Abstract
The data presented here are related to our research article entitled “miR-132/212 is induced by stress and its dysregulation triggers anxiety-related behavior” (Aten et al., 2018). In this article, we utilize immunofluorescent techniques to examine the protein-level expression of two microRNA-132/212 target genes, Sirt1 and Pten, in miR-132 transgenic and miR-132/212 conditional knockout (cKO) mouse lines. Additionally, using immunohistochemistry, we detail the expression profile of Sirt1 and Pten in the hippocampus and amygdala of WT mice after a 15 day chronic restraint stress paradigm.
Abbreviations: miR, microRNA; WT, wild-type; cKO, conditional knockout; NGS, Normal Goat Serum; TRE, tetracycline-regulated element
Specifications table
| Subject area | Biology |
| More specific subject area | Neuroscience |
| Type of data | Image, figure, graph |
| How data was acquired | Immunofluorescent and Immunohistochemical labeling for Sirt1 and Pten |
| Data format | Raw and analyzed/quantified |
| Experimental factors | WT, microRNA-132/212 cKO, and microRNA-132 transgenic mice |
| Experimental features | Brain tissue from naïve WT, microRNA-132/212 cKO, and microRNA-132 transgenic mice was immunolabeled for Sirt1 or Pten and CFP (cyan fluorescent protein). Additionally, brain tissue from WT mice exposed to chronic restraint stress was immunolabled for Sirt1 and Pten |
| Data source location | Department of Neuroscience; Ohio State University; Columbus, OH, USA |
| Data accessibility | Data are available within this article |
| Related research article | Aten, S., Page, C.E., Kalidindi, A., Wheaton, K.L., Niraula, A., Godbout, J.P., Hoyt, K.R., Obrietan, K., 2018. miR-132/212 is induced by stress and its dysregulation triggers anxiety-related behavior. Neuropharmacology 2019; 144, 256-270 [1] |
Value of the data
-
•
The expression patterns of Sirt1 and Pten in miR-132 transgenic and miR-132/212 cKO mice could be used to further the understanding of the wide-ranging effects of dysregulated microRNAs within the forebrain.
-
•
These data depicting amygdalar and hippocampal protein-level expression of Sirt1 and Pten (two validated miR-132/212 target genes) after chronic stress could serve as a basis for the examination of additional miR-132/212 targets following different stress paradigms (acute stress, unpredictable stress, etc.).
-
•
The data can be used to provide a foundation for further investigation of miR-132/212 targets that are implicated in stress/anxiety-related behavioral states.
1. Data
The data presented in this article depict protein-level expression of Sirt1 and Pten, both of which are miR-132/212 target genes that have been implicated in stress/anxiety [2], [3], [4], [5]. Expression of both targets is shown within the hippocampus and amygdala of naïve WT, miR-132/212 conditional knockout, and miR-132 transgenic mice (Fig. 1).
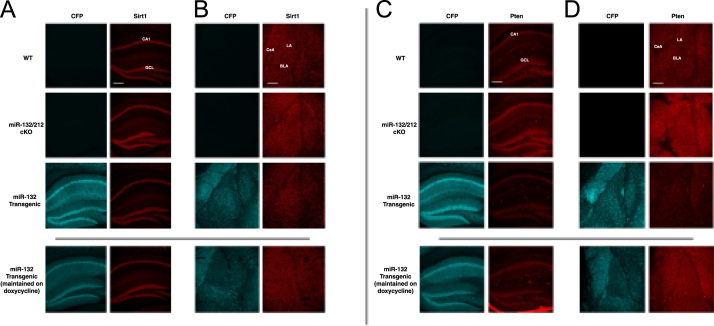
Fig. 1.
