Abstract
Snake venoms represent an enriched system for investigating the evolutionary processes that lead to complex and dynamic trophic adaptations. It has long been hypothesized that natural selection may drive geographic variation in venom composition, yet previous studies have lacked the population genetic context to examine these patterns. We leverage range-wide sampling of Mojave Rattlesnakes (Crotalus scutulatus) and use a combination of venom, morphological, phylogenetic, population genetic, and environmental data to characterize the striking dichotomy of neurotoxic (Type A) and hemorrhagic (Type B) venoms throughout the range of this species. We find that three of the four previously identified major lineages within C. scutulatus possess a combination of Type A, Type B, and a ‘mixed’ Type A + B venom phenotypes, and that fixation of the two main venom phenotypes occurs on a more fine geographic scale than previously appreciated. We also find that Type A + B individuals occur in regions of inferred introgression, and that this mixed phenotype is comparatively rare. Our results support strong directional local selection leading to fixation of alternative venom phenotypes on a fine geographic scale, and are inconsistent with balancing selection to maintain both phenotypes within a single population. Our comparisons to biotic and abiotic factors further indicate that venom phenotype correlates with fang morphology and climatic variables. We hypothesize that links to fang morphology may be indicative of co-evolution of venom and other trophic adaptations, and that climatic variables may be linked to prey distributions and/or physiology, which in turn impose selection pressures on snake venoms.
Introduction
Studying the context and geographic scale of local adaptation is critical to understanding the processes that shape the evolution of adaptive traits in natural populations1–3. Snake venom represents an excellent model for studying variation across populations as a result of stochastic and deterministic evolutionary processes. For example, strong selection is expected to act on snake venom due to its important ecological role in feeding and defense4–7, and because it is composed of numerous components that together manifest as distinct venom phenotypes between populations and species4,8. Additional dimensions involved in venom evolution are the physiological factors that must co-evolve with venom to allow for its production and prevent auto-toxicity9–12. Thus, snake venom provides opportunity to discern the causes of variation that have direct consequences for individual fitness at multiple scales.
There are several intriguing examples of phenotypic polymorphism in venom within rattlesnake species5,13,14, with the most well-studied example existing in Mojave Rattlesnakes (Crotalus scutulatus). In C. scutulatus, venom composition among individuals generally takes one of two forms, either a highly neurotoxic ‘Type A’ venom, or a hemorrhagic ‘Type B’ venom4,15–20. The venom of Type A individuals is dominated by a neurotoxin formed from a heterodimeric phospholipase A2 (PLA2) called Mojave Toxin (MTX) and has little snake venom metalloproteinase (SVMP) activity. Type B individuals lack MTX and tend to have high expression of SVMPs in their venom. Individuals possessing both phenotypes (i.e., Type A + B individuals) have also been found, though less frequently than strictly Type A and Type B individuals21,22.
Mojave Rattlesnakes are distributed in the deserts of North America, the Central Mexican Plateau, and the volcanic lowlands of south-central Mexico (Fig. 1). While previous proteomic16,19,23,24 and transcriptomic25 studies have shown that Mojave Rattlesnakes possess high intraspecific venom variation, venom composition has not been well characterized for the majority of the geographic distribution of these snakes. Type A, Type B, and Type A + B phenotypes are known from several distinct locations throughout the species distribution, and the spatial assortment of these phenotypes suggests that they are geographically structured15,19,21–23,26–28.
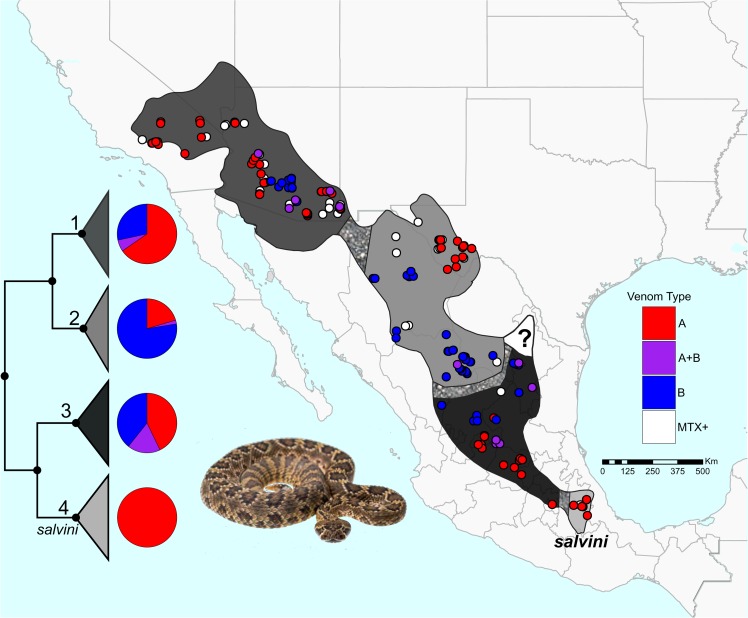
Figure 1.
Distribution and sampling of Mojave Rattlesnakes collected from throughout their range. Red, purple, and blue represent Type A, Type A + B, and Type B venom, respectively, in the pie charts and the sampling points. White points are individuals that were positive for Mojave Toxin but we could not distinguish between Type A and Type A + B. Mottling in the distribution are areas of gene flow between lineages. Pie charts represent the proportion of each venom type collected from each lineage based on samples with venom. Cladogram in lower left of the four lineages from Fig. 3 corresponding to those in Schield et al.38 numbered: 1 - Sonoran lineage, 2 - Chihuahuan lineage, 3 - Central Mexican Plateau lineage, 4 - salvini lineage, ?-not sampled/unknown lineage. Inset photo: Crotalus scutulatus by Travis Fisher.
Several selection-driven hypotheses have been proposed to explain the dichotomy between neurotoxic and hemorrhagic snake venom phenotypes because venom is expected to be locally adapted to prey at a very fine geographic scale7,29–32. These hypotheses have centered on potential differences in the digestive efficiency of the two venom types33; Type B venoms likely provide more efficient digestion of prey at lower temperatures or when temperature fluctuations are pronounced8,34, although evidence for an increase in digestive efficiency in Type B venoms remains controversial (e.g.33,35,36). Alternatively, neurotoxic venoms may be particularly advantageous when the chance of prey escaping is high34, as would be expected in metabolically active ectothermic prey in warm areas. This is because Type A venoms rapidly subdue prey through neuromuscular paralysis, but do not convey the potential digestive benefits that high SVMP activity does4. An additional selection pressure to consider is the types of prey found in regions with alternative venom types, as neurotoxic venom components have very specific targets and are likely effective against select prey items, while Type B venoms may have broad biological effects across more taxonomically diverse prey4.
While natural selection is thought to be a major driver of fine-scale geographic differences in venom phenotypes, little attention has been given to the potential role of other evolutionary forces (e.g., genetic drift, gene flow, population structure) in the evolution of the A/B dichotomy in the venom phenotypes of rattlesnakes. Recently, Dowell et al.37 proposed that Type A and Type B venom polymorphism in rattlesnakes might be maintained by balancing selection, and also that the presence of individuals with Type A + B venom may be the result of introgression between populations dominated by either Type A and Type B phenotypes. Schield et al.38 found evidence for four distinct geographically structured lineages within C. scutulatus, and for gene flow between these structured populations. Although sampling has been limited to date, previous studies have demonstrated that all three (i.e., Type A, Type B, and Type A + B) venom phenotypes occur in the three northern C. scutulatus lineages19,21,22,39,40, whereas C. scutulatus salvini (the most southern lineage) is thought to be exclusively Type A15,23. A number of questions remain, including what evolutionary mechanisms may underlie variation in venom phenotypes, and to what degree venom phenotypes are structured across the range of C. scutulatus.
In this study, we characterize venom phenotypes and genotypes throughout the entire range of C. scutulatus. We then link these data on venom variation with data on morphological variation, population genetic structure, and environmental variables to identify evolutionary factors that may underlie the polymorphic venom phenotypes in Mojave Rattlesnakes. Our results indicate that balancing selection is unlikely to explain venom variation, but instead, strong directional local selection for different venom phenotypes appears to drive range-wide venom variation. We also find evidence that venom phenotypes are correlated with annual environmental temperature, suggesting that the primary drivers of selection on venom phenotype variation include factors related to environmental temperature, such as snake digestive physiology, prey distribution, or prey physiology.
Methods
Ethics statement
Scientific collecting permits in the United States were issued by the State of Arizona Game and Fish Department (SP628489, SP673390, SP673626, SP715023), the California Department of Fish and Wildlife (SC-12985), the New Mexico Department of Game and Fish (3563, 3576) and Texas Parks and Wildlife (SPR-0390-029). In Mexico, collection permits were issued by the Secretaria de Medio Ambiente y Recursos Naturales of the Estados Unidos Mexicanos (SEMARNAT: SGPA/DGVS/03562/15, SGPA/DGVS/01090/17, and FAUT-0015). Interactions with animals were approved by the University of Central Florida’s (UCF) Institutional Animal Care and Use Committee under protocol 13–17 W and followed the American Society of Ichthyologists and Herpetologists ethical guidelines.
Sample collection and DNA extraction
We collected representatives of all previously identified lineages38 of Mojave Rattlesnakes from throughout their distribution. For most samples collected in the field, we obtained venom and tissue. When possible, voucher specimens were created and deposited (Supplemental Table 1 with abbreviations following41). Tissues were stored in 95% ethanol or RNAlater and venom was collected and vacuum dried, frozen in liquid nitrogen, and/or stored at −80 °C. We collected a total of 216 individuals: 114 of these had tissue and venom, 34 had only venom, and 68 had only tissue (Supplemental Table 1). Whole genomic DNA was extracted from samples using the Serapure bead extraction protocol of Rohland and Reich42 following modifications in Faircloth43.
