Abstract
Background & Aims:
Intestinal bacteria can modify the composition of bile acids and bile acids, which are regulated by the farnesoid X receptor (FXR), affect the survival and growth of gut bacteria. We studied the effects of obeticholic acid (OCA), a bile acid analog and FXR agonist, on the intestinal microbiomes of humans and mice.
Methods:
We performed a phase 1 study in 24 healthy volunteers given OCA (5 mg, 10 mg, or 25 mg per day, for 17 days). Fecal and plasma specimens were collected at baseline (day 0) and on days 17 (end of dosing) and 37 (end of study). The fecal specimens were analyzed by shotgun metagenomic sequencing. A Uniref90 high stringency genomic analysis was used to assign specific genes to the taxonomic signature of bacteria whose abundance was associated with OCA. Male C57BL/6 mice were gavaged daily with water, vehicle, or OCA (10 mg/kg) for 2 weeks. Small intestine luminal contents were collected by flushing with saline and fecal pellets were collected at baseline and day 14. Mouse samples were analyzed by 16S tagged sequencing. Culture experiments were performed to determine taxonomic-specific effects of bile acids and OCA on bacterial growth.
Results:
Suppression of endogenous bile acid synthesis by OCA in subjects led to a reversible induction of Gram-positive bacteria that are found in the small intestine and are components of diet and the oral microbiota. We found that bile acids reduced proliferation of these bacteria in minimum inhibitory concentration assays. In these organisms, there was an increase in the representation of microbial genomic pathways involved in DNA synthesis and amino acid metabolism with OCA treatment of subjects. Consistent with these findings, mice fed OCA had reduced endogenous bile acid levels and an increased proportion of Firmicutes, specifically in the small intestine, compared to mice fed water or vehicle.
Conclusions:
In studying the effects of OCA in humans and mice, we found evidence for interactions between bile acids and features of the small intestinal microbiome. These findings indicate that FXR activation alters the intestinal microbiota and could provide opportunities for microbiome biomarker discovery or new approaches to engineering the human microbiome. Clinical trial information: ClinicalTrials.gov no: NCT01933503.
Keywords: nuclear hormone receptor, genetics, gene regulation, metabolism
Introduction:
Although very little is known about the composition of the human small intestinal microbiota, its structure has been assumed to be very dynamic for several reasons. The relatively low bacterial biomass1, and the low level of diversity make the small intestinal community less resilient to environmental stressors. In addition, the intermittent delivery of food as well as the concomitant influx of both biliary and pancreatic secretions also likely have a robust effect on the composition of the proximal- and mid-small intestinal microbiota. Few studies have characterized the microbiota of the human small intestine by 16S tagged sequencing of samples obtained by esophagogastroduodenoscopy (EGD), nasoduodenal catheters, and ileostomy output. Despite technical limitations, such as the potential for contamination via instrumentation and the exposure of bacteria to oxygen in ileostomy samples, a number of studies have shown predominance of Streptococcus spp.,1, which accounted for 19% of 454 pyrosequencing reads with a high intra-genus and intra-species genetic diversity in samples obtained from ileostomy patients2. Other predominant genera include Veillonella spp., Prevotella spp., Rothia spp., Haemophilus spp., Actinobacillus spp., Escherichia spp., and Fusobacterium spp. The high abundance of Streptococcus, Veillonella, and Prevotella species, prominent members of the oral microbiota but present at relatively low abundances in the fecal microbiota3, supports the notion that the diet and/or the mouth can be a significant inoculation source for the small intestine.
The production of bile acids by the liver and their secretion into the proximal small intestine play an important role in the emulsification, solubilization, and absorption of lipids4. Bile acids are endogenous agonists for the nuclear hormone receptor FXR and the G-protein coupled receptor TGR5. Based on preclinical and clinical studies, FXR activation regulates multiple aspects of mammalian physiology, and the effects demonstrated in humans include inhibition of bile acid synthesis as a mechanism for the treatment of liver disease5. There is an important bidirectional relationship between bacteria and bile acids, whereby bacteria regulate the composition of luminal bile acids while bile acids have an impact on the survival and growth of bacteria6. A unique feature of bacteria that inhabit the mammalian gut is the expression of bile salt hydrolases (BSH) capable of deconjugating bile acids, where they are the rate limiting step in the conversion of primary bile acids, such as cholate and chenodeoxycholate, to secondary bile acids, such as deoxycholate and lithocholate, through 7 α-dehydroxylation by intestinal bacteria via the expression of genes in the bile acid inducible operons (BAI)6.
Although the concentration of bile acids is extremely high in the small intestine, levels in the colon are much lower due to the absorption of over 90% of luminal bile acids in the terminal ileum as part of enterohepatic circulation4. One might expect, therefore, that the interaction between bile acids and the gut microbiota would be more robust in the mammalian small intestine than the colon. The impact of bile acids on bacteria is complex but a few generalizations can be made. Gram-positive bacteria are generally more sensitive to inhibition by the toxic effects of bile than Gram-negative bacteria6. This observation has practical utility, as bile acids are added to McConkey agar to inhibit the growth of Gram-positive bacteria in clinical microbiology laboratories. However, bile acid tolerance can be a strain-specific trait, preventing generalizations about the sensitivity of bacteria to bile acids at the genus and species level6. The mechanisms of bile acid toxicity on bacteria are multifactorial and include membrane effects, DNA damage, oxidative stress, alterations to RNA structure, and protein denaturation6. Furthermore, there are many factors in addition to the expression of BSHs that may impact bacterial bile acid tolerance, including components of the outer membrane, such as lipopolysaccharide, multidrug efflux pumps, and various two-component systems6.
Obeticholic acid (OCA), a semi-synthetic bile acid derivative of chenodeoxycholic acid7, is a potent and selective agonist for FXR, a nuclear receptor expressed in the liver and intestine. FXR is a key regulator of bile acid, inflammatory, fibrotic, and metabolic pathways. FXR activation decreases the intracellular hepatocyte concentrations for bile acids by suppressing de novo synthesis from cholesterol, as well as, by increasing transport of bile acids out of the hepatocytes7. These mechanisms limit the overall size of the circulating bile acid pool while promoting choleresis, thus reducing hepatic exposure to bile acids. OCA, originally described for its anticholestatic and hepatoprotective properties7, has been approved for the treatment of primary biliary cholangitis7, and has shown promising results in the treatment of nonalcoholic steatohepatitis7.