Expression of Sirt1 and Pten within the forebrain of WT, miR-132/212 cKO, and miR-132 transgenic animals (top three rows). Representative 10X immunofluorescent images of Sirt1 in the hippocampus (A) and amygdala (B). Representative 10X immunofluorescent images of Pten in the hippocampus (C) and amygdala (D). Immunolabeling in miR-132 transgenic animals maintained on doxycycline is also shown (bottom row). Note the increased and decreased target gene expression in miR-132/212 cKO and miR-132 transgenic animals, respectively, relative to WT mice. Also note the decreased expression of CFP in the miR-132 transgenic animals maintained on doxycycline, in addition to the subtle increase in Sirt1 and Pten expression in miR-132 transgenic animals on doxycycline relative to miR-132 transgenic animals not maintained on doxycycline. CFP is a marker for the miR-132 transgenic mouse line, and as such, immunofluorescence was not observed in WT and miR-132/212 cKO animals. Scale bar = 300 μm for both hippocampal and amygdalar images. Abbreviations: CA1-cornu ammonis 1 hippocampal subfield; GCL-granule cell layer; LA-lateral amygdala; BLA-basolateral amygdala; CeA-central amygdala.
We also present data detailing relative Sirt1 (Fig. 2) and Pten (Fig. 3) protein levels within the hippocampus and amygdala of WT animals that have been exposed to 15 days of chronic restraint stress.
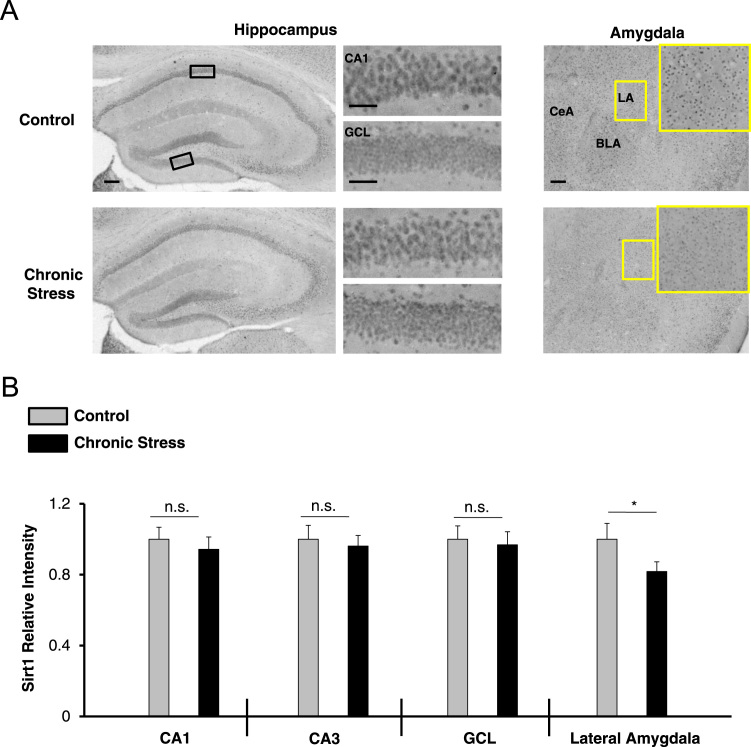
Fig. 2.
Expression of Sirt1 after chronic stress. (A) Representative immunohistochemical labeling for Sirt1 in the hippocampus and amygdala of control and chronically stressed WT mice. The boxed regions in the low-magnification images approximate the locations of the regions that are depicted in the high-magnification panels (to the right for the hippocampus, and inset for the amygdala). (B) Quantification of Sirt1 immunolabeling in three hippocampal regions (the CA1, CA3, and GCL) and in the lateral amygdala. Note the reduced expression of Sirt1, only within the amygdala of chronically stressed animals. For each brain region, the control condition was set equal to a value of one, and the stressed condition was normalized to this value. Scale bar = 50 μm for the low magnification images (i.e., whole hippocampus and amygdala) and 30 μm for high magnification images (i.e., CA1 cell layer and GCL-lower blade). Data were analyzed using the Student׳s t-tests and are presented as the mean ± SEM. *: p = 0.0424; n.s.: not significant (p > 0.05). N = 7 mice per condition. Abbreviations: CA1-cornu ammonis 1 hippocampal subfield; CA3-cornu ammonis 3 hippocampal subfield; GCL-granule cell layer; LA-lateral amygdala; BLA-basolateral amygdala; CeA-central amygdala.
