequency determination.
In conclusion, we demonstrate a previously unrecognized role for CDK1 in regulating tumor initiation. We show that MHC Ihi cells from the PDX tumors of melanoma, colon cancer, and pancreatic cancer possess a higher tumor initiating capacity compared to MHC Ilo cells, and that this variance in tumor initiation is due to the difference in CDK1 expression between these cell populations. We demonstrate that CDK1 interacts with Sox2 and promotes tumor initiation in human melanoma, and blockade of CDK1 leads to the downregulation of pluripotent stem cell genes governed by Sox2. Because the interaction of CDK1 and Sox2 is also observed in colon cancer, CDK1 may have similar effects on Sox2’s activity and function in other cancers. These data suggest a novel strategy in cancer treatment by interrupting CDK1’s function and its protein-protein interaction.
Supplementary Material
Statement of Significance: Findings uncover CDK1 as a new regulator of Sox2 during tumor initiation and implicate the CDK1-Sox2 interaction as a potential therapeutic target in cancer.
Acknowledgements
We thank the University of Colorado Denver (UCD) Skin Cancer Repository (Carol Amato) for providing human tumors, UCCC Grant (P30CA046934), SDRC Grant (P30AR057212) for helping with FACS sorting and histology, ALM Core (supported by CCTSI Grant UL1TR001082) for helping imaging experiments. We also thank Ms. Angelina Baroffio (Dermatology, UCD) for technical assistance, Dr. Natalie Ahn (Chemistry and Biochemistry, UCB) for helpful comments, and Ms. Joanne Domenico (Dermatology, UCD) for editing the manuscript.
Grant Support: This work was supported by an NIH/NCI R01CA197919 (to M. Fujita), Veterans Affairs Merit Review Award 5I01BX001228 (to M. Fujita), Cancer League of Colorado (to M. Fujita) and Tadamitsu Cancer Research Fund (to M. Fujita).
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Meacham CE and Morrison SJ, Tumour heterogeneity and cancer cell plasticity. Nature, 2013. 501(7467): p. 328–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medema JP, Cancer stem cells: the challenges ahead. Nat Cell Biol, 2013. 15(4): p. 338–44. [DOI] [PubMed] [Google Scholar]
- 3.Boyle SE, et al. , CD271 Expression on Patient Melanoma Cells Is Unstable and Unlinked to Tumorigenicity. Cancer Res, 2016. 76(13): p. 3965–77. [DOI] [PubMed] [Google Scholar]
- 4.Quintana E, et al. , Phenotypic heterogeneity among tumorigenic melanoma cells from patients that is reversible and not hierarchically organized. Cancer Cell, 2010. 18(5): p. 510–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quintana E, et al. , Efficient tumour formation by single human melanoma cells. Nature, 2008. 456(7222): p. 593–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen N, et al. , Understanding melanoma stem cells. Melanoma Manag, 2015. 2(2): p. 179–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishizawa K, et al. , Tumor-initiating cells are rare in many human tumors. Cell Stem Cell, 2010. 7(3): p. 279–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo Y, et al. , ALDH1A isozymes are markers of human melanoma stem cells and potential therapeutic targets. Stem Cells, 2012. 30(10): p. 2100–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boiko AD, et al. , Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature, 2011. 466(7302): p. 133–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar A and Hochedlinger K, The sox family of transcription factors: versatile regulators of stem and progenitor cell fate. Cell Stem Cell, 2013. 12(1): p. 15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu J, et al. , Induced pluripotent stem cell lines derived from human somatic cells. Science, 2007. 318(5858): p. 1917–20. [DOI] [PubMed] [Google Scholar]
- 12.Giorgetti A, et al. , Generation of induced pluripotent stem cells from human cord blood using OCT4 and SOX2. Cell Stem Cell, 2009. 5(4): p. 353–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rudin CM, et al. , Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat Genet, 2012. 44(10): p. 1111–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bass AJ, et al. , SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nat Genet, 2009. 41(11): p. 1238–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santini R, et al. , SOX2 regulates self-renewal and tumorigenicity of human melanoma-initiating cells. Oncogene, 2014. 33(38): p. 4697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Basu-Roy U, et al. , Sox2 maintains self renewal of tumor-initiating cells in osteosarcomas. Oncogene, 2012. 31(18): p. 2270–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holt LJ, et al. , Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science, 2009. 325(5948): p. 1682–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malumbres M and Barbacid M, Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer, 2009. 9(3): p. 153–66. [DOI] [PubMed] [Google Scholar]
- 19.Lim S and Kaldis P, Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development, 2013. 140(15): p. 3079–93. [DOI] [PubMed] [Google Scholar]