Herein, using shotgun metagenomic sequencing of fecal samples collected in a controlled human study where healthy subjects were given three different doses of OCA for 17 days followed by a 17-day washout period, we examined the impact of FXR activation by OCA on endogenous bile acid levels and gut microbiome composition.
As shown in the present study, the suppression of bile acid synthesis by OCA treatment, quantified by the reduction in plasma levels of 7α-hydroxy-4-cholesten-3-one (C4), a bile acid precursor, led to a reversible and highly significant increase in specific Gram-positive facultative anaerobic bacterial taxa. These were characterized as organisms found in both food and in the small intestine, such as S. thermophilus, L. casei, and L. lactis.
We provide three lines of evidence that support the notion that the increased abundance of these bacterial taxa is due to the suppression of small intestinal bile acid levels by OCA. First, cross species genomic pathway analysis for these three taxa showed a consistent enrichment of pathways associated with OCA treatment that were indicative of bacterial proliferation, such as DNA and amino acid synthesis. Second, in vitro experiments revealed significant growth inhibition of all three taxa in response to conjugated endogenous bile acids at the physiological levels observed in the human small intestine that were not observed with pharmacological doses of OCA. Finally, treatment of mice with OCA led to a significant induction of Firmicutes, specifically in the small intestine, which was concurrent with a significant reduction in conjugated bile acid levels. Thus, our results provide evidence for the important role of bile acids in the modulation of the human small intestinal microbiome through the activation of FXR. In addition to the opportunity for a biomarker predicting clinical response to FXR agonists, these data also point to novel opportunities to modulate the small intestinal microbiome to prevent and/or treat disease.
Materials and Methods:
Human Subjects.
The Phase 1 study “An Open Label, Randomized, Single Dose and Multiple Dose Trial to Assess the Pharmacokinetics of Obeticholic Acid (OCA)” was performed at a single center in healthy volunteers (ClinicalTrials.gov Identifier: NCT01933503). The trial protocol and the informed consent form were approved by the Chesapeake institutional review board (#133-07), and informed consent was obtained from all subjects. Stool specimens were collected at three time points. The first (day 1) stool specimens were collected on day 1 before OCA treatment. The second (day 16) were collected on day 15, 16, or 17; one subject in the 10 mg group was unable to provide a sample during these days. The last (day 37) were collected up to 2 days prior to submitting them to study staff on day 37. Plasma C4 concentration was measured via LC-MS/MS on Days 1 through 44 only in subjects receiving 5 mg and 10 mg of OCA (seven and six subjects completed the day 44 assessments, respectively). All authors had access to the study data and reviewed and approved the final manuscript.
16S rRNA marker gene and shotgun metagenomic sequencing.
16S rRNA sequencing8 and shotgun metagenomic sequencing9 were performed as previously described (see Supplementary Information).
Functional profiling.
6085 pathways from 233 species were identified from HUMAnN2 (involving 371 unique pathways). Quantile normalization was performed, and 2,603 pathways with low (<10%) present rate were removed. A total of 3,482 pathways (from 134 species, involving 311 unique pathways) were kept for further analysis.
Statistical analysis.
The generalized estimation equation (GEE) analysis was applied to study the correlation of C4 and species over time. Both C4 concentration and the relative abundance of species were log10 transformed when fitting the model: log10(Species) ~ log10(C4). The R function “geeglm” from package “geepack” was used. For each of the most varying genes, the repeated-measure ANOVA was performed to study OCA’s time effect, dose effect, and time:dose interaction effect on genes: log10(gene abundance) ~ day + dose + day:dose. For each identified pathway, the repeated-measure ANOVA was performed to study OCA’s time effect, dose effect, and time:dose interaction effect on pathways: log10(pathway abundance) ~ day + dose + day;dose. Samples were classified as OCA treated (from day 16) or un-treated (from day 1, and 37). The log10 value of relative abundance of species, C4 concentration, or transposase abundance was used, separately, as predictor. Logistic regression model was used to determine their discriminatory ability to the outcome (OCA treated or un-treated). The discriminatory power was assessed by receiver operating characteristic (ROC) curves and the area under the ROC curve (AUC) values, by varying the predictive probability of being OCA treated.
In vitro microbiology studies.
Inhibition of bacterial growth by bile acids was determined by the microbroth dilution method10. Growth was measured via optical density at 630 nm, and measurements were normalized against wells containing the appropriate bile acid level with no bacteria.
Animal Studies.
All animal experiments were performed according to Institutional Animal Care and Use Committee-approved protocols and adhere to the standards articulated in the Animal Research: Reporting of In Vivo Experiments (ARRIVE) guidelines. Three groups of male C57BL6 mice at 8 weeks of age (2-3 mice per cage) from Jackson Laboratory on Open Standard Diet (D11112201, Research Diets) were housed in small barrier cages with Bed- o’Cobs (1/8”, Andersons Lab Bedding Products, Maumee, OH) with nestlets and were gavaged daily (during the light cycle) with water, vehicle (0.5% methylcellulose, Sigma-Aldrich, St. Louis, MO; n=10) or obeticholic acid (OCA, 10 mg/kg dissolved in 0.5% methylcellulose; n=10) for two weeks. Mice were euthanized within 3-5 hours of the last gavage. The small intestine was divided into proximal and distal segments, and the luminal contents were collected by flushing each segment with 1 mL of sterile 1X PBS. Fecal pellets were collected at baseline (Day 0) and Day 14.
Results:
Treatment with OCA inhibits synthesis of endogenous bile acids and increases the relative abundance of several low level Gram-positive bacterial taxa detectable in human feces.
Twenty-four healthy subjects were randomly assigned to one of three dose groups (5 mg, 10 mg, or 25 mg OCA per day), with each dose group comprising eight subjects (four women and four men). A single oral OCA dose of 5 mg, 10 mg, or 25 mg tablets, depending on the treatment assignment, was administered on Day 1. In the multiple-dose phase, a single oral OCA dose of 5 mg, 10 mg, or 25 mg tablets, in accordance with the assigned treatment, was administered orally once daily for 14 days from Days 4-17. The patients remained at the study site until Day 30, and were followed up until the final visit on Day 44 (Supplementary Figure S1A). Fecal specimens were collected at baseline (Day 0), at the end of the multiple-dose phase (Day 17), and at the end of the study (Day 37). Pharmacokinetic blood samples were assessed pre-dose and on Days 1 – 3, 17, 35, 37, 39, and 44.
