Abstract
The relationships between HIV-1 DNA copy number, proviral transcriptional activity, and residual plasma viremia in individuals off and on ART are not well defined. To address this, we performed a cross-sectional study of 12 viremic donors and 23 ART-treated virologically suppressed (plasma HIV-1 RNA<20 copies/mL) donors. We report a strong association between HIV-1 DNA copy number and HIV-1 transcriptional activity in blood that persists on suppressive ART, but not between transcriptional activity and the levels of persistent viremia on ART. The latter finding contrasts with that in viremic donors and suggests that most HIV transcription in donors on suppressive ART does not result in virion production. This uncoupling of proviral transcription and viremia warrants closer investigation.
Keywords: HIV-1 DNA, HIV-1 RNA, HIV-1 viremia, HIV-1 reservoir, HIV-1 persistence, HIV-1 cure
Introduction
Although HIV-1 replication is suppressed during combination antiretroviral therapy (ART), replication-competent HIV-1 proviruses persist in latently-infected CD4+ T-cells (CD4), and their remarkably long half-life of 44 months prevents a “sterilizing” cure from ART alone (Finzi et al., 1997; Siliciano et al., 2003). In addition, various forms of cell-associated (CA) HIV-1 RNA continue to be synthesized during ART (Chun et al., 2003; Fischer et al., 2000; Lassen et al., 2004), and ultrasensitive PCR assays can detect HIV-1 virions in the plasma (Maldarelli et al., 2007; Palmer et al., 2008), indicating that HIV-1 proviral transcription and small amounts of virion production persist on ART.
With the growing interest in controlling or eliminating HIV-1 reservoirs, plasma HIV-1 RNA, along with total HIV-1 DNA and unspliced RNA, have been used as virologic markers of HIV-1 persistence to assess the therapeutic effect of new pharmacological and immunological approaches to achieve a functional cure of HIV-1 (Archin et al., 2014; Elliotet al., 2014; Rasmussen et al., 2014). Molecular markers offer reliable and inexpensive ways to monitor changes in the number and transcriptional activity of HIV-1-infected cells. This approach is potentially valuable because the gold standard of measuring the HIV-1 latent reservoir – the quantitative viral outgrowth assay (QVOA) – is expensive and time consuming, and other approaches to improving or replacing QVOA still require ex vivo cell culture (Laird et al., 2013; Procopio et al., 2015). Molecular markers of HIV-1 have also been evaluated extensively, and a number of observational studies have characterized the dynamics of their decay as a result of ART (Besson et al., 2014; Furtado et al., 1999; Koelsch et al., 2008; Palmer et al., 2008). Although some researchers have questioned the value of molecular markers because they do not correlate with the latent reservoir measured by QVOA (Eriksson et al., 2013), others have suggested that HIV-1 DNA and RNA could provide important information on the time to virological relapse after the interruption of ART (Li et al., 2016; Pasternak et al., 2009; Sneller et al., 2017; Williams et al., 2014).
HIV-1 DNA, unspliced RNA and plasma viral RNA represent three distinct stages in the lifecycle of HIV-1 replication, i.e., infection of CD4+ T-cells, transcription of proviruses and production of virions, respectively. It is therefore not surprising that these molecular markers are correlated during untreated viremia (Furtado et al., 1999). However, it is still unclear how suppressive ART alters these correlations. To explore their relationships on ART, we conducted a cross-sectional study of molecular markers in virologically suppressed donors on stable ART and compared them to viremic donors off ART. Although previous studies have explored associations between HIV-1 DNA and CA HIV-1 RNA (Malatinkova et al., 2015; Procopio et al., 2015), HIV-1 DNA and plasma viremia (Chun et al., 2011; Mexas et al., 2012), or CA HIV-1 RNA and plasma viremia (Procopio et al., 2015), the current study examines each of these correlations within the same set of samples; and importantly, compares viremic patients to those suppressed on ART.
Materials and Methods
Clinical specimens
The study was approved by the University of Pittsburgh Institutional Review Board. Study participants were recruited from the University of Pittsburgh AIDS Center for Treatment and provided written informed consent. Two groups of participants were enrolled: (1) viremic donors with plasma HIV-1 RNA >1,000 copies/ml (Roche COBAS AmpliPrep/COBAS TaqMan, v2.0) who were not currently receiving ART (some participants had a history of ART exposure); and (2) virologically suppressed donors who had been on stable suppressive ART for at least 6 months with plasma HIV-1 RNA <20 copies/ml (Roche). In a subset of donors, samples were collected at two timepoints (5 of 12 viremic donors and 19 of 23 virologically suppressed donors).
Sample specimens were collected through either large-volume phlebotomy (100 –180 ml) or leukapheresis, and then processed to peripheral blood mononuclear cells (PBMC) by Ficoll-Paque density gradient centrifugation (Sigma-Aldrich, USA) within 4 hours of collection. The cells were cryopreserved in 5 –10 million aliquots and stored in liquid nitrogen before analysis. Plasma was harvested from whole blood by double centrifugation (400 g × 10 min, followed by 1,350 g × 15 min) and stored at −80°C before analysis.
