Abstract
Adeno-associated virus (AAV) vectors have become one of the most widely used gene transfer tools in human gene therapy. Considerable effort is currently being focused on AAV capsid engineering strategies with the aim of developing novel variants with enhanced tropism for specific human cell types, decreased human seroreactivity, and increased manufacturability. Selection strategies based on directed evolution rely on the generation of highly variable AAV capsid libraries using methods such as DNA-family shuffling, a technique reliant on stretches of high DNA sequence identity between input parental capsid sequences. This identity dependence for reassembly of shuffled capsids is inherently limiting and results in decreased shuffling efficiency as the phylogenetic distance between parental AAV capsids increases. To overcome this limitation, we have developed a novel codon-optimization algorithm that exploits evolutionarily defined codon usage at each amino acid residue in the parental sequences. This method increases average sequence identity between capsids, while enhancing the probability of retaining capsid functionality, and facilitates incorporation of phylogenetically distant serotypes into the DNA-shuffled libraries. This technology will help accelerate the discovery of an increasingly powerful repertoire of AAV capsid variants for cell-type and disease-specific applications.
Keywords: AAV, library, directed evolution, codon optimization, DNA shuffling, capsid
Introduction
The recent approval of the gene and cell therapy products Glybera,1, 2 Luxturna,3 and Strimvelis4 marks a pivotal point in the maturation of the gene therapy field. Significantly, all of these products share a common technological underpinning: the use of recombinant viral vectors to safely deliver therapeutic transgenes into target cells, an indication of the importance of vector engineering for the future of gene therapy. One of the most powerful gene transfer vectors used in current research and clinical studies is based on recombinant adeno-associated virus (rAAV).5 AAV is a single-stranded DNA virus first employed as a gene transfer vector in 1984.6 Since AAV vectors entered the clinic in 1994 there have been more than 160 phase I, II, and/or III gene therapy trials utilizing rAAV, with the number initiated each year increasing 3-fold since 2011 (http://www.abedia.com/wiley/index.html). This accelerated adoption of rAAV technology reflects therapeutic efficacy in preclinical disease models and exciting early success in human clinical trials, most notably in the liver, eye, and CNS. A number of technical advances, such as discovery of vector pseudo-serotyping, the development of scalable high-yield production strategies,7, 8, 9 and the demonstration that rAAV can be used for both gene addition and gene correction,10, 11, 12 have also contributed to increased uptake of the AAV vector system. However, one of the leading challenges that remains to be addressed, in order to achieve the full therapeutic potential of rAAV, is limited efficiency in the functional transduction of primary human cells. Given that the tropism of AAV vectors is determined by the capsid,13 significant effort has been focused on AAV capsid engineering to develop novel variants with enhanced human tropism.14, 15
Multiple different approaches have been developed to facilitate identification of novel AAV capsid variants with improved transduction potential, ranging from searching for naturally occurring AAV variants in multiple different species, reconstructing ancestral variants,16, 17, 18 rational design based on custom modification of AAV properties by domain swapping,19 incorporation of tissue-specific ligands on the vector surface,20 through to directed evolution technologies.21 In contrast with rational design approaches, which require extensive knowledge of the relationships between capsid structure and function and AAV-target cell receptor interactions, directed evolution technologies are based on better understood AAV biology and aim to mimic the natural viral evolution process in a controlled laboratory environment.22 In contrast with sequential accumulation of mutations and modifications during natural evolution, directed evolution platforms rely on in vitro generation of large pools of AAV variants, which can be used in in vitro or in vivo selection schemes in the presence of specific selection pressure(s). One of the most commonly utilized molecular biology techniques to generate highly complex AAV capsid libraries is based on DNA-family shuffling, first adapted for use in the AAV system by Grimm et al.21
Shuffled AAV libraries have been successfully used by multiple groups to identify novel AAV capsid variants with improved properties. Using an AAV library based on eight parental AAV capsids in an in vitro selection system, Grimm et al.21 identified AAV-DJ, a variant with superior transduction efficiency in multiple cell types in vitro. Subsequently, similarly created libraries have been used to perform transduction-based selection on primary human hepatocytes in a xenograft mouse model of human liver, leading to the identification of AAV-LK0314 and more recently AAV-NP59,15 two novel human hepatotropic variants. Because library production by DNA-family shuffling involves enzymatic digestion of the parental AAV capsid sequences, followed by primerless PCR reassembly of DNA fragments to create novel capsid gene (cap) variants, shuffling efficiency relies on high degrees of sequence identity between parental serotypes at the DNA level.23 This identity dependence makes parental variants with higher sequence identity more efficient shuffling substrates and results in their overrepresentation in the final library, whereas DNA fragments from more distant capsid variants anneal less efficiently and are underrepresented (unpublished data). This identity-based bias reduces library diversity and may affect the likelihood of selecting optimal capsid variants.
In this study we describe an AAV shuffling strategy based on a novel codon-optimization approach developed to increase the sequence identity between parental capsid sequences while increasing the probability of retaining functionality. This strategy, designated localized codon-optimization (LCO), was used to design novel lcoAAV parental capsid variants with enhanced sequence identity for utilization in AAV shuffling applications. Use of these new parental variants facilitated more efficient incorporation of evolutionarily distant, and thus more sequence-divergent, variants, thereby confirming the utility and power of this new AAV engineering tool.
Results
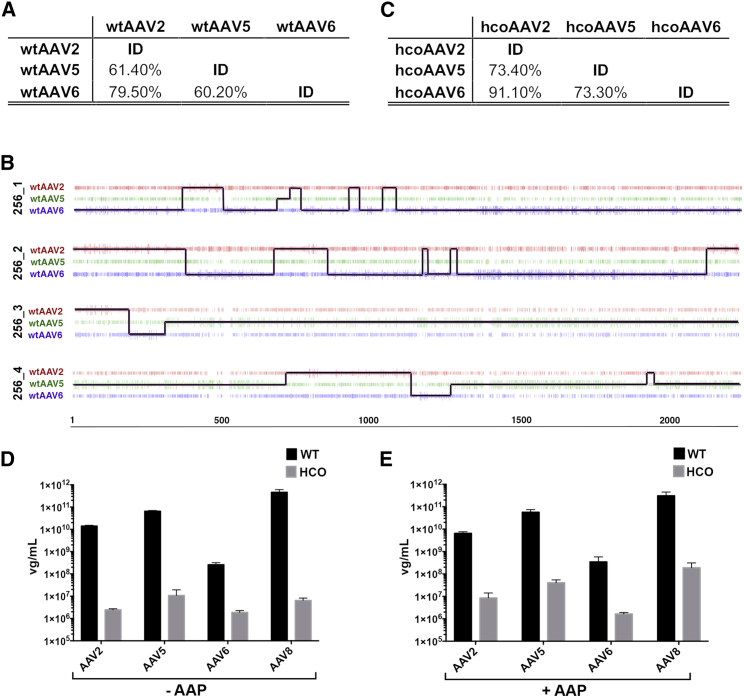
Wild-Type AAV Capsid Genes Are Suboptimal Substrates for DNA Shuffling
The efficiency of parental cap gene shuffling during the generation of AAV shuffled capsid libraries relies on stretches of sequence identity between the input sequences at the nucleotide level.23, 24 The level of sequence identity of the capsid genes between 12 natural AAV isolates used for library generation varied significantly, with AAV1 and AAV6 having the highest identity (97.1%) and AAV5 and AAV12 the lowest (55.1%). We hypothesized that such wide-ranging identities at the DNA level would directly influence the efficiency of shuffling of the individual input sequences and bias the final library, leading to overrepresentation of sequences contributed by closely related donor AAVs. To test this hypothesis and gain insight into the sequence identity-dependent likelihood of input parental capsids contributing sequences to individual clones within the highly complex library, we performed an initial shuffling experiment using capsid genes from three AAV serotypes belonging to Clade-A (AAV6), Clade-B (AAV2), and a distant clonal isolate (AAV5). The percent identity between these three capsid genes varied from 60.2% to 79.5%, with cap5 being the most distant variant, having identity to cap2 and cap6 of 61.4% and 60.2%, respectively (Figure 1A). Analysis of individual clones (n = 36) from the final shuffled library, AAVLib256, revealed that overall library diversity (number of unique clones within the pool) as well as clonal diversity (level of sequence shuffling within individual clones) were low. Specifically, the final library contained a considerable percentage of clones (25%, n = 9/36) that were 100% identical to parental AAV5 (data not shown). In addition, the analysis confirmed our hypothesis that more distant AAV donors, such as AAV5, do not shuffle efficiently with other variants and thus contribute poorly to shuffled clones (clones 256_1 and 256_2 in Figure 1B). In addition, a large percentage of the clones (36%) was >80% identical to AAV5 (clones 256_3 and 256_4 in Figure 1B), providing further evidence of inefficient shuffling of parental variants sharing a lesser degree of sequence identity. This almost certainly reflects the lower sequence identity between fragments of AAV5 and other DNA fragments present during the PCR reassembly step. Thus, fragments of AAV5 anneal to each other more efficiently than to DNA fragments from other variants, leading to preferential reassembly of the full-length or large fragments of the AAV5 capsid gene. This observation suggests that libraries generated using DNA-family shuffling that include more distant parental AAV donors are more likely to contain a significant proportion of full-length wild-type (WT) variants. This is especially problematic when attempting to create more diverse libraries with novel, often evolutionary distant, variants such as Anc8016 or mAAV-EVE118 included as parental donors. Our data indicate that these distant variants would not readily contribute to individual clones and thus would not increase complexity at the level of individual variants within the library, but could adversely affect the quality of the final library through reassembly of full-length parental sequences.
Figure 1.
Local Codon Optimization Increases DNA Shuffling but Negatively Affects Packaging Function
(A) Percent identity between capsid genes of indicated wild-type (WT) AAV serotypes. (B) Parental sequence contribution analysis of sample clones isolated from AAVLib256. Parental sequences are represented by red (AAV2), green (AAV5), and blue (AAV6) lines. Size of vertical bars represents probability that individual residue was contributed by given parental vector. Black line represents the most probable composition of individual shuffled clones based on the longest sequence of identity to parental variants in a 5′ to 3′ direction. (C) Percent identity between variants codon-optimized using human codon-usage table (hco). (D and E) rAAV yield comparison using helper plasmids encoding wild-type cap genes (wtAAV2, wtAAV5, wtAAV6, and wtAAV8) and corresponding hcoAAVs in the absence (D) or presence (E) of serotype-specific AAP provided in trans. vg = vector genomes. Values in (D) and (E) are shown as mean ± SD (n = 3).