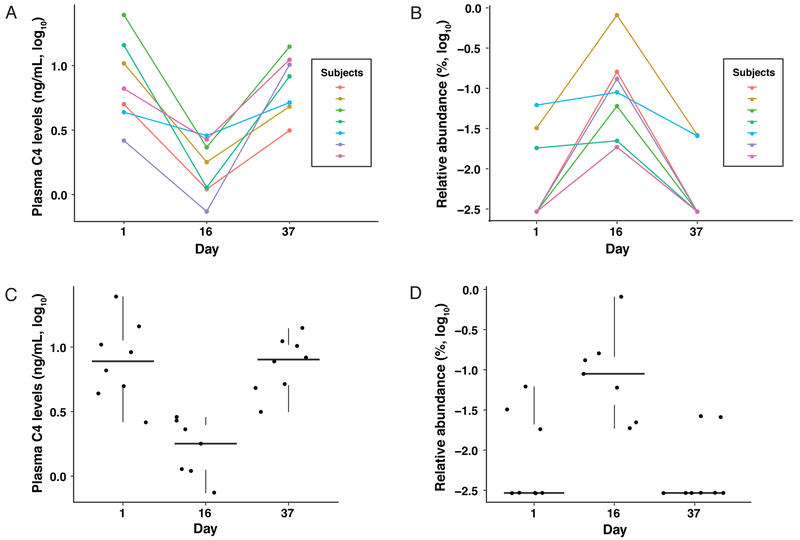
Serial quantification of plasma C4, an intermediate in the synthesis of bile acids from cholesterol11, showed a time-dependent reduction (repeated measure ANOVA, p=4.77×10−5) in response to OCA treatment (Supplementary Figure S1B). Therefore, plasma C4 levels are a reliable dynamic indicator of the host response to FXR activation by OCA that leads to the suppression of endogenous bile acid synthesis12. Features of the gut microbiome were matched to the variations in plasma levels of C4 that were maximally suppressed on Day 16 of the study relative to Day 0 (pre-OCA dose) and returned to normal by Day 37 (maximal time off OCA) (Figures 1A and C). We identified, in the 10 mg OCA group, 15 bacterial species that showed a time-dependent correlation with C4 levels (generalized estimation equation (GEE), p<0.05; Supplementary Table S1); five of these species remained statistically significant after correction for multiple comparisons (FDR<0.05). The time-dependent effect of OCA on Streptococcus thermophilus, the species showing the greatest increase, was striking, as this is a relatively low abundance taxon in the fecal microbiota, which, in most subjects, was undetectable without OCA treatment (Figures 1B and D). With two exceptions, Gram-positive bacterial genera showed increased abundances following OCA treatment. Meanwhile, the abundances of all Gram-negative bacteria decreased (Supplementary Table S1). Similar statistically-significant results were also observed for the 5 mg and 25 mg treatment groups (data not shown).
Figure 1.
Correlation between plasma C4 levels and the relative abundance of Streptococcus thermophilus in response to OCA treatment. Linear and box and whisker plots of: (A,C) plasma C4 levels and (B,D) S. thermophilus relative abundance in the 10 mg OCA group. The correlation between plasma C4 levels and the relative abundance of S. thermophilus were highly significant (FDR = 2.30e-05) based on a time-dependent GEE model.
Genomic representation of bacteria induced by treatment with OCA identifies a signature dominated by Streptococcus thermophilus and Lactococcus lactis consistent with bacterial proliferation.
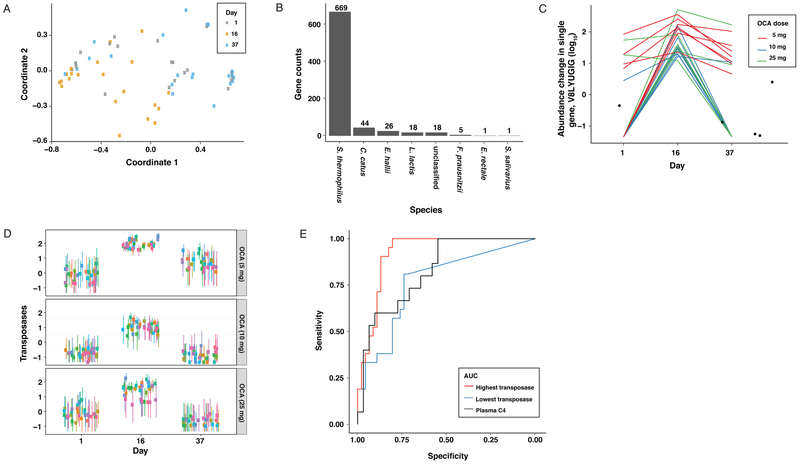
A Uniref90 high stringency genomic analysis was used to assign specific genes to the taxonomic signature of bacteria whose abundance was associated with OCA treatment and identified 782 genes assigned to eight bacterial species with a significant time-dependent effects in response to OCA treatment (Figures 2A and B). Nearly 86% of these belonged to Streptococcus thermophilus. The largest single category of genes was associated with S. thermophilus and L. lactis bacterial transposases (hypergeometric test, p=4.071×10−18) (Supplementary Table S2 and Figure 2C), enzymes that are important for the movement of mobile DNA elements throughout the genome and amongst the most abundant and ubiquitous class of genes in nature13. This family of genes showed a robust increase in representation at all three OCA doses (Figure 2D). The abundance of transposases can be used to predict OCA treatment with higher accuracy than plasma C4 levels based on a Receiver Operating Characteristics (ROC) curve analysis (Figure 2E).
Figure 2:
Genomic signature of the fecal microbiome associated with OCA administration. (A) A multidimensional scaling (MDS) plot of samples based on the Kendall rank correlation coefficient derived from 782 genes with a time-dependent effect in response to OCA administration based (repeated measure ANOVA, FDR<0.01). (B) Distribution of the 782 genes by bacterial taxonomy. (C) The abundance of a selected transposase (V8LYU6, from S. thermophilus) over time. (D) Out of 394 total transposases identified in the samples, 32 transposases had significant time-dependent responses to each of the three OCA doses (shown as log10 mean abundance and 95% confidence interval across samples). (E) ROC curves for transposases and plasma C4 (Red: transposase with highest AUC (0.917); Blue: transposase with lowest AUC (0.789); Black: plasma C4 levels (AUC = 0.828)).