Isolation and quantification of HIV-1 DNA and unspliced HIV-1 RNA
Total HIV-1 DNA and unspliced HIV-1 RNA levels in PBMC were quantified as reported (Hong et al., 2016b). Briefly, total nucleic acid (TNA) was isolated from 2.5 million PBMCs using Proteinase K/Guanidinium HCl/Guanidinium Thiocyanate lysis and isopropanol precipitation. TNA was split and one half used for HIV-1 DNA quantitation in triplicate by qPCR and one half treated with Dnase and then used for CA-HIV-1 RNA quantitation in triplicate by RT-qPCR. The target for both assays is the 3’ end of pol. The limit of detection for both HIV-1 DNA and RNA was one copy per reaction, as determined by limiting dilution analysis of DNA and RNA standards. Nucleic acid input of 700 (DNA) or 300 (RNA) ng was used for each qPCR (DNA) or RT followed by qPCR (RNA) reaction. The number of cell equivalents was estimated by qPCR targeting CCR5 according to a published protocol (Malnati et al., 2008), and was used to normalize HIV-1 DNA and unspliced RNA per million cells. The results were further normalized, as appropriate, by the percentage of CD4+ T-cells, i.e., HIV-1 DNA copies/million CD4+ T-cells was calculated by dividing HIV-1 DNA copies/million cells by CD4+ T-cell percent. For those donors from which samples were obtained at two timepoints (as above), the mean values from duplicate assays are reported.
Quantification of plasma HIV-1 RNA
Plasma HIV-1 RNA was quantified with single copy sensitivity (single copy assay; SCA) using qRT-PCR targeting a highly conserved region in pol (Cillo et al., 2014). The limit of detection for HIV-1 RNA was one copy per reaction as determined by limiting dilution analysis of HIV-1 transcripts. For those donors from which samples were obtained at two timepoints (as above), the mean values from duplicate assays are reported.
Flow cytometric analysis
CD4+ T-cell percentage in PBMC was determined by flow cytometry either by clinical assay or in-house. Briefly, cells were fixed using BD Cytofix buffer and analyzed for surface expression of T-cell markers using CD3-V450, CD4-APC-H7 and CD8-PE using an LSRII cytometer with FACSDiva software (BD Biosciences). All antibodies were obtained from BD Biosciences.
Statistical analysis
Plasma HIV-1 RNA (SCA) levels below the detection threshold were analyzed as half of the corresponding limit of detection. HIV-1 DNA and CA HIV-1 RNA levels per million CD4+ T-cells were calculated as HIV-1 DNA and CA HIV-1 RNA copies/106 PBMCs, respectively, and where appropriate were divided by the corresponding fraction of CD4+ T-cells in PBMC (which was determined by flow cytometry, as described above, or by clinical assay). Correlations between CA HIV-1 RNA, HIV-1 DNA, CA HIV-1 RNA/DNA ratio and SCA level were assessed using Spearman rank-based correlation coefficients.
Results
Thirty-five donors were studied, including 12 viremic and 23 virologically suppressed individuals on ART. Table 1 displays baseline characteristics including age, sex, CD4+ T-cell count and plasma HIV-1 RNA from before ART. At the time of the initial sample collection, the median CD4+ T-cell count was 240 cells/mm3 (range: 17; 1,053) for the viremic group and 642 cells/mm3 (range: 386; 1,667) for the virologically suppressed group after a median of 5.7 years of suppressive ART. Median CD4+T-cell nadir before ART was 227 cells/mm3 (range: 23, 578) in the virologically suppressed group.
Table 1.
Baseline characteristics
| GROUP | ||||