Reverse-phased High Performance Liquid Chromatography (RP-HPLC) to determine venom type
All venom was vacuum dried or lyophilized prior to use, resuspended in Millipore-filtered water, and centrifuged to remove insoluble debris. We determined protein concentration on a Qubit 3.0 Fluorometer (ThermoFisher Scientific) using the Qubit Protein Assay (ThermoFisher Scientific) following the manufacturer’s protocol. To determine the venom type (A, B, or A + B) of each individual, we used Reverse-phased High Performance Liquid Chromatography (RP-HPLC) based on the protocol in Margres et al.44. For RP-HPLC, we injected 100 μg of venom onto a Jupiter C18 column (250 × 2 mm; Phenomenex, Torrence, California, USA) using two solvents: 1 = 0.1% trifluoroacetic acid (TFA) in water and 2 = 0.075% TFA in acetonitrile. We used a Beckman System Gold HPLC (Beckman Coulter, Fullerton, California, USA) located in the Florida State University (FSU) Department of Biological Science Analytical Lab. The gradient started with 95% A and 5% B for 5 minutes followed by a 1% per minute linear gradient to 25% B, followed by a 0.25% per minute linear gradient to 55% B, a 2% per minute linear gradient to 75% B, a 14% per minute linear gradient to 5% B and then 5 minutes at the initial conditions all at a 0.2 mL/min flow rate. Run time was 180 minutes for each sample and the effluent was monitored at 220 and 280 nm45. Venoms were assayed using RP-HPLC for 148 individuals based on the presence of both subunits of MTX (Type A) and the presence of SVMPs (Type B) based on previous RP-HPLC profiles in C. scutulatus24,46 under these conditions5. When venom was available, RP-HPLC was used as the primary means of determining venom type.
Mojave toxin assay
To confirm venom type from RP-HPLC and to determine venom type when venom was unavailable, we used PCR assays for both subunits of Mojave Toxin (MTXA and MTXB). We amplified two fragments for each subunit using the primers designed by Zancolli et al.47. These primers were developed to determine MTX presence in C. scutulatus individuals from Arizona and New Mexico, USA and have also been shown to successfully amplify these fragments from individual C. scutulatus from Mexico22. PCR amplification was conducted on each DNA sample under the following conditions per 10 μL reactions: 3.5 μL PCR water, 1 μL 10X Sigma Buffer (Sigma-Aldrich, St. Louis, MO, USA), 1 μL of 25 mM MgCL2 (Sigma-Aldrich), 1.3 μL of 2.5 mM each dNTPs, 0.5 μL of each primer at 10 μM, 0.2 μL of Taq Polymerase (Sigma-Aldrich), and 2 μL DNA. PCR was conducted on PTC200 Thermal Cycler (Bio-Rad, Hercules, CA, USA): 3.5 minutes at 94 °C, 35 cycles of 30 seconds at 94 °C, 1 minute at 57 °C, and 1 minute at 72 °C, and a final extension at 72 °C for 5 minutes. PCR product amplification was evaluated on a 2% agarorose gel using GelRed dye (Biotium, Fremont, CA, USA) to determine if the subunits were present. If one assay was positive, the individual was considered to have MTX and be Type A because all available data suggest that if these loci are present, they are also expressed in the venom22,47. We were unable to distinguish Type A and Type A + B individuals using the PCR assay because we could not determine the presence or absence of SVMPs without a venom sample.
Venom phylogeography of C. scutulatus
To determine if venom type correlated with phylogenetic lineage, we PCR amplified and sequenced the mitochondrial NADH4 (ND4) gene for any individual in our dataset not already sequenced in Schield et al.38. As outgroups, we included one sample each from C. viridis, C. cerberus, and C. oreganus, the sister species complex to C. scutulatus48 (Supplemental Table 1). We used the primers ND4 and Leu to sequence the partial ND4 gene as well as the adjacent His, Ser, and Leu tRNA genes49. PCR was conducted under the following conditions per 10 μL reaction: 3.8 μL PCR water, 1 μL 10X Sigma Buffer (Sigma-Aldrich), 1 μL of 25 mM MgCL2 (Sigma-Aldrich), 0.8 μL of 2.5 mM each dNTPs (Invitrogen, Waltham, Massachusetts, USA), 0.5 μL of each primer at 10 μM, 0.4 μL of Taq Polymerase (Sigma-Aldrich) and 2 μL DNA. PCR was conducted on a PTC200 Thermal Cycler (Bio-Rad): 3.5 minutes at 94 °C, 35 cycles of 30 seconds at 94 °C, 1 minute at 53 °C, and 1 minute at 72 °C, and a final extension at 72 °C for 5 minutes.
Amplicons were purified by adding 0.5 μL FastAP (ThermoFisher Scientific #EF0651), 0.05 μL Exonuclease 1 (ThermoFisher Scientific #EN0581), and 7.45 μL PCR water to each 30 μL reaction and then placed on the thermal cycler for 30 minutes at 37 °C followed by 15 minutes at 85 °C. Sequencing was done in both directions at Eurofins Scientific (St. Charles, Missouri, USA) on an ABI 3730 genetic analyzer (Applied Biosystems, Waltham, MA, USA). Sequences were assembled and edited in Geneious v 10.1.2 (Biomatters Ltd., Auckland, New Zealand). Alignments were created with the MAFFT v 7.22 alignment algorithm50 implemented with default parameters in Geneious. We verified alignments by eye and trimmed low quality nucleotides and also checked to ensure there were no frameshift mutations. Our final alignment was 884 nucleotides for 190 ingroup and three outgroup taxa.
We used PartitionFinder 2.1.151 to determine the best-fit model of evolution for the ND4 alignment, partitioned by codon position and between protein-coding and tRNA regions. We used the “greedy” search algorithm, Bayesian Information Criterion (BIC), linked branch lengths, and only tested models available in MrBayes. The starting tree was generated using PhyML v 3.052. These analyses were run in the UCF Advanced Research Computing Center (ARCC) on the Stokes High Performance Computer (SHPC). Best fit models from PartitionFinder2 were HKY + I for the first codon position and the tRNA together, HKY for the second codon position, and HKY + Γ for the third codon position.
To determine the mitochondrial lineage of new samples in reference to Schield et al.38, we used Bayesian Inference (BI) in MrBayes v 3.2.653. Four independent MCMC runs were conducted for 107 generations and samples taken every 500 generations. The first 25% of each run was discarded as burnin and all runs were checked in Tracer v 1.654 to ensure stationarity was reached and that all ESS values for parameters from the individual and combined runs were ≥200. We combined the runs and generated a 50% majority rule tree.
Azocasein metalloproteinase assay
To determine if there are differences within and among venom types among populations, we performed an azocasein metalloproteinase assay on 146 samples in triplicate46. These assays were performed by incubating 20 μg of venom with 1 mg of azocasein substrate in buffer composed of 50 mM HEPES and 100 mM NaCl at a pH of 8.0 for 30 minutes at 37 °C. We stopped the reaction with 250 μL of 0.5 M trichloroacetic acid, vortexed, and brought it to room temperature. We then centrifuged it at 2000 rpm for 10 minutes. Sample absorbance was read at 342 nm and reported in Δ342nm/min/mg of venom protein46. To determine the statistical significance of differences among samples at an α of 0.05, we used a Kruskal-Wallis test with venom type and lineage as factors implemented in R v. 3.4.355. If there was significance globally, we used a Nemenyi post hoc test implemented in the R package PMCMR to determine pairwise significance56.
Kallikrein-like serine protease assay
To test if there are differences within venom types for other toxin classes, we performed a kallikrein-like serine protease assay on 60 samples following the protocol of Mackessy57. This assay was conducted by adding 0.8 μg of whole venom to 373 μL of the same buffer as that used for azocasein metalloporoteinase assay described above. Samples were incubated for 3 minutes at 37 °C and then 50 μL of substrate (Bz-ProPheArg-pNA; Bachem, Torrance, CA, USA), the sample was vortexed and placed back at 37 °C for three minutes. The reaction was stopped with 50% acetic acid. Sample absorbance was read at 405 nm and the specific activity was calculated based on a standard curve of p-nitroaniline and reported as nanomoles of product produced per minute per mg of venom46. We used a Kruskal-Wallis test with venom type and lineage as factors in R to test for significant differences at an α of 0.05. If there was significance globally, we used a Nemenyi post hoc test implemented in the R package PMCMR to determine pairwise significance56.
SDS-PAGE protein gel electrophoresis
To characterize overall venom peptide diversity, we performed SDS-PAGE protein gel electrophoresis on 110 samples following the protocol of Smith and Mackessy46. We loaded 20 μg of whole venom into wells of a NuPAGE Novex bis-tris 12% acrylamide mini gel (Life Technologies, Grand Island, NY, USA) and elecrophoresed in MES buffer at 175 volts for 45 minutes. To estimate the molecular weight, we used 7 μL of Mark 12 standard. We stained gels overnight on a gentle shake with 0.1% Coomassie brilliant blue R-250 in 50% and 20% acetic acid (v/v). Gels were destained for approximately two hours in 30% methanol and 7% glacial acetic acid (v/v) in water until bands were clearly visible. Gels were gently shaken overnight at room temperature in 7% acetic acid (v/v) storage solution and imaged the following day using an HP Scanjet 4570c scanner.
Head morphology analysis
To determine if there are head morphological differences, we measured morphological characters for 57 individuals from Arizona, USA. We followed Margres et al.13 and measured SVL (snout to vent length), HL (head length), HW (head width), IF (interfang distance), and FL (fang length) for both fangs when not broken and then averaged them. We measured SVL with a tailor’s tape from the tip of the snout to the posterior end of the cloaca to the nearest 1 mm. We used IP54 digital calipers (iGaging, San Clemente, CA, USA) to measure HL, HW, IF, and FL to the nearest 0.01 mm. Head length was measured from the tip of the snout to the articular-quadrate joint, HW was the widest point behind the eye, IF was the distance between the two fang maxillae, and FL was from the top of the maxilla to the tip of the fang while folded. Average FL was determined if neither fang was broken. If one fang was broken, then the unbroken fang was used as the sole measurement; if both fangs were broken, that individual was not included in analyses of FL. All measurements were natural-log-transformed so they met the assumptions of normality and homoscedasticity. To size correct the data, we used the lnSVL value and subtracted each other value from it (ex. lnHL-lnSVL) to generate new columns for HL, HW, IF, and FL that were standardized based on the length of the snake. Using these data, we first compared all individuals based on presence or absence of Mojave Toxin which allowed us to include all samples. We then excluded animals in which we could not distinguish between Type A and Type A + B and compared individuals of the three venom types. All comparisons were done using Kruskal-Wallis tests at an α of 0.05. All transformations and analyses of morphological data were done in R v. 3.4.355. If there was significance globally, we used a Nemenyi post hoc test implemented in the R package PMCMR to determine pairwise significance56.