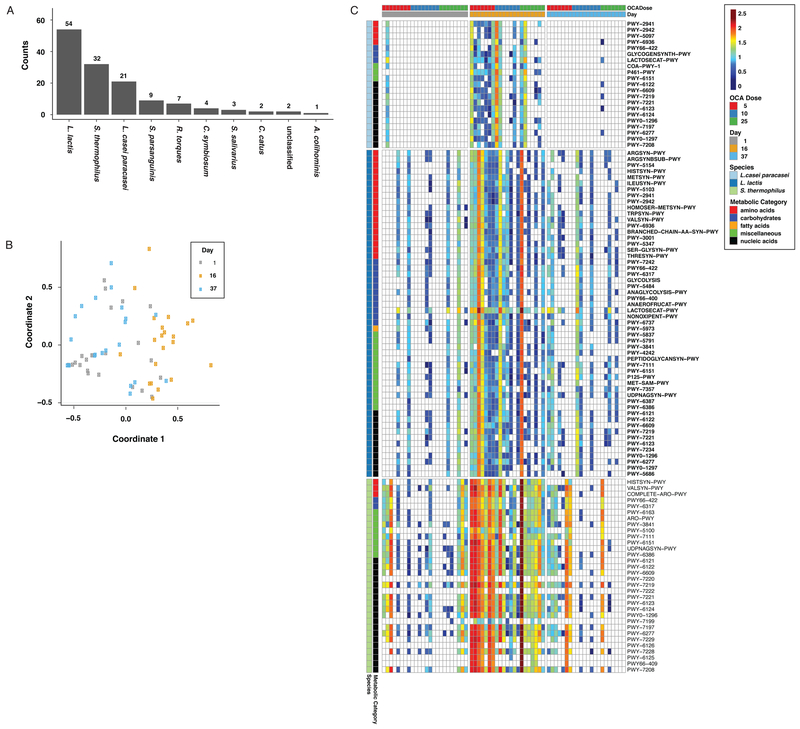
We also performed an analysis to identify bacterial metabolic pathways associated with OCA treatment. A repeated measure ANOVA identified 135 MetaCyc pathways with significant time effects (FDR<0.01). The majority of the 135 pathways showing a significant association belonged to three bacterial species: Lactococcus lactis, Streptococcus thermophilus, and Lactobacillus casei/paracasei (Figure 3A); these are Gram-positive bacteria that increased significantly with OCA treatment (Supplementary Table S1). A composite view of these pathways, visualized in a multidimensional scaling plot (Figure 3B), shows the effect of OCA and its reversibility, as the pathway abundances on Days 1 and 37 are more similar to each other compared with Day 16 (Figure 3C). For any given species, representation of a particular gene and/or pathway is likely to simply reflect the relative abundance of that taxon in the community, so functional relevance is unclear. We believe this to be the case for the association between transposases and OCA treatment (Figures 2C-E and Supplementary Table S2). However, pathways conserved across several species in response to OCA would likely indicate a functional interaction. Supporting this hypothesis, a heatmap of the statistically significant pathways for the top three bacterial taxa shows a robust time-dependent response to OCA; the associations are greatest at the lowest tested dose of OCA (Figure 3C). Common gene pathways between these three species, as well as other taxa identified in Figure 3A (Supplementary Table S3), show that pathways associated with nucleotide and amino acid biosynthesis are enriched by OCA treatment; this is consistent with bacterial proliferation.
Figure 3:
Bacterial metabolic pathways associated with OCA administration. (A) 135 metabolic pathways were significantly associated with OCA administration (repeated measure ANOVA, FDR < 0.01), categorized by bacterial taxa. (B) MDS plot of samples based on the Kendall rank correlation coefficient derived from the 135 metabolic pathways that were significantly associated with OCA administration. (C) Heatmap of significantly altered metabolic pathways from three major bacterial species sorted by time and dose. Metabolic pathways identified by Metacyc annotations were grouped into five classes (indicated by color on the y-axis).
Physiologically-relevant levels of endogenous intestinal bile acids lead to significant inhibition of bacterial growth under both aerobic and anaerobic conditions.
Gram-positive bacteria are generally more sensitive to growth inhibition by bile acids than Gram-negative bacteria6, and our genomic analysis is consistent with the notion that specific Gram-positive taxa become proportionally more abundant during OCA administration due to enhanced proliferation. Therefore, we hypothesized that FXR-dependent inhibition of endogenous bile acid synthesis by OCA may reduce the growth inhibitory effects on Grampositive bacterial species that are normally sensitive to bile acids. To determine if the three bacterial species with the greatest representation of pathways associated with OCA (Figure 3A) are sensitive to bile acids, we determined the species-specific minimal inhibitory concentrations (MIC) of the two most predominant bile acids in the human small intestine, glycochenodeoxycholic acid (GCDCA) and glycocholic acid (GCA) (Figure 4A). Since the environment of the small intestinal lumen may transition from an aerobic to anaerobic state along its length14, MICs were determined under both aerobic and anaerobic conditions. Although each species showed variable growth inhibition to the two bile acids under both aerobic and anaerobic conditions, the growth of all three species was significantly inhibited at the physiologically relevant concentrations (micromolar range) of bile acids normally found in the small intestine15. Although these results show a high level of consistency, it’s important to note that bile acid sensitivity can exhibit variability at the strain level as demonstrated by the greater level of growth inhibition of L. casei 393 relative to L. casei CP (Figure 4A).
Figure 4.
Minimal inhibitory concentrations (MICs) of the two Gram-position bacterial species most strongly associated with the use of OCA in response to two endogenous bile acids and OCA. (A) MICs of the species in response to treatment with the two dominant conjugated primary bile acids found in the human small intestine, glycochenodeoxycholic acid (GCDCA) and glycocholic acid (GCA), under both aerobic and anaerobic conditions. N=3 per measurement. Blue = Physiologically relevant concentrations of bile acids in the human small intestine. (B) MICs of the same bacterial taxa in response to treatment with OCA. N=3 per measurement. Blue = Estimated human small intestinal concentrations of OCA.