|---|---|---|---|---|
|
| ||||
| Total | Viremic | Suppressed | ||
| N = 35 | N = 12 | N = 23 | ||
| Age | Min, Max | 24, 66 | 24, 58 | 26, 66 |
| Median (Q1, Q3) | 50 (44, 54) | 49 (35, 53) | 51 (45, 57) | |
| Sex | M | 28 (80%) | 11 (92%) | 17 (74%) |
| F | 7 (20%) | 1 (8%) | 6 (26%) | |
| Race | African American | 22 (63%) | 10 (83%) | 12 (52%) |
| Caucasian | 12 (34%) | 1 (8%) | 11 (48%) | |
| Hispanic | 1 (3%) | 1 (8%) | 0 (0%) | |
| Nadir CD4 cell count (cells/mm3)* | N | 21 | ||
| Min, Max | 23, 578 | |||
| Median (Q1, Q3) | 227 (89, 314) | |||
| ART Duration at time of screening (years) | <4 | 9 (39%) | ||
| >4 | 11 (48%) | |||
| missing | 3 (13%) | |||
| Pre-ART HIV-1 RNA level (log10 (copies/mL)* | N | 20 | ||
| Min, Max | 3.7, 6.7 | |||
| Median (Q1, Q3) | 5.0 (4.3, 5.1) | |||
| Pre-ART HIV-1 RNA level (copies/mL)* | ≤ 10K | 2 (9%) | ||
| 10K – 100K | 11 (48%) | |||
| ≥ 100K | 7 (30%) | |||
| missing | 3 (13%) | |||
| Screening CD4 cell count (cells/mm3) | N | 35 | 12 | 23 |
| Min, Max | 17, 1,667 | 17, 1,053 | 386, 1,667 | |
| Median (Q1, Q3) | 628 (392, 835) | 240 (102, 810) | 642 (486, 835) | |
| Screening CD4 cell count (%) | N | 33 | 11 | 22 |
| Min, Max | 3, 55 | 3, 45 | 24, 55 | |
| Median (Q1, Q3) | 32 (25, 40) | 17 (8, 31) | 35 (30, 43) | |
Data not available for the Viremic group
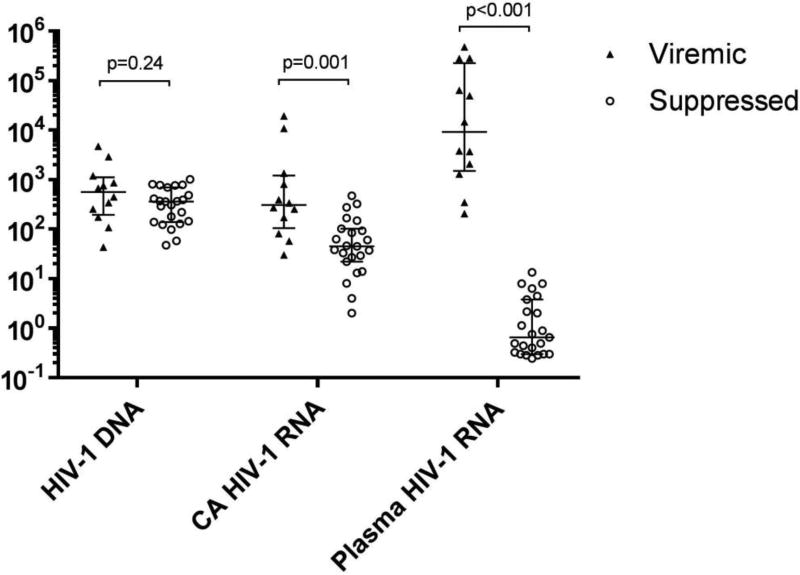
Figure 1 and Supplemental Table 1 summarize the HIV-1 DNA and CA HIV-1 RNA in PBMC and plasma HIV-1 RNA in the viremic and virologically suppressed groups. The median plasma HIV-1 RNA level in the viremic group was 9,189 copies/ml (range: 206; 474,211), ~10,000-fold higher than in the virologically suppressed group (median 0.7 copies/ml; range: 0.2; 13.4; 5 of 23 below limit of detection). The median HIV-1 DNA for the viremic and virologically suppressed groups was 557 (range: 43; 4,680) and 357 (range: 47; 1,005) copies/106 PBMCs, respectively, and the median CA HIV-1 RNA for the two groups was 305 (range: 30; 19,172) and 45 (range: 2; 471) copies/106 PBMCs, respectively. The sixfold lower CA HIV-1 RNA levels in donors on ART is small compared with the > 4 log10 difference in plasma viremia between the two groups, revealing a major dissociation between HIV-1 transcription and viremia on ART.
Fig. 1. Summary of results for HIV-1 DNA and RNA in PBMCs and HIV-1 RNA in plasma.
Long horizontal lines indicate median values; short horizontal lines denote first- and third-quartile values. Each symbol within a group represents results from a single donor. CA HIV-1 RNA and plasma HIV-1 RNA levels below the detection threshold are represented as one-half of the corresponding limit of detection. HIV-1 DNA (copies/106 PBMCs); CA (cell-associated) HIV-1 RNA (copies/106 PBMCs); Plasma HIV-1 RNA as determined by single-copy assay (copies/ml). Y axis scale is log10.
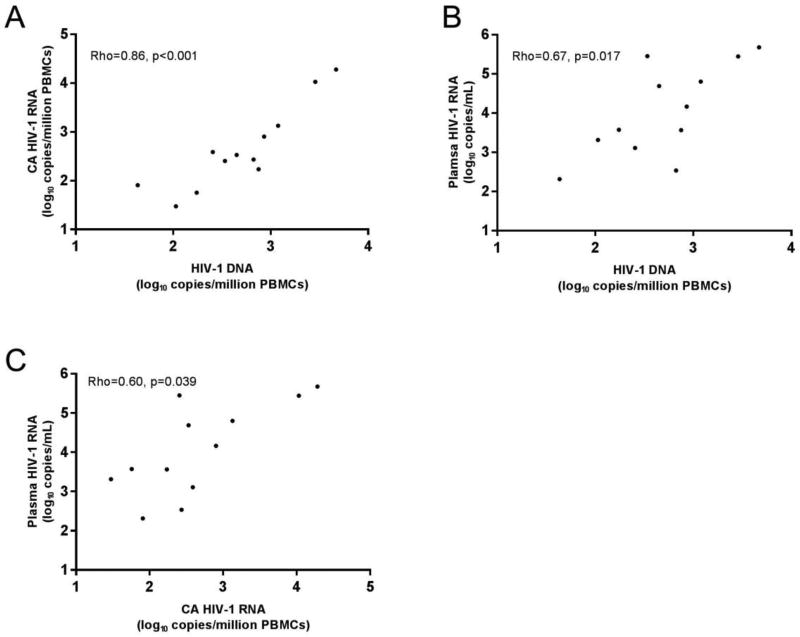
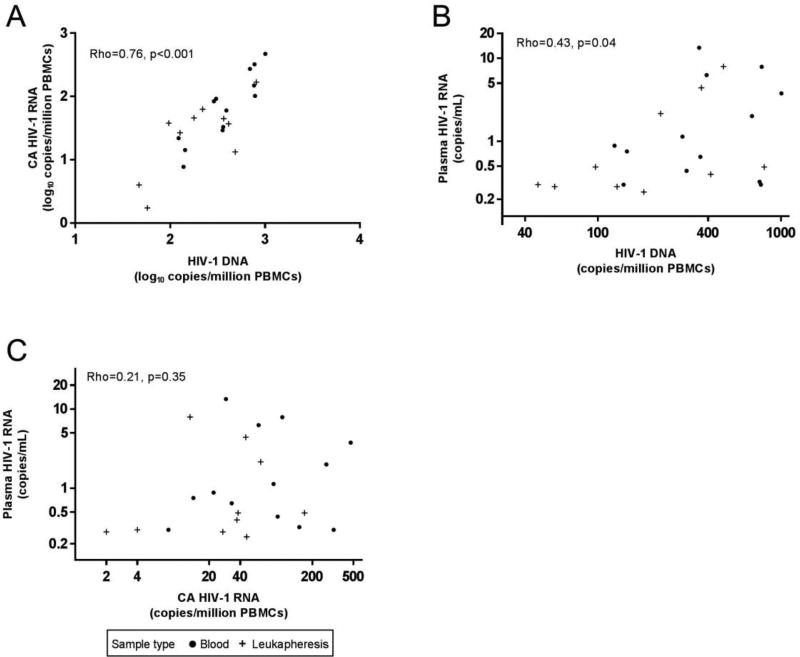
A strong, positive correlation was found between HIV-1 DNA and CA HIV-1 RNA in both the viremic (rho = 0.86, p < 0.001, Figure 2A) and virologically suppressed groups (rho = 0.76, p < 0.001, Figure 3A), indicating that the levels of proviral transcription in HIV-1-infected individuals is proportional to the number of cells containing HIV-1 DNA. Normalizing HIV-1 DNA copies per million CD4+ T-cells did not alter the correlations, and may have strengthened them (viremic: rho = 0.87, p < 0.001, Figure S1A; suppressed: rho = 0.85, p < 0.001, Figure S2A).