Niche modeling of venom type
To examine whether differences in venom type were correlated with differences in environmental variables, we constructed ecological niche models (ENMs) for the occurrence of Type A and Type B venom across the range of C. scutulatus. We used geographic localities for 81 type A and 68 type B C. scutulatus. Individuals that did not have venom but were positive for MTX were excluded from the analysis because we could not differentiate Type A vs Type A + B. ENMs were generated using MAXENT v 3.4.158 implemented in the R package dismo59. MAXENT uses occurrence records and a user-provided suite of environmental variables to predict the suitability of habitat and likelihood of occurrence across a landscape60. To limit the effect of sampling bias in the construction of EMS, we subsampled the total set of occurrence records to the same resolution as the our environmental data (30 arc seconds). This reduced the dataset to 72 and 54 representative Type A and Type B C. scutulatus occurrence records, respectively. For environmental data we used the 19 climatic variables collected in the WorldClim dataset v 1.461 as well as elevation, slope, and aspect with a 30 arc second resolution. To avoid biasing model fitting through inclusion of highly correlated inputs62, we removed 8 variables (BIO3, BIO5, BIO7, BIO10, BIO11, BIO13, BIO16, and BIO17) with a pair-wise Pearson’s correlation coefficient >0.90. BIO3 and BIO 7 were removed because they are derivatives of other bioclim variables and were correlated with them. BIO5 and BIO10 were removed because they were correlated with elevation. Elevation was kept because it was a variable specific to one of our hypotheses. BIO6 was removed in favor of BIO11 because temperature of the coldest month is less general than temperature of the coldest quarter. BIO13 and BIO16 were removed in favor of BIO12, the was more general. Lastly BIO14 was removed and BIO17 was kept, once again because BIO17 was more general. This left 14 remaining environmental variable characterizing southwestern North America. MAXENT models were run with 20 replicates and average model performance was evaluated by determining Area Under the Curve (AUC) for each model. ENMs were then generated for the Sonoran, Chihuahuan, and Central Mexican Plateau lineages independently following the same methods except only 10 replicates were used due to decreased sample size per lineage. This was done to further examine the correlations between environmental variables and venom types at finer geographic scales.
To test niche equivalency and niche similarity of Type A and Type B individual geographic distributions, we used the pseudoreplicate simulation method of Warren et al.63 as implemented in the R package phyloclim64. Both tests were run with 99 replicates to build a null distribution against which to test values of Schoener’s D and Warren’s I inferred from the full data models. To test niche equivalency, the occurrence points of two populations (e.g., individuals with Type A versus Type B venom) are combined and randomly partitioned into two datasets with sizes equal those of Type A and Type B. ENMs are generated for each dataset (e.g., a Type A model and a Type B model) and their similarity values (D and I) are calculated. This process is repeated to build a null distribution against which we test actual values of D and I inferred from each of the two full data models. The pseudoreplicate simulation method tests the null hypothesis that the two models (Type A and Type B) are not significantly different. In contrast, the test of niche similarity compares the ENMs of one venom type to an ENM of randomly selected subset of background cells (which include both presence and absence locations) of the other species. This is replicated 99 times and is then reversed such that the models for Type A venom are tested against background models for Type B venom and models for Type B venom are tested against background models for Type A venom. Comparison of the true D and I statistics to the pseudoreplicate distributions tests the hypothesis that one venom type’s niche model predicts the occurrences of the other venom type more than expected by chance. In addition, to more directly test for presence and absence differences as relating to specific environmental variables, we used logistic regression on the 14 variables used for ENMs. This approach was used to distinguish variables that may be important for C. scutulatus’ ecology, but may not be correlated with differences between venom types. Niche equivalency, niche similarity, and logistic regression of the model variables were then estimated for the Sonoran, Chihuahuan, and Central Mexican Plateau lineages independently to determine if the global pattern was the same at finer geographic scales.
Accession codes
Results
Venom type in C. scutulatus
For the 114 samples from which both venom and blood were sampled, the protein-based RP-HPLC venom type assay and the PCR-based MTX assay were in agreement in either detecting or not detecting MTX; no individuals that lacked MTX in the RP-HPLC profile had a positive result in the PCR assays (see representative examples in Fig. 2). All samples that had both subunits of MTX in their venom protein profile were positive for MTX in at least one PCR assay, and all but seven individuals were positive for all four MTX PCR assays. For the 68 samples that only had DNA available, all but five were unanimous for venom type. A total of 12 samples, likely due to DNA quality, were not positive for all four assays: two were positive for three of four assays, seven were positive for two of four, and three were positive for one assay. In total, 144 samples were positive for Mojave Toxin and 72 were negative for Mojave Toxin (Supplemental Table 1). Excluding samples in which Type A and Type A + B could not be distinguished, we had 86 Type A, 51 Type B, and 11 Type A + B (7.43% Type A + B). For the 47 individuals that were positive for the PCR MTX assay, we could not differentiate between Type A and Type A + B. We indicate when these are included in our analyses and when they are excluded.
Figure 2.
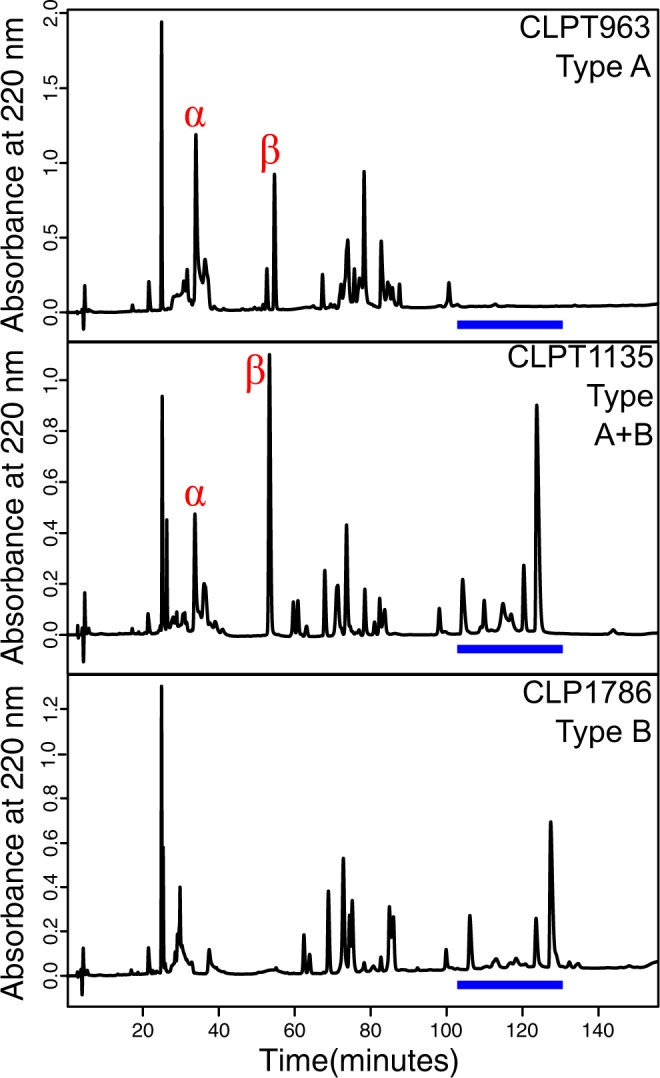
Representative Reverse-phased High Performance Liquid Chromatography (RP-HPLC) profiles of Type A (top), Type A + B (middle), and Type B (bottom) venom of Mojave Rattlesnakes. The acidic (α) and basic subunit (β) peaks for Mojave toxin are marked and the region where snake venom metalloproteinases elute is marked with a blue bar. Type B individuals lack both subunits of Mojave toxin.
Population genetic structure
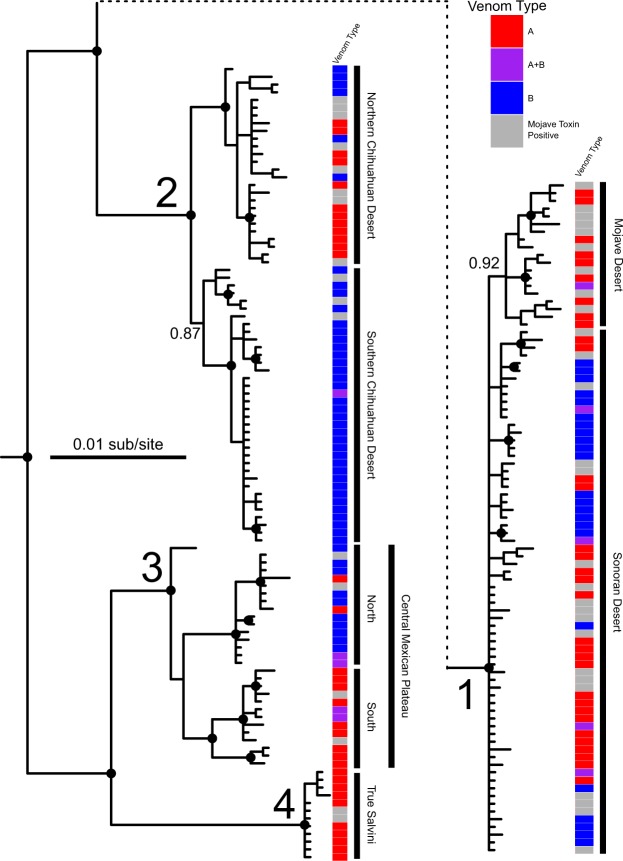
As expected, our mitochondrial ND4 phylogeny of individuals was highly similar to that of Schield et al.38 in recovering four main clades of haplotypes within C. scutulatus (Fig. 3). Because of the addition of more samples from the Central Plateau of Mexico, we identified potential substructure in this region (Fig. 3). Based on where each individual was in our phylogeny, we were able to assign each individual to a population and test if there were difference in venom characteristics that corresponded to shared ancestry. For our analyses, we used the three lineages with both Type A and Type B venom to compare populations and venom type. Only the lineage corresponding to C. scutulatus salvini was monotypic for venom type (Fig. 3).