As bile acid deconjugating enzymes have been shown to enhance bile acid tolerance6, we sought to determine if the strain(s) of S. thermophilus that increased in abundance with OCA treatment lack genomic representation of this enzyme family. Due to the low relative abundance of this species in our data set, it was not possible to assemble complete genomes directly from the sequence data. Instead, we compiled a custom database of all protein sequences annotated as “bile salt hydrolase” or “choloylglycine hydrolase” in bacterial genomes from NCBI RefSeq, and conducted a protein BLAST search against all available S. thermophilus genomes. This search yielded no matches. As a positive control, we applied the same method to genomes of Streptococcus equinus, which harbor bile acid deconjugating genes, and recovered thousands of matches. Thus, we could confirm that bile acid deconjugating enzymes were absent from all available reference genomes of S. thermophilus, confirming previous reports for this species as well as L. lactis16 and consistent with the bile acid sensitivity observed in vitro (Figure 4a).
Since OCA is a bile acid analogue possibly capable of inhibiting bacterial growth, we determined the MICs of OCA for the same bacterial species (Figure 4B). Although OCA was also able to inhibit growth of all three bacterial species, there was minimal to no inhibition of growth at OCA concentrations calculated to be reached in the human small intestine (~40 μM at a 10 mg/day dose)16, findings consistent with previous in vitro studies17. These findings support the notion that OCA treatment leads to the increased growth of bile acid sensitive bacterial taxa by suppressing endogenous bile acid synthesis.
OCA treatment in mice inhibits endogenous luminal bile acid levels and leads to increased Gram-positive bacteria, specifically in the small intestine.
The bacterial species that increase most prominently upon OCA treatment have been reported to represent a significant proportion of the small intestinal microbiota, but are very minor constituents in stool. For example, Streptococci represent as much as 20% of the human small intestinal microbiota by 16S tagged sequencing1. Some of these bacteria are environmental organisms used in the manufacturing of food introduced into the small intestine with diet, including Streptococcus thermophilus, Bifidobacterium breve, Lactobacillus casei, and Lactococcus lactis1.
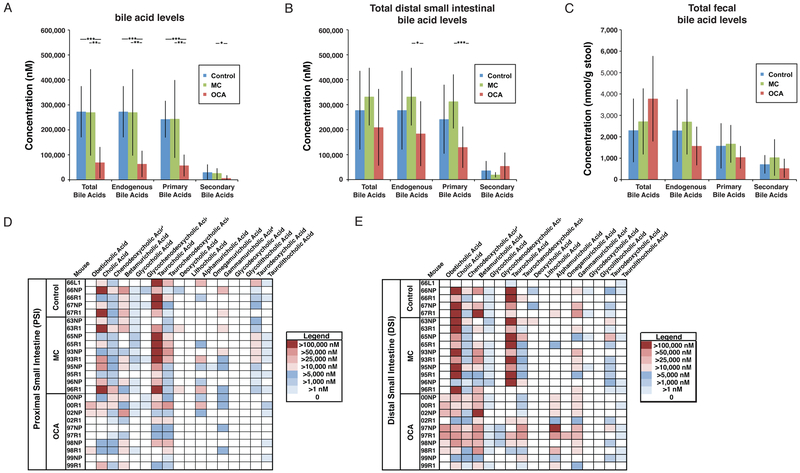
To determine whether OCA treatment can alter the gut microbiota composition specifically in the small intestine, we treated mice with OCA for 14 days, characterized the microbiota composition in the proximal and distal small intestine, as well as the stool, and quantified bile acids. Since OCA was prepared in methylcellulose, an additional methylcellulose control group was included due to its previously described effect on fecal bile acid levels18. Quantification of luminal bile acid concentrations revealed a significant reduction of endogenous primary bile acids that was greatest in the proximal small intestine but was also observed in the distal small intestine, with no effect in the feces (Figures 5A-C). The largest consistent decreases were observed in the primary bile acids cholic and taurocholic acids, with smaller and less consistent decreases in several of the less abundant bile acids (Figures 5D and E). Importantly, OCA was detectable in the small intestinal lumen of treated mice, where it contributed minimally to the total concentration of bile acids (Figures 5A-E). Also notable was the quantitative difference in the absolute levels of luminal bile acids in the small intestine (approximately 100-fold greater than in the feces, Figures 5A-C); this is consistent with enterohepatic reabsorption of over 95% of bile acids in the terminal ileum of the small intestine4.
Figure 5.
Effect of OCA administration on luminal bile acid concentrations in the murine small intestine and feces. Total (endogenous bile acids and OCA), total endogenous, total primary, and total secondary bile acids in the (A) lumen of the proximal small intestine; (B) lumen of the distal small intestine, and; (C) feces of mice following 14 days of gavage with either water (control, N=5), 0.5% methylcellulose (MC, N=10), or 0.5% methylcellulose with 10 mg/kg obeticholic acid (OCA, N=10). Mean±SE, *p<0.05, **p<0.01, ***p<0.001. Heatmaps of luminal bile acid concentrations in the proximal (D) and distal (E) small intestine.
The overall composition of the small intestinal microbiota in the OCA group, analyzed by 16S tagged sequencing, was different from both control groups. This was due to the increased abundance in the Clostridiaceae family of Firmicutes in both the proximal and distal small intestine of OCA-treated mice, but not in the feces (Figure 6 and Supplementary Figure S2). However, small intestinal and fecal bacterial load, as quantified by 16S copy number, showed no difference between the three treatment arms, revealing that OCA did not induce small intestinal bacterial overgrowth (Supplementary Figure S3). Collectively, these results demonstrate that treatment with OCA in mice leads to a significant reduction in luminal bile acid concentrations specifically in the small intestine, where there is a concurrent increase in Grampositive bacteria. Neither of these alterations was observed in fecal samples, showing that the activation of FXR by OCA and the resulting inhibition of endogenous bile acid synthesis impacts the small intestinal microbiota. These results, combined with the effects of OCA treatment on the abundance of bacterial taxa known to predominate in the small intestine, provide compelling evidence for the role of FXR in the dynamics of the human small intestinal microbiota.
Figure 6.
Effect of OCA on the proximal and distal small intestinal (PSI and DSI, respectively), as well as the fecal, microbiota composition in mice based on 16S tagged sequencing following 14 days of gavage with water (control), 0.5% methylcellulose (MC), or 0.5% methylcellulose with 10 mg/kg obeticholic acid (OCA).