Fig. 2. Correlations between levels of HIV-1 DNA or RNA in PBMC and plasma HIV-1 RNA among viremic donors.
(A) Correlation between HIV-1 DNA and CA HIV-1 RNA. (B) Correlation between HIV-1 DNA and plasma HIV-1 RNA. (C) Correlation between CA HIV-1 RNA and plasma HIV-1 RNA. Each symbol within the panels represents data from a unique donor (n=12). Axis scales are log10.
Fig. 3. Correlations between levels of HIV-1 DNA or RNA in PBMC and plasma HIV-1 RNA among virologically suppressed donors.
(A) Correlation between HIV-1 DNA and CA HIV-1 RNA. Axis scales are log10. (B) Correlation between HIV-1 DNA and plasma HIV-1 RNA. Axis scales are log2. (C) Correlation between CA HIV-1 RNA and plasma HIV-1 RNA. Axis scales are log2. Each symbol within a panel represents data from a unique donor (n=23).
Recent studies have reported that HIV-1 DNA and CA HIV-1 RNA reach a steady state after 4 years of suppressive ART (Besson et al., 2014; Gandhi RT et al., 2016). We therefore repeated the analysis on individuals who had been receiving ART for more than 4 years. Despite the small sample size, HIV-1 DNA and CA HIV-1 RNA remained strongly correlated in PBMC (rho = 0.78, p = 0.003, n = 11) and when corrected for CD4+ T-cells (rho = 0.89, p < 0.001, n = 10), which indicates that even among individuals on long-term effective ART, there is a strong proportional relationship between the number of HIV-infected cells and the levels of unspliced HIV-1 RNA.
Among the viremic group, there was a strong correlation between the levels of plasma HIV-1 RNA and HIV-1 DNA in PBMC (rho = 0.67, p = 0.017, Figure 2B) and when corrected for CD4+ T-cells (rho = 0.71, p = 0.009, Figure S1B), indicating that the production of HIV-1 virions is directly proportional to the number of HIV-infected cells. Surprisingly, this correlation persisted, albeit less strongly, after virologically suppressive ART (PBMC: rho = 0.43, p = 0.04, Figure 3B; corrected for CD4+ T-cells: rho = 0.52, p = 0.012, Figure S2B). Positive correlations were also found in the viremic group between the levels of plasma HIV-1 RNA and CA HIV-1 RNA, both in PBMC (rho = 0.60, p = 0.039, Figure 2C) and when corrected for CD4+ T-cells (rho = 0.85, p < 0.001, Figure S1C). This finding is consistent with previous reports (Furtado et al., 1999), and indicates that the level of transcription of HIV-1 proviruses is associated with the production of virions during uncontrolled viremia. By contrast, after ART suppression, the correlation between plasma and CA HIV-1 RNA is no longer evident (PBMC: rho = 0.21, p = 0.35, Figure 3C; corrected for CD4+ T-cells: rho = 0.30, p = 0.18, Figure S2C), suggesting that most of the HIV-1 transcription in the virologically suppressed group does not lead to virus production or viremia. When data were stratified by time on ART (<4 yrs, n=11 or >4 yrs, n=9) a non-significant trend toward a direct correlation was noted in those on ART <4 years (PBMC: rho = 0.57, p = 0.12, Figure S3A), but not in those on ART >4 yrs (PBMC: rho = 0.10, p = 0.78, Figure S3B). The reason for the dissociation between CA HIV-1 RNA and persistent viremia with longer-term ART is unknown, but may be related to the progressive clearance of virus producing cells and not those cells that are transcriptionally active without producing viral proteins or virions. With regard to clearance of virus producing cells on ART, longitudinal studies have shown decay of persistent viremia with long-term suppression on ART (Maldarelli et al., 2007; Riddler et al., 2016). No association was found between the CA HIV-1 RNA/DNA ratio and plasma HIV-1 RNA in either the viremic (rho = 0.33, p = 0.30) or virologically suppressed groups (rho = −0.06, p = 0.8).
Conclusions
This study explored quantitative relationships between the number of HIV-infected cells, HIV-1 transcription, and viremia in both viremic and virologically suppressed donors on ART using identical primer and probe sets to quantify total HIV-1 DNA, unspliced CA HIV-1 RNA and viral genomes in plasma, respectively. We found a strong positive correlation between total HIV-1 DNA and unspliced HIV-1 RNA in both groups, indicating proportionality between the number of infected cells and transcriptional activity, independent of ART. Our finding that there is a positive association between HIV-1 DNA and HIV-1 RNA regardless of ART status is somewhat unexpected, given that others have found that most proviruses persisting on ART are defective and thus incapable of infectious virus production (Eriksson et al., 2013; Fourati et al., 2012; Ho et al., 2013). The fraction of proviruses that are intact (i.e., can produce replication-competent virus) is small (<5%) and varies greatly from donor to donor, which has led to the assumption that total HIV-1 DNA is not a useful measure (Bruner et al., 2016; Ho et al., 2013). The inability to produce infectious virions, however, may not preclude proviral transcription; along these lines, recent reports indicate that defective or hyper-mutated proviruses can transcribe HIV-1 RNA (Thomas et al., 2016).