Conventional Codon-Optimization Affects Function of Individual Input Variants
To address the limitations of DNA shuffling efficiency using phylogenetically distant AAV serotypes, we attempted to increase the level of sequence identity between individual parental variants using a conventional codon optimization strategy (see Materials and Methods). Because standard AAV packaging protocols utilize human HEK293T cells, we decided to codon-optimize the cap genes of AAV2, AAV5, and AAV6 (subsequently referred to as “hcoAAV”) based on human codon-usage data (see Materials and Methods) (see Figure S3 for sequence of hcoAAV cap genes). The rep gene major splice acceptor site, start codons of VP2 and assembly-activating protein (AAP) were not codon optimized and were maintained as in the WT counterparts. This modification increased the DNA sequence identity between individual variants, with hcoAAV5 being 73.4% identical to hcoAAV2 and 73.3% to hcoAAV6 (Figure 1C). Because the function of variants selected from a library is directly linked to the functionality of the input variants, we performed functional analysis of hcoAAVs. To test the packaging efficiency, we used the three hcoAAV cap genes to generate pAAV packaging constructs containing the AAV2 rep gene. The resultant constructs, designated pAAV-hcoAAV2, pAAV-hcoAAV5, and pAAV-hcoAAV6, were used to package the AAV cassette expressing GFP under the control of a liver-specific promoter (LSP) (AAV-LSP-GFP) in parallel single-dish productions (n = 3) using the WT counterparts (pAAV2, pAAV5, and pAAV6) as controls. Because codon-optimization of the first open reading frame (ORF) affected sequences of individual aap25 genes encoded on the second reading frame (data not shown), it was not surprising that sequence optimization had a dramatic negative effect on all individual variants leading to significantly lower vector yields (Figure 1D). Because AAP26 is known to play an important role as an essential accessory protein for efficient assembly of functional AAV capsids,27, 28 we examined whether packaging of the hcoAAVs could be rescued by providing serotype-specific AAPs in trans during vector packaging. Interestingly, AAP complementation failed to substantially restore packaging efficiency (Figure 1E), indicating that codon-optimization affected processes and elements other than those related to AAP activity. To test whether the observed decrease in packaging efficiency after codon-optimization affects other AAVs, we generated and evaluated hcoAAV8 using the same approach with essentially the same outcome (Figures 1D and 1E). Collectively these data support the conclusion that conventional codon-optimization of cap gene sequences is an inadequate methodology for increasing nucleotide identity between input AAV sequences for DNA-family shuffling applications.
Novel Codon-Optimization Method Increases Capsid Sequence Identity while Retaining Function
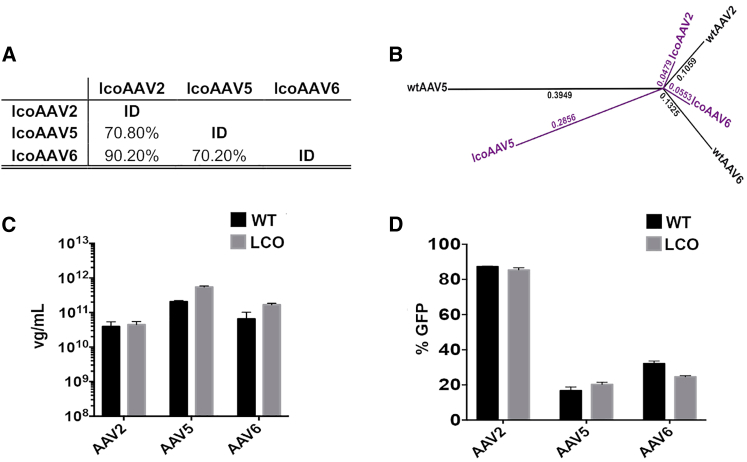
The dramatic loss of vector packaging efficiency after conventional codon-optimization (hcoAAVs) suggests that AAV capsid gene sequences, in addition to encoding structural capsid proteins and AAP, may contain other currently unknown elements important for cap gene function and AAV assembly. Accordingly, an alternative method of codon optimization was developed and evaluated for the ability to increase identity between individual AAV cap genes while maintaining the functionality of individual variants. We hypothesized that one way to preserve functional elements of the cap genes, while increasing identity between input genes, would be to only utilize codons used by input variants at each individual optimized position. To achieve this, we developed a novel codon-optimization method for improving the level of identity between serotypes while potentially maintaining capsid functionality. This localized codon-optimization (LCO) strategy increases identity between parental sequences by performing localized optimization at each codon independently of the rest of the gene sequence. Specifically, by generating a codon usage frequency table for each of the amino acid positions and then applying this local codon usage table to optimize individual sequences, the LCO algorithm minimizes the arbitrary changes that conventional codon optimization approaches introduce to the sequence (see Materials and Methods and Figure S1 for details).
To test the LCO algorithm, we performed localized codon-optimization of capsid genes from AAV2, AAV5, and AAV6 (subsequently referred to as lcoAAV2, lcoAAV5, and lcoAAV6). This modification increased the average identity between the three parental AAV caps by 10.03% when compared with wtAAV cap sequences (Figures 1A and 2A) and decreased the frequency of nucleotide substitutions per site (Figure 2B). In silico analysis of lcoAAVs confirmed that the essential and therefore homologous regions of the AAV genomes, such as the splice acceptor site (residue 2,228 in the wtAAV2 genome, GenBank: NC_001401.2), the VP2 and VP3 start codons (residues 2,614 and 2,809 in the wtAAV2 genome, GenBank: NC_001401.2), and the AAP start codon (CTG, residue 2,729 in the wtAAV2 genome, GenBank: NC_001401.2) were preserved. An important aspect was to confirm whether optimizing the VP proteins, encoded in the first ORF, would have a negative effect on the AAP ORF encoded in the second reading frame and thus affect capsid function.
Figure 2.
Functional Evaluation of AAVs Modified Using Localized Codon-Optimization Algorithm
(A) Percent identity at the DNA level between sequences of optimized AAV2, AAV5, and AAV6 capsid genes (lcoAAV2, lcoAAV5, and lcoAAV6, respectively). (B) DNA identity tree between cap genes from wtAAVs (wtAAV2, wtAAV5, and wtAAV6, in black) and LCO cap genes (lcoAAV2, lcoAAV5, and lcoAAV6, in purple). The numbers indicate the frequency of nucleotide substitutions per site. (C) rAAV yield comparison from three independent crude productions using helper plasmids encoding optimized capsids (lcoAAV2, lcoAAV5, and lcoAAV6) (gray bars: LCO). Packaging constructs encoding wild-type cap genes (wtAAV2, wtAAV5, wtAAV6) served as controls (black bars: WT). (D) Functional comparison between recombinant AAV2, AAV5, and AAV6 (WT: black bars) and lcoAAV2, lcoAAV5, and lcoAAV6 (LCO: gray bars). HuH7 cells were transduced at an MOI = 5.0 × 103 for AAV2 and AAV6 variants and at an MOI = 5.0 × 104 for AAV5 variants, with vectors encoding EGFP under the control of a liver-specific promoter (LSP). FACS analysis of EGFP was performed 72 hr after transduction. Values in (C) and (D) are shown as mean ± SD (n = 3).
In order to test functionality of the lcoAAVs, AAV packaging plasmids encoding optimized capsid sequences of AAV2, AAV5, and AAV6 were generated and used to package the AAV-LSP-GFP cassette in independent transfections (n = 3 per capsid), using the corresponding WT packaging constructs as controls. In marked contrast with the results obtained with hcoAAVs (Figure 1D), no substantial effect on lcoAAV vector yield was observed when compared with the wtAAV counterparts (Figure 2C). Western blot analysis revealed that, in contrast with hcoAAV2, steady-state levels of VP proteins from lcoAAV2 were indistinguishable from those detected for wtAAV2 control (Figure S5). Addition of AAP in trans had no effect on vector yield (data not shown), indicating that each of the lcoAAVs encoded functional AAP. Detailed analysis of lcoAAV2 revealed that codon optimization introduced an early stop codon in the AAP2 ORF leading to loss of two amino acids at the C terminus. Of the remaining 202 amino acids, 165 (81.68%) were fully conserved, 7 were conservative substitutions (3.47%), 5 semi-conservative (2.48%), and the remainder (25 amino acids [aa], 12.37%) non-conservative. Importantly, none of the non-conservative mutations occurred on AAP’s hydrophobic region (HR) nor on its conserved core (CC), the two essential domains recently described to mediate AAP-VP interactions in the context of AAV1.29 In fact, similar analysis on AAP1 revealed that the two essential regions are also conserved, thus providing further support to the hypothesis that local codon optimization preserves AAP functionality (Figure S4).
Sequence analysis also revealed that codon-optimization introduced a premature STOP codon in the X gene in lcoAAV2. Protein X has been implicated in AAV2 DNA replication, and mutations inhibiting X gene expression have been shown to cause a decrease in AAV replication and yield.30 However, our data show that truncation of the C terminus of protein X in lcoAAV2 had no effect on vector yield (Figure 2C), indicating that activity of protein X has not been affected or that protein X is dispensable. Most importantly, transduction studies performed using HuH7 cells showed that the transduction efficiency of lcoAAVs was similar to the wtAAV counterparts (Figure 2D), indicating that codon optimization using the LCO algorithm preserved vector yield and function.