Figure 3.
Bayesian Inference phylogeny based on ND4 sequence from 190 C. scutulatus. The dashed line indicates where the Sonoran and Mojave Desert lineage was moved from and no size adjustments occurred. Venom type as a discrete character is mapped onto the phylogeny. Dots on nodes represent significant posterior probability values of ≥0.95. The four primary clades are numbered as in Fig. 1 and subpopulations are denoted by bars and labeled.
Geographic and phylogenetic distribution of venom types
We find that venom types are variable within three of the four major lineages of C. scutulatus, and that the Type A phenotype is fixed only in the C. scutulatus salvini lineage (Figs 1 and 3). Within these major lineages, geographically-defined populations tend to be nearly fixed or fixed for a single venom type (i.e., Type A or Type B), and there is very little evidence of populations with highly polymorphic venom composition (Table 1 and Figs 1 and 3). Populations containing individuals with mixed Type A + B venom tend to occur in regions that effectively represent hybrid zones based on Schield et al.38, as these are regions were estimated to share substantial gene flow. Thus, the populations that exhibit phenotypic polymorphism in venom tend to be those that occur at the interface of populations that exchange gene flow. Otherwise, venom type tends to be fixed in a particular population outside of these zones (Table 1). Intriguingly, even within regions inferred to be panmictic based on nuclear data from Schield et al.38 (e.g., populations in Arizona; Fig. 1), we observe geographic subpopulations that are differentially fixed for one venom type or the other, and find little evidence for multiple venom types within the same geographically-distinct population (Figs 1 and 3). For example, the northern Chihuahuan Desert (Texas, USA) is fixed for Type A and all but one of the southern Chihuahuan Desert individuals is Type B (Table 1). Where these two lineages come into contact, there is a transition and both venom types occur. Additionally, all but one individual in the Mojave Desert (California and NW Arizona, USA) was Type A (Figs 1 and 3).
Table 1.
Percentage and number of each venom type for the four lineages as well as the subpopulations within the first three lineages.
| Type A | Type A + B | Type B | MTX+ | ||||
|---|---|---|---|---|---|---|---|
| % | No. | % | No. | % | No. | ||
| Lineage 1 | 65.43 | 53 | 6.17 | 5 | 28.4 | 23 | 28 |
| Sonoran | 53.45 | 31 | 6.9 | 4 | 39.66 | 23 | 19 |
| Mojave | 95.65 | 22 | 4.35 | 1 | 0 | 0 | 9 |
| Lineage 2 | 20.41 | 10 | 2.04 | 1 | 77.55 | 38 | 13 |
| Northern Chihuahuan | 62.5 | 10 | 0 | 0 | 37.5 | 6 | 10 |
| Southern Chihuahuan | 0 | 0 | 3.03 | 1 | 96.97 | 32 | 3 |
| Lineage 3 | 42.86 | 12 | 17.86 | 5 | 39.29 | 11 | 4 |
| North | 7.14 | 1 | 14.29 | 2 | 78.57 | 11 | 1 |
| South | 78.57 | 11 | 21.43 | 3 | 0 | 0 | 3 |
| Lineage 4 (salvini) | 100 | 11 | 0 | 0 | 0 | 0 | 2 |
Additionally, the number of individuals that were positive for the Mojave Toxin (MTX+) assay but could not be distinguished between Type A and Type A + B venom are indicated.
Venom characterization and activity assays
Metalloproteinase activity was significantly different between venom types (χ2 = 112.54, df = 2, p < 0.01) with Type A lower than both Type A + B (p < 0.01) and Type B (p < 0.01) and Type A + B and Type B not different (p = 0.11) using the Nemenyi post hoc test. Owing to the higher proportion of Type B individuals in the Chihuahuan Lineage, there was a significant difference among lineages (χ2 = 24.158, df = 2, p < 0.01) with the Chihuahuan Lineage being higher than the Sonoran (p < 0.01) and Central Mexican Plateau Lineage (p < 0.01) but the latter two not being different from each other (p = 0.94). Metalloproteinase activity was not different across lineages for Type A (χ2 = 1.33, df = 2, p = 0.515), Type A + B (χ2 = 0.54, df = 2, p = 0.76), or Type B (χ2 = 0.79, df = 2, p = 0.67) venom (Fig. 4). Type A venom metalloproteinase activity ranged from 0 to 0.668 Δ342nm/min/mg (n = 86, Average = 0.052, Median = 0.020), Type A + B from 0.483 to 1.275 Δ342nm/min/mg (n = 10, Average = 0.816, Median = 0.919), and Type B from 0.208 to 2.043 Δ342nm/min/mg (n = 50, Average = 0.945, Median = 0.919). Eleven of the Type A individuals had metalloproteinase activity values above 0.1 including six that had higher activity than the lowest Type B individual (CLP1834 = 0.208) but did not have SVMP peaks in their RP-HPLC profiles. These individuals may be functionally more similar to Type A + B individuals.
Figure 4.
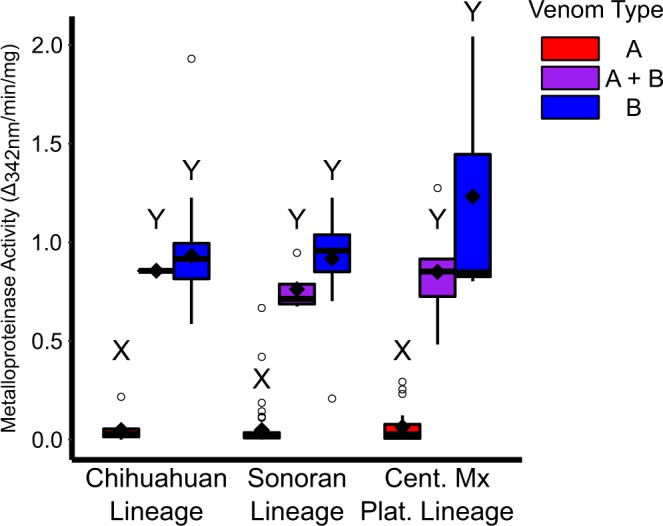
Metalloproteinase activity levels for venom types within each lineage. Type A venom had significantly less metalloproteinase activity regardless of lineage. Type B and A + B were not significantly different from each other in any comparison. Letters above bars (X and Y) indicate significance at an α of 0.05.
We only had data from the Sonoran and Chihuahuan lineages for Kallikrein-like activity (Fig. 5). Kallikrein-like serine protease activity was highly variable overall and was significantly different among venom types (χ2 = 8.45, df = 2, p = 0.01) with Type B being significantly lower than Type A (p = 0.01) but no different that Type A + B (p = 0.66) and Type A + B and Type A not being different (p = 0.66). There was no significant difference between the two lineages (χ2 = 0.24, df = 1, p = 0.62). Within the Chihuahuan lineage, there was significant differences among venom types (χ2 = 10.05, df = 2, p < 0.01; Fig. 5) with Type B being lower than Type A (p = 0.02) but not significantly different than Type A + B (p = 0.07). Type A and Type A + B were also not different (p = 0.899). Within the Sonoran Lineage, there was no difference among venom types (χ2 = 3.22, df = 2, p = 0.20; Fig. 5). Overall, Type A values ranged from 132.77 to 733.20 nmol product/min/mg (n = 28, Average = 386.58, Median = 380.10), Type A + B from 216.87 to 382.77 nmol product/min/mg (n = 5, Average = 301.54, Median = 298.30), and Type B from 70.69 to 469.24 nmol product/min/mg (n = 27, Average = 277.231, Median = 285.86).
Figure 5.
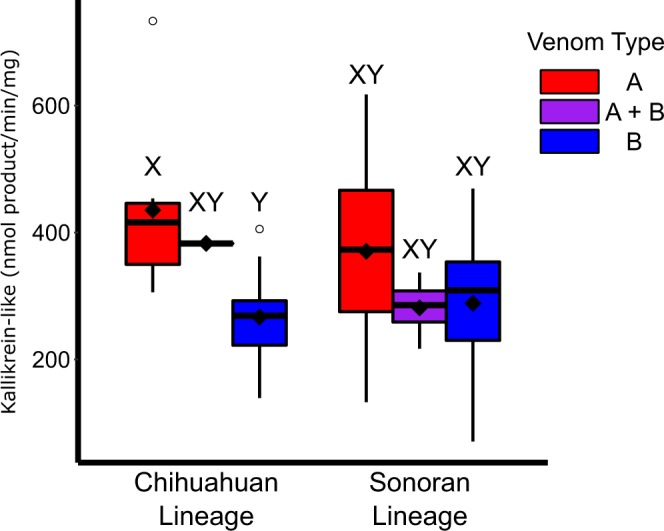
Kallikrein-like serine protease activity between the Chihuahuan and Sonoran lineages. Type B venom had significantly lower activity than Type A venom in the Chihuahuan lineage but there were no differences in the same venom type between the two populations and Type A + B venom was not significantly different from Type A or Type B venom. Letters above bars (X and Y) indicate significance at an α of 0.05.
SDS-PAGE confirmed venom type for all samples. The basic subunit of MTX was clear at 14 kD in Type A and Type A + B individuals and absent in Type B individuals (Figs S1 and S2). Additionally, SVMPs were clearly identifiable in Type B and Type A + B individuals at 55 kD and ~22 kD and absent in Type A individuals. Myotoxins at 6 kD were found in 56 of 110 individuals tested and were generally linked with Type A individuals in the Sonoran lineage and Type B individuals in the Chihuahuan lineage. SDS-PAGE illustrated additional diversity in C-type lectins and non MTX Phospholipase A2s as well as the uniformity in snake venom serine proteases and cysteine rich secretory proteins (Figs S1 and S2).