Compared with viremic donors, levels of HIV-1 DNA were less than twofold lower in virologically suppressed donors, and levels of unspliced CA HIV-1 RNA were approximately sixfold lower, which was surprising given the ~10,000-fold lower level of viremia. This finding suggested that the levels of HIV-1 transcription in donors with stable virologic suppression on ART are not related to viremia, and that most HIV-1 transcription is non-productive for virions. Indeed, we found no correlation between CA HIV-1 RNA levels and persistent viremia on ART. By contrast, CA HIV-1 RNA was directly and strongly correlated with the level of viremia in donors not receiving ART.
The lack of correlation between plasma HIV-1 RNA and unspliced CA HIV-1 RNA after virological suppression may be related, in part, to the progressive loss of productively-infected, virus producing cells after the initiation of ART, which is manifested by large declines (~85%) of HIV-1 DNA and the rapid decline of HIV-1 viremia (99.999%) (Besson et al., 2014). The infected cells that persist on ART are thus either transcriptionally silent (i.e., latent) or remain transcriptionally active but produce few virions or no virions. It appears then that following virologic suppression on ART, CA HIV-1 RNA largely becomes a measure of non-productive proviral transcription. Quantitative PCR-based assays targeting a specific CA HIV-1 RNA sequence can amplify a number of viral RNA forms containing the target sequences. The bulk of these viral RNA forms are likely transcriptional products of defective HIV-1 proviruses, given that >95% of proviruses in participants on ART are defective as a consequence of large deletions, point insertions or deletions, and G-to-A hypermutation. As noted above, recent findings indicate that defective proviruses can be expressed, and that transcripts containing some intact open reading frames have the potential to be translated, although virion production is improbable (Imamichi et al., 2016). Another source of defective CA HIV-1 RNA may be chimeric readthrough transcripts that are initiated at host promoters (Bullen et al., 2014), although their contribution is likely minor among virologically suppressed participants (Pasternak et al., 2015). A number of studies have also suggested that HIV-1 latency could be maintained by post-transcriptional blocks even when HIV-1 mRNAs are produced from integrated proviruses, such as by nuclear retention of multiply spliced HIV-1 mRNAs (Lassen et al., 2006) or by cellular mRNAs inhibiting HIV-1 expression (Huang et al., 2007). Indeed, a recent study by Yukl, et al. reported that most transcription products from proviruses in suppressed patients contain a variety of short and elongated but unprocessed RNA species, suggesting that incomplete transcripts result from both blocks in transcript elongation and post-elongation processing (polyadenylation and multiple splicing). Along these lines, because our qPCR assay targets the 3’end of pol transcripts it should not detect most short transcripts resulting from elongation blocks. By contrast, our assay should detect the substantial fraction of RNA transcripts that face post-elongation blocks, which could contribute to the lack of correlation between transcriptional activity (CA-HIV RNA) and virion production (plasma HIV-1 RNA by SCA) (Yukl et al., 2018).
It is important to point out that our results seem to contrast the finding that CA-RNA correlates with plasma viremia in suppressed patients by Kiselinova et al. (Kiselinova et al., 2016). However, in that study, residual viremia in suppressed patients was correlated with CA-RNA only at the baseline measurement, whereas the correlation diminished in subsequent measurements. This finding is very similar to the results of our analyses stratified by time on ART.
It has recently been shown that HIV-infected cells can undergo clonal expansion; a substantial fraction of HIV transcripts can be found in these clonally expanded cells, and transcriptionally active clones can persist over time (Wiegand et al., 2017). In some instances, the sequences of transcripts in clonally expanded cells match the rebound virus after the cessation of ART, indicating that transcriptionally active infected cell clones may have intact proviruses (Kearney et al., 2015). However, such cells are expected to be infrequent, because most proviruses are defective. Indeed, most of the transcripts in clonally expanded cells do not match an infectious virus that can be recovered from the same cell population (Hong F et al., 2016a; Kearney et al., 2015). The relative proportion of defective vs. intact proviruses that is transcriptionally active is not known; this is the topic of ongoing investigations. New methods are required to address this question: the CA HIV-1 RNA assay used in the current study and in most other studies of CA HIV-1 RNA (Hong F et al., 2016a; Kearney et al., 2015; Li et al., 2016; Thomas et al., 2016) measures HIV-1 transcription in infected cell populations, but cannot determine the fraction of HIV-1-infected cells that is actively transcribing HIV-1 mRNA. Assays that can assess transcription in single infected cells are thus required and have recently been reported (Wiegand et al., 2017).
The apparent disassociation between plasma HIV-1 RNA and unspliced CA HIV-1 RNA in suppressed patients calls into question the utility of using CA HIV-1 RNA as an endpoint in clinical trials that aim to evaluate the efficacy of latency-reversing agents on HIV-1 persistence. While helpful in monitoring the disturbance of proviral transcription, our data indicate that even a large increase in CA HIV-1 RNA may have uncertain significance without a concomitant and measurable increase in plasma HIV-1 RNA. Indeed, several studies of histone deacetylase inhibitors have reported increased CA HIV-1 RNA levels without statistically significant changes in either HIV-1 DNA or plasma HIV-1 RNA (Archin et al., 2014; Elliott et al., 2014), which highlights the importance of using a combination of assays to measure latency reversal.