A robust taxonomic signature for FXR activation in the human gut microbiome.
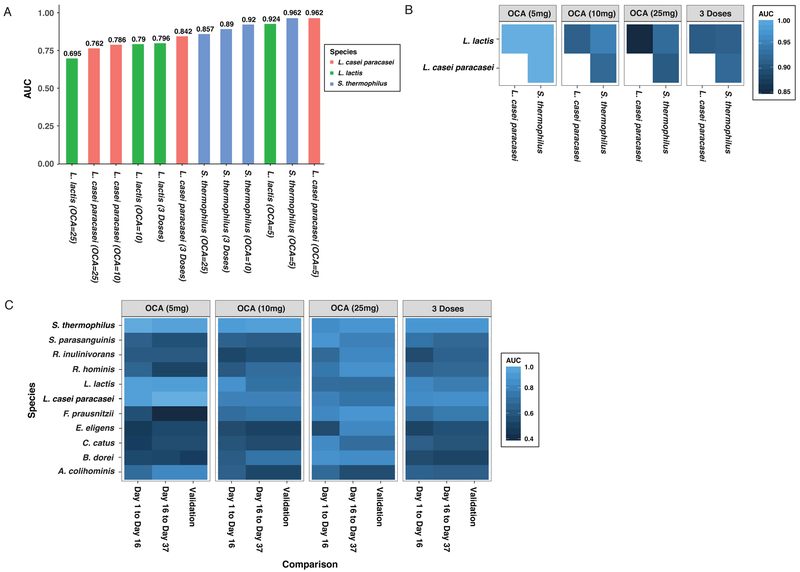
Growing evidence suggests that the composition of the gut microbiome might have value as a biomarker for drug metabolism19 and diet20 for personalized medicine, and discriminatory indices have been developed to categorize specific disease processes involving infections21, liver disease22, and inflammatory bowel disease23. The robust and reversible response of bacterial taxa to OCA treatment suggests that specific bacterial species, alone or in combination, might predict FXR-dependent inhibition of bile acid synthesis. ROC curves were generated and the area under the ROC curve (AUC) from logistic regression was calculated for each species characterized in our shotgun metagenomic dataset. The taxa that most accurately predicted treatment with the three doses of OCA, individually or combined, were L. casei-paracasei, L. lactis, and S. thermophilus (Figure 7A). For each of these species, the highest AUCs were observed at the OCA 5 mg dose. A combination of any two of these three species results in an AUC close to “1” for the 5 mg OCA dose (Figure 7B). This taxonomic signature exceeds the performance of C4 as a predictor of OCA administration at the 5 mg dose (Supplementary Figure S4). Finally, ROC analysis was performed for these three species, as well as a number of additional taxa, to determine the AUC at 5, 10, 25 mg doses, both independently and combined. To provide an assessment of the predictability of the models, data from Day 0 to 17 were used a training set, and data from Day 17 to 37 as a pseudo-validation set (Figure 7C). In general, the AUCs between these two intervals were very similar, which demonstrates the robust and reproducible nature of these associations. However, it should be noted that this reproducibility was not based on independent data sets since the data measured over three time points from the same set of individuals were used in the ROC analyses. Although the three species described in Figure 7A all had the highest AUCs at the 5 mg dose of OCA, two species showed a dose-dependent increase in AUC. Faecalibacterium prausnitzii and Bacteroides dorei both had AUCs of approximately 0.5 at the 5 mg dose, 0.7 at the 10 mg dose, and 0.85 at the 25 mg dose. Ultimately, taxonomic features that are maximally sensitive to the lowest dose of OCA, combined with others that show a more dynamic dose-dependent association, may prove useful in predicting clinical responses induced by OCA, and likely other FXR agonists.
Figure 7:
The power of the relative abundance of bacterial species to discriminate OCA treatment (day 16) vs. non-treatment (days 1 and 37) as assessed by logistic regression models. (A) The three species with the highest AUC values based on a ROC analysis of the three OCA doses. (B) AUC values based on a ROC analysis using the combination of any two of the three species with the highest AUC values. (C) AUC values based on separate ROC analyses for Day 1 vs. Day 16 and Day 37 vs. Day 16 based on logistic regression analysis. Species with high AUC values (AUC > 0.8) from at least one model are shown. The pseudo-validation AUCs were obtained by applying the logistic model derived from the Day 1 vs. Day 16 dataset (i.e., training set) to the Day 16 vs. Day 37 dataset (i.e., validation set).
Discussion:
Despite a large and growing literature characterizing the composition of the human fecal microbiome and its association with health and disease, only a few studies have analyzed the microbiome in the small intestine due to its technical accessibility. This is unfortunate, because the characterization of the small intestinal microbiome is highly relevant to understanding normal human physiologic responses to diet and nutrient absorption1, development of the mucosal immune system and the dynamics of bile acid biology4. The human small intestinal microbiota also plays a fundamentally important role in the pathogenesis of disease, such as bacterial overgrowth24, celiac disease, and irritable bowel syndrome, among many others1.
The effect of bile acids on bacteria has been the topic of intense investigation for decades and there are a number of studies in murine model systems examining the effect of exogenously-delivered bile acids on the composition of the gut microbiota. However, there are no studies in humans showing that modulation of endogenous bile acid production has an effect on the gut microbiome. Studies characterizing the gut microbiome in humans with cirrhosis infer that the highly significant discriminatory function to classify patients may be due to a reduction in bile acid synthesis but direct evidence is lacking25. Herein, we used treatment with OCA as a probe to characterize FXR-dependent dynamics of the small intestinal microbiome in response to alterations in endogenous bile acid concentrations. The high levels of bile acids in the small intestine relative to the colon and the differential sensitivity of bacterial species to the growth inhibitory effects of bile acids allowed us to discriminate a small intestinal from a colonic microbiome signature in response to FXR activation. This was confirmed in a mouse model where alterations in bile acids in response to treatment with OCA and their effects on the microbiota were observed specifically in the small intestine and not in the feces.