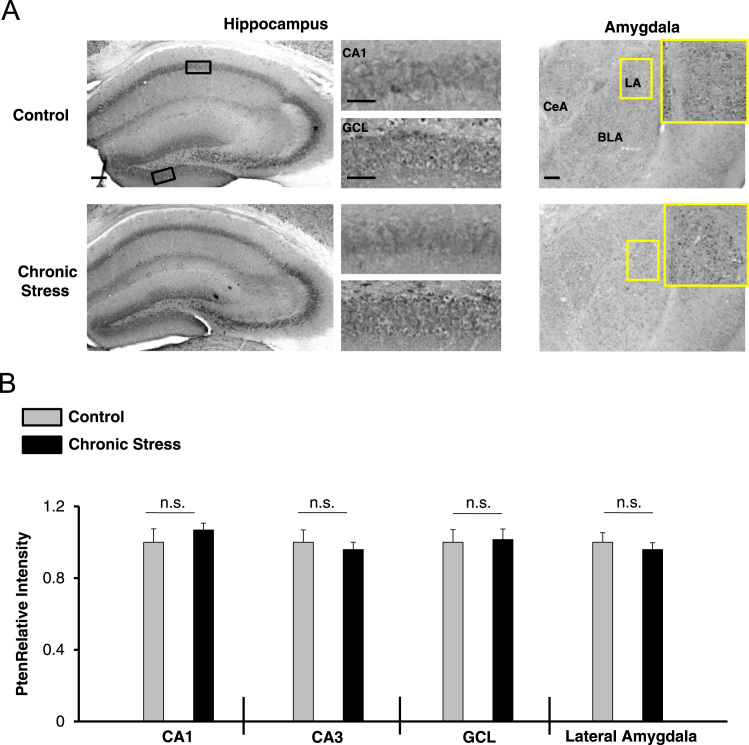
Fig. 3.
Expression of Pten after chronic stress. (A) Representative immunohistochemical labeling for Pten in the hippocampus and amygdala of control and chronically stressed WT mice. The boxed regions in the low-magnification images approximate the locations of the regions that are depicted in the high-magnification panels (to the right for the hippocampus, and inset for the amygdala). (B) Quantification of Pten immunolabeling in three hippocampal regions (the CA1, CA3, and GCL) and in the lateral amygdala. For each brain region, the control condition was set equal to a value of one, and the stressed condition was normalized to this value. Scale bar = 50 μm for the low magnification images (i.e., whole hippocampus and amygdala) and 30 μm for high magnification images (i.e., CA1 cell layer and GCL-lower blade). Data were analyzed using the Student׳s t-tests and are presented as the mean ± SEM. n.s.: not significant (p > 0.05). N = 7 mice per condition. Abbreviations: CA1-cornu ammonis 1 hippocampal subfield; CA3-cornu ammonis 3 hippocampal subfield; GCL-granule cell layer; LA-lateral amygdala; BLA-basolateral amygdala; CeA-central amygdala.
2. Experimental design, materials, and methods
2.1. miR-132 transgenic and miR-132/212 conditional knockout mice
Details related to the generation of the miR-132 transgenic and miR-132/212 knockout mouse lines can be found in the related research article [1] and in previous papers from our lab [6], [7], [8]. Briefly, in the ‘miR-132/212 cKO’ mouse line, the miR-132/212 locus is selectively excised within excitatory neurons of the forebrain (e.g. cortex, amygdala, hippocampus). ‘miR-132 transgenic’ animals were created by crossing miR-132/212 cKO mice with a tetracycline-regulated bidirectional miR-132/cyan fluorescent protein (CFP) transgenic mouse line driven by CaMKII:tTA. Hence, in this quadruple transgenic mouse line, the effects of endogenous miR-132/212 deletion and transgenic miR-132 over-expression can be observed within the same neuronal cell populations.
2.2. Doxycycline treatment
Doxycycline (0.40 μg/mL) was used to titer the level of tetracycline-inducible miR-132 transgene expression in miR-132 transgenic animals. Details of doxycycline administration can be found in our related research article [1].
2.3. Tissue processing
For all experiments (Fig. 1, Fig. 2, Fig. 3), brains were cut into 600 µm sections using a vibratome and were then fixed in 4% paraformaldehyde for six hours (4 ° C). Tissue was then incubated in 30% sucrose (in 1X PBS: Phosphate Buffered Saline) overnight, and coronal sections containing the hippocampus and amygdala were subsequently thin cut to 40 µm on a freezing microtome.