Localized Codon-Optimization of Parental Sequences Increases Identity and Improves DNA Shuffling of Distant Serotypes
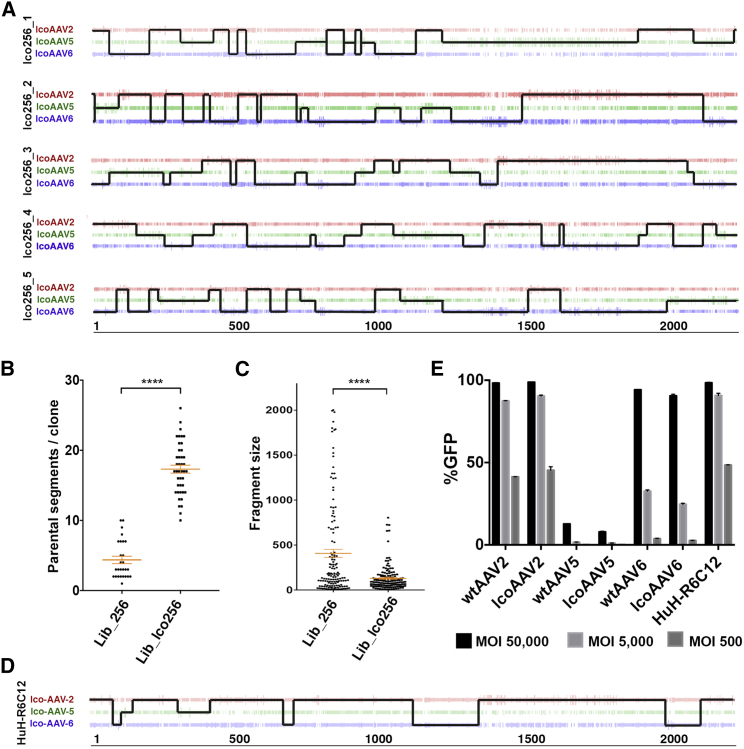
To evaluate the effect of sequence optimization on the efficiency of capsid shuffling, we used codon-optimized capsid genes (lcoAAV2, lcoAAV5, and lcoAAV6) as input sequences to create a shuffled library (AAVLiblco256). Random clones (n = 42) from the final library were fully sequenced to evaluate the presence of reassembled parental capsids and level of capsid shuffling. Detailed analysis of the sequenced clones revealed a marked increase in complexity at the level of individual variants within the library (Figure 3A). In comparison with the library generated using capsid sequences from wtAAV2, wtAAV5, and wtAAV6, which on average contained 4.37 ± 2.68 individual parental sequences per clone, clones in the new library contained on average 17.3 ± 3.70 individual fragments from parental donors (Figure 3B). Furthermore, the average size of each donor sequence segment decreased from 407 bp in AAVLib256 to 120 bp in AAVLiblco256 (Figure 3C), consistent with improved shuffling efficiency as a direct consequence of increased identity at the DNA level between input sequences. Furthermore, while all of the sequenced clones from the AAVLiblco256 library contained shuffled sequences from lcoAAV5, the percentage contribution ranged from 6.5% to 68%, and only 11.6% were >50% identical to AAV5 (data not shown), indicating that the use of lcoAAVs in shuffling protocol lowered the frequency of full AAV5 capsid reassembly.
Figure 3.
AAV Shuffled Library Based on lcoAAV2, lcoAAV5, and lcoAAV6
(A) Sequence analysis of representative clones from AAVLiblco256 showing increased shuffling efficiency at the DNA level. (B) Number of individual parental segments contributing to clones from shuffled AAV library based on capsids from wtAAV2, wtAAV5, and wtAAV6 (AAVLib256) (4.37 ± 0.53, mean ± SEM) and library generated using localized codon-optimization (LCO) algorithm optimized caps (AAVLiblco256) (17.31 ± 0.57, mean ± SEM). (C) Size distribution and mean (red line) size of parental fragments contributing to clones (n = 145) from AAVLib256 (407.2 ± 44.91, mean ± SEM) and AAVLiblco256 (129.5 ± 11.6, mean ± SEM). (D) Sequence composition of clone HuH-R6C12 isolated from AAVLiblco256 library during selection screen on HuH7 cells. (E) Functional analysis of HuH-R6C12 clone. HuH7 cells were transduced with iodixanol preparations of rAAV-HuH-R6C12, control wild-type (WT) vectors (rAAV2, rAAV5, and rAAV6), and lcoAAV vectors (lcoAAV2, lcoAAV5, and lcoAAV6) at MOIs indicated. FACS analysis of EGFP was performed 72 hr after transduction. Values are shown as mean ± SD (n = 3). ****p < 0.0001.
To test functionality, we cloned the novel highly shuffled library into a replication-competent recipient construct containing inverted terminal repeats (ITRs) and the rep gene (ITR-Rep-Caplibrary-ITR). The library was efficiently packaged using a standard transfection protocol and yielded 2 × 102 vector particles per packaging cell (2 × 1010 total particles per five 15-cm dishes). The library was used to perform six rounds of selection on HuH7 cells in the presence of wtAd5 virus, to facilitate library replication. Starting from round 4, at each round n = 20 clones were sequenced to track progression of the selection process. After round 6 of selection, a dominant AAV variant, clone 12 (designated HuH-R6C12), represented 40% of the analyzed pool of clones examined. Sequence analysis of AAV-HuH-R6C12 revealed that this novel variant was composed of sequences contributed by parental lcoAAV2, lcoAAV5, and lcoAAV6 (Figure 3D), providing a direct confirmation that lcoAAVs allow generation of shuffled clones that would be difficult or impossible to obtain using standard, unoptimized, AAV capsid sequences.
In order to evaluate the transduction potential of the selected HuH-R6C12 clone on HuH7 cells, the HuH-R6C12 cap gene was recovered using PCR, cloned into a standard AAV-helper construct containing the rep2 gene, and used to package the AAV-LSP-GFP construct. Packaging constructs expressing WT and lcoAAVs (pAAV2, pAAV5, pAAV6 and pAAV-lcoAAV2, pAAV-lcoAAV5, pAAV-lcoAAV6) were used in parallel to package the same AAV cassette and served as controls. The AAV-HuH-R6C12 vector functionally transduced 48.4% of HuH7 cells at MOI 500 (Figure 3E), validating the shuffling of lcoAAVs as a new addition to the AAV engineering toolbox.
lcoAAVs Enable Generation of Highly Diverse Libraries from Multiple Phylogenetically Distant Capsids
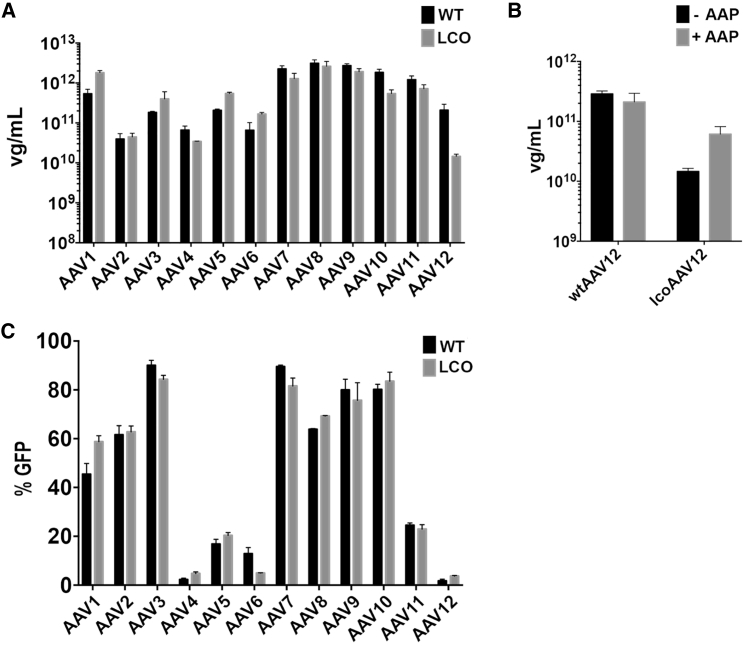
We next sought to validate the new codon optimization strategy by generating shuffled libraries using a larger number of input AAV cap sequences. To achieve this, we first used the LCO algorithm to optimize sequences of 12 parental wtAAVs (AAV1–12). To perform functional analysis of lcoAAV1–12, each lcoAAV capsid sequence (excluding the previously cloned lcoAAV2, lcoAAV5, and locAAV6) was used to generate the corresponding pAAV-lcoAAV packaging construct, which was subsequently used to test the ability to assemble into AAV particles. Packaging tests, performed in parallel with constructs encoding wtAAV cap sequences in independent packaging reactions (n = 3), revealed that the majority of optimized capsids retained packaging ability (Figure 4A). From the AAV panel tested, only lcoAAV12 showed a substantial decrease to 5.06% of wtAAV12 titer, which was partially rescued to 29.08% of wtAAV12 by providing AAP12 in trans (Figure 4B). Functional analysis on HuH7 cells (Figure 4C) revealed that codon optimization did not affect function of individual vectors when compared with wtAAV controls.
Figure 4.
Functional Evaluation of AAV1–12 Optimized Using Localized Codon Optimization Method
(A) Evaluation of lcoAAVs cap genes based on packaging efficiency. rAAV vectors produced using AAV-helper plasmids encoding corresponding wild-type cap genes were used as controls and were assigned a value of 100%. For clarity and convenience, we show again data for AAV2, AAV5, and AAV6 from Figure 2C. (B) Evaluation of the effect of AAP12 on packaging efficiency of lcoAAV12. (C) Functional assessment of rAAV vectors produced using lcoAAV and wtAAV cap genes for their ability to functionally transduce HuH7 cells. All values are shown as mean ± SD (n = 3).