Using features in the fecal gut microbiome that matched the pattern of the host physiologic response to FXR activation by OCA, namely the reversible inhibition of bile acid synthesis reflected by plasma C4 levels, we identified a very specific and robust induction of a limited number of Gram-positive bacteria. There are several common features amongst most prominent species in this group: they are nearly all Gram-positive, many are facultative anaerobes, and a number are organisms consumed in the diet, including S. thermophilus, L. casei/paracasei, B. breve, and L. lactis.1 Importantly, all these bacterial species have been described to be members of the human small intestinal microbiota, are observed only at very low levels in the feces, and have been described as probiotics with potential health benefits1.
In aggregate, these features suggest that the suppression of endogenous bile acid synthesis by OCA led to the increased representation of bile acid-sensitive bacterial species specifically in the human small intestine. Indeed, we show that bile salt hydrolases (BSH), unique to bacteria resident in the mammalian gut where they may enhance bile acid tolerance6, are missing from the genome of S. thermophilus, which is induced by OCA. We support this hypothesis by showing that these species are sensitive to growth inhibition in vitro when exposed to physiologically-relevant concentrations of conjugated bile acids found in the small intestine. However, growth inhibition was not observed upon exposure to OCA, which is consistent with previous data26 and suggests that FXR-dependent inhibition of endogenous bile acid secretion is responsible for the observed change in the gut microbiota and is not a direct effect of OCA. We provide additional support to the concept of small intestinal species expansion by characterizing the effect of OCA on the small intestinal and fecal bile acid compositions in mice and correlating these changes with the composition of the gut microbiota.
There have been a limited number of studies in rodent model systems examining the effect of exogenously-delivered bile acids or OCA on the composition of the fecal microbiota. OCA treatment of cirrhotic rats inhibited intestinal inflammation and bacterial translocation and led to alterations of the mucosa-associated ileal microbiota that varied depending on whether the rats were healthy or had liver cirrhosis26. In the latter group, there was a decrease in Proteobacteria and an increase in Gram-positive bacteria belonging to the Firmicutes phylum, which in part, is consistent with our findings in a disease model. By contrast, rats fed high levels of the weak FXR agonist cholic acid exhibited dramatic increases in the levels of both primary and secondary bile acids in the feces as well as phylum level alterations in gut microbiota composition, namely an increase in Firmicutes at the expense of Bacteroidetes27. However, the relevance of these observations to our study is unclear since we did not observe any significant alteration to the composition of the fecal microbiota or the levels of fecal bile acids in mice fed OCA.
A genomic and pathway analysis of the species induced by OCA revealed robust and consistent findings that not only serve to validate our taxonomic analyses, but also infer possible functionality. The largest single category of genes positively correlated with the administration of OCA were transposases, enzymes important for the movement of mobile DNA elements within the genome of an organism and between bacterial species via lateral gene transfer13. Although it is possible that bile acids are important for regulating mechanisms of lateral gene transfer in bacteria, it is much more likely that this association reflects the relative abundance of these specific taxa, since transposases and transposons are well annotated features in the genomic databases of bacterial strains13. However, many additional bacterial genes significantly associated with OCA administration were part of a sizable number of biological pathways belonging to three bacterial species, L. lactis, S. thermophilus, and L. casei/paracasei. The increased abundance of pathways associated with nucleic acid and amino acid synthesis, after OCA administration and shared by all three species, is consistent with the growth and persistence of these taxa by reduced bile acid inhibition.
These strong taxonomic and genomic microbiome associations with OCA administration may have practical value related to precision medicine, where there is growing evidence that the gut microbiome plays a role in the pharmacokinetic and pharmacodynamic responses to small molecule drugs19. Features of the gut microbiome have been shown to be of value in the development of personalized diets to improve post-prandial glycemic responses20. Moreover, drugs such as metformin can have a significant effect on the composition of the gut microbiome28. With these notions in mind, we show that the induction of transposases is a better marker of OCA administration than plasma C4 levels. Additionally, the abundance of L. lactis, S. thermophilus, and L. casei/paracasei, individually or in combination, have a very high AUCs in a ROC analysis, indicating that these taxa have high value as a binary classifier of OCA use. Some of these features respond in a dose-dependent fashion. As additional studies characterizing the gut microbiome are performed whereby the effects of FXR agonists such as OCA are correlated with clinical outcomes, features of the gut microbiome may become useful biomarkers.
In addition to providing the first glimpse into the dynamics of the human small intestinal microbiome in response to bile acid modulation, our results suggest potential opportunities related to disease treatment. Numerous studies on the use of probiotics, live microorganisms that confer a health benefit to the host when consumed, have shown benefit in the treatment of a wide variety of diseases associated with the liver29. However, clinical efficacy studies have so far not been sufficiently robust to allow currently available probiotics to achieve regulatory approval as a medically-indicated therapeutic modality possibly in part because they are unable to achieve a sufficient biomass via engraftment to exert a meaningful physiologic effect on the host. Indeed, studies have failed to demonstrate detectable levels of probiotic bacteria in fecal samples when delivered either in fermented food products or in capsule form30. However, since many of these probiotics may reside at higher levels in the small intestine, where they have shown an impact on intestinal gene expression30, a more robust beneficial host response might be observed if their effects focused on mechanisms involving the small intestine. Thus, the enhanced engraftment and/or bacterial load of probiotics targeting the small intestine via the FXR-dependent mechanisms we have demonstrated with OCA might have significant value. Such an approach might involve “engineering” the composition of the small intestinal microbiota. Indeed, we have shown that the composition and function of the gut microbiota can reliably be reconfigured in mice for the treatment of hyperammonemia31, an approach that we have recently shown might be clinically translatable to humans8.
In conclusion, the activation of FXR by OCA and its subsequent effects on the small intestinal microbiota via bile acid-dependent mechanisms has revealed a range of novel opportunities, not only to improve precision medicine regarding the administration of small molecule agonists, but also to develop more reliable biomarkers and utilize currently available and future probiotics targeting the small intestine for the prevention and/or treatment of a variety of diseases.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by a research grant from Intercept Pharmaceuticals (GDW), the PennCHOP Microbiome Program (GDW), P30 DK050306 Human-Microbial Analytic and Repository Core of the Center for Molecular Studies in Digestive and Liver Disease (GDW), NIH grant R01AA026302-01, K08-AA021424, R01GM123056, Robert Wood Johnson Foundation, Harold Amos Medical Faculty Development Award 7158, and IDOM DRC Pilot Award P30 DK019525 (RMC); and in part by NIH P30-DK050306.