Morphological analysis
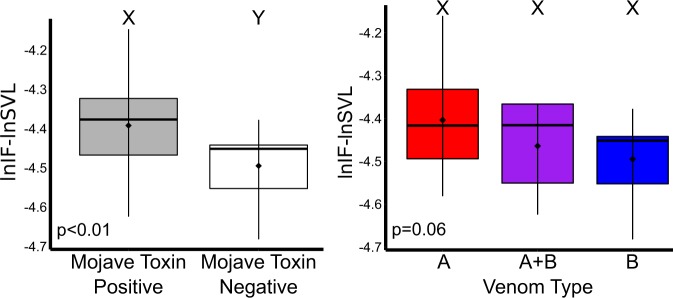
All 57 individuals used in this analysis were from the Sonoran lineage. Thus, they are all genetically similar and provide the best comparison of potential morphological differences associated with the presence of MTX and venom type. We removed eight individuals that were salvaged when comparing venom type because we could not determine if they were Type A or Type A + B. We did not find significant differences between head width based on the presence of MTX (χ2 = 0.72, df = 1, p = 0.40) or based on venom type (χ2 = 0.09, df = 2, p = 0.96). Head length was also not significant based on MTX presence (χ2 = 1.52, df = 1, p = 0.22) or venom type (χ2 = 1.53, df = 2, p = 0.46). We did find a significant difference in interfang distance based on the presence of MTX (χ2 = 7.12, df = 1, p < 0.01) but only marginal significance based on venom type (χ2 = 5.59, df = 2, p = 0.06; Fig. 6). Mojave Toxin positive individuals had a larger distance between the fangs than Mojave Toxin negative individuals. Additionally, there was a trend for MTX positive individuals to have longer fangs than MTX negative individuals (χ2 = 2.02, df = 1, p = 0.15) and for Type A individuals to have longer fangs than Type A + B and Type B individuals (χ2 = 3.97, df = 2, p = 0.14).
Figure 6.
Comparison of Interfang Distance (IF) based on presence and absence of Mojave Toxin (left) and by venom type (right) in the Sonoran lineage of Mojave Rattlesnakes. Snout-vent Length (SVL) was used to control for different sizes among animals. Smaller values on the y-axis (more negative) are smaller measurements. Mojave Toxin positive individuals had significantly wider distances between their fangs compared to Mojave Toxin negative individuals. After excluding animals in which Type A and Type A + B could not be distinguished because they were salvaged, there was marginal significance of interfang distance based on venom type with a trend of Type A individuals having a wider distance between fangs. Letters above bars (X and Y) indicate significance at an α of 0.05.
ENMs between venom type
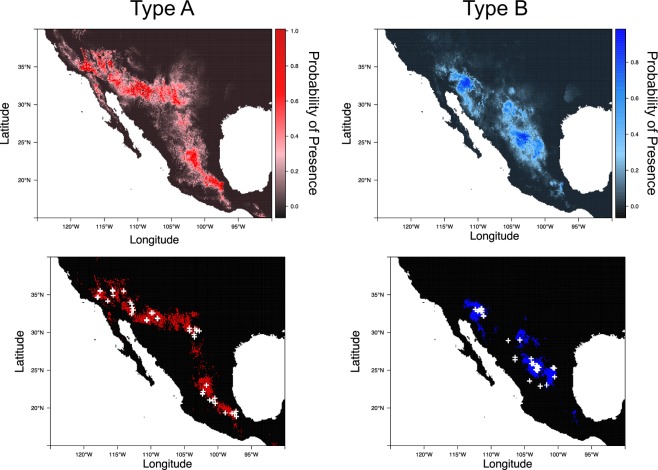
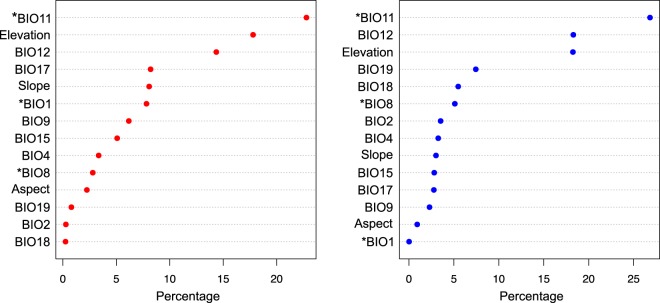
We found significant differences between the variables, particularly in regard to temperature for the ecological niche models (ENM) created for Type A and Type B individuals (Table 2 and Fig. 7). The ENM for Type A and Type B were not equivalent to each other (Fig. S3) but they were more similar than would be expected by chance (Fig. S7). Area under the curve (AUC) from comparison of the model and the Type B (AUC = 0.987) C. scutulatus was comparable to AUC’s from Type A test data (AUC = 0.907), indicating similarity in predictive power. The WorldClim bioclimatic variable BIO11 (Minimum temperature of the coldest quarter) explained the most variation for both the Type A and Type B models and was significantly different between the two models (Fig. 8 and Table 2). The other variables that were significantly different were BIO1 (annual mean temperature) and BIO8 (mean temperature of wettest quarter); the contribution of these variables differed between models (Fig. 8 and Table 2).
Table 2.
Logistic regression comparison of the 14 variables used in the Type A and Type B models for C. scutulatus.
| Independent Variable | All Samples | Sonoran | Chihuahuan | Cent. MX Plat. | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Df | F-value | P-value | Df | F-value | P-value | Df | F-value | P-value | Df | F-value | P-value | |
| BIO1 | 124 | −4.988 | <0.001 | 56 | −3.691 | <0.001 | 39 | −2.622 | 0.008 | 19 | 1.852 | 0.064 |
| BIO2 | 124 | −1.583 | 0.114 | 56 | −1.637 | 0.102 | 39 | −0.435 | 0.663 | 19 | 0.637 | 0.524 |
| BIO4 | 124 | 1.383 | 0.167 | 56 | −1.748 | 0.081 | 39 | 1.264 | 0.206 | 19 | −2.257 | 0.024 |
| BIO8 | 124 | −3.411 | <0.001 | 56 | −2.480 | 0.013 | 39 | −0.285 | 0.775 | 19 | 0.416 | 0.677 |
| BIO9 | 124 | −0.780 | 0.435 | 56 | −3.212 | 0.001 | 39 | −2.766 | 0.006 | 19 | 1.912 | 0.056 |
| BIO11 | 124 | −5.045 | <0.001 | 56 | −3.696 | <0.001 | 39 | −3.272 | 0.001 | 19 | 2.107 | 0.035 |
| BIO12 | 124 | 0.654 | 0.513 | 56 | −2.226 | 0.026 | 39 | 1.395 | 0.163 | 19 | 1.782 | 0.075 |
| BIO15 | 124 | −0.515 | 0.607 | 56 | 3.276 | 0.001 | 39 | −2.604 | 0.009 | 19 | 2.115 | 0.034 |
| BIO17 | 124 | −0.882 | 0.378 | 56 | −3.132 | 0.002 | 39 | 3.368 | <0.001 | 19 | −1.282 | 0.200 |
| BIO18 | 124 | −1.296 | 0.195 | 56 | −0.879 | 0.379 | 39 | 0.383 | 0.701 | 19 | 0.781 | 0.435 |
| BIO19 | 124 | 1.167 | 0.243 | 56 | −2.460 | 0.014 | 39 | 1.064 | 0.287 | 19 | −2.190 | 0.029 |
| Aspect | 124 | 1.369 | 0.171 | 56 | −0.995 | 0.320 | 39 | 2.444 | 0.015 | 19 | −0.250 | 0.802 |
| Elevation | 124 | 0.575 | 0.565 | 56 | 3.308 | <0.001 | 39 | −1.039 | 0.299 | 19 | −1.309 | 0.190 |
| Slope | 124 | 0.115 | 0.909 | 56 | 2.172 | 0.030 | 39 | 0.183 | 0.855 | 19 | −0.60 | 0.549 |
We used a full model and then compared the three lineages for which Type A and Type B individuals were identified. Bolded P-values were significantly different between the two models in a comparison. BIO1–BIO9 correspond to measures related to temperature and BIO12-BIO19 correspond to measures related to precipitation.
Figure 7.
Ecological Niche Models generated in MAXENT58 using 72 Type A (left) and 54 Type B (right) Mojave Rattlesnakes scaled by probability of presence (pp). Lower maps display model distributions where each venom type is expected to occur based on a threshold point where model sensitivity and specificity are highest (pp > 0.48 for A’s, pp > 0.24 for B’s).
Figure 8.
Model response to variables included in the Ecological Niche Models from MAXENT58 for Type A (left) and Type B (right) venoms. Asterisks (*) indicate variables that were significantly different between the two models. BIO1 = Annual Mean Temperature, BIO2 = Mean Diurnal Range (Mean of monthly (max temp − min temp)), BIO4 = Temperature Seasonality (standard deviation *100), BIO8 = Mean Temperature of Wettest Quarter, BIO9 = Mean Temperature of Driest Quarter, BIO11 = Mean Temperature of Coldest Quarter, BIO12 = Annual Precipitation, BIO15 = Precipitation Seasonality (Coefficient of Variation), BIO17 = Precipitation of Driest Quarter, BIO18 = Precipitation of Warmest Quarter, BIO19 = Precipitation of Coldest Quarter.
Lineage-specific ENMs varied in both the differentiation of models and in the variables that differed between venom types (Figs S3–S16, Table 2). Lineage-specific tests of correlation between environmental variables showed high variation in the variables found to be significantly different between venom types and the direction of their effects (Table 2). However, BIO11 was significant in all models and BIO1 was found to be highly significant across the full model, the Sonoran lineage model, and the Chihuahuan lineage and marginally significant for the Central Mexican Plateau lineage (Table 2).