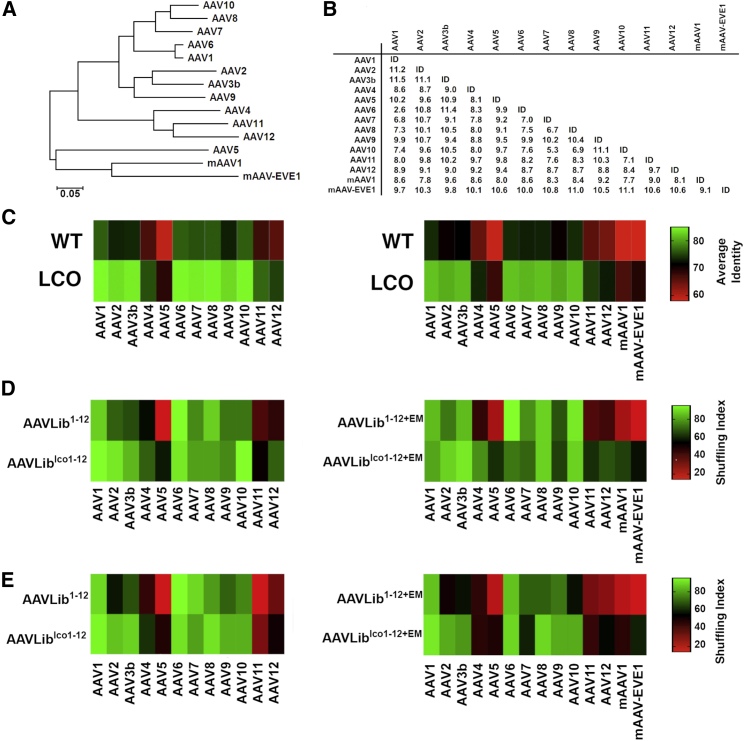
Next, lcoAAV1–12 were used to perform AAV capsid shuffling and library generation. In order to test the ability of capsid shuffling based on lcoAAV parental inputs to enhance shuffling of variants with lower identity at the DNA sequence level, we also included two novel, phylogenetically distant, contemporary AAV variants isolated from contemporary Australian marsupials (mAAV1) and an ultra-ancient AAV-derived endogenous viral element found within the genome of multiple marsupial species (mAAV-EVE1) (Figure 5A). Because the mAAV-EVE1 and mAAV1 variants had not been vectorized using the canonical AAV2 genome (Rep/ITR) (data not shown), the LCO algorithm’s built-in option was utilized to perform localized codon-optimization of mAAV-EVE1 and mAAV1 using sequence input from AAV1–12 only, without using mAAV-EVE1 and mAAV1 as input (Figure S6). As expected, localized codon-optimization increased identity between individual variants by up to 11.5% when compared with WT sequences (Figure 5B), with the identity range between variants increasing from 55%–75% to 75%–85% (Figure 5C).
Figure 5.
Parental Capsid Phylogeny, LCO-Based Enhancement of Sequence Identity, and Increased Shuffling Efficiency
(A) Phylogenetic relationship among the 14 AAV variants used in the study. (B) Relative increase in percent identity between wtAAV cap genes and cap genes optimized using the LCO algorithm. Values = %homologyLCO − %homologyWT. (C) Percent identity between individual wtAAV cap genes and lcoAAV cap genes. (D and E) Changes in shuffling index between plasmid libraries (D) and packaged AAV libraries (E) generated using wtAAV cap and lcoAAV cap, with green color indicating higher and red indicating lower shuffling index. n = 40 clones per library were used in the analysis.
Having optimized all 14 input sequences (AAV1–12, mAAV1, and mAAV-EVE1), we next performed DNA shuffling and generated libraries composed of lcoAAV1–12 only as well as lcoAAV1–12 including mAAV-EVE1 and mAAV1 (referred to as E and M in library names). In parallel, control libraries were generated using the same combination of parental unoptimized sequences. Thus, four shuffled capsid libraries were generated and designated AAVLiblco1–12 and control AAVLib1-12, as well as AAVLiblco1–12+EM and control AAVLib1–12+EM.
Sequence analysis of individual clones from each of the libraries (n = 40 per library) revealed that while the average number of individual sequence segments contributing to the fully reassembled capsid genes did not change considerably, the number of individual parental variants contributing to shuffled clones did increase for libraries based on lcoAAVs (Table 1). We also observed a moderate decrease in the average length of donor sequence segments for library AAVLiblco1–12+EM when compared with control AAVLib1–12+EM library (Table 1).
Table 1.
Quantitative Comparison of Shuffling Efficiency between AAV Libraries Generated Using Standard and lcoAAV Inputs
| AAVLib1-12 | AAVLiblco1–12 | AAVLib1–12+EM | AAVLiblco1–12+EM | |
|---|---|---|---|---|
| Plasmid Library | ||||
| Number of parental segments per clone | 17.77 ± 4.77 | 16.97 ± 3.09 | 14.57 ± 4.71 | 16.77 ± 3.21 |
| Average length of donor segments (bp) | 118.9 | 122.12 | 140.8 | 126 |
| Number of parental variants contributing | 8.03 ± 1.14 | 9.73 ± 1.12 | 7.87 ± 1.43 | 10.17 ± 1.57 |
| Packaged AAV Library | ||||
| Number of parental segments per clone | 19.66 ± 3.87 | 15.91 ± 3.78 | 16.50 ± 3.84 | 17.00 ± 4.93 |
| Average length of donor segments (bp) | 130.32 | 138.43 | 124.61 | 135.44 |
| Number of parental variants contributing | 7.17 ± 1.52 | 9.01 ± 1.38 | 6.41 ± 1.93 | 9.30 ± 1.87 |
The effects of using lcoAAV in capsid shuffling protocols became clear when the shuffling index (SI), expressed as percentage of individual clones containing at least one fragment (>15 bp) from a given parental donor, was calculated (Figures 5D and 5E). Specifically, the percentage of clones containing sequences from each of the individual parental donors in AAVLib1-12 and AAVLib1-12+EM paralleled the level of identity between individual donor sequences (Figure 5C), with the less identical variants (AAV5, AAV11, AAV12, mAAV1, and mAAV-EVE1) contributing to fewer shuffled clones. In striking contrast, the shuffling was more uniform between individual variants (higher SI) for libraries based on lcoAAVs (AAVLiblco1–12 and AAVLiblco1–12+EM) (Figures 5D and 5E), and again paralleled the identity between individual lcoAAVs (Figure 5C). Importantly, the more efficient shuffling was observed at the DNA level (Figure 5D) and in the final packaged AAV library (Figure 5E), confirming that cap variants containing sequences from more phylogenetically distant parental donors are functional and can assemble into AAV particles. These data clearly demonstrate that the novel system reported herein allows the generation of libraries containing distantly related family members and validates sequences optimized using LCO as a powerful tool for AAV shuffling applications.
Discussion
There is immense and growing excitement around AAV-based gene therapy in both the academic and commercial sectors. Leading disease indications based on evidence of therapeutic efficacy involve targeting the eye,31, 32, 33 CNS (reviewed in Hocquemiller et al.34), and liver (reviewed in Baruteau et al.35 and Nathwani et al.36), with diseases in multiple other target tissues at various stages of preclinical and clinical development.37, 38, 39, 40, 41, 42 To date, most AAV-based vectors used in human studies have been naturally occurring variants, such as AAV2, AAV5, AAV8, AAV9, and AAVrh10. Accumulating preclinical and clinical data indicate that many of the currently utilized AAV serotypes are less efficient at functionally transducing primary human cells than would be predicted from preclinical data obtained in in vitro and in vivo models. For example, based on analysis of the results of a hemophilia B clinical trial, where 2 × 1012 vg/kg of self-complementary (sc)AAV2/8 was used to deliver a codon-optimized human factor IX (hFIX) cDNA,36 one can estimate that only a few percent of human hepatocytes were functionally transduced and contributed to AAV-mediated hFIX transgene expression, with resultant peak FIX activity of 10%–12% of physiological levels. The same vector (scAAV2/8-LP1-hFIXco) tested at the same dose in non-human primates (NHP) led to transduction of 96%–99% hepatocytes with an average of 88–142 transgene copies per cell and peak hFIX levels between 399% ± 58% and 580% ± 34% of normal,43 while a similar dose of 1.2 × 1010 vg/mouse of ssAAV8 vector transduces the majority of mouse hepatocytes with average vector copy number of ∼1 per diploid genome.44 This highlights two critically important technological limitations that need to be addressed in order for AAV vectors to reach their full clinical potential: first, the failure of commonly used preclinical models to reliably predict human therapeutic utility; and second, the inadequate transduction efficiency of contemporary AAV capsid variants to treat many disease phenotypes potentially amenable to gene therapy in important target tissues, such as the human liver. AAV directed evolution has been successfully used to generate novel AAV variants with improved transduction of target cells/tissues. During this process, selection pressure is applied to an AAV library containing a large number of random variants of the capsid gene, each with unique sequence composition, and thus potentially unique tropism. One of the commonly utilized methods to generate AAV cap libraries is through DNA-family shuffling, a technique reliant on random fragmentation of capsid genes and PCR-based reassembly, facilitated by stretches of high sequence identity between different input capsid variants. Of note, the first synthetic AAV variant, AAV-LK03,14 selected using a directed evolution strategy for the ability to transduce primary human hepatocytes, has recently reached the clinic in the context of a gene therapy trial using AAV vector encoding human factor VIII (hFVIII) for hemophilia A.45 Based on the level of hFVIII activity, it is reasonable to conclude that the transduction efficiency achieved with the AAV-LK03 capsid is almost certainly higher than with any other vector tested in a clinical setting at the same dose, and supports preclinical predictions made using the FRG (Fah−/−/Rag2−/−/Il2rg−/−) human liver xenograft mouse model.10
Despite the power of directed evolution to select highly functional novel AAV variants, the main limitation of this approach is the efficiency with which parental capsid sequences can be shuffled, a critical step in library preparation that relies on stretches of DNA sequence identity between individual input capsid sequences. Because individual AAV serotypes differ to various degrees from one another, this has the potential to limit library complexity and cause unintentional bias in library composition. The predicted outcome of such bias would be underrepresentation of variants with lower sequence identity and overrepresentation of sequences from parental variants with higher sequence identity. Our results obtained with an AAV library generated by shuffling cap 2, 5, and 6 (AAVLib256) validate these concerns. Sequences from AAV5, which shares the lowest level of sequence identity among these capsids, proved more likely to recombine with other AAV5-derived fragments, leading to a high percentage of clones containing over 80% AAV5 genome and 25% of clones that were 100% AAV5. The likely explanation is that as a consequence of lower sequence identity with AAV2- and AAV6-derived DNA fragments present during the PCR reassembly step, AAV5-derived fragments annealed to each other more efficiently than to DNA fragments from AAV2 and AAV6, leading to preferential reassembly of the AAV5 cap gene or substantial proportions thereof. This effect can be reduced by introducing additional sequences into the reaction mixture, as in the case of AAVLib1-12 or AAVLib1–12+EM, generated from 12 and 14 parental donors, respectively, in which full-length AAV5 cap was not detected. This is attributable to the fact that in a library based on a larger number of parental donors, the chance of individual DNA fragments from the same parental donor interacting with one another is reduced, decreasing the probability of full-length capsid reassembly. Furthermore, the distribution of sequence identities between individual fragments within the PCR mixture is more normally distributed in the context of larger libraries, increasing the probability that fragments with lower degrees of identity will encounter another fragment with sufficiently high identity to allow annealing.