Abbreviations:
- OCA
obeticholic acid
- FXR
farnesoid X receptor
- GEE
generalized estimation equation
- ROC
receiver operating characteristics
- AUC
area under the curve
- GCDCA
glycochenodeoxycholic acid
- GCA
glycocholic acid
- MIC
minimum inhibitory concentration
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Intercept Pharmaceuticals performed the phase 1 clinical study and provided clinical metadata as well as shotgun metagenomic sequencing data for analysis by investigators at Penn and the Children’s Hospital of Philadelphia.
Conflict of interest statement: LA is a consultant for and has stock options in Intercept Pharmaceuticals. FB and JE are employees and shareholders of Intercept Pharmaceuticals. GDW and RMC receive research funding from Intercept Pharmaceuticals. The remaining authors declare no competing interests.
References
- 1.Derrien M, van Hylckama Vlieg JE. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol 2015;23:354–66. [DOI] [PubMed] [Google Scholar]
- 2.van den Bogert B, Erkus O, Boekhorst J, et al. Diversity of human small intestinal Streptococcus and Veillonella populations. FEMS Microbiol Ecol 2013;85:376–88. [DOI] [PubMed] [Google Scholar]
- 3.Charlson ES, Bittinger K, Haas AR, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med 2011;184:957–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schneider KM, Albers S, Trautwein C. Role of bile acids in the gut-liver axis. Journal of Hepatology 2018;68:1083–1085. [DOI] [PubMed] [Google Scholar]
- 5.Carr RM, Reid AE. FXR agonists as therapeutic agents for non-alcoholic fatty liver disease. Curr Atheroscler Rep 2015;17:500. [DOI] [PubMed] [Google Scholar]
- 6.Begley M, Gahan CGM, Hill C. The interaction between bacteria and bile. FEMS Microbiology Reviews 2005;29:625–651. [DOI] [PubMed] [Google Scholar]
- 7.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological 1995;57:289–300. [Google Scholar]
- 8.Ni J, Shen TD, Chen EZ, et al. A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci Transl Med 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lewis JD, Chen EZ, Baldassano RN, et al. Inflammation, Antibiotics, and Diet as Environmental Stressors of the Gut Microbiome in Pediatric Crohn’s Disease. Cell Host Microbe 2015;18:489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelkirk PG, Duben-Engelkirk JL. Laboratory diagnosis of infectious diseases: essentials of diagnostic microbiology: Lippincott Williams & Wilkins, 2008. [Google Scholar]
- 11.Galman C, Arvidsson I, Angelin B, et al. Monitoring hepatic cholesterol 7alpha- hydroxylase activity by assay of the stable bile acid intermediate 7alpha-hydroxy-4- cholesten-3-one in peripheral blood. J Lipid Res 2003;44:859–66. [DOI] [PubMed] [Google Scholar]
- 12.Hirschfield GM, Mason A, Luketic V, et al. Efficacy of obeticholic acid in patients with primary biliary cirrhosis and inadequate response to ursodeoxycholic acid. Gastroenterology 2015;148:751–61 e8. [DOI] [PubMed] [Google Scholar]
- 13.Aziz RK, Breitbart M, Edwards RA. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res 2010;38:4207–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He G, Shankar RA, Chzhan M, et al. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc Natl Acad Sci U S A 1999;96:4586–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Northfield TC, McColl I. Postprandial concentrations of free and conjugated bile acids down the length of the normal human small intestine. Gut 1973;14:513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, LaCerte C, Kansra S, et al. Comparative potency of obeticholic acid and natural bile acids on FXR in hepatic and intestinal in vitro cell models. Pharmacol Res Perspect 2017;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Verbeke L, Farre R, Verbinnen B, et al. The FXR agonist obeticholic acid prevents gut barrier dysfunction and bacterial translocation in cholestatic rats. Am J Pathol 2015;185:409–19. [DOI] [PubMed] [Google Scholar]
- 18.Cox LM, Cho I, Young SA, et al. The nonfermentable dietary fiber hydroxypropyl methylcellulose modulates intestinal microbiota. FASEB J 2013;27:692–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klaassen CD, Cui JY. Review: Mechanisms of How the Intestinal Microbiota Alters the Effects of Drugs and Bile Acids. Drug Metab Dispos 2015;43:1505–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeevi D, Korem T, Zmora N, et al. Personalized Nutrition by Prediction of Glycemic Responses. Cell 2015;163:1079–94. [DOI] [PubMed] [Google Scholar]
- 21.Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015;517:205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loomba R, Seguritan V, Li W, et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab 2017;25:1054–1062 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barber GE, Yajnik V, Khalili H, et al. Genetic Markers Predict Primary Non-Response and Durable Response To Anti-TNF Biologic Therapies in Crohn’s Disease. Am J Gastroenterol 2016;111:1816–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouhnik Y, Alain S, Attar A, et al. Bacterial populations contaminating the upper gut in patients with small intestinal bacterial overgrowth syndrome. Am J Gastroenterol 1999;94:1327–31. [DOI] [PubMed] [Google Scholar]
- 25.Qin N, Yang F, Li A, et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 2014;513:59–64. [DOI] [PubMed] [Google Scholar]
- 26.Ubeda M, Lario M, Munoz L, et al. Obeticholic acid reduces bacterial translocation and inhibits intestinal inflammation in cirrhotic rats. J Hepatol 2016;64:1049–1057. [DOI] [PubMed] [Google Scholar]
- 27.Islam KB, Fukiya S, Hagio M, et al. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology 2011;141:1773–81. [DOI] [PubMed] [Google Scholar]
- 28.Forslund K, Hildebrand F, Nielsen T, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015;528:262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen TD, Pyrsopoulos N, Rustgi VK. Microbiota and the liver. Liver Transpl 2018;24:539–550. [DOI] [PubMed] [Google Scholar]
- 30.Flach J, van der Waal MB, Kardinaal AFM, et al. Probiotic research priorities for the healthy adult population: A review on the health benefits of Lactobacillus rhamnosus GG and Bifidobacterium animalis subspecies lactis BB-12. Cogent Food & Agriculture 2018;4. [Google Scholar]
- 31.Shen T, Albenberg L, Bittinger K, et al. Engineering the gut microbiota to treat hyperammonemia. J Clin Invest. 2015;125:2841–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.