Discussion
The venom phenotype dichotomy exhibited in Mojave Rattlesnakes was first described in the 1930’s based on different symptoms of snakebite65. Subsequent studies of the distribution of venom types led to the conclusion that C. scutulatus has neurotoxic venom through the majority of their range15,21,27,37. Our data indicate that this century-long generalization is not accurate; both Type A and Type B phenotypes occur throughout the distribution of this species, and the geographic pattern of venom composition is much more complex than previously thought (Fig. 1). In this study, we provide the first range-wide analyses of venom composition variation in C. scutulatus, and our integration of venom characterization with new data on population structure have synergistically increased our ability to understand the patterns of evolution underlying and perpetuating the venom dichotomy in this species.
Previous studies examining the distribution of venom types in C. scutulatus lacked any population genetic context with which to interpret range-wide variation in venom. Schield et al.38 identified four major lineages within C. scutulatus, each of which shares gene flow with adjacent lineages, and within which there is inferred near-panmixia. In this study, we have demonstrated that all but one of the major phylogeographic lineages identified by Schield et al.38 possess Type A, Type A + B, and Type B individuals (Table 1 and Figs 1 and 3); the C. scutulatus salvini lineage appears fixed for Type A venom based on 13 individuals sampled. Combining our venom characterizations with population genetic patterns in C. scutulatus highlights two key features of venom variation within this system. The first is that, in the three lineages that possess each of the venom types, individuals with Type A + B venom tend to occur in regions where Type A and Type B venom types come into contact (Fig. 1). In one case, this corresponds to an area of introgression where the Chihuahuan and Central Mexican Plateau lineages contact one another and exchange genes38 (Fig. 1). This region contains a high concentration of Type A + B individuals. Collectively, these patterns suggest that Type A + B venom may represent a product of introgression between venom types rather than local selection for both types. The second key feature is that populations identified in Schield et al.38 appear to contain geographically segregated subpopulations that are differentially fixed for either Type A or Type B venom (Table 1 and Figs 1 and 3).
Since the discovery of the C. scutulatus venom system, different mechanisms have been speculated to drive the observed geographic variation in venom15–18. One mechanism that has been proposed to explain this apparently maintained Type A and Type B venom dichotomy is balancing selection (e.g., Dowell37). Our results provide convincing evidence against balancing selection in favor of differential local selection driving venom variation in C. scutulatus. While balancing selection should lead to frequent occurrences of observing both Type A and Type B individuals in a single population66–68, our data highlight a very different pattern in which geographically segregated subpopulations are largely fixed for a single venom type (i.e., Type A or Type B populations; Table 1). This pattern is instead most consistent with strong directional local selection that differs across populations by favoring one venom type over the other (Fig. 1), and that differential local selection may occur at a very fine geographic scale. We also find evidence that directional selection likely counteracts introgression resulting in mixed venom phenotypes, despite substantial gene flow within and between major C. scutulatus lineages. This is supported by the apparent rarity of the mixed venom phenotype across the broad range of C. scutulatus (i.e., only 11 of 148 individuals with Type A + B venom based on proteomic data). This pattern suggests that selection may be quite efficient in maintaining not only one venom type over the other, but also efficient in limiting introgression of genomic regions underlying the mixed venom phenotype.
A primary goal of this study was to identify biotic and abiotic factors that may predict the geographic variation in venom composition. We found evidence for correspondence between the presence or absence of Mojave Toxin and other trophic factors (e.g., inter-fang distance) within the Sonoran lineage (Fig. 6), suggesting possible selection on the integrated components of trophic function (e.g., Margres et al.13) to accommodate different prey dimensions and possibly different species. This suggests co-evolution, and potentially local and differential selection on trophic function that incorporates both venom and fang morphology. However, because we were only able to test this in one lineage, additional comparisons between these features in other C. scutulatus lineages, or other rattlesnake lineages, would be useful for testing our hypothesis and determining if there are broad patterns of co-evolution between venom and other trophic phenotypes or if other evolutionary mechanisms, such as drift, are acting on head morphology. If so, these data would further suggest that venom phenotype, along with other trophic adaptations, may be driven by local selection for different prey types or feeding behaviors.
We also found evidence for correlations between environmental variables, especially those related to temperature, (Table 2 Fig. 8) and venom phenotype. A correspondence between venom phenotype and climate is also recapitulated by the surprising evidence for distinct ecological niches for Type A and Type B phenotypes (Fig. 7). Moreover, the spatial distinction between Type A and Type B animals remained clear for Sonoran and Chihuahuan lineages. Though the specific variables of greatest contribution and significant difference between venom types varied across lineage specific models, this likely reflects the broad environmental heterogeneity across the range of C. scutulatus. The consistent recovery of temperature related variables, especially annual mean temperature and minimum temperature of the coldest quarter, as differing between venom types despite the broader environmental differences underscores its apparent relevance to the venom phenotype dichotomy (Table 2). While the precise link between environmental temperature parameters and venom composition is not clear, these environmental variables may be predictive of other important factors driving selection for different venom phenotypes. For example, annual temperature in different regions throughout the range of C. scutulatus may be a strong predictor of prey species distributions, behaviors, and physiological activities. These features of local prey populations may in turn impose substantial selection pressures on the venom of C. scutulatus populations29,30,47. A second but non-exclusive possibility is that venom composition and temperature are linked because Type B venom may be beneficial in cooler areas due to its potential digestive benefits8,33, including more rapid digestion by these ectothermic predators of prey in cooler conditions.
Perhaps one of the most surprising findings in our study is the apparent rarity of Type A + B venom in C. scutulatus, despite evidence for the otherwise strong selection for different venom types in different geographic regions. This rarity is intriguing because both major components in this dichotomy each have positive benefits4,6,69, and the presence of both could potentially have additive benefits for securing prey. The finding that Type A + B individuals are rare suggests, instead, that there are negative fitness consequences when both neurotoxic and hemorrhagic components are present and expressed within the venom of same individual. This conclusion is strengthened by consideration of the broad trend in venom phenotypes within the entire clade of rattlesnakes4,8. Across Sistrurus and Crotalus species, venom phenotype is either type I or type II (analogous to Type B and Type A in C. scutulatus venoms, respectively), with essentially no intermediate examples8. Why might this be? Neonate rattlesnakes often show an ontogenetic shift in composition from a type II venom to a type I venom as adults34, and type II venoms persisting into adulthood (C. oreganus concolor, C. scutulatus, C. tigris etc.) have been suggested to result from paedomorphosis70. This provides an intriguing new perspective on venom evolution that is often not considered. Specifically, while patterns of selection with respect to venom evolution are often viewed in the context of positive selection, our results, in light of the above trends, indicate that negative selection against “intermediate” or “mixed” venoms also play an important role in the evolution of venom composition.
Conclusion
Our characterization of C. scutulatus venom phenotypes throughout its range has provided among the most complete appraisals of venom variation in this species to date, and provides new interesting questions for the study of venom variation and evolution overall. We found that the geographic pattern of the three venom phenotypes in C. scutulatus is much more complex than previously hypothesized, and were able to rule out the fixation of venom phenotypes in major C. scutulatus lineages. Instead, fixation of venom phenotypes occurs at a much more fine geographic scale. The integration of venom phenotype data with recent data on population structure and gene flow provides evidence that venom variation in C. scutulatus is not likely the product of balancing selection, but instead likely representative of strong directional local selection acting on distinct venom loci (PLA2 and SVMP gene clusters) in geographically highly differentiated ways. We also find evidence that the mixture of venom phenotypes is likely the result of introgression between populations fixed for Type A and Type B venom, and that this admixture is rare. Links between venom phenotypes, fang morphology, and climatic variables represent intriguing areas for further study to determine the broad relevance of these features in shaping venom evolution.
Electronic supplementary material
Acknowledgements
For support and assistance in the field, J.L.S. would like to thank J. Aceves, J. Adams, I. Ahumada, L. Badillo, M. Bernard, B. Bloom, T. Burkhardt, J. Castañeda-Gaytán, B. Chambers, H. Dahn, D. Deem, T. Dimler, B. Eaton, R. Engeldorf, E. Fanti, M. Feldner, T. Fisher, E. Flores, H. Franz, F. García, R. Govreau, C. Grünwald, A. Gutierrez, F. Hernandez, M. Holding, J. Houck, W. Howell, C. Howell, T. Jones, J. Jones, T. LaDuc, P. Lindsey, M. Linsalata, J. Lock, C. Mallery, M. Margres, C. May, S. May, R. Mayerhofer, E. McCormick, C. McMartin, J. McNally, C. Moreno, D. Ortiz, A. Owens, T. Petty, M. Price, A. Quillen, G. Quintero, R. Ramirez, R. Rautsaw, E. Rivas, M. Rodriguez, B. Rodriguez, A. Salas, G. Salmon, J. Slone, R. Solis, D. Speckin, S. Stevens, R. Swanson, K. Swanson, E. Swanson, S.S. Sweet, G. Territo, B. Townsend, C. Trumbower, S. Valenzuela, K. VanSooy, I. Villalobos, C. Vratil, D. Weber, K. Wray, W. Wüster, G. Zancolli, and G. Zavala. C.L.S. would like to thank J. Jesús Sigala Rodríguez, Robert Bryson Jr., R. Hernández Árciga, E. Pérez-Ramos, C. A. Hernández Jiménez, L. Felipe Vázquez Vega, L. Fernando Dávila Galavíz, E. Alejandro García Trejo, G. E. Quintero Díaz, V. Bernal, C. Edith Esparza Estrada, J. Lenin Lara Galván, R. Rosales García, J. José Ayala Rodríguez, A. Guadalupe López Campos, C. Estefanía López Campos, A. Martínez Méndez, G. Contreras Cobos, and A. Álvarez Trillo (and his students from the Herpetário del Zoológico de Zacango, Estado de México) for assistance in the field collecting and/or providing access to live animals and venom. We thank M. Hogan, M. Margres, J. McGivern, and M. Ward from the Rokyta Lab and M. Seavy (FSU Department of Biological Science Analytical Lab) for assistance with RP-HPLC. Comments and suggestions were given by E. Hofmann, E. Hoffman, R. Rautsaw, and A. Savage which greatly improved the quality of this manuscript. We thank the following museums for providing tissues samples and/or cataloging specimens used in this study: Angelo State Natural History Collections, Arizona State University Natural History Collections, California Academy of Sciences, Museo de Zoología de la Facultad de Ciencias, Museum of Vertebrate Zoology, University of Texas at Arlington Amphibian and Reptile Diversity Center, and Yale Peabody Museum. Financial support was generously provided by the National Science Foundation to C.L.P. (DUE 1161228 which funded J.L.S. and A.J.M., DEB 1638879, and DEB 1822417), to D.R.R. (DEB 1145987 and DEB 1638902), to T.A.C. and S.P.M. (DEB 1655571), and to D.R.S. and T.A.C. (DDIG Grant DEB-1501886). Additional support was provided by UC MEXUS-CONACYT to C.L.S. and O.F.V. (CN-11-548), Clemson University and University of Texas at Arlington through faculty startup to C.L.P. and T.A.C., respectively, Consejo Nacional de Ciencia y Tecnologia to M.B. (CONACYT 247437), Consejo de Ciencia y Tecnología del Estado de Durango for support with project COCYTED 2018 01 15088 to M.B. and G.C.G., Phi Sigma Beta Phi Chapter (D.R.S.), Prairie Biotic Research Inc. (J.L.S.), Sigma Xi Grants-in-aid-of-research (J.L.S.), SnakeDays Research Grant (J.L.S.), the Southwestern Association of Naturalists McCarley Research Grant (J.L.S., A.J.M. and M.B.), and the Theodore Roosevelt Memorial Fund through the American Museum of Natural History (A.J.M. and J.L.S.).