In order to minimize this bias introduced during the DNA shuffling step by variants with lower sequence identities, we initially performed conventional codon optimization to increase pairwise identities between individual variants. Utilization of a single codon for each amino acid throughout the sequence, while having the most significant positive effect on the level of sequence identity, proved to have a negative effect on expression of capsid VP proteins (hcoAAV2 in Figure S5), leading to a dramatic decrease in vector titers. More interesting, however, was the fact that addition of corresponding AAPs in trans did not increase the total level of VP protein detected and did not rescue vector packaging. This result implies that AAV capsid genes encode additional currently unknown factors, or motifs, that serve important functions during cap expression and/or capsid assembly. Notably, the novel enhancer-promoter element recently identified in the 3′ UTR of AAV246 further supports the possibility that in order to fully utilize the limited genome size, AAV could use all six DNA reading frames and thus contain additional coding or functional regions that remain to be identified. Should additional data become available in support of this hypothesis, the known level of genomic complexity of this “simple” virus would increase significantly and many of the commonly accepted assumptions related to AAV biology and vectorology would need to be reevaluated. Alternative sophisticated algorithms and methods to codon-optimize DNA sequences for protein engineering have been described;23, 24, 47, 48 however, to the best of our knowledge, they currently remain untested in the context of large structural viral proteins.
By exploiting individual triplet codons utilized by parental input variants, the new LCO algorithm presented here allows identity between individual input AAV variants to be increased (Figure 5B) while minimizing the risk of loss of vector function (Figure 4; Figure S5). The increased identity between individual input parental AAV cap genes led to more efficient shuffling as measured by decreased average size and increased number of individual fragments contributing to fully reassembled cap variants within shuffled AAVLiblco256 (Figures 3B and 3C). As expected, this effect was less dramatic in the context of libraries generated using a larger number of input cap genes (AAV1–12 + mAAV-EVE1 and mAAV1) (Table 1), due to the fact that many of the variants were over 70% identical to one another even before the codon optimization step, allowing efficient shuffling between such variants. Importantly, despite the fact that in more complex libraries the average size and number of individual parental fragments contributing to reassembled cap variants did not change substantially between LCO and WT libraries (Table 1), the number of contributing parental variants per clone (Table 1) and the percentage of clones containing sequences from each parental donor (Figure 5E) increased when lcoAAVs were used as input sequences. These results indicate that the LCO algorithm facilitates more efficient shuffling and increases the frequency of individual clones containing fragments from more distant variants.
The shuffling of more distantly related sequence fragments, as facilitated by LCO, could potentially increase the proportion of individual variants within an AAV capsid library that are incapable of efficiently packaging an ITR-flanked DNA cargo, or are otherwise functionally impaired owing to structural incompatibility of capsid domains during assembly or packaging. Furthermore, diverse capsid contributions, even among clones that do package efficiently, will not necessarily translate into functional superiority of selected variants. Variants containing capsid gene elements derived from diverse parental capsids could also fail to successfully cross-complement the ITRs and Rep derived from AAV2, which would further complicate the manufacture of vectors utilizing such capsids. Accepting this uncertainty, the important positive consequence of increased library diversity is the otherwise unavailable prospect of identifying novel functional variants that would not exist in a less phylogenetically diverse library.
Out of the 12 AAV serotypes tested (Figure 4A), only AAV12 showed a decrease in vector packaging efficiency when the capsid sequence was optimized using the LCO algorithm. Expression of AAP12 in trans partially rescued vector packaging, providing further evidence supporting the hypothesis that AAP may not be the only accessory protein involved in AAV packaging. Interestingly, a number of AAV serotypes (AAV1, AAV3, AAV5, and AAV6) appear to be more efficient at packaging following LCO modification (Figure 4A). Although this may be related to the availability of specific tRNAs in HEK293T cells, it might also indicate that codon optimization on the first ORF as part of LCO affected cap gene functional elements and/or led to generation of more functional variants of accessory proteins involved in AAV packaging or replication.
In summary, AAV capsid shuffling based on lcoAAVs can be used to enhance currently utilized AAV shuffling at the DNA level, thereby providing a powerful addition to the AAV engineering toolbox. It is important to point out that the AAV cap shuffling based on lcoAAVs is a complementary method, not a replacement, to currently utilized shuffling methods. The authors do not claim that lcoAAV libraries are superior to libraries based on wtAAV or will necessarily lead to identification of more functional variants. The properties of selected variants depend on the library and selection model, and to this end, shuffling based on lcoAAVs offers an efficient way to generate libraries composed of highly shuffled clones, which otherwise have an extremely low probability of being present in libraries based on wtAAVs. Furthermore, based on the functional data obtained with lcoAAV capsids, the localized codon-optimization algorithm could be applied more generically for DNA shuffling of other genes to increase the identity of input parental sequences with reduced likelihood of impairing function. Importantly, this new technology can enhance other techniques based on homologous recombination, such as staggered extension process (StEP), random chimeragenesis on transient templates (RACHITT), and nucleotide exchange and excision technology (NExT), making localized codon-optimization a powerful new tool with potential applicability and significant impact on the broader field of bioengineering.
Materials and Methods
Standard Codon Optimization
Conventional human codon optimization was performed using Geneious (version 9.1.5) (https://www.geneious.com/).49 The nucleotide sequence identity between individual parental variants was increased using the Codon Adaptation Index (CAI) of a gene sequence as the geometric mean of the relative adaptiveness of the codons in the sequence, as defined in Sharp and Li.50 Specifically, the human codon frequency table was used to identify the codon most commonly used for each of the individual amino acids. Subsequently, the cap genes were reverse translated, applying the single most commonly used codon for each of the amino acids, so that the final codon-optimized variants were represented by the most likely non-degenerate coding sequence, and thus have the highest CAI.
Localized Codon-Optimization Algorithm
The LCO algorithm was written in Java as a native Geneious (version 9.1.5) (https://www.geneious.com/)49 plugin (the plugin and instructions are available for free download at https://github.com/CMRI-TVG/AAVcodons). The resulting enhanced-identity sequences are exportable to a FASTA format. The algorithm performs a specific multiple sequence alignment (MSA) on the target sequences using ClustalW2.51 In order to preserve the amino acid sequence of each variant, the algorithm uses translation MSA where nucleotide sequences are translated to amino acid sequences while saving positional references. The resulting amino acid sequences are then aligned and re-converted back to nucleotide sequences by using the positional references. Using the nucleotide MSA as input, the algorithm identifies individual codons, translates them, and identifies positions with 100% amino acid conservation. For those positions, the algorithm creates a local codon-usage table and selects the most common codon in all the variants in the alignment. In the case where two codons are used with equal frequency (50:50) to encode the same amino acid, the position is assigned to the codon of the first capsid used in the alignment, in our case, AAV1. In regions with indels, the algorithm ignores the sequences that have a gap and performs local codon optimization for all other sequences following the same method (Figure S1). To increase flexibility, the algorithm allows selection of input variant(s) that will not be included in the calculation of the most common codon, but will undergo codon optimization. This feature is important when including unverified or incomplete parental variant sequences. Furthermore, this feature allows users to perform codon optimization of novel parental sequences at a later point, without affecting previously optimized variants.
Sequence Contribution Analysis and Graphical Representation
To allow convenient contribution analysis of shuffled capsids, a Sequence Origin Depiction (SOD) plugin was created for Geneious (https://www.geneious.com/)49 (the plugin and instructions are available for free download at https://github.com/CMRI-TVG/AAVcodons). In contrast to a commonly used Xover tool (http://qpmf.rx.umaryland.edu/xover.html),52 the SOD allows the user to zoom in on the output sequence to perform detailed analysis at the nucleotide level. The tool also displays the crossover number, number of point mutations, Levenshtein distances to all parental sequences, effective mutation, as well as mean, minimum, and maximum size of contributing fragments. For convenience, the SOD graphical output can be exported in a number of file formats. Specifically, the graphical output for SOD depicts horizontal lines corresponding to parental sequences augmented with bars representing proportional likelihood of a nucleotide coming from a given sequence and a polygonal line depicting the most likely donor of individual fragments in the resulting sequence. To achieve this, ClustalW2 is used to align parental sequences and the sequence being analyzed, each represented as a horizontal line, with a bar at each position where the nucleotide on the corresponding donor sequence(s) matches residue(s) in the analyzed sequence. The height of the bar is proportional to the percentage likelihood that the given residue contributes to the novel sequence. Gaps between the likelihood bars indicate DNA stretches not present in the analyzed clone. The contribution line is calculated as the longest sequence of identity in a 5′ to 3′ direction. Implementation was done in Java as a Geneious Plugin. This offers the opportunity to choose alternative MSAs or perform manual alignment adjustments.