A recent study showed that HIV-1 DNA was predictive of disease progression as well as plasma viral rebound after ART cessation in participants with primary HIV-1 infection (Williams et al., 2014), suggesting that HIV-1 DNA is a clinically relevant biomarker. Interestingly, other studies have also suggested that unspliced CA HIV-1 RNA is an important biomarker of therapy outcomes among patients with undetectable plasma HIV-1 RNA (Pasternak et al., 2009) and of virological rebound after treatment cessation (Li et al., 2016). These associations between HIV-1 DNA and RNA and time to viral rebound after ART interruption may appear to contradict our data and others indicating that most HIV-1 DNA is defective and that CA HIV-1 RNA is not associated with viremia on ART. However, a small fraction of proviruses is intact and may be transcriptionally active, leading to rebound viremia. The likelihood that such intact, transcriptionally active proviruses are present may be proportional to the total number of infected cells and the total number of HIV-1 transcripts, both of which are strongly correlated. Such proportionality between total and intact proviruses and total and intact transcripts could explain the reported associations between HIV-1 DNA and RNA and time to viral rebound after stopping ART. Importantly, compartments other than blood may contribute to residual and/or rebound plasma viremia, and inclusion of data from these compartments could provide further insight.
In summary, we describe a strong and positive correlation between HIV-1-infected cells and the level of HIV-1 proviral transcription. We show that this relationship cannot be extended to residual viremia in individuals on long-term suppressive ART, especially the link between plasma viral RNA and unspliced CA HIV-1 RNA. The later finding is consistent with that reported in a different study in virologically suppressed donors (Gandhi et al., 2017). Additional studies are needed to better define the role of HIV-1 DNA and RNA as biomarkers of response to experimental interventions. Our findings highlight the importance of developing new assays that can differentiate HIV-1 DNA and CA HIV-1 RNA derived from intact vs. defective proviruses for the study of HIV-1 persistence and the evaluation of agents aimed at reducing HIV-1 reservoirs.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the volunteers for participating in this study. We thank Kelley Friel and Lorraine Pollini for editorial assistance.
Funding Information:
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number UM1AI106701 and to the Statistical and Data Management Center of the AIDS Clinical Trials Group (UM1 Ai068634). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure:
J.W.M. is a consultant to Gilead Sciences and holds share options in Cocrystal Pharmaceuticals, Inc. No other authors report disclosures.
References
- Archin NM, Bateson R, Tripathy MK, Crooks AM, Yang KH, Dahl NP, Kearney MF, Anderson EM, Coffin JM, Strain MC, Richman DD, Robertson KR, Kashuba AD, Bosch RJ, Hazuda DJ, Kuruc JD, Eron JJ, Margolis DM. HIV-1 expression within resting CD4+ T cells after multiple doses of vorinostat. J Infect Dis. 2014;210:728–735. doi: 10.1093/infdis/jiu155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson GJ, Lalama CM, Bosch RJ, Gandhi RT, Bedison MA, Aga E, Riddler SA, McMahon DK, Hong F, Mellors JW. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis. 2014;59:1312–1321. doi: 10.1093/cid/ciu585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho YC, Richman DD, Deeks SG, Siliciano JD, Siliciano RF. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nature medicine. 2016 doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nature medicine. 2014;20:425–429. doi: 10.1038/nm.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Justement JS, Lempicki RA, Yang J, Dennis G, Jr, Hallahan CW, Sanford C, Pandya P, Liu S, McLaughlin M, Ehler LA, Moir S, Fauci AS. Gene expression and viral prodution in latently infected, resting CD4+ T cells in viremic versus aviremic HIV-infected individuals. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1908–1913. doi: 10.1073/pnas.0437640100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun TW, Murray D, Justement JS, Hallahan CW, Moir S, Kovacs C, Fauci AS. Relationship between residual plasma viremia and the size of HIV proviral DNA reservoirs in infected individuals receiving effective antiretroviral therapy. J Infect Dis. 2011;204:135–138. doi: 10.1093/infdis/jir208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cillo AR, Vagratian D, Bedison MA, Anderson EM, Kearney MF, Fyne E, Koontz D, Coffin JM, Piatak M, Jr, Mellors JW. Improved single-copy assays for quantification of persistent HIV-1 viremia in patients on suppressive antiretroviral therapy. Journal of clinical microbiology. 2014;52:3944–3951. doi: 10.1128/JCM.02060-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott JH, Wightman F, Solomon A, Ghneim K, Ahlers J, Cameron MJ, Smith MZ, Spelman T, McMahon J, Velayudham P, Brown G, Roney J, Watson J, Prince MH, Hoy JF, Chomont N, Fromentin R, Procopio FA, Zeidan J, Palmer S, Odevall L, Johnstone RW, Martin BP, Sinclair E, Deeks SG, Hazuda DJ, Cameron PU, Sekaly RP, Lewin SR. Activation of HIV transcription with short-course vorinostat in HIV-infected patients on suppressive antiretroviral therapy. PLoS Pathog. 