Author Contributions
J.L.S., C.L.S., O.F.V., S.P.M., T.A.C., D.R.R. and C.L.P. designed the study. J.L.S., M.B., G.C.G., C.L.S., L.L.S., A.T., N.M.B., G.C.G., O.F.V., D.A.R. and S.P.M. collected samples. J.L.S., C.F.S., A.J.M., D.R.S., M.B., S.P.M., D.R.R. and C.L.P. generated data. J.L.S., C.F.S. and A.J.M. analyzed the data. C.L.S., O.F.V., S.P.M., T.A.C., D.R.R. and C.L.P. provided materials, reagents, computational resources, and lab space. J.L.S, A.J.M., D.R.S., T.A.C., D.R.R. and C.L.P. wrote the manuscript. All authors edited, reviewed, and approved the final manuscript.
Data Availability
New ND4 sequences generated in this study were deposited in GenBank under accession numbers MH883648-MH883754. Specimen vouchers were deposited in the appropriate museums based on permit requirements. All specimen data including morphology and assay data are listed in Supplemental Table 1.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-35810-9.
References
- 1.Tigano A, Friesen VL. Genomics of local adaptation with gene flow. Molecular Ecology. 2016;25:2144–2164. doi: 10.1111/mec.13606. [DOI] [PubMed] [Google Scholar]
- 2.Pinho C, Hey J. Divergence with gene flow: models and data. Annual Review of Ecology, Evolution, and Systematics. 2010;41:215–30. doi: 10.1146/annurev-ecolsys-102209-144644. [DOI] [Google Scholar]
- 3.Cox CL, Davis Rabosky AR. Spatial and temporal drivers of phenotypic diversity in polymorphic snakes. The American Naturalist. 2013;182:E40–E57. doi: 10.1086/670988. [DOI] [PubMed] [Google Scholar]
- 4.Mackessy, S. P. Venom composition in rattlesnakes: trends and biological significance. In Hayes, W. K., Beaman, K. R., Cardwell, M. D. & Bush, S. P. (eds) The Biology of Rattlesnakes, 495–510 (Loma Linda University Press, Loma Linda, CA, 2008).
- 5.Rokyta DR, Wray KP, McGivern JJ, Margres MJ. The transcriptomic and proteomic basis for the evolution of a novel venom phenotype within the Timber Rattlesnake (Crotalus horridus) Toxicon. 2015;98:34–48. doi: 10.1016/j.toxicon.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 6.Casewell NR, Wüster W, Vonk FJ, Harrison RA, Fry BG. Complex cocktails: the evolutionary novelty of venoms. Trends in ecology & evolution. 2013;28:219–29. doi: 10.1016/j.tree.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 7.Daltry JC, Wüster W, Thorpe RS. Diet and snake venom evolution. Nature. 1996;379:537–540. doi: 10.1038/379537a0. [DOI] [PubMed] [Google Scholar]
- 8.Mackessy SP. Evolutionary trends in venom composition in the Western Rattlesnakes (Crotalus viridis sensu lato): toxicity vs. tenderizers. Toxicon. 2010;55:1463–74. doi: 10.1016/j.toxicon.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 9.Munekiyo SM, Mackessy SP. Presence of peptide inhibitors in rattlesnake venoms and their effects on endogenous metalloproteases. Toxicon. 2005;45:255–263. doi: 10.1016/j.toxicon.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Mackessy SP, Baxter LM. Bioweapons synthesis and storage: The venom gland of front-fanged snakes. Zoologischer Anzeiger - A Journal of Comparative Zoology. 2006;245:147–159. doi: 10.1016/j.jcz.2006.01.003. [DOI] [Google Scholar]
- 11.Rokyta DR, Lemmon AR, Margres MJ, Aronow K. The venom-gland transcriptome of the Eastern Diamondback Rattlesnake (Crotalus adamanteus) BMC genomics. 2012;13:312. doi: 10.1186/1471-2164-13-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kerkkamp Harald M. I., Casewell Nicholas R., Vonk Freek J. Evolution of Venomous Animals and Their Toxins. Dordrecht: Springer Netherlands; 2015. Evolution of the Snake Venom Delivery System; pp. 1–11. [Google Scholar]
- 13.Margres MJ, et al. Phenotypic integration in the feeding system of the Eastern Diamondback Rattlesnake (Crotalus adamanteus) Molecular Ecology. 2015;24:3405–3420. doi: 10.1111/mec.13240. [DOI] [PubMed] [Google Scholar]
- 14.Glenn J, Straight R, Wolt T. Regional variation in the presence of canebrake toxin in Crotalus horridus venom. Comparative Biochemistry and Physiology Part C: Pharmacology, Toxicology and Endocrinology. 1994;107:337–346. doi: 10.1016/0305-0491(94)90057-4. [DOI] [PubMed] [Google Scholar]
- 15.Glenn JJL, Straight RC, Wolfe MMC, Hardy DDL. Geographical variation in Crotalus scutulatus scutulatus (Mojave Rattlesnake) venom properties. Toxicon. 1983;21:119–130. doi: 10.1016/0041-0101(83)90055-7. [DOI] [PubMed] [Google Scholar]
- 16.Glenn J, Straight R. Mojave Rattlesnake Crotalus scutulatus scutulatus venom: variation in toxicity with geographical origin. Toxicon. 1978;16:81–84. doi: 10.1016/0041-0101(78)90065-X. [DOI] [PubMed] [Google Scholar]
- 17.Weinstein SA, Minton SA, Wilde CE. The distribution among ophidian venoms of a toxin isolated from the venom of the Mojave Rattlesnake (Crotalus scutulatus scutulatus) Toxicon. 1985;23:825–844. doi: 10.1016/0041-0101(85)90014-5. [DOI] [PubMed] [Google Scholar]
- 18.Wooldridge B, et al. Mojave Rattlesnakes (Crotalus scutulatus scutulatus) lacking the acidic subunit DNA sequence lack Mojave toxin in their venom. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology. 2001;130:169–179. doi: 10.1016/S1096-4959(01)00422-5. [DOI] [PubMed] [Google Scholar]
- 19.Borja M, et al. Mojave Rattlesnake (Crotalus scutulatus scutulatus) with Type B venom from Mexico. Copeia. 2014;2014:7–13. doi: 10.1643/OT-12-041. [DOI] [Google Scholar]
- 20.Aird SD, Kruggel WG, Kaiser II. Amino acid sequence of the basic subunit of mojave toxin from the venom of the mojave rattlesnake (Crotalus s. scutulatus) Toxicon. 1990;28:669–673. doi: 10.1016/0041-0101(90)90255-6. [DOI] [PubMed] [Google Scholar]
- 21.Wilkinson JA, Glenn JL, Straight RC, Sites JW. Distribution and genetic variation in venom A and B populations of the Mojave Rattlesnake (Crotalus scutulatus scutulatus) in Arizona. Herpetologica. 1991;47:54–68. [Google Scholar]
- 22.Borja M, et al. Biological and proteolytic variation in the venom of Crotalus scutulatus scutulatus from Mexico. Toxins. 2018;10:35. doi: 10.3390/toxins10010035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dobson, J. et al. Rattling the border wall: pathophysiological implications of functional and proteomic venom variation between Mexican and US subspecies of the desert rattlesnake Crotalus scutulatus. Comparative Biochemistry and Physiology Part C: Toxicology & Pharmacology (2017). [DOI] [PMC free article] [PubMed]
- 24.Massey DJ, et al. Venom variability and envenoming severity outcomes of the Crotalus scutulatus scutulatus (Mojave Rattlesnake) from southern Arizona. Journal of Proteomics. 2012;75:2576–2587. doi: 10.1016/j.jprot.2012.02.035. [DOI] [PubMed] [Google Scholar]
- 25.Strickland J, Mason A, Rokyta D, Parkinson C. Phenotypic variation in Mojave Rattlesnake (Crotalus scutulatus) venom is driven by four toxin families. Toxins. 2018;10:135. doi: 10.3390/toxins10040135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez M, Rael ED, Maddux NL. Isolation of a hemorrhagic toxin from Mojave Rattlesnake (Crotalus scutulatus scutulatus) venom. Toxicon. 1990;28:685–694. doi: 10.1016/0041-0101(90)90257-8. [DOI] [PubMed] [Google Scholar]
- 27.Glenn JL, Straight RC. Intergradation of two different venom populations of the Mojave Rattlesnake (Crotalus scutulatus scutulatus) in Arizona. Toxicon. 1989;27:411–8. doi: 10.1016/0041-0101(89)90203-1. [DOI] [PubMed] [Google Scholar]
- 28.Bieber AL, Tu T, Tu AT. Studies of an acidic cardiotoxin isolated from the venom of Mojave Rattlesnake (Crotalus scutulatus) Biochimica et Biophysica Acta (BBA) - Protein Structure. 1975;400:178–188. doi: 10.1016/0005-2795(75)90139-7. [DOI] [PubMed] [Google Scholar]
- 29.Smiley-Walters SA, Farrell TM, Gibbs HL. Evaluating local adaptation of a complex phenotype: reciprocal tests of Pigmy Rattlesnake venoms on treefrog prey. Oecologia. 2017;184:739–748. doi: 10.1007/s00442-017-3882-8. [DOI] [PubMed] [Google Scholar]
- 30.Smiley-Walters SA, Farrell TM, Gibbs HL. The importance of species: Pigmy Rattlesnake venom toxicity differs between native prey and related non-native species. Toxicon. 2018;144:42–47. doi: 10.1016/j.toxicon.2018.01.022. [DOI] [PubMed] [Google Scholar]
- 31.Holding ML, Drabeck DH, Jansa SA, Gibbs HL. Venom resistance as a model for understanding the molecular basis of complex coevolutionary adaptations. Integrative and Comparative Biology. 2016;56:1032–1043. doi: 10.1093/icb/icw082. [DOI] [PubMed] [Google Scholar]