Shuffled AAV Capsid Plasmid Library Generation
AAV libraries were generated as previously described14 with minor modifications. The capsid genes from WT AAV serotypes AAV1 (GenBank: NC_002077.1), AAV2 (GenBank: NC_001401.2), AAV3 (GenBank: AF028705.1), AAV4 (GenBank: NC_001829.1), AAV5 (amplified from pXR5 plasmid, which differs at two positions, G1268C and C2131G, from GenBank: NC_006152.1), AAV6 (GenBank: AF028704.1), AAV7 (GenBank: AF513851.1), AAV8 (GenBank: AF513852.1), AAV9 (GenBank: AY530579.1), AAV10 (GenBank: AY631965.1), AAV11 (GenBank: AY631966.1), AAV12 (GenBank: DQ813647.1), mAAV-EVE1_modified (GenBank: MG657004), and mAAV1 (GenBank: MK026553) were cloned into the plasmid p-RescueVector (pRV 1–12), a construct based on the pGEM-T Easy Vector System (catalog [Cat] #A1360; Promega) modified to harbor trimethoprim resistance and randomized ends flanking the capsids, for optimal Gibson Assembly (GA). Individual clones were Sanger sequenced (Garvan Molecular Genetics). Codon-optimized AAV capsid genes were synthesized de novo (Genewiz) and cloned into the same system (pRV 1–12). Capsid genes (serotypes 1–12, mAAV-EVE1 and mAAV1 for Lib_1-12 and Lib_1-12EM, and serotypes 2, 5, and 6 for libraries 2/5/6) were excised using SwaI and NsiI (NEB), mixed at 1:1 molar ratios, and digested with 1:10 prediluted DNaseI (Cat #M030S; NEB) for 2–5 min. The pool of fragments was separated on a 1% (w/v) agarose gel and fragments ranging from 200 to 1,000 bp (for AAVLib1–12+EM, AAVLib1–12, AAVLiblco1–12+EM, AAVLiblco1–12) and from 200 to 500 bp (for AAVLib256 and AAVLiblco256) were recovered using the Zymoclean Gel DNA Recovery Kit (Cat #D4001T; Zymogen). For each primer-less PCR reassembly reaction, 500 ng of gel-extracted fragments was used and fully reassembled capsids were amplified in a second PCR with primers (Shuffling_Rescue-F: 5′-GTCGGAAAGCATATGCCGCG-3′, Shuffling_Rescue-R: 5′-GACGTCGCATGCAACTAGTAT-3′) binding the cap gene and carrying overlapping ends to pRV plasmids. A GA reaction was performed by mixing an equal volume of 2 × GA Master Mix (Cat #E2611L; NEB) with 1 pmoL PCR-amplified and DpnI-treated pRV (BB_GAR-F: 5′-ACTTGTTCACTTTGATGGCGAGG-3′, BB_GAR-R: 5′-CTGCACACGACATGACATCACG-3′) and 1 pmol of the recovered shuffled capsids, at 50°C for 30 min. DNA was ethanol precipitated and electroporated into SS320 electro-competent E. coli (Cat #60512-2; Lucigen). The total number of transformants was calculated by preparing and plating five 10-fold serial dilutions of the electroporated bacteria. The pool of transformants was grown overnight in 250 mL of Luria-Bertani media supplemented with trimethoprim (10 μg/mL). Total pRV library plasmids were purified with an EndoFree Maxiprep Kit (Cat #12362; QIAGEN). Thirty individual clones were picked and Sanger sequenced to sample library variability. pRV-based libraries were then digested overnight with SwaI and NsiI, and 1.4 μg of insert was ligated at 16°C with T4 DNA ligase (Cat #M0202; NEB) for 16 hr into 1 μg of a replication-competent AAV2-based plasmid platform (p-Replication-Competent [p-RC]) containing ITR-2 and rep2, and unique SwaI and NsiI sites flanking a 1-kb randomized stuffer [ITR2-rep2-(SwaI)-stuffer-(NsiI)-ITR2]. Ligation reactions were concentrated by using ethanol precipitation, electroporated into SS320 electro-competent bacteria, and grown as described above. Total pRC library plasmids were purified with an EndoFree Maxiprep Kit (Cat #12362; QIAGEN).
Production of rAAV Crude Lysates
All rAAV stocks were prepared by polyethylenimine (PEI) (Cat #239662; Polysciences) triple transfection (2:1 PEI:DNA ratio) of adherent HEK293T cells (Cat #CRL-3126; ATCC) with pAd5 helper plasmid,53 AAV transfer vector expressing GFP under the control of LSP (ssAAV-LSP1-GFP-WPRE-BGHpA),54 and an AAV-helper plasmid encoding rep2 and the capsid of interest at 2:1:1 molar ratios. When indicated, pAAP1-12 plasmids expressing FLAG-tagged AAP proteins under the control of the human cytomegalovirus immediate early (CMV-IE) enhancer-promoter28 were co-transfected to provide AAP in trans. Cells were seeded 18 hr prior to transfection into 15-cm tissue culture (TC)-treated dishes to obtain 90% confluency at the time of transfection. Cells were harvested 72 hr posttransfection and centrifuged for 20 min at 5,250 × g. Media were either discarded or used for qPCR titration following DNaseI treatment to remove free plasmid DNA. The cell pellet was resuspended in 1 mL of Benzonase Buffer (50 mM Tris [pH 8.5] with 2 mM MgCl2) and subjected to three freeze-thaw cycles. Genomic and free plasmid DNA was removed by incubating with Benzonase (Merck KGaA, Cat #1.101695.0002; EMD Chemicals) 200 U/mL at 37°C for 1 hr. Cellular debris was removed by centrifugation for 30 min at 5,250 × g. Supernatant was further cleared by adding 1 M CaCl2 to a final concentration of 25 mM and incubated on ice for 1 hr followed by centrifugation at 5,250 × g for 30 min at 4°C. Supernatant, 1 mL in total, was then transferred into a sterile cryotube and stored at −80°C.
Production of Replication-Competent AAV Libraries
Recombinant AAV capsid libraries were packaged following the same transfection protocol as described above, with the exception that only two plasmids were used: the pAd5 helper plasmid and the pRC libraries containing ITR-rep2-Caplibrary-ITR at a 1:1 molar ratio. Cell lysates from five 15-cm dishes were pooled and purified using iodixanol-based density gradients as previously described.55 Amicon Ultra-4 Centrifuge Filter Units with Ultracel-100 kDa membrane (Cat #UFC810024; EMD Millipore) were used to perform a buffer exchange (PBS, 50 mM NaCl, 0.001% Pluronic F68 [v/v]; Cat #24040-032; LifeTech) and concentration step. Based on the efficiency of transformation (total number of individual bacterial colonies), the variability of the library was estimated to have the upper limit of 2.72 × 107 variants for AAVLiblco256, 2.4 × 106 for AAVLib1–12, 3.9 × 106 for AAVLiblco1–12, 9.8 × 106 for AAVLib1–12+EM, and 6.9 × 106 for AAVLiblco1–12+EM.
AAV Titration by Real-Time qPCR
Vector preparations were titrated by real-time qPCR as previously described56 using the following primers: GFP-F: 5′-TCAAGATCCGCCACAACATC-3′ and GFP-R: 5′-TTCTCGTTGGGGTCTTTGCT-3′ for vectors encoding the LSP-GFP cassette, and rep2-F: 5′-AAGGATCACGTGGTTGAGGT-3′ and rep2-R: 5′-CCCACGTGACGAGAACATTT-3′ for replication-competent library preparations. Serial dilutions of linearized plasmid were used to generate a standard curve.
Replication-Competent AAV Library Selection in HuH7 Cells
Human hepatoma HuH7 cells were maintained as monolayer cultures in DMEM (Cat #D579; Sigma) supplemented with 10% (v/v) fetal bovine serum (Cat #F8192; Sigma), 100 μg/mL penicillin, and 100 μg/mL streptomycin. 1 × 105 cells were seeded per well in two 24-well TC dishes 16 hr prior to infection with AAV library. Four 10-fold dilutions of the AAV library were added to the media in duplicate plates. Cells were washed with 1× PBS 24 hr after infection and fresh media added. To facilitate AAV library replication, we added WT human Adenovirus 5 (hAd5) (Cat #VR-1516; ATCC) at an MOI of 0.42 (based on 7-day median TC infective dose [TCID50]) to one of the plates. The plate without hAd5 served as a qPCR control. Cells were harvested 72 hr after hAd5 infection and lysed by three freeze-thaw cycles. Cellular debris was removed by centrifugation and supernatant-containing AAV particles analyzed for AAV amplification by qPCR. Eight microliters of the matched library dilutions (+/− hAd5) was treated with DNaseI (Cat #M030S; NEB) at 37°C for 1 hr, and the enzyme was heat inactivated at 75°C for 10 min. qPCR with rep2-specific primers was used after each round to confirm library replication (Figure S2) and to select the library dilution to be moved into subsequent rounds of selection. At each step the highest library dilution that resulted in no less than a 2 log increase in AAV signal was selected to minimize cross-packaging of multiple vectors in single packaging cells and thus increase the stringency of selection. The library dilution selected for subsequent rounds of amplification was incubated at 65°C for 30 min to inactivate hAd5, and iterative selection was performed using the above described conditions.
Vectorization of Evolved AAV Capsids
After each round of selection, AAV capsid sequences were recovered from the media by PCR using primers flanking the capsid region (CapRescue-F: 5′-CCCTGCAGACAATGCGAGAGAATGAATCAGAATTCAAATATCTGC-3′, CapRescue-R: 5′-ATGCATATGGAAACTAGATAAGAAAGAAATACG-3′). PCR-amplified cap genes were cloned by GA in-frame downstream of the rep2 gene in a recipient pHelper packaging plasmid opened by PCR amplification using the following primers (pHelper-F: 5′-CGCATTGTCTGCAGGGAAACAGCATC-3′, pHelper-R: 5′-TTTCTTTCTTATCTAGTTTCCATATGCATGTAGATAAGTAGCATGGCGGG-3′) and DpnI treated. Twenty individual clones were sequenced to track progress of the selection process.