2014;10:e1004473. doi: 10.1371/journal.ppat.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Graf EH, Dahl V, Strain MC, Yukl SA, Lysenko ES, Bosch RJ, Lai J, Chioma S, Emad F, Abdel-Mohsen M, Hoh R, Hecht F, Hunt P, Somsouk M, Wong J, Johnston R, Siliciano RF, Richman DD, O'Doherty U, Palmer S, Deeks SG, Siliciano JD. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog. 2013;9:e1003174. doi: 10.1371/journal.ppat.1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science. 1997;278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- Fischer M, Gunthard HF, Opravil M, Joos B, Huber W, Bisset LR, Ott P, Boni J, Weber R, Cone RW. Residual HIV-RNA levels persist for up to 2.5 years in peripheral blood mononuclear cells of patients on potent antiretroviral therapy. AIDS research and human retroviruses. 2000;16:1135–1140. doi: 10.1089/088922200414974. [DOI] [PubMed] [Google Scholar]
- Fourati S, Lambert-Niclot S, Soulie C, Malet I, Valantin MA, Descours B, Ait-Arkoub Z, Mory B, Carcelain G, Katlama C, Calvez V, Marcelin AG. HIV-1 genome is often defective in PBMCs and rectal tissues after long-term HAART as a result of APOBEC3 editing and correlates with the size of reservoirs. J Antimicrob Chemother. 2012;67:2323–2326. doi: 10.1093/jac/dks219. [DOI] [PubMed] [Google Scholar]
- Furtado MR, Callaway DS, Phair JP, Kunstman KJ, Stanton JL, Macken CA, Perelson AS, Wolinsky SM. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N Engl J Med. 1999;340:1614–1622. doi: 10.1056/NEJM199905273402102. [DOI] [PubMed] [Google Scholar]
- Gandhi RT, McMahon DK, Bosch RJ, Lalama CM, Cyktor JC, Macatangay BJ, Rinaldo CR, Riddler SA, Hogg E, Godfrey C, Collier AC, Eron JJ, Mellors JW, Team AA. Levels of HIV-1 persistence on antiretroviral therapy are not associated with markers of inflammation or activation. PLoS Pathog. 2017;13:e1006285. doi: 10.1371/journal.ppat.1006285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DI, Lai J, Blankson JN, Siliciano JD, Siliciano RF. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell. 2013;155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong F, Spindler J, Musick A, Cillo AR, Bale Michael, Shao W, Coffin JM, Mellors JW, Kearney MF. ART reduces cellular HIV RNA but not the fraction of proviruses transcribing RNA; Conference on Retroviruses and Opportunistic Infections (CROI); Boston, MA. 2016a. [Google Scholar]
- Hong F, Aga E, Cillo A, Yates AL, Besson G, Fyne E, Koontz DL, Jennings C, Zheng L, Mellors JW. Novel assays to measure total cell-associated HIV-1 DNA and RNA. Journal of clinical microbiology. 2016b doi: 10.1128/JCM.02904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4+ T lymphocytes. Nature medicine. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- Imamichi H, Dewar RL, Adelsberger JW, Rehm CA, O'Doherty U, Paxinos EE, Fauci AS, Lane HC. Defective HIV-1 proviruses produce novel protein-coding RNA species in HIV-infected patients on combination antiretroviral therapy. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:8783–8788. doi: 10.1073/pnas.1609057113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearney MF, Wiegand A, Shao W, Coffin JM, Mellors JW, Lederman M, Gandhi RT, Keele BF, Li JZ. Origin of Rebound Plasma HIV Includes Cells with Identical Proviruses that are Transcriptionally Active Before Stopping Antiretroviral Therapy. Journal of virology. 2015 doi: 10.1128/JVI.02139-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiselinova M, De Spiegelaere W, Buzon MJ, Malatinkova E, Lichterfeld M, Vandekerckhove L. Integrated and Total HIV-1 DNA Predict Ex Vivo Viral Outgrowth. PLoS Pathog. 2016;12:e1005472. doi: 10.1371/journal.ppat.1005472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelsch KK, Liu L, Haubrich R, May S, Havlir D, Gunthard HF, Ignacio CC, Campos-Soto P, Little SJ, Shafer R, Robbins GK, D'Aquila RT, Kawano Y, Young K, Dao P, Spina CA, Richman DD, Wong JK. Dynamics of total, linear nonintegrated, and integrated HIV-1 DNA in vivo and in vitro. J Infect Dis. 2008;197:411–419. doi: 10.1086/525283. [DOI] [PubMed] [Google Scholar]
- Laird GM, Eisele EE, Rabi SA, Lai J, Chioma S, Blankson JN, Siliciano JD, Siliciano RF. Rapid quantification of the latent reservoir for HIV-1 using a viral outgrowth assay. PLoS Pathog. 2013;9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen KG, Bailey JR, Siliciano RF. Analysis of human immunodeficiency virus type 1 transcriptional elongation in resting CD4+ T cells in vivo. Journal of virology. 2004;78:9105–9114. doi: 10.1128/JVI.78.17.9105-9114.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassen KG, Ramyar KX, Bailey JR, Zhou Y, Siliciano RF. Nuclear retention of multiply spliced HIV-1 RNA in resting CD4+ T cells. PLoS Pathog. 2006;2:e68. doi: 10.1371/journal.ppat.0020068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Etemad B, Ahmed H, Aga E, Bosch RJ, Mellors JW, Kuritzkes DR, Lederman MM, Para M, Gandhi RT. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. Aids. 2016;30:343–353. doi: 10.1097/QAD.0000000000000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatinkova E, De Spiegelaere W, Bonczkowski P, Kiselinova M, Vervisch K, Trypsteen W, Johnson M, Verhofstede C, de Looze D, Murray C, Kinloch-de Loes S, Vandekerckhove L. Impact of a decade of successful antiretroviral therapy initiated at HIV-1 seroconversion on blood and rectal reservoirs. Elife. 