- 20.Dai M, et al. , CDK4 regulates cancer stemness and is a novel therapeutic target for triple-negative breast cancer. Sci Rep, 2016. 6: p. 35383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonuccelli G, et al. , Targeting cancer stem cell propagation with palbociclib, a CDK4/6 inhibitor: Telomerase drives tumor cell heterogeneity. Oncotarget, 2017. 8(6): p. 9868–9884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu T, et al. , CDK4/6-dependent activation of DUB3 regulates cancer metastasis through SNAIL1. Nat Commun, 2017. 8: p. 13923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uphoff CC and Drexler HG, Detecting Mycoplasma contamination in cell cultures by polymerase chain reaction. Methods Mol Med, 2004. 88: p. 319–26. [DOI] [PubMed] [Google Scholar]
- 24.Luo Y, et al. , Side population cells from human melanoma tumors reveal diverse mechanisms for chemoresistance. J Invest Dermatol, 2012. 132(10): p. 2440–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanjana NE, Shalem O, and Zhang F, Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods, 2014. 11(8): p. 783–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto M, et al. , Constitutively active inflammasome in human melanoma cells mediating autoinflammation via caspase-1 processing and secretion of interleukin-1beta. J Biol Chem, 2010. 285(9): p. 6477–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vassilev LT, et al. , Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci U S A, 2006. 103(28): p. 10660–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linares JF, et al. , Phosphorylation of p62 by cdk1 controls the timely transit of cells through mitosis and tumor cell proliferation. Mol Cell Biol, 2011. 31(1): p. 105–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Forloni M, et al. , NF-kappaB, and not MYCN, regulates MHC class I and endoplasmic reticulum aminopeptidases in human neuroblastoma cells. Cancer Res, 2010. 70(3): p. 916–24. [DOI] [PubMed] [Google Scholar]
- 30.Dejardin E, et al. , Regulation of major histocompatibility complex class I expression by NF-kappaB-related proteins in breast cancer cells. Oncogene, 1998. 16(25): p. 3299–307. [DOI] [PubMed] [Google Scholar]
- 31.Byth KF, et al. , AZD5438, a potent oral inhibitor of cyclin-dependent kinases 1, 2, and 9, leads to pharmacodynamic changes and potent antitumor effects in human tumor xenografts. Mol Cancer Ther, 2009. 8(7): p. 1856–66. [DOI] [PubMed] [Google Scholar]
- 32.Enserink JM and Kolodner RD, An overview of Cdk1-controlled targets and processes. Cell Div, 2010. 5: p. 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.L’Italien L, et al. , Unmasking the redundancy between Cdk1 and Cdk2 at G2 phase in human cancer cell lines. Cell Cycle, 2006. 5(9): p. 984–93. [DOI] [PubMed] [Google Scholar]
- 34.Ubersax JA, et al. , Targets of the cyclin-dependent kinase Cdk1. Nature, 2003. 425(6960): p. 859–64. [DOI] [PubMed] [Google Scholar]
- 35.Van Hoof D, et al. , Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell, 2009. 5(2): p. 214–26. [DOI] [PubMed] [Google Scholar]
- 36.Masui S, et al. , Pluripotency governed by Sox2 via regulation of Oct3/4 expression in mouse embryonic stem cells. Nat Cell Biol, 2007. 9(6): p. 625–35. [DOI] [PubMed] [Google Scholar]
- 37.Nantajit D, et al. , Cyclin B1/Cdk1 phosphorylation of mitochondrial p53 induces anti-apoptotic response. PLoS One, 2010. 5(8): p. e12341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cukier IH, Li Y, and Lee JM, Cyclin B1/Cdk1 binds and phosphorylates Filamin A and regulates its ability to cross-link actin. FEBS Lett, 2007. 581(8): p. 1661–72. [DOI] [PubMed] [Google Scholar]
- 39.Kim HH, et al. , Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev, 2008. 22(13): p. 1804–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hunter T and Karin M, The regulation of transcription by phosphorylation. Cell, 1992. 70(3): p. 375–87. [DOI] [PubMed] [Google Scholar]
- 41.Jeong CH, et al. , Phosphorylation of Sox2 cooperates in reprogramming to pluripotent stem cells. Stem Cells, 2010. 28(12): p. 2141–50. [DOI] [PubMed] [Google Scholar]
- 42.Ouyang J, et al. , Cyclin-dependent kinase-mediated Sox2 phosphorylation enhances the ability of Sox2 to establish the pluripotent state. J Biol Chem, 2015. 290(37): p. 22782–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang WW, et al. , Cdk1 is required for the self-renewal of mouse embryonic stem cells. J Cell Biochem, 2011. 112(3): p. 942–8. [DOI] [PubMed] [Google Scholar]
- 44.Neganova I, et al. , CDK1 plays an important role in the maintenance of pluripotency and genomic stability in human pluripotent stem cells. Cell Death Dis, 2014. 5: p. e1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwan KY, Shen J, and Corey DP, C-MYC transcriptionally amplifies SOX2 target genes to regulate self-renewal in multipotent otic progenitor cells. Stem Cell Reports, 2015. 4(1): p. 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boumahdi S, et al. , SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature, 2014. 511(7508): p. 246–50. [DOI] [PubMed] [Google Scholar]
- 47.Leis O, et al. , Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene, 2012. 31(11): p. 1354–65. [DOI] [PubMed] [Google Scholar]
- 48.Zhu F, et al. , SOX2 Is a Marker for Stem-like Tumor Cells in Bladder Cancer. Stem Cell Reports, 2017. 9(2): p. 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cesarini V, et al. , Sox2 is not required for melanomagenesis, melanoma growth and melanoma metastasis in vivo. Oncogene, 2017. 36(31): p. 4508–4515. [DOI] [PubMed] [Google Scholar]
- 50.Schaefer SM, et al. , Sox2 is dispensable for primary melanoma and metastasis formation. Oncogene, 2017. 36(31): p. 4516–4524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.