Western Blot Analysis
For western blot analysis of AAV VP and Rep protein, HEK293T cells were transfected with pAd5 plasmid, pAAV-LSP-GFP plasmid, and AAV packaging plasmid expressing Rep2 and Cap of corresponding WT or codon-optimized AAV following the PEI transfection protocol described above. Cells were harvested 72 hr posttransfection, and total proteins were extracted using radioimmunoprecipitation assay (RIPA) buffer (Cat #89900; Pierce, Thermo Fisher, Rockford, IL, USA) supplemented with protease inhibitors (Cat #04693116001; cOmplete Tablets, Roche, Mannheim, Germany) and quantified by Bradford assay. Total protein (10 μg) was separated by polyacrylamide gel electrophoresis using 4%–12% NuPAGE BisTris gels (Cat #NP0322; Life Technologies, Carlsbad, CA, USA) followed by transfer to nitrocellulose membranes and blocking in PBS−/−, 5% (w/v) milk powder, and 0.1% (v/v) Tween 20. Detection of VP1+VP2+VP3 proteins was performed using rabbit polyclonal primary antibody (1:300; Cat #03-61084; American Research Products, Waltham, MA, USA) and secondary antibody (goat anti-rabbit IgG-HRP; sc-2004; Santa Cruz Biotechnology, Dallas, TX, USA) while detection of AAV Rep proteins was performed using anti-AAV Replicase antibody (mouse monoclonal anti-Rep, 303.9, 1:100; Cat #03-61069; American Research Products, Waltham, MA, USA) and secondary antibody (goat anti-mouse IgG-HRP; Cat #P044701-2; Agilent Dako, Santa Clara, CA, USA). Vinculin (primary antibody: mouse monoclonal hVIN-1; Cat #V9131; Sigma-Aldrich, Saint Louis, MO, USA) and matching secondary antibody (goat anti-mouse IgG-HRP; Cat #P0447; Agilent Dako, Santa Clara, CA, USA) were used as an endogenous loading control. Signal was detected using SuperSignal West Pico Chemiluminescent Substrate (Cat #34080; Thermo Fisher, Rockford, IL, USA) and a FujiFilm Luminescent Image Analyzer system (LAS-4000).
In Vitro Transduction Analysis
For transduction studies, 2 × 105 HuH7 cells were plated per well into 24-well TC plates in complete media (DMEM with 10% [v/v] FBS). Four hours later, the vector stock was diluted into 0.5 mL of complete media and added to the cells at an indicated MOI (see figure legends for details). Data shown in Figure 4C were generated using the following MOIs for each vector pair (WT and LCO): AAV1 MOI = 3.2 × 104, AAV2 MOI = 2.7 × 103, AAV3 MOI = 1.0 × 104, AAV4 MOI = 1.4 × 104, AAV5 MOI = 5.2 × 104, AAV6 MOI = 1.6 × 104, AAV7 MOI = 4.4 × 105, AAV8 MOI = 2.0 × 105, AAV9 MOI = 6.3 × 105, AAV10 MOI = 3.6 × 105, AAV11 MOI = 2.8 × 105, and AAV12 MOI = 4.0 × 104. The cells were harvested 72 hr after transduction using TrypLE Express (Cat #12604021; Thermo Fisher) and resuspended in 200 μL of fluorescence-activated cell sorting (FACS) buffer (PBS−/− with 5% [v/v] FBS and 5 mM EDTA). EGFP expression was quantified using a BD Fortessa flow cytometer (Westmead Research Hub Flow Cytometry Facility, Westmead, NSW, Australia), and the data were analyzed using FlowJo 7.6.1.
Phylogenetic Analysis of Parental AAV Sequences
The 14 parental AAV sequences were aligned using MUSCLE version 3.8.31.57 The phylogenetic relationship among the sequences was inferred by using the maximum likelihood method based on the Tamura-Nei model.58 The tree with the highest log likelihood (−21,217.32) is depicted (Figure 5A). Initial trees for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with superior log likelihood value. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. There were a total of 2,400 positions in the final dataset. Evolutionary analyses were conducted in MEGA X.59
Statistical Analyses
Statistical analyses were conducted using GraphPad Prism (GraphPad Software, La Jolla, CA, USA) version 7.0a. Analysis of statistical significance between two groups in Figures 3B and 3C was performed using the two-tailed nonparametric Kolmogorov-Smirnov test.
Code Availability
The custom plugin used in this study to perform codon optimization and analysis can be obtained without restrictions from https://github.com/CMRI-TVG/AAVcodons.
Author Contributions
M.C.-C., S.L.G., A.K.A., S.H.Y.L., and L.L. designed the experiments. M.C.-C., S.L.G., A.W., C.V.H., A.K.A., S.H.Y.L., J.W., K.L.D., A.R., A.C.F., and L.L. generated reagents, algorithms, and protocols; performed experiments; and analyzed data. H.N. provided AAP constructs. M.C.-C., A.K.A., A.W., C.V.H., and L.L. wrote the manuscript and generated the figures. All authors reviewed, edited, and commented on the manuscript.
Conflicts of Interest
L.L., I.E.A., and A.J.T. have commercial affiliations. L.L. and I.E.A. have consulted on technologies addressed in this paper. L.L. and I.E.A. have stock and/or equity in companies with technology broadly related to this paper. All other authors declare no conflicts of interest.
Acknowledgments
We thank CMRI Vector and Genome Engineering Facility for help in vector preparation. This work was supported by a Project grant to L.L. from Australian National Health and Medical Research Council (NHMRC) (APP1108311) and a Discovery grant to I.E.A. from the Australian Research Council (ARC) (DP150101253). A.J.T. is a Wellcome Trust Principal Research Fellow. All research at Great Ormond Street Hospital NHS Foundation Trust and UCL Great Ormond Street Institute of Child Health is made possible by the NIHR Great Ormond Street Hospital Biomedical Research Centre. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. H.N. was supported by a grant (R01 NS088399) from the National Institutes of Health (NIH).
Footnotes
Supplemental Information includes six figures and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.10.016.
Supplemental Information
References
- 1.Bryant L.M., Christopher D.M., Giles A.R., Hinderer C., Rodriguez J.L., Smith J.B., Traxler E.A., Tycko J., Wojno A.P., Wilson J.M. Lessons learned from the clinical development and market authorization of Glybera. Hum. Gene Ther. Clin. Dev. 2013;24:55–64. doi: 10.1089/humc.2013.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ylä-Herttuala S. Endgame: glybera finally recommended for approval as the first gene therapy drug in the European union. Mol. Ther. 2012;20:1831–1832. doi: 10.1038/mt.2012.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(2018). FDA approves hereditary blindness gene therapy. Nat. Biotechnol. 36, 6. [DOI] [PubMed]
- 4.Schimmer J., Breazzano S. Investor outlook: rising from the Ashes; GSK’s European Approval of Strimvelis for ADA-SCID. Hum. Gene Ther. Clin. Dev. 2016;27:57–61. doi: 10.1089/humc.2016.29010.ind. [DOI] [PubMed] [Google Scholar]
- 5.Lisowski L., Tay S.S., Alexander I.E. Adeno-associated virus serotypes for gene therapeutics. Curr. Opin. Pharmacol. 2015;24:59–67. doi: 10.1016/j.coph.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 6.Hermonat P.L., Muzyczka N. Use of adeno-associated virus as a mammalian DNA cloning vector: transduction of neomycin resistance into mammalian tissue culture cells. Proc. Natl. Acad. Sci. USA. 1984;81:6466–6470. doi: 10.1073/pnas.81.20.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robert M.A., Chahal P.S., Audy A., Kamen A., Gilbert R., Gaillet B. Manufacturing of recombinant adeno-associated viruses using mammalian expression platforms. Biotechnol. J. 2017;12:e1600193. doi: 10.1002/biot.201600193. [DOI] [PubMed] [Google Scholar]
- 8.Kotin R.M., Snyder R.O. Manufacturing clinical grade recombinant adeno-associated virus using invertebrate cell lines. Hum. Gene Ther. 2017;28:350–360. doi: 10.1089/hum.2017.042. [DOI] [PubMed] [Google Scholar]
- 9.Kondratov O., Marsic D., Crosson S.M., Mendez-Gomez H.R., Moskalenko O., Mietzsch M., Heilbronn R., Allison J.R., Green K.B., Agbandje-McKenna M., Zolotukhin S. Direct head-to-head evaluation of recombinant adeno-associated viral vectors manufactured in human versus insect cells. Mol. Ther. 2017;25:2661–2675. doi: 10.1016/j.ymthe.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirata R., Chamberlain J., Dong R., Russell D.W. Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat. Biotechnol. 2002;20:735–738. doi: 10.1038/nbt0702-735. [DOI] [PubMed] [Google Scholar]
- 11.Miller D.G., Wang P.R., Petek L.M., Hirata R.K., Sands M.S., Russell D.W. Gene targeting in vivo by adeno-associated virus vectors. Nat. Biotechnol. 2006;24:1022–1026. doi: 10.1038/nbt1231. [DOI] [PubMed] [Google Scholar]
- 12.Russell D.W., Hirata R.K. Human gene targeting by viral vectors. Nat. Genet. 1998;18:325–330. doi: 10.1038/ng0498-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grimm D., Pandey K., Nakai H., Storm T.A., Kay M.A. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J. Virol. 2006;80:426–439. doi: 10.1128/JVI.80.1.426-439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sebastiano V., Zhen H.H., Haddad B., Bashkirova E., Melo S.P., Wang P., Leung T.L., Siprashvili Z., Tichy A., Li J. Human COL7A1-corrected induced pluripotent stem cells for the treatment of recessive dystrophic epidermolysis bullosa. Sci. Transl. Med. 2014;6:264ra163. doi: 10.1126/scitranslmed.3009540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paulk N.K., Pekrun K., Zhu E., Nygaard S., Li B., Xu J., Chu K., Leborgne C., Dane A.P., Haft A. Bioengineered AAV capsids with combined high human liver transduction in vivo and unique humoral seroreactivity. Mol. Ther. 2018;26:289–303. doi: 10.1016/j.ymthe.2017.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zinn E., Pacouret S., Khaychuk V., Turunen H.T., Carvalho L.S., Andres-Mateos E., Shah S., Shelke R., Maurer A.C., Plovie E. In silico reconstruction of the viral evolutionary lineage yields a potent gene therapy vector. Cell Rep. 2015;12:1056–1068. doi: 10.1016/j.celrep.2015.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santiago-Ortiz J., Ojala D.S., Westesson O., Weinstein J.R., Wong S.Y., Steinsapir A., Kumar S., Holmes I., Schaffer D.V. AAV ancestral reconstruction library enables selection of broadly infectious viral variants. Gene Ther. 2015;22:934–946. doi: 10.1038/gt.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith R.H., Hallwirth C.V., Westerman M., Hetherington N.A., Tseng Y.S., Cecchini S., Virag T., Ziegler M.L., Rogozin I.B., Koonin E.V. Germline viral “fossils” guide in silico reconstruction of a mid-Cenozoic era marsupial adeno-associated virus. Sci. Rep. 2016;6:28965. doi: 10.1038/srep28965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen X., Storm T., Kay M.A. Characterization of the relationship of AAV capsid domain swapping to liver transduction efficiency. Mol. Ther. 2007;15:1955–1962. doi: 10.1038/sj.mt.6300293. [DOI] [PubMed] [Google Scholar]