2.4. Immunofluorescent labeling and confocal microscopy
For Sirt1 immunofluorescent labeling in naïve WT, miR-132/212 cKO, and miR-132 transgenic animals, sections containing the hippocampus and amygdala were blocked for one hour in 10% NGS (Normal Goat Serum) in 1X PBS containing 1% Triton X-100 (PBST). Note that sections were washed three times (5 min/wash) in PBST between each labeling step. Sections were then incubated (3 hours at room temperature) in the following primary antibodies: chicken anti-GFP (1:2500 dilution; Abcam, Cat # ab13970) to detect the TRE-regulated CFP transgene in the miR-132 transgenic animals and rabbit anti- Sirt1/Sir2α (1:500 dilution; Millipore, Cat# 09-845). Next, sections were incubated in Alexa Fluor 488 goat anti-chicken IgY (1:1000 dilution; Invitrogen, Carlsbad, CA) and Alexa Fluor 594 goat anti-rabbit IgY (1:1000 dilution) for two hours at room temperature. Finally, tissue was mounted and coverslipped with Fluoromount-G (SouthernBiotech; Cat # 0100-01, Birmingham, AL). 10X fluorescent images of the hippocampus and amygdala were captured with a Leica TSC SP8 confocal microscope. Acquisition parameters were as follows for both hippocampus and amygdala images, and these settings were not changed between each genotype: 1024 ×1024 scan format, 100 Hz scan speed; 16 line average; 4.0 AU pinhole. Note that immunofluorescent labeling was conducted to obtain representative images only; statistical analysis was not performed.
For Pten immunofluorescent staining, we utilized a biotinylated tyramide labeling protocol, as previously described by our lab [9]. In brief, sections containing the hippocampus and amygdala were washed three times in PBST, followed by a 30 minute incubation in 0.3% hydrogen peroxide diluted in PBST. Tissue was blocked for one hour in 10% NGS diluted in PBST. Sections were then incubated in rabbit anti-Pten primary antibody in PBST containing 5% NGS (1: 2500 dilution; Cell Signaling Technology) and chicken anti-GFP (1: 2,500 dilution; Abcam) for three hours at room temperature. Tissue was then incubated for two hours at room temperature in goat anti-rabbit HRP-conjugated secondary antibody (1:1,000 dilution; Perkin Elmer, Cat # NEF812001EA) in PBST containing 5% NGS. Subsequently, tissue was incubated in tyramide 488 (1:800 dilution in 0.0015% H2O2 in PBST) for 30 minutes at room temperature. Preparation of the tyramide solution is outlined in [10]. Next, tissue was incubated in Alexa Fluor 594 goat anti-chicken IgY (1:1,000 dilution; Invitrogen) for two hours at room temperature. Images were acquired as described above, and imaging settings were held constant for all genotypes. As with Sirt1 immunofluorescent experiments, Pten immunofluorescent images were taken for representative photomicrographs only; statistical analysis was not conducted.
2.5. Chronic restraint stress paradigm
Details of the chronic restraint paradigm can be found in our related research paper [1]. Briefly, male WT mice were restrained in 50 mL conicals/tubes for two hours per day throughout a 15 day period. Control mice were handled daily. Animals were sacrificed on day 16 (24 h after the final restraint session), and tissue was processed for immunohistochemical analysis.
2.6. Immunohistochemistry and brightfield microscopy
For Sirt1 and Pten immunohistochemical labeling (performed on tissue from control and chronically stressed WT animals), sections (two per animal) containing the hippocampus and amygdala were incubated for 20 minutes in 0.3% hydrogen peroxide in PBST. Sections were washed three times in PBST between each labeling step. Next, tissue was blocked for one hour in PBST containing 10% NGS, followed by incubation at 4 °C (for eight hours) with rabbit primary antibodies against Sirt1/Sir2α (1:1000 dilution) or Pten (1:100 dilution). After secondary antibody incubation (for two hours at room temperature), sections were processed using the ABC labeling method (Vector Labs) followed by visualization with nickel-intensified DAB (3,3′-diaminobenzidine) substrate. After mounting with Permount Mounting Medium (Fisher Chemical), 4X bright-field images of the amygdala and hippocampus were acquired. A 16-bit digital camera (Micromax YHS 1300; Princeton Instruments) on a Leica DMIR microscope with Metamorph software (MetaMorph Microscopy Automation and Image Analysis Software) were used to capture the images. The average intensity for each region (after background subtraction) was calculated for each animal, and the group average was displayed as the mean ± SEM.