2015;4:e09115. doi: 10.7554/eLife.09115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldarelli F, Palmer S, King MS, Wiegand A, Polis MA, Mican J, Kovacs JA, Davey RT, Rock-Kress D, Dewar R, Liu S, Metcalf JA, Rehm C, Brun SC, Hanna GJ, Kempf DJ, Coffin JM, Mellors JW. ART suppresses plasma HIV-1 RNA to a stable set point predicted by pretherapy viremia. PLoS Pathog. 2007;3:e46. doi: 10.1371/journal.ppat.0030046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnati MS, Scarlatti G, Gatto F, Salvatori F, Cassina G, Rutigliano T, Volpi R, Lusso P. A universal real-time PCR assay for the quantification of group-M HIV-1 proviral load. Nat Protoc. 2008;3:1240–1248. doi: 10.1038/nprot.2008.108. [DOI] [PubMed] [Google Scholar]
- Mexas AM, Graf EH, Pace MJ, Yu JJ, Papasavvas E, Azzoni L, Busch MP, Di Mascio M, Foulkes AS, Migueles SA, Montaner LJ, O'Doherty U. Concurrent measures of total and integrated HIV DNA monitor reservoirs and ongoing replication in eradication trials. Aids. 2012;26:2295–2306. doi: 10.1097/QAD.0b013e32835a5c2f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Maldarelli F, Wiegand A, Bernstein B, Hanna GJ, Brun SC, Kempf DJ, Mellors JW, Coffin JM, King MS. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3879–3884. doi: 10.1073/pnas.0800050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak AO, DeMaster LK, Kootstra NA, Reiss P, O'Doherty U, Berkhout B. Minor Contribution of Chimeric Host-HIV Readthrough Transcripts to the Level of HIV Cell-Associated gag RNA. Journal of virology. 2015;90:1148–1151. doi: 10.1128/JVI.02597-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak AO, Jurriaans S, Bakker M, Prins JM, Berkhout B, Lukashov VV. Cellular levels of HIV unspliced RNA from patients on combination antiretroviral therapy with undetectable plasma viremia predict the therapy outcome. PLoS One. 2009;4:e8490. doi: 10.1371/journal.pone.0008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procopio FA, Fromentin R, Kulpa DA, Brehm JH, Bebin AG, Strain MC, Richman DD, O'Doherty U, Palmer S, Hecht FM, Hoh R, Barnard RJ, Miller MD, Hazuda DJ, Deeks SG, Sekaly RP, Chomont N. A Novel Assay to Measure the Magnitude of the Inducible Viral Reservoir in HIV-infected Individuals. EBioMedicine. 2015;2:874–883. doi: 10.1016/j.ebiom.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen TA, Tolstrup M, Brinkmann CR, Olesen R, Erikstrup C, Solomon A, Winckelmann A, Palmer S, Dinarello C, Buzon M, Lichterfeld M, Lewin SR, Ostergaard L, Sogaard OS. Panobinostat, a histone deacetylase inhibitor, for latent-virus reactivation in HIV-infected patients on suppressive antiretroviral therapy: a phase 1/2, single group, clinical trial. Lancet HIV. 2014;1:e13–21. doi: 10.1016/S2352-3018(14)70014-1. [DOI] [PubMed] [Google Scholar]
- Riddler SA, Aga E, Bosch RJ, Bastow B, Bedison M, Vagratian D, Vaida F, Eron JJ, Gandhi RT, Mellors JW, Team AAP. Continued Slow Decay of the Residual Plasma Viremia Level in HIV-1-Infected Adults Receiving Long-term Antiretroviral Therapy. J Infect Dis. 2016;213:556–560. doi: 10.1093/infdis/jiv433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siliciano JD, Kajdas J, Finzi D, Quinn TC, Chadwick K, Margolick JB, Kovacs C, Gange SJ, Siliciano RF. Long-term follow-up studies confirm the stability of the latent reservoir for HIV-1 in resting CD4+ T cells. Nature medicine. 2003;9:727–728. doi: 10.1038/nm880. [DOI] [PubMed] [Google Scholar]
- Sneller MC, Justement JS, Gittens KR, Petrone ME, Clarridge KE, Proschan MA, Kwan R, Shi V, Blazkova J, Refsland EW, Morris DE, Cohen KW, McElrath MJ, Xu R, Egan MA, Eldridge JH, Benko E, Kovacs C, Moir S, Chun TW, Fauci AS. A randomized controlled safety/efficacy trial of therapeutic vaccination in HIV-infected individuals who initiated antiretroviral therapy early in infection. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aan8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas A, Mellors J, Gandhi R, McMahon D, Eron J, Bosch R, Lalama C, Cytkor J, Walker B, Jones B. Only T-cell responses to Nef/Tat/Rev correlate with infected cell frequencies on ART; Conference on Retroviruses and Opportunistic Infections (CROI); Boston, MA. 2016. [Google Scholar]
- Wiegand A, Spindler J, Hong FF, Shao W, Cyktor JC, Cillo AR, Halvas EK, Coffin JM, Mellors JW, Kearney MF. Single-cell analysis of HIV-1 transcriptional activity reveals expression of proviruses in expanded clones during ART. Proceedings of the National Academy of Sciences of the United States of America. 2017;114:E3659–E3668. doi: 10.1073/pnas.1617961114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JP, Hurst J, Stohr W, Robinson N, Brown H, Fisher M, Kinloch S, Cooper D, Schechter M, Tambussi G, Fidler S, Carrington M, Babiker A, Weber J, Koelsch KK, Kelleher AD, Phillips RE, Frater J, Investigators SP. HIV-1 DNA predicts disease progression and post-treatment virological control. Elife. 2014;3:e03821. doi: 10.7554/eLife.03821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukl SA, Kaiser P, Kim P, Telwatte S, Joshi SK, Vu M, Lampiris H, Wong JK. HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci Transl Med. 2018;10 doi: 10.1126/scitranslmed.aap9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.