- 20.Bartlett J.S., Kleinschmidt J., Boucher R.C., Samulski R.J. Targeted adeno-associated virus vector transduction of nonpermissive cells mediated by a bispecific F(ab’gamma)2 antibody. Nat. Biotechnol. 1999;17:181–186. doi: 10.1038/6185. [DOI] [PubMed] [Google Scholar]
- 21.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimm D., Zolotukhin S. E pluribus unum: 50 years of research, millions of viruses, and one goal—tailored acceleration of AAV evolution. Mol. Ther. 2015;23:1819–1831. doi: 10.1038/mt.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moore G.L., Maranas C.D. eCodonOpt: a systematic computational framework for optimizing codon usage in directed evolution experiments. Nucleic Acids Res. 2002;30:2407–2416. doi: 10.1093/nar/30.11.2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maheshri N., Schaffer D.V. Computational and experimental analysis of DNA shuffling. Proc. Natl. Acad. Sci. USA. 2003;100:3071–3076. doi: 10.1073/pnas.0537968100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sonntag F., Schmidt K., Kleinschmidt J.A. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc. Natl. Acad. Sci. USA. 2010;107:10220–10225. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sonntag F., Köther K., Schmidt K., Weghofer M., Raupp C., Nieto K., Kuck A., Gerlach B., Böttcher B., Müller O.J. The assembly-activating protein promotes capsid assembly of different adeno-associated virus serotypes. J. Virol. 2011;85:12686–12697. doi: 10.1128/JVI.05359-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grosse S., Penaud-Budloo M., Herrmann A.K., Börner K., Fakhiri J., Laketa V., Krämer C., Wiedtke E., Gunkel M., Ménard L. Relevance of assembly-activating protein for adeno-associated virus vector production and capsid protein stability in mammalian and insect cells. J. Virol. 2017;91 doi: 10.1128/JVI.01198-17. e01198-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Earley L.F., Powers J.M., Adachi K., Baumgart J.T., Meyer N.L., Xie Q., Chapman M.S., Nakai H. Adeno-associated virus (AAV) assembly-activating protein is not an essential requirement for capsid assembly of AAV serotypes 4, 5, and 11. J. Virol. 2017;91 doi: 10.1128/JVI.01980-16. e01980-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tse L.V., Moller-Tank S., Meganck R.M., Asokan A. Mapping and engineering functional domains of the assembly-activating protein of adeno-associated viruses. J. Virol. 2018;92 doi: 10.1128/JVI.00393-18. e00393-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao M., You H., Hermonat P.L. The X gene of adeno-associated virus 2 (AAV2) is involved in viral DNA replication. PLoS ONE. 2014;9:e104596. doi: 10.1371/journal.pone.0104596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maguire A.M., Simonelli F., Pierce E.A., Pugh E.N., Jr., Mingozzi F., Bennicelli J., Banfi S., Marshall K.A., Testa F., Surace E.M. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2240–2248. doi: 10.1056/NEJMoa0802315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bainbridge J.W., Smith A.J., Barker S.S., Robbie S., Henderson R., Balaggan K., Viswanathan A., Holder G.E., Stockman A., Tyler N. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N. Engl. J. Med. 2008;358:2231–2239. doi: 10.1056/NEJMoa0802268. [DOI] [PubMed] [Google Scholar]
- 33.Cideciyan A.V., Aleman T.S., Boye S.L., Schwartz S.B., Kaushal S., Roman A.J., Pang J.J., Sumaroka A., Windsor E.A., Wilson J.M. Human gene therapy for RPE65 isomerase deficiency activates the retinoid cycle of vision but with slow rod kinetics. Proc. Natl. Acad. Sci. USA. 2008;105:15112–15117. doi: 10.1073/pnas.0807027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hocquemiller M., Giersch L., Audrain M., Parker S., Cartier N. Adeno-associated virus-based gene therapy for CNS diseases. Hum. Gene Ther. 2016;27:478–496. doi: 10.1089/hum.2016.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baruteau J., Waddington S.N., Alexander I.E., Gissen P. Delivering efficient liver-directed AAV-mediated gene therapy. Gene Ther. 2017;24:263–264. doi: 10.1038/gt.2016.90. [DOI] [PubMed] [Google Scholar]
- 36.Nathwani A.C., Tuddenham E.G., Rangarajan S., Rosales C., McIntosh J., Linch D.C., Chowdary P., Riddell A., Pie A.J., Harrington C. Adenovirus-associated virus vector-mediated gene transfer in hemophilia B. N. Engl. J. Med. 2011;365:2357–2365. doi: 10.1056/NEJMoa1108046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gafni Y., Pelled G., Zilberman Y., Turgeman G., Apparailly F., Yotvat H., Galun E., Gazit Z., Jorgensen C., Gazit D. Gene therapy platform for bone regeneration using an exogenously regulated, AAV-2-based gene expression system. Mol. Ther. 2004;9:587–595. doi: 10.1016/j.ymthe.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 38.Ulrich-Vinther M. Gene therapy methods in bone and joint disorders. Evaluation of the adeno-associated virus vector in experimental models of articular cartilage disorders, periprosthetic osteolysis and bone healing. Acta Orthop. Suppl. 2007;78:1–64. [PubMed] [Google Scholar]
- 39.Brown N.J., Hirsch M.L. Adeno-associated virus (AAV) gene delivery in stem cell therapy. Discov. Med. 2015;20:333–342. [PubMed] [Google Scholar]
- 40.Jaén M.L., Vilà L., Elias I., Jimenez V., Rodó J., Maggioni L., Ruiz-de Gopegui R., Garcia M., Muñoz S., Callejas D. Long-term efficacy and safety of insulin and glucokinase gene therapy for diabetes: 8-year follow-up in dogs. Mol. Ther. Methods Clin. Dev. 2017;6:1–7. doi: 10.1016/j.omtm.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wells D.J. Systemic AAV gene therapy close to clinical trials for several neuromuscular diseases. Mol. Ther. 2017;25:834–835. doi: 10.1016/j.ymthe.2017.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santiago-Ortiz J.L., Schaffer D.V. Adeno-associated virus (AAV) vectors in cancer gene therapy. J. Control. Release. 2016;240:287–301. doi: 10.1016/j.jconrel.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nathwani A.C., Rosales C., McIntosh J., Rastegarlari G., Nathwani D., Raj D., Nawathe S., Waddington S.N., Bronson R., Jackson S. Long-term safety and efficacy following systemic administration of a self-complementary AAV vector encoding human FIX pseudotyped with serotype 5 and 8 capsid proteins. Mol. Ther. 2011;19:876–885. doi: 10.1038/mt.2010.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakai H., Fuess S., Storm T.A., Muramatsu S., Nara Y., Kay M.A. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J. Virol. 2005;79:214–224. doi: 10.1128/JVI.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.George L.A., Ragni M.V., Samelson-Jones B.J., Cuker A., Runoski A.R., Cole G., Wright F., Chen Y., Hui D.J., Wachtel K. Spk-8011: preliminary results from a phase 1/2 dose escalation trial of an investigational AAV-mediated gene therapy for hemophilia a. Blood. 2017;130:604. [Google Scholar]
- 46.Logan G.J., Dane A.P., Hallwirth C.V., Smyth C.M., Wilkie E.E., Amaya A.K., Zhu E., Khandekar N., Ginn S.L., Liao S.H.Y. Identification of liver-specific enhancer-promoter activity in the 3′ untranslated region of the wild-type AAV2 genome. Nat. Genet. 2017;49:1267–1273. doi: 10.1038/ng.3893. [DOI] [PubMed] [Google Scholar]
- 47.Moore G.L., Maranas C.D., Lutz S., Benkovic S.J. Predicting crossover generation in DNA shuffling. Proc. Natl. Acad. Sci. USA. 2001;98:3226–3231. doi: 10.1073/pnas.051631498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milligan J.N., Garry D.J. Shuffle optimizer: a program to optimize DNA shuffling for protein engineering. Methods Mol. Biol. 2017;1472:35–45. doi: 10.1007/978-1-4939-6343-0_3. [DOI] [PubMed] [Google Scholar]
- 49.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sharp P.M., Li W.H. The codon Adaptation Index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 52.Huang W., Johnston W.A., Boden M., Gillam E.M. ReX: a suite of computational tools for the design, visualization, and analysis of chimeric protein libraries. Biotechniques. 2016;60:91–94. doi: 10.2144/000114381. [DOI] [PubMed] [Google Scholar]
- 53.Parmiani G. Immunological approach to gene therapy of human cancer: improvements through the understanding of mechanism(s) Gene Ther. 1998;5:863–864. doi: 10.1038/sj.gt.3300692. [DOI] [PubMed] [Google Scholar]
- 54.Cunningham S.C., Dane A.P., Spinoulas A., Logan G.J., Alexander I.E. Gene delivery to the juvenile mouse liver using AAV2/8 vectors. Mol. Ther. 2008;16:1081–1088. doi: 10.1038/mt.2008.72. [DOI] [PubMed] [Google Scholar]
- 55.Strobel B., Miller F.D., Rist W., Lamla T. Comparative analysis of cesium chloride- and iodixanol-based purification of recombinant adeno-associated viral vectors for preclinical applications. Hum. Gene Ther. Methods. 2015;26:147–157. doi: 10.1089/hgtb.2015.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q., Dong B., Firrman J., Roberts S., Moore A.R., Cao W., Diao Y., Kapranov P., Xu R., Xiao W. Efficient production of dual recombinant adeno-associated viral vectors for factor VIII delivery. Hum. Gene Ther. Methods. 2014;25:261–268. doi: 10.1089/hgtb.2014.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tamura K., Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 59.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.