2.7. Experimental design and statistical analysis
Statistical analysis was conducted using GraphPad Prism 7.0 (Graphpad Prism) software. All data are presented as the mean ± SEM, and significance for all experiments was set at *p < 0.05. Comparisons between ‘control’ and ‘chronic stress’ groups were performed using Student׳s two-tailed t-tests within each hippocampal/amygdalar region. Additionally, Grubb׳s test was conducted on data sets within each group; animals that were found to be statistically significant outliers (p < 0.05) were removed from analysis. Grubb׳s test was used to exclude one control animal from the Sirt1 immunolabeling experiment (Fig. 2).
Acknowledgments
This work was supported by National Institutes of Health; Grant code: MH103361, NS066345, NS091302 and National Science Foundation; Grant code: 1354612.
Footnotes
Transparency data associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.11.042.
Transparency document. Supplementary material
Supplementary material
.
References
- 1.Aten S., Page C.E., Kalidindi A., Wheaton K.L., Niraula A., Godbout J.P., Hoyt K.R., Obrietan K. miR-132/212 is induced by stress and its dysregulation triggers anxiety-related behavior. Neuropharmacology. 2019;144:256–270. doi: 10.1016/j.neuropharm.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Libert S., Pointer K., Bell E.L., Das A., Cohen D.E., Asara J.M., Kapur K., Bergmann S., Preisig M., Otowa T., Kendler K.S., Chen X., Hettema J.M., van den Oord E.J., Rubio J.P., Guarente L. SIRT1 activates MAO-A in the brain to mediate anxiety and exploratory drive. Cell. 2011;147:1459–1472. doi: 10.1016/j.cell.2011.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim H.D., Hesterman J., Call T., Magazu S., Keeley E., Armenta K., Kronman H., Neve R.L., Nestler E.J., Ferguson D. SIRT1 Mediates depression-like behaviors in the nucleus accumbens. J. Neurosci. 2016;36:8441–8452. doi: 10.1523/JNEUROSCI.0212-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kwon C.H., Luikart B.W., Powell C.M., Zhou J., Matheny S.A., Zhang W., Li Y., Baker S.J., Parada L.F. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou J., Blundell J., Ogawa S., Kwon C.H., Zhang W., Sinton C., Powell C.M., Parada L.F. Pharmacological inhibition of mTORC1 suppresses anatomical, cellular, and behavioral abnormalities in neural-specific Pten knock-out mice. J. Neurosci. 2009;29:1773–1783. doi: 10.1523/JNEUROSCI.5685-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aten S., Hansen K.F., Snider K., Wheaton K., Kalidindi A., Garcia A., Alzate-Correa D., Hoyt K.R., Obrietan K. miR-132 couples the circadian clock to daily rhythms of neuronal plasticity and cognition. Learn. Mem. 2018;25:214–229. doi: 10.1101/lm.047191.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen K.F., Sakamoto K., Wayman G.A., Impey S., Obrietan K. Transgenic miR132 alters neuronal spine density and impairs novel object recognition memory. PLoS One. 2010;5:e15497. doi: 10.1371/journal.pone.0015497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hansen K.F., Sakamoto K., Aten S., Snider K.H., Loeser J., Hesse A.M., Page C.E., Pelz C., Arthur J.S.C., Impey S., Obrietan K. Targeted deletion of miR-132/-212 impairs memory and alters the hippocampal transcriptome. Learn Mem. 2016;23:61–71. doi: 10.1101/lm.039578.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karelina K., Liu Y., Alzate-Correa D., Wheaton K.L., Hoyt K.R., Arthur J.S.C., Obrietan K. Mitogen and stress-activated kinases 1/2 regulate ischemia-induced hippocampal progenitor cell proliferation and neurogenesis. Neuroscience. 2015;285:292–302. doi: 10.1016/j.neuroscience.2014.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vize P.D., McCoy K.E., Zhou X. Multichannel wholemount fluorescent and fluorescent/chromogenic in situ hybridization in Xenopus embryos. Nat. Protoc. 2009;4:975–983. doi: 10.1038/nprot.2009.69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material