- 32.Pomento AM, Perry BW, Denton RD, Gibbs HL, Holding ML. No safety in the trees: local and species-level adaptation of an arboreal squirrel to the venom of sympatric rattlesnakes. Toxicon. 2016;118:149–155. doi: 10.1016/j.toxicon.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 33.Thomas RG, Pough FH. The effect of rattlesnake venom on digestion of prey. Toxicon. 1979;17:221–228. doi: 10.1016/0041-0101(79)90211-3. [DOI] [PubMed] [Google Scholar]
- 34.Mackessy SP. Venom ontogeny in the pacific rattlesnakes Crotalus viridis helleri and C. v. oreganus. Copeia. 1988;1988:92–101. doi: 10.2307/1445927. [DOI] [Google Scholar]
- 35.McCue MD. Prey envenomation does not improve digestive performance in Western Diamondback Rattlesnakes (Crotalus atrox) Journal of Experimental Zoology Part A: Ecological Genetics and Physiology. 2007;307:568–577. doi: 10.1002/jez.411. [DOI] [PubMed] [Google Scholar]
- 36.Chu C-W, Tsai T-S, Tsai I-H, Lin Y-S, Tu M-C. Prey envenomation does not improve digestive performance in Taiwanese pit vipers (Trimeresurus gracilis and T. stejnegeri stejnegeri) Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 2009;152:579–585. doi: 10.1016/j.cbpa.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 37.Dowell NL, et al. Extremely divergent haplotypes in two toxin gene complexes encode alternative venom types within rattlesnake species. Current Biology. 2018;28:1016–1026. doi: 10.1016/j.cub.2018.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schield DR, et al. Cryptic genetic diversity, population structure, and gene flow in the Mojave Rattlesnake (Crotalus scutulatus) Molecular Phylogenetics and Evolution. 2018;127:669–681. doi: 10.1016/j.ympev.2018.06.013. [DOI] [PubMed] [Google Scholar]
- 39.Dagda RK, Gasanov S, De La Oiii Y, Rael ED, Lieb CS. Genetic basis for variation of metalloproteinase-associated biochemical activity in venom of the Mojave Rattlesnake (Crotalus scutulatus scutulatus) Biochemistry Research International. 2013;2013:251474. doi: 10.1155/2013/251474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dagda RK, Gasanov SE, Zhang B, Welch W, Rael ED. Molecular models of the Mojave Rattlesnake (Crotalus scutulatus scutulatus) venom metalloproteinases reveal a structural basis for differences in hemorrhagic activities. Journal of Biological Physics. 2014;40:193–216. doi: 10.1007/s10867-013-9339-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabaj, M. Standard symbolic codes for institutional resource collections in herpetology and ichthyology: an online reference. Version 6.5 (16 August 2016). Online athttp://www.asih.org/, (American Society of Ichthyologists and Herpetologists, Washington, DC, 2016).
- 42.Rohland N, Reich D. Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Research. 2012;22:939–46. doi: 10.1101/gr.128124.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faircloth, B. Protocol: preparation of an AMPure XP substitute (AKA Serapure). Online at, 10.6079/j9mw2f26 (2014).
- 44.Margres MJ, et al. Linking the transcriptome and proteome to characterize the venom of the Eastern Diamondback Rattlesnake (Crotalus adamanteus) Journal of Proteomics. 2014;96:145–158. doi: 10.1016/j.jprot.2013.11.001. [DOI] [PubMed] [Google Scholar]
- 45.Rokyta DR, Wray KP, Lemmon AR, Lemmon EM, Caudle SB. A high-throughput venom-gland transcriptome for the Eastern Diamondback Rattlesnake (Crotalus adamanteus) and evidence for pervasive positive selection across toxin classes. Toxicon. 2011;57:657–671. doi: 10.1016/j.toxicon.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 46.Smith CF, Mackessy SP. The effects of hybridization on divergent venom phenotypes: characterization of venom from Crotalus scutulatus scutulatus × Crotalus oreganus helleri hybrids. Toxicon. 2016;120:110–123. doi: 10.1016/j.toxicon.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 47.Zancolli G, et al. Is hybridization a source of adaptive venom variation in rattlesnakes? A test, using a Crotalus scutulatus × viridis hybrid zone in southwestern New Mexico. Toxins. 2016;8:188. doi: 10.3390/toxins8060188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blair C, Sánchez-Ramírez S. Diversity-dependent cladogenesis throughout western mexico: Evolutionary biogeography of rattlesnakes (viperidae: Crotalinae: Crotalus and Sistrurus) Molecular Phylogenetics and Evolution. 2016;97:145–154. doi: 10.1016/j.ympev.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 49.Arevalo E, Davis SK, Sites JW. Mitochondrial DNA sequence divergence and phylogenetic relationships among eight chromosome races of the Sceloporus grammicus Complex (Phrynosomatidae) in Central Mexico. Systematic Biology. 1994;43:387–418. doi: 10.1093/sysbio/43.3.387. [DOI] [Google Scholar]
- 50.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution. 2016;34:msw260. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 52.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 53.Ronquist F, et al. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Systematic Biology. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rambaut, A., Suchard, M. A., Xie, D. & Drummond, A. J. Tracer v1.6 (2010).
- 55.R Core Team. R: A Language and Environment for Statistical Computing. http://www.R-project.org/ (R Foundation for Statistical Computing, Vienna, Austria, 2017).
- 56.Pohlert, T. The Pairwise Multiple Comparison of Mean Ranks Package (PMCMR), https://CRAN.R-project.org/package=PMCMR, R package (2014).
- 57.Mackessy SP. Fibrinogenolytic proteases from the venoms of juvenile and adult Northern Pacific Rattlesnakes (Crotalus viridis oreganus) Comparative Biochemistry and Physiology – Part B: Comparative Biochemistry. 1993;106:181–189. doi: 10.1016/0305-0491(93)90025-Z. [DOI] [PubMed] [Google Scholar]
- 58.Phillips, S. J., Dudik, M. & Schapire, R. E. Maxent software for modeling species niches and distributions (2018).
- 59.Hijmans, R. J., Phillips, S., Leathwick, J. & Elith, J. dismo: Species distribution modeling. R package version 1.1-4 (2017).
- 60.Phillips SJ, Anderson RP, Schapire RE. Maximum entropy modeling of species geographic distributions. Ecological Modelling. 2006;190:231–259. doi: 10.1016/j.ecolmodel.2005.03.026. [DOI] [Google Scholar]
- 61.Fick SE, Hijmans RJ. WorldClim 2: new 1-km spatial resolution climate surfaces for global land areas. International Journal of Climatology. 2017;37:4302–4315. doi: 10.1002/joc.5086. [DOI] [Google Scholar]
- 62.Dormann CF, et al. Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography. 2013;36:27–46. doi: 10.1111/j.1600-0587.2012.07348.x. [DOI] [Google Scholar]
- 63.Warren DL, Glor RE, Turelli M. Environmental niche equivalency versus conservatism: quantitative approaches to niche evolution. Evolution. 2008;62:2868–83. doi: 10.1111/j.1558-5646.2008.00482.x. [DOI] [PubMed] [Google Scholar]
- 64.Heibl, C. & Calenge, C. phyloclim: Integrating phylogenetics and climatic niche modeling (2015).
- 65.Githens T, George I. Comparative studies on the venoms of certain rattlesnakes. Bulletin of the Antivenin Institute of America. 1931;5:31–34. [Google Scholar]
- 66.Charlesworth D. Balancing selection and its effects on sequences in nearby genome regions. PLoS Genetics. 2006;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Llaurens V, Whibley A, Joron M. Genetic architecture and balancing selection: the life and death of differentiated variants. Molecular Ecology. 2017;26:2430–2448. doi: 10.1111/mec.14051. [DOI] [PubMed] [Google Scholar]
- 68.Fijarczyk A, Babik W. Detecting balancing selection in genomes: limits and prospects. Molecular Ecology. 2015;24:3529–3545. doi: 10.1111/mec.13226. [DOI] [PubMed] [Google Scholar]
- 69.LaBonte J, Welch K, Suarez R. Digestive performance in neonatal Southern Pacific Rattlesnakes (Crotalus oreganus helleri) Canadian Journal of Zoology. 2011;89:705–713. doi: 10.1139/z11-034. [DOI] [Google Scholar]
- 70.Mackessy SP, Williams K, Ashton KG. Ontogenetic variation in venom composition and diet of Crotalus oreganus concolor: A case of venom paedomorphosis? Copeia. 2003;2003:769–782. doi: 10.1643/HA03-037.1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
New ND4 sequences generated in this study were deposited in GenBank under accession numbers MH883648-MH883754. Specimen vouchers were deposited in the appropriate museums based on permit requirements. All specimen data including morphology and assay data are listed in Supplemental Table 1.